DOI:10.32604/cmc.2022.022701

| Computers, Materials & Continua DOI:10.32604/cmc.2022.022701 |  |
| Article |
Intelligent Classification Model for Biomedical Pap Smear Images on IoT Environment
1Department Electronics and Instrumentation Engineering, V. R. Siddhartha Engineering College, Vijayawada, 520007, India
2Department of Computer Applications, Government Arts & Science College, Kanyakumari, 629401, India
3Faculty of Science, AL-Azhar University, Cairo, 11651, Egypt
4Faculty of Computers and Information Technology, University of Tabuk, 47512, Saudi Arabia
5Department of Computer Science, College of Science & Art at Mahayil, King Khalid University, 62529, Saudi Arabia & Faculty of Computer and IT, Sana'a University, 31220, Yemen
6Department of Biomedical Engineering, College of Engineering, Princess Nourah bint Abdulrahman University, 11564, Saudi Arabia
7Department of Computer and Self Development, Preparatory Year Deanship, Prince Sattam bin Abdulaziz University, Alkharj, 16278, Saudi Arabia
*Corresponding Author: Fahd N. Al-Wesabi. Email: fwesabi@gmail.com
Received: 16 August 2021; Accepted: 16 September 2021
Abstract: Biomedical images are used for capturing the images for diagnosis process and to examine the present condition of organs or tissues. Biomedical image processing concepts are identical to biomedical signal processing, which includes the investigation, improvement, and exhibition of images gathered using x-ray, ultrasound, MRI, etc. At the same time, cervical cancer becomes a major reason for increased women's mortality rate. But cervical cancer is an identified at an earlier stage using regular pap smear images. In this aspect, this paper devises a new biomedical pap smear image classification using cascaded deep forest (BPSIC-CDF) model on Internet of Things (IoT) environment. The BPSIC-CDF technique enables the IoT devices for pap smear image acquisition. In addition, the pre-processing of pap smear images takes place using adaptive weighted mean filtering (AWMF) technique. Moreover, sailfish optimizer with Tsallis entropy (SFO-TE) approach has been implemented for the segmentation of pap smear images. Furthermore, a deep learning based Residual Network (ResNet50) method was executed as a feature extractor and CDF as a classifier to determine the class labels of the input pap smear images. In order to showcase the improved diagnostic outcome of the BPSIC-CDF technique, a comprehensive set of simulations take place on Herlev database. The experimental results highlighted the betterment of the BPSIC-CDF technique over the recent state of art techniques interms of different performance measures.
Keywords: Biomedical imaging; pap smear images; internet of things; deep learning; cervical cancer; disease diagnosis
Cervical cancer is the most dangerous and rapidly developing cancer that affects the lives of many females globally. As per the report of WHO, cervical cancers are dramatically increasing amongst Indian females that occur around 1 in 53 females than 1 in 100 females suffer from these ailments globally. The most frequent and common symptom examined almost all suffered persons was uncommon bleeding or discharge from vagina. For medicinal diagnosis and treatment, Pap smear tests are adapted for the detection of abnormalities existing in cervical cells such as changes in cell size, mucus, cell disruption, cell color, and so on [1]. Regular Pap smear screening is one of the effective and successful approaches in medicinal practices for facilitating the earlier screening and detection of cervical cancers. But, the manual analyses of Pap smear images are error prone, laborious, and time-consuming of 100 sub-images within an individual slide should be observed in a microscope using a trained cytopathologist for all patients screened [2]. In order to conquer the limitation related to the computer assisted Pap smear analyses system, manual analyses of Pap smear images using machine learning (ML) and image processing methods have been presented by various authors.
Wireless communication technology and Advanced ML methods have allowed improving a comprehensive medical diagnosis which is capable of operating in realtime, without human interaction, and accurately [3]. Still, there are numerous problems which must be resolved, e.g., robust algorithm to handle a number of variants in data, packet loss at the time of transmissions, and higher bandwidth requirements for medical video data transmission. In order to tackle some of these problems, edge based cloud computing has been presented for IoT [4], cloud based architecture and detecting voice pathology was accomplished, deep learning (DL) for recognizing emotions, edge based transmission, and a disease monitoring scheme. Computer aided system in cancerous cell recognition were employed in the works several times. In breast cancer detection, distinct feature extraction approaches like Laplacian Gaussian filter, histogram of gradient orientation, and local binary pattern have been applied [5]. Local texture analyses were employed for diagnosing pulmonary nodule. In order to study dermoscopy images for skin tumors, color component, and direction filter feature. An approach for detecting voice pathology with distinct input modals. In recent times, DL approach has carried a major development in accuracy in various applications. Because of its higher accuracy in various fields, it has turn into an advanced machine learning (ML) method.
The DL method, as a significant approach to artificial intelligence, was extensively employed in image detection [6]. Furthermore, the CNN attains outstanding results in image classification amongst the many DL methods. The network is capable of directly processing the original image, avoid the requirement for difficult preprocessing of an image. It integrates the 2 factors of weight sharing, pooling, and local receptive field, which significantly decrease the training parameter of neural network [7]. Consequently, study on using CNN for medicinal image diagnoses is growing.
This paper devises a new biomedical pap smear image classification using cascaded deep forest (BPSIC-CDF) model on Internet of Things (IoT) environment. The BPSIC-CDF technique enables the IoT devices for pap smear image acquisition. In addition, the pre-processing of pap smear images takes place using adaptive weighted mean filtering (AWMF) technique. Moreover, sailfish optimizer with Tsallis entropy (SFO-TE) manner has been executed for the segmentation of pap smear image. Furthermore, a deep learning based Residual Network (ResNet50) approach was implemented as a feature extractor and CDF as a classifier to determine the class labels of the input pap smear images. In order to showcase the improved diagnostic outcome of the BPSIC-CDF technique, a comprehensive set of simulations take place on Herlev database.
Dong et al. [8] presented a cell classification approach which integrates artificial and Inception v3 features that efficiently enhances the precision of cervical cells detection. Additionally, to tackle the underfitting problems and perform an efficient DL training with a comparatively smaller number of medicinal data, this study inherits the robust learning capacity from TL approach and attains effective and accurate cervical cell image classifications depends on Herlev dataset. Alyafeai et al. [9] developed a fully automatic pipeline for detecting cervical and cervix cancers classification from cervigram image. The presented pipeline includes 2 pre-rained DL methods for automated cervical tumor classification and cervix detection. Self-extracted features are employed using the succeeding method for classifying the cervix cancers. Such features are learned by 2 lightweight methods according to CNN approach.
Ghoneim et al. [10] proposes cervical cancer cells classification and detection systems on the basis of CNN method. The cell image is fed to CNN models for extracting deep learned features. Following, an ELM based classifier categorizes the input image. CNN models are employed through fine-tuning and TL approach. Also, Alternates to the ELM, MLP, and AE based classifiers are examined. Khamparia et al. [11] presented a fresh IoHT driven DL architecture for classification and detection of cervical cancer in Pap smear images with the idea of Tl technique. Next TL, CNN method was integrated with distinct traditional ML methods such as KNN, NB, LR, RF, and SVM. In this study, FE from cervical images is implemented by pretrained CNN methods such as VGG19, InceptionV3, ResNet50, and SqueezeNet that are fed to fattened and dense layers for abnormal and normal cervical cell classifications.
Chandran et al. [12] proposed 2 DL CNN frameworks for detecting cervical cancer via the colposcopy images; CYENET and VGG19 (TL) method. In the CNN framework, VGG19 is adapted as a TL for the research. A novel method is proposed and called as CYENET method for classifying cervical cancer from colposcopy image manually. The sensitivity, accuracy, and specificity are evaluated for the presented method. Zahir et al. [13] presented an inexpensive, portable, and automated breast cancer recognition on the basis of histopathological images with the help of DL method. The DL methods are developed with the help of CNN approach. The study relates the efficiency of the CNN method with the help of Tl method using a pretrained method (VGG16) and the efficiency of a CNN method without TL model.
In Wang et al. [14], an adoptive pruning deep TL method (PsiNet-TAP) is presented to Pap smear image classifications. Also, they developed a new network for classifying Pap smear images. Because of the constrained amount of images, they adapted TL model for obtaining the pretrained method. Next, it is optimized by adapting the convolutional layer and pruning few convolutional kernels which might interfere with the targeted classification tasks.
The working principle of the presented approach is demonstrated in Fig. 1. Initially, the images from iPad devices are obtained using camera, and users get informed for an image election. Users can choose framewise image or each image gradually. When an image got elected from the users, it would show distinguished classes, viz., abnormal and normal, to train cervical cells. This represents image is effectively loaded in scheme afterward preprocessing. When image loading is performed, image is transmitted to deep cervical predictive Web application that interacts with IoT scheme for extracting features, prediction, and training of abnormal and normal images/cells. The transmitted image consists of target labels and file names recognized using API prediction. The extracted feature is categorized by CRF classifier and forecasts the recall/accuracy rate of image classifications. Similarly, the JSON file saves computational time, viz., testing, and training needed using pretrained classifiers and models for predicting the result. For testing the system generalisability, an input test image has been provided to API predictions for detecting classes of Pap smear cells. When the classes are recognized, the result and hash code are saved or returned to smartphone/device via JSON network.
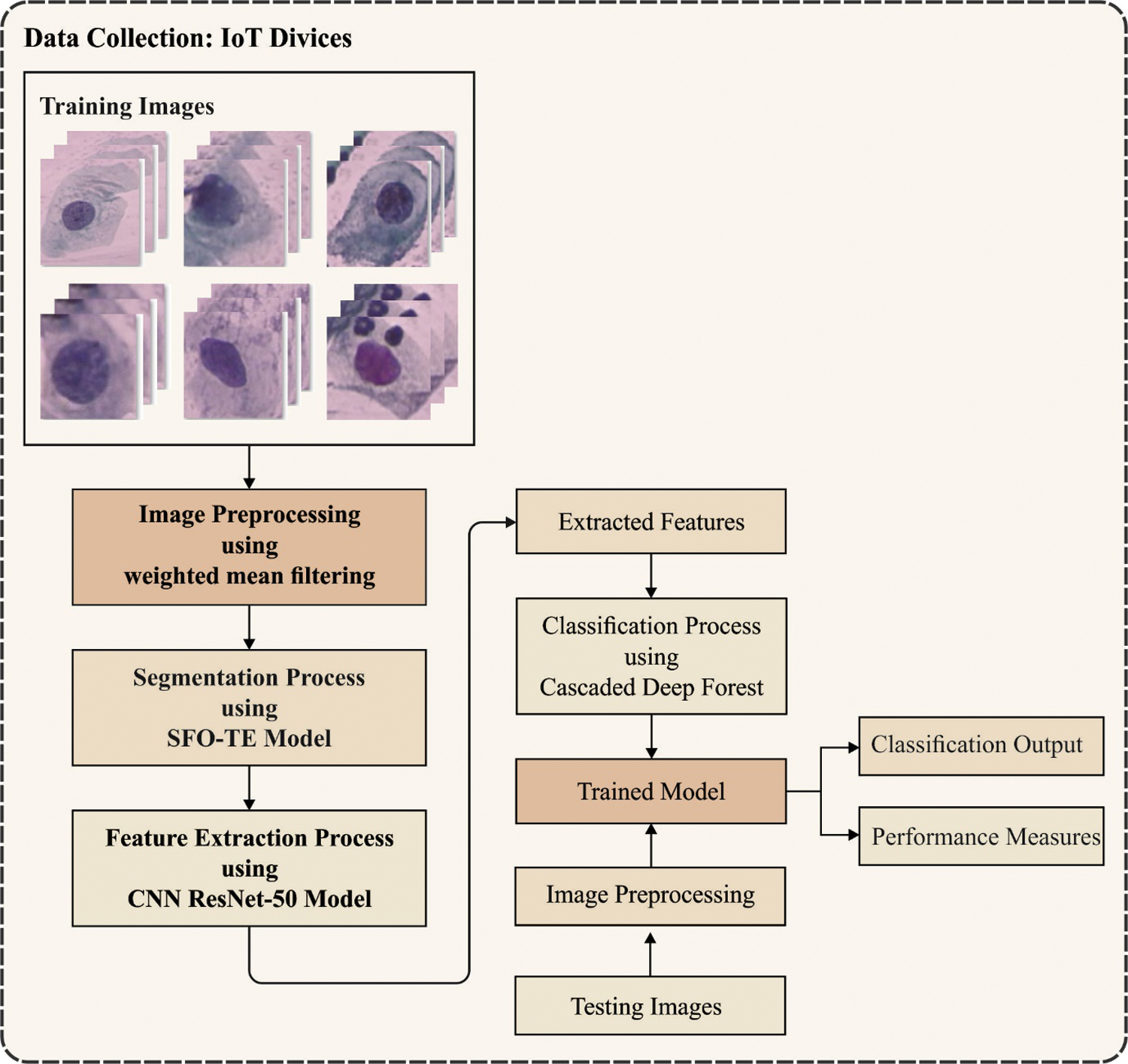
Figure 1: Overall process of proposed method
The traditional mean filter (MF) has an appropriate window size and can be employed for noise removal. But the AWMF technique makes use of a variable window size, which is distinct depending upon the minimal as well as maximal pixel values. In case, the intermittent pixel value in the window is similar to boundary values, the intermittent pixels are receiving the mean weighted value of chosen window. When the intermittent pixel values are distinct, the intensities remain same [15]. Here, the input image has a
The noisy level can be computed by
The pre-processed pap smear images are segmented using the SFO-TE technique. The entropy is associated with the chaos measure in a scheme. Initially, Shannon deliberated the entropy for measuring the ambiguity based on the data content of the scheme. The Shannon described that: if physical systems are divided into 2 statistically free sub-systems A & B, afterward the entropy value could be stated as follows:
According to Shannon's concept, a non-extensive entropy model was developed by Tsallis [16] i.e., determined by:
Whereas T represents the possible system, q indicates the entropic index, and
The Tsallis entropy could be deliberated for finding an optimum threshold of an image. Assume an image with L gray levels in the interval of
In which
In the multilevel thresholding procedure, it can be needed to define the optimum threshold value T that maximize the objective function
SFO [17] is a population based meta-heuristic technique that was simulated from the attack-alternation approach of set of hunting sailfish that hunt a school of sardines. This hunting approach provides upper hand for hunters by giving them the chance of soring their energy. It assumes that 2 populations: sailfishes as well as sardines populations. The sailfishes were assumed that candidate solution and these issues variables have been places of sailfishes from the search space. This technique attempts for randomizing the effort of search agents (both sailfish as well as sardine) more feasible. The sailfishes are regarded as scattered from the search space, but the places of sardines with use to finding optimum solutions from the search spaces.
The sailfish with optimum fitness value was known as ‘elite’ sailfish and their place at
where
where
where
where
where
where v implies the amount of variables and
3.3 ResNet Based Feature Extraction
In the feature extraction method, the ResNet50 method was implemented as a feature extractor for deriving the set of feature vectors. It is a developing network that resolves accuracy degradation problems once the network depths are improved, and their precision obtains saturate and again degrade quickly. The residual block utilizes residual function with stacked NN layer and attained topmost validation error rate of 3.57% i.e., superior to another deep network contributed in ILSVRC 2015. It is highly robust networks comprise of fifty deep layers i.e., capable of classifying thousand objects in single iteration. These networks avoid gradient vanishing problems that occur in all deep network and are complex for optimizing the classification task and interrelated features. Google Brain team integrated residual block method using GoogleNet and made innovative Inception–ResidualV2 method that decreases error rate to 3.1% that are farther superior to the outcomes attained with another pretrained method. The robustness of ResNet is the summary of skip net connections in which input is also included in the output of convolutional block in network.
Generally, training of CNN from scratch needed huge number of data, however, it is complex sometimes for gathering huge number of data for classification purposes [18]. But, in terms realtime problems, it is highly complex sometimes for matching testing and training data. It results in the summary of TL concept. It is most innovative ML models that learned the knowledge needed for solving a challenge and reutilize and employ similar knowledges for solving another challenge of any related field. On certain tasks, context networks are trained by appropriate datasets and the tasks are transmitted to certain domain i.e., trained using targeted datasets. The TL method includes pretrained problem size, similarity, and method selection. The method selections are highly applicable to the targeted problems i.e., related to the context task or problem; When the size of targeted datasets is similar/smaller to the source set, it will results in data overfitting. But, when the size of datasets is greater, afterward pretrained models require only finetuning irrespective of the step implemented that is model developments from scratch.
Finally, the feature vectors are categorized using CDF technique to allot appropriate class labels. The inspiration for taking into account the cascade Deep Forest model (CDF) in the presented cervical cancer subtype classification method is that traditional supervised machine learning classifiers usually operate with labelled data and neglect a significant number of data with inadequate data. As a result, smaller sample size of training data limits the growth in developing suitable classifiers. Furthermore, various problems might restrict the application of traditional machine leaning methods like RF and Support Vector Machine (SVM), to the tasks of cancer subtype classification. The abovementioned problem strengthens the threat of overfitting in training, i.e., considered with the help of smaller sample size and higher dimension of biology multiomics data. In addition, class imbalance is a quite usual scenario in multiomics data that increases the difficulty of module learning with the threat of weakening the capability of module approximation for larger sequence bias.
Even though many methods were lately emerged for addressing the aforementioned problems [19], constrained alternates are presented with validated methods for small scale multiomics data. Furthermore, robust and more accurate approaches are yet needed additional growths to achieve precise cervical cancer subtype classification. Alternatively, in comparison with the normal structure of convolution deep neural networks (DNN) with various fully connected and convolutional layers, also the DNN is extremely prone to overfitting, with high possibilities of convergence to local optimum, while offering relatively/imbalanced small size training data. But, regularization and dropout approaches are extensively used for alleviating that challenge, still, overfitting is an unavoidable challenge for DNN. Therefore, the advanced suggested the CDF method as an effective alternate for DNN to learn hyper level representation in a highly optimized manner. The CDF method fully exploits the characteristic of ensemble and DNN methods. The CDF method learns features of class distribution by assembling decision tree based forest when managing the input, instead of the overhead of employing backward and forward propagation approaches for learning hidden parameters as in DNNs.
The cascade forests follow a supervised learning system depending on the layers that employ ensemble RF for obtaining a class distribution of features which leads to accurate classification. The significance features in the CDF models aren't considered between multiple layers at the time of feature depiction training. Consequently, the obtained predictive accuracy is extremely influenced by several DTs in every forest, particularly with imbalanced/small scale data, as it can be crucial in the making of DT, in which the discriminative feature is employed for deciding split nodes. Fig. 2 displays the framework of applied CDF. As taking into account the employed CDF method, every level of the cascade includes 2 CRF (the yellow blocks) and 2 RF (the blue blocks). Hence, assume there is n subclass for predicting, every forest needs to output an n-dimension class vector, i.e., later connected to represent the original input.
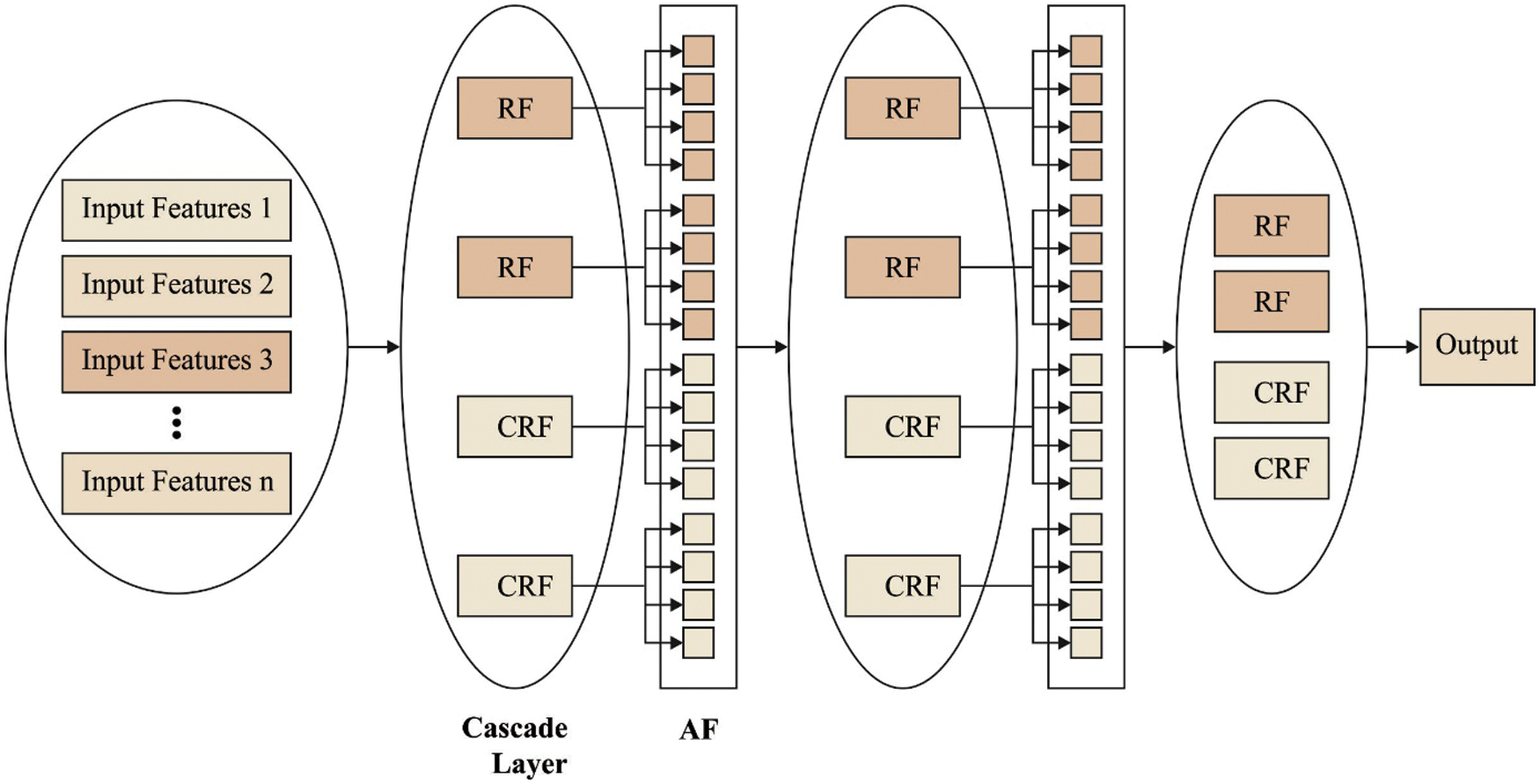
Figure 2: Structure of cascaded deep forest
This section investigates the performance of the proposed technique on Herlev database. It contains a set of 918 instances with 7 class labels. A set of 3 class labels comes under normal class (242 images) and remaining 4 classes belong to abnormal class (675 images).
The normal class includes 74 images into Superficial Squamous Epithelial (Class-1), 70 images into Intermediate Squamous Epithelial (Class-2), and 98 images into Columnar Epithelial (Class-3). In addition, 182 images fall into Mild Squamous Non-Keratinizing Dysplasia (Class-4), 146 images under Moderate Squamous Non-Keratinizing Dysplasia (Class-5), 197 images into Severe Squamous Non-Keratinizing Dysplasia (Class-6), and 151 images into Squamous Cell Carcinoma In Situ Intermediate (Class-7).
Fig. 3 depicts the confusion matrix offered by the BPSIC-CDF technique on the applied training/testing set of 80:20. The figure shown the BPSIC-CDF technique has classified a set of 73 images into class 0, 68 images into class 1, 97 images into class 2, 179 images into class 3, 142 images into class 4, 197 images into class 5, and 148 images into class 6.
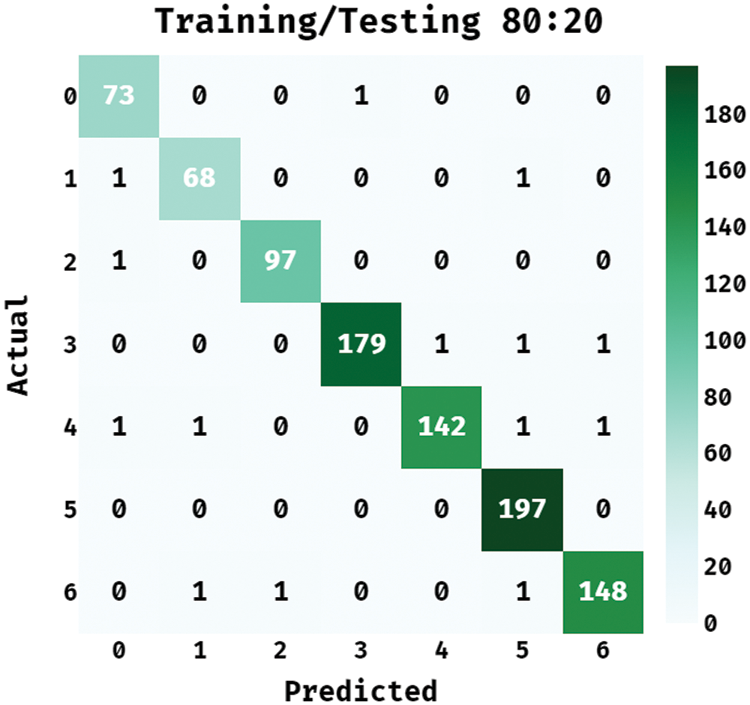
Figure 3: Confusion matrix of BPSIC-CDF model on training/testing (80:20)
Tab. 1 illustrates the classification results analysis of the BPSIC-CDF technique on the applied training and testing set of (80:20). The figure has shown that the BPSIC-CDF technique has resulted in increased classification accuracy on all the applied classes. For instance, the BPSIC-CDF technique has classified class 0 with the maximum accuracy of 0.9956, precision of 0.9605, and recall of 0.9865. Besides, the BPSIC-CDF method has classified class 2 with the superior accuracy of 0.9978, precision of 0.9898, and recall of 0.9898. Similarly, the BPSIC-CDF algorithm has classified class 4 with increased accuracy of 0.9946, precision of 0.9930, and recall of 0.9726. Moreover, the BPSIC-CDF methodology has classified class 6 with the superior accuracy of 0.9946, precision of 0.9867, and recall of 0.9801.
Fig. 4 showcases the ROC analysis of the BPSIC-CDF technique on the applied 80:20 dataset. The figure demonstrated that the BPSIC-CDF technique has showcased effective outcomes with the increased ROC of 99.8906.
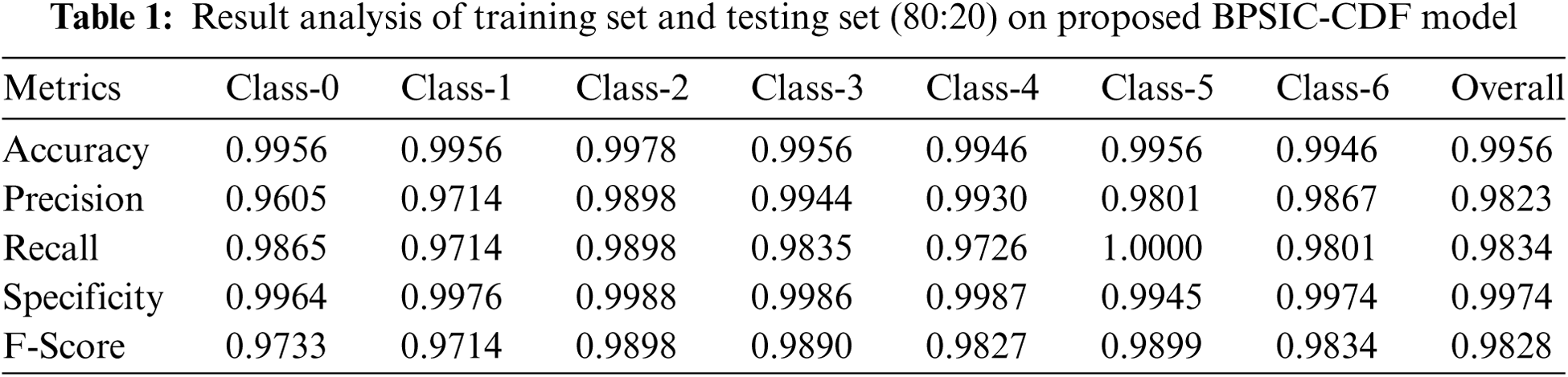
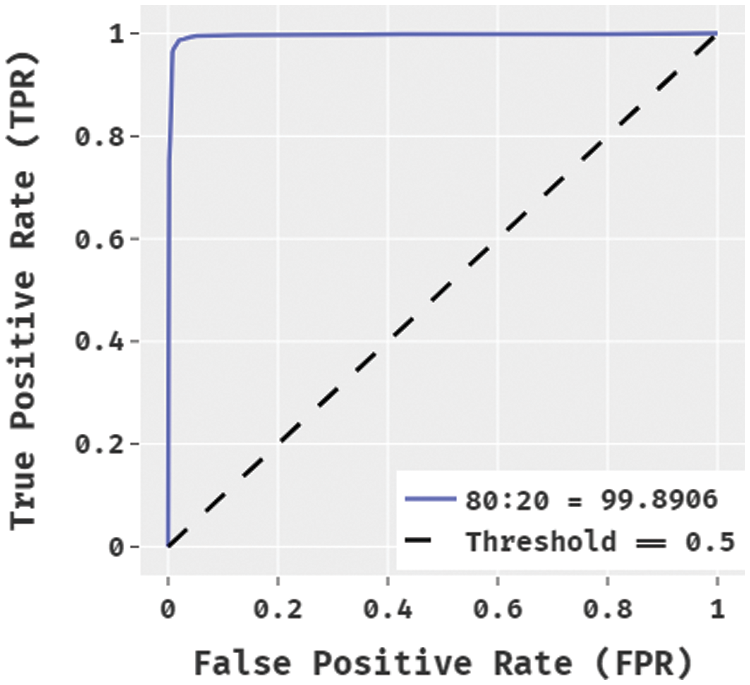
Figure 4: ROC analysis of BPSIC-CDF model on 80:20 dataset
Fig. 5 showcases the confusion matrix offered by the BPSIC-CDF algorithm on the applied training/testing set of 70:30. The figure outperformed the BPSIC-CDF approach has classified a set of 68 images into class 0, 66 images into class 1, 94 images into class 2, 177 images into class 3, 140 images into class 4, 194 images into class 5, and 148 images into class 6.
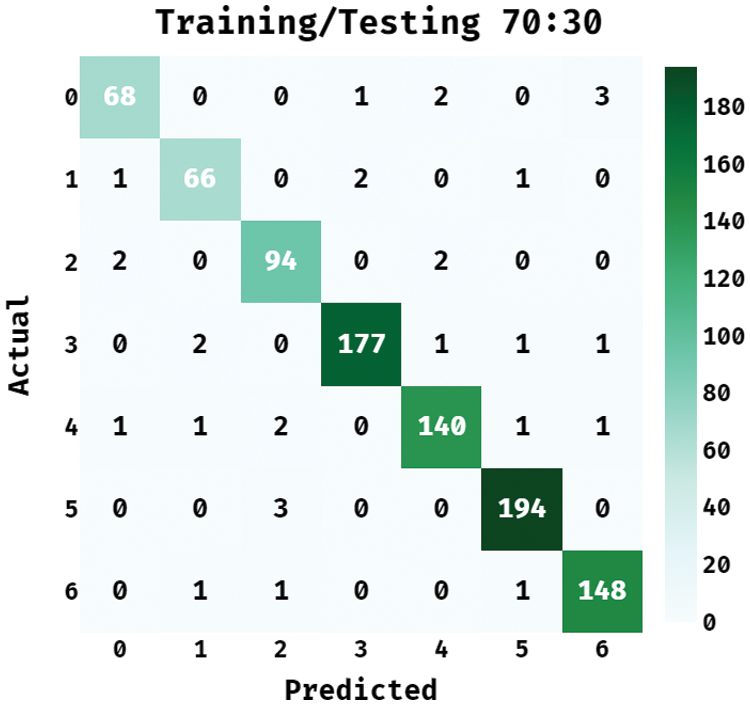
Figure 5: Confusion matrix of BPSIC-CDF model on training/testing (70:30)
Tab. 2 depicts the classification results analysis of the BPSIC-CDF algorithm on the applied training and testing set of (70:30). The figure exhibited that the BPSIC-CDF method has resulted in higher classification accuracy on all the applied classes. For instance, the BPSIC-CDF approach has classified class 0 with the maximal accuracy of 0.9891, precision of 0.9444, and recall of 0.9189. In line with, the BPSIC-CDF method has classified class 2 with the increased accuracy of 0.9891, precision of 0.94, and recall of 0.9592. Likewise, the BPSIC-CDF methodology has classified class 4 with the superior accuracy of 0.988, precision of 0.9655, and recall of 0.9589. In addition, the BPSIC-CDF manner has classified class 6 with increased accuracy of 0.9913, precision of 0.9673, and recall of 0.9801.
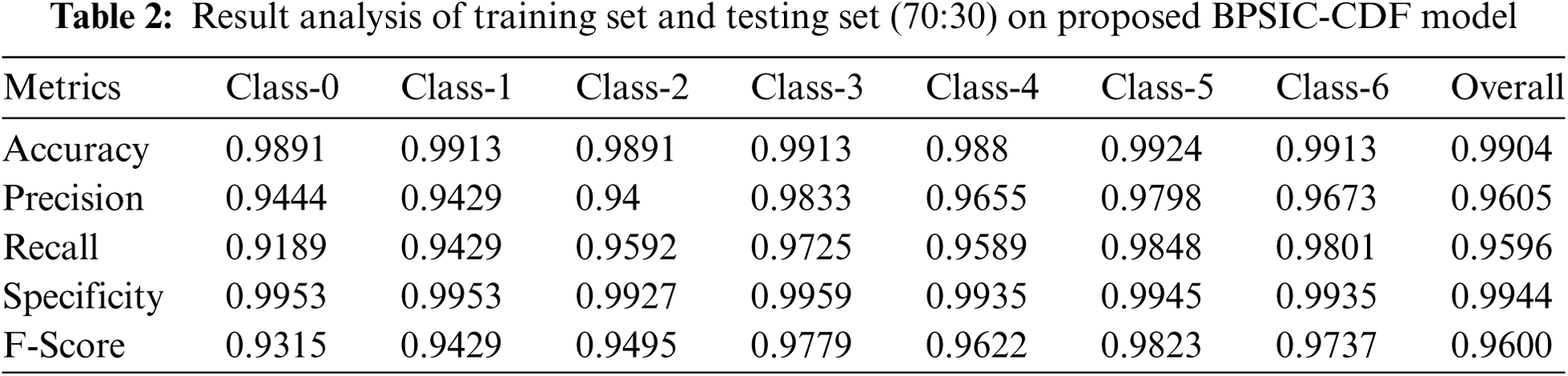
Fig. 6 demonstrated the ROC analysis of the BPSIC-CDF manner on the applied 70:30 dataset. The figure exhibited that the BPSIC-CDF method has outperformed effectual outcomes with the higher ROC of 99.6498.
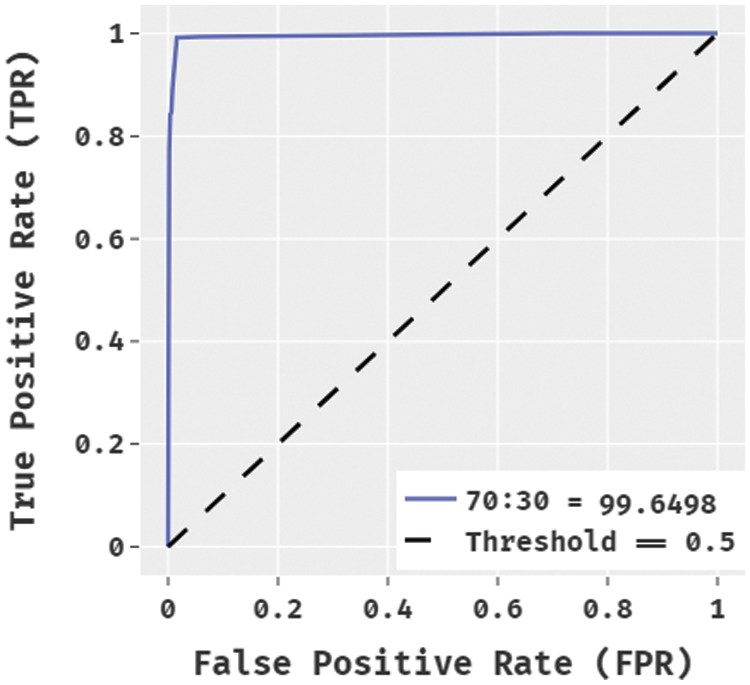
Figure 6: ROC analysis of BPSIC-CDF model on 70:30 dataset
Fig. 7 illustrates the confusion matrix accessible by the BPSIC-CDF technique on the applied training/testing set of 60:40. The figure demonstrated the BPSIC-CDF manner has classified a set of 68 images into class 0, 65 images into class 1, 88 images into class 2, 177 images into class 3, 140 images into class 4, 190 images into class 5, and 140 images into class 6.
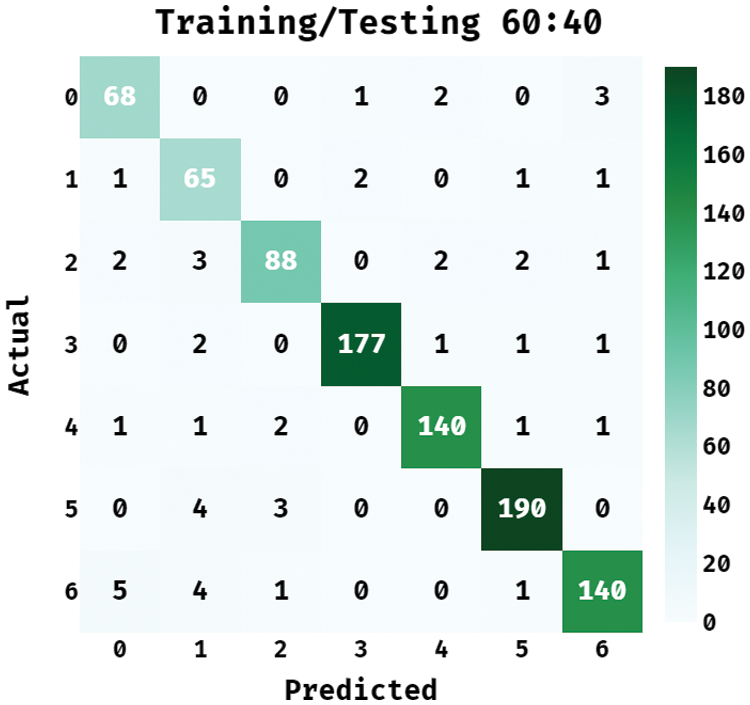
Figure 7: Confusion matrix of BPSIC-CDF model on training/testing (60:40)
Tab. 3 demonstrates the classification outcomes analysis of the BPSIC-CDF approach on the applied training and testing set of (60:40). The figure has exhibited that the BPSIC-CDF manner has resulted in improved classification accuracy on all the applied classes. For instance, the BPSIC-CDF method has classified class 0 with the maximal accuracy of 0.9837, precision of 0.8831, and recall of 0.9189. Moreover, the BPSIC-CDF system has classified class 2 with the superior accuracy of 0.9826, precision of 0.9362, and recall of 0.898. Also, the BPSIC-CDF method has classified class 4 with the maximum accuracy of 0.988, precision of 0.9655, and recall of 0.9589. Finally, the BPSIC-CDF approach has classified class 6 with higher accuracy of 0.9804, precision of 0.9524, and recall of 0.9272.
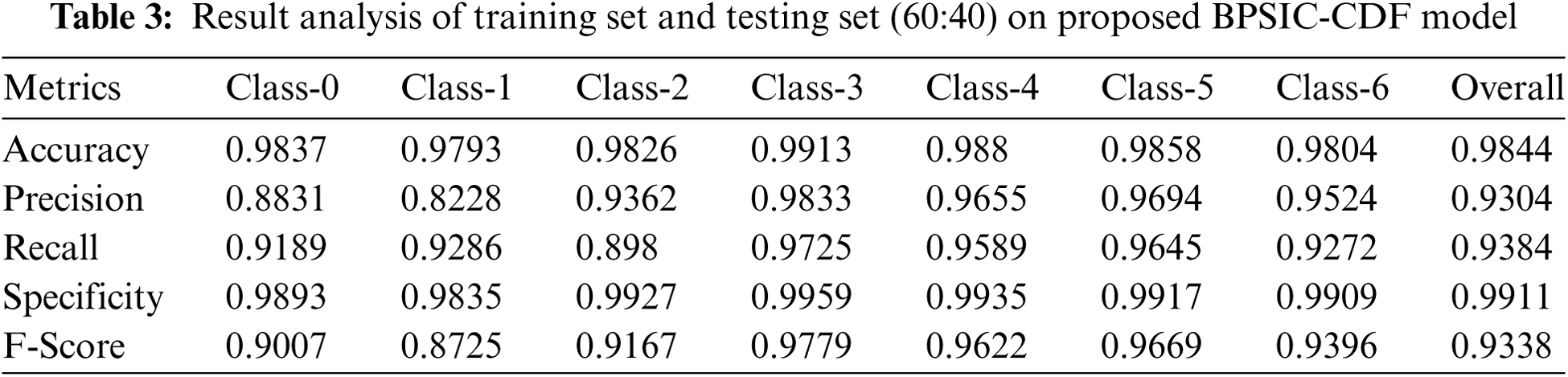
Fig. 8 illustrates the ROC analysis of the BPSIC-CDF manner on the applied 60:40 datasets. The figure outperformed that the BPSIC-CDF method has depicted effective results with the higher ROC of 99.6290.
For demonstrating the enhanced performance of the BPSIC-CDF technique, a comparative analysis is made in Tab. 4 [20]. The accuracy and specificity analysis of the BPSIC-CDF technique with state-of-art approaches exhibited that the 7-class DT technique has exhibited ineffectual outcomes with an accuracy of 91.45% and specificity of 95.01%. In addition, the 7-class K-NN technique has gained slightly improved outcomes with an accuracy of 91.68% and specificity of 95.15%. Moreover, the 2-class DT and 5-class DT techniques have resulted in a moderate accuracy of 92.66% and 92.92% respectively. Furthermore, the 5-class K-NN technique has gained somewhat considerable outcome with an accuracy of 93.31% whereas the 2-class K-NN and 7-class RF approaches have resulted in a reasonable performance with an accuracy of 94.24% and 94.76% respectively. Concurrently, the 5-class RF and 2-class RF approaches have showcased competitive accuracy of 95.67% and 98.42% respectively. However, the BPSIC-CDF technique has accomplished an increased accuracy and sensitivity of 99.01% and 99.43%.
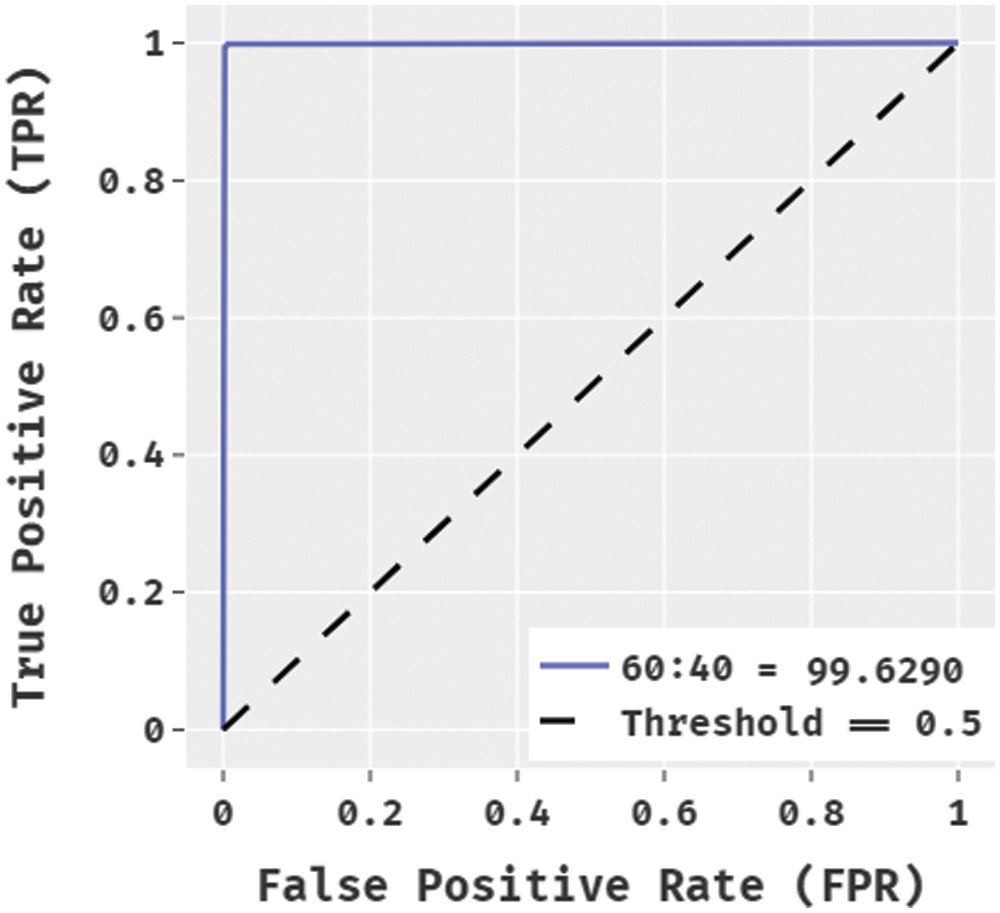
Figure 8: ROC analysis of BPSIC-CDF model on 60:40 dataset
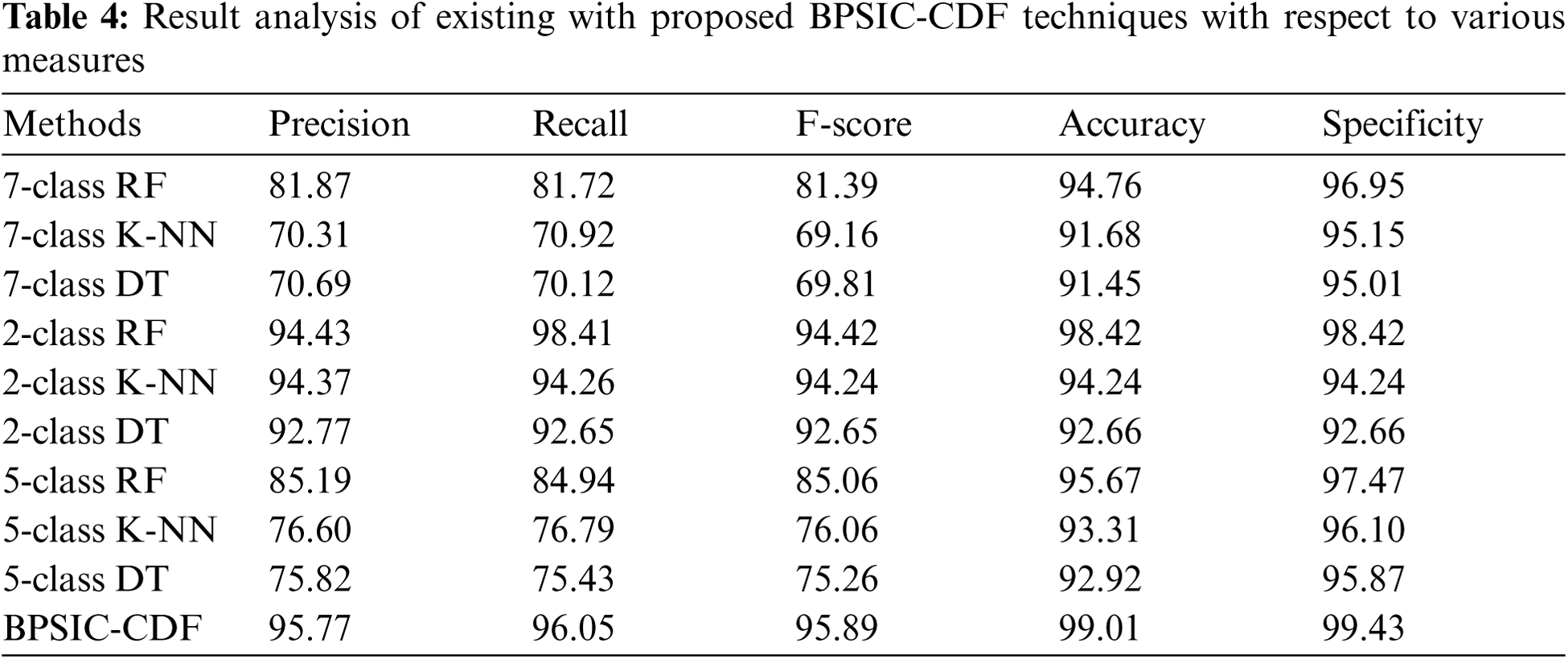
The precision, recall, and F-score analysis of the BPSIC-CDF approach with recent methods showcased that the 7-class K-NN manner has displayed ineffectual outcomes with the precision of 70.31%, recall of 70.92%, and F-score of 69.16%. Likewise, the 7-class DT approach has reached a slightly higher outcome with the precision of 70.69%, recall of 70.12%, and F-score of 69.81%. Additionally, the 5-class DT and 5-class K-NN methodologies have resulted in a moderate precision of 76.60%, recall of 76.79%, and F-score of 76.06% respectively. Besides, the 7-class F and 7-class RF techniques have resulted in a reasonable efficiency with the precision of 81.87%, recall of 81.72%, and F-score of 81.39% correspondingly. Simultaneously, the 5-class RF and 2-class DT approaches have showcased competitive precision of 85.19%, recall of 84.94%, and F-score of 85.06% respectively. But, the BPSIC-CDF manner has accomplished an improved precision of 95.77%, recall of 96.05%, and F-score of 95.89%.
In this study, an intelligent cervical cancer diagnosis model using BPSIC-CDF technique is designed using pap smear images in the IoT healthcare environment. The BPSIC-CDF technique involves IoT based image acquisition AWMF based preprocessing, SFO-TE based segmentation, ResNet50 based feature extraction, and CDF based classification. In order to showcase the improved diagnostic outcome of the BPSIC-CDF technique, a comprehensive set of simulations take place on Herlev database. The experimental results highlighted the betterment of the BPSIC-CDF technique over the recent state of art techniques interms of different performance measures. As a part of future extension, the performance of the BPSIC-CDF technique can be extended by the design of advanced DL architectures and effective feature reduction approaches.
Funding Statement: The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work under grant number (RGP 2/209/42). This research was funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-Track Path of Research Funding Program.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. W. William, A. Ware, A. H. B. Ejiri and J. Obungoloch, “Cervical cancer classification from Pap-smears using an enhanced fuzzy C-means algorithm,” Informatics in Medicine Unlocked, vol. 14, pp. 23–33, 2019. [Google Scholar]
2. T. Vaiyapuri, V. S. Parvathy, V. Manikandan, N. Krishnaraj, D. Gupta et al., “A novel hybrid optimization for cluster-based routing protocol in information-centric wireless sensor networks for iot based mobile edge computing,” Wireless Personal Communications, pp. 1–24, 2021. https://doi.org/10.1007/s11277-021-08088-w. [Google Scholar]
3. D. N. Le, V. S. Parvathy, D. Gupta, A. Khanna, J. J. P. C. Rodrigues et al., “IoT enabled depthwise separable convolution neural network with deep support vector machine for COVID-19 diagnosis and classification,” International Journal of Machine Learning and Cybernetics, vol. 12, pp. 3235–3248, 2021. [Google Scholar]
4. M. S. Hossain and G. Muhammad, “Emotion recognition using secure edge and cloud computing,” Information Sciences, vol. 504, pp. 589–601, 2019. [Google Scholar]
5. K. Shankar, E. Perumal, M. Elhoseny and P. T. Nguyen, “An IoT-cloud based intelligent computer-aided diagnosis of diabetic retinopathy stage classification using deep learning approach,” Computers, Materials & Continua, vol. 66, no. 2, pp. 1665–1680, 2021. [Google Scholar]
6. R. Bharathi, T. Abirami, S. Dhanasekaran, D. Gupta, A. Khanna et al., “Energy efficient clustering with disease diagnosis model for IoT based sustainable healthcare systems,” Sustainable Computing: Informatics and Systems, vol. 28, pp. 100453, 2020. [Google Scholar]
7. M. Elhoseny, M. M. Selim and K. Shankar, “Optimal deep learning based convolution neural network for digital forensics face sketch synthesis in internet of things (iot),” International Journal of Machine Learning and Cybernetics, vol. 12, pp. 3249–3260, 2020. [Google Scholar]
8. N. Dong, L. Zhao, C. H. Wu and J. F. Chang, “Inception v3 based cervical cell classification combined with artificially extracted features,” Applied Soft Computing, vol. 93, pp. 106311, 2020. [Google Scholar]
9. Z. Alyafeai and L. Ghouti, “A fully-automated deep learning pipeline for cervical cancer classification,” Expert Systems with Applications, vol. 141, pp. 112951, 2020. [Google Scholar]
10. A. Ghoneim, G. Muhammad and M. S. Hossain, “Cervical cancer classification using convolutional neural networks and extreme learning machines,” Future Generation Computer Systems, vol. 102, pp. 643–649, 2020. [Google Scholar]
11. A. Khamparia, D. Gupta, V. H. C. de Albuquerque, A. K. Sangaiah and R. H. Jhaveri, “Internet of health things-driven deep learning system for detection and classification of cervical cells using transfer learning,” The Journal of Supercomputing, vol. 76, no. 11, pp. 8590–8608, 2020. [Google Scholar]
12. V. Chandran, M. G. Sumithra, A. Karthick, T. George, M. Deivakani et al., “Diagnosis of cervical cancer based on ensemble deep learning network using colposcopy images,” BioMed Research International, vol. 2021, pp. 1–15, 2021. [Google Scholar]
13. S. Zahir, A. Amir, N. A. H. Zahri and W. C. Ang, “Applying the deep learning model on an iot board for breast cancer detection based on histopathological images,” Journal of Physics: Conference Series, vol. 1755, no. 1, pp. 012026, 2021. [Google Scholar]
14. P. Wang, J. Wang, Y. Li, L. Li and H. Zhang, “Adaptive pruning of transfer learned deep convolutional neural network for classification of cervical pap smear images,” IEEE Access, vol. 8, pp. 50674–50683, 2020. [Google Scholar]
15. P. Yugander, C. H. Tejaswini, J. Meenakshi, K. S. kumar, B. V. N. S. Varma et al., “MR image enhancement using adaptive weighted mean filtering and homomorphic filtering,” Procedia Computer Science, vol. 167, pp. 677–685, 2020. [Google Scholar]
16. V. Rajinikanth, S. C. Satapathy, S. L. Fernandes and S. Nachiappan, “Entropy based segmentation of tumor from brain MR images – a study with teaching learning based optimization,” Pattern Recognition Letters, vol. 94, pp. 87–95, 2017. [Google Scholar]
17. M. G. A. Nassef, T. M. Hussein and O. Mokhiamar, “An adaptive variational mode decomposition based on sailfish optimization algorithm and Gini index for fault identification in rolling bearings,” Measurement, vol. 173, pp. 108514, 2021. [Google Scholar]
18. I. Z. Mukti and D. Biswas, “Transfer learning based plant diseases detection using resnet50,” in 2019 4th Int. Conf. on Electrical Information and Communication Technology (EICTKhulna, Bangladesh, pp. 1–6, 2019. [Google Scholar]
19. Y. Guo, S. Liu, Z. Li and X. Shang, “BCDForest: A boosting cascade deep forest model towards the classification of cancer subtypes based on gene expression data,” BMC Bioinformatics, vol. 19, no. S5, pp. 118, 2018. [Google Scholar]
20. D. N. Diniz, M. T. Rezende, A. G. C. Bianchi, C. M. Carneiro, D. M. Ushizima et al., “A hierarchical feature-based methodology to perform cervical cancer classification,” Applied Sciences, vol. 11, no. 9, pp. 4091, 2021. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |