DOI:10.32604/cmc.2022.023178

| Computers, Materials & Continua DOI:10.32604/cmc.2022.023178 |  |
| Article |
Hybridization of CNN with LBP for Classification of Melanoma Images
1Faculty of Information Technology, University of Central Punjab, Lahore, Pakistan
2Department of Computer Sciences, Bahria University Lahore Campus, Pakistan
*Corresponding Author: Saeed Iqbal. Email: saeediqbalkhattak@gmail.com
Received: 30 August 2021; Accepted: 14 October 2021
Abstract: Skin cancer (melanoma) is one of the most aggressive of the cancers and the prevalence has significantly increased due to increased exposure to ultraviolet radiation. Therefore, timely detection and management of the lesion is a critical consideration in order to improve lifestyle and reduce mortality. To this end, we have designed, implemented and analyzed a hybrid approach entailing convolutional neural networks (CNN) and local binary patterns (LBP). The experiments have been performed on publicly accessible datasets ISIC 2017, 2018 and 2019 (HAM10000) with data augmentation for in-distribution generalization. As a novel contribution, the CNN architecture is enhanced with an intelligible layer, LBP, that extracts the pertinent visual patterns. Classification of Basal Cell Carcinoma, Actinic Keratosis, Melanoma and Squamous Cell Carcinoma has been evaluated on 8035 and 3494 cases for training and testing, respectively. Experimental outcomes with cross-validation depict a plausible performance with an average accuracy of 97.29%, sensitivity of 95.63% and specificity of 97.90%. Hence, the proposed approach can be used in research and clinical settings to provide second opinions, closely approximating experts’ intuition.
Keywords: Skin cancer; convolutional neural network; feature extraction; local binary pattern; classification
The skin being largest organ of the human body performs critical tasks of providing protective barrier against mechanical, thermal and physical injury, exposure to hazardous substances and light and maintaining body temperature. While performing these, the skin cells are regularly shed and new cells replace these. However, this systematic process may go awry and produce cells when these are not required. This malignant ailment of the skin, most commonly melanoma, bears the highest mortality and timely detection and treatment are thus imperative to lessen the concomitant adverse effects [1]. The underlying reason of melanoma is not fully understood, however, genetic predisposition, exposure to ultraviolet radiation and contact with hazardous materials are identified as the most common causes [2]. Melanoma is the malignant transformation of pigment producing melanocytes and therefore, these tumors are mostly brown or black albeit observed cases with pink, red or purple color when melanomas do not produce melanin [1–3].
According to the Global Cancer Statistics reported by International Agency for Research on Cancer (IARC), approximately 19.3 million new cancer patients and around 10 million deaths have been reported in 2020 [1,4]. The report further states incidence of almost 2–3 million non-melanoma and >130,000 melanoma skin cancer cases annually, worldwide. Europe and North America have significantly larger numbers of melanoma skin cancer patients according to the report of the IARC [1] and as shown in Fig. 1. In the United State of America alone, there have been 93,583 new cases (52,412 males and 41,171 female) reported in the year 2020, out of which 9,320 died due to the disease [3]. The leading causative factor reported is increased exposure to ultraviolet (UV) radiation due to depletion of the ozone layer and an additional 300,000 non-melanoma and 4,500 melanoma skin cancer cases may occur with further 10% decrease in ozone levels [5,6].
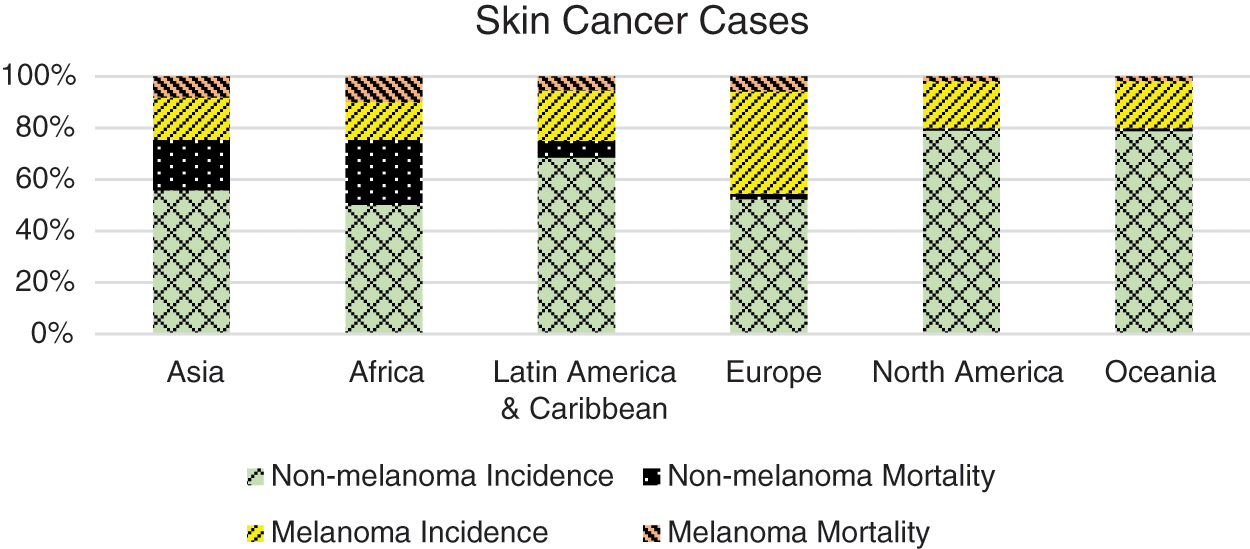
Figure 1: Continent wise ratio of skin cancer cases [1]. Europe and North America are reported with the highest number of incidences
Nowadays, computer-based technology has played a vital role in almost every field of human life. Computers are aiding humans in diverse fields where it is difficult for humans to work efficiently and identify and detect things of interest. Same is the case with detecting diseases like cancer where a significant impediment is that the early diagnosis of melanoma, even by seasoned specialists, is a hard-core process. Therefore, using a method for simplifying the diagnosis can be helpful for the specialists identifying diseases like cancer at an early stage is very important for the proper diagnosis and timely treatment [7]. Using these techniques increases the speed and accuracy of diagnostics and decreases probability of errors of physicians and pathologists.
Skin cancer occurs because of the abnormal behavior of cells or growth of anomalous cells which may be treatable if detected in earlier stages. A little ignorance may lead to death because if it is not treated timely, it starts infiltrating lymphatic system or circulatory system and reach other parts of the body [1,2]. There are many types of skin cancers out of which some are deadly and some are not deadly, thus detecting the specific type of cancer is another challenge [8]. Following list presents the main types of skin cancers deemed significant in relevant literature:
a) Actinic Keratosis (AK): It is recognized as a solar keratosis and has a very hard outlier e.g., cranky skin, dry and flaking skin. The main cause of this type of skin cancer is exposure to UV radiation.
b) Atypical moles: It is a benign mole (noncancerous) but it looks like melanoma.
c) Basal Cell Carcinoma (BCC): It is uncontrolled and abnormal growth or lesion that appear in basal cells. It looks like red patches, pink growths. Each year four million cases of basal cell carcinoma are diagnosed in the United States of America [9].
d) Melanoma: Benign melanoma is like a mole on skin usually of brown color, tan or black spot. It is may be rounded or oval whereas Malignant Melanoma looks like a sore on the skin which causes bleeding. Malignant Melanoma is the most dangerous form of skin cancer.
e) Merkel Cell Carcinoma: It is a limited, destructive skin cancer that mostly affects elderly Caucasians [9].
f) Squamous Cell Carcinoma (SCC): It is an uncontrolled and abnormal growth cells appear in the squamous cell [9].
Deep learning is a complete black box model. It presents data in a hierarchical way. Deep learning has several frameworks/models to implement a real time detection and recognition of objects like TensorFlow proposed and developed by [10], Keras proposed and developed by [11], Theano proposed and developed [12] and MXNET proposed and developed [11]. It uses Convolutional Neural Network to convolve the input image/s into several layers and predict class (object category) at the output layer. For convolution, we have used the Eq. (1), where ω represents weight and X is the value of the node of a layer. First layer's outputs are passed to the next subsequent layer and so on up to the output layer. The backbone of a training model is Gradient descent as known as Backpropagation in Neural Network. In back propagation, it processes the input images, computes the error values and compares with the ground truth values/labels using cost/error functions. Convolutional neural networks consist of convolutional layers to make an architecture which are Convolutional (1D, 2D and 3D), Pooling, Dense, Normalization and fully connected layers. In convolution layer, filtering is used in certain conditions–filter is a rectangular grid or a cubic block of neurons which slides over an image from top left to downside right. Weighted volume of the pixels is determined by Eq. (1) to get a neuron. It is used when we want to maintain the size of an image.
where * shows the discrete convolution mathematical operation on input image with filter (F). In a Neural network, after convolution layers we apply a pooling layer to reduce the size of the input image. The purpose of a fully connected layer is to reduce the spatial information, which is composed of the previously connected nodes.
The rest of the paper is organized as follows: In Section 2, we explain the basic idea of skin cancer detection. Section 3 explains the proposed framework/model using CNN, results and discussion are presented in Section 4 and in Section 5, we conclude the paper and future work.
Recently, it has been observed that cancer rates are increasing rapidly due to life-style of people; there are different type of cancers, like Actinic Keratoses, Basal cell carcinoma, Squamous cell carcinoma and Melanoma. Early detection of these cancers is curable and can save life. There are numerous skin cancer detectors made by using different techniques involving computer vision, machine learning and image processing.
Using convolutional neural networks, cancer classification is developed by Esteva et al. [13] to achieve good results. The system is designed by Iyatomi et al. [14] to classify melanomas using an automatic way. Recently Khan et al. [15] proposed a concept for multi-classification of skin cancer. The authors used a DarkNet19 with fine-tuning parameters and Multi-layered feed forward neural network and achieved plausible results. While Dorj et al. [16] proposed a system for classification of four kinds of melanoma. It presents a unified method for histopathological image representation learning, visual analysis interpretation and automatic classification of skin histopathology images as either having basal cell carcinoma or not. The authors used methodology in this paper are (1) collected images from datasets, (2) Apply preprocessing like cropping images and k-fold cross validation, (3) Using the features of convolutional neural network, they classify into different categories and finally used ECOC-SVM classifier, training and testing are performed. Using K-Means clustering and Support Vector Machine (SVM), Almansour et al. [17] proposed a method for melanoma identification. Another approach is demonstrated by Abbas et al. [9] and Capdehourat et al. [18] to recognize the dermoscopy images using Ada-boost MC. The Ruiz et al. [19] and Giotis et al. [4] developed a method for decision support system to utilize the basic image processing methods and algorithms (visual diagnostic attributes, degree of damage, texture and color analysis for melanoma) of neural networks. Automatic system for diagnosis melanoma is proposed by Isasi et al. [20] and an Android application is developed by Ramlakhan et al. [21] for its classification. They have developed an intelligent system using convolutional neural network to classify the melanoma.
Klautau et al. [22] proposed a method using deep learning and image representation learning that combines different layers of neural network (convolutional, auto encoder) and use soft-max classifier to detect cancer from melanoma images and interpret of visual analysis.
The novel approach is inspired from image feature representation learning and deep learning, proposed by Klautau et al. [22] and yields a deep learning architecture that combines an auto encoder learning layer, a convolutional layer and a soft-max classifier for cancer detection and representation of visual analysis. Different strategies are used for image representations like wavelet coefficients, Graph Representations and Discrete Cosine Transform (DCT). These strategies are combined with deep learning architecture to form multiple linear and non-linear data transformations by Capdehourat et al. [18]. To diagnose Basal Cell Carcinoma (BCC), Cruz et al. [23] proposed auto-encoder deep learning architecture for histopathology image classification. In this method, the interpretation layer recognizes the most contributed tissues. A unified deep learning model has different layers for image feature representation, image classification and prediction recording output. Using these approaches, authors visualize performance of classification.
Malignant melanoma is the most war-like and existence-danger of skin cancer. It is an uncontrolled and rapid growth of skin cells and it is easy to recover if diagnosed in its early stages. The stage of a cancer decides the survival rate of malignant melanoma. If malignant melanoma gets diagnosed in its early stages, then it could reduce the risk of death [24].
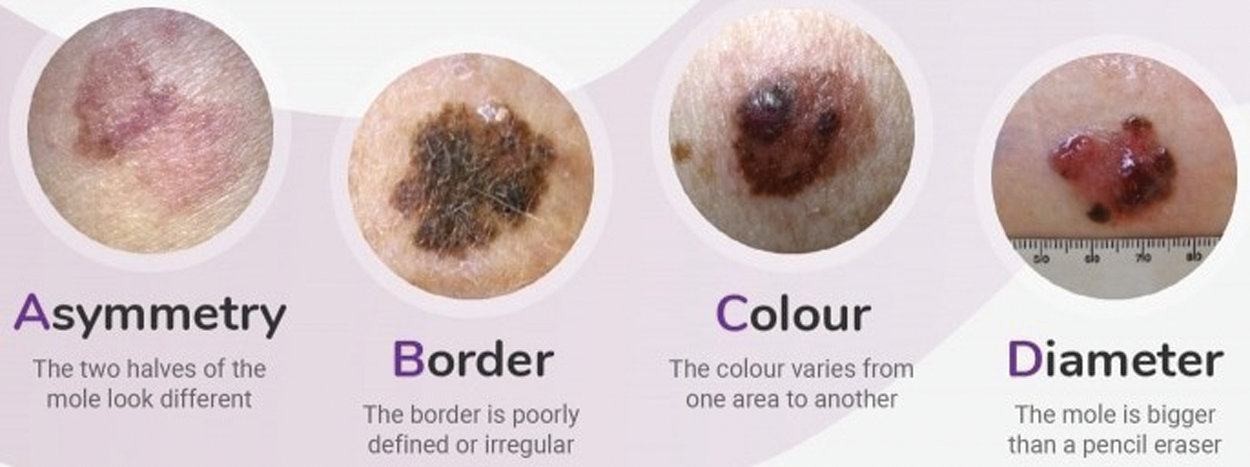
Figure 2: ABCD parameters: Asymmetry, Boundary irregularity, Color and skin lesion Diameter. [25]
For early detection of skin lesions different methodologies of Computer Vision and Machine Learning are used. The Intelligence system helps us to differentiate between malignant melanoma and benign lesions (non-cancerous). Fig. 2 depicts four types of parameters asymmetry, color, boundary irregularity and skin lesion diameter which is known as ABCD analysis depicted in [20]. Due to malignant melanoma, the anatomical structure of skin cells is agitated and the skin pattern is disrupted. As a feature set, disorder of the skin pattern is used to find out whether it is malignant or benign skin lesion. In previous procedures, the normal White Light Clinical (WLC) image is used to extract the skin pattern by high-pass filtering explored by [19] and the skin line direction and for lesion classification, the skin line intensity was used by processing a small image set proposed by [26].
These newly explored features have not been composed with ABCD (asymmetry, boundary irregularity, color and diameter of skin lesion) till now. All new features should be combined to enhance classification performance. If we combine the skin pattern feature and ABCD features, then that will be good to increase lesion discrimination as compared to the use of each feature alone. First of all, two features will be extracted from skin pattern and then the computational algorithms will be used to calculate the ABCD features and later, the lesion classification was conducted. The classifications using individual or combined the features and in the end, the results will be declared [24].
Boundary information is used for shape analysis of skin lesion classification. Shape classifies skin lesion images to be benign (non-melanoma) or malignant. Shape descriptor describes a given shape by some set of features vectors. In literature, Zhang et al. [8] proposed methods, that are used to describe shape i.e., Chain code to extract the boundary images, region, contour-based shape descriptor, curvature scale space is described [25] and shape signatures are developed by Davies et al. [26]. The author used Fourier descriptors to extract shape features from skin lesion images. For skin lesion shape analysis, it extract the coordinates of all possible points of the skin lesion image and forward them to a set of (xn,yn) (where N indicates the number of points on the boundary, n = 0, 1, 2, 3, …, N).
The most explored/advanced areas in a Computer Science discipline is Artificial Intelligence or Machine Learning and Computer Vision. In Machine Learning, we trained our framework/model using datasets to make them a decision-able system. At the end, the system will provide the most reliable output against inputs. Sometimes the outputs are not according to our requirements [27]. Computer Scientists have resolved this issue using deep learning and several algorithms (Convolutional Neural Network, Fully Convolutional Residual Network (FCRN) [28], fully convolutional network [29] and U-NET [30]). Deep learning has a hierarchical learning method which uses communication pattern and biological nervous system using extensive hardware's (GPU & TPU) and optimized algorithms (TensorFlow, Theano).
Several research communities have done good work on melanoma detection in dermoscopy images using Convolutional Neural Networks. Different methodologies and techniques are used to classify skin lesion images, [24] analyzed texture in skin images and found good results using baseline statistical methods. El Abbadi et al. [31] Wiener filter is used to remove noise and thresholding is used to segment the whole images. Abd ElGhany et al. [32] have used CNNs and different Fine-tuning methodologies for preprocessing and segmentation to achieve a plausible result.
Reportedly, millions of people are diagnosed with skin cancer on a yearly basis and late diagnosis becomes a major reason for death in many cases. To improvise the detection and ensure timely diagnosis, computer aided diagnostic mechanisms are designed through the use of deep learning techniques. Thomas et al. [3] have worked on most common types of skin cancer through the use of semantic segmentation in order to interpret full context of skin tissue types. The proposed method has achieved a high classification accuracy of 93.6%.
Another study by Tougaccar et al. [33] has proposed the classification of malignant and benign skin tumors. The data set rebuilt by auto-encoder model was fed to MobileNetV2 model for training. The study claims to achieve the classification success rate of 95.27%. Adegun et al. [34] have also presented a comprehensive survey of state-of-the-art techniques employed for skin cancer detection. The survey was aimed analyze the performance of the readily available techniques and their performance in order to allow the researchers to design more efficient and competitive models for skin lesion classification.
Pacheco et al. [35] have presented their work on ISIC 2019 dataset and used different approaches to handle the skin classification problem. Initially, they have used 13 different pretrained CNN models and all were fine-tuned with Adam optimizer. They have applied ensemble technique on the pretrained models to consider the majority voting, average and maximum probability. For outlier classification, they used hierarchical and entropy estimation. At the end, for exploration and classification of the meta-data of ISIC2019, they used histogram method. They have just used pretrained models with fine-tuned and Adam optimizer along with ensemble classifiers.
Tan et al. [36] proposed the concept of intelligent decision support system for skin cancer detection. They have combined different structure features such as color, border irregularity and texture features such as Local Binary Patterns and Histogram of Oriented Gradients (HoG) operators. After feature extraction, they have applied Particle Swarm Optimization (PSO) techniques to enhance the feature optimization. The dataset was acquired from UCI and ALL-IDB2 for experimentation and evaluation of the model. Although they have used different handcrafted features extraction methodologies with Partical Swarm Optimization (PSO) approach for analyzing skin lesions, however, it did not incorporate the customized and hybrid Convolutional Neural Network for classifications of skin lesions.
Srinivasu et al. [37] have used ISIC-2019 dataset (HAM10000) and applied pretrained CNN model (MobileNet V2) and LSTM. The advantage of MobileNet is that it requires less computational resources as compared to traditional CNN models. They have used statistical approach Gray-Level Occurrence Matrix (GLCM) for assessing continuous growth of skin lesion. It assesses and analyses the association between pixel texture and pixel intensity. The pretrained model (MobileNet V2) and LSTM is evaluated on ISIC2019 dataset and evaluate performance on different measurement metrics. Although they have used different pretrained models with ensemble classifier and used statistical approach for assessing skin lesion, however, it did not incorporate the local and discriminative features of skin lesion.
The thorough and detailed literature review suggests that hybrid Convolutional Neural Network techniques with Local Binary Pattern for detection and multi-classification of skin lesions have not been employed previously for the extraction of local and atomic features. Especially in skin lesions classification, local features consist of the inner details of skin lesions. For the precise and accurate classification of medical images (especially deadly diseases), we require appropriate and pertinent features from the dataset in a shorter time with the usage of minimum computational resources. Thus, we propose a hybridized Convolutional Neural Network with Local Binary Patterns to acquire the local and atomic features from the ISIC dataset to classify the skin lesion at multiple levels. In this study, we apply data augmentation techniques, preprocessing to remove hair (combination of different filters with morphological operations and inpainting techniques) from the skin lesion images and contrast enhancements using top-hat and bottom-hat approaches. After preprocessing, we have applied our LBPCNN model with different filter sizes to acquire plausible results (accuracy, sensitivity and specificity) and compared them with pretrained models.
In convolutional neural networks, layers are adjacent and connected to each other. Each node is connected to nodes of the adjacent layer. Input layers have pixel intensities as values. We have been using 16,384 input neurons for the 128 × 128-pixel images. When input values are passed to adjacent layer neurons, it contains random weighted values and are then moved to the next adjacent (hidden) layer and finally results are received by output neurons.
The main theme of this paper is to classify skin lesion images using convolutional neural network and classify according to Skin Cancer types (Basel Cell Carcinoma (BCC), Actinic Keratoses (AK), Melanoma and Squamous Cell Carcinoma (SCC)).
The results are obtained from 11529 images (which are downloaded from ISIC dataset (Combination of ISIC 2017, 2018 and 2019 (HAM10000)–327 of Actinic Keratoses, 379 of Basel Cell Carcinoma (BCC) and 153 of Squamous Cell Carcinoma (SCC))). Tab. 1 depicted the number of testing and training images for four types of Skin Cancer.

A convolutional neural network is a special type of network. In terms of machine learning, it is a class of deep or even may be shallow as argued by some, feed forward artificial neural networks that is applied to analyze visual imagery. CNN's were inspired by the biological process of the visual cortex. CNNs require relatively little preprocessing as compared to other image classification algorithms. A CNN like any other neural network consists of an input and an output layer and a single or multiple hidden layers. The hidden layers of a CNN typically consist of convolutional layers, pooling layers, fully connected layers and normalization layers. Convolutional neural networks are best for inputs which have some relation with other inputs. Due to it images are very suitable for CNN's as in an image, almost every given pixel has a relation to a pixel in its neighborhood. In Convolution Layers, filtering is used in certain conditions. A filter or Kernel is a rectangular grid or a cubic block of neurons which is slide over an image from top left to downside right. The number of steps or the number of jumps to convolve a filter or Kernel with an image is called striding. Padding means adding extra layer pixels on the edges of an image. It is a modified convolution which applies to input data with defined gaps – 1, 2 and 3, depending upon your image size. During convolution, it skipped the zero-pixel value from the light blue pixels. The receptive field is 7 × 7 and 15 × 15 when dilated convolution is 2 and 4 respectively. MaxPooling is a sample-based discretization process. Its objective is to down sample the input which in case of CNN's are images. Applying max pooling on an image reduces its dimensionality. It reduces the number of parameters within the model, to improve the speed of the network and generalizes the results from a convolutional filter. Dropout is defined as a regularization technique for reducing over-fitting in neural networks training data. The dense layer is a fully connected or a regular neural network layer which means that all the neurons in one layer are connected to those in the next layer. Flattening layer is the process of converting all two-dimensional arrays or a matrix into a single linear vector. The flattening step is needed because a fully connected layer is like a regular neural network layer so the input needs to be a vector.
In this paper, we propose a hybridized convolutional neural network framework to explore the main preprocessing task of skin cancer images collected from ISIC 2017, 2018 and 2019 – HAM10000). Before using convolutional neural networks, we have used the basic image processing steps depicted in the Fig. 3. Following steps are used for feature extraction of skin cancer.
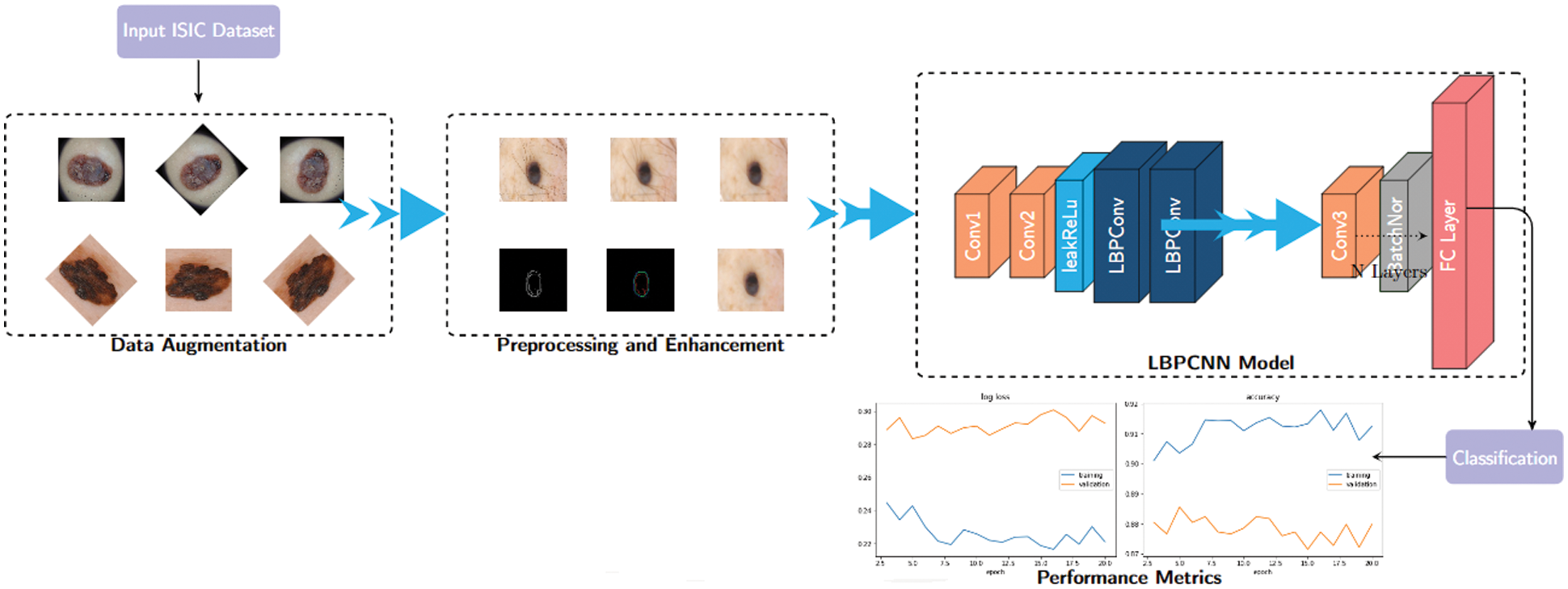
Figure 3: The basic structure of our proposed hybridized CNN–we have defined non-trainable filters with help of Bernoulli distribution to determine difference map and bitmap. Our proposed LBPCNN model have 17 layers (Convolutional layers with Rectified Linear Units, max pooling layers [38] (2 × 2), Dropout layer and Batch Normalization layers) including fully connected layer with stride one and padding value is zero (`SAME'). We have set the batch size of 50 and trained our network with the most successful classifier ADAM [39]
a) Image Acquisition: We have prepared a dataset collected images from ISIC 2017, 2018 and 2019 dataset. The total high quality RGB images of skin cancer are 11529 (Dimensions (pixels) 1022 × 767) with (Basel Cell Carcinoma have 2829, Squamous Cell Carcinoma have 2776, Actinic Keratoses have 2710 and Melanoma have 3214 images). Normally some images have noise such as organs, hairs or tools. These images are further passed to preprocessing steps (remove hair using filters and resizing). These images have four classes of skin cancers (Melanoma, Squamous Cell Carcinoma, Actinic Keratoses and Basal Cell Carcinoma) and are saved in different directories for testing and training.
b) Image Preprocessing: In this step, we adjust the size of each image to 128 × 128 pixels. We are using color images to detect skin cancer, color plays an essential role in detection of skin lesions, because it varies in colors (brown, blue, black and pink). We presented a full color image (RGB) to an array of pixels. Background color is determined by averaging the lesion image and subtracting it from the whole image. Different research methods (histogram analysis, lesion/object feature space) are used to analyze color skin lesions. We have used a histogram of color analysis for finding relevance among four categories (Melanoma, Squamous Cell Carcinoma, Actinic Keratoses and Basal Cell Carcinoma).
Further, we have removed noise (hair and unwanted objects) by using several filtering algorithms (Gaussain – Eq. (2), 3 – CImg(x,y) is a Gaussian filtered image, ck(k, l) is a kernel and ColImg(x,y) is a colored image).
For filtering, we have used the input image as shown in Fig. 4 and applied Gaussian and median filtering with a 7 × 7 kernel. Median filters and different combinations of morphological operations can help to remove all possible from the skin image. We apply black-hat and closing morphological operations on the hairy images. After morphological operations, we prepare and intensify the hair contour to apply inpaint methods to remove the hairs from the skin lesion images. Fig. 4 presents the removal of all possible hairs from the skin lesion images.
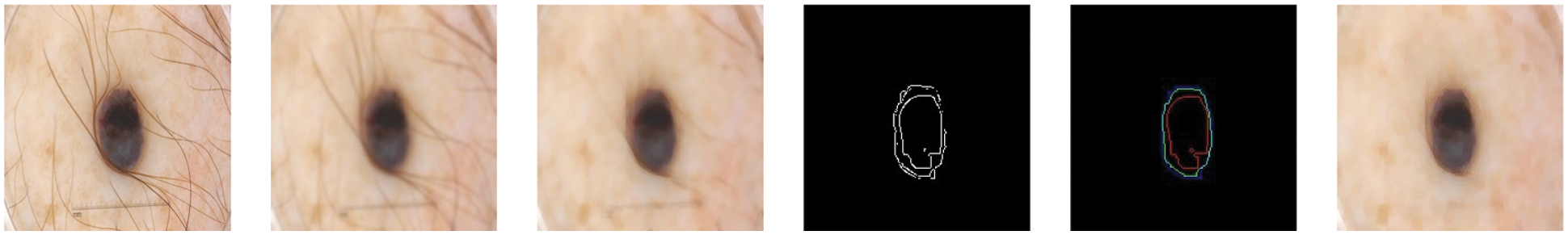
Figure 4: For filtering, we have used the hairy image (1st image) and Gaussian (2nd image) and Median filtering (3rd image) with 7 × 7 kernel. Median filters remove all possible hairs from the skin lesion images and give plausible results. After filtering, different morphological approaches applied with Canny (4th image) and Laplacian (5th image) operations and performed to find proper shape of a lesion (6th image)
After filtering, we used several edge detection algorithms (Canny and Laplacian). The Laplacian algorithm gave us positive results as compared to others (Canny). After edge detection (IMed(x,y)), we multiply the edge detected image with image that contains shape of lesion (IMLes(x,y)) using Eq. (4). It deletes the unwanted information (unwanted parts of skin lesions).
After deleting the unwanted information, the next state is morphological operation to close the open shape of skin lesion using a disk-shaped structuring element with radius-2.
c) Image Enhancement: After removing hairs from the datasets, we apply Top-hat and Bottom-hat morphological operations for contrast enhancement and background equalization. In top-hat operation, we apply opening operation on an input image with Structural Element (SE) and then subtract from an input image to get the bright object from the original image. In bottom-hat morphological operation, we apply closing operation on an input image with SE and subtract from the original image to extract a darkened object. For image enhancement (equation depicted in 7), we subtract top-hat from bottom-hat and apply addition operation with an input image to get a smooth and visually enhanced image. Eq. (6), 5 depict the Bottom and Top-Hat problem where ι is an input image and κ is a Structural Element (SE). In Eq. (7), we get the enhanced image, where ηimg presents Top-Hat and τimg is Bottom-Hat morphological operations.
d) Local Binary Pattern (LBP): Ideally for texture recognition, we use Local Binary Pattern (LBP) to detect/recognize or classify abnormality in skin lesions. Medical image analysis has two sub-problems (1) the best feature extraction method (2) and a dedicated classifier. Even the best classification methods fail when poor feature extraction methods are used. Local Binary Pattern (LBP) is normally used in computer vision and image analysis applications. It has simple implementation and low cost of computations. It requires two user-defined parameters, P (# of neighbors) and R (radius of comparisons).
where gpand gcrepresent the neighbor and central value of the gray pixel respectively. P and R are total neighbor pixel values and radius of the neighborhood respectively. The sign function s () is used to find the movement direction. For origin coordinates (0, 0), the coordinates are found by sign function (-r sin(2πn/p), r cos(2πn/p)) where r and p present as radius and neighborhood pixel respectively. We compared the center values with the neighbor pixel values and if the center value is less than the neighbor value then assigns it 1 else assign it a 0 value. After that the complete bit string is converted into decimal value on base 2 and make a vector (v = [10001101] = [1*27 + 0*26 + 0*25 + 0*24 + 1*23 + 1*22 + 0*21 + 1*20]). The main crux behind LBP is the activation function (Heaviside step function 9) is used to achieve the results. But it fails in high ordered complex CNN to achieve non-linearity, because the gradient descent would not be achieved when the derivative of x is zero.
Following are the basic LBP parameter to extract features from the patch of an image:
• Base: It takes any real values for weights to encode the LBP descriptor.
• Pivot: For comparing the patched intensity of the pixel, it chooses the center value as pivot. Choosing different intensity values in the patch gives us different local textures.
• Ordering: It partially encodes/preserves the local texture for a specific order (clockwise or anti-clock) by choosing different neighborhood sizes (P = 8, 12 and 16) and pivot values.
The LBP equation returns a minimum value of circularly rotated. The circular right rotation is represented by ROR (x, i) of bit sequence x by i steps.
We are extracting super pixels to capture the local clue (color, gradient pattern and texture) of an image. To handle different levels of magnification, we are using different sizes of kernels (3 × 3, 5 × 5 and 7 × 7) with striding and padding value set as 1; pooling layer is max-pooling (with stride = 2) – The network is translation invariant and extracts the most pertinent features like edges and we have used three fully connected layers. For each dimension the number of output feature of a layer is calculated by the following Eq. (11) where foutis number of output features, finis number of input features, s is strid size, k is kernel/filter size and p is padding size.
In our experiments the function used is LeakyRelu in all layers where activation is required. This caters for negative activations from neurons which are otherwise set to 0 when the ReLU is used. In our experiments, we have a set value of ψ = 0.01. It is noteworthy that although the ReLU and its modification, LeakyReLU, activation functions minimize the problem of vanishing gradients observed with Sigmoid activation functions, these functions are discontinuous and undefined when ϕ = 0. Therefore, these have piecewise derivatives. We have used LBP with our proposed novel hybrid CNN model that is depicted in Tab. 2. Using deep convolutional neural network, we have extracted features from datasets that are classified into four classes i.e., Basel Cell Carcinoma (BCC), Actinic Keratoses (AK), Melanoma and Squamous Cell Carcinoma (SCC). The proposed convolution model has 17 layers (with different filter sizes 3 × 3, 5 × 5, 7 × 7 and 11 × 11) with padding and stride values as 1, with three fully connected layers and pooling layer is max-pooling (with stride 2). For rectification of non-linearity, we have used ReLu in all hidden layers.

Input Layer: We have passed skin cancer raw images with a size of 128 × 128 that are composed of prominent features. For convolution filter size is 64, stride is one and padding is zero (‘SAME’). Total number of parameters is 4864 ((FilterHeight * FilterWidth * InputImageChannels + 1) * NumberOfFilters). Similarly, we can calculate the total number of parameters with a minor change that is InputImageChannels = NumberOfFilters (from the previous layer).
Local Binary Pattern Convolutional Layer: We use Local Binary Pattern Convolutional Neural Network to achieve highly efficient statistical and computational results. It is a hybrid combination of constant and variable (learned) weights. The concept of LBPCNN layers with difference maps, bit maps and feature maps. During the calculation difference map, we have used non-trainable kernels and used a set of learn-able linear weights. The non-trainable kernels comprise values from −1, 0, 1 using Bernoulli distribution. We apply 1 × 1 Convolution layer to obtain a feature map. The loss function of LBPCNN layers is back propagated as exactly the same method they are used for learnable linear weights. However, during training, 1 × 1 kernels are updated and the anchor weights are immune. The size of anchor weights is dependent upon the number of input channels (i), number of spatial sizes (x × y) and number of intermediate channels (k). Initially, we determine sparsity level and randomly assign non-zeros value −1, 1 according to the percentage of the weights with the help of Bernoulli distribution.
The feature maps are obtained from the proposed LBPCNN layer which is a linear combination of different intermediate bitmaps and anchor weights. Each patch of a bitmap is composed by convolving the input map of image with a predefined kernel and intermediate channel (k) and non-linear activation functions. The respective feature map is extracted from multiplication of intermediate bitmap with convolutional kernels and parameters (t1, t2, t3, …, tn). The process is depicted as:
where F is a matrix of number of input channels (i), number of spatial size (x × y) and number of intermediate channels (k) and imagex is a smaller patch of an image.
Batch Normalization: For reducing the problem of vanishing gradient, to regularize the model and achieve a high learning rate, we used batch normalization.
The minimization of covariance shift is achieved by the batch normalization layers in the network. If we consider B as set of mini batch of x1 ⋅⋅⋅xmtraining samples, the batch normalization uses second central statistical moment of σ2 to normalize the mini batch as given in Eq. (18) which is, the batch normalization subjected to scale and shift using Eq. (19).
where μ is the arithmetic mean of x1 ⋅⋅⋅xm samples within the mini-batch β and ε is a constant for numerical stability [33]. The normalized activation xi^ are internal to the transformation and the γ is for scaling and β for shift are used in the linear transformation given in Eq. (19).
In literature review, there have used different Convolutional Neural Network with diversified configurations that have been used filter size of 11 × 11 with stride 4 [40] and 7 × 7 with stride 2 to 3 in achieve plausible result. We have used all other filters (as mentioned above) and choose 3 × 3 in our proposed network with local binary pattern. We have used smaller filter for getting information from the most prominent features that have been captured in 3 × 3 filter size and others missed in larger filters.
For demonstration and evaluation of the LBPCNN model, we have used a Lenovo System with 8GB RAM, processor is Intel Core i7-7700 CPU @ 3.60 GHz × 8 and Graphic Card is GeForce GTX 1050 Ti/PCIe/SSE2. For implementation, we have used pyTorch and other python libraries for preprocessing of ISIC dataset.
We have applied various data augmentation techniques to achieve In-Distribution Generalization: generating examples that are novel but drawn from the same distribution as the training set. We have used different data augmentation techniques such as sequential rotation (45◦), shearing with a factor of 0.2, horizontal and vertical flipping in our proposed LBPCNN model. The same data augmentation techniques are used for pretrained models. Using data augmentation, we achieve rotation invariant and resolve transformation issues and avoid over-fitting problems in ISIC dataset. We also performed In-Distribution generalization on the ISIC dataset and class imbalance problem in order to achieve better performance.
For training of our proposed model and pretrained models, we have used pyTorch with other python based libraries. Our proposed model comprises of a combination of convolution, Local Binary Pattern (LBPCNN) layers with ReLu and Batch Normalization layers. The approach of LBPCNN layers is depicted in Fig. 5 that comprises a non-trainable filter and Bernoulli distribution to find the values of bitmap and difference map. The proposed model performance is evaluated using 5 fold cross validation with different kernel size. K-folding enlarges the available data for training and testing models to assess the performance. It's a pretty straight forward method to divide the dataset into various equal size chunks. We repeat the training and testing experimentation up to 5 times as shown in Fig. 6. The purpose of the k-folding is to shuffle the dataset and optimize the performance of CNN models and properly handle the unseen data.
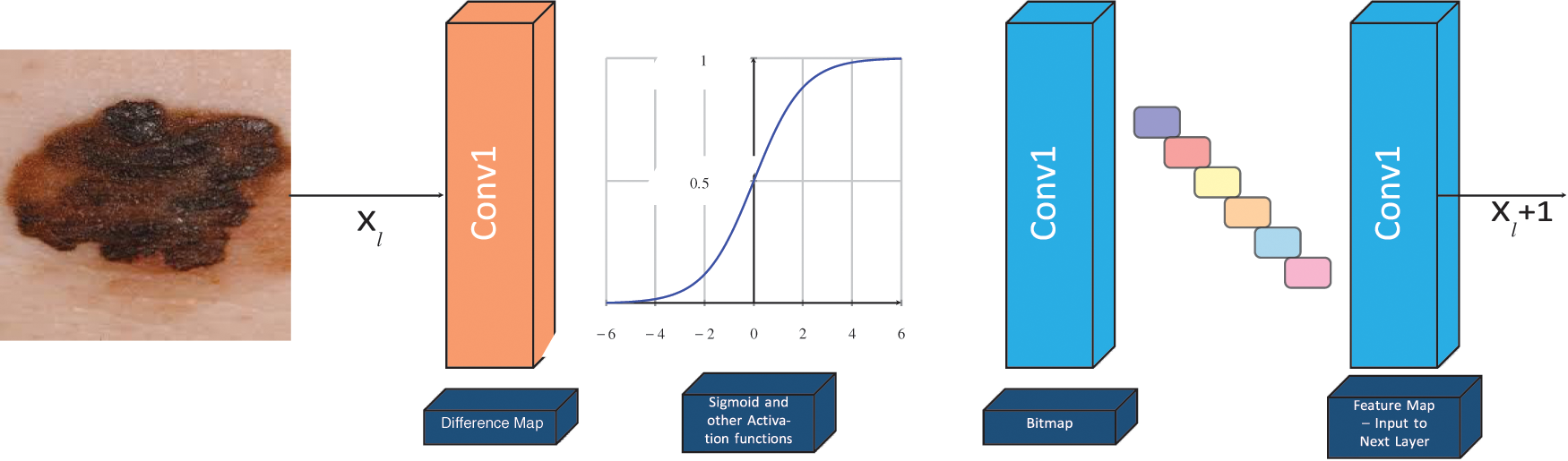
Figure 5: The basic structure of LBPCNN -- we have defined non-trainable filters with help of Bernoulli distribution to determine difference map and bitmap
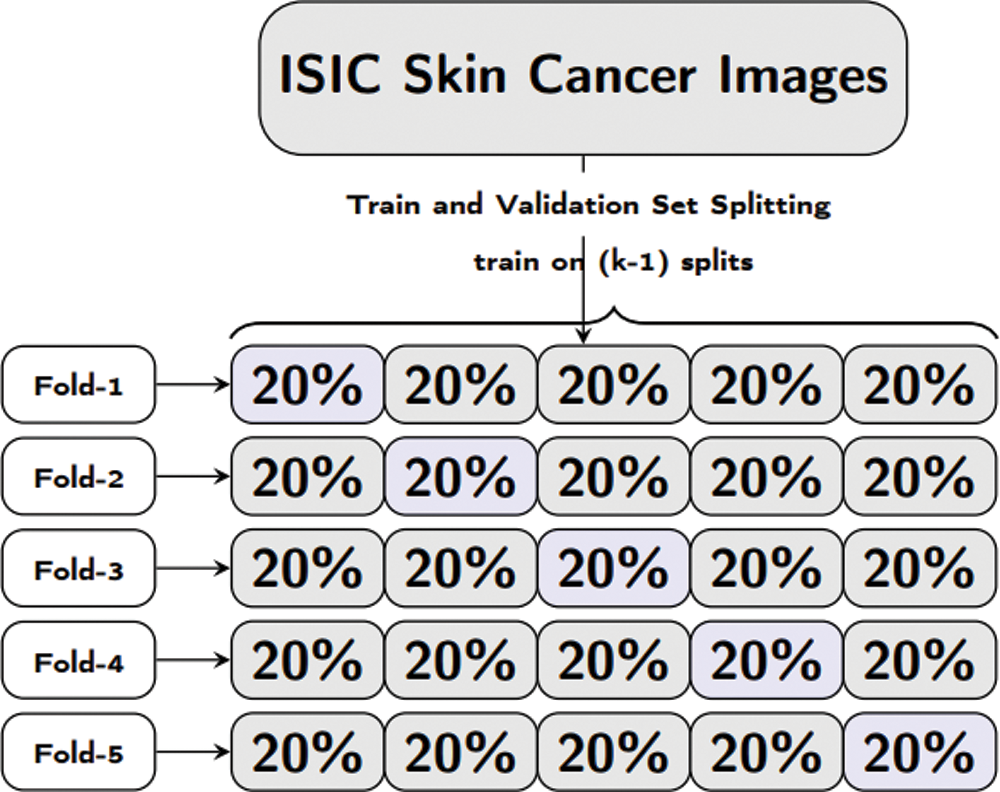
Figure 6: Formal representation of training and validation procedure employed in the 5-fold cross validation
We have used Local Binary Pattern Convolutional Neural Network (LBPCNN) with our proposed network model (similar to VGG Network) with hidden layers, ReLu, Dropout, Batch Normalization and Flatten Layers. Our LBPCNN model is depicted in Tab. 2.
Using LBPCNN and above mentioned preprocessing steps we achieved plausible results. We visually analyzed the separation and region-growing performance of the proposed LBPCNN, Figs. 7 and 8 presents an example of classification results. The white irregular line represents the segmentation of lesions in fair skins. We have cropped the original skin lesion dataset and resized it into 128128 for training and validation/testing. We adopted mirror and rotation operations to enlarge and× get the maximum results from the dataset. We have used local binary pattern (finding rotational invariance using the Eq. (10)) with convolutional neural network (LBPCNN) and then contour of those pixels to get the category of a lesion. LBPCNN has 17 layers (Convolutional layers with Rectified Linear Units, max pooling layers [38] (2 × 2), Dropout layer and Batch Normalization layers) including fully connected layer with stride one and padding value is zero (‘SAME’). We have set the batch size of 50 and trained our neural network with the most successful classifier ADAM [39]. Overfitting problem is catered by dropout [40] layer with a factor of 0.2 and 0.5. The ReLu layer is used to vanish gradient problems in convolutional layers.
We have passed 128 × 128 images to the input layer and convolved with 17 different layers (4 LBP and 10 convolutional layers) using ReLu to rectify the gradient problem. To overcome the covariance shift and to speed up the training process, we used batch normalization in LBPCNN that are widely used in deep learning frameworks/models e.g., Inception and ResNet [32]. Tab. 2 depicts our proposed Convolutional Neural Network model.
The main crux of LBPCNN is to extract local (neighborhood) features based on the current pixel and expeditiously acquire the local spatial patterns. After thorough and detailed research on the ISIC dataset, we finalized that LBPCNN is the best approach for detection and classification of skin cancer. It has the ability to extract local features with the help of radius (R) and neighborhood (P). P and R are the total neighbors of pixel values and radius of the neighborhood respectively. We set 3 × 3 neighborhood values to extract the atomic features from the ISIC dataset.
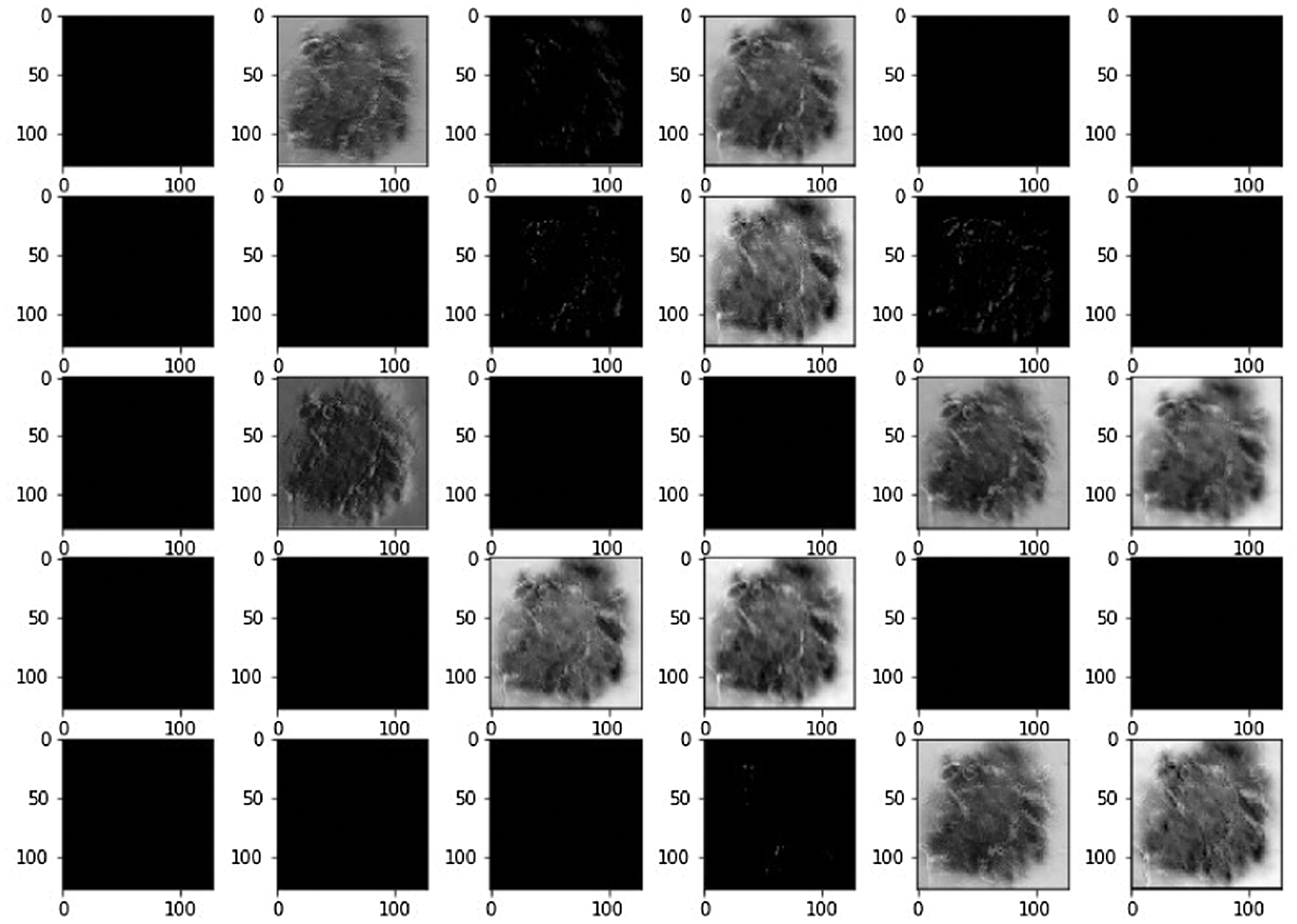
Figure 7: Systematically visualize the feature map of our proposed model LBPCNN. The idea behind the visualization of a feature map is what important features are to be highlighted during training for classification detection of cancer
For classification of skin lesions, we required the local features of an image. Generally, LBP has been used for local feature extraction. Mostly the image has only a single lesion, so from a single lesion image, the LBPCNN have acquired plausible results. Using LBPCNN, we can customize the P and R values to extract the different levels of discriminative features.
There is no specific approach for extraction of local features and describing atomic-scale appearance using LBPCNN in the previous research. The mentioned pretrained models such as AlexNet, ResNet, DenseNet169, InceptionV3, VGG16 and Xception have used different combinations of Convolution layers with different hyper parameters to achieve accuracy. Using LBPCNN, we use lesser computational resources as compared to other pretrained models and acquire discriminative and most prominent features for detection and classifications of skin lesions.
We have trained our proposed novel LBPCNN model with filters of three different sizes (depicted in Tab. 2). As shown in Fig. 6, we repeatedly perform the experiments up to five times. For all k-folding, dataset is sliced and wrapped for optimized validation and testing accuracy and loss. The overall and k-fold training, validation and testing accuracy are depicted in Fig. 9 and k-fold training, validation and testing loss are depicted in Fig. 10. The confusion matrix depicted in Fig. 13 and it has 8035 total images for training (Basel Cell Carcinoma have 2113, Squamous Cell Carcinoma have 1864, Actinic Keratoses have 1967 and Melanoma have 2091 images) on dataset (ISIC 2017 and 2018) for validation/testing we have used 3494 images (Basel Cell Carcinoma have 716, Squamous Cell Carcinoma have 912, Actinic Keratoses have 743 and Melanoma have 1123 images) other than training images. Fig. 13 upper row depicts the training confusion matrix and bottom row has testing confusion matrix of our proposed novel hybrid CNN (LBPCNN) model respectively.

Figure 8: Output Layer Result: It represents the output features belonging to a specific class with classification accuracy values using categorical cross-entropy function. The higher classification accuracy denotes that the image belongs to this class with high value
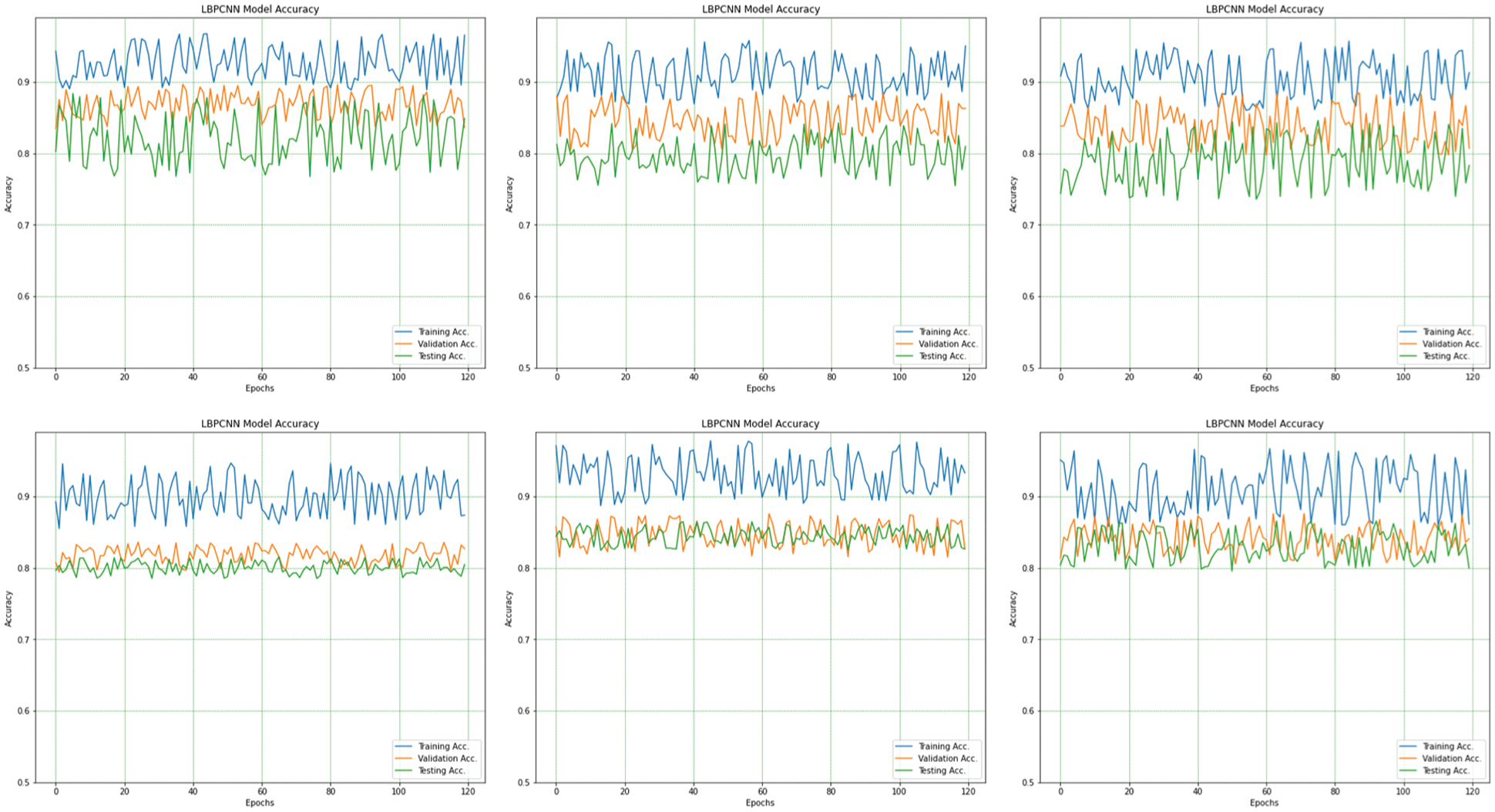
Figure 9: The overall (1st plot) and 5-fold accuracy (2nd to 6th plots) of our proposed novel hybrid CNN model training, testing and validation accuracy are obtained for multiclass classification problem of skin cancer (Basel Cell Carcinoma (BCC), Squamous Cell Carcinoma (SCC), Actinic Keratoses (AK) and Melanoma)
Our proposed novel hybrid LBPCNN model ROC's and confusion matrices are depicted in Figs. 12 and 11. Fig. 11 expressed the ROC of multi-class classification (Basel Cell Carcinoma (BCC), Squamous Cell Carcinoma (SCC), Actinic Keratoses (AK) and Melanoma) of ISIC 2017 and 2018 dataset that initially has 3 × 3 kernel has higher FPR than other kernel sizes, however, it then outperforms the 5 × 5 and 7 × 7 kernel higher values of TPR at different classification threshold. The area under curve (AUC) with the 3 × 3 kernel is 0.97 while it is 0.95 and 0.96 with the other kernels used in experiments. This is probably due to the finer image features extraction using Local Binary Pattern Convolutional Network.
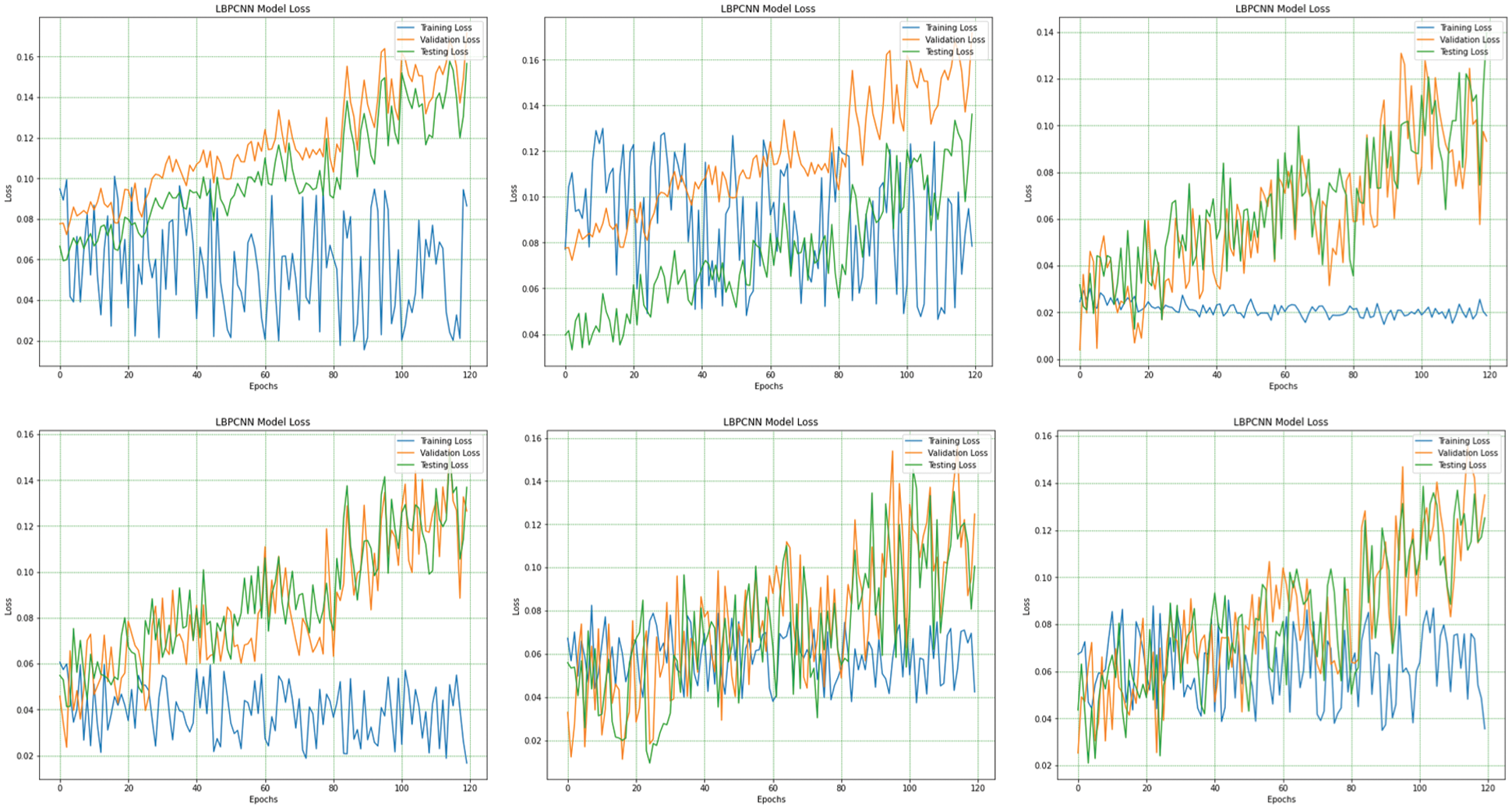
Figure 10: The overall (1st plot) and 5-fold loss (2nd to 6th plots) of our proposed novel hybrid CNN model training, testing and validation loss are obtained for multiclass classification problem of skin cancer (Basel Cell Carcinoma (BCC), Squamous Cell Carcinoma (SCC), Actinic Keratoses (AK) and Melanoma)
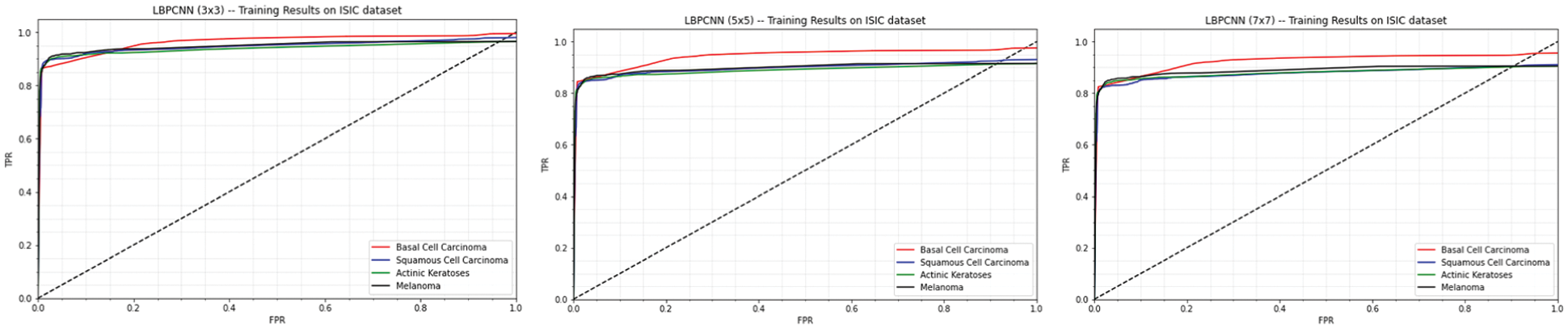
Figure 11: Our proposed novel hybrid CNN model (LBPCNN) is trained with kernel size 3 × 3 (1st plot), 5 × 5 (2nd plot) and 7 × 7 (3rd plot) have achieved 0.97, 0.96 and 0.95 training accuracy respectively
We tweak the filter size of LBPCNN model to train and test with the same datasets. Confusion matrix of training and testing are depicted in Fig. 13 and ROC of 5 × 5 filter size is depicted in Fig. 11 (2nd plot). The result of filter size 3 × 3 is much better than filter size 5 × 5.
During third time training/testing, we modify the filter size (7 × 7) for training, validation and testing with the same dataset. Confusion matrix of training and testing are depicted in Fig. 13 and ROC of 7 × 7 filter size is depicted in Fig. 11 (3rd plot).
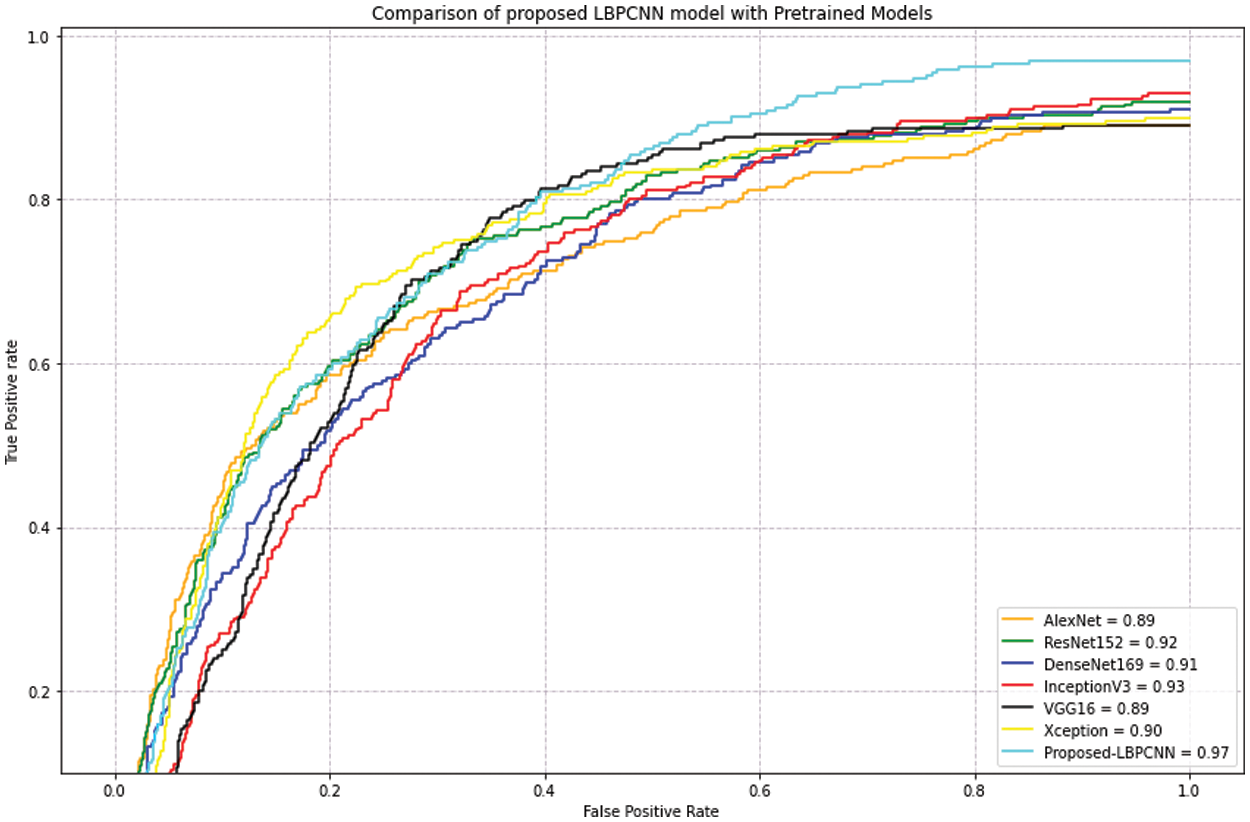
Figure 12: Our proposed novel hybrid CNN model (LBPCNN) is compared with pretrained models on dataset ISIC 2017 – some classes (327 of Actinic Keratoses, 379 of Basel Cell Carcinoma (BCC) and 153 of Squamous Cell Carcinoma (SCC)) images acquired from ISIC 2018 and 2019. The acquired accuracyof pretrained models are AlexNet (0.89), ResNet (0.92), DenseNet169 (0.91), InceptionV3 (0.93), VGG16 (0.89) and Xception (0.90)
The performance of multi-class classification of the proposed novel hybrid CNN (LBPCNN) model has been measured for each fold. It has been observed that 3 × 3 filter is given better accuracy as compared to other filters such as 5 × 5 and 7 × 7. Tab. 3 depicts performance (accuracy, precision, recall, specificity and sensitivity) of Basil Cell Carcinoma category during training and testing. We present the performance of Squamous Cell Carcinoma in Tab. 3. The tables show the results of all filters (3 × 3, 5 × 5 and 7 × 7) that we have used in our proposed LBPCNN model. Tab. 3 presents the result of Actinic Keratoses, Melanoma category of different filters respectively. In health care, early diagnosis and treatment can save a life of someone's and medical imaging plays a vital role for early diagnosis. Our proposed novel hybrid CNN (LBPCNN) model can detect the localization of the lesion and these types of model can play a vital role in identifying skin lesions in early stages. In previous studies, different approaches and models such as ResNet and VGG are generally used for skin lesion detection. However, the proposed novel hybrid CNN uses Local Binary Pattern methods to locate local patterns of skin lesions and achieve plausible results. It extracts the multi-dimensional micro-features and anisotropy features from different orientations which is more constructive and spatial for training. For maintaining consistency and classification loss and accuracy, we used 5-fold cross validation. During training, the LBPCNN layer reduces the learnable parameters from 9xto 169x. The convolutional layer weights further reduce the computational cost and memory complexity during training and testing phase. Small to medium size datasets, LBPCNN is prevented from overfitting. From the experimental studies, we summarize that LBPCNN with filter size (3 × 3) performs better to classify the skin lesion in multi-classification problems.
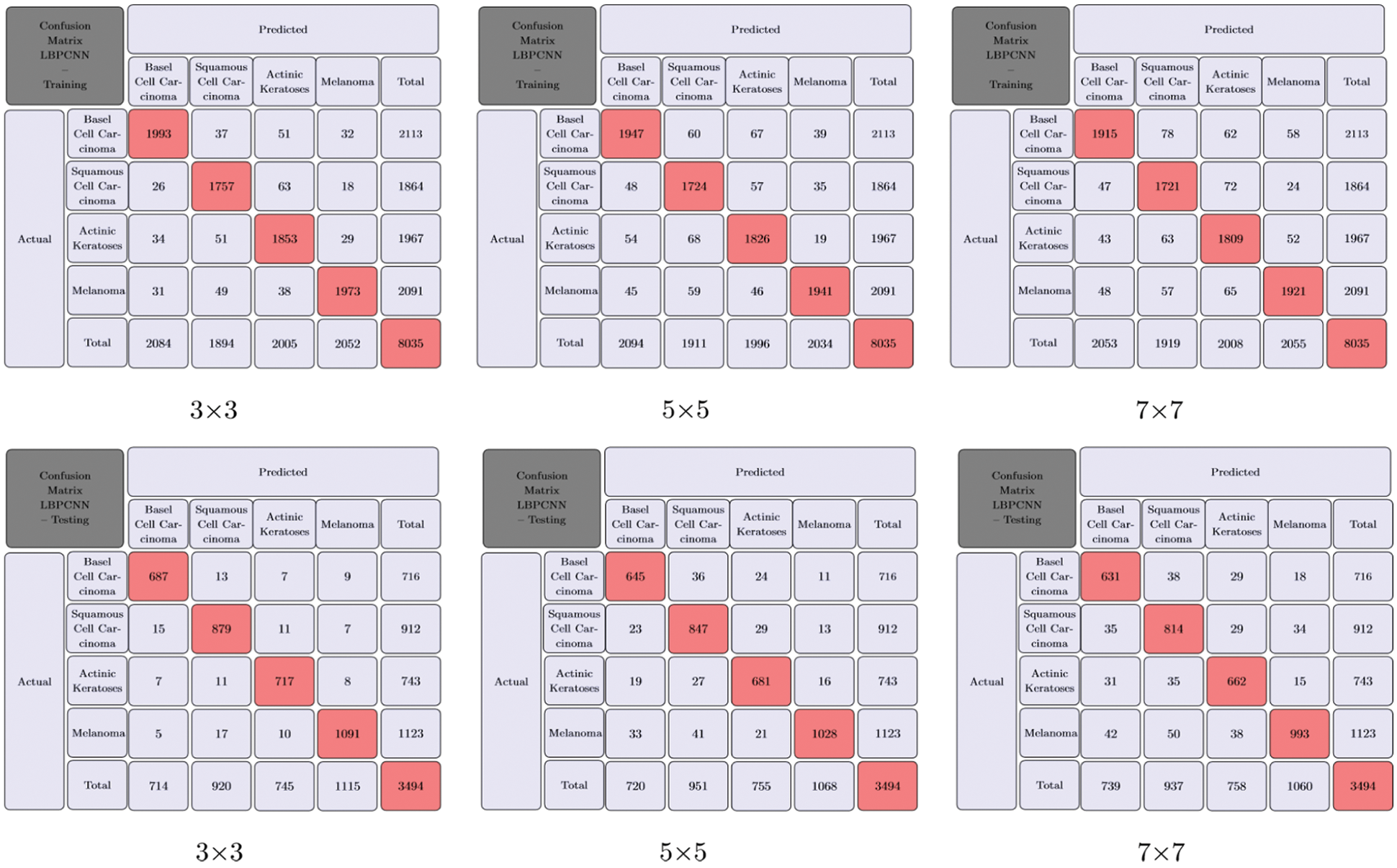
Figure 13: Our proposed novel hybrid CNN model (LBPCNN) Training and Testing with different filter (BCC), Squamous Cell Carcinoma (SCC), Actinic Keratoses (AK) and Melanoma). The upper row has size is 3 × 3, 5 × 5 and 7 × 7 for multiclass classification problem of skin cancer (Basel Cell Carcinoma training confusion matrix and bottom row has testing confusion matrix. It provides direct comparison of values such as True Positive (TP), True Negative (TN), False Positive (FP) and Flase Negative (FN) and visualize predictive analytics such as accuracy, sensitivity and specificity
For evaluation of our proposed hybrid LBPCNN model, we apply pretrained models such as AlexNet, ResNet, DenseNet169, InceptionV3, VGG16 and Xception on ISIC dataset and achieved results depicted on Tab. 4. After detailed evaluation and comparison, we summarize that our proposed model achieves better results in lesser and with minimum computational resources than pretrained models due to hybridized layers (LBPCNN).
We have compared our proposed framework/model result with other existing previous work [14,16–18,20,21,35,41] and [19]. Tab. 5 depicts the results of previous research work. We have achieved high accuracy and specificity than other existing framework/model, the sensitivity result of [14,16] is higher than our proposed framework/model.



In this paper, a local binary pattern with convolutional neural network is presented to classify the melanoma and non-melanoma skin lesions. We have used different R and P operators of LBP to get different textures using ROI. The result presents an outstanding performance for LBPCNN during training and testing with an accuracy 0.97 and 0.98 and sensitivity of 0.95 and 0.96 respectively. The main crux of LBPCNN is to extract local (neighborhood) features based on the current pixel and expeditiously acquire the local spatial patterns.
In future work, we plan to extract features with the help of handcrafted feature extraction algorithms such as Maximally Stable Extremal Regions (MSER) and Speeded-Up Robust Features (SURF) and embed with custom layers in Convolutional Neural Network and classify with traditional machine learning algorithms or ensemble classifiers. We also plan to improve the real time (using smartphone camera) classification of skin lesions (melanoma or non-melanoma). The developed model will be placed in a remote cloud to automate the detection of skin lesions and to help the affected patients immediately and it will reduce the clinic manual workload. After the detailed and necessary tests are done, we will deploy the developed and tested model in local clinics and hospitals for immediate screening and diagnosis.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. The Global Cancer Observatory, World Health Organization (WHO“International agency for research on cancer. melanoma cancer,” 2020. [Online] Available: https://gco.iarc.fr/today/data/factsheets/cancers/16-Melanoma-of-skin-fact-sheet.pdf. [Google Scholar]
2. K. Bashir, “Melanoma and tanning: A case study of sun safety knowledge and practices among 15 Canadian university women,” MA Thesis, School of Human Kinetics, University of Ottawa, Canada, 2013. [Google Scholar]
3. S. M. Thomas, J. G. Lefevre, G. Baxter and N. A. Hamilton, “Interpretable deep learning systems for multi-class segmentation and classification of non-melanoma skin cancer,” Medical Image Analysis, vol. 68, pp. 101915, 2021. [Google Scholar]
4. I. Giotis, N. Molders, S. Land, M. Biehl, M. F. Jonkman et al. “Med-node: A computer-assisted melanoma diagnosis system using non-dermoscopic images,” Expert Systems with Applications, vol. 42, no. 19, pp. 6578–6585, 2015. [Google Scholar]
5. C. Chen, D. Carlson, Z. Gan, C. Li and L. Carin, “Bridging the gap between stochastic gradient MCMC and stochastic optimization,” in Artificial Intelligence and Statistics, vol. 51, pp. 1051–1060, 2016. [Google Scholar]
6. S. A. ElGhany, M. R. Ibraheem, M. Alruwaili and M. Elmogy, “Diagnosis of various skin cancer lesions based on fine-tuned resnet50 deep network,” CMC-Computers Materials & Continua, vol. 68, no. 1, pp. 117–135, 2021. [Google Scholar]
7. N. Zhang, Y. Cai, Y. Y. Wang, Y. T. Tian, X. L. Wang et al., “Skin cancer diagnosis based on optimized convolutional neural network,” Artificial Intelligence in Medicine, vol. 102, pp. 101756–101763, art no. 101756, 2020. [Google Scholar]
8. D. Zhang and G. Lu, “A comparative study of three region shape descriptors,” DICTA2002: Digital Image Computing Techniques and Applications, vol. 2122, pp. 86–91, 2002. [Google Scholar]
9. Q. Abbas, M. E. Celebi, C. Serrano, I. F. GarcíA and G. Ma, “Pattern classification of dermoscopy images: A perceptually uniform model,” Pattern Recognition, vol. 46, no. 1, pp. 86–97, 2013. [Google Scholar]
10. M. Abadi, P. Barham, J. Chen, Z. Chen, A. Davis et al., “Tensorflow: a system for largescale machine learning,” in 12th USENIX Symp. on Operating Systems Design and Implementation (OSDI-16), Savannah, GA, USA, pp. 265–283, 2016. [Google Scholar]
11. F. Chollet, “Introduction to Keras and Tensorflow,” in Deep Learning with Python, 2nd ed., vol. 1, Manning Publications Co., Mountain View, CA, USA, 2020. [Google Scholar]
12. J. Bergstra, O. Breuleux, P. Lamblin, R. Pascanu, O. Delalleau et al., “Theano: Deep learning on gpus with python,” in Neural Information Processing Systems (NIPS), BigLearning Workshop, vol. 3, pp. 1–48, 2011. [Google Scholar]
13. A. Esteva, B. Kuprel, R. A. Novoa, J. Ko, S. M. Swetter et al., “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, no. 7639, pp. 115–118, 2017. [Google Scholar]
14. H. Iyatomi, H. Oka, M. E. Celebi, K. Ogawa, G. Argenziano et al., “Computer-based classification of dermoscopy images of melanocytic lesions on acral volar skin,” Journal of Investigative Dermatology, vol. 128, no. 8, pp. 2049–2054, 2008. [Google Scholar]
15. M. A. Khan, T. Akram, M. Sharif, S. Kadry and Y. Nam, “Computer decision support system for skin cancer localization and classification,” CMC-Computers Materials & Continua, vol. 68, no. 1, pp. 1041–1064, 2021. [Google Scholar]
16. U. O. Dorj, K. K. Lee, J. Y. Choi and M. Lee, “The skin cancer classification using deep convolutional neural network,” Multimedia Tools and Applications, vol. 77, pp. 9909–9924, 2018. [Google Scholar]
17. E. Almansour and M. A. Jaffar, “Classification of dermoscopic skin cancer images using color and hybrid texture features,” International Journal of Computer Science and Network Security, vol. 16, no. 4, pp. 135–139, 2016. [Google Scholar]
18. G. Capdehourat, A. Corez, A. Bazzano, R. Alonso and P. Musé, “Toward a combined tool to assist dermatologists in melanoma detection from dermoscopic images of pigmented skin lesions,” Pattern Recognition Letters, vol. 32, no. 16, pp. 2187–2196, 2011. [Google Scholar]
19. D. Ruiz, V. Berenguer, A. Soriano and B. SáNchez, “A decision support system for the diagnosis of melanoma: A comparative approach,” Expert Systems with Applications, vol. 38, no. 12, pp. 15217–15223, 2011. [Google Scholar]
20. A. G. Isasi, B. G. Zapirain and A. M. Zorrilla, “Melanomas non-invasive diagnosis application based on the abcd rule and pattern recognition image processing algorithms,” Computers in Biology and Medicine, vol. 41, no. 9, pp. 742–755, 2011. [Google Scholar]
21. K. Ramlakhan and Y. Shang, “A mobile automated skin lesion classification system,” in Tools with Artificial Intelligence (ICTAI23rd IEEE Int. Conf. on, Boca Raton, FL, USA, pp. 138–141, 2011. [Google Scholar]
22. A. Klautau, N. Jevtić and A. Orlitsky, “On nearest-neighbor error-correcting output codes with application to all-pairs multiclass support vector machines,” Journal of Machine Learning Research, vol. 4, pp. 1–15, 2003. [Google Scholar]
23. A. A. Cruz-Roa, J. E. A. Ovalle, A. Madabhushi and F. A. G. Osorio, “A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection,” in Int. Conf. on Medical Image Computing and Computer-Assisted Intervention-MICCAI. Lecture Notes in Computer Science, vol. 8150, Springer, Berlin, Heidelberg, pp. 403–410, 2013. [Google Scholar]
24. A. G. Pacheco, C. S. Sastry, T. Trappenberg, S. Oore and R. A. Krohling, “On out-of-distribution detection algorithms with deep neural skin cancer classifiers,” in Proc. of the IEEE/CVF Conf. on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, pp. 732–733, 2020. [Google Scholar]
25. Z. She and P. S. Excell, “Skin pattern analysis for lesion classification using local isotropy,” Skin Research and Technology, vol. 17, no. 2, pp. 206–212, 2011. [Google Scholar]
26. E. R. Davies, “Basic Image Filtering Operations,” in Computer and Machine Vision: Theory, Algorithms and Practicalities, 4th ed., London, UK, Elsevier, pp. 38–78, 2012. [Google Scholar]
27. S. Ioffe and C. Szegedy, “Batch normalization: accelerating deep network training by reducing internal covariate shift,” in Int. Conf. on Machine Learning, Proc. of Machine Learning Research, Lille, France, vol. 37, pp. 448–456, 2015. [Google Scholar]
28. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Las Vegas, Nevada, USA. pp. 770–778, 2016. [Google Scholar]
29. J. Long, E. Shelhamer and T. Darrell, “Fully convolutional networks for semantic segmentation,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Boston, Massachusetts, USA, pp. 3431–3440, 2015. [Google Scholar]
30. O. Ronneberger, P. Fischer and T. Brox. “U-net: convolutional networks for biomedical image segmentation,” in 18th Int. Conf. on Medical Image Computing and Computer Assisted Interventio (MICCAI 2015Part III, LNCS 9351, Springer, Munich, Germany, pp. 234–241, 2015. [Google Scholar]
31. E. Abbadi, N. Khdhair and A. H. Miry, “Automatic segmentation of skin lesions using histogram thresholding,” Journal of Computer Science, vol. 10, no. 4, pp. 632–639, 2014. [Google Scholar]
32. S. A. ElGhany, M. R. Ibraheem, M. Alruwaili and M. Elmogy, “Diagnosis of various skin cancer lesions based on fine-tuned resnet50 deep network,” CMC-Computers Materials & Continua, vol. 68, no. 1, pp. 117–135, 2021. [Google Scholar]
33. M. Toğaçar, Z. Cömert and B. Ergen, “Intelligent skin cancer detection applying autoencoder, mobilenetv2 and spiking neural networks,” Chaos, Solitons & Fractals, vol. 144, pp. 110714, 2021. [Google Scholar]
34. A. Adegun and S. Viriri, “Deep learning techniques for skin lesion analysis and melanoma cancer detection: A survey of state-of-the-art,” Artificial Intelligence Review, vol. 54, no. 2, pp. 811–841, 2021. [Google Scholar]
35. A. G. Pacheco, C. S. Sastry, T. Trappenberg, S. Oore and R. A. Krohling, “On out-of-distribution detection algorithms with deep neural skin cancer classifiers,” in Proc. of the IEEE/CVF Conf. on Computer Vision and Pattern Recognition Workshops, Seattle, WA, United States, pp. 732–733, 2020. [Google Scholar]
36. T. Y. Tan, L. Zhang and C. P. Lim, “Intelligent skin cancer diagnosis using improved particle swarm optimization and deep learning models,” Applied Soft Computing, vol. 84, pp. 105725, 2019. [Google Scholar]
37. P. N. Srinivasu, J. G. SivaSai, M. F. Ijaz, A. K. Bhoi, W. Kim et al., “Classification of skin disease using deep learning neural networks with mobilenet v2 and lstm,” Sensors, vol. 21, no. 8, pp. 2852, 2021. [Google Scholar]
38. A. Giusti, D. C. Ciresan, J. Masci, L. M. Gambardella and J. Schmidhuber, “Fast image scanning with deep max-pooling convolutional neural networks,” in 20th IEEE Int. Conf. on Image Processing (ICIP), Melbourne, Australia, pp. 4034–4038, 2013. [Google Scholar]
39. A. Krizhevsky, I. Sutskever and G. E. Hinton, “Imagenet classification with deep convolutional neural networks,” in Advances in Neural Information Processing Systems-Conf. on Neural Information Processing Systems, Nevada, USA, pp. 1097–1105, 2012. [Google Scholar]
40. V. Nair and G. E. Hinton, “Rectified linear units improve restricted boltzmann machines,” in Proc. of the 27th International Conf. on Machine Learning (ICML-10), Haifa, Israel, pp. 807–814, 2010. [Google Scholar]
41. M. Anas, K. Gupta and S. Ahmad, “Skin cancer classification using k-means clustering,” International Journal of Technical Research and Applications, vol. 5, no. 1, pp. 62–65, 2017. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |