DOI:10.32604/cmc.2022.024556

| Computers, Materials & Continua DOI:10.32604/cmc.2022.024556 |  |
| Article |
Automated Multi-Document Biomedical Text Summarization Using Deep Learning Model
1Department of Information Systems, College of Computer and Information Sciences, Prince Sultan University, Saudi Arabia
2Department of Computer Science, College of Science and Arts at Mahayil, King Khalid University, Saudi Arabia
3Faculty of Computer and IT, Sana'a University, Sana'a, Yemen
4Department of Computer Science, College of Computing and Information System, Umm Al-Qura University, Saudi Arabia
5Department of Computer and Self Development, Preparatory Year Deanship, Prince Sattam bin Abdulaziz University, AlKharj, Saudi Arabia
6Department of Natural and Applied Sciences, College of Community-Aflaj, Prince Sattam bin Abdulaziz University, Saudi Arabia
*Corresponding Author: Manar Ahmed Hamza. Email: ma.hamza@psau.edu.sa
Received: 22 October 2021; Accepted: 03 December 2021
Abstract: Due to the advanced developments of the Internet and information technologies, a massive quantity of electronic data in the biomedical sector has been exponentially increased. To handle the huge amount of biomedical data, automated multi-document biomedical text summarization becomes an effective and robust approach of accessing the increased amount of technical and medical literature in the biomedical sector through the summarization of multiple source documents by retaining the significantly informative data. So, multi-document biomedical text summarization acts as a vital role to alleviate the issue of accessing precise and updated information. This paper presents a Deep Learning based Attention Long Short Term Memory (DL-ALSTM) Model for Multi-document Biomedical Text Summarization. The proposed DL-ALSTM model initially performs data preprocessing to convert the available medical data into a compatible format for further processing. Then, the DL-ALSTM model gets executed to summarize the contents from the multiple biomedical documents. In order to tune the summarization performance of the DL-ALSTM model, chaotic glowworm swarm optimization (CGSO) algorithm is employed. Extensive experimentation analysis is performed to ensure the betterment of the DL-ALSTM model and the results are investigated using the PubMed dataset. Comprehensive comparative result analysis is carried out to showcase the efficiency of the proposed DL-ALSTM model with the recently presented models.
Keywords: Biomedical; text summarization; healthcare; deep learning; lstm; parameter tuning
Automatic text processing tool plays a vital role in efficient knowledge acquisition in massive source of textual data in the field of health care and life science, namely, clinical guidelines/electronic health records and scientific publications [1]. Automatic text summarization has been subregion of text mining and Natural Language Processing (NLP) which intends for producing a condensed form of more than one input documents by extracting the more important contents [2,3]. Text summarization tool could assist clinicians and researchers’ resources saving and time by manually presenting and identifying the key concepts within long document, with no need to read the entire text [4]. Initially, text summarization is based on frequency features to recognize the most relevant contents of textual documents. After, several summarization tools have integrated a broad range of heuristics and features into the procedure of content selection. The most commonly utilized feature includes the lengths of sentences, the position of sentences, keywords extracted from the texts, the existence of cue phrases, the title words, the centroid-based cohesion, the co-occurrence feature, the existence of arithmetical contents, etc [5].
In order to resolve this limitation, other strands of study investigated the evolution of technology which utilizes source of knowledge domain for mapping the texts into concept-based representations [6]. It allows measuring the useful content of the text regarding the semantics and context behindhand the sentence, instead of shallow features. But there are few difficulties in utilizing biomedical knowledge sources in text analyses, especially in summarization [7]. Maintaining, utilizing, and Building knowledge basis could be challenging. A massive amount of automatic annotation is required to widely determine the entities and concepts and to capture the relationships among them. The selection of relevant sources of knowledge domain is challenging which might seriously affect the performances of bio-medical summarization [8]. Another problem is how to measure the useful content of sentences relying on qualitative relationships among methods. Deep neural network (DNN) based language methods [9] could be used to tackle most of the problems related to knowledge domain in context aware bio-medical summarization. In deep language (DL) algorithm is pre-trained on massive quantities of text information and learns how to characterize 4 units of text, mostly words, in a vector space [10]. The pre-trained embedding could be finetuned on down-stream tasks or straightly utilized as an arithmetical feature.
[11] proposed a deep-reinforced, abstractive summarization method which can able to read bio-medical publication abstract and produce summary by means of a title or one sentence headline. They present a new reinforcement learning (RL) reward metrics based biomedical expert systems, namely MeSH and UMLS Metathesaurus also shows that this method can able to produce abstractive, domain-aware summaries. [12] presented a new text summarization method for documents with Deep Learning Modifier Neural Network (DLMNN) classification. It produces an enlightening summary of the document-based entropy values. The introduced DLMNN architecture contains 6 stages. Initially, the input document is preprocessed. Next, the feature can be extracted with preprocessed information. Then, the most relevant feature can be elected by the improved fruit fly optimization algorithm (IFFOA). The entropy values for each selected feature are calculated. In [13], the major focuses on application of ML methods in 2 distinct sub-regions that are associated with medical industry. The initial applications are Sentiment Analysis (SA) of user narrated drug reviews and the next is engineering in food technology. Since ML and AI methods enforce the limitations of scientific drug detection, ML methods are chosen as another technique for 2 main factors. Initially, ML method includes distinct learning approaches and also its feasibility for numerous NLP operations. Next, its inherent capacity to model various features that capture the features of sentiment in text.
[14] trying to address this limitation by suggesting a novel method with topic modelling, unsupervised neural networks, and documents clustering for building effective document representations. Initially, a novel document clustering method with the Extreme learning machine (ELM) method is implemented on massive text collection. Next, topic modelling is employed in the document collection for identifying the topic existing in all the clusters. Then, all the documents are characterized in a concept space using a matrix in which column represents the cluster topics and row represents the document sentence. The created matrix can be trained by numerous ensemble learning algorithms and unsupervised neural networks for building abstract representations of the document in the topic space. [15] designed a new biomedical text summarization method which integrates 2 commonly used data mining methods: Frequent itemset and clustering mining. Biomedical paper can be stated as a group of biomedical topics with the UMLS metathesaurus. The K-means method is utilized for clustering analogous sentences. Subsequently, the Apriori method is employed for discovering the frequent itemsets amongst the clustered sentences. Lastly, the relevant sentence from all the clusters is elected for building the summary with the detected frequent itemset.
[16] integrated frequent itemsets and sentence clustering mining for building an individual bio-medical text summarization model. A bio-medical document is denoted as a set of UMLS topics. The generic concept is rejected. The vector space method is applied for representing the sentence. The K-means clustering method is employed for semantically clustering analogous sentences. The frequent itemset is extracted between the global clusters. The detected frequent itemset is applied for calculating the score of sentence. The topmost N high scoring sentences are elected for representing the last summary. [17] resolve this problem in terms of biomedical text summarization. They measure the efficiency of a graph-based summarizer with distinct kinds of contextualized and context-free embeddings. The word representation is generated by pretraining neural language methods on massive amount of bio-medical texts. The summarizer modes the input texts as graphs where the strength of relationships among the sentences are evaluated by the domain specific vector representation.
The objective of this study is to design novel deep learning based Multi-document Biomedical Text Summarization model with hyperparameter tuning process.
This paper presents a Deep Learning based Attention Long Short Term Memory (DL-ALSTM) Model for Multi-document Biomedical Text Summarization. The proposed DL-ALSTM model initially performs data preprocessing to convert the available medical data into a compatible format for further processing. Then, the DL-ALSTM model gets executed to summarize the contents from the multiple biomedical documents. In order to tune the summarization performance of the DL-ALSTM model, chaotic glowworm swarm optimization (CGSO) algorithm is employed. Extensive experimentation analysis is performed to ensure the betterment of the DL-ALSTM model and the results are investigated using the PubMed dataset.
The remaining sections of the paper are arranged as follows. Section 2 offers the proposed DL-ALSTM model and Section 3 discusses the performance validation. Finally, Section 4 draws the conclusion of the study.
2 The Proposed Biomedical Text Summarization Technique
In this study, an effective DL-ALSTM model has been presented for Multi-document Biomedical Text Summarization. The proposed DL-ALSTM model performs pre-processing, summarization, and hyperparameter optimization. The detailed working of these processes is offered in the following sections.
The summarization procedure begins with running a pre-processed step. Initial, individual's part of an input document which is discarded to inclusion under the overview have been removed, and the essential text was taken. The redundant parts contain the title, abstract, keywords, author's data, headers of sections and subsections, figures and tables, and bibliography section [18]. It can be assumed that parts are unnecessary as it doesn't perform under the technique summary which is utilized for evaluating the amount of outline. The removal phase is customized dependent upon the framework of an input text and user's preference. When it can be chosen for including the title of section and subsection under the summary, further data has been saved together with all the sentences for specifying the section of text which sentence goes to.
As the input of feature extraction scripts of BERT are text files where all sentences perform in distinct lines, and all sentences are tokenized, the pre-processed step remains with splitting an input text as to distinct sentences and tokenization step. Utilizing the Natural Language ToolKit (NLTK), the summarizers split the essential text into groups of sentences, and signify all the sentences as groups of tokens. Afterward these pre-processed functions, an input sentence is ready that mapped as to contextualized vector representation.
2.2 Biomedical Text Summarization
The preprocessed data is fed into the DL-ALSTM model to summarize the multi-document biomedical text. The LSTM cell is comprised of 5 essential components: Input gate i, forget gate
Based on the subsequent explains the fundamental calculations in LSTM cell:
1. Gates
2. Input transform
3. State Update
The trained stage purposes for learning the parameters
In this case, it can utilize stacked LSTM layer on the vertical way in which input of present LSTM layers utilizes resultant of preceding layer. Fig. 1 illustrates the structure of LSTM model.
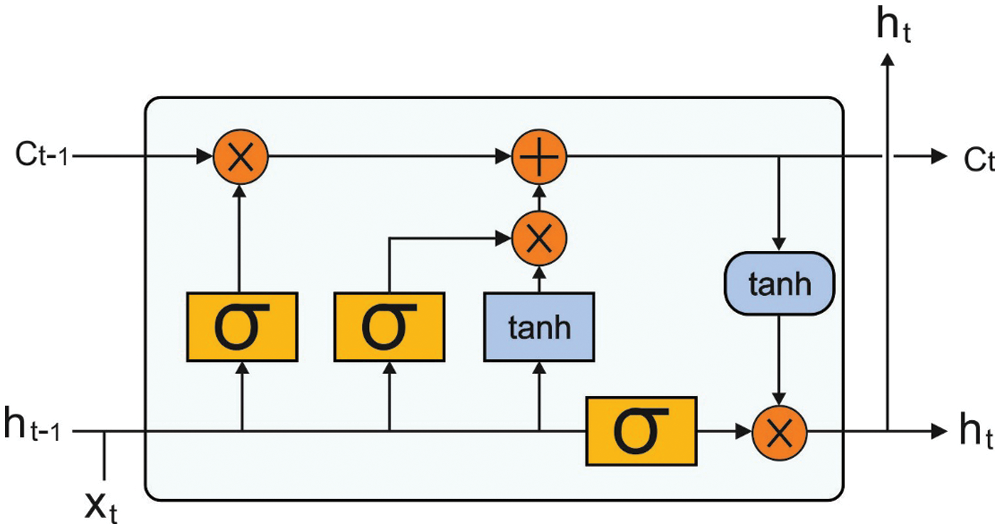
Figure 1: LSTM structure
Noticeably, this technique is exchanging classical RNN by LSTM unit. Besides the initial layer, this technique carries out passing the hidden state of preceding layer
This method has been simulated in Google neural machine translation method. It has 3 modules: Encoded, decoded, and attention networks. During the PCA-LSTM technique, the encoded utilizes stacked LSTM layer that contains 1
The value of hidden state significance score
where
where f generally utilizes
where g represents the GRU units. During this encoder-decoder structure dependent upon PCA-LSTM approach, the calculating of created paraphrase order
2.3 Design of CGSO Based Hyperparameter Optimization
In order to effectually adjust the hyperparameters involved in the DL-ALSTM model, the CGSO algorithm is utilized [20]. In the fundamental GSO, the n firefly (FF) individuals are arbitrarily distributed from
Fluorescein update:
where
Probability selection:
In particular, the region set is
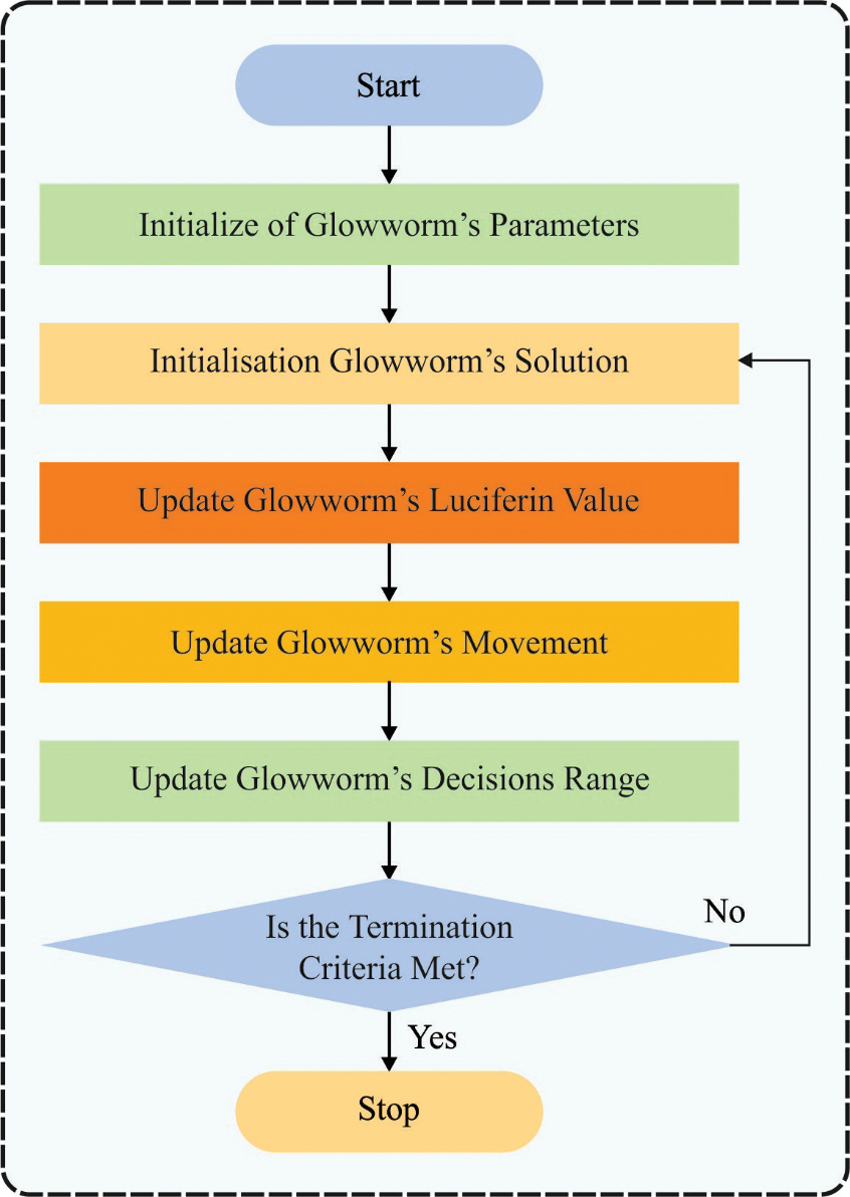
Figure 2: Flowchart of GSO
Location update:
where s represents the moving step.
The dynamic decision area radius upgrade:
The GSO technique contains a primary distribution of FFs, luciferase upgrade, FF progress, and decision-making area upgrade. To improve the performance of the GSO algorithm, the CGSO algorithm is derived by integrating the concepts of chaotic theory.
The chaos method is a subdivision of mathematics that performs on nonlinear dynamic process. Nonlinear denotes that it can be inconceivable for predicting the system's response with respect to the input, and dynamic mean alters from the system in one state to another over time. The chaos purposes signify the dynamical system using deterministic formula. However, based on the initial condition, chaotic function is divergent feature performances and generated wildly unpredictable. Thus, the chaos functions are improving the diversification and intensification of enhanced methods i.e., avert local optimum solution and alter neighboring global optimum. This purpose follows easier principle and has some interconnecting portions; but, in all iterations, the created value was depending on the primary condition and earlier values.
In this case, it is performed 3 different chaotic maps such as iterative mapping, tent mapping, and logistic mapping with power exponents (p) and sensory modality (c) computation from the BOA. The chaos purpose has been determined to exhibit high efficacy associated with another chaos purpose.
Logistic map:
Now,
Iterative map:
During the iterative map, the values of P are chosen among zero and one, as well the result
Tent map:
The tent map has 1D map which is same as logistic map. Now, the result
The proposed DL-ALSTM model has been validated using PubMed dataset [21], which comprises the instances in json format. The abstract, sections, and body are all sentence tokenized. The json objects includes several parameters such as: article_id, abstract_text, article_text, section_names, and sections.
The accuracy graph of the LSTM model is depicted in Fig. 3. The figure reported that the LSTM model has attained increased training and validation accuracies with an increase in epoch count. At the same time, it is noticed that the LSTM model has resulted in higher validation accuracy compared to training accuracy.
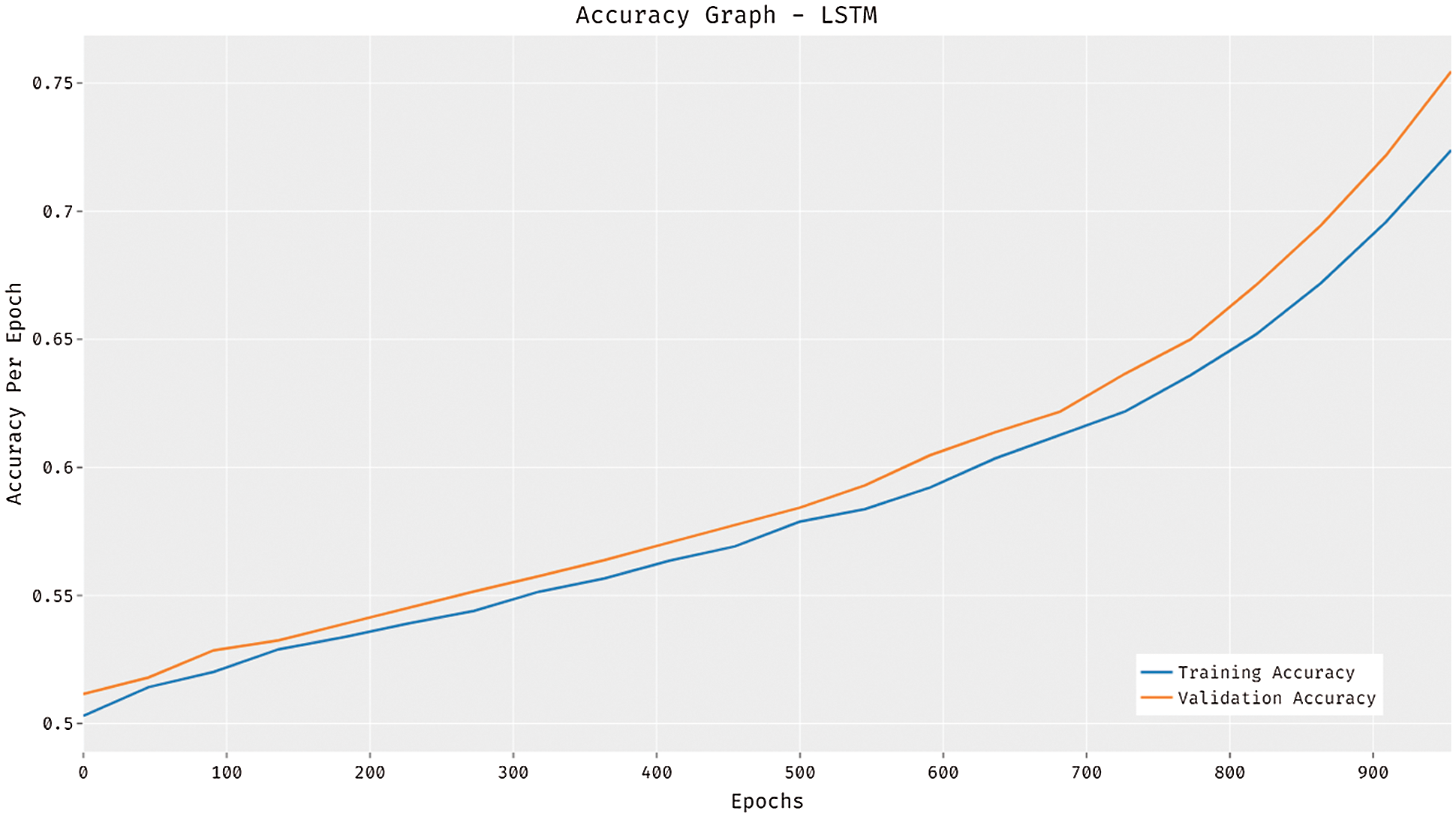
Figure 3: Accuracy analysis of LSTM model
Fig. 4 reports the loss graph analysis of the LSTM model on the test dataset applied. The figure showcased that the LSTM model has accomplished reduced loss with a rise in epoch count. It is observed that the LSTM model has resulted to reduced validation loss compared to training loss.
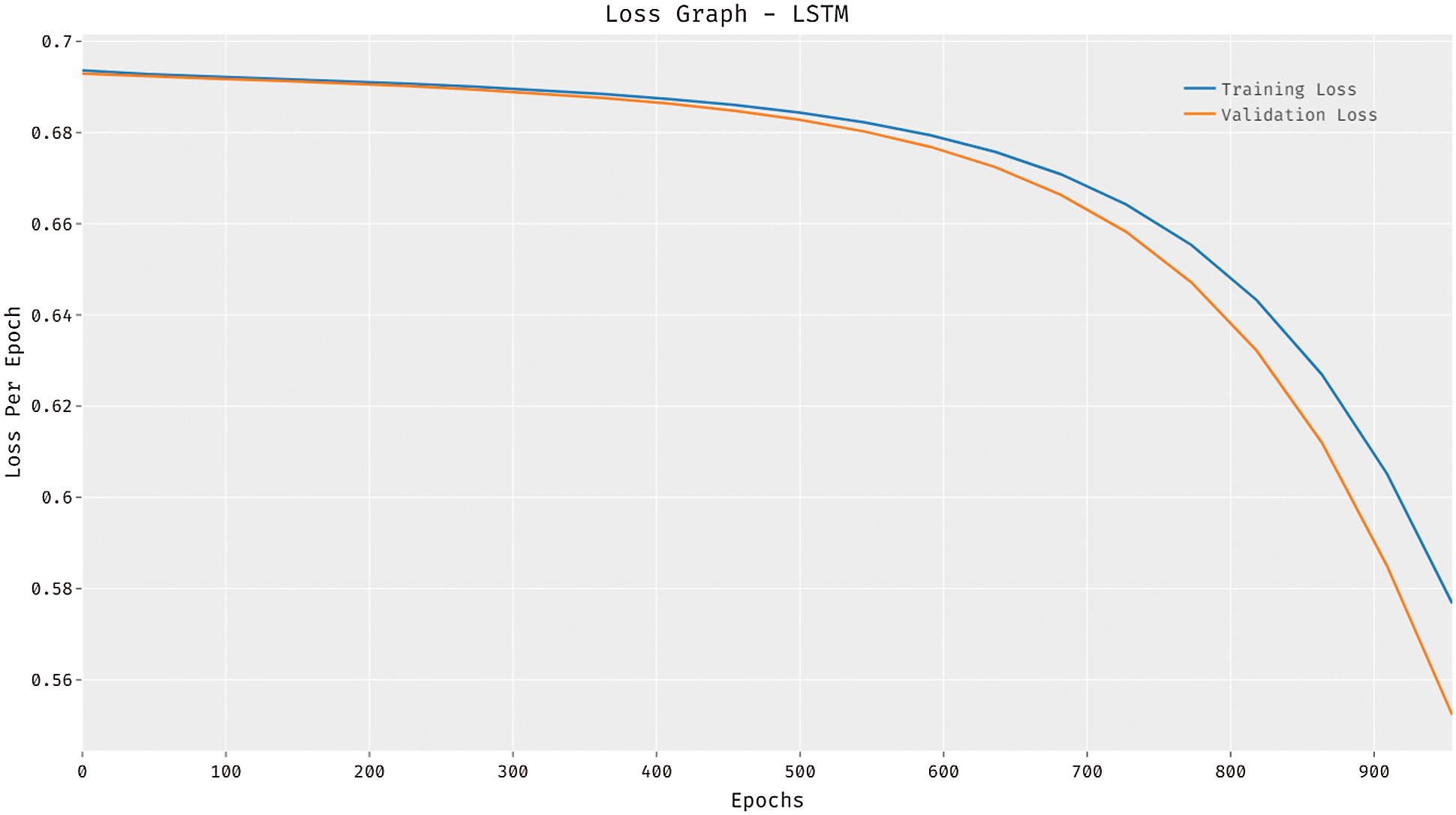
Figure 4: Loss analysis of LSTM model
The accuracy graph of the DL-ALSTM manner is demonstrated in Fig. 5. The figure stated that the DL-ALSTM technique has attained improved training and validation accuracies with a higher epoch count. Besides, it can be clear that the DL-ALSTM approach has resulted in superior validation accuracy compared to training accuracy.
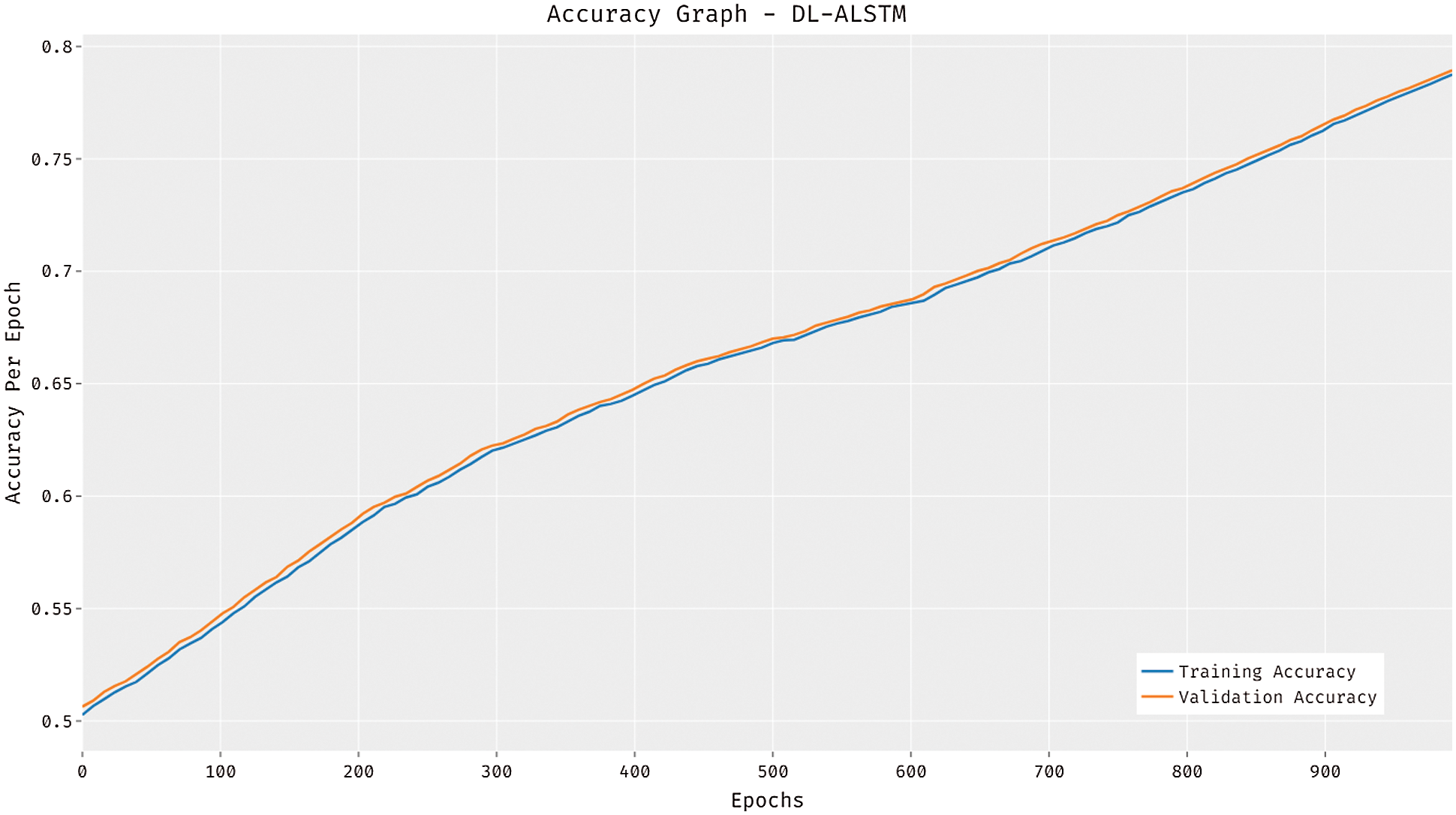
Figure 5: Accuracy analysis of DL-ALSTM model
Fig. 6 illustrates the loss graph analysis of the DL-ALSTM system on the test dataset applied. The figure outperformed that the DL-ALSTM technique has accomplished lesser loss with an increase in epoch count. It is demonstrated that the DL-ALSTM approach has resulted in decreased validation loss related to training loss.
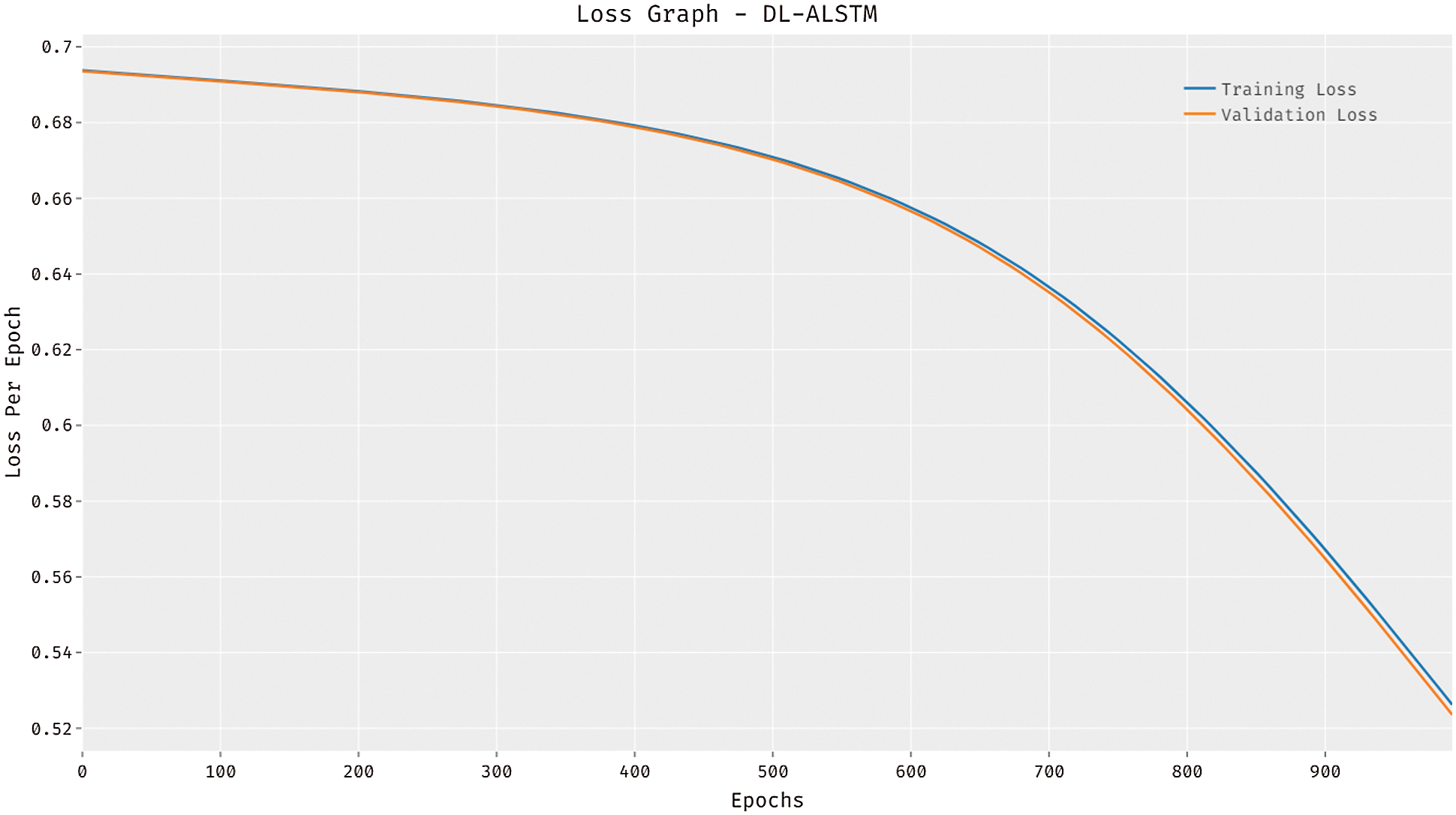
Figure 6: Loss analysis of DL-ALSTM model
Fig. 7 showcases the Rouge-1 analysis of the LSTM model on the applied dataset. The figure shows that that the LSTM model has attained moderately considerable ROUGE-1 values. For instance, the LSTM model has attained Rouge-1 of 0.7471 under the execution run-1. In addition, the LSTM Model has resulted in a Rouge-1 of 0.7354 under the execution run-2. Similarly, the LSTM technique has resulted in a Rouge-1 of 0.7470 under the execution run-4. Along with that, the LSTM manner has resulted in a Rouge-1 of 0.7528 under the execution run-8. Finally, the LSTM algorithm has resulted in a Rouge-1 of 0.7304 under the execution run-10.
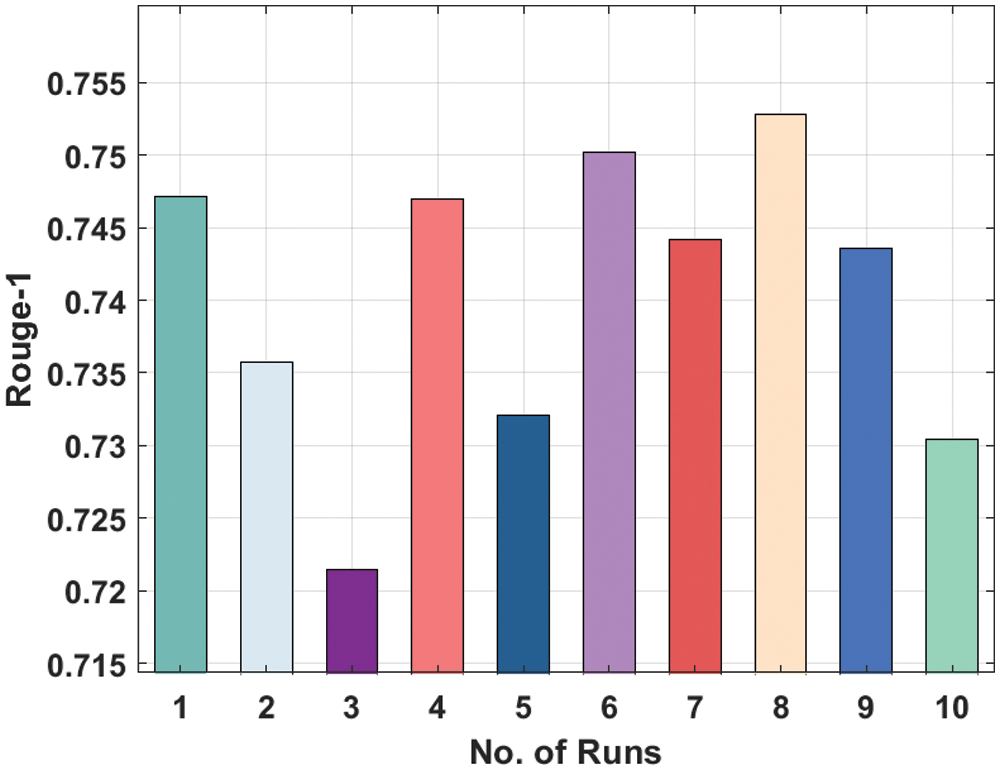
Figure 7: Result analysis of LSTM model in terms of Rouge-1
Fig. 8 depicts the Rouge-2 analysis of the LSTM manner on the applied dataset. The figure displayed that the LSTM manner has reached moderately considerable ROUGE-2 values. For instance, the LSTM technique has gained Rouge-2 of 0.3449 under the execution run-1. Similarly, the LSTM Model has resulted in a Rouge-2 of 0.3223 under the execution run-2. Likewise, the LSTM approach has resulted in a Rouge-2 of 0.3216 under the execution run-4. In addition, the LSTM methodology has led to a Rouge-2 of 0.3307 under the execution run-8. At last, the LSTM Model has resulted in a Rouge-2 of 0.3562 under the execution run-10.
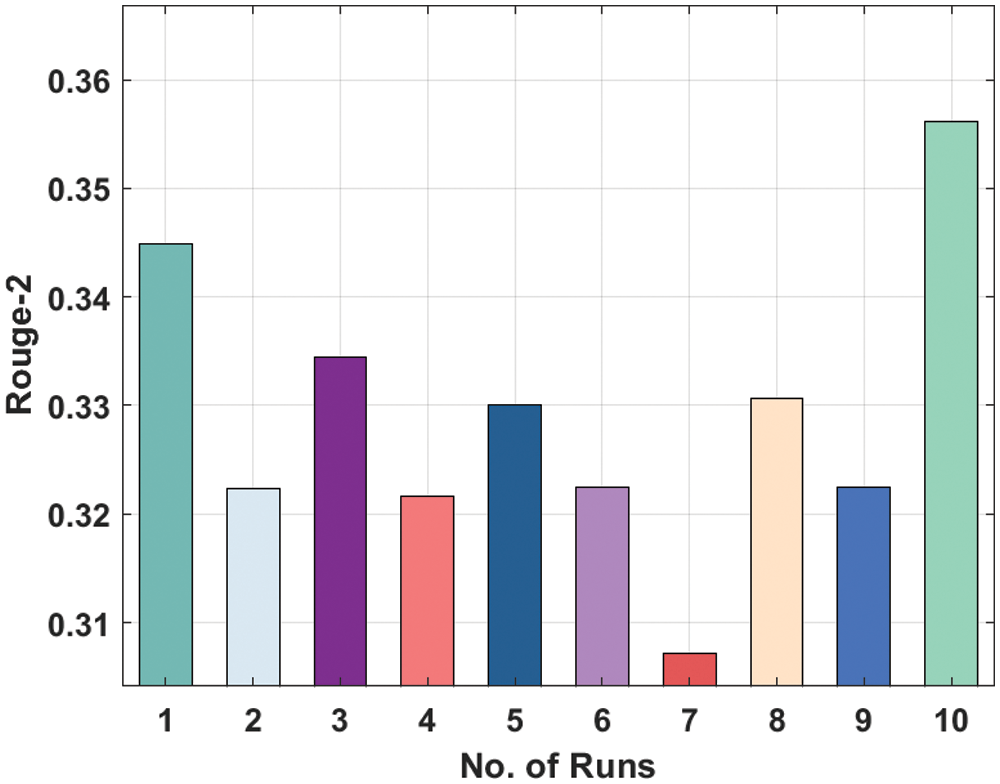
Figure 8: Result analysis of LSTM model in terms of Rouge-2
Tab. 1 illustrates the result analysis of LSTM and DL-ALSTM models with different runs.
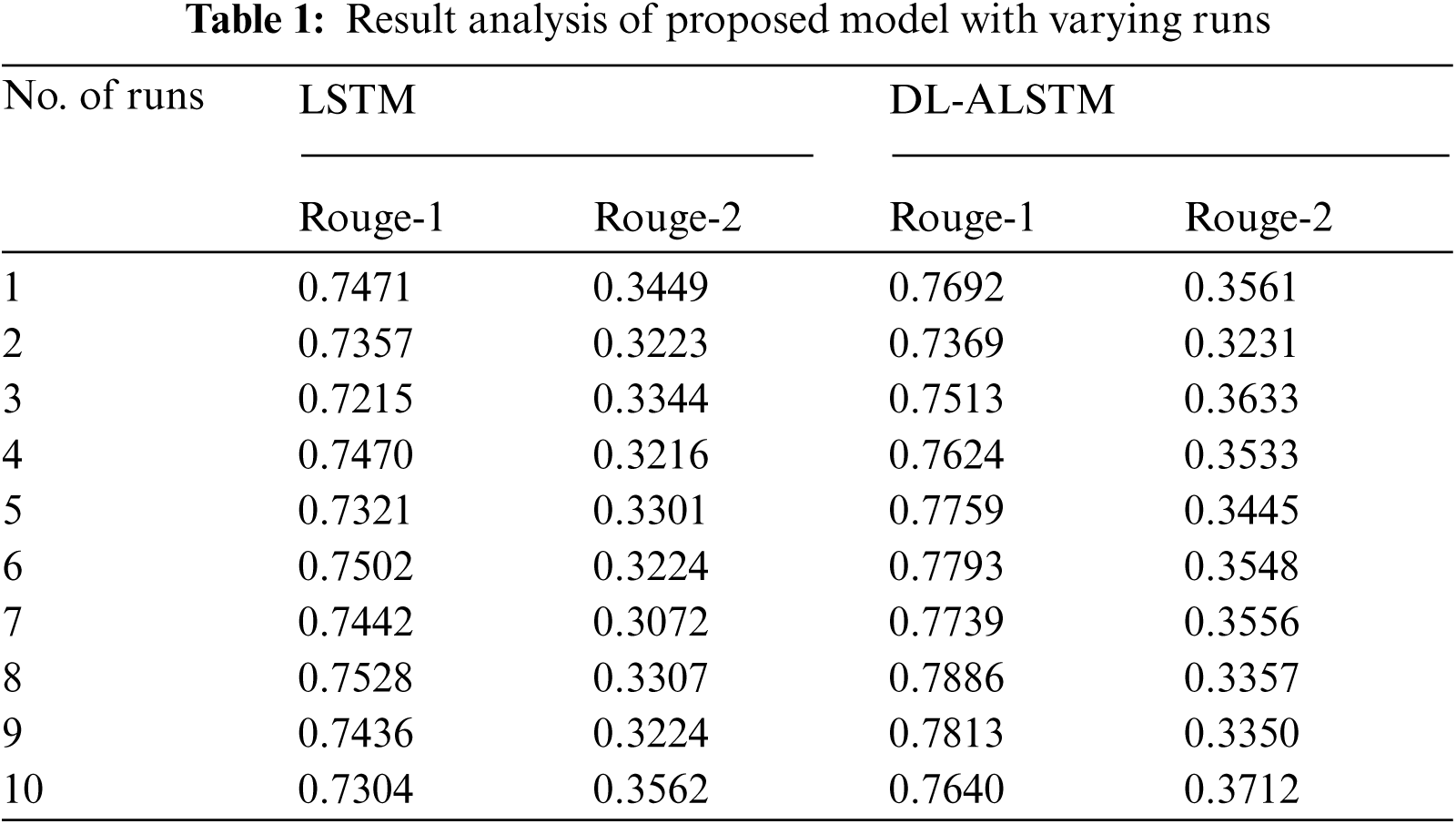
Fig. 9 portrays the Rouge-1 analysis of the DL-ALSTM manner on the applied dataset. The figure exhibited that the DL-ALSTM approach has reached moderately considerable ROUGE-1 values. For instance, the DL-ALSTM manner has reached Rouge-1 of 0.7692 under the execution run-1. Followed by, the DL-ALSTM approach has resulted in a Rouge-1 of 0.7369 under the execution run-2. At the same time, the DL-ALSTM technique has led to a Rouge-1 of 0.7624 under the execution run-4. In line with, the DL-ALSTM algorithm has resulted in a Rouge-1 of 0.7886 under the execution run-8. Eventually, the DL-ALSTM methodology has resulted in a Rouge-1 of 0.7640 under the execution run-10.
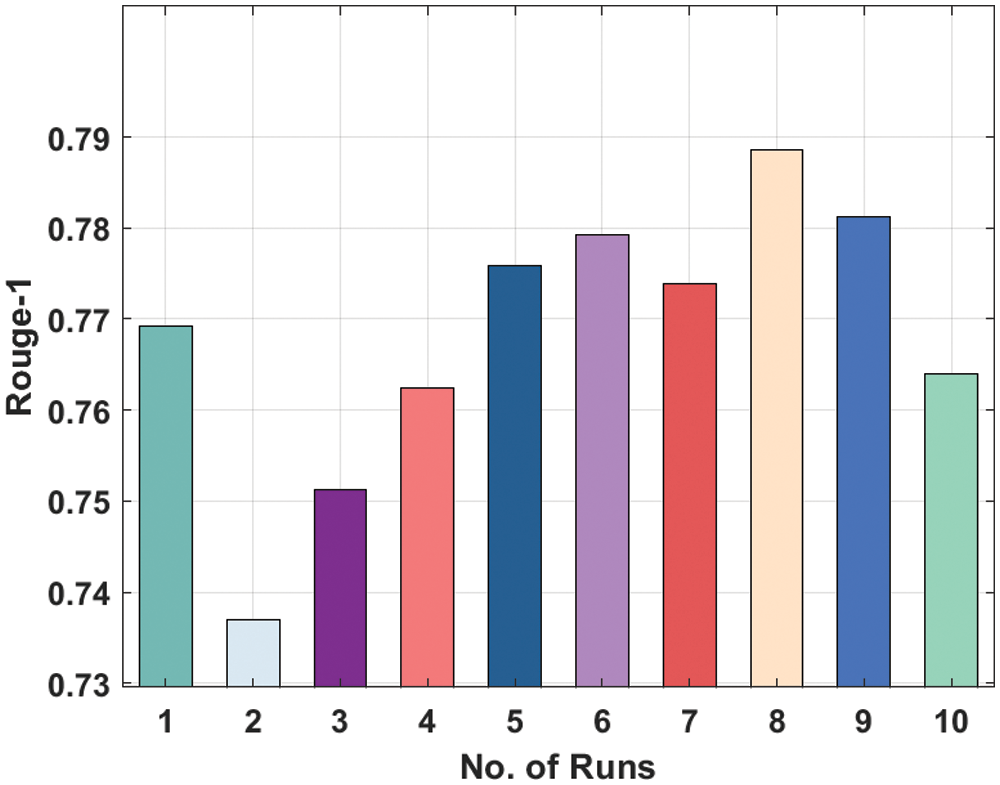
Figure 9: Result analysis of DL-ALSTM model in terms of Rouge-1
Fig. 10 displays the Rouge-2 analysis of the DL-ALSTM method on the applied dataset. The figure outperformed that the DL-ALSTM model has attained moderately considerable ROUGE-2 values. For instance, the DL-ALSTM model has attained Rouge-2 of 0.3561 under the execution run-1. Besides, the DL-ALSTM manner has resulted in a Rouge-2 of 0.3231 under the execution run-2. In the meantime, the DL-ALSTM algorithm has resulted in a Rouge-2 of 0.3533 under the execution run-4. Simultaneously, the DL-ALSTM technique has resulted in a Rouge-2 of 0.3357 under the execution run-8. Finally, the DL-ALSTM approach has resulted in a Rouge-2 of 0.3712 under the execution run-10.
Figure 10: Result analysis of DL-ALSTM model in terms of Rouge-2
A brief comparative results analysis of the DL-ALSTM model with recent approaches takes place in Tab. 2. Fig. 11 investigates the Rouge-1 analysis of the DL-ALSTM model with existing techniques. The figure shows that the Bayesian BS, BERT-Base and SUMMA approaches have obtained reduced Rouge-1 values of 0.7288, 0.7257, and 0.7098. At the same time, the BioBERT-pubmed, CIBS, and BioBERT-pmc techniques have obtained slightly increased Rouge-1 values of 0.7376, 0.7345, and 0.7309 respectively. Moreover, the LSTM and BERT-Large techniques have resulted in reasonable Rouge-1 values of 0.7528 and 0.7504 respectively. However, the proposed DL-ALSTM technique has accomplished superior performance with the maximum Rouge-1 of 0.7886.
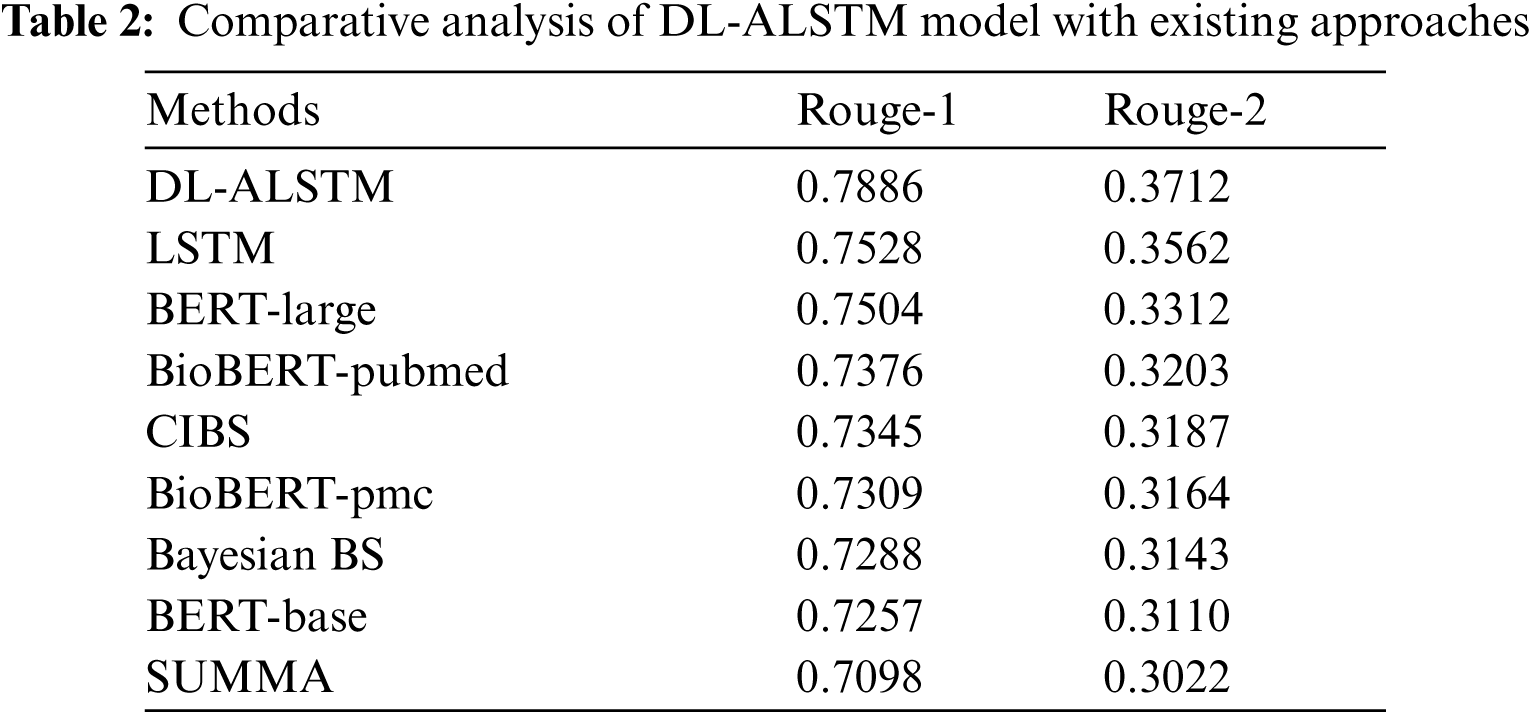
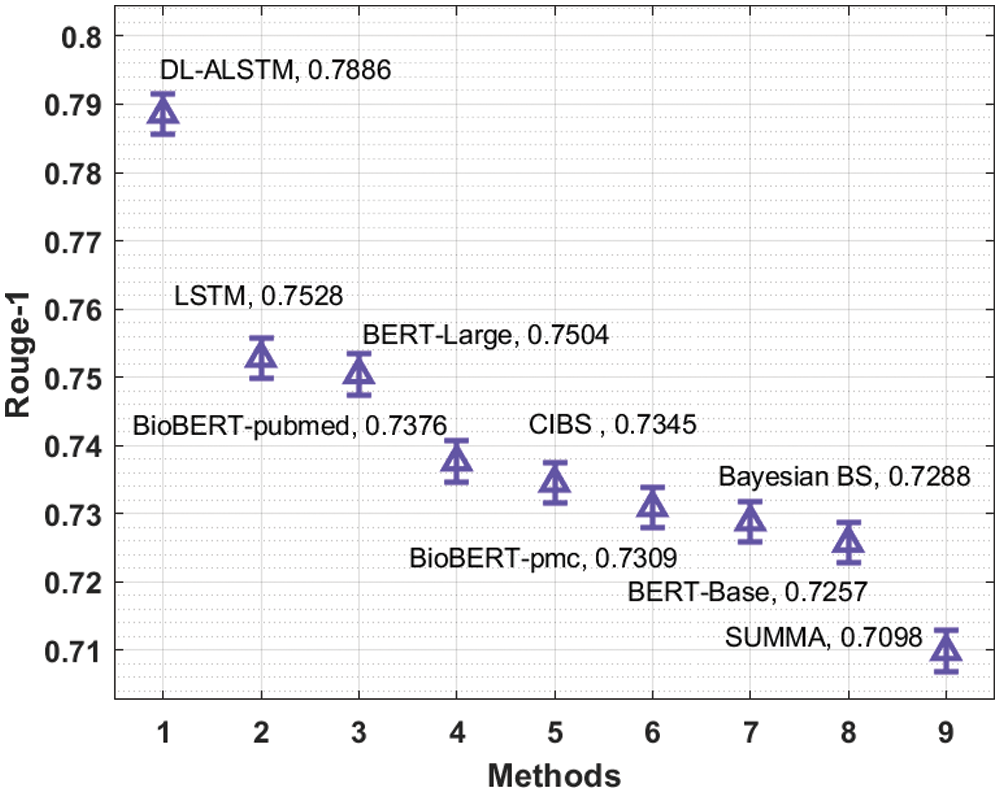
Figure 11: Comparative analysis of DL-ALSTM model interms of Rouge-1
Fig. 12 examines the Rouge-2 analysis of the DL-ALSTM manner with existing algorithms. The figure outperformed that the Bayesian BS, BERT-Base and SUMMA manners have reached minimal Rouge-2 values of 0.3143, 0.3110, and 0.3022. Likewise, the BioBERT-pubmed, CIBS, and BioBERT-pmc algorithms have gained slightly enhanced Rouge-2 values of 0.3203, 0.3187, and 0.3164 correspondingly. Moreover, the LSTM and BERT-Large techniques have resulted in reasonable Rouge-2 values of 0.3562 and 0.3312 correspondingly. But, the presented DL-ALSTM technique has accomplished higher efficiency with the maximal Rouge-2 of 0.3712.
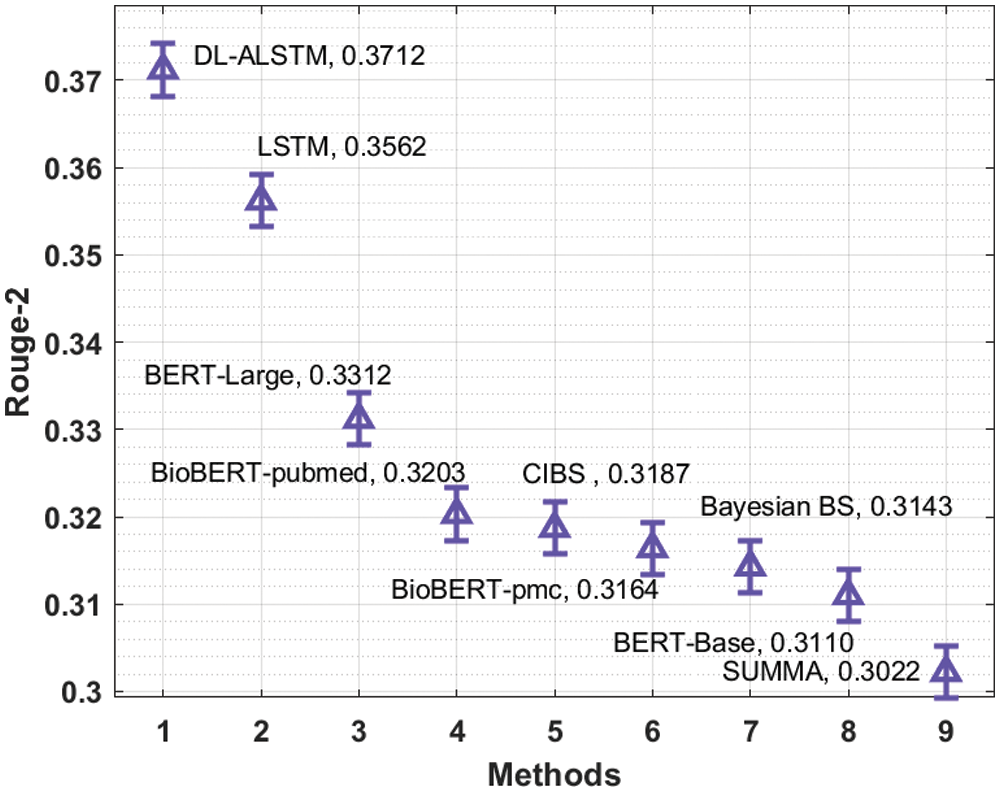
Figure 12: Comparative analysis of DL-ALSTM model interms of Rouge-2
By looking into the above mentioned tables and figures, it is apparent that the DL-ALSTM technique is found to be an effective tool for biomedical text summarization process.
In this study, an effective DL-ALSTM model has been presented for Multi-document Biomedical Text Summarization. The proposed DL-ALSTM model initially performs data preprocessing to convert the available medical data into a compatible format for further processing. Then, the DL-ALSTM model gets executed to summarize the contents from the multiple biomedical documents. In order to tune the summarization performance of the DL-ALSTM model, CGSO algorithm is employed. Extensive experimentation analysis is performed to ensure the betterment of the DL-ALSTM model and the results are investigated using the PubMed dataset. Comprehensive comparative result analysis is carried out to showcase the efficiency of the proposed DL-ALSTM model with the recently presented models. In future, the performance of the DL-ALSTM model can be improvised by the use of advanced hybrid metaheuristic optimization techniques.
Acknowledgement: The authors would like to acknowledge the support of Prince Sultan University for paying the Article Processing Charges (APC) of this publication.
Funding Statement: This work is funded by Deanship of Scientific Research at King Khalid University under Grant Number (RGP 1/279/42). https://www.kku.edu.sa.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. R. Mishra, J. Bian, M. Fiszman, C. R. Weir, S. Jonnalagadda et al., “Text summarization in the biomedical domain: A systematic review of recent research,” Journal of Biomedical Informatics, vol. 52, pp. 457–467, 2014. [Google Scholar]
2. Y. Shang, Y. Li, H. Lin and Z. Yang, “Enhancing biomedical text summarization using semantic relation extraction,” PLoS ONE, vol. 6, no. 8, pp. e23862, Aug. 2011. [Google Scholar]
3. R. Paulus, C. Xiong and R. Socher, “A deep reinforced model for abstractive summarization,” arXiv preprint arXiv:1705.04304v3, Nov. 2017. [Google Scholar]
4. C. Yongkiatpanich and D. Wichadakul, “Extractive text summarization using ontology and graph-based method,” in 2019 IEEE 4th Int. Conf. on Computer and Communication Systems (ICCCS), Singapore, pp. 105–110, 2019. [Google Scholar]
5. T. Xie, Y. Zhen, T. Li, C. Li and Y. Ge, “Self-supervised extractive text summarization for biomedical literatures,” in 2021 IEEE 9th Int. Conf. on Healthcare Informatics (ICHI), Victoria, BC, Canada, pp. 503–504, 2021. [Google Scholar]
6. M. Moradi, “Frequent itemsets as meaningful events in graphs for summarizing biomedical texts,” in 2018 8th Int. Conf. on Computer and Knowledge Engineering (ICCKE), Mashhad, Iran, pp. 135–140, 2018. [Google Scholar]
7. Z. Li, H. Lin, C. Shen, W. Zheng, Z. Yang et al., “Cross2self-attentive bidirectional recurrent neural network with bert for biomedical semantic text similarity,” in 2020 IEEE Int. Conf. on Bioinformatics and Biomedicine (BIBM), Seoul, Korea (Southpp. 1051–1054, 2020. [Google Scholar]
8. S. Candemir, S. Antani, Z. Xue and G. Thoma, “Novel method for storyboarding biomedical videos for medical informatics,” in 2017 IEEE 30th Int. Symp. on Computer-Based Medical Systems (CBMS), Thessaloniki, Greece, pp. 127–132, 2017. [Google Scholar]
9. Z. Gero and J. C. Ho, “Uncertainty-based self-training for biomedical keyphrase extraction,” in 2021 IEEE EMBS Int. Conf. on Biomedical and Health Informatics (BHI), Athens, Greece, pp. 1–4, 2021. [Google Scholar]
10. A. Rai, S. Sangwan, T. Goel, I. Verma and L. Dey, “Query specific focused summarization of biomedical journal articles,” in 2021 16th Conf. on Computer Science and Intelligence Systems (FedCSIS), Sofia, Bulgaria, pp. 91–100, 2021. [Google Scholar]
11. F. Gigioli, N. Sagar, A. Rao and J. Voyles, “Domain-aware abstractive text summarization for medical documents,” in 2018 IEEE Int. Conf. on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, pp. 2338–2343, 2018. [Google Scholar]
12. B. Muthu, C. Sivaparthipan, P. M. Kumar, S. Kadry, C. H. Hsu et al., “A framework for extractive text summarization based on deep learning modified neural network classifier,” ACM Transactions on Asian and Low-Resource Language Information Processing, vol. 20, no. 3, pp. 1–20, Jul. 2021. [Google Scholar]
13. H. Fouad, “A special section on machine learning in biomedical signal and medical image processing,” Journal of Medical Health Informatics, vol. 9, no. 3, pp. 480–481, 2019. [Google Scholar]
14. N. Alami, M. Meknassi, N. E. Nahnahi, Y. E. Adlouni and O. Ammor, “Unsupervised neural networks for automatic Arabic text summarization using document clustering and topic modeling,” Expert Systems with Applications, vol. 172, pp. 114652, 2021. [Google Scholar]
15. O. Rouane, H. Belhadef and M. Bouakkaz, “Combine clustering and frequent itemsets mining to enhance biomedical text summarization,” Expert Systems with Applications, vol. 135, pp. 362–373, 2019. [Google Scholar]
16. O. Rouane, H. Belhadef and M. Bouakkaz, “A new biomedical text summarization method based on sentence clustering and frequent itemsets mining,” in Int. Conf. on the Sciences of Electronics, Technologies of Information and Telecommunications, Springer, Cham, pp. 144–152, 2018. [Google Scholar]
17. M. Moradi, M. Dashti and M. Samwald, “Summarization of biomedical articles using domain-specific word embeddings and graph ranking,” Journal of Biomedical Informatics, vol. 107, pp. 103452, Jul. 2020. [Google Scholar]
18. M. Moradi, G. Dorffner and M. Samwald, “Deep contextualized embeddings for quantifying the informative content in biomedical text summarization,” Computer Methods and Programs in Biomedicine, vol. 184, pp. 105117, Feb. 2020. [Google Scholar]
19. E. Egonmwan and Y. Chali, “Transformer and seq2seq model for paraphrase generation,” in Proc. of the 3rd Workshop on Neural Generation and Translation, Hong Kong, pp. 249–255, 2019. [Google Scholar]
20. M. Marinaki and Y. Marinakis, “A glowworm swarm optimization algorithm for the vehicle routing problem with stochastic demands,” Expert Systems with Applications, vol. 46, pp. 145–163, 2016. [Google Scholar]
21. Dataset: https://github.com/armancohan/long-summarization, 2021. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |