DOI:10.32604/cmc.2022.024367

| Computers, Materials & Continua DOI:10.32604/cmc.2022.024367 |  |
| Article |
Optimal Deep Learning Based Inception Model for Cervical Cancer Diagnosis
1Computer Science Department, Faculty of Information Technology, Al Hussein bin Talal University, Ma'an, 71111, Jordan
2MIS Department, College of Business Administration, University of Business and Technology, Jeddah, 21448, Saudi Arabia
3Faculty of Science, Computer Science Department, Northern Border University, Arar, 91431, Saudi Arabia
*Corresponding Author: Bassam A. Y. Alqaralleh. Email: b.alqaralleh@ubt.edu.sa
Received: 15 October 2021; Accepted: 20 December 2021
Abstract: Prevention of cervical cancer becomes essential and is carried out by the use of Pap smear images. Pap smear test analysis is laborious and tiresome work performed visually using a cytopathologist. Therefore, automated cervical cancer diagnosis using automated methods are necessary. This paper designs an optimal deep learning based Inception model for cervical cancer diagnosis (ODLIM-CCD) using pap smear images. The proposed ODLIM-CCD technique incorporates median filtering (MF) based pre-processing to discard the noise and Otsu model based segmentation process. Besides, deep convolutional neural network (DCNN) based Inception with Residual Network (ResNet) v2 model is utilized for deriving the feature vectors. Moreover, swallow swarm optimization (SSO) based hyperparameter tuning process is carried out for the optimal selection of hyperparameters. Finally, recurrent neural network (RNN) based classification process is done to determine the presence of cervical cancer or not. In order to showcase the improved diagnostic performance of the ODLIM-CCD technique, a series of simulations occur on benchmark test images and the outcomes highlighted the improved performance over the recent approaches with a superior accuracy of 0.9661.
Keywords: Median filtering; convolutional neural network; pap smear; cervical cancer
Cervical cancer is worldwide disease cancer that is very popular amongst females, and simultaneously, this is a curable and preventable cancer [1]. Human Papilloma Virus (HPV) or so-called cervical disease is a very common kind of female cancer around the world [2]. The Papanicolaou research is becoming the keystone of cervical screening for the previous sixty years. The Papanicolaou test, also known as the Pap smear/Pap test, was conducted by Georgios Papanikolaou in the year of 1940. It comprises exfoliating cells from the cervix development to enable microscopic estimation of these cells to track precancerous/cancerous caries. The multiple aspects are due to prescription medication, body immune suppression of cells, and cigarettes. Screening smear test appears that time consuming and frequently outcomes are incorrect. Depiction contains dead blood/disproportionate on a cell, overlapped as well as irregular patterning [3]. Trend analyses and computerized tracking of the smear test are becoming an impressive function of screening images.
Up till now, different kinds of studies have focused on preventive HPV deoxyribonucleic acid (DNA) testing, HPV vaccine, Pap smear problems, and other recommendations for the preventive of cervical cancer. Prevention of secondary screening still remains a significant portion because screening plays an important part in HPV vaccine since the vaccine does not fairly recompense for higher risk HPV [4]. Cervical cancer is common cancer amongst females worldwide however during that time, it is the furthest treatable and preventable cancer. Mostly, cervical cancer initiates by pre-cancerous variations and grows relatively slow [5]. Clinicians in the medical centre have a problem recognizing the cancer cells since the nucleus of the cells is occasionally really difficult for observing with bare eyes. Conversely, the challenges to recognizing the precise information of the cancer stage. Many persons get the consequence that they are on stage 2, afterward re-testing, it can be essentially on stage 4 that the possibility to cure is lower [6]. This occurs since the clinician cannot detect the precise sample and balance data exactly. Currently, the method of computerized image analyses utilized for assisting artificial diagnoses of or tumors/cell abnormalities in histopathology/cytopathology could offer precise and objective assessment of nuclear morphology [7]. But, the proficient clinician would have distinct perceptions regarding the cancer stage in accordance with image screening.
This paper designs an optimal deep learning based Inception model for cervical cancer diagnosis (ODLIM-CCD) using pap smear images. The proposed ODLIM-CCD technique incorporates median filtering (MF) based pre-processing to discard the noise and Otsu model based segmentation process. Besides, deep convolutional neural network (DCNN) based Inception with Residual Network (ResNet) v2 model is utilized for deriving the feature vectors. Moreover, swallow swarm optimization (SSO) based hyperparameter tuning process is carried out for the optimal selection of hyperparameters. Finally, recurrent neural network (RNN) based classification process is done to determine the presence of cervical cancer or not. In order to showcase the enhanced diagnostic performance of the ODLIM-CCD technique, a series of simulations occur on benchmark test images and the outcomes highlighted the improved performance over the recent approaches.
In Moldovan [8], the cervical cancer diagnoses can be advanced with machine learning (ML) technique where the features are chosen by linear relation, and the information is categorized into the support vector machine (SVM) method. The hyperparameter of the SVM is chosen by chicken swarm optimization (CSO) model. The technique is validated and tested on the open-source cervical cancer (Risk Factor) Dataset from the UCI ML Repository. In Karim et al. [9] a method with ensemble method using SVM as the base classifiers are considered. The ensemble approach using Bagging method attained a precision of 98.12% with higher f-measure, accuracy, and recall values. In Li et al. [10], they presented a deep learning (DL) architecture for the precise recognitions of Low-Grade Squamous Intraepithelial Lesion (LSIL) (includes cervical cancer and Cervical intraepithelial neoplasia (CIN)) with time-lapsed colposcopic image. The presented architecture includes 2 major modules, viz., feature fusion network and key frame feature encoding network. In many fusion methods are related, each of them outperforms the present automatic cervical cancer diagnoses system with an individual time slot. A graph convolutional network with edge features (E-GCN) is established that an accurate fusion method, because of its outstanding explainability reliable with medical training.
In Erkaymaz et al. [11], cervical cancer is recognized by 4 fundamental classifications: Naïve Bayes (NB), K-nearest neighbor (KNN), multilayer perceptron (MLP), and decision tree (DT) methods and random subspace ensemble model. The Gain Ratio Attribute Evaluation (GRAE) feature extraction is employed for contributing to classification accuracy. The classification result attained by each dataset and reduced dataset is related based on efficiency criteria like Specificity performance criteria, accuracy, root mean square error (RMSE), and sensitivity. William et al. [12] proposed a summary of an advanced publication focuses on automatic diagnoses and classification of cervical cancer from pap-smear image. It analyses 30 journal papers attained automatically by 4 systematic datasets examined by 3 sets of keywords: (1) Pap-smear Images, Automated, Segmentation; (2) Cervical Cancer, Segmentation, Classification; (3) Machine Learning, pap-smear Images, Medical Imaging. The analysis establishes that few models are utilized often when compared to another model: e.g., KNN, filtering, and thresholding are the commonly utilized methods for preprocessing, classification, and segmentation of pap-smear images.
In Hemalatha et al. [13], the frequently employed neural networks. Dimensionally reduced Cervical Pap smear Datasets with Fuzzy Edge Recognition technique is taken into account for classifier. The 4 NN are related and the appropriate networks for classifying the datasets are estimated. Huang et al. [14] suggest an approach of cervical biopsy tissue image classification depends upon ensemble learning-support vector machine (EL-SVM) and least absolute shrinkage and selection operator (LASSO) methods. With the LASSO method for FS, the average optimization time has been decreased by 35.87 s when guaranteeing the exactness of the classification, later serial fusion has been carried out. The EL-SVM classification has been utilized for identifying and classifying 468 biopsy tissue images, and the error and ROC curves are utilized for evaluating the generalization capacity of the classification.
Haryanto et al. [15] focus on creating the classifier method of Cervical Cell Images with the convolutional neural network (CNN) method. The dataset utilized is the image datasets SIPaKMeD. The CNN method has been employed with the AlexNet framework and non-padding system. Nithya et al. [16] aim at detecting cervical cancer and the datasets utilized in the study comprising imbalanced target classes, missing values, and redundant features. Therefore, this work aims at handling this problem via incorporated FS method for attaining an optimum feature subset. The subset obtained by this combined method is applied in increased predictive tasks. The optimal and best feature subsets are according to the efficacy of the classifier in forecasting the outcomes. In Rahaman et al. [17], they offer a complete analysis of an advanced method depends on DL method for the analyses of cervical cytology images. Initially, present DL and its simplified framework which is employed in this region. Next, discusses the open-source cervical cytopathology dataset and assessment metrics for classification and segmentation method. Next, a complete study of the current growth of DL method for the classification and segmentation of cervical cytology images has been proposed. Lastly, examine the current method and appropriate methods for the analyses of pap smear cell.
In this study, a novel ODLIM-CCD technique is derived to classify cervical cancer using pap smear images. The proposed ODLIM-CCD technique incorporates MF based pre-processing, Otsu based segmentation, Inception with ResNet v2 model based feature extraction, SSO based hyperparameter tuning, and RNN based classification process is done to determine the presence of cervical cancer or not. Fig. 1 showcases the overall process of ODLIM-CCD model.
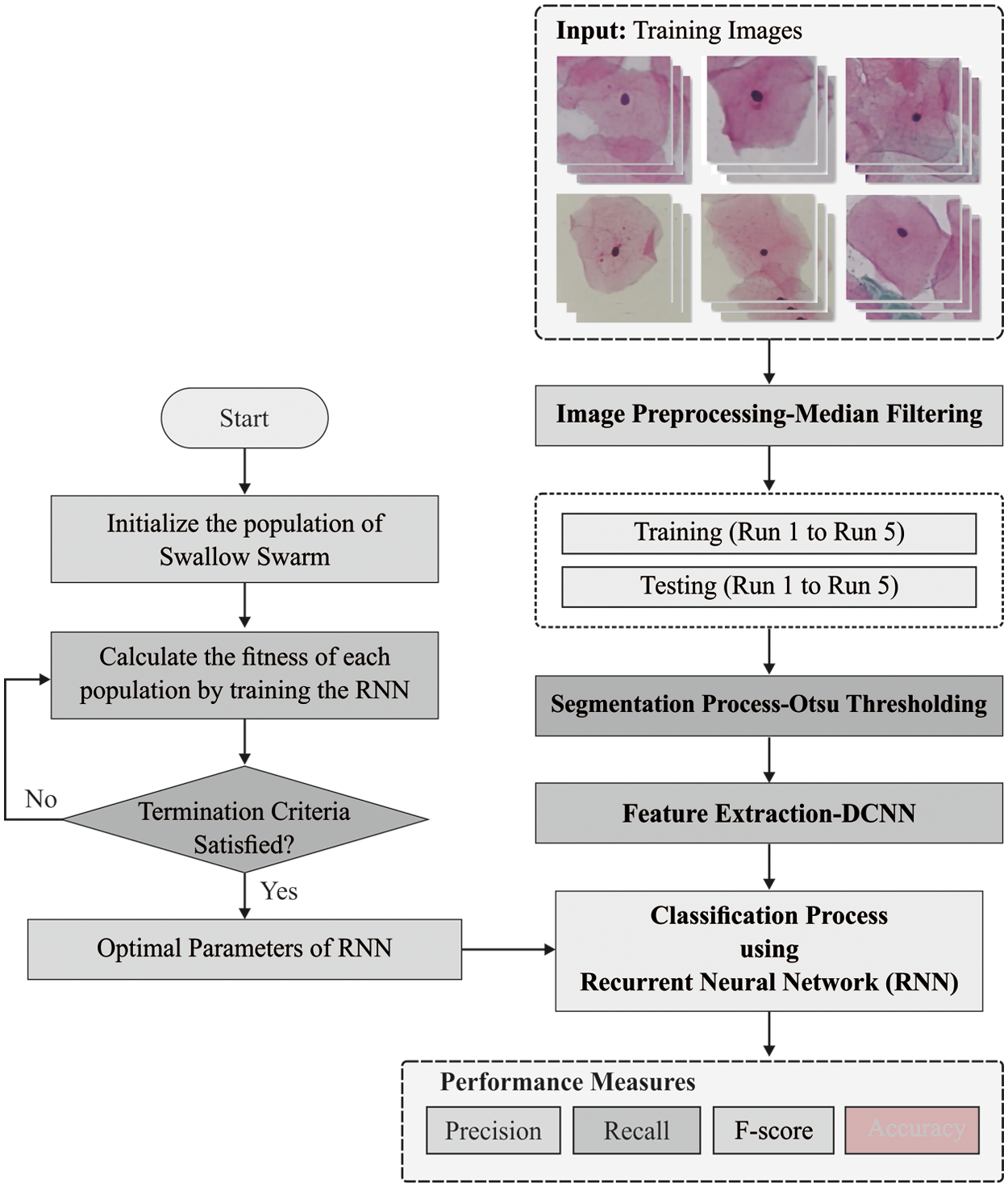
Figure 1: Overall process of ODLIM-CCD model
The MF has been non-linear signal modeling technologies dependent upon statistics. A noisy value of digital images or order has been exchanged with median values of region (masks). The pixel of masks have been ordered under the sequence of their gray level, and the median value of fixed has been stored for replacing the noisy values. The MF resultant has
The pre-processed images are segmented using Otsu technique to determine the affected regions. Otsu (1979) is a segmentation method utilized for finding an optimum threshold value of the images depending on increasing the between class variances. These approaches are utilized for finding the threshold optimal value which separates the images into several classes [18]. These methods identify
In which
Whereas
It is essential to detect the average intensity levels
It is noted that
Let
In the equation,
In which
whereas i denotes a certain class,
for mean values:
3.3 Inception with ResNet v2 Based Feature Extraction
CNN is certain type of neural network in which the weight is learned for the application of a sequence of convolution on the input image, being the filter weight shared over a similar convolution layer. This design and related learning mechanism is thoroughly discussed.
CNN replaces fully connected (FC) affine layer
Convolution layer j is defined as a set
All these convolution layers are followed by non-linear pointwise function and the spatial size
As above mentioned, the amount of algorithmic developments have been presented in the last few years. e.g., the execution of
Inception-ResNet-V2 (IRV2) proposed by Google Company is employed by an advanced method to classify mammograms. It is extremely a fusion of GoogLeNet (Inception) and ResNet. Inception has been popular network using a similar layer framework utilized in GoogleNet. Currently, Inception v1–v4 is a common method of GoogleNet. Therefore, the residual learning based ResNet has efficient when compared to ILSVRC 2015 that drives deeper to the 152 layers. The previous network framework is determined by a non-linear conversion of input, while Highway Network permits only certain output from the conventional network layer that should be often trained. Furthermore, the real input enhancement is transmitted to the successive layer. Simultaneously, ResNet safeguards the information by straightforward data forwarding of an output.
In Residual-Inception model, the Inception is employed since it is made up of low processing difficulty than the original Inception model. The amount of layers in this technique for each module is 5, 10, and 5, respectively. According to the conventional research, IRV2 has determined from that maps the original costs of Inception-v4 model. Now, methodological variations between non-residual and residual Inceptions are Inception-ResNet, i.e., denoted as BN algorithm was employed at the traditional layers.
3.4 SSO Based Hyperparameter Tuning
For optimally adjusting the hyperparameter involved in the Inception with ResNet v2 model, the SSO algorithm is utilized. The SSO method stimulated from the communal motion of swallows and the interactions between flock members have attained better outcomes. This method has proposed a metaheuristic model on the basis of the specific features of swallows, include intelligent social relations, fast flight, and hunting skills. Now, the method is equivalent to particle swarm optimization (PSO) algorithm; however, it has exclusive features that could not be established in an equivalent method, include the usage of 3 kinds of particles: Leader Particles
1. Location of the local leader (LL).
2. Location of the global leader (GL).
3. The optimal individual experience alongside the path.
4. The preceding path.
The particle uses the succeeding formula to search and continue the path:
Eq. (13) illustrates the velocity vector undeer the path of the global leader.
Eqs. (14) and (15) estimates the acceleration co-efficient
Eqs. (4) and (5) estimate the acceleration co-efficient
The
At the last stage, the feature vectors are provided into the RNN model, which identifies the presence or absence of cervical cancer. The RNN is a kind of feed forward neural networks (FFNN). The RNN has been desired to model orders on FFNN as it has a cyclic connection. The letters X, H, and y utilized for indicating an input order, a hidden vector order, and resultant vector order correspondingly.
where
At this point,
For accommodating a variable-length order input, the RNN has been employed to back-propagation training time (BPTT). This technique was primarily trained to utilize training data in back-propagation (BP) trained period, and the resultant error gradient has been saved to any time step. The RNN was extremely tough for training, however, it causes the gradient for bursting/disappearing with the entire trained with BPTT technique. This p has been declared. Fig. 2 illustrates the structure of RNN.
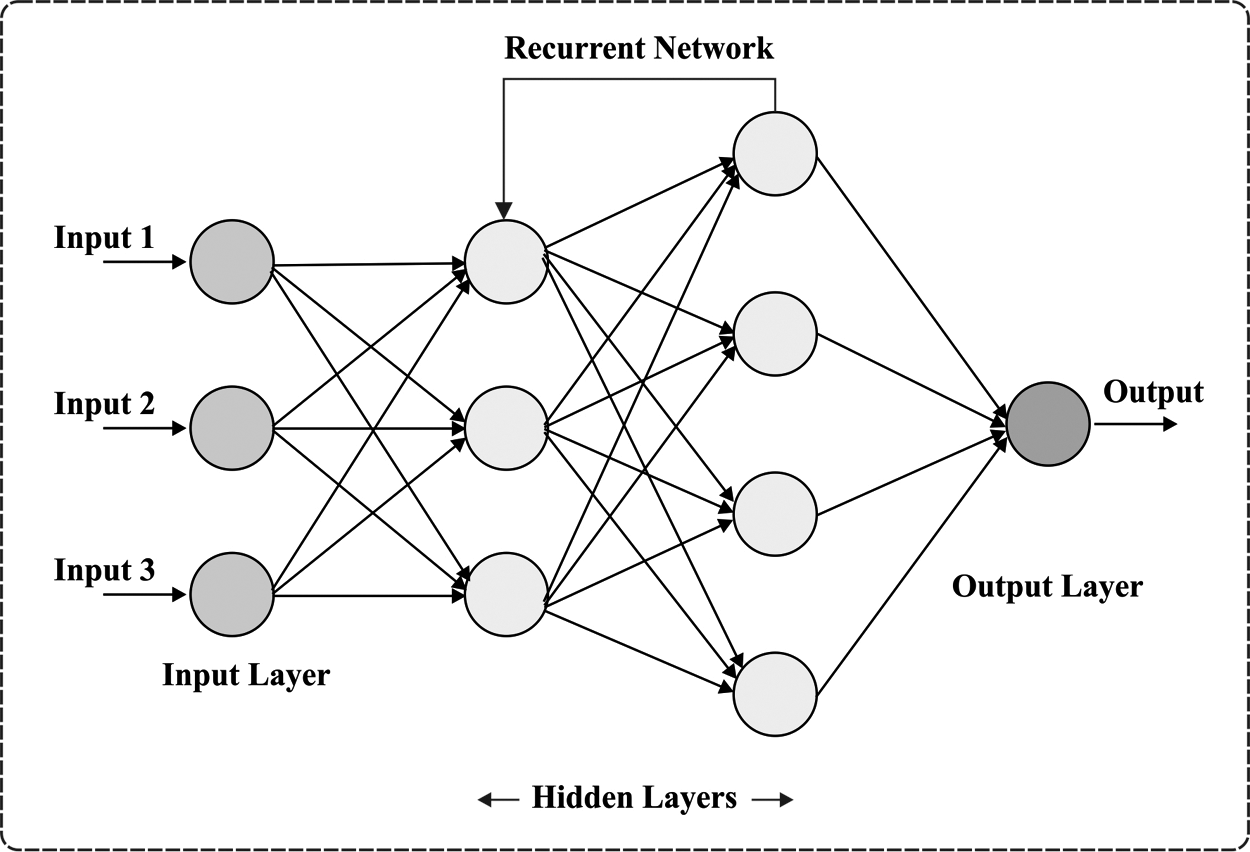
Figure 2: RNN structure
The performance validation of the ODLIM-CCD technique takes place using benchmark Herlev pap smear image dataset, which contains 918 images into normal and abnormal classes. Fig. 3 illustrates a few sample images.
Tab. 1 offers a detailed comparative analysis of the ODLIM-CCD model with other ones. Fig. 4 and Fig. 5 demonstrated that the ODLIM-CCD technique has outperformed the existing techniques interms of different measures under varying runs. For instance, the ODLIM-CCD technique has gained effective outcomes with an average precision of 96.68% whereas the MLP, random forest (RF), and SVM models have obtained lower average precision of 96.59%, 96.07%, and 95.42% respectively. Moreover, the ODLIM-CCD approach has reached effective outcomes with an average recall of 97.39% whereas the MLP, RF, and SVM techniques have attained lower average recall of 95.61%, 95.38%, and 95.10% correspondingly. Furthermore, the ODLIM-CCD approach has gained effective outcomes with an average accuracy of 96.61% whereas the MLP, RF, and SVM techniques have obtained reduced average accuracy of 96.12%, 95.18%, and 94.90% respectively. Likewise, the ODLIM-CCD methodology has obtained effective outcomes with an average F-score of 97.17% whereas the MLP, RF, and SVM systems have gained minimum average F-score of 95.85%, 95.34%, and 95.20% correspondingly.
Figure 3: Sample images
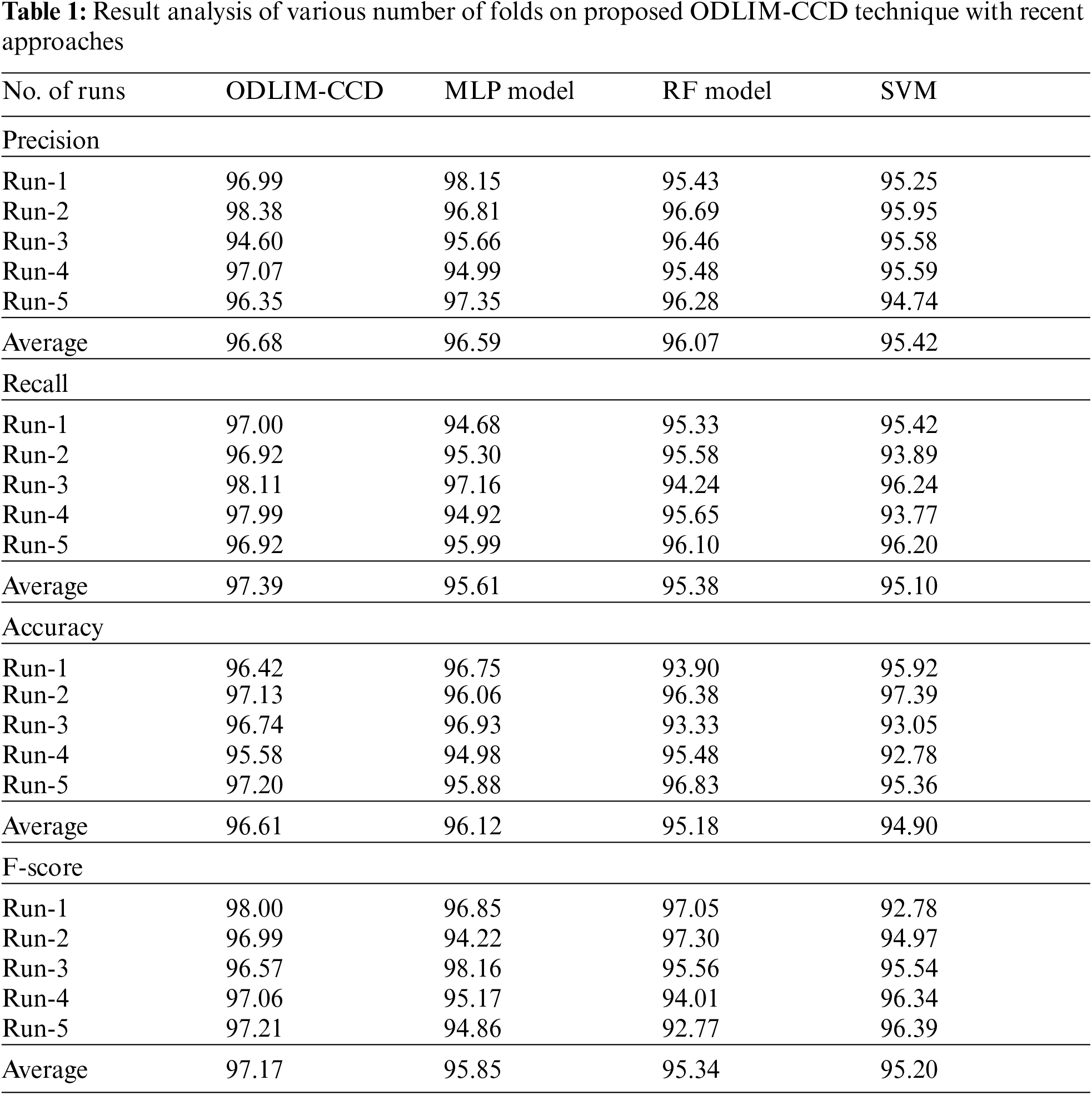

Figure 4: Precision analysis of different models under varying runs

Figure 5: Accuracy analysis of different models under varying runs
In order to ensure the enhanced cervical cancer classification outcome of the ODLIM-CCD technique, a detailed comparative analysis is made in Tab. 2.
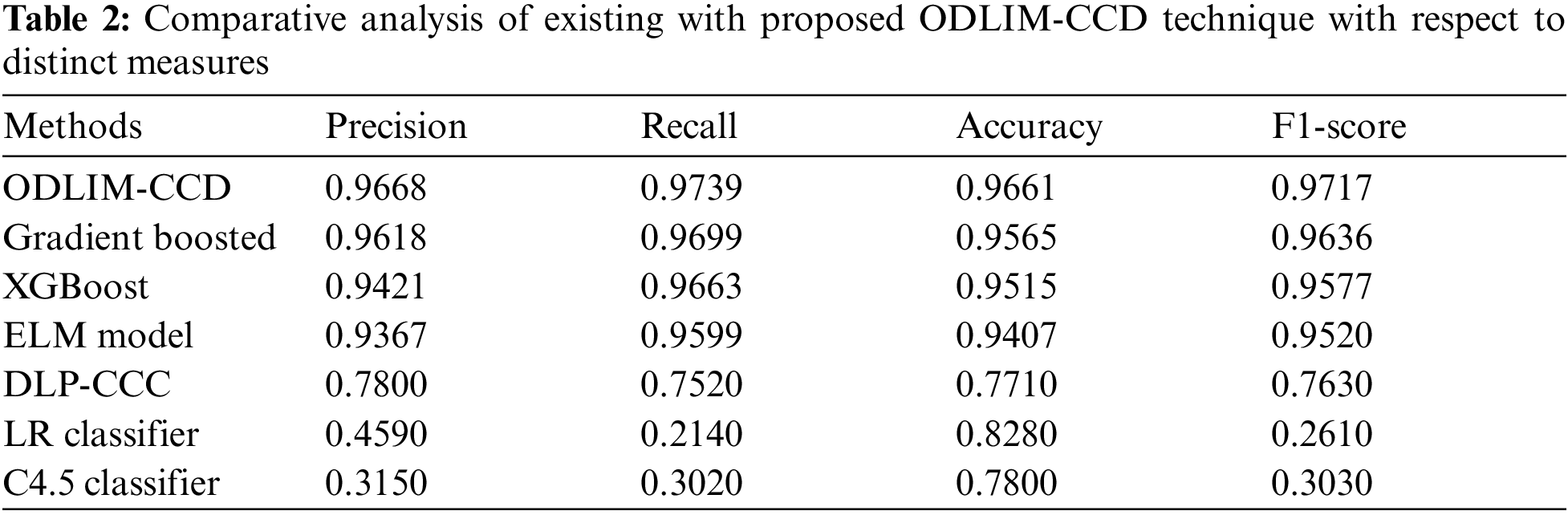
Fig. 6 portrays the comparative analysis of the ODLIM-CCD system with other ones in terms of precision. The figure has shown that the C4.5 and logistic regression (LR) classifiers have obtained reduced precision of 0.315 and 0.459 respectively. Besides, the sss has gained slightly increased precision of 0.78 whereas the extreme learning machine (ELM), extreme gradient boosting (XGBoost), and Gradient Boosting models have accomplished near optimal precision of 0.9367, 0.9421, and 0.9618 respectively. However, the ODLIM-CCD technique has showcased better outcomes with a higher precision of 0.9668.

Figure 6: Comparative precision analysis of ODLIM-CCD technique with recent methods
Fig. 7 show cases the comparative analysis of the ODLIM-CCD technique with other ones with respect to recall. The figure has shown that the C4.5 and LR classifiers have obtained reduced recall of 0.302 and 0.214 respectively. Besides, the DLP-CC model has gained slightly increased recall of 0.752 whereas the ELM, XGBoost, and Gradient Boosting models have accomplished near optimal recall of 0.9599, 0.9663, and 0.9699 respectively. However, the ODLIM-CCD technique has showcased better outcomes with a higher recall of 0.9739.

Figure 7: Comparative recall analysis of ODLIM-CCD technique with recent methods
Fig. 8 depicts the comparative analysis of the ODLIM-CCD technique with other ones in terms of accuracy. The figure exhibited that the C4.5 and LR classifiers have obtained reduced accuracy of 0.780 and 0.828 respectively. In addition, the DLP-CC model has gained somewhat higher accuracy of 0.771 whereas the ELM, XGBoost, and Gradient Boosting models have accomplished near optimal accuracy of 0.9407, 0.9515, and 0.9565 correspondingly. At last, the ODLIM-CCD technique has showcased better results with a superior accuracy of 0.9661.
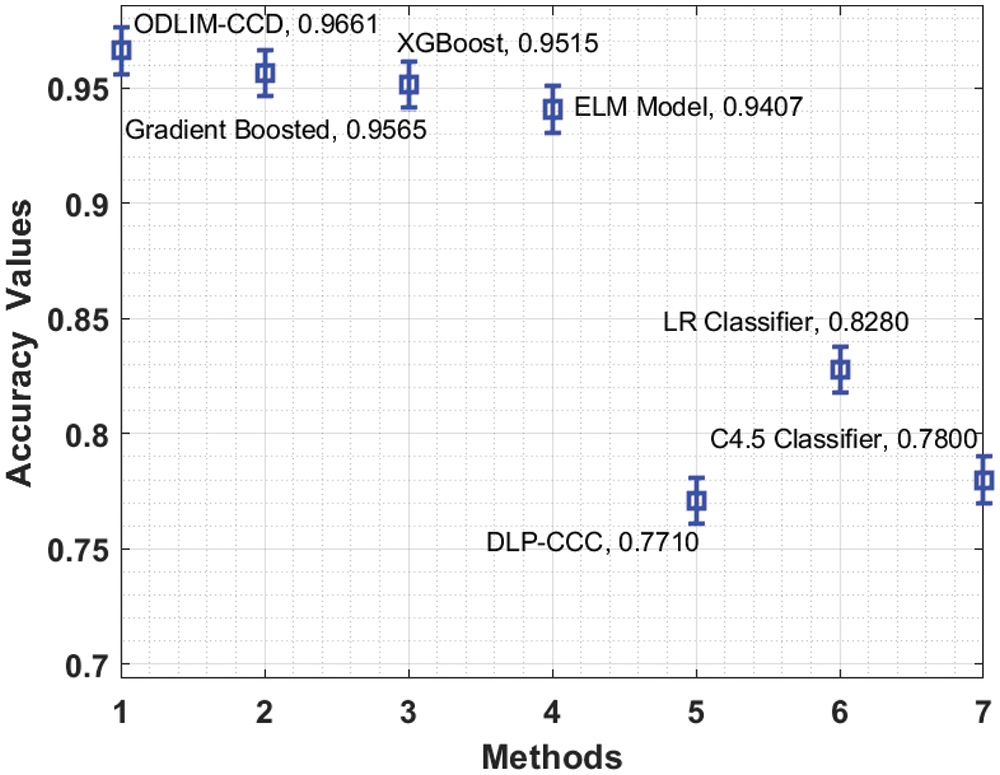
Figure 8: Comparative accuracy analysis of ODLIM-CCD technique with recent methods
Fig. 9 portrays the comparative analysis of the ODLIM-CCD technique with other ones with respect to F1-score. The figure depicted that the C4.5 and LR classifiers have obtained lower F1-score of 0.763 whereas the ELM, XGBoost, and Gradient Boosting approaches have accomplished near optimal F1-score of 0.952, 0.9577, and 0.9636 correspondingly. Finally, the ODLIM-CCD technique has outperformed optimum outcome with the maximum F1-score of 0.9717.

Figure 9: Comparative F-score analysis of ODLIM-CCD technique with recent methods
In this study, a novel ODLIM-CCD technique is derived to classify cervical cancer using pap smear images. The proposed ODLIM-CCD manner incorporates MF based pre-processing, Otsu based segmentation, Inception with ResNet v2 model based feature extraction, SSO based hyperparameter tuning and RNN based classification process. Moreover, SSO based hyperparameter tuning process is carried out for the optimal selection of hyperparameters. Finally, RNN based classification process is done to determine the presence of cervical cancer or not. In order to showcase the enhanced diagnostic efficiency of the ODLIM-CCD technique, a series of simulations occurs on benchmark test images and the outcomes highlighted the improved performance over the recent approaches. In future, the ODLIM-CCD technique can be executed on the cloud server for remote healthcare monitoring applications.
Funding Statement: This Research was funded by the Deanship of Scientific Research at University of Business and Technology, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. M. Schiffman, P. E. Castle, J. Jeronimo, A. C. Rodriguez and S. Wacholder, “Human papillomavirus and cervical cancer,” Lancet, vol. 370, no. 9590, pp. 890–907, 2007. [Google Scholar]
2. S. K. Shankar, L. P. Prieto, M. J. R. Triana and A. RuizCalleja, “A review of multimodal learning analytics architectures,” in 2018 IEEE 18th Int. Conf. on Advanced Learn. Techn. (ICALT), Mumbai, India, pp. 212–214, 2018. [Google Scholar]
3. S. Dimopoulos, C. E. Mayer, F. Rudolf and J. Stelling, “Accurate cell segmentation in microscopy images using membrane patterns,” Bioinformatics, vol. 30, no. 18, pp. 2644–2651, 2014. [Google Scholar]
4. J. Uthayakumar, N. Metawa, K. Shankar and S. K. Lakshmanaprabu, “Intelligent hybrid model for financial crisis prediction using machine learning techniques,” Information Systems and e-Business Management, vol. 18, no. 4, pp. 617–645, 2020. [Google Scholar]
5. R. Sparks and A. Madabhushi, “Explicit shape descriptors: Novel morphologic features for histopathology classification,” Medical Image Analysis, vol. 17, no. 8, pp. 997–1009, 2013. [Google Scholar]
6. M. Elhoseny, K. Shankar and J. Uthayakumar, “Intelligent diagnostic prediction and classification system for chronic kidney disease,” Scientific Reports, vol. 9, no. 1, pp. 9583, 2019. [Google Scholar]
7. E. Bengttson, “Recognizing signs of malignancy-the quest for computer assisted cancer screening and diagnosis systems,” in 2010 IEEE Int. Conf. on Computational Intelligence and Computing Research, Coimbatore, India, pp. 1–6, 2010. [Google Scholar]
8. D. Moldovan, “Cervical cancer diagnosis using a chicken swarm optimization based machine learning method,” in 2020 Int. Conf. on e-Health and Bioengineering (EHB), Iasi, Romania, pp. 1–4, 2020. [Google Scholar]
9. E. Karim and N. Neehal, “An empirical study of cervical cancer diagnosis using ensemble methods,” in 2019 1st Int. Conf. on Advances in Science, Engineering and Robotics Technology (ICASERT), Dhaka, Bangladesh, pp. 1–5, 2019. [Google Scholar]
10. Y. Li, J. Chen, P. Xue, C. Tang, J. Chang et al., “Computer-aided cervical cancer diagnosis using time-lapsed colposcopic images,” IEEE Transactions on Medical Imaging, vol. 39, no. 11, pp. 3403–3415, 2020. [Google Scholar]
11. O. Erkaymaz and T. Palabaş, “Classification of cervical cancer data and the effect of random subspace algorithms on classification performance,” in 2018 26th Signal Processing and Communications Applications Conf. (SIU), Izmir, Turkey, pp. 1–4, 2018. [Google Scholar]
12. W. William, A. H. B. Ejiri, J. Obungoloch and A. Ware, “A review of applications of image analysis and machine learning techniques in automated diagnosis and classification of cervical cancer from pap-smear images,” Computer Methods and Programs in Biomedicine, vol. 164, pp. 15–22, 2018. [Google Scholar]
13. K. Hemalatha and K. U. Rani, “An optimal neural network classifier for cervical pap smear data,” in 2017 IEEE 7th Int. Advance Computing Conf. (IACC), Hyderabad, India, pp. 110–114, 2017. [Google Scholar]
14. P. Huang, S. Zhang, M. Li, J. Wang, C. Ma et al., “Classification of cervical biopsy images based on LASSO and EL-SVM,” IEEE Access, vol. 8, pp. 24219–24228, 2020. [Google Scholar]
15. T. Haryanto, I. S. Sitanggang, M. A. Agmalaro and R. Rulaningtyas, “The utilization of padding scheme on convolutional neural network for cervical cell images classification,” in 2020 Int. Conf. on Computer Engineering, Network, and Intelligent Multimedia (CENIM), Surabaya, Indonesia, pp. 34–38, 2020. [Google Scholar]
16. B. Nithya and V. Ilango, “Machine learning aided fused feature selection based classification framework for diagnosing cervical cancer,” in 2020 Fourth Int. Conf. on Computing Methodologies and Communication (ICCMC), Erode, India, pp. 61–66, 2020. [Google Scholar]
17. M. M. Rahaman, C. Li, X. Wu, Y. Yao, Z. Hu et al., “A survey for cervical cytopathology image analysis using deep learning,” IEEE Access, vol. 8, pp. 61687–61710, 2020. [Google Scholar]
18. L. Xiao, H. Ouyang and C. Fan, “An improved otsu method for threshold segmentation based on set mapping and trapezoid region intercept histogram,” Optik, vol. 196, pp. 163106, 2019. [Google Scholar]
19. Q. Li, C. Li and H. Chen, “Incremental filter pruning via random walk for accelerating deep convolutional neural networks,” in Proc. of the 13th Int. Conf. on Web Search and Data Mining, Houston TX USA, pp. 358–366, 2020. [Google Scholar]
20. A. Kaveh, T. Bakhshpoori and E. Afshari, “An efficient hybrid particle swarm and swallow swarm optimization algorithm,” Computers & Structures, vol. 143, pp. 40–59, 2014. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |