DOI:10.32604/cmc.2022.026131

| Computers, Materials & Continua DOI:10.32604/cmc.2022.026131 |  |
| Article |
Artificial Intelligence Based Prostate Cancer Classification Model Using Biomedical Images
1Department of Computer Science, Faculty of Computing and Information Technology, King Abdulaziz University, Jeddah, 21589, Saudi Arabia
2Department of Computer Science, College of Computers and Information Technology, Taif University, Taif, 21944, Saudi Arabia
3Department of Computer Science, College of Science & Art at Mahayil, King Khalid University, Saudi Arabia
4Department of Computer Science, College of Computer and Information Sciences, Prince Sultan University, Saudi Arabia
5Department of Computer and Self Development, Preparatory Year Deanship, Prince Sattam bin Abdulaziz University, AlKharj, Saudi Arabia
*Corresponding Author: Fahd N. Al-Wesabi. Email: falwesabi@kku.edu.sa
Received: 16 December 2021; Accepted: 22 February 2022
Abstract: Medical image processing becomes a hot research topic in healthcare sector for effective decision making and diagnoses of diseases. Magnetic resonance imaging (MRI) is a widely utilized tool for the classification and detection of prostate cancer. Since the manual screening process of prostate cancer is difficult, automated diagnostic methods become essential. This study develops a novel Deep Learning based Prostate Cancer Classification (DTL-PSCC) model using MRI images. The presented DTL-PSCC technique encompasses EfficientNet based feature extractor for the generation of a set of feature vectors. In addition, the fuzzy k-nearest neighbour (FKNN) model is utilized for classification process where the class labels are allotted to the input MRI images. Moreover, the membership value of the FKNN model can be optimally tuned by the use of krill herd algorithm (KHA) which results in improved classification performance. In order to demonstrate the good classification outcome of the DTL-PSCC technique, a wide range of simulations take place on benchmark MRI datasets. The extensive comparative results ensured the betterment of the DTL-PSCC technique over the recent methods with the maximum accuracy of 85.09%.
Keywords: MRI images; prostate cancer; deep learning; medical image processing; metaheuristics; krill herd algorithm
Prostate cancer is one of the common forms of cancer that is accountable for 26% of cancer diagnoses for American men [1]. So far, prostate cancer is detected by systematic biopsies that contain millions of specimens taken from the prostate through a core needle. A systematic biopsy is invasive and has lower sensitivity; furthermore, it creates a possibility of bleeding, infection, and sepsis [2]. A non-invasive imaging technique to diagnose prostate cancer at an earlier stage might enhance prostate cancer treatment and diagnosis. For a new ultrasound technique, the Contrast Enhanced Ultrasound (CEUS) could offer suitable modality to visualize the dynamic pattern of the blood flows, allowing clinical experts to diagnose angiogenesis for cancer detection [3,4]. Until now, numerous studies of the CEUS based prostate cancer diagnosis are accomplished by measuring distinct parameters of the time intensity curve (TIC). Machine learning (ML) is a subdivision of artificial intelligence (AI) which is depending on the concept of the system learning patterns from a largescale dataset through statistical and probabilistic mechanisms and also make predictions or decisions on the new information [5]. In medical imaging sector, computer-aided diagnosis and detection (CAD), that is an integration of ML classification and imaging feature engineering, has shown promising results in supporting radiotherapists for precise diagnoses, reducing the time and cost of diagnosis [6].
Conventional feature engineering approaches are depending on quantitative imaging feature extraction [7] like intensity, texture, volume, shape, and different statistical features from imaging data as well as ML classifiers like Decision Tree (DT), Support Vector Machines (SVM), and Adaboost. The deep learning (DL) method has shown effective results in different kinds of computer vision (CV) tasks like object-detection, segmentation, and classification. Nonetheless, in order to attain effective implementation, a precise fine-tuning of hyperparameter and optimum structures and combinations of the layer are needed. This remains one of the key challenges of DL-based approaches while employed in distinct sectors like medical imaging [8–10]. With convolutional neural network (CNN) promising result in the fields of computer vision (CV), medical imaging researchers have changed their interest towards DL-based approaches to design CAD systems for the diagnosis of cancer.
This study develops a novel Deep Transfer Learning based Prostate Cancer Classification (DTL-PSCC) model using MRI images. The presented DTL-PSCC technique encompasses EfficientNet based feature extractor for the generation of a set of feature vectors. In addition, the fuzzy k-nearest neighbour (FKNN) model is utilized for classification process where the class labels are allotted to the input MRI images. Moreover, the membership value of the FKNN model can be optimally tuned by the use of krill herd algorithm (KHA) which results in improved classification performance. In order to demonstrate the enhanced classification outcome of the DTL-PSCC technique, a wide range of simulations take place on benchmark MRI datasets.
The rest of the study is organized as follows. Section 2 offers the related works, Section 3 discusses the proposed model and Section 4 provides the experimental validation. Lastly, Section 5 draws the conclusion.
Zhang et al. [11] integrated a GrowCut and Zernik feature extraction and extreme learning machine (ELM) approaches for lesion segmentation in MRI and prostate cancer diagnosis. They utilize GrowCut approach for the segmentation of the suspicious cancer region and the integration of ML models in ensemble learning to diagnose prostate cancer. De Vente et al. [12] developed a neural network (NN) system that grades and detects cancer tissue in end-to-end manner simultaneously. It is medically applicable when compared to the classifier goals of the ProstateX-2 challenge. They utilized the data set for testing and training. Also, employed a two-dimensional U-Net with MRI as input and lesion segmentation map which encodes the Gleason Grade Group (GGG), a measurement for the aggressiveness of cancer, as output.
Ye [13] designed an AI-based method (called AI-biopsy) for the earlier diagnoses of prostate cancer through MRI labelled with histopathology data. The DL method is designed to differentiate 1) higher-risk tumors from lower-risk tumors and 2) benign from cancerous tumors. Alkadi et al. [14] trained a deep convolution encoder-decoder framework for segmenting the malignant lesions, the prostate, and anatomical structure. To integrate the 3D contextual spatial data given by the MRI, we present a three-dimensional sliding window model that preserves two-dimensional domain difficulty when utilizing three-dimensional data.
Feng et al. [15] introduced a DL architecture for diagnosing prostate cancer in the CEUS image. The presented approach extracts feature uniformly from temporal and spatial dimensions by carrying out 3D convolutional operation that captures dynamic data of the perfusion encoded in many adjacent frames. The DL model was validated and trained against expert's delineation through the CEUS image recorded by 2 kinds of contrast agents. In [16], a strong DL based convolutional neural network (DL-CNN) approach is applied by means of transfer learning (TL) method. The outcomes are compared to several ML approaches. Cancer MRI databases are employed for training ML classifiers and GoogleNet, different features like Entropy based, Morphological, Texture, Elliptic Fourier Descriptors, and Scale Invariant Feature Transform (SIFT) is extracted.
In this study, an effective DTL-PSCC technique has been developed to classify prostate cancer using MRI images. The proposed DTL-PSCC technique involves several subprocesses namely preprocessing, EfficientNet based feature extraction, FKNN based classification, and KHA based parameter tuning. The membership value of the FKNN model can be optimally tuned by the use of KHA which results in improved classification performance.
The ground truth given by PROSTATEx-2 challenge is coordinate point at centre of lesion [17]. The region of interest (ROI) of size 65 × 65 nearby the ground truth has cropped in T2W image and ROI of size 21 × 21 are collected to remove 2D GLCM features. The size of ROI has been selected then a manual inspection so that the maximum tumor amongst the provided data set is suitable inside the ROI. The Lloyd-max quantization was executed on ROI with the amount of gray level fixed to baseline parameter 32 that minimized mean square error (MSE) to provide the amount of quantization levels.
3.2 Feature Extraction: EfficientNet Model
CNN is a typical DL approach which could produce cutting-edge results for almost all the classification problems [18]. CNN achieved good results on image classification, however, it could yield better accuracy on text data. CNN is Mainly utilized for automatically extracting the feature from the input data set, in addition ML method, where the user requirements to elect the feature 2D, and 3D CNN is employed for video and image data, correspondingly, while 1D CNN is applied to text classification. The CNN framework employs a sequence of convolution layers for extracting features from the input data set. Max pooling layer afterward every convolution layer and the dimension of extracted feature is decreased. In the convolution layer, the size of the kernel performs an important role in feature extraction. The model's hyperparameter represents the kernel size and number of filters.
This layer translates the word into a vector space module based on how frequently words appear closer to another word. The embedding layers use random weight to learn embedding for each term in the trained data set. The softmax layer is utilized as the classification layer that could achieve better results for the multi-class problems. The softmax function contains N units, in which the N represents the amount of units. All the units are connected fully with preceding layer and compute the likelihood of every class on N as follows
The pooling as well as convolution layers were utilized to remove the feature. These layers were utilized to remove the visual feature and recognize the difficult nature of image. But, the nature of skin cancer lesions has highly complex, and increasing an automated analysis model utilizing DL was stimulating. For alleviating this issue, TL was employed. Fig. 1 illustrates the structure of EfficientNet technique. During the current study, EfficientNetB3 was utilized for skin cancer recognition. An EfficientNetB3 is a recent, cost-efficient, and robust method established by scaling 3 parameters like depth, width, and resolution [19]. An EfficientNetB3 method with noisy-student weight has been utilized from scenarios I and III to TL method, but “isicall_eff3_weights” weights were utilized as pre-training to scenarios II and IV. The amount of parameters are decreased. Besides, the rectified linear unit (RELU) activation function was employed with 3 dense and 2 dropout layers. The resultant layer has several outcome units to multiclass classifier utilizing the softmax activation function.
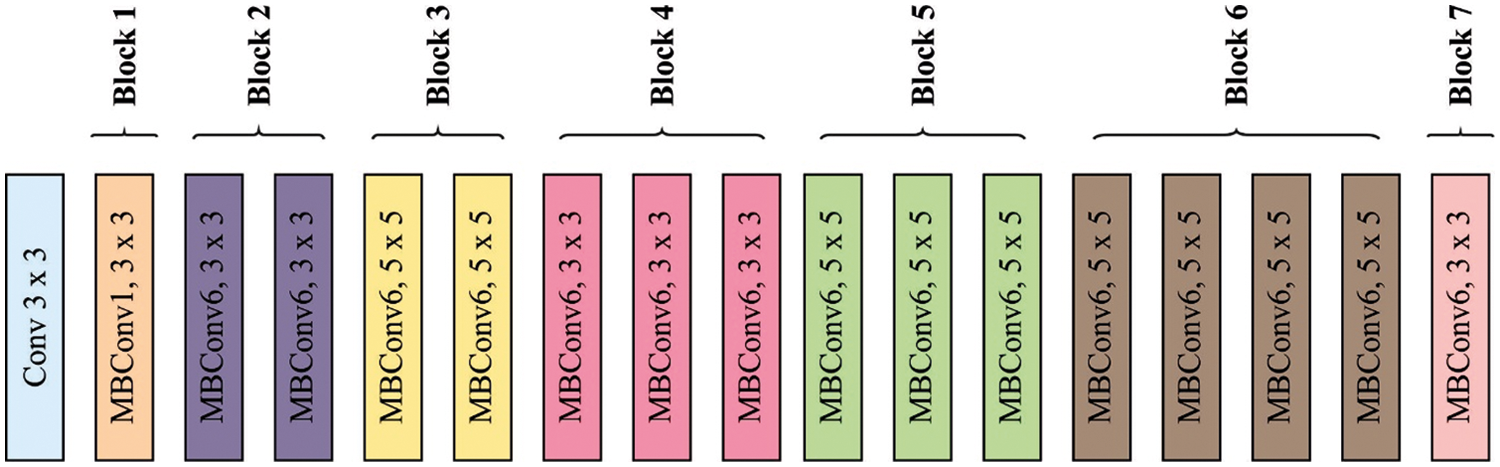
Figure 1: Framework of EfficientNet
3.3 Optimal Fuzzy KNN Based Classification
In this section, the FKNN classifier to detect and classify different classes of prostate cancer is applied. In the FKNN model, the fuzzy membership values of the instances are allocated to distinct class labels as given below [20].
while
where
Once the membership values are calculated, it gets allocated to the classes with maximum degree of membership, i.e.,
For tuning the parameters involved in the FKNN model, the KHA is applied. Antarctic krill is the main animal species on Earth. The capability to procedure huge swarm is most important feature of this species. An individual krill gets out from the herd once predators namely whales seals attack the krill. This attack decreases the density of krill herd (KH). The restructuring of KH then predation was influenced by several parameters. An essential purpose of herding performance of the krill individuals was improving krill density and attaining the food. The KH technique utilizes this multiobjective herding to resolve global optimized issues. Thus, the outcome, the krill individual transfers near an optimum solution once its searches for maximum density of herd as well as food. This performance generates the KH nearby the global minimal of optimized issue.
The time-dependent place of individual krill from
1. Effort induced by another krill individual;
2. Foraging motion
3. Physical or random diffusion
The subsequent Lagrangian method was generalizing to n-dimension decision space:
While
The effort of all krill individuals is determined as:
where
where
where rand stands for the arbitrarily created number amongst zero and one, I refers the actual iteration number and
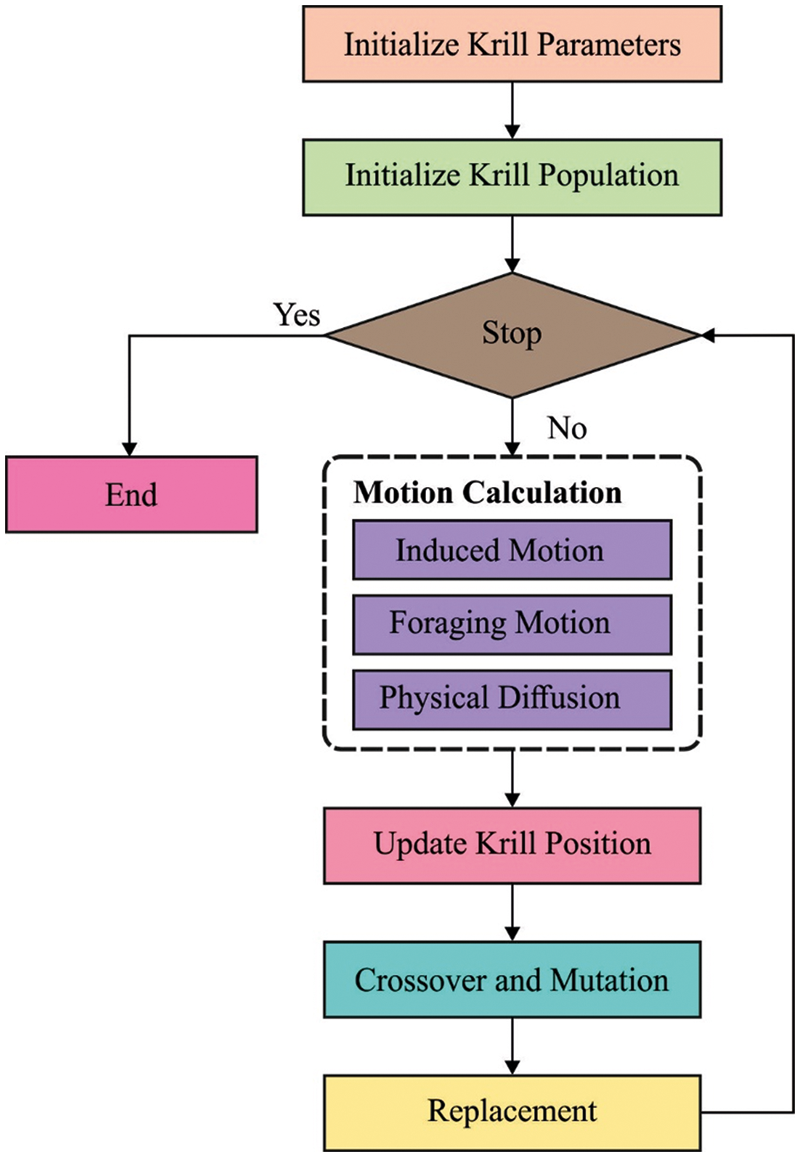
Figure 2: Flowchart of KH
The performance validation of the DTL-PSCC technique takes place using the PROSTATEx-2 Challenge dataset [22], which holds a set of 162 MRI images with 5 class labels namely transaxial T2W, sagittal T2W, ADC, DW, and Ktrans. In this study, the five classes are represented by targets. The results are examined under varying ratios of training and testing data. A few sample images are demonstrated in Fig. 3.
Figure 3: Sample images
Tab. 1 and Fig. 4 offer a brief prostate cancer classification result analysis of the DTL-PSCC technique under the training/testing dataset of 80:20. The results show that the DTL-PSCC technique has obtained effective performance. For instance, the DTL-PSCC technique has classified the images into target 1 with
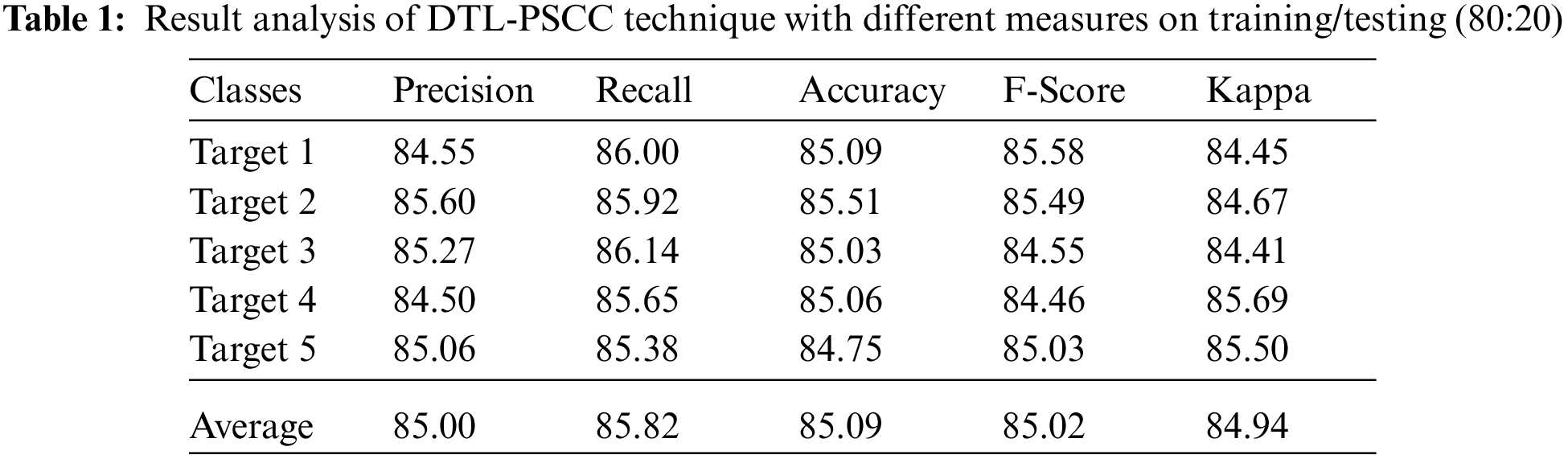
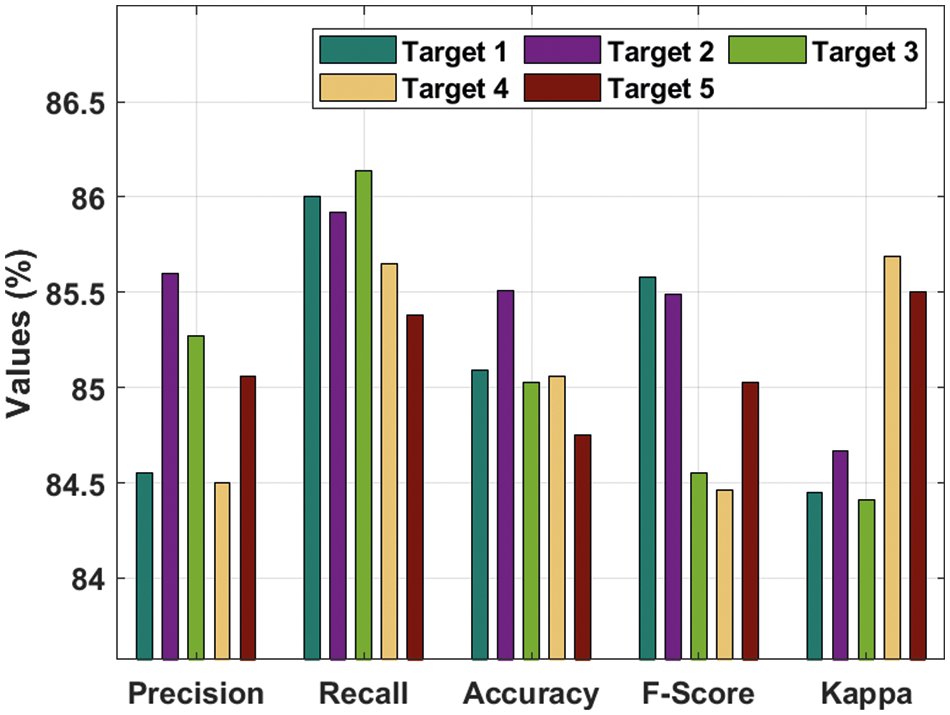
Figure 4: Result analysis of DTL-PSCC technique on training/testing (80:20)
Moreover, the DTL-PSCC technique has categorized the images into target 3 with
Fig. 5 demonstrates the average prostate cancer detection results of the DTL-PSCC technique under the training/testing dataset of 80:20. The figure reported that the DTL-PSCC technique has accomplished improved classification performance with the average
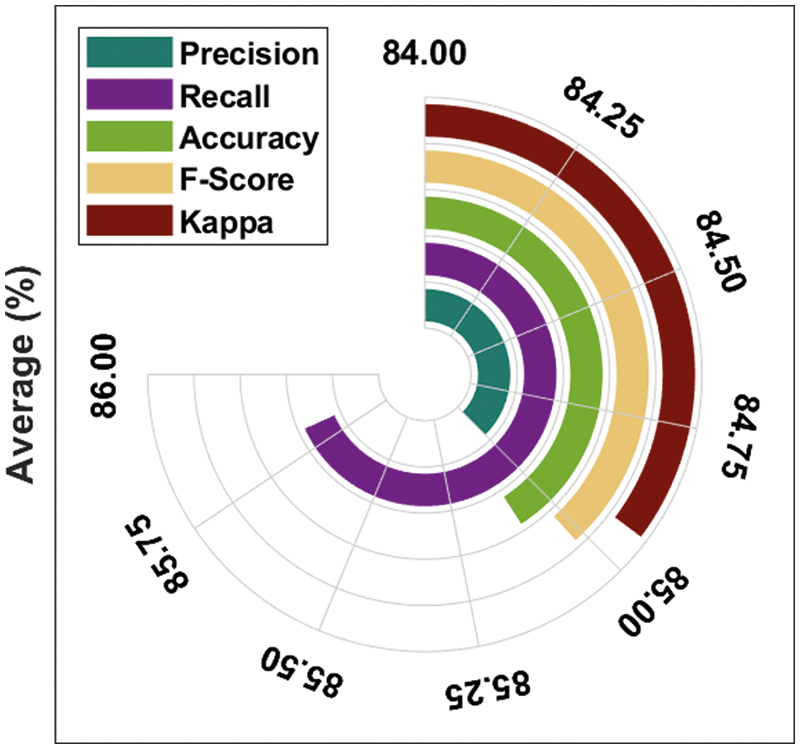
Figure 5: Average analysis of DTL-PSCC technique on training/testing (80:20)
Fig. 6 illustrates the accuracy analysis of the DTL-PSCC methodology on training and testing (80:20) dataset. The outcomes exhibited that the DTL-PSCC approach has accomplished increased efficiency with higher training and validation accuracy. It can be demonstrated that the DTL-PSCC manner has reached increased validation accuracy over the training accuracy.
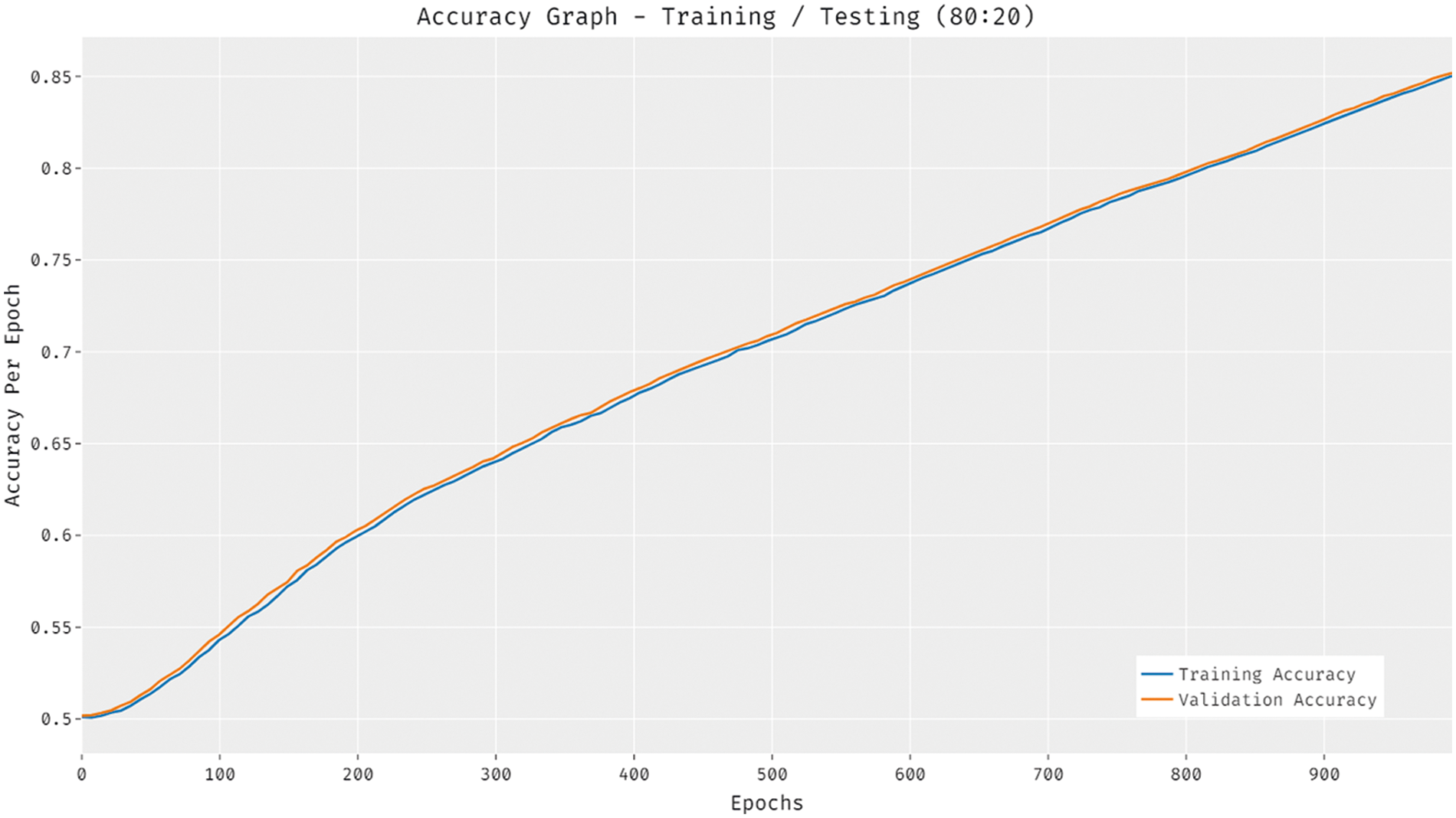
Figure 6: Accuracy analysis of DTL-PSCC technique on training/testing (80:20)
Fig. 7 showcases the loss analysis of the DTL-PSCC methodology on training and testing (80:20) dataset. The results established that the DTL-PSCC approach has resulted in a proficient outcome with the decreased training and validation loss. It can be stated that the DTL-PSCC technique has lower validation loss over the training loss.

Figure 7: Loss analysis of DTL-PSCC technique on training/testing (80:20)
Tab. 2 and Fig. 8 provide a detailed prostate cancer classification result analysis of the DTL-PSCC approach under the training/testing dataset of 70:30. The outcomes outperformed that the DTL-PSCC method has obtained effective performance. For instance, the DTL-PSCC algorithm has classified the images into target 1 with the
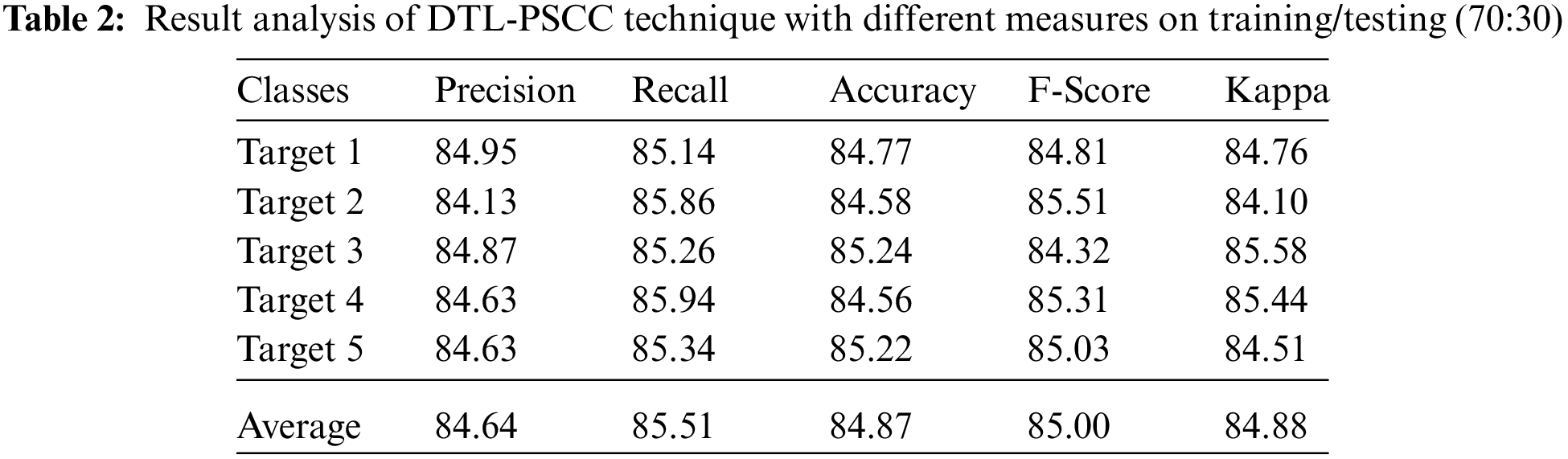
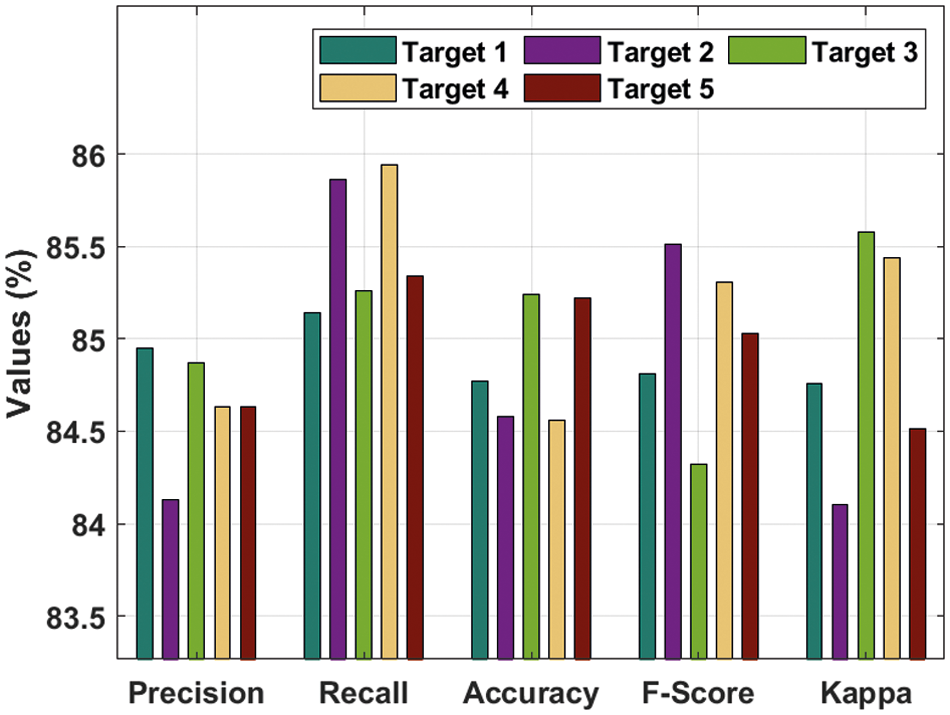
Figure 8: Result analysis of DTL-PSCC technique on training/testing (70:30)
Fig. 9 depicts the average prostate cancer detection outcomes of the DTL-PSCC methodology under the training/testing dataset of 70:30. The figure stated that the DTL-PSCC technique has accomplished increased classification performance with average
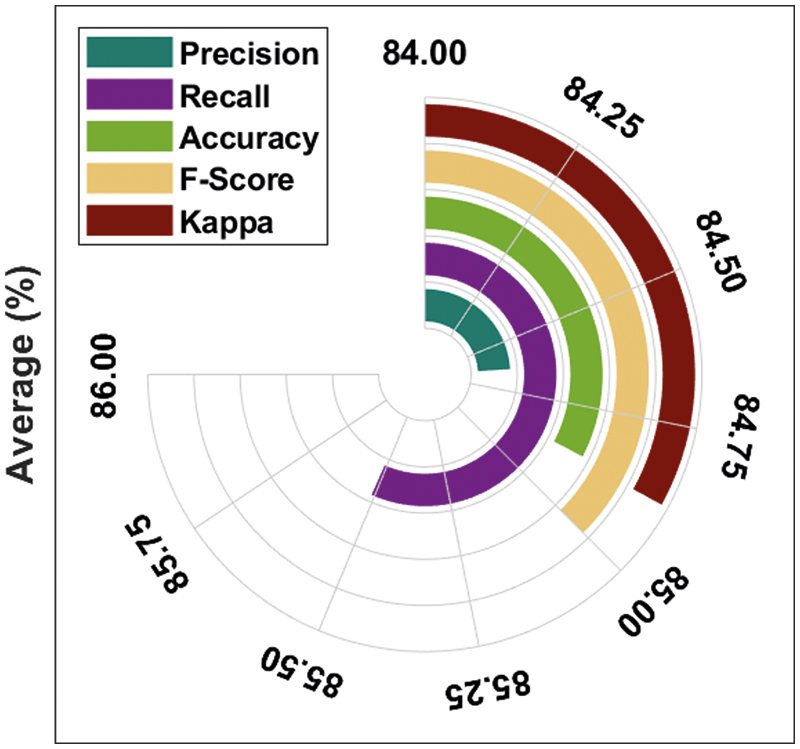
Figure 9: Average analysis of DTL-PSCC technique on training/testing (70:30)
Fig. 10 portrays the accuracy analysis of the DTL-PSCC approach on training and testing (70:30) dataset. The results proved that the DTL-PSCC methodology has achieved improved results with increased training and validation accuracy. It is noticed that the DTL-PSCC technique has gained improved validation accuracy over the training accuracy.
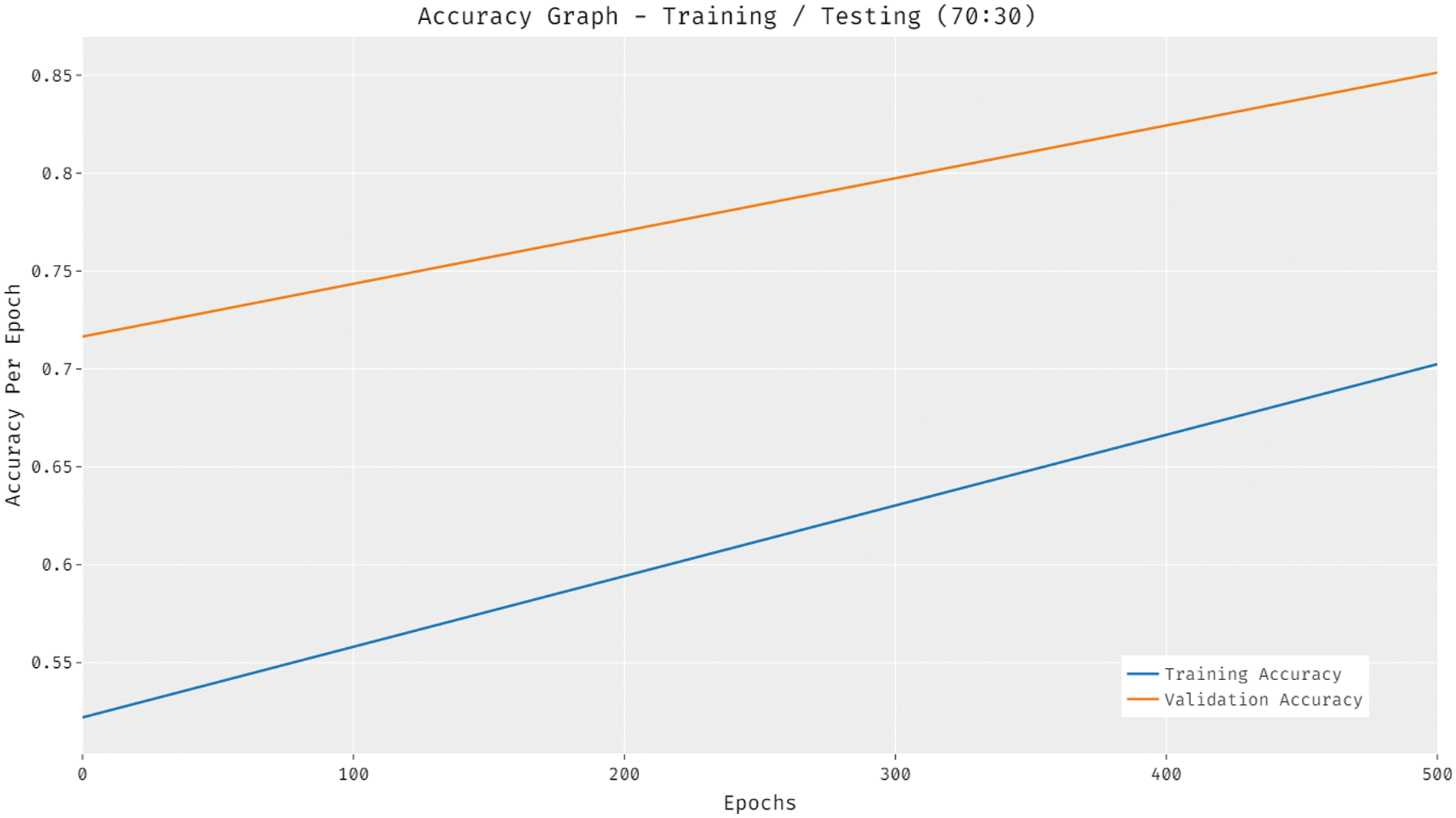
Figure 10: Accuracy analysis of DTL-PSCC technique on training/testing (70:30)
Fig. 11 depicts the loss analysis of the DTL-PSCC algorithm on training and testing (70:30) dataset. The outcomes established that the DTL-PSCC system has resulted in a proficient outcome with the decreased training and validation loss. It can be stated that the DTL-PSCC methodology has obtainable minimal validation loss over the training loss.
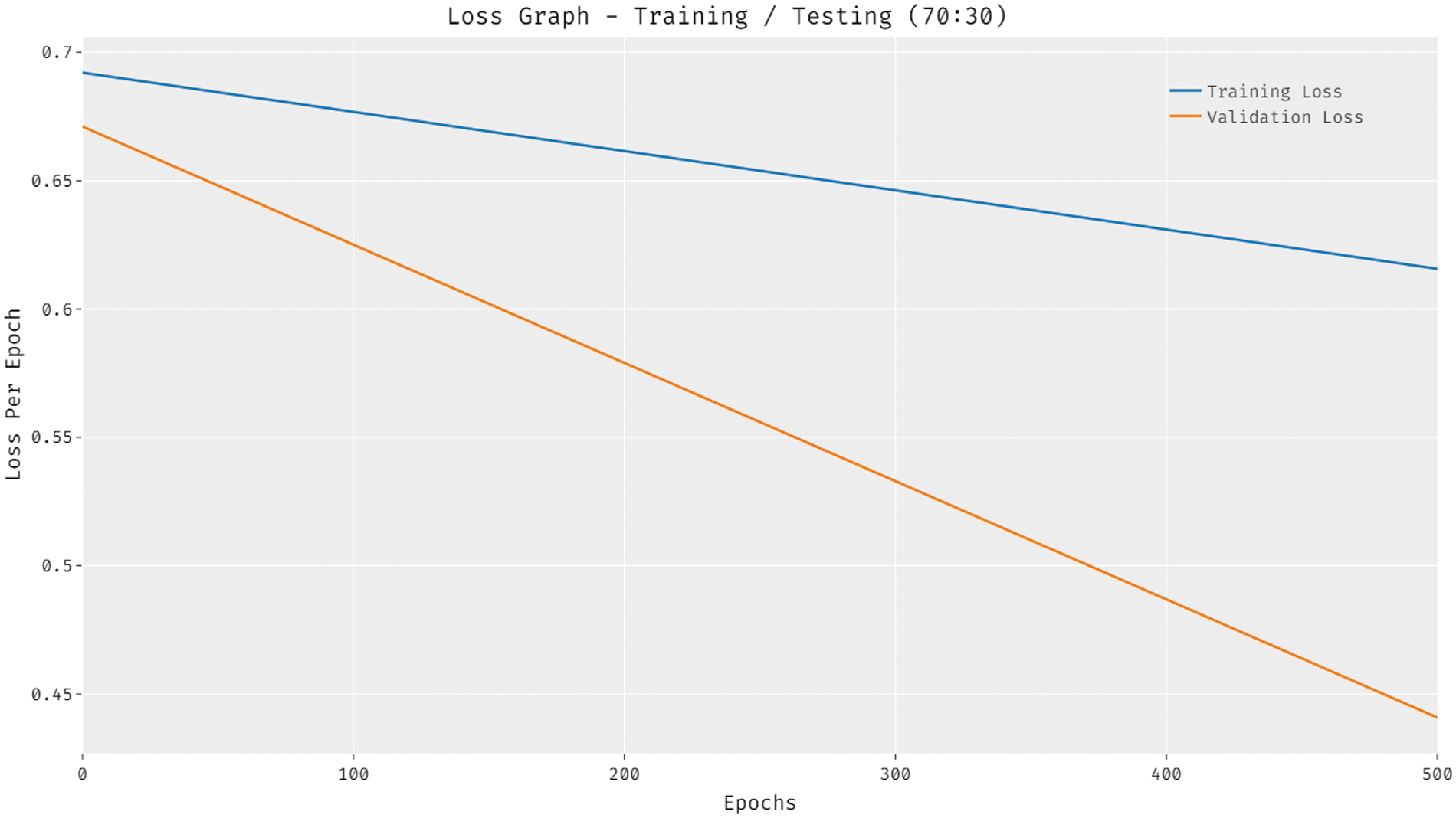
Figure 11: Accuracy analysis of DTL-PSCC technique on training/testing (70:30)
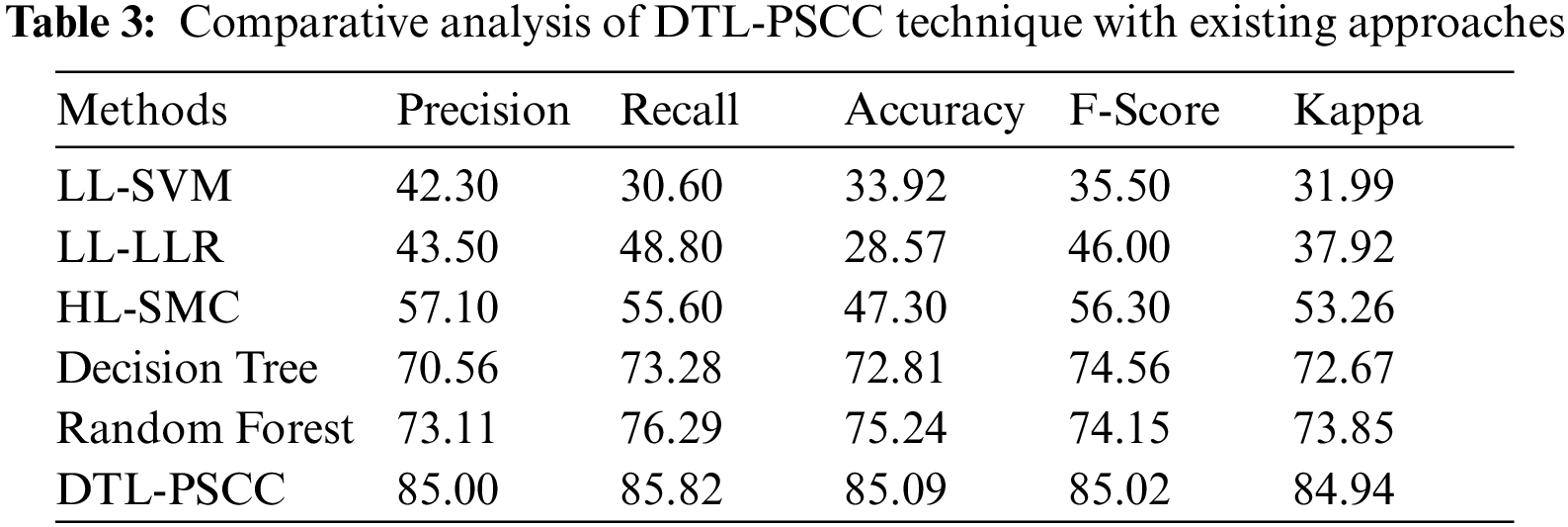
Lastly, a detailed comparative analysis of the DTL-PSCC technique with recent methods is offered in Tab. 3 and Fig. 12. The results demonstrated that the LL-support vector machine (SVM), LL-logistic regression with L1 penalty (LLR) (LLR), and HL-SMC methods have obtained least prostate classification performance with
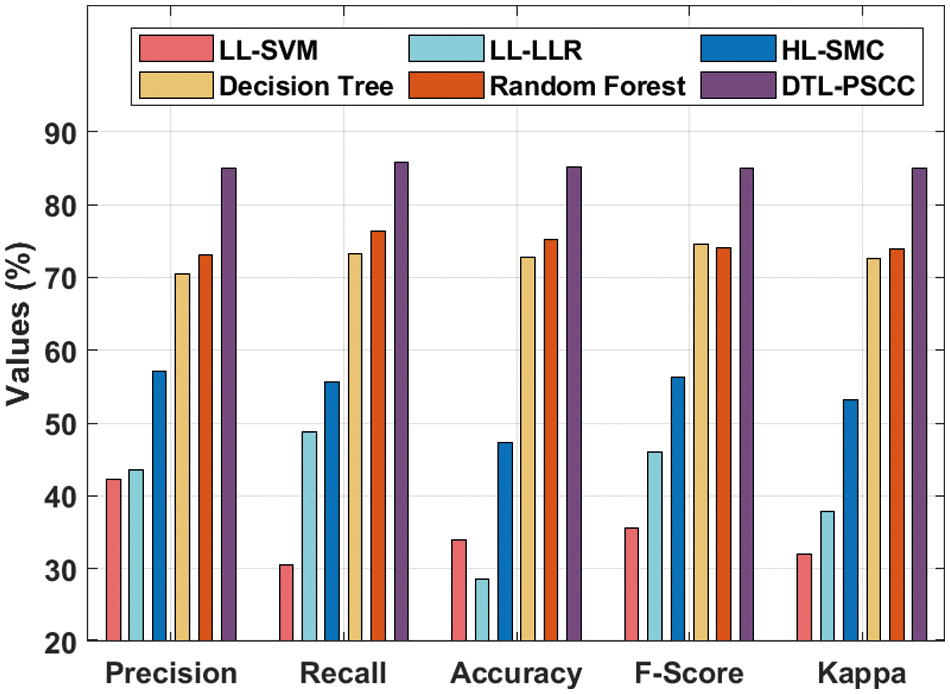
Figure 12: Comparative analysis of DTL-PSCC technique with existing approaches
In this study, an effective DTL-PSCC technique has been developed to classify prostate cancer using MRI images. The proposed DTL-PSCC technique involves several subprocesses namely preprocessing, EfficientNet based feature extraction, FKNN based classification, and KHA based parameter tuning. The membership value of the FKNN model can be optimally tuned by the use of KHA which results in improved classification performance. In order to demonstrate the enhanced classification outcome of the DTL-PSCC system, a wide range of simulations takes place on benchmark MRI datasets. The extensive comparative results ensured the advancement of the DTL-PSCC system over the recent methods with the higher accuracy of 85.09%. Hence, the DTL-PSCC technique has appeared as a proficient approach for prostate cancer classification and detection. In future, deep learning based segmentation techniques can be derived to improve the efficiency of the DTL-PSCC technique
Acknowledgement: The authors would like to acknowledge the support of Prince Sultan University for paying the Article Processing Charges (APC) of this publication.
Funding Statement: The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work under grant number (RGP 2/25/43). Taif University Researchers Supporting Project Number (TURSP-2020/346), Taif University, Taif, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Z. Wang, C. Liu, D. Cheng, L. Wang, X. Yang et al., “Automated detection of clinically significant prostate cancer in mp-mri images based on an end-to-end deep neural network,” IEEE Transactions on Medical Imaging, vol. 37, no. 5, pp. 1127–1139, 2018. [Google Scholar]
2. X. Zhong, R. Cao, S. Shakeri, F. Scalzo, Y. Lee et al., “Deep transfer learning-based prostate cancer classification using 3 tesla multi-parametric MRI,” Abdom Radiol, vol. 44, no. 6, pp. 2030–2039, 2019. [Google Scholar]
3. A. Chaddad, M. Kucharczyk and T. Niazi, “Multimodal radiomic features for the predicting gleason score of prostate cancer,” Cancers, vol. 10, no. 8, pp. 249, 2018. [Google Scholar]
4. N. Aldoj, S. Lukas, M. Dewey and T. Penzkofer, “Semi-automatic classification of prostate cancer on multi-parametric MR imaging using a multi-channel 3D convolutional neural network,” European Radiology, vol. 30, no. 2, pp. 1243–1253, 2020. [Google Scholar]
5. J. Li, Z. Weng, H. Xu, Z. Zhang, H. Miao et al., “Support vector machines (SVM) classification of prostate cancer gleason score in central gland using multiparametric magnetic resonance images: A cross-validated study,” European Journal of Radiology, vol. 98, pp. 61–67, 2018. [Google Scholar]
6. S. Mehralivand, J. H. Shih, S. Harmon, C. Smith, J. Bloom et al., “A grading system for the assessment of risk of extraprostatic extension of prostate cancer at multiparametric MRI,” Radiology, vol. 290, no. 3, pp. 709–719, 2019. [Google Scholar]
7. C. Jensen, J. Carl, L. Boesen, N. C. Langkilde and L. R. Østergaard, “Assessment of prostate cancer prognostic gleason grade group using zonal-specific features extracted from biparametric MRI using a KNN classifier,” Journal of Applied Clinical Medical Physics, vol. 20, no. 2, pp. 146–153, 2019. [Google Scholar]
8. R. Cuocolo, M. B. Cipullo, A. Stanzione, L. Ugga, V. Romeo et al., “Machine learning applications in prostate cancer magnetic resonance imaging,” European Radiology Experimental, vol. 3, no. 1, pp. 35, 2019. [Google Scholar]
9. R. Hao, K. Namdar, L. Liu, M. A. Haider and F. Khalvati, “A comprehensive study of data augmentation strategies for prostate cancer detection in diffusion-weighted MRI using convolutional neural networks,” Journal of Digital Imaging, vol. 34, no. 4, pp. 862–876, 2021. [Google Scholar]
10. X. Yang, C. Liu, Z. Wang, J. Yang, H. L. Min et al., “Co-trained convolutional neural networks for automated detection of prostate cancer in multi-parametric MRI,” Medical Image Analysis, vol. 42, pp. 212–227, 2017. [Google Scholar]
11. L. Zhang, L. Li, M. Tang, Y. Huan, X. Zhang et al., “A new approach to diagnosing prostate cancer through magnetic resonance imaging,” Alexandria Engineering Journal, vol. 60, no. 1, pp. 897–904, 2021. [Google Scholar]
12. C. de Vente, P. Vos, M. Hosseinzadeh, J. Pluim and M. Veta, “Deep learning regression for prostate cancer detection and grading in bi-parametric MRI,” IEEE Transactions on Biomedical Engineering, vol. 68, no. 2, pp. 374–383, 2021. [Google Scholar]
13. Z. Ye, “Editorial for “A deep learning approach to diagnostic classification of prostate cancer using pathology-radiology fusion,” Journal of Magnetic Resonance Imaging, vol. 54, no. 2, pp. 472–473, 2021. [Google Scholar]
14. R. Alkadi, F. Taher, A. E. baz and N. Werghi, “A deep learning-based approach for the detection and localization of prostate cancer in t2 magnetic resonance images,” Journal of Digital Imaging, vol. 32, no. 5, pp. 793–807, 2019. [Google Scholar]
15. Y. Feng, F. Yang, X. Zhou, Y. Guo, F. Tang et al., “A deep learning approach for targeted contrast-enhanced ultrasound based prostate cancer detection,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, vol. 16, no. 6, pp. 1794–1801, 2019. [Google Scholar]
16. A. A. Abbasi, L. Hussain, I. A. Awan, I. Abbasi, A. Majid et al., “Detecting prostate cancer using deep learning convolution neural network with transfer learning approach,” Cognitive Neurodynamics, vol. 14, no. 4, pp. 523–533, 2020. [Google Scholar]
17. B. Abraham and M. S. Nair, “Computer-aided classification of prostate cancer grade groups from MRI images using texture features and stacked sparse autoencoder,” Computerized Medical Imaging and Graphics, vol. 69, pp. 60–68, 2018. [Google Scholar]
18. H. Gunasekaran, K. Ramalakshmi, A. R. M. Arokiaraj, S. D. Kanmani, C. Venkatesan et al., “Analysis of DNA sequence classification using CNN and hybrid models,” Computational and Mathematical Methods in Medicine, vol. 2021, pp. 1–12, 2021. [Google Scholar]
19. G. Marques, D. Agarwal and I. de la Torre Díez, “Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network,” Applied Soft Computing, vol. 96, pp. 106691, 2020. [Google Scholar]
20. Z. Cai, J. Gu, C. Wen, D. Zhao, C. Huang et al., “An intelligent Parkinson”s disease diagnostic system based on a chaotic bacterial foraging optimization enhanced fuzzy KNN approach,” Computational and Mathematical Methods in Medicine, vol. 2018, pp. 1–24, 2018. [Google Scholar]
21. A. H. Gandomi and A. H. Alavi, “Krill herd: A new bio-inspired optimization algorithm,” Communications in Nonlinear Science and Numerical Simulation, vol. 17, no. 12, pp. 4831–4845, 2012. [Google Scholar]
22. J. Epstein, L. Egevad, M. Amin, B. Delahunt, J. Srigley et al., “The 2014 international society of urological pathology (isup) consensus conference on gleason grading of prostatic carcinoma,” American Journal of Surgical Pathology, vol. 40, no. 2, pp. 244–252, 2016. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |