DOI:10.32604/cmc.2022.029000

| Computers, Materials & Continua DOI:10.32604/cmc.2022.029000 |  |
| Article |
A Novel Hybrid Machine Learning Approach for Classification of Brain Tumor Images
1Radiological Sciences Department, College of Applied Medical Sciences, Najran University, Najran, 61441, Saudi Arabia
2Department of Computer Science, COMSATS University Islamabad, Sahiwal Campus, Sahiwal, 57000, Pakistan
3College of Engineering, Najran University, Najran, 61441, Saudi Arabia
*Corresponding Author: Tariq Ali. Email: Tariqali@cuisahiwal.edu.pk
Received: 22 February 2022; Accepted: 25 March 2022
Abstract: Abnormal growth of brain tissues is the real cause of brain tumor. Strategy for the diagnosis of brain tumor at initial stages is one of the key step for saving the life of a patient. The manual segmentation of brain tumor magnetic resonance images (MRIs) takes time and results vary significantly in low-level features. To address this issue, we have proposed a ResNet-50 feature extractor depended on multilevel deep convolutional neural network (CNN) for reliable images segmentation by considering the low-level features of MRI. In this model, we have extracted features through ResNet-50 architecture and fed these feature maps to multi-level CNN model. To handle the classification process, we have collected a total number of 2043 MRI patients of normal, benign, and malignant tumor. Three model CNN, multi-level CNN, and ResNet-50 based multi-level CNN have been used for detection and classification of brain tumors. All the model results are calculated in terms of various numerical values identified as precision (P), recall (R), accuracy (Acc) and f1-score (F1-S). The obtained average results are much better as compared to already existing methods. This modified transfer learning architecture might help the radiologists and doctors as a better significant system for tumor diagnosis.
Keywords: Brain tumor; magnetic resonance images; convolutional neural network; classification
A healthy brain can be affected by many diseases. A brain tumor is one of them. As an initial signs of brain tumor its formation is due to the evolution of unusual cells in the brain. The initial signs are specific and general in terms of signs. As the general sign it create pressure on brain or spinal cord, while the specific signs indicate when the specific part of the brain is not working well. In the benign tumor tissues, the growth is sluggish as matched to the malignant tumor. Although it is not metastatic [1]. A malignant tumor is cancerous cells that cause a malignant tumor. The speed of this kind of tumor is quick and destroys the neighboring cells immediately. It is metastatic as compared to a benign tumor. The cure of the brain tumor hinge on upon its type and position [2]. Medical field technology is emerging, and different techniques are used in the medical field for different medical diagnoses. These techniques have helped radiologists to diagnose and detection of tumors [3]. In the case of brain tumors, magnetic resonance imaging (MRI) is the modality of choice. MRI is one of the innovative techniques for imaging in the field of radiology having several sequences (T1, T2 weighted and fluid-attenuated inversion recovery images). It gives a detailed image and an inside view of the brain and other parts [4]. Several methods and algorithms were offered by scholars and researchers. As compared to other algorithms, the use of deep learning is increasing day by day due to its better accuracy results when large data is trained. It can learn and seek the data having dominant attributes step by step. It provides better accuracy results [5].
Deep learning can seek and learn from the data which is disorganized even without any label. For refining the results, this method repeats the tasks again and again through little modification. Deep learning has shown a great influence on problem-solving techniques like the classification of images, processing, and recognition of speech. A neural network having many additional layers is called a deep neural network. If compared to other neural networks, the framework of a deep neural network has contrast due to the existence of many hidden layers. Just like neurons in the human brain deep neural networks also have interrelated components and elements. These elements assign the network's tasks. Several groups are present in the network in the form of layers like input, hidden and output layers. Separately every layer has a link with the other. Weights are assigned to each connection between layers. In other words, we can say that it work as a feed-forward neural network that has several hidden layers [6]. A standout amongst the most well-liked kind of deep neural architecture is acknowledged as convolutional neural networks (CNN). It is one of the prominent deep learning architectures which provide the best accuracy results in supervised and unsupervised classification [7]. Every year almost 11,000 people from the population are diagnosed with a brain tumor. This population includes 500 youngsters and children. Each year 5000 people lost their lives [8]. Its proper diagnosis, detection, and grading with high accuracy are still challenging in the medical and research field as its characteristics vary from patient to patient.
In this research work, three robust automated approach based on CNN, multi-level CNN, ResNet-50 based multi-level CNN model are applied for the classification of tumor and healthy patients by using MRI for attaining good accuracy results and making the right treatment decisions. Real-time data is collected from local hospitals for the experiments.
The key objective of this work is to detect and classify brain tumor-affected patients. For classification purposes, three approaches are used. Following are the contributions of this work:
1. Collection of normal and brain tumor-affected patient data from local hospitals and online repository.
2. Using state-of-the-art machine learning techniques, we have used three different models known as CNN, multi-level CNN, ResNet-50 based multi-level CNN for the classification of affected patients.
3. This model will help the radiologist to better understanding of low-level feature of MRI.
4. Might help the radiologists as a better significant system for tumor diagnosis.
5. The obtained average results of the used model are much better as compared to already existing methods.
The reaming paper is prepared as the combination of four sections known as: related work provides most recent work in the field, methodology explores the architecture of proposed model, results and discussion provide the results and finally the conclusion section explains the real purpose of proposed work.
Bhaiya et al. [9] used 3 dimensional MRI images for detecting the tumors. For tumor segmentation, a random forest was applied. For the classification, two properties of the images were used like local binary pattern in three orthogonal planes and histogram of oriented gradients. Kumari [10] proposed a method for classification in which for preprocessing of images principle component analysis was used. In this methodology, three types of neural networks were supervised: back propagation algorithm, learning vector quantization (LVQ), and radial basis function (RBF). The training time of RBF was more as compared to LVQ because more time was spent in LVQ. The support vector machine demonstrated in [11] contained the feature extraction using the gray level scoring matrix. This technique with other kernel functions like radial and linear basis was used for classifying the images on types with an accuracy of 98%. Shree et al. [12] showed a technique for classifying the tumor using MRI through discrete wavelet transform along with a three-dimensional gray level dependence matrix for extracting the features. For minimizing the size of features simulated annealing was used and for removing the over-fitting stratified k-fold cross-validation was used. In work done by Naik et al. [13], preprocessing was done for noise removal. For extracting the coefficient of wavelet, 2-D discrete wavelet transform was used. A probabilistic neural network (PNN) was used for classifying tumors and GLMC was used for feature extraction.
In [14], by using the median filter, the preprocessing of the images was done. Through the rules of a decision tree, the classification was done. A hybrid method is applied for the classification of the images for MRIs from the SICAS Repository. The features of the discrete wavelet transform the specific and required features of the images were extracted [15]. Another approach is used for multi-level deep CNN to classify the images and to maximize the accuracy [16]. Through a genetic algorithm, the features which were extracted were reduced and then the classification of the images was done by using a support vector machine. The combination of two methods like principal component analysis and Gaussian model was proposed for automatically extracting the essential and required features in [17]. By using the real data sets, improved classification results were shown in [18]. For segmenting the tumor region k-means were used and a two-tier method was used for classification. For feature extraction a discrete wavelet transform was used and for training of these features a neural network was used. For training purposes, the k-nearest neighbor was used. Aamir et al. [19] designed a method through creating the pixel matrix, the specific features were obtained using GLCM. Knapsack problem was considered for genetic algorithm implementation. The fitness function was used for checking the solution's quality [20]. Removing the over-fitting decision tree along with random forest was used for better training. Firstly, changing the input MRI images into the grayscale images were done by using matplotlib Library in python. For extracting the required features discrete wavelet transform was applied. The BPNN was used for classification of images. Neural network was adjusted and used for better classification in [21].
Alfonse et al. [22] designed a technique for segmenting and to classify the MRIs of the brain tumor using BWT and SVM. For better detection skull was stripped for getting the high accuracy level. In [23], a SVM was used for classifying the tumor images and for extracting the features fast Fourier transform was used, and for achieving the required features maximal relevancy method was used. In the end accuracy of 98% was achieved. MRI of the brain has two types of regions having normal and abnormal cells. For extracting the desired region of the tumor, these two regions need to be separated for further technique implementation [24]. A new methodology using the techniques of ANN along with features of the gray wolf technique was proposed in [25] for providing better classification results on MRI of the brain. Comparisons were made for analyzing the proposed method with other neural networks and results were provided showing better classification accuracy.
In this section a brief illustration of the technique used for MRI brain tumor classification is presented. The current machine learning methods have the minimum number of kernels and learning parameters and huge time complexity. To handle this problem, a multi-level convolutional neural network is proposed. The proposed model is divided into two levels: One for the detection of tumor and the second level is for the classification of the tumor. The first level uses the 17 CNN layers for detection and the second level uses the 10 CNN layers for the classification of malignant and benign tumor types [26]. A brief summary of the convolutional neural network is presented in Fig. 1.

Figure 1: Generic architecture of convolutional neural network
3.1 Level-I Convolutional Neural Network Architecture
The layers present in the CNN architecture have their working and specificity. In our proposed methodology at the level-I architecture of CNN only detection of tumor and the normal patient is done. This architecture uses 17 layers for the detection of the tumor. The first one layer is the image input layer the size of the image is specified by adjusting the weight, height, and size of the channel. Filter size, the learning rate of the network, and stride are specified using the convolution layer. The formula for a convo layer that we have used is presented in Eq. (3). To speed up the network a batch normalization layer is added to the framework. An activation function is added in the form of the ReLU layer as shown in Eq. (1). Input is divided into several pooling regions and extreme value is calculated of each region through the max-pooling layer. Layers used in this study are presented in Tab. 1 and Fig. 2 with different parameters and filter sizes that differ from 8 to 32 and the learning rate is set to 0.0001 with epoch value 30. A fully connected layer combines all the outputs of previous layers in other words it combines all the learned features of the layers for the recognition of the immense patterns as shown in Eq. (2). The softmax layer works as an activation function for normalization. Utilizing the probabilities, the classification layer classifies it into specified classes.

Here, f(x) is the ReLU function and x is the target value as shown in Eq. (1).
In Eq. (2),
Here Eq. (3) is the convolutional layer,
3.2 Level-II Convolutional Neural Network
In our used methodology, at level II, we have utilized a total of ten layers of CNN with different constraints and filters for the classification of normal, malignant, and benign tumors. The first layer is the image input layer having parameters weight, height, and size of the channel. In the convolutional layer number of filters, size and stride are defined while the kernel values are specified from 8 to 32. For the training of the network the learning rate is set to 0.0001 with an epoch value of 30. The detailed layers description is given in Tab. 2 and workflow is also shown in Fig. 2.

Figure 2: Workflow of multi-level CNN model

3.3 Feature Extractor ResNet-50 Model
The ResNet-50 model consists of residual connections between different convolutional layers. The residual connections indicates that the results of the residual net is convolution of its input. By using these connections, the residual network maintains the minimum loss, attain the maximum knowledge gain and integrate the higher performance during the training phase for brain tumor analysis. The ResNet-50 technique contains of five steps with convo and identity block. Individually a convo block contains 3 convolutional layers and a block identity. This model consists of almost 23 billion trainable parameters. The block diagram of pre-trained ResNet-50 is shown in Fig. 3.

Figure 3: Architecture of Pre-trained ResNet-50
3.4 ResNet-50 Based Multi-Level CNN Model
During the transfer learning (TL) of pre-trained ResNet model all the layers remain the same in all the stages of the pre-trained model. After the training of pre-trained ResNet model the output flow continued as a layer wise manner. Initially all the layers are in the frozen state with all the parameters. When the residual network forward output after pre-trained to the fine-tuning network, then a block of extracted features created. After the unfreezing of all the pre-trained model layers, a new layer of classifier is introduced to get the required results. The freezing and new classifier block is also shown in Fig. 4.

Figure 4: Architecture of proposed pre-trained ResNet-50 with multi-level CNN
We have analyzed results through Python language by using supportive libraries; numpy array (np), operating system (os), keras models, layers and utils, matplotlib, sklearn utils and metrics, tensorflow models, pandas, tqdm, and seaborn. These libraries support the development of machine-learning applications. The proposed model was evaluated on the system with an Intel(R) Core i7 7th generation, Nvidia GeForce GTX 1060 (6 GB) and 16 GB of RAM.
4.1 Dataset Preparation and Preprocessing
The total number of 2043 MRIs patient's data was used for this study. The used dataset is not publically available and it's the private property of a hospital. In this dataset each category of malignant benign and normal consists of different number of MRI images respectively for training and testing purpose. Well-reputed Neurosurgeons and Radiologist helped in labeling the images manually by using their expertise. Collected images are in DICOM file format and using PmsDView 3.0 software they are viewed and saved in JPG format. The dimension (width and height) of the images is set to 120 × 100. All the images are converted into RGB three-channel images. Important information and detail of the image are saved by clearing up the images. The dataset is distributed into testing and training folders. The fractions of training and testing on the dataset is 85% and 15% correspondingly, as shown in Tab. 3. Different types of tumors are shown in Fig. 5.
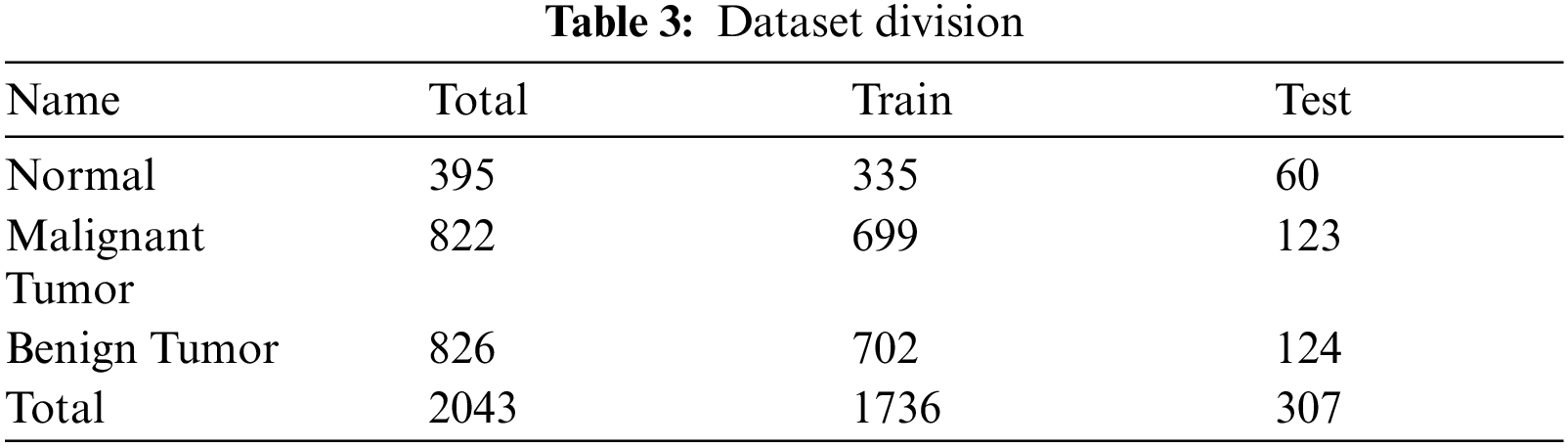

Figure 5: Tumor types
The performance correlation of our presented methodology for MRI brain tumor classification describes accuracy (Acc). On average, the gain of Acc is 95%. An epoch depicts several times the algorithm grasps the whole informational index. Thus, every time the calculation viewed all examples of the database, an epoch has finished. The confusion matrix is created to show the results of the recognition rate of our defined three classes (Normal, malignant, and benign Tumor). These measurements are often known a true positive rate and false negative (fn) rate, separately as shown in Fig. 5. The mathematical results are measured with the following statistical equations from Eqs. (4) to (7):
True positive (tp) denotes correctly classified values for images and true negative (tn) shows incorrectly classified images.
4.3 Results with Convolutional Neural Network
This model is applied to get the required results for brain tumor. The testing confusion matrix of normal, benign and malignant tumors is presented below in Fig. 6. The boxes in the confusion matrix shows the amount of images that are used in the testing process. The average statistical evaluation of convolutional neural network (CNN) is presented in Tab. 4 as precision (P) 0.88%, recall (R) 0.88%, F1-score (F1-S) 0.87% and accuracy 0.88%. The Fig. 7 shows the illustration of training and validation accuracy and loss in graphical form.

Figure 6: CNN model confusion matrix of 3-classes for the test dataset


Figure 7: CNN illustration of training and validation accuracy and loss
4.4 Results with Multi-Level CNN Model
The multi-level CNN model is applied to get the required results with its pre-defined structure. While the testing confusion matrix of all the tumor categories as normal, benign and malignant tumors is shown below in Fig. 8. The different color boxes in the confusion matrix shows the total number of images that are used to get the testing results. The calculation for the average statistical params is shown in Tab. 4 given below. The statistical values for multi-level CNN model are precision (P) 0.87%, recall (R) 0.86%, F1-score (F1-S) 0.87% and accuracy 0.88%. The Fig. 9 shows the illustration of training and validation accuracy and loss in graphical form.

Figure 8: Multi-level CNN Model confusion matrix of 3-classes for the test dataset
4.5 Results with ResNet-50 Based Multi-Level CNN Model
The multi-level CNN model with pre-trained ResNet-50 is used to get the better results. For this purpose, different categories of brain tumors from datasets are used known as normal, benign, and malignant tumors shown in confusion matrix in Fig. 10. The number of images used for testing process are shown in the confusion matrix with different colors as shown in Fig. 11. The parameters for the transfer learning are set to execute to get the different statistical values for the testing process. The average statistical evaluation of proposed multi-level CNN model of pre-trained ResNet-50 is presented in Tab. 4 as precision (P) 0.94%, recall (R) 0.94%, F1-score (F1-S) 0.93% and accuracy 0.95%. The Fig. 10 shows the demonstration of training and validation accuracy and loss in graphical form.

Figure 9: Multi-level CNN illustration of training and validation accuracy and loss

Figure 10: Multi-level CNN with ResNet-50 model confusion matrix of 3-classes for the test dataset

Figure 11: Multi-level CNN with ResNet-50 model demonstration of training and validation accuracy and loss
The results presented in Tab. 4, show the assessment calculation of the proposed methods on the MRI images database. To get the accurate results three methods are applied. First one is the CNN model consist of multiple convo layers and filters. Second one is the multi-level CNN model which is divided into two levels: first one for the detection of tumor and the second level is for the classification of the tumor. The first level uses the 17 CNN layers for detection and the second level uses the 10 CNN layers for the classification of malignant and benign tumor types. The average statistical evaluation of multi-level convolutional neural network is presented in Tab. 4 on the complete dataset.
The third model is the multi-level CNN model with pre-trained ResNet-50 model applied to calculate the dataset results. This model is applied to get the required results with its pre-defined structure. This model consists of residual connections among different convolutional layers. The residual connections specifies that the results of the residual net is convolution of its input. By using these connections, the residual network maintains the minimum loss, attain the maximum knowledge gain, and integrate the higher performance during the training phase for brain tumor analysis. The applied model comprises of five phases with convo and identity block. Individually a convo block contains 3 convolutional layers and every identity block also consists of 3 convolutional layers. When the training of pre-trained ResNet-50 model is completed then the output flow continued as a layer wise manner. At starting stage all the layers are in the frozen state with all the parameters. When the residual network forward output after pre-trained to the network, then a block of extracted features created. After the unfreezing of all the pre-trained model layers, a new layer of classifier is introduced to get the required results.
Tab. 5 displays the comparison of accuracy values with state-of-the-art techniques. The three proposed techniques attain an accuracy percentage as 0.88 for CNN model, 0.91 for multi-level CNN, and 0.95 for multi-level CNN with ResNet-50 model which is the maximum one as matched to the previously presented techniques and among our applied techniques. The highest value of accuracy is due to the transfer architecture of pre-trained ResNet-50 model, because it uses a block of extracted features to get the better results.

We have proposed a brain tumor recognition technique utilizing a factual methodology on MRI images as the input. The identification results of brain tumor disorder for every MRI image are either it's healthy or tumor affected (Benign, Malignant). In our method, images are preprocessed by converting them into one common format and channel for feature extraction by utilizing CNN architecture. We have worked with three models CNN, multi-level CNN, and multi-level CNN with pre-trained ResNet-50 for detection of brain tumors for contribution in this research. The proposed transfer learning methodology for MRI brain tumor classification provides results concerning different statistical values known as precision (P) 0.94%, recall (R) 0.94%, F1-score (F1-S) 0.93% and accuracy 0.95%. The results reveal that this technique is much better in each constant as compared to the other state-of-the-art techniques. As a future directions the applied research may help the radiologist to accurately diagnosis the brain tumors.
Funding Statement: Authors would like to acknowledge the support of the Deputy for Research and Innovation-Ministry of Education, Kingdom of Saudi Arabia for funding this research through a project (NU/IFC/ENT/01/014) under the institutional funding committee at Najran University, Kingdom of Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. J. Amin, M. Sharif, M. Yasmin and S. L. Fernandes, “Big data analysis for brain tumor detection: Deep convolutional neural networks,” Future Generation Computer Systems, vol. 87, no. 1, pp. 290–297, 2018. [Google Scholar]
2. H. Dong, G. Yang, F. Liu, Y. Mo and Y. Guo, “Automatic brain tumor detection and segmentation using u-net based fully convolutional networks,” in Proc. Annual Conf. on Medical Image Understanding and Analysis, Edinburgh, UK, pp. 506–517, 2017. [Google Scholar]
3. J. Amin, M. Sharif, M. Yasmin and S. L. Fernandes, “A distinctive approach in brain tumor detection and classification using MRI,” Pattern Recognition Letters, vol. 139, no. 1, pp. 118–127, 2017. [Google Scholar]
4. P. Sapra, R. Singh and S. Khurana, “Brain tumor detection using neural network,” International Journal of Science and Modern Engineering, vol. 1, no. 9, pp. 2319–6386, 2013. [Google Scholar]
5. “Data science, machine learning and deep learning,” datos.gob.es, 05-Apr-2018. [Online]. Available: https://datos.gob.es/en/noticia/data-science-machine-learning-and-deep-learning. [Accessed: 02-Dec-2021]. [Google Scholar]
6. Houseofbots.com. [Online]. Available: https://www.houseofbots.com/news-detail/1443-1-what-is-deep-learning-and-neural-network. [Accessed: 02-Dec-2021]. [Google Scholar]
7. Missinglink.ai. [Online]. Available: https://missinglink.ai/guides/convolutional-neural-networks/convolutional-neural-networks-image-classification/. [Accessed: 02-Dec-2021]. [Google Scholar]
8. S. Abbasi and F. Tajeripour, “Detection of brain tumor in 3D MRI images using local binary patterns and histogram orientation gradient,” Neurocomputing, vol. 219, no. 1, pp. 526–535, 2017. [Google Scholar]
9. L. P. Bhaiya and V. K. Verma, “Classification of MRI brain images using neural network,” Network, vol. 2, no. 5, pp. 751–756, 2012. [Google Scholar]
10. R. Kumari, “SVM classification an approach on detecting abnormality in brain MRI images,” International Journal of Engineering Research and Applications, vol. 3, no. 4, pp. 1686–1690, 2013. [Google Scholar]
11. A. Kharrat, M. B. Halima and M. B. Ayed, “MRI brain tumor classification using support vector machines and meta-heuristic method,” in Proc. 15th Int. Conf. on Intelligent Systems Design and Applications, Marrakech, Morocco, pp. 446–451, 2015. [Google Scholar]
12. N. V. Shree and T. Kumar, “Identification and classification of brain tumor MRI images with feature extraction using DWT and probabilistic neural network,” Brain Informatics, vol. 5, no. 1, pp. 23–30, 2018. [Google Scholar]
13. J. Naik and S. Patel, “Tumor detection and classification using decision tree in brain MRI,” International Journal of Computer Science and Network Security, vol. 14, no. 6, pp. 87–91, 2014. [Google Scholar]
14. S. Kumar, C. Dabas and S. Godara, “Classification of brain MRI tumor images: A hybrid approach,” Procedia Computer Science, vol. 122, no. 1, pp. 510–517, 2017. [Google Scholar]
15. M. Aamir, T. Ali, A. Shaf, M. Irfan and M. Q. Saleem, “ML-DCNNet: Multi-level deep convolutional neural network for facial expression recognition and intensity estimation,” Arabian Journal for Science and Engineering, vol. 45, no. 12, pp. 10605–10620, 2020. [Google Scholar]
16. A. Chaddad, “Automated feature extraction in brain tumor by magnetic resonance imaging using Gaussian mixture models,” Journal of Biomedical Imaging, vol. 2015, no. 1, pp. 8–18, 2015. [Google Scholar]
17. V. Anitha and S. Murugavalli, “Brain tumour classification using two-tier classifier with adaptive segmentation technique,” Institution of Engineering and Technology Computer Vision, vol. 10, no. 1, pp. 9–17, 2016. [Google Scholar]
18. E. S. B. J. a. M. Ravikumar, “Human brain tumor classification using genetic algorithm approach,” International Journal of Emerging Technology in Computer Science & Electronics, vol. 25, no. 4, pp. 86–89, 2018. [Google Scholar]
19. M. Aamir, T. Ali, M. Irfan, A. Shaf, M. Z. Azam et al., “Natural disasters intensity analysis and classification based on multispectral images using multi-layered deep convolutional neural network,” Sensors, vol. 21, no. 28, pp. 2648–2661, 2021. [Google Scholar]
20. B. M. Zahran, “Classification of brain tumor using neural network,” Computers and Software, vol. 9, no. 4, pp. 673–678, 2014. [Google Scholar]
21. N. B. Bahadure, A. K. Ray and H. P. Thethi, “Image analysis for MRI based brain tumor detection and feature extraction using biologically inspired BWT and SVM,” International Journal of Biomedical Imaging, vol. 2017, no. 1, pp. 9749108–9749121, 2017. [Google Scholar]
22. M. Alfonse and A. -B. M. Salem, “An automatic classification of brain tumors through MRI using support vector machine,” Egyptian Computer Science Journal, vol. 40, no. 3, pp. 1–11, 2016. [Google Scholar]
23. G. Coatrieux, H. Huang, H. Shu, L. Luo and C. Roux, “A watermarking-based medical image integrity control system and an image moment signature for tampering characterization,” IEEE Journal of Biomedical and Health Informatics, vol. 17, no. 6, pp. 1057–1067, 2013. [Google Scholar]
24. H. M. Ahmed, B. A. Youssef, A. S. Elkorany, A. A. Saleeb and F. A. El-Samie, “Hybrid gray wolf optimizer–artificial neural network classification approach for magnetic resonance brain images,” Applied Optics, vol. 57, no. 7, pp. 25–31, 2018. [Google Scholar]
25. M. Aamir, M. Irfan, T. Ali, G. Ali, A. Shaf et al., “An adoptive threshold-based multi-level deep convolutional neural network for glaucoma eye disease detection and classification,” Diagnostics, vol. 10, no. 8, pp. 602–616, 2020. [Google Scholar]
26. X. Chen, Y. Xu, D. W. K. Wong, T. Y. Wong and J. Liu, “Glaucoma detection based on deep convolutional neural network,” in Proc. 37th Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society, Milan, Italy, pp. 715–718, 2015. [Google Scholar]
27. R. Asaoka, H. Murata, A. Iwase and M. Araie, “Detecting preperimetric glaucoma with standard automated perimetry using a deep learning classifier,” Ophthalmology, vol. 123, no. 9, pp. 1974–1980, 2016. [Google Scholar]
28. A. A. Salam, T. Khalil, M. U. Akram, A. Jameel and I. Basit, “Automated detection of glaucoma using structural and non structural features,” Springerplus, vol. 5, no. 1, pp. 1–21, 2016. [Google Scholar]
29. M. D. L. Claro, L. D. M. Santos, W. Lima e Silva, F. H. D. De Araújo, N. H. De Moura et al., “Automatic glaucoma detection based on optic disc segmentation and texture feature extraction,” Latin American Center for Informatics Electronic Journal, vol. 19, no. 2, pp. 1–10, 2016. [Google Scholar]
30. Q. Abbas, “Glaucoma-deep: Detection of glaucoma eye disease on retinal fundus images using deep learning,” International Journal of Advanced Computer Science and Applications, vol. 8, no. 6, pp. 41–46, 2017. [Google Scholar]
31. M. Al Ghamdi, M. Li, M. Abdel-Mottaleb and M. A. Shousha, “Semi-supervised transfer learning for convolutional neural networks for glaucoma detection,” in Proc. IEEE Int. Conf. on Acoustics, Speech and Signal Processing, Brighton, UK, pp. 3812–3816, 2019. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |