DOI:10.32604/cmc.2022.029073

| Computers, Materials & Continua DOI:10.32604/cmc.2022.029073 |  |
| Article |
Automated Machine Learning for Epileptic Seizure Detection Based on EEG Signals
1University of Science and Technology Beijing, Beijing, 100083, China
2Hwa Create Co., Ltd, Beijing, 100193, China
3Amphenol Global Interconnect Systems, San Jose, CA 95131, US
*Corresponding Author: Xiang Wang. Email: wangxiang@ustb.edu.cn
Received: 23 February 2022; Accepted: 31 March 2022
Abstract: Epilepsy is a common neurological disease and severely affects the daily life of patients. The automatic detection and diagnosis system of epilepsy based on electroencephalogram (EEG) is of great significance to help patients with epilepsy return to normal life. With the development of deep learning technology and the increase in the amount of EEG data, the performance of deep learning based automatic detection algorithm for epilepsy EEG has gradually surpassed the traditional hand-crafted approaches. However, the neural architecture design for epilepsy EEG analysis is time-consuming and laborious, and the designed structure is difficult to adapt to the changing EEG collection environment, which limits the application of the epilepsy EEG automatic detection system. In this paper, we explore the possibility of Automated Machine Learning (AutoML) playing a role in the task of epilepsy EEG detection. We apply the neural architecture search (NAS) algorithm in the AutoKeras platform to design the model for epilepsy EEG analysis and utilize feature interpretability methods to ensure the reliability of the searched model. The experimental results show that the model obtained through NAS outperforms the baseline model in performance. The searched model improves classification accuracy, F1-score and Cohen’s kappa coefficient by 7.68%, 7.82% and 9.60% respectively than the baseline model. Furthermore, NAS-based model is capable of extracting EEG features related to seizures for classification.
Keywords: Deep learning; automated machine learning; EEG; seizure detection
According to World Health Organization (WHO) statistics, epilepsy is the most common neurological disease second only to stroke [1] and 2.4 million people are diagnosed with epilepsy each year [2]. Seizures are caused by an abnormal discharge of neurons in the brain [3,4]. During the seizure, the patient will lose consciousness. Some parts of the body or the whole body will even twitch [5], which can last from a few seconds to a few minutes [6]. Sudden unprovoked seizures can make patients unable to protect themselves in time and even cause fainting and life-threatening [7,8]. Electroencephalography (EEG) has become the standard technology for epilepsy detection due to its high spatial and temporal resolution, non-invasiveness and low cost [9]. It collects electrical signals during brain activity through electrodes placed in different positions of the brain, so that it can detect abnormal brain potentials such as spikes and spikes in patients during epileptic seizures in real time [6,10]. In traditional clinical diagnosis, EEG-based seizure detection relies on well-trained professionals to complete. Since epileptic seizures are random and sudden, doctors and other hospital staff need to face EEG data recorded for several hours or even days, which will undoubtedly consume a lot of time and energy for professionals to complete this work. At the same time, the large patient population will also put pressure on them [11]. Moreover, nearly three-quarters of epilepsy patients come from countries below the middle income, and the high cost of artificial seizure detection will make them prohibitive [12].
Considering the limitations of artificial epilepsy detection, many researchers begin to focus on the research of automatic epilepsy detection technology based on EEG [13–15]. Feature extraction is the key to the automatic detection algorithms for epilepsy EEG and the current feature extraction methods can be divided into manual design features and automatic feature extraction. Manual methods utilize prior knowledge to model the characteristics of epilepsy EEG, which usually extract temporal and frequency domain features related to seizures as the basis for epilepsy detection. In the time domain analysis, the authors in reference [16] apply sample entropy and approximate entropy as epileptic features and designed a corresponding classifier. In the field of frequency domain analysis, reference [17] uses Fourier transform to convert EEG epilepsy features into frequency domain representation for automatic epilepsy detection. Similarly, discrete wavelet transform method to extract features of epilepsy EEG signals is employed in [18]. In addition, the authors in article [19] extract time domain and frequency domain features simultaneously to realize an automatic epilepsy detection algorithm. The hand-crafted approaches combined with the experience of professionals are capable of boosting the performance of the automatic epilepsy detection system in certain scenarios, but they face many challenges in actual application scenarios. Firstly, the EEG signal of epilepsy is non-stationary, so the features extracted from the same patient at different times of epileptic seizures are quite different [20]. It is difficult to extract information from the features of all patients with epilepsy EEG episodes using manual design methods, which will cause information loss in the feature extraction stage to a certain extent, and reduce the accuracy of the EEG-based automatic epilepsy detection system. Secondly, due to the low signal-to-noise ratio of the EEG, the methods of manually extracting features are easily affected by noise or artifacts, resulting in inaccurate feature extraction. Therefore, it is difficult for hand-crafted approaches to adapt to the real EEG acquisition scene, which is complex and changeable.
The method of automatically extracting epilepsy EEG features based on deep learning has gradually attracted attention. Deep learning technology can automatically learn the feature extraction mode of related tasks from the provided data through the deep structure of the neural network, which has achieved excellent results in multiple EEG analysis tasks including epilepsy EEG detection and exceeds the manual feature extraction method on performance [21]. Discriminative models, representative models and hybrid models are the frequently applied architectures in deep learning based methods. In the area of discriminative models, convolutional neural networks (CNN) are capable of utilizing convolutional structures to extract local features of multiple channels of EEG at the same time and are widely used in automatic epilepsy detection algorithms [22,23]. The authors in reference [24] designs a 13-layer convolutional neural network to automatically detect epileptic seizures. As for the representative models, deep belief networks (DBN) [25] and authoencoder (AE) [26] also have many applications in automatic epilepsy detection and have achieved good recognition accuracy. Hybrid models can apply the advantages of different deep learning models to improve the system performance. A combination of CNN and long short term memory (LSTM) is proposed in [27] to build an automatic epilepsy EEG detection system. Although these deep learning-based automatic detection methods for epilepsy EEG have achieved good results in their respective data sets and experimental environments, the collection environment and EEG equipment of different medical institutions are different. It will lead to differences in the signal-to-noise ratio of EEG signals and the number of acquisition channels for different data sets, so it is not clear whether these automatic detection algorithms can adapt to these changes and achieve qualified performance. Furthermore, the design of deep learning architecture for epilepsy EEG detection is time-consuming and difficult, which requires the cooperation of medical institutions and professionals with computer-related backgrounds. The high threshold of algorithm design limits the application of EEG-based automatic epilepsy detection systems in countries below the middle income.
Deep learning is playing an increasingly important role in medical-related fields [28–30], especially in brain research [31–36]. With the development of computing power, Automated Machine Learning (AutoML) provides the possibility of using machine learning algorithms to solve problems for people without relevant knowledge [37]. In the task of automatic detection for epilepsy EEG, Neural Architecture Search (NAS) of AutoML can quickly build a detection model that adapts to specific epilepsy EEG signals and users only need to provide the corresponding data set. It will undoubtedly save much time and cost of algorithm development and accelerate the deployment of the epilepsy EEG automatic detection system. NAS methods can be divided into three types, namely NAS based on deep reinforcement learning [38,39], NAS based on evolutionary algorithm [40,41], and NAS based on Bayesian [34]. NAS has been attempted to be applied in EEG analysis, where the application areas include emotion recognition [42], motor imagery EEG [43] and state evaluation [44]. Moreover, feature interpretability tools are very important for deep learning-based EEG analysis, as the tools can ensure the reliability of black-box models like CNNs. Analysis of model weights [45], model activations [46] and the correlation of input and output [47] are the frequently employed methods for the inspection of neural networks in EEG signals classification.
In this paper, we apply the Bayesian-based NAS algorithm of the open source platform AutoKeras [37] to implement the search of the convolutional neural network for the automatic detection of epilepsy EEG. We compare the searched model with EEGNet that can perform EEG analysis across EEG paradigms [48]. Then deep learning feature interpretability approaches are utilized to verify the reliability of the searched model. The contributions of this work can be categorized as:
• We study the possibility of automatic machine learning in the field of automatic epilepsy EEG detection. The experimental results show that the searched model achieves an accuracy of 76.61% in the test set.
• We use the deep learning feature interpretability method to analyze the NAS-based model. The analysis results show that the model extracts the EEG features related to epileptic seizures, which ensures the reliability of automatic machine learning algorithms.
The organization of this paper is as follows. Section 2 introduces the dataset, NAS algorithm and experimental protocol. The classification results and the analysis of feature explainability are given in Section 3. Section 4 presents discussion and the conclusion is illustrated in Section 5.
The experimental data we use comes from [49] and here is a brief description of dataset. This dataset contains five sets (represented as A-E), and each set has a total of 100 pieces of EEG data. The EEG signals of sets A and B are extracranial data collected from five healthy subjects. The difference is that the set A is recorded with eyes open and the signals in set B are closed eyes. Sets C, D and E are intracranial data acquired from five epilepsy patients, in which the epileptogenic zone of the patients has been diagnosed through resection. EEG in sets C and D are acquired when the epilepsy patient had no seizures, where the data collection location of set C is located in the epileptic area and that of set D is set in the hippocampal formation of the opposite hemisphere of the brain. The EEG recorded during the epileptic seizure is included in set E. All analog EEG signals are converted to digital signals at a sampling rate of 173.61 Hz and saved as single-channel data. Each EEG signal has been screened for artifacts to avoid the influence of noise and has passed the weak stationarity criterion. Then the original data is divided into EEG segments with a duration of 23.6 seconds [49].
2.2 Neural Architecture Search Method
We use AutoKeras (AK) to implement automatic neural architecture search, which is one of the most widely used open-source AutoML system [37]. Network morphism, a technique that changes the structure of a neural network but maintains its function, is adopted to improve search efficiency in this system [50,51]. Moreover, AK applies Bayesian optimization to choose the most prospective operation of network morphism each time, thus guiding through the neural architecture search space [37]. The flowchart of the NAS algorithm is shown in Fig. 1.
The purpose of the NAS algorithm is to search for the best performing model on a given epilepsy EEG dataset. Here, we define the epilepsy EEG dataset as
where
In order to make the NAS space meet the hypothesis of Gaussian process, AK propose an edit-distance kernel function for neural networks and it can be written as [37],
where
where
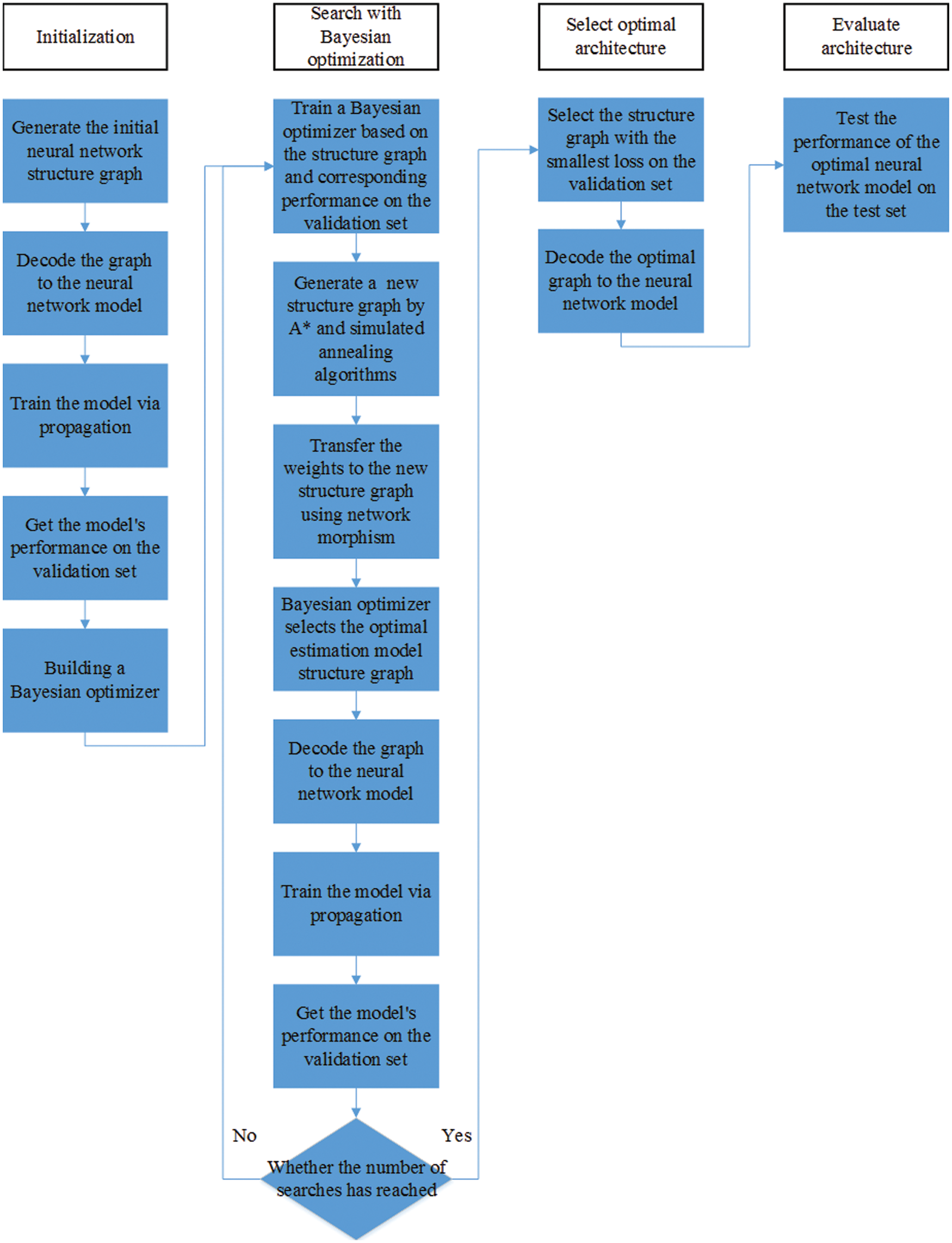
Figure 1: The flowchart of the NAS algorithm
Similarly,
where
Then AK choose upper-confidence bound (UCB) [52] as acquisition function and propose a novel approach combining simulated annealing [53] and A* search [54] to optimize UCB on tree-structured space [37].
The acquisition function UCB can be written as,
where
where

Moreover, AK introduce a graph-level network morphism to maintain the consistency of the intermediate tensor shape [37].
In the preprocessing, we apply the Butterworth filter to filter all segments from 0.43 to 100 Hz. Then the data of each segments is divided into 23 epochs containing 178 sampling points, and the duration of each epoch is approximately 1.025 s. The epochs are z-normalized so that the mean is equal to zero and the standard deviation is equal to one, where the z-normalization is one of the common and useful preprocess procedures for time series classification [55]. Finally, we randomly selected epochs according to the ratio of 3:1:1 to form the training set, test set, and validation set. The experimental procedure of epilepsy EEG detection algorithm is shown in the Fig. 2.
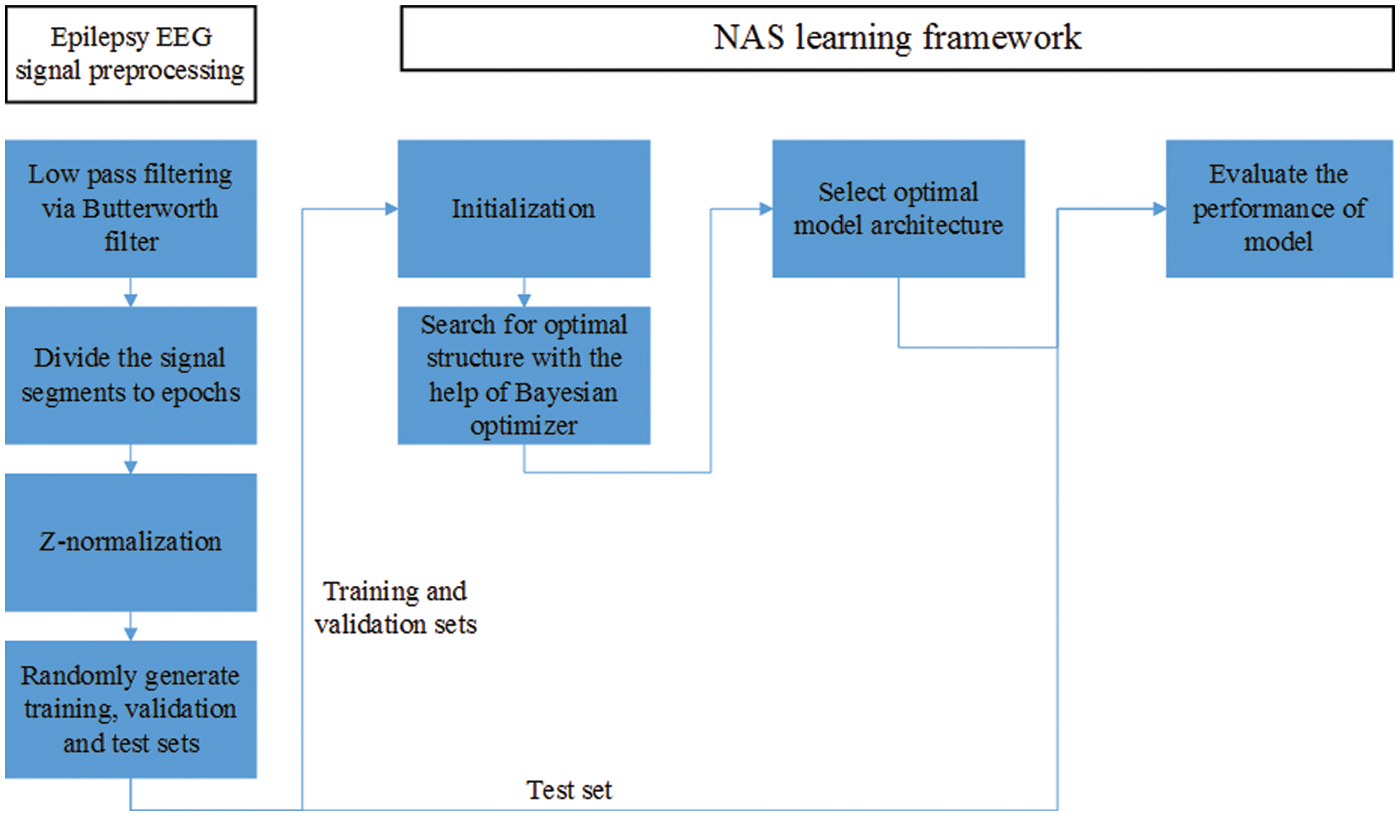
Figure 2: The experimental procedure of epilepsy EEG detection algorithm
When using AK to automatically search the neural network structure, we regarded the single-channel epilepsy EEG signal as a special type of image signal, and configure the ImageClassifier in AK to customize the search space. Specifically, we set the block type parameter in CnnModule to regular convolutions, which means that AK is capable of finding the model with the best performance by searching the structure composed of raw convolution. We set different search times and conduct multiple experiments. It should be noted that in the NAS algorithm, the number of searches is equal to the number of generated neural networks. In the search process, only the training set and the validation set were used. After the search was completed, the searched best model was trained for 100 more training iterations. Then we saved the model with the smallest cross-entropy loss on the validation set and tested its performance on the test set.
EEGNet [48] was chosen as the baseline model in our experiment, which is a deep convolutional neural network that can extract EEG features and classify signals across EEG paradigms. The previous classification results showed that EEGNet achieved good results on several EEG classification tasks, not limited to a specific EEG paradigm. Depthwise convolutions and separable convolutions are employed to reduce the amount of model parameters to adapt to different sizes of EEG datasets in EEGNet [48]. Moreover, depthwise convolutions are also used as spatial filters to extract the frequency-specific spatial features of EEG, and separable convolutions are applied for compression and correlation extraction of high-dimensional feature maps [48].
Three performance metrics are introduced to evaluate the classification results in our experiment, including classification accuracy, F1-score and Cohen’s kappa coefficient [56]. The accuracy is the ratio of the number of samples correctly classified by the model to the total number of samples for a given test set. And it can be expressed as,
where tps and tns is the true positives and true negatives, respectively. In addition, fps and fns denotes the false positives and false negatives.
The calculation of the F1-score takes into account the Precision and Recall, which can reflect the performance of the algorithm to a certain extent, especially for unbalanced data. F1-score can be written as,
Cohen’s kappa coefficient is used to measure the degree of consistency between model classification and manual labeling. The larger the value, the better the model performance. The calculation of the Cohen’s kappa coefficient can be expressed as,
where
We apply one-way analysis of variance to perform probability testing, modeling classification indicators as response variables and different approaches as factors.
Deep learning interpretability methods, as an important tool for testing the reliability of black box models such as deep convolutional neural networks, is very necessary for the application of algorithms in medical scenarios such as epilepsy detection. For the deep learning model obtained using the NAS method, since there is no prior knowledge and artificial experience involved in the structural design, it is difficult to ensure the reliability of neural networks only from the performance metrics on test set. Therefore, we use the feature explainability approaches to analyze the features extracted by the model, verifying that the neural networks extract features related to epilepsy EEG for classification instead of noise or artifacts.
We apply t-distributed stochastic neighbor embedding (t-SNE) [57] algorithm to reduce the dimensions of the high-dimensional features extracted by the searched model to three dimensions for visualization. The t-SNE algorithm is a machine learning algorithm widely used in nonlinear dimensionality reduction. Through dimensionality reduction and visualization of high-dimensional data, we can intuitively see the differences of extracted features between the classes, thus evaluating the model's ability to extract task-related features.
We employ the ‘Gradient * Input’ method [58] in the DeepExplain framework [59] to calculate the correlation between EEG input features and model decisions. This method is based on the gradient and the forward-backward iterations of the neural network to obtain the influence of each input sample on the neurons in the model decision layer, so as to determine which features will activate the neurons related to the right decision and which will interfere with the activation of the correct neurons in the model. We can visualize the main basis for the analysis of the searched model using this approach, and verify that the model extracts the relevant features of epilepsy EEG for classification.
Fig. 3 shows the performance of the convolutional neural network on the test set under different trial settings of the search algorithm, where the number of trials is the number of deep learning models tried by the NAS and error bars represent two standard errors of the mean. It can be clearly seen from the figure that the performance metrics (accuracy, F1-score and Cohen’s kappa coefficient) of the searched model reach the best when the search algorithm has the largest number of trials (150). And the classification accuracy of this searched model on the test set is 76.61%. When the number of models tried by the search algorithm is less than 100, the performance of the searched model is basically the same, and there is no obvious improvement. However, when the number of trials of the search algorithm is greater than 100, the performance of the final searched model on the test set boosts as the number of trials increases.
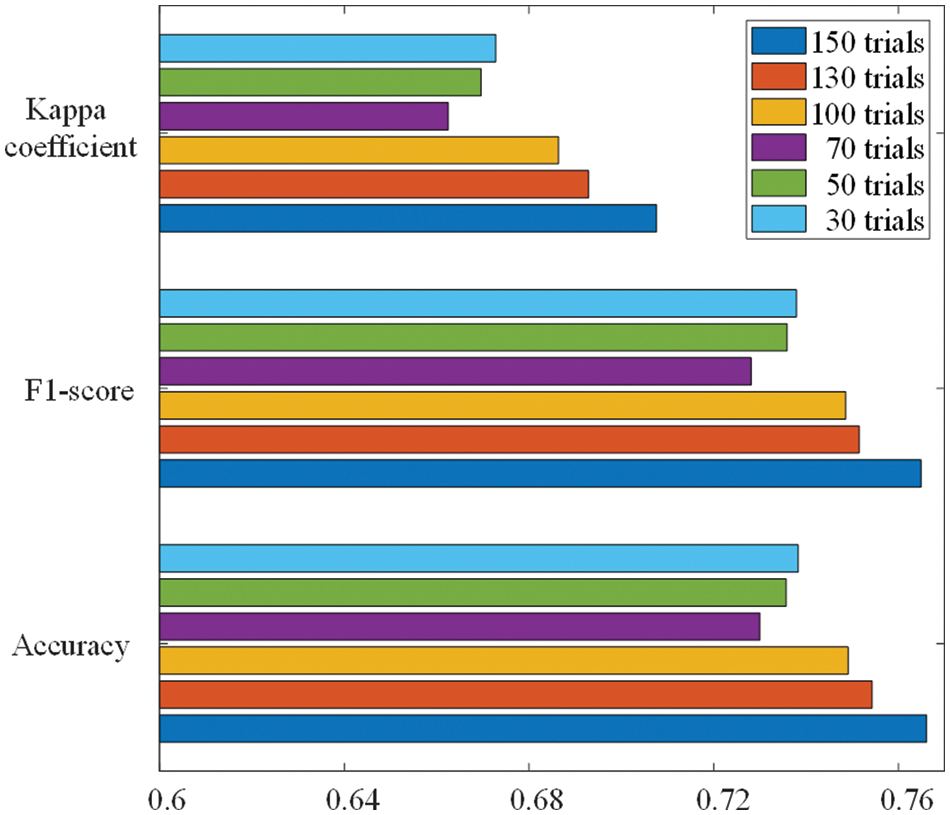
Figure 3: Classification performance of models under different trial settings of NAS
Through the experimental results, we can find that the performance of the optimal model grows faster after the NAS algorithm evaluation exceeds 100 trials, indicating that the Bayesian optimizer plays an important role in generating new neural network architectures. At the beginning of the NAS, the Bayesian optimizer is continuously trained by the performance of the model on the test set and the corresponding model structure to build the relationship between the model and the accuracy. In the later stage of the NAS algorithm, the trained Bayesian optimizer is capable of evaluating the accuracy of the neural network on the test set to find a model architecture with better performance, thus reducing the search time and improving the efficiency of the NAS.
We use the t-SNE algorithm to reduce the dimensionality of the high-dimensional feature vectors output by the global average pooling layer and visualize it in a 3-dimensional space. The visualization results are shown in the Fig. 4, where the data is randomly selected from the test set, and the color represents the classes of the EEG feature. We can see that the features extracted by the convolutional neural network are clustered together according to the corresponding classes, reflecting the class difference of the high-dimensional features. The characteristics of the data collected from healthy subjects are the most similar, while the features of EEG acquired from patients with epilepsy are quite different. This indicates that trained model obtained by NAS has the feature extraction ability to classify epilepsy EEG.
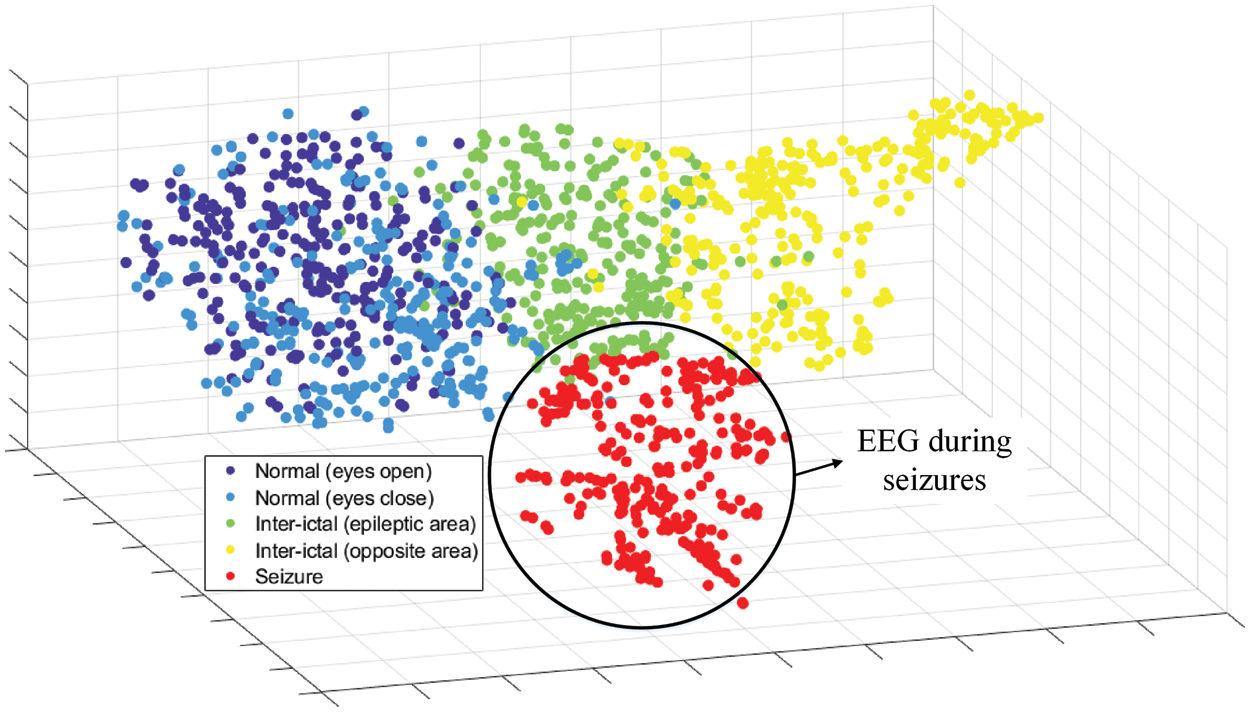
Figure 4: t-SNE of high-dimensional features from global average pooling layer
Through the feature representation extracted by the model searched by NAS, we can see that the model is good at extracting the features of epilepsy EEG signals and classifying epilepsy EEG and signals with other labels. In addition, it can be clearly observed that it is difficult for the NAS model to extract distinguishable features from the EEG collected in normal people with eyes open and eyes closed, which means that the signals with these two labels have high similarity. Boosting the classification performance of the NAS model on these two labels is one of our follow-up works.
Fig. 5 shows the relevance between EEG data with epilepsy class and model decisions calculated using the “Gradient * Input” [58] method in the Deep Explain framework [59]. We randomly select high-confidence and low-confidence data in test set for analysis, where the time domain signal displayed is processed by z-normalization. It can be seen from the figure that regardless of the level of confidence, the significant changes in the amplitude of the input original EEG signal are highly correlated with the model decision relevance, which indicates that the model has extracted abnormal discharge features of epileptic EEG for seizure detection. Furthermore, we can see that the signal part with sudden amplitude changes has a high degree of consistency with the model for high-confidence data, which means that the model basically does not miss the extraction of the characteristics of each abnormal discharge. But for low-confidence data, the feature of abnormal discharge has a low degree of matching with the model decision-making correlation. The model misses the extraction of some epilepsy features in the original input EEG signal, which may easily cause classification errors.
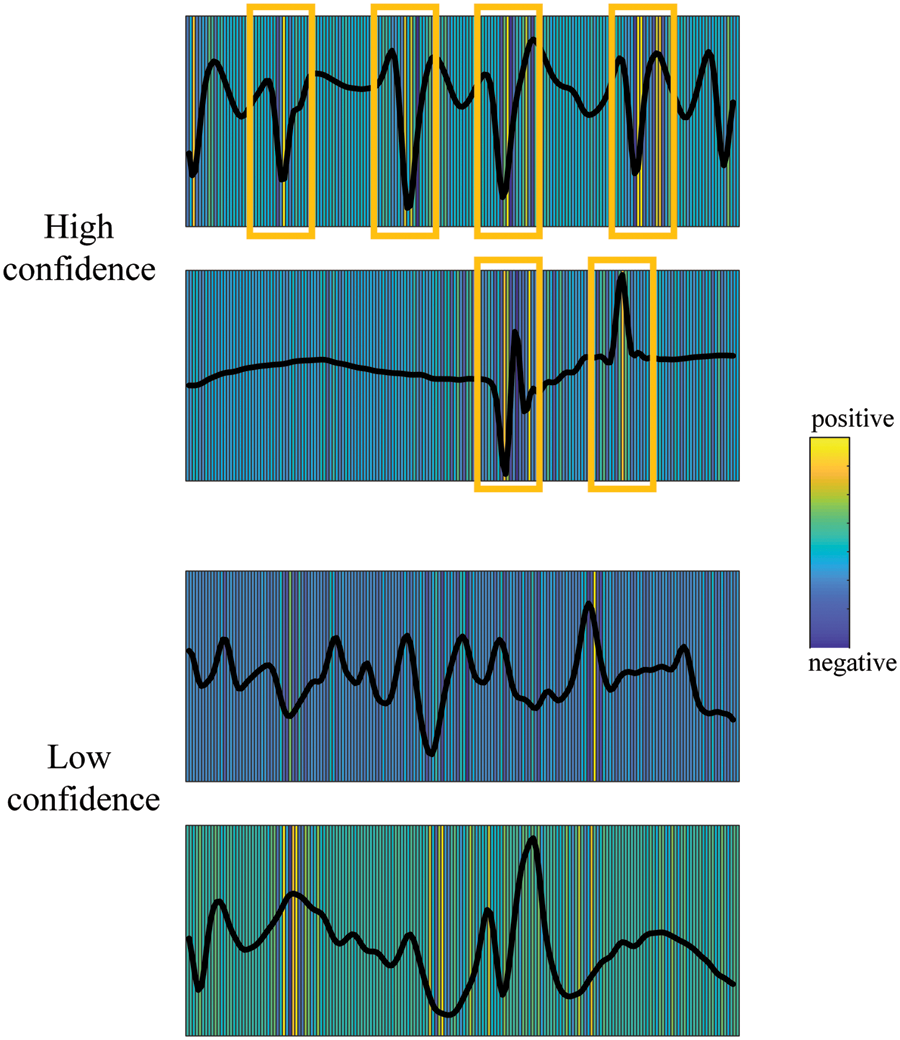
Figure 5: EEG feature relevance for high-confidence and low-confidence data in test set
The performance comparison between the searched deep convolutional neural network using NAS and EEGNet is illustrated in the Fig. 6. We can see that the performance metrics (accuracy, F1-score and Cohen’s kappa coefficient) of searched model on the test set are better than that of baseline model (p < 0.05), which reflects the great potential of the NAS algorithm in realizing the automatic detection system for epilepsy EEG. In terms of classification accuracy, the model searched by NAS can reach 76.61% when that of EEGNet is only 68.93%. A similar situation occurs in the evaluation of F1-score. The NAS model achieves 76.49% while the F1-score of baseline model is 68.68%. EEGNet reaches 61.17% in Kappa coefficient, and the neural network model searched by NAS achieves 70.76%. Using NAS to implement epilepsy EEG detection algorithms can reduce the dependence of algorithm design on professional experience and reduce development costs, thereby promoting the popularization of epilepsy EEG automatic detection system.
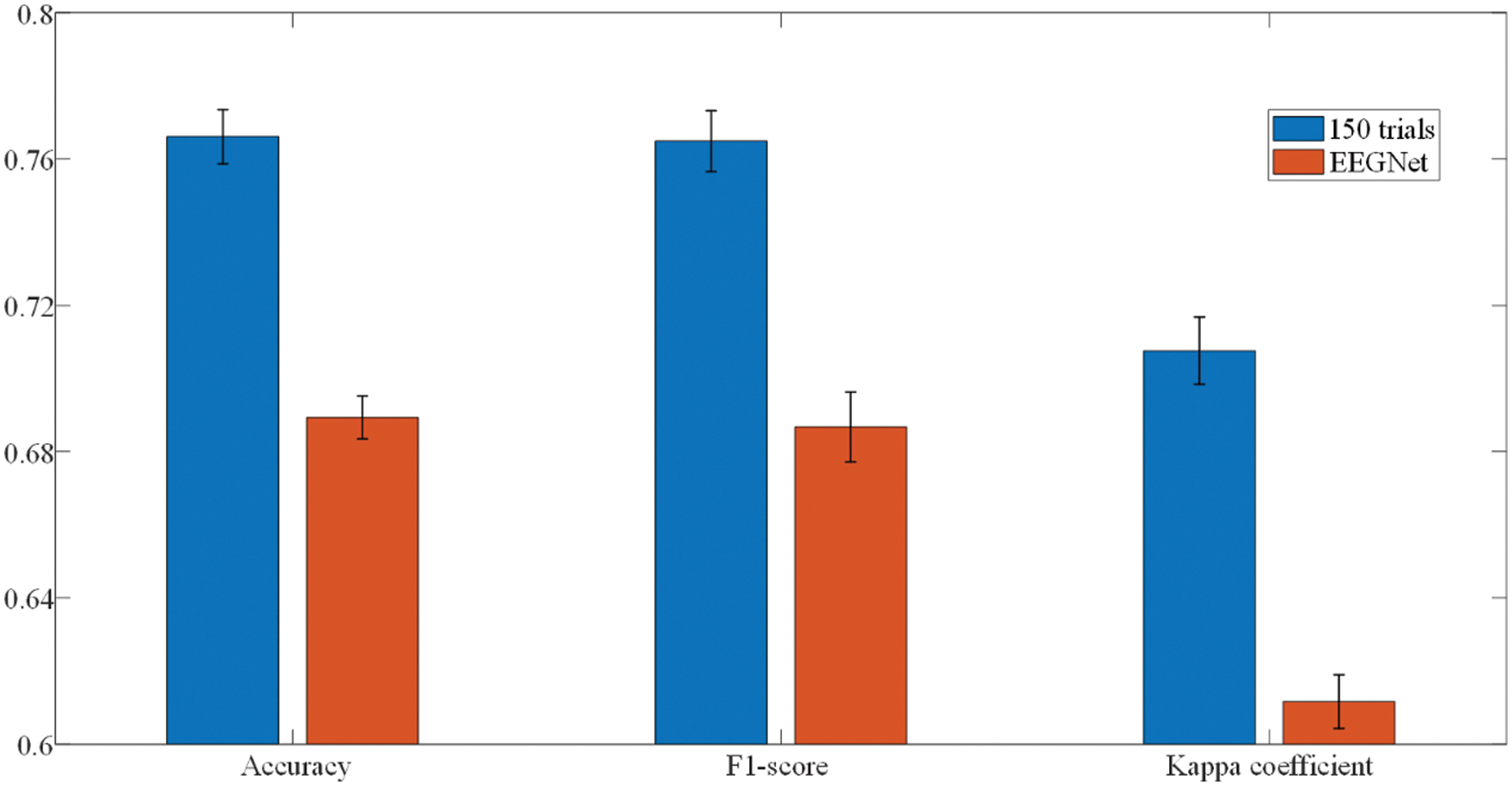
Figure 6: Classification performance of EEGNet and NAS-searched model
4.2 The Architect of Searched Model
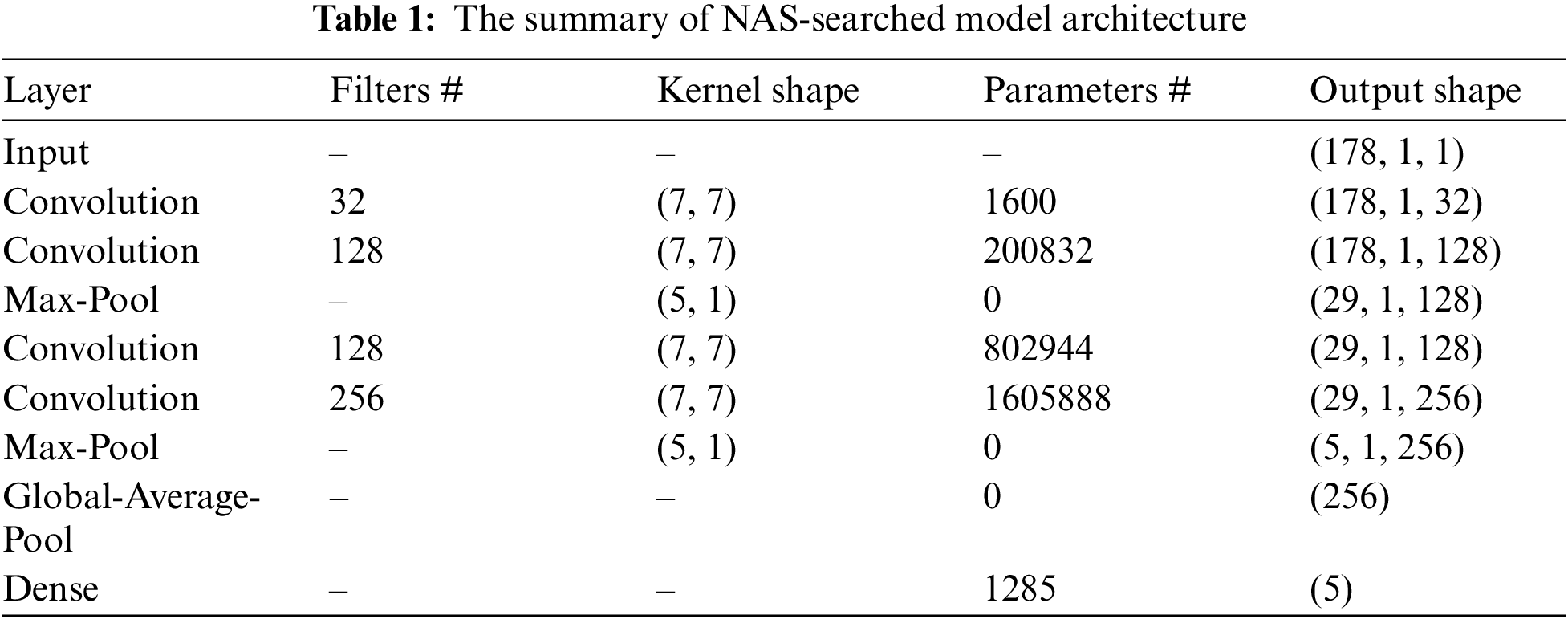
Fig. 7 and Tab. 1 shows the architecture of the convolutional neural network with the best performance obtained by NAS. It can be seen from the figure that the feature extraction part of this model consists of two convolution-max-pooling blocks, where each convolution-max-pooling block contains two convolutional layers and one maximum pooling layer. The convolutional layers are used to extract the local features of epilepsy EEG signals, and the stacking of two convolutional layers are capable of making the extracted EEG features more high-dimensional. The maximum pooling layers are added to compress the dimensionality of the feature map, thus avoiding the model from overfitting due to excessive parameters and improving the model’s ability to resist noise. The feature extraction part of this model is similar to the design of DeepConvNet [60] and convolutional neural network in [61] for EEG analysis. Then the model extracts global EEG features through the global average pooling layer and employs dropout technology as a regularization measure.
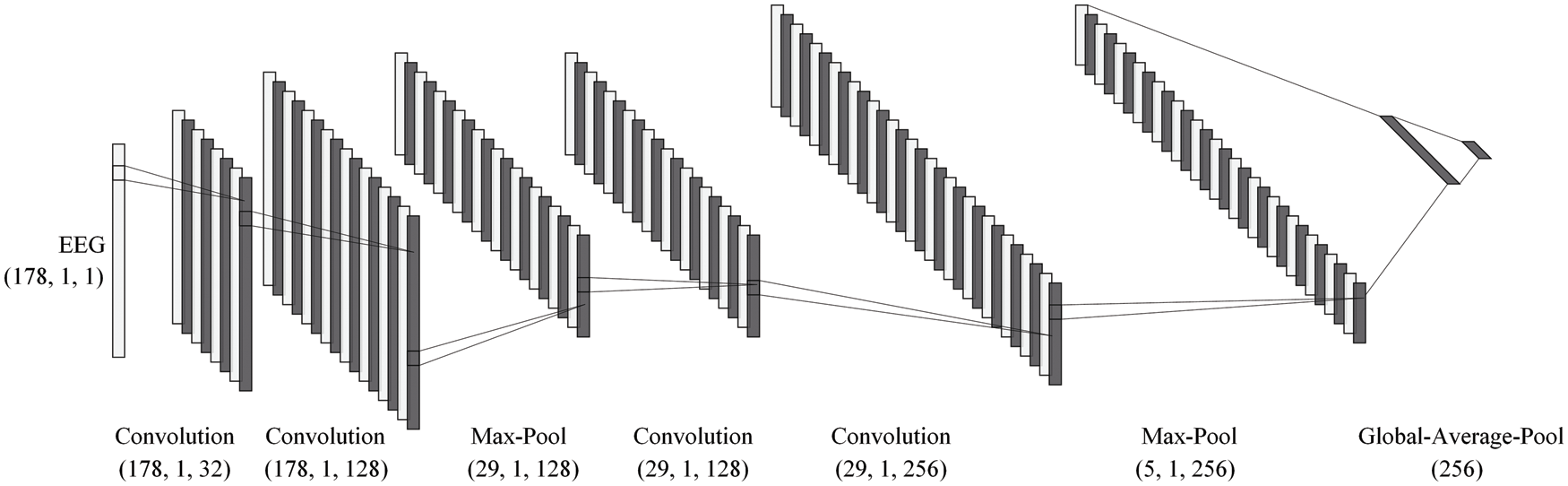
Figure 7: The model architecture of NAS-searched model
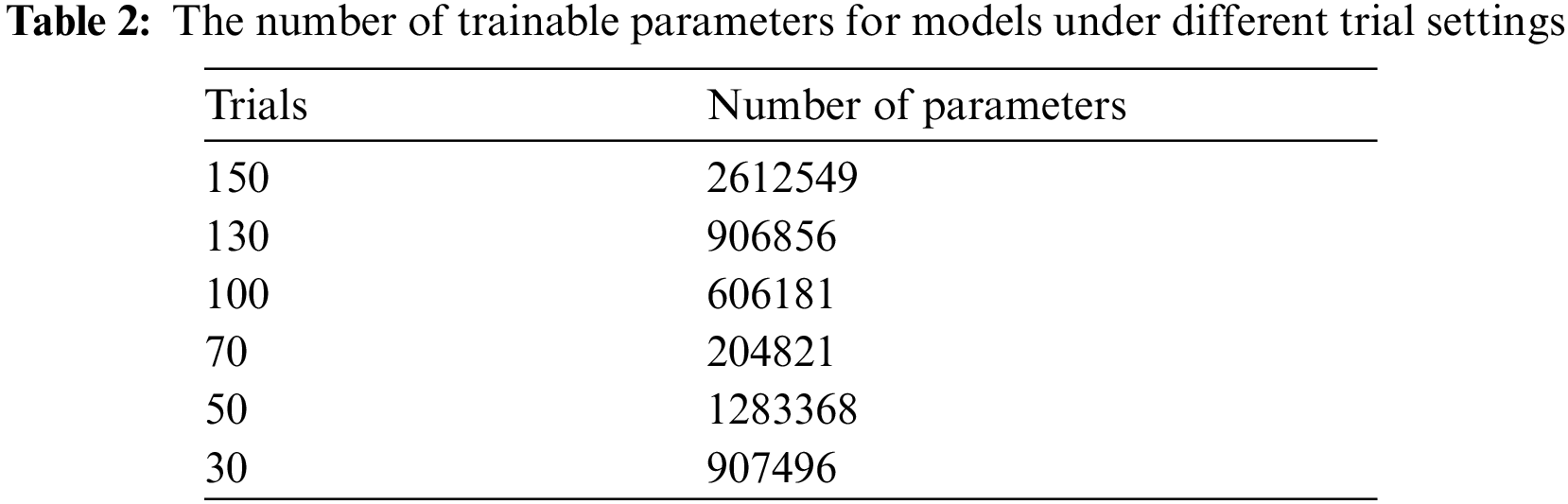
The number of searched model parameters under different trial settings is shown in Tab. 2. Since the AutoKeras platform applies network morphism, the model built by NAS gradually changes from a simple model to a complex one. With the increase in the number of trials, the amount of model parameters is increasing. It can be seen from the classification results that the corresponding feature extraction capabilities of searched model are also improving. This provides practical experience for the subsequent application of NAS to realize epilepsy EEG detection algorithms. A model with great performance requires sufficient number of trials for NAS.
In this work, we explore the possibility of AutoML playing a role in the task of automatic epilepsy EEG detection. We employ the neural architecture search algorithm based on network morphism in the AutoKeras platform to realize the design of the automatic epilepsy EEG detection model. The experimental results show that the multiple performance metrics (accuracy, F1-score and Cohen’s kappa coefficient) of the model obtained by NAS are better than the baseline model on the test set. In addition, deep learning feature interpretability methods are applied to analyze the feature extraction of the model, ensuring the reliability of the algorithm. However, there are still some limitations in this work. First, the search efficiency of the NAS algorithm needs to be improved. Second, the model generated by NAS has a large number of model parameters, which limits the response speed of practical applications. In future works, we will further boost the search efficiency of the NAS algorithm and promote the NAS to optimize the amount of parameters in the generated neural networks. Moreover, we intend to improve the ability of the model to analyze EEG signals in different states, and deploy it to the actual epilepsy EEG detection system.
Funding Statement: This work is supported by Fundamental Research Funds for the Central Universities (Grant No. FRF-TP-19-006A3).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. A. Ahmadi, M. Behroozi, V. Shalchyan and M. R. Daliri, “Classification of epileptic EEG signals by wavelet based CFC,” in Proc. EBBT, Tepekent, Turkey, IAU, TUR, pp. 1–4, 2018. [Google Scholar]
2. World Health Organization, Epilepsy, 2017. [Online]. Available: http://www.who.int/mediacentre/factsheets/fs999/en/. [Google Scholar]
3. American Epilepsy Society, Facts and Figures, https://www.aesnet.org/for_patients/facts_figures. 2017. [Google Scholar]
4. Harvard Health Publications, Harvard Medical School, Seizure Overview, 2014. [Online]. Available at: http://www.health.harvard.edu/mind-and-mood/seizure-overview. [Google Scholar]
5. M. S. Hossain, S. U. Amin, M. Alsulaiman and G. Muhammad, “Applying deep learning for epilepsy seizure detection and brain mapping visualization,” ACM Transactions on Multimedia Computing, Communications and Applications, vol. 15, no. 1, pp. 1–17, 2019. [Google Scholar]
6. A. Ahmed and M. Bayoumi, “A deep learning approach for automatic seizure detection in children with epilepsy,” Frontiers in Computational Neuroscience, vol. 15, no. 1, pp. 15–29, 2021. [Google Scholar]
7. T. Yan, X. Bi, M. Zhang, W. Wang, Z. Yao et al., “Age-related oscillatory theta modulation of multisensory integration in frontocentral regions,” NeuroReport, vol. 27, no. 11, pp. 796–801, 2016. [Google Scholar]
8. A. Y. Mutlu, “Detection of epileptic dysfunctions in EEG signals using Hilbert vibration decomposition,” Biomedical Signal Processing and Control, vol. 40, no. 4, pp. 33–40, 2018. [Google Scholar]
9. P. Thodoroff, P. Joelle and L. Andrew, “Learning robust features using deep learning for automatic seizure detection,” Machine Learning for Healthcare Conference, vol. 56, pp. 178–190, 2016. [Google Scholar]
10. Y. Ye, G. Xun, K. Jia and A. Zhang, “A multi-view deep learning framework for EEG seizure detection,” IEEE Journal of Biomedical and Health Informatics, vol. 23, no. 1, pp. 83–94, 2018. [Google Scholar]
11. R. Hussein, H. Palangi, R. Ward and Z. J. Wang, “Epileptic seizure detection: A deep learning approach,” arXiv preprint arXiv:1803.09848, 2018. [Google Scholar]
12. F. Mormann, R. G. Andrzejak, C. E. Elger and K. Lehnertz, “Seizure prediction: The long and winding road,” Brain, vol. 130, no. 2, pp. 314–333, 2007. [Google Scholar]
13. U. R. Acharya, S. L. Oh, Y. Hagiwara and J. H. Tan, “Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals,” Computers in Biology and Medicine, vol. 100, no. 21, pp. 270–278, 2018. [Google Scholar]
14. U. R. Acharya, S. V. Sree, G. Swapna, R. J. Martis and J. S. Suri, “Automated EEG analysis of epilepsy: A review,” Knowledge-Based Systems, vol. 45, no. 1, pp. 147–165, 2013. [Google Scholar]
15. O. Faust, U. R. Acharya, H. Adeli and A. Adeli, “Wavelet-based EEG processing for computer-aided seizure detection and epilepsy diagnosis,” Seizure, vol. 26, no. 3, pp. 56–64, 2015. [Google Scholar]
16. Y. D. Song, J. Crowcroft and J. Zhang, “Automatic epileptic seizure detection in EEGs based on optimized sample entropy and extreme learning machine,” Journal of Neuroscience Methods, vol. 210, no. 2, pp. 132–146, 2012. [Google Scholar]
17. K. Samiee, K. Peter and G. Moncef, “Epileptic seizure classification of EEG time-series using rational discrete short-time Fourier transform,” IEEE transactions on Biomedical Engineering, vol. 62, no. 2, pp. 541–552, 2014. [Google Scholar]
18. A. Hamad., E. H. Houssein, A. E. Hassanien and A. A. Fahmy, “A hybrid EEG signals classification approach based on grey wolf optimizer enhanced SVMs for epileptic detection,” in Proc. Int. Conf. on Advanced Intelligent Systems and Informatics, Cham, Springer, pp. 108–117, 2017. [Google Scholar]
19. A. T. Tzallas, M. G. Tsipouras and D. I. Fotiadis, “Automatic seizure detection based on time-frequency analysis and artificial neural networks,” Computational Intelligence and Neuroscience, vol. 2007, no. 4, pp. 1–13, 2007. [Google Scholar]
20. T. McShane, “A clinical guide to epileptic syndromes and their treatment,” Archives of Disease in Childhood, vol. 89, no. 6, pp. 591–591, 2004. [Google Scholar]
21. X. Zhang, L. Yao, X. Wang, J. Monaghan and D. McAlpine, “A survey on deep learning-based non-invasive brain signals: Recent advances and new frontiers,” Journal of Neural Engineering, vol. 18, no. 3, pp. 1–44, 2021. [Google Scholar]
22. K. Wang, Y. Zhao, Q. Xiong, M. Fan and G. Sun, “Research on healthy anomaly detection model based on deep learning from multiple time-series physiological signals,” Scientific Programming, vol. 2016, pp. 1–9, 2016. [Google Scholar]
23. T. S. Robin, G. Lukas, E. Katharina, H. Frank and B. Tonio, “Deep learning with convolutional neural networks for decoding and visualization of eeg pathology,” arXiv e-prints, arXiv-1708, 2017. [Google Scholar]
24. U. R. Acharya, S. L. Oh, Y. Hagiwara, J. H. Tan and H. Adelid, “Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals,” Computers in Biology and Medicine, vol. 100, no. 21, pp. 270–278, 2018. [Google Scholar]
25. J. T. Turner, A. Page, T. Mohsenin and T. Oates, “Deep belief networks used on high resolution multichannel electroencephalography data for seizure detection,” in Proc. 2014 AAAI Spring Sym. Series, California, America, pp. 75–81, 2014. [Google Scholar]
26. M. P. Hosseini, H. Soltanian-Zadeh, K. Elisevich and D. Pompili, “Cloud-based deep learning of big EEG data for epileptic seizure prediction,” in Proc. 2016 IEEE Global Conf. on Signal and Information Processing, Washington, America, pp. 1151–1155, 2016. [Google Scholar]
27. A. M. Abdelhameed, H. G. Daoud and M. Bayoumi, “Deep convolutional bidirectional LSTM recurrent neural network for epileptic seizure detection,” in Proc. 2018 16th IEEE Int. New Circuits and Systems Conf., Montreal, Canada, pp. 139–143, 2018. [Google Scholar]
28. X. R. Zhang, X. Sun, W. Sun, T. Xu and P. P. Wang, “Deformation expression of soft tissue based on BP Neural network,” Intelligent Automation & Soft Computing, vol. 32, no. 2, pp. 1041–1053, 2022. [Google Scholar]
29. X. R. Zhang, J. Zhou, W. Sun and S. K. Jha, “A lightweight CNN based on transfer learning for COVID-19 diagnosis,” Computers, Materials & Continua, vol. 72, no. 1, pp. 1123–1137, 2022. [Google Scholar]
30. D. Shen, G. Wu and H. Suk, “Deep learning in medical image analysis,” Annual Review of Biomedical Engineering, vol. 19, no. 1, pp. 221–248, 2017. [Google Scholar]
31. T. Jeslin and J. A. Linsely, “Agwo-cnn classification for computer-assisted diagnosis of brain tumors,” Computers, Materials & Continua, vol. 71, no. 1, pp. 171–182, 2022. [Google Scholar]
32. R. Rajaragavi and S. P. Rajan, “Optimized u-net segmentation and hybrid res-net for brain tumor mri images classification,” Intelligent Automation & Soft Computing, vol. 32, no. 1, pp. 1–14, 2022. [Google Scholar]
33. R. Muthaiyan and D. M. Malleswaran, “An automated brain image analysis system for brain cancer using shearlets,” Computer Systems Science and Engineering, vol. 40, no. 1, pp. 299–312, 2022. [Google Scholar]
34. H. A. Mengash and H. A. Hosni Mahmoud, “Brain cancer tumor classification from motion-corrected mri images using convolutional neural network,” Computers, Materials & Continua, vol. 68, no. 2, pp. 1551–1563, 2021. [Google Scholar]
35. E. Gothai, A. Baseera, P. Prabu, K. Venkatachalam, K. Saravanan et al., “Machine learning technique to detect radiations in the brain,” Computer Systems Science and Engineering, vol. 42, no. 1, pp. 149–163, 2022. [Google Scholar]
36. Y. Du, M. Yin and B. Jiao, “InceptionSSVEP: A multi-scale convolutional neural network for steady-state visual evoked potential classification,” in Proc. 2020 IEEE 6th Int. Conf. on Computer and Communications, Chengdu, China, pp. 2080–2085, 2020. [Google Scholar]
37. H. Jin, Q. Song and X. Hu, “Auto-keras: An efficient neural architecture search system,” in Proc. of the 25th ACM SIGKDD Int. Conf. on Knowledge Discovery & Data Mining, Anchorage, America, pp. 1946–1956, 2019. [Google Scholar]
38. B. Baker, O. Gupta, N. Naik and R. Raskar, “Designing neural network architectures using reinforcement learning,” arXiv preprint arXiv:1611.02167, 2016. [Google Scholar]
39. Z. Zhao, J. Yan and C. Liu, “Practical network blocks design with q-learning,” arXiv preprint arXiv:1708.05552, 2017. [Google Scholar]
40. H. Liu, K. Simonyan, O. Vinyals, C. Fernando and K. Kavukcuoglu, “Hierarchical representations for efficient architecture search,” arXiv preprint arXiv:1711.00436, 2017. [Google Scholar]
41. E. Real, A. Aggarwal, Y. Huang and Q. V. Le, “Regularized evolution for image classifier architecture search,” Proceedings of the AAAI Conference on Artificial Intelligence, vol. 33, no. 01, pp. 4780–4789, 2019. [Google Scholar]
42. C. Li, Z. Zhang, R. Song, J. Cheng, Y. Liu et al., “EEG-based emotion recognition via neural architecture search,” IEEE Transactions on Affective Computing, vol. 1, no. 1, pp. 1–12, 2021. [Google Scholar]
43. E. Rapaport, O. Shriki and R. Puzis, “Eegnas: Neural architecture search for electroencephalography data analysis and decoding,” International Workshop on Human Brain and Artificial Intelligence, Macao, China (Conference Proceedingspp. 3–20, 2019. [Google Scholar]
44. Y. Yang, Z. Gao, Y. Li and H. Wang, “A CNN identified by reinforcement learning-based optimization framework for EEG-based state evaluation,” Journal of Neural Engineering, vol. 18, no. 4, pp. 1–12, 2021. [Google Scholar]
45. W. L. Zheng, W. Liu, Y. Lu, B. L. Lu and A. Cichocki, “Emotionmeter: A multimodal framework for recognizing human emotions,” IEEE Transactions on Cybernetics, vol. 49, no. 3, pp. 1110–1122, 2019. [Google Scholar]
46. Y. Zhong and J. Zhang, “Cross-session classification of mental workload levels using EEG and an adaptive deep learning model,” Biomedical Signal Processing and Control, vol. 33, no. 1, pp. 30–47, 2017. [Google Scholar]
47. K. G. Hartmann, R. T. Schirrmeister and T. Ball, “Hierarchical internal representation of spectral features in deep convolutional networks trained for EEG decoding,” in Int. Conf. on Brain-Computer Interface, Jeongseon, South Korea, pp. 1–6, 2018. [Google Scholar]
48. V. J. Lawhern, A. J. Solon, N. R. Waytowich, S. M. Gordon and C. P. Hung, “EEGNet: A compact convolutional neural network for EEG-based brain-computer interfaces,” Journal of Neural Engineering, vol. 15, no. 5, pp. 1–18, 2018. [Google Scholar]
49. R. G. Andrzejak, K. Lehnertz, F. Mormann, C. Rieke, P. David et al., “Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state,” Physical Review E, vol. 64, no. 6, pp. 1–8, 2001. [Google Scholar]
50. T. Chen, I. Goodfellow and J. Shlens, “Net2net: Accelerating learning via knowledge transfer,” arXiv preprint arXiv:1511.05641, 2015. [Google Scholar]
51. T. Wei, C. Wang, Y. Rui and C. W. Chen, “Network morphism,” in Int. Conf. on Machine Learning. PMLR, New York, America, pp. 564–572, 2016. [Google Scholar]
52. P. Auer, N. Cesa-Bianchi and P. Fischer, “Finite-time analysis of the multiarmed bandit problem,” Machine Learning, vol. 47, no. 2, pp. 235–256, 2016. [Google Scholar]
53. D. Bertsimas and J. Tsitsiklis, “Simulated annealing,” Statistical Science, vol. 8, no. 1, pp. 10–15, 1993. [Google Scholar]
54. R. E. Korf, “Depth-first iterative-deepening: An optimal admissible tree search,” Artificial Intelligence, vol. 27, no. 1, pp. 97–109, 1985. [Google Scholar]
55. A. Bagnall, J. Lines, A. Bostrom, J. Large and E. Keogh, “The great time series classification bake off: A review and experimental evaluation of recent algorithmic advances,” Data Mining and Knowledge Discovery, vol. 31, no. 3, pp. 606–660, 2017. [Google Scholar]
56. J. Cohen, “A coefficient of agreement for nominal scales,” Educational and Psychological Measurement, vol. 20, no. 1, pp. 37–46, 1960. [Google Scholar]
57. L. V. der Maaten and G. Hinton, “Visualizing data using t-SNE,” Journal of Machine Learning Research, vol. 9, no. 11, pp. 2579–2605, 2008. [Google Scholar]
58. A. Shrikumar, P. Greenside, A. Shcherbina and A. Kundaje, “Not just a black box: Learning important features through propagating activation differences,” arXiv preprint arXiv:1605.01713, 2016. [Google Scholar]
59. M. Ancona, E. Ceolini, C. Öztireli and M. Gross, “Towards better understanding of gradient-based attribution methods for deep neural networks,” arXiv preprint arXiv:1711.06104, 2017. [Google Scholar]
60. R. T. Schirrmeister, J. T. Springenberg, L. D. J. Fiederer, M. Glasstetter, K. Eggensperger et al., “Deep learning with convolutional neural networks for EEG decoding and visualization,” Human Brain Mapping, vol. 38, no. 11, pp. 5391–5420, 2017. [Google Scholar]
61. S. Zhao, J. Yang and M. Sawan, “Energy-efficient neural network for epileptic seizure prediction,” IEEE Transactions on Bio-medical Engineering, vol. 69, no. 1, pp. 401–411, 2021. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |