DOI:10.32604/cmc.2022.028935

| Computers, Materials & Continua DOI:10.32604/cmc.2022.028935 |  |
| Article |
Efficient Segmentation Approach for Different Medical Image Modalities
1Department Electronics and Electrical Communications, Faculty of Electronic Engineering, Menoufia University, Menouf, 32952, Egypt
2Security Engineering Laboratory, Department of Computer Science, Prince Sultan University, Riyadh, 11586,Saudi Arabia
3Department of Information Technology, College of Computer and Information Sciences, Princess Nourah Bint Abdulrahman University, Riyadh, 11671, Saudi Arabia
4Department of Computer Sciences, College of Computer and Information Sciences, Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia
*Corresponding Author: Amel A. Alhussan. Email: aaalhussan@pnu.edu.sa
Received: 21 February 2022; Accepted: 24 March 2022
Abstract: This paper presents a study of the segmentation of medical images. The paper provides a solid introduction to image enhancement along with image segmentation fundamentals. In the first step, the morphological operations are employed to ensure image detail protection and noise-immunity. The objective of using morphological operations is to remove the defects in the texture of the image. Secondly, the Fuzzy C-Means (FCM) clustering algorithm is used to modify membership function based only on the spatial neighbors instead of the distance between pixels within local spatial neighbors and cluster centers. The proposed technique is very simple to implement and significantly fast since it is not necessary to compute the distance between the neighboring pixels and the cluster centers. It is also efficient when dealing with noisy images because of its ability to efficiently improve the membership partition matrix. Simulation results are performed on different medical image modalities. Ultrasonic (Us), X-ray (Mammogram), Computed Tomography (CT), Positron Emission Tomography (PET), and Magnetic Resonance (MR) images are the main medical image modalities used in this work. The obtained results illustrate that the proposed technique can achieve good results with a short time and efficient image segmentation. Simulation results on different image modalities show that the proposed technique can achieve segmentation accuracies of 98.83%, 99.71%, 99.83%, 99.85%, and 99.74% for Us, Mammogram, CT, PET, and MRI images, respectively.
Keywords: Image segmentation; ultrasonic; mammogram; CT; PET; MRI; morphological operations; FCM; active contours
Image modalities differ as they reflect the internal anatomy of the different body organs. Morphology refers to the description of the shape and structure of the object in a specific image. It depends on the set theory and relies more on the relative arrangement of pixels instead of on their numerical values. The important parameters for structuring elements are shape, size, and origin. The structuring element shape depends on the arrangement of ones and zeros in the matrix. The structuring element acts as a window over which the interaction takes place. It also helps to characterize the image objects or features. The mechanism of structuring element operation is similar to that of masks used in spatial filtering. A structuring element is moved over the whole image [1].
Image segmentation [2–4] is the process of splitting an image into several non-overlapping segments (sets of pixels, also known as image objects). The success of the image analysis process depends on the accuracy of the segmentation process, but a successful segmentation of an image is generally a difficult problem. The input is an image during an image pre-processing operation, and the output is an enhanced high-quality image. Image segmentation is also an important tool in various medical imaging applications. It simplifies delineating boundaries or edges of structures, organs, or other regions of interest. Several techniques have been proposed for image segmentation. The active contour is the most well-known technique because of its efficiency and speediness. This paper will focus on medical image segmentation based on morphological operations, FCM, and active contour models.
Tab. 1 summarizes some of the related work [5–17] in medical image segmentation and the metrics of segmentation evaluation. This paper aims to develop a segmentation approach for different medical image modalities. In particular, the main target is the segmentation of the tumor region of interest (ROI). The proposed segmentation approach uses FCM combined with morphological operations and the active contour model to delineate the tumor region in the medical image and classify it as benign or malignant in future work. The motivation of this work is to enhance patient medical insurance by providing greater and more accurate information for medical diagnosis. Furthermore, this paper aims to detect, analyze and solve important and relevant medical problems.
The main contributions of this paper can be summarized as:
• It presents an efficient segmentation technique for different medical image modalities.
• It combines FCM with morphological operations, so that the tumor region segmentation is represented by the membership functions of the tissues derived from the minimization of the energy in the FCM.
• It adds both the global and local spatial information into the membership function to decrease the sensitivity problem to the noise and intensity inhomogeneity in the image data.
• It employs the active contour model to delineate the ROI, which can be extracted.
This paper is organized as follows. Section 2 illustrates the proposed segmentation process. The simulation results on Us, Mammogram, CT, PET, and MR images are given in Section 3. Section 4 handles the effect of different noise types on the segmentation process. Finally, Section 5 presents the conclusions and future work.
2 The Proposed Image Segmentation Technique
The proposed image segmentation technique consists of four steps:
Step 1. Pre-processing and Enhancement using Histogram Equalization.
Step 2. Morphological Operations and FCM.
Step 3. Active Contour Segmentation.
Step 4. ROI Extraction.
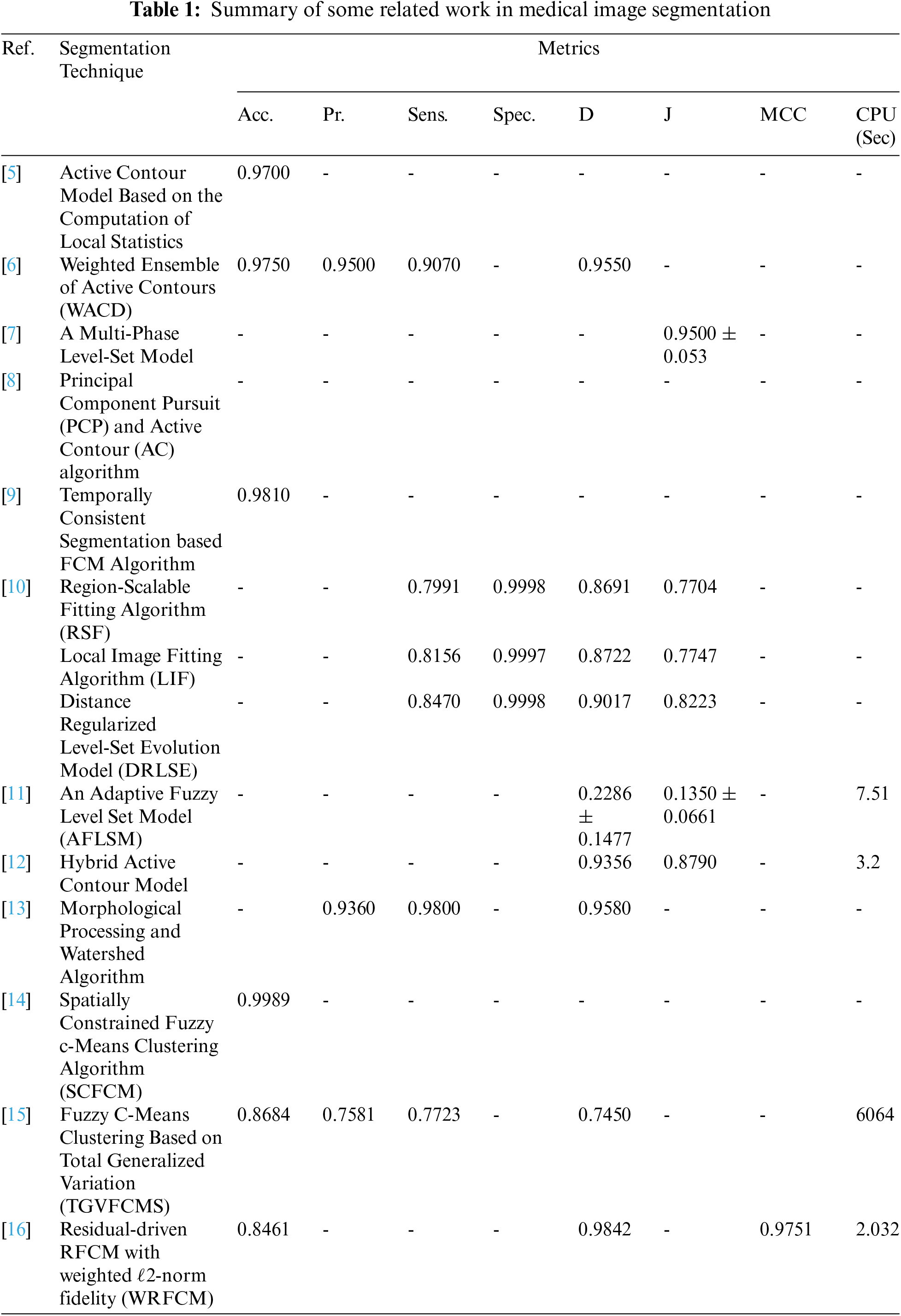
The steps of the segmentation process are shown in Fig. 1. The proposed work is implemented on different medical image modalities. Fig. 1a is a sample of Us breast image, Fig. 1b is a sample of X-ray (mammogram) image, Fig. 1c is a sample of CT chest image, Fig. 1d is a sample of PET brain image and Fig. 1e is a sample of MR brain image. All steps will be explained in the following steps.
Figure 1: Steps of the proposed segmentation technique
Step 1. Pre-processing and Image Enhancement using Histogram Equalization
Image pre-processing actually means that the input image is fed to a particular algorithm to obtain a better quality image. Image enhancement is known to be the digital image adjustment so that the results are easier and simpler for analysis and display. Histogram equalization [18–24] is one of the pixel brightness transformations techniques. It is a well-known contrast enhancement technique due to its performance on almost all types of images.
Step 2. Morphological Operations and FCM
The main target of step 2 is removing the image imperfections by accounting for the form and structure of the image using a collection of non-linear operations related to the shape or morphology of features, and finding the cluster centers that maximize a similarity function or minimize the dissimilarity function. This combination of morphological operation and FCM clustering algorithm produces better segmentation results due to their advantages.
Step 3. Active Contour Segmentation
Segmentation methods based on the active contour model [25–27] outperform many of the traditional image segmentation methods introduced in the literature. The Chan-Vese algorithm is based on an energy minimization problem, which can be reformulated to be the easiest way to solve the problem.
3 Simulation Results and Discussions
Simulation results were carried out using MATLAB R2019a on a Dell machine, Core i5 processor, 8 Gbytes RAMs, and 320 Gbytes hard disk. Simulation results are conducted on five different examples of the scanned image (Us, Mammogram, CT, PET, and MRI) [28,29].
To reveal the efficiency of the proposed technique, a sample of each different modality of medical images is tested in the experiments. To assess the segmentation performance of the proposed technique, the accuracy and similarity indices are calculated: the segmentation Accuracy (Acc.), Recall or Sensitivity (Sens.), Precision (Pr.), Matthews Correlation Coefficient (MCC), Dice coefficient (D), also known as Dice similarity index (F1 score), Jaccard (J) and Specificity (Spec.) [30]:
where TP is the true positive, FP is the false positive, TN is the true negative, FN is the false negative.
Fig. 2 shows samples of the Us breast images, Fig. 2b shows the Us images after pre-processing stage, Fig. 2c shows the FCM and morphological stage output images, Fig. 2d shows the segmentation results of the ROI produced by active contour model (the tumor is marked by a red contour), and Fig. 2e shows the resultant ROI images. Fig. 3a shows samples of the mammogram original images, Fig. 3b shows the mammogram images after pre-processing stage, Fig. 3c shows the FCM and morphological stage output images, Fig. 3d shows the segmentation results of the ROI produced by the active contour model, and Fig. 3e shows the resultant ROI images. Fig. 4a shows a sample of the CT chest images, Fig. 4b shows the CT images after pre-processing stage, Fig. 4c shows the FCM and morphological stage output images, Fig. 4d shows the segmentation results of the ROI produced by active contour model, and Fig. 4e shows the resultant ROI images.
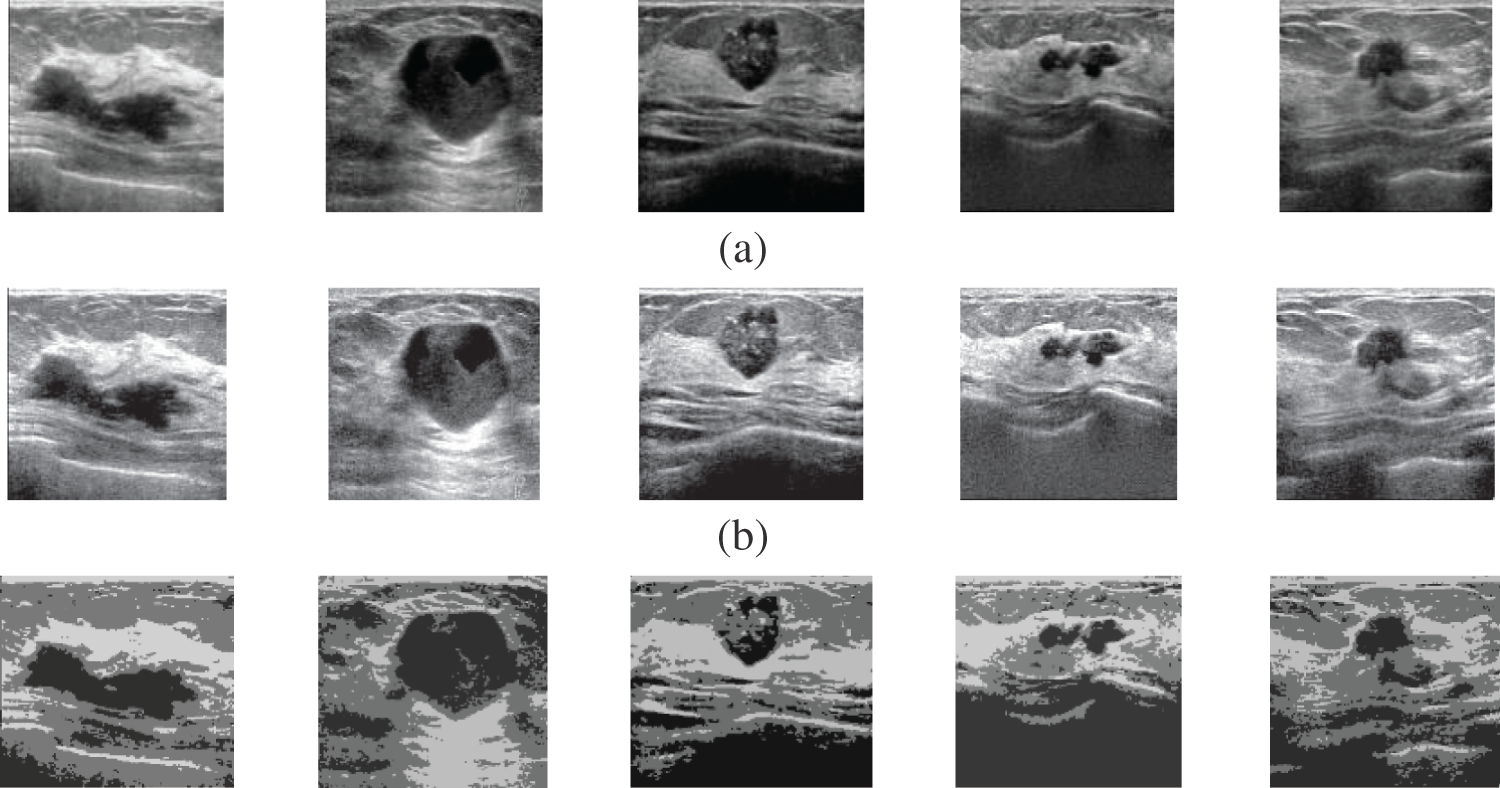
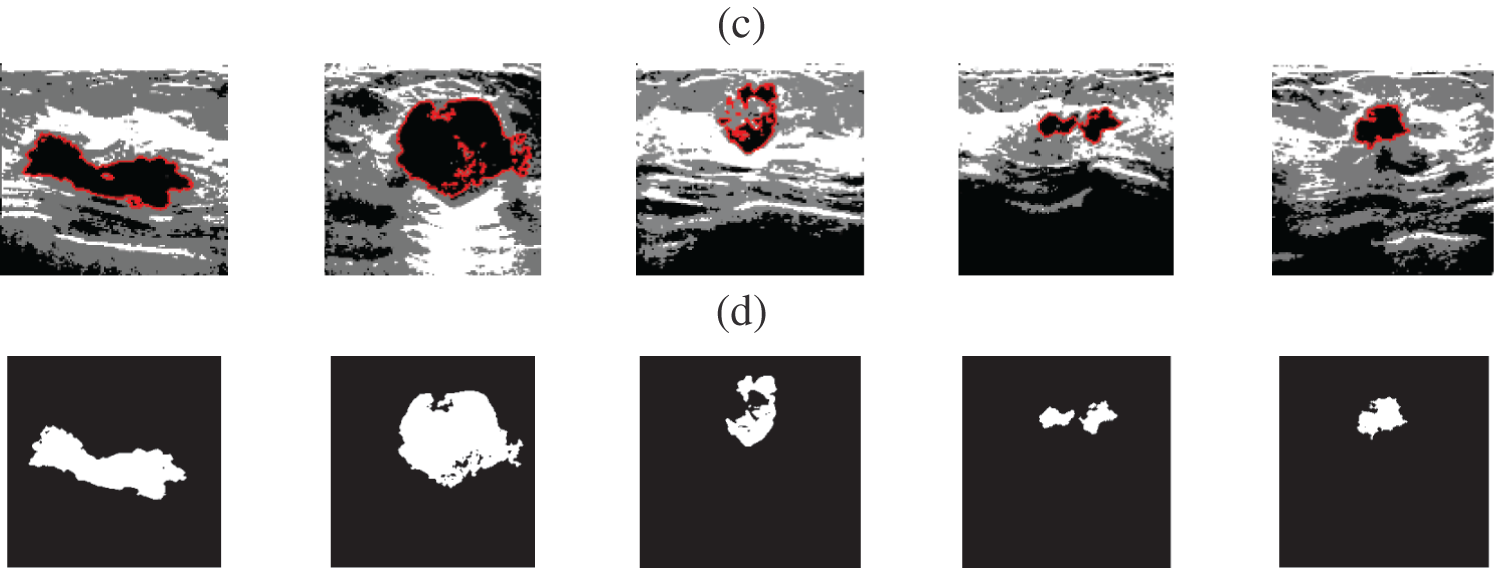
Figure 2: A sample of the Us medical images (a) Us images, (b) Us images after preprocessing stage, (c) FCM and morphological operation output images, (d) Segmented images after active contour, and (e) The extracted tumor images
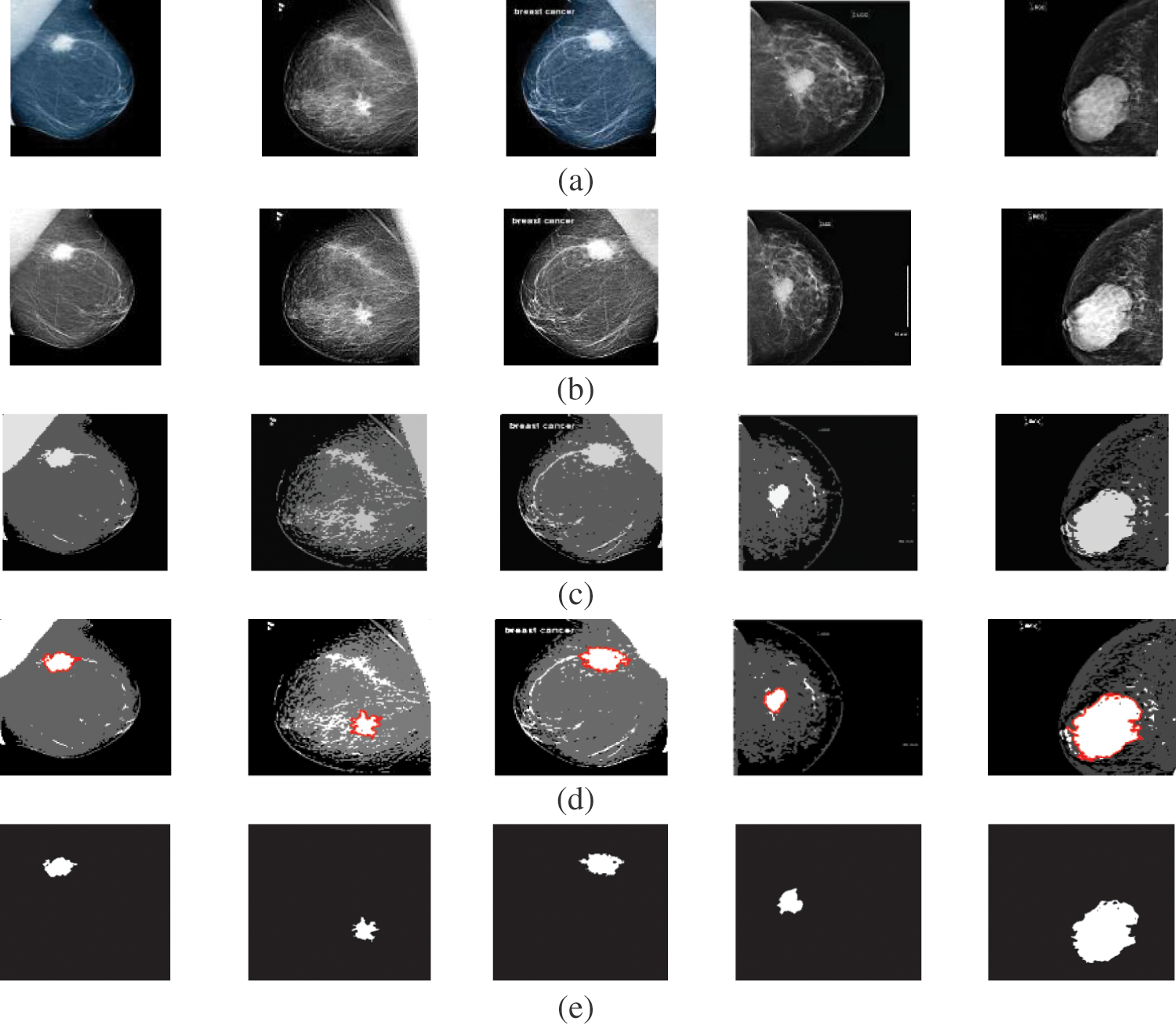
Figure 3: A sample of the Mammogram medical images (a) Mammogram images, (b) Mammogram images after preprocessing stage, (c) FCM and morphological operation output images, (d) Segmented images after active contour, and (e) The extracted tumor images
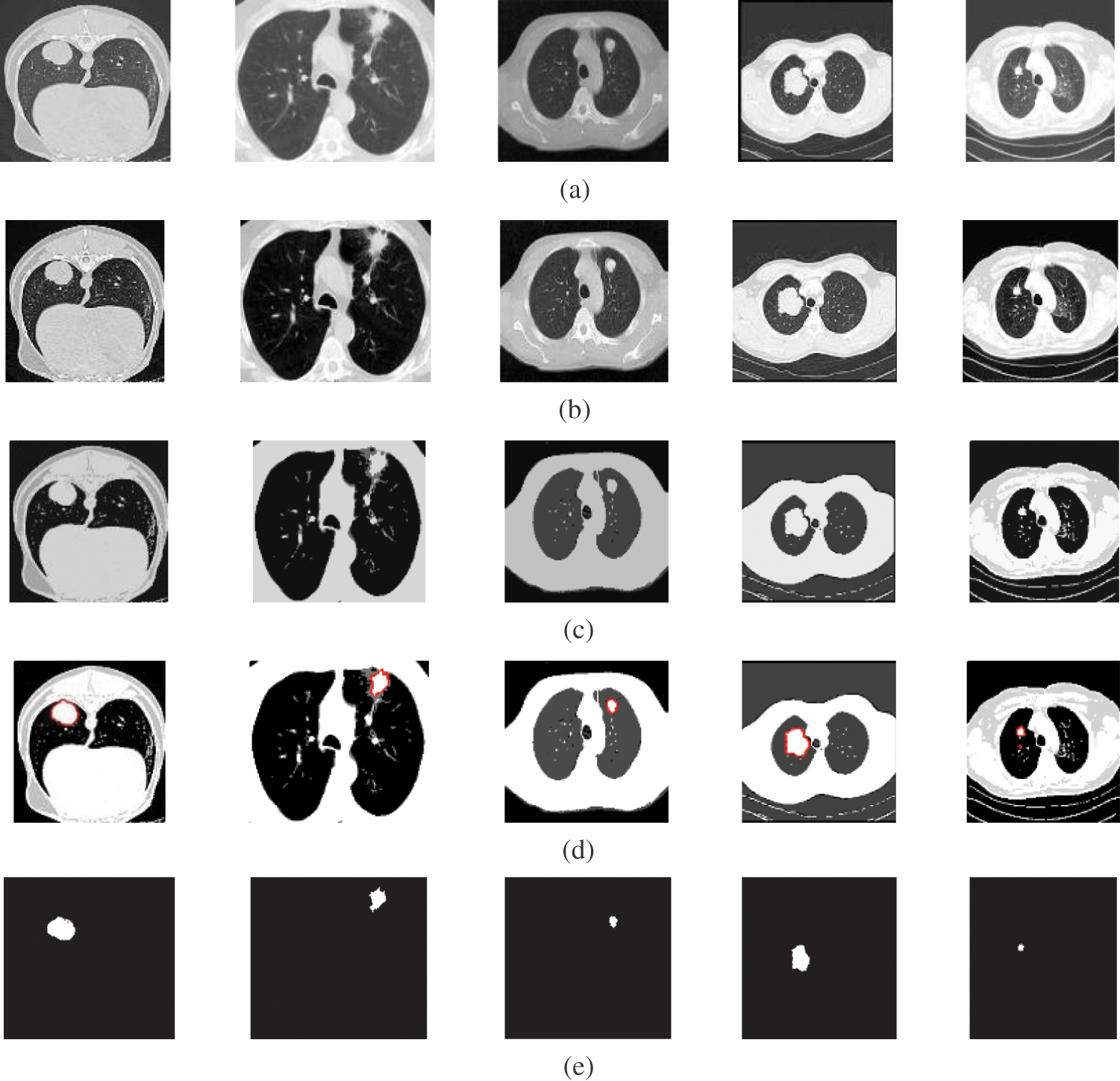
Figure 4: A sample of the CT medical images (a) CT images, (b) CT images after preprocessing stage, (c) FCM and morphological operation output images, (d) Segmented images after active contour, and (e) The extracted tumor images
Fig. 5a shows samples of the PET brain images, Fig. 5b shows the PET images after pre-processing stage, Fig. 5c shows the FCM and morphological stage output images, Fig. 5d shows the segmentation results of the ROI produced by active contour model, and Fig. 5e shows the resultant ROI images. Fig. 6a shows samples of the MRI brain images, Fig. 6b shows the MRI images after pre-processing stage, Fig. 6c shows the FCM and morphological stage output images, Fig. 6d shows the segmentation results of the ROI produced by active contour model, and Fig. 6e shows the resultant ROI images. Tabs. 1–5 show the segmentation Accuracy (Acc.), Sensitivity (Sens.), Precision (Pr.), F-Measure (FM), Matthews Correlation Coefficient (MCC), Dice (D), Jaccard (J), and Specificity (Spec.) numerical results on different image modalities.
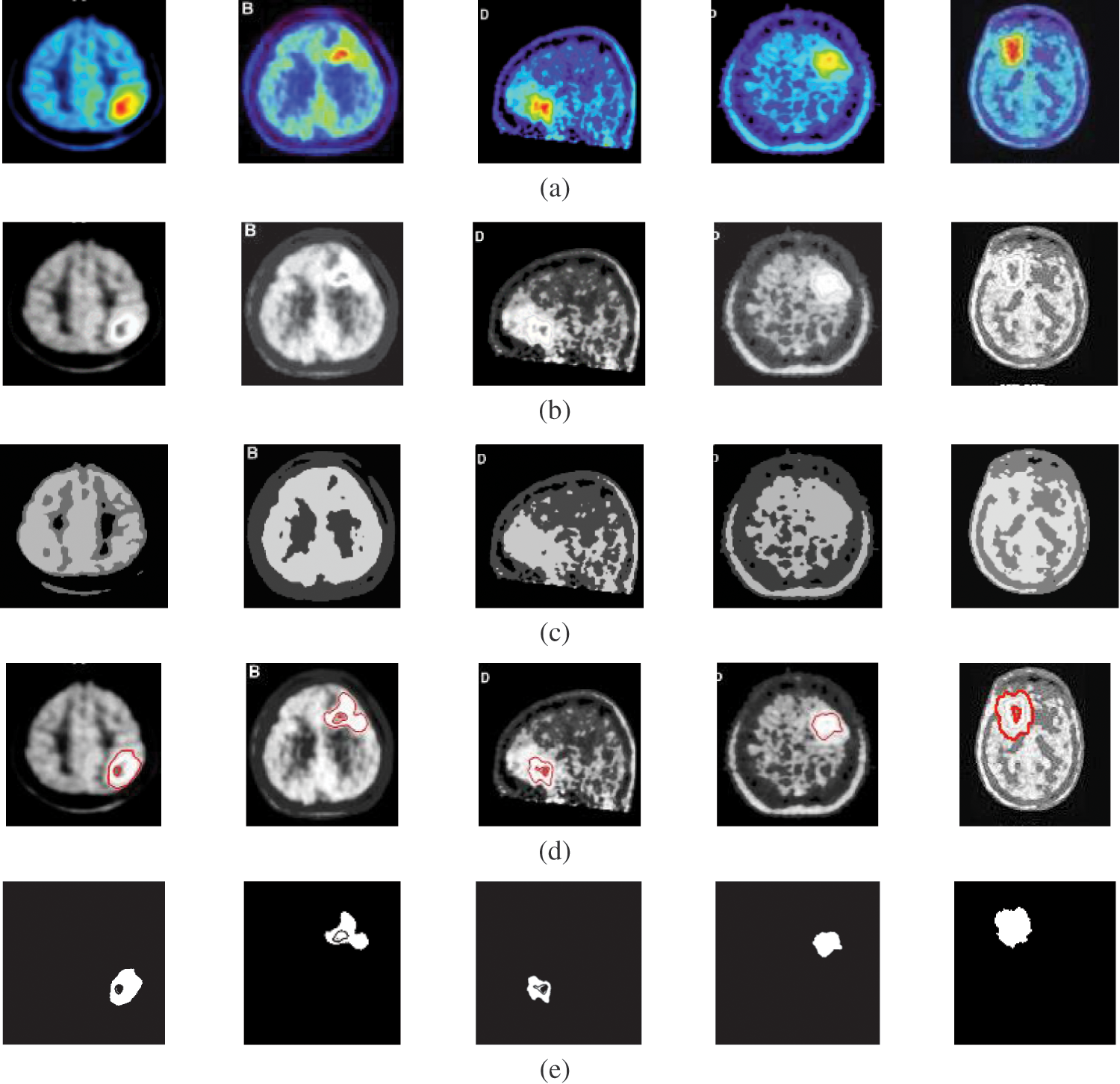
Figure 5: A sample of the PET medical images (a) PET images, (b) PET images after preprocessing stage, (c) FCM and morphological operation output images, (d) Segmented images after active contour, and (e) The extracted tumor images
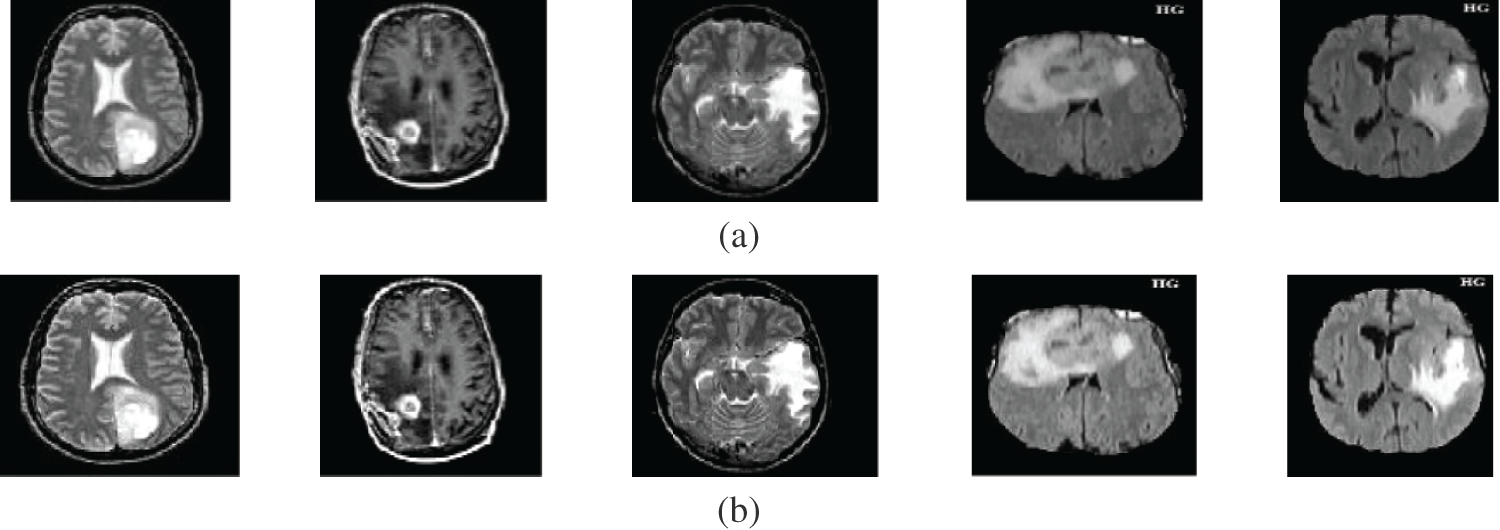
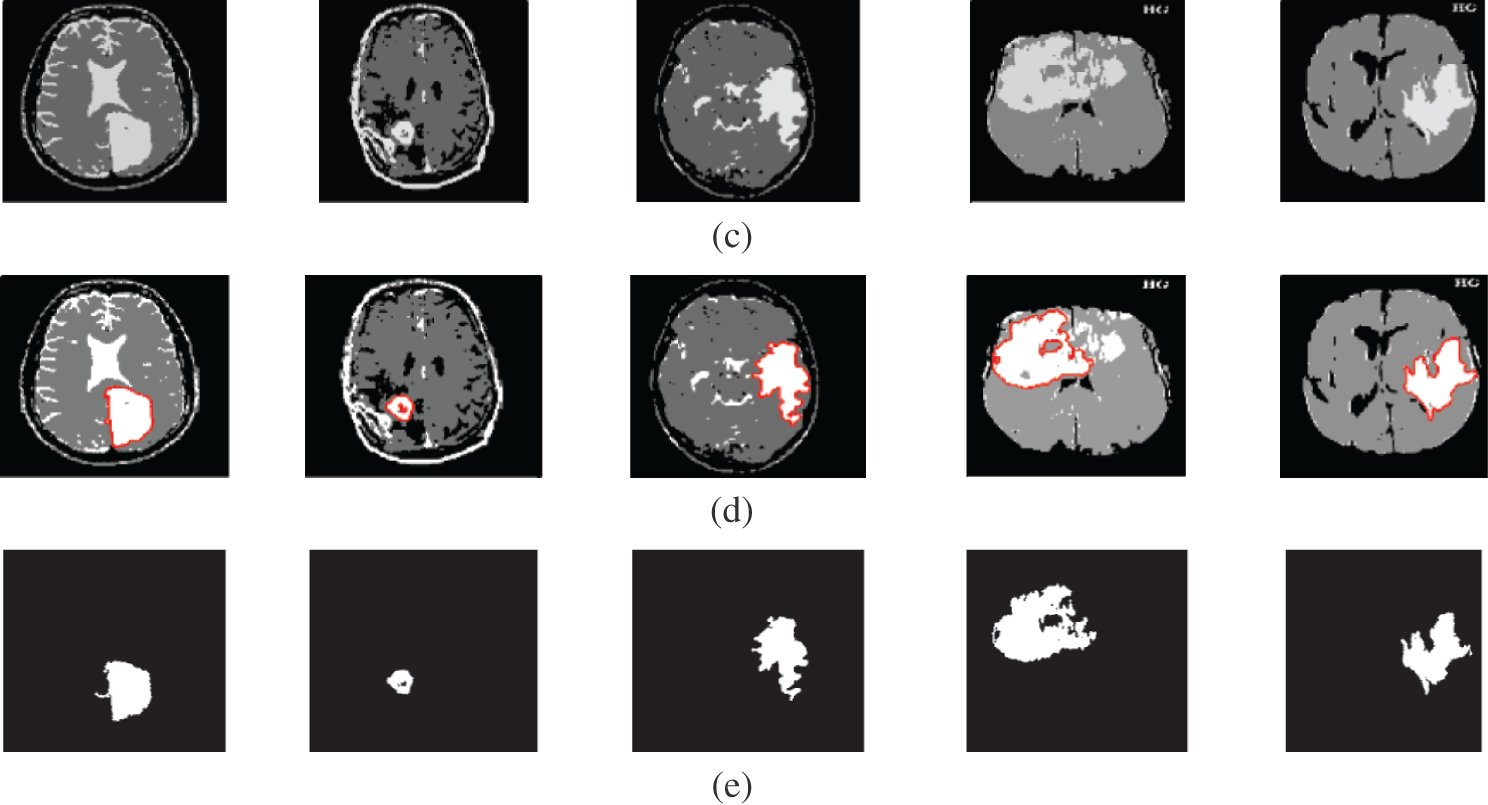
Figure 6: A sample of the MR medical images (a) MR images, (b) MR images after preprocessing stage, (c) FCM and morphological operation output images, (d) Segmented images after active contour, and (e) The extracted tumor images
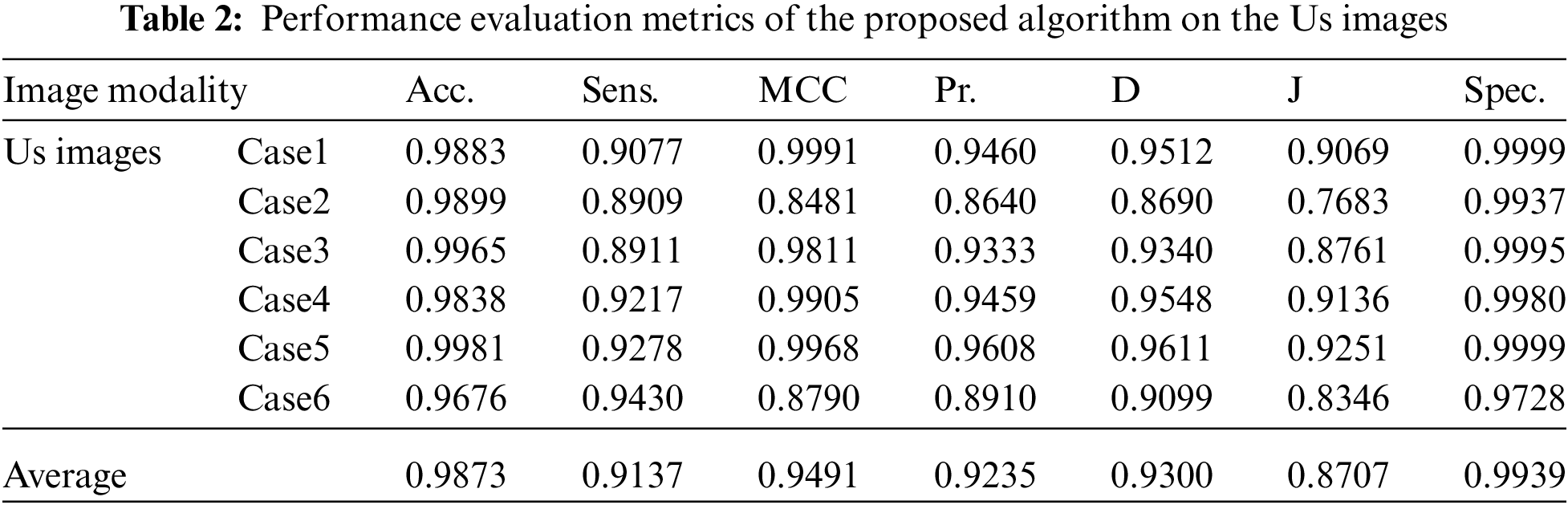
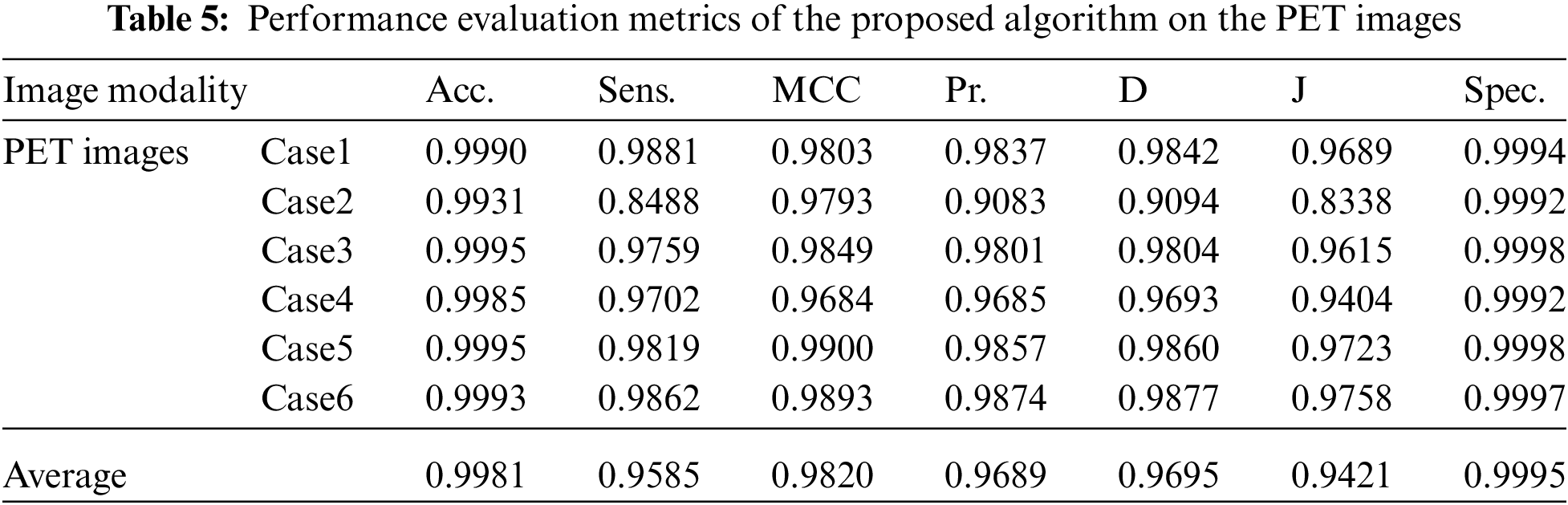
Visual results of the Us breast image, shown in Fig. 2, reveal the importance of each step in the segmentation approach. According to Fig. 2b, the preprocessing step output images, it is apparent that the Us images are sharpened and histogram equalized. This process increases the image contrast and helps to obtain accurate segmentation. The morphological operations and FCM, shown in Fig. 2c, are considered the first stage of segmentation. The active contour model segmentation delineates the tumor region of interest, as shown in Fig. 2d. These results are the second stage of segmentation. The delineated tumors are then extracted, as shown in Fig. 2e. The quantitative results for the Us images are given in Tab. 2. The algorithm is applied to six cases of Us breast images. Finally, the average value of each metric is calculated. These results show that the proposed approach can achieve an average segmentation accuracy of 0.9873.
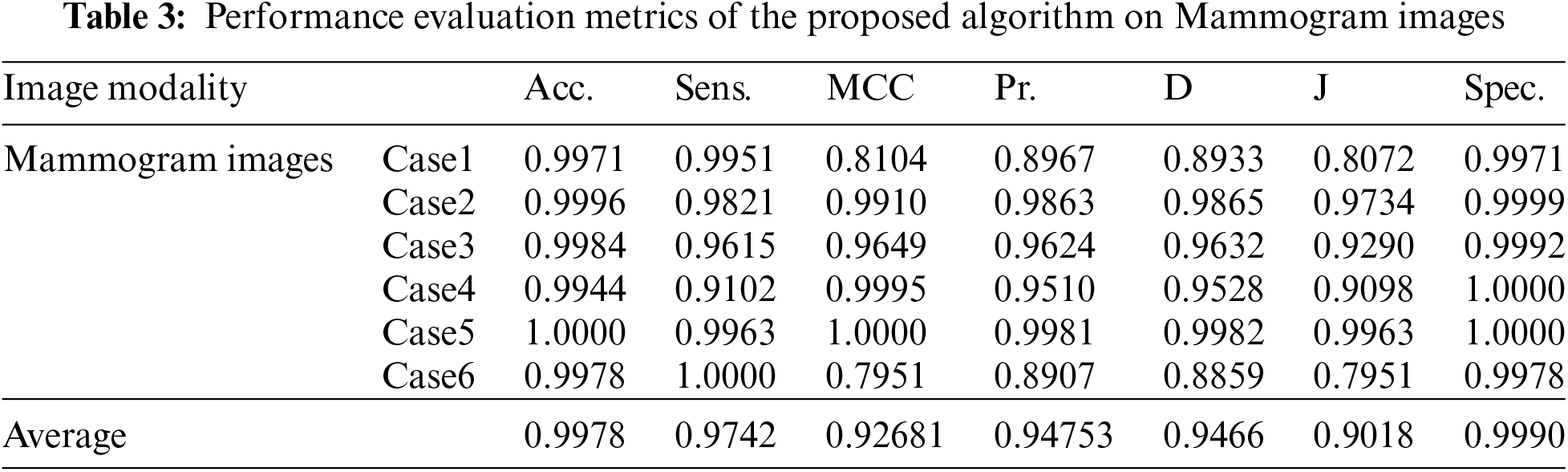
Fig. 3 shows the visual results of the mammogram images. The enhancement step increases the image contrast and improves the image quality, and bright pixels are enhanced significantly. The results of the morphological operations and FCM, shown in Fig. 3c, segments the FCM clusters, and the active contour outlines the tumor region of interest, as shown in Fig. 3d. Finally, the tumors are extracted, as shown in Fig. 3e. The quantitative results for the mammogram images are given in Tab. 3. The algorithm is applied to six cases of mammogram images. The average value of each metric is calculated. The proposed segmentation approach can achieve a segmentation accuracy of 0.9990 on the six cases.
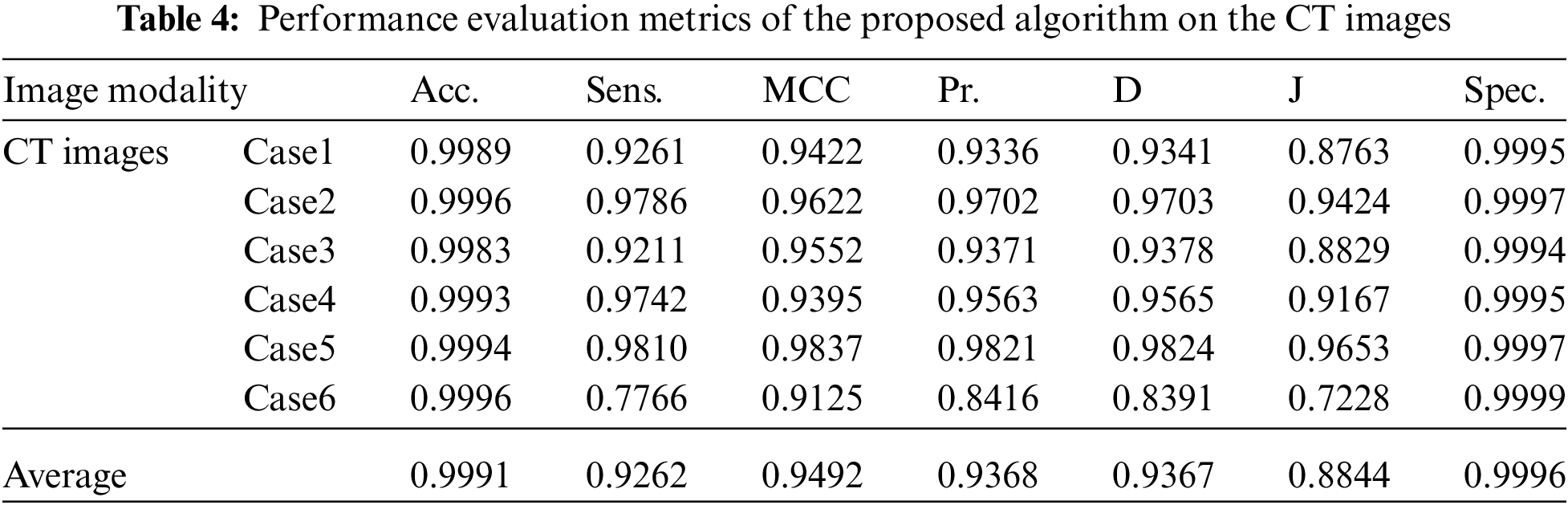
Fig. 4 illustrates the results of the proposed segmentation approach applied to the CT chest images. The enhancement results, shown in Fig. 4b, considerably enhances and improves the image contrast. The results of the morphological operations and FCM, shown in Fig. 4c, divide the images into adjacent regions as shown in Fig. 4d. Finally, the proposed approach extracts the tumors in each image, as shown in Fig. 4e. The segmentation metrics for the CT images are given in Tab. 4, and the average segmentation accuracy reached 0.9991.
The fourth modality is the PET brain images, shown in Fig. 5. The PET image quality is very low. Firstly, the images are converted into grayscale, and then the proposed approach is applied. The preprocessing step, shown in Fig. 5b, does not improve image quality. Therefore, the segmentation process may not give the desired results. Subsequently, the output of step 3 shown in Fig. 5c, hardly results in good results due to the very low quality of the acquired PET original images. As shown in Figs. 5d and 5e, the active contour model segmentation delineates and extracts the tumor region. The quantitative results for the PET images are given in Tab. 5. The proposed approach can achieve an average segmentation accuracy of 0.9981.
Fig. 6 gives the results of the proposed segmentation approach applied to the MRI brain images. As shown in Fig. 6b, the enhancement results considerably enhance and improve the image contrast. The results of the morphological operations and FCM, shown in Fig. 6c, splits the images into sub-regions as shown in Fig. 6d. Finally, the proposed approach extracts the tumors in each image, as shown in Fig. 6e. The segmentation metrics for the MRI brain images are given in Tab. 6, and the obtained average segmentation accuracy is 0.9978. The proposed approach shows excellent performance for delineating and extracting the tumor boundaries with good quantitative and qualitative results.
4 Noise Effect on the Segmentation Process
It is well known that the histogram can describe the distribution characteristics of image data. It is simple to perform segmentation if the histogram has a number of peaks. Speckle noise with 0.05 variance was added to the Us images, and Gaussian noise with 0 mean and 0.02 variance was added to the other image modalities. The original image histogram has one or two obvious peaks on contrary to the noisy image histogram, which has no obvious peaks. Fig. 7 shows a comparison of distribution characteristics of image data for the original image and noisy image.
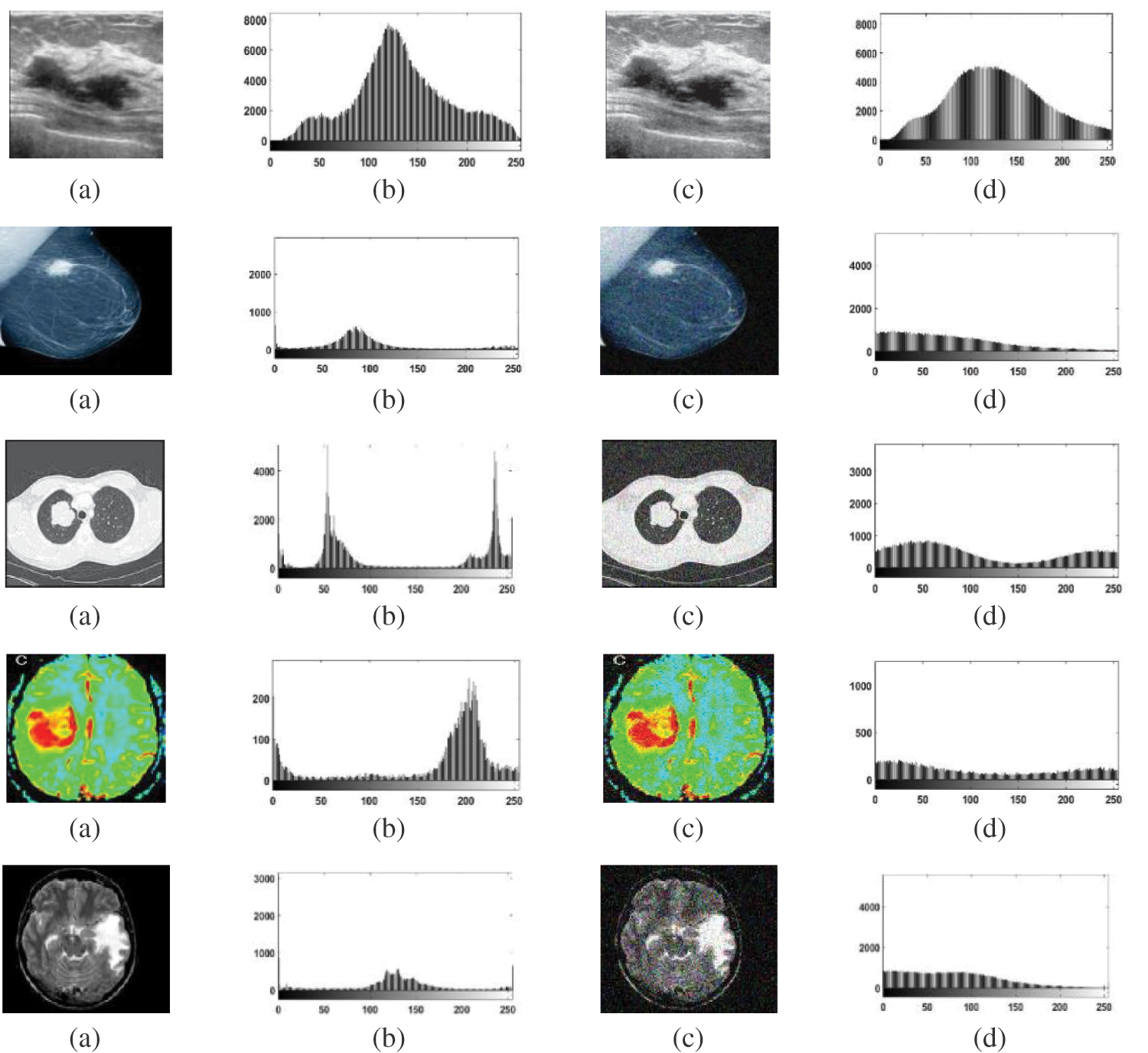
Figure 7: Comparison of distributions of data for original images and noisy images (a) Original images (b) The original images histograms, (c) Noisy images, and (d) Noisy images histograms
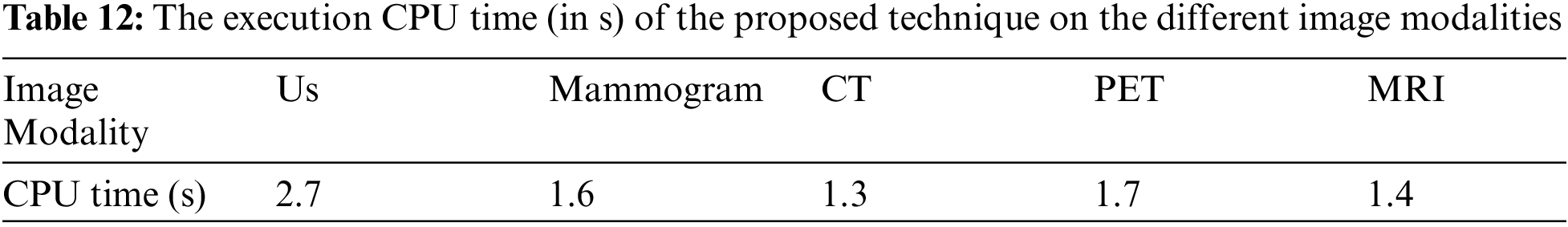
Tabs. 7 to 11 give the performance evaluation metrics of the proposed technique on different image modalities in the presence of noise. Tab. 12 illustrates the execution time (in seconds) of the proposed technique on the different image modalities.
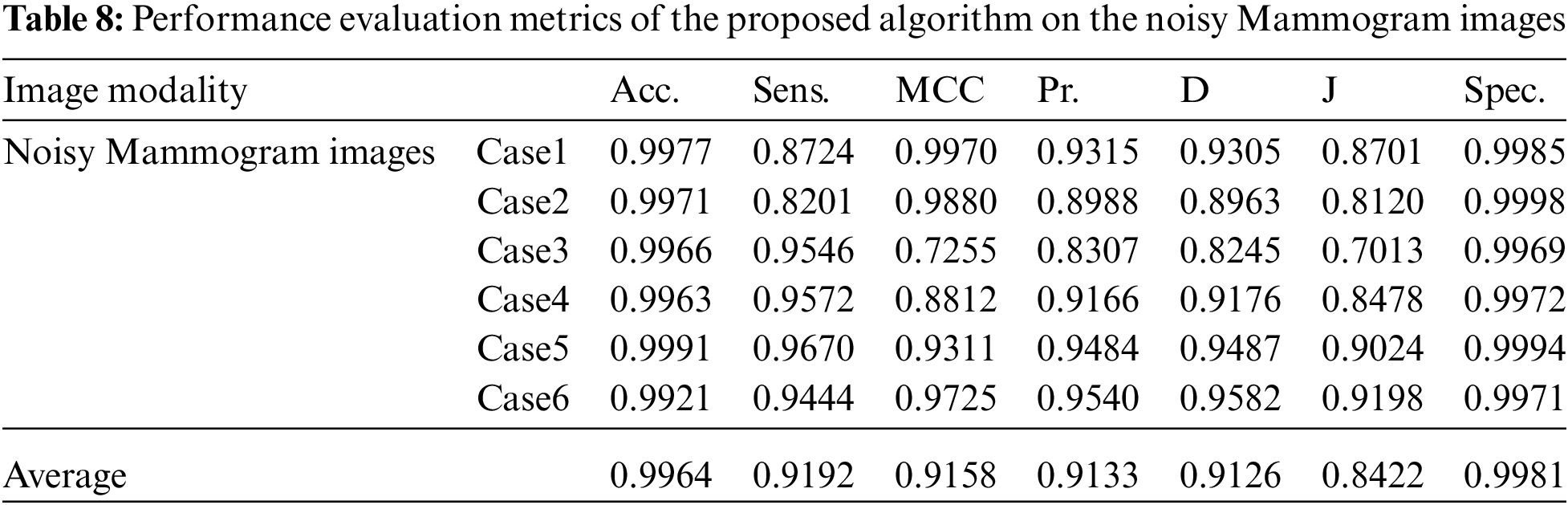
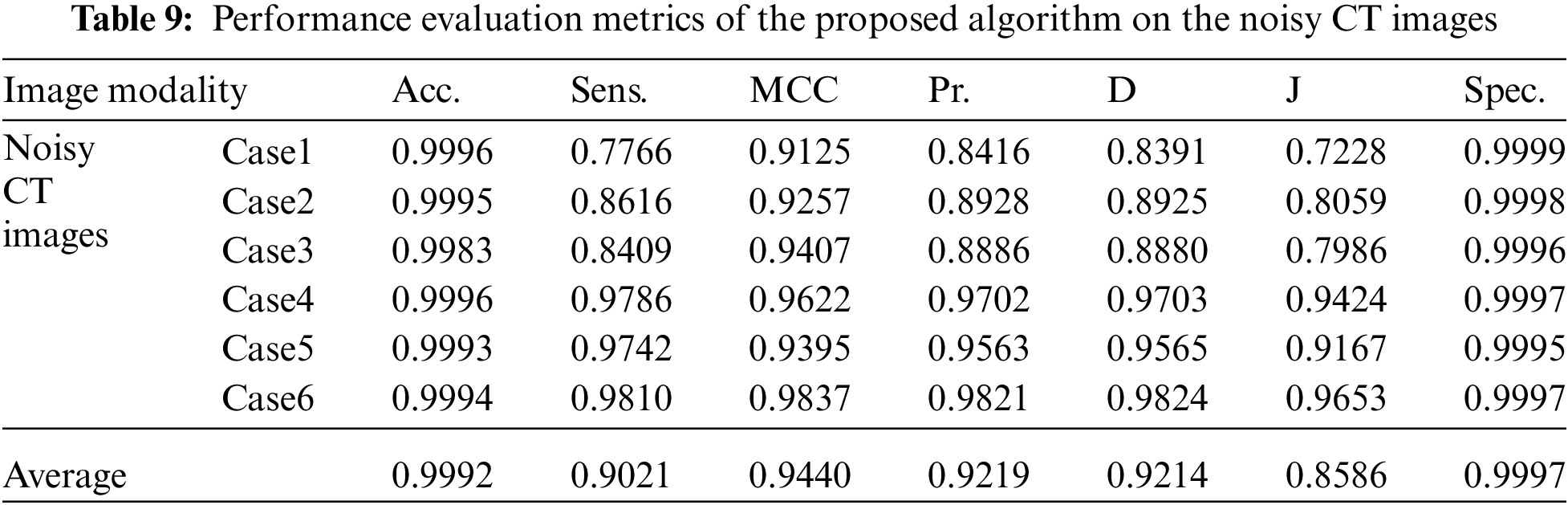
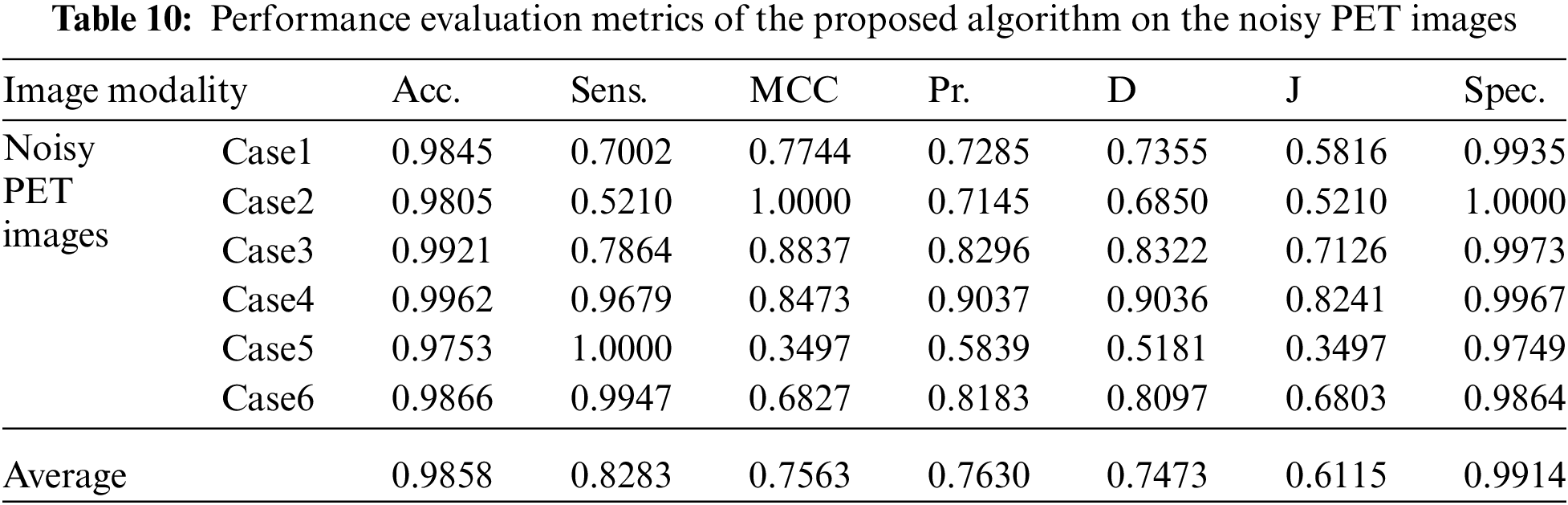
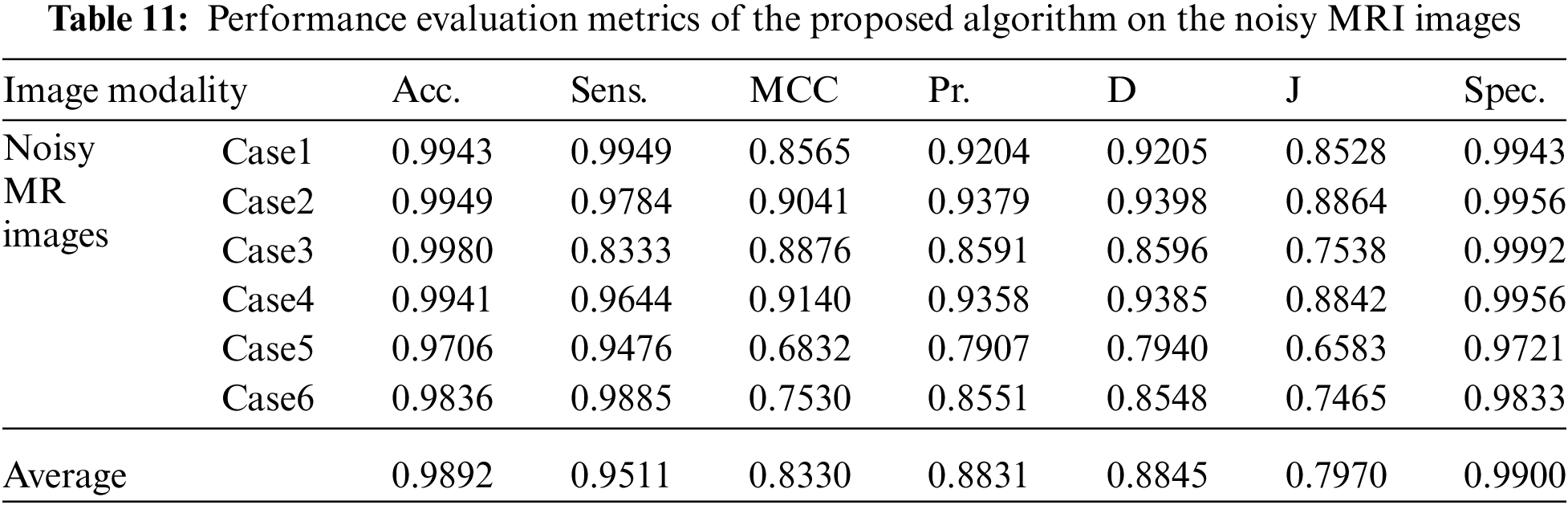
It is shown that the proposed approach can achieve average segmentation accuracies of 0.9813, 0.9964, 0.9992, 0.9858, and 0.9892 for Us, mammogram, CT, PET, and MRI images. Furthermore, results show good accuracy and similarity results for the noise-free and noisy images. Obviously, combining morphological operation with the FCM algorithm can improve the distribution of the image data and facilitate the segmentation process. In addition, morphological operations can maintain the object boundaries and eliminate the noise effect. The proposed technique has low complexity, as shown in Tab. 12. It is easy to obtain good results for image segmentation with the introduction of membership and morphological operations filtering. The illustrated results show that the proposed technique can segment and extract the tumor region successfully even in the presence of different noise types.
This paper introduced an efficient image segmentation framework to reduce the influence of noise and improve the segmentation quality. By introducing morphological operations, the local spatial information of the image is utilized to improve the segmentation effect. Because it is possible to suppress noise while preserving the contour of objects, a trade-off has easily been achieved between noise suppression and detail preservation. Moreover, morphological operations are able to provide good reconstructed results for images corrupted by different types of noise. Furthermore, the FCM membership filtering is employed to exploit the local spatial constraints. The obtained outcomes illustrated that the proposed technique gives good segmentation findings without tuning parameters for different modalities of medical images. The future work will focus on presenting an efficient segmentation stage and a classification stage for tumors in different multi-modality medical images as an effective tool for identifying malignant ultrasonic breast tumors from benign ones.
Acknowledgement: Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R66), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Funding Statement: Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R66), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. T. Lei, X. Jia, Y. Zhang, L. He, H. Meng et al., “Significantly fast and robust fuzzy c-means clustering algorithm based on morphological reconstruction and membership filtering,” IEEE Transactions on Fuzzy Systems, vol. 3, no. 3, pp. 1–15, 2018. [Google Scholar]
2. G. Li, Y. Zhao, L. Zhang, X. Wang, Y. Zhang et al., “Entropy-based global and local weight adaptive image segmentation models,” IEEE Tsinghua Science and Technology, vol. 25, no. 1, pp. 1–12, 2020. [Google Scholar]
3. W. El-Shafai, S. Abd El-Nabi, E. El-Rabaie, A. Ali and F. Soliman, “Efficient deep-learning-based autoencoder denoising approach for medical image diagnosis,” Computers, Materials and Continua, vol. 70, no. 3, pp. 6107–6125, 2022. [Google Scholar]
4. W. El-Shafai, A. Mohamed, E. El-Rabaie, A. Ali and F. Soliman, “Automated COVID-19 detection based on single-image super-resolution and CNN models,” Computers, Materials and Continua, vol. 69, no. 3, pp. 1141–1157, 2021. [Google Scholar]
5. A. Soudani and E. Zagrouba, “Adaptive region based active contour model for image segmentation,” in Proc. of IEEE/ACS 14th Int. Conf. on Computer Systems and Applications (AICCSA), Kuala Lumpur, Malaysia, pp. 210–219, 2017. [Google Scholar]
6. W. Xu, X. Yue, Y. Chen and M. Reformat, “Ensemble of active contour-based image segmentation,” in Proc. IEEE Int. Conf. on Image Processing (ICIP), Beijing, China, pp. 113–118, 2017. [Google Scholar]
7. F. Alqahtani, M. Amoon and W. El-Shafai, “A fractional Fourier based medical image authentication approach,” Computers, Materials and Continua, vol. 70, no. 2, pp. 3133–3150, 2022. [Google Scholar]
8. W. El-Shafai, F. Khallaf, E. El-Rabaie and F. Abd El-Samie, “Robust medical image encryption based on DNA-chaos cryptosystem for secure telemedicine and healthcare applications,” Journal of Ambient Intelligence and Humanized Computing, vol. 1, no. 5, pp. 1–29, 2021. [Google Scholar]
9. W. El-Shafai, A. Algarni, G. El Banby, F. El-Samie and N. Soliman, “Classification framework for COVID-19 diagnosis based on deep CNN models,” Intelligent Automation and Soft Computing, vol. 30, no. 3, pp. 1561–1575, 2022. [Google Scholar]
10. P. Yugander and G. R. Reddy, “Segmentation of noisy images using improved distance regularized level set evolution,” in Proc. IEEE Int. Conf. on Circuit, Power and Computing Technologies (ICCPCT), Kollam, India, pp. 1–5, 2017. [Google Scholar]
11. Z. Zhang and J. Song, “An adaptive fuzzy level set model with local spatial information for medical image segmentation and bias correction,” IEEE Access, vol. 7, pp. 27322–27338, 2019. [Google Scholar]
12. Y. Cao, “A novel hybrid active contour model for intracranial tuberculosis mri segmentation applications,” IEEE Access, vol. 8, pp. 149569–149585, 2020. [Google Scholar]
13. C. Shi-Gang, L. Heng, W. Xing-Li, Z. Yong-Li and H. Lin, “Study on segmentation of lettuce image based on morphological reorganization and watershed algorithm,” in Proc. IEEE Chinese Control and Decision Conf. (CCDC), Shenyang, China, pp. 6595–6597, 2018. [Google Scholar]
14. X. Li, Z. Qu and X. Yang, “Spatially constrained fuzzy c-means clustering algorithm for image segmentation,” Journal of Physics Conference Series, vol. 1237, no. 3, pp. 1–22, 2019. [Google Scholar]
15. X. Zhang, W. Pan, Z. Wu, J. Chen, Y. Mao et al., “Robust image segmentation using fuzzy c-means clustering with spatial information based on total generalized variation,” IEEE Access, vol. 8, pp. 95681–95697, 2020. [Google Scholar]
16. C. Wang, W. Pedrycz, Z. Li and M. Zhou, “Residual-driven fuzzy c-means clustering for image segmentation,” IEEE/CAA Journal of Automatica Sinica, vol. 8, no. 4, pp. 876–889, 2020. [Google Scholar]
17. A. Mahmoud, M. El-Rabaie, E. Taha, A. Elfishawy, O. Zahran et al., “Medical image segmentation techniques, a literature review, and some novel trends,” Menoufia Journal of Electronic Engineering Research, vol. 27, no. 2, pp. 23–58, 2018. [Google Scholar]
18. N. Soliman, S. Abd-Alhalem, W. El-Shafai, S. Abdulrahman and F. Abd El-Samie, “An improved convolutional neural network model for DNA classification,” Computers, Materials and Continua, vol. 70, no. 3, pp. 5907–5927, 2022. [Google Scholar]
19. C. Ding, X. Pan, X. Gao, L. Ning and Z. Wu, “Three adaptive sub-histograms equalization algorithm for maritime image enhancement,” IEEE Access, vol. 8, pp. 147983–147994, 2020. [Google Scholar]
20. L. Bai, W. Zhang, X. Pan and C. Zhao, “Underwater image enhancement based on global and local equalization of histogram and dual-image multi-scale fusion,” IEEE Access, vol. 8, pp. 128973–128990, 2020. [Google Scholar]
21. M. F. Khan, D. Goyal, M. M. Nofal, E. Khan, R. Al-Hmouz et al., “Fuzzy-based histogram partitioning for bi-histogram equalisation of low contrast images,” IEEE Access, vol. 8, pp. 11595–11614, 2020. [Google Scholar]
22. Y. Yao, C. Xia, J. Li and Q. Li, “Head CT image convolution feature segmentation and morphological filtering for densely matching points of IoTs,” IEEE Access, vol. 8, pp. 11617–11633, 2020. [Google Scholar]
23. Y. Li and J. Wang, “Novel binary adaptive morphological operators,” in Proc. 12th IEEE Int. Conf. on Anti-Counterfeiting, Security, and Identification (ASID), Xiamen, China, pp. 132–135, 2018. [Google Scholar]
24. F. Hu, H. Chen and X. Wang, “An intuitionistic kernel-based fuzzy c-means clustering algorithm with local information for power equipment image segmentation,” IEEE Access, vol. 8, pp. 11203–11229, 2020. [Google Scholar]
25. S. Soomro and K. N. Choi, “Robust active contours for mammogram image segmentation,” in Proc. IEEE Int. Conf. on Image Processing (ICIP), Beijing, China, pp. 2149–2153, 2017. [Google Scholar]
26. X. Yang, X. Jiang, L. Zhou, Y. Wang and Y. Zhang, “Active contours driven by local and global region-based information for image segmentation,” IEEE Access, vol. 8, pp. 11220–11240, 2020. [Google Scholar]
27. T. Dietenbeck, M. Alessandrini, D. Friboulet and O. Bernard, “Creaseg: A free software for the evaluation of image segmentation algorithms based on level-set,” in Proc. IEEE Int. Conf. on Image Processing (ICIP), Hong Kong, China, pp. 665–668, 2010. [Google Scholar]
28. W. El-Shafai and F. Abd El-Samie, “Extensive COVID-19 X-ray and CT chest images dataset,” Mendeley Data, v3, http://dx.doi.org/10.17632/8h65ywd2jr.3, 2020. [Google Scholar]
29. A. Algarni, W. El-Shafai, G. El Banby, F. El-Samie and N. Soliman, “An efficient CNN-based hybrid classification and segmentation approach for COVID-19 detection,” Computers, Materials and Continua, vol. 70, no. 2, pp. 4393–4410, 2022. [Google Scholar]
30. W. El-Shafai, A. Mahmoud, E. El-Rabaie, T. Taha and F. El-Samie, “Efficient deep CNN model for COVID-19 classification,” Computers, Materials and Continua, vol. 70, no. 3, pp. 4373–4391, 2022. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |