DOI:10.32604/csse.2022.022305

| Computer Systems Science & Engineering DOI:10.32604/csse.2022.022305 |  |
| Article |
A Robust 3-D Medical Watermarking Based on Wavelet Transform for Data Protection
1Engineering Research Center of Digital Forensics, Ministry of Education, Jiangsu Engineering Center of Network Monitoring, School of Computer and Software, Nanjing University of Information Science & Technology, Nanjing, 210044, China
2Jiangsu Collaborative Innovation Center of Atmospheric Environment and Equipment Technology (CICAEET), Nanjing University of Information Science & Technology, Nanjing, 210044, China
3Wuxi Research Institute, Nanjing University of Information Science & Technology, Wuxi, 214100, China
4IT Fundamentals and Education Technologies Applications, University of Information Technology and Management in Rzeszow, Rzeszow, Voivodeship, 100031, Poland
*Corresponding Author: Xiaorui Zhang. Email: zxr365@126.com
Received: 03 August 2021; Accepted: 13 September 2021
Abstract: In a telemedicine diagnosis system, the emergence of 3D imaging enables doctors to make clearer judgments, and its accuracy also directly affects doctors’ diagnosis of the disease. In order to ensure the safe transmission and storage of medical data, a 3D medical watermarking algorithm based on wavelet transform is proposed in this paper. The proposed algorithm employs the principal component analysis (PCA) transform to reduce the data dimension, which can minimize the error between the extracted components and the original data in the mean square sense. Especially, this algorithm helps to create a bacterial foraging model based on particle swarm optimization (BF-PSO), by which the optimal wavelet coefficient is found for embedding and is used as the absolute feature of watermark embedding, thereby achieving the optimal balance between embedding capacity and imperceptibility. A series of experimental results from MATLAB software based on the standard MRI brain volume dataset demonstrate that the proposed algorithm has strong robustness and make the 3D model have small deformation after embedding the watermark.
Keywords: 3-D medical watermarking; robust watermarking; PCA; BF-PSO
In the current Internet of Medical Things, the emergence of new diagnosis and treatment modes such as telemedicine ensures that doctors can use CT, MRI, ultrasound or other medical equipment to diagnose diseases [1,2]. With the huge increase of 5G technology in communication speed, more and more medical images are being presented to doctors and patients in the form of 3D models. By performing operations such as scaling, rotating, and moving on these models [3,4], doctors can more intuitively find out whether there are lesions in tissues or organs, and further conduct comprehensive pathological inquiry, technical communication, demonstration and surgery simulation [5]. In order to ensure the safe transmission and storage of these medical images, watermark technology [6–9] is applied to medical data to solve the possible malicious tampering or attack in the process of medical data transmission.
As a new information hiding technology, medical watermarking embeds watermark information into digital media to protect the original carrier [10]. Through certain watermarking algorithms, watermark information such as patient's name, gender, age, and symptoms, etc., is embedded into medical data to prevent from being tampered in the process of transmission and storage. Also, this operation can protect the legitimate rights and interests of doctors and patients, and provide an effective way to solve the increasingly critical doctor-patient relationship [11]. The emerging remote consultation system can establish a communication bridge between patients, local medical personnel, and non-local experts, by which severe diseases are further accurately judged. In remote consultation, medical watermarking is an emerging technology that can help medical institutions to securely store or transmit medical information and ensure that the data is not attacked by illegal users [12].
3D watermarking technology is one of the rapid developing technologies in the field of multimedia information security. It as a new technology can protect copyright and authentication sources as well as integrity in an open internet environment. In recent years, it has attracted great attention and become a research hotspot. There are many ways to implement 3D medical models. Li et al. [13] proposed a new mechanism of digital watermarking. By replacing some vertex coordinates of 3D mesh models and embedding watermarked images, high-capacity watermark information can be embedded. Liu et al. [14] proposed a 3D point cloud model watermarking algorithm based on feature point location The watermark is embedded into the vertices with large average curvature by modifying the azimuth of the feature vertices. Hao et al. [15] proposed an adaptive embedding watermarking algorithm for 3D mesh models, which has a high embedding capacity by adaptive threshold to select the vertices to be embedded. Yang et al. [16] adopted the edge embedding technique in integer wavelet transform. It uses the amplitude information and sub-band category of wavelet coefficients to achieve the balance between the capacity and the visual invisibility. Ali et al. [17] used genetic algorithm to select the coefficients in integer wavelet transform and embedded the optimal watermark depth in the selected pixels. As can be seen from the literatures [18–20], some watermark authentication schemes may cause obvious deformation of the original carrier and cannot be effectively restored to the original version. However, in the medical field, any distortion of the original medical data is unacceptable. Therefore, these irreversible watermarking schemes which lead to the distortion of the original carrier are not suitable for the integrity protection of medical data.
To address the above challenge, a 3D medical watermarking algorithm based on wavelet transform is proposed in this paper, where the watermark is embedded by selecting the feature points whose normal vector changes greatly after dimension reduction. These points have a good invariance of rotation and translation, so that the embedded watermark can resist the attacks from translation, rotation, and scaling. At the same time, the multi-level wavelet transform is used to embed the watermark into the intermediate frequency region to maximize the visual invisibility of the original carrier. This study proposes a bacterial foraging algorithm based on particle swarm optimization, which combines the optimal solution with watermark pixels to achieve an optimal balance between embedding capacity and imperceptibility. The extraction process does not need the original 3D model and can realize a reversible embedding on the premise of protecting the integrity and security of medical data, which avoids the limitation of existing 3D watermarking technologies in low robustness and poor visual invisibility.
This paper studies the watermarking algorithm of a 3D medical model based on the wavelet transform. Firstly, the algorithm uses principal component analysis (PCA) to reduce the dimension of the 3D model, where, the data are projected to the low-dimensional subspace to reduce the complexity of data processing. At the same time, the vertices with large changes in the normal direction were selected as the feature points of the 3D model, and the bacterial foraging algorithm based on particle swarm optimization was used to adjust the embedding depth during the watermarking process. This part mainly introduces several corresponding key techniques used in the algorithm and their relationship each other, as shown in Fig. 1. These techniques and their improvements will be introduced in detail below.
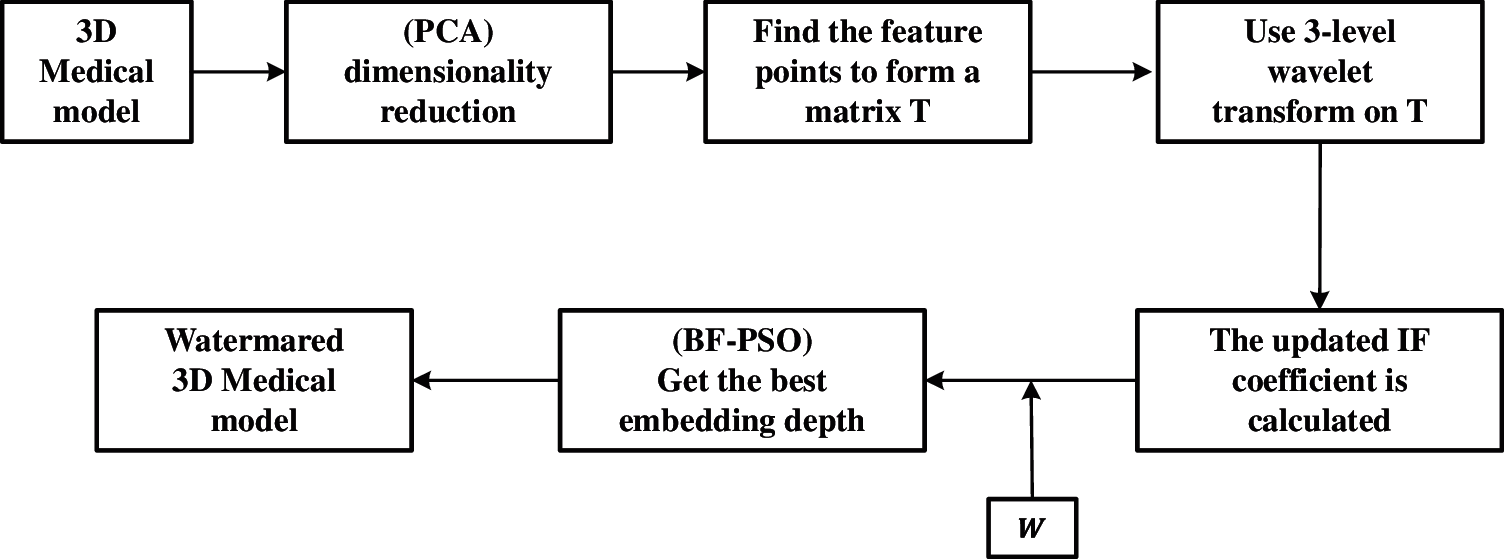
Figure 1: Watermark frame diagram of medical model
2.1 Principal Component Analysis
Dimension reduction is an effective preprocessing method of high-dimensional feature data, which can not only remove noise and unimportant features of high-dimensional data, but also improve data processing speed subsequently. Principal component analysis (PCA) can map high-dimensional data to low-dimensional space through linear projection to realize the dimensionality reduction [21]. The variance of the data is the largest in the projected dimension, which makes it possible to use less data dimensions and retain the characteristics of more original data points. It can reduce the dimensions of the processed data and minimize the loss of information after dimension reduction.
The generation methods of the 3D model include 3D modeling, 3D reconstruction and 3D scanning. Also, the obtained 3D model is relatively arbitrary in size, position, and direction. Therefore, it is necessary to place the 3D model in a standard coordinate system, that is, normalization. It includes three steps: translation normalization, rotation normalization, and scale normalization. The normalization process makes the model with the same shape maintain the same spatial state after translation, rotation or scaling operation.
Suppose the 3D model is composed of three sides in a triangle, let (vi = (xi, yi, zi)T ∈ R3, i = 1, 2, …n) to be a vertex set, T = {t1, t2, …tm} to be a set of triangular faces. The 3D model was processed by PCA. Let vp be the geometric center of the 3D model, and the covariance matrix Cv of the vertices of the 3D model is obtained by Eq. (1) and Eq. (2).
Calculate the corresponding eigenvector e1, e2, e3 based on the formula Cvek = λkek(║ek║ = 1, k = 1, 2, 3), where Cv is a symmetric matrix, and the eigenvectors of the matrix are orthogonal to each other. The spatial coordinate system, namely PCA coordinate system, can be constructed according to the eigenvectors. PCA transformation will not change the position of the 3D center point.
Wavelet transform [22] has a powerful ability in data analysis and processing, which is developed to overcome the shortcomings of the Fourier transform. It has been successfully applied in many fields, such as signal processing, image processing, pattern recognition, and so on. In 1989, according to the theory of multi-resolution analysis, Mallat proposed a fast decomposition and reconstruction algorithm based on an orthogonal wavelet, namely Mallat algorithm [23], where the processed signal is firstly filtered and then the filtered results are down-sampled to realize fast wavelet decomposition. For multi-level wavelets, the decomposition algorithm is implemented in a cascade manner, in which the wavelet transform of each layer is derived from the low-frequency components generated by the previous decomposition. Also, the fast wavelet reconstruction algorithm is the inverse process of the corresponding decomposition algorithm. The basic operation of discrete wavelet transform is given by Eq. (3).
In order to ensure the relationship between inverse wavelet transform (IDWT) and wavelet transform (DWT), the orthogonal condition is indicated by Eq. (4).
The implementation of 2D-DWT is shown in Fig. 2.
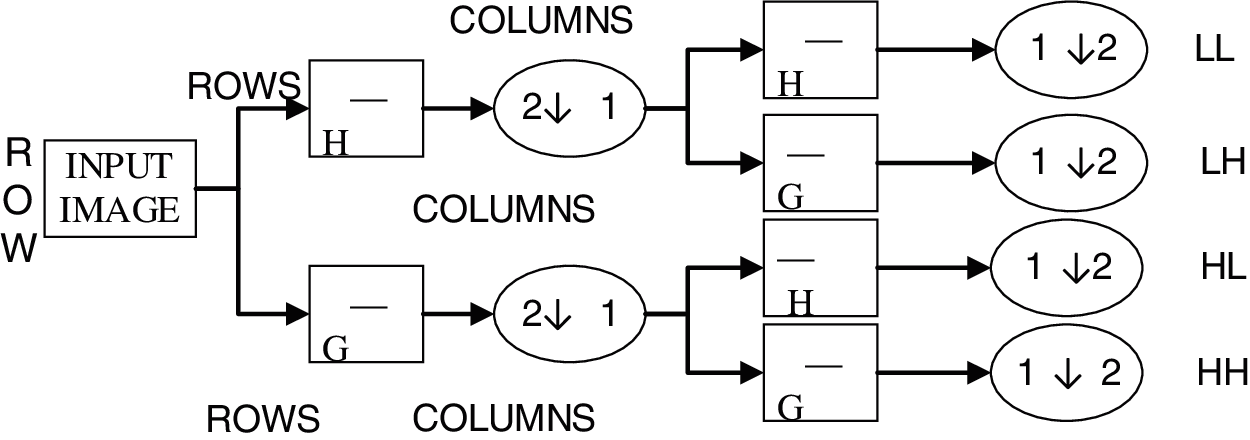
Figure 2: Implementation of 2D-DWT
After wavelet transform, the important features of the image are summarized as follows:
(1) The transformed low-frequency coefficients can maintain the frequency characteristics well, and the high-frequency information has strong spatial selectivity;
(2) Wavelet transform coefficients have the characteristics of horizontal, vertical, and diagonal selectivity, and these characteristics are consistent with the characteristics of the human visual system;
(3) The energy in the image is mainly concentrated in the low-frequency wavelet coefficients of wavelet transform;
(4) The low-pass sub-band of the image has a strong correlation with the corresponding direction. The sub-band coefficient in the horizontal direction has a great correlation with that in the vertical direction, as are the vertical and diagonal sub-band coefficients.
2.3 Bacterial Foraging Optimization(BFO) Based on Particle Swarm Optimization(PSO)
Particle swarm optimization (PSO) [24] draws lessons from the concepts of population learning and self-learning in bird behavior. It, as a global optimization evolutionary algorithm, searches for the global optimum by following the currently searched optimal value. Bacterial foraging optimization (BFO) realizes optimization through competition and cooperation among bacterial populations. It is a search technology based on bacterial populations [25]. The detailed problem-solving process of BFO algorithm is as follows: generate the initial solution group, calculate the value of fitness function, and use the interaction mechanism of the group for iterative optimization. The three main operators of chemotaxis, reproduction, and migration are performed in a loop to obtain the optimal or quasi-optimal solution. Due to the randomness of chemotactic behavior of bacteria, the standard BFO algorithm leads to the slow chemotactic speed and low accuracy of optimization solution. Therefore, in order to improve the traditional BFO algorithm, researchers have proposed various improved schemes, including adjusting the algorithm parameters and integrating other intelligent algorithms. In this study, a bacterial foraging algorithm based on particle swarm optimization (BF-PSO) is adopted, where the BFO algorithm is improved by improving the chemotaxis operator.
The main steps of implementing the BF-PSO algorithm are described as follows.
(1) Parameter initialization. Initialize parameters Ned (number of migration), Nre (number of reproduction), Nch (number of chemotaxis), Ped (basic migration probability), S (number of bacterial scale), Ns (number of swims), ω, C1, C2, α, and β.
(2) Initialize bacterial location. Eq. (5) is used to generate the initialization position and initialize the fitness function value X of bacteria.
(3) Rotate operation. Migration cycle is E = 1: Ned, reproductive cycle is R = 1: Nre, chemotactic cycle is C = 1: Nch. The information of bacteria i is expressed as
(4) Perform bacterial chemotaxis cycle.
(a) Flip operation.
All bacteria are divided into two categories according to the fitness function value. Those superior to the average fitness function value are searched locally by tracking the direction of the optimal bacteria; on the contrary, the global search is carried out by tracking the central position of the bacterial population.
If neither of the above methods is satisfied, update the location of bacteria according to Eq. (6) and Eq. (7).
where C(i) > 0 represents the unit step of forward swimming; Δ(i) ∈ [ − 1, 1] represents any direction vector generated by direction change; φ(j) represents the unit step vector in a random direction selected after rotation.
(b) Swimming operation. If the flipped fitness function value is improved, swim in the flipped direction until the fitness function value is no longer improved or reaches the predetermined maximum number of walks Ns.
P(j, k, g) = {θi(j, k, g)|i = 1, 2, …, S} indicates the location of the bacterial population, J(i, j, k, g) represents the fitness function value of bacteria after the i-th chemotaxis operation, the k-th reproduction operation and the g-th migration operation are performed. The influence value Jcc of signal transmission between population bacteria is calculated according to Eq. (8).
where datt is the depth of attraction, ωatt is the width of attraction, hrep represents the height of repulsion force and ωrep represents the width of repulsion force. After performing a chemotaxis operation, the fitness function value of bacteria is updated according to Eq. (9).
(5) Reproductive cycle. After the chemotactic cycle is completed, the fitness of each bacterium in its life cycle is accumulated to obtain bacterial energy, and then the fitness is sorted according to the obtained bacterial energy. During reproductive cycle, half of the bacteria with poor energy acquisition ability are eliminated, and half of the bacteria with the strong energy acquisition ability are self-replicated to generate new same bacteria as the original bacteria. The new bacteria have the same location as the original bacteria, that is, they have the same foraging ability.
(6) Migration cycle. After the propagation operation is completed, a random probability is generated and compared with the basic migration probability Ped. If the generated probability is less than Ped, the individual dies. Initialize in the solution space according to Eq. (5) to generate a new bacterial individual, which will have different positions and different foraging abilities of the extinct individual. The new bacterial individual may be closer to the global optimal solution, which is conducive to jumping out of the local optimal solution and finding the global optimal solution.
(7) Judge the ending condition of the cycle. If the conditions of the above three loops are met, it will end and output the result.
3D medical model data such as CT and MRI, are different from 2D images. When they are used for disease analysis and diagnosis, the amount of data is large and the analysis is difficult [26]. During network transmission, in order ensure real-time diagnosis, fast transmission speed, as well as secure and robust watermarking, we propose a robust watermarking method in the wavelet domain. The dimension of the 3D model is reduced to lower the complexity of the data processing.
In the process of watermark embedding and extraction, the quality of the formed and extracted watermark image largely depends on the watermark embedding coefficient. The low value of the embedding coefficient will reduce the robustness of the watermark, while the higher value will minimize the invisibility of the watermark. Therefore, it is necessary to select an optimal value for embedding coefficient to balance the invisibility and robustness of the watermark.
The vertex coordinate
The vertices with large changes in the extracted normal vector are taken as the feature points of the medical model. These points have a good rotation and translation invariance, so that the embedded watermark can resist geometric attacks such as translation, rotation, and scaling. Find out each vertex connected with vi to form a set N(vi), and vp is the geometric center of the medical model.
Determine the discrete normal vector of vertex vi:
Estimate the area where the normal direction changes sharply:
The sequence D(vi) is obtained according to the feature point extraction method, and the first 8S×8S results of the sequence are arranged in descending order. The corresponding vertices are taken as feature points, and then the ri values corresponding to the feature points are composed into a matrix T of 8S×8S, where S×S is the size of the watermark image.
A series of multi-resolution sub-bands {HLi, LHi, HHi}, i = 1, 2, 3 and approximate sub-bands LL3 are obtained by three-level wavelet transform of matrix T. Then, HL3 is divided into non-overlapping 2 × 2 sub blocks and recorded as Bk, k = 1, 2, …; The watermark bits are embedded into non-overlapping block Bk by wavelet transform. For each sub block,
This study proposes a bacterial foraging algorithm based on particle swarm optimization (BF-PSO) to obtain the best watermark embedding depth. Inspired by the historical optimal idea of particle position in PSO algorithm, the chemotaxis operator in BF algorithm is improved. When the chemotactic operator is executed, the bacteria whose fitness values are better than the average value are searched by tracking the optimal bacteria; on the contrary, searched by tracking the central position of the bacterial population. If the above two methods fail, the random search method of standard BFO algorithm is adopted. By evaluating the bacterial energy after reproduction and the fitness of new bacterial individuals after the migration cycle, the local optimal solution and global optimal solution are obtained. The embedding process of the watermark is shown in Fig. 3, and the extraction process is the inverse process of watermark embedding.
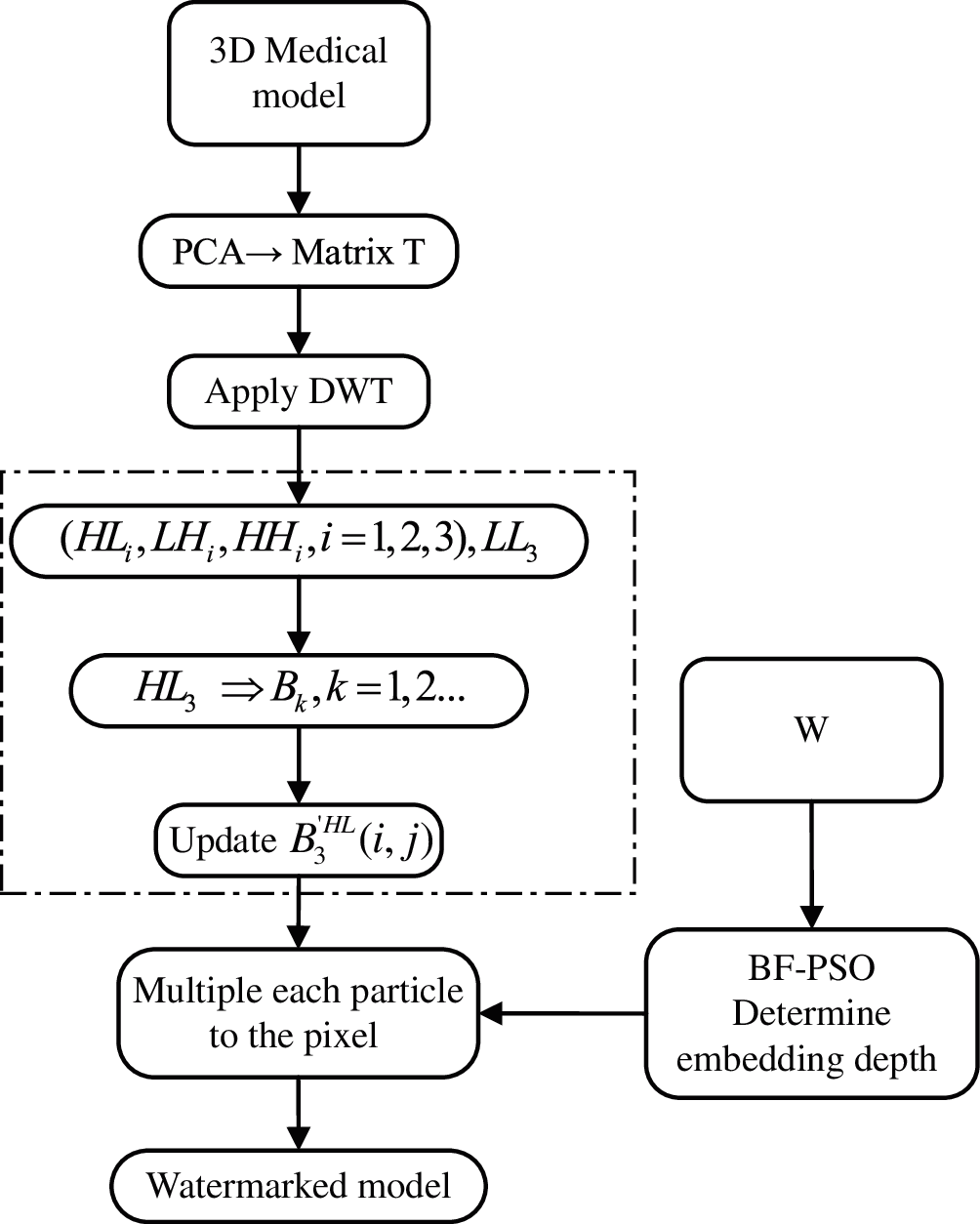
Figure 3: Watermark embedding flowchart
Step1. Due to the characteristics of 3D model data, it is necessary to obtain the reduced dimensional 3D model after preprocessing, and select ri in the vertex coordinate si in 3D model as the input of wavelet transform;
Step2. Process ri to form the 8S × 8S matrix T, and the wavelet coefficients are obtained by three-level wavelet transform, in which the obtained HL3 is divided into 2 × 2 non-overlapping sub-blocks and recorded as Bk, k = 1, 2, …
Step3. Embed the watermark according to watermark embedding formula coef′ = coef + a*w, where coef′ is the wavelet coefficient of ri in spherical coordinates, a is the watermark embedding coefficient. Note that a is also the scaling factor and used to adjust the embedding depth;
Step4. The optimal watermark embedding depth is obtained by using the bacterial foraging algorithm based on particle swarm optimization (BF-PSO);
Step5. coef′ is transformed by inverse wavelet transform, and the 3D model containing watermark can be obtained.
The process of the watermark extraction algorithm is the inverse process of the corresponding watermark embedding algorithm. The specific steps of the algorithm are as follows.
Step1. The model dimension reduction operation proposed in this algorithm is performed for the 3D model, and the ri value under the spherical coordinates of the vertices of the 3D model is calculated;
Step2. The ri value is processed and transformed into a matrix, and the wavelet coefficients are obtained by wavelet transform;
Step3. Restore watermark image;
Step4. Evaluate the performance of the watermarking algorithm.
This section will evaluate the performance of the proposed algorithm. Simulation experiments use various attacks on the 3D model embedded with watermark information to verify the robustness and invisibility of the watermark algorithm in the paper. The experimental results show that the 3D model can still successfully extract the watermark information after being attacked, which shows the high robustness and visual invisibility of our algorithm. The watermark system is evaluated by Peak Signal-to-Noise Ratio (PSNR):
The original 3D model data [27] used for the test is the standard MRI brain data from MATLAB R2019a software. Fig. 4 shows the standard MRI brain slice image, and Fig. 5 shows the MRI brain model and watermark image.
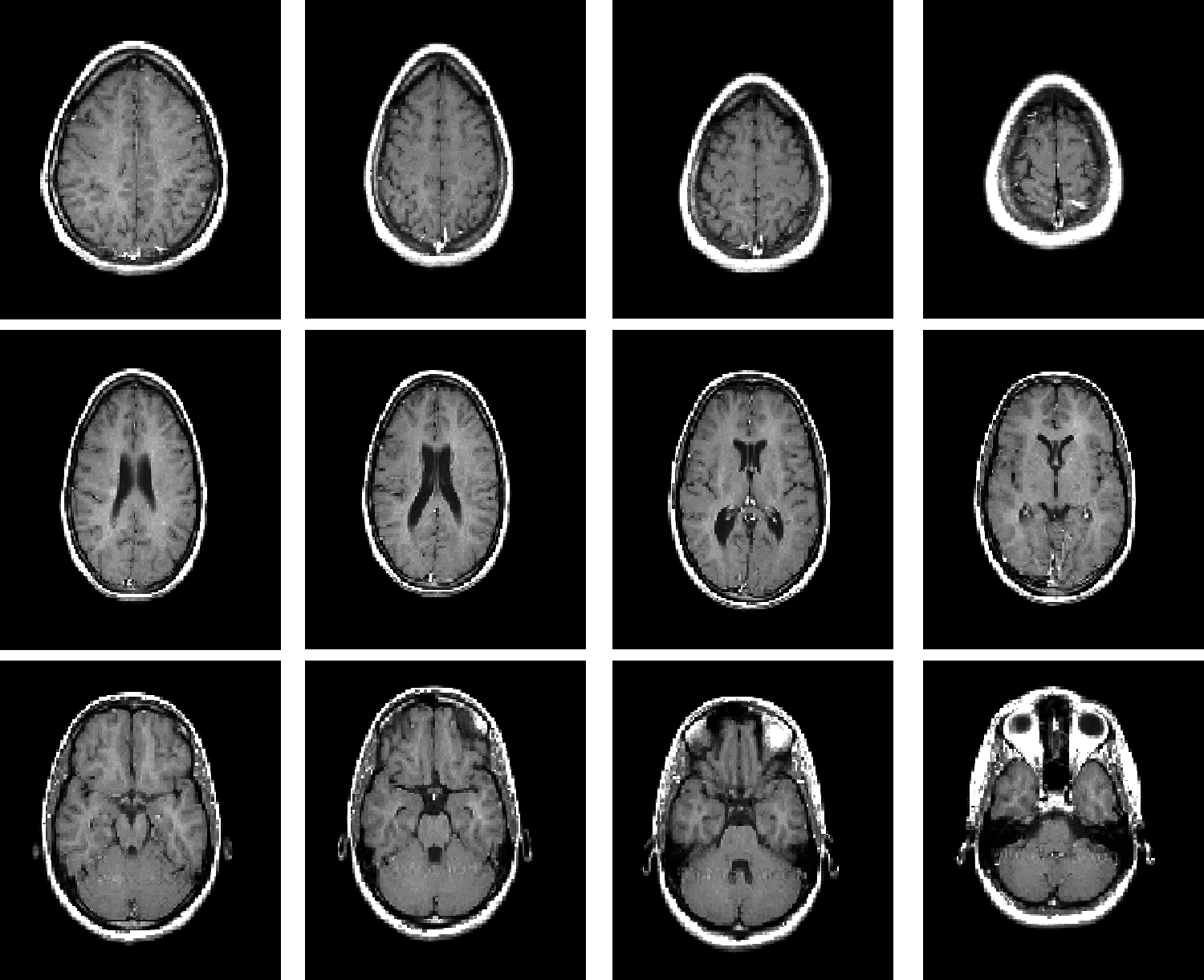
Figure 4: Standard MRI brain slices
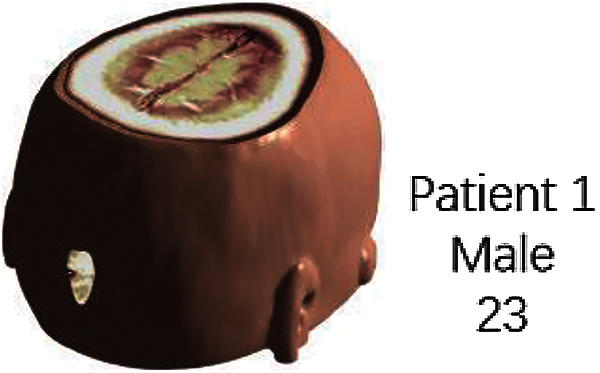
Figure 5: MRI brain model and watermark image
Gaussian noise attacks with different intensity are applied to medical data. Fig. 6 shows the medical model and brain slice image under the interference of three kinds of different Gaussian noise. It can be seen that the greater the intensity of Gaussian noise, the worse the quality of the slice image after being attacked. Tab. 1 shows the calculated PSNR and Normalized Correlation (NC) values under three kinds of Gaussian noise attacks with different intensities. It can be seen from the data that although the model is attacked, the quality of the extracted watermark image is still good, indicating that our algorithm has strong robustness to Gaussian noise.
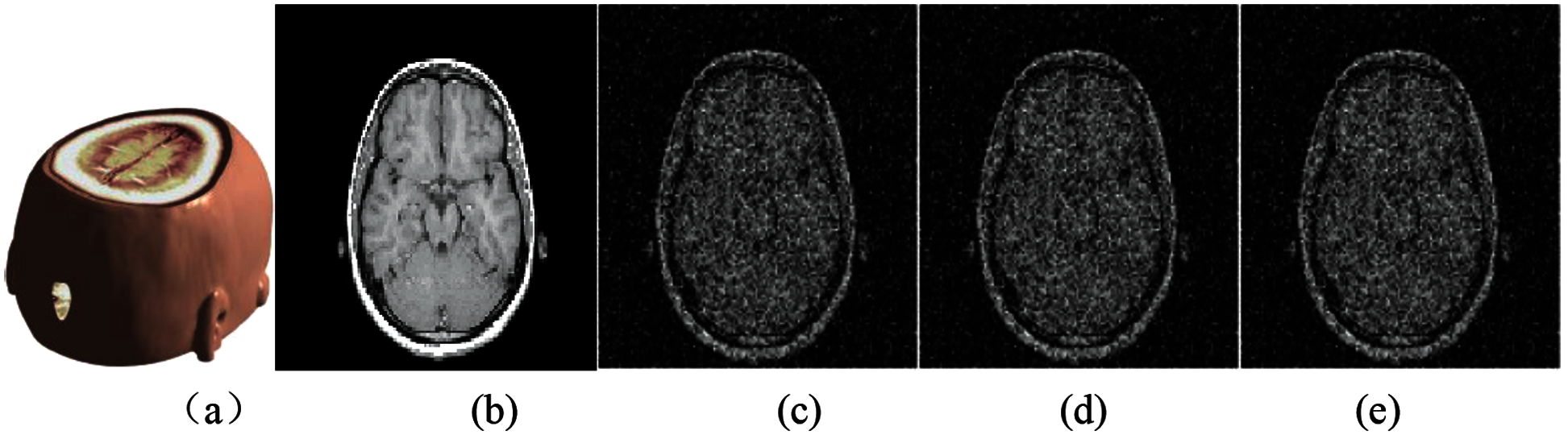
Figure 6: The slice images of the watermarked brain data under Gaussian noise attacks with different intensities: (a) Watermarked brain volume data; (b) Slice of MRI brain data; (c) Slice attacked by Gaussian noise with 0.01variance; (d) Slice attacked by Gaussian noise with 0.15 variance; (e) Slice attacked by Gaussian noise with 0.40 variance
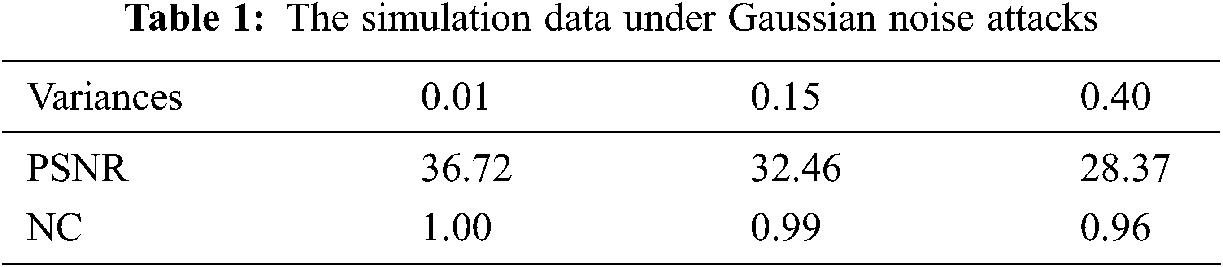
Fig. 7 shows the medical model and brain slice images when attacked by the JEPG compression under different intensities. It can be seen that the greater the compression intensity, the more obvious the blocking effect. Tab. 2 shows the calculated PSNR and NC values under three JEPG compression attacks with different intensities. Seen from the data, the calculated NC value is still close to 1.0, indicating that the invisibility of the watermark is good.
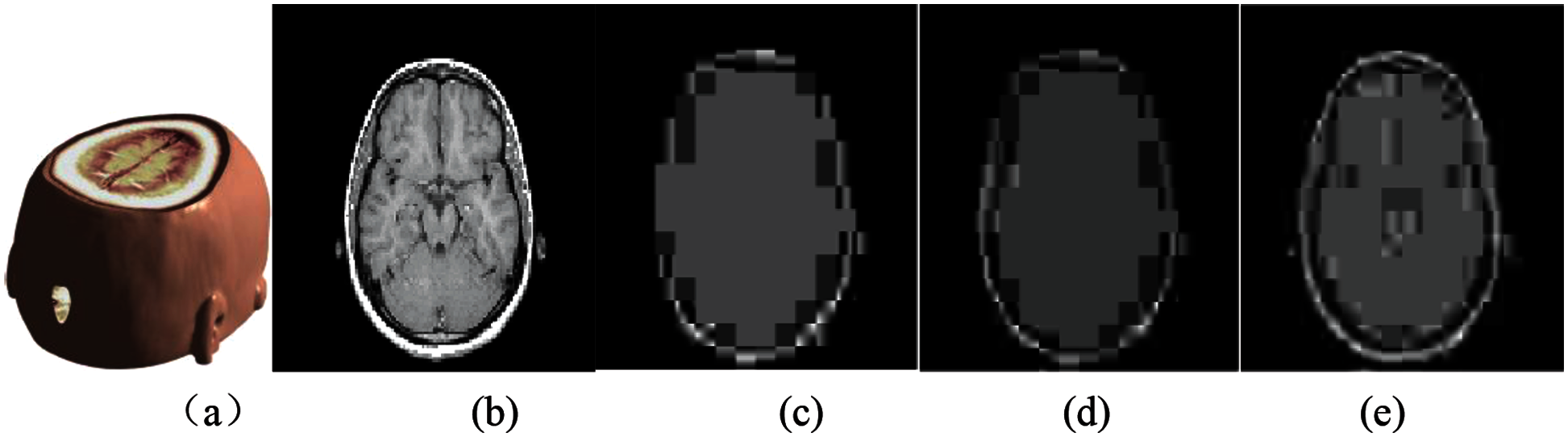
Figure 7: The slice images of the watermarked brain volume data under JPEG compression attacks with different intensities: (a) Watermarked brain volume data; (b) Slice of volume data; (c) Slice attacked by JEPG compression with 0.01 variance; (d) Slice attacked by JEPG compression with 0.03 variance; (e) Slice attacked by JEPG compression with 0.06 variance
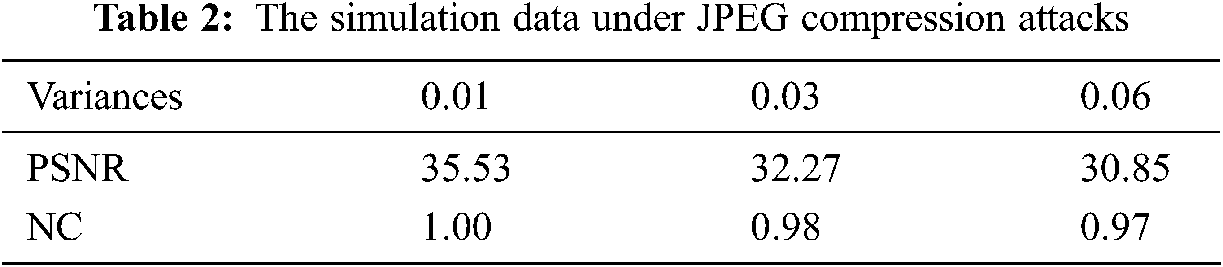
Fig. 8 shows the comparison of NC values between proposed algorithm and other algorithms. The comparison results in show that the NC value of the proposed algorithm is slightly higher than that proposed by Liu et al. [28] and loan et al. [29]. The algorithm proposed by loan et al. embedded the watermark by selecting and setting the difference and range of the intermediate frequency coefficient in advance, which effectively managed the intermediate frequency coefficient of the image. The algorithm proposed by Liu et al. generated a watermark for the region of interest and the region of non-interest separately, a recovery function of the region of interest is designed to ensure that the watermark can be extracted effectively. It is worth mentioning that although the proposed algorithm has no obvious change in scaling attack, compared with other algorithms, it remains the optimal value of 1.0. In addition, under a low scaling factor, the smaller the size of the image, the more obvious its robustness. Especially when the scale factor is as low as 0.1, the watermark can still be extracted completely, so it is obviously better than other algorithms in Fig. 8a. Seen from JEPG compression and clockwise rotation attacks, the overall performance of the proposed algorithm is better than other existing algorithms.
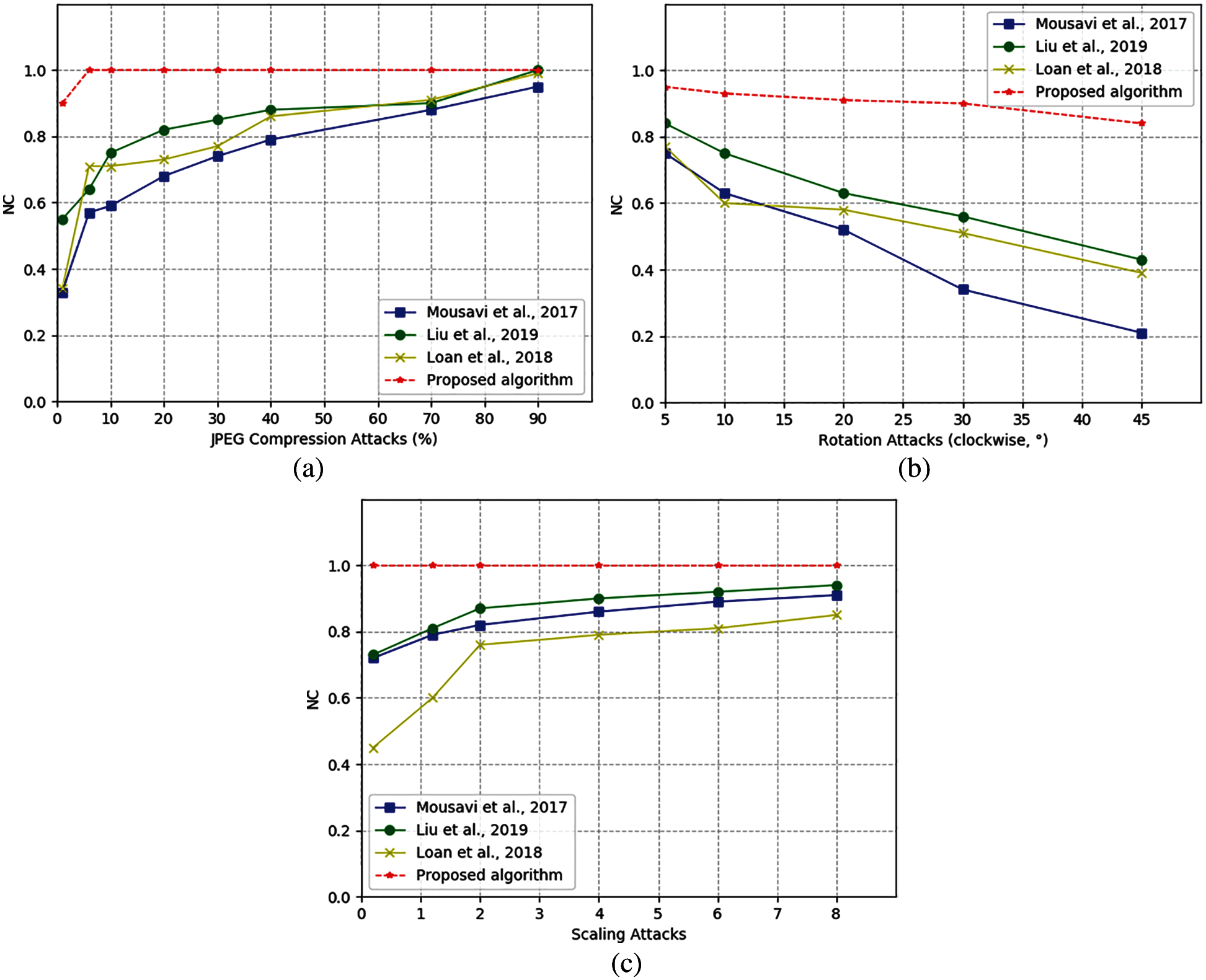
Figure 8: Algorithm comparison chart under different attacks: (a) by JPEG compression attacks; (b) By Clockwise rotation attacks; (c) By Scaling attacks
This paper proposes a 3D medical watermark based on wavelet domain, which embeds the watermark information into the vertices of the significant region in the 3D model, so as to ensure the invisibility of the 3D model. The bacterial foraging algorithm model based on particle swarm optimization (BF-PSO) uses the correlation between adjacent pixels in the image to achieve the optimal balance between embedding capacity and imperceptibility and further find the optimal wavelet coefficients for embedding. In telemedicine, the receiver needs undistorted medical data for diagnosis, and the watermark cannot destroy the readability of the original data. Our scheme realizes the invisibility of the watermark, guarantees the undistorted original data, and does not need any additional information in the extraction process. Simulation results show that our method has good imperceptibility and strong robustness, which can resist geometric attacks such as rotation, scaling and clipping. In future work, we will continue to study the watermark embedding capacity of the model, and find the best balance between capacity and robustness.
Funding Statement: This work was supported, in part, by the Natural Science Foundation of Jiangsu Province under grant numbers BK20201136, BK20191401; in part, by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) fund.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. K. L. Bagot, N. Moloczij and K. Barclay-Moss, “Sustainable implementation of innovative, technology-based health care practices: A qualitative case study from stroke telemedicine,” Journal of Telemedicine and Telecare, vol. 26, no. 1, pp. 79–91, 2020. [Google Scholar]
2. J. W. Kim, J. Friedman and S. Clark, “Implementation of a pediatric emergency telemedicine program,” Pediatric Emergency Care, vol. 36, no. 2, pp. e104–e107, 2020. [Google Scholar]
3. M. A. Lago, C. K. Abbey and M. P. Eckstein, “Foveated model observers for visual search in 3D medical images,” IEEE Transactions on Medical Imaging, vol. 40, no. 3, pp. 1021–1031, 2020. [Google Scholar]
4. B. Aubert, C. Vazquez and T. Cresson, “Toward automated 3D spine reconstruction from biplanar radiographs using CNN for statistical spine model fitting,” IEEE Transactions on Medical Imaging, vol. 38, no. 12, pp. 2796–2806, 2019. [Google Scholar]
5. J. H. Cho and J. A. Fessler, “Regularization designs for uniform spatial resolution and noise properties in statistical image reconstruction for 3-d x-ray CT,” IEEE Transactions on Medical Imaging, vol. 34, no. 2, pp. 678–89, 2015. [Google Scholar]
6. B. Wang, W. Kong, W. Li and N. N. Xiong, “A dual-chaining watermark scheme for data integrity protection in internet of things,” Computers, Materials & Continua, vol. 58, no. 3, pp. 679–695, 2019. [Google Scholar]
7. B. Wang and P. Zhao, “An adaptive image watermarking method combining SVD and wang-landau sampling in DWT domain,” Mathematics, vol. 8, pp. 691, 2020. [Google Scholar]
8. R. Chen, L. Pan, Y. Zhou and Q. H. Lei, “Image retrieval based on deep feature extraction and reduction with improved CNN and PCA,” Journal of Information Hiding and Privacy Protection, vol. 2, no. 2, pp. 9–18, 2020. [Google Scholar]
9. J. Liu, J. Li, Y. Chen, X. Zou, J. Cheng et al., “A robust zero-watermarking based on sift-dct for medical images in the encrypted domain,” Computers, Materials & Continua, vol. 61, no. 1, pp. 363–378, 2019. [Google Scholar]
10. L. L. Cui and Y. B. Xu, “Research on copyright protection method of material genome engineering data based on zero-watermarking,” Journal on Big Data, vol. 2, no. 2, pp. 53–62, 2020. [Google Scholar]
11. A. Anand, A. K. Singh and Z. Lv, “Compression-then-encryption based secure watermarking technique for smart healthcare system,” IEEE Multimedia, vol. 27, no. 4, pp. 133–143, 2020. [Google Scholar]
12. X. R. Zhang, W. F. Zhang and W. Sun, “A robust watermarking scheme based on ROI and IWT for remote consultation of COVID-19,” Computers, Materials & Continua, vol. 64, no. 3, pp. 1435–1452, 2020. [Google Scholar]
13. L. Li, H. Li, W. Yuan, J. Lu and X. Feng, “A watermarking mechanism with high capacity for 3D mesh objects using integer planning,” IEEE Multimedia, vol. 25, no. 3, pp. 49–64, 2018. [Google Scholar]
14. J. Liu, Y. Yang, D. Ma, Y. Wang and Z. Pan, “A watermarking method for 3D models based on feature vertex localization,” IEEE Access, vol. 6, pp. 56122–56134, 2018. [Google Scholar]
15. L. Hao, B. Yan, J. Pan, N. Chen and M. A. Acquah, “Adaptive unified data embedding and scrambling for 3D mesh models,” IEEE Access, vol. 7, pp. 162366–162386, 2019. [Google Scholar]
16. C. S. Chan, “An image authentication method by applying Ham-ming code on rearranged bits,” Pattern Recognition Letters, vol. 32, no. 14, pp. 1679–1690, 2011. [Google Scholar]
17. G. Xuan, “Y. Q. Shi,” in C. Yang, Y. Zheng and D. Zou, “Lossless data hiding using integer wavelet transform and threshold embedding technique,” in Proc. ICME, Amsterdam, Netherlands, pp. 1520–1523, 2005. [Google Scholar]
18. H. M. Al-Otum, “Semi-fragile watermarking for gray scale image authentication and tamper detection based on an adjusted expanded-bit multi-scale quantization-based technique,” Image Represent, vol. 25, no. 5, pp. 1064–1081, 2014. [Google Scholar]
19. T. Bianchi and A. Piva, “Secure watermarking for multimedia content protection: A review of its benefits and open issues,” IEEE Signal Processing Magazine, vol. 30, no. 2, pp. 87–96, 2013. [Google Scholar]
20. T. Rahim, S. Khan and M. Arslan, “Exploiting de-noising convolutional neural networks DnCNNs for an efficient watermarking scheme: A case for information retrieval,” IETE Technical Review, vol. 38, no. 2, pp. 245–255, 2020. [Google Scholar]
21. C. Lu, J. Feng and Y. Chen, “Tensor robust principal component analysis with a new tensor nuclear norm,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 42, no. 4, pp. 925–938, 2020. [Google Scholar]
22. V. A. Pimpalkhute, R. Pagea and A. Kotharib, “Digital image noise estimation using DWT coefficients,” IEEE Transactions on Image Processing, vol. 30, pp. 1962–1972, 2021. [Google Scholar]
23. H. Bi, L. Xu and X. Cao, “Polarimetric SAR image semantic segmentation with 3D discrete wavelet transform and markov random field,” IEEE Transactions on Image Processing, vol. 29, pp. 6601–6614, 2020. [Google Scholar]
24. P. Ghamisi and J. A. Benediktsson, “Feature selection based on hybridization of genetic algorithm and particle swarm optimization,” IEEE Geoscience & Remote Sensing Letters, vol. 12, no. 2, pp. 309–313, 2014. [Google Scholar]
25. Z. Yalan, “Research and application on bacteria foraging optimization algorithm,” Computer Engineering & Applications, vol. 46, no. 20, pp. 16–21, 2010. [Google Scholar]
26. W. Zhang, J. Liu and J. Yao, “Mesenteric vasculature-guided small bowel segmentation on 3-d CT,” IEEE Transactions on Medical Imaging, vol. 32, no. 11, pp. 2006–2021, 2013. [Google Scholar]
27. X. R. Zhang, H. L. Wu, W. Sun and K. J. Sunil, “A fast and accurate vascular tissue simulation model based on point primitive method,” Intelligent Automation & Soft Computing, vol. 27, no. 3, pp. 873–889, 2021. [Google Scholar]
28. X. Liu, J. Lou and H. Fang, “A novel robust reversible watermarking scheme for protecting authenticity and integrity of medical images,” IEEE Access, vol. 7, pp. 76580–76598, 2019. [Google Scholar]
29. N. A. Loan, N. N. Hurrah and S. A. Parah, “Secure and robust digital image watermarking using coefficient differencing and chaotic encryption,” IEEE Access, vol. 6, pp. 19876–19897, 2018. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |