DOI:10.32604/csse.2023.024295

| Computer Systems Science & Engineering DOI:10.32604/csse.2023.024295 |  |
| Article |
Brain Tumor Segmentation through Level Based Learning Model
1Department of Electronics and Communication Engineering, Adhi College of Engineering and Technology, 631 605, India
2Department of Electronics and Communication Engineering, Infant Jesus College of Engineering, Vallanadu, Thoothukudi, 628 851, India
*Corresponding Author: K. Dinesh Babu. Email: dinesh.comm@gmail.com
Received: 12 October 2021; Accepted: 17 January 2022
Abstract: Brain tumors are potentially fatal presence of cancer cells over a human brain, and they need to be segmented for accurate and reliable planning of diagnosis. Segmentation process must be carried out in different regions based on which the stages of cancer can be accurately derived. Glioma patients exhibit a different level of challenge in terms of cancer or tumors detection as the Magnetic Resonance Imaging (MRI) images possess varying sizes, shapes, positions, and modalities. The scanner used for sensing the location of tumors cells will be subjected to additional protocols and measures for accuracy, in turn, increasing the time and affecting the performance of the entire model. In this view, Convolutional Neural Networks deliver suitable models for efficient segmentation and thus delivered promising results. The previous strategies and models failed to adhere to diversity of sizes and shapes, proving to be a well-established solution for detecting tumors of bigger size. Tumors tend to be smaller in size and shape during their premature stages and they can easily evade the algorithms of Convolutional Neural Network (CNN). This proposal intends to furnish a detailed model for sensing early stages of cancer and hence perform segmentation irrespective of the current size and shape of tumors. The size of networks and layers will lead to a significant weightage when multiple kernel sizes are involved, especially in multi-resolution environments. On the other hand, the proposed model is designed with a novel approach including a dilated convolution and level-based learning strategy. When the convolution process is dilated, the process of feature extraction deals with multiscale objective and level-based learning eliminates the shortcoming of previous models, thereby enhancing the quality of smaller tumors cells and shapes. The level-based learning approach also encapsulates the feature reconstruction processes which highlights the sensing of small-scale tumors growth. Inclusively, segmenting the images is performed with better accuracy and hence detection becomes better when compared to that of hierarchical approaches.
Keywords: Glioma detection; segmentation; smaller tumour; growth; machine learning; feature analysis
There are various forms of cancers which originate from the brain, and the most common form of cancer is known as glioma. This type of cancer is prominently found in glial cells and based on their intensity, texture, and mass, they are categorized into to low-level and high-level glioma. With the latter variant of glioma, a patient is deemed to alive for a span of 15 months’ maximum. The former variant, before proceeding to higher level glioma, can be divided into benign for malignant tumour. Eventually, they will become higher level glioma leading to fatality. The commonly known diagnosis and treatment includes radiotherapy, chemotherapy and obviously surgery [1]. The purpose of any diagnosis plan is to mitigate the effects and delay the chances of fatality. One primary way of doing it is to detect the presence of tumour cells before they become malignant. Segmentation is a process where medical professionals and expert doctors will cut out the tumour affected cells, tissues, and neighbouring tissues for proper examination. This study will ultimately lead them to plan and deliver a proper treatment, understand the progression of the disease, and hence recommend further therapies for saving the lives of patients [2]. Segmentation is performed after a thorough analysis of the brain tissues from different imaging devices and mechanism. The prominent ways of imaging a brain for sensing the tumour cells are positron emission tomography, computed tomography, and Magnetic resonance imaging [3]. The commonly preferred methodology is to perform an MRI owing to high contrast, clarity, and resolution through non-invasive methods.
Medical experts, in the traditional methodology, prefer to investigate the three-dimensional images for proper diagnosis. The 3D images are preferred since they exhibit better visibility towards the tumours’ size, shape, and mass. It is not as easy as it sounds but a time-consuming process and requires great expertise to sense the stages of tumours accordingly. These difficulties demanded the processes of automatic segmentation [4], apart from reducing the workload and human error from the equation, the chances of accuracy have been greatly increased. Better prediction delivered better treatment plants and hence becomes a better and safer option for doctors to opt computational methodologies [5]. There are other common forms of cancers, such as meningioma, carcinoma etc. which are much easier to be detected and localized for segmentation. The discussed variants, glioma and glioblastoma increases the challenges due to its smaller sizes, shapes, and masses. In terms of delineation, gliomas are harder to be detected since the level of contrast and colour are too subtle and they easily hide behind the natural features of the brain [6]. There are chances for any computational model to neglect a glioma tumour, considering that to be a normal and healthy brain tissue. Since there is no standard localization found for brain tumours, they can be distributed anywhere across different brain tissues. Different sizes, shapes, masses, scale, and ranges of different voxel-based parameters of brain areas captured on an MRI can easily hide the tumour cells. Adding to the difficulties, the brain imaging devices differ according to different activation protocols [7] and hence it varies to different medical institutions.
The brain tumour will be continuously monitored with different images devices, assuming that the patients are affected and there are multiple models involved. According to the investigative model of MRI, the brain structure is visualized into T1 weighted, T2 weighted, post contract T1 weighted and Flair [8]. These are the different modalities which are better than one another in terms of segmentation and hence complement each other. Process of manually segmenting the image is time consuming and challenging even for professionals, making the processes of automated models mandatory. Tumours must be segmented as the first and foremost process before planning for any diagnosis. Proper interpretations after proper segmentations will yield a better and effective treatment plan. Machine learning models have been introduced for segmentation owing to the benefits that they are more reliable and accurate even in different scenarios [9]. There is also a challenge that machine learning algorithms, and their predictions cannot be relied upon completely, medical experts and professionals will be cross checking the predictions. Extensively, machine learning has been substituted by deep learning models, convolutional neural networks for betterments with respect to accuracy and reliability. Comparatively, deep learning established an improvised version of machine learning to optimize the results and provide meaningful insights over the analysis. One of the standard deep learning models will be using the U Net and Fully Conventional networks for better results. In terms of accuracy, U Net have yielded accuracy as well as with optimal performance. The model was based out on a U symmetrical feature, where the model was separated into two windows, in a window on the left–processes of encoding was taken care of and the right window responsible for decoding [10]. The process was also extended beyond the encoder and decoder, where a feature map would be generated to distinguish low level and high-level features extracted from the input images.
The information retrieved from different levels of brain segments or slices are used for further integration of different layers to understand the tumours much better. Magnetic resonance images are used in a three dimensional and a two-dimensional model of segmentation. When three dimensional segmentations are preferred [11], the levels are provided with suitable labels and the model has undergone sufficient training to enhance the accuracy of predictions. The technical difficulty involved in three-dimensional segmentation process is that the amount of network parameters required is huge and there is a lot of memory requirements associated [12]. This problem was addressed by building a multi part convolutional neural network with two dimensional slices of the brain extracted from a Magnetic Resonance Imaging. Just like any other model of machine learning, the classes of input were filled with numerous imbalances. The CNN model [13] also included two training steps to process the classes of inputs and hence remove the imbalance issues. This model was extended into a fully convolutional neural network model, by considering the boundary of brain and supporting tissues based on which the segmentation was carried out. A model named the Deep Medic [14] was introduced to extract the features from different scales, levels and finally integrate them in a two-part architecture either locally or globally.
Because a machine learning model or a deep learning model should not involve huge dependency over time and space complexity [15], it is advised that the special features should be generated with minimal intervention. Likewise, location sensitive information is potentially important for a detailed analysis and hence can be used for solving various problems related to brain segmentation [16]. The architectures which we have discussed so far, are subjected to these limitations, and hence can affect the quality of predictions or the time taken for predictions [17]. In this context, the proposed model addresses the limitations along with analysing and defining the impact of early-stage cancer.
1. Cancer cells or tumours of any size can be detected with the proposed model, and this is specialized with localizing small sized distributions of tumour cells.
2. Networks which were defined as heavyweight are reduced to process at simpler levels analysis dependency over resources and time.
3. A model of segmentation is proposed with dilated convolution and different levels of learning to overcome the deficiencies of hierarchical analysis.
Image processing has been extensively used in several domains for analysing the images and delivering promising outcomes. Especially in the medical domain, images captured from various medical equipment are subjected to analysis for helping medical experts to conclude a treatment plan. Radiologists rely on different medical imaging techniques, analysis methods to detect, plan and deliver a diagnostic approach [18]. Computer aided diagnosis are the final outcomes of medical analysis and hence delivered to the medical institutions for further utilization. The time consumed for analysis and to arrive at a treatment plan can considerably be reduced with automated techniques. Machine learning has been a potential candidate of computer aided diagnosis techniques, specifically when it comes to tumour detection, nodule detection, classification, segmentation, isolation, image retrieval and annotation processes [19]. Before the introduction of machine learning techniques, these processes were carried out through a naked eye and with sufficient experience of medical professionals. These techniques were further extended into deep learning models for enhanced features detection and engineering [20]. Sensitive information from these images is the features required for analysis, which are the outcomes of a neural network.
Concentrating on the segmentation of medical images, in the detection of brain tumour, organs capturing and segmentation, ventricles and vessels segmentation, deep learning models have been proven to deliver better outcomes than ordinary machine learning algorithms. With better accuracy, the chances of survival and the treatment plan to work are higher [21]. The features are extracted based on sizes, shapes, textures, volumes, localization, and presence of any abnormal features in the targeted area. Otsu thresholding approach [22] acts is the foundational method for all segmentation processes in the medical domain. The captured image cannot directly be processed in a machine learning algorithm and are subjected to various pre-processing stages. Presence of noise, weak edges and other parameters affect the accuracy of the classification model. Considering all these factors, a model must acknowledge all these deficiencies before applying the automatic segmentation techniques [23]. In a technique proposed in recent times, fuzzy clustering was implemented to highlight constraint-based along with edge detection algorithms distributed across a spatial image for segmentation. The approach was tested on synthetic images and later applied with real images to ensure the level of accuracy [24]. The conventional methods of brain tumour segmentation involved several parameters such as edges, regions of interest, atlas, clustering features and classification features and inevitably thresholds. The level of human intervention was also higher, despite the fact of demanding huge computational and time resources. Yet again, these models had to include multiple modalities for enhanced performance and accuracy. In this regard, various methods and approaches have been introduced. The Atlas [25] obtained for a brain tumour image was compared against healthy patients’ brain image for identifying the difference and hence classifying the grade of tumour. It was later identified that even healthy part of brain may seem deformed and cannot be considered as a standard threshold for the said approach. Moreover, tumour affected cells resulted in in deformed healthy tissues in the neighbourhood regions and the algorithm classified benign into malignant tumour. The incorrect registration resulted in in demand for better models that monitors tumour growth, compares, and classifies the tumours accordingly.
Multi Atlas models work proposed with a suitable searching algorithm to detect the presence of a signature patch of deformations [26]. The same models were extended to consider active and passive contour approaches along with symmetrical factors, alignment practice found in the left and right brain. The brain cannot be aligned into a proper structure especially with a large tumour in the image. This raised the need of voxel-based morphometry parameters upon which support vector machines and random forests were implemented for delivering better accuracy. The information about neighbouring voxel, faraway voxel including the symmetry of the brain assisted in classifying magnetic resonance images. Recent debates convolutional neural networks are modelled to process magnetic resonance images and registered promising results. The available models can be categorised into one of the three classes where the first class is based out on Deep learning approaches for analysing two dimensional images. The second class concentrates on three-dimensional images and features, eliminating the analysis of two-dimensional image inputs, thereby resulting in higher computational time and cost [27]. The final class included both three dimensional and two-dimensional images and were dependent on volumetric parameters. U Net architectures were the other kinds of approaches which concentrated over and encoder and decoder-based architectures. A fully functional and a full resolution-based mask was available in the U Net architectures which emphasized on convolutional layers and max pooling layers [28]. The method of dilated convolution was recommended in several techniques for reducing the computational cost incurred in the previous methods. Owing to a fact that three-dimensional segmentation model demanded heavy processes over training data inform of labelled or sliced information. This heaviness increased the preference over two dimensional models since they were optimal and required lesser resources for functioning [29]. The other models included a patch-based approach for determining the features and distinguish between normal and tumour affected cells or tissues. Any patch-based model would include pre-processing, modelling, classification of the models and finally the post processing stages. Interim processes like modelling and classification resulted in higher computation time, which eliminated the process of patch-based models [30].
The tumours, their structures, and substructures are found to be stratified. The whole tumour may not deliver the accuracy during segmentation since the enhanced tumour characteristics are found within the tumour Core and can be only determined after the segmentation process. The efficiency of Brain Tumour segmentation processes was supposed to be improved and hence some authors presented a model of cascading approach [31]. Various layers were included in CNN networks to isolate the tumour core from the mass of tumour cells to get a better perspective of the brain tumour. Combining different models together and training them as one also promised better outcomes when compared to that of a model working alone. End to end networks such as modified U Net architectures depended on two-dimensional image, avoiding the three-dimensional special features which were significant measures of accuracy. Parcellation of brains’ tissues [32] and cells was introduced along with patch-based neural networks for enhancing the performance of U Net architectures. These architectures for able to reverse the process of individual layers and can be altered for backpropagation.
When search architectures were combined with multi-level learning strategies of deep learning, the accuracy of brain tumour segmentation has significantly increased. The performance of these networks was effective when the processes of encoding and decoding models were defined for better reconstruction of input images. Once the models have learnt from the training data sets the process of classification has been introduced as an auxiliary approach. After the reconstruction process is done, the segmentation process is carried out despite aligning to heavy computational cost and huge memory requirements [33]. The feature resolutions can be down sampled for reducing the computation cost but is necessary for detecting objects of smaller sizes [34]. A pixel to pixel-based methodology is implemented for enhancing the accuracy of segmentation and this demanded three-dimensional image as an input. The presented architecture addresses all these deficiencies and hence proposes a model of detection and classification of small size tumour cells [35]. Multistage learning increases the chances of detection of minor features and hence can be helpful for classification processes.
This section will be covering the fundamentals of the proposed method starting from the data sets to the algorithms which have been applied the proposed model for enhancing the accuracy of predictions and segmentation.
This model includes the standard data bases which are available open source namely Brain Tumor Segmentation Challenge (BraTS) 2017 and 2018 respectively. BraTS 2017 data set comprises of nearly 285 patients affected by glioma, where are these images are classified into High-Grade Gliomas (HGG) and Lower Grade Glioma (LGG) categories accordingly. There are a few patients whose condition cannot be determined, and segmentation cannot be performed. From the available information, the ground truth is computed for comparison with affected patients. The ground truth is a signature element which are derived by experts and doctors. The ground truth will later be used for annotations and the model can only be generated upon the server of BraTS. The labels of annotation are described as follows, increasing tumour, necrosis, edema or healthy tissue. All the processes are applicable to both the data sets and the number of inputs vary in 2018 data set accordingly. The following Fig. 1 illustrates the ground truth and other information of the obtained datasets.
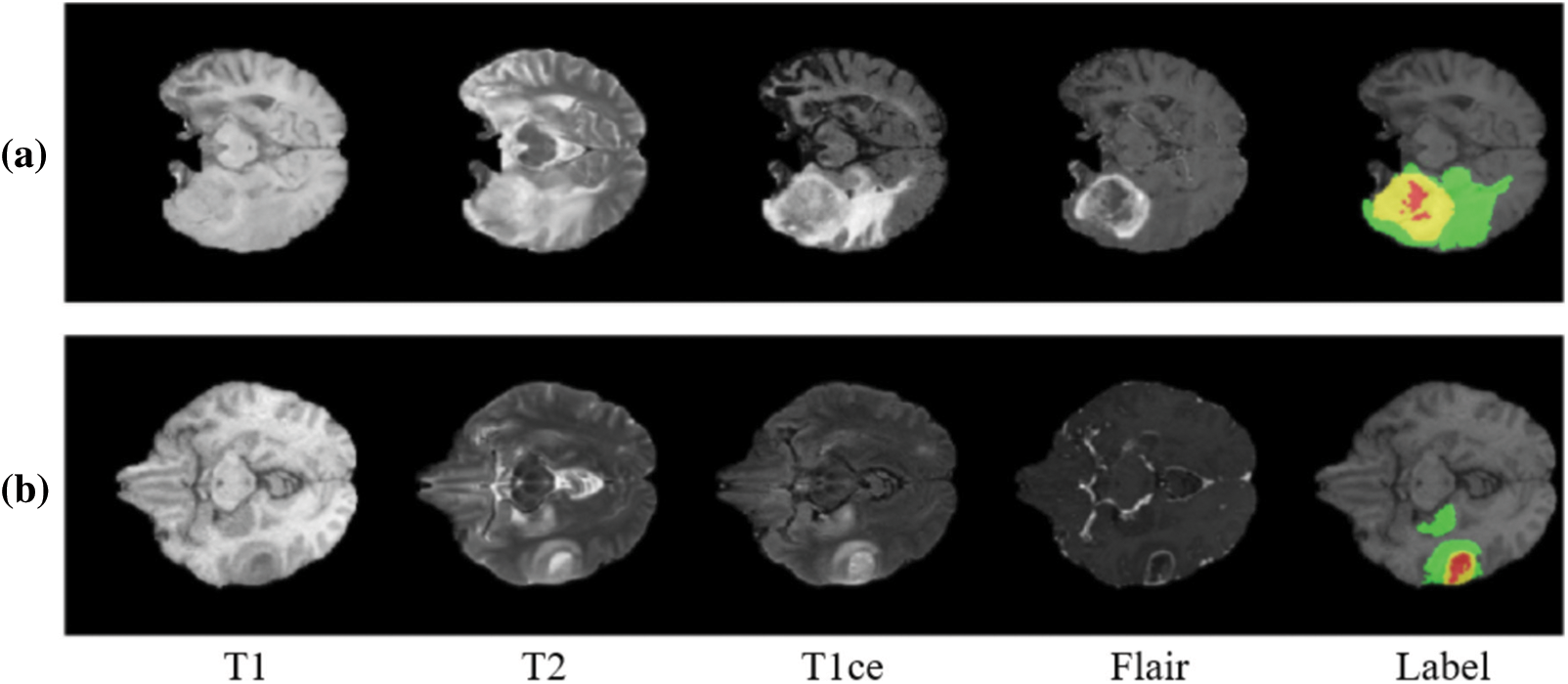
Figure 1: Stages of brain tumour according to Brain Tumour Segmentation Challenge (BraTS) datasets
It is understood that the images obtained from any open-source data set cannot be directly processed and a deep learning model is sensitive to noise and other disturbances present on the images. To enhance the quality of prediction and segmentation it is advised to that; all images are subjected to pre-processing using a standard algorithm. In this case, these images are applied with N4ITK algorithm for bias rectification and converting all the images into a standard template. From the survey of literature, it is presumed that N4ITK algorithm is an independent and reliable technique for pre-processing the brain images. The output images retrieved from the algorithm are normalised to unit variance adhering to 0 mean.
In the proposed deep learning model, there is no significant difference between the layers available in the architecture such as Shallow or Deep. These layers are carefully connected through a bridge of information to enhance the features available globally and locally. The following Fig. 2 illustrates the architecture of the proposed model, and it works best with images of the resolution 128 × 128. The output images will be obtained in the same dimensions. This model is also assisted by a multistage technique for learning and enhancing the features during the reconstruction process after down sampling is applied. The overall architecture is divided into two segments, one for encoding and the other for decoding along with some layers for padding. The architecture of the encoder is further divided into two convolutional layers and a dropout layer blocks for assuring a max pooling process. The output of every layer is processed to buy the next layer using a Conv2D Transpose a methodology before it is concatenated into one output image in the Decoder block. The final block present in the decoder adds another convolutional layer for filtering tumours of the least size available. Contracted images are enhanced by the Decoder blocks for better analysis and segmentation of small sized tumours. The following equations represent the activation and normalisation function respectively.
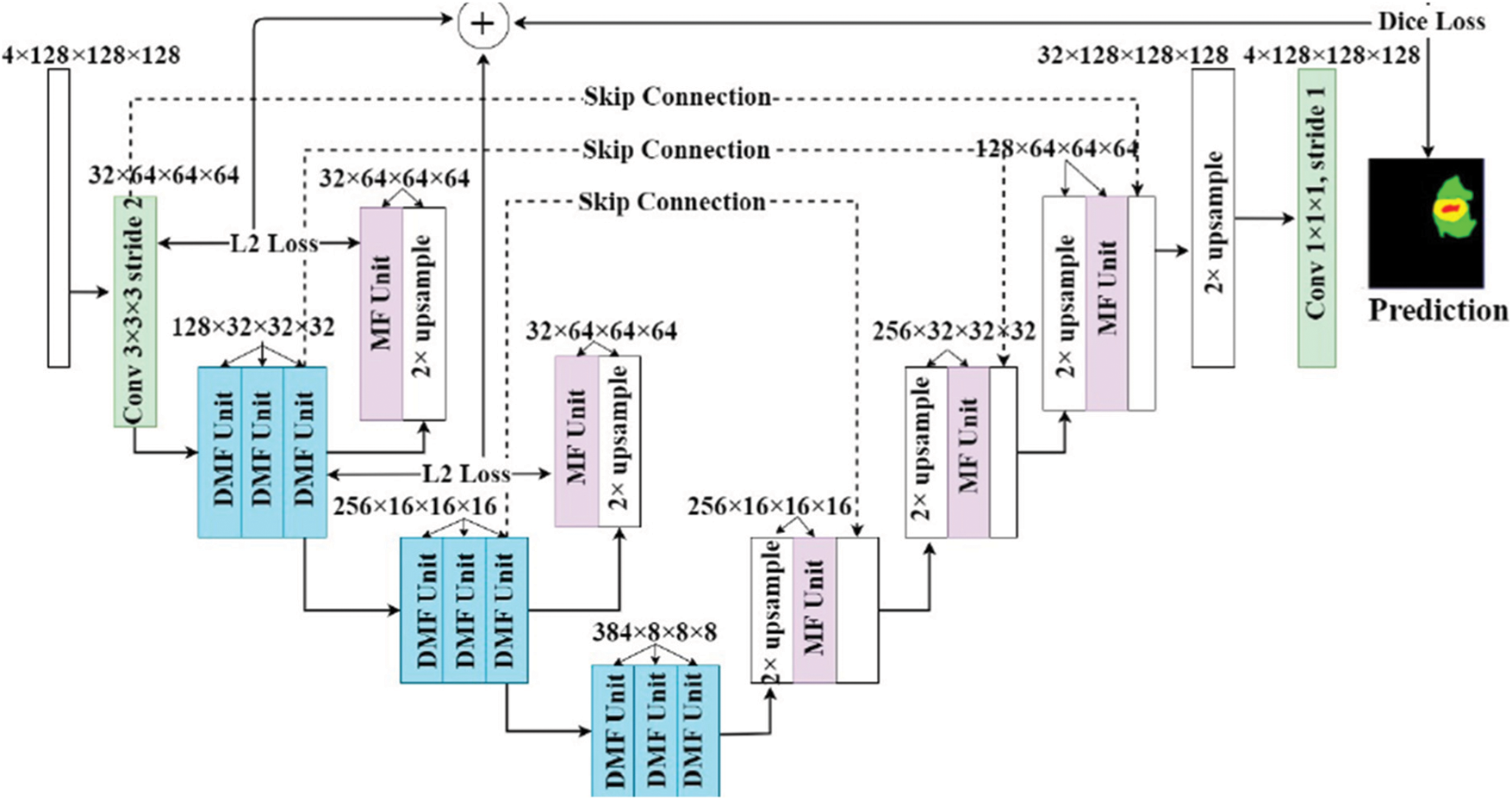
Figure 2: Proposed architecture with deep learning and multistage learning technique
The middle layers of the architecture responsible for filtering the input images, and a Dilated Multi-Fiber (DMF) unit is implemented as part of the encoder and decoder. The DMF Unit abbreviated as dilated multi fibre network access the information Bridge between the encoder and decoder. Since this unit it consumes lesser energy, requires lesser memory, and exhibits optimal computational cost. The information between encoder and Decoder are routed in most optimal way when DMF unit is implemented. This process greatly assists the model of the proposed architecture to reduce the computation cost during the segmentation process. Multi fibre and dilated multi fibre networks are also suitable for concatenating different resolution features within the encoder. Having said that the batch normalization and a linear function is applied before every convolution operation, the information between different layers of the proposed architecture must be passed without any damage. The proposed architecture during the observation’s phases recorded the benefits of cascading conditional layers and by optimizing the hyperparameters. The hyperparameters were set to the maximum tuning to control the Drop out ratio to 0.27, which is the best dropout ratio for the given network architecture. Adam optimizer was implemented to deliver the specialized loss function and the learning rate was defined as 0.01.
Intermediate level layers were responsible for extracting the medium to low level features and preserving the quality of input images without discarding sensitive parameters. Regions found with cancer cells have displayed huge size variations and formed a shape accordingly. After eliminating the huge shapes, the intention of the proposed model is to predict the presence of small size tumours. Deeper layers are assigned to perform a contextual aggregation process over different scales of images. This ensures that the outcome is scale invariant. As an outcome, the proposed architecture delivers better segmentation results. Similarly, on the other side of the Decoder network, two convolutional layers are implemented for filtering and decoding respectively. The outcomes are in form of N × 1 images and are found to possess great equality towards segregating the features and enhance the accuracy. Different combinations were tested out for extracting the features and almost the same result was yielded in any combination. The output of the decoder was clear enough to segregate the different classes of tumour. During the process of aggregation, which happens at the layers of transition, the segmented regions are found to be reconstructed with greater accuracy.
As we mentioned in the previous sections, pre-processing is important for brain segmentation processes and similarly balancing the class data is equally important. BraTS is a standard asset and the distribution of different classes in the training data range from 98.47% for the healthy tissue, 1.02 for the edema tissue, 0.26 for increasing tumour and 0.23 for not increasing tumour. The difference understood from this statistic due to the segmentation accuracy. Problem of class imbalances can be removed with the help of loss function. The following Eq. (3) explain the need of dice loss coefficient and it is expressed mathematically.
The performance is detailed as a quantitative and qualitative measure of segmentation accuracy. The dice score is a combination of two objective functions, where one is responsible for identifying the maximum level of overlapping conditions of the ground truth and the other determines the quality of prediction and segmentation. Combining these two objective functions delivers the dice loss coefficient. One objective function it is also considered to be a cross entropy estimation which portrays the classification accuracy of affected tissues shown in Figs. 3a, 3b. The proposed model has computed over 230 images, where 80% of the dataset was used for training the model and remaining 20 for validating the outcomes and this is the standard measure of any machine learning or deep learning algorithms. The measured outcomes are listed in the Tab. 1 below and are found to have similar computational cost, dependency on optimizers and other factors which affect the performance such as pre-processing quality. The proposed model with multistage learning technique has yielded nearly 8% percent improvisation when compared to that of the conventional architectures Fig. 4. The loss function and coefficient has delivered utmost betterment in terms of performance and segmentation quality when compared to that of the standard methodologies. Tab. 1 demonstrates the comparative results of classifying increasing tumours with other U Net architectures. There is a significant improvement as much as 0.72% for differentiating the increasing tumour and core tumour.
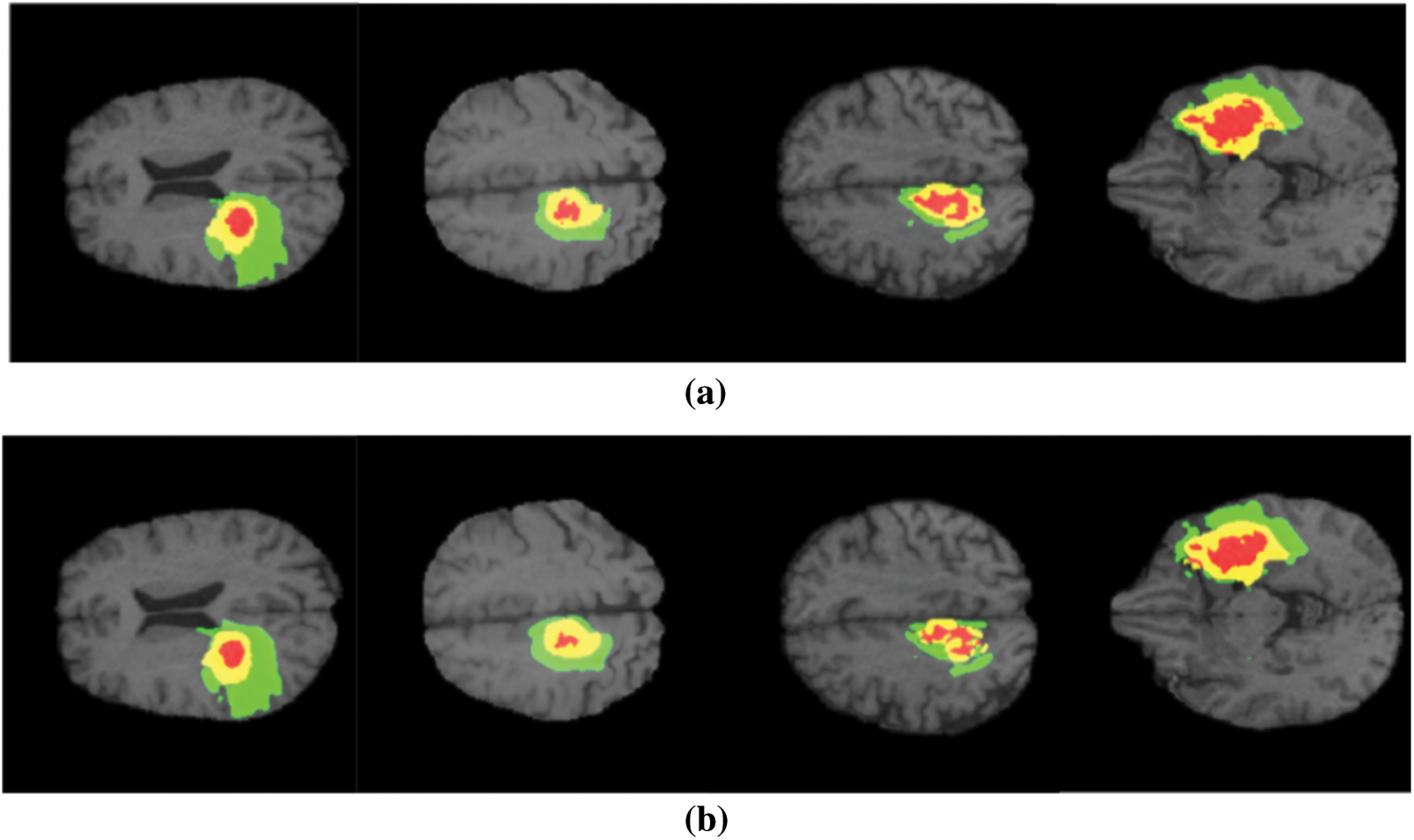
Figure 3: (a) Conventional U net performance (b) Proposed architecture performance (colour variations to demonstrate different categories of tumour, Red–Necrosis, Green–Edema tissue, Yellow–increasing tumour)
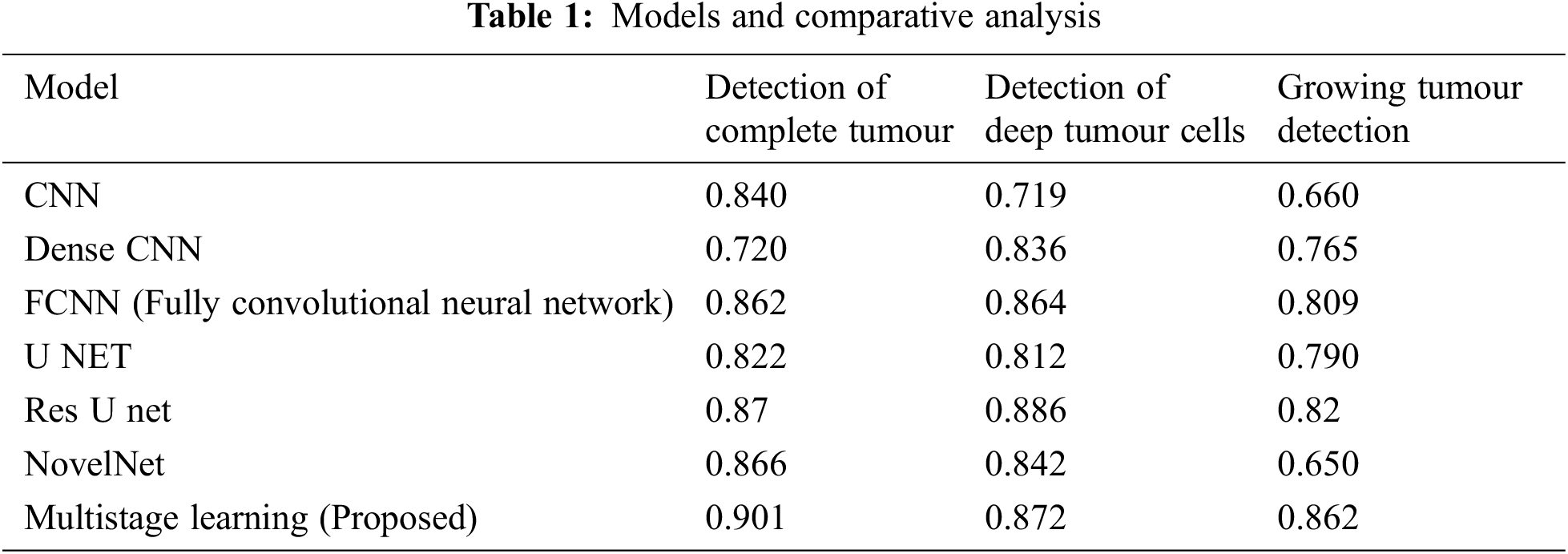
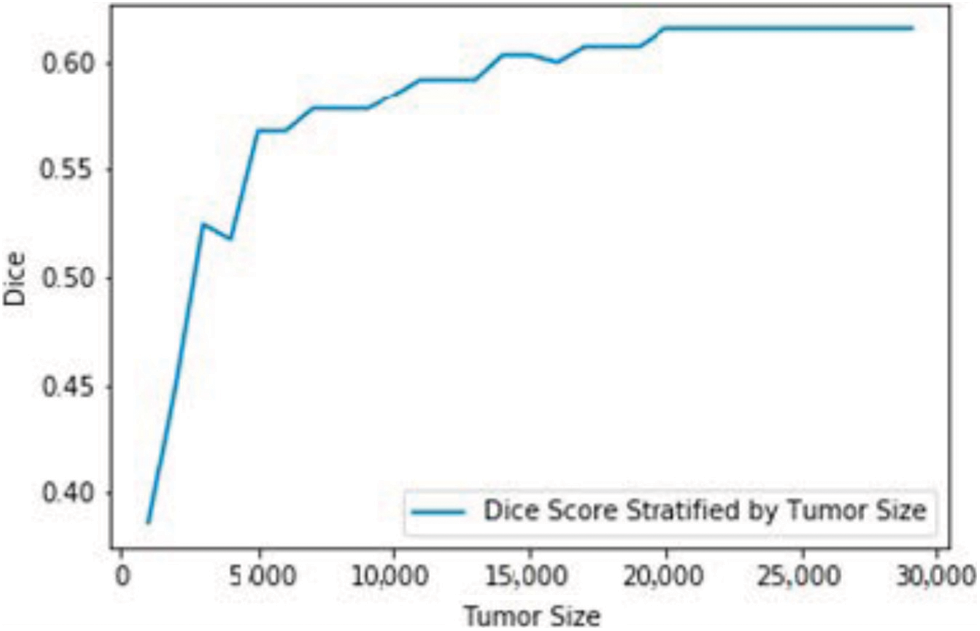
Figure 4: Dice loss coefficient for different tumour sizes
The second part of the analysis is to determine the qualitative difference between the models of segmentation, in which the results are furnished in form of box plots and MRI images. The level of ground truth and segmented regions based on predictions of segmented regions. There are other models which implements the Dilated Multifibre (DMF) Unit, and they fail to detect tumours of smaller sizes. But the proposed model exhibits better accuracy in terms of detecting smaller tumours with better accuracy. The Tab. 2 delivers the comparative results of models in terms of detecting different sizes of tumours.
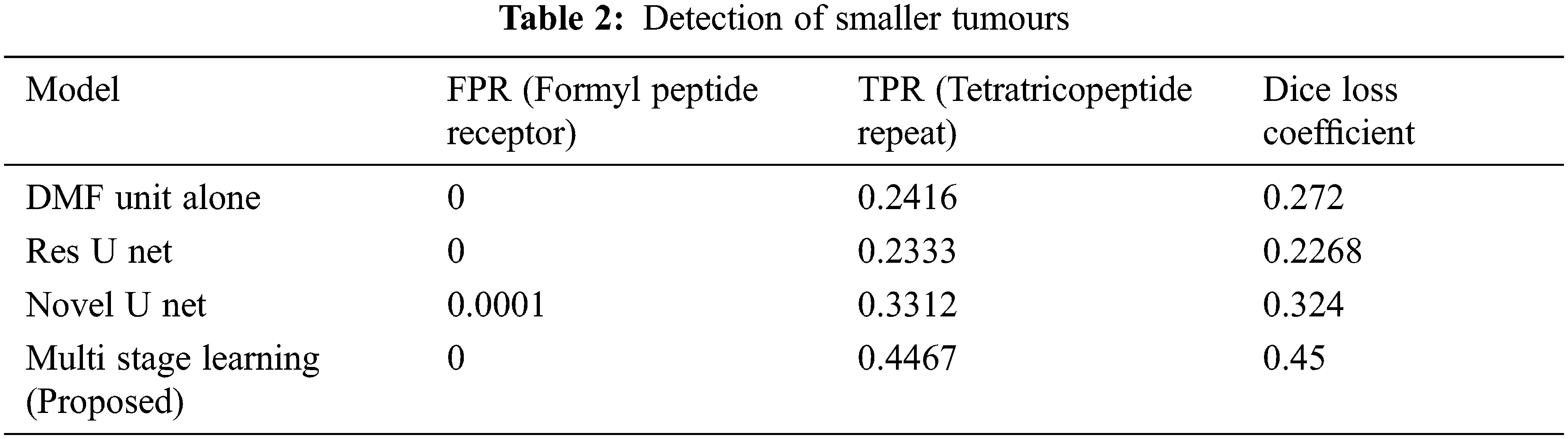
The model proposed in the article demonstrates a technique for brain tumour segmentation from brain MRI images. The multistage learning technique was enforced with a DMF Unit and acted as an information bridge between the encoder and decoder units in the deep learning model. The presented model could detect smaller tumour cells, which can be easily missed in other methodologies. Feature resolution was reduced in other standard architectures during down sampling and that issue was addressed by tweaking the encoder operations. The performance of the model was tested with standard datasets such as BraTS 2017 and 2018 respectively. From the investigative results, it is evident that our model has outperformed other conventional architectures. It has shown a significant 7% increase in segmentation accuracy when compared to other standard models. The process of multistage learning assisted in delivering better features during reconstruction and to preserve the structure of small tumours. Overall, the proposed model has shown significant improvements over other segmentation techniques.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare they have no conflicts of interest to report regarding this study.
1. A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury et al., “PyTorch: An imperative style, high-performance deep learning library,” in Advances in Neural Information Processing Systems, Cambridge, MA, USA: MIT Press, pp. 8026–8037, 2019. [Google Scholar]
2. C. Zhou, S. Chen, C. Ding and D. Tao, “Learning contextual and attentive information for brain tumour segmentation,” in Proc. of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Lecture Notes in Computer Science, Berlin/Heidelberg, Germany: Springer International Publishing, pp. 497–507, 2019. [Google Scholar]
3. S. Cui, L. Mao, J. Jiang, C. Liu, S. Xiong et al., “Automatic semantic segmentation of brain gliomas from MRI images using a deep cascaded neural network,” Journal of Healthcare Engineering, vol. 2018, no. 4940593, pp. 1–14, 2018. [Google Scholar]
4. S. Pavlov, A. Artemov, M. Sharaev, A. Bernstein, E. Burnaev et al., “Weakly supervised fine-tuning approach for brain tumor segmentation problem,” in 2019 18th IEEE Int. Conf. on Machine Learning and Applications (ICMLA), Boca Raton, FL, USA, pp. 1600–1605, 2019. [Google Scholar]
5. F. Hoseini, A. Shahbahrami and P. Bayat, “Adapt ahead optimization algorithm for learning deep CNN applied to MRI segmentation,” Journal of Digital Imaging, vol. 32, no. 1, pp. 105–115, 2019. [Google Scholar]
6. H. Li, A. Li and M. Wang, “A novel end-to-end brain tumor segmentation method using improved fully convolutional networks,” Computational Biol. Med, vol. 108, pp. 150–160, 2019. [Google Scholar]
7. K. Hu, Q. Gan, Y. Zhang, S. Deng, F. Xiao et al., “Brain tumor segmentation using multi-cascaded convolutional neural networks and conditional random field,” IEEE Access, vol. 7, pp. 92615–92629, 2019. [Google Scholar]
8. L. Chen, Y. Wu, A. M. DSouza, A. Z. Abidin, A. Wismüller et al., “MRI tumor segmentation with densely connected 3D CNN,” in Proc. of the Medical Imaging 2018, Image Processing, Houston, TX, USA, no. 10574, pp. 10–15, 2018. [Google Scholar]
9. P. Mlynarski, H. Delingette, A. Criminisi and N. Ayache, “Deep learning with mixed supervision for brain tumor segmentation,” Journal Medical Imaging, vol. 6, no. 34002, pp. 1–14, 2019. [Google Scholar]
10. W. Chen, B. Liu, S. Peng, J. Sun, X. Qiao et al., “S3d-UNet: Separable 3D U-net for brain tumor segmentation,” in Proc. of the Int. MICCAI Brain Lesion Workshop, Granada, Spain, pp. 358–368, 2019. [Google Scholar]
11. S. Devunooru, A. Alsadoon, P. Chandana and A. Beg, “Deep learning neural networks for medical image segmentation of brain tumours for diagnosis-A recent review and taxonomy,” J. Ambient Intell. Human. Comput., vol.12, no. 1, pp. 455–483, 2020. [Google Scholar]
12. L. Wang, S. Wang, R. Chen, X. Qu, Y. Chen et al., “Nested dilation networks for brain tumor segmentation based on magnetic resonance imaging,” Frontiers in Neuroscience, vol. 13, no. 285, pp. 1–14 2019. [Google Scholar]
13. F. Isensee, P. Kickingereder, W. Wick, M. Bendszus, K. H. Maier-Hein et al., “No new-net,” in Proc. of the Int. MICCAI Brain Lesion Workshop, Granada, Spain, pp. 234–244, 2018. [Google Scholar]
14. N. Nuechterlein and S. Mehta, “3D-ESPNet with pyramidal refinement for volumetric brain tumor image segmentation,” in Proc. of the Int. MICCAI Brain Lesion Workshop, Granada, Spain, pp. 245–253, 2018. [Google Scholar]
15. L. C. Chen, G. Papandreou, I. Kokkinos, K. Murphy, A. L. Yuille et al., “Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 40, pp. 834–848, 2017. [Google Scholar]
16. X. Zhao, Y. Wu, G. Song, Z. Li, Y. Zhang et al., “A deep learning model integrating FCNNs and CRFs for brain tumor segmentation,” Med. Image Anal., vol. 43, pp. 98–111, 2018. [Google Scholar]
17. H. Liu, X. Shen, F. Shang, F. Ge and F. Wang, “CU-Net: Cascaded U-net with loss weighted sampling for brain tumor segmentation,” in Proc. of the Int. Workshop, MFCA 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, pp. 102–111, 2019. [Google Scholar]
18. R. Brügger, C. F. Baumgartner and E. Konukoglu, “A partially reversible U-net for memory-efficient volumetric image segmentation,” in Proc. of the Int. Conf. on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Shenzhen, China, pp. 429–437, 13–17, 2019. [Google Scholar]
19. X. Wang, R. Girshick, A. Gupta and K. He, “Non-local neural networks,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, pp. 7794–7803, 18–23, 2018. [Google Scholar]
20. H. Zhao, Y. Zhang, S. Liu, J. Shi, C. Change Loy et al., “Psanet: Point-wise spatial attention network for scene parsing,” in Proc. of the European Conf. on Computer Vision (ECCV), Munich, Germany, pp. 267–283, 8–14, 2018. [Google Scholar]
21. Z. Meng, Z. Fan, Z. Zhao and F. Su, “ENS-Unet: End-to-end noise suppression U-net for brain tumor segmentation,” in Proc. of the 2018 40th Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, pp. 5886–5889, 2018. [Google Scholar]
22. J. Liu, F. Chen, C. Pan, M. Zhu, X. Zhang et al., “A cascaded deep convolutional neural network for joint segmentation and genotype prediction of brainstem gliomas,” IEEE Transactions on Biomedical Engineering., Eng., vol. 65, pp. 1943–1952, 2018. [Google Scholar]
23. S. Pereira, A. Pinto, J. Amorim, A. Ribeiro, V. Alves et al., “Adaptive feature recombination and recalibration for semantic segmentation with fully convolutional networks,” IEEE Transactions on Medical Imaging, vol. 38, no. 12, pp. 2914–2925, 2019. [Google Scholar]
24. C. Chen, X. Liu, M. Ding, J. Zheng, J. Li et al., “3D dilated multi-fiber network for real-time brain tumor segmentation in MRI,” in Proc. of the Int. Conf. on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Shenzhen, China, pp. 184–192, 2019. [Google Scholar]
25. M. U. Rehman and K. T. Chong, “DNA6mA-MINT: DNA-6mA modification identification neural tool,” Genes, 2020, vol. 11, no. 898, pp. 1–12, pp. 2020. [Google Scholar]
26. K. Muhammad, S. Khan, J. D. Ser and V. H. C. de Albuquerque, “Deep learning for multigrade brain tumor classification in smart healthcare systems: A prospective survey,” IEEE Transactions on Neural Networks and Learning Systems, vol. 32, no. 2, pp. 507–522, 2020. [Google Scholar]
27. T. Ilyas, A. Khan, M. Umraiz and H. Kim, “SEEK: A framework of superpixel learning with CNN features for unsupervised segmentation,” Electronics, vol. 9, no. 383, pp. 1–15, 2020. [Google Scholar]
28. W. L. Geon and K. K. Hong, “Multi-task learning U-net for single-channel speech enhancement and mask-based voice activity detection,” Applied Science, vol. 10, no. 3230, pp. 1–15, 2020. [Google Scholar]
29. T. He, J. Hu, Y. Song, J. Guo, Z. Yi et al., “Multi-task learning for the segmentation of organs at risk with label dependence,” Medical Image Analysis, vol. 61, no. 101666, pp. 1–16, 2020. [Google Scholar]
30. A. Ghani, C. H. See, V. Sudhakaran, J. Ahmad, R. Abd-Alhameed et al., “Accelerating retinal fundus image classification using artificial neural networks (ANNs) and reconfigurable hardware (FPGA),” Electronics, vol. 8, no. 1522, pp. 1–17, 2019. [Google Scholar]
31. J. Nalepa, M. Marcinkiewicz and M. Kawulok, “Data augmentation for brain-tumor segmentation: A review,” Frontiers in Computational Neuroscience, vol. 13, no. 83, pp. 1–18, 2019. [Google Scholar]
32. L. Sun, S. Zhang, H. Chen and L. Luo, “Brain tumor segmentation and survival prediction using multimodal mri scans with deep learning,” Frontiers in Neuroscience, vol. 13, no. 810, pp. 1–9, 2019. [Google Scholar]
33. A. Banan, A. Nasiri and A. Taheri-Garavand, “Deep learning-based appearance features extraction for automated carp species identification,” Aquacultural Engineering, vol. 89, no. 102053, pp. 1–34, 2020. [Google Scholar]
34. W. Alam, S. D. Ali, H. Tayara and K. To Chong, “A CNN-based RNA N6-methyladenosine site predictor for multiple species using heterogeneous features representation,” IEEE Access 2020, vol. 8, pp. 138203–138209, 2020. [Google Scholar]
35. S. Markkandan, S. Sivasubramanian, J. Mulerikkal, N. Shaik, B. Jackson et al., “Massive MIMO codebook design using Gaussian mixture model-based clustering,” Intelligent Automation & Soft Computing, vol. 32, no. 1, pp. 361–375, 2022. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |