DOI:10.32604/iasc.2021.017021

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2021.017021 |  |
| Article |
Chinese Q&A Community Medical Entity Recognition with Character-Level Features and Self-Attention Mechanism
1School of Management, Nanjing University of Posts & Telecommunications, Nanjing, 210023, China
2Jiangsu Provincial Key Laboratory of Data Engineering and Knowledge Service, Nanjing, 210023, China
3School of Information Management, Nanjing University, Nanjing, 210093, China
4School of Business and Economics, Loughborough University, Leicestershire, LE11 3TU, United Kingdom
5School of Basic Medical Sciences, Nanjing Medical University, Nanjing, 210029, China
*Corresponding Author: Xiaoyan Li. Email: xylihappy@njmu.edu.cn
Received: 18 January 2021; Accepted: 15 March 2021
Abstract: With the rapid development of Internet, the medical Q&A community has become an important channel for people to obtain and share medical and health knowledge. Online medical entity recognition (OMER), as the foundation of medical and health information extraction, has attracted extensive attention of researchers in recent years. In order to further improve the research progress of Chinese OMER, LSTM-Att-Med model is proposed in this paper to capture more external semantic features and important information. First, Word2vec is used to generate the character-level vectors with semantic features on the basis of the unlabeled corpus in the medical domain and open domain respectively. Then, the two character-level vectors are embedded into BiLSTM-CRF as features to construct LSTM-Wiki and LSTM-Med models. Finally, Self-Attention mechanism is introduced into LSTM-Med model, and the performance of the model is validated by using the self-labeled data. The 10-fold cross-validation experiment shows that LSTM-Att-Med with Self-Attention mechanism introduced achieves the best performance and the F-value can be up to 91.66%, which is 0.72% higher than that of BiLSTM-CRF. In addition, the experiment result demonstrates that the improvements of F-value are inconsistent for different corpora based on LSTM-Att-Med. The paper also analyzes the recognition performance and error results of different medical entities.
Keywords: Q &A community; deep learning; online medical entity recognition; external semantic features; self-attention mechanism
With the rapid development of Internet, the medical Q&A community has become an important channel for people to acquire and share medical and health knowledge. A Pew Research Center survey found that 72% American adults had searched medical and health information online [1]. In a 2016 survey, about 195 million Chinese people stated that they had used online medical services [2]. In recent years, a large amount of medical and health Q&A data has been accumulated on the Internet. In this context, the medical and health community has become an important data source for patient requirements analysis [3,4], epidemiological surveillance [5–7], adverse drug reaction detection [8], and disease prediction [9,10].
Compared with electronic medical records (EMRs) and medical literatures, user-generated content in the medical Q&A community is more arbitrary, and the use of terminology therein is more irregular. In addition, the descriptions of the same disease from ordinary users and professional doctors are always inconsistent. Therefore, how to effectively extract medical entities from the online Q&A community is very challenging. In this study, we proposed a method by combining self-attention mechanism and deep learning model. The method effectively improved the performance of medical entity recognition (MER) for the Q&A community by capturing more external semantic features and important information.
Generally, medical entities include diseases, symptoms, medications, examinations, and treatments. OMER, as the foundation of medical and health information extraction, is also a critical step of NLP. Despite lots of attention it has been paid, there still exist many challenges in the study of OMER. Firstly, there is no reference standard for naming medical entities in the Q&A community. Secondly, abbreviations, acronyms, and variations are extremely common in the medical field. In addition, medical entities usually contain more complex word structures. These problems are the main obstacles to MER.
Due to the importance of its application, OMER has attracted considerable attention. Based on patient descriptions in online medical forums, medical entities were extracted through the lexico-syntactic patterns [11]. Jimeno-Yepes et al. [12,13] developed Micromed, an OMER tool for online texts, which was more effective than the MetaMap and Stanford NER for the general texts. Using Twitter data, Magumba et al. [14] comparatively analyzed the performance of different deep learning models on disease recognition. Yao et al. [15] used CRF to recognize medical entities. Yang et al. [16] extracted medical entities from Chinese online health consultations using deep neural networks. Liu et al. [17] found that BiLSTM-CRF could achieve an optimal performance for OMER.
Recent studies have shown that embedding features in neural networks can effectively extract medical entities and classify entity relationships. Luo [18] verified the effects of word embeddings in relationship classifications based on LSTM model and found that word embeddings from the medical domain were better than those from the open domain. Using i2b2/VA, DrugBank, and Medline datasets, Unanue et al. [19] found that character-level embeddings could improve the performance of MER. By embedding the randomly initialized word vectors and pretrained word vectors of biomedical texts, Cho et al. [20] found that the pretrained embeddings of biomedical texts performed best in MER.
In recent years, attention mechanism has been widely used in NLP tasks, especially in deep learning models. In 2014, attention mechanism was firstly added to recurrent neural network for image classification [21]. Subsequently, Bahdanau et al. [22] applied attention mechanism to machine translation. In 2017, self-attention mechanism was used to learn text representations [23]. Meanwhile, a new approach for MER which combines attention mechanism with neural network model was proposed. This approach exhibited state-of-the-art results in MER studies. Li et al. [24] introduced attention mechanism into the BiLSTM-CRF model, and F-values on EMRs of CCKS1 2017 and CCKS 2018 reached 90.48% and 86.11% respectively. For entity recognition on EMRs, researchers found that the experiment performance could be improved by introducing attention mechanism into neural network model [25]. Based on existing research, it can be found that BiLSTM-CRF is the best model on sequence tasks. For better use of external semantics and important information, this paper embedded the character vectors generated from the external corpora into BiLSTM-CRF. Moreover, self-attention mechanism was introduced to capture potential semantic features to further improve the performance of OMER.
3.1 Data Source and Preprocessing
The Q&A data used in this study consists of communications between doctors and patients. Since most patients have no medical background and medical knowledge, they usually use colloquial language to describe their problems. For a more comprehensive experiment comparison, we obtained data from 39ask.net2 and qiuyi.cn.3 The two websites are well-known Chinese medical Q&A communities. On 39ask.net, patients’ expressions are less colloquial, doctors’ answers are more professional. In contrast, the descriptions of entities on qiuyi.cn are more casual, especially in the patient question section. In total, 1,197 questions and 4,651 answers from 39ask.net and 831 questions and 2,398 answers from qiuyi.cn were crawled. Hereafter, a rule-based method was used for text extraction, data cleaning and sentence segmentation.
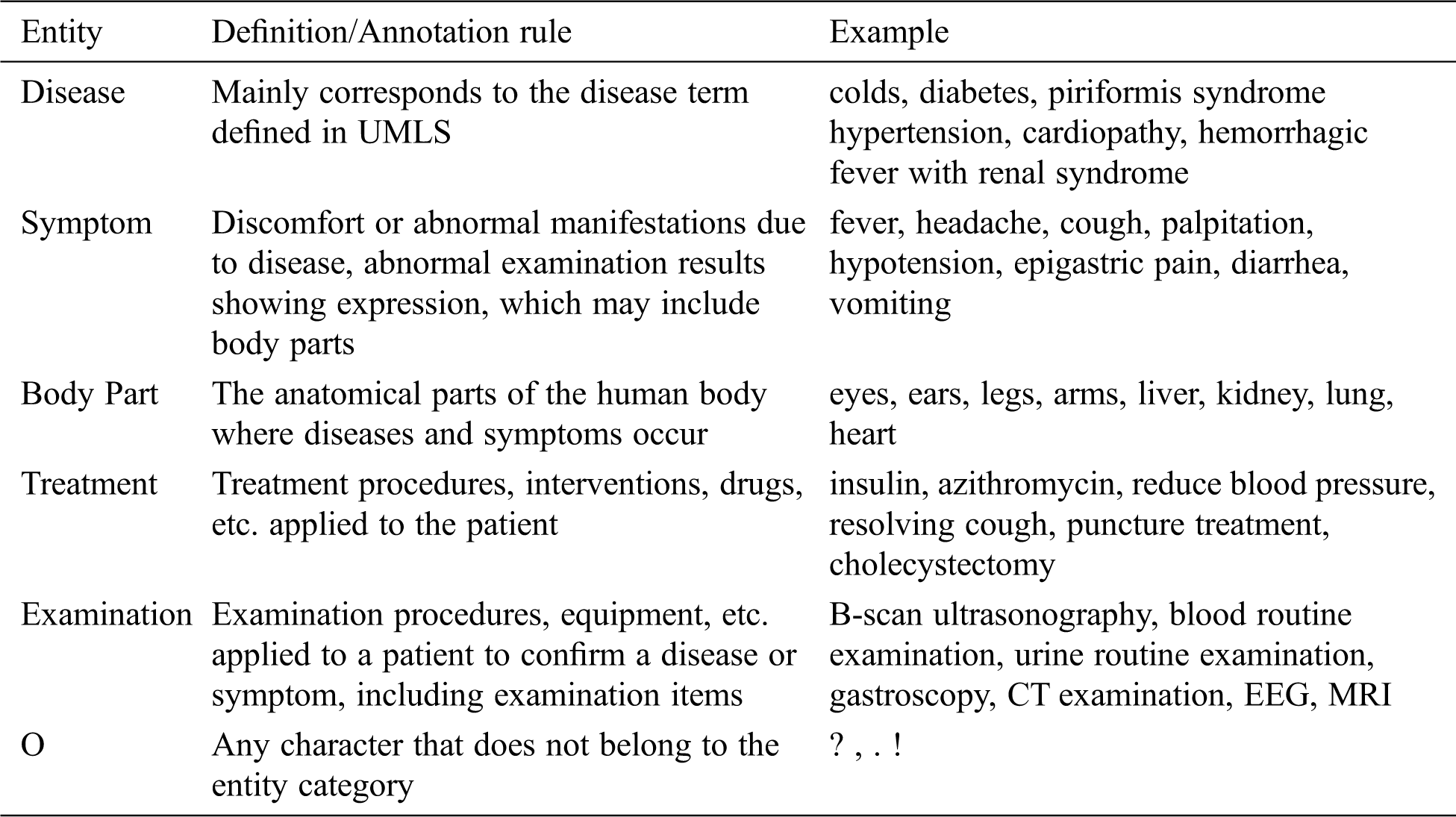
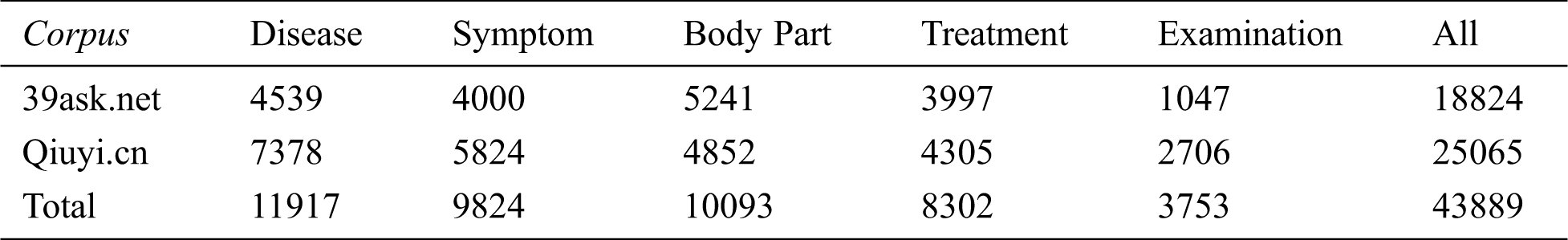
This paper referred to the definition of medical entity in UMLS; followed the principles of non-overlapping, non-nesting between entities, and no punctuation in entities. Moreover, the BIO labeling system was used to annotate entities. The specific format is B-X, I-X, and O, where X represents the type of entity, B-X is used to mark the beginning of entity X, I-X is used to mark the interior of entity X, and O is used to indicate a non-entity. Referring to previous researches, we annotated disease, symptom, body part, treatment, and examination entities. The definition and annotation rules as well as examples of corresponding entities are presented in Tab. 1. The quantities of different entities are shown in Tab. 2.
Table 1: The definition/annotation rules and corresponding examples of five entities
To improve efficiency and ensure annotation quality, the labeling tool YEDDA4 was used [26]. YEDDA, a text span annotation tool with lightweight collaboration, is commonly used to label natural language text. In addition, YEDDA is highly effective for manually annotating text.
Table 2: Entity statistics of two corpora
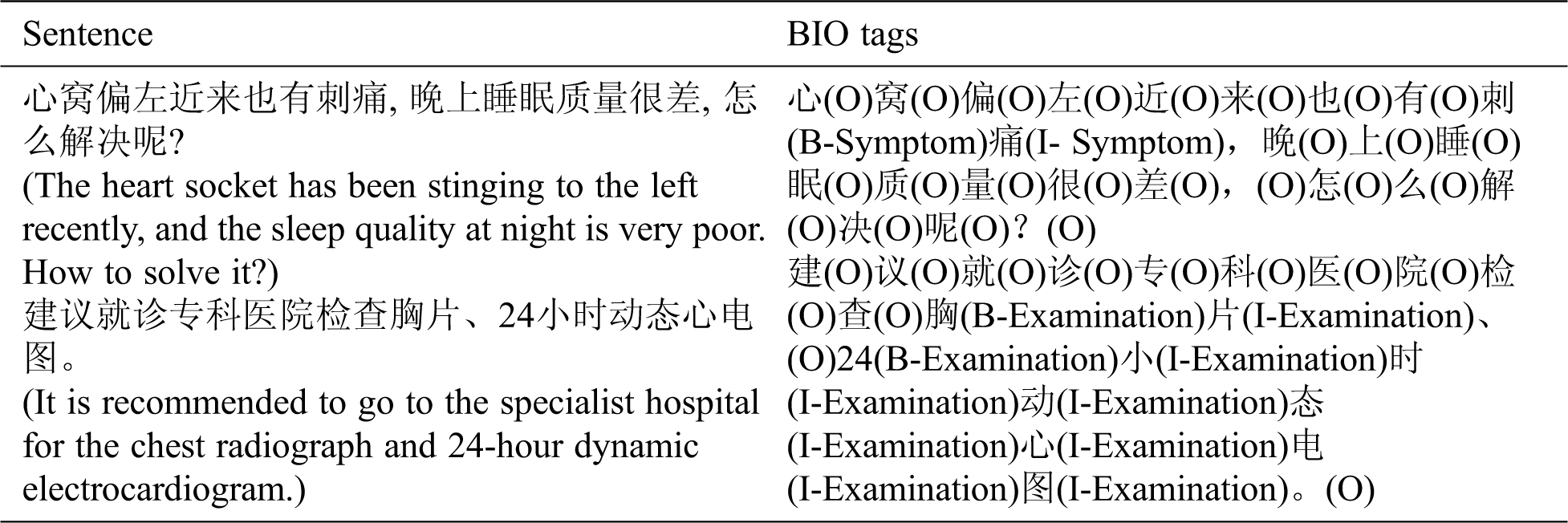
The data was annotated in multiple rounds to ensure the quality of the corpora. In addition, the consistency of annotation had been checked. One example of BIO annotation is listed in Tab. 3.
Table 3: One example of BIO annotation
In this section, we verified the influence of character embeddings from different fields on OMER. Specifically, Word2vec [27] was used to train character vectors and then the vectors were converted into character-level embeddings. Hereafter, the skip-gram model was chosen to generate 100-dimensional character vectors. In addition, the window length was set to 5, min_count was set to 5, and the remaining parameters were set to the default values.
In the experiment, a total of 2.2 GB open-domain texts were crawled from Chinese Wikipedia website, while 815 MB medical-domain texts were crawled from the two Chinese Q&A communities for training character vectors. The scale of open-domain vectors is greater than that of medical-domain vectors.
The architecture of this study is shown in Fig. 1. Firstly, the crawled data was preprocessed by text extraction and deduplication. Secondly, medical entities were annotated with YEDDA. Thirdly, CRF, BiLSTM-CRF, LSTM-Wiki (the BiLSTM-CRF with character embeddings from Chinese Wikipedia), and LSTM-Med (the BiLSTM-CRF with character embeddings from medical data) were adopted to conduct comparative experiments. Finally, LSTM-Att-Med with self-attention mechanism introduced was constructed to further improve the performance of OMER.
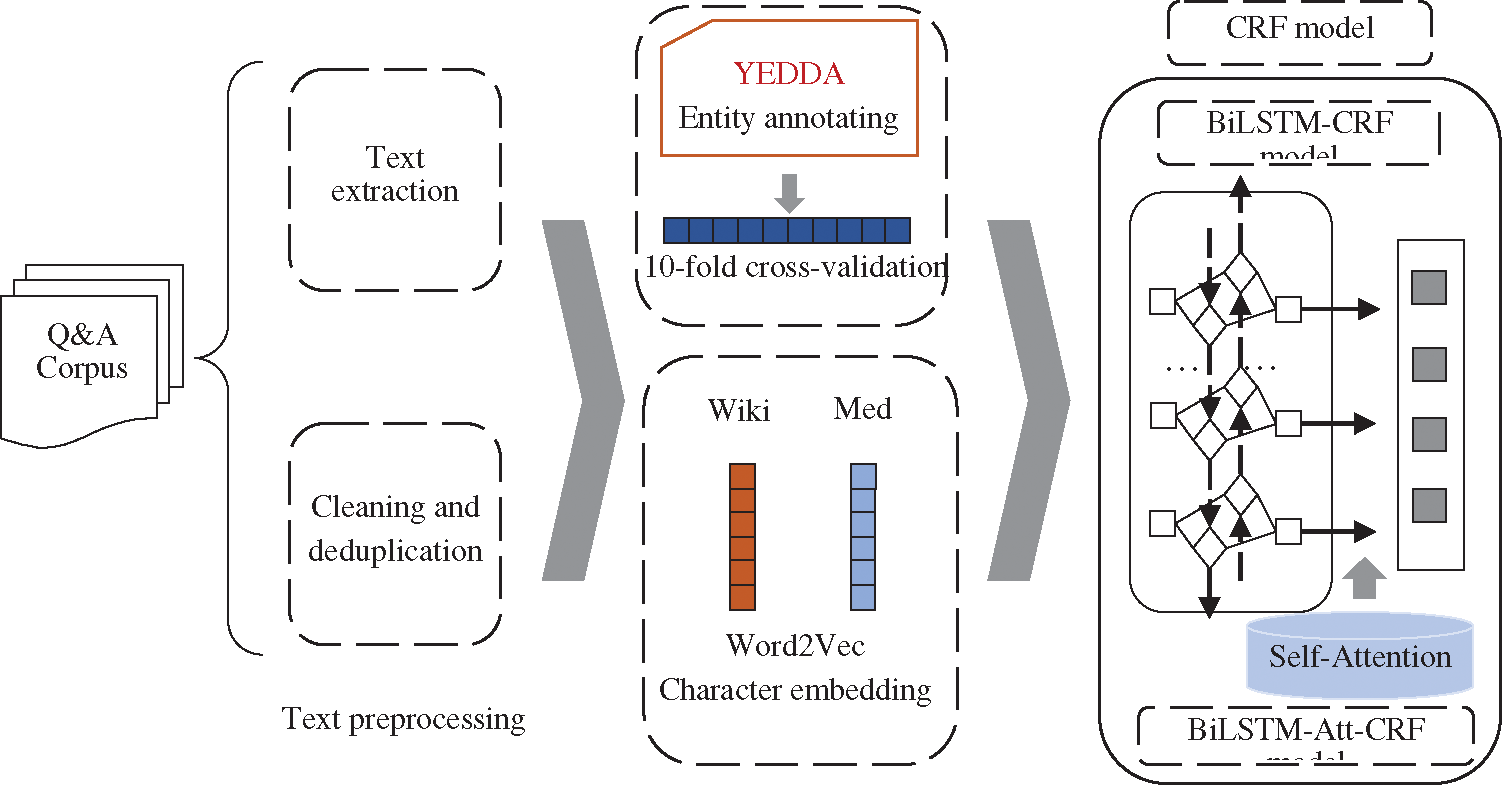
Figure 1: The architecture of this research
RNN is a recurrent neural network used to process sequence data, such as automatic speech recognition and machine translation. The structure of RNN is displayed in Fig. 2. xt is the input at time t, and ht and ot are the hidden layer and output layer respectively, corresponding to time t.
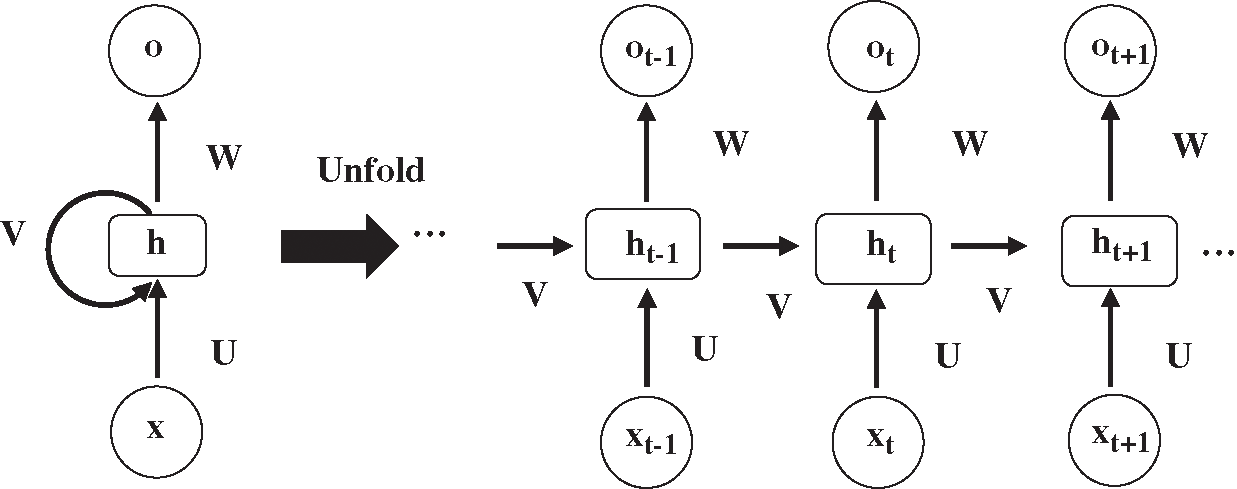
Figure 2: The structure of recurrent neural network
The RNN structure dictates that the output value of RNN at the next moment is affected by the input values at multiple previous moments. However, the output value will be affected by the input value of the later moment. For example, for the text “小明最近感冒了, 今天准备去医院检查一下____, 看看中性粒细胞是否在正常范围内” (“Ming has caught a cold recently, and today he is going to the hospital to take ____ to check if the number of neutrophils is within the normal range”), it may be impossible to accurately determine the specific examination item by simply analyzing the content before the space. Therefore, it is necessary to learn representations from future time steps in order to better understand the context. Bidirectional RNN (BiRNN) could learn representations from future time steps [28]. The structure of BiRNN is shown in Fig. 3.
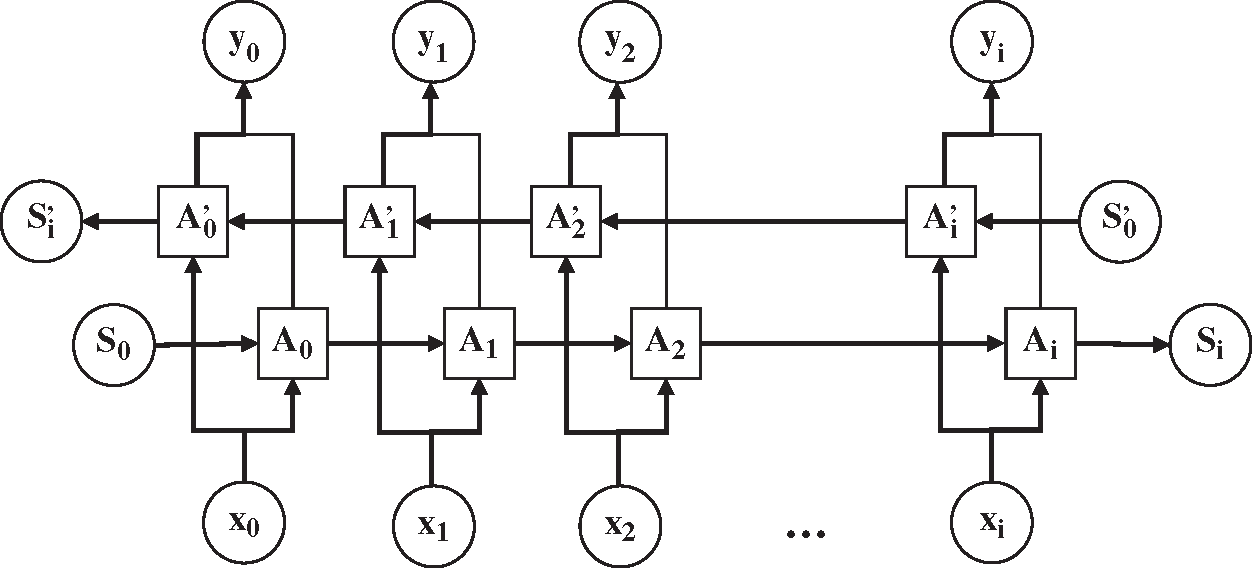
Figure 3: The structure of bidirectional recurrent neural network
In theory, RNN can better capture long-term dependencies. However, RNN cannot learn long-term dependencies of the sequences for the affection of the latest vectors in practical applications [29], indicating that RNN has the problem of gradient disappearance or gradient explosion. More concretely, RNN has an insufficiency in storage capacity such that it cannot solve the problem of long-term dependencies. Accordingly, LSTM is designed to overcome this problem. The difference between LSTM and RNN lies in the state of the cell. The specific structure of a LSTM neuron is shown in Fig. 4, where C is the memory information stored by the cell. In LSTM, three gate structures (the input gate, forget gate, and output gate) are used to selectively forget part of the historical information, add part of the current input information, and finally integrate all the information to generate the output.
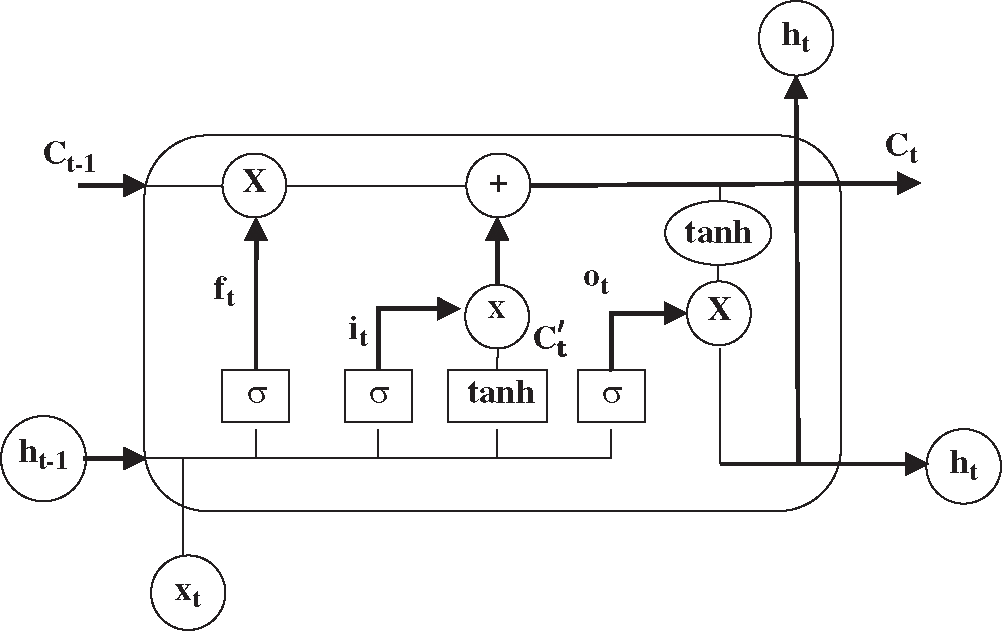
Figure 4: The structure of a LSTM neuron
Forget gate:
Input gate:
Calculate updated value:
Update cell status:
Output gate:
Calculate output value:
In the above formulas, W and b represent the weight matrix and offset vector. The network structure of BiLSTM is similar to BiRNN. The BiLSTM is composed of a forward LSTM and a backward LSTM to ensure the forward and backward information stored in the cell at the same time. Correspondingly, BiLSTM [30] can learn long-term dependencies and simultaneously capture bidirectional information about characters. However, BiLSTM cannot directly use the predicted labels, which leads to invalidity of the predicted label sequence. For example, in predicted result, the label “I-Treatment” will be followed by the label “B-Disease”. In addition, CRF has a transfer feature which uses the order of the output labels to ensure the rationality of predictions. The combination of BiLSTM and CRF can not only effectively save the information of the entire sentence but also can use contextual information to achieve highly accurate sequence labeling.
The long-term dependency phenomenon is extremely common in the text of medical Q&A communities, especially in patient-contributed content. Although BiLSTM can take advantage of long-term contextual information, it is prone to local optimization rather than global optimization. This problem can be solved by CRF. In light of this, BiLSTM-CRF was adopted for OMER. The architecture of BiLSTM-CRF is shown in Fig. 5. Moreover, a dropout layer was added between the embedding layer and the BiLSTM layer to improve the generalizability. The red arrow in Fig. 5 indicates that the dropout layer has been used.
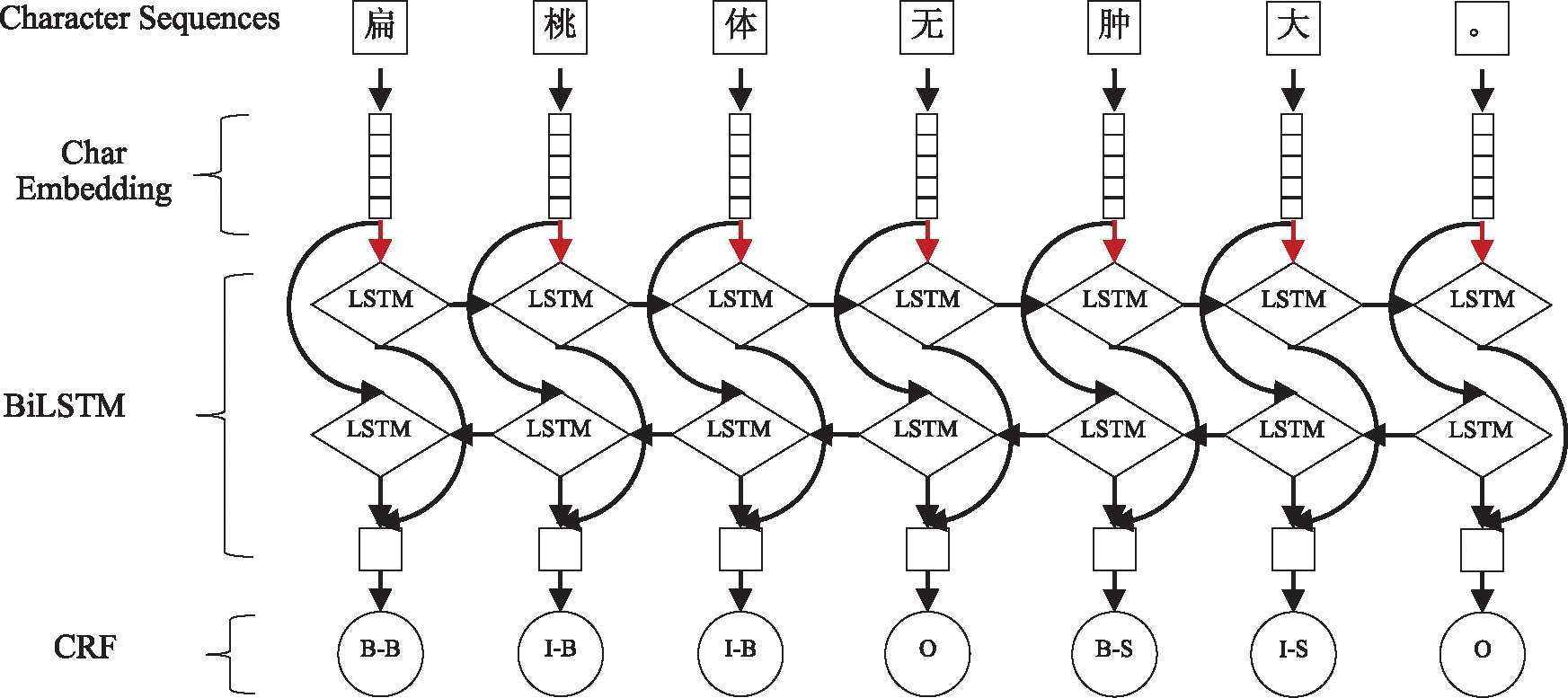
Figure 5: The main architecture of BiLSTM-CRF model
3.4.2 Self-Attention Mechanism
As a selection mechanism, attention mechanism focuses on more important information in semantic information [31]. Self-attention is a type of attention mechanism [23], and it is used to capture the relationship between weight and sequence position when calculating the same sequence representation. Specifically, attention focuses on the weight of input to output, but self-attention focuses on the weight within itself. Thus, self-attention can better capture the semantic features of words in a sentence [32]. The advantage of self-attention mechanism is that it can directly calculate dependent relationships between words, learn the internal structure of the sentence, and obtain long-term dependencies regardless of the distance between words.
Although LSTM can retain long-term information through the gate mechanism, it still shows inconsistencies in labels of long sentences [22]. For long-term dependencies, LSTM need to accumulate information step by step. The greater the distance, the less likely LSTM is to capture features effectively. Therefore, aiming to directly calculate dependencies between words regardless of distance, this research adopted a deep learning model named BiLSTM-Att-CRF combining BiLSTM-CRF neural network with self-attention mechanism. Specifically, the self-attention layer is added between the BiLSTM layer and the CRF layer. The model’s framework is shown in Fig. 6.
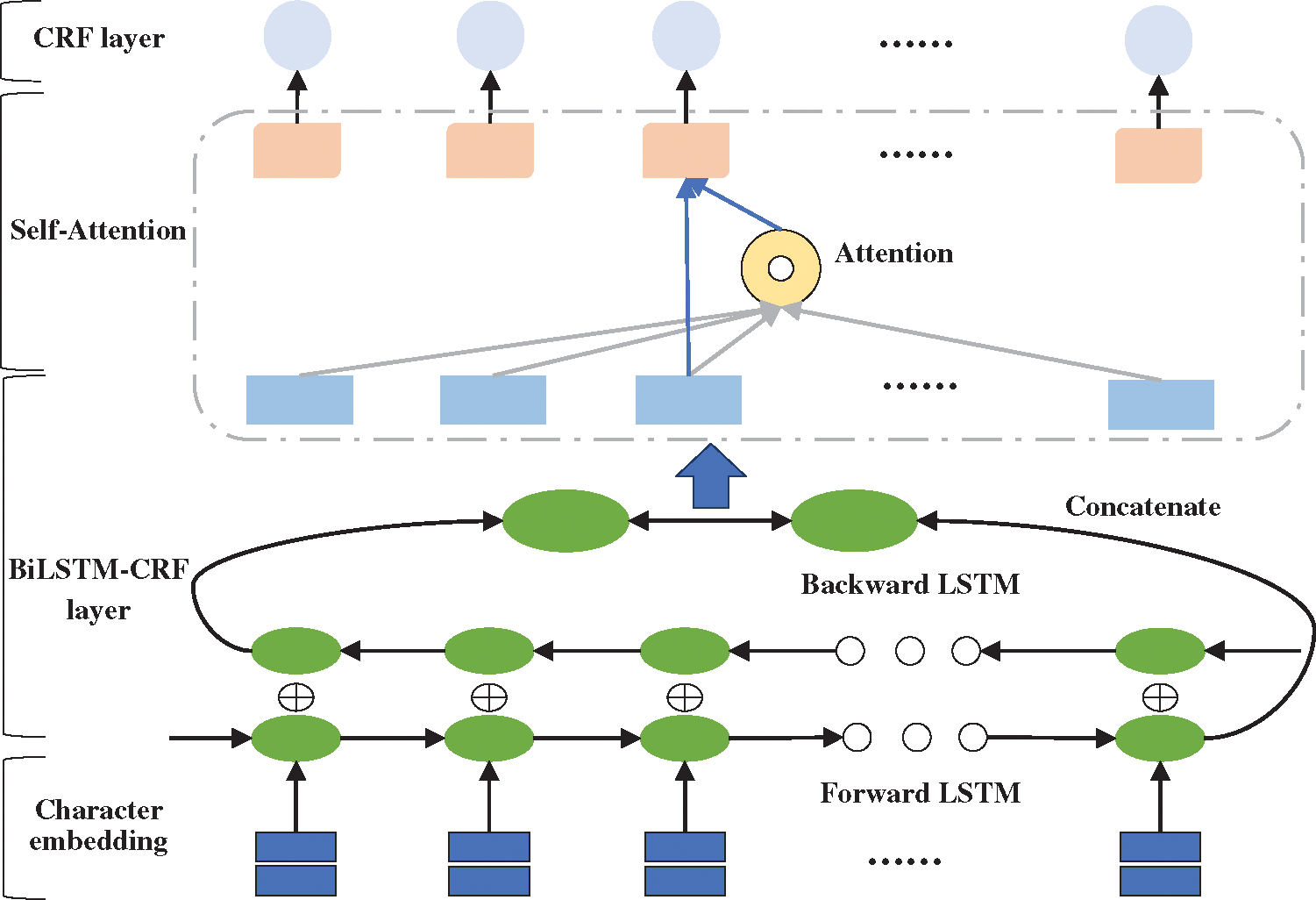
Figure 6: The framework of BiLSTM-Att-CRF model
In BiLSTM-Att-CRF, the input sentence is represented as a vector sequence X (x1, x2, x3, …, xn) through the pretrained character embedding table, where n is the length of the sentence. What’s more, the vector is used as the input for the BiLSTM layer. The BiLSTM can obtain a representation ht′ at each time step t and combine another representation
In formula (7),
The attention weight is used to generate a weighted sum for each time step:
In formula (9),
Finally, a CRF layer is added to decode the best marked path among all possible marked paths. The score from state i to j is represented by the probability transfer matrix
In the experiment, 10-fold cross-validation, a commonly used method in the small-scale dataset, was used to reduce the impact of insufficient data. This method is commonly used in the small-scale dataset [33–35]. The data was divided into 10 parts, and the cross-validation was repeated 10 times. Training, testing, and model selection were carried out on the datasets, and the average result of 10 times was taken as the final experiment result. Specifically, after the model training was completed, the test set was used to test the model, and then the parameters were adjusted according to the results. However, there is a potential problem in deep learning models. The more times the test set is evaluated, the higher the risk of overfitting is. Hence, it is necessary to add a validation set to assist the construction of model. Subsequently, the weights were trained on the training set, and the training effect of the model was evaluated on the validation set. This process is shown in Fig. 7.
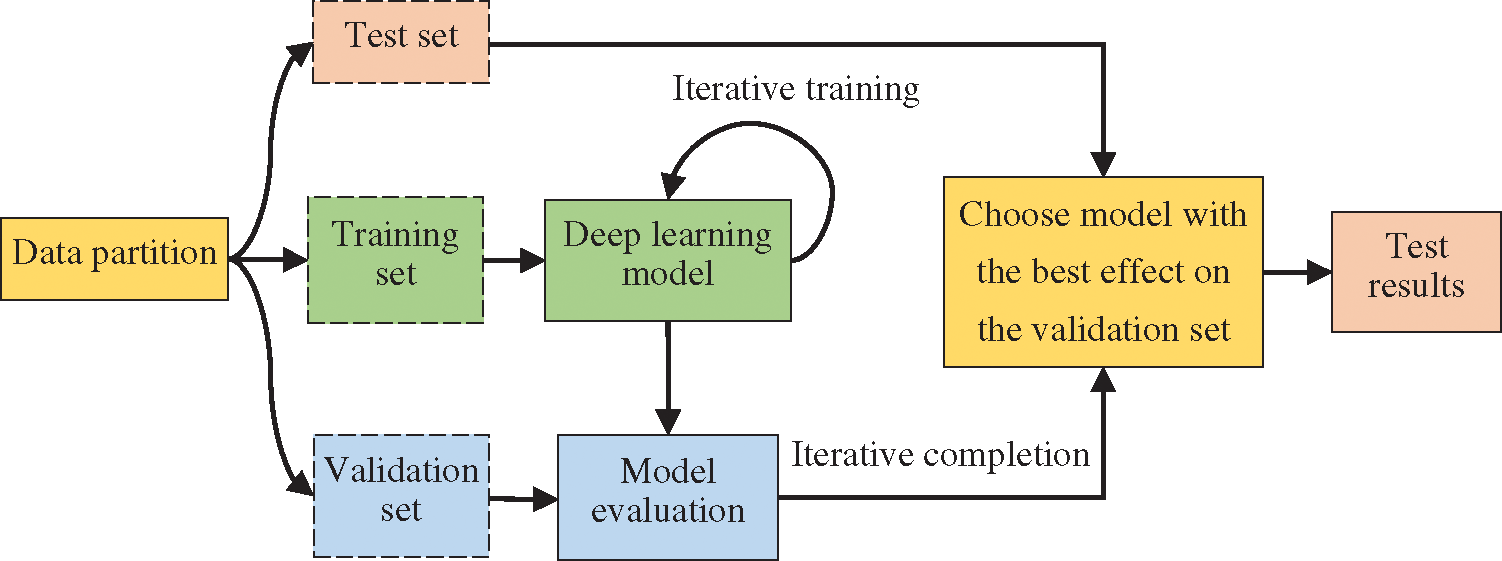
Figure 7: Training flowchart
In the experiment, the data used for the CRF model was randomly divided into training and test sets according to the ratio of 7:3, and the data used for the BiLSTM-CRF model was randomly divided into training, test, and validation sets according to the ratio of 7:2:1.
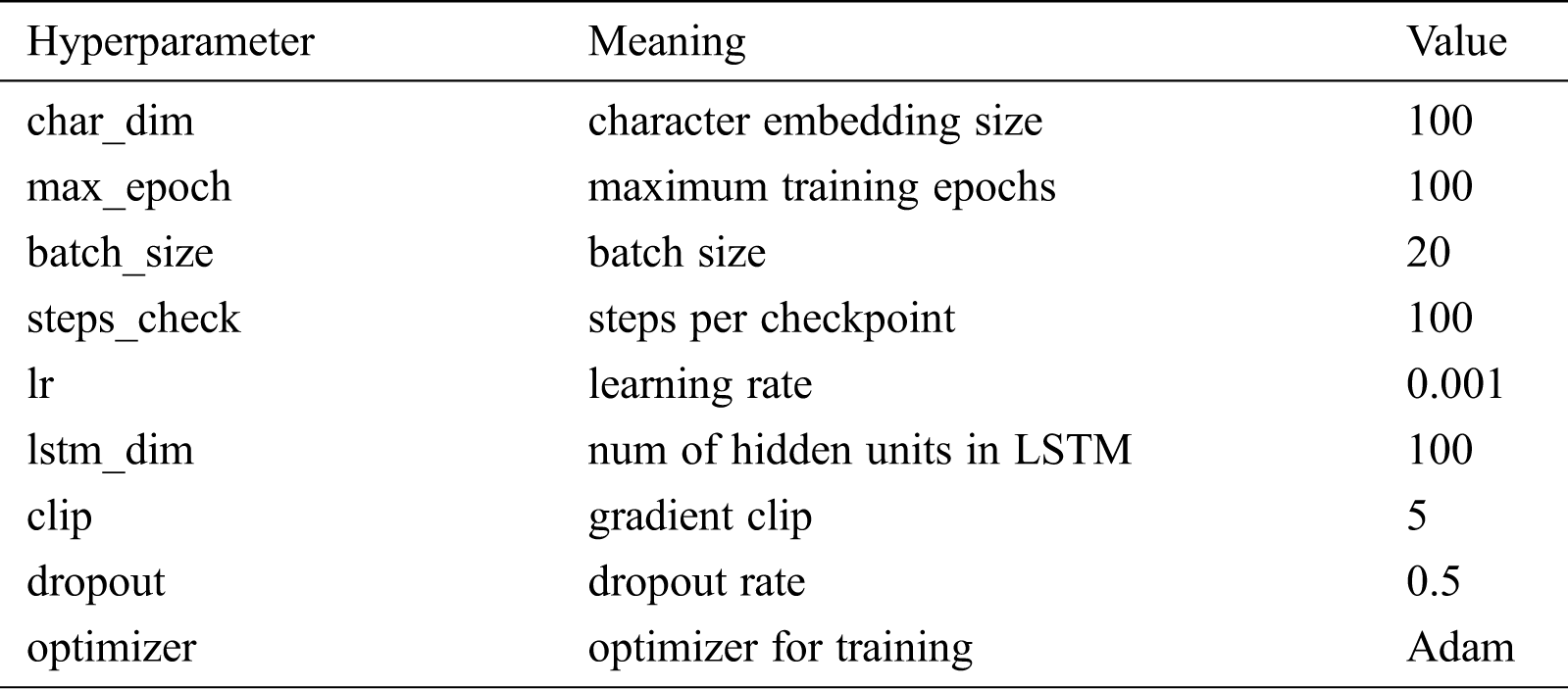
All neural network models in this paper were implemented on the Windows operating using Python version 3.5 and the TensorFlow framework version 1.2.1. The open-source Python tool CRF++ version 0.58 was used to construct the CRF model. The hardware environment is as follows: Intel i5 CPU, 8 GB memory, and NVIDIA GeForce MX150 graphics card. Tab. 4 shows the hyperparameter settings of the deep learning models in the experiment.
Table 4: The hyperparameter settings of deep learning models
Referring to existing research, precision (P), recall (R), and F-value were used in the experiments. In OMER, there are four possible performance classifications: a true positive (TP) means that a positive label is predicted as positive (i.e., correctly identified); a false positive (FP) means that a negative label is predicted as positive (i.e., incorrectly identified); a false negative (FN) means that a positive label is predicted as negative (i.e., incorrectly rejected); and a true negative (TN) means that a negative label is predicted as negative (i.e., correctly rejected). Based on these four classification cases, the calculation formulas for precision, recall, and F-value are defined as follows:
Since the combination of precision and recall can fully reflects the effectiveness of model, the F-value is used as the main evaluation criterion.
Existing researches indicate that BiLSTM-CRF has excellent performance in sequence tasks. Therefore, BiLSTM-CRF was constructed as a comparison with the CRF model. In this paper, three main issues were explored. The first one is the performance of an OMER model that integrates the external semantic features. The second one is the impact of corpora from different sources on OMER. The last one is the difference in improvement by introducing self-attention mechanism into the model. In view of the above three subjects, the CRF model was used as a baseline and three control experiments were designed as follows.
Experiment 1: Verify the performance of four models and the external semantic features. CRF, BiLSTM-CRF, LSTM-Wiki, and LSTM-Med were used to conduct a control experiment.
Experiment 2: Verify the impact of corpora from different sources on OMER. Using medical Q&A data from 39ask.net and qiuyi.cn, we compared the experiment results of four models and analyzed the influence of corpora from different sources on the experiment results.
Experiment 3: Verify the performance of self-attention mechanism. We introduced self-attention mechanism into the optimal performance model, conducted the experiments on different corpora, and analyzed the experiment results.
4.4.1 Comparison with Benchmark Model: CRF
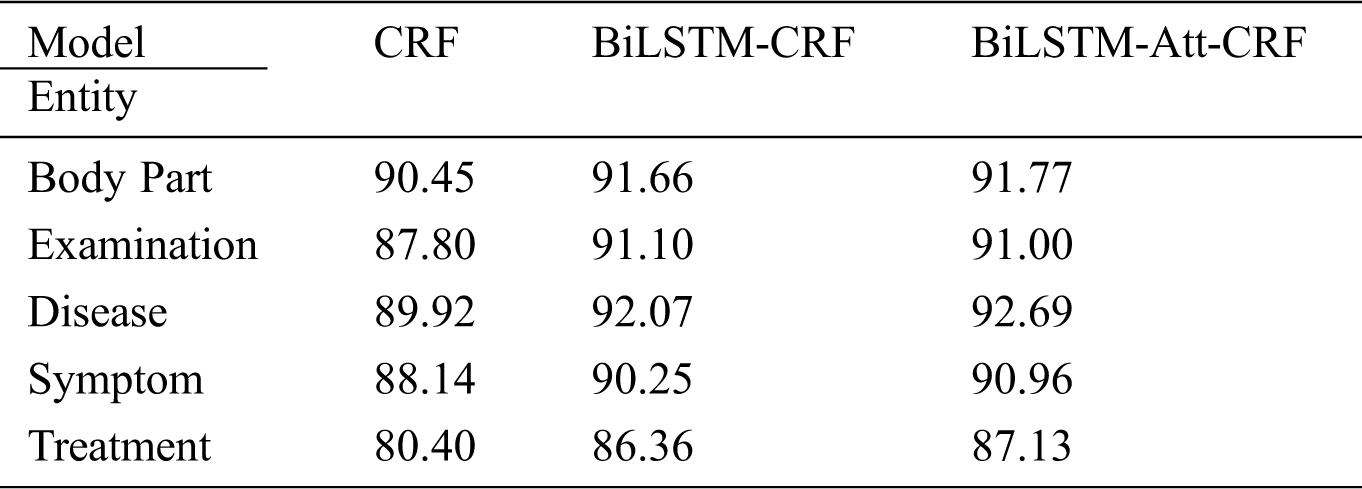
As shown in Tab. 5, the overall F-value of CRF reaches 89.62%, indicating that the quality of experiment data is generally higher after multiple rounds of annotations. In four models, BiLSTM-CRF performs slightly better than CRF; LSTM-Wiki and LSTM-Med with external semantic features further improve the performance of OMER. LSTM-Med achieves the best performance with F-value of 90.17%, 0.55% higher than the benchmark model. Similarly, LSTM-Wiki has an F-value of 0.44% higher than CRF, showing that embedding external semantic vectors can improve the performance of OMER.
Table 5: F-values of four models for various entities (unit: %)
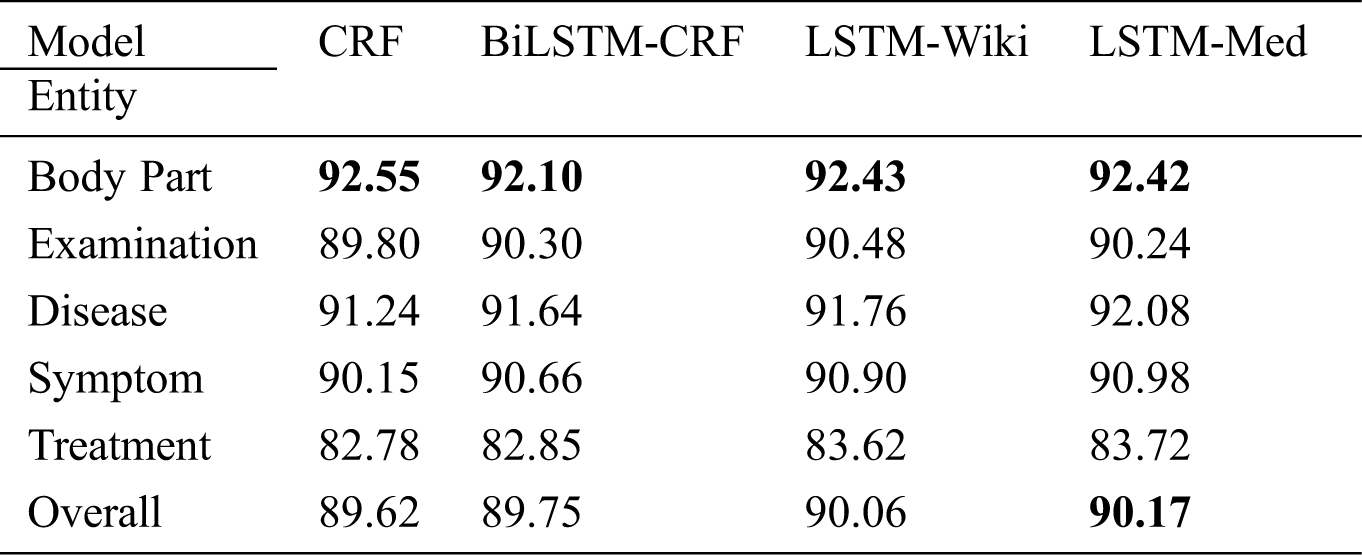
Tab. 5 also indicates that LSTM-Med performs slightly better than LSTM-Wiki. It is worth noting that the scale of medical-domain vectors is smaller than that of open-domain vectors, meaning that the embeddings from same field have a better performance. In addition, the result also shows that embedding external semantic vectors can not only help the model learn similarities between input characters but also can capture more contextual information. Therefore, the introduction of external semantic features from the same field is more effective for OMER.
In order to more intuitively present the experiment results, Fig. 8 shows the F-values of five entities. In deep learning models, the F-values of body part, disease, symptom, and examination entities all exceed 90%; F-value of the treatment entity is the lowest. Further analysis of the experiment corpus indicates that body part, disease, and examination entities in the Q&A community generally with the fixed expressions and the descriptions of symptoms are less colloquial, while the treatment entities involve many characters and complex structures. These differences result in the recognition of treatment entities is more difficult than other types. The average character length of the treatment entity is 3.80, and the longest entity contains 21 characters. Besides, the important semantic features of treatment entity are fewer than other entities. Furthermore, the treatment entity can be easily interfered by other entities. The phenomenon can cause entity boundary recognition errors and entity type recognition errors. For example, the quantity of “局部麻醉扁桃体切除术” (“Treatment”) is small so that it is easily affected by marked entities, and it will be recognized as “局部麻醉” (“Treatment”) \ “扁桃体” (“Body part”) \ “切除” (“Treatment”) \ “术” (“O”). These phenomena may have a negative effect on the experiment results.
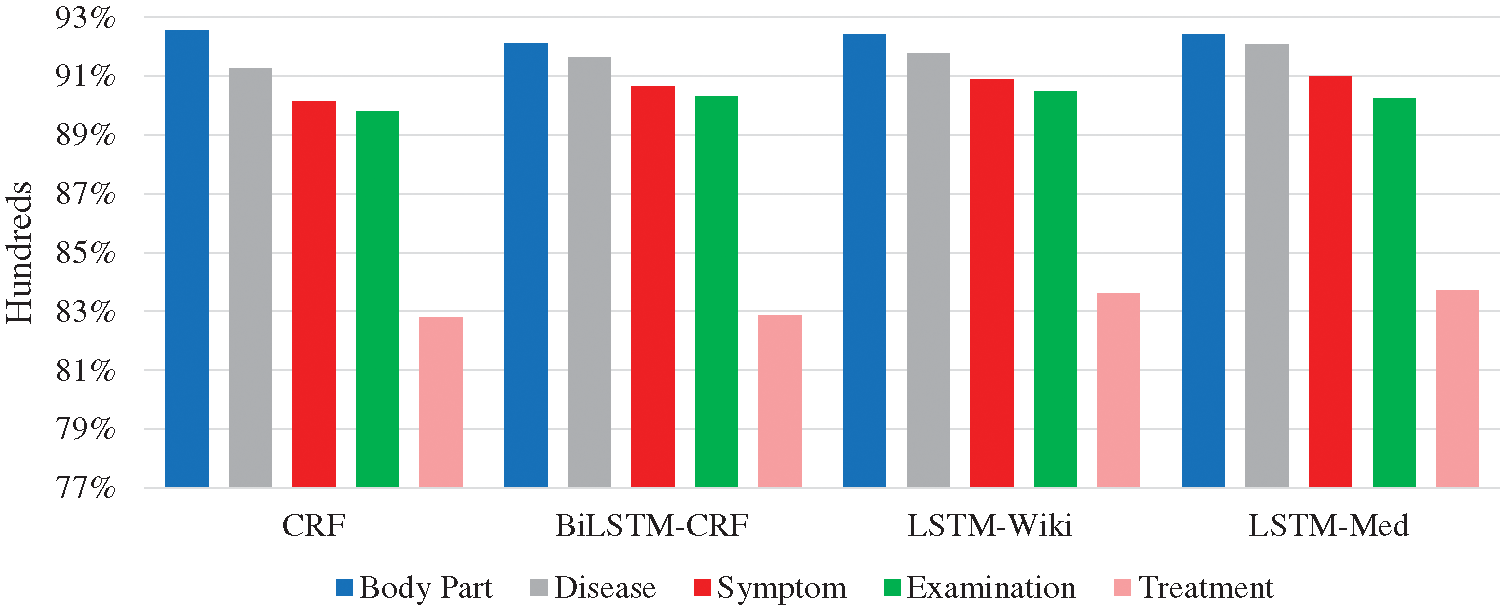
Figure 8: F-values of five entities in four models
4.4.2 Comparison with Different Sources of Corpora
As shown in Tab. 6, LSTM-Med achieves the best performance with F-value of 91.64% on 39ask.net and 90.98% on qiuyi.cn respectively, and followed by LSTM-Wiki. This is consistent with the results of the merged corpus.
Table 6: Experiment results of four models on different corpora (unit: %)
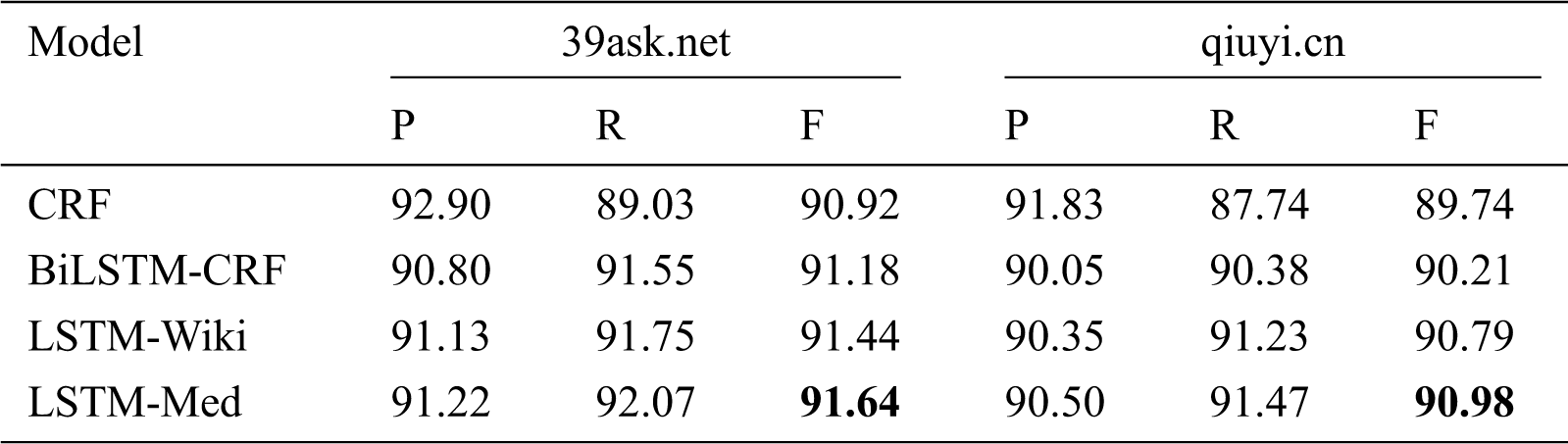
In addition, as can be seen in Tab. 6, CRF has the highest precision and lowest recall among four models, illustrating that the CRF model can be used for tasks with high precision requirements. In contrary, three deep learning models have higher recall than precision. In addition, it can be found that the precision and recall of three deep learning models exceed 90%. LSTM-Med has the best overall performance, indicating that deep learning models have better overall effects for OMER.
As shown in Fig. 9, the experiment results of four models on 39ask.net are better than those on qiuyi.cn. According to the previous analysis of two corpora, descriptions on 39ask.net are more professional, and the experiment result of 39ask.net is better than qiuyi.cn. In addition, the improvement effect is more obvious on qiuyi.cn. The F-value of LSTM-Med is 1.24% higher than that of CRF on qiuyi.cn, while the F-value of LSTM-Med is 0.72% higher than that of CRF on 39ask.net. Further analysis finds that qiuyi.cn contains lots of colloquial sentences, abbreviations and irregular statements. Through comparative analysis, it can be inferred that more information will be learned from the colloquial corpora by embedding character vectors from the same field.
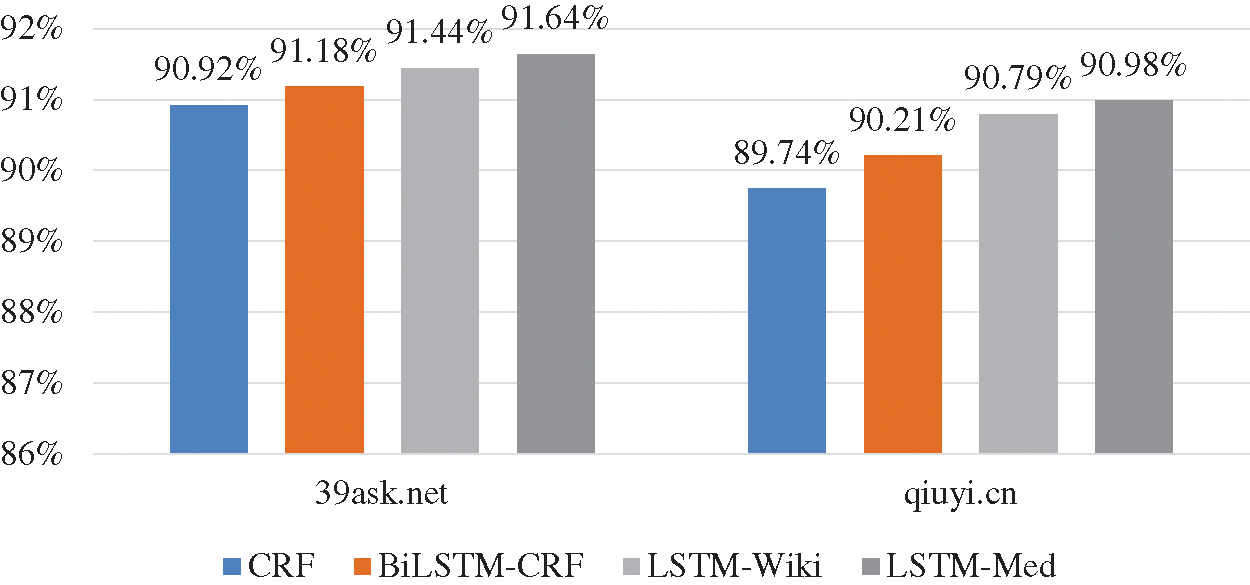
Figure 9: F-values of four models on two corpora
4.4.3 Performance of Self-Attention Mechanism
Through the previous experiments, it is showed that LSTM-Med with medical character embeddings has the best performance for OMER. Hence, self-attention mechanism is introduced into BiLSTM-CRF and LSTM-Med to construct BiLSTM-Att-CRF and LSTM-Att-Med models.
To compare the information storage capabilities of CRF, BiLSTM-CRF, and BiLSTM-Att-CRF, the recall is used for analysis in this section. A higher recall indicates that the model can store more detailed information about the entities. As shown in Fig. 10, BiLSTM-Att-CRF has the optimal recall, with the highest improvement achieved on qiuyi.cn.
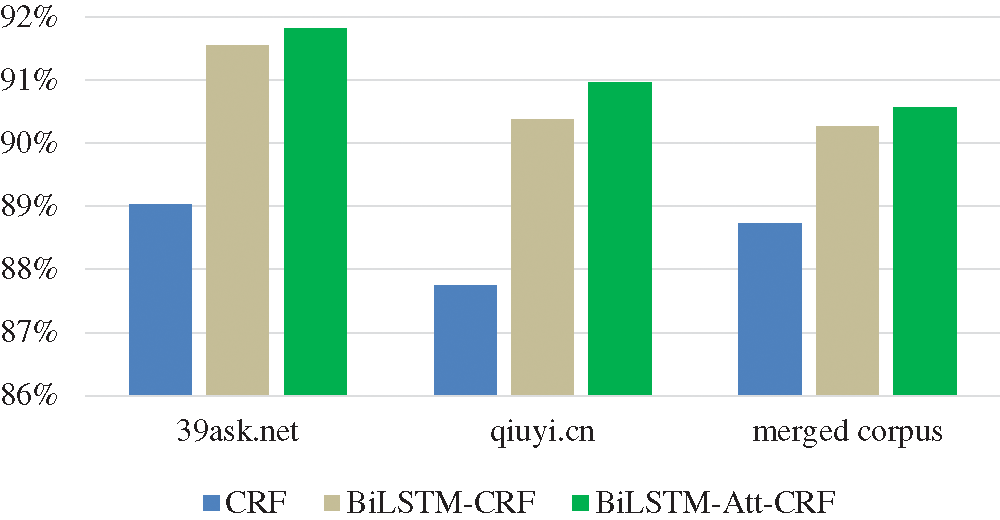
Figure 10: The recall of three models on three corpora
Tab. 7 shows the recall of three basic models without any additional features on qiuyi.cn. BiLSTM-Att-CRF has significantly improved the recall, especially for treatment entity. Compared with other entities, treatment entity usually contains more characters and complex structures, which can easily lead to recognition errors, such as “行结肠镜下息肉切除术” (“undergo polypectomy under colonoscopy”), “3%盐水和1:5000呋喃西林溶液反复漱口”(“rinse mouth repeatedly with 3% saline and 1:5000 Furacilin solution”), and so on. However, there are fixed rules for the descriptions in treatment entities. For example, some regular boundary characters often appear on both sides of treatment entity, such as “行…” (“undergo…”) and “…术/手术” (“…surgery”). If the boundary character is close to the entity, three models can predict the entity, but CRF cannot capture the contextual information at a long distance. Although BiLSTM can solve the problem of long-term dependencies through the gate control mechanism, the latter word is more dominant than the former word in terms of semantics, which makes it difficult to recognize long entities. Consequently, the greater the distance between the boundary character and the keyword of the entity, or the more characters an entity contains, the better the effect of introducing self-attention mechanism.
Table 7: The recall of three models on qiuyi.cn (unit: %)
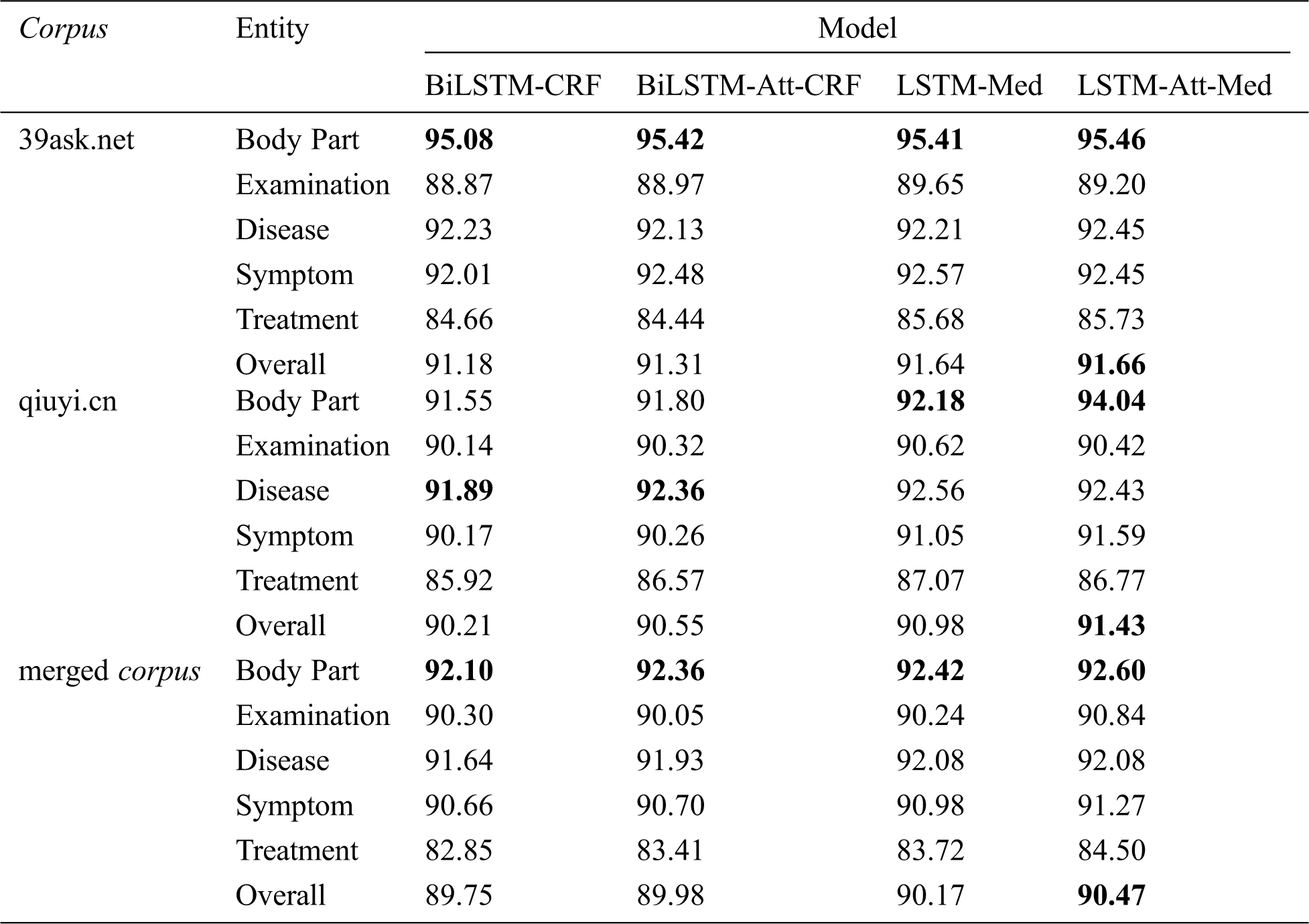
Tab. 8 displays the F-values of five entities before and after the introduction of self-attention mechanism into BiLSTM-CRF and LSTM-Med. It can be seen that LSTM-Att-Med achieves the best performance. The overall F-values of LSTM-Att-Med are 91.66%, 91.43%, and 90.47% on 39ask.net, qiuyi.cn, and the merged corpus respectively. The experiment results show that LSTM-Att-Med has an F-value improvement of 0.45% compared with LSTM-Med on qiuyi.cn. However, the improvements are 0.02% and 0.30% on 39ask.net and the merged corpus respectively. The main reason for the result is that there are many long sentences in qiuyi.cn. After introducing self-attention mechanism, the distance between characters in long sentences can be ignored, and correlations will be automatically learned so as to obtain more potential features. The expressions in 39ask.net are more professional. Accordingly, the experiment result tends to be stable, and the improvements are not obvious. Furthermore, it is found that the F-value on the merged corpus is lower than that on 39ask.net and qiuyi.cn. The main reason might be the complex contexts in the merged corpus. Subsequently, the interference of entities will appear.
As shown in Tab. 8, F-value of body part is the highest among five entities on 39ask.net. However, the performance of body part is improved most obviously after the introduction of self-attention mechanism on qiuyi.cn, with F-value reaching 94.04%—the best result among five entities. As we can see that LSTM-Att-Med can obtain long-term dependencies, learn the sentence structure, and capture the relationship between the labels to improve the experiment result in a colloquial corpus.
Table 8: F-values of four models on different corpora (unit: %)
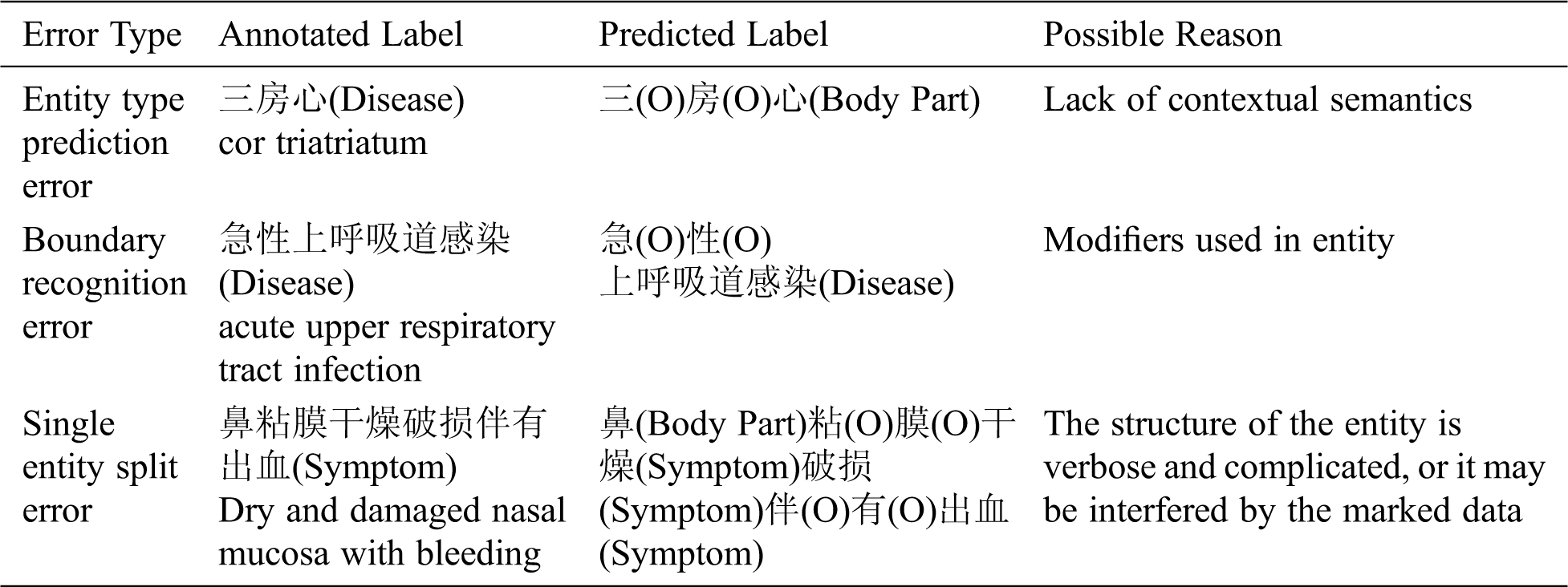
In order to deeply understand the experiment results, the error cases are analyzed and summarized, and the hidden reasons are explored. Tab. 9 presents the types and examples of errors as well as the possible reasons.
Table 9: Examples of online medical entity recognition errors
Through the analysis of results, errors are summarized as follows.
(1) Entity type prediction error: This error is mainly caused by lack of contextual semantics. The data was randomly divided by 10-fold cross-validation, which will result in the unreasonable separation of sentences, as shown in Tab. 9.
(2) Boundary recognition error: For example, “阵发性痉挛性咳嗽” (“paroxysmal spastic cough”) should be predicted as a symptom entity, but the model only predicts “咳嗽” (“cough”) as a symptom entity.
(3) Single entity split error: For example, “行结肠镜下息肉切除术” (“undergo polypectomy under colonoscopy”) should be predicted as a treatment entity, but “结肠镜” (“colonoscopy”) and “息肉切除术” (“polypectomy”) are split and predicted as an examination entity and a treatment entity respectively.
In this paper, LSTM-Att-Med model is proposed to extract disease, symptom, body part, treatment, and examination entities from Chinese Q&A communities. Firstly, in order to verify the impact of introducing external semantic features on OMER, the character vectors with semantic features from open domain and medical domain are embedded in BiLSTM-CRF respectively. Secondly, the differences in OMER for two sources of Q&A data are compared and analyzed. Finally, LSTM-Att-Med model is constructed to further improve the performance of OMER. The research finds that:
(1) Deep learning models embedded with external semantic feature vectors can improve the performance of OMER, and small-scale embeddings from medical domain are more effective than large-scale embeddings from open domain.
(2) The professionalism of Q&A community expressions has a significant impact on the experiment results. For more colloquial corpora, the neural network model embedded with external semantic feature vectors shows more prominent improvement in the performance.
(3) The proposed LSTM-Att-Med model can further improve the performance of OMER. This improvement effect is more obvious in the colloquial corpora, indicating that the model can obtain long-term dependencies and learn sentence structure to capture the semantic associations between different labels after introducing self-attention mechanism.
In this study, although our models achieved comparatively high F-values compared with existing studies, there are still some limitations. For instance, large-scale annotated data is necessary in deep learning models. However, in this experiment, the scale of annotated data from the online Q&A community is relatively small. In order to achieve better performance, transfer learning and semi-supervised learning methods will be used to reduce reliance on manual annotation.
Acknowledgement: The authors would like to thank all anonymous reviewers for their constructive comments.
Funding Statement: This work is supported by the National Social Science Foundation of China “Research on entity semantics mining in health field under big data environment” (No. 17CTQ022).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1China Conference on Knowledge Graph and Semantic Computing.
4https://github.com/jiesutd/YEDDA.
1. S. Fox and M. Duggan, “Health online 2013. Washington, DC, USA: Pew Internet & American Life Project, 2013. [Online]. Available at: http://www.pewinternet.org/2013/11/26/the-diagnosis-difference. [Google Scholar]
2. The 39th Statistical Report on Internet Development in China.China Internet Network Information Center, 2017. [Online]. Available at: http://www.cnnic.cn/gywm/xwzx/rdxw/20172017/201701/t20170122_66448.htm. [Google Scholar]
3. Y. Zhang, “Contextualizing consumer health information searching: An analysis of questions in a social Q&A community,” in Proc. of the 1st ACM Int. Health Informatics Sym., Arlington, Virginia, USA, pp. 210–219, 2010. [Google Scholar]
4. D. Demner-Fushman, Y. Mrabet and A. B. Abacha, “Consumer health information and question answering: helping consumers find answers to their health-related information needs,” Journal of the American Medical Informatics Association, vol. 27, no. 2, pp. 194–201, 2020. [Google Scholar]
5. J. Ginsberg, M. H. Mohebbi, R. S. Patel, L. Brammer, M. S. Smolinski et al., “Detecting influenza epidemics using search engine query data,” Nature, vol. 457, no. 7232, pp. 1012–1014, 2009. [Google Scholar]
6. A. Culotta, “Towards detecting influenza epidemics by analyzing Twitter messages,” in Proc. of the first workshop on social media analytics, New York, NY, USA, pp. 115–122, 2010. [Google Scholar]
7. E. Diazaviles and A. Stewart, “Tracking Twitter for epidemic intelligence: Case study: EHEC/HUS outbreak in Germany, 2011,” in Proc. of the 4th annual ACM web science conf., New York, NY, USA, pp. 82–85, 2012. [Google Scholar]
8. M. Liu, E. R. M. Hinz, M. E. Matheny, J. C. Denny, J. S. Schildcrout et al., “Comparative analysis of pharmacovigilance methods in the detection of adverse drug reactions using electronic medical records,” Journal of the American Medical Informatics Association, vol. 20, no. 3, pp. 420–426, 2013. [Google Scholar]
9. M. A. Khan, S. Abbas, A. Atta, A. Ditta, H. Alquhayz et al., “Intelligent cloud based heart disease prediction system empowered with supervised machine learning,” Computers, Materials & Continua, vol. 65, no. 1, pp. 139–151, 2020. [Google Scholar]
10. B. Yan, X. Tang, J. Wang, Y. Zhou and G. Zheng, “An improved method for the fitting and prediction of the number of Covid-19 confirmed cases based on LSTM,” Computers, Materials & Continua, vol. 64, no. 3, pp. 1473–1490, 2020. [Google Scholar]
11. S. Gupta, D. Maclean, J. Heer and C. D. Manning, “Induced lexico-syntactic patterns improve information extraction from online medical forums,” Journal of the American Medical Informatics Association, vol. 21, no. 5, pp. 902–909, 2014. [Google Scholar]
12. A. Jimeno-Yepes, A. MacKinlay and B. Han, “Investigating Public Health Surveillance using Twitter,” in Proceedings of BioNLP, Beijing, China, pp. 164–170, 2015. [Google Scholar]
13. A. Jimeno-Yepes, A. MacKinlay, B. Han and Q. Chen, “Identifying diseases, drugs, and symptoms in Twitter,” Studies in Health Technology and Informatics, vol. 216, pp. 643–647, 2015. [Google Scholar]
14. M. A. Magumba, P. Nabende and E. Mwebaze, “Ontology boosted deep learning for disease name extraction from Twitter messages,” Journal of Big Data, vol. 5, no. 1, pp. 6, 2018. [Google Scholar]
15. C. Yao, Y. Qu, B. Jin, L. Guo, C. Li et al., “A convolutional neural network model for online medical guidance,” IEEE Access, vol. 4, pp. 4094–4103, 2016. [Google Scholar]
16. H. Yang and H. Gao, “Toward sustainable virtualized healthcare: extracting medical entities from Chinese online health consultations using deep neural networks,” Sustainability, vol. 10, no. 9, pp. 3292, 2018. [Google Scholar]
17. X. Liu, Y. Zhou and Z. Wang, “Recognition and extraction of named entities in online medical diagnosis data based on a deep neural network,” Journal of Visual Communication and Image Representation, vol. 60, no. 6, pp. 1–15, 2019. [Google Scholar]
18. Y. Luo, “Recurrent neural networks for classifying relations in clinical notes,” Journal of Biomedical Informatics, vol. 72, no. 1, pp. 85–95, 2017. [Google Scholar]
19. I. J. Unanue, E. Z. Borzeshi and M. Piccardi, “Recurrent neural networks with specialized word embeddings for health-domain named-entity recognition,” Journal of Biomedical Informatics, vol. 76, no. 5, pp. 102–109, 2017. [Google Scholar]
20. M. Cho, J. Ha, C. Park and S. Park, “Combinatorial feature embedding based on CNN and LSTM for biomedical named entity recognition,” Journal of Biomedical Informatics, vol. 103, no. 5, pp. 103381, 2020. [Google Scholar]
21. V. Mnih, N. Heess, A. Graves and K. kavukcuoglu, “Recurrent models of visual attention,” in Twenty-eighth Conf. on Neural Information Processing Systems (NIPS 2014Montreal, QC, Canada, pp. 2204–2212, 2014. [Google Scholar]
22. D. Bahdanau, K. Cho and Y. Bengio, “Neural machine translation by jointly learning to align and translate,” arXiv preprint arXiv: 1409. 0473, 2014. [Google Scholar]
23. A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones et al., “Attention is all you need,” in Thirty-first Conf. on Neural Information Processing Systems (NIPS 2017Long Beach, California, USA, pp. 5998–6008, 2017. [Google Scholar]
24. L. Li, J. Zhao, L. Hou, Y. Zhai, J. Shi et al., “An attention-based deep learning model for clinical named entity recognition of Chinese electronic medical records,” BMC Medical Informatics and Decision Making, vol. 19, no. 5, pp. 395, 2019. [Google Scholar]
25. B. Ji, R. Liu, S. Li, J. Yu, Q. Wu et al., “A hybrid approach for named entity recognition in Chinese electronic medical record,” BMC Medical Informatics and Decision Making, vol. 19, no. 2, pp. 64, 2019. [Google Scholar]
26. J. Yang, Y. Zhang, L. Li and X. Li, “YEDDA: A lightweight collaborative text span annotation tool,” in Meeting of the Association for Computational Linguistics, Melbourne, Australia, pp. 31–36, 2018. [Google Scholar]
27. T. Mikolov, I. Sutskever, K. Chen, G. S. Corrado and J. Dean, “Distributed representations of words and phrases and their compositionality,” in Twenty-seventh Conf. on Neural Information Processing Systems (NIPS 2013Lake Tahoe, Nevada, USA, pp. 3111–3119, 2013. [Google Scholar]
28. M. Schuster and K. K. Paliwal, “Bidirectional recurrent neural networks,” IEEE Transactions on Signal Processing, vol. 45, no. 11, pp. 2673–2681, 1997. [Google Scholar]
29. Y. Bengio, P. Simard and P. Frasconi, “Learning long-term dependencies with gradient descent is difficult,” IEEE Transactions on Neural Networks, vol. 5, no. 2, pp. 157–166, 1994. [Google Scholar]
30. L. Ding, L. Li, J. Han, Y. Fan and D. Hu, “Detecting domain generation algorithms with Bi-lLSTM,” Computers, Materials & Continua, vol. 61, no. 3, pp. 1285–1304, 2019. [Google Scholar]
31. Z. Bo, W. Haowen, L. Jiang, Y. Shuhan and L. Meizi, “A novel bidirectional lstm and attention mechanism based neural network for answer selection in community question answering,” Computers, Materials & Continua, vol. 62, no. 3, pp. 1273–1288, 2020. [Google Scholar]
32. Y. Shen, Y. Li, J. Sun, W. Ding, X. Shi et al., “Hashtag recommendation using lstm networks with self-attention,” Computers, Materials & Continua, vol. 61, no. 3, pp. 1261–1269, 2019. [Google Scholar]
33. S. Hassanpour and C. P. Langlotz, “Information extraction from multi-institutional radiology reports,” Artificial Intelligence in Medicine, vol. 66, no. 2, pp. 29–39, 2016. [Google Scholar]
34. I. Haapala, M. Karjalainen, A. Kontunen, A. Vehkaoja, K. Nordfors et al., “Identifying brain tumors by differential mobility spectrometry analysis of diathermy smoke,” Journal of Neurosurgery, vol. 133, no. 1, pp. 1–7, 2019. [Google Scholar]
35. Y. M. Chan, E. Y. K. Ng, V. Jahmunah, J. E. W. Koh, O. S. Lih et al., “Automated detection of glaucoma using optical coherence tomography angiogram images,” Computers in Biology and Medicine, vol. 115, no. 8, pp. 103483, 2019. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |