DOI:10.32604/iasc.2021.017898

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2021.017898 |  |
| Article |
COVID-19 Diagnosis Using Transfer-Learning Techniques
1College of Applied Computer Sciences, King Saud University, Riyadh, Saudi Arabia
2Turabah University College, Taif University, Taif, Saudi Arabia
3College of Computer and Information Science, King Saud University, Riyadh, Saudi Arabia
*Corresponding Author: Mohammed Faisal. Email: Mfaisal@ksu.edu.sa
Received: 16 February 2021; Accepted: 07 April 2021
Abstract: COVID-19 was first discovered in Wuhan, China, in December 2019 and has since spread worldwide. An automated and fast diagnosis system needs to be developed for early and effective COVID-19 diagnosis. Hence, we propose two- and three-classifier diagnosis systems for classifying COVID-19 cases using transfer-learning techniques. These systems can classify X-ray images into three categories: healthy, COVID-19, and pneumonia cases. We used two X-ray image datasets (DATASET-1 and DATASET-2) collected from state-of-the-art studies and train the systems using deep learning architectures, such as VGG-19, NASNet, and MobileNet2, on these datasets. According to the validation and testing results, our proposed diagnosis systems achieved excellent results with the VGG-19 architecture. The two-classifier diagnosis system achieved high sensitivity for COVID-19, with 99.5% and 100% on DATASET-1 and DATASET-2, respectively. The three-classifier diagnosis system achieves high sensitivity for COVID-19, with 98.4% and 100% on DATASET-1 and DATASET-2, respectively. The high sensitivity of these diagnostic systems for COVID-19 will significantly improve the speed and precision of COVID-19 diagnosis.
Keywords: Covid-19; deep learning; diagnosis system; VGG-19; NASNet; MobileNet2
Since preventive or experimental vaccine therapy for extreme acute respiratory syndrome coronavirus (COVID-19) is not available, its early detection is of paramount importance in enabling infected persons to gain rapid immunity and to reduce the risk of infection for the healthier population. Key diagnosis methods for COVID-19 are reverse transcription-polymerase chain (RT-PCR) and gene sequencing of respiratory or blood samples [1]. However, an overall positive RT-PCR average of 30%–60% is obtained by analyzing throat swab samples, which results in having undiagnosed patients with COVID-19 who may contagiously infect a large, healthy population [2]. Given the high prevalence of COVID-19 and the shortage of qualified radiologists, automatic methods for detecting COVID-19 can assist the diagnostic process and improve high-precision diagnosis at an early stage. Artificial intelligence (AI) and machine learning (ML) techniques are effective tools that can be used to develop methods for the early diagnosis of COVID-19. In this regard, using chest X-ray images, we use an end-to-end deep learning (DL) framework to classify COVID-19. Unlike traditional AI/ML techniques that use a two-stepped process (i.e., the manual extraction of features, which is followed by image recognition) for classifying medical images, we have developed DL-based systems that explicitly predict COVID-19 from raw images without requiring feature extraction. Recently, in most machine vision and medical image processing activities, deep-level learning models, specifically convolution neural networks (CNNs), have outperformed conventional AI models and have been used for several tasks, including image grouping, image segmentation, facial recognition, super-resolution, and image improvement [3–5]. In this study, we train three CNNs, including VGG-19, MobileNetV2, and NASNet, which have achieved promising results in several tasks, and we evaluate their success on COVID-19 X-ray-datasets for the detection of COVID-19.
The application of sophisticated AI techniques combined with radiological imaging can help to reliably diagnose COVID-19 and compensate for the shortage of trained physicians in remote areas. DL models have been widely used for many tasks, including classification. In the detection of patient details through image segmentation and lesion, the time between the onset of initial symptoms and the imaging test may be an important factor in the reliability of X-ray results. Although X-rays showed no symptoms of the disease within the first three days of the onset of coughing and fever, these symptoms were more evident in the next 10 to 12 days after the initial 3 days [6]. Consensus guidelines for imaging for children with COVID-19 [7] are now in place. According to these guidelines, if an infant is suspected of developing COVID-19 and shows mild to serious signs of an acute respiratory disorder, X-ray tests should be conducted. Repeated X-ray tests may be necessary to track the progression of the disease if original chest X-ray images yield concrete symptoms of COVID-19, and this will also be warranted, if a patient’s state of health deteriorates. With the global prevalence of the COVID-19 pandemic, chest X-ray radiographs for healthcare services should be considered as a valuable method for identifying COVID-19 [8]. The above findings indicate that DL with X-ray imaging can provide significant COVID-19 diagnosis results.
The rapid identification of confirmed COVID-19 cases at an early stage is necessary for quarantine and medical care, as well as the initial diagnosis, management, and public health safety of patients. However, with current conditions, medical staff, especially radiologists, are under tremendous pressure due to the large number of suspected patients having to go through CT scanning; in addition, the lack of scanning devices will increase the chance of failure to detect minor lesions due to visual exhaustion of radiologists. As the primary emerging AI technology used for medical imaging, ML has been effective in the automated detection of lung diseases [9–11]. An image classification project achieved human-level success with a further one million training photographs in 2015 [12] and achieved genuinely exciting results on lung cancer testing in 2019 [13]. Most ML techniques for diagnosing diseases enable lesions to be observed, particularly for identifying CT-related diseases.
The motivations of this research are (1) to contribute to overcoming the COVID-19 pandemic; (2) to introduce an automated and fast COVID-19 diagnosis system as a simple alternative diagnosis technique to mitigate the spread of COVID-19; (3) to enhance the accuracy of existing diagnosis systems (see Section 7, Comparison and Discussions) by introducing an accurate COVID-19 diagnosis system. Therefore, in this study, we aim to introduce an accurate COVID-19 diagnosis system using state-of-the-art CNN architectures and transfer-learning (TL) techniques. In this study, we investigated two TL scenarios. In the first scenario, we froze pretrained layers in a VGG-19 model, except for the last four layers; in the second scenario, we fine-tuned all layers of the Mobilenetv2 and NASNet models.
The rest of this paper is structured as follows. Section 2 presents the literature review. Section 3 presents the proposed methodology, with a description of the selected CNN architecture. Section 4 describes the two datasets used in this study, and Section 5 proposes two COVID-19 diagnosis systems; Section 6 describes the results of the two proposed diagnosis systems in different metrics; Section 7 presents a comparison and discussion of the proposed diagnosis systems and reference studies, and Section 8 provides the conclusion.
Currently, there is an extreme shortage of radiologists, and radiologists are exhausted by the large number of COVID-19 casualties and cases due to the rapid spread of COVID-19. In this situation, a poorly controlled diagnosis of COVID-19 is possible and can be harmful. A patient’s temperature level is one of the easiest detection markers of the disease. COVID-19 belongs to the 2B beta-coV category and has at least 70% resemblance to the genetic sequence of SARS-CoV [13], and it is the 7th RNA coronavirus family member to have infected humans [14]. COVID-19 infection symptoms include respiratory signs, fever, cough, and pneumonia [15]. Its diagnosis has become a major concern in hospitals due to the lack of nucleic acid detection boxes. CT and radiology have proven effective for the early detection and diagnosis of COVID-19 [16–18], but due to the insufficient number of radiologists [18], specialized computer-aided lung CT diagnosis systems are required to reliably validate suspected COVID-19 cases, scan patients, and conduct virus supervision.
DL is a common AI research field that enables the development of end-to-end models to achieve promising results with input data, without the need for manual extraction of features [19]. There are several recent studies on COVID-19 where they employ various DL models with X-ray images for COVID-19 detection [20]. They used a DL-based model with a total of 16,756 X-ray images with a multiclass classification (three classes) and a proposed dedicated dataset of COVID-19 X-ray images named COVIDx [21]. Narin et al. [22] proposed a support vector machine (SVM) model that classified characteristics obtained from various CNN models using X-ray images (25 COVID-19 positive and 25 healthy patients). The study claims that ResNet50 with the SVM classifier produces great results. Apostolopoulos et al. [23] used three different CNN models (ResNet50, InceptionV3, and InceptionResNetV2) using 50 open access COVID-19 X-ray images from Joseph Cohen and 50 typical images from a Kaggle repository. El-Din Hemdan et al. [24] deployed DL models to diagnose patients with COVID-19 using chest X-rays. It proposed a COVIDx-Net model that includes seven CNN models with 50 Chest X-Ray images (25 COVID-19 positives, 25 healthy). Ozturk et al. [25] proposed a model based on the DarkNet method that is completely automated with an end-to-end structure without the need for manual feature extraction. The authors used 1,125 images (125 COVID-19 positives, 500 Pneumonia images, and 500 NoFindings images) to experiment with their developed model. In 2020, Al-Waisy et al. [26] proposed a hybrid COVID-19 detection system called COVIDCheXNet. They used the contrast-limited adaptive histogram equalization to enhance the data instead of fussing the results of two pretrained DL models (ResNet34 and high-resolution network model). COVIDCheXNet system achieved an accuracy of 99.99% and a sensitivity of 99.98%. In 2021, Al-Waisy et al. [27] proposed a hybrid COVID-19 detection system called COVID-DeepNet system based on ML. The proposed COVID-DeepNet system was used to diagnose patients into two classes (COVID-19 and healthy). In 2021, Abdulkareem et al. [28] proposed a model based on ML techniques (Naive Bayes, random forest, and SVM) and the Internet of Things (IoT) to diagnose patients with COVID-19 in smart hospitals. The proposed system achieved an accuracy of 95% with the SVM model. Ismael et al. [29] used several DL approaches to classify COVID-19 using healthy chest X-ray images. For feature extraction, they used an SVM classifier with linear, quadratic, cubic, and Gaussian functions, and for fine-tuning procedures, they used ResNet18, ResNet50, ResNet101, VGG-16, and VGG-19. They used a dataset containing 180 COVID-19 and 200 healthy chest X-ray images. The proposed system achieved a maximum accuracy of 94.7% with the ResNet50 model and SVM classifier with the linear kernel function. Jain et al. [30] used DL-based CNN models ( Inception V3, Xception, and ResNeXt) to detect the COVID-19 on chest X-ray images. They used a dataset of 6,432 chest X-ray image samples, and 5,467 samples were used for training and 965 for validation. The Xception model achieved the highest accuracy of 97.97%. Tab. 1 illustrates several COVID-19 classification methods, the accuracy achieved, and the size of the dataset used for their training and validation.
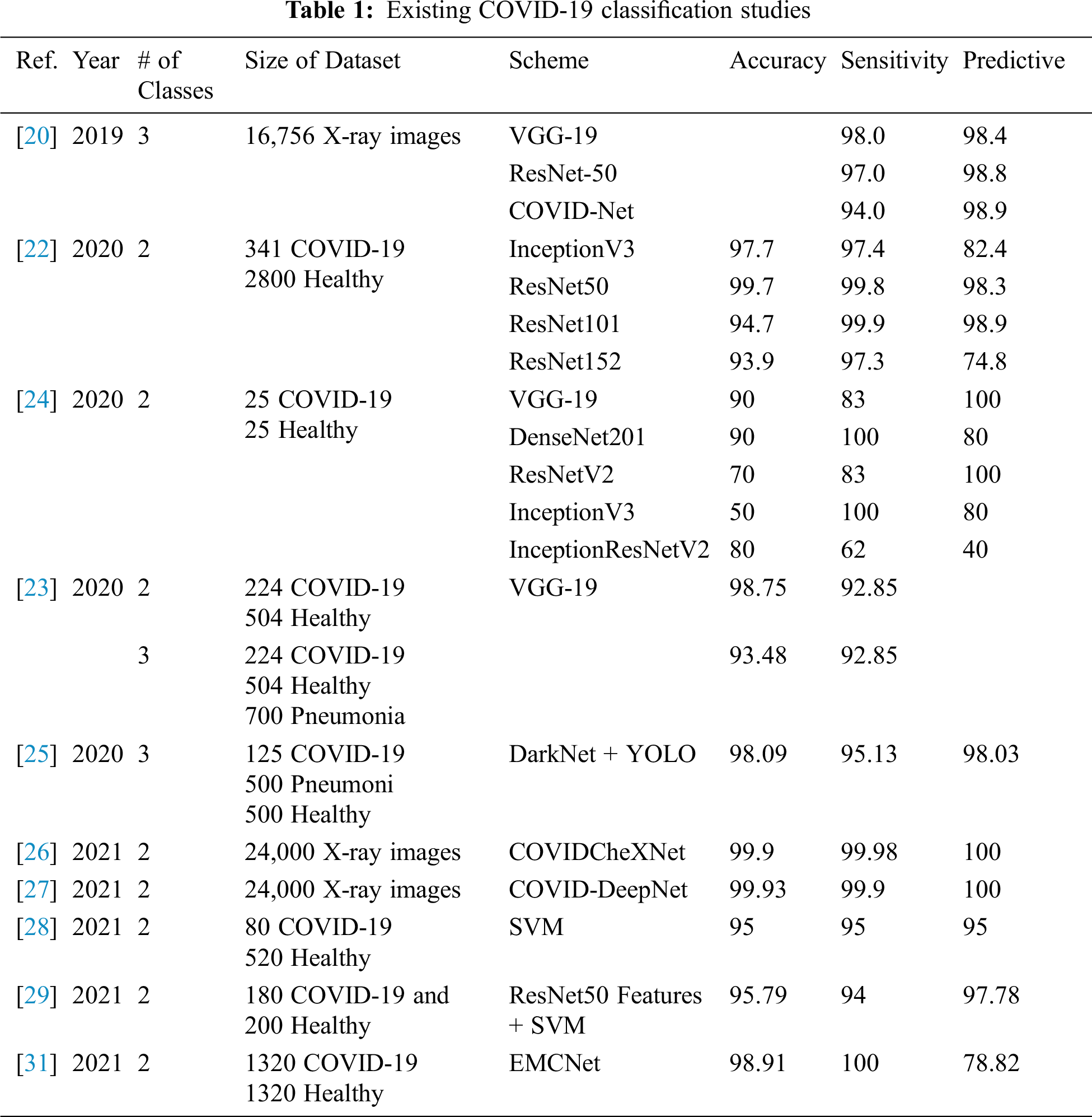
As can be seen in Tab. 1, the existing COVID-19 diagnosis systems have many disadvantages such as only using two classes for classification, having limited positive cases, or achieving low accuracy and sensitivity.
Usually, deep neural networks with a larger dataset perform better than those with smaller datasets. For applications where the dataset is not large, TL can be effective. The TL concept uses a well-trained model from large datasets, such as ImageNet, and applies it with comparatively small datasets. This eliminates the need for large datasets, thus decreasing the amount of training that the DL algorithm needs when built from scratch.
In this study, our system is trained, evaluated, and tested with three well-known CNNs—VGG-19 [32], NASNet [33], and MobileNet2 [34]. VGG is developed with minimum preprocessing to identify graphic patterns from pixel images. The ImageNet project has been configured for the detection of visual objects. A VGG network is characterized by its simplicity and is build using only 3 × 3 convolutional layers stacked on top of each other in increasing depth. The reducing volume size is handled by max-pooling. Two fully connected layers, each with 4,096 nodes, are then followed by a softmax classifier. NASNet is a google ML model that generates small neural networks. Google first revealed in May 2017 that its AutoML project represents a significant step forward in the use of state-of-the-art ML models to identify images. The accuracy of the validation system is 82.7%, which is an improvement on all previous models created by the team.
MobileNetV2 is a 53-layer profound CNN in which a retrained network trained on more than one million images from the ImageNet dataset can be loaded. Consequently, the network represents a wide variety of images. The image input size in the network is 224 × 224.
In this study, we investigated two scenarios of TL. In the first scenario, the pretrained layers in the VGG-19 model, except for the last 4 layers, are frozen; in the second scenario, all layers of Mobilenetv2 and NASNet models are fine-tuned. In VGG-19, we freeze all layers except for the last 4 layers. Then, we added the following layers:
• Global average pooling
• Dropout (0.3)
• Dense (128)
• Dense (64)
• Softmax (2/3 classes)
In Mobilenetv2/NASNet, we did not freeze any layer from the pretrained model. Then, we added the following layers:
• Global average pooling
• Dense (1024)
• Batch Healthyization
• Dense (1024)
• Softmax (2/3 classes)
We used two datasets, which comprise a collection of healthy, pneumonia, and COVID-19 infection X-ray images from a state-of-the-art study [23,35]. The following three datasets were used to train our system with the pretrained CNN models: VGG-19, NASNet, and MobileNetV2. We compared the performance of our system with those of the studies from which these datasets are obtained.
DATASET-1 [35] comprises post-to-antheral chest X-ray (CXR) images as this vision is commonly used in pneumonia diagnosis by radiologists. DATASET-1 comprises four subdatasets; two subdatasets were constructed by the authors, and the other two were collected from two repositories, Kaggle and GitHub, which are publicly accessible. The first subdataset is collected from the “Italian Society of Medical and Interventional Radiology (SIRM) COVID-19 DATABASE” and comprises 330 positive COVID-19 radiography CT and CXR images with different resolutions, where 70 images are CXR and 250 images are CT images of the lung. The second subdataset is collected from “Novel Corona Virus 2019 Dataset” and comprises 179 radiography images of COVID-19, Middle East respiratory syndrome, severe acute respiratory syndrome, and ARDS from written papers and web resources, created by Lan Dao, Joseph Paul Cohen, and Paul Morrison in GitHub46. The third subdataset is collected from “COVID-19 positive CXR images from different articles.” Datasets collected from GitHub have motivated researchers to study the literature, and as a result, in less than two months, more than 1200 articles were published. The fourth subdataset is a Kaggle CXR dataset, comprising 5247 CXR images with resolutions ranging from 400 to 2,000 pixels for regular, viral, and bacterial pneumonia. Of these, 3,906 are pneumonia-affected pictures from multiple subjects (25,561 bacterial pneumonia pictures and 1,345 virus pneumonia pictures) and 1,341 are from healthy subjects. Fig. 1 illustrates some samples of DATASET-2.
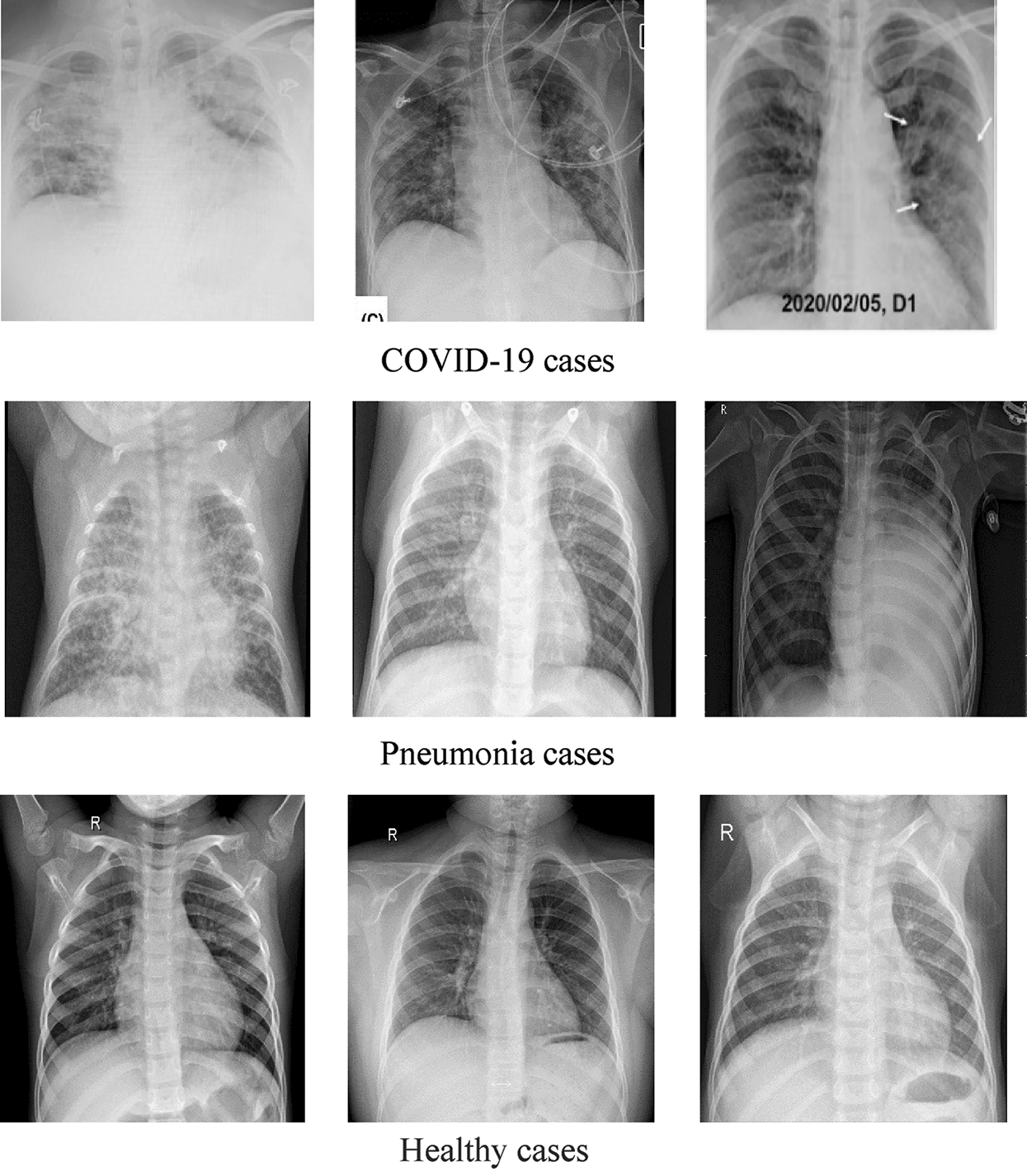
Figure 1: Sample X-ray images from DATASET-1 showing COVID-19, viral pneumonia, and healthy cases
DATASET-2 [23] comprises three subdatasets. For the first subdataset, GitHub was explored for similar datasets. The authors took a set of X-ray images from Cohen [36]. For the second subdataset, the North American Radiological Society, Radiopaedia, and SIRM were closely examined. These sets can be found online [37]. The third subdataset was complemented with a series of common X-ray bacterial-pneumonia scans to enable the CNNs to differentiate COVID-19 from common pneumonia. Kermany et al. [38] made this collection available on the Internet. A total of 700 identified severe pneumonia images, 224 identified COVID-19 images, and 504 healthy condition images were included in DATASET-2 [23]. Fig. 2 illustrates some samples of DATASET-2.
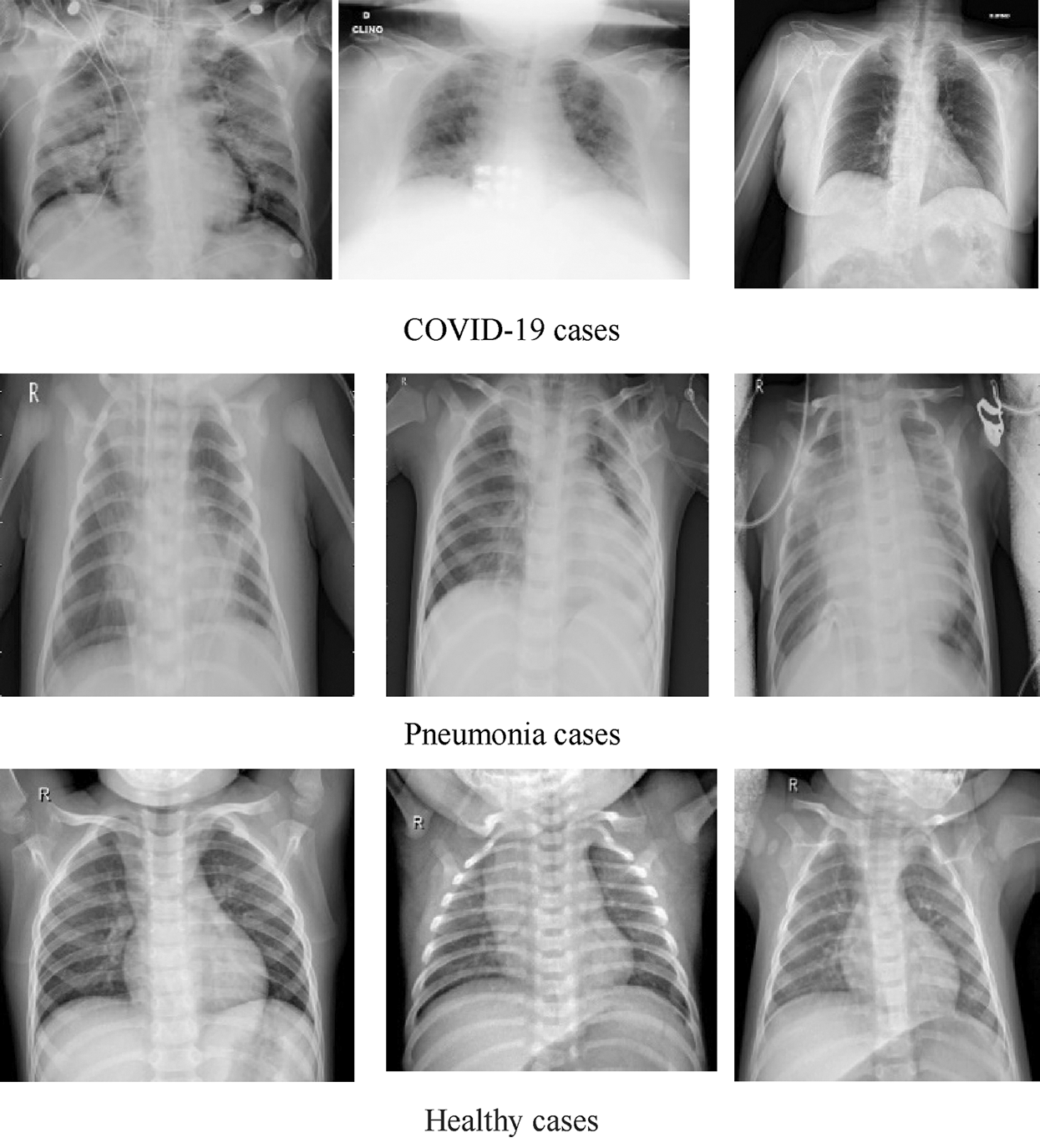
Figure 2: Sample X-ray image from DATASET-2 shows COVID-19, viral pneumonia, and healthy cases
This paper proposes diagnosis systems that will classify COVID-19 infection into two and three classes using DL techniques. The two-classifier system can classify X-ray images into two classes, i.e., healthy and COVID-19 positive cases. The three-classifier system can classify X-ray images into three classes, i.e., healthy, pneumonia, and COVID-19 positive cases.
The two-classifier diagnosis system uses an end-to-end DL framework to classify COVID-19 from the CXR images. Compared to traditional techniques for classifying medical images, which use a two-step process (manual extraction of features and image recognition), our two-classifier ML system explicitly predicts COVID-19 from raw images, without requiring feature extraction, and classifies X-ray images into two categories: COVID-19 and healthy cases. As illustrated in Fig. 3, we started by collecting the image data (thousands of X-ray images) of patients having COVID-19 and healthy persons. Then, we classified the images (dataset) into two types (COVID-19 and healthy cases) and augmented the images by resizing them according to the standard size of their respective CNN models. After that, we divided the dataset into training and test sets and applied the pretrained CNN models (VGG 19, NASNet, and MobileNetV2 ) to generate the diagnosis, i.e., detect whether a patient is a COVID-19 patient or a healthy person.
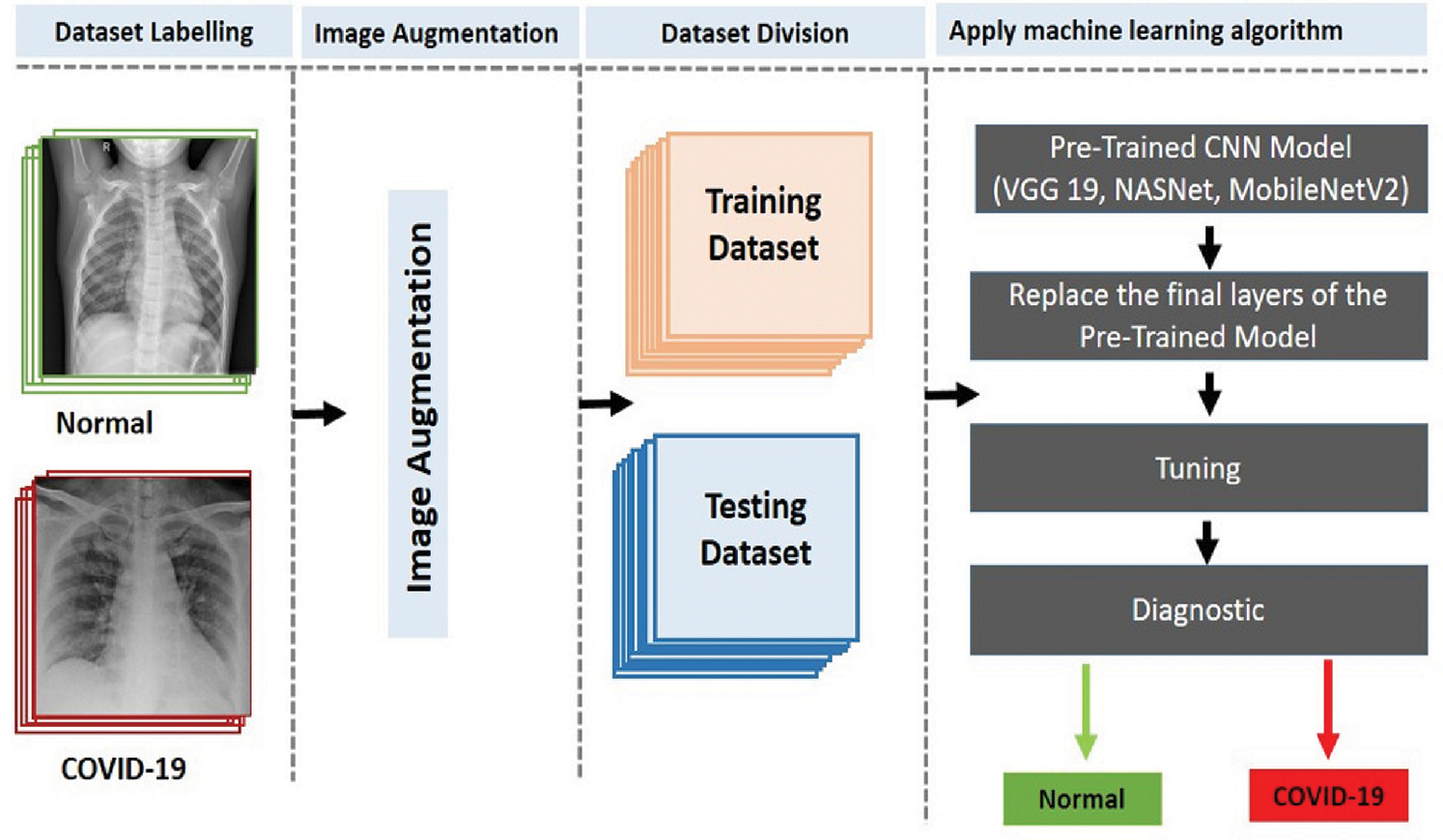
Figure 3: Proposed two-classifier diagnosis system
The three-classifier diagnosis system can classify CXR images into three classes, i.e., COVID-19, pneumonia, or healthy cases, as illustrated in Fig. 4, using the pretrained CNN models (VGG 19, NASNet, and MobileNetV2).
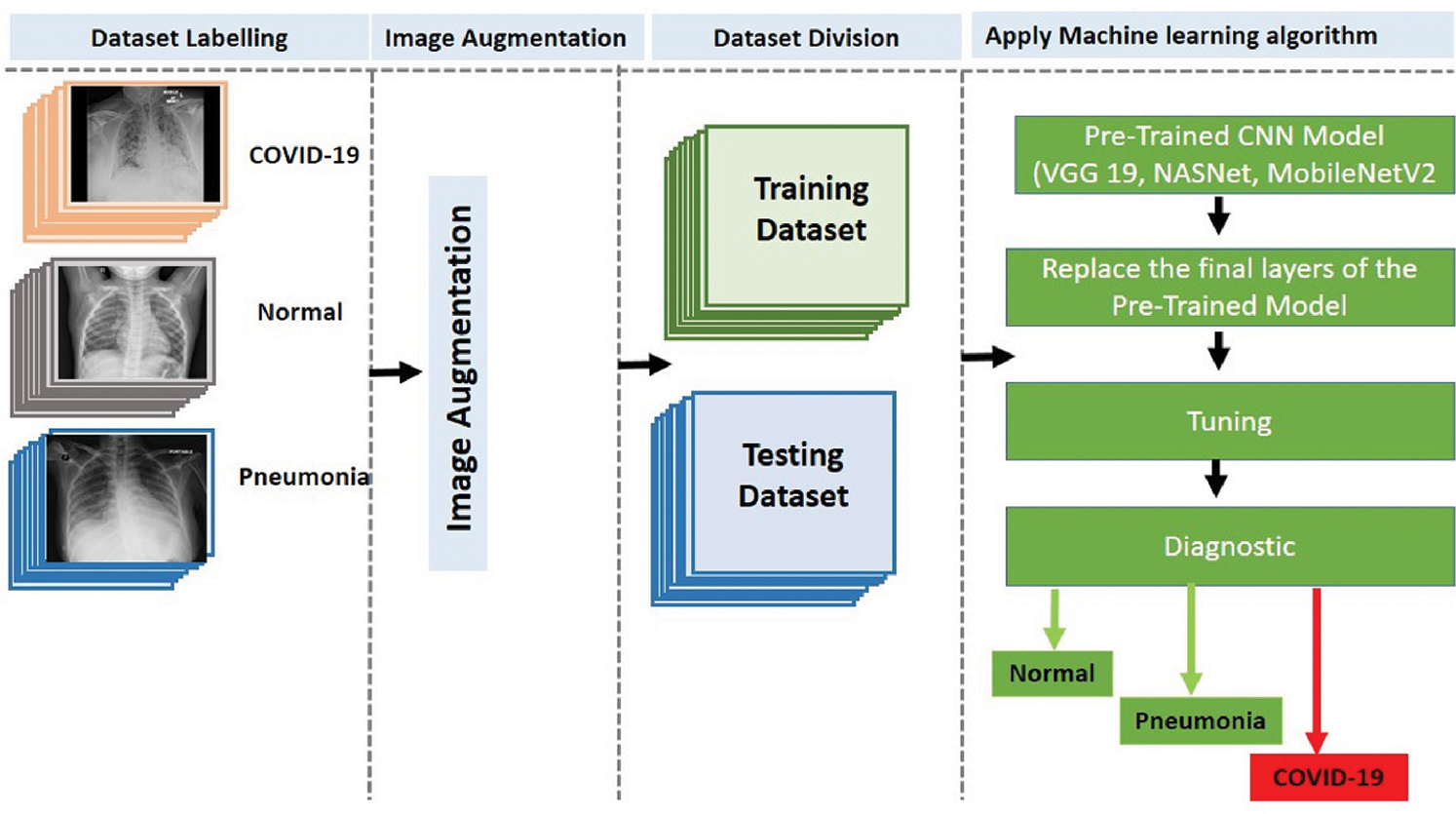
Figure 4: Proposed three-classifier diagnosis system
In this study, three well-known pretrained DL CNNs––VGG-19, NASNet, and MobileNet2––were retrained, evaluated, and tested using the KERAS framework to classify the X-ray images. The training of the models was conducted on a machine equipped with the Intel © i9-9880H core @2.3 GHz processor, 16 GB RAM, 2 GB graphics card, and 64-bit Windows 10 as the operating system. We used the ImageData Generator with the following parameters (rotation range = 40, width shift range = 0.2, height shift range = 0.2, shear range = 0.2, and zoom range = 0.2) for augmentation. In addition, we resized all images to 224 × 224 pixels to meet the requirement of the pretrained models. Furthermore, we used five-fold cross-validation for training and testing with the following training parameters: batch size = 16, the number of epochs = 30, and ADAM optimizer with the learning rate = 0.0001.
We evaluated the CNN architectures using two public datasets: DATASET-1 [35] and DATASET-2 [23]. For each dataset, we tested the three CNN models for the two- and three-classifier scenarios. Different well-known metrics such as the accuracy, F1 score, precision, recall, and confusion matrix were used to evaluate the models and compare them with other obtained results. The following specific metrics were documented in the classification work of the CNNs: (a) accurately categorized disease cases (true positive, TP), (b) falsely categorized disease cases (false positive, FP), (c) correctly identified healthy cases (true negatives, TN), and (d) incorrectly classified healthy cases (false negative, FN). TP = the correctly diagnosed COVID-19 cases, FP = pneumonia or healthy cases that were diagnosed as COVID-19, TN = pneumonia or healthy cases that were diagnosed as non-COVID-19 cases, and FN = the COVID-19 cases that were diagnosed as non-COVID-19. We calculated the performance metrics using the following equations (references):
Furthermore, we used five-fold cross-validation for all experiments and took the overall average of all results. To test the models (GG-19, NASNet, and MobileNet2), we tested the trained models on a new test dataset (20% of the total dataset) and generated results using the same configuration and parameters that were used in training and validating the models.
Tab. 2 summarizes the performance of the CNN models tested on the two different datasets. For DATASET-1, VGG-19 outperformed the other models with the following performance metrics: 99.5% accuracy, 99.2% F1 score, 97.6% sensitivity for COVID-19 cases, and 1% sensitivity for healthy cases.
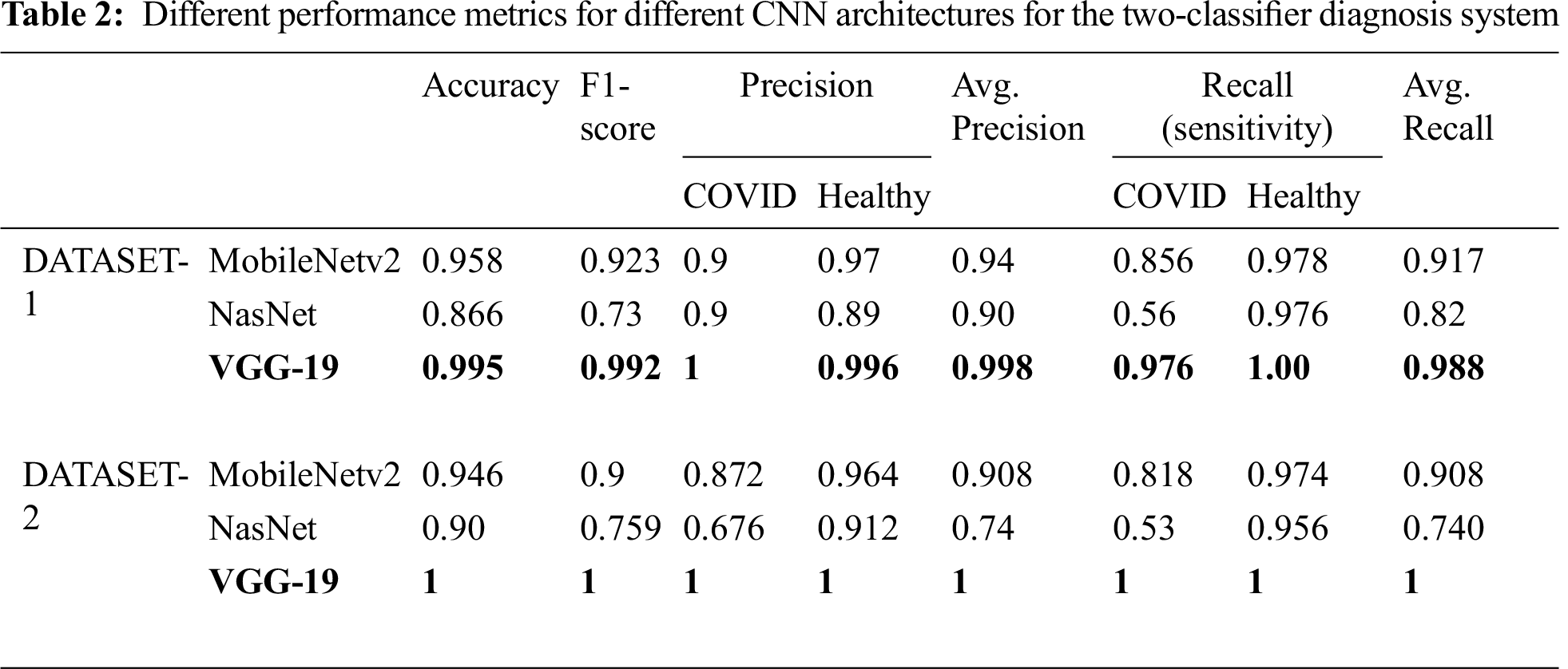
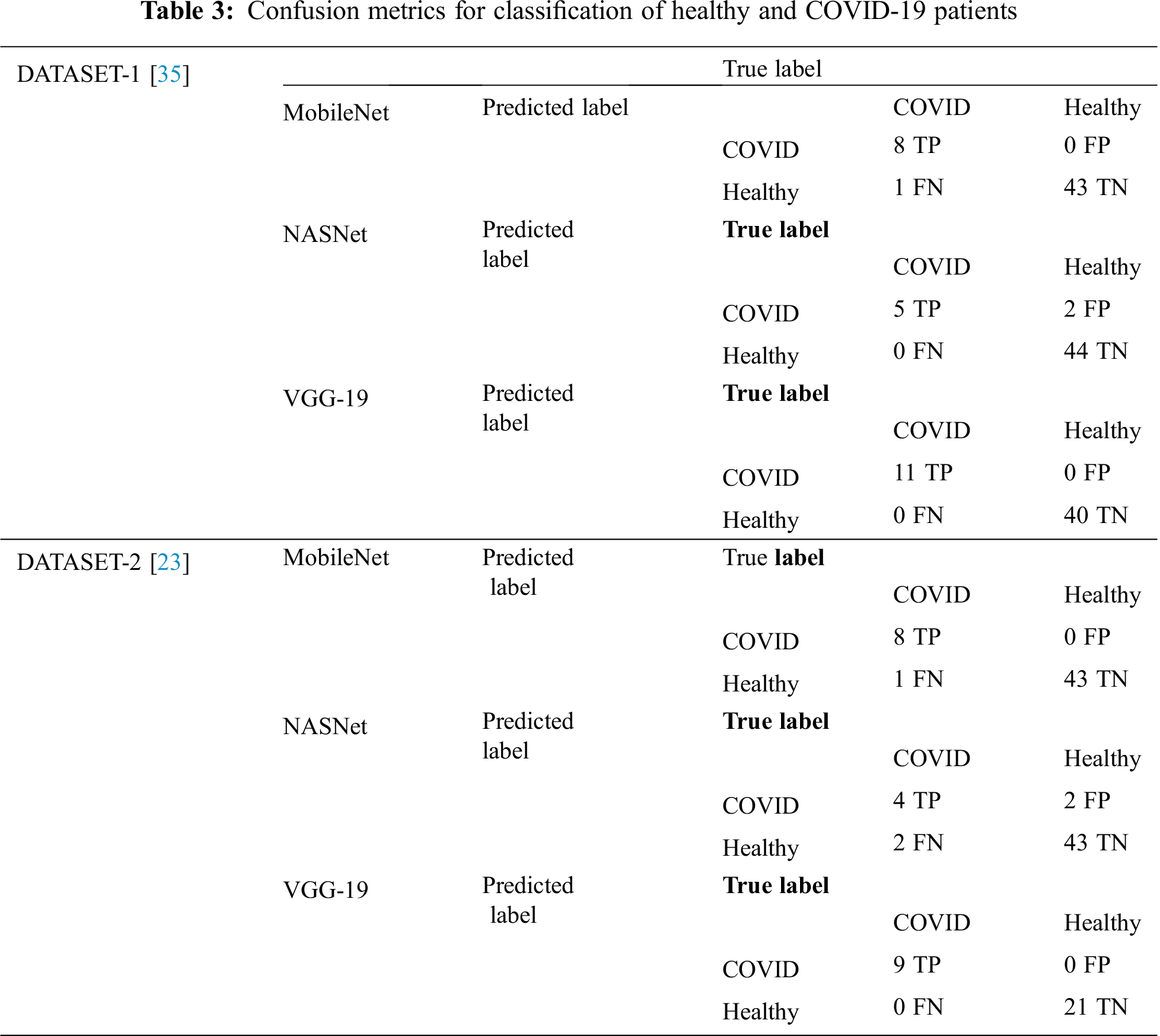
In DATASET-2, we achieved a similar result, i.e., VGG-19 outperformed the other models with 1% accuracy, 1% F1 score, 1% sensitivity for COVID-19 cases, and 1% sensitivity for healthy cases, as shown in Tab. 3, which also shows the confusion matrix for MobileNet, NASNet, and VGG 19 for the best fold for the two-classifier system, on both datasets.
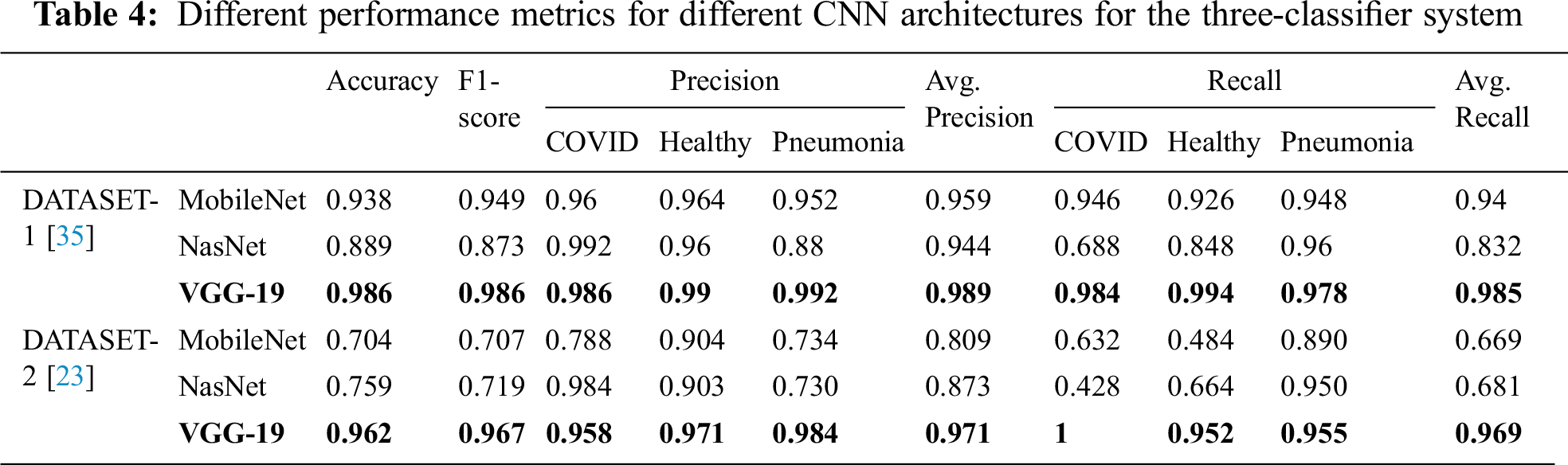
Tab. 4 encapsulates the performance metrics for the different CNN architectures tested for the three-classifier system. VGG-19 outperforms the other models with the following performance metrics in DATASET-1: 98.6% accuracy, 98.6% F1 score, 98.4% sensitivity for COVID cases, 99% for healthy cases, and 99.2% sensitivity for pneumonia cases. In DATASET-2, VGG-19 achieved excellent performance with 96.2% accuracy, 96.7% F1 score, 95.8% sensitivity for COVID cases, 97.1% sensitivity for healthy cases, and 98.4% sensitivity for pneumonia cases, as shown in Tab. 5.
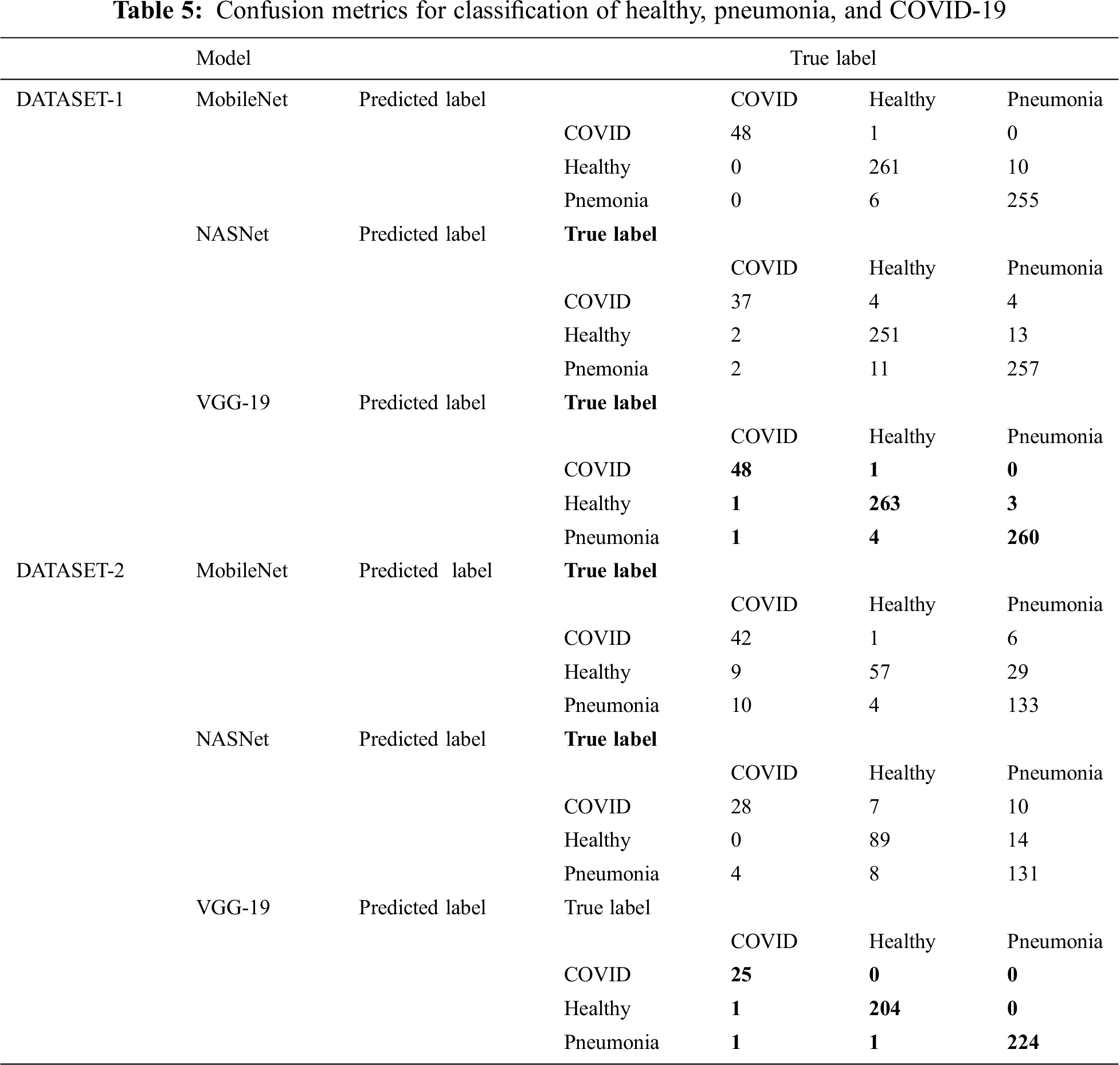
Tab. 5 shows the confusion matrix for MobileNet, NASNet, and VGG 19 for the best fold for the three-classifier system for the two datasets.
We performed five-fold cross-validation with 30 epochs for both classifier systems for the VGG-19, MobileNet, and NASNet models and took the overall average of the results. Figs. 5–8 illustrate the learning performance accuracy of VGG-19 in single-fold cross-validation, with 30 epochs of the two-classifier and three-classifier systems for DATASET-1 [35] and DATASET-2 [23].
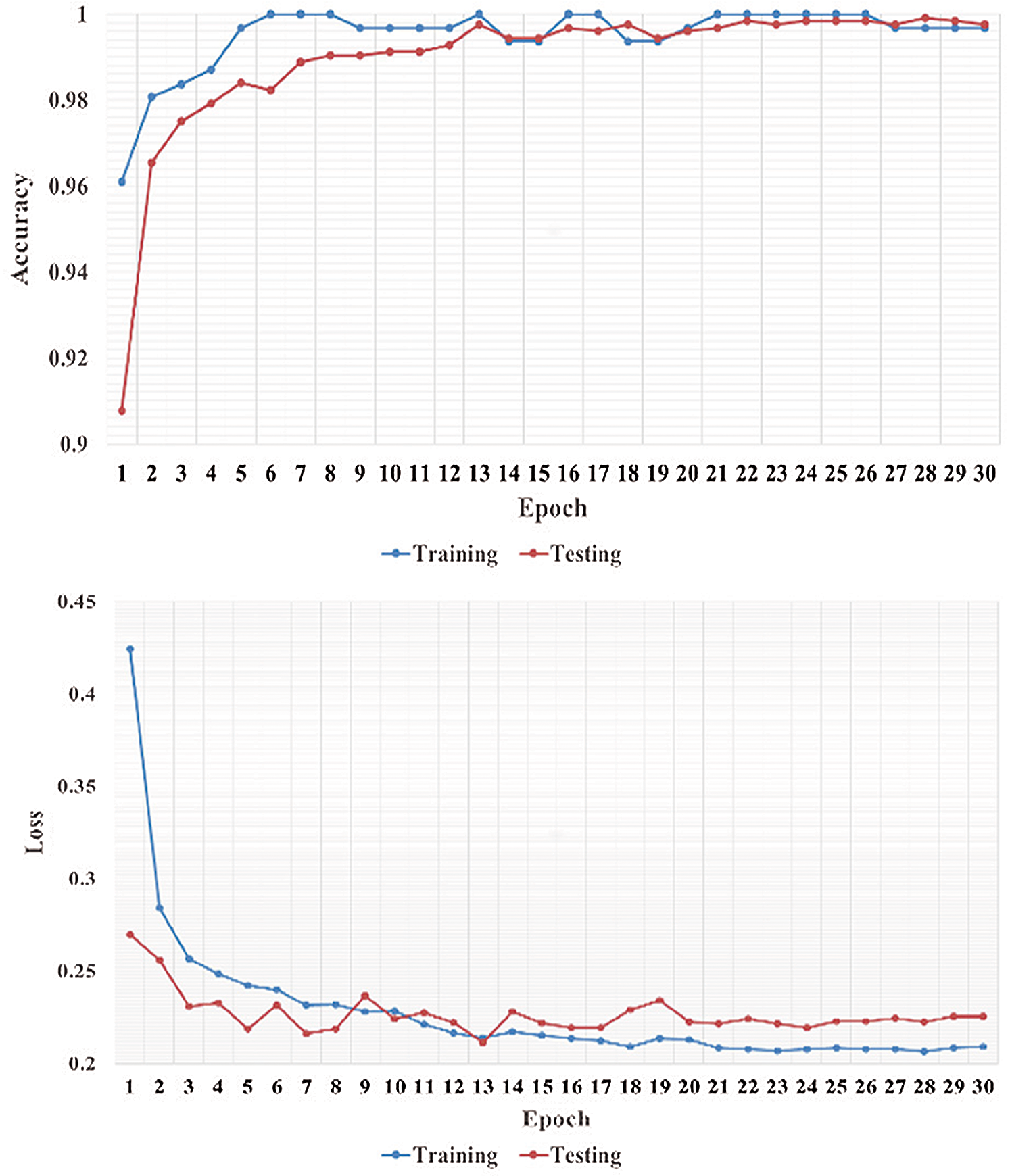
Figure 5: Learning performance accuracy and training and validation learning curves of VGG-19 for the two-Classifier system in one fold with 30 epochs for DATASET-1 [35]
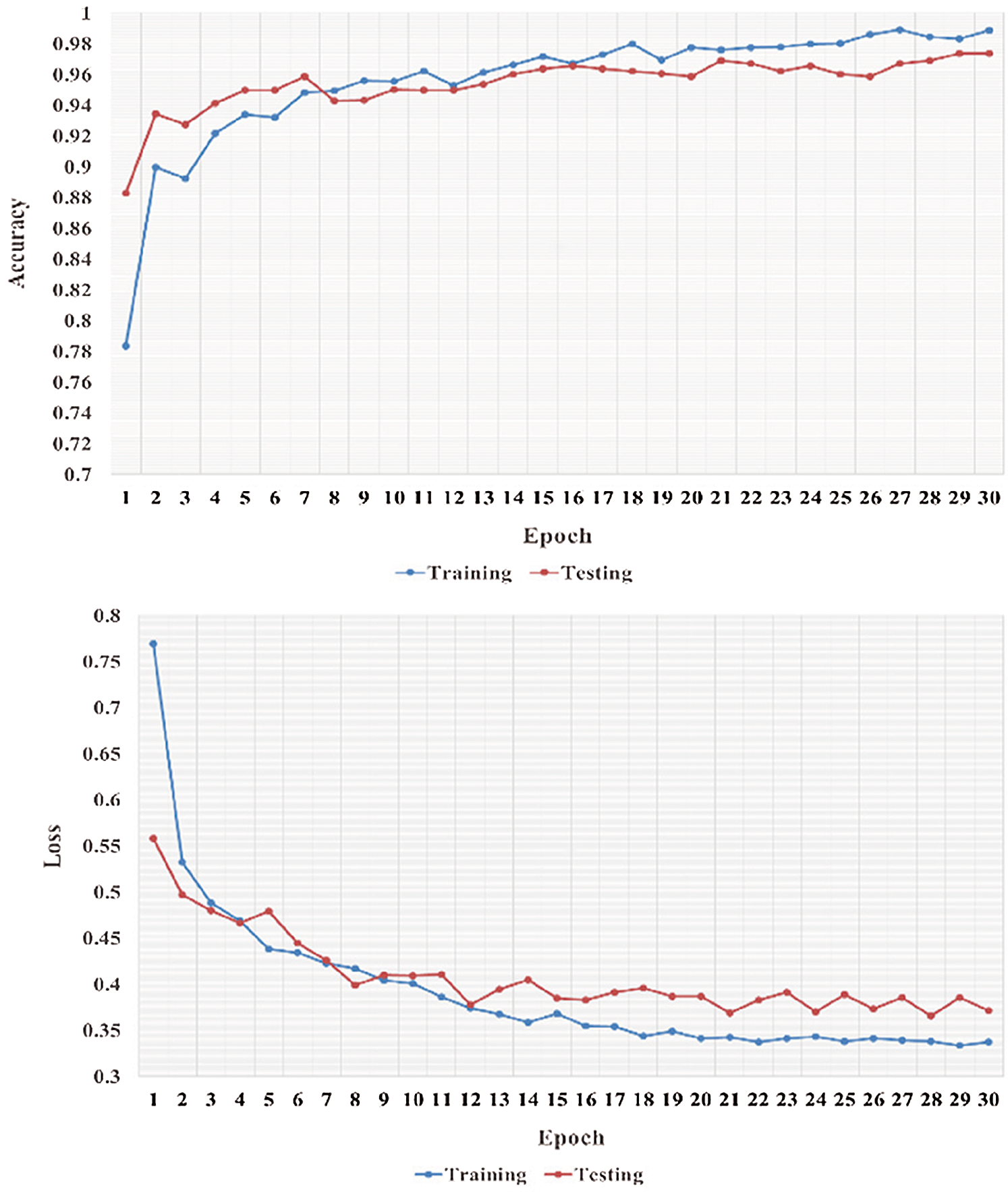
Figure 6: Learning performance accuracy and training and validation learning curves of VGG-19 for the tree-Classifier system in one fold with 30 epochs for DATASET-1 [35]
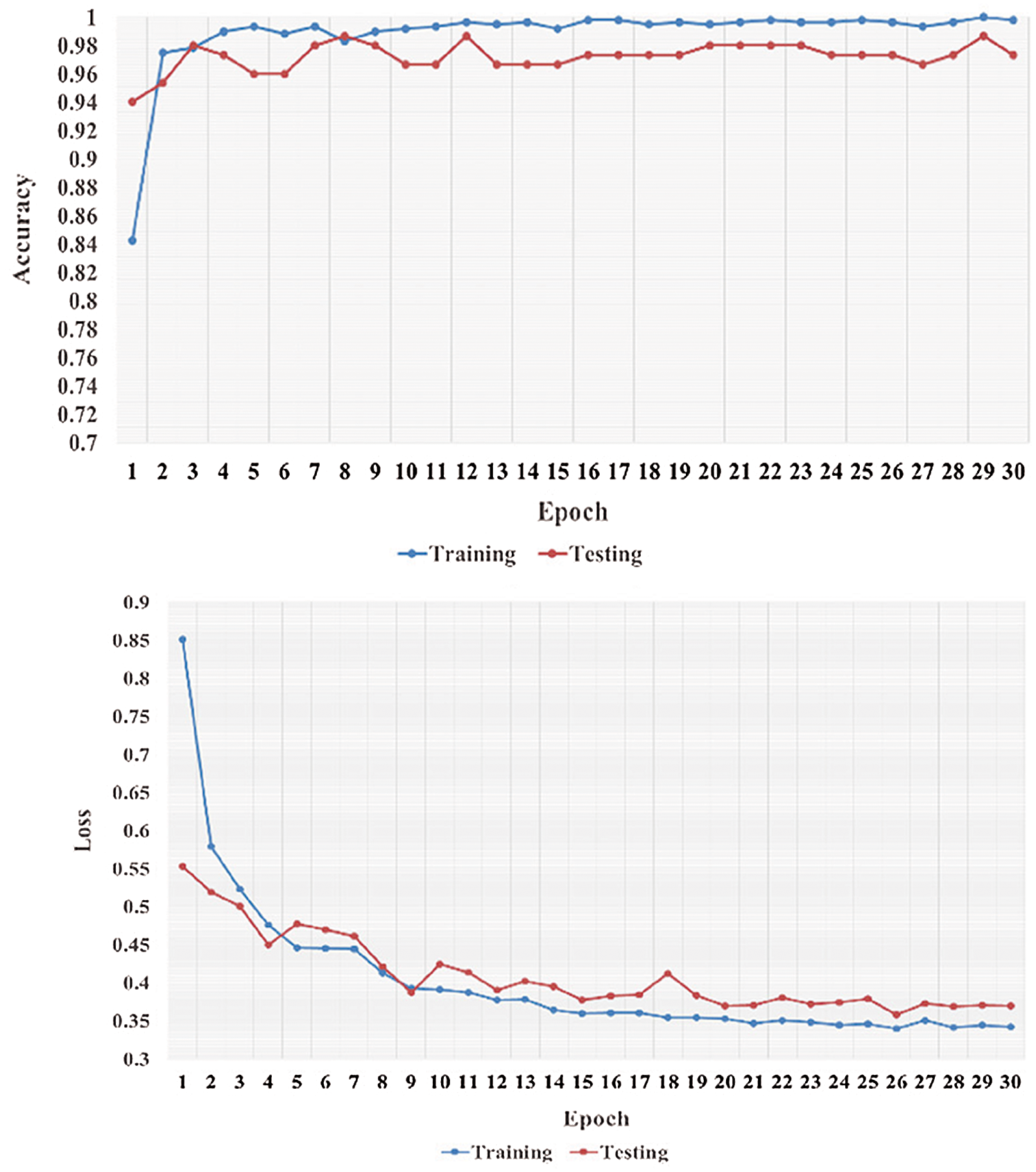
Figure 7: Learning performance accuracy and training and validation learning curves of VGG-19 for the two-Classifier system in one fold with 30 epochs for DATASET-2 [23]
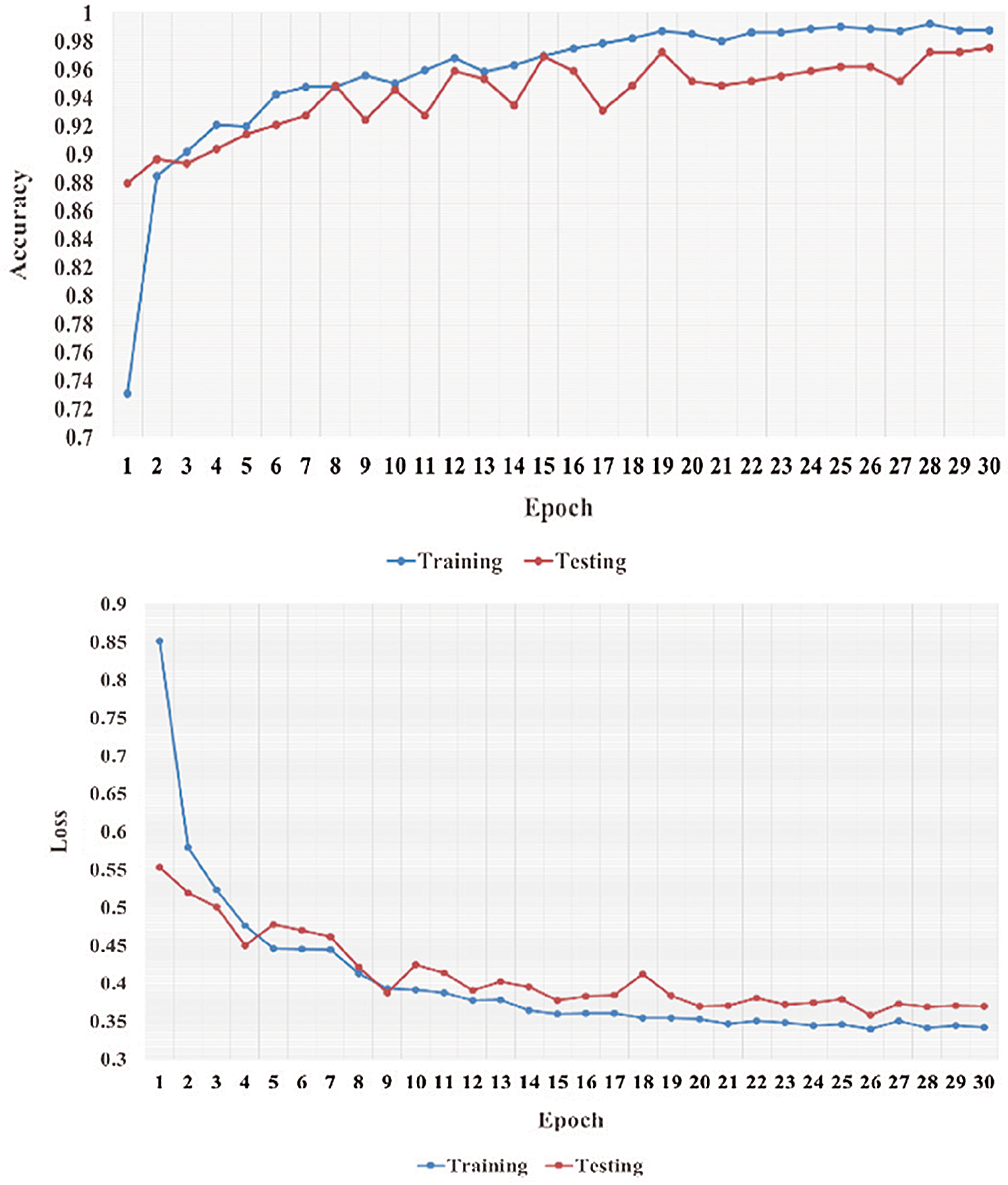
Figure 8: Learning performance accuracy and training and validation learning curves of VGG-19 for the three-Classifier system in one fold with 30 epochs for DATASET-2 [23]
In Figs. 5–8, training and validation accuracies show that there is no overfitting; for example, VGG-19 has an excellent fit and stable performance. The training and validation loss decreased to the point of stability with a minimal gap between the two final loss values in all folds.
In this section, we compare the performance metrics of the models in our study with those of the reference studies that used the same datasets (DATASET-1 and DATASET-2).
7.1 Two-Classifier System (DATASET-1)
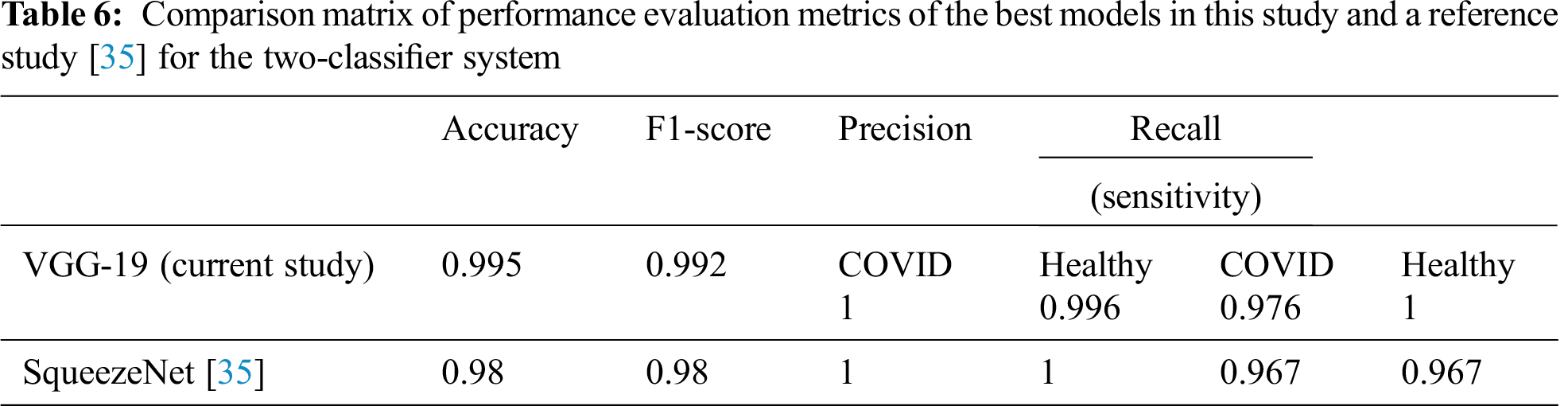
Tab. 6 summarizes the performance evaluation comparison matrix of the best results of the models in this study and the reference study [35] for the two-classifier system. In this study, VGG-19 outperforms and shows outstanding results with different performance metrics on DATASET-1: 99.5% accuracy, 99.2% F1 score, 100% sensitivity for COVID cases, and 99.6% sensitivity for healthy cases. In contrast, in the reference study [35], four CNN models were used to train their system, where SqueezeNet shows the best results with different performance metrics on DATASET-1: 98.3% accuracy, 98.3% F1 score, 96.7% sensitivity for COVID cases, and 96.7% sensitivity for healthy cases.
7.2 Three-Classifier System (DATASET-1)
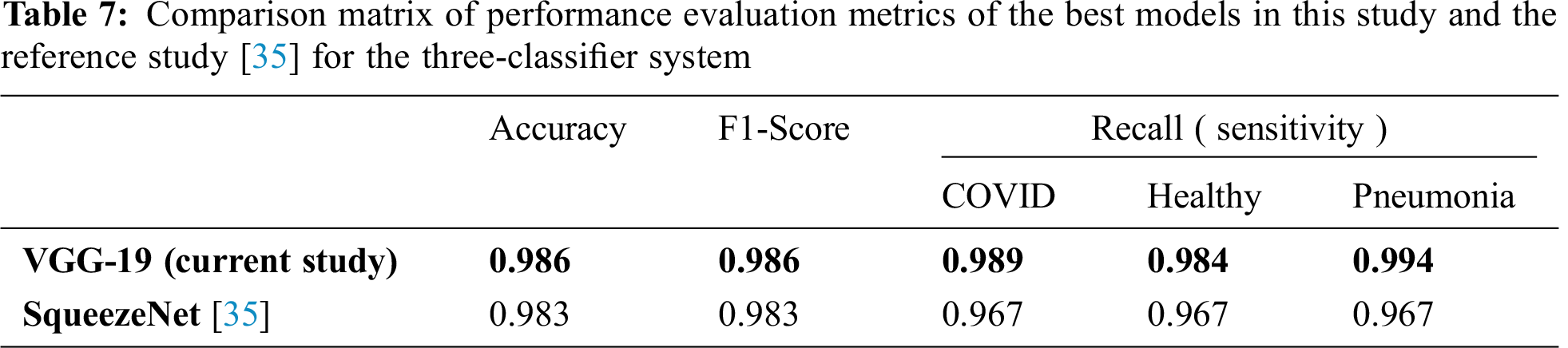
Tab. 7 compares the performance evaluation metrics of the best models in this study and the reference study [35] for the three-classifier system. In this study, VGG-19 outperformed and showed outstanding results with different performance metrics on DATASET-1; it showed 98.6% accuracy, 98.6% F1 score, 98.9% sensitivity for COVID cases, 98.4% sensitivity for healthy cases, and 99.4% for pneumonia cases. In contrast, in the reference study [35], four CNN models were used to train their system and SqueezeNet showed the best results for different performance metrics; it showed 98.3% accuracy, 98.3% F1 score, 96.7% sensitivity for COVID cases, 96.7% sensitivity for healthy cases, and 96.7% for pneumonia cases on DATASET-1.
7.3 Two- and Three-Classifier Systems (DATASET-2)
Tab. 8 summarizes the performances of the best models using the two- and three-classifier systems of this study and a state-of-the-art reference study [23] on the same dataset, which is DATASET-2. For the two-classifier system in our study, the VGG-19 model outperformed the other CNN models with 100% accuracy and 100% sensitivity, while the reference study [23] exhibited 98.75% accuracy and 92.85% sensitivity. Furthermore, for the three-classifier system, the VGG-19 model in our study exhibited 96.2% accuracy and 96.7% sensitivity, while the reference study [23] exhibited 93.48% accuracy and 92.85% sensitivity.
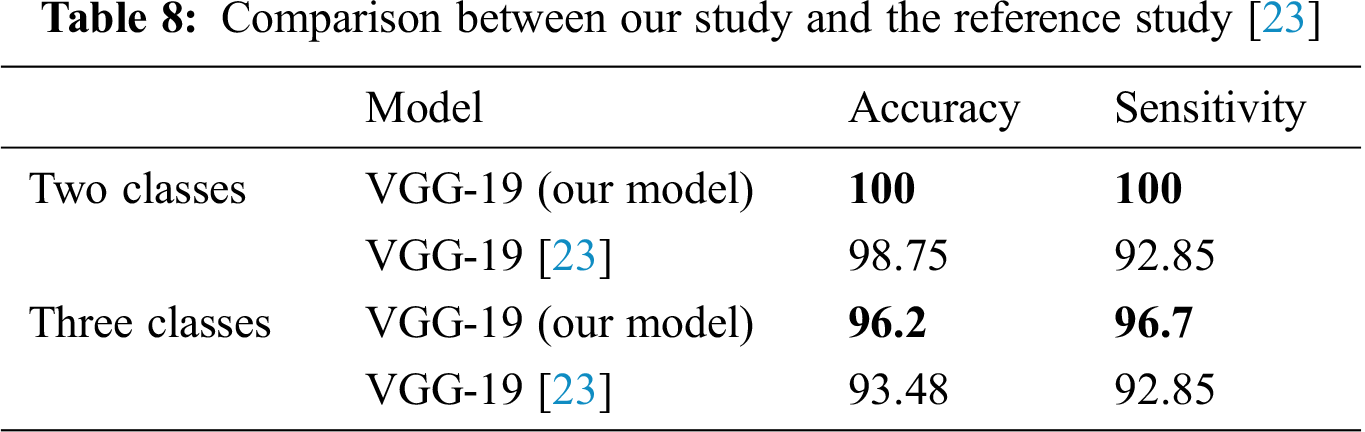
Tab. 8 shows that in the proposed DL-based two- and three-classifier systems, the VGG-19 model achieves accurate results and can be useful for diagnosing COVID-19 cases.
In this paper, we proposed DL-based systems for the automatic identification of COVID-19 in CXR images by retraining three pertained CNN models (VGG 19, MobileNetV2, and NASNet ) on two datasets (DATASET-1 and DATASET-2). CXR and X-ray images were classified into two and three classes (healthy, COVID-19, and pneumonia cases) using the proposed two- and three-classifier systems. In particular, the two-classifier system classified X-ray images into COVID-19 or healthy cases, and the three-classifier system classified CXR images into COVID-19, healthy, or pneumonia cases. We conducted a detailed theoretical study to determine the efficiency of each of the three CNN models and found that the VGG-19 model outperforms the other CNN models. In addition, a detailed comparison study was conducted by comparing the results of this study with those of reference studies, and we found that the proposed classifier systems of this study outperform the related existing systems of the reference studies. In the present study, the VGG-19 model outperformed all other CNN models for the two classification systems. On DATASET-1, the VGG-19 model achieved 99.5% accuracy, 99.2% F1 score, 97.6% sensitivity for COVID-19 cases, and 100% sensitivity for healthy cases; on DATASET-2, it achieved 100% accuracy, 100% F1 score, 100% sensitivity for COVID-19 cases, and 100% sensitivity for healthy cases. Furthermore, in the three-classifier system, the VGG-19 model outperformed the other models. On DATASET-1, the model achieved 98.6% accuracy, 98.6% F1 score, 98.4% sensitivity for COVID-19 cases, 99.4% for healthy cases, and 97.8% sensitivity for pneumonia cases; on DATASET-2, the model achieved 96.2% accuracy, 96.7% F1 score, 100% sensitivity for COVID-19 cases, 95.2% sensitivity for healthy cases, and 95.5% sensitivity for pneumonia cases.
In future work, we plan to extend the dataset by adding more COVID-19 cases and including different types of images like CT scanning images. In addition, we plan to use more epochs.
Funding Statement: This work was supported by the Deanship of Scientific Research (DSR) with King Saud University, Riyadh, Saudi Arabia, through a research group program under Grant RG-1441-503.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. W. Wang, Y. Xu, R. Gao, R. Lu, K. Han et al., “Detection of SARS-CoV-2 in different types of clinical specimens,” JAMA, vol. 323, no. 18, pp. 1843–1844, 2020. [Google Scholar]
2. Y. Yang, M. Yang, C. Shen, F. Wang, J. Yuan et al., “Laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections,” Innovation, vol. 1, no. 3, pp. 100061, 2020. [Google Scholar]
3. Y. LeCun, L. Bottou, Y. Bengio and P. Haffner, “Gradient-based learning applied to document recognition,” Proceedings of the IEEE, vol. 86, no. 11, pp. 2278–2324, 1998. [Google Scholar]
4. V. Badrinarayanan, A. Kendall and R. Cipolla, “SegNet: A deep convolutional encoder-decoder architecture for image segmentation,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 39, no. 12, pp. 2481–2495, 2017. [Google Scholar]
5. S. Minaee, A. Abdolrashidi, H. Su, M. Bennamoun and D. Zhang, “Biometric recognition using deep learning: A survey,” arXiv preprint arXiv:1912.00271, 2019. [Google Scholar]
6. H. Y. F. Wong, H. Y. S. Lam, A. H. T. Fong, S. T. Leung, T. W. Y. Chin et al., “Frequency and distribution of chest radiographic findings in COVID-19 positive patients,” Radiology, vol. 296, no. 2, pp. E72–E78, 2020. [Google Scholar]
7. A. Chen, J. X. Huang, Y. Liao, Z. Liu, D. Chen et al., “Differences in clinical and imaging presentation of pediatric patients with COVID-19 in comparison with adults,” Radiology: Cardiothoracic Imaging, vol. 2, no. 2, pp. e200117, 2020. [Google Scholar]
8. B. Sarkodie, K. Osei-Poku and E. Brakohiapa, “Diagnosing COVID-19 from chest X-ray in resource limited environment-case report,” Journal of Medical Case Reports, vol. 6, pp. 135, 2020. [Google Scholar]
9. D. Ardila, A. P. Kiraly, S. Bharadwaj, B. Choi, J. J. Reicher et al., “Author correction: End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography,” Nature Medicine, vol. 25, no. 8, pp. 1319, 2019. [Google Scholar]
10. K. Suzuki, “Overview of deep learning in medical imaging,” Radiological Physics and Technology, vol. 10, no. 3, pp. 257–273, 2017. [Google Scholar]
11. N. Coudray, P. S. Ocampo, T. Sakellaropoulos, N. Narula, M. Snuderl et al., “Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning,” Nature Medicine, vol. 24, no. 10, pp. 1559–1567, 2018. [Google Scholar]
12. K. He, X. Zhang, S. Ren and J. Sun, “Delving deep into rectifiers: Surpassing human-level performance on imagenet classification,” in Proc. of the IEEE Int. Conf. on Computer Vision, Santiago, Chile, pp. 1026–1034, 2015. [Google Scholar]
13. I. A. E. HuiDS, T. Madani, F. Ntoumi, R. Koch and O. Dar, “The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health: The latest 2019 novel coronavirus outbreak in Wuhan, China,” International Journal of Infectious Diseases, vol. 91, no. 4, pp. 264–266, 2020. [Google Scholar]
14. N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang et al., “A novel coronavirus from patients with pneumonia in China, 2019,” New England Journal of Medicine, vol. 382, no. 8, pp. 727–733, 2020. [Google Scholar]
15. World Health Organisation, “Coronavirus disease (COVID-2019) situation reports,” 2020. [Online]. Available: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/. [Google Scholar]
16. J. Lei, J. Li, X. Li and X. Qi, “CT imaging of the 2019 novel coronavirus (2019-nCoV) pneumonia,” Radiology, vol. 295, no. 1, pp. 18, 2020. [Google Scholar]
17. H. Shi, X. Han and C. Zheng, “Evolution of CT manifestations in a patient recovered from 2019 novel coronavirus (2019-nCoV) pneumonia in Wuhan, China,” Radiology, vol. 295, no. 1, pp. 20, 2020. [Google Scholar]
18. F. Song, N. Shi, F. Shan, Z. Zhang, J. Shen et al., “Emerging 2019 novel coronavirus (2019-nCoV) pneumonia,” Radiology, vol. 295, no. 1, pp. 210–217, 2020. [Google Scholar]
19. A. Krizhevsky, I. Sutskever and G. E. Hinton, “Imagenet classification with deep convolutional neural networks,” Advances in Neural Information Processing Systems. Vol. 25, pp. 1097–1105, 2012. [Google Scholar]
20. L. Wang and A. Wong, “COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images, arXiv preprint arXiv:2003.09871, 2020. [Google Scholar]
21. P. K. Sethy and S. K. Behera, “Detection of coronavirus disease (COVID-19) based on deep features,” Preprints, 2020030300, 2020. [Google Scholar]
22. A. Narin, C. Kaya and Z. Pamuk, “Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks,” arXiv preprint arXiv:2003.10849, 2020. [Google Scholar]
23. I. D. Apostolopoulos and T. A. Mpesiana, “Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks,” Physical and Engineering Sciences in Medicine, vol. 43, no. 2, pp. 635–640, 2020. [Google Scholar]
24. E. El-Din Hemdan, M. A. Shouman and M. E. Karar, “COVIDX-Net: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images,” arXiv, p. arXiv: 2003.11055, 2003. [Google Scholar]
25. T. Ozturk, M. Talo, E. A. Yildirim, U. B. Baloglu, O. Yildirim et al., “Automated detection of COVID-19 cases using deep neural networks with X-ray images,” Computers in Biology and Medicine, vol. 121, no. 7798, pp. 103792, 2020. [Google Scholar]
26. A. S. Al-Waisy, S. Al-Fahdawi, M. A. Mohammed, K. H. Abdulkareem, S. A. Mostafa et al., “COVID-CheXNet: Hybrid deep learning framework for identifying covid-19 virus in chest X-rays images,” Soft Computing, vol. 21, pp. 1–16, 2020. [Google Scholar]
27. A. Al-Waisy, M. A. Mohammed, S. Al-Fahdawi, M. Maashi, B. Garcia-Zapirain et al., “COVID-DeepNet: Hybrid multimodal deep learning system for improving covid-19 pneumonia detection in chest X-ray images,” Computers, Materials & Continua, vol. 67, no. 2, 2021. [Google Scholar]
28. K. H. Abdulkareem, M. A. Mohammed, A. Salim, M. Arif, O. Geman et al., “Realizing an effective COVID-19 diagnosis system based on machine learning and IOT in smart hospital environment,” IEEE Internet of Things Journal, 2021. [Google Scholar]
29. A. M. Ismael and A. Şengür, “Deep learning approaches for covid-19 detection based on chest X-ray images,” Expert Systems with Applications, vol. 164, no. 4, pp. 114054, 2021. [Google Scholar]
30. R. Jain, M. Gupta, S. Taneja and D. J. Hemanth, “Deep learning based detection and analysis of COVID-19 on chest X-ray images,” Applied Intelligence, vol. 51, no. 3, pp. 1690–1700, 2021. [Google Scholar]
31. P. Saha, M. S. Sadi and M. M. Islam, “EMCNet: Automated COVID-19 diagnosis from X-ray images using convolutional neural network and ensemble of machine learning classifiers,” Informatics in Medicine Unlocked, vol. 22, no. 3, pp. 100505, 2021. [Google Scholar]
32. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” arXiv preprint arXiv: 1409.1556, 2014. [Google Scholar]
33. M. Sandler, A. Howard, M. Zhu, A. Zhmoginov and L. C. Chen, “Mobilenetv2: Inverted residuals and linear bottlenecks,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, pp. 4510–4520, 2018. [Google Scholar]
34. B. Zoph, V. Vasudevan, J. Shlens and Q. V. Le, “Learning transferable architectures for scalable image recognition,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, pp. 8697–8710, 2018. [Google Scholar]
35. M. E. Chowdhury, T. Rahman, A. Khandakar, R. Mazhar, M. A. Kadiret et al., “Can AI help in screening viral and COVID-19 pneumonia?,” arXiv preprint arXiv: 2003.13145, 2020. [Google Scholar]
36. J. P. Cohen, P. Morrison and L. Dao, “COVID-19 image data collection,” arXiv preprint arXiv: 2003.11597, 2020. [Google Scholar]
37. Larxel, “COVID-19 X-rays dataset,” 2020. [Online]. Available: https://www.kaggle.com/andrewmvd/convid19-x-rays. [Google Scholar]
38. D. S. Kermany, M. Goldbaum, W. Cai, C. C. Valentim, H. Liang et al., “Identifying medical diagnoses and treatable diseases by image-based deep learning,” Cell, vol. 172, no. 5, pp. 1122–1131, 2018. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |