DOI:10.32604/iasc.2022.020508

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2022.020508 |  |
| Article |
Automated Deep Learning of COVID-19 and Pneumonia Detection Using Google AutoML
Department of Mathematical Sciences, Faculty of Science and Technology Universiti Kebangsaan Malaysia, 43600, Bangi, Malaysia
*Corresponding Author: Saiful Izzuan Hussain. Email: sih@ukm.edu.my
Received: 27 May 2021; Accepted: 08 July 2021
Abstract: Coronavirus (COVID-19) is a pandemic disease classified by the World Health Organization. This virus triggers several coughing problems (e.g., flu) that include symptoms of fever, cough, and pneumonia, in extreme cases. The human sputum or blood samples are used to detect this virus, and the result is normally available within a few hours or at most days. In this research, we suggest the implementation of automated deep learning without require handcrafted expertise of data scientist. The model developed aims to give radiologists a second-opinion interpretation and to minimize clinicians’ workload substantially and help them diagnose correctly. We employed automated deep learning via Google AutoML for COVID-19 X-ray detection to provide an automated and faster diagnosis. The model is employed on X-ray images to detect COVID-19 based on several binary and multi-class cases. It consists of three scenarios of binary classification categorized as healthy, pneumonia, and COVID-19. The multi-classification model based on these three labels is employed to differentiate between them directly. An investigational review of 1125 chest X-rays indicates the efficiency of the proposed method. AutoML enables binary and multi-classification tasks to be performed with an accuracy up to 98.41%.
Keywords: COVID-19; deep learning; AutoML; pneumonia
Coronavirus (COVID-19) is reported to officially start on December 31, 2019, with the unknown causes of pneumonia recorded in Wuhan, Hubei Province, China, and becomes a pandemic a few weeks later [1,2]. The disease is called as COVID-19, which is scientifically termed from serious acute respiratory syndrome coronavirus (SARS-CoV-2). This new virus aggressively effects the human respiratory system and spreads over 30 days from Wuhan to a large part of China [3]. There are only first seven cases reported on January 20, 2020, but due to its rapid transmission, the number of cases surpassed more than 300,000 by April 5, 2020 [4]. Coronaviruses mostly affect animals, but because of their zoonotic nature, they can also be transmitted to humans. SARS-CoV and COVID-19 have caused extreme human breathing disease and death. As of September 21, 2020, there are 30.9 million cases with more than 950,000 deaths around the world [5]. COVID-19 is usually clinically characterized by respiratory illness with flu-like symptoms such as cough, headache, fever, sore throat, fatigue, body aches, and shortness of breath [6].
In addition to early detection diagnostic tests for care and insulation of the disease, radiological imagery is a critical factor in the COVID-19 epidemic management process. Radiological images from COVID-19 cases provide valuable diagnostic details. The most popular research technique to identify COVID-19 is by applying a reverse transcription-polymerase chain reaction (RT-PCR) in real time. The early diagnosis and treatment of chest radiology using X-rays and computed tomography (CT) play a vital role in this process [7].
Researchers also made important discoveries through imaging studies of COVID-19. An RT-PCR sensitivity of 60% to 70% enables the analysis of patients’ radiological images to detect symptoms [8,9]. A few studies also demonstrated the changes in CT and X-ray chest images before symptoms of COVID-19 start [10,11]. Ground-glass opacification (GGO) signs and specific patterns for peripheral and bilateral consolidations can be seen through images taken. Multifocal or peripheral focal GGO affects 50% to 75% of a patient’s lungs as stated in Kanne et al. [8]. In a similar vein, Yoon et al. [12] reported a single nodular opacity found in the lower left lung region of one out of the three patients studied, while the other two had four and five abnormal opacities in both lungs. In another study, Zhao et al. [13] reported to not only find GGO but also identify consolidation and vascular lesion dilation. Li et al. [14] reported common characteristics in COVID-19 patients including interlobular septal thickening, GGO and consolidation, and air bronchograms with or without vascular expansion.
The use of automated diagnostic methods in the health area has recently gained prominence by being a supplementary instrument for clinicians [15–17]. Machine learning (ML) approach allows the development of end-to-end models for better predicted results without the need to extract the features manually [18]. The functionality of ML framework is based on the basic unit of computation that operates on data and illustrates the theory of learning from experience and errors in a probabilistic manner. ML techniques specifically via deep learning have been used successfully for a number of problems including detection of arrhythmia [19–21], classification of skin cancer [22,23], detection of breast cancer [24,25], classification of gastric carcinoma [26], prediction of colorectal cancer [27], classification of brain diseases [28], detection of pneumonia in via X-ray images [29], and segmentation of the lung [30,31]. The rapid growth of the COVID-19 epidemic demands expertise in this area. All of this showed an increased interest in developing artificial intelligence-based automatic detection systems.
CT is a sensitive approach to be considered better than X-rays as an RT-PRC screening for identification of COVID-19 based on the previous studies. However, there are several disadvantages to consider in using X-ray images. The number of CT scan units is limited in hospitals. X-ray images can be used extensively since they are not as costly as CT procedure and do not involve extensive patient planning in order to perform an examination.
There is an increase of study using radiology images for the identification of COVID-19. In X-ray image area, Hemdan et al. [32] employed COVIDX-Net to identify COVID-19 and suggested that deep learning has the potential to be implemented in hospitals for screening process. In a similar study, Wang et al. [33] obtained 92.4% accuracy in the classification of labels between normal, pneumonia, and COVID-19. Luján-García et al. [34] proposed the use of deep learning model with pretrained weights on ImageNet. The deep learning model established by Ioannis et al. [35] by using 224 images verified as COVID-19 reached success rates of 98.75% and 93.48% in two and three groups, respectively. Similarly, Narin et al. achieved 98% of COVID-19 detection accuracy with the execution with ResNet50 model [36]. Sethy et al. [37] detected several features from different neural network models with X-ray and reported the best performance belongs to the ResNet50 model with SVM classification. The study by Ozturk et al. [38] achieved 98.08% accuracy in binary scenarios and 87.02% accuracy in multi-class situations. Brunese et al. [39] have shown the efficiency of deep learning method with an average accuracy of 97% and 2.5 seconds as detection time of the disease. The results indicate that deep learning demonstrates correct prediction features of this disease. To conclude, all the studies show the potential of using X-ray images in providing high accuracy in COVID-19 detection process. The model was able to distinguish GGOs, aggregation areas, and nodular opacity in the pathognomonic results of COVID-19 patients at high accuracy.
While ML algorithms demonstrate high COVID-19 identification potential, it has been a daunting task to create an adaptive deep learning model through a conventional procedure. This is due not just to the limited availability of high-quality X-ray images related to COVID-19 image dataset but also to the difficulty of understanding the environment and architecture of deep learning algorithms [40]. The development of the deep leaning model for COVID-19 models requires an experienced and high-knowledge data scientist or expertise in programming language to design the model carefully and validate [41,42]. However, with the rapid evolution of deep learning technology, this learning barrier has been reduced recently [43,44]. AutoML offers methods and processes for selecting a suitable model automatically, optimizing its hyperparameters, and analyzing its results. All of these make the process of creating a model easy and improve the accuracy of the model through optimization and extensive search at the same time. AutoML is appealing in combination with the integration of cloud computing such as Google Cloud Platform and AWS [45,46] with the ability to take full advantage of scalable and flexibility cloud infrastructures and resources.
Deep learning models are proven and adaptive to COVID-19 cases, and the probability diagnostic accuracy rate is high. The highlight of this study is to explore and evaluate the performance of using advanced AutoML technology during the process of COVID-19 identification. The research expands previous research in this field by employing and assessing an investigational deep learning model based on Google Cloud AutoML Vision [46] rather than a customized deep learning environment and architecture from a sketch. The next section begins with a description of the methodology and public dataset. Early diagnosis of the COVID-19 is critical in order to avoid disease transmission and provide timely care. AutoML model has been proven as a valuable approach in recognizing early stages of COVID-19 patients. In the last section, this study concludes with remarks on the purpose of the study, the limitation during the procedure, and suggestions for future studies.
Artificial intelligence is transformed by the emergence of deep learning technology. A convolutional neural network (CNN) model based on deep learning is developed by taking one or more layers with the internal parameters that will be optimized to achieve a certain task, for example, for recognition or classification of objects [28]. The structure known as convolution comes from a mathematical operator. The word intensity depends on the size and number of layers of this network.
CCN is quite difficult to be implemented as it needs a few understanding and insights of layers to be optimized. A better approach to tackle this problem is to build a model with already established models rather to develop a deep model from sketch. AutoML is identified as the preliminary step for the deep learning model to be applied in this paper.
In this article, we suggest a binary and triple classification using AutoML:
i) To diagnose the relationship of chest X-rays between a healthy patient (no finding) and related pulmonary disease (pneumonia and COVID-19),
ii) To differentiate between related pulmonary disease and COVID-19,
iii) To differentiate between healthy patient, pneumonia, and COVID-19 using triple classification.
The model performance is evaluated by three metrics: sensitivity, specificity, and accuracy. These measurements are widely used for the description of a diagnostic test in ML area. These have been used to quantify and understand how reliable and suit a model is. Sensitivity is used to indicate how good a positive disease can be detected. It shows how much diseases are correctly predicted based on the abnormal cases. High sensitivity demonstrates a low level of probability in patients treated as normal because of misdiagnosis. Specificity estimates the likelihood of correct exclusion of patients without disease. The number shown might be high, but it is less important compared to the sensitivity in the medical diagnosis. Accuracy indicates how the total labels are correctly predicted with the condition. For this study, it showed how good diseases and nondiseases are correctly predicted. The accuracy from the test with prevalence can be measured based on specificity and sensitivity.
True positive (TP), true negative (TN), false negative (FN), and false positive (FP) are terms needed to be understood to measure sensitivity, specificity, and accuracy. If a patient has a confirmed illness, the diagnostic test also reveals that the disease occurs; hence, the predicted results are called TP. Similar to a TN situation, if a patient has been found to be absent of any disease, the diagnostic test indicates that the disease is absent as well. A clear outcome between the predicted results and the real condition (also known as the norm of truth) is both TP and TN. However, there could be probability misdiagnoses for some medical examination. When the diagnostic test shows that a patient does not have such a disease where it is not true in the real condition, it is called FP. Similarly, when the diagnostic test results show that a patient is misdiagnosed with a disease, the test outcome is known as FN. Both, FP and FN, suggest that the findings of the experiments are contradictory to the real situation.
The following shows how the terms TP, TN, FN, and FP are used to describe sensitivity, specificity, and accuracy:
Sensitivity = TP/(TP + FN) = (Total of true predicted positive assessment)/(Total of all positive assessment)
Specificity = TN/(TN + FP) = (Total of negative predicted assessment)/(Total of all negative assessment)
Accuracy = (TN + TP)/(TN + TP + FN + FP) = (Total of correct predicted assessment)/Total of all assessment).
In this section, we present the process selection of datasets used for this research, concept of Google AutoML, how the model is built, and the performance measures of models. A comparison between models based on previous studies is made at the end of the section.
Public database is used to evaluate the proposed approach. All datasets concerned are collections of chest X-rays from the source of [38]. These datasets (Fig. 1) randomly used 500 healthy and 500 pneumonia class X-ray images. Dataset for COVID-19 originally are from Cohen JP [47] who developed these X-ray images with images of different open-access sources. The collection includes 125 X-ray images diagnosed with COVID-19. A full metadata is not given for all patients in this dataset. The images for healthy and pneumonia cases are from Wang et al. [48].

Figure 1: Example of images that have been used with labels: (a) and (b) healthy patient; (c) and (d) pneumonia; (e) and (f) COVID-19. The source of image is originally from the dataset of [47,48]
AutoML is an AI algorithm based on a deep learning structure. Deep learning approach is an algorithmic variant of ML. In this study, Google Cloud AutoML Vision is chosen as our main platform to construct our model for COVID-19 detection. AutoML Vision is an application deployment for image recognition and object detection on Google Cloud Platform. AutoML Vision’s core capabilities include allowing users with limited knowledge to develop high-quality personalized deep learning models based on unique tasks. AutoML Vision is established on standardized and pretrained and ready-to-use AI applications like the Vision API in the selection of Google AI/ML offers [46].
Google’s AutoML Vision helps a non-ML professional run an ML model according to a unique criteria, with limited ML knowledge. Cloud-based models are supported with the latest GPUs that allow users to execute a customized task in a few hours (depending on the dataset size). AutoML splits the dataset into three different sets by default for the preparation of the model to be designed: 80% preparation, 10% validation, and 10% off-box checking. The default setting can be changed at any time.
The introduction of an ML model into the pathology workflow is a difficult work. ML is a source of computational engineering with a complex computation behind it. In order to construct an image processing model in ML, you may need a high level of coding language knowledge such as Python and MATLAB, image recognition computer vision techniques, data processing, and predictive analysis. In regular clinical workflows, it could be hard for a pathologist or digital pathologist to learn and develop these skills either to employ a graduate specialist or to work together with an engineering community.
Task pace and computer hardware are other obstacles that need to be taken into consideration. ML models are very hard to run on local machine channels computationally. The batch work takes days to run on a personal computer without installed GPUs even for basic image classification. The design and development of an ML model is another obstacle to feature engineering and data preprocessing. Function engineering is the ability to remove specific elements such as data cleaning, context and noise filtration, and item blockage and segmentation. The quality of the model depends on the performance of the functional engineering task and on the related features used in software implementation. Irregular object limits and moderate tissue stain strength are the variables that make pathologists confused when measuring heterogeneity. Often, the human eye cannot perceive objects beyond a certain distance as a normal restriction. To resolve these issues, skills and training should be required in feature engineering and in ML fields, which are rare in pathology laboratories. AutoML is a feasible option to overcome these considerations and exclude this field from engineering fields.
The following steps are taken to develop our experimental COVID-19 model. A few procedures need to be followed before the model is trained. The process also includes to upload our expanded dataset in Google Cloud Storage:
1. Prepare the collection of images that will be uploaded to cloud storage route. The process requires a minimum of 100 image examples per category or label. The probability of classification increases with the number of good-quality images.
2. Import the images to the Google Cloud AutoML Vision UI to create a dataset. All data need to be labeled based on the category to be classified.
3. Train the data and select the process.
AutoML Vision produces a training process that works automatically. The training dataset process has a default setting of preparation (80%), validation (10%), and evaluation (10%). It will try to identify COVID-19 models to find the best performing algorithms in the training data collection. AutoML Vision employs validation and test process on datasets to evaluate model performance.
We conducted tests for the identification and classification of COVID-19 using X-ray images in two separate models: binary and triple classification. The binary model is used to train the automatic deep learning model to detect and discern two groups. It distinguishes between healthy and diseases (pneumonia and COVID-19) in the first case. In the second scenario, the model differentiates the data between healthy and COVID-19. In the third scenario, pneumonia is separated from COVID-19. The multi-class classification model is employed to classify X-ray images into three categories: healthy, pneumonia, and COVID19. All the execution is carried out via AutoML.
The efficiency of the proposed method for binary as well as triple classification problems is evaluated. Eighty percent of the X-ray pictures are used for preparation, 10% for analysis, and 10% for testing. In this paper, we select a confidence threshold of 0.5. Confidence threshold is used by the machine as decision threshold. In a simple term, the confidence threshold showed how much confidence level of the model is with its predicted probability to assign a chosen category.
The process of evaluation consists the predictive accuracy of the test samples from 0.0 to 1.0 at different levels with a compatibility threshold. Fig. 2 presents a precision-recall curve produced based on the previous computation. At a given confidence threshold, every point in the curve denotes a value for the pair of precision and recall.
Figs. 2 and 3, present an area under precision-recall curve and precision-recall curve for the training models.

Figure 2: Area under precision-recall curve and precision-recall curve for the binary model. (a) and (b) healthy vs. disease; (c) and (d) healthy vs. COVID-19; (e) and (f) pneumonia vs. COVID-19

Figure 3: Area under precision-recall curve and precision-recall curve for healthy vs. pneumonia vs. COVID-19
The average accuracy of the model is computed by the accuracy of the area under the precision-recall curve (AuPRC). As we refer to the AutoML model for binary cases in scenario 1 in Fig. 2a, an AuPRC score of 81.42% is achieved. Precision variation and reminder with confidence threshold change. The precision increases as the confidence threshold increases with the possibility of the recall to decrease.
To further characterize the performance, confusion matrix analyses were conducted on both models. The findings of confusion matrices for the binary and multi-class classification problem are shown in Fig. 4.

Figure 4: Confusion matrix for (a) healthy vs. disease; (b) healthy vs. COVID-19; (c) pneumonia vs. COVID-19; and (d) healthy vs. pneumonia vs. COVID-19
For all models, all classes were predicted into the correct categories with a probability of at least greater than 76%. For model 1, the process between healthy and disease classifications is the hardest. Disease classifications were most likely to be misclassified as healthy at 24% and the healthy to be misclassified as disease at 12%. Classification for model 1 scenario 2 and scenario 3 seems to provide a higher accuracy. COVID-19 classifications were most likely to be misclassified as healthy at 8% and the healthy to be misclassified as COVID-19 at 0% as shown in Fig. 4b. COVID-19 classifications were most likely to be misclassified as pneumonia at 0% and the pneumonia to be misclassified as COVID-19 at 2% as shown in Fig. 4c.
The confusion matrix for the validation of Fig. 4d is used to separate between healthy, pneumonia, and COVID-19 classifications. Based on the triple classification model, 8% is marked as healthy when there is COVID-19, 12% as pneumonia when there is healthy, and 2% as healthy when there is pneumonia. The classification results in terms of precision and recall from AutoML for both models are shown in Tab. 1. Precision is defined as items that are actually correct and correctly predicted. A high precision model could associate with less false positives. A high recall model could associate with fewer false negatives.
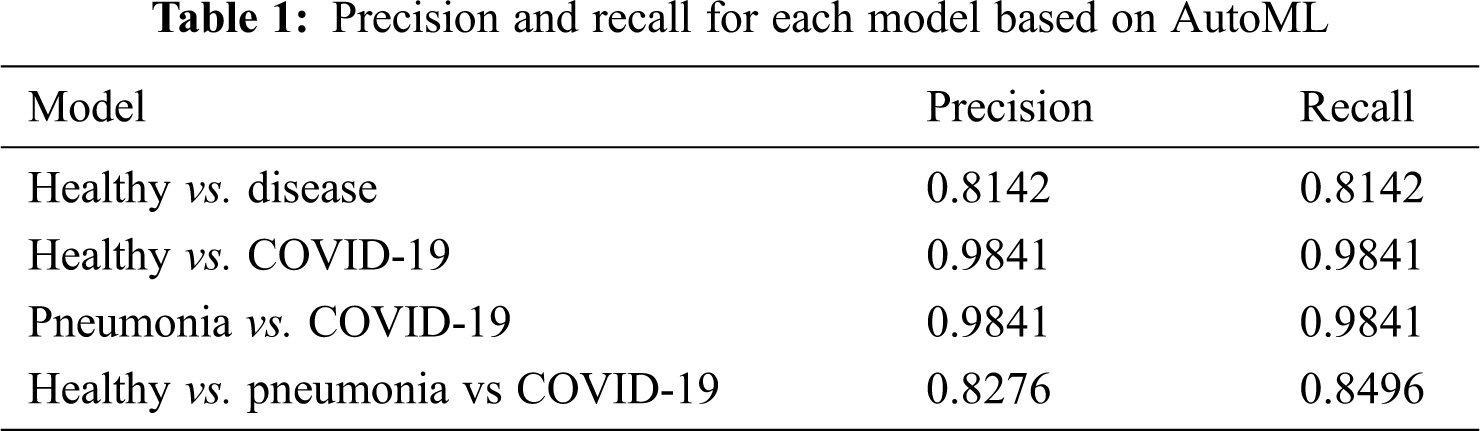
Further, sensitivity, specificity precision, and accuracy performance for the binary and multi-classification models are computed and presented in Tab. 2 for a standardized understanding as the results from AutoML are presented in terms of precision and recall.
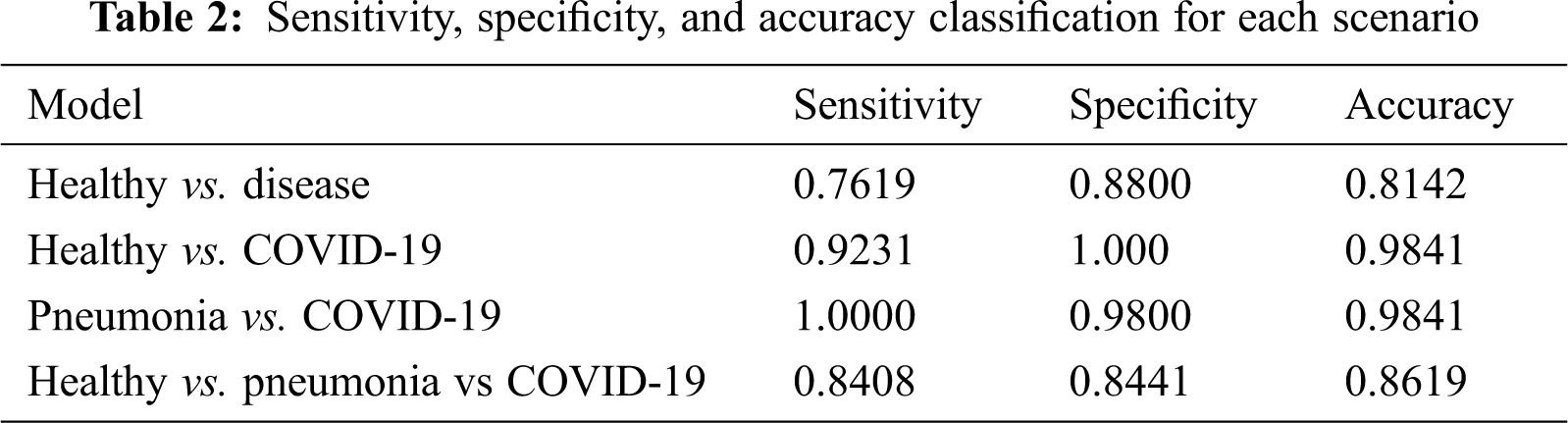
As far as accuracy is concerned, the first scenario for binary healthy vs disease achieves an accuracy of 81.42% and the other two binary scenarios achieve a value of 98.41%. The model was able to distinguish pneumonia and COVID-19 well for scenario 2 with a higher accuracy. The sensitivity is also almost perfect as 100%. The accuracy for triple classification also presents a sign potential of the model to be employed at 86.19%.
With the results, able to produce accuracy better than 80% as for the overall scenario. At some scenario, accuracy was able to achieve 98.41%. The results obtained showed the potential of the model to be employed in hospitals or far health centers. The waiting period for the radiologist to screen the images could be reduced which cost few hours and days. More energy could be concentrated to isolate the suspect in the faster process. The process of spread of this disease may therefore be greatly decreased. This model works as an alternative to reduce the workload of clinicians. If the potential patient is classified as positive, patients could obtain a second opinion and details faster.
The performance metrics in terms of accuracy between the models from previous publications and AutoML model for further evaluation is considered and compared. Tab. 3 lists the findings of a model comparison that was used in terms of the average accuracy of the running deep learning model.
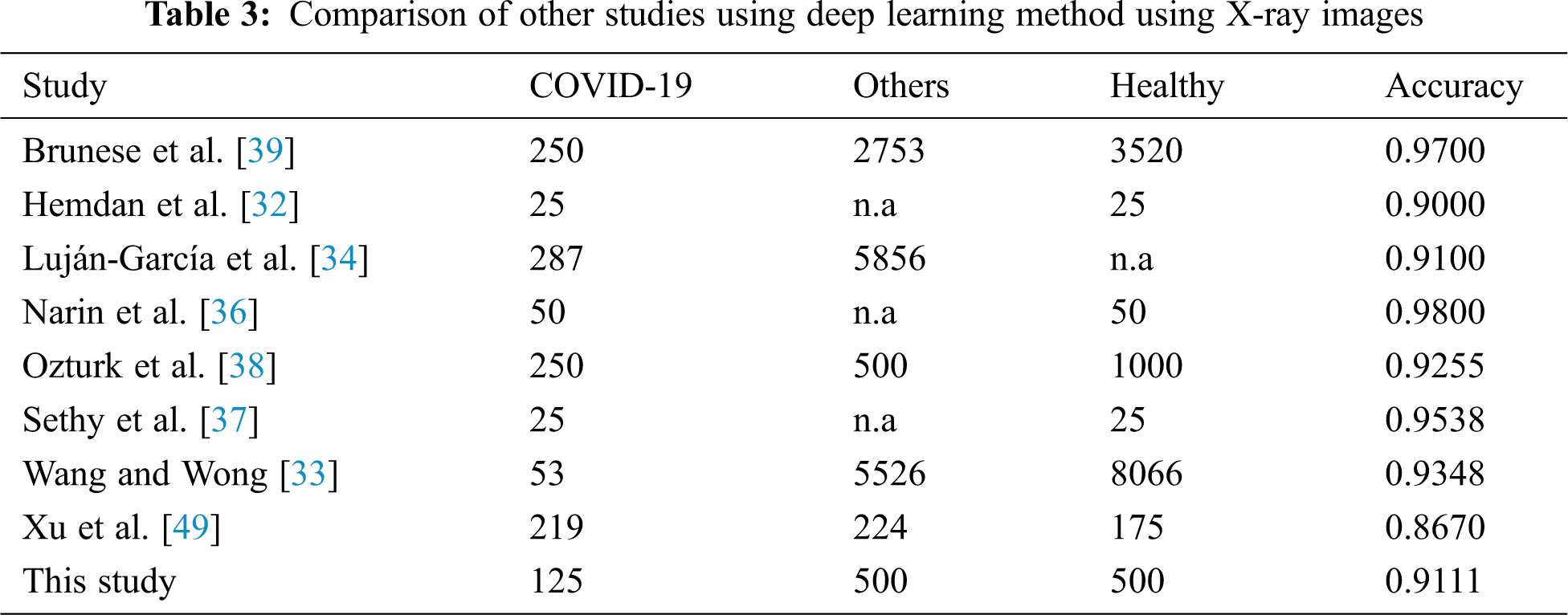
The average accuracy of model is 0.9111 performed better than half of the previous studies. If we compare the average of accuracy based on the scenario employed, it seems that the AutoML is only able to provide better prediction than a few studies only. However, as we compare how flexible the model is without the need of handcrafted deep learning expert, it still provides a great potential to be developed and explored. Despite AutoML Vision’s comprehensive search and optimization, the changes are still minimal and easy to be implemented by a non-code researcher. This is because previous models of human engineering may have been similar to an ideal model for the given dataset.
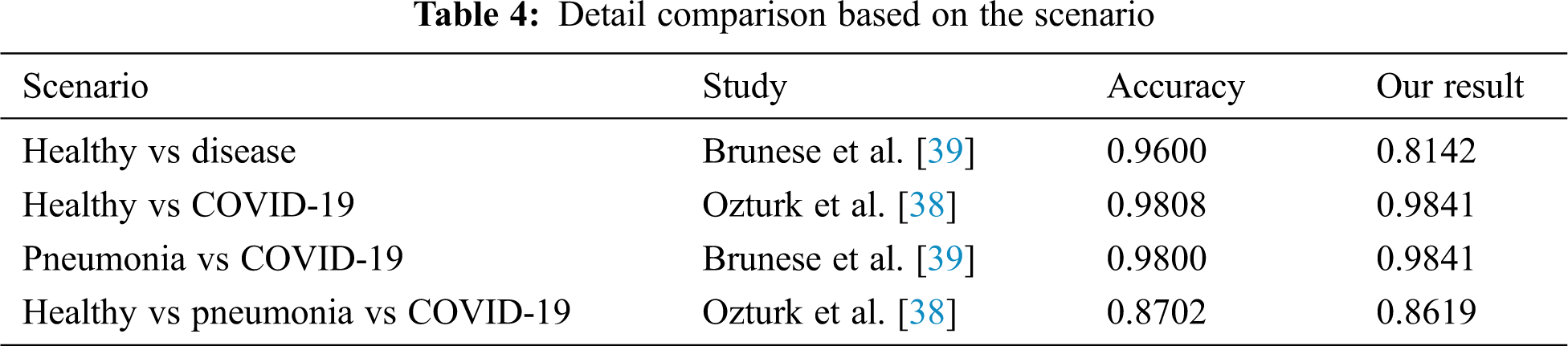
In Tab. 4, we compared and evaluated our results from the results of other previous studies in terms of situation which has the same scenario. Interestingly, AutoML Vision from this analysis was found to marginally outperform models reported earlier in both the assessment metrics and the non-tests in the same situation. In many cases, the prediction of greater than 98% was achieved. This model is able to separate the labels between healthy and COVID-19 or pneumonia and COVID-19 patients well in the binary cases. The same goes to triple classification where the results were able to produce almost the same as those of a handcrafted model by experts. By considering the results and how flexible this method is, it makes AutoML seem fit to be implemented into the real world and have greater potential to be developed. The present study contributes evidence that suggest the huge potential of AutoML to be applied in COVID-19 detection.
There is an important consideration with regard to the increasing number of population diagnosed with COVID-19. The high-risk patients must be detected at an early stage so they could receive timely care and to reduce the possibility of death.
Many models are designed to distinguish patients diagnosed with diseases linked to pneumonia (including COVID-19) from patients with healthy condition. The prediction methodology such as deep learning via ML can help with the diagnosis process of using X-ray images of COVID-19 patients. The images are useful since they are available and cost less compared to others. They are often used in health centers around the world throughout the pandemic and help to detect the symptoms in seconds. CT is an expensive procedure that is not readily available since it typically is only found in bigger areas of health centers or hospitals. At the same time, the patient absorbs more radiation as compared to X-rays.
A deep learning approach using X-ray imaging is therefore suggested because it is more accessible compared to CT. Chest X-rays with lower radiation demonstrated certain standard findings of COVID-19 in the lungs. The patients identified with positive COVID by the model can be assigned to the sophisticated confirmatory centers and are accompanied by immediate therapy. In addition, the PCR testing and occupation of health centers can be avoided and reduce the workload for patients.
This paper employed an automated deep learning model for the detection and classification of COVID-19 cases from radiographs using AutoML. This study employed a fully automated deep learning executed using Google Cloud Platform. Our built-in users can automate classification binary and multi-class tasks, respectively, and obtain an accuracy of 98.41%. Based on the results, the CNN deep learning has a major impact on the automatic detection and extra main features of X-ray linked to the COVID-19 diagnosis accurately. AutoML model is designed to differentiate COVID-19 automatically using X-ray images, without the need of handcraft extraction techniques of experienced data scientists. The model developed helps provide specialist radiologists in health centers with a second opinion and increase the accuracy. Our findings show AutoML can correctly differentiate between COVID-19 patients and healthy patients with an outstanding accuracy rate of 0.9841. This model has also been able to differentiate between patients infected COVID-19 and patients with bacteria-infected pneumonia with the same precision. It can greatly reduce clinicians’ workload and help them make an informed diagnosis in their day-to-day service. The model proposed was able to save time as the process of diagnostic only takes up to a few minutes and thus making specialists to concentrate more on important cases.
The collection of images related to COVID-19 could be a major limitation of the current study and may be difficult to overcome. A more detailed analysis especially in terms of the number of data of patients diagnosed with COVID-19 is needed and could be used in future research. Exploration of a more promising concept for future research would be also interesting to distinguish between patients having mild symptoms from pneumonia and patients having symptoms that may neither be visualized nor visualized specifically in radiation. As for our study, we intend to validate our model with additional images especially using COVID-19 images for better accuracy. At the same time, the model also could be improved in terms of demography by locally collecting radiology images from patients diagnosed with COVID-19. There will also be a possibility to deploy the model developed for screening in local hospitals. The model based on deep learning can be uploaded in a cloud storage to automatically diagnose and increase the efficiency of pandemic risk management. This should greatly reduce the workload of clinicians. Lastly, it is also interesting to explore the possibility of using CT images for COVID-19 process identification and compare the results obtained from this research for future studies.
Funding Statement: Part of this work was supported by the Universiti Kebangsaan Malaysia grant GGPM-2018-070.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. F. Wu, S. Zhao, B. Yu, Y. M. Chen, W. Wang et al., “A new coronavirus associated with human respiratory disease in China,” Nature, vol. 579, no. 7798, pp. 265–269, 2020. [Google Scholar]
2. C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao et al., “Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China,” Lancet, vol. 395, no. 10223, pp. 497–506, 2020. [Google Scholar]
3. Z. Wu and J. M. McGoogan, “Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese center for disease control and prevention,” JAMA, vol. 323, no. 13, pp. 1239–1242, 2020. [Google Scholar]
4. M. L. Holshue, C. DeBolt, S. Lindquist, K. H. Lofy, J. Wiesman et al., “First case of 2019 novel coronavirus in the United States,” New England Journal of Medicine, vol. 328, no. 10, pp. 929–936, 2020. [Google Scholar]
5. World Health Organization Coronavirus Disease, [Online]. Available: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (Accessed on 18 September 20202019. [Google Scholar]
6. T. Singhal, “A review of coronavirus disease-2019 (COVID-19),” Indian Journal of Pediatrics, vol. 87, no. 4, pp. 281–286, 2020. [Google Scholar]
7. Z. Y. Zu, M. D. Jiang, P. P. Xu, W. Chen, Q. Q. Ni et al., “Coronavirus disease 2019 (COVID-19A perspective from China,” Radiology, vol. 296, no. 2, pp. E15–E25, 2020. [Google Scholar]
8. J. P. Kanne, B. P. Little, J. H. Chung, B. M. Elicker and L. H. Ketai, “Essentials for radiologists on COVID-19: An update—Radiology scientific expert panel, pp. 113–114, 2020. [Google Scholar]
9. X. Xie, Z. Zhong, W. Zhao, C. Zheng, F. Wang et al., “Chest CT for Typical 2019-nCoV pneumonia: Relationship to negative RT-PCR testing,” Radiology, vol. 296, no. 2, pp. E41–E45, 2020. [Google Scholar]
10. J. F. W. Chan, S. Yuan, K. H. Kok, K. K. W. To and H. Chu, “A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster,” Lancet, vol. 395, no. 10223, pp. 514–523, 2020. [Google Scholar]
11. S. Simpson, F. U. Kay, S. Abbara, S. Bhalla, J. H. Chung et al., “Radiological society of north America expert consensus document on reporting chest CT findings related to COVID-19: Endorsed by the society of thoracic radiology, the American college of radiology, and RSNA,” Radiology: Cardiothoracic Imaging, vol. 2, no. 2, pp. 1–10, 2020. [Google Scholar]
12. S. H. Yoon, K. H. Lee, J. Y. Kim, Y. K. Lee, H. Ko et al., “Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID-19Analysis of nine patients treated in Korea,” Korean Journal of Radiology, vol. 21, no. 4, pp. 494–500, 2020. [Google Scholar]
13. W. Zhao, Z. Zhong, X. Xie, Q. Yu and J. Liu, “Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: A multicenter study,” American Journal of Roentgenology, vol. 214, no. 5, pp. 1072–1077, 2020. [Google Scholar]
14. Y. Li and L. Xia, “Coronavirus disease 2019 (COVID-19Role of chest CT in diagnosis and management,” American Journal of Roentgenology, vol. 214, no. 6, pp. 1280–1286, 2020. [Google Scholar]
15. G. Litjens, T. Kooi, B. E. Bejnordi, A. A. A. Setio, F. Ciompi et al., “A survey on deep learning in medical image analysis,” Medical Image Analysis, vol. 42, no. 13, pp. 60–88, 2017. [Google Scholar]
16. J. Ker, L. Wang, J. Rao and T. Lim, “Deep learning applications in medical image analysis,” IEEE Access, vol. 6, pp. 9375–9389, 2018. [Google Scholar]
17. D. Shen, G. Wu and H. I. Suk, “Deep learning in medical image analysis,” Annual Review of Biomedical Engineering, vol. 19, no. 1, pp. 221–248, 2017. [Google Scholar]
18. Y. LeCun, Y. Bengio and G. Hinton, “Deep learning,” Nature, vol. 521, no. 7553, pp. 436–444, 2015. [Google Scholar]
19. O. Yıldırım, P. Pławiak, R. S. Tan and U. R. Acharya, “Arrhythmia detection using deep convolutional neural network with long duration ECG signals,” Computers in Biology and Medicine, vol. 102, no. 4, pp. 411–420, 2018. [Google Scholar]
20. A. Y. Hannun, P. Rajpurkar, M. Haghpanahi, G. H. Tison, C. Bourn et al., “Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network,” Nature Medicine, vol. 25, no. 1, pp. 65–69, 2019. [Google Scholar]
21. U. R. Acharya, S. L. Oh, Y. Hagiwara, J. H. Tan, M. Adam et al., “A deep convolutional neural network model to classify heartbeats,” Computers in Biology and Medicine, vol. 89, no. 1, pp. 389–396, 2017. [Google Scholar]
22. A. Esteva, B. Kuprel, R. A. Novoa, J. Ko, S. M. Swetter et al., “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, no. 7639, pp. 115–118, 2017. [Google Scholar]
23. N. C. Codella, Q. B. Nguyen, S. Pankanti, D. A. Gutman, B. Helba et al., “Deep learning ensembles for melanoma recognition in dermoscopy images,” IBM Journal of Research and Development, vol. 61, no. 4/5, pp. 5:1–5:15, 2017. [Google Scholar]
24. Y. Celik, M. Talo, O. Yildirim, M. Karabatak and U. R. Acharya, “Automated invasive ductal carcinoma detection based using deep transfer learning with whole-slide images,” Pattern Recognition Letters, vol. 133, no. 7, pp. 232–239, 2020. [Google Scholar]
25. A. Cruz-Roa, A. Basavanhally, F. González, H. Gilmore, M. Feldman et al., “Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks, Medical Imaging 2014,” Digital Pathology, vol. 9041, pp. 904103, 2014. [Google Scholar]
26. H. Sharma, N. Zerbe, I. Klempert, O. Hellwich and P. Hufnagl, “Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology,” Computerized Medical Imaging and Graph, vol. 61, no. 2, pp. 2–13, 2017. [Google Scholar]
27. D. Bychkov, N. Linder N, R. Turkki, S. Nordling, P. E. Kovanen et al., “Deep learning based tissue analysis predicts outcome in colorectal cancer,” Scientific Reports, vol. 8, no. 1, pp. 85, 2018. [Google Scholar]
28. M. Talo, O. Yildirim, U. B. Baloglu, G. Aydin and U. R. Acharya, “Convolutional neural networks for multi-class brain disease detection using MRI images,” Computerized Medical Imaging and Graph, vol. 78, no. 1, pp. 101673, 2019. [Google Scholar]
29. P. Rajpurkar, J. Irvin, K. Zhu, B. Yang, H. Mehta et al., “Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning,” arXiv preprint arXiv:1711.05225, 2017. [Google Scholar]
30. G. Gaal, B. Maga and A. Lukacs, “Attention U-Net based adversarial architectures for chest X-Ray lung segmentation,” arXiv preprint arXiv:2003.10304, 2020. [Google Scholar]
31. J. C. Souza, J. O. B. Diniz, J. L. Ferreira, G. L. F. da Silva, A. C. Silva et al., “An automatic method for lung segmentation and reconstruction in chest X-ray using deep neural networks,” Computer Methods and Programs in Biomedicine, vol. 177, no. 5, pp. 285–296, 2019. [Google Scholar]
32. E. E. D. Hemdan, M. A. Shouman and M. E. Karar, “COVIDX-Net: A framework of deep learning classifiers to diagnose COVID-19 in X-Ray images,” arXiv preprint arXiv:2003.11055, 2020. [Google Scholar]
33. L. Wang and A. Wong, “COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images,” Scientific Reports, vol. 10, no. 1, pp. 1–12, 2020. [Google Scholar]
34. J. E. Luján-García, M. A. Moreno-Ibarra, Y. Villuendas-Rey and C. Yáñez-Márquez, “Fast COVID-19 and pneumonia classification using chest X-ray images,” Mathematics, vol. 8, no. 9, pp. 1423, 2020. [Google Scholar]
35. I. D. Apostolopoulos and T. A. Mpesiana, “Covid-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks,” Physical and Engineering Sciences in Medicine, vol. 43, no. 2, pp. 635–640, 2020. [Google Scholar]
36. A. Narin, C. Kaya and Z. Pamuk, “Automatic detection of coronavirus disease (COVID- 19) using X-ray images and deep convolutional neural networks,” Pattern Analysis and Applications, pp. 1–14, 2021. [Online]. Available: https://link.springer.com/article/10.1007/s10044-021-00984-y. [Google Scholar]
37. P. K. Sethy and S. K. Behera, “Detection of coronavirus disease (COVID-19) based on deep features and support vector machine,” 2020. [Google Scholar]
38. T. Ozturk, M. Talo, E. A. Yildirim, U. B. Baloglu, O. Yildirim et al., “Automated detection of COVID-19 cases using deep neural networks with X-ray images,” Computers in Biology and Medicine, vol. 121, no. 7798, pp. 103792, 2020. [Google Scholar]
39. L. Brunese, F. Mercaldo, A. Reginelli and A. Santone, “Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays,” Computer Methods and Programs in Biomedicine, vol. 196, pp. 105608, 2020. [Google Scholar]
40. X. Chen, L. Yao and Y. Zhang, “Residual attention U-Net for automated multi-class segmentation of COVID-19 chest CT images,” arXiv preprint arXiv:2004.05645, 2020. [Google Scholar]
41. W. Rawat and Z. Wang, “Deep convolutional neural networks for image classification: A comprehensive review,” Neural Computation, vol. 29, no. 9, pp. 2352–2449, 2017. [Google Scholar]
42. A. Janowczyk and A. Madabhushi, “Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases,” Journal of Pathology Informatics, vol. 7, no. 1, pp. 29, 2016. [Google Scholar]
43. A. Romano and A. Hernandez, “Enhanced deep learning approach for predicting invasive ductal carcinoma from histofigpathology images,” in 2nd Int. Conf. on Artificial Intelligence and Big Data, ICAIBD, Chengdu, China, pp. 142–148, 2019. [Google Scholar]
44. M. Heller, “Automated machine learning or autoML explained, InfoWorld,” 2019. [Online]. Available: https://www.infoworld.com/article/3430788/automated-machine-learning-or-automl-explained.html. [Google Scholar]
45. Amazon web service (AWS“Amazon SageMaker autopilot,” 2020. [Online]. Available: https://aws.amazon.com/sagemaker/autopilot/. [Google Scholar]
46. Google, Google cloud AutoML 2020. [Online]. Available: https://cloud.google.com/automl. [Google Scholar]
47. J. P. Cohen, “COVID-19 image data collection,” 2020. [Online]. Available: https://github.com/ieee8023/COVID-chestxray-dataset. [Google Scholar]
48. X. Wang, Y. Peng, L. Lu, Z. Lu, M. Bagheri et al., “Chestx-ray8: Hospital- scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Honolulu, USA, pp. 2097–2106, 2017. [Google Scholar]
49. X. Xu, X. Jiang, C. Ma, P. Du, X. Li et al., “A deep learning system to screen novel coronavirus disease 2019 pneumonia,” Engineering, vol. 6, no. 10, pp. 1122–1129, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |