DOI:10.32604/iasc.2022.021211

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2022.021211 |  |
| Article |
Smart and Automated Diagnosis of COVID-19 Using Artificial Intelligence Techniques
1Department of Computer Engineering, College of Computers and Information Technology, Taif University, Taif, 21944, Saudi Arabia
2Department of Information Technology, College of Computers and Information Technology, Taif University, Taif, 21944, Saudi Arabia
3Department of Electronics and Electrical Communications, Faculty of Electronic Engineering, Menoufia University, Menouf, 32952, Egypt
4Department of Electrical Engineering, Faculty of Engineering, Menoufia University, Shebin El-Kom, 32511, Egypt
5Department of Computer Science and Engineering, Faculty of Electronic Engineering, Menoufia University, Menouf, 32952, Egypt
6Department of Electronics and Electrical Communication Engineering, Al-Obour High Institute for Engineering and Technology, Al-Obour, 3036, Egypt
7Department of the Robots and Intelligent Machines, Faculty of AI, Kafrelsheikh University, Egypt
*Corresponding Author: Masoud Alajmi. Email: ms.alajmi@tu.edu.sa
Received: 27 June 2021; Accepted: 24 September 2021
Abstract: Machine Learning (ML) techniques have been combined with modern technologies across medical fields to detect and diagnose many diseases. Meanwhile, given the limited and unclear statistics on the Coronavirus Disease 2019 (COVID-19), the greatest challenge for all clinicians is to find effective and accurate methods for early diagnosis of the virus at a low cost. Medical imaging has found a role in this critical task utilizing a smart technology through different image modalities for COVID-19 cases, including X-ray imaging, Computed Tomography (CT) and magnetic resonance image (MRI) that can be used for diagnosis by radiologists. This paper combines ML with imaging analysis in an artificial deep learning approach for COVID-19 detection. The proposed methodology is based on convolutional long short term memory (ConvLSTM) to diagnose COVID-19 automatically from X-ray images. The main features are extracted from regions of interest in the medical images, and an intelligent classifier is used for the classification task. The proposed model has been tested on a dataset of X-ray images for COVID-19 and normal cases to evaluate the detection performance. The ConvLSTM model has achieved the desired results with high accuracy of 91.8%, 95.7%, 97.4%, 97.7% and 97.3% at 10, 20, 30, 40 and 50 epochs that will detect COVID-19 patients and reduce the medical diagnosis workload.
Keywords: COVID-19; deep learning; X-ray images; conv LSTM
In December 2019, the COVID-19 pandemic emergency was identified in China and is still spreading from human to human rapidly worldwide. In addition to its public health implications, it is a substantial financial burden for all countries on top of the problem surrounding the low efficiency of vaccinations and the difficulty of obtaining them. Therefore, the detection of COVID-19 at an early stage plays a vital role in controlling and transmitting the virus. According to the Chinese government guidelines [1], the virus diagnosis by gene sequencing for blood or respiratory samples is the main pointer for reverse transcription-polymerase chain reaction (RT-PCR). However, the RT-PCR process takes a long time to get results compared to the rapid spread rate of COVID-19, which means many infected patients go undetected. Several efforts have been made to search for alternative methods to mitigate the current inefficiency and scarcity of COVID-19 testing.
Over the past two decades, automatic disease diagnosis has been used in the medical sciences to provide fast and accurate results. Medical imaging has become one of the most important medical sciences which are based on visualization diagnosis. It has captured researchers’ attention through different modalities such as X-ray, CT, and MRI to diagnose different diseases. The mechanism of image acquisition plays a role in the correct diagnosis of the COVID-19 pandemic from the acquired images [2–5]. X-ray beams are directed towards the lung, as X-ray imaging depends on the interaction between photons and electrons, resulting in different absorption levels due to densities of tissues classes [6–11]. Hence, there is a difference between normal lung images, pneumonia images, and COVID-19 images, as illustrated in Fig. 1.
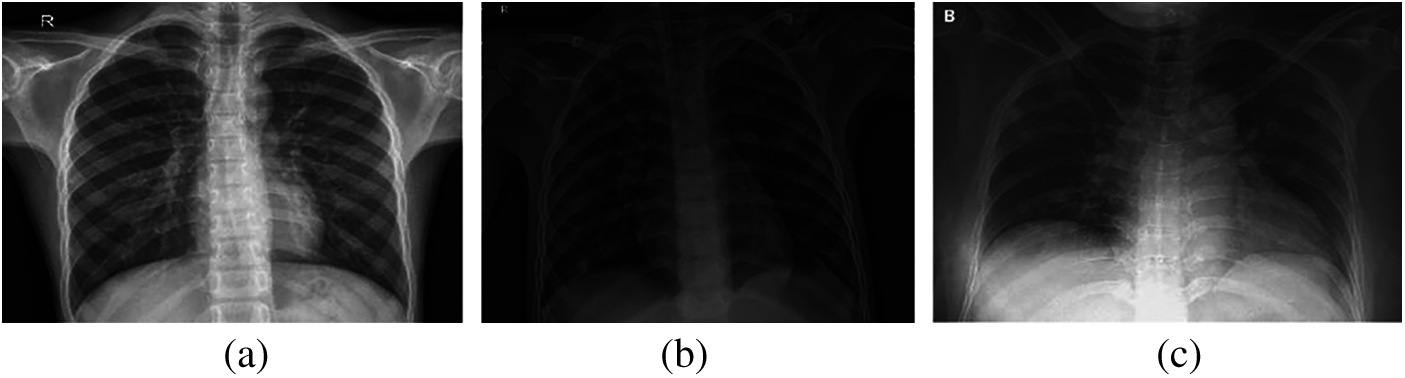
Figure 1: (a) Normal, (b) pneumonia, and (c) COVID-19 X-ray images
On the other hand, CT imaging was applied in COVID-19 detection cases to show ground glass patterned areas in the images. If these areas are determined accurately, this will help in the process of diagnosis [12–15]. Generally, CT images are generated by collecting several angular projections of X-ray images. The norma, pneumonia, and COVID-19 cases can be detected based on CT images, as illustrated in Fig. 2. Researchers have conducted several approaches by applying medical images using computer vision [16], machine and deep learning [17–19] to automatically diagnose several human body ailments for smart healthcare [20,21]. The recent advanced applications are based on machine learning and artificial intelligence concept to produce proper performance. Deep learning methods have improved the detection accuracy of tumor areas in the lungs, bone suppression by X-ray, diabetic retinopathy, prostate segmentation, skin lesions, and myocardial presence in coronary CT imaging [22–25]. Moreover, deep learning models are generally used for predicting time arrangement to obtain impressive results across diverse fields.

Figure 2: (a) Normal, (b) pneumonia, and (c) COVID-19 CT images
Therefore, several researchers have executed simulated intelligence models such as convolutional neural network (CNN), recurrent neural network (RNN) and LSTM to distinguish the medical images, which have proven their effectiveness and efficiency.
This paper focuses on the fully automatic identification of COVID-19 patients by deep learning techniques. The proposed strategy employs an enhanced deep learning algorithm ConvLSTM as a feature extract to accurately detect COVID-19 cases. The main role of the algorithm is based on images classification of X-rays images for Non-COVID-19 and COVID-19 cases with high accuracy. The proposed algorithm is evaluated to measure the classification model performance to ensure its validity.
The remaining sections are arranged as follows: Section 2 explores the related work. Section 3 presents the work methodology. Section 4 studies the proposed model with evaluation matrices. The strategy is concluded in Section 5.
The deep learning technique has performed classification tasks directly from images since 2006 to achieve an optimum modality for early detection. From this perspective, various approaches for COVID-19 detection are presented.
The authors in [26] presented a multi-channel scheme for pneumonia detection based on the utilization of two pre-trained convolutional neural networks: RetinaNet and Mask R-CNN. Each of them is used for the localization of affected pneumonia areas in chest X-ray images. Bounding boxes with different sizes are set to localize the regions of interest. Optimization of the bounding box size at each region of interest is performed depending on the histograms of the pixels in the bounding boxes. The pneumonia detection results obtained with this ensemble method were not good enough. Hence, the authors recommended the augmentation of frontal and lateral chest X-ray images for a more efficient diagnosis.
The authors in [27] adopted the concept of transfer learning and a deep residual network for the diagnosis of pediatric pneumonia from X-ray chest images. The transfer learning facilitates the diagnosis process because it depends on pre-trained neural networks, which saves the heavy training burden for the diagnosis of pediatric pneumonia. The residual network is used to capture the small changes that appear in the images due to pneumonia. The residual network avoids the deterioration of the fitting effect with the increase of the depth of the network. In the convolution processes adopted in the deep residual network, each neuron is selected as small as possible to the total receptive field to capture tiny changes in the images. Dilated convolution is also adopted to minimize the effect of each convolution layer on the subsequent one. This designed structure managed to achieve an accuracy level of 90% in the classification of pediatric pneumonia cases compared to 74%, 82%, 85% and 88% achieved by VGG16, DenseNet121, InceptionV3 and Xception networks, respectively.
The authors in [28] presented a hybrid deep learning approach for pneumonia from X-ray chest images based on the utilization of multiple pre-trained networks, including AlexNet, VGG-16, and VGG-19. Each of these networks gives 1000 deep features from each image at hand. A feature reduction strategy from 1000 to 100 features was adopted based on the minimum redundancy maximum relevance algorithm for each pre-trained network. Feature concatenation was also adopted in this algorithm to generate a feature vector composed of 300 features that can be used subsequently for classification with a traditional classifier. Comparisons among linear regression, linear discriminant analysis, k-nearest neighbors, decision tree, and support vector machine classifiers were presented in this study. An accuracy of 99.41% for pneumonia classification was achieved. This high level of accuracy is attributed to the utilization of multiple pre-trained networks and the feature reduction and concatenation strategy. It is expected that this strategy will succeed if applied for the classification of COVID-19 images.
The authors in [29] developed a deep learning approach for pneumonia and introduced a CNN called Mask-RCNN. The objective of this network is to perform pixel-wise segmentation considering both local and global features. The authors performed augmentation on the available data and images. In addition, they developed a post-processing strategy to merge different outputs in a multi-channel decision fusion process. They managed to localize opacities with bounding boxes but concluded that the chest image size is an important factor in determining pneumonia effects correctly. This may appear as a drawback of this method that needs further consideration.
The authors in [30] studied lung diseases from X-ray and CT scans as an abnormality detection process. Abnormalities incurred in either X-ray or CT images have been detected with two scenarios. The first one is a modified version of AlexNet by introducing a flattening layer, two fully connected layers and a support vector machine classifier for abnormality detection. The second scenario depends on merging the deep-learned features with hand-crafted features to generate more representative features to classify abnormal regions. The hand-crafted features include features extracted from the gray level co-occurrence matrix and Hu moments. Feature selection is performed to make the hand-crafted feature vector equal in size to the deeply extracted feature vector. After that, feature fusion is performed with the principle component analysis to generate unified feature vectors that can be valid for further classification of abnormalities. Simulation experiments with these scenarios on large lung datasets lead to accuracy levels of classification up to 97%.
The authors in [31] presented a study about the progression of COVID-19 patients through the utilization of high-resolution CT images. They divided the phases of the disease into four phases: early phase, progressive phase, severe phase, and dissipative phase based on the available CT images and the occurrence of lesions in lung images. The early-stage corresponds to the early days of infection. In this phase, a few glass opacities with thick blood vessel shadows appear in the lungs. In the progressive stage, multiple patchy glass opacities appear in the lungs. In addition, lesions reveal sub-pleural distribution with thickened vascular shadows. In the severe phase, multiple consolidations appear in the two lungs in addition to ground-glass opacities. In the dissipative phase, nodular consolidations appear in the lungs with the spread of lesions. In addition, some halo signs are seen around the nodules. From this study, it is clear that the phase of the disease can be known from the lung CT images with different distinguishing features. In addition, one of the criteria adopted for releasing patients from quarantine is chest image improvement. This improvement is represented in the reabsorption of the lesions. The authors of this study recommended using artificial intelligence to determine the phase of the disease to give the appropriate treatment and evaluate recovery based on lung chest images to allow patients to leave quarantine.
The authors in [32] studied the classification of bacterial pneumonia, COVID-19 disease, and normal incidents from X-ray images based on deep learning and transfer learning. They worked on pre-trained deep networks including VGG19, MobileNet v2, Inception, Xception and Inception ResNet v2 [22–25] and performed slight modifications of their parameters to classify between the three diseases. They worked on two datasets to test the pre-trained deep networks. The first one comprises 1427 X-ray images: 224 images for COVID-19, 700 images for bacterial pneumonia, and 504 images of normal cases. The second dataset comprises 224 images for COVID-19, 714 images for pneumonia, and 504 images for normal cases. The maximum accuracy of classification in this study was 96.78%, the maximum sensitivity was 98.66%, and the maximum specificity was 96.46%. These levels reveal that further enhancement is still possible through parameter optimization of the deep networks used to diagnose COVID-19 cases.
The authors in [33] demonstrated a practical methodology to detect the abnormalities related to the loss in local lung volume as an early indication of coronavirus disease. A crazy-paving pattern was found in ultra-high-resolution computed tomography (U-HRT) in addition to lobules that were smaller in the affected lungs. Six tested patients with COVID-19 were subjected to U-HRT imaging. The result showed that all patients had several bilateral lesions in the lungs with a crazy-paving pattern. The authors noticed that his CT volume decreases to around 75% for the patient who needs oxygen. Santosh discussed the importance of artificial intelligence tools to work as cross-population train/test models [34]. In [35], the authors review all available information about the epidemiology, diagnosis, isolation and treatments of COVID-19. In addition, the registered trials for investigating COVID-19 infection treatments, antibodies, convalescent plasma transfusion and vaccines are listed. In the COVID-19 global pandemic, smart city has great relevance, closely tied in with local resources availability. The role of the smart city during an outbreak is demonstrated in detail in [36] for the entire world population. The study in [37] has manipulated the data unavailability via two data augmentation strategies by using a set of simple image transformations and a data augmentation framework that distends the limited dataset of X-ray and CT images by 100%. It compared the work performance against traditional machine learning techniques employing the SVM and k-NN algorithms. In [38], the paper provides a COVID-19 detection system based on two deep learning modalities, CNN and ConvLSTM, using a set of CT images and a set of X-ray images. The simulation results proved that the proposed modalities could be adopted for the rapid detection of COVID-19.
3.1 Long Short-Term Memory (LSTM)
On image classification, CNN is considered the first choice because it extracts image components and learns the larger structures. Also, RNN learns to extract image features over time. However, RNN is practically unstable and causes gradient explosion through long time windows of back-propagating gradients. To avoid the long-term dependencies problem of RNN, the LSTM method is applied by using memory cells instead of the hidden units to store data until it is relevant. Therefore, LSTM is RNN architecture with a proper gradient-based learning algorithm presented to control information through the memory cell. Also, an LSTM network can learn over 1000 steps to bridge intervals in the noisy case without any loss in time and can connect previous information to the present obtained data [39,40].
An LSTM layer consists of memory blocks that act as differentiable memory chips in a digital computer. Every block has recurrently connected memory cells and three multiplicative gates or units; an input gate, a “forget” gate to provide the continuous analogues operations of cells through the switch of the gates, and an output gate as shown in Fig. 3. LSTM external inputs are x(t) the current input vector, previous cell state c(t−1) and h(t−1) the previous hidden state. The three gates using the nonlinear activation functionσ are calculated as [41]:
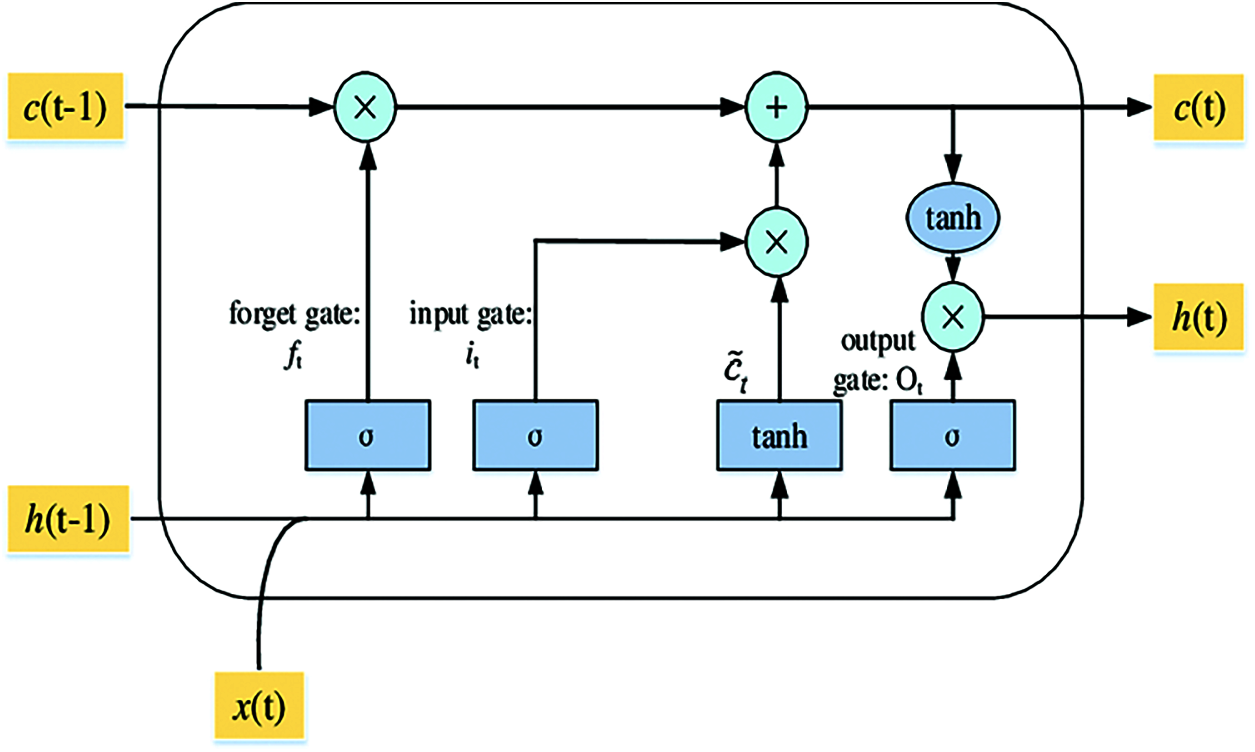
Figure 3: The structure of the LSTM unit
The sigmoid function is usually employed as an activation function for the gates using nonlinear (tanh). After that, an intermediate state C(t) inside the LSTM is generated, and the states of the memory cell and hidden are updated using multiplication operation for two vectors as:
The real dataset was prepared using various x-ray images of two different subsets to train and enable efficient diagnosis through the LSTM model, which obtained from [42,43]. Each subset contains a combined dataset as 304 images of COVID-19 and 304 normal images. Firstly, the dataset is transformed and coded into a proper interpretation of the LSTM model. Then, the model learns from the transformed samples by a function that maps the past observation sequence input to an output observation. The datasets are divided into 80% of data utilized for training and the remaining 20% utilized for the testing purpose.
4.1 LSTM Proposed Architecture
The development of the suggested smart and automated LSTM for the COVID-19 diagnosis framework is debated in three stages: pre-processing, feature extraction, and classification stages. In pre-processing stage, frame sampling is carried out on the input medical image to extract its frames. Then, the input medical images are resized to a unified size determined in the input layer. The stage of feature extraction has layers that enclose neurons arranged in 4 dimensions: the samples number, the input image width and height, and the filters number utilized for each convolution layer.
The proposed ConvLSTM modality comprises five layers: the LSTM, the fully connected, the Rectified Linear Unit (ReLU), the dropout, and the softmax layers, as shown in Fig. 4. The classification process was performed with the activation function in the softmax layer. The convolutional layer with a 3 × 3 kernels size is employed for feature extraction activated by the ReLU function using an input image frame size of 224 × 224.
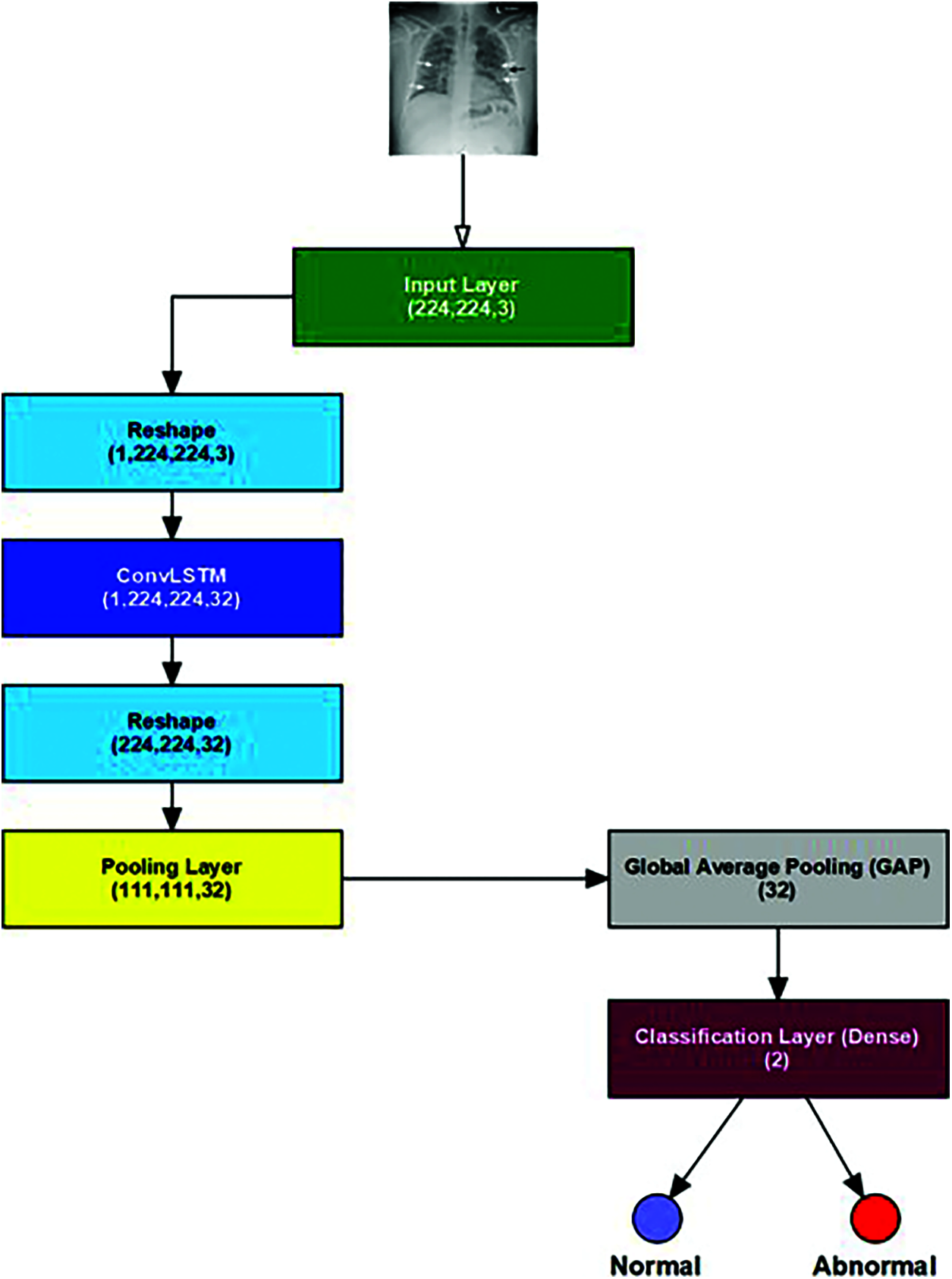
Figure 4: The proposed ConvLSTM architecture
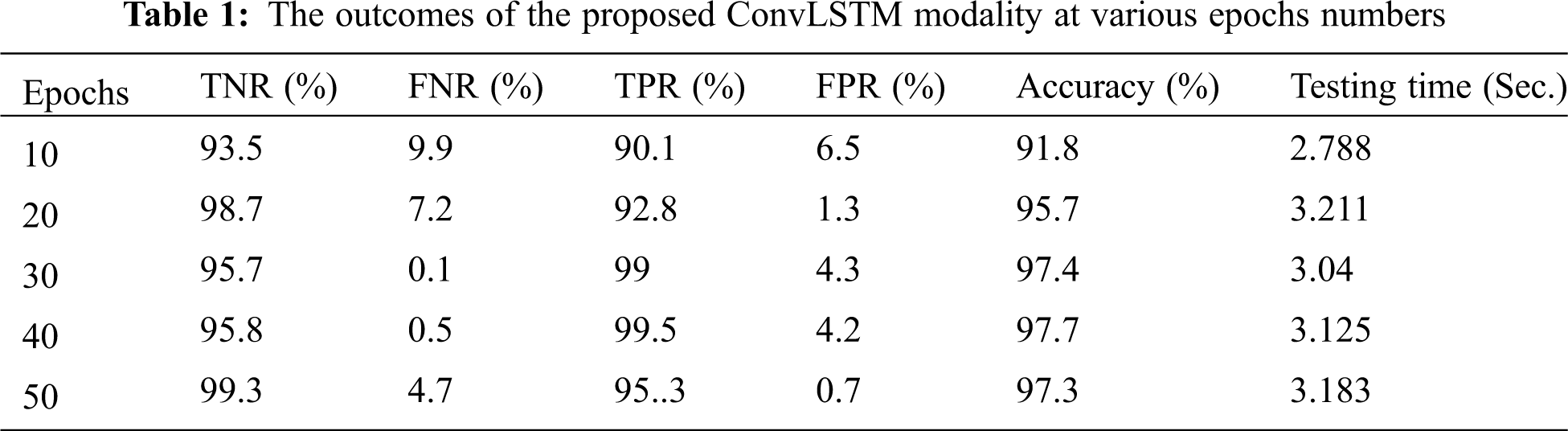
The proposed ConvLSTM modality is examined on the applied X-ray dataset to differentiate among normal and COVID-19 (abnormal) states. Figs. 5 and 6 depict the curves of accuracy and loss models for the training and validation of the proposed ConvLSTM modality. Tab. 1 depicts the outcomes of the proposed ConvLSTM modality at various numbers of epochs of 10, 20, 30, 40 and 40 for True Negative Ratio (TNR), False Negative Ratio (FNR), True Positive Ratio (TPR), False Negative Ratio (FPR), accuracy and testing time. The accuracy of the proposed ConvLSTM modality in detection can be calculated as:
where,
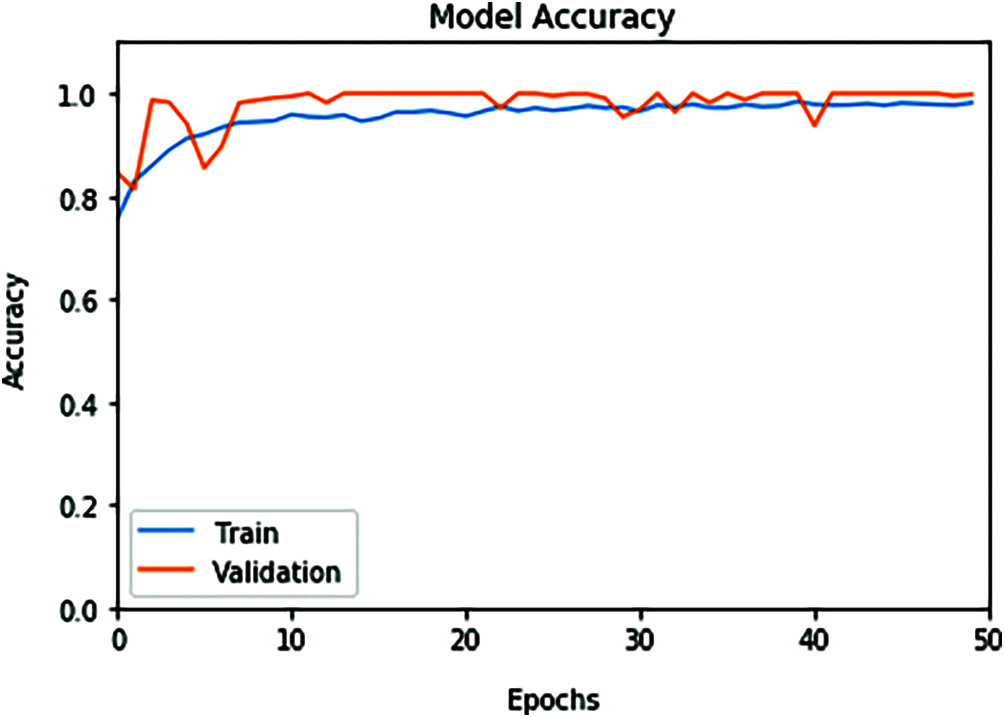
Figure 5: ConvLSTM accuracy model curve of the training and validation data
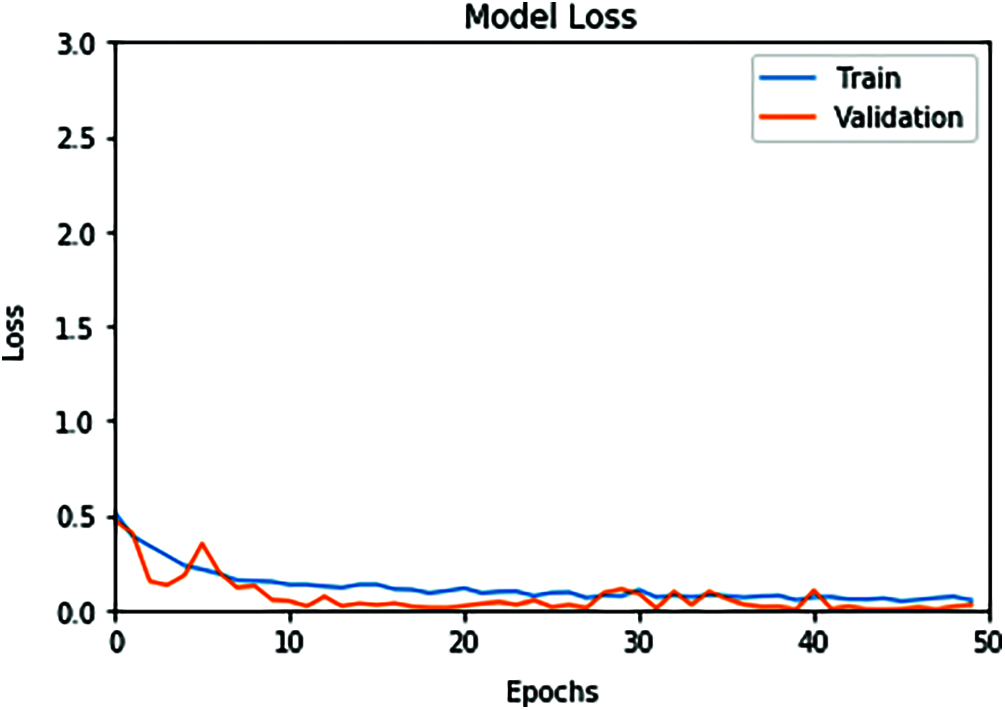
Figure 6: ConvLSTM loss model curve of the training and validation data
(TN) denotes the cases classified as healthy, and (TP) denotes the cases classified as patients.
(FN) denotes the cases classified incorrectly as healthy. (FP) denotes the cases classified incorrectly as patients.
The simulation outcomes demonstrate that the presented ConvLSTM modality has an accuracy of 91.8%, 95.7%, 97.4%, 97.7% and 97.3% at 10, 20, 30, 40 and 50 epochs respectively. In addition, TNR, FNR, TPR, FPR and the testing time of the ConvLSTM modality are estimated. It is indicated that the proposed model achieves high performance and can effectively detect COVID-19 patients with high accuracy using X-ray images.
This work presented an effective method for detecting COVID-19 cases based on the deep learning modality of ConvLSTM. The proposed ConvLSTM modality is tested on X-ray images dataset for COVID-19 and normal cases to evaluate detection performance. The proposed ConvLSTM modality achieved a high classification performance with an accuracy of 91.8%, 95.7%, 97.4%, 97.7% and 97.3% at 10, 20, 30, 40 and 50 epochs respectively. Hence, the proposed ConvLSTM can be considered and adopted for rapid detection of COVID-19.
Acknowledgement: The authors would like to thank the Deanship of Scientific Research, Taif University, Saudi Arabia, for funding the research project number 1–441–21.
Funding Statement: This research was funded by the Deanship of Scientific Research, Taif University, Saudi Arabia, under research project number 1–441–21.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. T. Ai, Z. Yang, H. Hou, C. Zhan, C. Chen et al., “Correlation of chest CT and RT-pCR testing in coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases,” Radiology, vol. 296, no. 2, pp. E32–E40, 2020. [Google Scholar]
2. T. Ozturk, M. Talo, E. Yildirim, A. Baloglu, U. Yildirim et al., “Automated detection of COVID-19 cases using deep neural networks with X-ray images,” Computers in Biology and Medicine, vol. 121, no. pp. 103792, pp. 1–11, 2020. [Google Scholar]
3. E. Dong, H. Du and L. Gardner, “An interactive web-based dashboard to track COVID-19 in real time,” Lancet Infectious Diseases, vol. 20, no. 5, pp. 533–534, 2020. [Google Scholar]
4. F. Shi, J. Wang, J. Shi, Z. Wu, Q. Wang et al., “Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19,” IEEE Reviews in Biomedical Engineering, vol. 14, pp. 4–15, 2021. [Google Scholar]
5. L. Li, L. Qin, Z. Xu, Y. Yin, X. Wang et al., “Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT,” Radiology, vol. 296, no. 2, pp. E67–E72, 2020. [Google Scholar]
6. S. Rao and J. Vazquez, “Identification of COVID-19 can be quicker through artificial intelligence framework using a mobile phone-based survey when cities and towns are under quarantine,” Infection Control & Hospital Epidemiology, vol. 41, no. 7, pp. 826–830, 2020. [Google Scholar]
7. R. Vaishya, M. Javaid, I. Khan and A. Haleem, “Artificial intelligence (AI) applications for COVID-19 pandemic,” Diabetes & Metabolic Syndrome: Clinical Research & Reviews, vol. 14, no. 4, pp. 337–339, 2020. [Google Scholar]
8. Z. Allam and D. Jones, “On the coronavirus (COVID-19) outbreak and the smart city network: Universal data sharing standards coupled with artificial intelligence (AI) to benefit urban health monitoring and management,” Healthcare, vol. 8, no. 1, 46, pp. 1–9, 2020. [Google Scholar]
9. K. C. Santosh, “AI-Driven tools for coronavirus outbreak: Need of active learning and cross-population train/test models on multitudinal/multimodal data,” Journal of Medical Systems, vol. 44, no. 93, pp. 1–5, 2020. [Google Scholar]
10. I. D. Apostolopoulos and T. A. Mpesiana, “COVID-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks,” Physical and Engineering Sciences in Medicine, vol. 43, pp. 635–640, 2020. [Google Scholar]
11. X. Xu, X. Jiang, C. Ma, P. Du, X. Li et al., “A deep learning system to screen novel coronavirus disease 2019 pneumonia,” Engineering, vol. 6, no. 10, pp. 1122–1129, 2020. [Google Scholar]
12. D. Singh, V. Kumar and M. Kaur, “Classification of COVID-19 patients from chest CT images using multi-objective differential evolution-based convolutional neural networks,” European Journal of Clinical Microbiology & Infectious Diseases, vol. 39, pp. 1379–1389, 2020. [Google Scholar]
13. L. Huang, R. Han, T. Ai, P. Yu, H. Kang et al., “Serial quantitative chest CT assessment of COVID-19: Deep-learning approach,” Radiology: Cardiothoracic Imaging, vol. 2, no. 2, pp. 1–8, 2020. [Google Scholar]
14. P. Belfiore, F. Urraro, R. Grassi, G. Giacobbe, G. Patelli et al., “Artificial intelligence to codify lung CT in COVID-19 patients,” Radiologica Medica, vol. 125, pp. 500–504, 2020. [Google Scholar]
15. M. Hasan, M. AL-Jawad, H. Jalab, H. Shaiba, W. Ibrahim et al., “Classification of COVID-19 coronavirus, pneumonia and healthy lungs in CT scans using Q-deformed entropy and deep learning features,” Entropy, vol. 22, no. 5, 517, pp. 1–15, 2020. [Google Scholar]
16. J. Thevenot, M. B. Lopez and A. Hadid, “A survey on computer vision for assistive medical diagnosis from faces,” IEEE Journal of Biomedical and Health Informatics, vol. 22, no. 5, pp. 1497–1511, 2018. [Google Scholar]
17. B. Jin, Y. Qu, L. Zhang and Z. Gao, “Diagnosing Parkinson disease through facial expression recognition: Video analysis,” Journal of Medical Internet Research, vol. 22, no. 7, e18697, pp. 1–12, 2020. [Google Scholar]
18. M. Islam, H. Iqbal, M. Haque and M. Hasan, “Prediction of breast cancer using support vector machine and K-nearest neighbors,” in IEEE Region 10 Humanitarian Technology Conf. (R10-HTCpp. 226–229, Dhaka, Bangladesh, 2017. [Google Scholar]
19. M. Haque, M. Islam, H. Iqbal, M. Reza and M. Hasan, “Performance evaluation of random forests and artificial neural networks for the classification of liver disorder,” in Int. Conf. on Computer, Communication, Chemical, Material and Electronic Engineering (IC4ME2pp. 1–5, Rajshahi, Bangladesh, 2018. [Google Scholar]
20. H. Naz and S. Ahuja, “Deep learning approach for diabetes prediction using PIMA Indian dataset,” Journal of Diabetes & Metabolic Disorders, vol. 19, pp. 391–403, 2020. [Google Scholar]
21. N. Tajbakhsh, J. Y. Shin, S. R. Gurudu, R. T. Hurst, C. B. Kendall et al., “Convolutional neural networks for medical image analysis: Full training or fine tuning,” IEEE Transactions on Medical Imaging, vol. 35, pp. 1299–1312, 2016. [Google Scholar]
22. A. Rahaman, M. Islam, M. Sadi and S. Nooruddin, “Developing IoT based smart health monitoring systems: A review,” Revue Intelligence Artificielle, vol. 33, no. 6, pp. 435–440, 2019. [Google Scholar]
23. M. Islam, A. Rahaman and M. Islam, “Development of smart healthcare monitoring system in IoT environment,” SN Computer Science, vol. 1, no. 185, pp. 1–11, 2020. [Google Scholar]
24. S. Lakshmanaprabu, S. Mohanty, K. Shankar, N. Arunkumar and G. Ramirez, “Optimal deep learning model for classification of lung cancer on CT images,” Future Generation Computer Systems, vol. 92, pp. 374–382, 2019. [Google Scholar]
25. Y. Chen, X. Gou, X. Feng, Y. Liu, G. Qin et al., “Bone suppression of chest radiographs with cascaded convolutional networks in wavelet domain,” IEEE Access, vol. 7, pp. 8346–8357, 2019. [Google Scholar]
26. I. Sirazitdinov, M. Kholiavchenko, T. Mustafaev, Y. Yixuan, R. Kuleev et al., “Deep neural network ensemble for pneumonia localization from a large-scale chest X-ray database,” Computers and Electrical Engineering, vol. 78, pp. 388–399, 2019. [Google Scholar]
27. G. Liang and L. Zheng, “A transfer learning method with deep residual network for pediatric pneumonia diagnosis,” Computer Methods and Programs in Biomedicine, vol. 187, no. 104964, pp. 1–9, 2020. [Google Scholar]
28. M. To˘gaçar, B. Ergen, Z. Cömert and F. Özyurt, “A deep feature learning model for pneumonia detection applying a combination of mRMR feature selection and machine learning models,” IRBM, vol. 41, no. 4, pp. 212–222, 2020. [Google Scholar]
29. A. Jaiswal, P. Tiwari, S. Kumar, D. Gupta, A. Khanna et al., “Identifying pneumonia in chest X-rays: A deep learning approach,” Measurement, vol. 145, pp. 511–518, 2019. [Google Scholar]
30. A. Bhandary, G. Prabhu, V. Rajinikanth, K. Thanaraj, S. Satapathy et al., “Deep-learning framework to detect lung abnormality-A study with chest X-ray and lung CT scan images,” Pattern Recognition Letters, vol. 129, pp. 271–278, 2020. [Google Scholar]
31. L. Mingzhi, L. Pinggui, B. Zeng, L. Zongliang, P. Yu et al., “Coronavirus disease (COVID-19Spectrum of CT findings and temporal progression of the disease,” Academic Radiology, vol. 27, no. 5, pp. 603–608, 2020. [Google Scholar]
32. I. Apostolopoulos and T. Mpesiana, “COVID-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks,” Physical and Engineering Sciences in Medicine, vol. 43, pp. 635–640, 2020. [Google Scholar]
33. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” in the 3rd Int. Conf. on Learning Representations (ICLRKuala Lumpur, Malaysia, pp. 1–14, 2015. [Google Scholar]
34. A. Howard, M. Zhu and B. Chen, “Mobile nets: efficient convolutional neural networks for mobile vision applications,” ArXiv preprint:1704.04861, pp. 1–9, 2017. [Google Scholar]
35. P. Zhai, Y. Ding, X. Wu, j. Long, Y. Zhong et al., “The epidemiology, diagnosis and treatment of COVID-19,” International Journal of Antimicrobial Agents, vol. 55, no. 5, no. 105955, pp. 1–13, 2020. [Google Scholar]
36. S. Yang and Z. Chong, “Smart city projects against COVID-19: Quantitative evidence from China,” Sustainable Cities and Society, vol. 70, no. 102897, pp. 1–9, 2021. [Google Scholar]
37. A. Sedik, A. Iliyasu, B. Abd El-Rahiem, M. Abdel Samea, A. Abdel-Raheem et al., “Deploying machine and deep learning models for efficient data-augmented detection of COVID-19 infections,” Viruses, vol. 12, no. 7, no. 769, pp. 1–29, 2020. [Google Scholar]
38. A. Sedik, M. Hammad, F. Abd El-Samie, B. Gupta and A. Abd El-Latif, “Efficient deep learning approach for augmented detection of coronavirus disease,” Neural Computing and Application, pp. 1–18, 2021. [Google Scholar]
39. A. Esteva, B. Kuprel, R. A. Novoa, J. Ko and S. M. Swetter, “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, pp. 115–118, 2017. [Google Scholar]
40. M. Nour, Z. Cömert and K. Polat, “A novel medical diagnosis model for COVID19 infection detection based on deep features and Bayesian optimization,” Applied Soft Computing, vol. 97, no. 106580, pp. 1–13, 2020. [Google Scholar]
41. K. Greff, R. Srivastava, J. Koutnik, B. Steunebrink and J. Schmidhuber, “LSTM: A search space odyssey,” IEEE Transactions on Neural Networks & Learning Systems, vol. 28, no. 10, pp. 2222–2232, 2015. [Google Scholar]
42. L. L. Wang, K. Lo, Y. Chandrasekhar, R. Reas, J. Yang et al., “CORD-19: The COVID-19 open research dataset,” arXiv:2004.10706v2, preprint, 2020. [Google Scholar]
43. J. Zhang, Y. Xie, Y. Li, C. Shen and Y. Xia, “COVID-19 screening on chest X-ray images using deep learning based anomaly detection.” arXiv:2003.12338, preprint, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |