DOI:10.32604/iasc.2022.023753

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2022.023753 |  |
| Article |
A Convolutional Neural Network for Skin Lesion Segmentation Using Double U-Net Architecture
1Institute of Southern Punjab, Multan, 32100, Pakistan
2College of Computer Science and Information Systems, Najran University, Najran, 61441, Saudi Arabia
3Department of Creative Technology, Air University, Islamabad, 44200, Pakistan
4Department of Computer Science College of Computers and Information Technology, Taif University, Taif, 21944, Saudi Arabia
5College of Computer Science and Engineering, University of South Florida, Tampa, 33620, United States
*Corresponding Author: Abdulmajeed Alqhatani. Email: aaalqhatani3@gmail.com
Received: 20 September 2021; Accepted: 20 December 2021
Abstract: Skin lesion segmentation plays a critical role in the precise and early detection of skin cancer via recent frameworks. The prerequisite for any computer-aided skin cancer diagnosis system is the accurate segmentation of skin malignancy. To achieve this, a specialized skin image analysis technique must be used for the separation of cancerous parts from important healthy skin. This procedure is called Dermatography. Researchers have often used multiple techniques for the analysis of skin images, but, because of their low accuracy, most of these methods have turned out to be at best, inconsistent. Proper clinical treatment involves sensitivity in the surgical process. A high accuracy rate is therefore of paramount importance. A generalized and robust model is needed to accurately assess and segment skin lesions. In this regard, a novel approach named Double U-Net has been proposed to provide necessary strength and Robustness. This process uses two U-Net architectures stacked upon each other with ASPP which is used to squeeze out a high resolution and redundant information. In this paper, we trained the proposed architecture on the PH2 dataset and the model was evaluated on the PH2 test, ISIC-2016 and HAM datasets. Evaluation of information shows the model achieved a DSC of 0.9551 on the PH2 test dataset, 0.8104 on ISIC-2016 and 0.7645 on the HAM dataset. Analyses show results comparable to the most recently available state-of-the-art techniques.
Keywords: Skin lesion; convolutional neural network; U-net; double U-net
The most common cause of death from diseases of the skin is malignant growth. This is generally seen when the cells that make up the skin follow a pattern of growth that is different from the normal. The growth takes the form of unnatural and rapid progress whereby some character of the native tissue may be retained or the new growth may be completely different from the native tissue. In Skin tumors, there are generally three types of malignant growth which are Squamous cell carcinoma, Basal cell carcinoma and Melanoma. The most common types of skin cancer are Squamous cell carcinoma and Basal cell carcinoma. These are also known as “Non-Melanoma skin cancer” [1]. The most dangerous form of these three types is Melanoma. If this is caught in a late-stage, or, if left untreated, Melanomas can spread to other body organs, increasing significantly, the chances of death. Non-Melanoma skin cancer is the 5th [2] most common tumor, whereas Melanoma of the skin is the 19th most common cancer worldwide. Fortunately, detection of skin cancer particularly Melanoma in its early stages can result in excellent outcomes [3]. With the help of proper diagnosis and treatment, survival rates from malignant skin conditions can easily be improved. The earlier the disease is diagnosed the better the management and the better the outcome. Computer-assisted technology in medical imaging has been of vital importance in the diagnosis and management of skin cancer [4]. This technique has been widely used as a tool for separating and identifying cancerous parts of the skin from normal skin [1]. Early detection is important for proper treatment to be programmed accordingly. Usually a macroscopic, followed by a microscopic examination is done [5]. Microscopic images are obtained through the use of a magnifying lens or a specialized microscope which is used to examine the compromised area more closely, a process called Dermatoscopy. This is a non-intrusive strategy for imaging used to capture skin lesions by removing the surface layers, to get to the deeper layers [6]. Different medical image analysis techniques have been proposed to help delineate the suspicious area with images obtained through dermatoscopy. For medical image segmentation, there are two types of methodologies, traditional and Semi-Automated and/or Automated. Traditional methods relate to a visual inspection of the suspicious area by a physician. Semi-automated and automated methods involve point-based pixel intensity operations [7,8], pixel clustering methods [9–11], level set methods [12], deformable models [13] and deep-learning-based methods [14].
In medical imaging, segmentation is a challenging and exigent task. Segmentation is used to label pixels of the diseased segment through medical images, a procedure that helps the clinician get detailed information of the affected area and to separate diseased cancerous parts from healthy skin [15]. Medical Image segmentation may prove to be very demanding and taxing as it has to deal with many variables such as difficulty in acquiring high-quality images, unavailability of annotated masks and variation in imaging data of different patients [16]. In Semantic Image Segmentation, machine learning plays a vital role, especially in the deep learning-based approaches [17]. For Robotic Medical Image segmentation, Convolution neural networks have proven to be the best-in-class execution [18]. In machine learning-based methodologies, pixel-wise prediction for picture segmentation is performed to fully chart out neural architecture, stressing the need for semantic segmentation of images [19]. Another recognized method used for segmentation is U-Net architecture. Accordingly, two main functionalities are performed by the two parts of U-Net architecture called Analysis and Synthesis. These are used for feature learning and segmenting based on inherent data. U-Net is a Fully Convolutional Network (FCN) that works with the concept of skipping layers in between parts which allows deep supervision to improve network performance [20]. Robustness and Generalization are two main objectives that are important across diverse biomedical applications. Generalization centers around the capacity of the model to perform on a free dataset that is utilized to prepare the model, whereas Robustness centers on the capacity of the model to play out the difficult assignment [21]. Working on this principle, another architecture named Double U-Net design has been proposed to provide a baseline for achieving these objectives. This technique uses modified U-Net and VGG19 as encoders in the network that allow deep learning networks to produce improved segmented masks which optimize the overall segmentation performance compared to U-Net alone.
Our endeavor, therefore, was to use the Double U-Net design to segment the cancerous area from normal skin to check the Generalization and Robustness of the architecture over U-Net. The role of this paper is to examine the accuracy of the proposed Double U-Net architecture in comparison with other methods to semantically segment skin lesions in medical images. This method has been benchmarked with other techniques via results through different statistics like Accuracy, Dice score and other performance indicators, that aid in measuring with other similar segmentation processes.
The rest of this paper is structured as follows. Section 2 presents related work. Section 3 focuses on materials and methods and Section 4 explores the implementation and performance evaluation of the proposed model. Section 5 presents the benchmark. Finally, Section 6 provides conclusions and future directions.
Many methods have been developed over the past few years to achieve better and higher accuracy in precisely identifying the segmented area of a skin lesion in Dermatoscopic images. Deep learning has played a vital role in Medical Image segmentation. Recently a new deep learning architecture has been proposed named NABLA-N. This provides a variety of combination procedures through deciphering units to help in better division assignments of dermatographic images. A blend of low-level and undeniable level feature maps is provided in this model. This ensures better feature representation for segmentation and essentially impacts with better qualitative and quantitative results when tested on International Skin Imaging Collaboration 2018 (ISIC-2018) [22,23]. Another system, which is a modified form of U-Net, has been proposed for semantic segmentation. This includes two branches known as Semantic and Detail branches that extract information from the Deep and Shallow layers. A Malblock module is used that capitalizes on the idea of collective knowledge and Class Activation MAP (CAM) to reduce the redundant feature [24]. Another modification in U-Net is made on paper [25]. This utilizes nearby binary Convolution on U-Net rather than standard Convolution and replaces the encoder part of the U-Net with Local Binary Convolutional Neural Network (LBCNN) layers to naturally learn and fragment highlights. Multi-scale context-guided architecture called Multi-Scale Context-Guidednet (MSCGnet) has been proposed to address the difficulties of low contrast and varieties of shading across space and fluffiness. Context space attention structures are used in the down-sampling part to reduce the loss. MSCGnet network with iteration has been proposed to boost performance in what is known as Iterative Multi-Scale Context-Guidednet (IMSCGnet) [26].
An efficient fully convolutional neural network, named DermoNet, an FCN, has been proposed to productively section skin cancer. This architecture reuses the data of past layers due to thickly associated Convolutional squares and Skip associations, with accurate extraction of the segmentation [27]. Another Framework named Dual Objective Networks (DONet) has been proposed. This utilizes two symmetric decoders using different loss functions for approaching different objectives. It is then aggregated to deliver a final prediction. Furthermore, the purpose of the Recurrent Context Encoding Module (RCEM) is to deal with challenges of size and shape [28]. TMD-UNET is the successor to U-Net architecture. This has been proposed with some modifications which include dilated Convolution instead of regular Convolution, coordination of multi-scale input highlights on the info side of the model and dense Skip association rather than customary Skip association [29]. Another architecture named I-Net has been proposed to get accurate semantic segmentation without losing any spatial information. This augments the responsive field by expanding the kernel size of Convolutional layers rather than down-sampling and by co-relating with the featured map of previous layers. It uses two overlapping max-pooling to extract the sharpest features. Multiple shortcuts can be added because of a fixed size and fixed number of channels [30]. Another modification has been made in U-Net which combines dilated Convolution and Pyramid Pooling Module. Dilated Convolution computes High Spatial Resolution feature maps and Pyramid Pooling obtains more contextual information [31]. For automatic Semantic segmentation, a method has been proposed called the DSNet. In this model, researchers utilized Profundity Shrewd Distinct Convolution to project learned features onto the pixel space of the encoder at various stages to make the architecture lightweight [32]. To add to this, a neutrosophic graph cut algorithm using an optimized clustering estimation algorithm for Dermatoscopic skin lesion segmentation has been proposed. Here, researchers have used Histogram Based Clustering Estimation (HBCE) to obtain the initial number of clusters with the corresponding centroid. Results of this novel approach when compared to traditional HBCE, have achieved an accuracy of 97% [33]. Another approach that uses a fully automatic deep learning ensemble method to segment the skin lesion boundary of demographic images and evaluates the performance on the ISIC-2017 data set has been completed. This has achieved a Jacquard index of 79.58% showing out-performance over Full Resolution Convolutional Network (FrCN), FCN, U-Net and SegNet by achieving an accuracy of 97.6% [34]. For automatic segmentation of skin lesions in dermatographic images, an architecture based on a fully deep Convolutional-deConvolutional Neural Network (CDNN) has been proposed that specifically focuses on effective network architecture and appropriate training strategies [35]. Similar work for skin lesion segmentation named FrCN has been accomplished. This methodology improves the segmentation of skin lesions by learning full resolution features of every pixel of every input image without considering the need for pre/post-processing. This methodology was tested on the International Symposium on Biomedical Imaging (ISBI 2017) and the PH2 dataset. Accuracy of 94.03% and 95% have been achieved respectively. This has outperformed FCN, U-Net and SegNet [36]. In 2019, a skin lesion segmentation approach along with deep CNN and named “You Only Look Once” (YOLO) and grab cut algorithm was established. This performed skin lesion segmentation in 4 steps including detection, segmentation, removal and post-processing. This was tested on the PH2 dataset and ISBI 2017. It achieved results similar to other deep learning methods in terms of the Dice coefficient and metrics of accuracy [34].
During the last few years, different deep learning methods have been proposed to segment skin lesions and have been tested on different publicly available datasets. The recent development of artificial intelligence systems requires both Generalizations, which is the model's ability to execute on independent datasets and Robustness, which is the model's ability to tackle challenging images, to provide standardization. It was therefore deemed crucial to develop a model that was both Robust and Generalizable [21]. An architecture was therefore introduced for image segmentation named Double U-Net architecture. This uses two-unit architectures stacked upon each other. The first one includes VGG–19 as the encoder. It also uses Atrous Spatial Pyramid Pooling (ASPP) for contextual information. This was tested on Medical Image Computing and Computer-Assisted Intervention (MICCAI) automatic polyp detection, CVC-clinic DB and Data Science Bowl 2018 boundary segmentation datasets. This has shown better results than U-Net architecture alone. In this paper, we have shown that the Double U-Net architecture can be a strong baseline for medical image segmentation as it provides both Generalization and Robustness [37].
Other researchers have also used techniques for image segmentation based on the CNN U-Net architecture, modified by ResNet in the decoder and Convolutional layer. This architecture has been applied to three scenarios of skin damage. Experimental results show an improvement in image segmentation with 89% accuracy compared to the original U-Net, which achieves 84% and 81% accuracy for the RelayNet. Another related work by Dash et al. [38] has reported effective performance for image segmentation in detecting the Psoriasis skin lesion using a modified U-Net-based full Convolutional network (PsLSNet). The reported results were measured by different evaluation metrics to show the novelty of their proposed method and achieved a Dice coefficient (DSC) of 93.03%, accuracy (ACC) of 94.80%, Jaccard index (JI) of 86.40%, Sensitivity of 89.60% (SE) and Specificity of 97.60% (SP). Mishra et al. [39] proposed CNN architecture for lesion segmentation based on U-Net and GPU acceleration and compared it with other research work in the ISBI challenge. Their proposed method achieved 84% of the JC index whereas; the other four researchers achieved an accuracy below 77% of the JC index.
2.1 Atrous Spatial Pyramid Pooling
Atrous Partial Pyramid Pooling (Fig. 1) known as ASPP [40] helps to extract a high-resolution feature map that will assist in the segmentation process [41]. It is used in obtaining multi-scale context information by applying parallel Convolutions with different Atrous rates which handle the segmentation of the object at different scales [42]. The parallel results are concatenated one by one. Convolution is applied to get the output. This process helps in Semantic Segmentation [43]. Researchers, which include Long et al. [44] have shown that multi-scale context information can be aggregated without the loss of resolution through Atrous Convolution, which is generally applied to one or two-dimensional input data x[i]. After categorizing, w[k], Output y[i] is attained as follows:
where i is the area of the pixels, the atrous convolution dilated value is r and the dimension of the Convolution kernel is k. A special Atrous Convolution with a dilated rate of 1 is standard Convolution. Diverse dilated rates can be set to change the scope of the amenable field. Additional time is required for training if the rate is slower, a more thorough division of the rough feature map is required. The stride can be further classified into the following for standard
1. The dimension of the feature map obtained by Convolution will diminish if S > 1 which represents down-sampling while performing Convolution.
2. S = 1, denotes the Convolution of the standard step size of 1.
3. 0 < S < 1, shows the fractional stride Convolution, which is comparable to up-sampling the illustration.

Figure 1: Atrous spatial pyramid pooling
The dimension of the feature map acquired by Convolution will show a rise, such as S = 0.5 which denotes putting an empty pixel at the back of each pixel of the illustration making the resulting feature map two times as large as the Convolution of S = 1 in the same circumstances. Alternatively, Atrous Convolution does not insert empty pixels between normal pixels but misses some of the native pixels, or retains the information, increasing weights of 0 to expand the receptive field of the Convolution kernel. Understandably, the Convolution with S > 1 has the same consequence but there is a down-sampling of the Convolution which reflects in a reduction in the dimension of the feature map. The size of the receptive field F can be achieved if the size of the Convolution nucleus is k and the void rate of the void Convolution is r.
Fig. 1 shows the Atrous Convolution layers in parallel with variable dilated rates in the pyramid model to secure multifaceted data. The higher rate relates to long-range pixels while the lower rate correlates to the closest pixels. Nevertheless, due to the image boundary effect, some of the remote boundary data cannot be captured correctly. This pattern is separate from the typical pooling output of the feature map recorded directly into the pyramid model by ICNet. More exhaustive information can be acquired to enhance segmentation accuracy even though criteria and training time are enhanced in this model.
Squeeze and Excitation blocks were introduced by Hu et al. [45]. The idea of Squeeze and Excitation blocks is to attain global information of every channel with the help of global average pooling. This squeezes the feature map into a single numeric value. Further on, information passes through the fully connected neural network, Relu and Sigmoid functions. The reason behind using the Squeeze and Excitation block is to reduce redundant information and pass out more relevant information [46].
To test the Generalizability and Robustness of Double U-Net, three different datasets relating to the skin lesion were used. The summary is given in Tab. 1.
• The first dataset is the PH2 dataset. Here, the information comprises 200 Dermatoscopic images obtained from hospital Pedro Hispano. These are 8-bit color images containing pictures of different types of Cancer. The database contains the annotated images of all the Dermatoscopic lesion images [47].
• The second dataset is the ISIC-2016 challenge dataset. The first part of the challenge is called lesion segmentation which contains 900 images of skin cancer and their related annotated masks for the training of the data. The training data here will be used for testing purposes [48].
• The third dataset used to test the architecture is the HAM dataset. This dataset contains 10015 images of common pigmented skin lesion disease with the ground truth of the images [49].
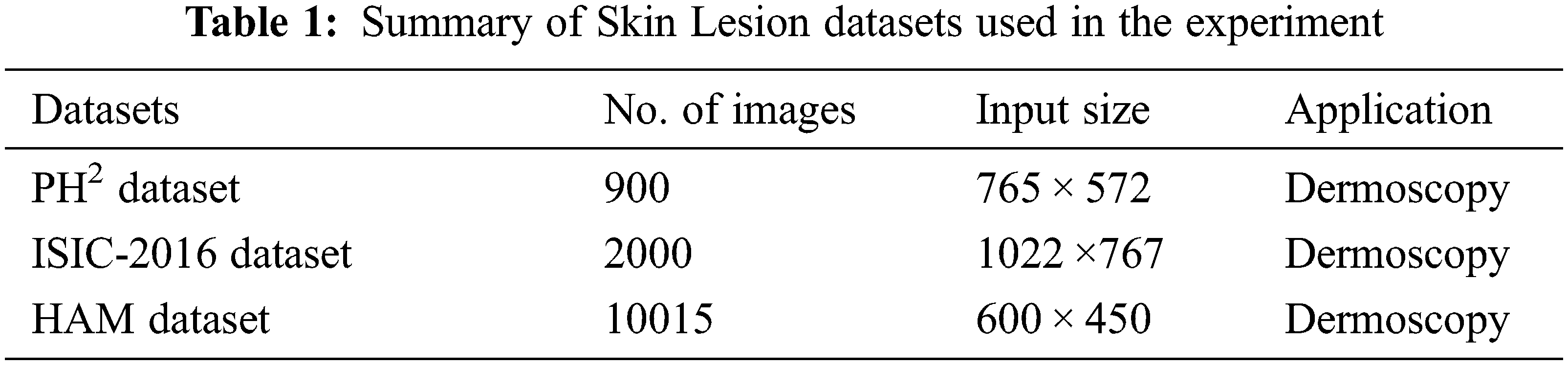
The proposed network architecture to segment skin cancer in Dermatoscopic images is the Double U-Net. This architecture contains two different U-Net architectures which are stacked upon each other to produce better results than a single U-Net architecture. This is shown in Fig. 2. An input image of the skin lesion is fed to the first part of architecture which performs necessary operations and produces a resulting image Fig. 3. This image is then combined with the input image and fed to the second part of the architecture as shown in Fig. 4. This produces the final segmented image. The displayed output contains four parts: the original image, annotated mask, the output produced by the first part of the Network and the output produced from the second part respectively. Fig. 2 shows the architecture of the Double U-Net. The architecture contains two U-Net networks, the first of which contains VGG-19 as an encoder that makes it different from the original U-Net architecture. It also includes ASPP which helps in extracting high-resolution feature maps that assist in the segmentation process and lead to better results. In addition to this, there is a Decoder block that includes a Squeeze and Excitation block. These blocks help to reduce redundant information and pass out more relevant information. The result produced from one network is then combined with the input image by element-wise multiplication. Following this, the information goes through the second architecture to produce the final segmented results.

Figure 2: Double U-Net architecture

Figure 3: Network I of proposed architecture

Figure 4: Network II of proposed architecture
Figs. 3 and 4 explain the proposed architecture in detail. Initially, the input image is fed to the encoder of Network 1. The encoder includes a Convolutional operation of 3 × 3 to program the information of the input image and then performs Batch normalization to regularize the model and reduce the Shift to internal co-variants. It then performs Rectified Linear Function (Relu) to introduce non-linearity and finally the Squeeze and Excitation block. Max pooling is then performed with steps 2 and 2 × 2 windows to decrease the window of the feature maps. Two decoders are used in each network. They provide up-sampling of 2 × 2 on given features that are used to widen the window of the feature map. Skip connections are then used from an encoder. In the first network, connection feature maps of the first encoder are used to the resulting feature map. In the second network, the skip connection feature map from both the encoders is used to enhance the quality of the image. After that, the Convolution operation is performed which is followed by batch normalization, activation functions and Squeeze and Excitation block respectively. Finally, the Convolutional layer is applied along with the Sigmoid Function to generate a mask for the modified networks. Understandably, it is essential to realize where the skin lesion boundaries exist. Once this is defined, the necessary object must be adjusted into this area. Concerning this, additional decoders for contour generation and distance map regression have to be accommodated. Here the multitask learning methodology makes the model aware of the boundaries of the affected area. Consequently, this has a bearing on the results as affected and healthy tissues are more clearly defined.
In this section, evaluation methods, experimental setup with configuration, data augmentation methods and comparisons are presented to test the Generalizability and Robustness of Double U-Net architecture.
The evaluation of Double U-Net architecture is performed on three datasets based on four statistics which include, Dice Sorensen coefficient (DSC), mean Intersection over Union (mIoU), Precision and Recall. A comparison between the datasets shows how well the model performs. Through this evaluation, the Generalizability and Robustness of the model are tested. Here, mIoU, is the official evaluation method from the official challenge site.
This model has been trained on the PH2 dataset which contains 900 images. Here, data augmentation is performed which includes cropping, rotation, transposition and transformation, etc, a process that modifies one image into 26 images. Work on this model was carried out utilizing the Keras Framework with tensor flow 2. 1. 0, as backend. This is pure Python work, the training of which was performed on Tesla T4 with the compute capability of 7.5, a core clock of 1.5 GHz and a core count of 40. It has a device memory size of 14.73 GiB and a device memory bandwidth of 298.08 GiBs. During training, the original image size for the smaller dataset, the PH2, ISIC-2016 and HAM segmentation dataset were used. The training time and complexity are optimized to help the Lesion Boundary segmentation challenge dataset once the images are resized to 384 × 512. The size of ETIS-Larib was amended accordingly to the PH2. The Nadam optimizer and its dimensions along with binary cross-entropy were also significant factors to be considered. For the lesion boundary segmentation and Nuclei segmentation datasets, where Dice loss and Adam optimizer performed slightly better, Batch sizes 8 and learning rate trained for 100 epochs with early stopping and ReduceLROnPlateau. The trained accuracy of this model is 0.99186. This outperforms DSC and 0.983868 in mIoU U-Net [50] and multi-ResUNet [50]. The image size accepted by the model is 192 × 256. Preprocessing is performed to resize the image to the required size.
In this section, the Generalizability and Robustness of the model are tested on different datasets to judge whether the model can be used as a baseline. Results are tabulated in Tab. 2 and are shown in both the statistic and image format.
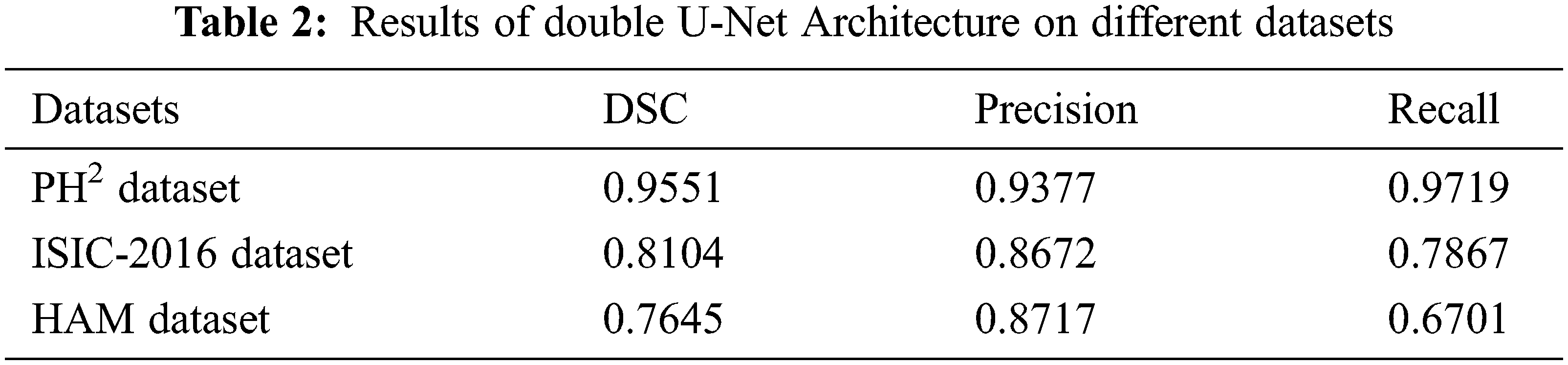
The proposed architecture has been trained on a PH2 dataset which contains 900 images. Augmentation is performed on images that change one image into 26 images through different operations like cropping, zooming and transposing, etc. A training accuracy of 0.9649 was achieved. Preprocessing is performed before feeding the images to the architecture that changes the image size to the required 192 × 256. Data augmentation is performed to eliminate the problem of high-quality images with labeled information.
As shown in Tab. 2, PH2 with a DSC of 0.9551 showed that it performed well on this particular data set and achieved better results. Fig. 5 shows the resulting images. The first image in Fig. 5 is input followed by ground truth, with outputs 1 and 2 respectively. Output 1 is the output of network 1 used in this architecture and output 2 is the final output after the second network. In Fig. 5, there are some resultant skin lesion images segmented through Double U-Net architecture. In the second and third pictures, the difference between output 1 and output 2 can be seen which justifies using two networks in this architecture.

Figure 5: Resulted images of PH2 dataset
ISIC-2016 and the HAM dataset containing 10050 images have been used for comparison Tab. 2.
5.1 Comparison with Other Frameworks
Various techniques have been employed for segmentation of the skin lesion. The expectation that the Double U-Net exploration technique would section the cancerous regions with higher accuracy when contrasted with other strategies has been justified. Tab. 3. shows the correlation of proposed architecture with different architectures. The outcomes show that the Double U-Net architectures performed better than the other best-in-class techniques.
5.2 Evaluation Comparison of PH2 Dataset
For the assessment of the Robustness and Generalizability of the proposed model, an evaluation of the outcomes on the PH2 dataset was made which compared it with other state-of-the-art methods. The outcomes are recorded in Tab. 4. Our technique accomplished promising results.

The second top-ranked participant Yuan et al. [55] obtained a DSC (0.920) by employing a double U-Net framework. Our technique achieved a DSC of 0.959 with the proposed technique stated earlier. Based on the results, our model performed better than existing techniques used in the associative field of study.
Skin cancer segmentation is a fundamental advance in building up a computer-aided diagnostic framework for skin malignancy. In this paper, we built a skin cancer segmentation architecture using CNN with two U-Net models stacked upon each other, improving the Dice coefficient and precision impressively. Our model engineering was tested against the PH2, ISIC-2016 and HAM datasets. The DSC gained was 0.9551, 0.8104 and 0.7645 separately. The proposed methodology achieved high accuracy results when compared to state-of-the-art strategies. Double U-Net has shown promising results. The performance of DoubleU-Net is significantly better when compared to baselines and UNet on all four datasets. Moreover, the proposed structural design is flexible which makes it possible to integrate other CNN blocks into the Double U-Net architecture. In the future, a new network model based on an improvement with interconnections of the nodes needs to be modified to take advantage of the segmentation process more effectively. In addition, research should focus more on designing low Dice coefficients on experimental cases by Triple U-Net with a multi-scale loss for skin lesion segmentation while retaining its ability.
Funding Statement: This research was made possible by a generous fund from the deanship of scientific research at Taif University, Taif, Saudi Arabia, under Taif University researchers supporting the project, Project No. TURSP-2020/344.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. D. L. Pham, C. Xu and J. L. Prince, “Current methods in medical image segmentation,” Annual Review of Biomedical Engineering, vol. 2, no. 2000, pp. 315–337, 2000. [Google Scholar]
2. N. C. Institute, “Melanoma of the skin—Cancer stat facts,” Seer, USA, (Accessed 26 November 20212021. Available: https://seer.cancer.gov/statfacts/html/melan.html. [Google Scholar]
3. R. B. Oliveira, M. E. Filho, Z. Ma, J. P. Papa, A. S. Pereira et al., “Computational methods for the image segmentation of pigmented skin lesions: A review,” Computer Methods and Programs in Biomedicine, vol. 131, pp. 127–141, 2016. [Google Scholar]
4. M. Silveira, J. C. Nascimento, J. S. Marques, A. R. S. Marçal, T. Mendonça et al., “Comparison of segmentation methods for melanoma diagnosis in dermoscopy images,” IEEE Journal of Selected Topics in Signal, vol. 3, no. 1, pp. 35–45, 2009. [Google Scholar]
5. K. Korotkov and R. Garcia, “Computerized analysis of pigmented skin lesions: A review,” Artificial Intelligence in Medicine, vol. 56, no. 2, pp. 69–90, 2012. [Google Scholar]
6. H. Kittler, H. Pehamberger, K. Wolff and M. Binder, “Diagnostic accuracy of dermoscopy,” Lancet Oncology, vol. 3, no. 3, pp. 159–165, 2002. [Google Scholar]
7. G. Schaefer, M. I. Rajab, M. Emre Celebi and H. Iyatomi, “Colour and contrast enhancement for improved skin lesion segmentation,” Computerized Medical Imaging and Graphics, vol. 35, no. 2, pp. 99–104, 2011. [Google Scholar]
8. C. Barata, M. Ruela, M. Francisco, T. Mendonc and J. S. Marques, “Two systems for the detection of melanomas in dermoscopy images using texture and color features,” IEEE Systems Journal, vol. 8, no. 3, pp. 965–979, 2014. [Google Scholar]
9. D. D. Gómez, C. Butakoff, B. K. Ersbøll and W. Stoecker, “Independent histogram pursuit for segmentation of skin lesions,” IEEE Transactions on Biomedical Engineering, vol. 55, no. 1, pp. 157–161, 2008. [Google Scholar]
10. M. E. Yüksel and M. Borlu, “Accurate segmentation of dermoscopic images by image thresholding based on type-2 fuzzy logic,” IEEE Transactions on Fuzzy Systems, vol. 17, no. 4, pp. 976–982, 2009. [Google Scholar]
11. F. Y. Xie, S. Y. Qin, Z. G. Jiang and R. S. Meng, “PDE-Based unsupervised repair of hair-occluded information in dermoscopy images of melanoma,” Computerized Medical Imaging and Graphics, vol. 33, no. 4, pp. 275–282, 2009. [Google Scholar]
12. M. Silveira, J. C. Nascimento, J. S. Marques, A. R. S. Marcal, T. Mendonca et al., “Comparison of segmentation methods for melanoma diagnosis in dermoscopy images,” IEEE Journal of Selected Topics in Signal Processing, vol. 3, no. 1, pp. 35–45, 2009. [Google Scholar]
13. Z. Ma and J. M. R. S. Tavares, “A novel approach to segment skin lesions in dermoscopic images based on a deformable model,” IEEE Journal of Biomedical and Health Informatics, vol. 20, no. 2, pp. 615–623, 2016. [Google Scholar]
14. Y. Li and L. Shen, “Skin lesion analysis towards melanoma detection using deep learning network,” Sensors, vol. 18, no. 2, pp. 1–16, 2018. [Google Scholar]
15. A. D. Brébisson and G. Montana, “Deep neural networks for anatomical brain segmentation.” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition Workshops, Boston, MA, USA, pp. 20–28, 2015. [Google Scholar]
16. F. Zhao and X. Xie, “An overview on interactive medical segmentation,” Annals of the British Machine Vision Association, vol. 2013, no. 7, pp. 1–22, 2013. [Google Scholar]
17. K. Abhishek, G. Hamarneh and M. S. Drew, “Illumination-based transformations improve skin lesion segmentation in dermoscopic images,” in Proc. of the IEEE/CVF Conf. on Computer Vision and Pattern Recognition Workshops, Seattle, USA, pp. 728–729, 2020. [Google Scholar]
18. D. Jha, M. A. Riegler, D. Johansen, P. Halvorsen and H. D. Johansen, “DoubleU-Net: A deep convolutional neural network for medical image segmentation,” in IEEE 33rd Int. Symp. on Computer-Based Medical Systems (CBMS), Rochester, USA, pp. 558–564, 2020. [Google Scholar]
19. X. Liu, Z. Deng and Y. Yang, “Recent progress in semantic image segmentation,” Artificial Intelligence Review, vol. 52, pp. 1089–1106, 2019. [Google Scholar]
20. L. Chen, P. Bentley, K. Mori, K. Misawa, M. Fujiwara et al., “DRINet for medical image segmentation,” IEEE Transactions on Medical Imaging, vol. 37, no. 11, pp. 2453–2462, 2018. [Google Scholar]
21. T. Roβ, A. Reinke, P. M. Full, M. Wagner, H. Kenngott et al., “Robust medical instrument segmentation challenge 2019,” Robust Medical Instrument Segmentation Challenge 2019, vol. 2003, no. 10299, pp. 1–55, pp. 2020. [Google Scholar]
22. M. Z. Alom, T. Aspiras, T. M. Taha and V. K. Asari, “Skin cancer segmentation and classification with nabla-n and inception recurrent residual convolutional networks,” arXiv, vol. 1904, no. 11126, pp. 1–7, 2019. [Google Scholar]
23. R. Arora, B. Raman, K. Nayyar and R. Awasthi, “Automated skin lesion segmentation using attention-based deep convolutional neural network,” Biomedical Signal Processing and Control, vol. 65, no. 102358, pp. 1–10, 2021. [Google Scholar]
24. Z. Cheng, A. Qu and X. He, “Contour-aware semantic segmentation network with spatial attention mechanism for medical image,” The Visual Computer, vol. 2021, no. 1, pp. 1–14, 2021. [Google Scholar]
25. O. Salih and S. Viriri, “Skin lesion segmentation using local binary convolution-deconvolution architecture,” International Society for Stereology & Image Analysis, vol. 39, no. 3, pp. 169–185, 2020. [Google Scholar]
26. Y. Tang, Z. Fang, S. Yuan, Y. Xing, J. T. Zhou et al., “IMSCGnet: Iterative multi-scale context-guided segmentation of skin lesion in dermoscopic images,” IEEE Access, vol. 8, pp. 39700–39712, 2020. [Google Scholar]
27. S. Baghersalimi, B. Bozorgtabar, P. Schmid-Saugeon, H. K. Ekenel and J. P. Thiran, “DermoNet: Densely linked convolutional neural network for efficient skin lesion segmentation,” EURASIP Journal on Image and Video Processing, vol. 2019, no. 1, pp. 1–10, 2019. [Google Scholar]
28. Y. Wang, Y. Wei, X. Qian, L. Zhu and Y. Yang, “DONet: Dual objective networks for skin lesion segmentation,” arXiv, vol. 2008, no. 8278, pp. 1–10, 2020. [Google Scholar]
29. S. T. Tran, C. H. Cheng, T. T. Nguyen, M. H. Le, D. G. Liu et al., “TMD-Unet: Triple-unet with multi-scale input features and dense skip connection for medical image segmentation,” Healthcare, vol. 9, No. no. 1, pp. 1–19, 2021. [Google Scholar]
30. W. Weng and X. Zhu, “INet: Convolutional networks for biomedical image segmentation,” IEEE Access, vol. 9, pp. 16591–16603, 2021. [Google Scholar]
31. B. Hafhouf, A. Zitouni, A. C. Megherbi and S. Sbaa. “A modified u-net for skin lesion segmentation.,” in 1st IEEE Int. Conf. on Communications, Control Systems and Signal Processing (CCSSP), El Oued, Algeria, pp. 225–228, 2020. [Google Scholar]
32. M. K. Hasan, L. Dahal, P. N. Samarakoon, F. I. Tushar and R. Martí, “DSNet: Automatic dermoscopic skin lesion segmentation,” Computers in Biology and Medicine, vol. 120, no. 103738, pp. 1–10, 2020. [Google Scholar]
33. A. R. Hawas, Y. Guo, C. Du, K. Polat and A. S. Ashour, “OCE-NGC: A neutrosophic graph cut algorithm using optimized clustering estimation algorithm for dermoscopic skin lesion segmentation,” Applied Soft Computing, vol. 86, no. 105931, pp. 1–13, 2020. [Google Scholar]
34. H. M. Ünver and E. Ayan, “Skin lesion segmentation in dermoscopic images with combination of yolo and grabcut algorithm,” Diagnostics, vol. 9, no. 3, pp. 72–94, 2019. [Google Scholar]
35. Y. Yuan, “Automatic skin lesion segmentation with fully convolutional-deconvolutional networks,” Computer Vision and Pattern Recognition, arXiv Preprint arXiv, vol. 1703, no. 5165, pp. 1–4, 2017. [Google Scholar]
36. M. A. Al-masni, M. A. Al-antari, M. T. Choi, S. M. Han and T. S. Kim, “Computer methods and programs in biomedicine skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks,” Computer Methods and Programs in Biomedicine, vol. 162, pp. 221–231, 2018. [Google Scholar]
37. I. Rasool, A. S. Mahboob, S. Shahbandegan and N. Baniasadi, “Skin lesion segmentation using convolutional neural networks with improved U-Net architecture,” in 6th Iranian Conf. on Signal Processing and Intelligent Systems (ICSPIS), Mashhad, Iran, pp. 1–5. IEEE, 2020. [Google Scholar]
38. M. Dash, N. D. Londhe, S. Ghosh, A. Semwal and R. S. Sonawane, “PsLSNet: Automated psoriasis skin lesion segmentation using modified U-Net-based fully convolutional network,” Biomedical Signal Processing and Control, vol. 52, pp. 226–237, 2019. [Google Scholar]
39. R. Mishra and O. Daescu, “Deep learning for skin lesion segmentation,” in IEEE Int. Conf. on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, USA, pp. 1189–1194, 2017. [Google Scholar]
40. L. C. Chen, G. Papandreou, S. Member, I. Kokkinos, K. Murphy et al., “DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolution and fully connected CRFs.” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 40, no. 4, pp. 834–848, 2017. [Google Scholar]
41. L. C. Chen, G. Papandreou, F. Schroff and H. Adam, “Rethinking atrous convolution for semantic image segmentation,” Computer Vision and Pattern Recognition, arXiv Preprint arXiv, vol. 1706, no. 5587, pp. 1–14, 2017. [Google Scholar]
42. J. Chen, C. Wang and Y. Tong, “AtICNet: Semantic segmentation with atrous spatial pyramid pooling in image cascade network,” EURASIP Journal on Wireless Communications and Networking, vol. 2019, no. 1, pp. 1–7, 2019. [Google Scholar]
43. F. B. K. Nasir, M. J. T. K. Nasir, M. Ziaratban, F. Bagheri, M. J. Tarokh et al., “Semantic segmentation of lesions from dermoscopic images using yolo-deeplab networks,” International Journal of Engineering, vol. 34, no. 2, pp. 458–469, 2021. [Google Scholar]
44. J. Long, E. Shelhamer and T. Darrell, “Fully convolutional networks for semantic segmentation,” in IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), Boston, USA, pp. 3431–3340, 2015. [Google Scholar]
45. J. Hu, L. Shen and G. Sun, “Squeeze-and-excitation networks,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, pp. 7132–7141. 2018. [Google Scholar]
46. J. F. Rodrigues, B. Brandoli and S. Amer-Yahia, “DermaDL: Advanced convolutional neural networks for automated melanoma detection,” in Proc. of IEEE Symposium on Computer-Based Medical Systems, Minnesota, USA, vol. 2020-July, pp. 504–509, 2020. [Google Scholar]
47. J. R. T. Mendonça, P. M. Ferreira, J. Marques and A. R. S. Marca, “PH2-A dermoscopic image database for research and benchmarking,” in 35th Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, pp. 5437–5440, 2013. [Google Scholar]
48. N. C. F. Codella, D. Gutman, M. E. Celebi, B. Helba, M. A. Marchetti et al., “Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (ISBIhosted by the international skin imaging collaboration (ISIC),” in Int. Symp. on Biomedical Imaging, Melbourne, Australia, vol. 2018-April, pp. 168–172, 2017. [Google Scholar]
49. P. Tschandl, C. Rosendahl and H. Kittler, “Data descriptor: The HAM10000 dataset, a large collection of multi-sources dermatoscopic images of common pigmented skin lesions,” Scientific Data, vol. 5, no. 1, pp. 1–9, 2018. [Google Scholar]
50. N. Ibtehaz and M. S. Rahman, “MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation,” Neural Networks, vol. 121, no. 1, pp. 74–87, 2020. [Google Scholar]
51. O. Ronneberger, P. Fischer and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” in Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, pp. 234–241, Springer, Cham, 2015. [Google Scholar]
52. J. Long, E. Shelhamer and T. Darrell, “Fully convolutional networks for semantic segmentation,” in IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), Boston, United States, vol. 39, no. 4, pp. 640–651, 2014. [Google Scholar]
53. H. Wen, “II-FCN for skin lesion analysis towards melanoma detection,” Computer Vision and Pattern Recognition, arXiv Preprint arXiv, vol. 1702, no. 8699, pp. 1–4, 2017. [Google Scholar]
54. M. Attia, M. Hossny, S. Nahavandi and A. Yazdabadi, “Spatially aware melanoma segmentation using hybrid deep learning techniques,” Computer Vision and Pattern Recognition, arXiv Preprint arXiv, vol. 1702, no. 7963, pp. 1–4, 2017. [Google Scholar]
55. Y. Yuan, M. Chao and Y. C. Lo, “Automatic skin lesion segmentation using deep fully convolutional networks with jaccard distance,” IEEE Transactions on Medical Imaging, vol. 36, no. 9, pp. 1876–1886, 2017. [Google Scholar]
56. K. Zafar, S. O. Gilani, A. Waris, A. Ahmed, M. Jamil et al., “Skin lesion segmentation from dermoscopic images using convolutional neural network,” Sensors, vol. 20, no. 6, pp. 1–14, 2020. [Google Scholar]
57. M. Taheri, M. Rastgarpour and A. Koochari, “A novel method for medical image segmentation based on convolutional neural networks with SGD optimization,” Journal of Electrical and Computer Engineering Innovations, vol. 9, no. 1, pp. 37–46, 2021. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |