DOI:10.32604/iasc.2023.019198

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2023.019198 |  |
| Article |
Analysis of Brain MRI: AI-Assisted Healthcare Framework for the Smart Cities
1Department Electronics and Electrical Communications Engineering, Faculty of Electronic Engineering, Menoufia University, Menoufia, 32952, Egypt
2Department of ROBOTICS and Intelligent Machines, Faculty of Artificial Intelligence, Kafrelsheikh University, Kafrelsheikh, Egypt
3Department of Computer Engineering, College of Computers and Information Technology, Taif University, Taif, 21944, Saudi Arabia
*Corresponding Author: Walid El-Shafai. Email: eng.waled.elshafai@gmail.com
Received: 06 April 2021; Accepted: 30 August 2021
Abstract: The use of intelligent machines to work and react like humans is vital in emerging smart cities. Computer-aided analysis of complex and huge MRI (Magnetic Resonance Imaging) scans is very important in healthcare applications. Among AI (Artificial Intelligence) driven healthcare applications, tumor detection is one of the contemporary research fields that have become attractive to researchers. There are several modalities of imaging performed on the brain for the purpose of tumor detection. This paper offers a deep learning approach for detecting brain tumors from MR (Magnetic Resonance) images based on changes in the division of the training and testing data and the structure of the CNN (Convolutional Neural Network) layers. The proposed approach is carried out on a brain tumor dataset from the National Centre of Image-Guided Therapy, including about 4700 MRI images of ten brain tumor cases with both normal and abnormal states. The dataset is divided into test, and train subsets with a ratio of the training set to the validation set of 70:30. The main contribution of this paper is introducing an optimum deep learning structure of CNN layers. The simulation results are obtained for 50 epochs in the training phase. The simulation results reveal that the optimum CNN architecture consists of four layers.
Keywords: Healthcare; smart cities; clinical automation; CNN; machine learning; brain tumor; medical diagnosis
Deep learning has been developed as a new tool for classifying images. It resembles in its operation the human visual system in acquiring a decision from batches in the images through convolution operations. Image classification into normal and abnormal images has captured the researchers interest as it is the first step towards automated diagnosis of diseases. The classification process can be performed through different disease representation methods such as medical images. The MRI is a type of medical imaging techniques, which relies on a powerful magnetic field. The image classification process can be performed using several techniques [1–4]. Deep learning is an efficient technique among these techniques [5]. The utilization of deep learning in medical image classification is an advanced step in the track of automated diagnosis [6–10].
In this paper, deep learning is used for sorting and diagnosis of brain tumors. The brain tumor is a collection or mass of abnormal cells in the brain. The invention of MRI played an important role in extracting, analyzing, and categorizing brain tumors [11]. The MRI imaging is widely used for brain tumor classification [12]. The obtained MR images have high quality for future processing. There are different types of MR images including T1, T2 and PD proton density weighted images [13]. The new trends of deep learning are appropriate for the classification of MR images [14].
Different approaches have been implemented for brain tumor detection. Segmentation of brain images along the X, Y, and Z-axis is performed using both MvNet and SPNet techniques [15]. These techniques were evaluated on BraTS 17 from the QTIM @ MGH (Quantitative Translational Imaging in Medicine Lab at the Martinos Center) [16], and they achieved an accuracy of 55% for survival prediction. Another technique was proposed in [17]. This technique is based on the transformation of 3D segmentation into atri-planar 2D CNN operation. This architecture achieved an accuracy of 88%. The authors in [18] proposed an automated segmentation algorithm for brain tumors based on DCNNs (Deep Convolutional Neural Networks). This algorithm has been carried out on BRATS 2013 dataset organized in conjunction with the MICCAI 2012 and 2013 conferences [19]. It achieved a sensitivity of 0.83. An automatic segmentation technique based on CNN and intensity normalization, as a pre-processing step, was proposed in [20]. This technique was compared with BraTS 2015 using the DSC (Dice Similarity Coefficient) evaluation metric. It achieved DSC values of 0.75, 0.65, and 0.78 for the enhanced, core, and complete regions.
Nie et al. [21] proposed an approach based on multi-channel data and an SVM (Support Vector Machine), as a supervised learning technique. This approach achieved an accuracy of 89.9%. Xiao et al. [22] proposed an approach based on deep learning and 3D images. It consists of three stages, pre-processing, a deep learning model, and post-processing. They achieved an accuracy of 98.44%. Chato et al. [23] proposed an approach to predict the number of survivors from glioma brain tumors, automatically. This approach is based on the classification of MRI images using ML (Machine Learning) algorithms, logistic regression, quadratic, and discriminant analysis. The feature extraction process is performed using CNNs. It has been carried out on BRATS2017 (163 samples). It achieved an accuracy of 68.8%. Shreyas et al. [24] proposed a simple FCN (Fully-Convolutional Network) for brain tumor segmentation. This approach has been carried out on the dataset provided for the BraTS challenge from the MICCAI (Medical Image Computing and Computer-Assisted Intervention) society. It achieved a sensitivity of 0.92. Balasooriya et al. [25] adopted a technique to recognize the tumor type based on a sophisticated CNN. The performance of this technique is based on the average F1-score evaluation metric with a value of 99.46%. In addition, an accuracy of 99.68% has been achieved. The work in [26] is based on CNN activations trained by an image net to extract features (13.3% active, and 4096 neurons). The system achieved an accuracy of 97.5% for classification and 84% for segmentation. The work in [27] is based on both 3D and 2D CNN to classify the CT (Computerized Tomography) brain images into 3 categories: AD (Alzheimer’s Disease), lesion (e.g., tumor) and normal. The accuracy levels are 95%, 76.7%, and 88.8% for normal, lesion and AD classes, respectively, and this led to an average accuracy of 86.8%.
The main contributions of the proposed work are as follows:
1. Presenting a deep learning approach for brain tumor detection from MR images.
2. Introducing the optimum deep learning structure of CNN layers.
3. Presenting extensive simulation tests on different images and different parameters of the CNN model.
This paper consists of five sections. Section 2 presents the process of MR image classification based on deep learning, while Section 3 introduces the suggested approach. Section 4 illustrates the dataset description and visualization. Section 5 shows the results of the proposed approach. Finally, the concluding remarks are introduced in Section 6.
2 Deep Learning Classification
The basic idea of deep learning is to use different convolution kernels to extract features from images. These kernels are 2D in nature and have different weights and orientations [28–31]. Features are captured, when the applied convolution kernel is matched to the scanned area [32]. Convolution kernels are arranged within different convolutional layers. Activation functions are used to get decisions at the outputs of the convolutional layers. Pooling layers are used as decision tools for the obtained feature maps from the convolutional layers. In addition, batch normalization is used as a tool for regularization to avoid overfitting [33–35]. The Fully-Connected (FC) layers are used for the classification task.
A. Convolutional (CNV) Layer
The convolutional layer is used on the brain images collected from patients to extract a sufficient set of features. The convolutional layer contains filters that perform 2-D convolution operations on the input images. The resultant characteristics of the convolutional layer vary in consistence with the used filters. This theory is very admirably suitable to the MR images for tumor detection. Fig. 1 indicates the convolutional layer operation.

Figure 1: Convolutional layer operation
There are three modes for convolution, when applied to images:
a) Valid mode: Filter never goes outside the input image, as shown in Fig. 2. If the input length equals N and the filter length equals K, then the output length equals N − K + 1.

Figure 2: Valid mode illustration
b) Full mode: Filter is allowed to go outside the input image, far enough, so that there is at least one overlapping element as shown in Fig. 3. If the input length equals N and the filter length equals K, then the output length equals N + K − 1.

Figure 3: Full mode illustration
c) Same mode: Padding is set such that the input length equals the output length. If the input length equals N = 5 and the filter length equals K = 3, then the padding length P = 1, as shown in Fig. 4.

Figure 4: Same mode illustration
B. Pooling Layer (PL)
Pooling strategy is a tool used to reduce the number of features. Different types of pooling can be used such as max and mean pooling. An example of max pooling is shown in Fig. 5

Figure 5: Maximum pooling process
C. Rectified Linear Unit (ReLU)
It is used for effective and fast training through converting negative values to zero and maintaining positive values. The output of the ReLU layer is fed to a linear activation function, and the output of the neuron is as shown in Eq. (1).
D. FC Layer
The FC layer is used for classification in order to obtain the final decision about states. The outputs are obtained in the form of probabilities as indicated by Eq. (2).
E. Optimization
Optimization techniques are used to minimize the cost function. The idea of the optimization is to bring down the error, while the cost function is very high. So, the weights must be adjusted in such a way and the cost function must be minimized. In the gradient descent technique, the rate of learning should be known and be optimum, because if there is a high learning rate, the cost would not be minimized. At some point, it will cross over to the side on the other hand. On the other hand, if we have a very low learning rate, then it might take a long time for convergence. Hence, we need to come up with the optimum learning rate, and once that is done, the error function is minimized. The formulas for the cost function and loss are given as follows:
3 The Proposed AI-Assisted Healthcare Approach
The proposed approach steps are illustrated in Fig. 6. It comprises five convolutional layers combined with five pooling layers giving maximum values, and finally a pooling layer giving average values. As shown in Fig. 6, the input image size is 224 × 224. These input images are passed through different convolutional layers, and finally passed through a dense layer to make a decision. A dense layer with a size of 2 is exploited for final classification, as demonstrated in Fig. 6.

Figure 6: Structure of the proposed deep learning layers
The suggested approach is performed on the National Centre of Image-Guided Therapy dataset [36]. This dataset includes about 4700 MR images of ten brain tumor cases in both normal and abnormal states. These cases have different tumor positions, as shown in Tab. 1. Fig. 7 shows samples of images included in this dataset. Fig. 8 shows data visualization of training and testing. In this paper, 70% of the data is used for training and 30% for testing during the CNN-based classification.
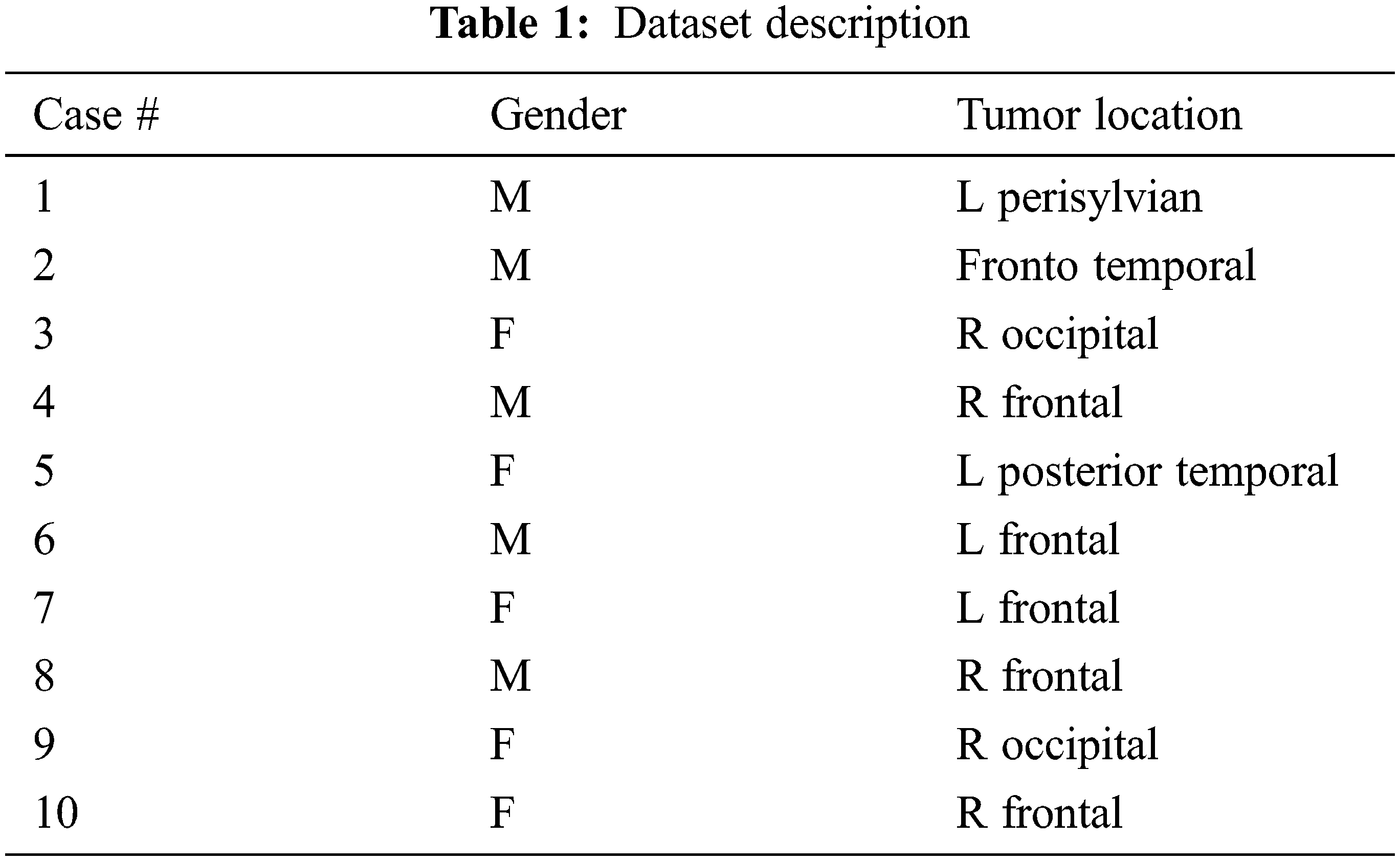

Figure 7: Samples of the dataset

Figure 8: Dataset visualization
The accuracy of the CNN model is given as:
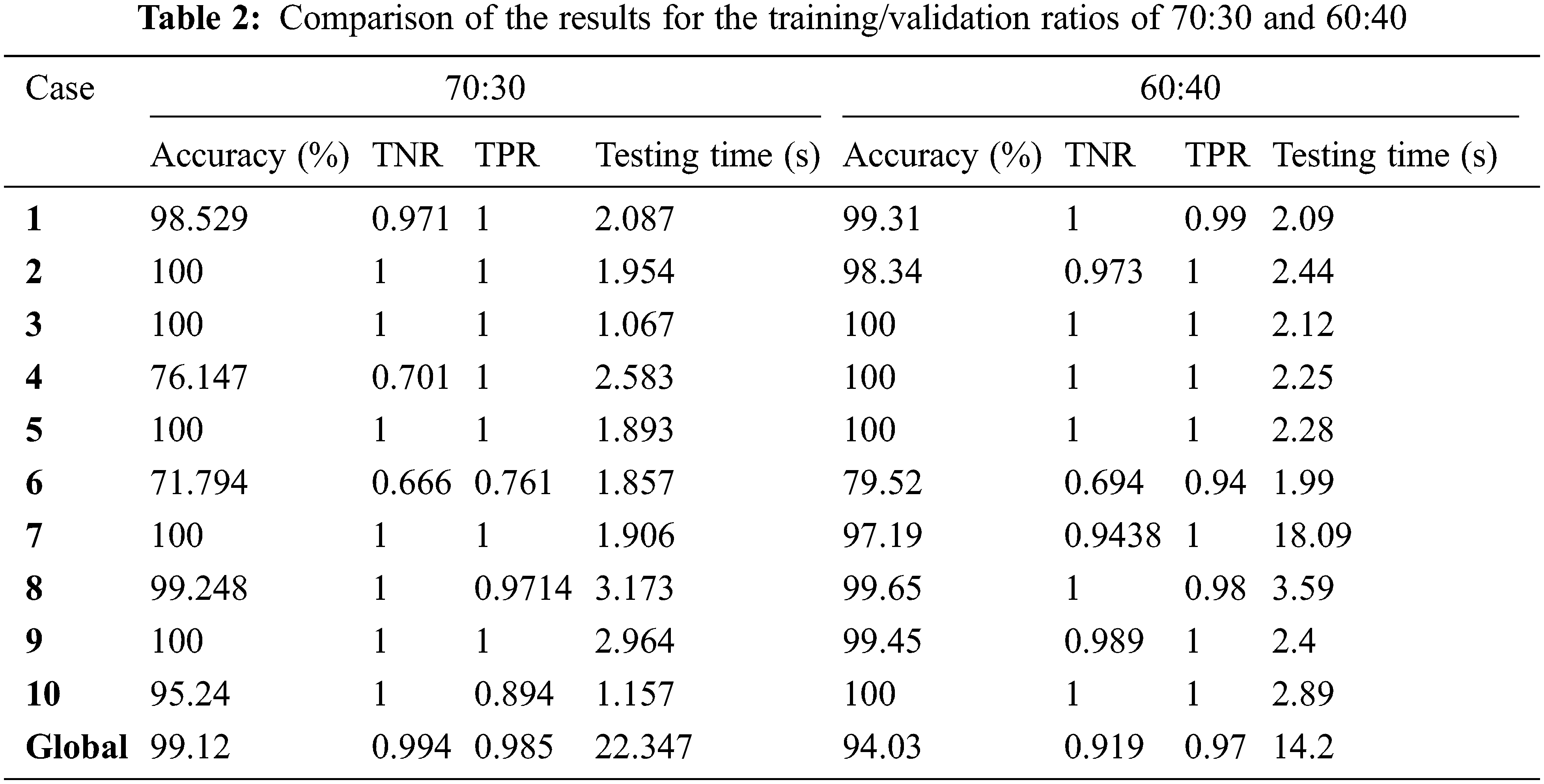
The suggested approach is divided into two parts: training and validation. The model consists of 6 convolutional layers followed by 6 max-pooling layers, a dense layer, a global average pooling layer, and the last decision SoftMax layer. Simulation findings are obtained using Python-based Pillow, TensorFlow, and Keras libraries [37–40]. In the simulation results, we compared different training and validation ratios of 60:40 and 70:30 at different epochs of 15, 25, and 50. Tab. 2 shows comparison results for the training/validation ratios of 70:30 and 60:40. It is noticed from the comparison results that the case of 70:30 is better than the case of 60:40 from the obtained accuracy, TPR (True Positive Rate), TNR (True Negative Rate), and testing time perspectives. So, the case of 70:30 training/testing ratio is considered for the rest of the experimental results.
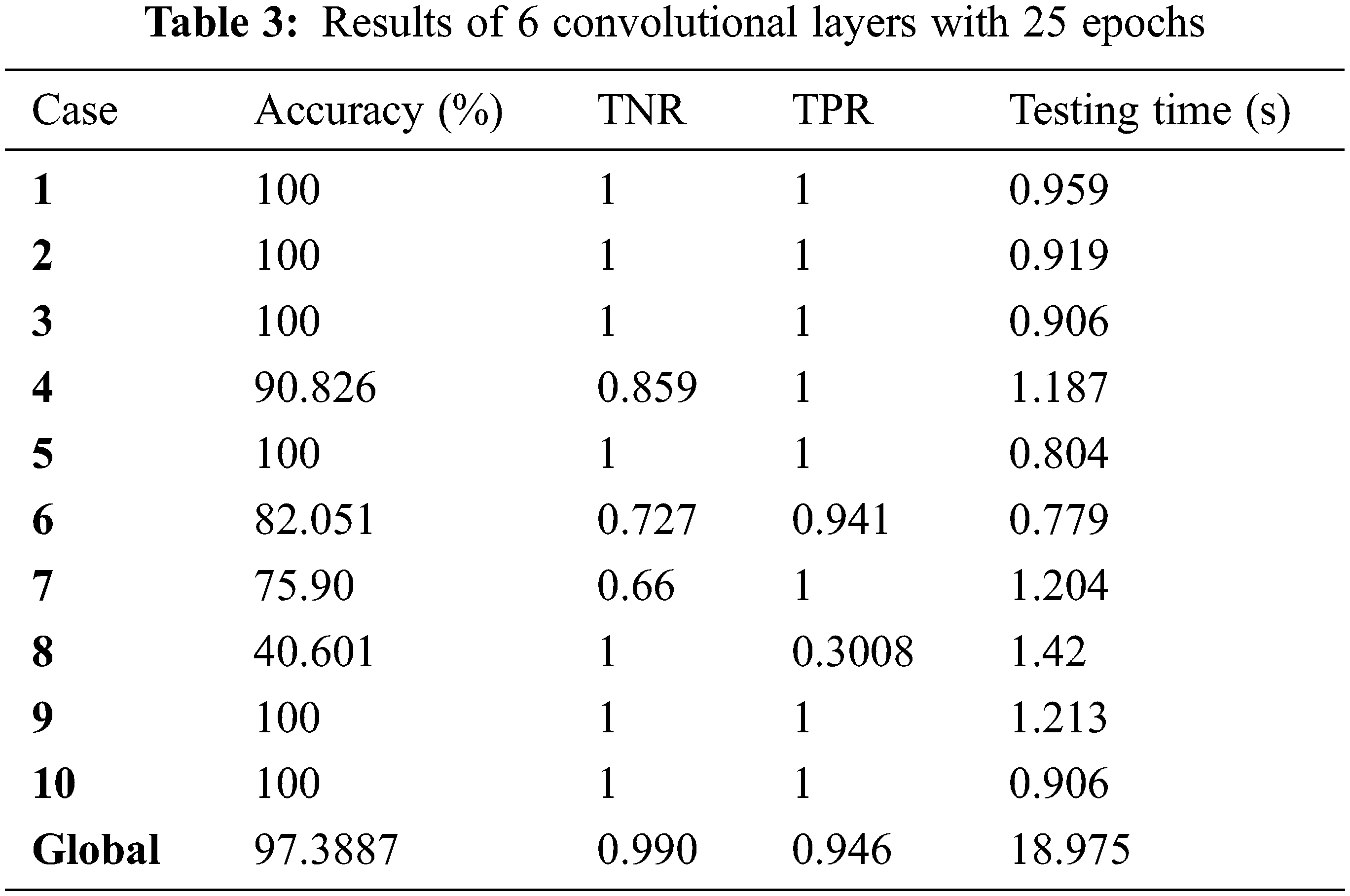
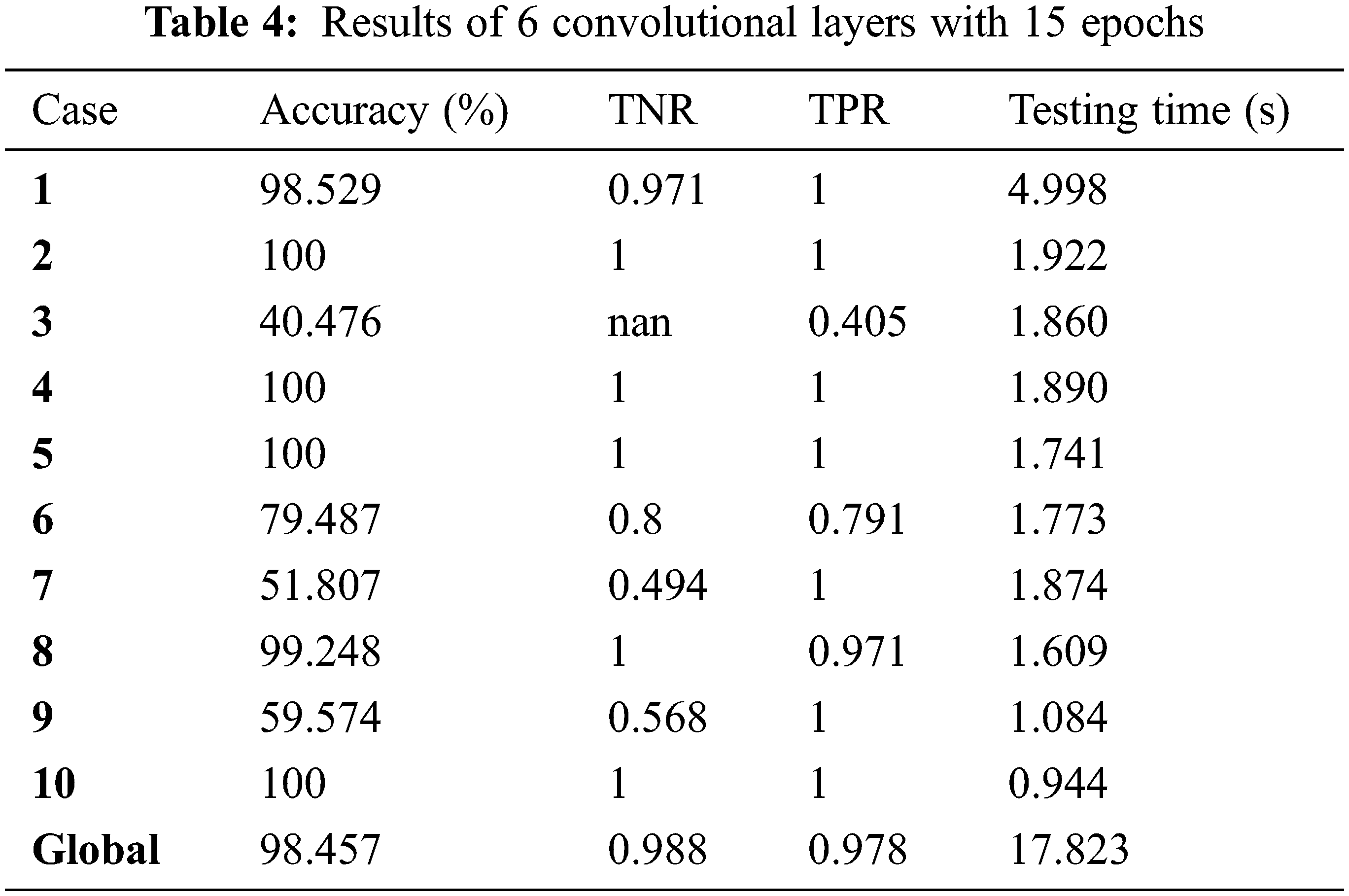
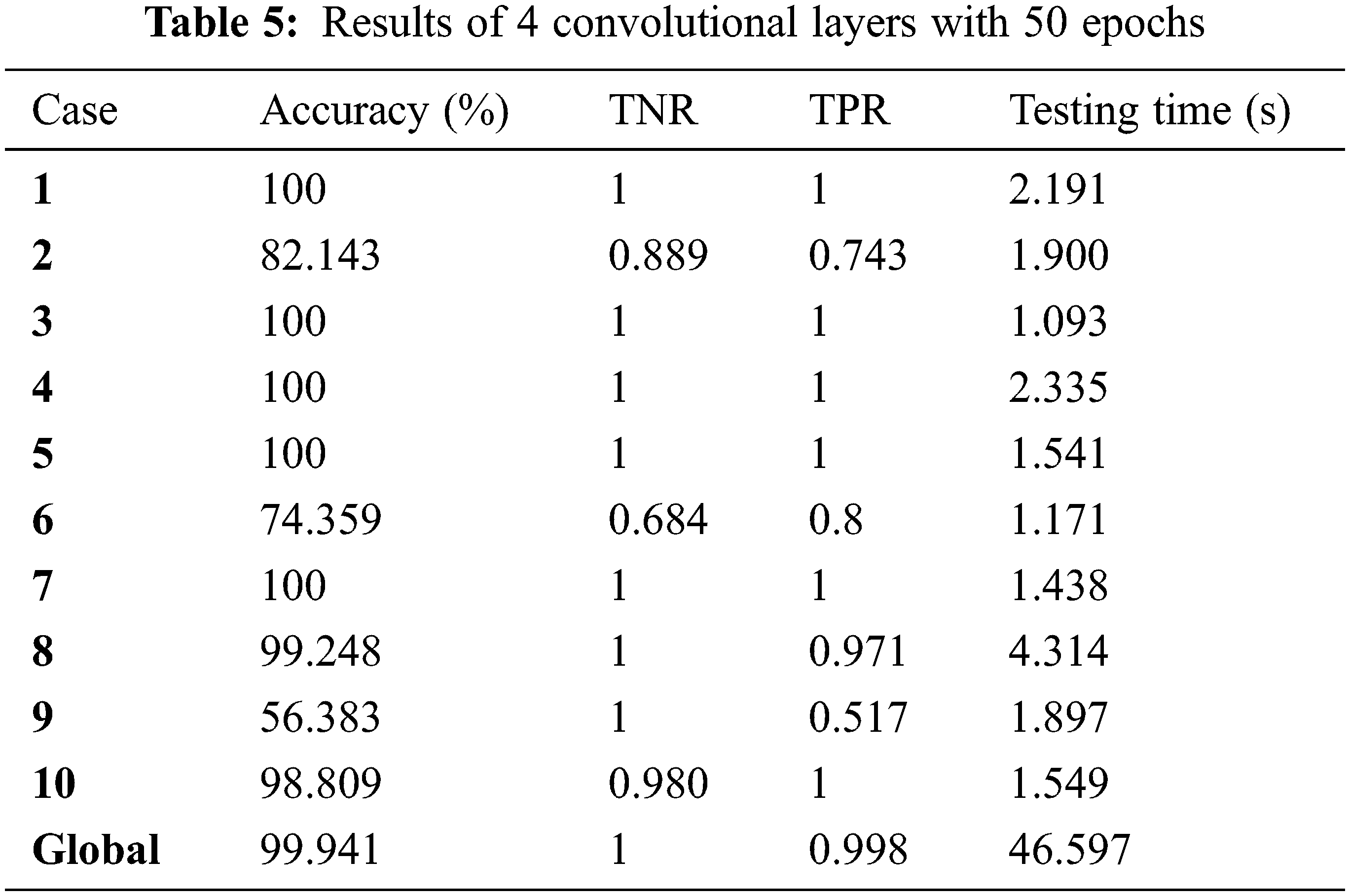
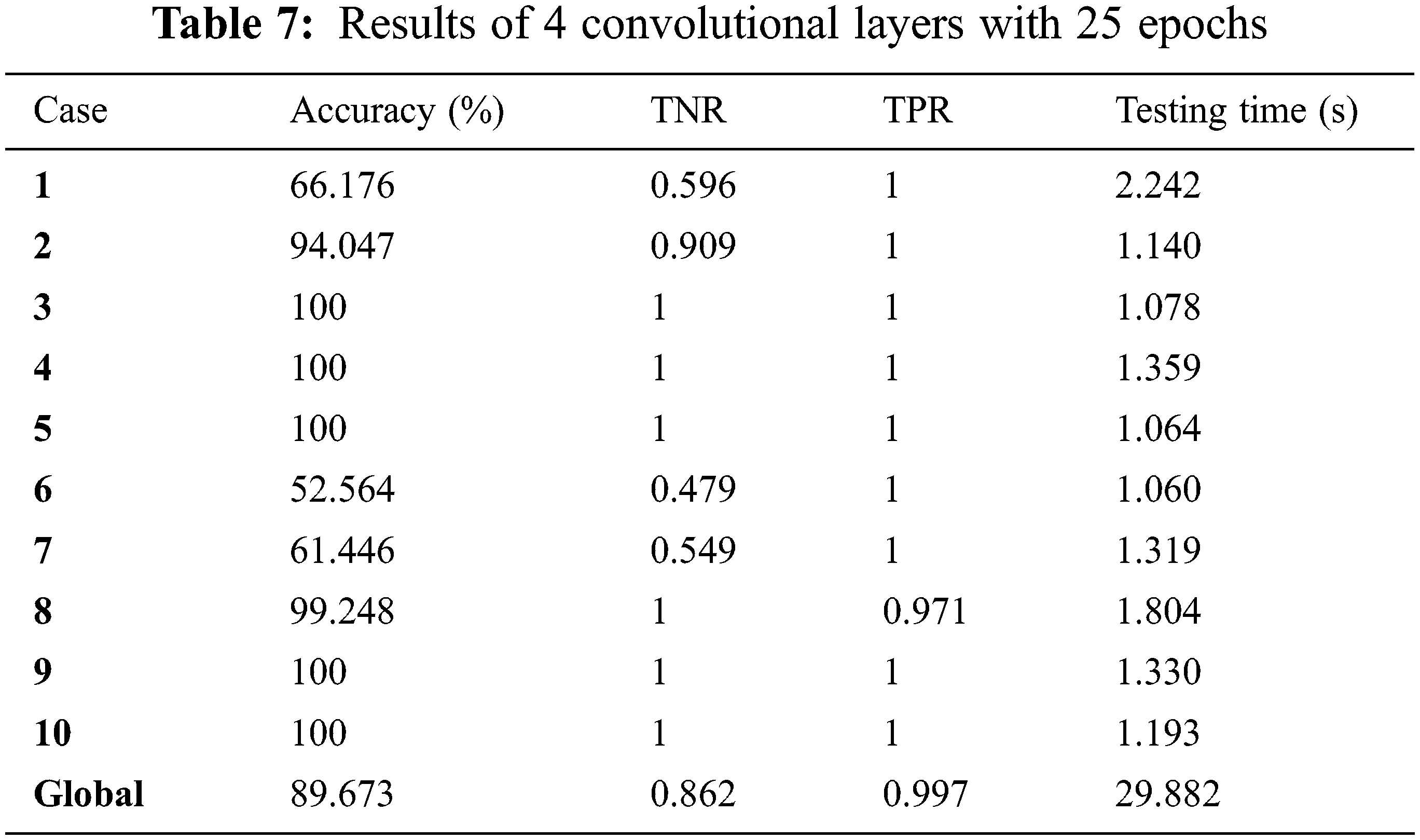
Tab. 3 shows the results of 6 convolutional layers with 25 epochs. Tab. 4 shows the results of 6 convolutional layers with 15 epochs. Tabs. 5 and 6 show the results and curves of 4 convolutional layers with 50 epochs. Tab. 7 shows the results and curves of 4 convolutional layers with 25 epochs. Tab. 8 shows the results and curves of 4 convolutional layers with 15 epochs. Tab. 9 presents a summary of comparison results for 6 and 4 convolutional layers with different numbers of epochs. Tab. 10 presents a comparison between the proposed approach and other traditional ones. It is observed from the obtained outcomes that the proposed approach is superior compared to other traditional ones. Also, it is demonstrated that the case of 4 convolutional layers with 50 epochs gives the best results.




This paper presented a brain tumor detection approach based on deep CNN. The proposed approach has been tested on a set of 10 cases in both patient-specific and global scenarios. It achieved an accuracy of 99.9% for four convolutional layers and 50 epochs. On the other hand, it achieved an accuracy of 98.5% in the global detection scenario. The simulation results revealed that the proposed approach is efficient for brain tumor detection as it achieves an acceptable accuracy in both patient-specific and global detection. In the future, we plan to use advanced deep learning techniques, especially convolutional neural networks in image fusion, to get images with high resolution and accuracy.
Acknowledgement: The authors would like to acknowledge the support received from Taif University Researchers Supporting Project Number (TURSP-2020/147), Taif University, Taif, Saudi Arabia.
Funding Statement: This work was funded and supported by the Taif University Researchers, Taif University, Taif, Saudi Arabia, under Project TURSP-2020/147.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. W. El Shafai, B. Hrušovský, M. El-Khamy and M. El-Sharkawy, “Joint space-time-view error concealment algorithms for 3D multi-view video,” in Proc. 18th IEEE Int. Conf. on Image Processing (ICIP), Brussels, Belgium, pp. 2201–2204, 2011. [Google Scholar]
2. O. Faragallah, M. Alzain, H. El-Sayed, J. Al-Amri, W. El-Shafai et al., “Block-based optical color image encryption based on double random phase encoding,” IEEE Access, vol. 7, pp. 4184–4194, 2018. [Google Scholar]
3. W. El-Shafai, “Pixel-level matching based multi-hypothesis error concealment modes for wireless 3D H. 264/MVC communication,” 3D Research, vol. 6, no. 3, pp. 1–11, 2015. [Google Scholar]
4. W. El-Shafai, E. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Efficient multi-level security for robust 3D color-plus-depth HEVC,” Multimedia Tools and Applications, vol. 77, no. 23, pp. 30911–30937, 2018. [Google Scholar]
5. W. El-Shafai, “Joint adaptive pre-processing resilience and post-processing concealment schemes for 3D video transmission,” 3D Research, vol. 6, no. 1, pp. 1–10, 2015. [Google Scholar]
6. K. Al-Afandy, W. El-Shafai, E. El-Rabaie, F. Abd El-Samie, O. Faragallah et al., “Robust hybrid watermarking techniques for different color imaging systems,” Multimedia Tools and Applications, vol. 77, no. 19, pp. 25709–25759, 2018. [Google Scholar]
7. W. El-Shafai, S. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Security of 3D-HEVC transmission based on fusion and watermarking techniques,” Multimedia Tools and Applications, vol. 78, no. 19, pp. 27211–27244, 2019. [Google Scholar]
8. W. El-Shafai, S. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Encoder-independent decoder-dependent depth-assisted error concealment algorithm for wireless 3D video communication,” Multimedia Tools and Applications, vol. 77, no. 11, pp. 13145–13172, 2018. [Google Scholar]
9. W. El-Shafai, S. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Efficient hybrid watermarking schemes for robust and secure 3D-MVC communication,” International Journal of Communication Systems, vol. 31, no. 4, pp. e3478, 2018. [Google Scholar]
10. W. El-Shafai, S. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Enhancement of wireless 3D video communication using color-plus-depth error restoration algorithms and Bayesian Kalman filtering,” Wireless Personal Communications, vol. 97, no. 1, pp. 245–268, 2017. [Google Scholar]
11. W. El-Shafai, S. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Recursive Bayesian filtering-based error concealment scheme for 3D video communication over severely lossy wireless channels,” Circuits Systems, and Signal Processing, vol. 37, no. 11, pp. 4810–4841, 2018. [Google Scholar]
12. K. Abdelwahab, S. Abd El-atty, W. El-Shafai, S. El-Rabaie and F. Abd El-Samie, “Efficient SVD-based audio watermarking technique in FRT domain,” Multimedia Tools and Applications, vol. 79, no. 9, pp. 5617–5648, 2020. [Google Scholar]
13. O. Faragallah, A. Afifi, W. El-Shafai, H. El-Sayed, E. Naeem et al., “Investigation of chaotic image encryption in spatial and FrFT domains for cybersecurity applications,” IEEE Access, vol. 8, pp. 42491–42503, 2020. [Google Scholar]
14. S. Gawande and V. Mendre, “Brain tumor diagnosis using deep neural network (DNN),” International Journal of Advanced Research in Electrical, Electronics and Instrumentation Engineering, vol. 5, no. 5, pp. 1–23, 2017. [Google Scholar]
15. Y. Li and L. Shen, “Deep learning based multimodal brain tumor diagnosis,” in Int. MICCAI Brain lesion Workshop, Cham, Springer, pp. 149–158, 2017. [Google Scholar]
16. J. Kalpathy-Cramer, J. Freymann, J. Kirby, P. Kinahan and F. Prior, “Quantitative imaging network: Data sharing and competitive algorithm validation leveraging the cancer imaging archive,” Translational Oncology, vol. 7, no. 1, pp. 147–152, 2014. [Google Scholar]
17. L. Zhao and K. Jia, “Deep feature learning with discrimination mechanism for brain tumor segmentation and diagnosis,” in Proc. IEEE Int. Conf. on Intelligent Information Hiding and Multimedia Signal Processing (IIH-MSP), Adelaide, SA, Australia, Australia, pp. 306–309, 2015. [Google Scholar]
18. S. Hussain, S. Muhammad and M. Majid, “Brain tumor segmentation using cascaded deep convolutional neural network,” in Proc. 39th IEEE Annual Int. Conf. of the Engineering in Medicine and Biology Society (EMBC), Jeju, South Korea, South Korea, pp. 1998–2001, 2017. [Google Scholar]
19. S. Reza and K. Iftekharuddin, “Multi-class abnormal brain tissue segmentation using texture,” Multimodal Brain Tumor Segmentation, vol. 38, pp. 38–42, 2013. [Google Scholar]
20. S. Pereira, A. Pinto, V. Alves and A. Silva, “Brain tumor segmentation using convolutional neural networks in MRI images,” IEEE Transactions on Medical Imaging, vol. 35, no. 5, pp. 1240–1251, 2016. [Google Scholar]
21. D. Nie, H. Zhang, E. Adeli, L. Liu and D. Shen, “3D deep learning for multi-modal imaging-guided survival time prediction of brain tumor patients,” in Proc. Int. Conf. on Medical Image Computing and Computer-assisted Intervention, Cham, Springer, pp. 212–220, 2016. [Google Scholar]
22. Z. Xiao, R. Huang, Y. Ding and T. Lan, “A deep learning-based segmentation method for brain tumor in MR images,” in Proc. IEEE 6th Int. Conf. on Computational Advances in Bio and Medical Sciences (ICCABS), Atlanta, GA, USA, pp. 1–6, 2016. [Google Scholar]
23. L. Chato and S. Latifi, “Machine learning and deep learning techniques to predict overall survival of brain tumor patients using MRI images,” in Proc. IEEE 17th Int. Conf. on Bioinformatics and Bioengineering, Washington, DC, USA, pp. 9–14, 2017. [Google Scholar]
24. V. Shreyas and V. Pankajakshan, “A deep learning architecture for brain tumor segmentation in MRI images,” in Proc. 19th IEEE Int. Workshop on Multimedia Signal Processing (MMSP), Luton, UK, pp. 1–6, 2017. [Google Scholar]
25. M. Balasooriya and D. Nawarathna, “A sophisticated convolutional neural network model for brain tumor classification,” in Proc. IEEE Int. Conf. on Industrial and Information Systems (ICIIS), Peradeniya, Sri Lanka, pp. 1–5, 2017. [Google Scholar]
26. Y. Xu, Z. Jia, Y. Ai, F. Zhang, M. Lai et al., “Deep convolutional activation features for large scale brain tumor histopathology image classification and segmentation,” in Proc. IEEE Int. Conf. on Acoustics, Speech and Signal Processing (ICASSP), South Brisbane, QLD, Australia, pp. 947–951, 2015. [Google Scholar]
27. W. Gao and R. Hui, “A deep learning based approach to classification of CT brain images,” in Proc. IEEE SAI Computing Conf., London, UK, pp. 28–31, 2016. [Google Scholar]
28. O. Faragallah, M. AlZain, H. El-Sayed, J. Al-Amri, W. El-Shafai et al., “Secure color image cryptosystem based on chaotic logistic in the FrFT domain,” Multimedia Tools and Applications, vol. 79, no. 3, pp. 2495–2519, 2020. [Google Scholar]
29. A. Sedik, H. Emara, A. Hamad, E. Shahin, N. El-Hag et al., “Efficient anomaly detection from medical signals and images,” International Journal of Speech Technology, vol. 22, no. 3, pp. 739–767, 2019. [Google Scholar]
30. H. El-Hoseny, W. El-Rahman, W. El-Shafai, E. El-Rabaie, K. Mahmoud et al., “Optimal multi-scale geometric fusion based on non-subsampled contourlet transform and modified central force optimization,” International Journal of Imaging Systems and Technology, vol. 29, no. 1, pp. 4–18, 2019. [Google Scholar]
31. W. El-Shafai, E. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Proposed adaptive joint error-resilience concealment algorithms for efficient colour-plus-depth 3D video transmission,” IET Image Processing, vol. 12, no. 6, pp. 967–984, 2018. [Google Scholar]
32. W. El-Shafai, S. El-Rabaie, M. El-Halawany and F. Abd El-Samie, “Performance evaluation of enhanced error correction algorithms for efficient wireless 3D video communication systems,” International Journal of Communication Systems, vol. 31, no. 1, pp. 1–22, 2018. [Google Scholar]
33. H. El-Hoseny, W. Abd El-Rahman, W. El-Shafai, G. El-Banby, E. El-Rabaie et al., “Efficient multi-scale non-sub-sampled shearlet fusion system based on modified central force optimization and contrast enhancement,” Infrared Physics & Technology, vol. 102, pp. 1–24, 2019. [Google Scholar]
34. N. El-Hag, A. Sedik, W. El-Shafai, H. El-Hoseny, A. Khalaf et al., “Classification of retinal images based on convolutional neural network,” Microscopy Research and Technique, vol. 84, no. 3, pp. 394–414, 2020. [Google Scholar]
35. A. Mahmoud, W. El-Shafai, T. Taha, E. El-Rabaie, O. Zahran et al., “A statistical framework for breast tumor classification from ultrasonic images,” Multimedia Tools and Applications, vol. 80, no. 4, pp. 1–20, 2020. [Google Scholar]
36. National centre of image-guided therapy dataset, last accessed on 20-11-2020. [Online]. http://2014.ncigt.org/publications/bitstream/download/6076. [Google Scholar]
37. Python library, last accessed on 20-11-2020. [Online]. https://www.python.org/downloads/release/python-350/. [Google Scholar]
38. Keras library, last accessed on 20-11-2020. [Online]. https://keras.io/. [Google Scholar]
39. Pillow library, last accessed on 20-11-2020. [Online]. https://pillow.readthedocs.io/en/3.0.x/handbook/tutorial.html. [Google Scholar]
40. Tensorflow library, last accessed on 20-11-2020. [Online]. https://www.tensorflow.org/. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |