DOI:10.32604/iasc.2023.025597

| Intelligent Automation & Soft Computing DOI:10.32604/iasc.2023.025597 |  |
| Article |
A Transfer Learning Based Approach for COVID-19 Detection Using Inception-v4 Model
1College of Computer Science and Information Systems, Najran University, Najran, 61441, Saudi Arabia
2Department of Computer Science and Information Technology, University of Sargodha, Sargodha, 40100, Pakistan
3School of Systems and Technology, University of Management and Technology, Lahore, 54782, Pakistan
4Department of Computer Science, COMSATS University Islamabad, Wah Campus, Islamabad, 47040, Pakistan
5Department of Medical Imaging, King Khaled Hospital, Najran, 61441, Saudi Arabia
6Electrical & Electronics Engineering Department, Universiti Teknologi, Petronas, 31750, Malaysia
7Department of Electrical Engineering, HITEC University, Taxila, 47080, Pakistan
*Corresponding Author: Muhammad Ramzan. Email: mramzansf@gmail.com
Received: 29 November 2021; Accepted: 13 January 2022
Abstract: Coronavirus (COVID-19 or SARS-CoV-2) is a novel viral infection that started in December 2019 and has erupted rapidly in more than 150 countries. The rapid spread of COVID-19 has caused a global health emergency and resulted in governments imposing lock-downs to stop its transmission. There is a significant increase in the number of patients infected, resulting in a lack of test resources and kits in most countries. To overcome this panicked state of affairs, researchers are looking forward to some effective solutions to overcome this situation: one of the most common and effective methods is to examine the X-radiation (X-rays) and computed tomography (CT) images for detection of Covid-19. However, this method burdens the radiologist to examine each report. Therefore, to reduce the burden on the radiologist, an effective, robust and reliable detection system has been developed, which may assist the radiologist and medical specialist in effective detecting of COVID. We proposed a deep learning approach that uses readily available chest radio-graphs (chest X-rays) to diagnose COVID-19 cases. The proposed approach applied transfer learning to the Deep Convolutional Neural Network (DCNN) model, Inception-v4, for the automatic detection of COVID-19 infection from chest X-rays images. The dataset used in this study contains 1504 chest X-ray images, 504 images of COVID-19 infection, and 1000 normal images obtained from publicly available medical repositories. The results showed that the proposed approach detected COVID-19 infection with an overall accuracy of 99.63%.
Keywords: COVID-19; transfer learning; deep learning; artificial intelligence; chest X-rays; machine learning
Professionals from Wuhan, China, reported in December 2019 that a number of patients with an unknown disease had common symptoms similar to those of pneumonia [1]. They adopted a surveillance mechanism to identify novel pathogens in a timely manner following the outbreak of Severe Acute Respiratory Syndrome (SARS) in 2003, leading to the identification of novel viral diseases referred to as COVID-19 [2]. A month later, the World Health Organization (WHO) declared the novel virus to be a pandemic, indicating that it could spread very quickly and could affect a large segment of the population worldwide. The pandemic is rapidly escalating, and approximately 800 million people have already been infected as shown in Fig. 1 [3]. Dry cough, fever, chills, shortness of breath, inflammation and lung infection are the main symptoms of COVID-19 [4,5]. Reverse transcription polymerase chain reaction (RT-PCR) is considered to be the standard for diagnosis of COVID-19 [6,7]. These test kits are expensive, and they also take more time to produce results. Testing material for RT-PCR is not widely available, and laboratory technicians are also not experienced in performing the test properly. As this disease can spread very quickly from one patient to another, extensive tests need to be carried out at a rapid pace so that all infected individuals can be traced and quarantined.
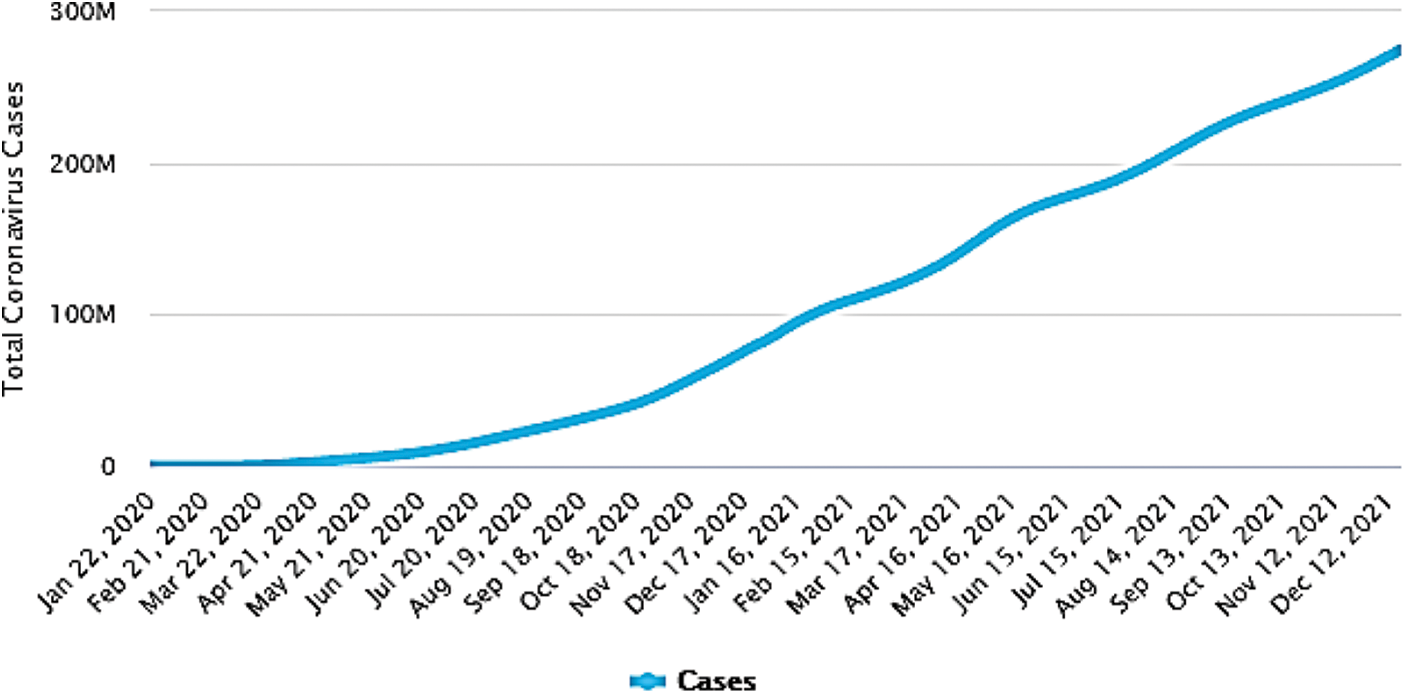
Figure 1: Number of COVID-19 infected cases worldwide [3]
Developing countries with wrecked health care systems cannot afford a large number of expensive RT-PCR tests. There is therefore a considerable difference between the number of COVID-19 tests carried out in developed and developing countries. Fig. 2 shows a comparison of the COVID-19 tests performed in different economies [8]. PT-PCR test suffers from serious learning curve problems during diagnosis. The RT-PCR tests for COVID-19 are believed to have a high specificity but show a sensitivity value as low as 60 to 70 percent [9]. Thus, diagnosis of COVID-19 requires a number of negative tests that are very difficult to perform with a short supply of test kits [10]. It takes at least twenty-four hours to produce a result. Time is crucial in this serious situation, and one cannot wait for an unexpected spread of infection. Therefore, experts are already recommending the use of radiological imaging techniques such as X-rays [11]. X-rays play an important role in the field of experimental medical science. X-rays are electromagnetic rays with wavelengths ranging from 0.01 to 10 nanometers. By inventing technology to combine 2D X-ray image traces into a 3D image, X-rays have improved the accuracy of bone breakage point detection, organ inflammation, tumor location, and many other internal disorders of the body. In clinical practice, a chest X-ray is a quick and easy way to identify COVID-19 based on clinical symptoms.

Figure 2: Number of COVID-19 tests conducted in different countries [8]
The major motivation behind this research was to develop a less expensive, reliable and effective diagnostic tool which may be used to assist the decision makers in medical diagnostic centers. Due to the very limited number of publicly available chest X-ray images related to COVID-19, the transfer learning approach [12] is the best technique for the training of complex deep learning models. In this study, we adopt a well-known transfer learning technique and a fine-tuned neural model, Inception-v4, to the best of the authors’ knowledge. It has not been explored yet for the detection of COVID-19 cases. The proposed model ImageNet pre-trained weights to extract the generic feature set from 15 million images. Moreover, the most challenging dataset is created by combining the two publicly available datasets, which consists of 1504 chest X-ray images, 504 confirmed COVID-19 images and 1000 normal images. To the best of authors’ knowledge, this is the largest dataset as it contains the largest number of X-rays images of confirmed Covid positive cases. The results show that the proposed approach is more effective than existing COVID-19 detection techniques and has achieved a higher accuracy of 99.63%. The model’s accuracy is superior to other existing tools, it may be used in real-world clinical environments for fast and reliable diagnostic tools.
The rest of the paper is divided into the following sections. A detailed overview of the exsiting literature is given in Section 2. Section 3 describes the proposed material and methods for the effective detection and classification of symptoms of COVID-19 by means of chest X-ray images. The results of the proposed approach are discussed briefly in Section 4. Finally, Section 5 summarizes this study and presents a brief conclusion and directions for future work.
Therapeutic treatment and vaccines for COVID-19 are not yet available, and so isolation of suspected individuals from a healthy population is an effective measure to reduce the transmission of the virus [13]. A timely diagnosis is therefore very important for the immediate isolation of infected persons. Machine learning approaches and artificial intelligence can play a significant role in helping front-line fighters to quickly and accurately detect positive cases of COVID-19. In recent years, deep learning has achieved tremendous success in the field of medical imaging and early diagnosis of various diseases [14]. Biomedicine [15], health information [16] and Magnetic Resonance Imaging (MRI) analysis [17] are used. In the medical field, deep learning is used more specifically in the segmentation, classification, and identification of various anatomical areas of interest [18]. Diseases such as brain tumors, skin cancer, lung cancer, breast cancer, and diabetes can be detected through medical imaging. Some deep-learning approaches have shown equivalent results or have even surpassed human performance in these applications [19].
Researchers are now making use of these technological advances in the automatic detection of COVID-19 cases [20]. Xu et al. [21] proposed a three-dimensional DCNN model. CT images were segregated into the following three categories: Covid-19, viral pneumonia, and normal cases. The candidate region segmentation technique was utilized for preprocessing of images to extract the pulmonary regions from the CT images. The proposed model showed an accuracy of 86.7% for classification. In order to propose a multi-classification DCNN model for the automatic detection of COVID-19, Ouchicha et al. used X-ray images. The proposed approach identified COVID-19 with an accuracy of 97.20% and showed an average accuracy of 96.69% for multi-classification [22]. Maghdid et al. [23] proposed a transfer learning approach to early diagnosis of the symptoms of COVID-19. The proposed framework used a simple Convolutional Neural Network (CNN) with a modified AlexNet pre-trained model. In this study, a dataset containing a combination of 120 X-ray images (60 with confirmed COVID-19 and 60 normal images) and 339 Computed Tomography (CT) scan images (191 images with COVID-19 and 147 normal images) was analyzed. The proposed approach identified COVID-19 cases with 98% accuracy. A CNN model with pre-trained weights has been used for a multi-classification task to classify normal/healthy, COVID-19, and pneumonia X-ray images. The results showed that the CNN based model achieved an average accuracy of 0.91 and an F1 score of 0.88 [24].
Since the outbreak of the novel COVID-19, researchers have been continuously trying to do their part to overcome this epidemic. Various machine learning and deep learning systems for automatic detection of Covid-19 cases have been developed based on CT scan images, X-ray images and clinical images. Rahimzadeh et al. [25] developed a deep CNN model based on the concatenation of 50 layers deep Residual Network version 2 (ResNet50V2) and extreme inception (Xception) models. The proposed system used a multi-classification approach to classify chest X-ray images in Covid-19 cases, viral pneumonia and normal cases with an overall accuracy of 91.4%. Afshar et al. [26] proposed a technique for automatic diagnosis of Covid-19 called COVID-CAPS. The proposed system was based on a neural Capsule network capable of working well with small datasets. It consisted of different capsules and convolution layers to reduce the problem of imbalanced data in different classes. The proposed system was evaluated on a Chest X-ray8 dataset [27] and demonstrated 95.7% accuracy, 90% sensitivity, 95.8% specificity, and 97% Area under the Receiver Operating Characteristic (AUROC) curve Covid and non-Covid classification. Ahuja et al. [28] proposed a transfer learning approach for the automatic detection of Covid-19 from CT scan images. The classification results of ResNet18, ResNet50, ResNet101, and SqueezeNet are compared. ResNet18 showed better performance among others with 99.4% classification accuracy.
Das et al. [29] developed a truncated inception Net model to classify Covid-19 positive X-ray images from non-Covid X-ray images. The proposed model achieved an accuracy of 99.96 percent. Five different models such as ResNet152, ResNet50, RestNet101, Inception-ResNetV2 and Inception-v3 were proposed by Narin et al. [30] for autonomous diagnosis of Covid-19. The Proposed models were tested on three different datasets named as the GitHub repository by Dr. Joseph Cohen et al. [31], Chest X-ray8 dataset [27], and Kaggle repository [32]. The pre-trained ResNet50 model outperformed other models with 96.1% accuracy, 99.5% accuracy, and 99.7% accuracy for the first, second, and third datasets.
Jain et al. [33] compared the performance of various deep CNN models such as Xception, ResNet, and Inception V3 classify of Covid positive X-ray images from healthy X-ray images. The Xception model showed better classification performance among other models with 97.97% accuracy. Ozturk et al. [34] proposed a real time object detection system named as you look only once (YOLO) for the automatic detection of Covid-19. The Darknet-19 model is used as classifier with the YOLO architecture. The proposed approach showed 87.02% accuracy for binary classification between Covid-19 and healthy X-ray images. The multi-classification model classified X-ray images into three classes such as Covid-19 positive, non-Covid, and pneumonia with 87.02% accuracy. Another deep learning technique for automatic detection of Covid-19 on the basis of X-ray images has been proposed by Jain et al. [35]. The proposed approach was tested on two datasets, such as image data collection [31] and the Kaggle repository [32] and achieved 97.77% accuracy. Saha et al. [36] proposed a deep CNN approach for autonomous classification of Covid-19 and achieved 98.91% accuracy.
Loey et al. [37] proposed a Generative Adversarial Network (GAN) with deep transfer learning for automatic detection of novel coronavirus. The GAN network has been used to generate X-ray images, and the deep learning model GoogLeNet was used classify of Covid-19 images from healthy images. The proposed system achieved an accuracy value of 80.6%. Khan et al. [38] proposed an automatic Covid-19 detection system named as CoroNet. The proposed system is based on Xception deep CNN model pre-trained on the ImageNet dataset. The proposed model has been implemented in three different scenarios to detect coronavirus from chest X-rays. The first scenario was multi-classification, which classified X-rays in to four classes — Normal, Pneumonia-viral Pneumonia-bacterial and COV19 positive cases — with an accuracy of 89.5%. In second scenario, the multi-classification model classified X-rays into three classes — Normal, Pneumonia and Covid-19 positive cases — with an accuracy of 94.59%. Binary classification was the third scenario that classify chest X-rays into Covid-19 positive and Normal cases with 99% accuracy. It achieved an overall accuracy of 89.6%.
Another system for automatic detection of Covid-19 was proposed by Islam et al. [39]. The proposed system was based on the combination of deep CNN and Long Short-Term Memory (LSTM) architecture for taking the advantages of both models. Deep CNN was used for feature extraction, while the LSTM model used these extracted features for classification. The proposed deep architecture consisted of 20 layers containing 12 convolutional, 5 pooling, one fully connected (FC) layer, one LSTM, and one output layer with the softmax activation function. The proposed CNN-LSTM architecture showed 97% accuracy. Wang et al. [40] proposed COVID-Net for automatic detection of Covid-19 cases from chest X-rays. An open-source dataset named as COVIDx was also created in this study. It showed an accuracy score of 93.1% for classification between Covid-19 and healthy chest X-ray images. Karhan et al. [41] proposed another system using chest X-ray for automatic detection of Covid-19 cases. The deep transfer learning model, ResNet50, was implemented in this research and achieved a classification accuracy score of 99.5%. A comprehensive list of recent studies related to deep learning-based diagnoses of Covid-19 cases is presented in Tab. 1. We have done a thorough study of existing techniques of coronavirus detection and found various research gaps and limitations in literature techniques, such as only a few studies are conducted on reliable data sources. Several authors stated the inconsistent quality of available data as their research’s limitation. Lacking of heterogeneous data to train neural networks reduces the soundness of produced results and deep learning algorithm. Various literature studies suffered from over fitting problem and could not produce good results against an unseen dataset. Covid-19 symptoms lead to multi- organ failure, but as best of the authors’ knowledge, no literature technique diagnoses the severity of lung engagement in Covid-19 subjects.

3 Proposed Research Methodology
This research proposes an automated COVID-19 detection system by applying state-of-the-art approaches to deep learning in the medical field. A detailed flowchart of steps included in the proposed research methodology is shown in Fig. 3. Each step is explained in detail in the sections below.

Figure 3: Framework showing steps of the proposed research methodology
3.1 Dataset Acquisition and Preparation
In this study, we used 1504 chest X-ray images from two publicly available datasets, COVID-19 Image Data Collection databases [31], and the Kaggle chest X-ray repository [44]. The first repository has recently been published and contains images from different publications on the novel COVID-19 Pandemic [31]. This includes a combination of CT scan images and COVID-19 chest X-ray images. As more images related to COVID-19 are continually being added, its size is gradually increasing. The experiments in this study included 504 X-ray images of the chest and 25 CT scan images. This dataset also includes certain metadata for patients with COVID-19, such as their patient identification number and gender. All the COVID-19 images used in this research are collected from this repository. Other than this, 1000 normal chest X-ray images have been collected from the Kaggle repository [44]. Figs. 4a and 4b show the chest X-rays of an infected COVID-19 patient and a healthy person, respectively.

Figure 4: Sample chest X-ray images from dataset: (a) COVID-19 infected chest X-ray [31]; (b) Normal/healthy chest X-ray [44]
3.2 Inception-v4 Model Construction
In this paper, we implemented the Inception-v4 framework to develop the proposed approach for the classification of COVID-19 and normal chest X-ray images. This deep neural framework has evolved from Inception-v1 and GoogLeNet [45]. The architecture of Inception-v4 is more uniform and simplified due to several inception modules. Inception-v4 is used in this study to predict COVID-19 cases due to its exceptional performance in ILSVRC 2012, resulting in an error of 3.08% in three residual networks [46]. The model has also achieved top-1 accuracy of 80%, which is better than Inception v3 and GoogLeNet. The Inception-v4 model has been progressed from Inception-v1, Batch Normalization (BN) Inception, and factorization. The overall schema of Inception-v4 is shown in Fig. 5. Stem refers to the initial operation performed before the Inception block. Inception-v4 consists of three inception modules A, B and C. Reduction blocks are used between inception modules to change the height and width of the grid [47,48].

Figure 5: Overall schema showing pure Inception-v4 network
3.3 Applying Transfer Learning to Inception-v4
Transfer learning [12] as shown in Fig. 6, is an important practice in machine learning to solve the problem of limited training data. In the case of transfer learning, a neural network model trained in one situation can be replicated in another related situation. For example, an image classification model can be trained on ImageNet (a large image repository containing millions of labeled images) to start learning on a limited dataset for the Covid-19 image classification. In this way, the model parameters are initialized with good initial values, so only small changes are needed to achieve better results in a new task.

Figure 6: Learning process of transfer learning
There are two variants of the transfer learning process: fine-tuning and a fixed feature extractor. In the fixed feature extractor variant, the model is firstly trained on a large dataset, and it learns the common features i.e., edges shapes from the available dataset and then only the last classification layer is trained with small amount of dataset from a specific domain. Whereas in fine-tuning variant, only the final layer is not trained in this technique, but selectively few of the previous layers are also trained. The initial layers mostly capture the generic features, and the last layers capture the specific features of the specific problem In the proposed approach, we used a fine-tuning variant to establish a more reliable, robust and effective medical detection system.
The mathematical description of transfer learning as fine-tuning variant of [49] is as follows:
where χ is the feature space and P(X) is the marginal distribution, X = {xi, …, xn}, xi Є χ.
For a specific task T and domain D,
where у is the targeted labels, and ƞ is the predictive function.
for each vector/label set (xi, yi), xi Є χ, yi Є у:
end for
if fine-tune then
end if
Return f
ƞ is an initial feature set that is obtained when the model is initially trained on the ImageNet dataset. Г is a learner function that takes training set of examples of the specific problem and pretrained feature set,
3.4 Model Fine-Tuning and Predicting Output
In this step, the convolutional layers are initialized with pre-trained weights that have been trained through1000 ImageNet dataset classes. In order to improve the generalization of the model, the last layers are randomly initialized, switching from ImageNet images to chest X-ray images. The last layer is changed from 1000 output classes to two output classes: COVID-19 and normal/healthy. The pre-trained inception v4 consists of an input layer that accepts 299 × 299 × 3 input shape, Inception-A, Reduction-B, Inception-B, Inception-C, Reduction-A and Reduction-C modules, where the last three layers are used for prediction purposes. It consists of the AveragePooling2D layer, the Dropout layer, the Flattens layer and the Dense layer. The last layer is modified by the number of output classes. The Stochastic Gradient Descent (SGD) Optimizer is used with 0.001 learning rate, 0.9 momentum and 10-6 decay weighting. Cross-entropy loss is used for equal weighting across classes during model training. The model has been trained over 100 epochs. The output layer classifies the input chest X-ray images as a normal image or a COVID-19 effected image.
Inception-v4 evaluated chest X-rays for the detection of a novel coronavirus. We have implemented a fine-tuned CNN with 1356 images. We used an SGD optimizer that predicts images from normal and COVID-19 affected classes compared to ground truth labels. The model is fit for training at 100 epochs with early stopping, and the model training stops after approximately 90 epochs. The training and validation loss/accuracy graphs for both modules are shown in Fig. 7. In Fig. 7a, the model accuracy curve gradually increases as the epochs are increasing, similarly the model loss curve is decreases as the epochs are increasing. Despite the limited dataset, the model has achieved 99.63% accuracy. There exist other performance metrics for measuring the reliability of the classification model, such as specificity, sensitivity, F1 score, kappa score (κ) and precision. Detailed results obtained for different performance metrics are shown in Tab. 2. The specificity and sensitivity values depict that the model is reliable for detection COVID-19.

Figure 7: Training and validation graphs of Inception-v4 model: (a) training accuracy graph; (b) training loss graph

The results of the classification show that the proposed model has achieved a κ-score of 0.99, which means that the model has an excellent distinguishing capacity between classes and an inter-rater categorical property agreement. The AUROC graph is plotted with a few other multi-class extensions such that a one-to-one scheme is used to uniquely compare the pairwise combination of classes with the reported micro and macro average.
The AUC is approximately 1 as shown in Fig. 8, which means the model has a good class separation capability. The confusion matrix, along with the classification results, is shown in Fig. 9. In comparison with the method [23], which uses the same datasets has achieved 100% sensitivity on X-rays images, and 90% sensitivity is achieved on CT-images, whereas our model has achieved 98.5 sensitivity on both image types. However, the model was only trained for 20 epochs with a small batch size of 10, which may result into quick convergenc. Our model is trained for approximately 90 epochs with 25 batch sizes.

Figure 8: AUROC curve of Inception-v4 fine-tuned CNN

Figure 9: Confusion matrix of Inception-v4 fine-tuned CNN
A comparison of results from the proposed approach to existing studies is shown in Tab. 3. The proposed approach appears to have produced better results compared to previous studies.

A sufficient number of chest X-ray images of COVID-19 patients are not yet available for training of machine learning models for the automatic detection of COVID-19 symptoms. Therefore, in order to overcome this problem, we implemented a transfer learning approach based on Inception-v4 to distinguish between a Covid-19 X-ray image and a normal X-ray image. This research can help developing countries to identify COVID-19 cases cost-effectively and support their fight against the novel disease outbreak. The proposed approach has achieved promising results in the automatic detection of COVID-19 cases with an accuracy of 99.63%, a sensitivity of 98.55%, a specificity of 100%, an F1 score of 99.26%, and a κ-score of 0.99%. However, the proposed model has achieved higher accuracy, sensitivity and specificity but it doesn’t assure a production-ready solution with this limited number of datasets. As a potential future research perspective, this research can be extended to identify various stages of the COVID-19 virus, such as mild, moderate, and severe infections. Researchers can collect more data samples from COVID-19 to make their research more reliable and accurate. Multiclass models can be implemented to identify different types of pneumonia. More attributes of COVID-19 patients, such as phenotypes, CT scan images, history, can be analyzed in order to produce more accurate results. Location attention mechanisms can be put in place to improve the accuracy of COVID-19 predictions. The ability of the detection system to identify the level or day of the COVID-19 virus in a patient can be enhanced.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang et al., “A novel coronavirus from patients with pneumonia in China,” 2019 New England Journal of Medicine, vol. 382, no. 8, pp. 723–733, 2020. [Google Scholar]
2. Q. Li, M. Med, X. Guan, P. Wu, X. Wang et al., “Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia,” New England Journal of Medicine, vol. 382, no. 13, pp. 1199–1207, 2020. [Google Scholar]
3. Worldometer, “Coronavirus worldwide graphs,” 2021. (Accessed 21 Nov 2021). [Online]. Available: https://www.worldometers.info/coronavirus/coronavirus-cases/. [Google Scholar]
4. Y. Shi, Y. Wang, C. Shao, J. Huang, J. Gan et al., “COVID-19 infection: The perspectives on immune responses,” Cell Death & Differentiation, vol. 27, no. 5, pp. 1451–1454, 2020. [Google Scholar]
5. Z. Xu, L. Shi, Y. Wang, J. Zhang, L. Huang et al., “Pathological findings of COVID-19 associated with acute respiratory distress syndrome,” The Lancet Respiratory Medicine, vol. 8, no. 4, pp. 420–422, 2020. [Google Scholar]
6. V. M. Corman, O. Landt, M. Kaiser, R. Molenkamp, A. Meijer et al., “Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR,” Eurosurveillance, vol. 25, no. 3, pp. 2000045, 2020. [Google Scholar]
7. W. H. K. Chiu, V. Vardhanabhuti, D. Poplavskiy, P. L. H. Yu, R. Du et al., “Detection of COVID-19 using deep learning algorithms on chest radiographs,” Journal of Thoracic Imaging, vol. 35, no. 6, pp. 369–376, 2020. [Google Scholar]
8. Our World in Data, “Our world in data: Total COVID-19 tests,” 2021. (Accessed 21 Nov 2021). [Online]. Available: https://ourworldindata.org/grapher/full-list-total-tests-for-covid-19. [Google Scholar]
9. Y. Fang, H. Zhang, J. Xie, M. Lin, L. Ying et al., “Sensitivity of chest CT for COVID-19: Comparison to RT-PCR,” Radiology, vol. 296, no. 2, pp. 200432, 2020. [Google Scholar]
10. J. P. Kanne, B. P. Little, J. H. Chung, B. M. Elicker and L. H. Ketai, “Essentials for radiologists on COVID-19: An update—radiology scientific expert panel,” Radiological Society of North America, vol. 296, no. 2, pp. 200527, 2020. [Google Scholar]
11. V. I. Mikla and V. V. Mikla, “Advances in imaging from the first X-ray images,” in Medical Imaging Technology, 1st ed., vol. 1. Amsterdam, Netherlands: Elsevier, pp. 1–22, 2014. [Google Scholar]
12. C. Tan, F. Sun, T. Kong, W. Zhang, C. Yang et al., “A survey on deep transfer learning,” in Int. Conf. on Artificial Neural Networks. Rhodes, Greece, pp. 270–279, 2018. [Google Scholar]
13. WHO, “Coronavirus disease 2019 (COVID-19) situation report–72,” 2020. (Accessed 10 Nov 2021). [Online]. Available: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200401-sitrep-72-COVID-19.pdf. [Google Scholar]
14. J. Ker, L. Wang, J. Rao and T. Lim, “Deep learning applications in medical image analysis,” IEEE Access, vol. 6, no. 1, pp. 9375–9389, 2017. [Google Scholar]
15. M. Wainberg, D. Merico, A. Delong and B. J. Frey, “Deep learning in biomedicine,” Nature Biotechnology, vol. 36, no. 9, pp. 829–838, 2018. [Google Scholar]
16. D. Ravi, C. Wong, F. Deligianni, M. Berthelot, J. Andreu-Perez et al., “Deep learning for health informatics,” IEEE Journal of Biomedical and Health Informatics, vol. 21, no. 1, pp. 4–21, 2016. [Google Scholar]
17. J. Liu, Y. Pan, M. Li, Z. Chen, L. Tang et al., “Applications of deep learning to MRI images: A survey,” Big Data Mining and Analytics, vol. 1, no. 1, pp. 1–18, 2018. [Google Scholar]
18. M. Bakator and D. Radosav, “Deep learning and medical diagnosis: A review of literature,” Multimodal Technologies and Interaction, vol. 2, no. 3, pp. 47, 2018. [Google Scholar]
19. A. Esteva, B. Kuprel, R. A. Novoa, J. Ko, S. M. Swetter et al., “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, no. 7639, pp. 115–118, 2017. [Google Scholar]
20. A. Makris, I. Kontopoulos and K. Tserpes, “COVID-19 detection from chest X-ray images using deep learning and convolutional neural networks,” in Hellenic Conf. on Artificial Intelligence, Athens, Greece, pp. 60–66, 2020. [Google Scholar]
21. X. Xu, X. Jiang, C. Ma, P. Du, X. Li et al., “A deep learning system to screen novel coronavirus disease 2019 pneumonia,” Engineering, vol. 6, no. 1, pp. 1122–1129, 2020. [Google Scholar]
22. C. Ouchicha, O. Ammor and M. Meknassi, “CVDNet: A novel deep learning architecture for detection of coronavirus (Covid-19) from chest X-ray images,” Chaos, Solitons & Fractals, vol. 140, no. 1, pp. 110245, 2020. [Google Scholar]
23. H. S. Maghdid, A. T. Asaad, K. Z. Ghafoor, A. S. Sadiq and M. K. Khan, “Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms,” in Proc. of Multimodal Image Exploitation and Learning, Orlando, Florida, USA, pp. 26, 2021. [Google Scholar]
24. J. E. Luján-García, M. A. Moreno-Ibarra, Y. Villuendas-Rey and C. Yáñez-Márquez, “Fast COVID-19 and pneumonia classification using chest X-ray images,” Mathematics, vol. 8, no. 9, pp. 1423, 2020. [Google Scholar]
25. M. Rahimzadeh and A. Attar, “A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2,” Informatics in Medicine Unlocked, vol. 19, no. 1, pp. 100360, 2020. [Google Scholar]
26. P. Afshar, S. Heidarian, F. Naderkhani, A. Oikonomou, K. N. Plataniotis et al., “Covid-caps: A capsule network-based framework for identification of covid-19 cases from X-ray images,” Pattern Recognition Letters, vol. 138, no. 1, pp. 638–643, 2020. [Google Scholar]
27. X. Wang, Y. Peng, L. Lu, Z. Lu, M. Bagheri et al., “ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Honolulu, HI, USA, pp. 2097–2106, 2017. [Google Scholar]
28. S. Ahuja, B. K. Panigrahi, N. Dey, V. Rajinikanth and T. K. Gandhi, “Deep transfer learning-based automated detection of COVID-19 from lung CT scan slices,” Applied Intelligence, vol. 51, no. 1, pp. 571–585, 2021. [Google Scholar]
29. D. Das, K. C. Santosh and U. Pal, “Truncated inception net: COVID-19 outbreak screening using chest X-rays,” Physical and Engineering Sciences in Medicine, vol. 43, no. 3, pp. 915–925, 2020. [Google Scholar]
30. A. Narin, C. Kaya and Z. Pamuk, “Automatic detection of coronavirus disease (covid-19) using X-ray images and deep convolutional neural networks,” Pattern Analysis and Applications, vol. 24, no. 1, pp. 1207–1220, 2021. [Google Scholar]
31. J. P. Cohen, P. Morrison, L. Dao, K. Roth, T. Q. Duong et al., “Covid-19 image data collection: Prospective predictions are the future,” Machine Learning for Biomedical Imaging, vol. 1, no. 1, pp. 18272, 2020. [Google Scholar]
32. P. Patel, “Chest X-ray (Covid-19 & pneumonia),” 2021. (Accessed 17 Jan 2021). [Online]. Available: https://www.kaggle.com/prashant268/chest-xray-covid19-pneumonia. [Google Scholar]
33. R. Jain, M. Gupta, S. Taneja and D. J. Hemanth, “Deep learning based detection and analysis of COVID-19 on chest X-ray images,” Applied Intelligence, vol. 51, no. 3, pp. 1–11, 2020. [Google Scholar]
34. T. Ozturk, M. Talo, E. A. Yildirim, U. B. Baloglu, O. Yildirim et al., “Automated detection of COVID-19 cases using deep neural networks with X-ray images,” Computers in Biology and Medicine, vol. 121, no. 1, pp. 103792, 2020. [Google Scholar]
35. G. Jain, D. Mittal, D. Thakur and M. K. Mittal, “A deep learning approach to detect Covid-19 coronavirus with X-ray images,” Biocybernetics and Biomedical Engineering, vol. 40, no. 4, pp. 1391–1405, 2020. [Google Scholar]
36. P. Saha, M. S. Sadi and M. M. Islam, “EMCNet: Automated COVID-19 diagnosis from X-ray images using convolutional neural network and ensemble of machine learning classifiers,” Informatics in Medicine Unlocked, vol. 22, no. 1, pp. 100505, 2021. [Google Scholar]
37. M. Loey, F. Smarandache and N. E. M. Khalifa, “Within the lack of chest COVID-19 X-ray dataset: A novel detection model based on GAN and deep transfer learning,” Symmetry, vol. 12, no. 4, pp. 651, 2020. [Google Scholar]
38. A. I. Khan, J. L. Shah and M. M. Bhat, “CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest X-ray images,” Computer Methods and Programs in Biomedicine, vol. 196, no. 1, pp. 105581, 2020. [Google Scholar]
39. M. Z. Islam, M. M. Islam and A. Asraf, “A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images,” Informatics in Medicine Unlocked, vol. 20, no. 1, pp. 100412, 2020. [Google Scholar]
40. L. Wang, Z. Q. Lin and A. Wong, “Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest X-ray images,” Scientific Reports, vol. 10, no. 1, pp. 1–12, 2020. [Google Scholar]
41. Z. Karhan and A. Fuat, “Covid-19 classification using deep learning in chest X-ray images,” in Proc. of Medical Technologies National Conf., Antalya, Turkey, pp. 1–4, 2020. [Google Scholar]
42. RSNA Pneumonia Detection Challenge, “kaggle,” 2021. (Accessed 20 Jan 2021). [Online]. Available: https://www.kaggle.com/c/rsna-pneumonia-detection-challenge. [Google Scholar]
43. D. S. Kermany, “Identifying medical diagnoses and treatable diseases by image-based deep learning,” Cell, vol. 172, no. 5, pp. 1122–1131, 2018. [Google Scholar]
44. P. Mooney, “Chest X-ray images (pneumonia),” 2021. (Accessed 20 Jul 2020). [Online]. Available: https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia. [Google Scholar]
45. C. Szegedy, W. Liu, Y. Jia and P. Sermanet, “Going deeper with convolutions,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Boston, MA, USA, pp. 1–9, 2015. [Google Scholar]
46. C. Szegedy, S. Ioffe, V. Vanhoucke and A. Alemi, “Inception-v4, inception-ResNet and the impact of residual connections on learning,” in Proc. of Thirty-first AAAI Conf. on Artificial Intelligence, San Francisco, California, USA, vol. 31, 2017. [Google Scholar]
47. S. -H. Tsang, “Review: Inception-v4—Evolved from GoogLeNet, merged with ResNet idea (Image classification),” 2020. (Accessed 25 Oct 2021). [Online]. Available: https://towardsdatascience.com/review-inception-v4-evolved-from-googlenet-merged-with-resnet-idea-image-classification-5e8c339d18bc. [Google Scholar]
48. T. -C. Pham, C. -M. Luong, M. Visani and V. -D. Hoang, “Deep CNN and data augmentation for skin lesion classification,” in Proc. of Asian Conf. on Intelligent Information and Database Systems, Đồng Hới, Quảng Bình, Vietnam, pp. 573–582, 2018. [Google Scholar]
49. D. Sarkar, “A comprehensive hands-on guide to transfer learning with real-world applications in deep learning,” 2020. (Accessed 20 Oct 2021). [Online]. Available: https://towardsdatascience.com/a-comprehensive-hands-on-guide-to-transfer-learning-with-real-world-applications-in-deep-learning-212bf3b2f27a. [Google Scholar]
50. British Society of Thoracic Imaging (BSTI“COVID-19 imaging database,” 2020. (Accessed 15 Oct 2021). [Online]. Available: https://www.bsti.org.uk/training-and-education/covid-19-bsti-imaging-database/. [Google Scholar]
51. I. Bickle, C. Hacking and A. Er, “Normal chest imaging examples,” 2020. (Accessed 15 Aug 2021). [Online]. Available: https://radiopaedia.org/articles/normal-chest-imaging-examples. [Google Scholar]
52. S. Minaee, R. Kafieh, M. Sonka, S. Yazdani and G. J. Soufi, “Deep-covid: Predicting covid-19 from chest X-ray images using deep transfer learning,” Medical Image Analysis, vol. 65, no. 1, pp. 101794, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |