DOI:10.32604/jai.2022.028092

| Journal on Artificial Intelligence DOI:10.32604/jai.2022.028092 |  |
| Article |
Classification of Bone Marrow Cells for Medical Diagnosis of Acute Leukemia
Bahria University Islamabad, Pakistan
*Corresponding Author: Samabia Tehsin. Email: stehseen.buic@bahria.edu.pk
Received: 02 February 2022; Accepted: 04 March 2022
Abstract: Leukemia is the cancer that starts in the blood cells due to the excess production of immature leucocytes that replace the cells with normal blood cells. Physicians rely on their experience to determine the type and subtype of Leukemia from the blood sample. Most people are misdiagnosed when it comes to its subtypes, the error rates can be up to 40% during the classification process. That too depends on the expertise of the physician. This research represents a Convolutional Neural Network based medical image classifier. The proposed technique can classify Leukemia and its five subtypes. State of the art deep learning and transfer learning techniques have been used, all of the network architectures are fine-tuned followed by feature extraction. This research can assist the pathologist in decision making or can be used for the process automation. The proposed methodology can give better results in terms of accuracy and is more effective than the hematologist's visual classification in terms of time.
Keywords: Convolutional neural network; transfer learning; medical imaging; acute leukemia
Leukemia is the most common cancer worldwide with 250,000–300,000 new cases every year, the percentage of death caused by Leukemia is 74% which is typically because of poor identification of types or subtypes of Leukemia or delay in diagnosis. Referring to the score given by INEGI-2006, leukemia ranked 5th as a reason of death in men (7%), 6th in women (5.8%) and 1st in children (48.5%) with cancer, respectively [1]. Leukemia begins in the bone marrow and bone marrow cells start to generate unusual WBC. Due to absence of defenses and normal WBC, body starts losing its fight against all sorts of infections and diseases. Fig. 1 [2] shows the image of healthy and Leukemia infected bone marrow samples. Early diagnosis of the disease can result in quick recovery in case of children.
Leukemia can mainly be classified as Acute Lymphoblastic Leukemia (ALL) & Acute Myeloid Leukemia (AML). Acute leukemia's grow fast and invade the body in a few weeks or months, whereas chronic leukemia's are slow growing but worsen over the time. The FAB classification categorizes ALL into three subtypes “L1-L2-L3” and AML into eight different subtypes “M0, M1, M2, M3, M4, M5, M6, M7” respectively. Fig. 2 shows L1 and L2 type Cells.
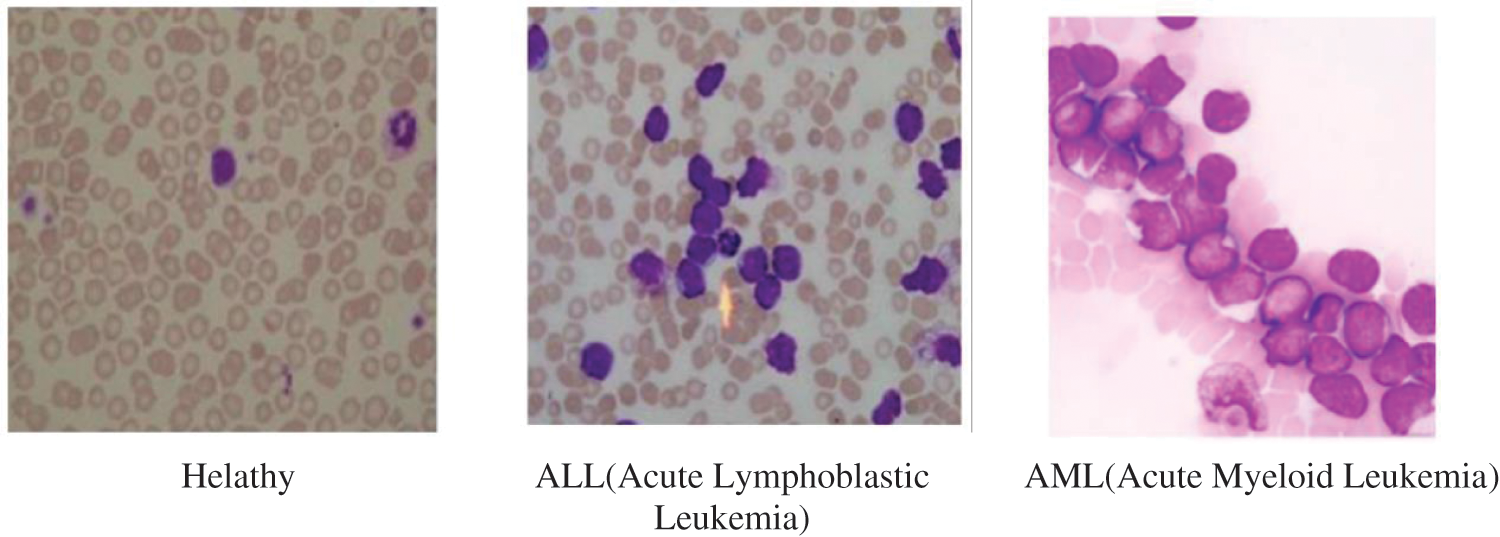
Figure 1: Healthy bone marrow and leukemia
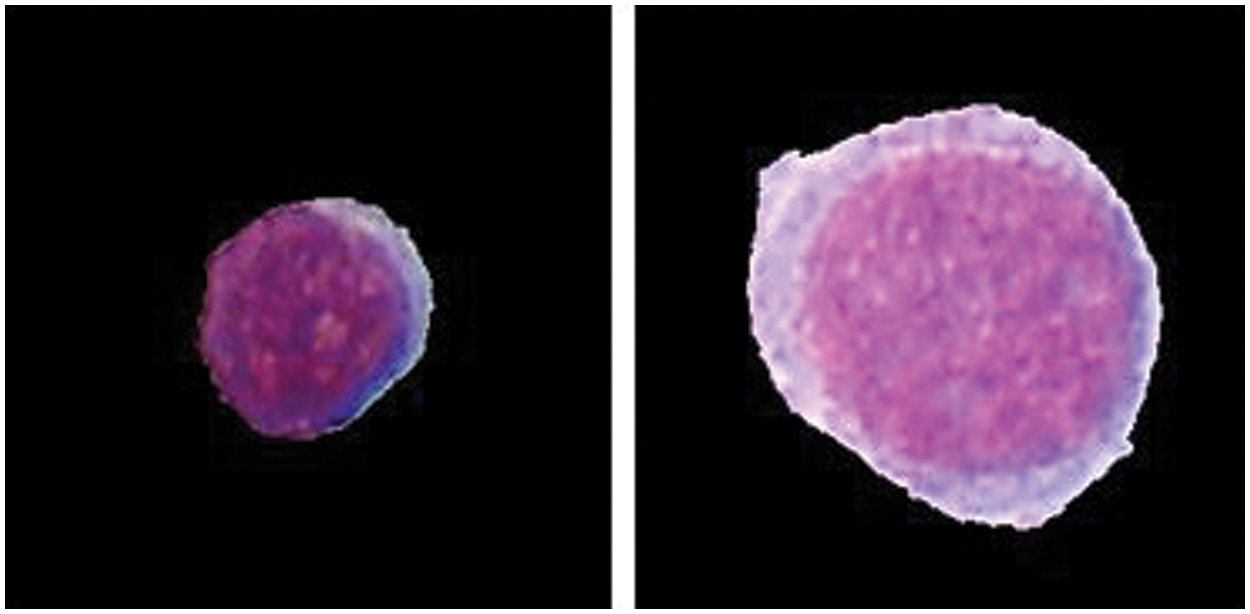
Figure 2: L1 and L2 type cell
1.1 Acute Lymphocytic Leukemia
Acute lymphocytic leukemia, additionally known as “acute lymphoblastic leukemia” and “acute lymphoid leukemia”, is a kind of blood cancer that occurs when unusual WBC aggregate in the bone substance. ALL advances quickly, supplanting solid cells that create healthy lymphocytes with leukemia cells that can't develop appropriately. Leukemia cells are transferred in the blood to various structures, cells and tissues. Over production and propagation of these infected cells might lead to various conceivable side effects.
ALL is predictably links to getting extra B lymphatic cells as compared to T cells. T and B cells undertake dynamic jobs in keeping the body from contaminations and germs and decimating cells that have just turned out to be tainted. B cells especially prevent from contaminating the body, however T cells eliminate infected blood cells. ALL can strike at any stage of life; however acute lymphocytic leukemia occurrences happen most often in individuals younger than 15 or beyond 45 years old. Albeit ALL makes up the biggest level of leukemia analyze in youngsters younger than 15 (mostly amid in the age of 2–4 years), it is uncommon for grown-ups to have this ailment.
ALL Treatment may incorporate radiation/chemotherapy, chemotherapy with undeveloped cell transplant, radiation therapy. The incorporated group of leukemia specialists will answer the inquiries and suggest treatment options reliant on his specialization in findings. Chemotherapy for ALL typically starts with enlistment chemotherapy, in which a blend of medications is utilized to destroy the malignant cells.
AML is the deadliest kind of Leukemia as it progresses quickly and gives us a very small window to its cure. It is also the highest acknowledged form of leukemia. It occurs once the bone marrow starts to form cells those are not yet totally developed, the cells are abnormal and affect the normal cells. And the body loses its defense mechanism. In AML, the bone marrow likewise make anomalous RBCs and platelets. The quantity of these unusual cells increments quickly, and the anomalous cells start to swarm out the ordinary WBCs, RBCs and platelets that the body requires. One of the fundamental things that separate AML from the other principle types of leukemia is that it has eight diverse subtypes.
“Myeloblastic (M0)-on special analysis, Myeloblastic (M1)-without maturation, Myeloblastic (M2)-with maturation, Promyeloctic (M3), Myelomonocytic (M4), Monocytic (M5), Erythroleukemia (M6), Megakaryocytic (M7)”. Fig. 3 represents the structure of different AML types. AML Treatment might incorporate chemotherapy, radiation treatment, or stem cell transplant. Pathologist will prescribe treatment options dependent on the subtype of Leukemia according to his expertise. Which may also we wrong and time taking, which will lead to a very ill-timed treatment.
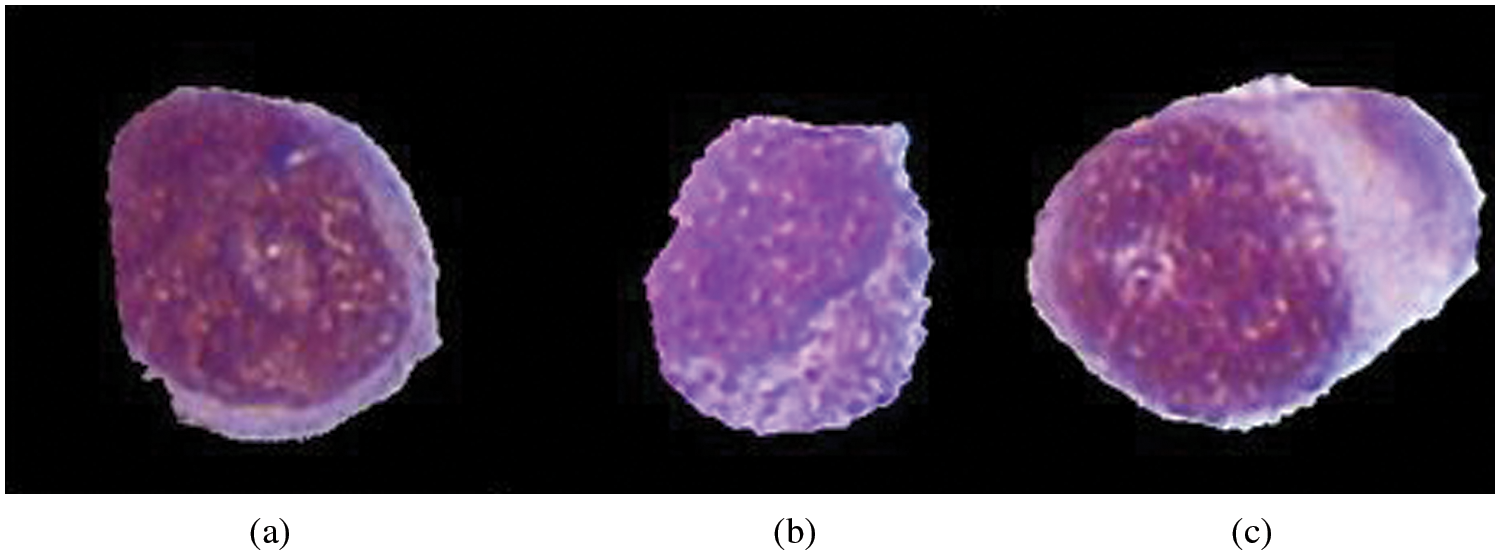
Figure 3: Categorization of AML according to FAB. (a) M2-ALL, (b) M3-ALL, (c) M5-AML
There is an adequate need to set up a quick diagnosis because the classification process only depends upon the expertise of the hematologist and his experience, which can lead to many problems like mis diagnosis or delay in the diagnosis our proposed system will give the hematologist another machine learning based opinion which will give him an insight on the classification problem.
The accompanying disadvantages may fundamentally hamper manual minute morphological examination of bone marrow cells:
• Poorly arranged or recolored blood smears.
• Time utilization. Despite the fact that an accomplished master plays out the PB spread examination, it can't be considered as a quick procedure, where the administrator needs to complete a watchful report on the impact cells morphology (estimate, shape, core chromatin structure) with the end goal to turn out with a right diagnosis and guarantee that the correct treatment will be given.
• The technique is vulnerable to human mistake.
1.4 Bone Marrow Aspirate Morphological Examination
Bone Marrow is an extraordinary greasy tissue containing immature microorganisms, situated inside a couple of vast bones. These foundational microorganisms can change into WBCs, RBCs and platelets that have different jobs. Inside this uncommon tissue, youthful stems cells dwell, alongside additional iron. Undeveloped cells stay undifferentiated until irregular, debilitated, or harmed cells should be supplanted. This is the main procedure through which cells get supplanted to keep up a solid body. Doctors normally utilize the 5-year survival rates to quantify illness result. Survival rates incorporate patients who survive 5 years after diagnosis, whether disappearing, i.e., in a state amid which the symptoms of the infection are lessened.
Key contributions for the proposed study includes the classification of diseased and healthy samples. It also proposed a methodology for the classification of sub-types as well. The proposed techniques are focused on classification of ALL and AML including their respective subtypes, using state of the art pre trained models. Rest of the article is organizes as: Section 2 provides analysis of state-of-the-art of the field, Section 3 provides the proposed methodology, Section 4 explains the results of the proposed methodology and Section 5 concludes the research article. Before moving further, Tab. 1 list down the abbreviations used in this study for better understanding
This section is solely dedicated to the conclusions of the literature reviews presented. Some of the most prevalent and significant transfer learning techniques are presented and reviewed which have been previously used for the classification purposes, CNNs have been the least employed for the classification, so are transfer learning techniques, in the previous literature there is lot of work done for the segmentation and classification of Leukemia using Digital images of blood cells whereas bone marrow images were narrowly used. We have listed some notable work from previous studies to get an insight on the research problem. Various procedures are suggested by the research community to spot on these issues and to get the improved sample image for better segmentation.
This section represents state of the art of the domain. The overview of techniques is given and in-depth analysis is also presented. In this section, a summary of important achievements for the categorization of acute leukemia and its sub-categories, are discussed. Numerous machine learning procedures are explained and assessed in the presented research work, for grouping of healthy and infected cells. The vast rise of machine learning tools and usage gets the question of choosing between different methods in different scenarios. Empirical investigation can give the most suitable choice for this. In this section, we have discussed different classification methods along with their suitability in different scenarios.
Vincent et al. [1] used Histogram Equalization, morphological filtering of ALL sample-images, followed by fuzzy C-Means for segmentation of WBCs. After employing Gabor filter and feature extraction, SVM is used for the grouping of infected cells. This technique is capable to get 0.90 accuracy. Singh et al. [3] had improved the accuracy to 92% by using LDA. SVM is utilized for the categorization of Leukemia-infected and healthy cells. Harjoko et al. [4] attained an accuracy of 93.57% by deploying their recommended approach. Histogram Equalization, Median filter are utilized for image quality enhancement. After that, different features including shape and statistical features were obtained from the segmented lymphocytes. Moreover, a strong ensemble classifier using backpropagation momentum was used to classify infected and non-infected cells which led to an accuracy of 93.57%. Lim et al. [5] used histogram equalization and median filtering to enhance contrast and noise removal. HIS color space is used for improved segmentation results. After extracting geometrical based features like pixel-count, boundary-length, diameter and obliqueness features, they used SVM which improved the accuracy to 97.55%. Jagadev et al. [6] acquired accuracy of 92% by using K-Means clustering, Marker controlled Watershed method, HSV color based segmentation, after extracting features like cell area, average intensity value, Entropy, and few other statistical features. SVM is used for the classification. Fatma et al. [7] proposed K means clustering, Image Acquisition, Linear Contrast Enhancement, HSI Color Modelling, Median Filter segmentation techniques for white blood. Classifiers like Perceptron, LDAs, and SVM are used and attained the accuracy of 91%. Thanh et al. [8] achieved the accuracy of 96.43% by using Clinical decision support system classifiers. Reference [9] presented new techniques for classification like genetic algorithm, simulated annealing and hill climbing, and concluded that simulated annealing is the best for detection of leukemia, they received around an accuracy of 97.22%, but they only classified between M3 and other subtypes.
EPMs [10] provides a cheap alternative to a better diagnosis of acute leukemia, EPMs requires no human intervention, it automatically selects machine learning classifiers, It gave a result of 97.68% for two types of leukemia [11] They used different machine learning algorithms for the segmentation and classification for types and subtypes of leukemia, And even came up with a diagnostic algorithm which gave an accuracy of 95%.
After investigating the prior approaches, we can conclude that there is a need of improving the classification results of blasted and non-blasted cells for Leukemia detection [12–14]. The accurate diagnosis of such fatal disease is a very critical aspect. For reliability and robustness of diagnostic system, classification results must be further enhanced. The improved system can be used to assist the human diagnosis process or it can be used for the process automation as well. This is an extremely challenging job, as the blood cells are greatly overlying making those difficult to segregate. It is also analyzed that classification of ALL and AML sub-categorization is overlooked by the research community. It is quite much a challenging task because of high inter-class resemblance and intra-class inconsistency. These sub-categories are tough to categorize but can be essential in detection and cure of the disease.
In the recent years, manual classification of Leukemia has been done by the hematologist that was totally based on their expertise. In modern era, machine learning plays a vital role in the classification. Many algorithm like Random Forest, SVM have been used in the classification process. Deep learning is very much used for the extraction of features. It gives the researcher an easy way of extracting features, as compare to hand-crafted features. This section describes the proposed procedure for categorization of Leukemia and its subtypes. CNN is employed for the feature extraction. Detailed architectural details are available in the section. Experiments are carried out by various pre-trained models on our custom dataset. Annotation of the segmented images is done by the expert hematologist for ground truth labeling.
In this study, we deployed the pre-trained CNN's for classification of ALL, AML and its subtypes. In the recent years, this manual extraction of features (normally known has hand-crafted features) is substituted by machine-learned features using deep neural network, particulary by CNNs. We also employ CNNs in our study.
CNN basic architecture is presented first, followed by a description that how it is employed in the current problem. In our study, we deploy transfer learning techniques to classify Cellular Elements Nucleus cellula and then Leukemia and its subtypes. We have employed few well-established pre-trained CNNs applying the transfer learning techniques. The employed networks are briefly outlined in this section following the system overview. Training classifiers to recognize images requires labeled images so we have been manually generating and labeling the training data which was supervised by a hematologist. Proposed architecture has two convolutional layers with 24 (9 × 9) and 48 (7 × 7) filter respectively. The convolutional layers are followed by ReLU and the max-pooling layers. The last layer is fully connected for classification purpose. The networks are trained using the medical images of the bone marrow cells.
This study also employs Alex Net, VGG-16, inception V3 and a mixed architecture of inception V3 & ResNets 101 as pretrained networks. Network parameters for system training are: Batch-size: 30, # of Epochs: 20, Momentum: 0.9, Base learning rate: 0.001.
The database of Images used in this study was created in collaboration with physicians and the Mexican Social Security Institute in order to implement a system for the automatic morphological identification of acute leukemia's from bone marrow cells images. Image set is comprised of 633 bone marrow leukemia cells images. Different color staining techniques are used in the dataset collection. All images were digitalized by Carl Zeiss optical microscope. Hence the image resolution was the same. For the classification of subtypes of ALL and AML, these images were labelled to L1, L2 and M2, M3, M5 by expert oncologist who labelled each image into ALL and AML subtype manually. Fig. 4 shows images from the dataset. Training and testing ratio is kept at 0.75 and 0.25, respectively. It means 75% data is being used to update the system weights and train the model for the focused problem and results of the trained model are tested on remaining 25% of data. Our main objective of the work was to classify the subtypes of ALL which were mostly neglected in the previous literature because they are difficult to classify due to their inter class similarity and intra class variability. Equations should be flushed to the left of the column.
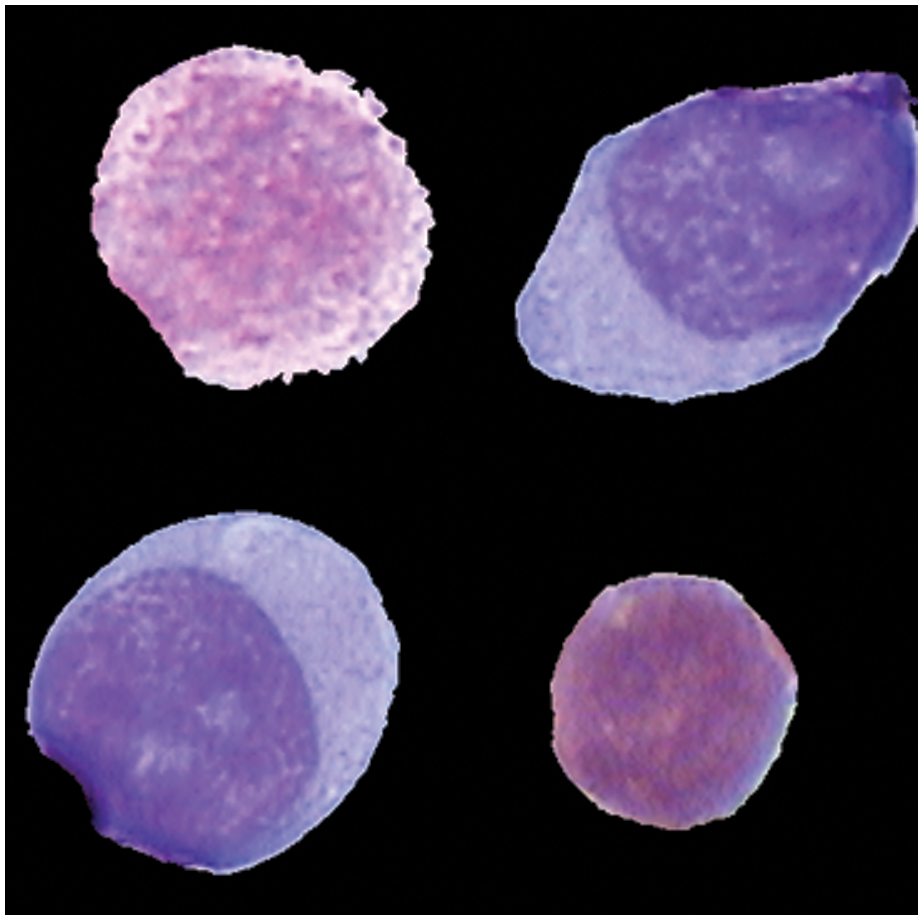
Figure 4: Different subtype cells of leukemia
Due to scarcity of reliable medical data, data augmentation is used. It will help in proper training of network architectures. Image spin and reflection is used to improve the size of training data. Images are spined for three angles including, ninety, hundred and eighty and two hundred seventy. The size of data becomes four-times, after applying the data augmentation. After augmentation, the size of dataset becomes 2532.
3.4 Convolutional Neural Networks
Convolutional neural systems are great ANNs, and are employed primarily to group images, cluster them by comparability, and execute target identification within scenes. These are the algorithms which can be used to distinguish among road signs, people, faces and numerous different parts of visual information. CNNs are especially valuable for discovering designs in pictures to perceive articles, scenes, and appearances. They gain straightforwardly from picture data, utilizing patterns to arrange or classify pictures and wiping out the requirement for manual element extraction [15]. The adequacy of convolutional network in an image recognition is one of the major cause for why the universe has turned out the viability of deep learning.
They are charging significant priors in computer vision, that have the evident operations for mechanical technology, drones, self-driving cars, medications, drones, and security for visually debilitated. A CNN can have many layers in which every layer figures out how to expose or identify distinctive components or attributes of a picture. CNN have filters which are connected by every training picture on various firmness or decisions, and an output of every picture is utilized for contribution with the upcoming layer. Like other neural network, a CNN is made out of an output layer, an input layer, and many invisible layers in between input and output layers. These layers accomplish assignments that modify the weights with the goal of learning features particular to the data. There are various types of layers introduced in the literature. The most widely used layers are as given below.
Convolution puts the input pictures through an arrangement of convolutional filters, every one of which initiates certain features from the pictures.
3.4.2 Rectified Linear Unit (ReLu)
ReLu takes into account quicker and more powerful training. It truncates the negative values to zero, while keeping the positive values unchanged. This is in some cases referred to as activation, as just the activated features are conveyed forward into the following layer.
Pooling is a use to rearranges an output through executing nonlinear down sampling, and lessening the parameter's quantity which is required to study by network. These tasks are rehashed more than tens or many layers, with each layer figuring out how to distinguish diverse features.
Whenever training of neural network is required for any specific task on a given data, we start to train from the scratch with the random set of weights. Once the system is fully trained we record the weights of the neural network that we use somewhere and we can make it available for people to use as well. But when we have to work on a different problem we definitely would want to save time and not repeat the whole training process from scrape, So we might consider to use the already trained weights and augment those for the new data set, This process is also known as fine tuning. Medical data is difficult to collect, so in order to cater this data scarcity, the model can be pre-trained on some large dataset and fine-tune on the medical data. And then by removing the weights of fully connected layers then attaching the new layers with randomized weights. Pre-training comes with many advantages firstly it is really beneficial for producing good results on small dataset, saving time as well. As for large datasets they are always not available and cannot be easily trained as well. We will describe feature extraction & fine tuning along with their architectures.
One way to deal with get around this issue is to first pre-train a deep net on a huge scale dataset, as ImageNet. At that point, given another dataset, we can begin with these pre-trained weights when training on our new task. This procedure is generally called fine-tuning. There are various varieties of fine-tuning. Now and then, the underlying neural network is utilized just as a feature extractor. That implies that we solidify each layer preceding the output layer and essentially learn another output layer [16,17]. To fine-tune a network, we should initially replace the last completely associated layer with another one that yields the coveted number of classes. We introduce its weights arbitrarily. At that point we keep preparing as ordinary. Now and then it's normal to utilize a smaller learning rate dependent on the instinct that we may already be close a good outcome. The task of finetuning a network is to change the parameters of an officially prepared network with the goal that it adjusts to the new task at hand. The starting layers learn extremely broad features and as we go higher up the system, the layers tend to learn patterns more particular to the task it is being prepared/trained on. In this way, for fine-tuning, we need to keep the underlying layers flawless (or freeze them) and retrain the later layers for our task.
Feature Extraction uses a CNN that is trained so that the features are computed for the following picture. The features that are extracted from the hidden layer or many other types of layers in the presented picture. In the presented features the trained classifiers can accomplish aggressive outcomes, in some cases leaving behind the features that are engineered by humans. However, the examinations demonstrate the hyper parameters, ought to be chosen for better execution [18,19].
The Feature extraction doesn't modify the real network but it enables the undertakings so that it could gain benefits from the features that are complex and gaining from the past undertakings [20,21]. However, the features present were not particularly assigned for the generally new task and can be repeatedly enhanced using the fine tuning. In this research, five pre-trained models are empirically evaluated. Feature extraction task is accomplished by the combination of convolution, pooling and activation layers. Five-class classification task is achieved by the fully connected and Softmax layers.
In this section, we begin discussing the results of the experiments performed. At First we will discuss the dataset, then we will briefly describe the proposed methodology and the network parameters on which we have trained the system. Results of fine tuning and feature extraction will be presented. Finally, we will discuss system evaluation and comparing our results with the other work. The implementation of the methods is mostly discussed in Section 3.
4.1 Results for Classification of Cellular Elements
A pre-trained CNN was used for categorization of ALL, AML and its subtypes. We investigated various outcomes attained from segmented data. For the dataset ALL & AML detection accuracy was good but subtypes classification accuracy was somehow higher than the previous work. As before the classification was carried out on small datasets or individually on just AML OR ALL.
We first present the overall classification results for Nucleus and cellula in the Tab. 2. In the first experiment classified the Cellular Elements with fine tuning, and truncated the weights of last layer according to our problem with the same Network parameters discussed earlier. Mixed architecture had 825 layers, we froze 822 and used the last three layers using the same Network Parameters. The classification was carried out using Convolution Neural Networks, we were able to obtain the best results of classification of cellular elements with inception V3. We achieved a result of 85.3% for feature Extraction and 68.04% for fine tuning.
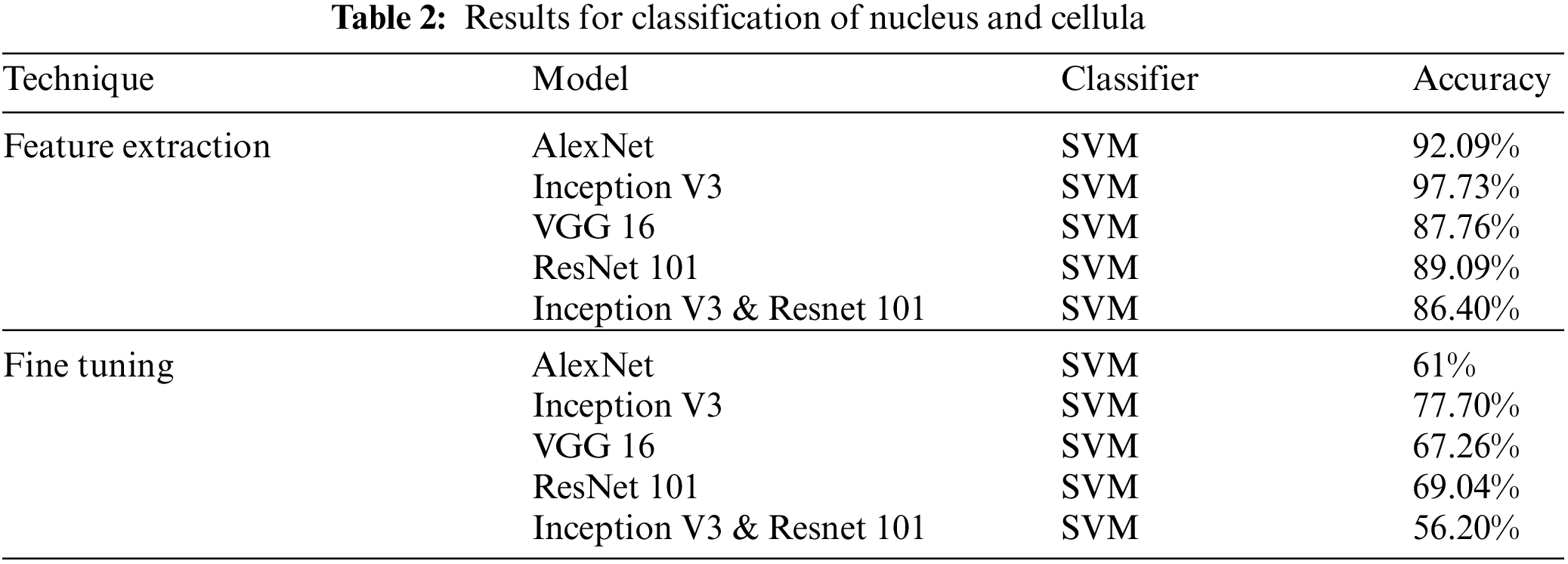
Classification Results for Leukemia and its subtypes are presented in Tab. 3.
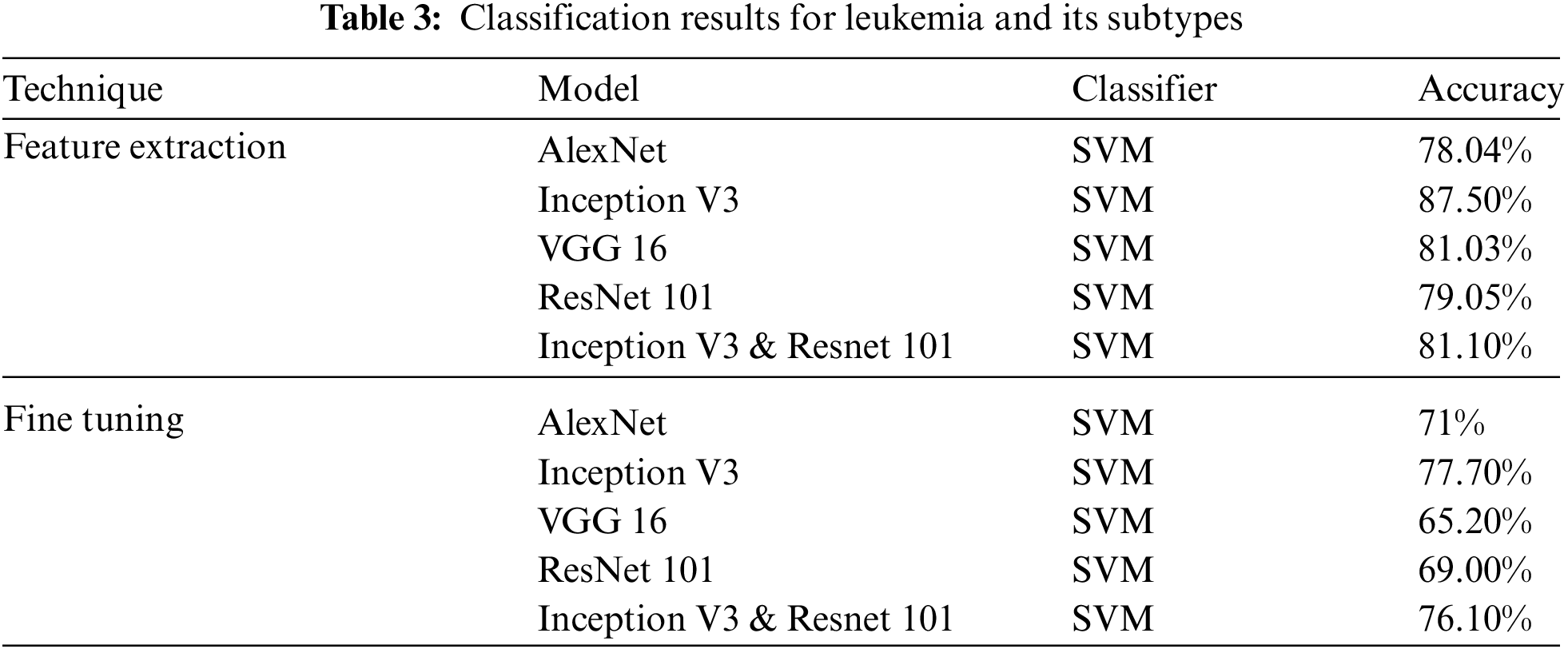
In the first experiment classified the Cellular Elements with fine tuning, and truncated the weights of last layer according to our problem with the same Network parameters discussed earlier. Mixed architecture had 825 layers, we froze 822 and used the last three layers using the same Network Parameters. The classification was carried out using Convolution Neural Networks, we were able to obtain the best results of classification of cellular elements with inception V3. Classification Results & Evaluations using transfer learning are summarized in Tab. 3. Alex Net & VGG 16 have three fully connected layers ALL. FC 6, FC 7 & FC 8 & we employed these for feature extraction.
All classifiers reported a higher result for feature extraction on FC-6 layer. This shows that CNNs were able to classify more accurately over the machine learning classifiers i.e., Random Forest or SVM. It is also worth mentioning that this is the first time these experiments are carried out on Five subtypes of leukemia altogether with a large dataset. Comparing the Five trained networks, it can be seen that Inception V3 out performs every other network. Tab. 4 shows comparison of different classifiers outcome.
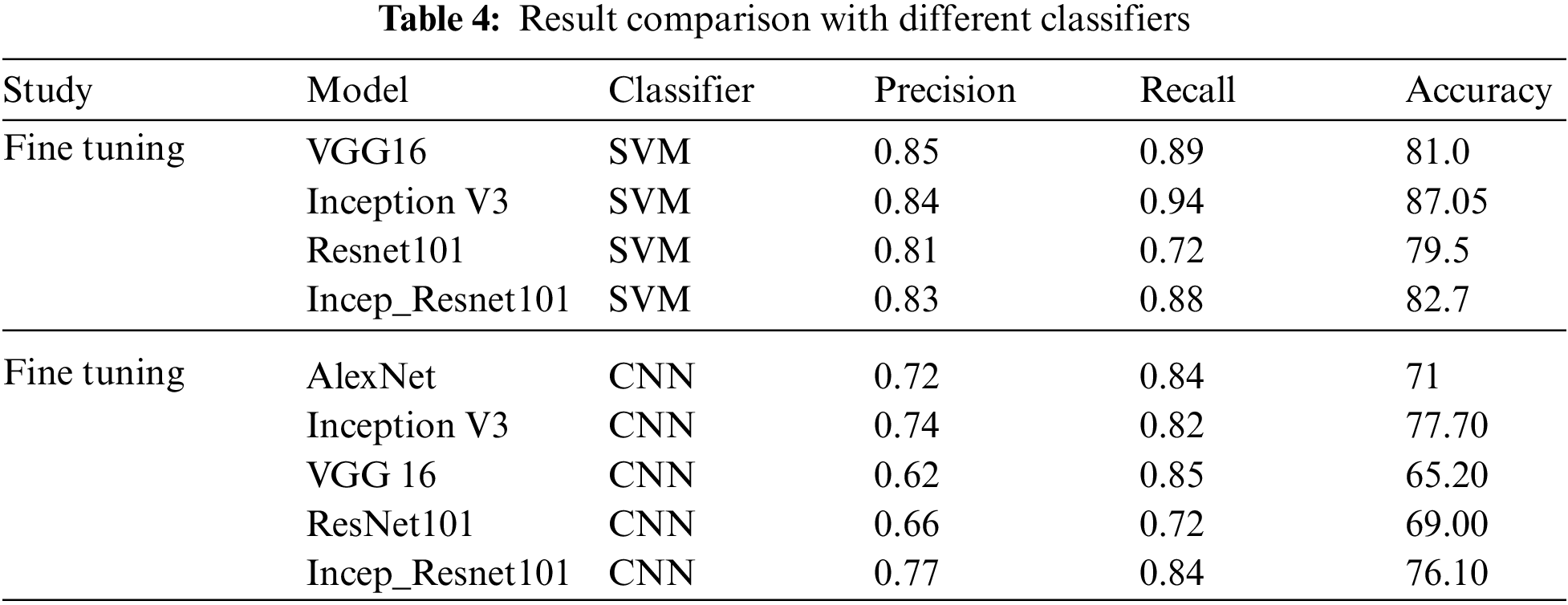
The second experimental scenario was to Classify the medical images for leukemia and its subtypes, System was trained on a total of 2352 images and the features used for classification purposes was Nucleus and cellula. Comparing the Pre trained networks, it can be seen that Inception V3 outperforms other networks in all the scenarios. Classification process was carried out using SVM for Feature Extraction because it was able to obtain the highest results as per old studies. The criteria for evaluating the models to find out overall percentage of correct classifications is done by Precision, Recall. Regarding precision, the results were promising. We showed how a system actually trained for common image data can be applied to classify Leukemia and its subtypes. The system gave a promising classification accuracy of 81.57% for feature extraction and 71.8% for fine tuning.
4.3 Comparison of Results with Notable Studies
This research presented detection for acute leukemia and its subtypes using bone marrow cells. The classification process was supported by extricating the descriptive features from nucleus and cellula, they classified on the basis of descriptive features such as shape geometry, eigen values, and machine learning classifiers (random forest, SVM, RC, IBk, SL & SMO, were used to classify the cellular elements, and their results inferred that use of descriptive features gives better results than from its cellular elements, The classification process was carried out after cell segmentation techniques, They used Wolds composition method for segmentation of the cells. They also came up with a diagnostic algorithm that diagnosed leukemia and its subtypes. They achieved an accuracy of 0.84 for the lymphoblastic sub-categories, and 0.92 for the myeloblastic sub-categories. But the catch was they have used two class or three class problems like L1 vs. L2 they got a precision of 81% by using Simple Logistic classifier, and for three class problem like M2 vs. M3 vs. M5 they got an accuracy of 78% by using Random committee and random forest as classifiers. They evaluated their classifiers by presenting TPR and TNR. Recall was missing from their results altogether. Our work was less time consuming and it classified five classes simultaneously instead of classifying two or three classes. It can be derived the CNN's are more successful in classifying problems more efficiently than just machine learning algorithms [14].
Despite of technology innovation, microscopic analysis of blood-sample even now continues as standard and hence cost-effective method for leukemia diagnosis. However it's really not an adequate solution to efficiently be the only factor deciding the diagnosis of leukemia. Also, less advances have been made to categorize the subtypes of Leukemia as it is equally important to categorize the Leukemia in different types, in order to do recommend an exclusive treatment. All these approaches of manually identifying the disease depends on the expertise of hematologist. So, there should be some efficient and robust automated system for classification of leukemia through which accuracy of classification is increased hence the diagnosis scores can be substantially improved without human involvement. Moreover, computer-based systems as compare to human diagnosis, can improve the accuracy and quick diagnosis. Quick diagnosis may lead to better treatment and early recovery from the fatal disease. This research endeavor can also be a great hope for the rural areas, where medical experts are not available. A lot of acute Leukemia cases are discovered in under-privileged people, who have a remote access to hospitals, which allows the disease to progress where it is hard to diagnose. The main purpose of our work is that we will be capable of delivering a method for detection of Leukemia earlier and with reliable precision, our research will focus on classification of Leukemia and its subtypes. This method will provide pathologists with a second opinion which will lead to a lesser chance of error and a quick tool to determine the type of disease, which will be helpful for suggesting a proper diagnostic, as before the treatment dangled on the experience of the pathologist and had a greater chance of error rate of 30%–40%, Which lead to the cause of high morality.
This work can be further enhanced by increasing the training and testing data. Although, data acquisition of medical data is a tedious task. The proposed method can be applied to other types of cancer detection as well.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. I. Vincent, K. R. Kwon, S. H. Lee and K. S. Moon, “Acute lymphoid leukemia classification using two-step neural network classifier,” in Proc. FCV, Mokpo, Korea, pp. 1–4, 2015. [Google Scholar]
2. N. Ahmed, A. Yigit, Z. Isik and A. Alpkocak, “Identification of leukemia subtypes from microscopic images using convolutional neural network,” Diagnostics, vol. 9, no. 3, pp. 1–11, 2019. [Google Scholar]
3. G. Singh, G. Bathla and S. Kaur, “A review to detect leukemia cancer in medical images,” in Proc. ICCCA, Greater Noida, India, pp. 1043–1047, 2016. [Google Scholar]
4. A. Harjoko, T. Ratnaningsih, E. Suryani, S. Palgunadi and N. P. T. Prakisya, “Classification of acute myeloid leukemia subtypes M1, M2 and M3 using active contour without edge segmentation and momentum backpropagation artificial neural network,” in Proc. MATEC Web of Conf., Yogyakarta, Indonesia, pp. 01041–01048, 2018. [Google Scholar]
5. H. N. Lim, E. U. Francis, M. Y. Mashor and R. Hassan, “Classification of bone marrow acute leukemia cells using multilayer perceptron network,” in Proc. ICED, Phuket, Thailand, pp. 486–490, 2016. [Google Scholar]
6. P. Jagadev and H. G. Virani, “Detection of leukemia and its types using image processing and machine learning,” in Proc. ICEI, Tirunelveli, India, pp. 522–526, 2017. [Google Scholar]
7. M. Fatma and J. Sharma, “Identification and classification of acute leukemia using neural network,” in Proc. MedCom, Greater Noida, India, pp. 142–145, 2014. [Google Scholar]
8. T. T. P. Thanh, C. Vununu, S. Atoev, S. H. Lee and K. R. Kwon, “Leukemia blood cell image classification using convolutional neural network,” International Journal of Computer Theory and Engineering, vol. 10, no. 2, pp. 54–58, 2018. [Google Scholar]
9. W. Ismail, R. Hassan, A. Payne and S. Swift, “The detection and classification of blast cell in leukaemia acute promyelocytic leukaemia (AML M3) blood using simulated annealing and neural networks,” in Proc. AIME, Bled, Slovenia, pp. 1–10, 2011. [Google Scholar]
10. T. TTP, G. N. Pham, J. H. Park, K. S. Moon, S. H. Lee et al., “Acute leukemia classification using convolution neural network in clinical decision support system,” CS & IT Conference Proceedings, vol. 7, no. 13, pp. 1–5, 2017. [Google Scholar]
11. P. S. Kumar and S. Vasuki, “Automated diagnosis of acute lymphocytic leukemia and acute myeloid leukemia using multi-SV,” Journal of Biomedical Imaging and Bioengineering, vol. 1, no. 1, pp. 1–5, 2017. [Google Scholar]
12. M. Ghaderzadeh, F. Asadi, A. Hosseini, D. Bashash, H. Abolghasemi and A. Roshanpour, “Machine learning in detection and classification of leukemia using smear blood images: A systematic review,” Scientific Programming, vol. 2021, Article ID 9933481, 2021. [Google Scholar]
13. H. J. Escalante, M. Montes-y-Gómez, J. A. González, P. Gómez-Gil, L. Altamirano et al., “Acute leukemia classification by ensemble particle swarm model selection,” Artificial Intelligence in Medicine, vol. 55, no. 3, pp. 163–175, 2012. [Google Scholar]
14. C. Reta, L. Altamirano, J. A. Gonzalez, R. Diaz-Hernandez, H. Peregrina et al., “Segmentation and classification of bone marrow cells images using contextual information for medical diagnosis of acute leukemias,” PLoS One, vol. 10, no. 6, pp. e0130805, 2015. [Google Scholar]
15. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” in Proc. ICLR, San Diego, CA, USA, pp. 1–14, 2015. [Google Scholar]
16. M. I. Khalil, S. Tehsin, M. Humayun, N. Z. Jhanjhi and M. A. AlZain, “Multi-scale network for thoracic organs segmentation,” Computers Materials & Continua, vol. 70, no. 2, pp. 3251–3265, 2022. [Google Scholar]
17. M. I. Khalil, M. Humayun, N. Z. Jhanjhi, M. N. Talib and T. A. Tabbakh, “Multi-class segmentation of organ at risk from abdominal CT images: A deep learning approach,” in Proc. ICTIDS, Malaysia, pp. 425–434, 2021. [Google Scholar]
18. N. Tajbakhsh, J. Y. Shin, S. R. Gurudu, R. T. Hurst, C. B. Kendall et al., “Convolutional neural networks for medical image analysis: Full training or fine tuning?,” IEEE Transactions on Medical Imaging, vol. 35, no. 5, pp. 1299–1312, 2016. [Google Scholar]
19. A. Kumar, J. Kim, D. Lyndon, M. Fulham and D. Feng, “An ensemble of fine-tuned convolutional neural networks for medical image classification,” IEEE Journal of Biomedical and Health Informatics, vol. 21, no. 1, pp. 31–40, 2017. [Google Scholar]
20. M. Humayun and A. Alsayat, “Prediction model for coronavirus pandemic using deep learning,” Computer Systems Science and Engineering, vol. 40, no. 3, pp. 947–961, 2022. [Google Scholar]
21. L. Gaur, U. Bhatia, N. Z. Jhanjhi, G. Muhammad and M. Masud, “Medical image-based detection of COVID-19 using deep convolution neural networks,” Multimedia Systems, vol. 20, no. 1, pp. 1–10, 2021. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |