

 | Molecular & Cellular Biomechanics |  |
DOI: 10.32604/mcb.2022.018958
ARTICLE
Nuclear Stress-Strain State over Micropillars: A Mechanical In silico Study
1LJAD, UMR CNRS 7351, Université Côte d’Azur, Nice, 06300, France
2Université Paris-Saclay, CentraleSupélec, UMR CNRS 8579, Laboratoire de Mécanique des Sols, Structures et Matériaux, Gif-sur-Yvette, 91190, France
*Corresponding Author: Rachele Allena. Email: rachele.allena@unice.fr
Received: 26 August 2021; Accepted: 20 October 2021
Abstract: Cells adapt to their environment and stimuli of different origin. During confined migration through sub-cellular and sub-nuclear pores, they can undergo large strains and the nucleus, the most voluminous and the stiffest organelle, plays a critical role. Recently, patterned microfluidic devices have been employed to analyze the cell mechanical behavior and the nucleus self-deformations. In this paper, we present an in silico model to simulate the interactions between the cell and the underneath microstructured substrate under the effect of the sole gravity. The model lays on mechanical features only and it has the potential to assess the contribution of the nuclear mechanics on the cell global behavior. The cell is constituted by the membrane, the cytosol, the lamina, and the nucleoplasm. Each organelle is described through a constitutive law defined by specific mechanical parameters, and it is composed of a fluid and a solid phase leading to a viscoelastic behavior. Our main objective is to evaluate the influence of such mechanical components on the nucleus behavior. We have quantified the stress and strain distributions in the nucleus, which could be responsible of specific phenomena such as the lamina rupture or the expression of stretch-sensitive proteins.
Keywords: Nuclear mechanics; micropillared substrate; in silico model
Cells continuously adapt themselves to their environment and stimuli they received (i.e., chemical, electrical, mechanical, …) [1–3]. More specifically, they are able to undergo large strains during confined migration through sub-cellular or sub-nuclear pores. During such a process, the nucleus, the most voluminous and the stiffest cellular organelle, plays a critical role [4,5]. Some cells such as cancerous cells can even undergo the rupture of the nuclear lamina to be able to migrate across healthy tissues [6,7]. Therefore, quantifying nucleus strains and stresses can be crucial to diagnose cancer and other pathologies in patients.
To do so, patterned microfluidic devices have been employed during the last few years in order to characterize the cell mechanical behaviour [8,9] and the nucleus self-deformations [10–13] and shape changes [14,15] induced by mechanical forces, which are due to the interaction between the cell and the topological surface. Assays on micropillared substrates involve successive steps: (i) contact between the cell and the pillars, (ii) adhesion of the cell on the pillars surface, (iii) cell spreading, (iv) cell polarization and (v) cell crawling.
A series of analytical and numerical models exist in the literature focusing on the interactions between the cell and a flat substrate. The former provides information on the spreading process with no excessive computational cost [16–19]. The latter, which may be discrete [20–22] or continuum [15,23–25], allows to investigate the intra-cellular rearrangement or to obtain quantitative results at the global or the local scale. In our previous paper [26], we have proposed an in silico two-dimensional (2D) model which simulates the first three steps (i.e., contact, adhesion and spreading) of the interaction between the cell and the micropillared substrate and provides insights on the mechanisms inducing nuclear deformation. We have been able to determine the role of the gravity and of the actin fibers above and beneath the nucleus responsible for a pushing and a pulling force, respectively.
In the present paper, we have adapted the model presented in [26] and we have focused on step one only (i.e., contact between the cell and the pillars). The cell and nucleus behaviours are described using specific mechanical tools (i.e., constitutive laws, mechanical properties, fluid and solid phase balance). By performing a sensibility study, our objective has been to determine the influence of these parameters on the interaction between the cell and underneath micropillared substrate. Then, the model provides the nucleus stress and strain fields over time and gives insights on the global cellular behavior that can be further explored experimentally.
In Section 2, we describe the mathematical framework of the model including the geometry (Section 2.1), the constitutive laws governing the behaviour of the cell and of its components (Section 2.2), the external forces applied to the cell (Sections 2.3 and 2.4) and the numerical implementation (Section 2.5). The results of the different simulations are presented in Section 3 and some conclusions and perspectives are drawn in Section 4.
Given the symmetry conditions, we consider here the cell
with
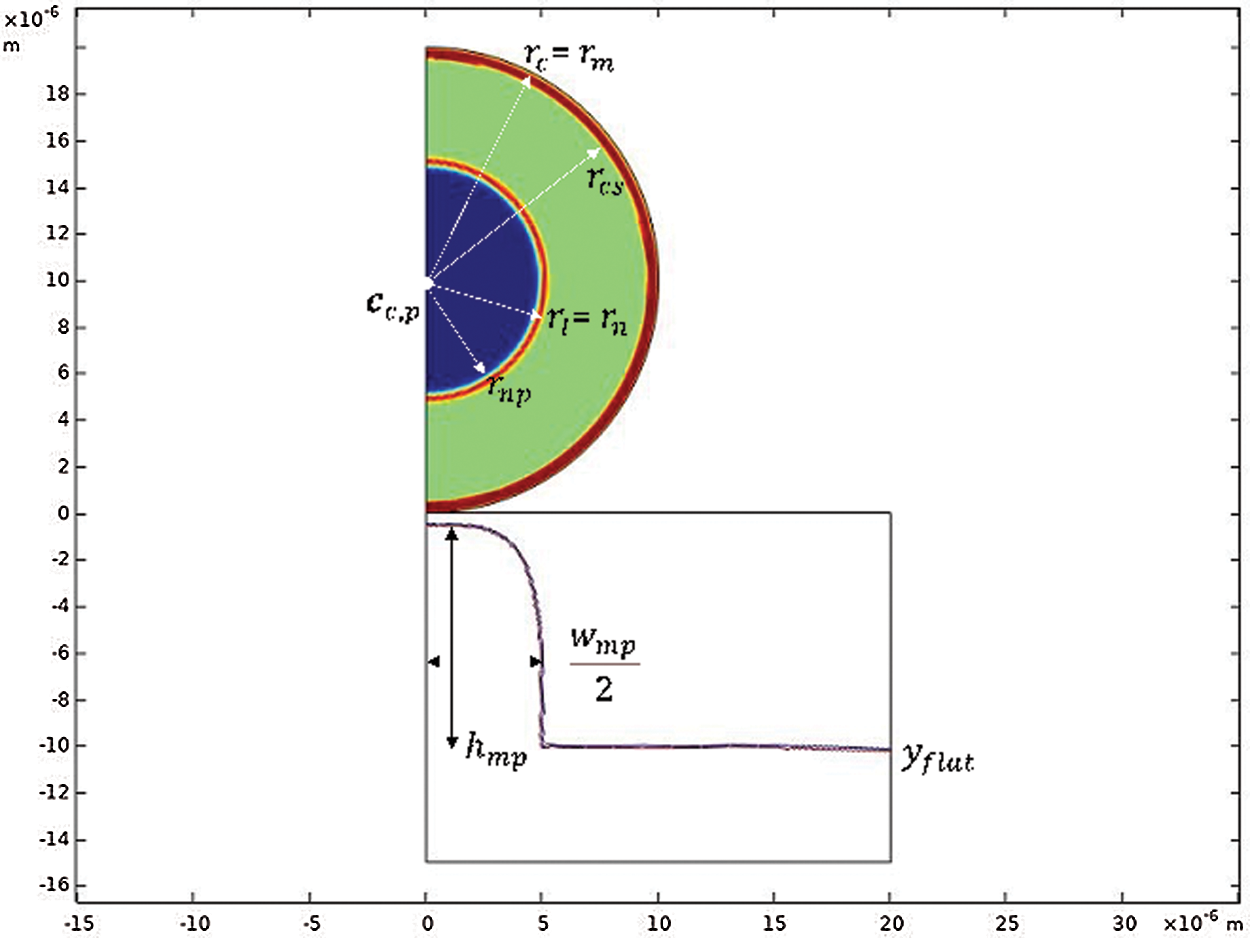
Figure 1: Cell and substrate geometry at t = 0 s. The cell has an external radius
Each cell component is composed of a solid and a fluid phase. To simplify the approach, we assume that both phases deform in parallel like in a Kelvin–Voigt model. Consequently, the overall stress S and the deformation F can be expressed as
where the subscripts s and f indicate the solid and the fluid phases, respectively, while the subscript i indicates a specific constitutive law for the solid phase, as described in the followings (Section 2.2.1), and
When describing the solid phase of an isotropic elastic material, for stability reasons, its strain energy must be poly-convex with respect to the three invariants
where
From a physical point of view,
1. A standard Saint-Venant material, which only depends on the first and second invariants
2. A Neo-Hookean compressible material, which depends on the first
3. The Mooney-Rivlin and Yeoh compressible materials which depend on the three invariants [25,35] and describe the deformations along lines, volumes and surfaces (i.e., the membrane and the lamina). Furthermore, the Yeoh material takes into account the successive softening and stiffening behavior of an elastic material made of fibers.
The second Piola Kirchhoff stress
– for the standard Saint-Venant material
with
– for the compressible neo-Hookean material
with
– for the Mooney-Rivlin material
with
– finally, for the Yeoh material
where
For the fluid phase of the cell, a Newtonian viscous fluid is considered but it must be defined in the Lagrangian configuration in order to ensure the compatibility with the solid phase. Thus, the Cauchy stress
where
with the superscript T indicating the transpose of a matrix.
Since
The cell is submitted to the gravity force
where
with t the time and
As the gravity is applied, the cell settles down and starts interacting with the underneath micropillared substrate, which is constituted by a micropillar and a flat region and it is defined by a characteristic function
where
Once the cell approaches the substrate, the contact force
where
The global equilibrium of the system in the initial configuration can be expressed as
with
In order to employ a classical finite element approach, we multiply each term of Eq. (30) by the kinematically admissible displacement test function w and we integrate over the cellular domain
where
Eq. (26) has been manually implemented using the weak form tool in COMSOL Multiphysics. The spatial discretization is obtained via quadratic polynomials for each isoparametric element of the mesh (mesh size between 0.3 µm and 1 µm). The time discretization is achieved via a second-order backward differentiation formula (BDF). The solution is computed using a nonlinear Newton scheme with a relative tolerance of 1% on the displacement error estimation.
The radii of the cell (
3.1 Cell Components Constitutive Law
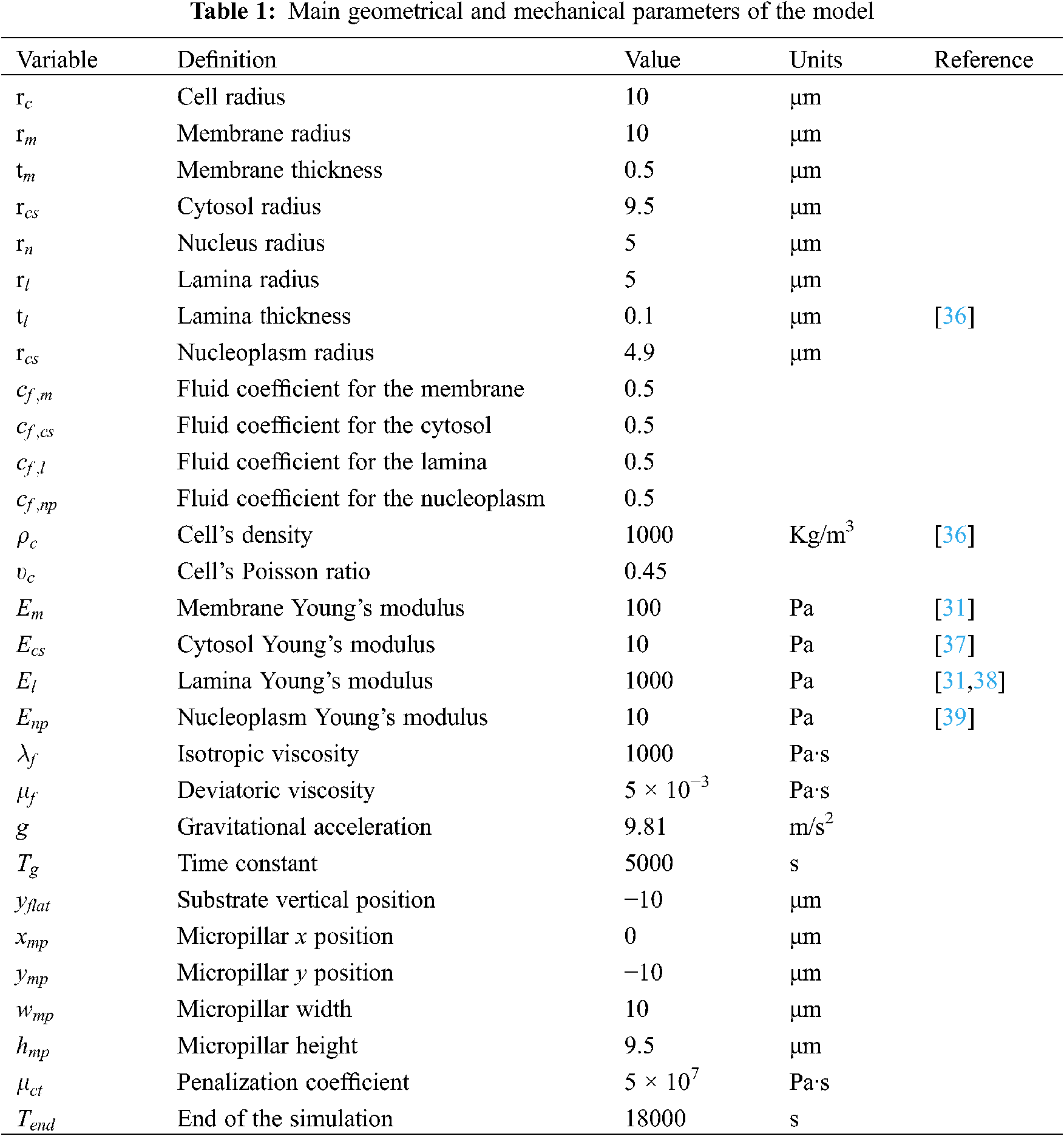
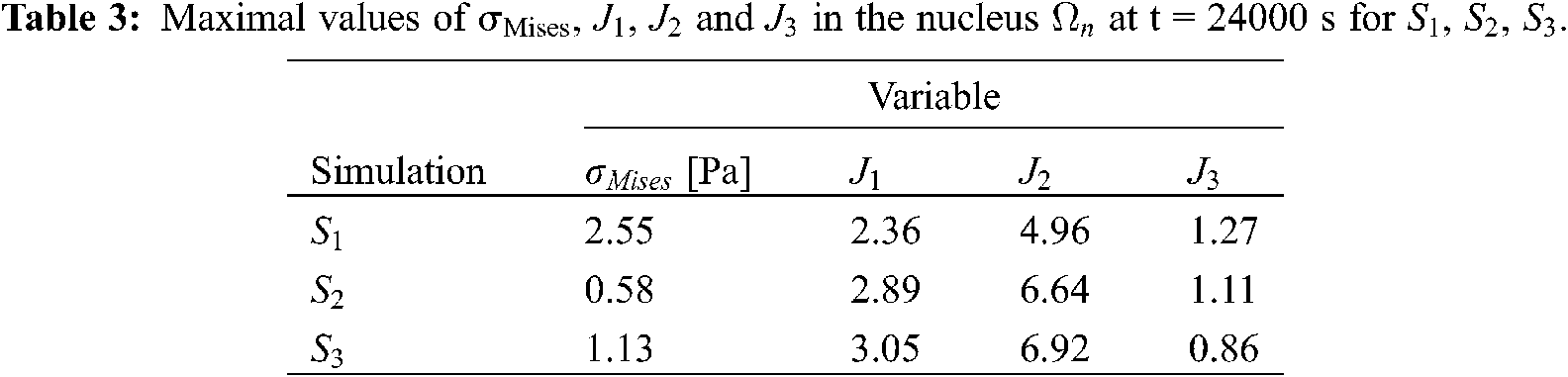
For the first series of simulations, we have tested different constitutive laws for the cell components. All the parameters defining each constitutive law are reported in Table 1. The fluid coefficients have been set equal to
The Saint-Venant material model is the simplest and mostly used in the literature, but it is not robust enough to describe very large deformations. The Neo-Hookean and the Mooney-Rivlin materials are efficient in considering large strains, but they do not exhibit typical stiffness-softening followed by hardening during deformation. Finally, the Yeoh model is theoretically the most consistent, but it is much more complex than the others due to a higher number of parameters to define (Eq. (21)).
The evolution as a function of time of the Mises stress
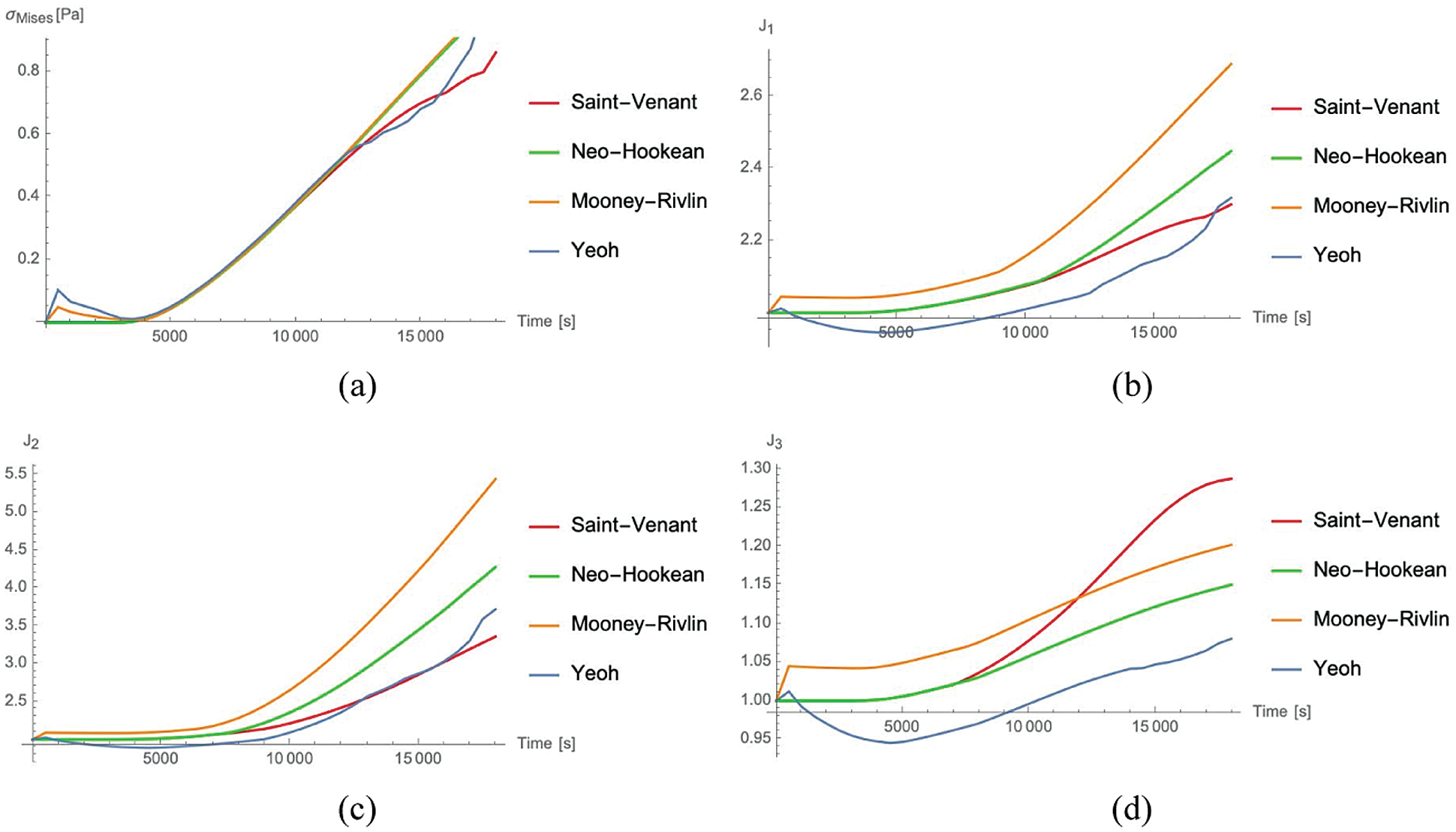
Figure 2: von Mises Cauchy stress
For the strains, the Mooney Rivlin material provides the maximum values of
We can estimate the contribution of the nucleus to the total cell deformation. In terms of fibers elongation (
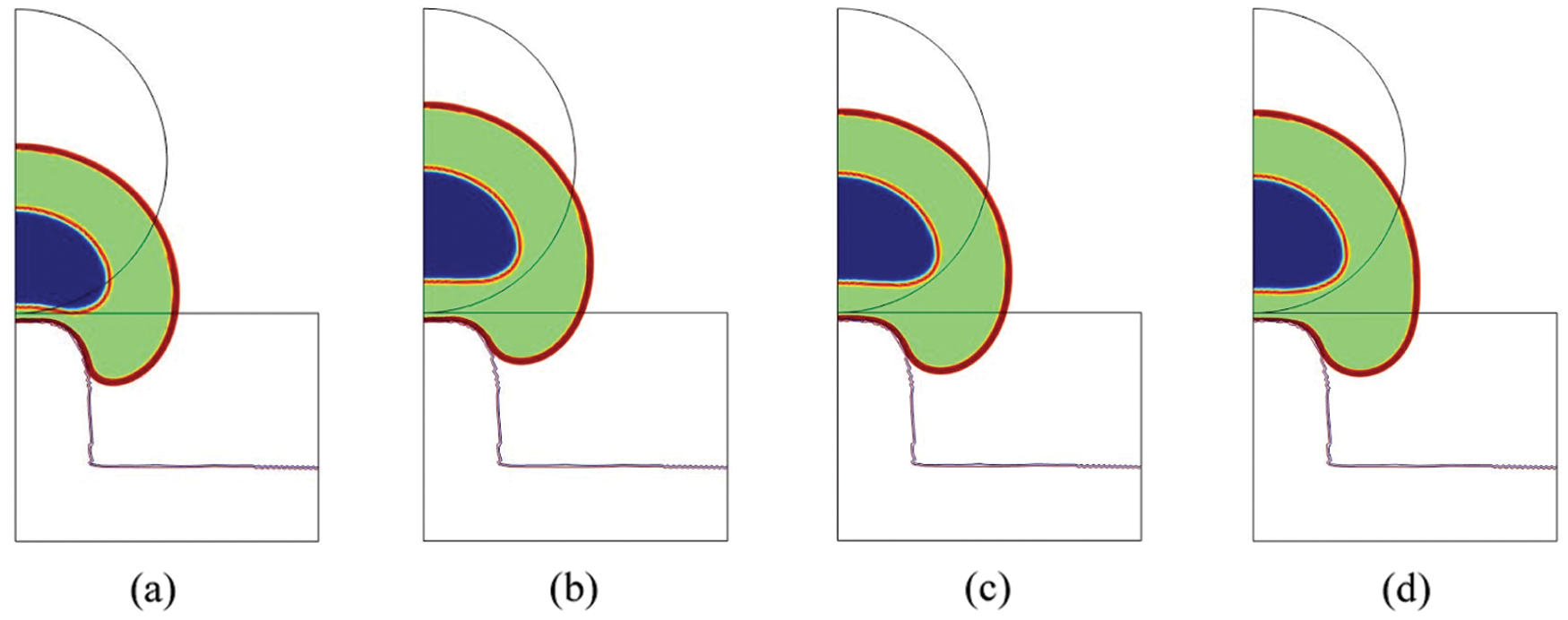
Figure 3: Total cell deformation at t = 18000 s for the Saint-Venant (a), the Neo-Hookean (b), the Mooney-Rivlin (c) and the Yeoh (d) material (blue = nucleoplasm, orange = lamina, green = cytosol, red = membrane)
Since the cell is composed by several organelles having different structures and geometries, there is a need for a constitutive macroscopic law able to describe the stresses associated to such heterogeneous strains. More specifically, taking into account these stresses implies low values of
3.2 Nucleus Fluid and Solid Phases
For the second series of simulations we have evaluated the influence of the fluid-solid phases of the lamina and of the nucleoplasm on the nucleus behaviour. Specifically, we have let vary
In Fig. 4, values of
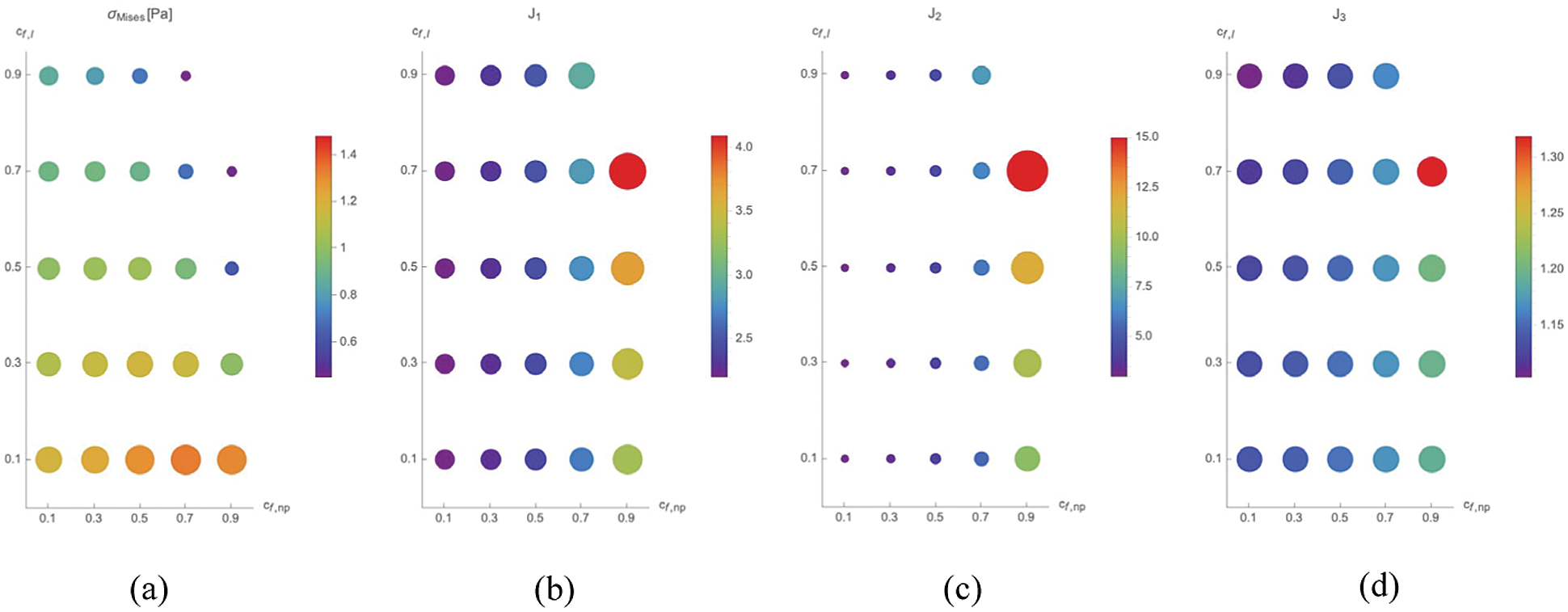
Figure 4: Maximal value of
On the one side, the maximal strains are found for
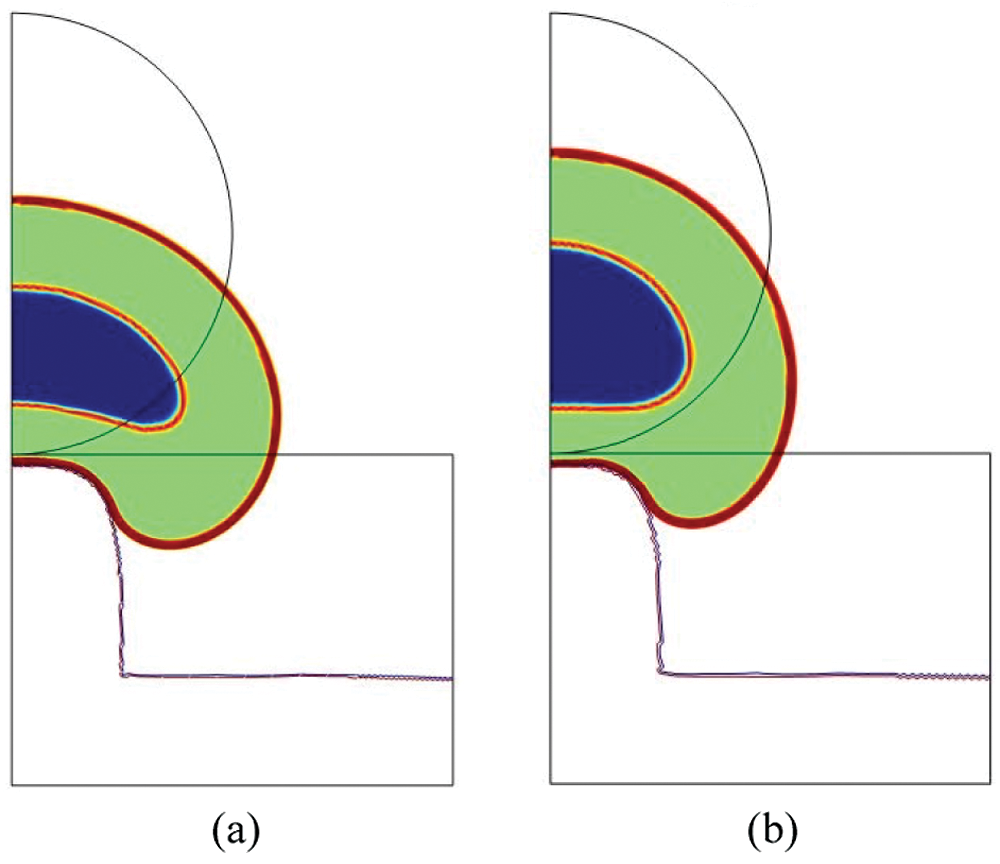
Figure 5: Total cell deformation at t = 18000 s for
On the other side, the lowest values of
3.3 Nucleus Mechanical Properties
For the last series of simulations, we have let vary the Young’s moduli of the nucleoplasm (
Fig. 6, we show the results in terms of Mises Cauchy stress
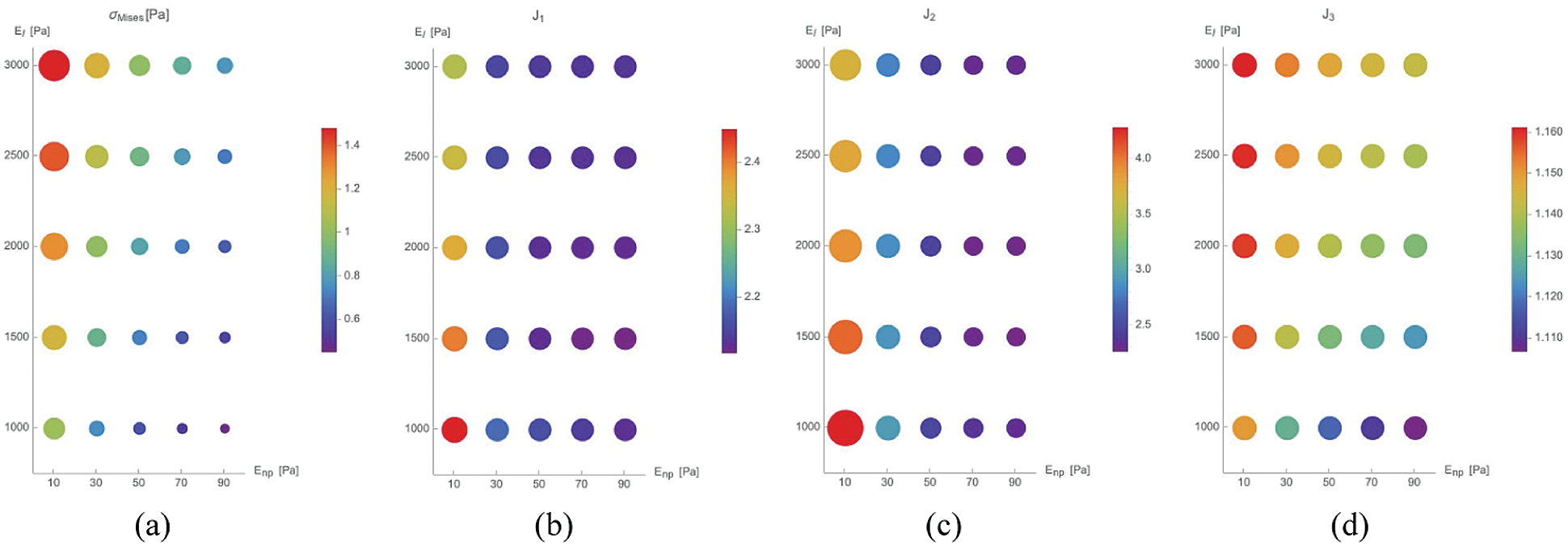
Figure 6: Maximal value of
3.4 Comparison with Experimental Data
In the previous series of simulations,
We have run three additional simulations (
From a quantitative point of view, the results in terms of maximal stress (
In Fig. 7, the total cell and nucleus deformation at t = 24000 s is shown. One can notice that for
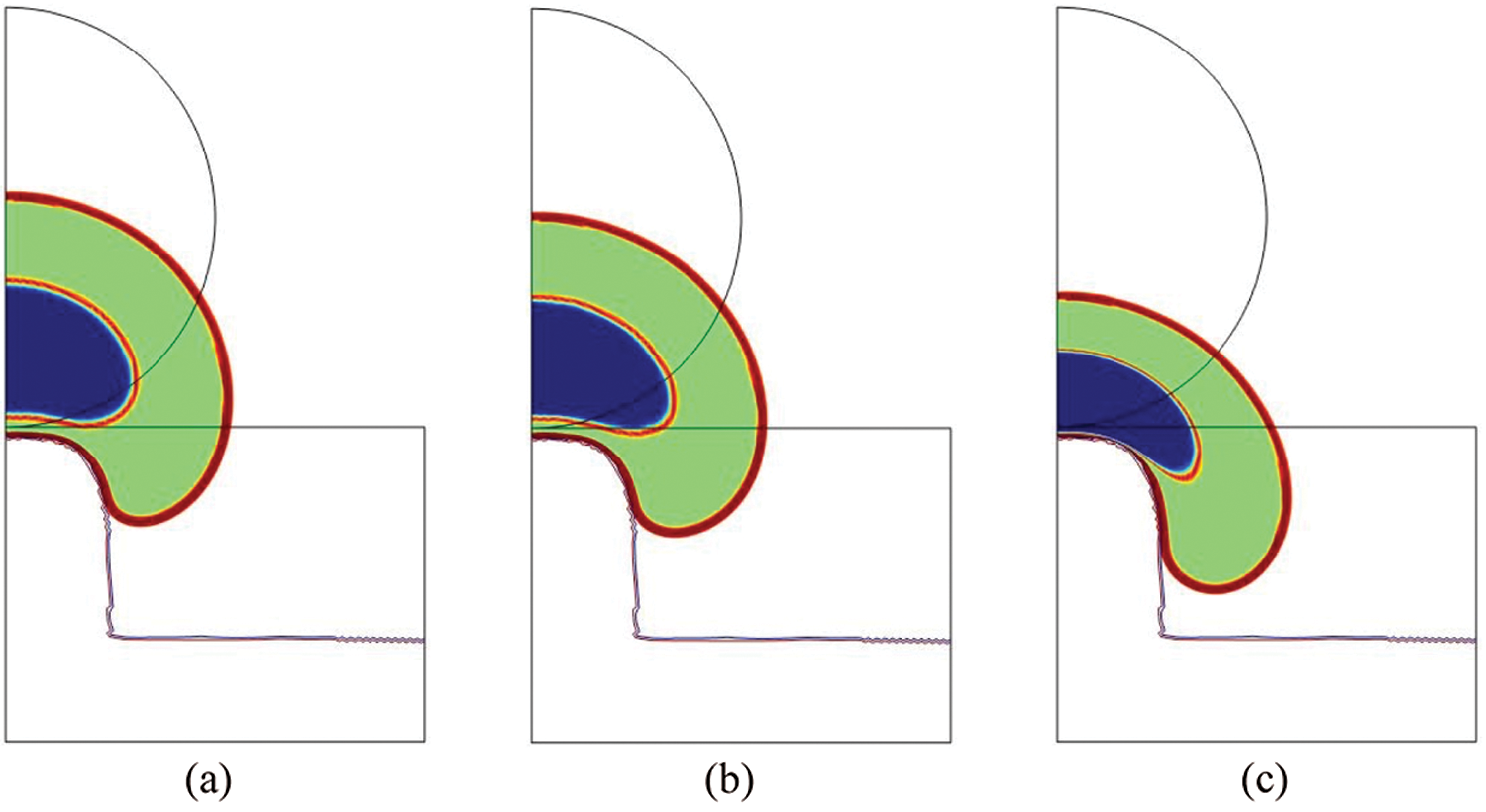
Figure 7: Total cell deformation at t = 24000 s for
In [12–14] a shape index (SI) parameter is used to quantify the nucleus self-deformation. SI is defined as follows:
where S and l are the nucleus surface and perimeter, respectively. SI is equal to 1 for a perfect circle (i.e., no deformation), whereas it is equal to 0 for a straight line. In [12–14], SI is measured in the plan perpendicular to the micropillars. This is not possible in our model since we are in 2D. Nonetheless, we believe that it is even more interesting to measure SI in the sagittal plan to quantify the nucleus self-deformation, but also its penetration. We found that SI is equal to 0.32, 0.26 and 0.21 for
In this paper we have presented a 2D computational model to investigate the interactions between the cell and the micropillared substrate. The cell is initially suspended and gently meets the pillar due to the gravity force. Then, the contact force between the cell and the pillar is applied over a very thin layer.
The model is equipped with specific mechanical tools, namely the constitutive laws describing the cell components, their mechanical properties, and the fluid and solid phase mixture for each component. Our main objective has been establishing a correlation between such elements and the nucleus stress-strain state. To do so, a thorough sensibility study has been performed. More specifically, three series of simulations have been run: (i) the first one involves different materials models to describe the cell behaviour, i.e., Saint-Venant, Neo-Hookean, Mooney-Rivlin and Yeoh materials, (ii) the second one focuses on the balance between the fluid and solid phases of the lamina and nucleoplasm, (iii) the third one takes into account the variation of the mechanical parameters (i.e., the Young modulus) of the lamina and the nucleoplasm.
Through the large spectrum of combinations that we have provided, biologists may identify the one corresponding to specific cellular phenotypes or behaviors. Additionally, our model could inspire further experimental investigations to explore the interactions between the cell and the micropillared substrate.
We have been able to quantify the stress and the strain in the nucleus
We have shown that the variation of the balance between the fluid and solid phase of the nucleus induces the maximum strains of the nucleus. In fact, we have found that varying
In terms of stress, even though the maximal values of the Mises Cauchy stress
Through some additional simulations, we have been able to compare our numerical results to the experimental observations on different cellular phenotypes. More specifically, we have shown that by increasing the simulation time, the nucleus undergoes significant self deformation and it acquires a ‘peanut’ shape by embracing the micropillar as it has been reported in [11–14]. Such a phenomenon is exacerbated when the nuclear lamina is ablated (i.e., reducing the Young modulus) and it confirms data according to which tumor cells are able to adjust their mechanical properties in order to invade healthy tissues [43].
To conclude, our model, which has been built only using mechanical tools, has drawn a large spectrum of scenarios to analyze and quantify the nucleus stress-strain state. It can help identifying the mechanical features responsible for specific nucleus responses and their impact on the global cell behavior. Although the interesting results, the present model could be improved in the different ways. First, a three-dimensional description of the system would allow to better evaluate the interactions between the cell and its surroundings and the nucleus role. Secondly, it could be interesting to quantify the pressure gradient in the cell in order to assess the proteins traffic flow to and from the nucleus. Finally, a precise description of the structure of the nuclear lamina could provide new information regarding its rupture and remodelling.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Mammoto, T., Ingber, D. E. (2010). Mechanical control of tissue and organ development. Development (Cambridge, England), 137(9), 1407–1420. DOI 10.1242/dev.024166. [Google Scholar] [CrossRef]
2. Versaevel, M., Grevesse, T., Gabriele, S. (2012). Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nature Communications, 3(1), 671. DOI 10.1038/ncomms1668. [Google Scholar] [CrossRef]
3. Swift, J., Ivanovska, I. L., Buxboim, A., Harada, T., Dingal, P. C. D. P. et al. (2013). Nuclear Lamin–A scales with tissue stiffness and enhances matrix-directed differentiation. Science, 341(6149), 1240104. DOI 10.1126/science.1240104. [Google Scholar] [CrossRef]
4. Friedl, P., Wolf, K., Lammerding, J. (2011). Nuclear mechanics during cell migration. Current Opinion in Cell Biology, 23(1), 55–64. DOI 10.1016/j.ceb.2010.10.015. [Google Scholar] [CrossRef]
5. Wolf, K., Te Lindert, M., Krause, M., Alexander, S., Te Riet, J. et al. (2013). Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. The Journal of Cell Biology, 201(7), 1069–1084. DOI 10.1083/jcb.201210152. [Google Scholar] [CrossRef]
6. Denais, C. M., Gilbert, R. M., Isermann, P., McGregor, A. L., Te Lindert, M.et al. (2016). Nuclear envelope rupture and repair during cancer cell migration. Science (New York, N.Y.), 352(6283), 353–358. DOI 10.1126/science.aad7297. [Google Scholar] [CrossRef]
7. Bell, E. S., Lammerding, J. (2016). Causes and consequences of nuclear envelope alterations in tumour progression. European Journal of Cell Biology, 95(11), 449–464. DOI 10.1016/j.ejcb.2016.06.007. [Google Scholar] [CrossRef]
8. Lu, H., Koo, L. Y., Wang, W. M., Lauffenburger, D. A., Griffith, L. G. et al. (2004). Microfluidic shear devices for quantitative analysis of cell adhesion. Analytical Chemistry, 76(18), 5257–5264. DOI 10.1021/ac049837t. [Google Scholar] [CrossRef]
9. Rosenbluth, M. J., Lam, W. A., Fletcher, D. A. (2008). Analyzing cell mechanics in hematologic diseases with microfluidic biophysical flow cytometry. Lab on a Chip, 8(7), 1062–1070. DOI 10.1039/b802931h. [Google Scholar] [CrossRef]
10. Davidson, P. M., Fromigué, O., Marie, P. J., Hasirci, V., Reiter, G. et al. (2010). Topographically induced self-deformation of the nuclei of cells: Dependence on cell type and proposed mechanisms. Journal of Materials Science. Materials in Medicine, 21(3), 939–946. DOI 10.1007/s10856-009-3950-7. [Google Scholar] [CrossRef]
11. Badique, F., Stamov, D. R., Davidson, P. M., Veuillet, M., Reiter, G. et al. (2013). Directing nuclear deformation on micropillared surfaces by substrate geometry and cytoskeleton organization. Biomaterials, 34(12), 2991–3001. DOI 10.1016/j.biomaterials.2013.01.018. [Google Scholar] [CrossRef]
12. Liu, X., Liu, R., Gu, Y., Ding, J. (2017). Nonmonotonic self-deformation of cell nuclei on topological surfaces with micropillar array. ACS Applied Materials & Interfaces, 9(22), 18521–18530. DOI 10.1021/acsami.7b04027. [Google Scholar] [CrossRef]
13. Liu, R., Yao, X., Liu, X., Ding, J. (2019). Proliferation of cells with severe nuclear deformation on a micropillar array. Langmuir, 35(1), 284–299. DOI 10.1021/acs.langmuir.8b03452. [Google Scholar] [CrossRef]
14. Pan, Z., Yan, C., Peng, R., Zhao, Y., He, Y. et al. (2012). Control of cell nucleus shapes via micropillar patterns. Biomaterials, 33(6), 1730–1735. DOI 10.1016/j.biomaterials.2011.11.023. [Google Scholar] [CrossRef]
15. Hanson, L., Zhao, W., Lou, H. Y., Lin, Z. C., Lee, S. W. et al. (2015). Vertical nanopillars for in situ probing of nuclear mechanics in adherent cells. Nature Nanotechnology, 10(6), 554–562. DOI 10.1038/nnano.2015.88. [Google Scholar] [CrossRef]
16. Cuvelier, D., Théry, M., Chu, Y. S., Dufour, S., Thiéry, J. P. et al. (2007). The universal dynamics of cell spreading. Current Biology, 17(8), 694–699. DOI 10.1016/j.cub.2007.02.058. [Google Scholar] [CrossRef]
17. Sarvestani, A. S., Jabbari, E. (2008). Modeling the kinetics of cell membrane spreading on substrates with ligand density gradient. Journal of Biomechanics, 41(4), 921–925. DOI 10.1016/j.jbiomech.2007.11.004. [Google Scholar] [CrossRef]
18. Nisenholz, N., Rajendran, K., Dang, Q., Chen, H., Kemkemer, R. et al. (2014). Active mechanics and dynamics of cell spreading on elastic substrates. Soft Matter, 10(37), 7234–7246. DOI 10.1039/C4SM00780H. [Google Scholar] [CrossRef]
19. Cao, X., Lin, Y., Driscoll, T. P., Franco-Barraza, J., Cukierman, E. et al. (2015). A chemomechanical model of matrix and nuclear rigidity regulation of focal adhesion size. Biophysical Journal, 109(9), 1807–1817. DOI 10.1016/j.bpj.2015.08.048. [Google Scholar] [CrossRef]
20. Ingber, D. E. (2003). Tensegrity I. Cell structure and hierarchical systems biology. Journal of Cell Science, 116(7), 1157–1173. DOI 10.1242/jcs.00359. [Google Scholar] [CrossRef]
21. Milan, J.-L., Lavenus, S., Pilet, P., Louarn, G., Wendling, S. et al. (2013). Computational model combined with in vitro experiments to analyse mechanotransduction during mesenchymal stem cell adhesion. European Cells & Materials, 25, 97–113. DOI 10.22203/eCM.v025a07. [Google Scholar] [CrossRef]
22. Fang, Y., Lai, K. W. C. (2016). Modeling the mechanics of cells in the cell-spreading process driven by traction forces. Physical Review E, 93(4), 042404. DOI 10.1103/PhysRevE.93.042404. [Google Scholar] [CrossRef]
23. Etienne, J., Duperray, A. (2011). Initial dynamics of cell spreading are governed by dissipation in the actin cortex. Biophysical Journal, 101(3), 611–621. DOI 10.1016/j.bpj.2011.06.030. [Google Scholar] [CrossRef]
24. Golestaneh, A. F., Nadler, B. (2016). Modeling of cell adhesion and deformation mediated by receptor-ligand interactions. Biomechanics and Modeling in Mechanobiology, 15(2), 371–387. DOI 10.1007/s10237-015-0694-9. [Google Scholar] [CrossRef]
25. Zeng, X., Li, S. (2011). Multiscale modeling and simulation of soft adhesion and contact of stem cells. Journal of the Mechanical Behavior of Biomedical Materials, 4(2), 180–189. DOI 10.1016/j.jmbbm.2010.06.002. [Google Scholar] [CrossRef]
26. Mondésert-Deveraux, S., Aubry, D., Allena, R. (2019). In silico approach to quantify nucleus self-deformation on micropillared substrates. Biomechanics and Modeling in Mechanobiology, 18(5), 1281–1295. DOI 10.1007/s10237-019-01144-2. [Google Scholar] [CrossRef]
27. Itskov, M., Aksel, N. (2004). A class of orthotropic and transversely isotropic hyperelastic constitutive models based on a polyconvex strain energy function. International Journal of Solids and Structures, 41(14), 3833–3848. DOI 10.1016/j.ijsolstr.2004.02.027. [Google Scholar] [CrossRef]
28. Bonet, J., Gil, A. J., Ortigosa, R. (2015). A computational framework for polyconvex large strain elasticity. Computer Methods in Applied Mechanics and Engineering, 283, 1061–1094. DOI 10.1016/j.cma.2014.10.002. [Google Scholar] [CrossRef]
29. Holzapfel, G. A. (2000). Nonlinear solid mechanics: A continuum approach for engineering. (1st ed.) Hoboken: Wiley. [Google Scholar]
30. Bonet, J., Wood, R. D. (2008). Nonlinear continuum mechanics for finite element analysis. (2nd ed.) Cambridge: Cambridge University Press. [Google Scholar]
31. Allena, R., Aubry, D. (2012). Run-and-tumble or look-and-run? A mechanical model to explore the behavior of a migrating amoeboid cell. Journal of Theoretical Biology, 306, 15–31. DOI 10.1016/j.jtbi.2012.03.041. [Google Scholar] [CrossRef]
32. Fan, H., Li, S. (2015). Modeling universal dynamics of cell spreading on elastic substrates. Biomechanics and Modeling in Mechanobiology, 14(6), 1265–1280. DOI 10.1007/s10237-015-0673-1. [Google Scholar] [CrossRef]
33. Jean, R. P., Chen, C. S., Spector, A. A. (2003). Analysis of the deformation of the nucleus as a result of alterations of the cell adhesion area. ASME 2003 International Mechanical Engineering Congress and Exposition, pp. 121–122. Washington, DC, USA. [Google Scholar]
34. Mokbel, M., Mokbel, D., Mietke, A., Träber, N., Girardo, S. et al. (2017). Numerical simulation of real-time deformability cytometry to extract cell mechanical properties. ACS Biomaterials Science & Engineering, 3(11), 2962–2973. DOI 10.1021/acsbiomaterials.6b00558. [Google Scholar] [CrossRef]
35. Wang, H., Biao, Y., Chunlai, Y., Wen, L. (2017). Simulation of AFM indentation of soft biomaterials with hyperelasticity. IEEE 12th International Conference on Nano/Micro Engineered and Molecular Systems, pp. 550–553. Los Angeles, CA, USA. [Google Scholar]
36. Righolt, C. H., Raz, V., Vermolen, B. J., Dirks, R. W., Tanke, H. J. et al. (2011). Molecular image analysis: Quantitative description and classification of the nuclear lamina in human mesenchymal stem cells. International Journal of Molecular Imaging, 2011(8), 1–11. DOI 10.1155/2011/723283. [Google Scholar] [CrossRef]
37. Crick, F. H. C., Hughes, A. F. W. (1950). The physical properties of cytoplasm. Experimental Cell Research, 1(1), 37–80. DOI 10.1016/0014-4827(50)90048-6. [Google Scholar] [CrossRef]
38. Caille, N., Thoumine, O., Tardy, Y., Meister, J. J. (2002). Contribution of the nucleus to the mechanical properties of endothelial cells. Journal of Biomechanics, 35(2), 177–187. DOI 10.1016/S0021-9290(01)00201-9. [Google Scholar] [CrossRef]
39. Vaziri, A., Lee, H., Mofrad, M. R. K. (2006). Deformation of the cell nucleus under indentation: Mechanics and mechanisms. Journal of Materials Research, 21(8), 2126–2135. DOI 10.1557/jmr.2006.0262. [Google Scholar] [CrossRef]
40. Lomakin, A. J., Cattin, C. J., Cuvelier, D., Alraies, Z., Molina, M. et al. (2020). The nucleus acts as a ruler tailoring cell responses to spatial constraints. Science, 370(6514), eaba2894. DOI 10.1126/science.aba2894. [Google Scholar] [CrossRef]
41. Enyedi, B., Jelcic, M., Niethammer, P. (2016). The cell nucleus serves as a mechanotransducer of tissue damage-induced inflammation. Cell, 165(5), 1160–1170. DOI 10.1016/j.cell.2016.04.016. [Google Scholar] [CrossRef]
42. Venturini, V., Pezzano, F., Castro, F. C., Häkkinen, H. M., Jiménez-Delgado, S. et al. (2020). The nucleus measures shape changes for cellular proprioception to control dynamic cell behavior. Science, 370(6514), eaba2644. DOI 10.1126/science.aba2644. [Google Scholar] [CrossRef]
43. Roberts, A. B., Zhang, J., Raj Singh, V., Nikolić, M., Moeendarbary, E. et al. (2021). Tumor cell nuclei soften during transendothelial migration. Journal of Biomechanics, 121(5), 110400. DOI 10.1016/j.jbiomech.2021.110400. [Google Scholar] [CrossRef]
44. Hatch, E. M. (2018). Nuclear envelope rupture: Little holes, big openings. Current Opinion in Cell Biology, 52, 66–72. DOI 10.1016/j.ceb.2018.02.001. [Google Scholar] [CrossRef]
45. Vargas, J. D., Hatch, E. M., Anderson, D. J., Hetzer, M. W. (2012). Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus, 3(1), 88–100. DOI 10.4161/nucl.18954. [Google Scholar] [CrossRef]
46. de Vos, W. H., Houben, F., Kamps, M., Malhas, A., Verheyen, F. et al. (2011). Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Human Molecular Genetics, 20(21), 4175–4186. DOI 10.1093/hmg/ddr344. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |