
 | Oncologie |  |
DOI: 10.32604/Oncologie.2021.015906
ARTICLE
TUG1 Indicate Unfavorable Prognosis of Gastric Cancer for Promoting Proliferation, Migration and Multidrug Resistance
1The Second Affiliated Hospital of Nanchang University, Jiangxi Key Laboratory of Clinical and Translational Cancer Research, Nanchang, 330006, China
2The Affiliated Hospital of Jiangxi University of Traditional Chinese Medicine, Nanchang, 330006, China
3The First People’s Hospital of Xiangyang, Xiangyang, 441000, China
*Corresponding Author: Anwen Liu. Email: awliu666@163.com
Received: 22 January 2021; Accepted: 25 February 2021
#First author
Abstract: Gastric cancer (GC) is the most common digestive system malignant tumor and second most common cause of cancer-related death. Even for the early gastric cancer, after radical operation, perioperative and postoperative chemotherapy the recurrence and metastasis are also high. 5-year overall survival of all GC patients is only 10–15%. Chemo-resistance still poses a major obstacle to successful treatment of GC. The aberrant expression of TUG1 (Taurine Upregulated Gene 1) was closely related to chemo-resistance and metastasis in many cancers. Here we over-expressed TUG1 in GC cell line SGC-7901 and MGC-803. We compared the migration ability and sensitivity of cisplatin (CDDP), doxorubicin(ADM), 5-fluorocrail(5-Fu) on SGC-7901, SGC-7901/TUG1 and MGC-803, MGC-803/TUG1. The TUG1 induced the multi-drugs resistant and promoted the mobility of GS cell. Furthermore, we investigate the clinical significance of TUG1 and evaluate its prognostic value in patients with GC. The expression of TUG1 was positive in GC tissues and was significantly correlated with tumor size and lymphatic metastasis. Our findings demonstrate the TUG1 may function as a protooncogene and potential biomarker of GC.
Keywords: Gastric cancer; TUG1; biomarker; multiple-drug resistance
For the aging population of lifestyle, the incidence of tumor increased year by year in China. Although the environmental risk factors and life style [1] changing affected the tendence of tumor morbidity, the gastric cancer still was the fourth most frequently diagnosed cancer and third leading cause of cancer deaths worldwide [2,3], almost 4.1 million people were diagnosed and approximately 3 million were cancer-related deaths annually. Gastric cancer is a relatively sensitive to chemotherapy [4,5]. The standard first curative treatment in early GC was radical surgery. However, 50% of advanced GC patients were unresectable at first diagnosed and benefited from chemotherapy, especially in combination chemotherapy. Multiple-drug resistance (MDR), is the bottleneck in the treatment of many cancers [6]. Only about 1 percent of genome code for proteins. Non-coding RNAs demonstrated their significant role in maintaining genomic stability including long and small noncoding RNA. Long non-coding RNAs (lncRNAs) are a vast family of long (>200nt) noncoding RNAs, taking part in diverse biological functions like chromatin modifications, post-transcriptional regulation, imprinting, etc. [7]. So more and more aberrantly expressed lncRNAs had been confirmed in a broad spectrum of cancers. TUG1 over-expressed in many tumor such as lung cancer [8], ovarian cancer [9], pancreatic cancer [10] and so on [11]. It is a multifunctional gene and diverses many biological processes: proliferation [12], apoptosis [13], EMT [14] for examples. Jiang reported that TUG1 regulated the chemo-sensitivity in esophageal squamous cell carcinoma [15]. How about the role of TUG1 in chemo-resistance of GC remained uncertain. Here we analyzed the expression of TUG1 in GC tissue by ISH and over-expressed the TUG1 in SGC-7901 and MGC-803 to explore its function.
The human GC cell lines SGC-7901 and MGC-803 were purchased from the Chinese Academy of Sciences Cell Bank, while SGC-7901/DDP and BGC-823/5-Fu were purchased from Fenghui Biotechnology Co., Ltd., China, and seeded in DMEM supplemented with 10% fetal bovine serum (FBS) and 100 units of penicillin/streptomycin (Invitrogen, San Diego, CA, USA) at 37°C in a humidified atmosphere with 5% CO2.
2.2 Over-Expressed TUG1 by Lentiviral Vector and Transfection
According to the UCSC database, we designed the primer to amplified the cDNA by PCR and recombinated into lentiviral vector pLv-GFP. The HEK293T cells was 80% in the10 cm culture dish for transient transfection with 2 μg packaging lentiviral vectors and 10 μg pLv-TUG1. 2 days later, viruses were gathered from the supernatants. After incubated with various concentrations of lentivirus for 24 h, the tumor cells were transformed to fresh culture medium DMEM with 10% FBS in it. Continuing culturing the cell for 24 h, the transfection efficiency was detected by Flow Cytometric Assay. The GFP was the positive marker.
2.3 Quantitative Real-Time PCR Assay
Total mRNA was extracted from the tumor cell using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The extracted RNA was reverse transcribed into cDNA using the SYBR Premix Ex Taq Kit (Takara Bio, Inc., Japan) according to the manual. The primers specific for TUG1 were forward, 5’-GCCATG AAG CCC TTT GAG-3’ and reverse, 5’-CGG AAG AGT TCA AGG TGT TG-3’. The primers of GAPDH were forward, 5’-GAAGGTGAAGGTCGGAGT-3’ and reverse, 5’-GAAGATGGTGATGGGATTTC-3’. The qRT-PCR protocol was as follows: 5 min at 95°C followed by 40 cycles at 95°C for 10 s and at 60°C for 45 s. TUG1 expression data were normalized to GAPDH expression from the same sample. The Ct value of each target gene was normalized against the Ct value of the reference gene (Ct (TUG1)-Ct (GAPDH)). Relative expression levels were determined using the 2−ΔΔCt method.
The SGC-7901, SGC-7901/TUG1 and MGC-803, MGC-803/TUG1 cells were placed in 96-well plates (4 × 103 cells/well) with different concentration of CDDP, ADM, 5-Fu. The chemo-sensitivity of CDDP, ADM, 5-Fu on both SGC-7901, SGC-7901/TUG1 and MGC-803, MGC-803/TUG1 cells was monitored using Cell Counting Kit-8 assay (CCK8, Dojindo Molecular Technologies, Inc., Kunamoto, Japan). After adding chemo-drugs for 24 h, the CCK8 solution was used to assess to measure cell viability according to the absorbance of each well at 450 nm. All experiments were performed in triplicate.
2.5 Scratch Wound Healing Assay
The logarithmic phase SGC-7901, MGC-803 cells and its pLv-TUG1 subtype cells were grown in 24 mm culture dishes at 80% confluence. 1-mm-diameter scrape was made across the confluent monolayer using a 10-μl tip. Afterward, the dishes were washed twice and incubated at 37°C in fresh DMEM containing 10% FBS. After 24 h, two arbitrary places were marked at the bottom of each dish where the width of the wound was measured with an inverted microscope (10× objective) (1X71; Olympus).
The SGC-7901/pLv-GFP, SGC-7901/TUG1 and MGC-803/pLv-GFP, MGC-803/TUG1 seeded in 8-μm transwell inserts to undergo transwell motility assays for 24 h. The migrated cells were fixed with 4% paraformaldehyde and stained with crystal violet. Photographs of five random fields were captured with an Olympus IX71 microscope. The cells were counted in 20 random fields at high power.
2.7 Patients and Tissue Samples
This study was approved by the Research Ethics Committee of the First People’s Hospital of Xiangyang. Written informed consent was obtained from all patients. Paraffin section samples were collected from 207 patients with GC who diagnosed based on histopathological evaluation and underwent surgery, chemotherapy at the First People’s Hospital of Xiangyang between 2009 and 2012. 5 years of clinical follow-up data of all patients were assessed retrospectively to evaluate the relationship of TUG1 level and clinicopathological characteristics of the patients.
2.8 In Situ Hybridization (ISH) Assay
TUG1 level was evaluated in formalin-fixed paraffin-embedded GC tissues sections by in situ hybridization. After being dewaxed, rehydrated, digested, fixed and dehydrated, 5-digoxigenin-labeled LNATM probes (Exiqon, Denmark) complementary to TUG1 was added and hybridized for 2 h at 55°C. After rigorous washing, a specific anti-digoxigenin antibody which is directly conjugated with alkaline phosphatase was used to recognize probe. Then, sections were stained with NBT/BCIP and nuclear fast red was used to counterstain. Positive staining were presented blue particles in the plasma. The staining intensity was scored as follows: 0 (negative); 1 (weak); 2 ( intermediate); 3(strong).
All computations were carried out using SPSS version 18.0 for Windows (SPSS, Inc., Chicago, IL). The data were expressed as the mean ± SD. The median score of TUG1 was used to stratify patients into two groups. Survival curves and survival rates were analyzed using the Kaplan-Meier method; statistically significant differences in survival were identified using the log-rank test. Differences were considered statistically significant when the p value was less than 0.05.
3.1 Over-Expressed TUG1 Induced the Multi-drugs Resistance in GC Cells
We compared TUG1 expression in four GC cell lines and found higher TUG1 expression in the drug-resistant cell lines (Fig. 1A). To explore the role of TUG1 in GC cells, we overexpressed TUG1 in SGC-7901 and MGC-803 cells and downregulated TUG1 in SGC-7901/DDP and BGC823/5-Fu cells. The transfection efficiencies of pLv-TUG1 and siRNA-TUG1 were detected by FCM after transfection of pLv-TUG1 or siRNA-TUG1 into GC cells for 24 h (Fig. 1B). The proportion of positively stained cells was more than 85%. Furthermore, we compared the difference between TUG1 expression before and after the above treatments by qRT-PCR analysis. The expression of TUG1 was 6.4- and 5.7- fold higher in SGC-7901/TUG1 and MGC-803/TUG1, respectively, than in their parent cells. With regard to the efficiency of siRNA-UTG1, we found that the expression levels of TUG1 in SGC-7901/DDP/siTUG1 and BGC823/5-Fu/siTUG were 23.1% and 25.7%, respectively (Fig. 1C).
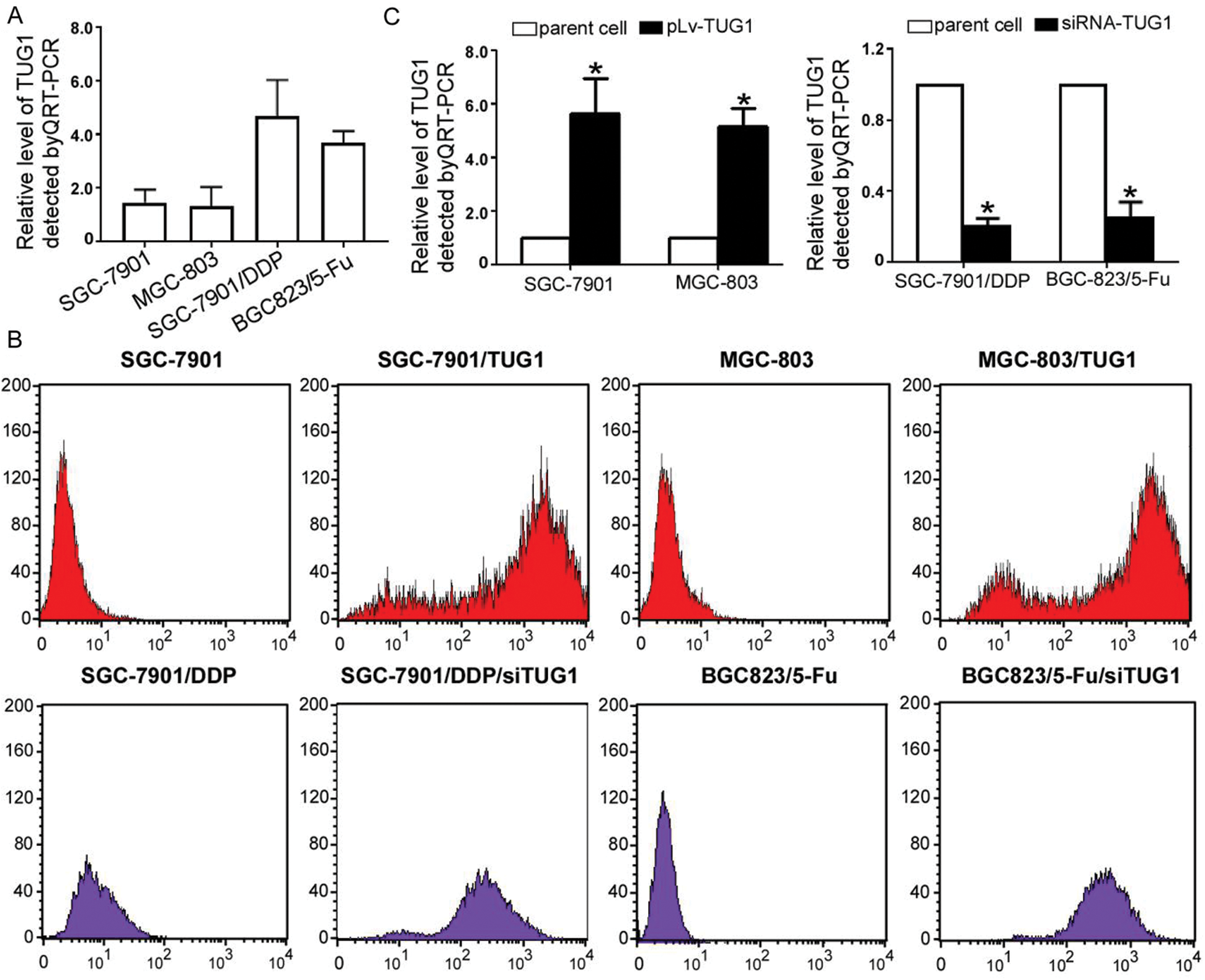
Figure 1: TUG1 expression in GC cells. (A) TUG1 expression was detected by qRT-PCR assay in four GC cell lines. (B) The transfection ratio of the pLv-TUG1 plasmid and siRNA-TUG1 in GC cell lines was detected by FCM. (C) TUG1 expression increased in pLv-TUG1-transfected cells by approximately 5 folds. In the siRNA-TUG1 group, TUG1 expression was less than 30% (*p < 0.01 compared with the parent cells)
3.2 TUG1 Closely Related to Multi-Drugs Resistance in GC Cells
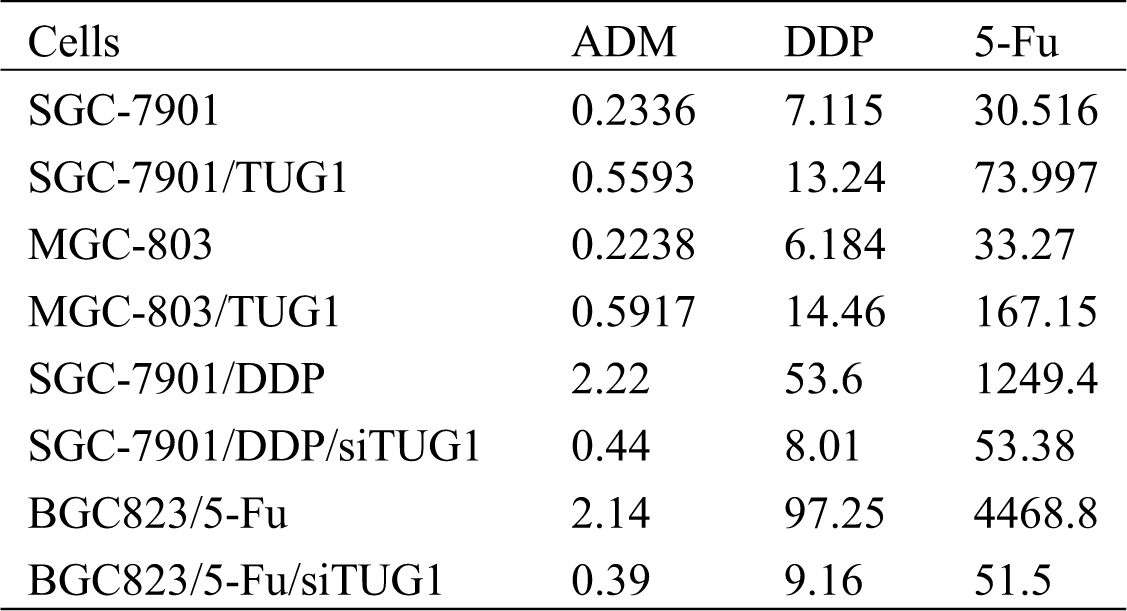
SGC-7901/TUG1, MGC-803/TUG1, SGC-7901/DDP/siTUG1, BGC823/5-Fu/siTUG1, and their parent cells were analyzed by the CCK-8 assay to assess their chemosensitivity to ADM, DDP, and 5-Fu (p < 0.001, Fig. 2A). Compared with SGC-7901/GFP and MGC-803/GFP, pLv-TUG1 markedly sensitized both TUG1-overexpressing GC subtype cell lines to multiple drugs. The siRNA-transfected GC subtype cells displayed significant resistance to multiple drugs (p < 0.05, Fig. 2B). The IC50 of DDP, ADM, and 5-Fu in each type of cells is shown in Tab. 1.
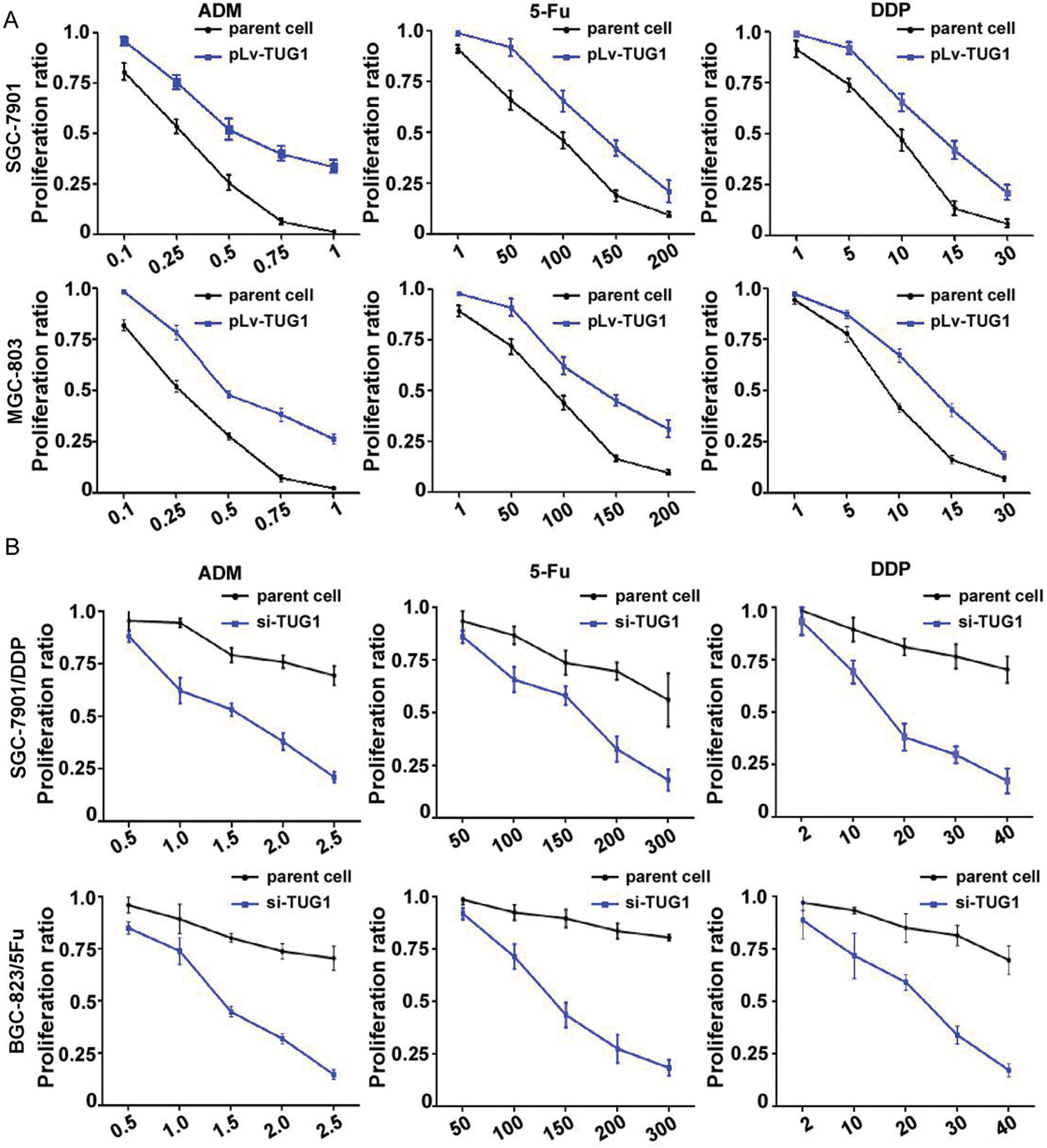
Figure 2: TUG1 regulated the drug resistance of GC cells. The CCK-8 assay was performed to measure cell proliferation. (A) Growth of SGC-7901 and MGC-803 cells and their pLv-TUG1 subclones was inhibited by ADM, DDP, and 5-Fu. (B) Viability of SGC-7901/DDP and BGC-823/5-Fu cells and their siRNA-TUG1 subclones was assessed after treatment with different concentrations of ADM, DDP, and 5-Fu. The dots represent the concentrations of chemotherapeutic drugs
Table 1: The IC50 of all drugs on the cell lines
3.3 Migration of GC Cells was Altered by TUG1 Regulation
In the wound healing assay, the percentage wound closure for SGC-7901/TUG1 and MGC-803//TUG1 cells was dramatically reduced compared with that for the respective parent cells, at 41.2% ± 7.63% vs. 69.4% ± 8.76% and 36.1% ± 13.15% vs. 66.1% ± 10.87%, respectively. Conversely, the percentage of wound closure for SGC-7901/DDP/siTUG1 and BGC823/5-Fu/siTUG1 cells was dramatically increased compared with that for the respective parent cells, at 78.61% ± 9.26% vs. 50.22% ± 6.07% and 80.33% ± 9.92% vs. 61.04% ± 8.39%, respectively (p < 0.01, Fig. 3A). The Transwell assay results (Fig. 3B) revealed dramatically increased invasion for SGC-7901/TUG1 and MGC-803/TUG1 cells compared with their respective parent cells. However, the opposite trend was found for SGC-7901/DDP/siTUG1 and BGC823/5-Fu/siTUG1 cells.
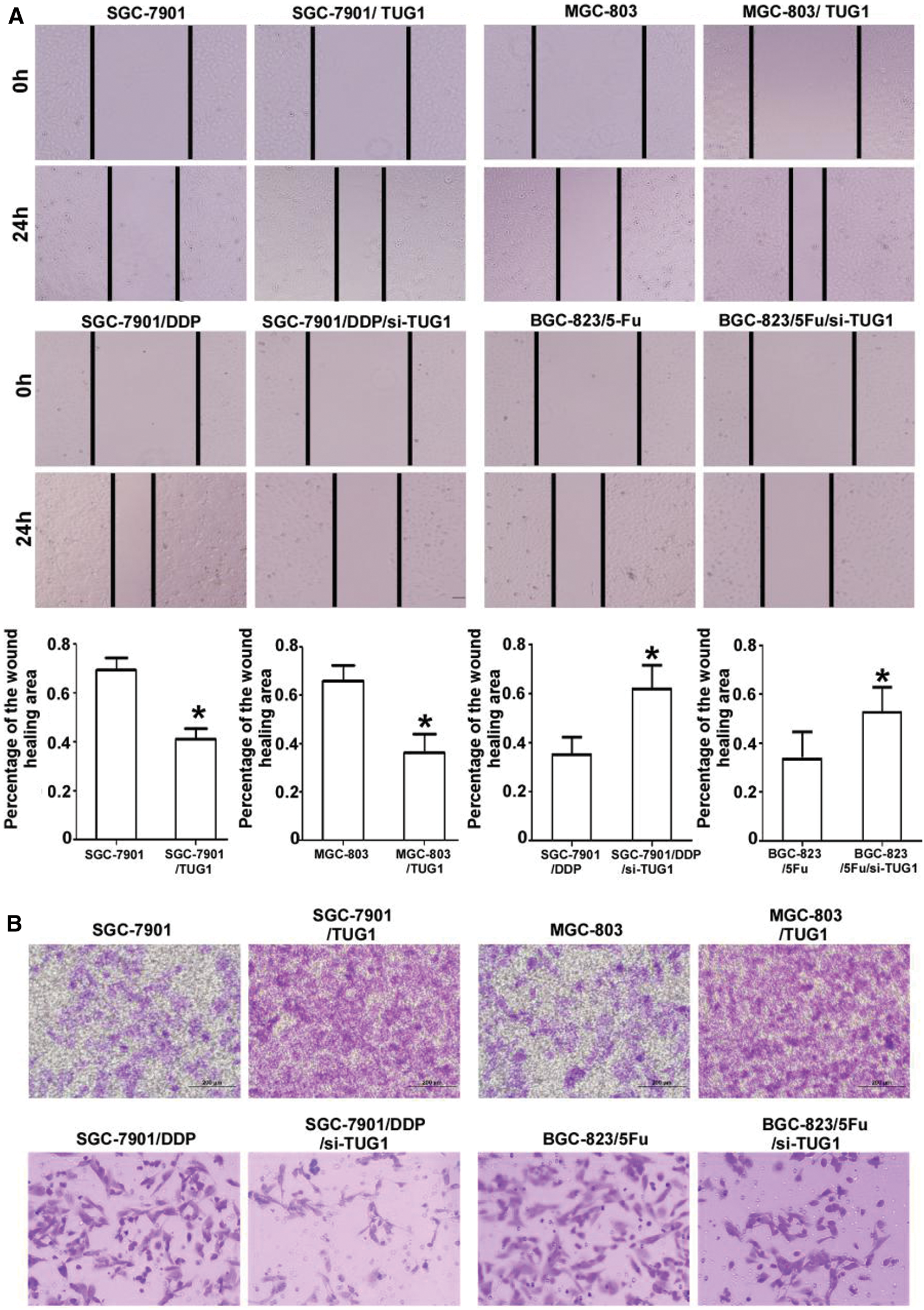
Figure 3: TUG1 affects the mobility of GC cells. A: Over-expression of TUG1 improved the mobility of SGC-7901 and MGC-803 cells, with wound healing areas only half of those for the parent cells. Compared with the parent cells, the mobility of SGC-7901/DDP/siTUG1 and BGC-823/5-Fu/siTUG1 decreased sharply after the siRNA-mediated silencing of TUG1. B: Transwell assays showed the same tendency as that observed in the wound healing assay. Representative photographs were captured at a 400× magnification
3.4 The Relationship between TUG1 Level and Clinicopathological Characteristics of GC Patients
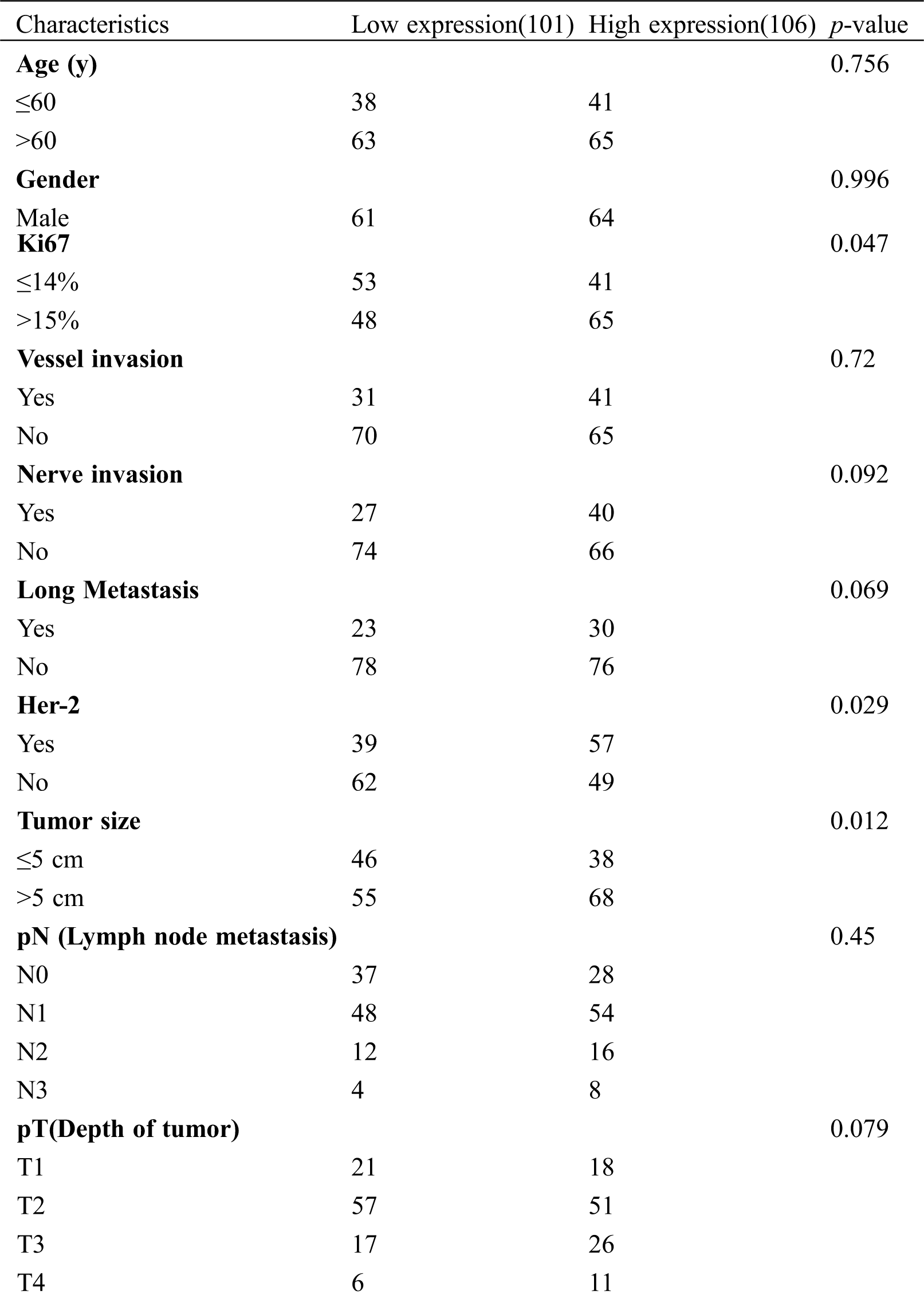
In total, 207 patients with GC were enrolled in this study, with an average age of 54.9 years of age (range: 24–72 years of age). The TUG1 staining score in the GC tissue was detected using ISH. As shown in Fig. 4A, TUG1-positive staining appeared as dark blue in the plasma of tumor cells. The patients were divided into two groups based on the median staining score, the low expression group (n = 101) and the high expression group (n = 106). From Tab. 2, it can be seen that high TUG1 expression was closely correlated with lymph node metastasis (p = 0.045), Ki67 (p = 0.047), the Her-2 positive ratio (p = 0.029), and tumor size (p = 0.012). There was also no association between TUG1 expression and other clinical factors, including age, sex, family history, nerve invasion, and histological grade (all p > 0.05).
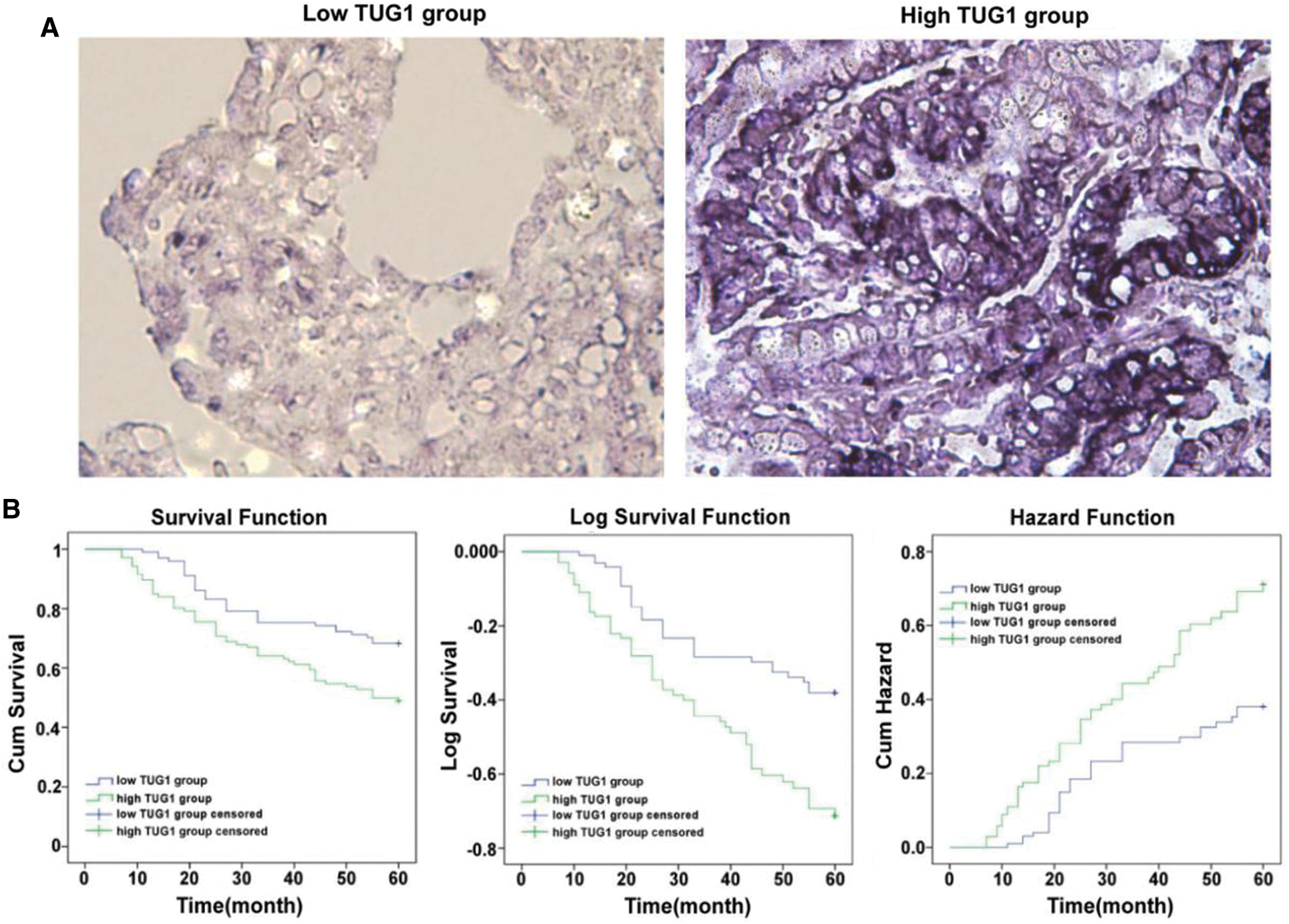
Figure 4: TUG1 expression in GC samples and prognostic analysis. A: TUG1 expression in GC tissue samples detected by ISH. Positive staining (blue granules) observed in the cytoplasm of tumor cells (×400). B: The correlation of TUG1 expression and overall survival (OS) in GC patients. The Kaplan-Meier method was used to estimate the probability of OS. It was presented in Cum Survival, Log Survival and Cum Hazard, respectively
Table 2: Patient characteristics and association of TUG1 expression with clinicopathological characteristics of GC patients (n = 207)
Using the Kaplan-Meier method, we analyzed differences in the 5-year OS rate between patients with GC in the high and low TUG1 groups. The cumulative survival, logarithmic survival, and cumulative hazard risk are shown in Fig. 4B. The log-rank test results showed that high expression of TUG1 was correlated with a poor prognosis of patients with GC (68.3% vs. 49.1%; p = 0.003).
Gastric cancer is one of the leading causes of cancer-related death accounting for 20% of the total worldwide especially in developing countries such as China. Each year more than 950,000 GC patients were new diagnoses [16]. Surgery is the only curative treatment for GC. But due to no census was held, many patients were inoperable disease at diagnosis, means AGC, who would benefit from conversion therapy or called neoadjuvant chemotherapy [17]. It is the main reason for recurrence that tumor-cell spilled to peritoneal dissemination in perioperative period or during surgery. As it is found from US 0116 trial that compared with surgery, chemo-radiotherapy could significantly reduce mortality and risk of tumor recurrence [18]. So it is necessary for GC patients with unfavorable prognostic factors undergo systemic chemotherapy. With the development of molecular biology, the complete sequencing of human genome had been done by the Human Genome Project. More and more carcinomas would be classified based on molecular subtypes. Response to cisplatin-based chemotherapy had been reported associated with mutations in various proto-oncogenes and tumoursuppressor genes such as MSI. During the past 20 years, the function of LncRNA had been a growing concern including regulating chemo-resistant of multiple types of cancer. Taurine Upregulated Gene 1 (TUG1) first cloned by Young et al. [19]. It is a spliced, polyadenylated RNA that does not encode any open reading frame greater than 82 amino acids in its full-length, 6.7 kilobase (kb) RNA sequence be named TUG1 on this account its first function regulating taurine, the necessary proper cysteine derivative for neural development. Soon high expression of TUG1 was verified in more and more tumor. The mechanism may due to TUG1 constitute chromatin-modifying complexes and plays roles in gene regulation. Through ribosomal fractionation van Heesch obtained ribosome-associated RNA pools and detected the systematic RNA sequencing. Then they compared the RNA content with nuclear and (non-ribosome bound) cytosolic RNA pools. It is TUG1 take in particular in ribosome complexes and have a function in cytoplasmic processes [20]. Xie reported TUG1 activated the Wnt/β-catenin pathway. Over-expression of TUG1 induced the bladder cancer (BC) cell line resistant to doxorubicin (Dox). Whereas knockdown of TUG1 inhibited the Dox resistance of BUC cells [21]. Niu also demonstrated that TUG1 affects cell growth and chemoresistance of SCLC [22]. In our study, TUG1 up-regulation also depressed the sensitivity of SGC-7901, MGC-803 to anticancer drugs, cisplatin (DDP), Adriamycin (ADM), and 5-Fu. The IC50 of SGC-7901/TUG1, MGC-803/TUG1 raised 3 to 5 times of their parent cells. It is the first time demonstrated that TUG1 impacts on the chemosensitivity of GC. Lin showed that miR-455-3p down-regulated with knocking-down of TUG1 which induces marked inhibition of cell migration, invasion, and glycolysis [23]. Zhao et al. reported TUG1 was overexpressed in PC tissues. Overexpressed TUG1 in Pancreatic Cancer cells triggered EMT formation and promoted cell proliferation, migration [14,24]. Our results also revealed over-expressed TUG1 promoted migration of GC cells. The scratched wound assay discovered healing area in TUG1 groups were almost half of control groups. Transwell migration assay showed migrated cells of TUG1 groups up-regulated two to three times of control groups. Wang investigated the prognostic and clinical significance of TUG1 in various types of cancers. This meta-analysis suggested overexpression of TUG1 was significantly correlated with unfavorable overall survival in patients with cancer. Here we found TUG1 level was statistically correlated with lymph node metastasis, Ki67, Her-2 positive ratio and tumor size.
In summary, TUG1 promoted the GC cell proliferation, migration and multi-drug resistance, which is correlated with poor prognosis. Our data suggested that TUG1 may serve as a novel predictive biomarker of GC. Further work is needed to deeply clarify the TUG1-related regulation mechanisms and establish an optimum drug delivery system. The diagnosis and therapeutic strategies based on it are still in an elementary stage and extensive studies are required. However, These findings provided us with new insights.
Availability of Data and Materials: The dataset supporting the conclusions of this article is included within this article and is available from the corresponding author upon request.
Author’s Contribution: Conception and design: Anwen Liu. Methodology, Data analysis and interpretation: Yuan Liu, Yun Jia, Bing Zhang. Manuscript writing: Yuan Liu. Final approval of manuscript: All authors.
Funding Statement: This work was supported by the Science and Technology Project of Jiangxi Administration of Traditional Chinese Medicine [Grant No. 2019A047].
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
1. Shams, A. Z., Haug, U. (2017). Strategies for prevention of gastrointestinal cancers in developing countries: A systematic review. Journal of Global Health, 7(2), 020405. DOI 10.7189/jogh.07.020405. [Google Scholar] [CrossRef]
2. Dong, J., Thrift, A. P. (2017). Alcohol, smoking and risk of oesophago-gastric cancer. Best Practice & Research Clinical Gastroenterology, 31(5), 509–517. DOI 10.1016/j.bpg.2017.09.002. [Google Scholar] [CrossRef]
3. Ruíz-García, E., Guadarrama-Orozco, J., Vidal-Millán, S., Lino-Silva, L. S., López-Camarillo, C. et al. (2017). Gastric cancer in Latin America. Scandinavian Journal of Gastroenterology, 53(2), 124–129. DOI 10.1080/00365521.2017.1417473. [Google Scholar] [CrossRef]
4. Wagner, A. D., Syn, N. L., Moehler, M., Grothe, W., Yong, W. P. et al. (2017). Chemotherapy for advanced gastric cancer. Cochrane Database of Systematic Reviews, 8, CD004064. DOI 10.1002/14651858. [Google Scholar] [CrossRef]
5. Cheng, X., Lu, Y. (2017). A review of capecitabine-based adjuvant therapy for gastric cancer in the Chinese population. Future Oncology, 14(8), 771–779. DOI 10.2217/fon-2017-0558. [Google Scholar] [CrossRef]
6. Zhang, H. H., Guo, X. L. (2016). Combinational strategies of metformin and chemotherapy in cancers. Cancer Chemotherapy and Pharmacology, 78(1), 13–26. DOI 10.1007/s00280-016-3037-3. [Google Scholar] [CrossRef]
7. Chen, Q. N., Wei, C. C., Wang, Z. X., Sun, M. (2017). Long non-coding RNAs in anti-cancer drug resistance. Oncotarget, 8(1), 1925–1936. DOI 10.18632/oncotarget.12461. [Google Scholar] [CrossRef]
8. Ou, C., Li, G. (2017). Long non-coding RNA TUG1: A novel therapeutic target in small cell lung cancer. Journal of Thoracic Disease, 9(7), E644–E645. DOI 10.21037/jtd.2017.06.94. [Google Scholar] [CrossRef]
9. Li, L., Gan, Z. H., Qin, L., Jiao, S. H., Shi, Y. (2017). AIB1 regulates the ovarian cancer cell cycle through TUG1. European Review for Medical and Pharmacological Sciences, 21(24), 5610–5617. [Google Scholar]
10. Qin, C. F., Zhao, F. L. (2017). Long non-coding RNA TUG1 can promote proliferation and migration of pancreatic cancer via EMT pathway. European Review for Medical and Pharmacological Sciences, 21(10), 2377–2384. [Google Scholar]
11. Li, N., Shi, K., Kang, X., Li, W. (2017). Prognostic value of long non-coding RNA TUG1 in various tumors. Oncotarget, 8(39), 65659–65667. DOI 10.18632/oncotarget.20025. [Google Scholar] [CrossRef]
12. Zhang, E. B., Yin, D. D., Sun, M., Kong, R., Liu, X. H. et al. (2014). P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death & Disease, 5(5), e1243. DOI 10.1038/cddis.2014.201. [Google Scholar] [CrossRef]
13. Liu, Y., Yang, S., Zhang, X. (2013). WITHDRAWN: Down-regulation of long non-coding RNATUG1suppresses melanoma cell proliferation and induces apoptosis via up-regulating microRNA-9. Biochemical and Biophysical Research Communications, S0006-291X(13)01521-0. DOI 10.1016/j.bbrc.2013.09.050. [Google Scholar] [CrossRef]
14. Zhao, L., Sun, H., Kong, H., Chen, Z., Chen, B. et al. (2017). The Lncrna-TUG1/EZH2 axis promotes pancreatic cancer cell proliferation, migration and EMTPhenotype formation through sponging Mir-382. Cellular Physiology and Biochemistry, 42(6), 2145–2158. DOI 10.1159/000479990. [Google Scholar] [CrossRef]
15. Jiang, L., Wang, W., Li, G., Sun, C., Ren, Z. (2016). High TUG1 expression is associated with chemotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. Cancer Chemotherapy and Pharmacology, 78(2), 333–339. DOI 10.1007/s00280-016-3066-y. [Google Scholar] [CrossRef]
16. van, C. E., Sagaert, X., Topal, B., Haustermans, K., Prenen, H. et al. (2016). Gastric cancer. Lancet, 388(10060), 2654–2664. DOI 10.1016/S0140-6736(16)30354-3. [Google Scholar] [CrossRef]
17. Oh, D. Y., Bang, Y. J. (2013). Adjuvant and neoadjuvant therapy for gastric cancer. Current Treatment Options in Oncology, 14(3), 311–320. DOI 10.1007/s11864-013-0238-4. [Google Scholar] [CrossRef]
18. Yeh, J. M., Tramontano, A. C., Hur, C., Schrag, D. (2017). Comparative effectiveness of adjuvant chemoradiotherapy after gastrectomy among older patients with gastric adenocarcinoma: A SEER–Medicare study. Gastric Cancer, 20(5), 811–824. DOI 10.1007/s10120-017-0693-x. [Google Scholar] [CrossRef]
19. Young, T. L., Matsuda, T., Cepko, C. L. (2005). The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Current Biology, 15(6), 501–512. DOI 10.1016/j.cub.2005.02.027. [Google Scholar] [CrossRef]
20. van, H. S., van, I. M., Jacobi, J., Boymans, S., Essers, P. B. et al. (2014). Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biology, 15(1), R6. DOI 10.1186/gb-2014-15-1-r6. [Google Scholar] [CrossRef]
21. Xie, D., Zhang, H., Hu, X., Shang, C. (2017). Knockdown of long non-coding RNA Taurine Up-Regulated 1 inhibited doxorubicin resistance of bladder urothelial carcinoma via Wnt/β-catenin pathway. Oncotarget, 8(51), 88689–88696. DOI 10.18632/oncotarget.20927. [Google Scholar] [CrossRef]
22. Niu, Y., Ma, F., Huang, W., Fang, S., Li, M. et al. (2017). Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Molecular Cancer, 16(1), 5. DOI 10.1186/s12943-016-0575-6. [Google Scholar] [CrossRef]
23. Lin, Y. H., Wu, M. H., Huang, Y. H., Yeh, C. T., Cheng, M. L. et al. (2018). Taurine up-regulated gene 1 functions as a master regulator to coordinate glycolysis and metastasis in hepatocellular carcinoma. Hepatology, 67(1), 188–203. DOI 10.1002/hep.29462. [Google Scholar] [CrossRef]
24. Wang, X., Chen, X., Zhang, D., Yang, G., Yang, Z. et al. (2017). Prognostic and clinicopathological role of long non-coding RNA taurine upregulated 1 in various human malignancies: A systemic review and meta-analysis. Tumor Biology, 39(7), 1010428317714361. DOI 10.1177/1010428317714361. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |