
 | Oncologie |  |
DOI: 10.32604/oncologie.2021.017786
ARTICLE
Overexpression of lnc-ERP44-3:6 Causes Cell Death and Sensitivity to Cisplatin in Breast Cancer Cell Lines
1Departamento de Bioquímica y Medicina Molecular, Facultad de Medicina, Universidad Autónoma de Nuevo León, Nuevo León, 64460, Mexico
2Centro de Biotecnología-FEMSA, Tecnológico de Monterrey, Nuevo León, 64849, Mexico
3Delee Corp, Mountain View, CA 94041, USA
4Department of Basic Science, School of Medicine, Universidad de Monterrey, Nuevo León, 66238, Mexico
5Department of General Surgery, Hospital Universitario, Dr. José Eleuterio González, Universidad Autónoma de Nuevo León, Nuevo León, 66460, Mexico
6Department of Pathology and Cytopathology, Facultad de Medicina, Dr. José Eleuterio Gonzáles, Universidad Atónoma de Nuevo León, Nuevo León, 66460, Mexico
7Clínica de Mama, Hospital Metropolitano, Dr. Bernardo Sepúlveda, Secretaría de Salud, Nuevo León, 66480, Mexico
*Corresponding Author: Carlos Córdova-Fletes. Email: carlos.cordovafl@uanl.edu.mx
Received: 06 June 2021; Accepted: 14 August 2021
Abstract: Breast cancer (BC) is one of the leading causes of death in women worldwide. A major challenge in BC is chemoresistance, which is often modulated by epigenetic regulators such as long non-coding RNAs (lncRNAs). Because these regulator lncRNAs may play diverse roles, determining their specific pathways and/or functions is crucial to identify possible biomarkers and/or therapeutic targets for BC. In this study, we used gene expression microarrays in order to identify lncRNAs related to the BC biology. We found, among six differentially expressed (DE) lncRNAs, that the expression of lnc-ERP44-3:6 was consistently down-regulated in all breast tumor tissues compared to normal adjacent tissues of BC patients from northeastern Mexico. Since the role of lnc-ERP44-3:6 remains unknown in BC, we induced its transient overexpression in three different BC cell lines (i.e., MCF10A, MCF-7, and HCC1395) by transfection in order to elucidate its potential downstream effects. Remarkably, our results showed a significant increase in cell death and chemosensitivity to cisplatin at 48 h post-transfection (p < 0.01). In addition, we observed that GAPDH and FAS were up-regulated following the overexpression of lnc-ERP44-3:6. As GAPDH and FAS promote apoptosis in cancer, our findings suggest that the lnc-ERP44-3:6 overexpression induces cell death possibly via up-regulation of one or both genes in BC cell lines. To the best of our knowledge, this is the first study evaluating the overexpression of lnc-ERP44-3:6 and providing insights about its role in BC cells, which suggests that this lncRNA may be an interesting candidate as a potential therapeutic target for BC in our population. However, further studies would be needed to clarify the mechanisms by which an overexpression of lnc-ERP44-3:6 causes both cell death and chemosensitivity to cisplatin.
Keywords: Breast cancer; lncRNA; lnc-ERP44-3:6; biomarker; therapeutic target; chemoresistance; tumor suppressor
BC is one of the major public health problems worldwide and is the most common cancer, with more than 2 million cases, 600 000 deaths, and an increase of 0.3% per year in the world. It is the second-leading cause of death—after lung cancer—among women younger than 50 years old [1,2]. Since BC is a heterogeneous disease, it has been classified into different subtypes according to overexpression or underexpression of different receptors (e.g., estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2)) [3,4]. A further highly aggressive and invasive BC subtype lacking hormone receptors expression is commonly known as triple-negative breast cancer (TNBC) subtype. The current therapies for BC rely on these subtypes [5,6]; yet, notwithstanding the progress in the management of BC patients, reaching a timely diagnostic and/or treatment is often not straightforward (especially for the TNBC subtype) and chemoresistance remains to be the foremost hurdle to overcome as it leads to metastasis and a poor prognosis in BC patients.
The observed heterogeneity in BC may partly be caused by aberrant epigenetic changes that lead to either the silencing of tumor suppressor genes or overexpression of oncogenes. These modifications can include CpG methylation, and hypoacetylation and hypermethylation of histones [7–9]. Recent discoveries have postulated that the lncRNAs may be involved in said changes [10–13]. LncRNAs are essential epigenetic regulators, span more than 200 nucleotides in length, locate mainly in the nucleus, transcribe by the RNA polymerase II, present post-transcriptional modifications, and their expression lower than mRNA is tissue-specific [11,14–18]. LncRNAs participate in multiple molecular processes such as chromatin remodeling, transcription enhancing or interfering, alternative splicing, protein stability, and as microRNAs decoys [19–21]. More recently, lncRNAs have been identified in various cellular and cancer-related processes, including stem cell pluripotency, cell cycle and proliferation, cell migration, lineage differentiation, and tissue invasion [14,15,22], and they may act as tumor suppressors (e.g., MEG3) [23], oncogenes (e.g., BCRT1) [24], and/or cell sensitizers to chemotherapeutic agents (e.g., SNORD3) [22]. Therefore, identifying new lncRNAs and their underlying functions would be essential to propose new biomarkers and/or therapeutic targets that allow for a more precise diagnosis and an improved treatment for BC.
In this study, we focused to find differentially expressed (DE) lncRNAs not yet reported in the literature to be related to BC or with unknown function. Thereby, we found six lncRNAs, among which the expression of lnc-ERP44-3:6 was consistently down-regulated in all BC tissue samples compared to adjacent normal tissue of patients from northeastern Mexico. lnc-ERP44-3:6 (also known as LOC101928438, lnc-ERP44-3:6, AL359710.1, lnc-ALG2-5, lnc-ALG2-1, and STX17-AS1) is an intergenic lncRNA that consists of 1917 bp and six exons and is located on chromosome 9 (genomic coordinates 99585324-99773612; hg38) (Fig. S1). Except for a mention in an abandoned patent [25], lnc-ERP44-3:6 has not yet been reported in other instances related to BC. Hence, we evaluate its effect on cell death and chemoresistance using a BC cellular model. Strikingly, we found that its overexpression in the model decreased cell viability and promoted cell death by apoptosis, as well as enhanced chemosensitivity to cisplatin in tumoral and non-tumoral cell lines.
The study was approved by the ethics committees of Hospital Universitario “Dr. José Eleuterio González” and Hospital Metropolitano “Dr. Bernardo Sepúlveda” with the protocol numbers of BI15-002 and DEISC-19 01 16 23, respectively. All the procedures performed were carried out according to the ethics committees and the latest version of the 1964 Helsinki Declaration.
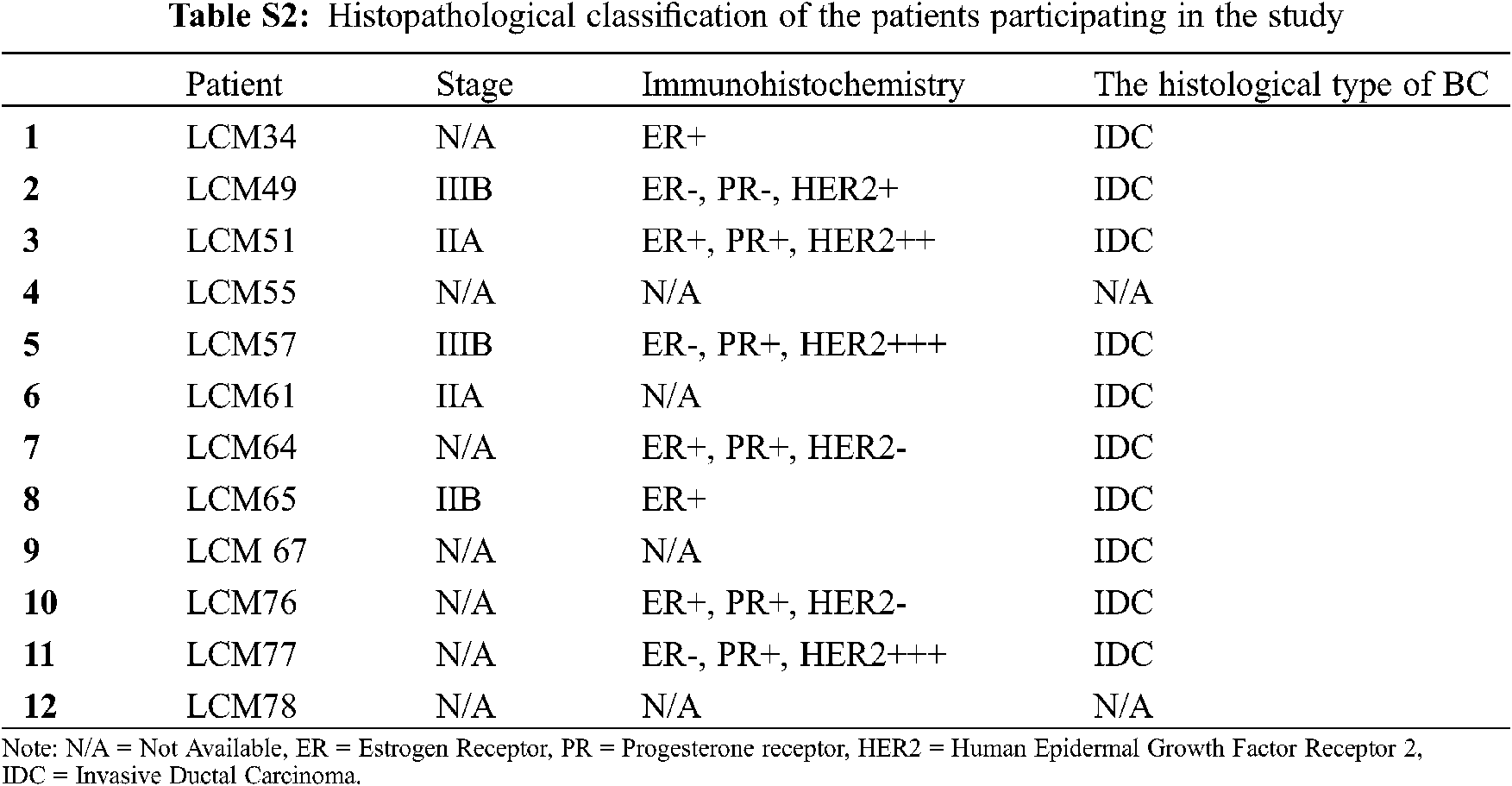
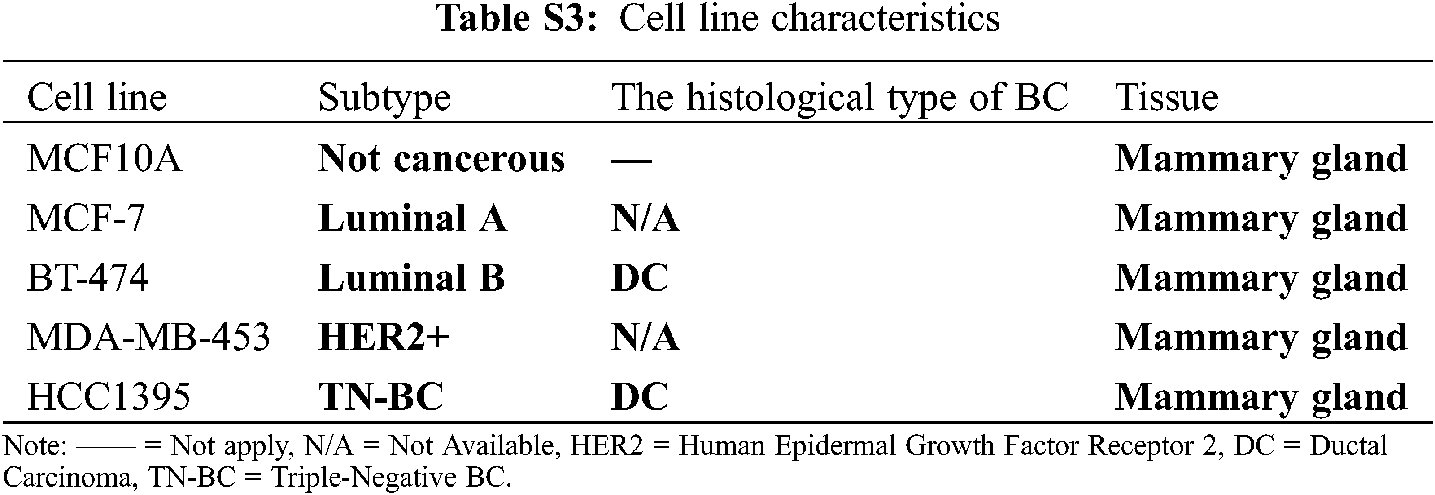
Twelve Mexican patients with untreated invasive ductal carcinoma (IDC) were recruited in the period 2015–2017 from the Hospital Universitario “Dr. José Eleuterio González” and Hospital Metropolitano “Dr. Bernardo Sepúlveda” (Monterrey, Nuevo León, México). Primary tumor samples from 12 biopsies (Luminal A, n = 2; Luminal B, n = 2 and HER2+, n = 4; and unidentified samples n = 4) and their corresponding 12 adjacent non-cancerous tissue samples (total = 24 biopsies, 5 × 5 × 5 mm) from these BC patients were dissected and preserved in RNA later solution (QIAGEN, USA). No TNBC subtype samples were found in this cohort. Total RNA was isolated and purified by the All Prep DNA/RNA mini tube kit (QIAGEN, USA) according to the manufacturer’s instructions. Purified RNA was later retrotranscribed and used for gene expression analysis by microarrays and real-time quantitative PCR (RT-qPCR). Next, we describe the experiments we conducted in this study to find and investigate the potential role of lnc-ERP44-3:6 in BC (Fig. 1A).
To perform a global differential gene expression analysis, the cDNA from each tumor and adjacent non-cancerous tissue sample was labeled with Cy3 dye and hybridized on the SurePrint G3 Human Gene Exp v3, 8x60K microarray (Agilent Technologies, USA), following the manufacturer’s instructions for the Agilent Low Input QuickAmp Labeling kit, one color protocol (Agilent Technologies, USA). GeneSpring software v14.5 (Agilent Technologies, USA) was used to process data, and a T-student test with Benjamin-Hochberg correction (p ≤ 0.01 and fold change ≥2.0) was performed to compare expression changes between tumor and non-cancerous tissues. Through this analysis, we focused on DE lncRNAs.
2.4 Design and Production of pCMV-lnc-ERP44-3:6-GFP-Vector
Once we got the lnc-ERP44-3:6 sequence from the LNCipedia database (v5.2), it was synthesized by GenScript (Piscataway, USA). This synthesized sequence was cloned into a pCMV-GFP expression vector (ATUM, USA) using the SapI restriction site and a T4 DNA ligase (New England Biolabs, USA). The vector is regulated by a T7 promoter and resistant to ampicillin. We transformed our vectors pCMV- lnc-ERP44-3:6-GFP (Fig. 1B) and pCMV-GFP using chemically competent (Molecular Cloning: A Laboratory Manual Sambrook) E.coli (strain TOP10). Then was selected the ampicillin-resistant clones and was confirmed through PCR the positive clones with the lnc-ERP44-3:6 sequence in the vector. Finally, we grow the bacterial culture and purify the plasmid DNA using the PureYieldTM Plasmid Maxiprep system (Promega, USA).
2.5 Cell Culture and Transfection
MCF10A (normal mammary epithelial) was grown in MEBM medium with 100 ng/ml of cholera toxin; MCF-7 (represents the luminal A subtype) was cultured in EMEM (Gibco, USA) medium with 0.01 mg/ml of human recombinant insulin (Gibco, USA) and 1 µM 4-hydroxytamoxifen (Sigma-Aldrich, USA); BT-474 (represents the luminal B subtype) was grown in DMEM/F-12 (Gibco, USA); MDA-MB-453 (represents the HER2+ subtype) was cultured in Leibovitz’s medium; and HCC1395 (represents the triple-negative subtype) was cultured in RPMI-1640 medium (Gibco, USA). All culture media were supplemented with 10% Fetal bovine serum (FBS; Gibco, USA), Amphotericin B (250 ng ml–1; Gibco, USA), Gentamicin (50 µg ml–1; Gibco, USA), L-Glutamine (20 µM ml–1; Sigma-Aldrich, USA), Penicillin and Streptomycin (10 units ml–1 and 10 µg ml–1, respectively; Gibco, USA). The cells were incubated at 37°C with 5% CO2, under sterile conditions. MCF10A, MCF-7, and HCC1395 were transfected with the pCMV-Inc ERP44-3:6-GFP or pCMV-GFP (control vector) plasmids, using Xfect transfection reagent (Takara Bio, USA), following the manufacturer’s instructions. Cells were then incubated at 24-, 48-, and 72-h post-transfection by technical triplicate. Statistical analysis was performed in GraphPad Prism 9 (v.9.2.0), using One-way ANOVA with a p-value < 0.05.
2.6 RNA Extraction and RT-qPCR
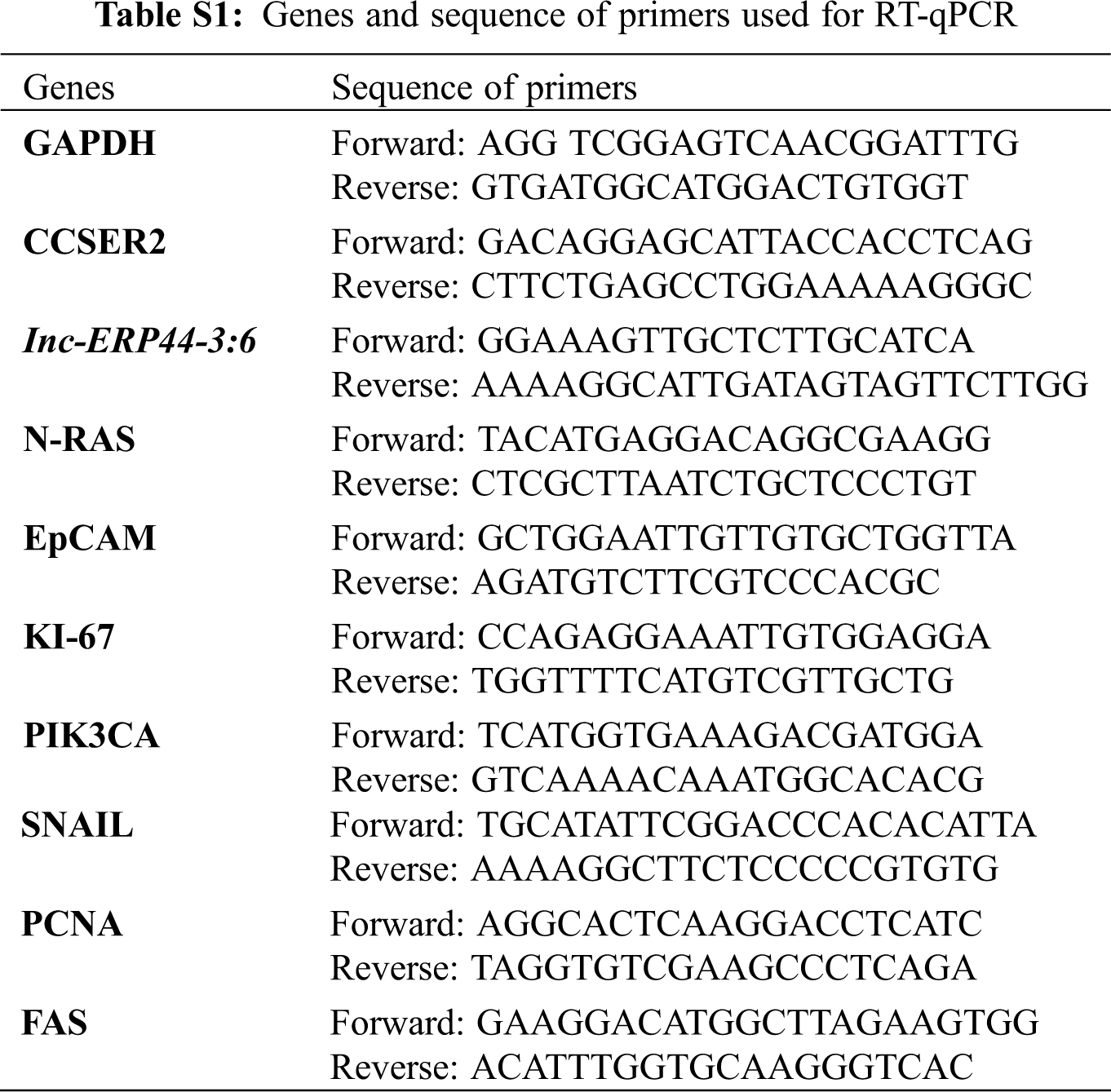
Trizol was used to extract total RNA according to the manufacturer’s instructions (Invitrogen, USA). The RNA pellet was resuspended in nuclease-free water (Corning, USA). Next, we performed RT-PCR using SuperScript III (Invitrogen, USA) to obtain the cDNA, following the manufacturer’s instructions. Later, Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent, USA) was used to process the samples on an Applied Biosystem 7,500 fast real-time PCR system (Thermo Fisher, USA). The lnc-ERP44-3:6 specific primers for RT-qPCR were designed using primer 3 plus [25]; their quality, specificity, and characteristics were analyzed and proved by OligoAnalyzer Tool (IDT) [26] and Amplify4 v1.0 software [27], respectively. CCSER2 was used as an endogenous gene to calculate relative levels of expression. GAPDH (metabolism and apoptosis), N-RAS (cell growth, motility, angiogenesis, and immune escape), EpCAM (EMT: epithelial-mesenchymal transition), KI-67 (cell proliferation), PCNA (cell proliferation), SNAIL (chemotherapeutic resistance, and EMT), and PIK3CA (cellular growth, proliferation, survival, angiogenesis, and motility), FAS (death receptor) genes were selected to determinate potential pathways affected after overexpressing lnc-ERP44-3:6 (Tab. S1). The RT-qPCR was performed by technical triplicate and the mean values were calculated to determine the relative expression using the 2−ΔΔCt method. T-student and one and two-way ANOVA tests were performed for statistical analysis with a p-value <0.05 using GraphPad Prism 9 (v.9.2.0).
2.7 Immunoassay with Transfected Cells
MCF10A, MCF-7, and HCC1395 cell lines were used to seed 10,000 cells per well on 96-well plates. The cells were fixed with paraformaldehyde and incubated for 10 min. The plate with the fixed cells was blocked with 200 µl of 5% skimmed milk in PBS (Phosphate Buffered Saline) and incubated for 1 h at room temperature; the next three washes with PBS containing 0.05% Tween-20TM were performed to remove waste. Then we added 100 µl of antibodies anti-GAPDH, anti-PCDNA, anti-KI-67, anti-PI3, anti-N-RAS, anti-EpCAM, and anti-SNAIL with a dilution of 1:2000 (Santa Cruz Biotechnology) and incubated for 1 h at room temperature with three subsequent washes using PBS containing 0.05% Tween-20TM. We used an antibody anti-mouse marked with HRP (100 µl, 1:10000 dilution; Pierce, Rockford IL, USA) was then incubated for 1 h at room temperature, followed by three washes with PBS containing 0.05% Tween-20TM. The HRP was then detected by adding 100 µl of 1-StepTM Ultra TMB-ELISA (Pierce, Rockford IL, USA) until a blue color was observed. The reaction was stopped by adding 100 µl 1M H2SO4, and the absorbance was measured at 450 nm in the Cytation 5 microplate reader (Biotek, USA). The immunoassay was performed by technical triplicate and GraphPad Prism 9 (v.9.2.0) and SPSS (v25) [28] were used for statistical analyses; a two-way ANOVA was performed with a p-value < 0.05.
2.8 Cell Viability, Death, and Cytotoxicity Assays
Sterile 96 well plates were used to seed 10,000 cells of MCF10A, MCF-7, or HCC1395 by technical triplicates and incubated for 24 h. Cells were transfected with pCMV-lncERP44-3:6-GFP and/or pCMV-GFP (empty vector, used as control) vector and incubated for 24, 48, and 72 h post-transfection. The cell viability was determined using 100 µl of CellTiter-Glo® 3D (Promega, USA) to each well, and luminescence was measured after 25 min of incubation at room temperature, following the manufacturer’s instructions. To detect cell death, we added 100 µl of Caspase-Glo® 3/7 Reagent to each well of 96-well plates with 10,000 cells of MCF10A, MCF-7, and HCC1395; the cells were incubated for 30 min at room temperature and luminescence was measured according to the manufacturer’s instructions. All assays were performed by technical triplicate and statistical analysis was performed in GraphPad Prism 9 (v.9.2.0), using two-way ANOVA with a p-value < 0.05.
Finally, the transfected cells with the lnc-ERP-443:6 or control vector were incubated with different concentrations of Cisplatin (2.5, 5, 10, and 20 µM) 24 h post-transfection and CellTiter-Glo® and Caspase-Glo® 3/7 reagents were used to determine viability cell and rate of apoptosis, respectively, at 48 h post-transfection. All assays were performed by technical triplicate and statistical analysis was performed in GraphPad Prism 9 (v.9.2.0), using two-way ANOVA with a p-value < 0.05.
3.1 lnc-ERP44-3:6 Expression in BC Patients’ Tissue and BC Cell Lines
Gene expression microarrays were performed to determine the global differential expression pattern from BC tissue (n = 12) compared to adjacent non-cancerous tissue (n = 12). Our results showed 99 DE entities (p < 0.01, unpublished data), among which, lnc-C9orf131-1, lnc-SLC25A21-2, and lnc-GLB1-1 were overexpressed, and lnc-ERP44-3, lnc-BBOX1-1, and lnc-SPRYD3-1 were down-regulated (Fig. 2 and data not shown). In this regard, lnc-ERP44-3:6 (fold change –3.719) was consistently down-regulated in all samples (and probes) of our study population (Figs. 2A and 2B, and data not shown); this result was validated by RT-qPCR assays (Fig. 2C). Given that the samples collected from patients were positive for either luminal A, B, or HER2+ subtype (Tab. S2), we evaluated the expression pattern of lnc-ERP44-3:6 in cell lines corresponding to such subtypes as well as in both a non-tumor cell line and a TNBC (Tab. S3). Thus, we determined its endogenous expression by RT-qPCR in all tumoral (i.e., MCF-7 (luminal A subtype), BT-474 (luminal B subtype), MDA-MB-453 (HER2+), and HCC1395 (TNC)) and normal (i.e., MCF10A) cell lines. The results showed no significant expression changes in the MCF-7 cell line compared to MCF10A; conversely, HCC1395 cell line showed the lowest expression of lnc-ERP44-3:6 compared to MCF10A, MCF-7, BT-474, and MDA-MB-453 (**** p < 0.0001) (Fig. 2D).

Figure 1: (A) Workflow we follow to determine the role of lnc-ERPP44-3:6, steps 1 to 7: 1. Gene expression analysis in BC patients and BC cell lines, 2. Overexpression of lnc-ERPP44-3:6 transfecting BC cell lines with the pCMV-lnc-ERP44-3:6-GFP vector, 3. RNA extraction from the cell lines transfected to perform the RT-qPCR, 4. Analysis of the gene expression, 5. Analysis of the protein profiles through immunoassays, 6. Viability and drug resistance following an overexpression of lnc- ERPP44-3:6. (B) Map of the pCMV-lnc-ERP44-3:6-GFP vector, which includes a CMV promoter and a GFP protein, and the sequence of lnc-ERPP44-3:6 inserted using the SapI restriction. (C) Prediction of the secondary structure of lnc-ERPP44-3:6 using RNAfold WebServer [29]

Figure 2: (A) Heat map showing in blue color the down-regulation of lnc-ERP44-3:6 compared to adjacent normal tissue in red color. (B) Fold change graphic denoting the lnc-ERP44-3:6 differential expression from microarray results. (C) Validation by RT-qPCR of the expression pattern of lnc-ERP44-3:6 in BC patients. (D) RT-qPCR results for the endogenous expression of lnc-ERPP44-3:6 in the BC cell lines. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001
3.2 Overexpression of lnc-ERP44-3:6 in BC Cell Lines
Three cell lines were selected to transiently overexpress lnc-ERP44-3:6 by the pCMV-GFP vector: MCF10A was used as a control cell line, while the MCF-7 and HCC1395 cell lines represented the least aggressive and the most aggressive subtypes of BC, respectively. The results showed the highest expression of lnc-ERP44-3:6 at 24 h, in all transfected cell lines (Figs. 3 and S2 in Annex B).
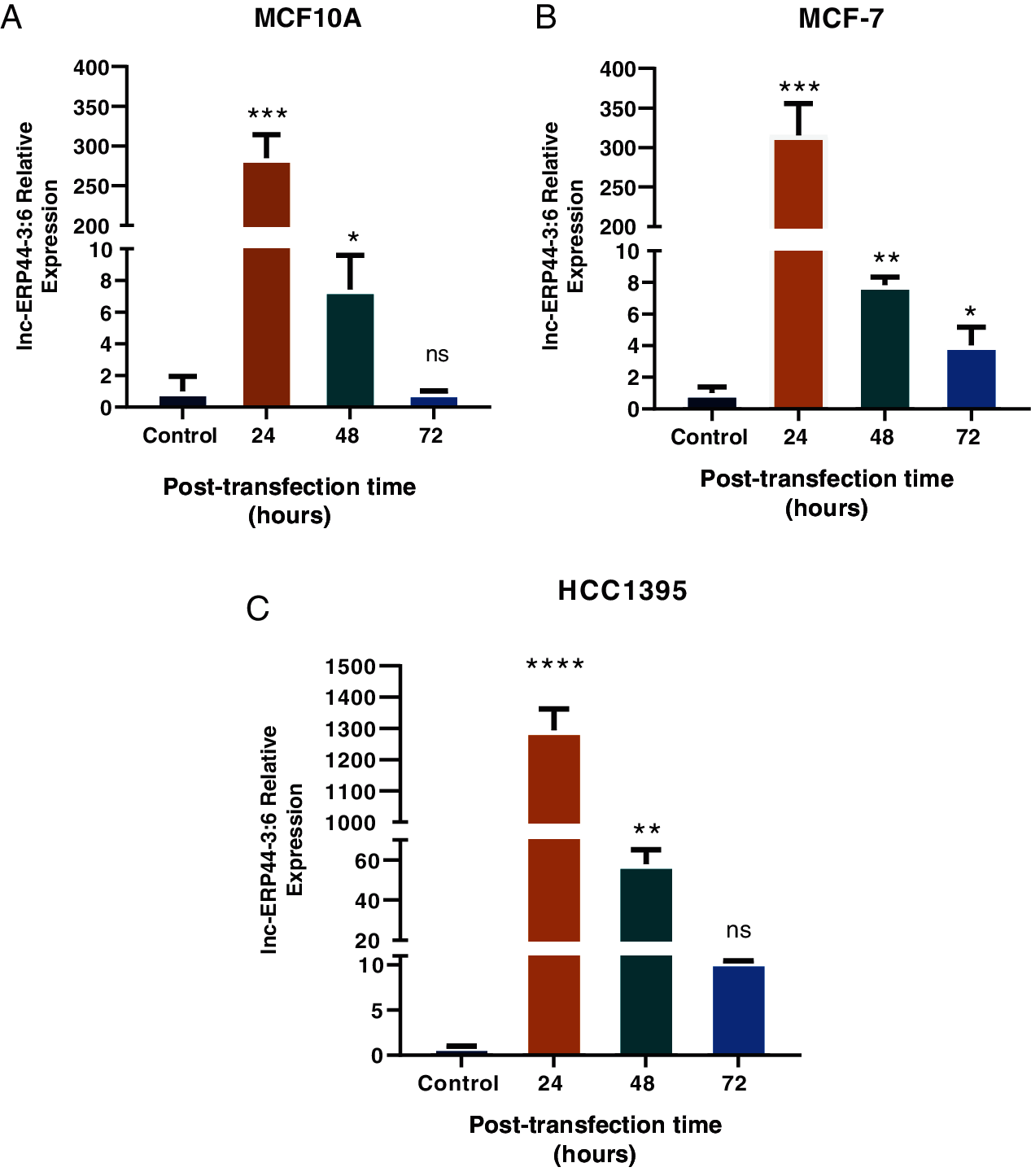
Figure 3: Expression of lnc-ERPP44-3:6 at 24, 48, and 72 h post-transfection with lnc-ERPP44-3:6 and control (empty vector) in (A) MCF10A, (B) MCF-7, and (C) HCC1395 cell lines. One-way ANOVA mean ± S.D. ns: not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001
3.3 Transcripts and Proteins Affected by lnc-ERP44-3:6 Overexpression
In order to characterize the functional role of this lncRNA, we analyzed the effect of the lnc-ERP44-3:6 up-regulation on the expression (at both mRNA and protein levels) of pivotal genes involved in cell proliferation (i.e., PCNA, KI-67, and PIK3CA), EMT (i.e., EpCAM and SNAIL), and oncogenicity (N-RAS) in our representative BC cell lines. Because GAPDH expression was observed to be altered by overexpressing lnc-ERP44-3:6, we used CCSER2 as an internal control for our RT-qPCR experiments. MCF10A, MCF-7, and HCC1395 cell lines showed the highest expression—at mRNA level—of GAPDH (****p < 0.0001), EpCAM (****p < 0.0001), and SNAIL (****p < 0.0001) at 48 h post-transfection with lnc-ERP44-3:6, whereas no significant changes in expression were observed in the remaining genes (Fig. S2 and 4A). The results were verified by immunoassays at 48 h post-transfection with lnc-ERP44-3:6 in MCF10, MCF-7, and HCC1395 cell lines, as we detected altered the expression (up-regulated) of GAPDH (**** p < 0.0001), EpCAM (*p ≤ 0.05), and SNAIL (**p ≤ 0.01) proteins in contrast to control cell lines. We also observed at 48 h post-transfection, that in the MCF-7 cell line both the KI-67 (*p < 0.05) and PCNA (*p < 0.05) proteins were overexpressed, whereas in the HCC1395 cell line, the expression of KI-67 (*p < 0.05), PIK3CA (*p < 0.05) and N-RAS (*p < 0.05) was up-regulated compared to the control cell line. In contrast, no significant changes in protein expression were observed for the remaining proteins (Fig. 4B).
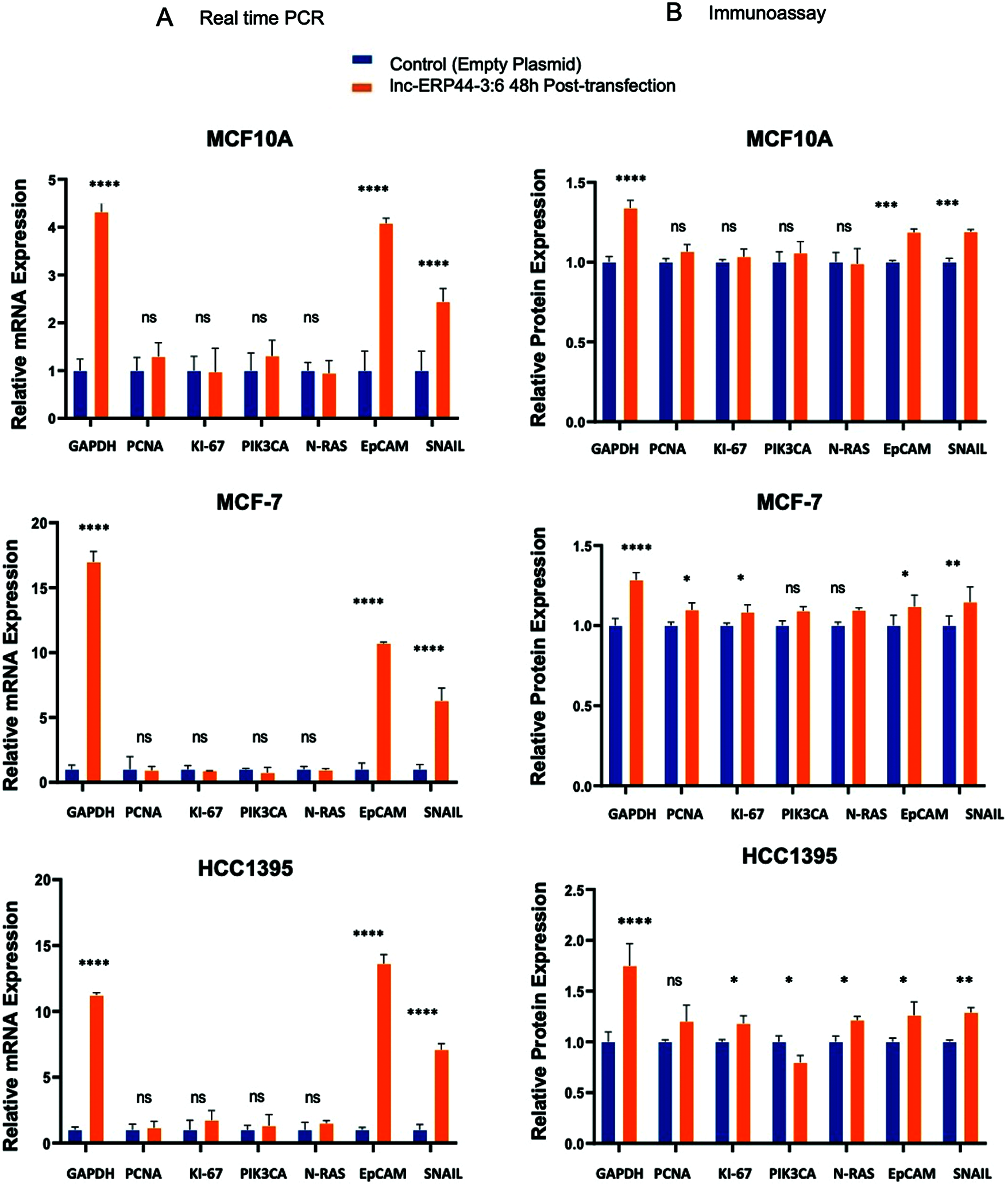
Figure 4: (A) mRNA and (B) proteins expression of GAPDH, PCNA, KI-67, PIK3CA, N-RAS, EpCAM and SNAIL in MCF10A, MCF-7 and HCC1395 cell lines at 48 h post-transfection with control vector and lnc-ERP44-3:6. Two-way ANOVA, mean ± S.D. ns: not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001
3.4 lnc-ERP44-3:6 Overexpression Promotes Cell Death and Chemosensitivity to Cisplatin
At 48 h post-transfection, MCF10A, MCF-7, and HCC1395 cell lines showed a significant decrease in cell viability and an increase in apoptosis rate compared to the control cell lines (p < 0.01) (Figs. 5A and 5B). After that, MCF10A, MCF-7, and HCC1395 cell lines, at 72 h post-transfection with lnc-ERP44-3:6, showed an increase in cell viability compared to 48 h post-transfection, which is consistent with a loss of lnc-ERP44-3:6 expression at this time point (Fig. 5A).
Moreover, we used cell lines at 48 h post-transfection to analyze if the increase in the apoptosis ratio after overexpression of lnc-ERP44-3:6 was linked to the activation of the death receptor FAS (cell surface death receptor), which activates the extrinsic apoptosis pathway. Then, we analyze the expression of FAS at the mRNA level in MCF10A, MCF-7, and HCC1395 cell lines by real-time PCR at 48 h post-transfection with lnc-ERP44-3:6 compared to control. We found a significant increase of FAS, which suggests us that FAS probably participates in the apoptosis after overexpression of lnc-ERP44-3:6 (Fig. 6).
Hence, we promptly inquired whether an overexpression of lnc-ERP44-3:6 would stimulate chemosensitivity to cisplatin (Fig. 7). Our results show that the cells treated with cisplatin at different concentrations and 48 h post-transfection with lnc-ERP44-3:6 exhibit a significant decrease in cell viability (Fig. 7A) and a significant increase in the apoptosis ratio (Fig. 7B) when compared to untransfected control cell lines. Indeed, the overexpression of lnc-ERP44-3:6 at 48 h post-transfection promoted chemosensitivity to cisplatin.
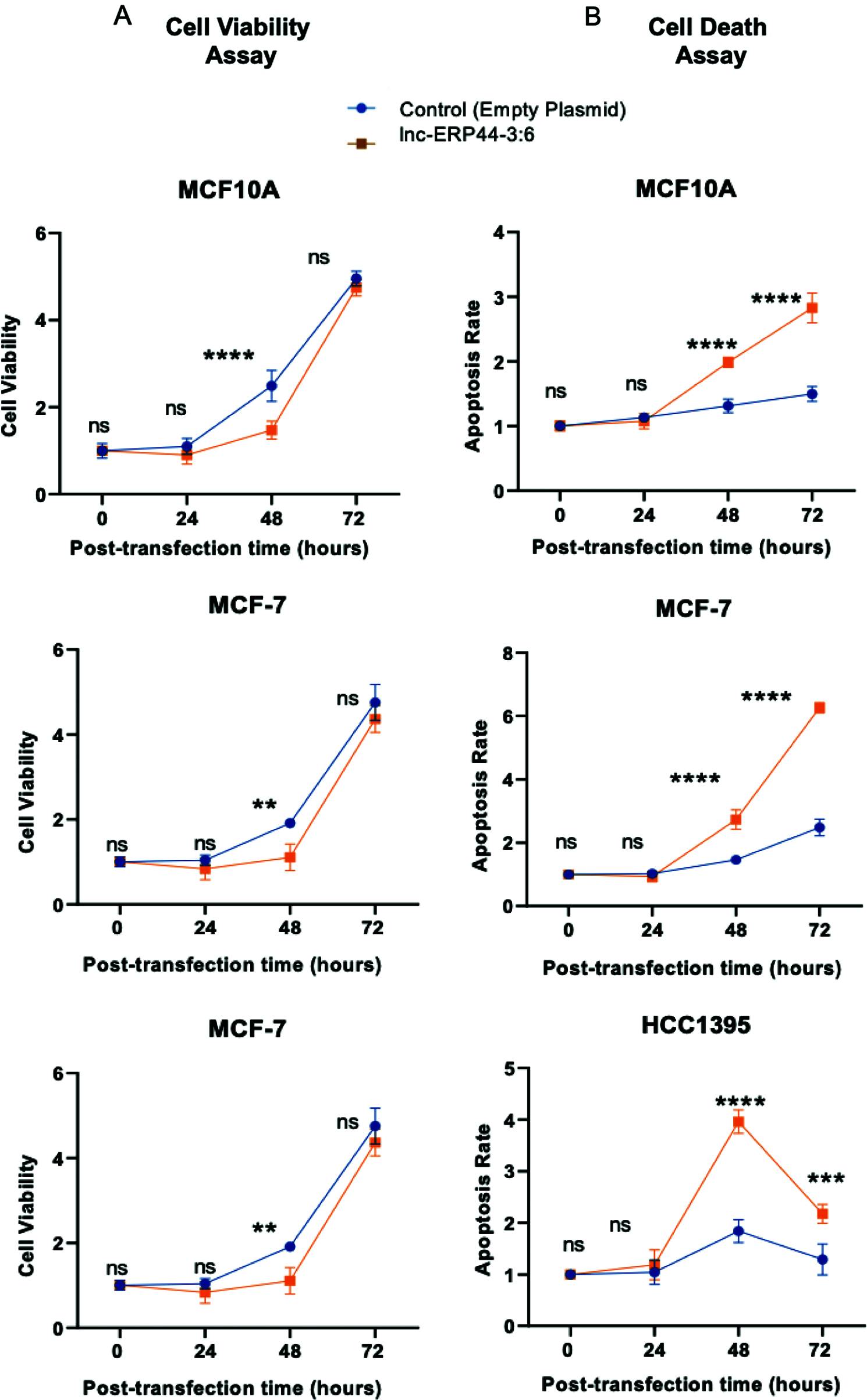
Figure 5: Effect of up-regulation of lnc-ERP44-3:6 at 0, 24, 48 and 72 h post-transfection on (A) cell viability and (B) apoptosis in MCF10A, MCF-7 and HCC1395 cell lines compared with control cell lines. Two-way ANOVA mean ± S.D. ns: not significant, ** p < 0.01, *** p < 0.001, **** p < 0.0001
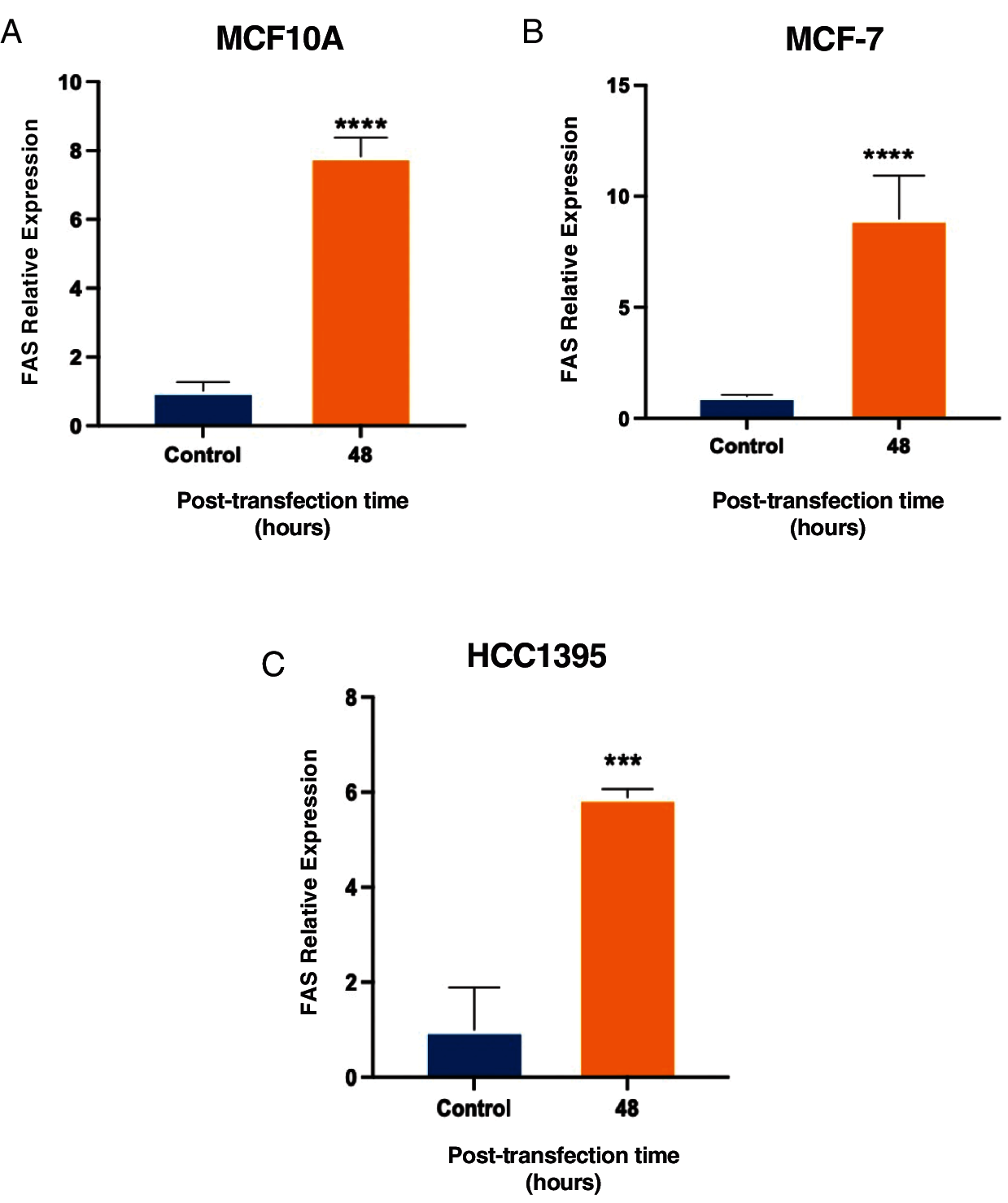
Figure 6: Increase in FAS expression at 48 h post-transfection with lnc-ERP44-3:6 in (A) MCF10A, (B) MCF-7 and (C) HCC1395 cell lines with respect to the control. T-student test mean ± S.D. *** p < 0.001, **** p < 0.0001
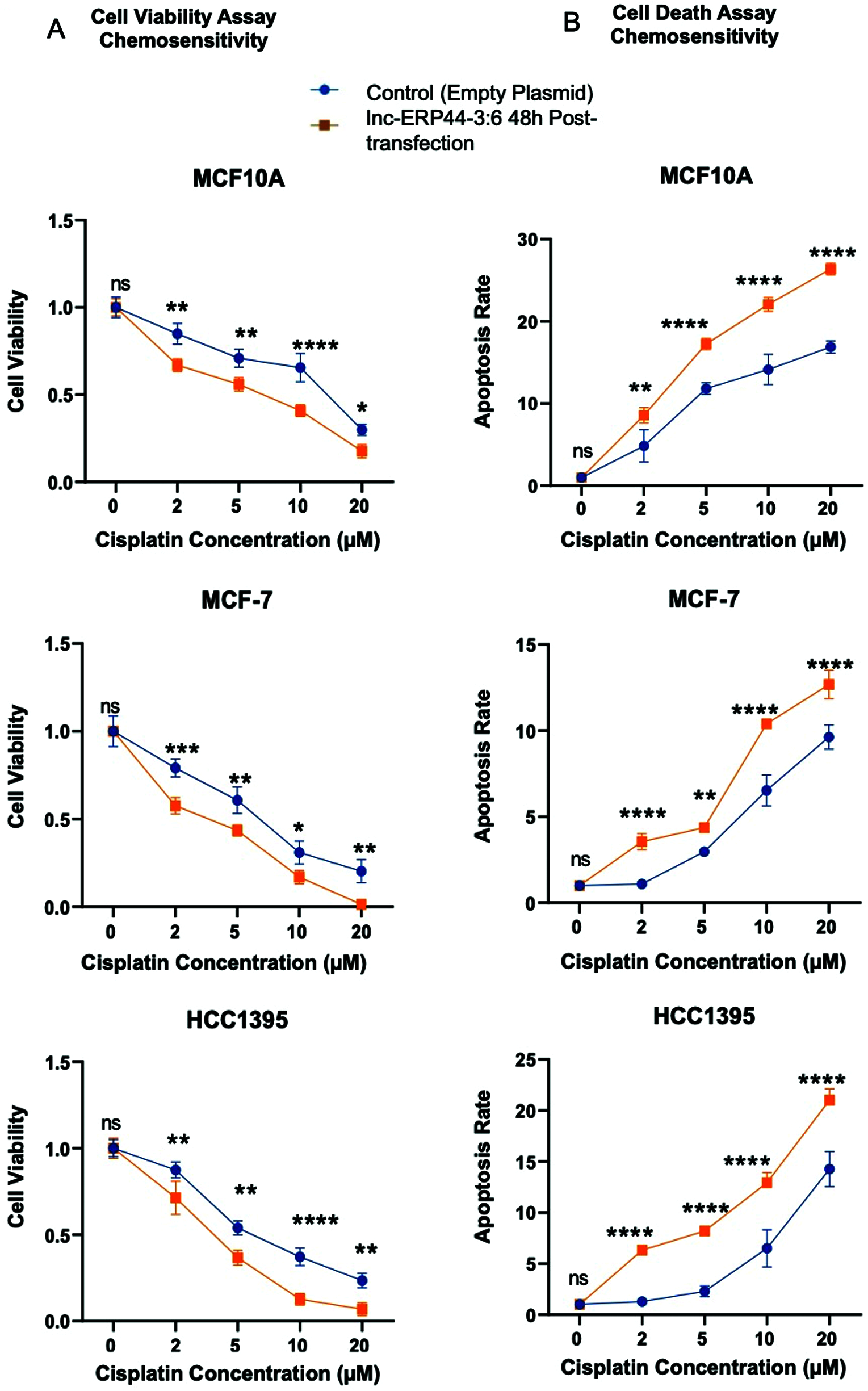
Figure 7: Impact of overexpression of lnc-ERP44-3:6 at 48 h post-transfection on chemosensitivity at different concentrations of cisplatin in MCF10A, MCF-7 and HCC1395 cell lines was evaluated by (A) cell viability and (B) cell death assays. Two-way ANOVA mean ± S.D. ns: not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001
LncRNAs play multiple roles in maintaining cell homeostasis and their aberrant expression in cancer cells has been widely associated with promoting tumor development, metastasis, and resistance to anticancer drugs [22,30]. In this study, we found six DE lncRNAs (i.e., lnc-C9orf131-1, lnc-SLC25A21-2, lnc-GLB1-1, lnc-ERP44-3:6, lnc-BBOX1-1, and lnc-SPRYD3-1) whose targets, pathways, and/or functions are largely unknown [31,32]. Among these, lnc-ERP44-3:6 presented a consistent down-regulated expression in all BC samples compared to their normal adjacent tissue (Figs. 2A–2C). Focusing on this lncRNA, we confirmed a decreased expression pattern ranging from non-significant to significant in BC cell lines here evaluated (i.e., MCF-/luminal, BT4-74/luminal B, MDA-MB-453/HER2+, and HCC1395/TNBC) compared to the non-tumoral one (i.e., MCF10A), which is partially compatible with the observed in our patients’ samples. Of note, the TNBC cell line exhibited the lowest expression of lnc-ERP44-3:6 (Fig. 2D). Moreover, our results agree with those mentioned by Khalil and Markowitz in their patent, in which states (though without providing additional details and/or references) that linc-ALG2-5 (gene expressing lnc-ERP44-3:6) is down-regulated in BC [33]. We also demonstrated that overexpression of lnc-ERP44-3:6—induced by CMV-GFP vector—reduces cell viability, promotes cell death, and enhances the chemosensitivity to cisplatin in MCF10A, MCF-7, and HCC1395 cell lines at 48 h post-transfection. Thus, our findings suggest that lnc-ERP44-3:6 could be a potential universal pro-apoptotic lncRNA, that is, it may act in both pathological and non-pathological states. Partially supporting this scenario, lnc-ERP44-3:6 presents certain pro-apoptotic characteristics seen in other lncRNAs such as GAS5 [34] and SNORD3A [22], including a low expression in tumor tissue to evade apoptosis, as well as that when overexpressed (by induction) in cancer cell lines, promote chemosensitivity and cell death [35]. These findings and observations underscore the value of lnc-ERP44-3:6 as a potential therapeutic target in BC, particularly in the TNBC subtype.
Although we did not evaluate the transcriptome of BC cells overexpressing lnc-ERP44-3:6, we obtained interesting expression data from some genes related to cell growth, motility, angiogenesis, and immune escape (i.e., N-RAS and/or PIK3CA), EMT (i.e., EpCAM and/or SNAIL), cell proliferation (i.e., KI-67 and PCNA) chemotherapeutic resistance (i.e., SNAIL), and apoptosis (i.e., GAPDH and FAS). In this regard, we identified two key events from our experiments: apoptosis and chemosensitivity. The more plausible explanation about lnc-ERP44-3:6 overexpression-related apoptosis could be linked to GAPDH and/or FAS, which were up-regulated in the cells here evaluated. For the former, Tarze et al. [36] observed that the exogenous addition of GAPDH or a high concentration of this protein induced mitochondrial membrane permeabilization, causing activation of the intrinsic apoptosis pathway and releasing of the pro-apoptotic proteins Cytochrome C and AIF (apoptosis-inducing factor). Similarly, FAS is involved in the extrinsic apoptosis pathway activating the initiator caspase 8, which spreads the apoptosis signal by directly processing downstream effector caspases such as caspase-3 [37,38]. Supporting these hypotheses, we indirectly observed a significant increase in the activity of caspase-3 and caspase-7 downstream effectors of both intrinsic and extrinsic apoptosis pathways [37,39] through a high apoptosis rate induced at 48 h post-transfection with lnc-ERP44-3:6 in the cell death assays (Fig. 5). On the other hand, the increased chemosensitivity to cisplatin observed in the cell lines that overexpressed lnc-ERP44-3:6 (Fig. 7), may be due to a “double pro-apoptotic hit” as the cytotoxic mechanism of cisplatin also induce the activation of both intrinsic and extrinsic apoptosis pathways [40].
As the cancer cells are in a constant internal fight between cell death and survival, they undergo multiple chaotic genomic and transcriptomic changes. For this reason, we often observe unpredictable changes and even opposite or dual roles from multiple genes in cancer cells exposed or not to drugs [41–45]. Here, we observed up-regulation of SNAIL and EpCAM at both mRNA and protein levels. SNAIL and EpCAM proteins are involved in promoting chemoresistance, aggressiveness, and progression in cancer cells [46,47]. Although we currently unknown the exact mechanisms and meaning of their overexpression, we speculate that they could have increased in response to the pro-apoptotic factors seemingly induced by the lnc-ERP44-3:6 overexpression (and probably cisplatin exposure). We envisage a similar scenario for KI-67, N-RAS, and PCNA, which were significantly (p < 0.05) up-regulated—but only at protein level—at 48 h post-transfection in the MCF-7 and/or HCC1395 cell lines compared to the control (Fig. 4). This apparent inconsistency between mRNA and protein levels of KI-67, N-RAS, and PCNA (Fig. 4) in such cell lines perhaps was due to a strong selective pressure to maintain the protein abundance even though the mRNA abundance diverges [48]. In this regard, protein expression levels depend on protein stability, translation, and post-transcriptional processes, but between 20–60% depend on mRNA levels, which may vary between organisms and conditions [48–51]. However, further studies would be needed to confirm these assumptions and ascertain if the expression patterns observed in our experiments actually depend on the exposure time to the pro-apoptotic factors, since we only evaluated an exposure to lnc-ERP44-3:6 overexpression of 48 h.
Within the limitations of our study, we are aware that remains to be evaluated the transcriptome of BC cells overexpressing lnc-ERP44-3:6, the effects of a stable overexpression of this lncRNA (as opposed to transient overexpression), as well as assess the expression patterns of DE genes in our experiments but at different times and after combining lnc-ERP44-3:6 overexpression and cisplatin. Nevertheless, our current work represents a preliminary framework to continue studying the pathways and/or functions of this and other lncRNAs found DE in the BC tissues from patients of our population.
In this study, we identified the down-regulation of lnc-ERP44-3:6 in BC tissues compared with their corresponding non-cancerous mammary tissues. According to our results of overexpression, lnc-ERP44-3:6 is a potential pro-apoptotic lncRNA. Furthermore, we found that combining lnc-ERP44-3:6 overexpression with cisplatin enhances the chemosensitivity of BC cells to this drug. Strikingly, we observed an exacerbated pro-apoptotic effect induced by this lncRNA on the TNBC cell line. We also found that GAPDH and FAS were up-regulated by lnc-ERP44-3:6 transient overexpression. We propose that the lnc-ERP44-3:6 overexpression-related apoptosis could be linked to one or both of these genes, which can activate both intrinsic and extrinsic apoptotic pathways. Supporting this, we indirectly observed a significant increase in the activity of downstream effectors caspase-3 and caspase-7 of both apoptosis pathways. To the best of our knowledge, this is the first study evaluating the overexpression of lnc-ERP44-3:6 and providing insights about its role in BC cells, which suggests that this lncRNA may be an interesting candidate as a potential therapeutic target for BC in our population. However, further studies would be needed to better understand the mechanisms by which an overexpression of lnc-ERP44-3:6 causes both cell death and chemosensitivity to cisplatin, as well as refine the pathways and/or functions of this and other lncRNAs.
Acknowledgement: The authors thank CONACYT for the scholarship (Scholarship No. 631205, for Elda A. Flores-Contreras). Likewise, we thank Sonia A Lozano-Sepúlveda, Gerardo Padilla, and Juan Francisco Islas for their support on technical and statistical issues.
Funding Statement: This research was funded by CONACYT (Grants INFRA-2013–204423 and S0008–2014–1-233212 https://www.conacyt.gob.mx/) and PAICYT 2019 (Grant SA761-19 http://investigacion.uanl.mx/paicyt-provericyt/) for Carlos Córdova-Fletes.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Harbeck, N., Gnant, M. (2017). Breast cancer. Lancet, 389(10074), 1134–1150. DOI 10.1016/S0140-6736(16)31891-8. [Google Scholar] [CrossRef]
2. Mondal, P., Meeran, S. M. (2020). Long non-coding RNAs in breast cancer metastasis. Non-Coding RNA Research, 5(4), 208–218. DOI 10.1016/j.ncrna.2020.11.004. [Google Scholar] [CrossRef]
3. Becker, S. (2015). A historic and scientific review of breast cancer: The next global healthcare challenge. International Journal of Gynecology & Obstetrics, 131(S1), S36–S39. DOI 10.1016/j.ijgo.2015.03.015. [Google Scholar] [CrossRef]
4. Elster, N., Collins, D. M., Toomey, S., Crown, J., Eustace, A. J. et al. (2015). HER2-family signalling mechanisms, clinical implications and targeting in breast cancer. Breast Cancer Research and Treatment, 149(1), 5–15. DOI 10.1007/s10549-014-3250-x. [Google Scholar] [CrossRef]
5. Masoud, V., Pagès, G. (2017). Targeted therapies in breast cancer: New challenges to fight against resistance. World Journal of Clinical Oncology, 8(2), 120–134. DOI 10.5306/wjco.v8.i2.120. [Google Scholar] [CrossRef]
6. Nagini, S. (2017). Breast cancer: Current molecular therapeutic targets and new players. Anti-Cancer Agents in Medicinal Chemistry, 17(2), 152–163. DOI 10.2174/1871520616666160502122724. [Google Scholar] [CrossRef]
7. Cheng, Y., He, C., Wang, M., Ma, X., Mo, F. et al. (2019). Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduction and Targeted Therapy, 4(1), 1–39. DOI 10.1038/s41392-019-0095-0. [Google Scholar] [CrossRef]
8. Fahrner, J. A., Eguchi, S., Herman, J. G., Baylin, S. B. (2002). Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Research, 62(24), 7213–7218. [Google Scholar]
9. Guo, M., Peng, Y., Gao, A., Du, C., Herman, J. G. (2019). Epigenetic heterogeneity in cancer. Biomarker Research, 7(1), 23. DOI 10.1186/s40364-019-0174-y. [Google Scholar] [CrossRef]
10. Campos, E. I., Reinberg, D. (2009). Histones: Annotating chromatin. Annual Review of Genetics, 43(1), 559–599. DOI 10.1146/annurev.genet.032608.103928. [Google Scholar] [CrossRef]
11. Flores-Contreras, E., Córdova-Fletes, C. (2019). Overview of long non-coding RNA and its role in breast cancer. Medicina Universitaria, 21(2), 63–73. DOI 10.24875/RMU.19000040. [Google Scholar] [CrossRef]
12. Kouzarides, T. (2007). Chromatin modifications and their function. Cell, 128(4), 693–705. DOI 10.1016/j.cell.2007.02.005. [Google Scholar] [CrossRef]
13. Vicentini, C., Galuppini, F., Corbo, V., Fassan, M. (2019). Current role of non-coding RNAs in the clinical setting. Non-Coding RNA Research, 4(3), 82–85. DOI 10.1016/j.ncrna.2019.09.001. [Google Scholar] [CrossRef]
14. Batista, P. J., Chang, H. Y. (2013). Long noncoding RNAs: Cellular address codes in development and disease. Cell, 152(6), 1298–1307. DOI 10.1016/j.cell.2013.02.012. [Google Scholar] [CrossRef]
15. Kondo, Y., Shinjo, K., Katsushima, K. (2017). Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Science, 108(10), 1927–1933. DOI 10.1111/cas.13342. [Google Scholar] [CrossRef]
16. Lee, J. T. (2012). Epigenetic regulation by long noncoding RNAs. Science, 338(6113), 1435–1439. DOI 10.1126/science.1231776. [Google Scholar] [CrossRef]
17. Soudyab, M., Iranpour, M., Ghafouri-Fard, S. (2016). The role of long non-coding RNAs in breast cancer. Archives of Iranian Medicine, 19(7), 508–517. DOI 0161907/AIM.0011. [Google Scholar]
18. Vikram, R., Ramachandran, R., Abdul, K. S. M. (2014). Functional significance of long non-coding RNAs in breast cancer. Breast Cancer, 21(5), 515–521. DOI 10.1007/s12282-014-0554-y. [Google Scholar] [CrossRef]
19. Marchese, F. P., Raimondi, I., Huarte, M. (2017). The multidimensional mechanisms of long noncoding RNA function. Genome Biology, 18, 206. DOI 10.1186/s13059-017-1348-2. [Google Scholar] [CrossRef]
20. Quinn, J. J., Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nature Reviews Genetics, 17(1), 47–62. DOI 10.1038/nrg.2015.10. [Google Scholar] [CrossRef]
21. Yoon, J. H., Abdelmohsen, K., Gorospe, M. (2013). Post-transcriptional gene regulation by long noncoding RNA. Journal of Molecular Biology, 425(19), 3723–3730. DOI 10.1016/j.jmb.2012.11.024. [Google Scholar] [CrossRef]
22. Luo, L., Zhang, J., Tang, H., Zhai, D., Huang, D. et al. (2020). LncRNA SNORD3A specifically sensitizes breast cancer cells to 5-FU by sponging miR-185-5p to enhance UMPS expression. Cell Death & Disease, 11(5), 1–12. DOI 10.1038/s41419-020-2557-2. [Google Scholar] [CrossRef]
23. Zhang, C., Yu, M., Li, X., Zhang, Z., Han, C. et al. (2017). Overexpression of long non-coding RNA MEG3 suppresses breast cancer cell proliferation, invasion, and angiogenesis through AKT pathway. Tumor Biology, 39(6), 1010428317701311. DOI 10.1177/1010428317701311. [Google Scholar] [CrossRef]
24. Liang, Y., Song, X., Li, Y., Chen, B., Zhao, W. et al. (2020). LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Molecular Cancer, 19, 85. DOI 10.1186/s12943-020-01206-5. [Google Scholar] [CrossRef]
25. https://primer3plus.com/. [Google Scholar]
26. OligoAnalyzer Tool-primer analysis | IDT. (2021). Integrated DNA Technologies. https://www.idtdna.com/pages/tools/oligoanalyzer. [Google Scholar]
27. Amplify4. (2021). https://engels.genetics.wisc.edu/amplify/. [Google Scholar]
28. SPSS Statistics-España | IBM. (2021). https://www.ibm.com/es-es/products/spss-statistics. [Google Scholar]
29. RNAfold Web Server. (2021). http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi. [Google Scholar]
30. Bhan, A., Soleimani, M., Mandal, S. S. (2017). Long noncoding RNA and cancer: A new paradigm. Cancer Research, 77(15), 3965–3981. DOI 10.1158/0008-5472.CAN-16-2634. [Google Scholar] [CrossRef]
31. Mak, A. C. Y., White, M. J., Eckalbar, W. L., Szpiech, Z. A., Oh, S. S. et al. (2018). Whole-genome sequencing of pharmacogenetic drug response in racially diverse children with asthma. American Journal of Respiratory and Critical Care Medicine, 197(12), 1552–1564. DOI 10.1164/rccm.201712-2529OC. [Google Scholar] [CrossRef]
32. Wain, L. V., Vaez, A., Jansen, R., Joehanes, R., van der Most, P. J. et al. (2017). Novel blood pressure locus and gene discovery using genome-wide association study and expression data sets from blood and the kidney. Hypertension. 10.1161/HYPERTENSIONAHA.117.09438. [Google Scholar] [CrossRef]
33. Khalil, A., Markowitz, S. (2017). Diagnostic and therapeutic targeting of DNMT-1 Associated RNA in human cancer. (USA Patent No. 20,170,268,008). USA Patent and Trademark Office. https://patents.google.com/patent/US20170268008A1/en. [Google Scholar]
34. Li, J., Li, L., Yuan, H., Huang, X. W., Xiang, T. et al. (2019). Up-regulated lncRNA GAS5 promotes chemosensitivity and apoptosis of triple-negative breast cancer cells. Cell Cycle, 18(16), 1965–1975. DOI 10.1080/15384101.2019.1635870. [Google Scholar] [CrossRef]
35. Su, Y., Wu, H., Pavlosky, A., Zou, L. L., Deng, X. et al. (2016). Regulatory non-coding RNA: New instruments in the orchestration of cell death. Cell Death & Disease, 7(8), e2333. DOI 10.1038/cddis.2016.210. [Google Scholar] [CrossRef]
36. Tarze, A., Deniaud, A., Le Bras, M., Maillier, E., Molle, D. et al. (2007). GAPDH, a novel regulator of the pro-apoptotic mitochondrial membrane permeabilization. Oncogene, 26(18), 2606–2620. DOI 10.1038/sj.onc.1210074. [Google Scholar] [CrossRef]
37. Aubrey, B. J., Kelly, G. L., Janic, A., Herold, M. J., Strasser, A. (2018). How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death & Differentiation, 25(1), 104–113. DOI 10.1038/cdd.2017.169. [Google Scholar] [CrossRef]
38. Fulda, S., Debatin, K. M. (2006). Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene, 25(34), 4798–4811. DOI 10.1038/sj.onc.1209608. [Google Scholar] [CrossRef]
39. Kulbacka, J., Saczko, J., Chwilkowska, A., Choromańska, A., Skołucka, N. (2012). Apoptosis, free radicals and antioxidant defense in antitumor therapy. Antioxidant enzyme. IntechOpen, 265–302. DOI 10.5772/50357. [Google Scholar] [CrossRef]
40. Tchounwou, P. B., Dasari, S., Noubissi, F. K., Ray, P., Kumar, S. (2021). Advances in our understanding of the molecular mechanisms of action of cisplatin in cancer therapy. Journal of Experimental Pharmacology, 13, 303–328. DOI 10.2147/JEP.S267383. [Google Scholar] [CrossRef]
41. Nitulescu, G. M., van de Venter, M., Nitulescu, G., Ungurianu, A., Juzenas, P. et al. (2018). The Akt pathway in oncology therapy and beyond (Review). International Journal of Oncology, 53(6), 2319–2331. DOI 10.3892/ijo.2018.4597. [Google Scholar] [CrossRef]
42. Sun, B., Dong, C., Lei, H., Gong, Y., Li, M. et al. (2019). Knockdown of inhibitor of differentiation 1 suppresses proliferation and induces apoptosis by inactivating PI3K/Akt/mTOR signaling in hemangioma-derived endothelial cells. Biomedicine & Pharmacotherapy, 111, 236–243. DOI 10.1016/j.biopha.2018.12.072. [Google Scholar] [CrossRef]
43. Vu, H. L., Aplin, A. E. (2016). Targeting mutant NRAS signaling pathways in melanoma. Pharmacological Research, 107, 111–116. DOI 10.1016/j.phrs.2016.03.007. [Google Scholar] [CrossRef]
44. Liu, K., Tang, Z., Huang, A., Chen, P., Liu, P. et al. (2017). Glyceraldehyde-3-phosphate dehydrogenase promotes cancer growth and metastasis through upregulation of SNAIL expression. International Journal of Oncology, 50(1), 252–262. DOI 10.3892/ijo.2016.3774. [Google Scholar] [CrossRef]
45. Henderson, V., Smith, B., Burton, L. J., Randle, D., Morris, M. et al. (2015). Snail promotes cell migration through PI3K/AKT-dependent Rac1 activation as well as PI3K/AKT-independent pathways during prostate cancer progression. Cell Adhesion & Migration, 9(4), 255–264. DOI 10.1080/19336918.2015.1013383. [Google Scholar] [CrossRef]
46. Tayama, S., Motohara, T., Narantuya, D., Li, C., Fujimoto, K. et al. (2017). The impact of EpCAM expression on response to chemotherapy and clinical outcomes in patients with epithelial ovarian cancer. Oncotarget, 8(27), 44312–44325. DOI 10.18632/oncotarget.17871. [Google Scholar] [CrossRef]
47. Ma, S. Y., Park, J. H., Jung, H., Ha, S. M., Kim, Y. et al. (2017). Snail maintains metastatic potential, cancer stem-like properties, and chemoresistance in mesenchymal mouse breast cancer TUBO‐P2J cells. Oncology Reports, 38(3), 1867–1876. DOI 10.3892/or.2017.5834. [Google Scholar] [CrossRef]
48. Laurent, J., Vogel, C., Kwon, T., Craig, S. A., Boutz, D. R. et al. (2010). Protein abundances are more conserved than mRNA abundances across diverse taxa. Proteomics, 10(23), 4209–4212. DOI 10.1002/pmic.201000327. [Google Scholar] [CrossRef]
49. Anderson, L., Seilhamer, J. (1997). A comparison of selected mRNA and protein abundances in human liver. Electrophoresis, 18(3–4), 533–537. DOI 10.1002/elps.1150180333. [Google Scholar] [CrossRef]
50. Lu, P., Vogel, C., Wang, R., Yao, X., Marcotte, E. M. (2007). Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nature Biotechnology, 25(1), 117–124. DOI 10.1038/nbt1270. [Google Scholar] [CrossRef]
51. Schrimpf, S. P., Weiss, M., Reiter, L., Ahrens, C. H., Jovanovic, M. et al. (2009). Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biology, 7(3), e48. DOI 10.1371/journal.pbio.1000048. [Google Scholar] [CrossRef]
Appendix A
Appendix B
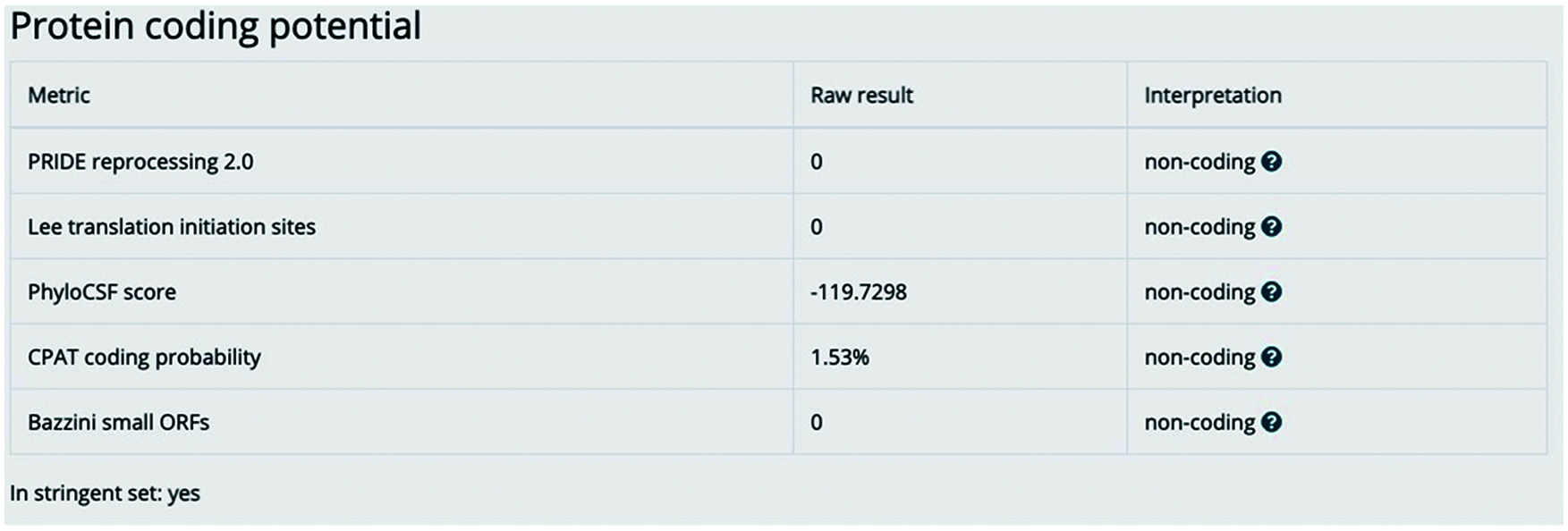
Figure S1: lnc-ERP44-3:6 has no encoding potential. The image represents the bioinformatic analysis by the LNCipedia version 5.2 database to determine the non-coding potential of lnc-ERP44-3:6 [1]
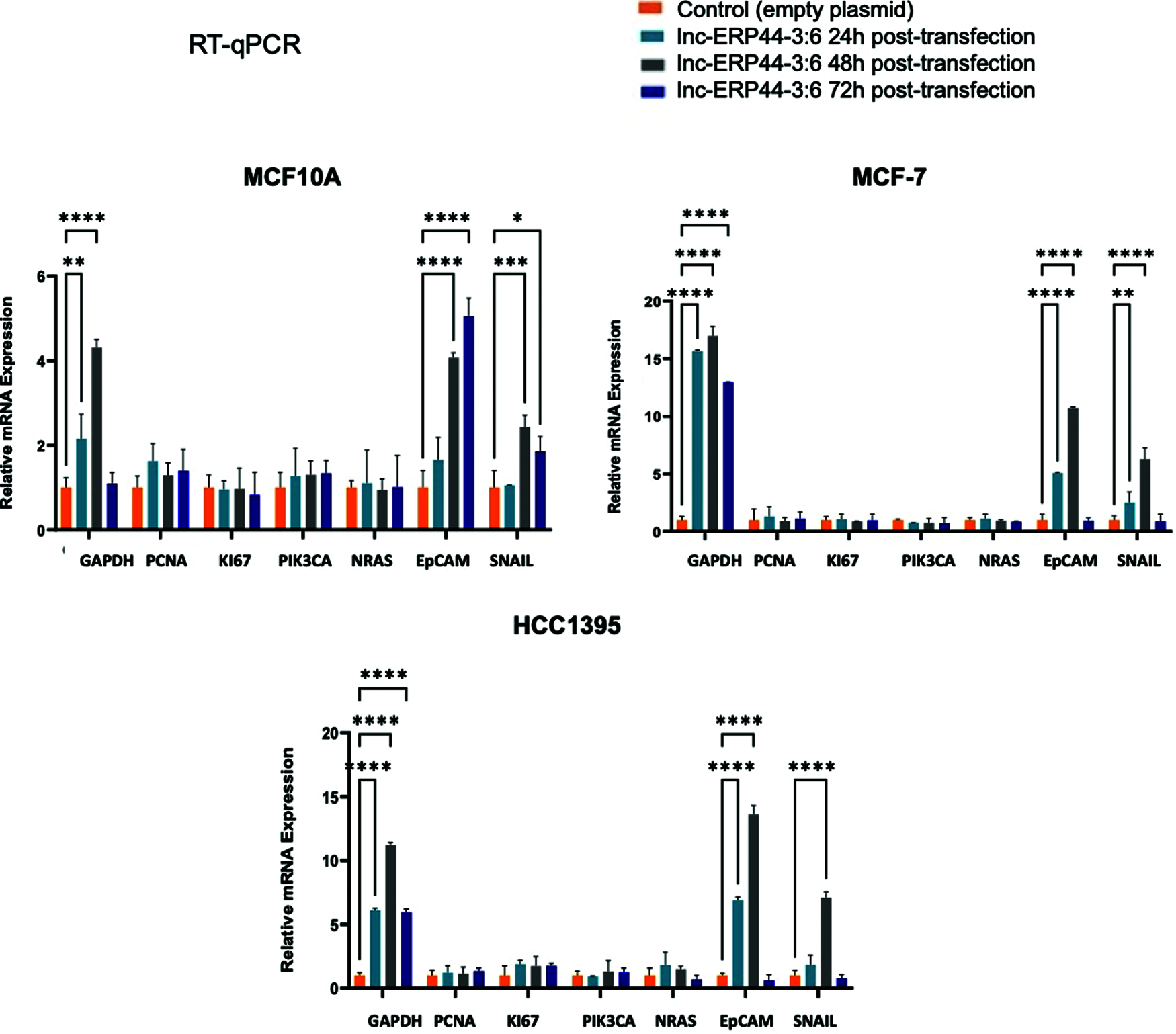
Figure S2: Expression pattern at 24, 48 and 72 h post-transfection of genes involved in BC. The relative mRNA expression of GAPDH, PCNA, KI-67, PIK3CA, N-RAS, EpCAM and SNAIL was measured in MCF10A, MCF-7 and HCC1395 cell lines at 24, 48 and 72 h post-transfection with control vector and lnc-ERP44-3:6. Two-way ANOVA, mean ± S.D. ns: not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001
References
1. Volders, P. J., Anckaert, J., Verheggen, K., Nuytens, J., Martens, L. et al. (2019). LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Research, 47(D1D135–D139. DOI 10.1093/nar/gky1031
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |