
 | Oncologie |  |
DOI: 10.32604/Oncologie.2021.016958
ARTICLE
PIWI Interacting RNA-651 Inhibition Transforms the Genetic Features of MCF-7 Breast Cancer Cells
1Medical Faculty, Department of Medical Biology and Genetics, Maltepe University, İstanbul, 34857, Turkey
2Medical Faculty, Departfiment of Biostatistics, Eskişehir Osmangazi University, Eskişehir, 26040, Turkey
*Corresponding Author: Çağrı Öner. Email: cagri.oner@maltepe.edu.tr
Received: 14 April 2021; Accepted: 22 June 2021
Abstract: piRNAs are novel members of small non-coding RNAs and have an impact on genetic and epigenetic mechanisms of cells. It was aimed to investigate the role of piR-651 on MCF-7 benign breast cancer cells by focusing on molecular characteristics. Anti-piR-651 was transfected and effects of piR-651 on proliferation, adhesion, and motility of MCF-7 cells were detected after the 24th, 48th, and 72nd hour. Gene expressions of piR-651, Ki-67, MMP-2, ERα, HIF-1α, and hTERT were determined by using RT-PCR. piR-651 inhibition caused the decrease of proliferation, adhesion (p < 0.001), and motility of MCF-7 cells. The efficiency of anti-piR-651 transfection supported the determination of the decrease of piR-651 expression after transfection after the 48th hour (p < 0.001). Anti-piR-651 transfection caused the downregulation of Ki-67, MMP-2, ERα, HIF-1α, and hTERT gene expressions after the 48th hour (p < 0.001). In MCF-7 cells, piR-651 inhibition can change both cellular characteristics and gene expressions which were related to these characteristics. piR-651 inhibition causes the decrease of both proliferation and adhesion of cells, which are especially important cellular marks of MCF-7 cells. These results have shed light on whether we can mitigate the effects of cancer cells via piRNA inhibition. The absence of piRNA expression caused the change of fate of benign breast cancer cells by decreasing malign features of MCF-7 cells.
Keywords: Adhesion; breast cancer; motility; PIWI interacting RNAs; piR-651; proliferation
Breast cancer is the most common cancer type in women and can be classified according to estrogen dependence. MCF-7 breast cancer cells are estrogen positive (especially ERα), benign, and adhesive breast cancer cells. It is known that high ERα expression is especially observed in hormone-dependent breast cancer cells [1,2]. According to the general characterization of cancer cells, cancer cells can have high proliferation rates and telomerase activity can modulate adhesion, migration, and invasion mechanisms, and can differentiate during carcinogenesis [3,4]. Recent studies show that ER is very important for controlling the development of ER-positive breast cancer cases [5]. ERα and ERβ are the types of ER in cells and studies have indicated that ERα modulates cellular mechanisms [5]. ERα acts as a ligand-activated oncogene and the upregulation of ERα in breast cancer causes more aggressive mannerisms [6,7].
PIWI-interacting RNAs (piRNAs) are the novel and the longest members of small non-coding RNAs. The main aim of piRNAs is to protect the genome against transposons [8]. They can regulate the cellular characteristics via epigenetic and genetic mechanisms by using transposable elements [9]. Transposable elements as mobile factors in the genome are widely distributed among all regions, causing deletions, gene breakdowns, and chromosomal regulations [10], and also lead to cancer initiation and development [11,12]. Furthermore, piRNAs can inhibit transcription and post-transcriptional mechanisms epigenetically [13,14]. Although initial research into piRNAs focused on germline and stem cells, the similarities of these cells and cancer cells canalized research to studying the relationship between cancer cells and piRNAs [14,15]. Key features of piRNAs are preserving DNA integrity, epigenetic regulation, differentiation during embryogenesis, and the occurrence and development of diseases [16]. piRNAs might have oncogenic or tumor suppressor functions in various cancers [16]. Some piRNAs, including piR-651, piR-20365, piR-4987, piR-20485, and piR-20582, are upregulated in breast cancer tissues and cell lines [17–19]. piR-651 expression especially studied in gastric cancer and expression pattern was shown as an oncogene previously [20]. piR‑651 can be upregulated and can be an oncogene in carcinogens of several cancer tissues and cell lines [21]. The upregulation of piR‑651 was detected in gastric, colon, lung, and breast cancer tissues and cell lines, compared with non‑cancerous tissues [21]. Furthermore, piR-651 upregulation has been demonstrated in cell lines of mesothelium, cervix, and liver cancer cell lines [17,18,22].
Ki-67 is the nucleus core protein that can direct the proliferation of cells, especially tumor cells [23]. From grade I breast cancer cells, such as MCF-7, to grade IV breast cancer cells, Ki-67 expression is the basic marker to understand proliferation potential [24]. In cell lines, Ki-67 expression is high in the G2 phase and mitosis of the cell cycle, and the accumulation of a transcription factor, which is called as E2F, is a result of high Ki-67 expression.
Matrix metalloproteinases (MMPs) are important for degrading extracellular matrix and basal membrane molecules. By affecting these structures, MMPs can control the invasion and metastasis in cancer cells [25]. Low enzyme levels of MMPs are identified in a benign type of breast cancer cells, such as MCF-7, compared to the malign types such as MDA-MB-231. MMP-2 is well studied and related to cancer cell migration and invasion [26]. MMP-2 gene expression is observed in early-stage breast cancer and gives the first signatures that are leading to tumor formation [27,28]. MMP-2 expression becomes higher with increasing tumor grade and MMP-2 activity has been found to be a key feature of decreased survival of breast cancer patients [27,29].
Hypoxia-inducible factor 1α (HIF-1α) is a transcription factor and has an important role in expressions of special gene regions, which are related to survival, proliferation, angiogenesis, and metastasis of cancer cells under hypoxic conditions [30]. The hypoxic tumor microenvironment is associated with the development and metastasis of solid tumors. HIF-1α expressions especially indicate the rate of transition from epithelial characteristics to mesenchymal characteristics in cancer cells [31]. Various studies with breast cancer patients have indicated a distinct survival advantage associated with decreased HIF-1α expression which is the regulatory subunit of HIF-1 [32]. During breast carcinogenesis, HIF-1α expression is a signature of cancer development and its expression pattern differentiates according to the grade of breast tumors [33]. HIF‑1α and its target genes expressions upregulate in breast cancer; upregulation of HIF‑1α expression can be used to predict early recurrence, metastasis, and poor clinical outcomes in breast cancer patients [31].
Telomeres are important for cells to protect themselves from DNA damages and recombination against chromosomal instability. In healthy cells, telomere length becomes shorter after each division, although this mechanism cannot be observed in cancerous cells. Human telomerase reverse transcriptase (hTERT) is a component of telomerase and during telomere elongation, hTERT makes it complex with telomerase RNA component (TERC) and other associated proteins. hTERT is the catalytic subunit of telomerase holoenzyme and its expression is crucial for the telomerase activity during carcinogenesis [34,35]. In cancer and stem cells, hTERT expression is upregulated [36]. The mechanisms which have an impact on hTERT expression in breast cancer cells are DNA methylation and hormonal regulation [37–40]. If tumor suppressor genes such as TP53, PTEN, and RB are suppressed and lose their functions in cells, overexpression of hTERT has been found to drive tumorigenesis and promote oncogenic functions [41].
In this study, we aimed to determine the possible molecular and cellular mechanisms of piR-651 in MCF-7 breast cancer cells. This study indicates that piR-651 has an important role in the proliferation, adhesion, and motility of MCF-7 cells. Moreover, as a genetic perspective, Ki-67, MMP-2, ERα, HIF-1α, and hTERT might be targets of piR-651 in non-invasive breast cancer cells. Our obtained data supports and verifies that anti-piR-651transfection cause to transform characteristics of non-invasive breast cancer cells. According to our knowledge, these results are the first results of the molecular mechanisms of piR-651 in breast cancer.
2.1 Cell Culture and Transfection
MCF-7 breast cancer cells were grown in a growth medium which contains Dulbecco’s Modified Eagle Medium (DMEM; Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Biowest, France) and 1% penicillin/streptomycin (Capricorn, Germany) until 5 days before transfection. The growth medium was removed and the experimental medium was used to inhibit the level of estrogen which was secreted from MCF-7 cells. The experimental medium was phenol red-free RPMI 1640 medium (Biowest, France) which was supplemented by 10% dextran-charcoal stripped fetal bovine serum (DC-FBS; Sigma, USA) and 1% penicillin/streptomycin (Capricorn, Germany). 5 days later, cells were ready to transfect with anti-piR-651 and non-target sequences (GenScript, USA) by using lipofectamine (TaKaRa, Japan). A non-target sequence (GenScript, USA) was used as a scrambled sequence and was used to determine the effect of the transfection agent. Anti-piR-651 and non-target sequences are shown in Tab. 1.
Before transfection, transfected sequences (anti-piR-651 and non-target) were mixed and incubated for 15 minutes at room temperature according to the manufacturer protocol of transfection reagent (TaKaRa, Japan). After incubation, anti-piR-651 and non-target sequences were transfected to MCF-7 cells with Opti-MEM (serum-free medium; Gibco, USA). After transfection, cell behavior assays and gene expressions were determined, respectively.

MCF-7 cells were seeded to a 96 well cell culture plate (Greiner, Germany) and transfected with non-target and anti-piR-651 as described previously. We obtained control, non-target, and anti-piR-651 transfected groups after transfections. The proliferation of MCF-7 cells was observed by using the XTT method (Biological Industries, Israel) at the 24th, 48th, and 72nd hour. The absorbance of cells at 450 nm was determined by using a microplate reader (Biotek, Japan).
MCF-7 cells were seeded to a 96 well cell culture plate and then these cells were transfected with non-target and anti-piR-651. As a result of transfections, control, non-target, and anti-piR-651 groups were obtained. Each group had two lines (2 × 7 wells) because one line of each group was washed with dPBS (Sigma, USA) three times before XTT treatment to understand the adhesion characteristics of groups. The adhesion of cells was detected by using the XTT method at the 24th, 48th, and 72nd hour. The absorbance of cells at 450 nm was determined by using a microplate reader (Biotek, Japan).
2.4 Wound Healing (Motility) Assay
The wound healing rate of MCF-7 cells was analyzed in Petri dishes (Greiner, Germany). 2 × 105 transfected cells were suspended in Petri dishes with a medium. After 24 h, the wound areas were generated at the base of the petri dish by the tip of a 1000 μl pipette. The width of wounded areas was measured following incubation for 0, 24, 48, and 72 h. The motility in this area was assessed using measurements performed at 7 different locations for each group and for each hour.
2.5 Total RNA Isolation and Real-Time Polymerase Chain Reaction (RT-PCR)
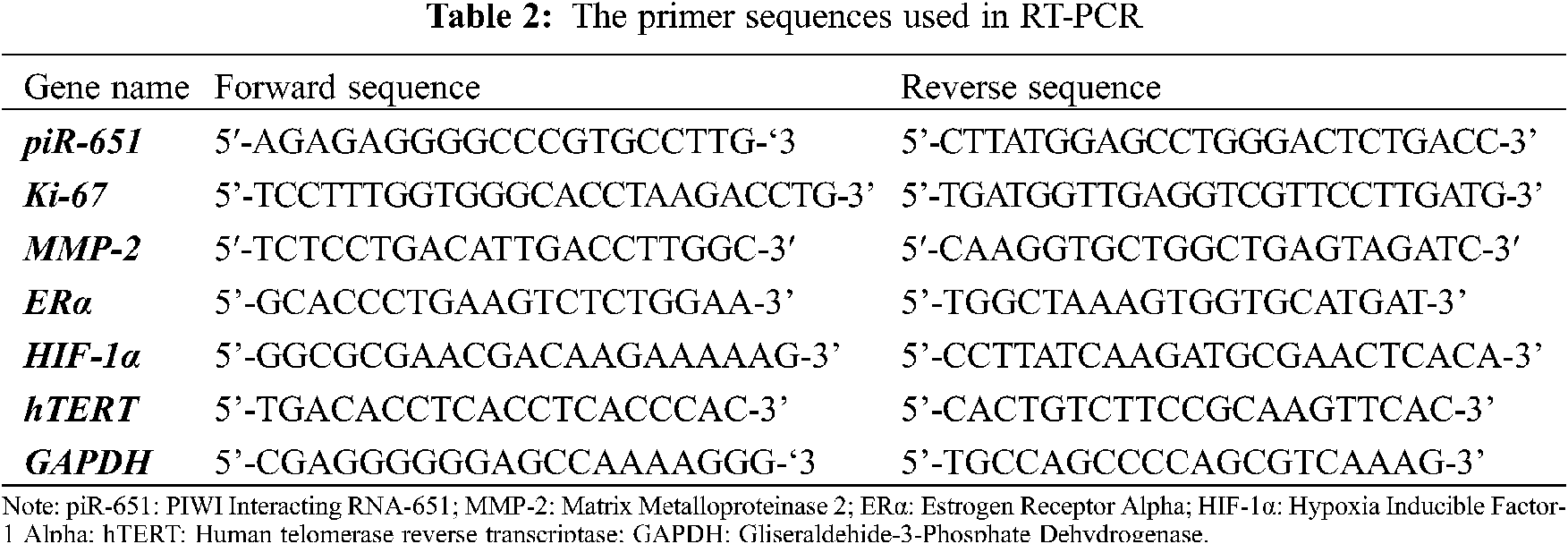
After transfection with non-target and anti-piR-651 sequences, total RNA was isolated according to the manufacturer’s procedure (Nucleospin RNA, Macherey-Nagel, GERMANY). The primers used in RT-PCR were obtained and synthesized from Oligcus, TURKEY, and are shown in Tab. 2. After total RNA isolation, RNAs were converted to cDNA by the reverse transcription mechanism. In the obtained cDNA, piR-651, Ki-67, MMP-2, ERα, HIF-1α, and hTERT expressions, RT-PCR was determined (Lighcycler96, Roche, USA). Gliseraldehide-3-Phosphate Dehydrogenase (GAPDH) was used as an internal control. Gene expressions of experimental groups were calculated by using the ΔΔCT formula.
The normal distribution of the continuous variables was enabled using the Kolmogorov-Smirnov suitability test. Comparisons between groups of normally distributed variables were evaluated using One-Way variance analysis (ANOVA). The Tukey HSD test was used for multiple comparisons of proliferation and adhesion assays. The comparisons between groups of variables that were not normally distributed were evaluated using the Kruskal-Wallis test. Multiple comparisons of gene expressions were compared using the Student t-test. All analyses were carried out using the IBM SPSS Statistics 21.0 software package. The obtained data were indicated as the mean ± standard deviation (sd). In the figures, only mean values have been shown.
To detect the efficiency of transfection, the piR-651 expression of anti-piR-651 transfected cells were compared to the control group. According to the obtained data, piR-651 expression was downregulating as statistically significant in anti-piR-651 transfected MCF-7 cells (–2.05 ± 0.0024) compared to the control group (0.397 ± 0.0017) after 48 h of anti-piR-651 transfection (Fig. 4A; P < 0.001).
3.2 Effect of Anti-piR-651 on the Proliferation of MCF-7 Cells
At the 24th, 48th, and 72nd hour of transfection, the proliferation of MCF-7 cells was determined. According to our obtained data, we could not observe any statistically significant changes at 24th and 72nd hour of anti-piR-651 transfected MCF-7 cells (P > 0.05, Figs. 1A and 1C). On the other hand, the impact of transfection on MCF-7 cells was determined statistically significant at the 48th hour (P < 0.001). The proliferation of anti-piR-651 transfected group (17,830 ± 1,214.68) decreased statistically significantly compared to control (21,665 ± 3,753.29) at the 48th hour (P < 0.001; Fig. 1B).
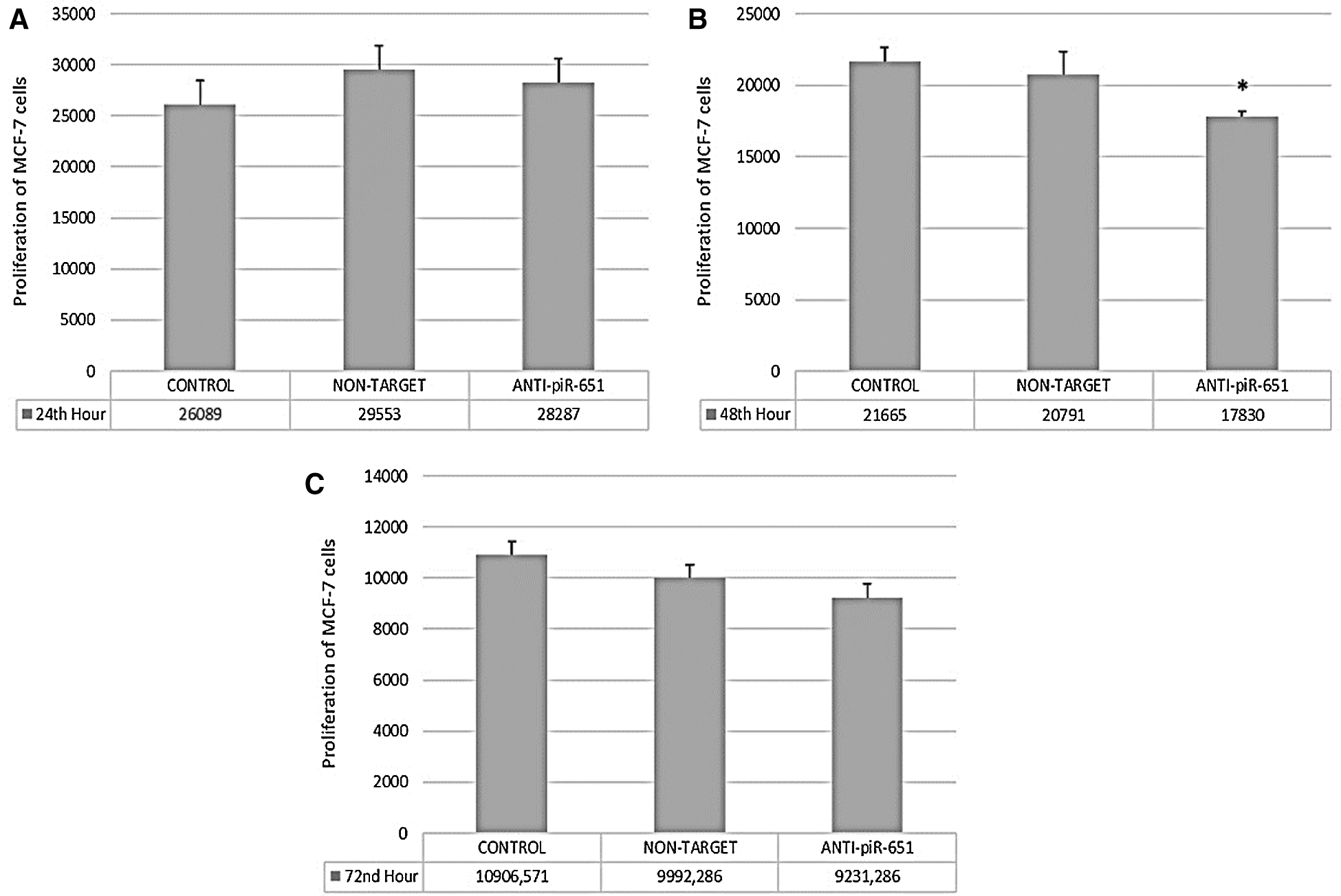
Figure 1: The proliferation of control, non-target and anti-piR-651 transfected MCF-7 cells at 24th, 48th and 72nd hours. A. The proliferation of control, non-target and anti-piR-651 transfected MCF-7 cells at 24th hour (p > 0.05). B. The proliferation control, non-target and anti-piR-651 transfected MCF-7 cells at 48th hour (p < 0.001). C. The proliferation control, non-target and anti-piR-651 transfected MCF-7 cells at 72nd hour (p > 0.05)
3.3 Effect of Anti-piR-651 on the Adhesion of MCF-7 Cells
According to our obtained data, the adhesion of the anti-piR-651 transfected group (0.729 ± 0.06) decreased statistically significantly compared to control (0,996 ± 0.06) after 24 h (P < 0.001; Fig. 2A). The largest decrease in adhesion was observed after 48 h in the anti-piR-651 transfected group (0,662 ± 0.091) compared to control (0,945 ± 0.091; P < 0.001; Fig. 2B). After 72 h, we could not observe any statistically significant difference between groups (P = 0.145; Fig. 2C).
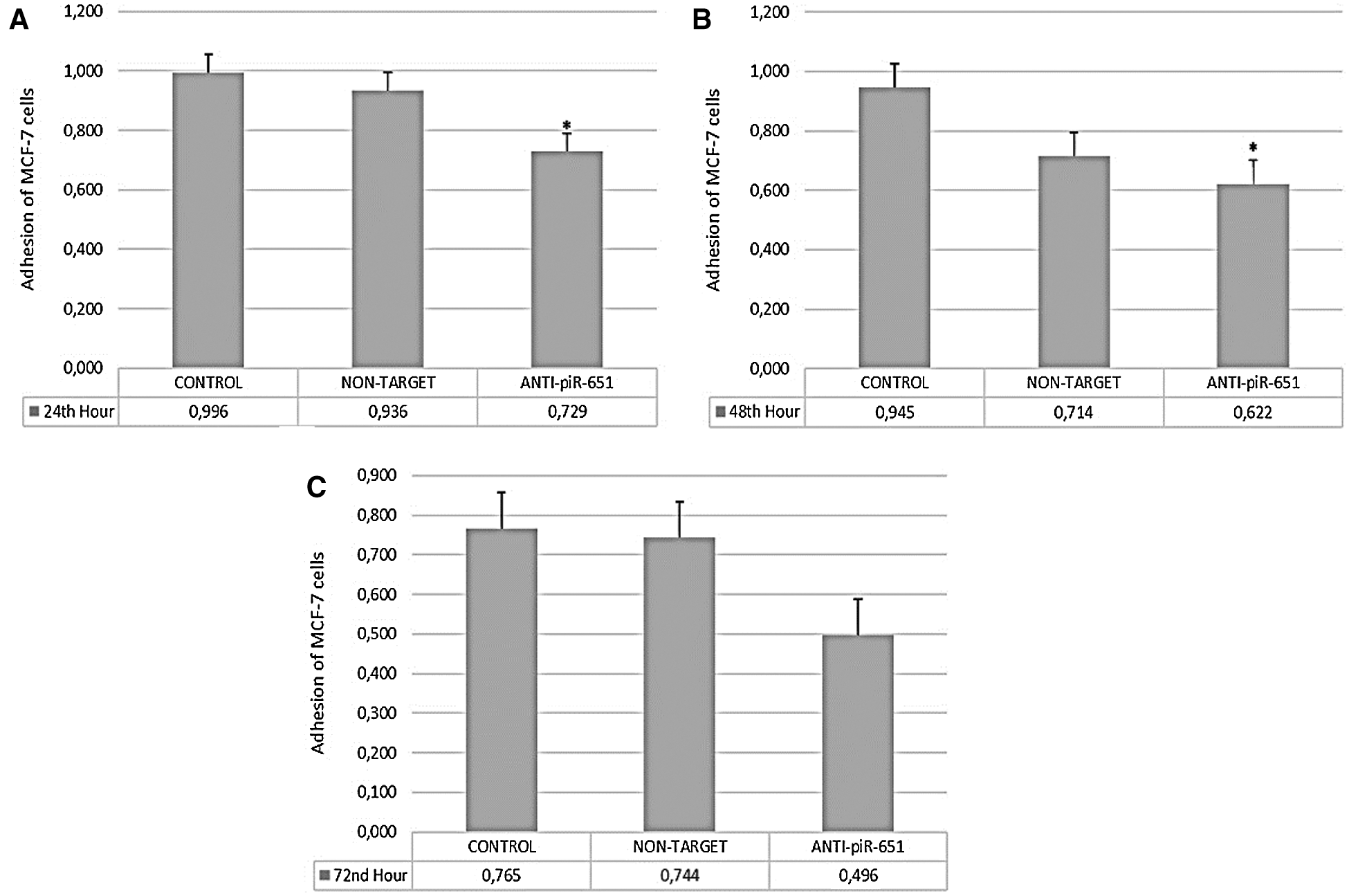
Figure 2: The adhesion of control, non-target and anti-piR-651 transfected MCF-7 cells at 24th, 48th and 72nd hours. A. The adhesion of control, non-target and anti-piR-651 transfected MCF-7 cells at 24th hour (p < 0.001). B. The adhesion of control, non-target and anti-piR-651 transfected MCF-7 cells at 48th hour (p < 0.001). C. The adhesion of control, non-target and anti-piR-651 transfected MCF-7 cells at 72nd hour (p > 0.05)
3.4 Anti-piR-651 Transfection Inhibits Motility of MCF-7 Cells
The wound healing of the control group was faster than non-target and anti-piR-651 transfected groups after 24 h (Fig. 3B). After 48 h, the motility of anti-piR-651 transfected cells was slower than the other groups (Fig. 3C). After 48 h, we could not observe any wound on the areas of non-target and control groups, although there was a wide wound area on the anti-piR-651 transfected MCF-7 cells (Fig. 3C).
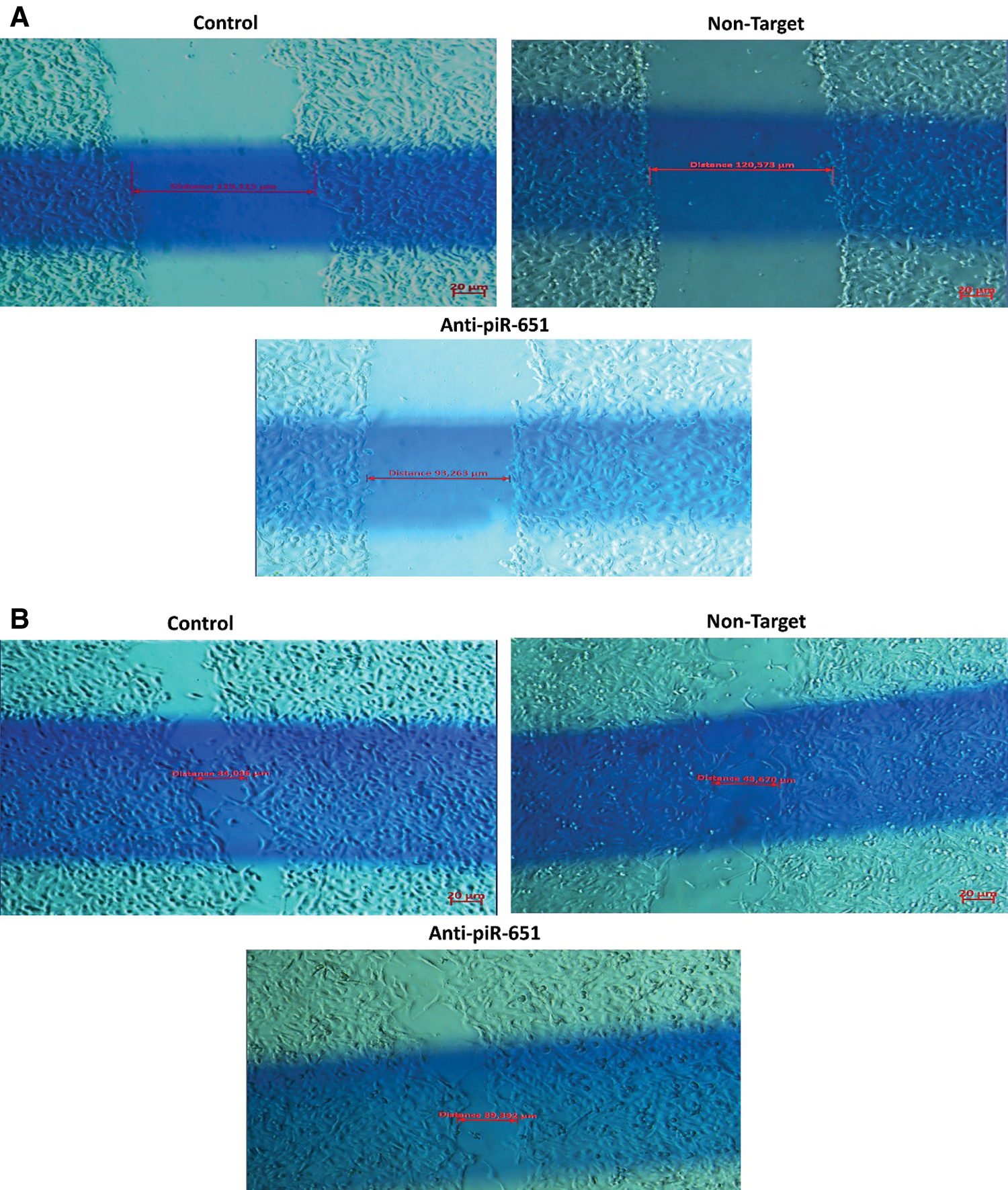
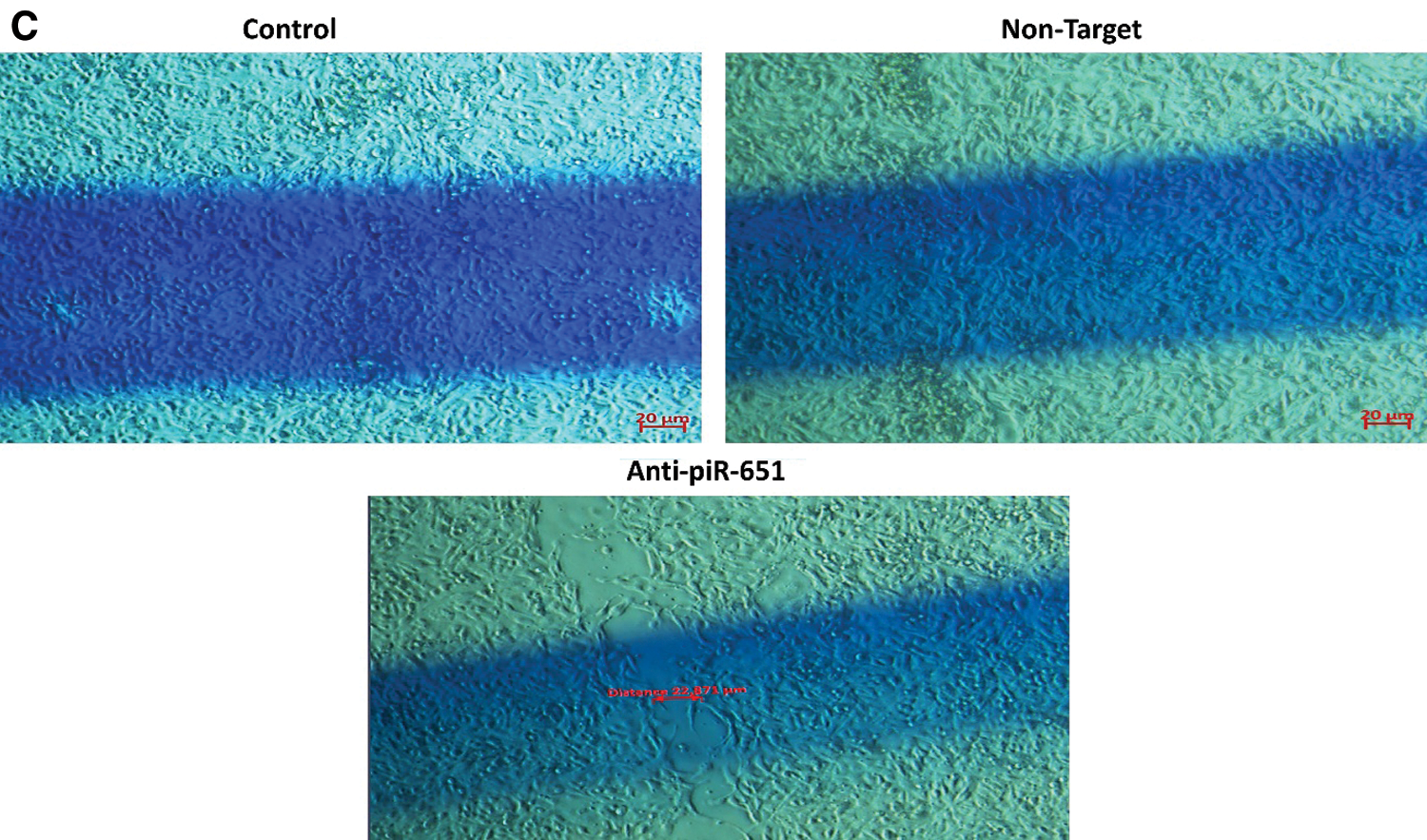
Figure 3: Wound healing images of control, non-target and anti-piR-651 transfected MCF-7 cells at 0, 24th and 48th hours. The wound healing of 48th hour is completely over in control and non-target groups while wound was still detected in anti-piR-651 transfected MCF-7 cells (Magnification Ratio: 20 μm). A indicates 0 hour; B indicates 24th hour; C indicates 48th hour
3.5 piR-651 Inhibition Impact on the Gene Expressions of Which are Related to the Molecular Characteristics of MCF-7 Cells by Using RT-PCR
Inhibition of piR-651 caused the decrease in Ki-67 expression in the transfected group (0,999 ± 0.0025) compared to control (1,21 ± 0.0036; P < 0.001, Fig. 4B). MMP-2 gene expression was decreased in the anti-piR-651 transfected group (–8,67 ± 0.0034) compared to control (–6,09 ± 0.0022; P < 0.001; Fig. 4C). ERα gene expression of the inhibited group (–5.14 ± 0.117) was downregulated compared to the control group (0.14 ± 0.032; P < 0.001; Fig. 4D). The differentiation of cells as a result of anti-piR-651 transfection was evaluated to observe the HIF-1α gene expression. We observed that the HIF-1α gene expression was downregulated in piR-651 inhibited MCF-7 cells (–1.34 ± 0.051) compared to the control (2.65 ± 0.079; P < 0.001; Fig. 4E). hTERT gene expression decreased statistically significantly in inhibited cells (−1.594 ± 0.058) compared to the control (1.517 ± 0.247; P < 0.001; Fig. 4F).
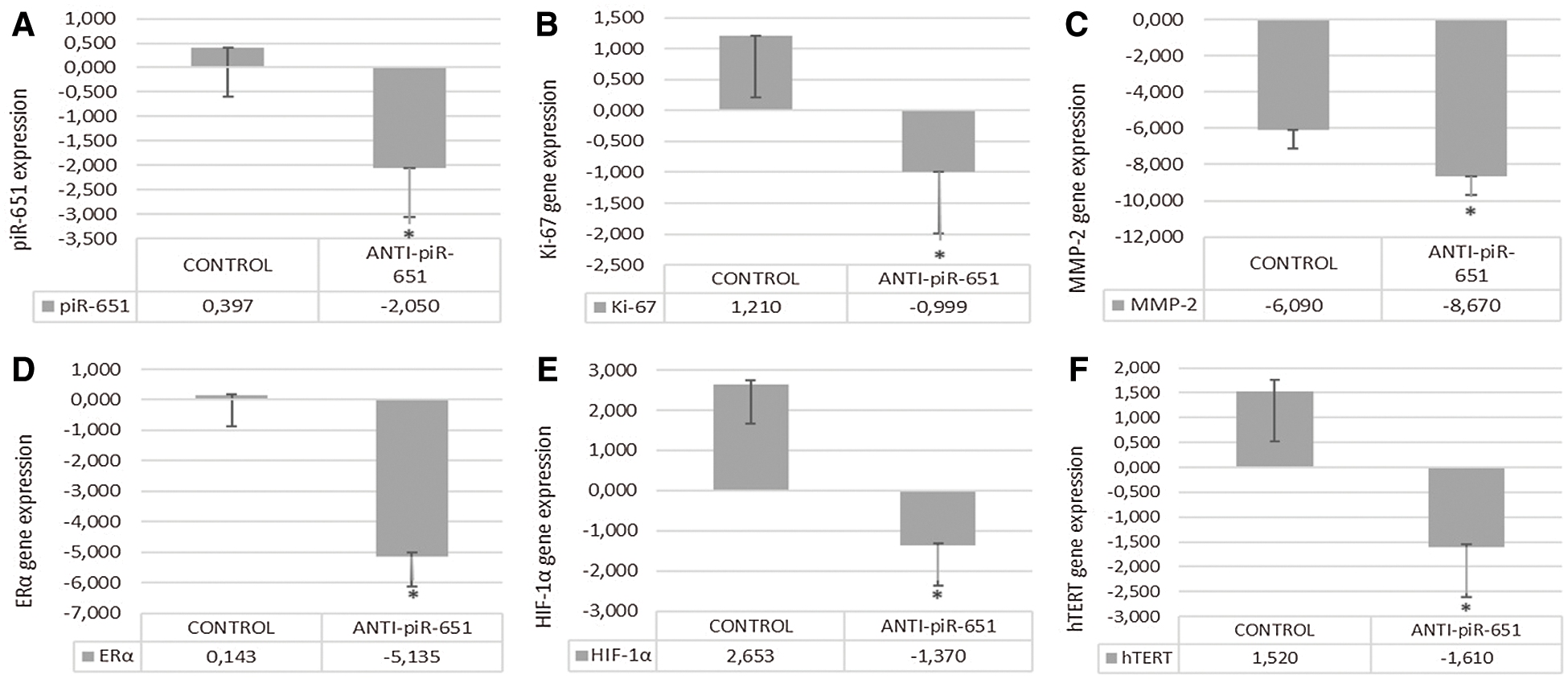
Figure 4: The effect of anti-piR-651 transfection on gene expressions of (A) piR-651; (B) Ki-67; (C) MMP-2; (D) ERα; (E) HIF-1α and (F) hTERT (p < 0.001)
Epigenetic mechanisms, especially hypermethylation, change the expression patterns of some genes which are related to adhesion (cadherins as CDH1, CDH13), DNA repair (BRCA1), proapoptotic functions (HOXA5, TMSF1) or the cell cycle (p16, p53, RASSF1A) in breast cancer [5,42]. piRNAs is a member of the epigenetic mechanisms, which are called non-coding RNAs, and are especially useful in controlling the methylations (DNMTs) and acetylation (HDAC, H3K27me3 methyltransferase, EZH2) mechanisms [43–45]. The preliminary studies of piRNAs indicate that the main targets of piRNAs are methyltransferases (DNMT1, 3a, and 3b) or histone methyltransferases like Enhancer of zeste homolog 2 (EZH2) or H3K27me3 methyltransferases [15,46–51]. By targeting the same molecules with other known mechanisms, we aimed to use the piRNA sequence (anti-piR-651) to determine its effects on molecular mechanisms such as proliferation, adhesion, and motility in benign breast cancer cells.
The first report about piR-651 and cancer suggested that the upregulation of piR-651 was associated with the tumor‑node-metastasis (TNM) in gastric cancer tissues [21]. Yao et al. [16] identified that inhibition of endogenous piR‑651 may prevent tumor development by inhibiting cell proliferation, migration, and invasion, and induces cell apoptosis in the NSCLC cell line. In another study involving piR-651 and lung cancer, it was determined that the upregulation of piR-651 caused a higher risk of death in NSCLC patients [11]. Previously, detecting piR-651 expression in MCF-7 cells after transfection was deemed essential for understanding the efficiency of transfection. As a result of transfection, piR-651 expression was decreased in anti-piR-651 transfected MCF-7 cells. After identifying the efficiency of transfection, the other gene expressions were detected in MCF-7 cells.
ERα is an important parameter for this study because MCF-7 cells are ERα positive and dependent cells. Therefore, we aimed to decrease the ERα gene expression of these cells in order to affect their proliferation. Furthermore, recent studies showed that the upregulation of ERα caused MCF-7 cells to be more aggressive. In a study regarding disruption of ER signaling in MCF-7 cells, polycomb repressors (like EZH2) and histone deacetylases (HDACs) were enhanced and this enhancement repressed ER target genes in a stable manner [52]. In our study, inhibition of piR-651 decreased ERα gene expression in ER-dependent breast cancer cells. We suggested that the ER-dependent breast cancer cells (MCF-7) cannot use this receptor to survive and proliferate; they should go apoptosis at that point. Targeting ERα by using anti-piR-651 transfection might be a useful treatment against benign breast cancer to control their proliferation.
Another perspective of the piR-651 effect on proliferation is that hTERT expression is an important key feature. Various studies have indicated that hTERT overexpression in mammary epithelial cells reduces their dependence on mitogens by regulating genes that are related to proliferation [53], while telomerase inhibition promoted apoptosis in ovarian cancer cells [54]. Li et al. [55] detected inhibition of hTERT, causing a reduction in cancer cell proliferation and growth without affecting telomere length. Hormone dependency and hormone receptor-mediated signaling pathways, such as estrogen receptor (ER)-signaling, can regulate TERT expression in hormone-dependent breast cancer. In ER-dependent breast cancer, high expression of hTERT is observed [38]. The other mechanism of hTERT depends on abnormal DNA methylation. DNA methyltransferase I (DNMT1) is the key feature of hTERT activity and in cancer cases, the expression of DNMT1 is overexpressed [37,56,57]. In a study, they indicated that the high DNMT1 expression is especially determined in MCF-7 cells and they suggested that DNA methylation, and also DNMT1, might be linked to the immortalization of cancer cells [37]. piRNA researchers have also known that one of the proteins, which is correlated with piRNA expression, is DNMT1. Leu et al. [52] determined the strong relationship between DNA methyltransferases and ERα in breast cancer cells. We suggested that ERα induction using anti-piR-651 caused hTERT gene expression to decrease in MCF-7 cells (Fig. 4F). For future studies, DNMTs might be one of the target molecules of ERα and an hTERT related proliferation pathway in benign breast cancer cells. Furthermore, as we observed, the downregulation of the hTERT gene might decrease the proliferation rate of MCF-7 cells (Fig. 1B). Given this point, we consider that cells cannot proliferate and some mortality mechanisms like apoptosis might occur in cells.
The third parameter which helps us to understand the piR-651 effect on MCF-7 cells is Ki-67. Previous studies have also indicated that Ki-67 expression generally indicates and gives information about the proliferation of cells, especially breast cancer cells. Sobecki et al. determined the extremely low Ki-67 gene expression on quiescent cells [23]. Low Ki-67 expression was observed in breast cancer cells, which have low proliferation rates [58]. A study that focused on the relationship between untreated early-stage breast cancer patients and the Ki-67 gene expression profile showed that there is a distinct correlation between early-stage breast cancer and reduced Ki-67 expression [59]. Moreover, it was determined that inhibition of the Ki-67 gene by using shRNA transfection caused decreased tumor growth in immunodeficient mice [60]. Not only Ki-67 gene expression but also proliferation assay was applied to determine the effect of piR-651 inhibition on the proliferation of MCF-7 cells. According to our results from proliferation assay and Ki-67 gene expression, inhibition of piR-651 caused MCF-7 cells to decrease proliferation after the 48th hour (Figs. 1B and 4B).
EZH2 is an important molecule to understand the relationship between MMPs and piR-651. MMPs and piR-651 both have an impact on carcinogenesis by affecting the EZH2 molecule. A study aimed to understand the potential effect of EZH2 on triple-negative breast cancer cells and according to their high-level data obtained, EZH2 caused an increase in the expression of MMP-2 in breast cancer cells [26]. In benign breast cancer cells like MCF-7, generally, low expressions of MMPs are observed. Moreover, Li et al. observed that MMP‑2 and MMP‑9 expression profiles in breast cancer tissues were correlated with lymph node metastasis and tumor grade [61]. High MMP‑2 and MMP‑9 expression cause degradation of type IV collagens and promotes the invasion and metastasis of tumor cells [62]. Previous studies have indicated that MMP‑2 and MMP‑9 expressions in breast cancer tissues were significantly higher than in fibrous adenomas and in patients with infiltrative breast cancer and lymph node metastasis compared to patients with non‑infiltrative and non‑lymph node metastasis [61,63]. Furthermore, Jones et al determined that MMP‑2 was mainly expressed in the cytosol of breast cancer cells, and a small amount of these cells were present in the normal breast duct and basal membrane [64]. Furthermore, Zhang et al. [17] have identified that piR-651 promotes invasion and metastasis of non-small cell lung cancer (NSCLC) cells. According to our data obtained, a downregulation of MMP-2 was determined after piR-651 inhibition. Moreover, we detected loss of adhesion of MCF-7 cells after piR-651 inhibition (Figs. 2A–2C). We observed that the motility of anti-piR-651 transfected MCF-7 cells decreased after the 48th hour. MMP-2 gene expression data also supports motility results and indicates that the downregulation of piR-651 prevents MCF-7 cells from spreading over a wide area (Fig. 3).
Hypoxic microenvironment mainly occurs in breast cancer and approximately 25%–40% of invasive breast cancer has hypoxic microenvironments [65]. Hypoxia can promote the development of breast cancer cells and epithelial-mesenchymal transition-mediated breast cancer cell migration, which has a negative impact on the survival rate of breast cancer patients with intratumoral hypoxia [66]. Li et al. [67] showed that HIF-1α downregulation caused the reversal of chemoresistance and inhibits proliferation, migration, and invasion of breast cancer cells in vitro. Furthermore, Wong et al. [68] indicated that observing metastatic formation during breast cancer development is a HIF-1α dependent situation. A study on the relationship between HIF-1α expression and MCF-7 cells supports our hypothesis and indicates that inhibition of HIF-1α decreases the tumorigenicity of MCF-7 cells [69]. Determining the expression patterns of HIF-1α is important due to the detection of the differentiation rate of cells after the transfection of anti-piR-651. According to our data obtained, inhibition of piR-651 caused an decrease of HIF-1α expression. This result indicates that piR-651 is functional in controlling the differentiation of MCF-7 benign breast cancer cells. Furthermore, the absence of piR-651 caused a decrease in HIF-1α expression, and low HIF-1α expression in MCF-7 cells showed that these breast cancer cells are early and benign types of breast cancer and the mechanisms of angiogenesis, migration, and invasion cannot be detected in that grade of breast cancer cells. Our hypothesis is piR-651 inhibition causes to reduce proliferation, adhesion, differentiation and motility of MCF-7 breast cancer cells. By this way, we can understand the impact of piR-651 in breast cancer. As a result of all our obtained data, inhibition of piR-651 affects various mechanism of cancer cells which were damaged during carcinogenesis and might transform these damaged mechanisms as previous conditions (Fig. 5).
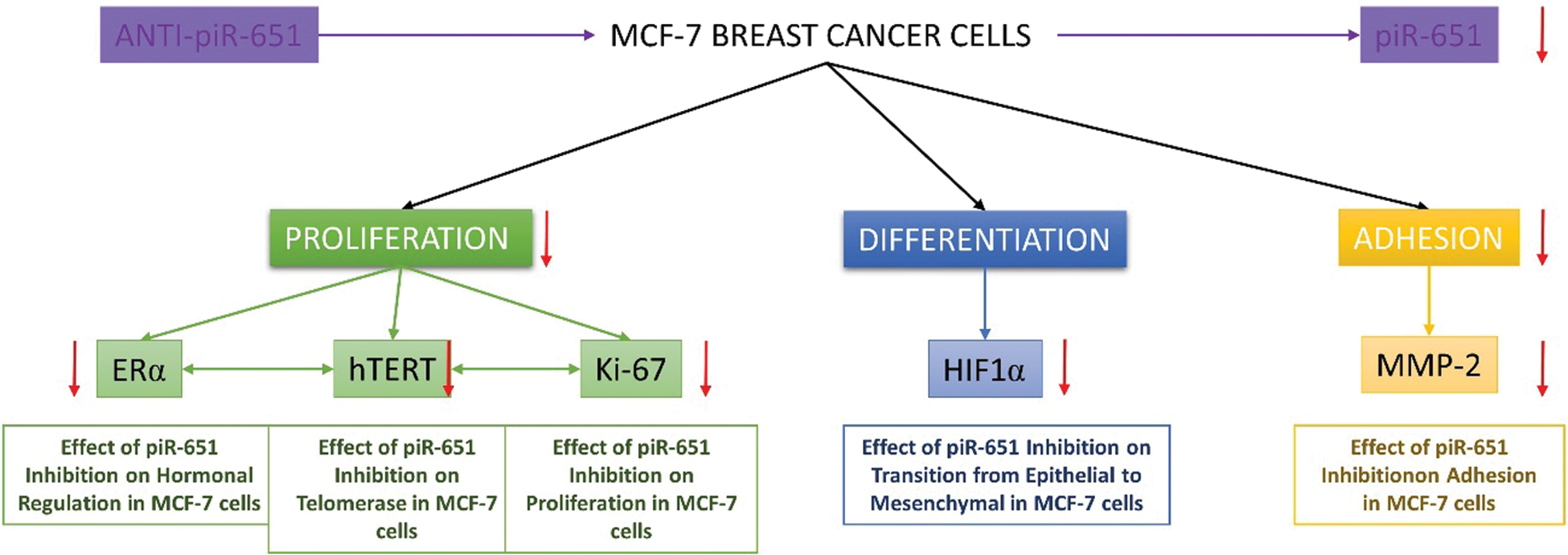
Figure 5: The impact of piR-651 on proliferation, differentiation and adhesion characteristics of MCF-7 breast cancer cells
The current study is subject to limitations. First of all, we could only observe the cellular characteristic changes of MCF-7 breast cancer cell line. The impact of piR-651 and other piRNAs should be detected in other breast cancer cell lines, tissue and blood samples. However, piRNA studies gain interest day by day, there are less knowledge about them. Furthermore, more studies are needed to identify the cellular mechanisms of various piRNAs.
This study indicates that piR-651 affects the main characteristics of benign breast cancer cells and it might be useful to transform benign cells into the healthy cells in the future. The relationship between piRNAs and ERα was determined and we suggest that hormonal regulation of breast cancer cells can be regulated by piRNAs as a result of the study results. To find a successful treatment for a disease, you should know the potential molecular mechanism of disease. By these results we cannot say that piR-651 is a good biomarker or target for gene treatment for now, but these results indicate that piR-651 might be a good candidate for treatment or early diagnosis of breast cancer. To the best of our knowledge, this study is the preliminary study of focusing on the relationship between piR-651 and breast cancer.
Acknowledgement: The authors thank Associate Prof. Dr. Bilge GÜVENÇ TUNA, Department of Biophysics, Yeditepe University School of Medicine, for kindly providing MCF-7 cells for this study.
Funding Statement: This study is funded by Maltepe University Scientific Research Committee (MUAR).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Mallick, A., Ahmed, S. (2019). Investigation of the effect of estradiol and subculturing on the receptor expression within hormone-dependent breast cancer cells. bioRxiv. DOI 10.1101/548271. [Google Scholar] [CrossRef]
2. Oner, C., Turgut Cosan, D., Colak, E. (2016). Estrogen and androgen hormone levels modulate the expression of PIWI interacting RNA in prostate and Breast Cancer. PLoS One, 11(7), e0159044. [Google Scholar]
3. Al-Aws, G. R. L., Chalap, E. D. (2019). A general overview of the genetic effects of extracellular polymers For Enterococcus faeciumin cancer cells. International Journal of Research in Pharmaceutical Sciences, 10(1), 436–443. [Google Scholar]
4. Harrington, K. J. (2016). The biology of cancer. Medicine, 44(1), 1–5. [Google Scholar]
5. Pathiraja, T. N., Stearns, V., Oesterreich, S. (2010). Epigenetic regulation in estrogen receptor positive breast cancer--role in treatment response. Journal of Mammary Gland Biology and Neoplasia, 15(1), 35–47. [Google Scholar]
6. Shao, W., Brown, M. (2004). Advances in estrogen receptor biology: Prospects for improvements in targeted breast cancer therapy. Breast Cancer Research, 6(1), 39–52. [Google Scholar]
7. Giacinti, L., Claudio, P. P., Lopez, M., Giordano, A. (2006). Epigenetic information and estrogen receptor alpha expression in breast cancer. Oncologist, 11(1), 1–8. [Google Scholar]
8. Grimson, A., Srivastava, M., Fahey, B., Woodcroft, B. J., Chiang, H. R. et al. (2008). Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature, 455(7217), 1193–1197. [Google Scholar]
9. Castaneda, J., Genzor, P., Bortvin, A. (2011). piRNAs, transposon silencing, and germline genome integrity. Mutation Research, 714(1–2), 95–104. [Google Scholar]
10. Levin, H. L., Moran, J. V. (2011). Dynamic interactions between transposable elements and their hosts. Nature Reviews Genetics, 12(9), 615–627. [Google Scholar]
11. Li, D., Luo, Y., Gao, Y., Yang, Y., Wang, Y. et al. (2016). piR-651 promotes tumor formation in non-small cell lung carcinoma through the upregulation of cyclin D1 and CDK4. International Journal of Molecular Medicine, 38(3), 927–936. [Google Scholar]
12. Chenais, B. (2013). Transposable elements and human cancer: A causal relationship? Biochimica et Biophysica Acta, 1835(1), 28–35. [Google Scholar]
13. Brennecke, J., Malone, C. D., Aravin, A. A., Sachidanandam, R., Stark, A. et al. (2008). An epigenetic role for maternally inherited piRNAs in transposon silencing. Science, 322(5906), 1387–1392. [Google Scholar]
14. Öner, Ç. (2019). Two different mechanisms of two different non-coding RNAs—MicroRNAs and PIWI-interacting RNAs: From origin to cancer, pp. 3–34. Academic Press. [Google Scholar]
15. Siddiqi, S., Matushansky, I. (2012). Piwis and piwi-interacting RNAs in the epigenetics of cancer. Journal of Cellular Biochemistry, 113(2), 373–380. [Google Scholar]
16. Yao, J., Wang, Y. W., Fang, B. B., Zhang, S. J., Cheng, B. L. (2016). piR-651 and its function in 95-D lung cancer cells. Biomedical Reports, 4(5), 546–550. [Google Scholar]
17. Zhang, S. J., Yao, J., Shen, B. Z., Li, G. B., Kong, S. S. et al. (2018). Role of piwi-interacting RNA-651 in the carcinogenesis of non-small cell lung cancer. Oncology Letters, 15(1), 940–946. [Google Scholar]
18. Huang, G., Hu, H., Xue, X., Shen, S., Gao, E. et al. (2013). Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clinical & Translational Oncology, 15(7), 563–568. [Google Scholar]
19. Zhang, H., Ren, Y., Xu, H., Pang, D., Duan, C. et al. (2013). The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surgical Oncology, 22(4), 217–223. [Google Scholar]
20. Cui, L., Lou, Y., Zhang, X., Zhou, H., Deng, H. et al. (2011). Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clinical Biochemistry, 44(13), 1050–1057. [Google Scholar]
21. Cheng, J., Guo, J. M., Xiao, B. X., Miao, Y., Jiang, Z. et al. (2011). piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clinica Chimica Acta, 412(17–18), 1621–1625. [Google Scholar]
22. Cheng, J., Deng, H. X., Xiao, B. X., Zhou, H., Zhou, F. et al. (2012). piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Letters, 315(1), 12–17. [Google Scholar]
23. Sobecki, M., Mrouj, K., Colinge, J., Gerbe, F., Jay, P. et al. (2017). Cell-cycle regulation accounts for variability in Ki-67 expression levels. Cancer Research, 77(10), 2722–2734. [Google Scholar]
24. Cuzick, J., Dowsett, M., Pineda, S., Wale, C., Salter, J. et al. (2011). Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. Journal of Clinical Oncology, 29(32), 4273–4278. [Google Scholar]
25. Gan, R. Y., Li, H. B., Sui, Z. Q., Corke, H. (2017). Absorption, metabolism, anti-cancer effect and molecular targets of epigallocatechin gallate (EGCGAn updated review. Critical Reviews in Food Science and Nutrition, 58(6), 924–941. [Google Scholar]
26. Chien, Y. C., Liu, L. C., Ye, H. Y., Wu, J. Y., Yu, Y. L. (2018). EZH2 promotes migration and invasion of triple-negative breast cancer cells via regulating TIMP2-MMP-2/-9 pathway. American Journal of Cancer Research, 8(3), 422–434. [Google Scholar]
27. Singer, C. F., Kronsteiner, N., Marton, E., Kubista, M., Cullen, K. J. et al. (2002). MMP-2 and MMP-9 expression in breast cancer-derived human fibroblasts is differentially regulated by stromal-epithelial interactions. Breast Cancer Research and Treatment, 72, 69–77. [Google Scholar]
28. Poulsom, R., Hanby, A. M., Pignatelli, M., Jeffery, R. E., Longcroft, J. M. et al. (1993). Expression of gelatinase A and TIMP-2 mRNAs in desmoplastic fibroblasts in both mammary carcinomas and basal cell carcinomas of the skin. Journal of Clinical Pathology, 46(5), 429–436. [Google Scholar]
29. Polette, M., Clavel, C., Cockett, M., Girod de Bentzmann, S. (1993). Detection and localization of mRNAs encoding matrix metalloproteinases and their tissue inhibitor in human breast pathology. Invasion Metastasis, 13(1), 31–37. [Google Scholar]
30. Semenza, G. L. (2012). Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends in Pharmacological Sciences, 33(4), 207–214. [Google Scholar]
31. Li, Q., Ma, R., Zhang, M. (2018). CoCl2 increases the expression of hypoxic markers HIF-1alpha, VEGF and CXCR4 in breast cancer MCF-7 cells. Oncology Letters, 15(1), 1119–1124. [Google Scholar]
32. Campbell, E. J., Dachs, G. U., Morrin, H. R., Davey, V. C., Robinson, B. A. et al. (2019). Activation of the hypoxia pathway in breast cancer tissue and patient survival are inversely associated with tumor ascorbate levels. BMC Cancer, 19(307), 1–13. [Google Scholar]
33. Gruber, G., Greiner, R. H., Hlushchuk, R., Aebersold, D. M., Altermatt, H. J. et al. (2004). Hypoxia-inducible factor 1 alpha in high-risk breast cancer: An independent prognostic parameter? Breast Cancer Research, 6(3), R191–198. [Google Scholar]
34. Ozturk, M. B., Li, Y., Tergaonkar, V. (2017). Current insights to regulation and role of telomerase in human diseases. Antioxidants (Basel), 6(17), 1–13. [Google Scholar]
35. Gardano, L., Pucci, F., Christian, L., Le Bihan, T., Harrington, L. (2013). Telomeres, a busy platform for cell signaling. Frontiers of Oncology, 3, 146. [Google Scholar]
36. Kim, N. W., Piatyszek, M. A., Prowse, K. R., Harley, C. B., West, M. D. et al. (1994). Specific association of human telomerase activity with immortal cells and cancer. Science, 266(5193), 2011–2015. [Google Scholar]
37. Zhao, C., Meng, L., Hu, H., Wang, X., Shi, F. et al. (2010). Spontaneously immortalised bovine mammary epithelial cells exhibit a distinct gene expression pattern from the breast cancer cells. BMC Cell Biology, 11, 82. [Google Scholar]
38. Calado, R. T., Yewdell, W. T., Wilkerson, K. L., Regal, J. A., Kajigaya, S. et al. (2009). Sex hormones, acting on the TERT gene, increase telomerase activity in human primary hematopoietic cells. Blood, 114(11), 2236–2243. [Google Scholar]
39. Orlando, C., Gelmini, S. (2001). Telomerase in endocrine and endocrine-dependent tumors. Journal of Steroid Biochemistry and Molecular Biology, 78(3), 201–214. [Google Scholar]
40. Leon-Blanco, M. M., Guerrero, J. M., Reiter, R. J., Pozo, D. (2004). RNA expression of human telomerase subunits TR and TERT is differentially affected by melatonin receptor agonists in the MCF-7 tumor cell line. Cancer Letters, 216(1), 73–80. [Google Scholar]
41. Boehm, J. S., Hession, M. T., Bulmer, S. E., Hahn, W. C. (2005). Transformation of human and murine fibroblasts without viral oncoproteins. Molecular Cell Biology, 25(15), 6464–6474. [Google Scholar]
42. Widschwendter, M., Jones, P. A. (2002). DNA methylation and breast carcinogenesis. Oncogene, 21(35), 5462–5482. [Google Scholar]
43. Huang, Y., Bai, J. Y., Ren, H. T. (2014). piRNA biogenesis and its functions. Russian Journal of Bioorganic Chemistry, 40(3), 293–299. [Google Scholar]
44. Ng, K. W., Anderson, C., Marshall, E. A., Minatel, B. C., Enfield, K. S. et al. (2016). Piwi-interacting RNAs in cancer: Emerging functions and clinical utility. Molecular Cancer, 15, 5. [Google Scholar]
45. Pandya, G. M., Ramani, U. V., Janmeda, M., Dangar, N. S., Tyagi, K. et al. (2014). piRNA: Basics and their Association with PIWI proteins. Current Trends in Biotechnology and Pharmacy, 8(3), 303–308. [Google Scholar]
46. Grimaud, C., Bantignies, F., Pal-Bhadra, M., Ghana, P., Bhadra, U. et al. (2006). RNAi components are required for nuclear clustering of Polycomb group response elements. Cell, 124(5), 957–971. [Google Scholar]
47. Horwich, M. D., Li, C., Matranga, C., Vagin, V., Farley, G. et al. (2007). The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Current Biology, 17(14), 1265–1272. [Google Scholar]
48. Siddiqi, S., Terry, M., Matushansky, I. (2012). Hiwi mediated tumorigenesis is associated with DNA hypermethylation. PLoS One, 7(3), e33711. [Google Scholar]
49. Simon, J. A., Lange, C. A. (2008). Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutation Research, 647(1–2), 21–29. [Google Scholar]
50. Tsang, D. P., Cheng, A. S. (2011). Epigenetic regulation of signaling pathways in cancer: Role of the histone methyltransferase EZH2. Journal of Gastroenterology and Hepatology, 26(1), 19–27. [Google Scholar]
51. Yan, H., Wu, Q. L., Sun, C. Y., Ai, L. S., Deng, J. et al. (2015). piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia, 29(1), 196–206. [Google Scholar]
52. Leu, Y. W., Yan, P. S., Fan, M., Jin, V. X., Liu, J. C. et al. (2004). Loss of estrogen receptor signaling triggers epigenetic silencing of downstream targets in breast cancer. Cancer Research, 64(22), 8184–8192. [Google Scholar]
53. Smith, L. L., Coller, H. A., Roberts, J. M. (2003). Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nature Cell Biology, 5(5), 474–479. [Google Scholar]
54. Saretzki, G., Ludwig, A., von Zglinicki, T., Runnebaum, I. B. (2001). Ribozyme-mediated telomerase inhibition induces immediate cell loss but not telomere shortening in ovarian cancer cells. Cancer Gene Therapy, 8(10), 827–834. [Google Scholar]
55. Li, S., Rosenberg, J. E., Donjacour, A. A., Botchkina, I. L., Hom, Y. K. et al. (2004). Rapid inhibition of cancer cell growth induced by lentiviral delivery and expression of mutant-template telomerase RNA and anti-telomerase short-interfering RNA. Cancer Research, 64(14), 4833–4840. [Google Scholar]
56. Saito, Y., Kanai, Y., Sakamoto, M., Saito, H., Ishii, H. et al. (2001). Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology, 33(3), 561–568. [Google Scholar]
57. Kanai, Y., Ushijima, S., Kondo, Y., Nakanishi, Y., Hirohashi, S. (2001). DNA methyltransferase expression and DNA methylation of CPG islands and peri-centromeric satellite regions in human colorectal and stomach cancers. International Journal of Cancer, 91(2), 205–212. [Google Scholar]
58. Marusyk, A., Tabassum, D. P., Altrock, P. M., Almendro, V., Michor, F. et al. (2014). Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature, 514(7520), 54–58. [Google Scholar]
59. Hurvitz, S., Abad, M. F., Rostorfer, R., Chan, D., Egle, D. et al. (2016). Breast cancer, early stage Interim results from neoMONARCH: A neoadjuvant phase II study of abemaciclib in postmenopausal women with HR+/HER2- breast cancer (BC). Annals of Oncology, 27, vi552. [Google Scholar]
60. Zheng, J. N., Ma, T. X., Cao, J. Y., Sun, X. Q., Chen, J. C. et al. (2006). Knockdown of Ki-67 by small interfering RNA leads to inhibition of proliferation and induction of apoptosis in human renal carcinoma cells. Life Sciences, 78(7), 724–729. [Google Scholar]
61. Li, H., Qiu, Z., Li, F., Wang, C. (2017). The relationship between MMP-2 and MMP-9 expression levels with breast cancer incidence and prognosis. Oncology Letters, 14(5), 5865–5870. [Google Scholar]
62. Parsons, S. L., Watson, S. A., Collins, H. M., Griffin, N. R., Clarke, P. A. et al. (1998). Gelatinase (MMP-2 and -9) expression in gastrointestinal malignancy. British Journal of Cancer, 78(11), 1495–1502. [Google Scholar]
63. Katunina, A. I., Gershtein, E. S., Ermilova, V. D., Tereshkina, I. V., Nazarenko, A. Y. et al. (2011). Matrix metalloproteinases 2, 7, and 9 in tumors and sera of patients with breast cancer. Bulletin of Experimental Biology and Medicine, 151(3), 359–362. [Google Scholar]
64. Jones, J. L., Glynn, P., Walker, R. A. (1999). Expression of MMP-2 and MMP-9, their inhibitors, and the activator MT1-MMP in primary breast carcinomas. Journal of Pathology, 189(2), 161–168. [Google Scholar]
65. Liu, Z. J., Semenza, G. L., Zhang, H. F. (2015). Hypoxia-inducible factor 1 and breast cancer metastasis. Journal of Zhejiang University–Science B, 16(1), 32–43. [Google Scholar]
66. Park, S. J., Kim, J. G., Kim, N. D., Yang, K., Shim, J. W. et al. (2016). Estradiol, TGF-beta1 and hypoxia promote breast cancer stemness and EMT-mediated breast cancer migration. Oncology Letters, 11(3), 1895–1902. [Google Scholar]
67. Li, S., Wei, Q., Li, Q., Zhang, B., Xiao, Q. (2015). Down-regulating HIF-1alpha by lentivirus-mediated shRNA for therapy of triple negative breast cancer. Cancer biology & Therapy, 16(6), 866–875. [Google Scholar]
68. Wong, C. C., Gilkes, D. M., Zhang, H., Chen, J., Wei, H. et al. (2011). Hypoxia-inducible factor 1 is a master regulator of breast cancer metastatic niche formation. Proceedings of the National Academy of Sciences of the United States of America, 108(39), 16369–16374. [Google Scholar]
69. Li, J., Shi, M., Cao, Y., Yuan, W., Pang, T. et al. (2006). Knockdown of hypoxia-inducible factor-1alpha in breast carcinoma MCF-7 cells results in reduced tumor growth and increased sensitivity to methotrexate. Biochemical and Biophysical Research Communications, 342(4), 1341–1351. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |