

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.014967
REVIEW
Non-Canonical Functions of the E2F/DP Pathway with Emphasis in Plants
1Facultad de Química, Departamento de Bioquímica, Universidad Nacional Autónoma de México, Avenida Universidad y Copilco, Ciudad de México, 04510, México
2Plant Development and (Epi) Genetics Group, Swammerdam Institute for Life Sciences, Universiteit van Amsterdam, Amsterdam, 1098XH, Netherlands
*Corresponding Author: Jorge M. Vázquez-Ramos. Email: jorman@unam.mx
Received: 12 November 2020; Accepted: 19 January 2021
Abstract: The E2F/DP pathway is a widely conserved regulatory mechanism in pluricellular organisms. The family of E2F and DP transcription factors was originally described having a role in the transition from the G1 to the S phase of the cell cycle. However, the discovery of hundreds of possible gene targets and their involvement in many other biochemical processes, soon showed that they participated in cell development and differentiation, chromatin remodeling, DNA repair and others. The E2F/DP transcription factors can act as either activators or repressors of transcription depending on their association to other regulatory proteins, particularly the retinoblastoma protein, or even depending to their protein structure that can define their role. In plants the E2F/DP pathway also regulates endoreduplication, a process present along the life cycle, from organ elongation to root differentiation and reproduction. These transcription factors also help plant cells to respond to environmental disturbances such as those caused by different types of radiation, or by pathogens. This review focuses on the “so called” non-canonical functions of the E2F/DP family proteins in animal and plant cells, that are in fact essential activities that connect regulatory circuits among multiple metabolic pathways by means of their atypical functions.
Keywords: E2F; DP; transcription factors; atypical functions; plant cells
Cell theory establishes that all biological organisms are composed of cells, the fundamental units of life, and postulates that all form of life originates from pre-existent life, i.e., every cell comes from another cell. Pluricellular organisms are composed of a great variety of cell types arising from a unique cell, or zygote, and thus have the same genetic information. Therefore, the diversity of cells in the human body is the result of a differential expression of the genome in the different cell lineages, expression that must be finely coordinated with cell proliferation [1]. The cell cycle is the process that will provide the new cells that will enter the distinct developmental programs for a cell to perform a determined role within an organism.
The E2F protein family is a group of transcription factors that have been widely studied, particularly for their role in the G1/S transition of the cell cycle. This review focuses on the non-canonical functions of the E2F family members, both in mammals and plants, highlighting the nodes in which these proteins connect regulatory circuits among multiple metabolic pathways by means of their atypical functions.
2 A Brief History of the E2F Family
Three decades have passed since the first reports on the E2F family of proteins. The presence of a nuclear factor in adenovirus infected HeLa cells was able to stimulate the early expression of the E2 viral gene, and thus was named E2 PROMOTER BINDING FACTOR (E2F) [2]. In parallel, Lee and Green (1987) identified a cell factor involved in the expression of diverse adenovirus genes, like E1A, E2A, E3 and E4 and was named E4F1 (E4 FACTOR 1) [3]. Later studies concluded that both groups characterized the same factor and that this was also present in non-infected cells, although in low proportion [4,5]. Two duplicated elements in cis were found in the promoter of the E2 gene that were necessary for its expression, and this depended on E2F [2,6]. Years later, it was found that the E2F factor was formed by two proteins. One of them, E2F1, was responsible for both gene transactivation and interaction with the hypophosphorylated form of the retinoblastoma (pRb) protein, whose function will be mentioned below; the second protein, DP (DRTF1/E2F-POLYPEPTIDE or DIMERIZATION PARTNER), was able to interact with E2F sites in the promoter of the E2 viral gene [7]. The DP human gene was cloned and the interaction of DP-E2F proteins was shown to potentiate both the transcriptional activity and the binding to pRb, demonstrating that the activity of this transcription factor depends on the cooperation of the E2F and DP subunits [8].
Initially, studies were directed to understand the influence of E2F-DP in the expression of genes required for entry and progression of the S phase of the cell cycle. However, most recent studies have shown that cell cycle regulation is not the only function as E2F-DP participates in multiple biological processes as mitosis, response to and repair of damaged DNA, differentiation, development and apoptosis [9–13].
3 The E2F/DP Family in Mammals
During the G1/S phase transition, the E2F-DP activity is regulated by proteins of the pocket family, integrated in mammals by the pRb, p107 and p130 proteins, that are efficient repressors of the E2F transcription factor (TF). In response to mitogenic stimuli, there is an increase in the kinase activity of complexes formed by the proteins named Cyclins and Cyclin Dependent Kinases (Cyc-CDK complexes) [12]. Pocket proteins are among the targets of these kinases that, when hyperphosphorylated, suffer structural changes, loosing affinity and releasing the E2F TF [14,15], thereby allowing activation of essential genes for the establishment and advance of the S phase of the cell cycle. Within the E2F family there are members related to either activation or repression of gene expression. In humans, E2F1, E2F2 and E2F3a have been characterized as activators, while E2F3b, E2F4, E2F5 and E2F6 are related to transcriptional repression and all of them form complexes with DP, a feature that identifies all of them as Typical E2Fs. A special type of E2Fs with transcriptional repression properties was described in Arabidopsis thaliana [16,17] and later homologues were found in animals. These E2Fs were named E2F7 and E2F8, and differently from the other members of the family, did not associate with DP, instead binding DNA as monomers, and were considered as Atypical E2Fs [18,19].
The typical E2F1-6 proteins share a similar domain organization. In the amino end they all possess a nuclear localization signal, in the central region a DNA binding domain and a DP heterodimerization domain in which a leucine zipper and a region called mark box are localized. The transactivation domain is found in the carboxyl end, where the capacity of gene expression regulation is found; the motif for interaction with the pocket proteins is embedded within this domain (DYXXXLXXXEGIXDLFD, where X represents any amino acid) [15,20]. The DP subunit of the complex consists of a nuclear localization signal in the amino end, in the central region a DNA binding domain and an E2F heterodimerization domain in the carboxyl end (Fig. 1). Atypical members E2F7 and E2F8 contain two DNA binding domains in their structure that confer a structural folding that allow them to contact the E2F motif in the DNA with high affinity, without the need of a DP subunit [21].
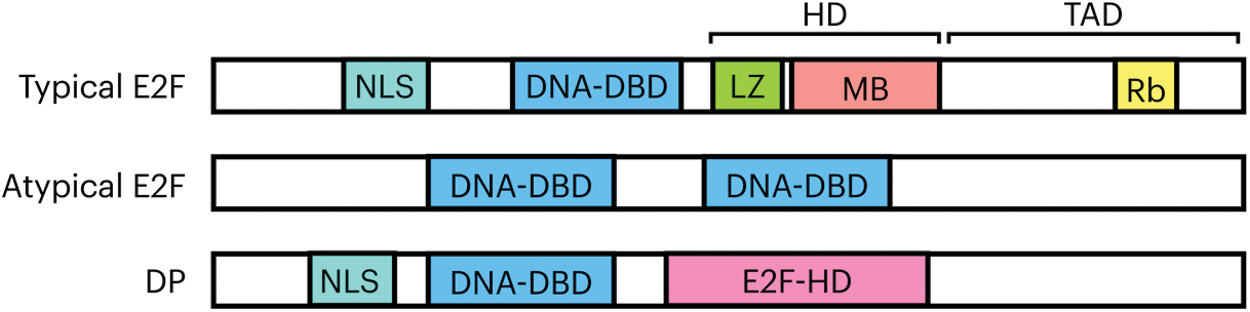
Figure 1: General organization of domains and motifs of E2F-DP proteins. The typical family members possess a nuclear localization signal (NLS); a DNA binding domain (DNA-DBD); a DP heterodimerization domain (HD) consisting of a leucine zipper (LZ) and the marked box (MB); and the transactivation domain (TAD), that contains within its sequence the motif of interaction with the Rb repressor. In the atypical E2F proteins only two main domains have been recognized and are those responsible for the binding to the DNA, (DNA-DBD 1 and 2). The DP structure comprises a nuclear localization signal, a DNA binding domain and a region that allows it to form complexes with E2F known as heterodimerization region (E2F-HD)
3.1 Opposing Interests within the E2F Family
As stated above, there are activators and repressors within the E2F family. E2F1, E2F2 and E2F3a induce proliferation in quiescent cells when overexpressed, while E2F3b, E2F4, E2F5, E2F6, E2F7 and E2F8 promote cell cycle arrest in serum-stimulated cell cultures [12] (Fig. 2A). However, when every member is analyzed individually, highly specialized and contrasting functions can be observed. As stated, E2F1-3a activates genes necessary for the S phase. However, high levels of E2F1 also promote apoptosis through two mechanisms. On one side, E2F1 induces the expression of pro-apoptotic genes as p14Arf (Arf) and caspases [22–25]; on the other side, it represses the expression of the anti-apoptotic gene Mcl-1 by binding to its promoter. In contrast, over-expression of the E2F4 repressor does not affect Mcl-1 [26], indicating target selectivity within the E2F family and opposing functions for the same protein (Fig. 2B). For E2F3 there are two isoforms coming from the same locus, E2F3a and E2F3b. The main difference between these isoforms is in the amino end since they are the product of the alternative use of promoters making the first exon different [27,28]. The E2f3 gene null mice dies soon after birth and cells with an interrupted E2f3 gene show proliferation defects due to a decrease in the activation of G1/S genes. The loss of E2F3b has no phenotype, whereas specifically silencing E2F3a causes a slight negative effect in cell proliferation, indicating that the two isoforms are partially redundant. However, silencing E2f3a in combination with E2f1 mutation causes severe proliferation defects, neonatal lethality, and defects in cartilage development, but not silencing of E2F3b, indicating a differential role for both isoforms [29–31].
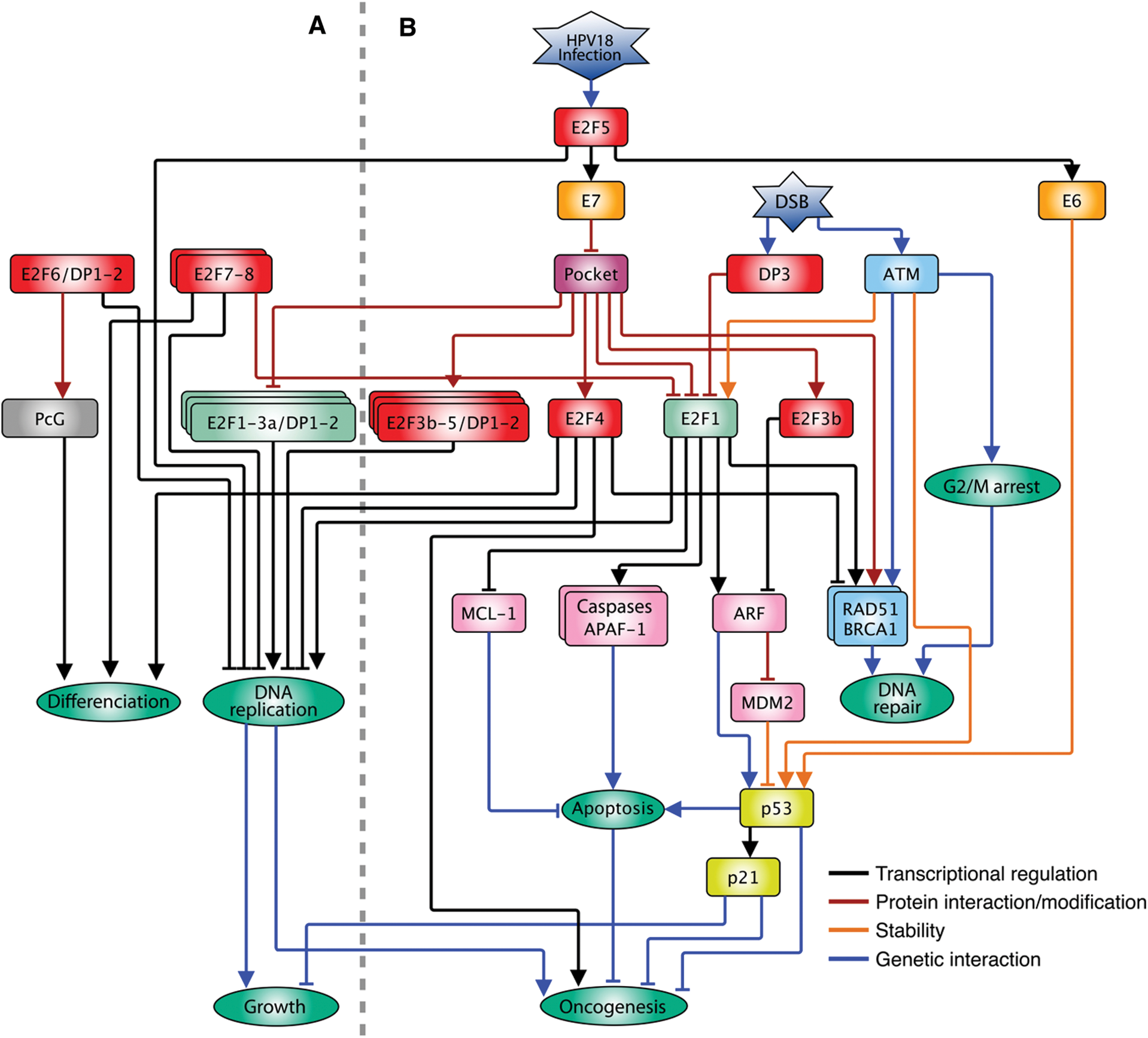
Figure 2: Interconnection of the E2F family with multiple pathways in animals. A. Participation of the E2F pathway in the G1/S transition of the cell cycle under normal conditions in which E2F1-3a positively regulates the expression of DNA replication while E2F3b-5 repressors negatively affect their expression in the absence of mitogenic stimuli and the pocket proteins modulate the function of the E2F family. B. When the cell perceives different types of stress such as viral infections or damage to its DNA (DSB), the cell cycle is arrested in both G1/S and G2/M by means of different pathways; the expression of DNA repair genes is stimulated and, if necessary, the cell death pathway is activated, involving atypical functions of E2F family members. The arrow heads indicate positive regulations, and the truncated heads indicate negative regulations. The color code of the connecting lines is indicated on the insert
The molecular mechanism by which E2F proteins repress gene expression is not known. However, they may function as scaffolding to recruit pocket proteins or associated co-repressors. Some of these co-repressors modify chromatin structure to silence transcription, as is the case of histone deacetylases (HDAC1 and HDAC2), histone demethylases (RBP2), DNA methyltransferases (DNMT1), nucleosome remodelers of the SWI/SNF type (Brg1, Brm), histone methyltransferases (Suv39, Suv4) and chromatin binding proteins like HP1 [32].
The predominant form of E2F3 is isoform b, and it has been considered a repressor since most of the time during the cell cycle is associated to pRb. Specific targets of the E2F3b-pRb complex are genes related to exit to cell differentiation and pro-apoptotic genes like Ink4/Arf, specifically the isoform p19Arf (murine homologue of Arf), that activates the p53-p21Cip1 pathway [33]. In this way, the repressive function of E2F3b allows cell cycle advance under normal conditions [34,35].
Similarly, E2F4 in complex with pocket proteins (mainly p130) represses the expression of G1/S genes during G0 (i.e., quiescence, senescence and differentiated states) and early G1, avoiding premature entrance into the cycle [36]. This TF possesses two nuclear export signals (NES) that prevent it from a stable permanence within the nucleus, and its residence in the nucleus, and repressive activity, depend on its association with pocket proteins [37]. E2F4, as other typical E2Fs, contains a transactivation domain; in vitro assays have shown that this protein can activate the expression of reporter genes, although weakly, and its overexpression does not allow entry into the S phase in quiescent cells. However, when co-expressed with DP1, this TF shows a higher capacity to stimulate cell cycle progression [38], but differently to E2F1, E2F4-DP1 overexpression (E2F4-DP1OE) does not induce neither the Arf expression nor the apoptosis process [39,40]. In contrast, in highly proliferating tissues as stem cells or tissue generating cells, E2F4 seems to substitute E2F1 as a primary activator, since it is the form predominantly expressed and it is localized in the cell nucleus, while E2F1 shows a more diffused expression pattern [41,42]. Loss of E2F4 reduces the rate of cell division in these tissues; nonetheless, it is not known if in this context, the E2F targets are cell cycle genes. In cancer models, E2F4 acts as an oncogene, probably through its non-canonical role as an activator and not as a repressor. Consistently, E2F4-DP1OE induces skin tumors in mice [43]. On the other hand, its function as repressor could also be related to cancer progression, since E2F4 increases the survival of transformed cells, repressing the expression of pro-apoptotic genes as E2f1 and Apaf-1 [44]. In addition, E2F4 in complex with p130 could promote genomic instability repressing genes essential for DNA damage repair by homologous recombination (HR) like BRCA1 and RAD51, increasing tumor cell survival [45,46]. These data indicate that E2F4 function, besides its interaction with pocket proteins, responds to other factors that depend on the cell context/ tissue in which it is expressed. Consistently, E2F4 overexpression, besides reducing the expression of certain genes, induces the expression of many others, indicating a dual function in gene expression regulation [47] (Fig. 2B).
E2F5, similarly to E2F4, associates to p107 and p130 to repress cell cycle genes transcription. Overexpression of E2F5 (E2F5OE) arrests cell cycle and can promote differentiation; however, the mechanism is still unknown [48–50]. The E2F pathway regulates not only adenovirus infection but also infection by other viruses as the human papillomavirus (HPV) [51]. The variety 18 (HPV18) is one of the more aggressive with a high probability of progression into carcinogenesis [52], and the high transformation of cells after infection resides in two viral proteins, E6 and E7, encoded in the same viral gene, and are a product of alternative splicing [53]. E6 protein stimulates p53 degradation via proteasome, while E7 dissociates pRb from E2F, thereby stimulating the cell cycle progression [54]. E6/E7 gene promoter is regulated by E2F through E2F sites bound by E2F2 and E2F5 proteins. Silencing of E2f1, E2f2 and E2f3 genes (individual or multiple) does not alter significantly the E6/E7 expression, but silencing E2f5 highly reduces expression [55]. Interestingly, this atypical E2F5 function as gene activator has only been described for cells infected by HPV18 and specifically on the E6/E7 promoter. The high rate of carcinogenesis after HPV18 infection could be because the high expression of E6/E7 (through E2F5) that produces a positive feedback loop, in which on one side stimulates the expression of the S phase genes by sequestering pRb and on the other side, halts the activation of apoptosis via p53 [55] (Fig. 2B).
Structure and function of the E2F6 protein differs from the other typical members of the E2F family due to the lack of the transactivation domain and of the pocket protein interaction motif, besides being an active repressor of transcription [56–58]. E2F6 overexpression (E2F6OE) differs from the activator E2Fs, as it induces no cell proliferation; on the contrary, the cell cycle is arrested independently of the association to pocket proteins. E2F6 is able to repress E2F target genes even under conditions in which E2F1-DP1 is overexpressed, reverting the high proliferation phenotype associated to this TF. Interestingly, E2F6OE cells are not arrested in the G1/S but in the S phase, indicating a possible role in the S phase exit, the opposite function to that of other members of the family [56] (Fig. 2A).
The main activity of this complex is the displacement of nucleosomes in the DNA to regulate transcription. Also, E2F6 is part of the POLYCOMB REPRESSIVE COMPLEX 1 (PRC1), and it associates to the core subunits of the MBI-1/Pcgf4 and RING1B E3 ubiquitin ligase. Another type of PcG complex related to E2F6 is E2F6PRC2, containing the catalytic subunit of PRC2, ENHANCER OF ZESTE 2 (EZH2), and the EPC1 protein, the anchoring protein. PRC2 trimethylates lysine 27 of histone H3 (H3K27me3), a mark related to transcription repression. The transcription repression activity of E2F6 has been linked to the function of the polycomb repressor complex (PcG), that is associated to chromatin structure. A possible mechanism to explain the repression of E2F1 target genes is through the interaction with the BRG1 subunit of the SWI/SNF remodeling complex [59]. During G1/S both E2F6 and BRG1 associate to promoters of E2F1 target genes, as E2f1 and PCNA [60]. PRC1 monoubiquitinates lysine 119 of histone H2A (H2AK119ub1), a chromatin mark related to gene expression silencing [61,62]. During G0, E2F6PRC2 associates to promoters of cell cycle genes and when cells are serum-stimulated to enter the cycle, the E2F6PRC2 complex becomes undetectable [63]. E2f6 null mice are viable and apparently healthy; however, they show homeotic transformations in the axial skeleton, similar to the phenotype of mice lacking the PcG components, indicating that this gene is part of the same genetic pathway [64].
The polycomb group proteins cannot bind DNA directly and so other TF could perform this function. This view is compatible with evidence that E2F6 can be localized in the same loci than other PcG proteins, both in G0 and in advanced stages of development and pattern formation, perhaps acting as a protein scaffolding to direct the PcG activity to specific loci [65,66].
The other two members of the E2F family, E2F7 and E2F8, known as atypical, are transcriptional repressors independent of pRb since they do not possess the interaction motif to pocket proteins. Overexpression of these TF represses the expression of cell cycle genes promoting its arrest [11,18,67]. Under normal conditions, expression of the E2f7 and E2f8 genes increases during G1/S via E2F1-3 and after the transition E2F7 and E2F8 proteins accumulate and repress the E2f1 expression, creating a transcriptional wave in G1/S by a negative feedback loop. Mutations in E2f7 or E2f8 do not show evident defects, indicating that they perform redundant functions; deletion of both genes causes an increase in apoptosis. This increase in cell death is in part due to a deregulated activity of E2F1 that, as mentioned before, stimulates the expression of proapoptotic genes. Consistently, E2F1 activity extended to the S phase exit and G2 promotes the activation of the p53 pathway and cell death [68]. Some of the factors that naturally limit E2F1 activity are association to pRb, phosphorylation by CycA/CDK2 in the S phase, ubiquitination and binding of E2F sites by repressor complexes associated to E2F6 [11,69,70]. Anyhow, none of these mechanisms is enough to counter the E2F activity in the absence of E2F7 and E2F8, pointing that the antagonistic role against E2F1 is not the only action they perform [68,71]. Another characteristic inherent to these atypical E2Fs is their capacity to form either homo or heterodimers among them. The predominant dimeric form is E2F7/E2F7, E2F7/E2F8 is the second most representative, and E2F8/E2F8 is the least common. Mice with genotype E2f7+/- E2f8-/- develop normally, while E2f7-/- E2f8+/- mice present multiple defects in postnatal development, indicating that their function is not at all that redundant [68]; the way this happens is not known although it is hypothesized that, since they all recognize the same response element on DNA, could compete for the sites with other E2Fs. Since E2F7 can form homodimers and both E2F7 and E2F8 can heterodimerize with E2F1, all these complexes are able to bind adjacent E2F sites on DNA in a cooperative manner [72]. As with DP [73], E2F1 interacts with E2F7/8 by the DNA binding domain. In vivo, E2F1 binds selectively to one of the two E2F sites in its own promoter, suggesting that during G1/S E2F1 activates its own expression, and when E2F7/8 accumulate, they can compete for the E2F sites forming a stable dimer with E2F1 on DNA displacing it. On the other hand, ChIP-reChIP experiments showed that E2F7/E2F7 can form complexes in the E2F sites with the transcriptional regulator CtBP2, whose silencing reverts the E2F7-dependent repression of the E2f1 gene [72]. This evidence shows that part of the repressive function of atypical E2Fs is related to competition for DNA sites and with chromatin remodeling of target genes via CtBP2 [74].
In human cells, the E2F dimerization partner (DP) subgroup is formed by three members, DP1, DP2/DP3 and TFDP3/DP4. Nomenclature can be confusing because there are multiple alternative splicing forms of DP2, and also, the murine homologue of DP2 in humans is known as DP3 [75]. When the third DP (TFDP3) was reported [76], another report appeared on a protein named DP4 [77], both referring to the same gene. So, from now on, we will refer to DP2/DP3 as DP2, a protein structurally and functionally related to DP1, while TFDP3/DP4 will be DP3, whose function is antagonistic to DP1 and DP2 [76]. DP1 shows the highest levels of expression of the family, followed by DP2 and then DP3; DP1 is considered as the main E2F interactor and its function is redundant with DP2; overexpression of DP1 stimulates entry to the S phase whereas DP3 promotes arrest in G1 [77]. All these proteins heterodimerize with E2F, but only DP1 and DP2 are related to activation of E2F target genes; meanwhile, DP3 inhibits both association to DNA and capacity of E2F activation [76,77]. Despite the low levels of DP3, abundance can be greatly increased after DNA damage by chemical agents that provoke double strand breaks (DSB), and its localization changes to the nucleus; this expression pattern is the opposite to that of DP1, whose levels decrease in response to DNA damage [78]. Overexpression of DP3 causes the displacement of DP1 from the complex with E2F1 substituting it, and so inhibiting the E2F function [76,78]. This unusual role can be seen as an inhibitory mechanism of p53-mediated E2F1 induced apoptosis [79]. However, Komori et al. [39] have reported that expression of the proapoptotic Arf gene is increased when E2F1 activity is deregulated by pRb inhibition as a consequence of the expression of the viral E1A protein. Interestingly, Arf expression or entry into apoptosis are not affected by silencing of the DP members, despite detention of the cell cycle in G1, indicating that E2F1 is acting independently of DP [39]. Evidence points to the interaction of E2F1 to CG repeats similar to the core E2F DNA motif (TTTCGCGCG), without the need of DP, defying the classical model of interaction as a heterodimer [73,80]. Besides Arf, expression of BIM, another proapoptotic gene, is increased when the E2F1 activity is deregulated [81]. Both genes lack canonical E2F sites in their promoter, but they contain CG repetitions, that are bound by this TF [39]. The classical model of E2F-induced apoptosis implies the existence of an E2F activity threshold that, when exceeded, more than inducing proliferation, stimulates entry into cell death. Experimental evidence challenges this model since in the absence of DP, cell cycle gene expression stops, while expression of proapoptotic genes continue. It is important to recall that the proapoptotic activity of E2F1 takes place mainly when the E2F pathway is altered, i.e., under conditions in which the pRb protein cannot repress its activity.
These functions of the atypical E2F proteins could work as a reserve mechanism to contend with unfavorable situations in which genome integrity is at risk, first, arresting the cell cycle and then, in case of major damage, activating processes as programmed cell death to maintain organism homeostasis.
3.2 Small Non-Coding RNAs as New Regulatory Nodes in the Cell Cycle Network
In recent years, the discovery of small non-coding RNAs (or miRNA) involved in cell cycle regulation opened an exciting new field in cell cycle research [82–84]. A prototypical miRNA, after several processing steps, consists of 21 to 24 nucleotide-long RNA; the sequence of a mature miRNA is partially complementary to the 3’ UTR of target mRNA(s) [85]. In cooperation with the RNA-induced silencing complex (RISC), miRNAs can direct either the cleavage of the target mRNA or interfere with the translation process, thus decreasing protein levels of their targets in a post-transcriptional manner [85]. miRNAs are transcribed by RNA pol II/III and they can exist both hosted by other genes (e.g., within introns), and as independent genetic units regulated by their own promoters [85]. miRNA function is crucial for cell cycle control, since they have the potential to decrease the levels of important proteins as oncogenes or tumor suppressors. For example, the CDK repressors p27kip1 and p57kip2 are targeted by miR221/222 and miR-181 [86–88]. Overexpression of the mir-221-222 cluster provokes the ectopic activation of CDK2 and enhanced tumor growth [89]. After mitogenic stimuli for cell cycle reentry, the activation of E2F-dependent transcription takes place; among the hundreds of E2F target genes there are several miRNA such as the mir-17-92 cluster which, paradoxically, target E2F1-3 mRNAs. This leads to a decrease in the activator-E2F levels, forming a negative feedback loop that can fine tune the abundance of E2F1-3 after the S phase establishment [90–92]. Other clusters like mir-15b-16-2, mir-106b-25, let-7a-d, and let-7i are also transcriptionally stimulated by E2F1-3. This miRNA clusters target central regulators of the G1/S transition like D-type cyclins, CDK4, CDK6, CDC25A, among others [93]. In the absence of these miRNAs the augmented E2F activity induces a strong DNA replication activity that results in replicative stress and DNA damage [93]. Altogether, these observations provide an exquisite model of regulation of E2F activity through several positive and negative feedback loops promoted by itself, supported in part by the activity of miRNAs, which in recent years have started to be regarded as an important tool for therapy in diseases such as cancer.
4 The E2F Family and the Green Ancestry
The first reports on the plant E2F/DP family were published 20 years ago. Cloning of maize cDNAs encoding pRb type proteins suggested conservation of the E2F/Rb pathway in plants [94,95]. The first plant E2F was reported in wheat [96,97] and then DP was also reported [98,99]. The discovery of the E2F pathway in plants was followed by studies in multiple species, including A. thaliana [17,100], tobacco [101], carrot [102], rice [103] and algae like Chlamydomonas reinhardtii [104] and Ostreococcus tauri [105], among others.
The E2F family of Arabidopsis has been the most widely studied; six E2F-type proteins and two DP-type proteins have been described. There are three typical E2Fs (i.e., E2Fa, E2Fb and E2Fc) and they have a domain organization similar to the human E2F1-5; it means, a nuclear localization signal, a DNA binding domain, a DP dimerization domain, a leucine zipper and a transactivation domain within which the RBR (Retinoblastoma-Related) motif is embedded, towards the carboxyl end [17,100]. Also, there are three atypical members that in plants are known as DEL (DP-E2F-LIKE), that include DEL1/E2Fe, DEL2/E2Fd and DEL3/E2Ff. Atypical E2Fs, similarly to animal E2F7/8, have two in tandem DNA binding domains, that allow them to bind DNA as monomers and do not contain DP interaction or transactivation domains [16]. DEL proteins are considered as transcriptional repressors [106,107]. Arabidopsis also has two DP-type proteins (DPa and DPb), that interact with E2Fa-c to form functional heterodimers with capacity to bind DNA [17,98]. These DP proteins have a nuclear localization signal, a DNA binding domain and an E2F dimerization domain. Finally, E2Fa and E2Fb present properties as transcriptional activators, while E2Fc works as a repressor [17].
In mammals, the consensus DNA binding sequence both for typical and atypical E2F is TTTSSCGS (S = nucleotides C or CG), regardless the direction with respect to the transcription initiation site [73,108,109]. Ramírez-Parra et al. [110] determined a slightly larger motif in Arabidopsis, TYTCCCGCC (where Y = T or C) [110], with a dinucleotide CG in positions 6 and 7 considered as indispensable for recognition by E2F/DP. Vandepoele et al. [111] reported a similar consensus, with some variations, WTTSSCSS (W = A or T) [111]. Consistent with the similarities observed for plant and animal DNA sequences, DNA binding domains of plant E2F, DEL and DP proteins in several plant species conserve the signature RRXYD (where X represents any amino acid) that allows them to contact the CG-rich core of the E2F motif [73,110,112–114]. The high conservation of the E2F DNA binding domain among eukaryotes is illustrated by its occurrence spanning major clades such as metazoan (616 sp.), viridiplantae (169 sp.) and fungi (17 sp.) when searched within the non-redundant DNA collection of NCBI (unpublished data).
5 Function of the Plant E2F/DP Family
In plants, as in animals, the E2F pathway regulates expression of cell cycle genes, mainly during the G1/S transition [115]. In Arabidopsis, E2Fa and E2Fb stimulate expression of genes related to start and advance of the S phase, while repressors E2Fc and DEL induce cell cycle arrest in G2 or G2/M, probably negatively regulating some of the E2F target genes for G1/S [107,116].
Besides the mitotic cell cycle, there is an alternative cell cycle that has been conserved from fungi to humans. This is known as endoreduplication and its main characteristic is that cells duplicate their genome in the absence of cell division, resulting in polyploid cells [117]. In humans, endoreduplication has been related to terminal differentiation and commonly occurs in highly specialized tissues such as hepatocytes or in the placenta [118]. Unlike animals, endoreduplication in plants is a process present along the life cycle. For instance, it is a strategy for organ elongation in leaves and hypocotyl and as part of the genetic program for root differentiation [119,120]. Polyploidy is also present during the reproductive stage.
5.1 Function of E2F in Endocycles
Flowering plants go through a process of double fertilization by which the embryo (2n) and a triploid endosperm are originated, which are the result of the fusion of the maternal (2n) and the paternal (n) nuclei [121]. Additionally, in some species as maize, the endosperm goes through several rounds of acytokinetic mitoses that form a multinucleated syncytium, followed by cellularization by mitosis [122–124]. Once this is finished, endosperm cells start synthesizing storage compounds and go through multiple rounds of endoreduplication, reaching a ploidy up to 192c, to finally enter the programmed cell death as part of the seed maturation process [122–125]. During this endoreduplicative stage, target genes of the E2F pathway as the subunits of the MCM2-7 replicative helicase, or the DNA polymerase processivity factor PCNA, greatly increase expression compared with the mitotic stage, pointing towards the participation of E2F in this process. On the other hand, silencing of one of the four genes coding for the maize pRb-type repressors, RBR1, the most expressed form, results in an increase in ploidy of endosperm cells, connecting the E2F/RBR pathway with control of ploidy in this developmental stage [125].
During mitosis (M), chromosomes condense and align with the mitotic spindle and then migrate (segregate) towards the poles to finally give way to cytokinesis creating two new daughter cells. Accumulation of the mitotic B-type cyclins (hereafter named as CycB) in the G2/M transition, and the increase in the associated kinase activity, leads to the establishment of the M phase; later, degradation of CycB is necessary for chromosome segregation and exit from M to happen. The anaphase promoting complex (APC) ubiquitinates mitotic cyclins, labelling them for degradation via 26S proteasome to allow M phase advance. The APC complex is composed of 11 subunits in vertebrates and 13 in yeast [126]; Arabidopsis APC has at least 13 subunits in the complex [127], of which the homologs of human CDH1 in Arabidopsis, CELL CYCLE SWITCH52 (CCS52) A and B, activate APC and show a peak of expression during late M and in G2/M, respectively [128].
The E2Fa/RBR1 complex binds to E2F sites in the CCS52A promoter to repress its expression during early G2, allowing CycB accumulation and the subsequent advance to the M phase [129]. Plants overexpressing E2Fa-DPa (E2Fa-DPaOE) show high levels of endoreduplication [130], probably the consequence of arrest in G2 due to the ectopic activation of CCS52A expression and the concomitant degradation of CycB. Additionally, arrest in G2/M could be enhanced by the increased expression of the E2Fc repressor protein in E2Fa-DPaOE plants [111,116]. RBR1 also plays an important role in the establishment of endoreduplication, since e2fa mutant plants, or plants expressing an E2Fa version lacking the RBR interacting motif (E2Fa∆RBR) are unable to repress CCS52A expression, increasing the arrest in G2 and endoreduplication [129].
The atypical E2F proteins also control the G2/M transition through repression of CCS52A expression, since DEL1OE plants show lower levels of ploidy, possibly due to arrest in the G1/S transition, promoting mitosis advance by regulating CCS52A. On the contrary, del1 mutant plants accumulate copies of the genome and ectopically express replication genes [106,107], that could direct entry into the endocycle. DEL proteins have no transactivation domain and therefore, it is a hypothesis that they antagonize the function of E2F-DP competing for the E2F site in target promoters, preventing its occupation by activator E2Fs [107]. However, most recent studies have shown that a subunit of an important expression regulating complex, MEDIATOR (MED), subunit 16 (MED16), genetically and physically interacts with DEL1. The joint activity of DEL1-MED16 stimulates transcriptional repression of CCS52A, and as a result, a decrease in ploidy, although the molecular mechanism has not been elucidated (Fig. 3) [131].
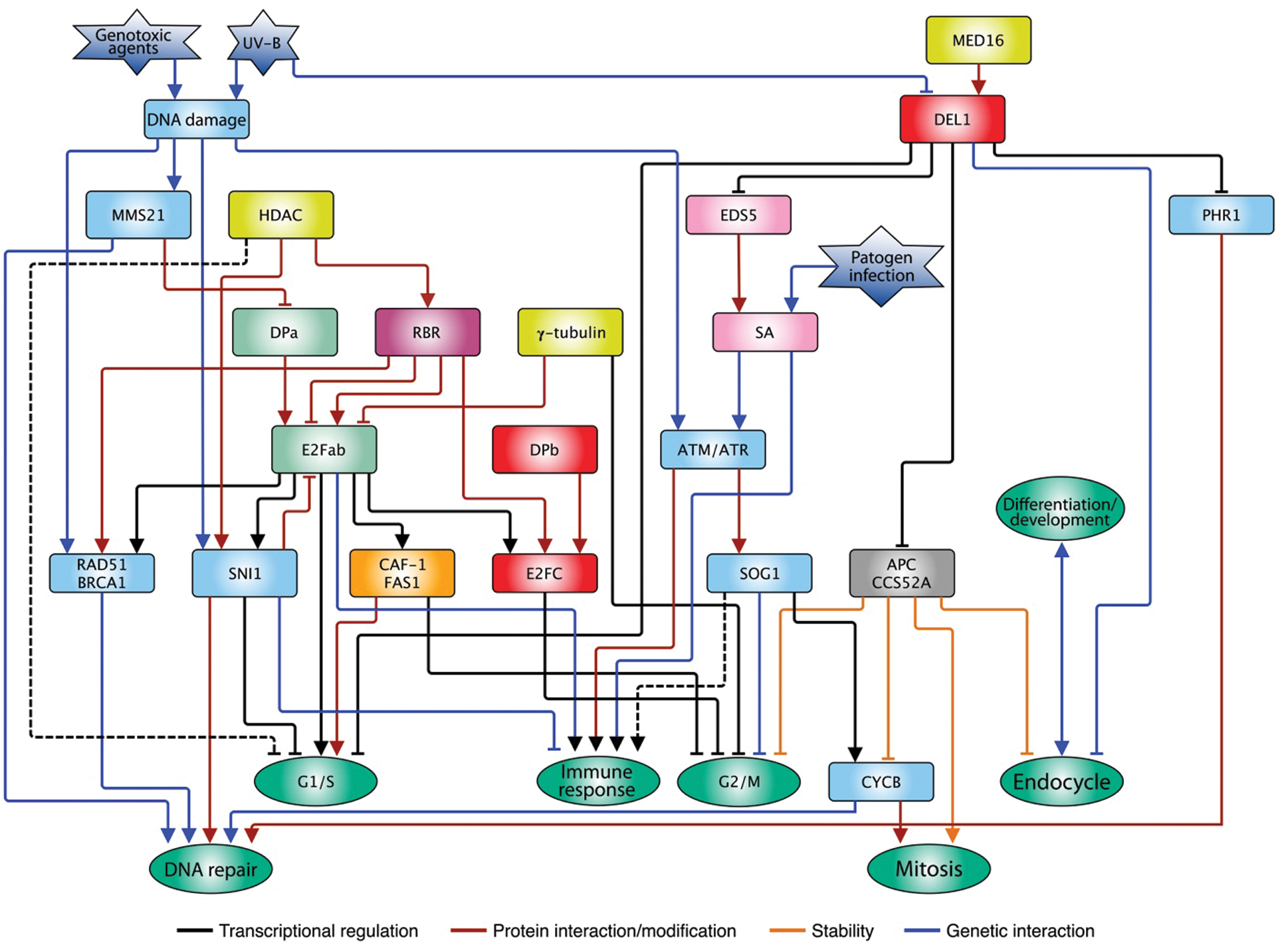
Figure 3: E2F family interconnection with multiple pathways in plants. Typical proteins E2Fa and b stimulate cell cycle progression in G1/S by the activation of replication genes, and their activity is counteracted by the RBR repressor. Besides, other repressor proteins control E2F function in response to stress such as the infection by pathogens or the DNA damage caused by environmental agents. The atypical protein DEL modulates multiple pathways such as cell cycle progression, immune response, DNA repair, endocycle, among others. The type of regulation exerted by the different genes/proteins is indicated in the insert. The arrows indicate a positive relationship, while the truncate ends indicate a negative relationship. The dotted lines represent hypothetical relationships
5.2 Function of E2F in Response to Environment
All living things are constantly exposed to agents that can compromise genome integrity. One of these agents, ultraviolet radiation (UV) is part of the solar light that, paradoxically, is the main energy source for photosynthetic organisms. The UV spectrum is divided in UV-C (λ < 280 nm), UV-B (λ 280 nm–315 nm) and UV-A (λ 315 nm–400 nm), of which, fraction UV-C is retained by the ozone layer in the stratosphere, while UV-B (5%–10%) and UV-A (>90%) reach the earth surface [132]. UV-C and UV-B are absorbed by DNA causing genotoxic damage in the form of pyrimidine-cyclobutanes (CPDs) [133], whereas UV-A can produce elevated levels of reactive oxygen species (ROS) when they impinge on biomolecules in cells [134]. The presence of CPDs on DNA can alter both replication and transcription and thus, cell cycle advance [135]. Arabidopsis plant leaves exposed to UV-B stress show a considerable increase in CPDs, besides an increase in ploidy. This damage can be repaired by photoreactivation for which the enzyme photolyase PHR1 is required, and its expression is stimulated by UV-B treatment. Under normal conditions, PHR1 expression is restricted by the association of DEL1 to its promoter, but UV-B decreases the abundance of DEL1 messenger RNA, releasing such repression [136]; since DEL1 is a negative regulator of endoreduplication, when its levels decrease, accumulation of DNA is favored due to the increase in CCS52A expression (Fig. 3).
Other genes in plants that could be influenced via the E2F pathway during DNA damage are those codifying for the three subunits of the chromatin assembly factor (CAF-1). The largest subunit, FASCIATA1 (FAS1), catalyzes the transference of histones H3/H4 to the recently synthesized DNA during replication and also participates in chromatin compaction and in HR [137]. The FAS1 promoter possesses two E2F sites that permit to increase or repress its expression, respectively. Under normal conditions, during G1/S, FAS1 forms part of the genes activated by E2Fa and E2Fb because of the transition to the S phase [130,138] and its expression diminishes in late S phase, maybe in response to the activity of E2Fc or DEL1 [139], allowing the advance of the cycle. Nonetheless, a replicative stress activates the expression of the G2 checkpoint genes and the subsequent cell cycle arrest. In a similar way, loss of the FAS1 gene provokes a delay of the S phase and the constitutive expression of DNA integrity checkpoint genes like RAD51 and BRCA1, possibly due to the increase in acetylation of histone H3 (H3ac) in their promoters, and also produces a systemic increase in ploidy [140]. DSB induction with chemical agents like zeocine, or others that cause replicative stress, partially phenocopy loss of FAS1 [140]. Altogether, these data connect the function of several members of the E2F family with the process of endoreduplication, either as part of the normal development of plants or in response to stress in the DNA [141].
In plants, the functional analog of the tumor suppressor p53 is known as SUPPRESSOR OF GAMMA RESPONSE 1 (SOG1). SOG1 is the TF responsible of integrating the signals arising from the DNA damage checkpoint kinases ATM or ATR when there is DNA damage to generate a transcriptional response [142]. Some of the genes whose expression is increased via SOG1 are related to cell cycle arrest like the CDK inhibitors of the SIAMESSE RELATED (SMR) group and of the KIP RELATED PROTEIN (KRP) group, besides inducing the expression of the CDK repressing kinase WEE1. SOG1 function also promotes an increase in ploidy [141], perhaps by stimulating the expression of WRKY25, a TF that associates to the promoter of DEL1 to repress its expression [143,144]. In parallel, SOG1 function promotes the expression of numerous genes concerned with DNA repair like CYCB1;1, RAD51 or BRCA1 [143].
E2F not only participates in endoreduplication regulation, but also directly acts in response to DNA damage, as evidenced by the transcriptional regulation of BRCA1, and the promotion of localization of RBR-BRCA1 in genomic sites where lesions accumulate [145], signaled by the presence of the phosphorylated form of histone H2A.X (γH2A.X), a marker of damage in DNA. On the contrary, while RAD51 expression can be regulated by E2Fa and RBR under normal conditions, its expression is independent of the E2F pathway when DNA damage is produced. However, the RBR protein, as with BRCA1, forms pools in genomic sites with lesions and co-localizes with RAD51, but in an E2F independent manner, as e2fa mutations illustrate, with an enhanced sensitivity to DNA damage, but unaffected RBR-RAD51 pools [146].
Other proteins not related to RBR can also repress the E2F activity, such is the case of SNI1, one of the eight subunits of the STRUCTURAL MAINTENANCE OF CHROMOSOMES SMC5/6 complex [147,148]. One of the best-known functions of this complex is the resolution of complex structures in DNA during DNA damage repair by HR, and during the blockage of replication forks during the S phase. It has recently been discovered that Arabidopsis SNI1 works similarly to RBR, masking the E2F transactivation domain and also recruiting histone deacetylases to repress expression of its replication target genes, including SNI1 itself [149]. This new function creates a link between the DNA integrity checkpoint and the cell cycle. On the other hand, it is known that pathogen-infected plants trigger the Salicylic Acid (SA) pathway to mediate the immune response, which apart from inducing expression of immunity genes, also promotes an increase in plant DNA damage, in the absence of genotoxic agents; accumulation of DNA damage, in turn, stimulates genes of the HR pathway as ATR and RAD17 [150]. SNI1 gene was discovered when sni1 mutants showed an increased immune response in the absence of stimuli, indicating that it was a repressor of the pathway. It is thought that SA-produced DNA damage contributes to signal and stimulate the systemic resistant pathway, since the resistance phenotype of sni1 plants is reverted by mutation of RAD17 and ATR sensors [150]. Additionally, e2fabc mutant plants are more susceptible to infections and are unable to express some of the immune response genes [151]. DEL1 also participates in the immune response directly regulating the expression of the EDS5 gene, a SA transporter (Fig. 3). Consistently, del1 plants show increased immunity in the absence of a pathogenic challenge [152], recalling the phenotype of SNI1 repressor absence. All this evidence shows a tight connection between the E2F pathway, the response to stress by pathogens and the DNA repair pathway.
In addition to SNI1, another protein of the SMC5/6 complex, SUMO E3 ligase MMS21, negatively regulates E2F activity. There are two mechanisms by which MMS21 regulates the cell cycle. The first consists in promoting dissociation of the E2F-DP complex by competing for DPa; the second is consequence of DP SUMOylation, potentiating heterodimer separation. DPa SUMOylation also decreases TF nuclear localization, provoking alterations in the regulation of its target genes. Therefore, mutant plants lacking MMS21 gene, a mutant showing an increased expression of G1/S E2F target genes, have higher expression of PCNA1, MCM3 and ORC2, implying that this E3 ligase could participate in the regulation of the S phase [153]. MMS21 mutation also affects expression of G2/M genes as CDKB1 and CYCB1 but, differently to G1/S genes, expression of G2/M genes decreases, arresting the cell cycle and causing an increase in ploidy. Overexpression of MMS21 attenuates the polyploid phenotype in E2Fa-DPaOE plants, suggesting an additional participation in the regulation of the cell cycle, independently of mediating the response to DNA damage (Fig. 3) [153,154].
During the cell cycle, the γ-tubulin cytoskeleton has a vital role in the reorganization of cell structures. In animal cells centrosomes are the organelles responsible of nucleating and polarizing microtubules starting from two centrioles that during interphase (probably in G1/S) are duplicated and help to organize cell organelles. During mitosis, these microtubules will form the mitotic spindle, the scaffolding in which chromosomes will anchor to be segregated later towards the opposite poles of the spindle; microtubules will also participate in the separation of the two new cells during cytokinesis. A structural role of γ-tubulin is not the only contribution to the cell cycle; apparently, it can also physically regulate the position of the genome within the nucleus in collaboration with the nuclear envelope and can also associate to transcriptional complexes, although these functions are not well understood [155].
There are no centrosomes in plants, however, microtubules form a structure known as microtubule organizing center (MTOC), whose function is like that of centrioles. In Arabidopsis, besides being part of the cytoskeleton, γ-tubulin associates to E2Fa-c in the nucleus and blocks their activity, but differently to MMS21, tubulin neither dissociates the E2F-DP complex nor blocks the DNA association. The ternary E2F-DP-tubulin complex, associated to chromatin, promotes cell cycle arrest, becoming another type of repressor complex for both G1/S and G2/M, preventing the increase in ploidy. This atypical function of γ-tubulin could coexist with the RBR pathway, since it interacts with the amino end and the DNA binding domain of E2Fa-c [156]. Finally, although in mammals γ-tubulin can interact with activating E2Fs (E2F1-3), ternary complex formation is circumscribed to G1 and G1/S, while in plants it also occurs in S and G2, having a unique role in endocycle regulation [157,158].
5.3 Seeds, Cell Cycle and the E2F Pathway
Seed development is a key innovation in the history of plant evolution that allowed them to colonize predominantly dry continental environments. Seeds can originate by ovule fertilization (sexual reproduction), or by apomixis (asexual reproduction), and are composed of three main structures: i) The zygote (2n), resulting from the fusion of a spermatic nucleus (n) with the arquegonium egg cell (n), ii) The endosperm (3n) coming from the fusion of another spermatic nucleus from pollen with two polar nuclei of the embryo sac central cell; and iii) Testa, a protecting layer of maternal origin [159].
In cereals, the development and growth of a seed is composed of three phases: Phase 1, starting with fertilization and includes cellularization of both the embryo and the endosperm. Phase 2, in which there is a notable increase in mass, attributed to cell elongation and massive accumulation of storage compounds; increase in ploidy of endosperm cells by endoreduplication also occurs in this phase. In Phase 3, or maturation phase, seeds enter a process of drying, loosing up to 80% water and in some species the dormancy process begins [160].
During embryogenesis, disturbing the G1/S CDKA;1 or CycA3 axis, or loss of APC/C components like HOBBIT (CDC27 homolog) slow down the cell cycle affecting the formation of embryo patterns, morphogenesis and tissue specification, like the root meristem [161–163]. Null cdka;1 mutations in Arabidopsis show a reduced number of cells, together with a drastic delay in embryogenesis, while rbr mutants reverse the post-embryonic defects of cdka;1 mutants, indicating the participation of both genes in the control of cell proliferation in the embryo [164]. Maize seeds with low levels of RBR1 show an increase in the expression of E2F/DP target genes, and an increase in the rate of cell division and DNA content in mature endosperm cells [125]. However, reserve material content and seed size are not affected, suggesting a functional redundancy with other members of the RBR pathway or other compensatory mechanisms [165].
In Arabidopsis, the seed maturation stage causes a reduction in nuclear size and a higher chromatin condensation. Nonetheless, there is expression of certain genes like those required for dormancy [166,167]. e2fa and e2fb mutant plants do not show alterations in cellularization during the embryo formation stage, but exhibit premature expression of seed maturation genes, suggesting that these E2Fs could participate in the balance between proliferation and differentiation during seed development performing the atypical function of maturation gene repression [168].
After maize seed maturation, during water imbibition, reactivation of the cell cycle would be necessary for proper germination to occur [169]. In maize, most of the cells in the embryo axis are blocked in the G1 phase of the cycle, and interestingly, they seem to have in store most of the components of the cell cycle machinery, like cyclins, CDKs, CDK inhibitors (ICK/KRP), active cyclin-CDK complexes, PCNA, among many others [170–178]. Recently, Sánchez-Camargo et al. [114] reported the existence of 12 genes of the E2F/DP pathway and similarly to other cell cycle players, mRNAs of 9 out of the 12 genes are present in embryo axes of dry seeds, and their levels fluctuate during germination. Detailed studies show that maize E2Fa/b1;1 can act as a transcriptional activator, while E2Fc does not have this capacity and evidence suggests that it is a transcriptional repressor. In many plant species E2Fc proteins have a truncated RBR interaction domain, whereas activating proteins have an additional motif towards the carboxyl end. Maize E2Fc could have lost this motif during evolution and therefore its transactivating function (Romero-Rodríguez and Sánchez-Camargo, data not published), thus becoming a repressor competing for E2F response elements or recruiting RBR to E2F target sites.
In dry maize seeds and during germination, replication genes like MCM3;1, RPA2 and PCNA1 show open chromatin marks as H3K9/K14ac and H3K4me3. However, their expression levels vary during germination. Immunoprecipitation experiments of chromatin associated E2F in dry seeds, indicate that the repressor E2Fc and RBR1 are bound to S phase promoters and after imbibition, these are replaced by activator E2Fa/b1;1, together with an increase in the expression of target genes [114]. At later germination times, when the first cell cycle round has finished [173], RBR1, E2Fa/b1;1 and E2Fc can be detected bound to chromatin, possibly forming repressor complexes in DNA. In this way, E2Fc-RBR1 could prevent the start of a premature cell cycle, and then RBR could regulate E2Fa/b1;1 function both for reentry to the next cell cycle and for exit to differentiation (Fig. 4) [114]. Evidence accumulated for years tends to indicate that seeds could possess some kind of biochemical memory formed, among other factors, by the Cyc/CDK/KRP signaling pathway, the E2F/DP/RBR factors and the chromatin state in the embryos. The proper communication among these pathways would allow the coordination of seed germination and the first round of cell cycles in the embryo axis, providing the physiological and environmental conditions are propitious.
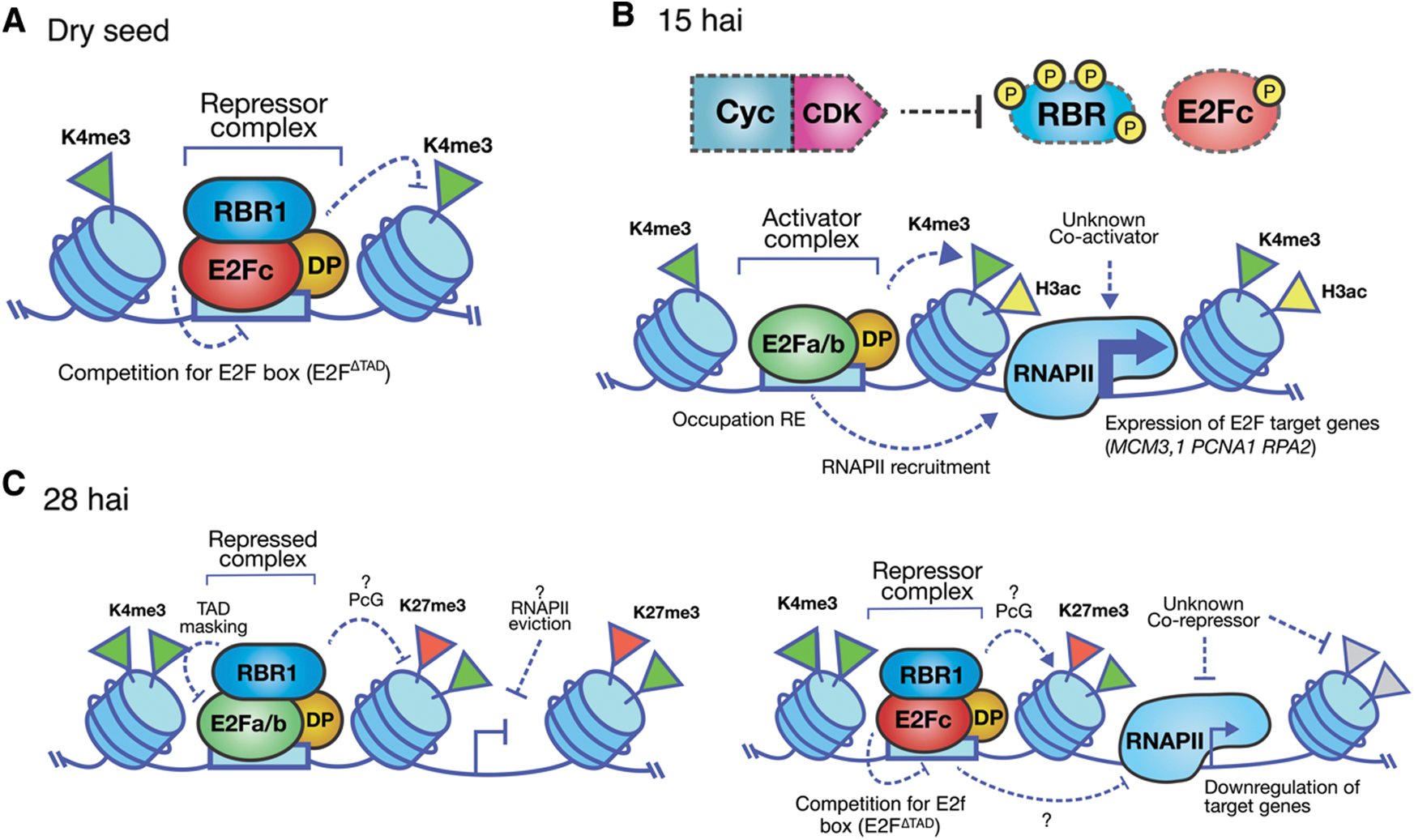
Figure 4: Model of the E2F/DP pathway during maize seed germination. A. In the dry seed the promoters of G1/S genes are decorated by accessible chromatin marks such as H3K4me3. However, they are occupied by the repressor complex E2Fc-RBR1, preventing from ectopic expression of the G1/S genes. B. After 15 h of imbibition (hai), the activity of Cyc/CDK complexes increase, being RBR and E2Fc some of their kinase substrates. At the same time, the open chromatin mark H3ac is enriched in the region of the transcription start site of the G1/S genes withthe concomitant increase in expression. C. Around 30 hai, the RBR1 protein co-localizes with E2Fa/b1;1 and E2Fc on G1/S promoters, accompanied by the appearance of the repressor mark H3K27me3 in these loci. The repressor function of these complexes could favor either the exit of the cycle or mark the beginning of a new round of cell division
Understanding cell cycle control has been of utmost interest for a long time since in multicellular organisms the precise balance between cell proliferation and differentiation is determinant for an adequate life cycle. Proof of that is the high incidence of oncogenesis in mammals when the cell cycle homeostasis is disturbed. For similar reasons, knowing the dynamics of the cycle in plants, with a genetic plasticity that causes amazement, can offer new perspectives to generate strategies, both with therapeutic purposes and with the purpose of increasing the productivity of agricultural products. Therefore, understanding the role of the E2F family as the core of transcriptional regulation of the cell cycle is imperative. Both, regulation of the E2F/RB pathway and its function illustrates the enormous complexity inherent to the coordination of proliferation, growth, and development. Likewise, the concept of E2F as a control module of multiple pathways apparently biochemically distant, like endoreduplication, DNA repair, immunity, cell death and even the dynamics of cell structure, is crucial to have an integral vision, that goes from understanding cell proliferation control to ascertain into the evolutionary processes that eukaryotes have gone through to travel from an unicellular form to the constitution of complex multicellular organisms, and how the environment has had an influence.
Funding Statement: This work was supported by Consejo Nacional de Ciencia y Tecnología [Grant No. CB220661, Postgraduate and Mobility grants to V.A.S.C. and SNI III grant to S.R.R.] and Universidad Nacional Autónoma de México [DGAPA-PAPIIT IN215316, PAIP 5000-9124, PAIP 5000-9130 and PAEP-UNAM grant to V.A.S.C.].
Conflicts of Interest: The authors declare that they have no conflicts of interest regarding this study.
1. Feher, J. J. (2017). Quantitative human physiology: An introduction. London, UK: Academic Press. [Google Scholar]
2. Kovesdi, I., Reichel, R., Nevins, J. R. (1987). Role of an adenovirus E2 promoter binding factor in E1A-mediated coordinate gene control. Proceedings of the National Academy of Sciences of the United States of America, 84(8), 2180–2184. DOI 10.1073/pnas.84.8.2180. [Google Scholar] [CrossRef]
3. Lee, K. A., Green, M. R. (1987). A cellular transcription factor E4F1 interacts with an E1a-inducible enhancer and mediates constitutive enhancer function in vitro. EMBO Journal, 6(5), 1345–1353. DOI 10.1002/j.1460-2075.1987.tb02374.x. [Google Scholar] [CrossRef]
4. Lee, K. A., Hai, T. Y., SivaRaman, L., Thimmappaya, B., Hurst, H. C. et al. (1987). A cellular protein, activating transcription factor, activates transcription of multiple E1A-inducible adenovirus early promoters. Proceedings of the National Academy of Sciences of the United States of America, 84(23), 8355–8359. DOI 10.1073/pnas.84.23.8355. [Google Scholar] [CrossRef]
5. Reichel, R., Kovesdi, I., Nevins, J. R. (1988). Activation of a preexisting cellular factor as a basis for adenovirus E1A-mediated transcription control. Proceedings of the National Academy of Sciences of the United States of America, 85(2), 387–390. DOI 10.1073/pnas.85.2.387. [Google Scholar] [CrossRef]
6. Hearing, P., Shenk, T. (1983). The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell, 33(3), 695–703. DOI 10.1016/0092-8674(83)90012-0. [Google Scholar] [CrossRef]
7. Girling, R., Partridge, J. F., Bandara, L. R., Burden, N., Totty, N. F. et al. (1993). A new component of the transcription factor DRTF1/E2F. Nature, 362(6415), 83–87. DOI 10.1038/362083a0. [Google Scholar] [CrossRef]
8. Helin, K., Wu, C. L., Fattaey, A. R., Lees, J. A., Dynlacht, B. D. et al. (1993). Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes & Development, 7(10), 1850–1861. DOI 10.1101/gad.7.10.1850. [Google Scholar] [CrossRef]
9. Bracken, A. P., Ciro, M., Cocito, A., Helin, K. (2004). E2F target genes: Unraveling the biology. Trends in Biochemical Sciences, 29(8), 409–417. DOI 10.1016/j.tibs.2004.06.006. [Google Scholar] [CrossRef]
10. Cam, H., Balciunaite, E., Blais, A., Spektor, A., Scarpulla, R. C. et al. (2004). A common set of gene regulatory networks links metabolism and growth inhibition. Molecular Cell, 16(3), 399–411. DOI 10.1016/j.molcel.2004.09.037. [Google Scholar] [CrossRef]
11. DeGregori, J., Johnson, D. G. (2006). Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Current Molecular Medicine, 6(7), 739–748. [Google Scholar]
12. Dimova, D. K., Dyson, N. J. (2005). The E2F transcriptional network: Old acquaintances with new faces. Oncogene, 24(17), 2810–2826. DOI 10.1038/sj.onc.1208612. [Google Scholar] [CrossRef]
13. Ren, B., Cam, H., Takahashi, Y., Volkert, T., Terragni, J. et al. (2002). E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes & Development, 16(2), 245–256. DOI 10.1101/gad.949802. [Google Scholar] [CrossRef]
14. Burke, J. R., Deshong, A. J., Pelton, J. G., Rubin, S. M. (2010). Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. Journal of Biological Chemistry, 285(21), 16286–16293. DOI 10.1074/jbc.M110.108167. [Google Scholar] [CrossRef]
15. Lee, C., Chang, J. H., Lee, H. S., Cho, Y. (2002). Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor. Genes & Development, 16(24), 3199–3212. DOI 10.1101/gad.1046102. [Google Scholar] [CrossRef]
16. Kosugi, S., Ohashi, Y. (2002). E2Ls, E2F-like repressors of Arabidopsis that bind to E2F sites in a monomeric form. Journal of Biological Chemistry, 277(19), 16553–16558. DOI 10.1074/jbc.M200913200. [Google Scholar] [CrossRef]
17. Mariconti, L., Pellegrini, B., Cantoni, R., Stevens, R. Bergounioux, C. et al. (2002). The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. Journal of Biological Chemistry, 277(12), 9911–9919. DOI 10.1074/jbc.M110616200. [Google Scholar] [CrossRef]
18. de Bruin, A., Maiti, B., Jakoi, L., Timmers, C., Buerki, R. et al. (2003). Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. Journal of Biological Chemistry, 278(43), 42041–42049. DOI 10.1074/jbc.M308105200. [Google Scholar] [CrossRef]
19. Maiti, B., Li, J., De Bruin, A., Gordon, F., Timmers, C. et al. (2005). Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. Journal of Biological Chemistry, 280(18), 18211–18220. DOI 10.1074/jbc.M501410200. [Google Scholar] [CrossRef]
20. Frolov, M. V., Dyson, N. J. (2004). Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. Journal of Cell Science, 117(11), 2173–2181. DOI 10.1242/jcs.01227. [Google Scholar] [CrossRef]
21. Morgunova, E., Yin, Y., Jolma, A., Dave, K., Schmierer, B. et al. (2015). Structural insights into the DNA-binding specificity of E2F family transcription factors. Nature Communications, 6(1), 1–8. DOI 10.1038/ncomms10050. [Google Scholar] [CrossRef]
22. Müller, H., Helin, K. (2000). The E2F transcription factors: Key regulators of cell proliferation. Biochimica et Biophysica Acta, 1470(1), M1–M12. [Google Scholar]
23. Nahle, Z., Polakoff, J., Davuluri, R. V., McCurrach, M. E., Jacobson, M. D. et al. (2002). Direct coupling of the cell cycle and cell death machinery by E2F. Nature Cell Biology, 4(11), 859–864. DOI 10.1038/ncb868. [Google Scholar] [CrossRef]
24. Qin, X. Q., Livingston, D. M., Kaelin, W. G., Adams, P. D. (1994). Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proceedings of the National Academy of Sciences of the United States of America, 91(23), 10918–10922. DOI 10.1073/pnas.91.23.10918. [Google Scholar] [CrossRef]
25. Shan, B., Lee, W. H. (1994). Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Molecular and Cell Biology, 14(12), 8166–8173. DOI 10.1128/MCB.14.12.8166. [Google Scholar] [CrossRef]
26. Croxton, R., Ma, Y., Song, L., Haura, E. B., Cress, W. D. (2002). Direct repression of the Mcl-1 promoter by E2F1. Oncogene, 21(9), 1359–1369. DOI 10.1038/sj.onc.1205157. [Google Scholar] [CrossRef]
27. He, Y., Armanious, M. K., Thomas, M. J., Cress, W. D. (2000). Identification of E2F-3B, an alternative form of E2F-3 lacking a conserved N-terminal region. Oncogene, 19(30), 3422–3433. DOI 10.1038/sj.onc.1203682. [Google Scholar] [CrossRef]
28. Leone, G., Nuckolls, F., Ishida, S., Adams, M., Sears, R. et al. (2000). Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Molecular and Cell Biology, 20(10), 3626–3632. DOI 10.1128/MCB.20.10.3626-3632.2000. [Google Scholar] [CrossRef]
29. Chong, J. L., Tsai, S. Y., Sharma, N., Opavsky, R., Price, R. et al. (2009). E2f3a and E2f3b contribute to the control of cell proliferation and mouse development. Molecular and Cell Biology, 29(2), 414–424. DOI 10.1128/MCB.01161-08. [Google Scholar] [CrossRef]
30. Chong, J. L., Wenzel, P. L., Sáenz-Robles, M. T., Nair, V., Ferrey, A. et al. (2009). E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature, 462(7275), 930–934. DOI 10.1038/nature08677. [Google Scholar] [CrossRef]
31. Danielian, P. S., Friesenhahn, L. B., Faust, A. M., West, J. C., Caron, A. M. et al. (2008). E2f3a and E2f3b make overlapping but different contributions to total E2f3 activity. Oncogene, 27(51), 6561–6570. DOI 10.1038/onc.2008.253. [Google Scholar] [CrossRef]
32. Talluri, S., Dick, F. A. (2014). Regulation of transcription and chromatin structure by pRB: Here, there and everywhere. Cell Cycle, 11(17), 3189–3198. DOI 10.4161/cc.21263. [Google Scholar] [CrossRef]
33. de Stanchina, E., McCurrach, M. E., Zindy, F., Shieh, S. Y., Ferbeyre, G. et al. (1998). E1A signaling to p53 involves the p19ARF tumor suppressor. Genes & Development, 12(15), 2434–2442. DOI 10.1101/gad.12.15.2434. [Google Scholar] [CrossRef]
34. Aslanian, A., Iaquinta, P. J., Verona, R., Lees, J. A. (2004). Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics. Genes & Development, 18(12), 1413–1422. DOI 10.1101/gad.1196704. [Google Scholar] [CrossRef]
35. Asp, P., Acosta-Alvear, D., Tsikitis, M., van Oevelen, C., Dynlacht, B. D. (2009). E2f3b plays an essential role in myogenic differentiation through isoform-specific gene regulation. Genes & Development, 23(1), 37–53. DOI 10.1101/gad.1727309. [Google Scholar] [CrossRef]
36. Popov, B., Chang, L. S., Serikov, V. (2005). Cell cycle-related transformation of the E2F4-p130 repressor complex. Biochemical and Biophysical Research Communications, 336(3), 762–769. DOI 10.1016/j.bbrc.2005.08.163. [Google Scholar] [CrossRef]
37. Gaubatz, S., Lees, J. A., Lindeman, G. J., Livingston, D. M. (2001). E2F4 is exported from the nucleus in a CRM1-dependent manner. Molecular and Cell Biology, 21(4), 1384–1392. DOI 10.1128/MCB.21.4.1384-1392.2001. [Google Scholar] [CrossRef]
38. Lukas, J., Bartkova, J., Rohde, M., Strauss, M., Bartek, J. (1995). Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Molecular and Cell Biology, 15(5), 2600–2611. DOI 10.1128/MCB.15.5.2600. [Google Scholar] [CrossRef]
39. Komori, H., Goto, Y., Kurayoshi, K., Ozono, E., Iwanaga, R. et al. (2018). Differential requirement for dimerization partner DP between E2F-dependent activation of tumor suppressor and growth-related genes. Scientific Reports, 8(1), 1–12. DOI 10.1038/s41598-017-17765-5. [Google Scholar] [CrossRef]
40. Wang, D., Russell, J. L., Johnson, D. G. (2000). E2F4 and E2F1 have similar proliferative properties but different apoptotic and oncogenic properties in vivo. Molecular and Cell Biology, 20(10), 3417–3424. DOI 10.1128/MCB.20.10.3417-3424.2000. [Google Scholar] [CrossRef]
41. Danielian, P. S., Hess, R. A., Lees, J. A. (2016). E2f4 and E2f5 are essential for the development of the male reproductive system. Cell Cycle, 15(2), 250–260. DOI 10.1080/15384101.2015.1121350. [Google Scholar] [CrossRef]
42. Deschênes, C., Alvarez, L., Lizotte, M., Vézina, A., Rivard, N. (2004). The nucleocytoplasmic shuttling of E2F4 is involved in the regulation of human intestinal epithelial cell proliferation and differentiation. Journal of Cellular Physiology, 199(2), 262–273. DOI 10.1002/jcp.10455. [Google Scholar] [CrossRef]
43. Wang, D., Russell, J., Xu, H., Johnson, D. G. (2001). Deregulated expression of DP1 induces epidermal proliferation and enhances skin carcinogenesis. Molecular Carcinogenesis, 31(2), 90–100. DOI 10.1002/mc.1044. [Google Scholar] [CrossRef]
44. Dingar, D., Konecny, F., Zou, J., Sun, X., von Harsdorf, R. (2012). Anti-apoptotic function of the E2F transcription factor 4 (E2F4)/p130, a member of retinoblastoma gene family in cardiac myocytes. Journal of Molecular and Cellular Cardiology, 53(6), 820–828. DOI 10.1016/j.yjmcc.2012.09.004. [Google Scholar] [CrossRef]
45. Bindra, R. S., Crosby, M. E., Glazer, P. M. (2007). Regulation of DNA repair in hypoxic cancer cells. Cancer and Metastasis Reviews, 26(2), 249–260. DOI 10.1007/s10555-007-9061-3. [Google Scholar] [CrossRef]
46. Hegan, D. C., Lu, Y., Stachelek, G. C., Crosby, M. E., Bindra, R. S. et al. (2010). Inhibition of poly (ADP-ribose) polymerase down-regulates BRCA1 and RAD51 in a pathway mediated by E2F4 and p130. Proceedings of the National Academy of Sciences of the United States of America, 107(5), 2201–2206. DOI 10.1073/pnas.0904783107. [Google Scholar] [CrossRef]
47. Lee, B. K., Bhinge, A. A., Iyer, V. R. (2011). Wide-ranging functions of E2F4 in transcriptional activation and repression revealed by genome-wide analysis. Nucleic Acids Research, 39(9), 3558–3573. DOI 10.1093/nar/gkq1313. [Google Scholar] [CrossRef]
48. Apostolova, M. D., Ivanova, I. A., Dagnino, C., D’Souza, S. J. A., Dagnino, L. (2002). Active nuclear import and export pathways regulate E2F-5 subcellular localization. Journal of Biological Chemistry, 277(37), 34471–34479. DOI 10.1074/jbc.M205827200. [Google Scholar] [CrossRef]
49. Reed, S. A., Ouellette, S. E., Liu, X., Allen, R. E., Johnson, S. E. (2007). E2F5 and LEK1 Translocation to the nucleus is an early event demarcating myoblast quiescence. Journal of Cellular Biochemistry, 101(6), 1394–1408. DOI 10.1002/jcb.21256. [Google Scholar] [CrossRef]
50. Sardet, C., Vidal, M., Cobrinik, D., Geng, Y., Onufryk, C. et al. (1995). E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle. Proceedings of the National Academy of Sciences of the United States of America, 92(6), 2403–2407. DOI 10.1073/pnas.92.6.2403. [Google Scholar] [CrossRef]
51. Wu, E. W., Clemens, K. E., Heck, D. V., Münger, K. (1993). The human papillomavirus E7 oncoprotein and the cellular transcription factor E2F bind to separate sites on the retinoblastoma tumor suppressor protein. Journal of Virology, 67(4), 2402–2407. DOI 10.1128/JVI.67.4.2402-2407.1993. [Google Scholar] [CrossRef]
52. Castellsague, X., De Sanjose, S., Albero, G., Ferrer, E., Byrne, S. (2007). HPV and cervical cancer in the world: 2007 report. Vaccine, 25(3), C1–C230. www.who.int/hpvcentre. [Google Scholar]
53. Tang, S., Tao, M., McCoy, J. P., Zheng, Z. M. (2006). The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16-or type 18-positive cervical cancer cell lines via translation reinitiation. Journal of Virology, 80(9), 4249–4263. DOI 10.1128/JVI.80.9.4249-4263.2006. [Google Scholar] [CrossRef]
54. Villa, L. L., Schlegel, R. (1991). Differences in transformation activity between HPV-18 and HPV-16 map to the viral LCR-E6-E7 region. Virology, 181(1), 374–377. DOI 10.1016/0042-6822(91)90507-8. [Google Scholar] [CrossRef]
55. Teissier, S., Pang, C. L., Thierry, F. (2010). The E2F5 repressor is an activator of E6/E7 transcription and of the S-phase entry in HPV18-associated cells. Oncogene, 29(36), 5061–5070. DOI 10.1038/onc.2010.246. [Google Scholar] [CrossRef]
56. Cartwright, P., MuÈller, H., Wagener, C., Holm, K., Helin, K. (1998). E2F-6: A novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene, 17(5), 611–623. DOI 10.1038/sj.onc.1201975. [Google Scholar] [CrossRef]
57. Morkel, M., Wenkel, J., Bannister, A. J., Kouzarides, T., Hagemeier, C. (1997). An E2F-like repressor of transcription. Nature, 390(6660), 567–568. DOI 10.1038/37507. [Google Scholar] [CrossRef]
58. Trimarchi, J. M., Fairchild, B., Verona, R., Moberg, K., Andon, N. et al. (1998). E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proceedings of the National Academy of Sciences of the United States of America, 95(6), 2850–2855. DOI 10.1073/pnas.95.6.2850. [Google Scholar] [CrossRef]
59. El Hassan, M. A., Yu, T., Song, L., Bremner, R. (2015). Polycomb repressive complex 2 confers BRG1 dependency on the CIITA locus. Journal of Immunology, 194(10), 5007–5013. DOI 10.4049/jimmunol.1403247. [Google Scholar] [CrossRef]
60. Leung, J. Y., Nevins, J. R. (2012). E2F6 associates with BRG1 in transcriptional regulation. PLoS One, 7(10), e47967. DOI 10.1371/journal.pone.0047967. [Google Scholar] [CrossRef]
61. Aranda, S., Mas, G., Di Croce, L. (2015). Regulation of gene transcription by Polycomb proteins. Science Advances, 1(11), e1500737. DOI 10.1126/sciadv.1500737. [Google Scholar] [CrossRef]
62. Blackledge, N. P., Fursova, N. A., Kelley, J. R., Huseyin, M. K., Feldmann, A. et al. (2020). PRC1 catalytic activity is central to Polycomb system function. Molecular Cell, 77(4), 857–874. DOI 10.1016/j.molcel.2019.12.001. [Google Scholar] [CrossRef]
63. Attwooll, C., Oddi, S., Cartwright, P., Prosperini, E., Agger, K. et al. (2005). A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation-specific manner. Journal of Biological Chemistry, 280(2), 1199–1208. DOI 10.1074/jbc.M412509200. [Google Scholar] [CrossRef]
64. Storre, J., Elsässer, H., Fuchs, M., Ullmann, D., Livingston, D. M. et al. (2002). Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2f6. EMBO Reports, 3(7), 695–700. DOI 10.1093/embo-reports/kvf141. [Google Scholar] [CrossRef]
65. Horard, B., Tatout, C., Poux, S., Pirrotta, V. (2000). Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Molecular and Cell Biology, 20(9), 3187–3197. DOI 10.1128/MCB.20.9.3187-3197.2000. [Google Scholar] [CrossRef]
66. Yu, M., Mazor, T., Huang, H., Huang, H. T., Kathrein, K. L. et al. (2012). Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Molecular Cell, 45(3), 330–343. DOI 10.1016/j.molcel.2011.11.032. [Google Scholar] [CrossRef]
67. Christensen, J., Cloos, P., Toftegaard, U., Klinkenberg, D., Bracken, A. P. et al. (2005). Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Research, 33(17), 5458–5470. DOI 10.1093/nar/gki855. [Google Scholar] [CrossRef]
68. Dyson, N. (1998). The regulation of E2F by pRB-family proteins. Genes & Development, 12(15), 2245–2262. DOI 10.1101/gad.12.15.2245. [Google Scholar] [CrossRef]
69. Helin, K. (1998). Regulation of cell proliferation by the E2F transcription factors. Current Opinion in Genetics & Development, 8(1), 28–35. DOI 10.1016/S0959-437X(98)80058-0. [Google Scholar] [CrossRef]
70. Li, J., Ran, C., Li, E., Gordon, F., Comstock, G. et al. (2008). Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Developmental Cell, 14(1), 62–75. DOI 10.1016/j.devcel.2007.10.017. [Google Scholar] [CrossRef]
71. Moon, N. S., Dyson, N. (2008). E2F7 and E2F8 keep the E2F family in balance. Developmental Cell, 14(1), 1–3. DOI 10.1016/j.devcel.2007.12.017. [Google Scholar] [CrossRef]
72. Liu, B., Shats, I., Angus, S. P., Gatza, M. L., Nevins, J. R. (2013). Interaction of E2F7 transcription factor with E2F1 and C-terminal-binding protein (CtBP) provides a mechanism for E2F7-dependent transcription repression. Journal of Biological Chemistry, 288(34), 24581–24589. DOI 10.1074/jbc.M113.467506. [Google Scholar] [CrossRef]
73. Zheng, N., Fraenkel, E., Pabo, C. O., Pavletich, N. P. (1999). Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes & Development, 13(6), 666–674. DOI 10.1101/gad.13.6.666. [Google Scholar] [CrossRef]
74. Dcona, M. M., Morris, B. L., Ellis, K. C., Grossman, S. R. (2017). CtBP-an emerging oncogene and novel small molecule drug target: Advances in the understanding of its oncogenic action and identification of therapeutic inhibitors. Cancer Biology & Therapy, 18(6), 379–391. DOI 10.1080/15384047.2017.1323586. [Google Scholar] [CrossRef]
75. Singh, S., Johnson, J., Chellappan, S. (2010). Small molecule regulators of Rb-E2F pathway as modulators of transcription. Biochimica et Biophysica Acta, 1799(10–12), 788–794. DOI 10.1016/j.bbagrm.2010.07.004. [Google Scholar] [CrossRef]
76. Qiao, H., Di Stefano, L., Tian, C., Li, Y. Y., Yin, Y. H. et al. (2007). Human TFDP3, a novel DP protein, inhibits DNA binding and transactivation by E2F. Journal of Biological Chemistry, 282(1), 454–466. DOI 10.1074/jbc.M606169200. [Google Scholar] [CrossRef]
77. Milton, A., Luoto, K., Ingram, L., Munro, S., Logan, N. et al. (2006). A functionally distinct member of the DP family of E2F subunits. Oncogene, 25(22), 3212–3218. DOI 10.1038/sj.onc.1209343. [Google Scholar] [CrossRef]
78. Ingram, L., Munro, S., Coutts, A. S., La Thangue, N. B. (2011). E2F-1 regulation by an unusual DNA damage-responsive DP partner subunit. Cell Death & Differentiation, 18(1), 122–132. DOI 10.1038/cdd.2010.70. [Google Scholar] [CrossRef]
79. Tian, C., Lv, D., Qiao, H., Zhang, J., Yin, Y. et al. (2007). TFDP3 inhibits E2F1-induced, p53-mediated apoptosis. Biochemical and Biophysical Research Communications, 361(1), 20–25. DOI 10.1016/j.bbrc.2007.06.128. [Google Scholar] [CrossRef]
80. Huber, H. E., Edwards, G., Goodhart, P. J., Patrick, D. R., Huang, P. S. et al. (1993). Transcription factor E2F binds DNA as a heterodimer. Proceedings of the National Academy of Sciences of the United States of America, 90(8), 3525–3529. DOI 10.1073/pnas.90.8.3525. [Google Scholar] [CrossRef]
81. Biswas, S. C., Liu, D. X., Greene, L. A. (2005). Bim is a direct target of a neuronal E2F-dependent apoptotic pathway. Journal of Neuroscience, 25(37), 8349–8358. DOI 10.1523/JNEUROSCI.1570-05.2005. [Google Scholar] [CrossRef]
82. Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S. et al. (2002). Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America, 99(24), 15524–15529. DOI 10.1073/pnas.242606799. [Google Scholar] [CrossRef]
83. Bottoni, A., Piccin, D., Tagliati, F., Luchin, A., Zatelli, M. C. et al. (2005). miR-15a and miR-16-1 down-regulation in pituitary adenomas. Journal of Cellular Physiology, 204(1), 280–285. DOI 10.1002/jcp.20282. [Google Scholar] [CrossRef]
84. Bonci, D., Coppola, V., Musumeci, M., Addario, A., Giuffrida, R. et al. (2008). The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nature Medicine, 14(11), 1271–1277. DOI 10.1038/nm.1880. [Google Scholar] [CrossRef]
85. O’Brien, J., Hayder, H., Zayed, Y., Peng, C. (2018). Overview of microRNA biogenesis, mechanisms of actions, and circulation. Frontiers in Endocrinology, 9, 402. DOI 10.3389/fendo.2018.00402. [Google Scholar] [CrossRef]
86. Kim, Y. K., Yu, J., Han, T. S., Park, S. Y., Namkoong, B. et al. (2009). Functional links between clustered microRNAs: Suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Research, 37(5), 1672–1681. DOI 10.1093/nar/gkp002. [Google Scholar] [CrossRef]
87. Visone, R., Russo, L., Pallante, P., De Martino, I., Ferraro, A. et al. (2007). MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocrine-Related Cancer, 14(3), 791–798. DOI 10.1677/ERC-07-0129. [Google Scholar] [CrossRef]
88. Wang, X., Gocek, E., Liu, C. G., Studzinski, G. P. (2014). MicroRNAs181 regulate the expression of p27Kip1 in human myeloid leukemia cells induced to differentiate by 1,25-dihydroxyvitamin D3. Cell Cycle, 8(5), 736–741. DOI 10.4161/cc.8.5.7870. [Google Scholar] [CrossRef]
89. Miller, T. E., Ghoshal, K., Ramaswamy, B., Roy, S., Datta, J. et al. (2008). MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. Journal of Biological Chemistry, 283(44), 29897–29903. DOI 10.1074/jbc.M804612200. [Google Scholar] [CrossRef]
90. Sylvestre, Y., De Guire, V., Querido, E., Mukhopadhyay, U. K., Bourdeau, V. et al. (2007). An E2F/miR-20a autoregulatory feedback loop. Journal of Biological Chemistry, 282(4), 2135–2143. DOI 10.1074/jbc.M608939200. [Google Scholar] [CrossRef]
91. Woods, K., Thomson, J. M., Hammond, S. M. (2007). Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. Journal of Biological Chemistry, 282(4), 2130–2134. DOI 10.1074/jbc.C600252200. [Google Scholar] [CrossRef]
92. O’Donnell, K. A., Wentzel, E. A., Zeller, K. I., Dang, C. V., Mendell, J. T. (2005). c-Myc-regulated microRNAs modulate E2F1 expression. Nature, 435(7043), 839–843. DOI 10.1038/nature03677. [Google Scholar] [CrossRef]
93. Bueno, M. J., Gómez de Cedrón, M., Laresgoiti, U., Fernández-Piqueras, J., Zubiaga, A. M. et al. (2010). Multiple E2F-induced microRNAs prevent replicative stress in response to mitogenic signaling. Molecular and Cellular Biology, 30(12), 2983–2995. DOI 10.1128/MCB.01372-09. [Google Scholar] [CrossRef]
94. Grafi, G., Burnett, R. J., Helentjaris, T., Larkins, B. A., DeCaprio, J. A. et al. (1996). A maize cDNA encoding a member of the retinoblastoma protein family: Involvement in endoreduplication. Proceedings of the National Academy of Sciences of the United States of America, 93(17), 8962–8967. DOI 10.1073/pnas.93.17.8962. [Google Scholar] [CrossRef]
95. Xie, Q., Sanz-Burgos, A. P., Hannon, G. J., Gutierrez, C. (1996). Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO Journal, 15(18), 4900–4908. DOI 10.1002/j.1460-2075.1996.tb00870.x. [Google Scholar] [CrossRef]
96. Elena, R. P., Xie, Q., Boniotti, M. B., Gutierrez, C. (1999). The cloning of plant E2F, a retinoblastoma-binding protein, reveals unique and conserved features with animal G1/S regulators. Nucleic Acids Research, 27(17), 3527–3533. DOI 10.1093/nar/27.17.3527. [Google Scholar] [CrossRef]
97. Sekine, M., Ito, M., Uemukai, K., Maeda, Y., Nakagami, H. et al. (1999). Isolation and characterization of the E2F-like gene in plants. FEBS Letters, 460(1), 117–122. DOI 10.1016/S0014-5793(99)01296-X. [Google Scholar] [CrossRef]
98. Magyar, Z., Atanassova, A., De Veylder, L., Rombauts, S., Inze, D. (2000). Characterization of two distinct DP-related genes from Arabidopsis thaliana. FEBS Letters, 486(1), 79–87. DOI 10.1016/S0014-5793(00)02238-9. [Google Scholar] [CrossRef]
99. Ramirez-Parra, E., Gutierrez, C. (2000). Characterization of wheat DP, a heterodimerization partner of the plant E2F transcription factor which stimulates E2F-DNA binding. FEBS Letters, 486(1), 73–78. DOI 10.1016/S0014-5793(00)02239-0. [Google Scholar] [CrossRef]
100. de Jager, S. M., Menges, M., Bauer, U. M., Murra, J. A. (2001). Arabidopsis E2F1 binds a sequence present in the promoter of S-phase-regulated gene AtCDC6 and is a member of a multigene family with differential activities. Plant Molecular Biology, 47(4), 555–568. DOI 10.1023/A:1011848528377. [Google Scholar] [CrossRef]
101. Chabouté, M. E., Clément, B., Sekine, M., Philipps, G., Chaubet-Gigot, N. (2000). Cell cycle regulation of the tobacco ribonucleotide reductase small subunit gene is mediated by E2F-like elements. Plant Cell, 12(10), 1987–1999. [Google Scholar]
102. Albani, D., Mariconti, L., Ricagno, S., Pitto, L., Moroni, C. et al. (2000). DcE2F, a functional plant E2F-like transcriptional activator from Daucus carota. Journal of Biological Chemistry, 275(25), 19258–19267. DOI 10.1074/jbc.M909390199. [Google Scholar] [CrossRef]
103. Kosugi, S., Ohashi, Y. (2002). E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant Journal, 29(1), 45–59. DOI 10.1046/j.1365-313x.2002.01196.x. [Google Scholar] [CrossRef]
104. Bisova, K., Krylov, D. M., Umen, J. G. (2005). Genome-wide annotation and expression profiling of cell cycle regulatory genes in Chlamydomonas reinhardtii. Plant Physiology, 137(2), 475–491. DOI 10.1104/pp.104.054155. [Google Scholar] [CrossRef]
105. Robbens, S., Khadaroo, B., Camasses, A., Derelle, E., Ferraz, C. et al. (2005). Genome-wide analysis of core cell cycle genes in the unicellular green alga Ostreococcus tauri. Molecular Biology and Evolution, 22(3), 589–597. DOI 10.1093/molbev/msi044. [Google Scholar] [CrossRef]
106. Lammens, T., Li, J., Leone, G., De Veylder, L. (2009). Atypical E2Fs: New players in the E2F transcription factor family. Trends in Cell Biology, 19(3), 111–118. DOI 10.1016/j.tcb.2009.01.002. [Google Scholar] [CrossRef]
107. Vlieghe, K., Boudolf, V., Beemster, G. T. S., Maes, S., Magyar, Z. et al. (2005). The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Current Biology, 15(1), 59–63. DOI 10.1016/j.cub.2004.12.038. [Google Scholar] [CrossRef]
108. Black, A. R., Azizkhan-Clifford, J. (1999). Regulation of E2F: A family of transcription factors involved in proliferation control. Gene, 237(2), 281–302. DOI 10.1016/S0378-1119(99)00305-4. [Google Scholar] [CrossRef]
109. Tao, Y., Kassatly, R. F., Cress, W. D., Horowitz, J. M. (1997). Subunit composition determines E2F DNA-binding site specificity. Molecular and Cell Biology, 17(12), 6994–7007. DOI 10.1128/MCB.17.12.6994. [Google Scholar] [CrossRef]
110. Ramirez-Parra, E., Frundt, C., Gutierrez, C. (2003). A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant Journal, 33(4), 801–811. DOI 10.1046/j.1365-313X.2003.01662.x. [Google Scholar] [CrossRef]
111. Vandepoele, K., Vlieghe, K., Florquin, K., Hennig, L., Beemster, G. T. S. et al. (2005). Genome-wide identification of potential plant E2F target genes. Plant Physiology, 139(1), 316–328. DOI 10.1104/pp.105.066290. [Google Scholar] [CrossRef]
112. Su, L. H., Pan, Y. J., Huang, Y. C., Cho, C. C., Chen, C. W. et al. (2011). A novel E2F-like protein involved in transcriptional activation of cyst wall protein genes in Giardia lamblia. Journal of Biological Chemistry, 286(39), 34101–34120. DOI 10.1074/jbc.M111.280206. [Google Scholar] [CrossRef]
113. Vandepoele, K., Raes, J., De Veylder, L., Rouzé, P., Rombauts, S. et al. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell, 14(4), 903–916. DOI 10.1105/tpc.010445. [Google Scholar] [CrossRef]
114. Sánchez-Camargo, V. A., Suárez-Espinoza, C., Romero-Rodríguez, S., Garza-Aguilar, S. M., Stam, M. et al. (2020). Maize E2F transcription factors. Expression, association to promoters of S-phase genes and interaction with the RBR1 protein in chromatin during seed germination. Plant Science, 296(1999), 110491. DOI 10.1016/j.plantsci.2020.110491. [Google Scholar] [CrossRef]
115. Inzé, D., De Veylder, L. (2006). Cell cycle regulation in plant development. Annual Review of Genetics, 40(1), 77–105. DOI 10.1146/annurev.genet.40.110405.090431. [Google Scholar] [CrossRef]
116. del Pozo, J. C., Diaz-Trivino, S., Cisneros, N., Gutierrez, C. (2006). The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell, 18(9), 2224–2235. DOI 10.1105/tpc.105.039651. [Google Scholar] [CrossRef]
117. Shu, Z., Row, S., Deng, W. M. (2018). Endoreplication: The good, the bad, and the ugly. Trends in Cell Biology, 28(6), 465–474. DOI 10.1016/j.tcb.2018.02.006. [Google Scholar] [CrossRef]
118. Fox, D. T., Duronio, R. J. (2012). Endoreplication and polyploidy: Insights into development and disease. Development, 140(1), 3–12. DOI 10.1242/dev.080531. [Google Scholar] [CrossRef]
119. Berckmans, B., Lammens, T., Van Den Daele, H., Magyar, Z., Bögre, L. et al. (2011). Light-dependent regulation of DEL1 is determined by the antagonistic action of E2Fb and E2Fc. Plant Physiology, 157(3), 1440–1451. DOI 10.1104/pp.111.183384. [Google Scholar] [CrossRef]
120. Bhosale, R., Boudolf, V., Cuevas, F., Lu, R., Eekhout, T. et al. (2018). A spatiotemporal DNA endoploidy map of the Arabidopsis root reveals roles for the endocycle in root development and stress adaptation. Plant Cell, 30(10), 2330–2351. DOI 10.1105/tpc.17.00983. [Google Scholar] [CrossRef]
121. Dante, R. A., Larkins, B. A., Sabelli, P. A. (2014). Cell cycle control and seed development. Frontiers in Plant Science, 5, 493. DOI 10.3389/fpls.2014.00493. [Google Scholar] [CrossRef]
122. Sabelli, P. A., Larkins, B. A. (2009). The contribution of cell cycle regulation to endosperm development. Sexual Plant Reproduction, 22(4), 207–219. DOI 10.1007/s00497-009-0105-4. [Google Scholar] [CrossRef]
123. Sabelli, P. A., Larkins, B. A. (2009). Regulation and function of retinoblastoma-related plant genes. Plant Science, 177(6), 540–548. DOI 10.1016/j.plantsci.2009.09.012. [Google Scholar] [CrossRef]
124. Sabelli, P. A., Larkins, B. A. (2009). The development of endosperm in grasses. Plant Physiology, 149(1), 14–26. DOI 10.1104/pp.108.129437. [Google Scholar] [CrossRef]
125. Sabelli, P. A., Liu, Y., Dante, R. A., Lizarraga, L. E., Nguyen, H. N. et al. (2013). Control of cell proliferation, endoreduplication, cell size, and cell death by the retinoblastoma-related pathway in maize endosperm. Proceedings of the National Academy of Sciences of the United States of America, 110(19), E1827– E1836. DOI 10.1073/pnas.1304903110. [Google Scholar] [CrossRef]
126. Castro, A., Bernis, C., Vigneron, S., Labbe, J. C., Lorca, T. (2005). The anaphase-promoting complex: A key factor in the regulation of cell cycle. Oncogene, 24(3), 314–325. DOI 10.1038/sj.onc.1207973. [Google Scholar] [CrossRef]
127. Eloy, N. B., Coppens, F., Beemster, G. T. S., Hemerly, A. S., Ferreira, P. C. G. (2006). The Arabidopsis anaphase promoting complex (APCRegulation through subunit availability in plant tissues. Cell Cycle, 5(17), 1957–1965. DOI 10.4161/cc.5.17.3125. [Google Scholar] [CrossRef]
128. Fülöp, K., Tarayre, S., Kelemen, Z., Horváth, G., Kevei, Z. et al. (2005). Arabidopsis anaphase-promoting complexes: Multiple activators and wide range of substrates might keep APC perpetually busy. Cell Cycle, 4(8), 4084–4092. DOI 10.4161/cc.4.8.1856. [Google Scholar] [CrossRef]
129. Magyar, Z., Horvath, B., Khan, S., Mohammed, B., Henriques, R. et al. (2012). Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO Journal, 31(6), 1480–1493. DOI 10.1038/emboj.2012.13. [Google Scholar] [CrossRef]
130. De Veylder, L., Beeckman, T., Beemster, G. T. S., de Almeida Engler, J., Ormenese, S. et al. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO Journal, 21(6), 1360–1368. DOI 10.1093/emboj/21.6.1360. [Google Scholar] [CrossRef]
131. Liu, Z., Chen, G., Gao, F., Xu, R., Li, N. et al. (2019). Transcriptional repression of the APC/C activator genes CCS52A1/A2 by the mediator complex subunit MED16 controls endoreduplication and cell growth in Arabidopsis. Plant Cell, 31(8), 1899–1912. DOI 10.1105/tpc.18.00811. [Google Scholar] [CrossRef]
132. Sage, E., Girard, P. M., Francesconi, S. (2012). Unravelling UVA-induced mutagenesis. Photochemical Photobiological Sciences, 11(1), 74–80. DOI 10.1039/C1PP05219E. [Google Scholar] [CrossRef]
133. Sutherland, J. C., Griffin, K. P. (1981). Absorption spectrum of DNA for wavelengths greater than 300 nm. Radiation Research, 86(3), 399–410. DOI 10.2307/3575456. [Google Scholar] [CrossRef]
134. Tyrrell, R. E. X. M., Pidoux, M. (1989). Singlet oxygen involvement in the inactivation of cultured human fibroblasts by UVA (334 nm, 365 nm) and near-visible (405 nm) radiations. Photochemistry and Photobiology, 49(4), 407–412. DOI 10.1111/j.1751-1097.1989.tb09187.x. [Google Scholar] [CrossRef]
135. Girard, P. M., Pozzebon, M., Delacôte, F., Douki, T., Smirnova, V. et al. (2008). Inhibition of S-phase progression triggered by UVA-induced ROS does not require a functional DNA damage checkpoint response in mammalian cells. DNA Repair, 7(9), 1500–1516. DOI 10.1016/j.dnarep.2008.05.004. [Google Scholar] [CrossRef]
136. Radziejwoski, A., Vlieghe, K., Lammens, T., Berckmans, B., Maes, S. et al. (2011). Atypical E2F activity coordinates PHR1 photolyase gene transcription with endoreduplication onset. EMBO Journal, 30(2), 355–363. DOI 10.1038/emboj.2010.313. [Google Scholar] [CrossRef]
137. Schönrock, N., Exner, V., Probst, A., Gruissem, W., Hennig, L. (2006). Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. Journal of Biological Chemistry, 281(14), 9560–9568. DOI 10.1074/jbc.M513426200. [Google Scholar] [CrossRef]
138. Sozzani, R., Maggio, C., Varotto, S., Canova, S., Bergounioux, C. et al. (2006). Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiology, 140(4), 1355–1366. DOI 10.1104/pp.106.077990. [Google Scholar] [CrossRef]
139. Del Pozo, J. C., Boniotti, M. B., Gutierrez, C. (2002). Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCFAtSKP2 pathway in response to light. Plant Cell, 14(12), 3057–3071. DOI 10.1105/tpc.006791. [Google Scholar] [CrossRef]
140. Ramirez-Parra, E., Gutierrez, C. (2007). E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiology, 144(1), 105–120. DOI 10.1104/pp.106.094979. [Google Scholar] [CrossRef]
141. Adachi, S., Minamisawa, K., Okushima, Y., Inagaki, S., Yoshiyama, K. et al. (2011). Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 108(24), 10004–10009. DOI 10.1073/pnas.1103584108. [Google Scholar] [CrossRef]
142. Yoshiyama, K. O., Kobayashi, J., Ogita, N., Ueda, M., Kimura, S. et al. (2013). ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Reports, 14(9), 817–822. DOI 10.1038/embor.2013.112. [Google Scholar] [CrossRef]
143. Bourbousse, C., Vegesna, N., Law, J. A. (2018). SOG1 activator and MYB3R repressors regulate a complex DNA damage network in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 115(52), E12453–E12462. DOI 10.1073/pnas.1810582115. [Google Scholar] [CrossRef]
144. O’Malley, R. C., Huang, S. C., Song, L., Lewsey, M. G., Bartlett, A. et al. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell, 165(5), 1280–1292. DOI 10.1016/j.cell.2016.04.038. [Google Scholar] [CrossRef]
145. Horvath, B. M., Kourova, H., Nagy, S., Nemeth, E., Magyar, Z. et al. (2017). Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO Journal, 36(9), 1261–1278. DOI 10.15252/embj.201694561. [Google Scholar] [CrossRef]
146. Biedermann, S., Harashima, H., Chen, P., Heese, M., Bouyer, D. et al. (2017). The retinoblastoma homolog RBR1 mediates localization of the repair protein RAD51 to DNA lesions in Arabidopsis. EMBO Journal, 36(9), 1279–1297. DOI 10.15252/embj.201694571. [Google Scholar] [CrossRef]
147. De Piccoli, G., Cortes-Ledesma, F., Ira, G., Torres-Rosell, J., Uhle, S. et al. (2006). Smc5–Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nature Cell Biology, 8(9), 1032–1034. DOI 10.1038/ncb1466. [Google Scholar] [CrossRef]
148. Wu, N., Yu, H. (2012). The Smc complexes in DNA damage response. Cell & Bioscience, 2(1), 5. DOI 10.1186/2045-3701-2-5. [Google Scholar] [CrossRef]
149. Wang, L., Chen, H., Wang, C., Hu, Z., Yan, S. (2018). Negative regulator of E2F transcription factors links cell cycle checkpoint and DNA damage repair. Proceedings of the National Academy of Sciences of the United States of America, 115(16), E3837–E3845. DOI 10.1073/pnas.1720094115. [Google Scholar] [CrossRef]
150. Yan, S., Wang, W., Marqués, J., Mohan, R., Saleh, A. et al. (2013). Salicylic acid activates DNA damage responses to potentiate plant immunity. Molecular Cell, 52(4), 602–610. DOI 10.1016/j.molcel.2013.09.019. [Google Scholar] [CrossRef]
151. Wang, S., Gu, Y., Zebell, S. G., Anderson, L. K., Wang, W. et al. (2014). A noncanonical role for the CKI-RB-E2F cell-cycle signaling pathway in plant effector-triggered immunity. Cell Host & Microbe, 16(6), 787–794. DOI 10.1016/j.chom.2014.10.005. [Google Scholar] [CrossRef]
152. Chandran, D., Rickert, J., Huang, Y., Steinwand, M. A., Marr, S. K. et al. (2014). Atypical E2F transcriptional repressor DEL1 acts at the intersection of plant growth and immunity by controlling the hormone salicylic acid. Cell Host & Microbe, 15(4), 506–513. DOI 10.1016/j.chom.2014.03.007. [Google Scholar] [CrossRef]
153. Liu, Y., Lai, J., Yu, M., Wang, F., Zhang, J. et al. (2016). The Arabidopsis SUMO E3 ligase AtMMS21 dissociates the E2Fa/DPa complex in cell Cycle regulation. Plant Cell, 28(9), 2225–2237. DOI 10.1105/tpc.16.00439. [Google Scholar] [CrossRef]
154. Ishida, T., Fujiwara, S., Miura, K., Stacey, N., Yoshimura, M. et al. (2009). SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell, 21(8), 2284–2297. DOI 10.1105/tpc.109.068072. [Google Scholar] [CrossRef]
155. Doxsey, S., Zimmerman, W., Mikule, K. (2005). Centrosome control of the cell cycle. Trends in Cell Biology, 15(6), 303–311. DOI 10.1016/j.tcb.2005.04.008. [Google Scholar] [CrossRef]
156. Kállai, B. M., Kourová, H., Chumová, J., Papdi, C., Trögelová, L. et al. (2020). γ-Tubulin interacts with E2F transcription factors to regulate proliferation and endocycling in Arabidopsis. Journal of Experimental Botany, 71(4), 1265–1277. [Google Scholar]
157. Binarová, P., Cenklová, V., Hause, B., Kubátová, E. Lysák, M. et al. (2000). Nuclear γ-tubulin during acentriolar plant mitosis. Plant Cell, 12(3), 433–442. [Google Scholar]
158. Höög, G., Zarrizi, R., von Stedingk, K., Jonsson, K., Alvarado-Kristensson, M. (2011). Nuclear localization of γ-tubulin affects E2F transcriptional activity and S-phase progression. FASEB Journal, 25(11), 3815–3827. DOI 10.1096/fj.11-187484. [Google Scholar] [CrossRef]
159. Reiser, L., Fischer, R. L. (1993). The Ovule and the embryo sac. Plant Cell, 5(10), 1291–1301. DOI 10.2307/3869782. [Google Scholar] [CrossRef]
160. Egli, D. B. (2006). The role of seed in the determination of yield of grain crops. Australian Journal of Agricultural Research, 57(12), 1237–1247. DOI 10.1071/AR06133. [Google Scholar] [CrossRef]
161. Hemerly, A. S., Ferreira, P. C., Van Montagu, M., Engler, G., Inzé, D. (2000). Cell division events are essential for embryo patterning and morphogenesis: Studies on dominant-negative cdc2aAt mutants of Arabidopsis. Plant Journal, 23(1), 123–130. DOI 10.1046/j.1365-313x.2000.00800.x. [Google Scholar] [CrossRef]
162. Yu, Y., Steinmetz, A., Meyer, D., Brown, S., Shen, W. H. (2003). The tobacco A-Type cyclin, Nicta; CYCA3;2, at the nexus of cell division and differentiation. Plant Cell, 15(12), 2763–2777. DOI 10.1105/tpc.015990. [Google Scholar] [CrossRef]
163. Blilou, I., Frugier, F., Folmer, S., Serralbo, O., Willemsen, V. et al. (2002). The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes & Development, 16(19), 2566–2575. DOI 10.1101/gad.237302. [Google Scholar] [CrossRef]
164. Nowack, M. K., Harashima, H., Dissmeyer, N., Zhao, X., Bouyer, D. et al. (2012). Genetic framework of cyclin-dependent kinase function in Arabidopsis. Developmental Cell, 22(5), 1030–1040. DOI 10.1016/j.devcel.2012.02.015. [Google Scholar] [CrossRef]
165. Leiva-Neto, J. T., Grafi, G., Sabelli, P. A., Dante, R. A., Woo, Y. et al. (2004). A dominant negative mutant of cyclin-dependent kinase A reduces endoreduplication but not cell size or gene expression in maize endosperm. Plant Cell, 16(7), 1854–1869. DOI 10.1105/tpc.022178. [Google Scholar] [CrossRef]
166. van Zanten, M.,Koini, M. A., Geyer, R., Liu, Y., Brambilla, V. et al. (2011). Seed maturation Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proceedings of the National Academy of Sciences of the United States of America, 108(50), 20219–20224. DOI 10.1073/pnas.1117726108. [Google Scholar] [CrossRef]
167. Zhao, W., Guan, C., Feng, J., Liang, Y., Zhan, N. et al. (2016). The Arabidopsis CROWDED NUCLEI genes regulate seed germination by modulating degradation of ABI5 protein. Journal of Integrative Plant Biology, 58(7), 669–678. DOI 10.1111/jipb.12448. [Google Scholar] [CrossRef]
168. Leviczky, T., Molnár, E., Papdi, C., Oszi, E., Horváth, G. V. et al. (2019). E2FA and E2FB transcription factors coordinate cell proliferation with seed maturation. Development, 146(22), dev179333. DOI 10.1242/dev.179333. [Google Scholar] [CrossRef]
169. Vázquez-Ramos, J. M., de la Paz Sánchez, M.,. (2003). The cell cycle and seed germination. Seed Science Research, 13(2), 113–130. DOI 10.1079/SSR2003130. [Google Scholar] [CrossRef]
170. Coello-Coutiño, P., García-Ramírez, E., Vázquez-Ramos, J. M. (1994). Preparation of an antibody against a maize DNA polymerase holoenzyme: Identification of the polymerase catalytic subunit. Canadian Journal of Botany, 72(6), 818–822. DOI 10.1139/b94-104. [Google Scholar] [CrossRef]
171. de la Paz Sanchez, M., Torres, A., Boniotti, M. B., Gutierrez, C., Vazquez-Ramos, J. M. (2002). PCNA protein associates to Cdk-A type protein kinases in germinating maize. Plant Molecular Biology, 50(2), 167–175. DOI 10.1023/A:1016029001537. [Google Scholar] [CrossRef]
172. Gutierrez, R., Quiroz-Figueroa, F., Vazquez-Ramos, J. M. (2005). Maize cyclin D2 expression, associated kinase activity and effect of phytohormones during germination. Plant and Cell Physiology, 46(1), 166–173. DOI 10.1093/pcp/pci007. [Google Scholar] [CrossRef]
173. Sanchez, MDLP., Gurusinghe, S. H., Bradford, K. J., Vazquez-Ramos, J. M. (2005). Differential response of PCNA and Cdk-A proteins and associated kinase activities to benzyladenine and abscisic acid during maize seed germination. Journal of Experimental Botany, 56(412), 515–523. DOI 10.1093/jxb/eri029. [Google Scholar] [CrossRef]
174. García, E., Quiroz, F., Uchiyama, Y., Sakaguchi, K., Vázquez-Ramos, J. M. (2006). Expression of a maize δ-type DNA polymerase during seed germination. Physiologia Plantarum, 127(2), 268–276. DOI 10.1111/j.1399-3054.2006.00659.x. [Google Scholar] [CrossRef]
175. Lara-Nunez, A., de Jesus, N., Vazquez-Ramos, J. M. (2008). Maize D4;1 and D5 cyclin proteins in germinating maize. Associated kinase activity and regulation by phytohormones. Physiologia Plantarum, 132(1), 79–88. DOI 10.1111/j.1399-3054.2007.00995.x. [Google Scholar] [CrossRef]
176. Garza-Aguilar, S. M., Lara-Nunez, A., Garcia-Ramirez, E., Vazquez-Ramos, J. M. (2017). Modulation of CycD3;1-CDK complexes by phytohormones and sucrose during maize germination. Physiologia Plantarum, 160(1), 84–97. DOI 10.1111/ppl.12537. [Google Scholar] [CrossRef]
177. Godinez-Palma, S. K., Rosas-Bringas, F. R., Rosas-Bringas, O. G., Garcia-Ramirez, E., Zamora-Zaragoza, J. et al. (2017). Two maize Kip-related proteins differentially interact with, inhibit and are phosphorylated by cyclin D-cyclin-dependent kinase complexes. Journal of Experimental Botany, 68(7), 1585–1597. DOI 10.1093/jxb/erx054. [Google Scholar] [CrossRef]
178. Garza-Aguilar, S. M., Axosco-Marin, J., Lara-Nunez, A., Guerrero-Molina, E. D., Lemus-Enciso, A. T. et al. (2019). Proliferating cell nuclear antigen associates to protein complexes containing cyclins/cyclin dependent kinases susceptible of inhibition by KRPs during maize germination. Plant Science, 280(9), 297–304. DOI 10.1016/j.plantsci.2018.12.020. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |