

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.012897
ARTICLE
Natarajania thailandica sp. nov. (Stilbosporaceae, Diaporthales) from Thailand
1Department of Plant Pathology, Agriculture College, Guizhou University, Guiyang, 550025, China
2Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai, 57100, Thailand
3Inovative Institute of Plant Health, Zhongkai University of Agriculture and Engineering, Guangzhou, 510225, China
*Corresponding Author: Yong Wang. Email: yongwangbis@aliyun.com
Received: 17 July 2020; Accepted: 10 October 2020
Abstract: A fungus similar to the monotypic genus Natarajania, isolated from dead wood and collected in Thailand, is reported. Analysis of partial ribosomal LSU and a protein coding gene (RPB2) demonstrated that the new isolate belonged to Stilbosporaceae, Diaporthales and genetically different from N. indica. It is unique in producing synnematous conidiophores, smooth-walled conidiogenous cells and a flared collarette but lacks an elongated collar-canal which is distinct in the type species. Therefore, sequence data and morphological traits are used to introduce the new species, Natarajania thailandica.
Keywords: Fungi; hyphomycete; one new species; saprobes; taxonomy
Link [1] introduced Stilbosporaceae to include Prosthecium with its asexual morph. Stilbosporaceae has been synonymized under different families [2–4]. Voglmayr et al. [5] established Stilbosporaceae in Diaporthales based on phylogenetic investigation of LSU sequence data and accommodated Stegonsporium and Stilbospora within the family, and synonymized Prosthecium under Stilbospora. The type species of Stilbospora, S. macrosperma, has been linked to its asexual morph Prosthecium ellipsosporum, the generic type of Prosthecium [6]. Natarajania was placed in Stilbosporaceae based on phylogenetic analyses of concatenated LSU, SSU, TEF and RPB2 sequence data by Maharachchikumbura et al. [7]. This is the only hyphomycetous taxon affiliated to the diaporthales which are known to have coelomycetous asexual morphs. Crinitospora, Natarajania, Stegonsporium and Stilbospora are presently placed within this family [8]. The reliability of available sequence data and identification of taxon require further investigation [2].
The monotypic dematiaceous hyphomycete genus, Natarajania, introduced by Pratibha and Bhat [9] is typified by N. indica Pratibha and Bhat. The genus is charactarized by mononemaotus, macronematous, erect, branched conidiophores, monophialidic, verrucose conidiogenous cells with a distinct collar-canal and dark-brown, slimy, smooth conidia [9]. This genus shares similar features of Cryphonectriaceae in Diaporthales which largely comprises coelomycetous asexual morphs [10].
We are carrying out inventories of fungi throughout Thailand where the diversity is proving to be extremely diverse with numerous new species [11,12]. The aim of the present paper is to introduce a second species of Natarajania with evidence from phylogenetic analyses of combined LSU and RPB2 sequence data and morphology. A comprehensive morphological description and illustrations are provided.
Fresh material was collected from Cha-Am, Phetchaburi Province, Thailand in August, 2017. Samples were labeled and brought to the laboratory in Zip lock plastic bags.
2.2 Incubation, Specimen Examination and Isolation
Specimen were incubated in plastic boxes with moistened sterilized tissue papers at room temperature over one week after which they were examined with a Motic SMZ 168 dissecting microscope for fungal fruiting bodies. Scrape mounts of the fungal structures were mounted in water on clean glass slides and stained with Melzer’s reagent or Indian ink or Congo red for microscopic studies and photomicrography. Micro-morphological structures of the fungus were examined and photographs were taken by Nikon ECLIPSE 80i compound microscope fitted with a Canon 600D digital camera. Measurements of photomicrographic structures were made with the Tarosoft®Image Frame Work version 0.9.7. program and images used for figures were assembled with Adobe Photoshop CS6 Extended version 13.0.1 software (Adobe Systems, USA). Isolates were made from single spores following the modified method of Chomnunti et al. [13]. Conidial suspensions were incubated at 25–28°C. Germinating conidia were transferred to potato dextrose agar (PDA) media. Pure cultures were obtained after sub-culturing. The cultural characteristics (mycelium color, shape, texture, and growth rate) were recorded [14]. Cultures and herbarium specimens of isolated fungi of this study were deposited in Mae Fah Luang University culture collection (MFLUCC) and Mae Fah Luang University Herbarium (Herb. MFLU) respectively.
2.3 DNA extraction, Polymerase Chain Reactions (PCR) and Sequencing
Total fungal DNA were obtained from fresh fungal mycelium grown on PDA media at 16–25°C for four weeks using Biospin Fungus Genomic DNA Extraction Kit (BioFlux®, China), (Hangzhou, P. R. China) following the instructions of the manufacturer. DNA amplifications were performed by polymerase chain reaction (PCR) using the primer pairs listed in Tab. 1. Amplifications were performed in 25 μl of PCR mixtures containing 12.5 μl of PCR Master Mix, 9.5 μl of ddH2O, 1 μl of DNA template and 1 μl of each primer set (10 μM). The polymerase chain reactions (PCR) for LSU and RPB2 were performed according to Senanayake et al. [2]. The PCR conditions for LSU was as follows: Initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 45 s, extension at 72°C for 90 s and a final extension at 72°C for 10 min and for RPB2 was: initial denaturation at 94°C for 120 s, followed by 35 amplification cycles of denaturation at 95°C for 45 s, annealing at 57°C for 50 s and extension at 72°C for 90 s. PCR products were visualized and confirmed on 1% agarose electrophoresis gels stained with green stain. Purification and sequencing of PCR products were carried out at Tsingke Company, Beijing, P.R. China. Newly generated DNA sequences were deposited in GenBank database.
Table 1: Partial gene regions and PCR primers used in this study

2.4 Molecular Phylogenetic Analyses
DNA sequences obtained in this study were analyzed and compared with other sequences retrieved from GenBank based on BLAST searches and recently published data. Sequence data were aligned by MAFFT [18,19] and manually improved with BioEdit v.7.2.5 [20]. Maximum likelihood (ML) and Bayesian Inference (BI) analyses of the combined LSU and RPB2 dataset were used.
Maximum-likelihood analysis was performed using the RAxML-HPC2 on XSEDE (v. 8.2.10) [21] in the CIPRES Science Gateway platform [22]. In this analysis nonparametric bootstrap iterations [23] was run in 1,000 replicates with the GTR model and a discrete gamma distribution [24]. Best-fit models for Bayesian and maximum likelihood analyses were selected using MrModeltest v. 2.2 [25] and the best model was GTR + I + G.
Bayesian analysis was performed using MrBayes v. 3.1.2 [26] to evaluate Posterior probabilities (PP) [27,28] by Markov Chain Monte Carlo sampling (MCMC). Six simultaneous Markov chains were run for 1000000 generations and trees were sampled at every 100th generation in two parallel runs. The first 20% of trees, representing the burn-in phase of the analyses were discarded. The remaining 80% trees were used to calculate PP in the majority rule consensus tree (Fig. 1). Phylograms were visualized with FigTree v1.4.0 program [29] and reorganized in Microsoft power point (2007) (Tab. 2).
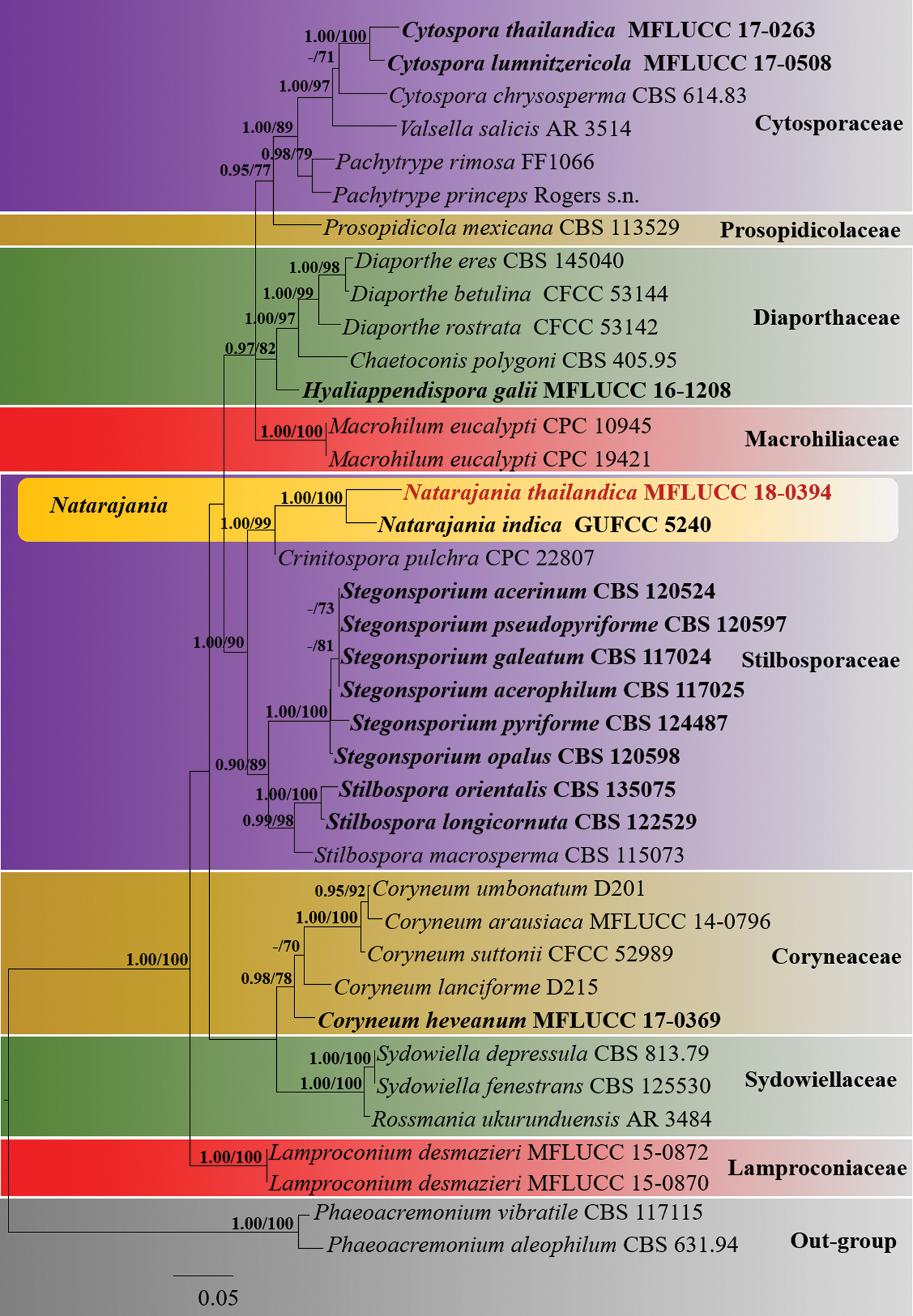
Figure 1: The Maximum likelihood phylogenetic tree based on a combined LSU and RPB2 sequence dataset which comprised 39 strains including Phaeoacremonium aleophilum (CBS 631.94) and P. vibratile (CBS 117115) as the outgroup taxa. Maximum likelihood bootstrap (ML) values >65% and Bayesian posterior probabilities (PP) >0.90 are indicated at the nodes. The ex-type strains are in bold black and the new isolate is in red bold
Table 2: GenBank accessions of taxa used in the phylogenetic analysis. Those of the novel taxon generated in this study are in blue bold and ex-types strains are in black bold


The combined LSU and RPB2 dataset belonging to Coryneaceae, Cytosporaceae, Diaporthaceae, Lamproconiaceae, Macrohilaceae, Prosopidicolaceae, Stilbosporaceae and Sydowiellaceae comprised 39 taxa with Phaeoacremonium aleophilum (CBS 631.94) and P. vibratile (CBS 117115) as the outgroup taxa. RAxML analysis of the combined dataset resulted a best tree with a final ML optimization likelihood value of −17816.176831 (Fig. 1). The matrix comprised 1304 distinct alignment patterns, with 51.11% of undetermined characters. Estimated base frequencies were as follows; gamma distribution shape parameter α = 0.226432; A = 0.240879, C = 0.269773, G = 0.278187, T = 0.211160; substitution rates AC = 1.679941, AG = 2.987693, AT = 1.497541, CG = 1.259527, CT = 7.992461, GT = 1.000000. Phylogenetic trees obtained from BI were similar in topology to the ML tree. Phylogenetic results indicated that the isolate of N. thailandica (MFLUCC18-0394) clustered with N. indica (GUFCC 5240) with strong support (100% ML, 1.00 PP) within the Stilbosporaceae (Fig. 1).
Natarajania J. Pratibha & Bhat, Kavaka 33: 129 (2006) [2005] amend.
Saprobic on dead wood or leaves. Sexual morph: Undetermined. Asexual morph: Colonies numerous, effuse, greyish or dark brown to black, hairy, velvety. Mycelium partly superficial, partly immersed in the substrate, composed of smooth, hyaline to pale brown, branched, septate, thick-walled hyphae. Conidiophores macronematous, mononematous or synnematous, septate, branched above, erect, straight or moderately flexuous, solitary or fasciculate, hyaline or brown to dark brown, smooth and thick-walled; when synnematous compactly intertwined in below half, producing spherical fertile heads at apex. Conidiogenous cells monophialidic, terminal, integrated, elongated, cylindrical, entirely smooth-walled or upper-hall distinctly verrucose and smooth below, pale brown, terminating in a collarette, sometimes with a narrow, elongated, cylindrical, smooth, colourless collar-canal. Conidia slimy, solitary, subglobose to ellipsoidal, dark brown, smooth walled, aseptate, straight or slightly curved, with a truncate base.
Natarajania thailandica Dayar & K.D. Hyde, sp. nov.
Index Fungorum Number: IF557508; Fig. 2

Figure 2: Natarajania thailandica (MFLU 18-0588, holotype). A Substrate. B Colonies on substrate. C-E Conidiophores. F Conidiogenous cells. G-J Conidia. K, L Culture on PDA (K upper, L lower). Scale bars: C, E = 500 μm, D = 100 μm, F–J = 20 μm
Etymology: Species epithet refers to the country, Thailand where it is originated.
Holotype: MFLU 18-0588
Saprobic on unidentified wood. Sexual morph: Undetermined. Asexual morph: Colonies on substrate hairy, numerous, effuse, dark brown to black, velvety. Conidiophores 250−950 × 8−16 μm (x̄ 560 × 14 μm, n = 25), macronematous, synnematous, compactly intertwined in below half, septate, branched above, erect, straight or moderately flexuous, producing spherical fertile heads at apex, 345−555 μm diam. (x̄ 485 μm, n = 20), brown to dark brown, smooth- and thick-walled. Conidiogenous cells 40−48 × 8−12 μm (x̄ 45 × 10 μm, n = 30), monophialidic, terminal, integrated, elongated, cylindrical, smooth-walled, pale brown, terminating in a collarette. Conidia 15−22 × 10−18 μm diam. (x̄ 18.5 × 14 μm, n = 30), solitary, subglobose to ellipsoidal, dark brown, smooth-walled, aseptate.
Culture characteristics: Colonies becoming 3 cm diam. on PDA withing 30 days at 25°C, circular, with smooth margin, white at the beginning, becoming yellowish white/cream after six weeks, flat from the surface, lacking aerial mycelium, reverse yellowish brown.
Material examined: THAILAND, Phetchaburi Province, Cha-Am district, on an unidentified terrestrial wood, 31 Aug. 2017, Dayarathne M.C., MCD 010 (MFLU 18-0588, holotype), ex-type living culture MFLUCC18-0394.
Notes: Natarajania thailandica is morphologically and phylogenetically closely related to N. indica (Figs. 1 and 2). In our phylogenetic analyses, Natarajania thailandica formed a sister lineage to N. indica with high statistical support (100% ML, 1.00 PP) (Fig. 1). Considering there are 2.06% base pair differences in the LSU region between N. thailandica and N. indica (18 bp out of 872 bp without gaps) we consider they are different species. However, RPB2 sequence data are unavailable for N. indica. Furthermore, Natarajania thailandica is clearly distinguished from N. indica by the characteristics of conidiophores, conidiogenous cells and conidia. Conidiophores of N. thailandica are synnematous and spirally arranged, while N. indica has mononematous conidiophores [9]. Additionally, conidiophores of N. thailandica are longer than those of N. indica (250−950 × 8−16 μm vs. 50−120 × 2−4.5 μm) [9]. Conidiogenous cells of N. thailandica are significantly different from those of N. indica in size (40−48 × 8−12 μm vs. 30−45 × 2−3 μm) and by the lack of a collar-canal. Further, the conidiogenous cells of N. indica are verrucose in the upper part whereas N. thailandica has smooth-walled conidiogenous cells. The conidia of N. thailandica are comparatively larger than those of N. indica (15−22 × 10−18 μm vs. 5−7.5 × 3−5 μm) [9]. Considering the morpho-molecular differences, we establish this species as a novel taxon within Natarajania.
Natarajania, typified by N. indica, is characterized by mononematous conidiophores, phialidic verrucose conidiogenous cells with a narrow, elongated, cylindrical, smooth, colorless collar-canal and apically flared collarette [9]. Our novel species N. thailandica widens the morphological diversity of the genus by having synnematous conidiophores and smooth conidiogenous cells without a collar-canal. Crinitospora pulchra B. Sutton & Alcorn showed close phylogenetic affinities to Natarajania in our phylogenetic analyses (Fig. 1). However, Crinitospora pulchra can be easily distinguished from Natarajania by being a coelomycete species with acervular conidiomata and median eu-septate conidia with divergent cellular appendages [30]. Furthermore, Natarajania was earlier confined to India [31], but our sampling from Thailand extends the geographical range to the ASEAN region. Natarajania indica has been reported from leaf litter of Antiaris toxicaria [9], while N. thailandica is isolated from dead wood.
Acknowledgement: Monica Dayarathne thanks Prof. D. Jayarama Bhat, Goa, India and Dr. Dhanushka Wanasinghe for advising and correcting the manuscript of this paper.
Funding Statement: Thailand Research Fund (TRF) grant no RSA5980068 (Biodiversity, phylogeny and role of fungal endophytes on above parts of Rhizophora apiculata) is acknowledged. Monika C. Dayarathne would like to acknowledge projects: National Natural Science Foundation of China (Nos. 31972222, 31560489), and Talent project of Guizhou Science and Technology Cooperation Platform ([2017]5788-5 and [2019]5641) and Guizhou Science, Technology Department International Cooperation Basic project ([2018]5806).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Link, H. F. (1826). Abhandlungen der königlichen Akademie der Wissenschaftenzu Berlin. Aus Dem Jahre, 1824, 145–194. [Google Scholar]
2. Senanayake, I. C., Crous, P. W., Groenewald, J. Z., Maharachchikumbura, S. S. N., Jeewon, R. et al. (2017). Families of Diaporthales based on morphological and phylogenetic evidence. Studies in Mycology, 86, 217–296. DOI 10.1016/j.simyco.2017.07.003. [Google Scholar] [CrossRef]
3. Senanayake, I. C., Jeewon, R., Chomnunti, P., Wanasinghe, D. N., Norphanphoun, C. et al. (2018). Taxonomic circumscription of Diaporthales based on multigene phylogeny and morphology. Fungal Diversity, 93(1), 241–443. DOI 10.1007/s13225-018-0410-z. [Google Scholar] [CrossRef]
4. Hyde, K. D., Norphanphoun, C., Maharachchikumbura, S. S. N., Bhat, D. J., Jones, E. B. G. et al. (2020). Refined families of Sordariomycetes. Mycosphere, 11(1), 305–1059. DOI 10.5943/mycosphere/11/1/7. [Google Scholar] [CrossRef]
5. Voglmayr, H., Jaklitsch, W. M. (2014). Stilbosporaceae resurrected: Generic reclassification and speciation. Persoonia, 33(1), 61–82. DOI 10.3767/003158514X684212. [Google Scholar] [CrossRef]
6. Voglmayr, H., Jaklitsch, W. M. (2008). Prosthecium species with Stegonsporium anamorphs on Acer. Mycological Research, 112(8), 885–905. DOI 10.1016/j.mycres.2008.01.020. [Google Scholar] [CrossRef]
7. Maharachchikumbura, S. S. N., Hyde, K. D., Jones, E. B. G., McKenzie, E. H. C. (2015). Towards a natural classification and backbone tree for Sordariomycetes. Fungal Diversity, 72, 199–301. [Google Scholar]
8. Wijayawardene, N. N., Hyde, K. D., Al-Ani, L. K. T., Tedersoo, L., Rajeshkumar, K. C. et al. (2020). Outline of Fungi and fungus-like taxa. Mycosphere, 11(1), 1060–1456. DOI 10.5943/mycosphere/11/1/8. [Google Scholar] [CrossRef]
9. Pratibha, J., Bhat, D. J. (2006). Natarajania indica gen. et sp. nov., a new dematiaceous hyphomycete from the forests of Western Ghats. India Kavaka, 3, 129–133. [Google Scholar]
10. Jiang, N., Fan, X., Tian, C., Crous, P. W. (2020). Reevaluating Cryphonectriaceae and allied families in Diaporthales. Mycologia, 112(2), 267–292. DOI 10.1080/00275514.2019.1698925. [Google Scholar] [CrossRef]
11. Hyde, K. D., Norphanphoun, C., Chen, J., Dissanayake, A. J., Doilom, M. et al. (2018). Thailand’s amazing diversity: Up to 96% of fungi in northern Thailand are novel. Fungal Diversity, 93(1), 215–239. DOI 10.1007/s13225-018-0415-7. [Google Scholar] [CrossRef]
12. Hyde, K. D., Tennakoon, D. S., Jeewon, R., Bhat, D. J., Maharachchikumbura, S. S. N. (2019). Fungal diversity notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity, 96(1), 1–242. DOI 10.1007/s13225-019-00429-2. [Google Scholar] [CrossRef]
13. Chomnunti, P., Hongsanan, S., Aguirre-Hudson, B., Tian, Q. Persoh, D. et al. (2014). The sooty moulds. Fungal Diversity, 66(1), 1–36. DOI 10.1007/s13225-014-0278-5. [Google Scholar] [CrossRef]
14. Rayner, R. W. (1970). A mycological colour chart. Kew, Surrey, England: CMI and British Mycological Society. [Google Scholar]
15. Vilgalys, R., Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology, 172(8), 4238–4246. [Google Scholar]
16. Rehner, S. A., Samuels, G. J. (1994). Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycological Research, 98(6), 625–634. DOI 10.1016/S0953-7562(09)80409-7. [Google Scholar] [CrossRef]
17. O’Donnell, K., Rooney, A. P., Proctor, R. H., Brown, D. W., McCormick, S. P. et al. (2013). Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genetics and Biology, 52, 20–31. DOI 10.1016/j.fgb.2012.12.004. [Google Scholar] [CrossRef]
18. Katoh, K., Rozewicki, J., Yamada, K. D. (2019). MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics, 20(4), 1160–1166. DOI 10.1093/bib/bbx108. [Google Scholar] [CrossRef]
19. Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780. DOI 10.1093/molbev/mst010. [Google Scholar] [CrossRef]
20. Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98. [Google Scholar]
21. Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30(9), 1312–1313. DOI 10.1093/bioinformatics/btu033. [Google Scholar] [CrossRef]
22. Miller, M. A., Pfeiffer, W., Schwartz, T. (2010). Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop, GCE(1), 1–8. [Google Scholar]
23. Stamatakis, A., Hoover, P., Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML Web Servers. Systematic Biology, 57(5), 758–771. DOI 10.1080/10635150802429642. [Google Scholar] [CrossRef]
24. Liu, Y. J., Whelen, S., Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Molecular Biology and Evolution, 16(12), 1799–1808. DOI 10.1093/oxfordjournals.molbev.a026092. [Google Scholar] [CrossRef]
25. Nylander, J. A. A. (2004). MrModeltest 2.0. Program distributed by the author. Uppsala University: Evolutionary Biology Centre. [Google Scholar]
26. Huelsenbeck, J. P., Rronquist, F. R. (2001). MrBayes: Bayesian inference of phylogenetics trees. Biometrics, 17, 754–755. [Google Scholar]
27. Rannala, B., Yang, Z. (1996). Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. Journal of Molecular Evolution, 43(3), 304–311. DOI 10.1007/BF02338839. [Google Scholar] [CrossRef]
28. Zhaxybayeva, O., Gogarten, J. P. (2002). Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genomics, 3(1), 5088. DOI 10.1186/1471-2164-3-4. [Google Scholar] [CrossRef]
29. Rambaut, A. (2012). Fig.Tree. Tree figure drawing tool. Version 1.4.0. http://tree.bio.ed.ac.uk/software/figtree/. [Google Scholar]
30. Sutton, B. C., Alcorn, J. L. (1985). Undescribed species of Crinitospora gen. nov., Massariothea, Mycoleptodiscus and Neottiosporina from Australia. Transactions of the British mycological Society, 84, 437–445. [Google Scholar]
31. Shenoy, B. D., Jeewon, R., Wang, H., Amandeep, K., Ho, W. H. et al. (2010). Sequence data reveals phylogenetic affinities of fungal anamorphs Bahusutrabeeja, Diplococcium, Natarajania, Paliphora, Polyschema, Rattania and Spadicoides. Fungal Diversity, 44(1), 161–169. DOI 10.1007/s13225-010-0059-8. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |