
International Journal of
Experimental Botany

 | Phyton- International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.012075
ARTICLE
A New Record of Aspergillus vadensis (Ascomycota) Isolated from Soil in Yunnan Province, China
1CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Science, Kunming, 650201, China
2Centre for Mountain Futures, Kunming Institute of Botany, Kunming, 654400, China
3Department of Chemistry, University of Science and Technology, Bannu, 28100, Pakistan
*Corresponding Authors: Jianchu Xu. jxu@mail.kib.ac.cn; Peter Mortimer. peter@mail.kib.ac.cn
Received: 13 June 2020; Accepted: 12 October 2020
Abstract: During a survey of soil fungi in Yunnan Province, several isolates of Aspergillus were obtained. Based on morphology and molecular analyses of internal transcribed spacer regions and intervening 5.8S nrRNA gene (ITS), β-tubulin and calmodulin (CaM) genes sequences, two isolates were identified as Aspergillus vadensis (section Nigri). A phylogenetic tree, detailed descriptions, illustrations and scanned electron microscopy morphology are provided for the new isolates. This is the first record of A. vadensis from China.
Keywords: Morphology; phylogeny; section Nigri; soil fungi
Aspergillus is a large and diverse fungal genus in the family Aspergillaceae, with over 400 recognized species [1] classified into six subgenera and 25 sections [2]. The genus Aspergillus is one of the most studied fungal genera due to its application in human and animal health, the food industry, enzyme technology, pharmacology, and as genetic model organisms [3–6].
Aspergillus sect. Nigiri, known as ‘black aspergilli’ [7], includes 27 accepted species [8] occurring predominantly in soil, but they have also been isolated from other sources [2,9–11]. Species in this section are versatile and are of industrial and biotechnological importance [12–14]. Aspergillus niger is used in the fermentation industry for the production of enzymes and citric acid and has been granted status as a GRAS (generally recognized as safe) organism [12,14]. Other members are exploited for the production of biopharmaceuticals, mycotoxins and a variety of extracellular enzymes [12,13]. Aspergillus tubingenesis was reported to be a degrader of polyurethane plastics, a promising discovery in the field of mycoremediation [15]. Some species in this section are reported to be pathogenic to both humans and animals [16,17] and are also known to produce ochratoxin, a mycotoxin that is toxic and carcinogenic [18].
Members of the section Nigri have been divided into two separate clades based on their conidiophore morphology: Biseriate species (e.g., A. carbonarius and A. niger complex) and uniseriate species (e.g., A. aculeatus and A. japonicas), which have been shown to exhibit differences in their sexual morphs, sclerotium formation and extrolite profiles [19].
Aspergillus vadensis belongs to the section Nigri and was first identified in 2004 [20]. This species displays several unique characteristics and differs from other members of sect. Nigri in its restriction fragment length polymorphism (RFLP) and production of secondary metabolites. Unlike other fungi of this section that predominantly produce black colonies, Aspergillus vadensis was found to produce olive-green to brown colonies when grown on Czapek Yeast Agar (CYA) and malt extract agar (MEA) media.
During a survey of soil microfungi in Yunnan Province, China, several axenic cultures of Aspergillus were obtained. The objectives of this study were (1) To sequence one non-translated loci and two protein-coding genes of the new isolates; (2) Identify phylogenetic relationships of the new isolates, with close affinities in Aspergillus by using ML, MP and BI analyses; and (3) Provide comprehensive morphological descriptions, micro-photographic illustration and discussions.
2.1 Soil Sampling and Fungal Isolation
The study was carried out in February 2018, utilizing abandoned (over 12 months) agricultural land in Kunming, Yunnan Province, China. First, surface litter was scraped away to obtain a uniformly thick slice of soil from the surface to the plough depth at each spot. Next, a V-shaped cut was made with a sterilized spade (using 70% ethanol) to remove a 1–2 cm slice of soil and placed into a clear plastic bag. After that, the soil was cleared of large objects such as stones, litter, wood pieces and plant roots by removing them with gloved hands. Using this method, samples were collected from five randomly selected spots within a 20 m × 20 m area. Later, these samples were pooled into a single sample. Aspergillus species were obtained from this pooled soil sample by using the soil plate method [21] on malt extract agar (MEA) media in petri dishes, with the incubation temperature at 28°C for seven days.
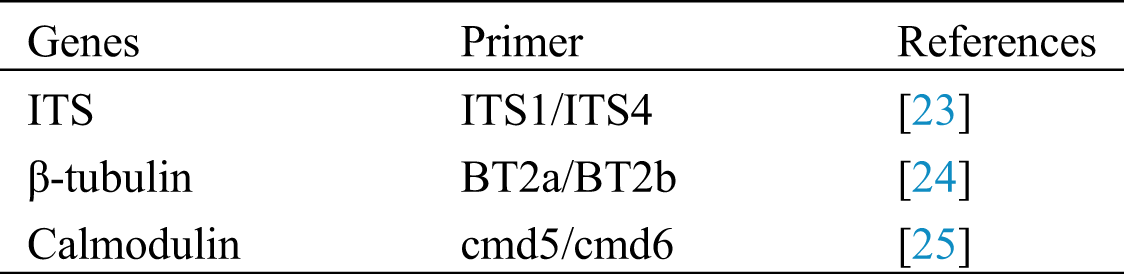
DNA was isolated from pure cultures (seven days old) grown on MEA. DNA was extracted using Biospin Fungal Genomic DNA isolation Kit–BSC14S1 (BioFlux, China). Polymerase chain reaction (PCR) was used to amplify partial gene regions of internal transcribed spacer regions and intervening 5.8S nrRNA gene (ITS), β-tubulin and calmodulin (CaM) gene regions as described by Tanikiwa et al. [22]. Primers used are shown in Tab. 1. PCR amplicons were separated on 1% agarose gel and sequenced by Sangon Biotech, Shanghai, China.
Table 1: List of genes and primers used in this study
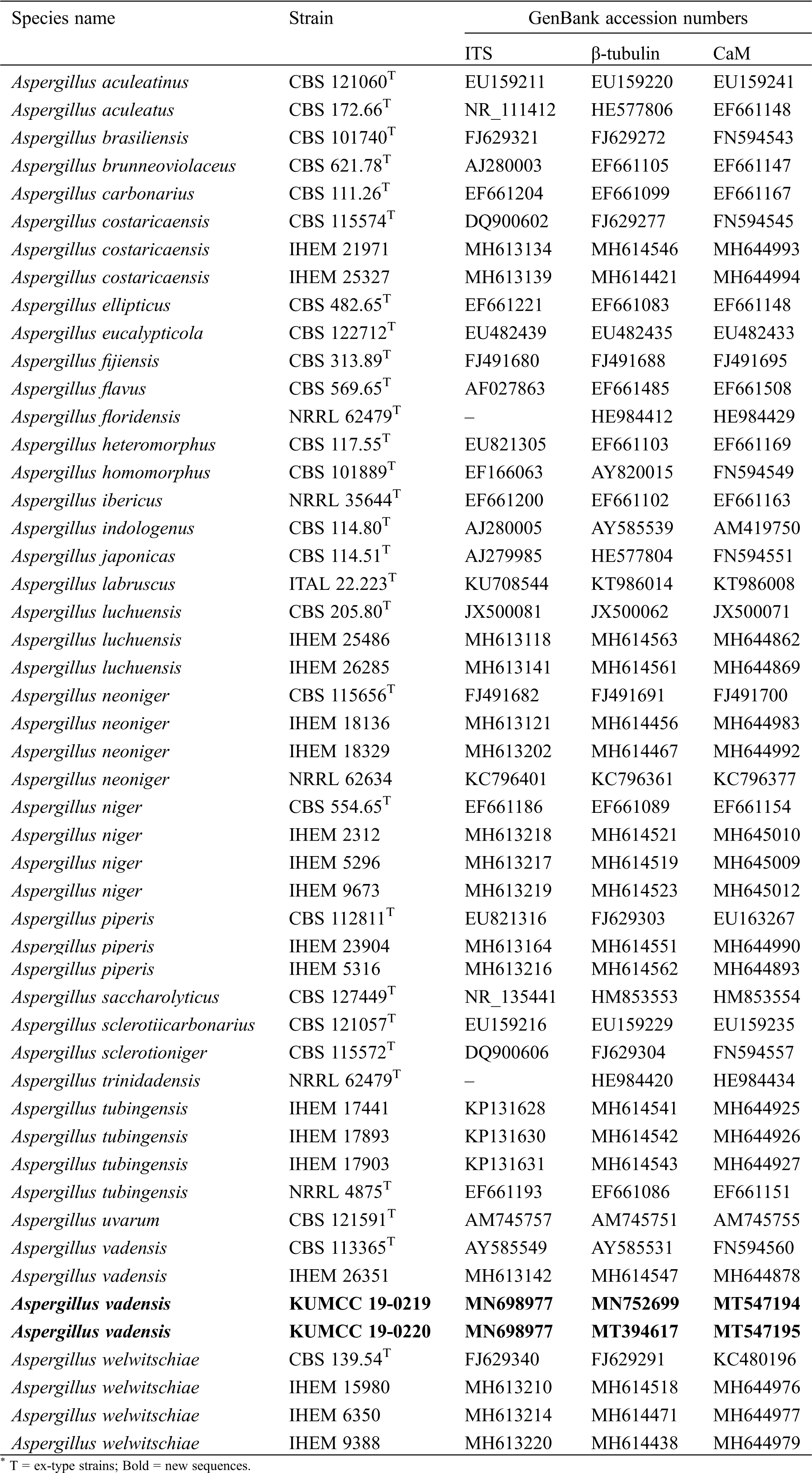
BLASTn searches were made using the newly generated sequences to determine the closest relatives for the taxonomic framework. Sequence data used for phylogenetic analyses in this study were downloaded from the NCBI database based on recently published data [7] (Tab. 2). Each locus was automatically aligned with MAFFT v. 7 [26] after which alignments were manually checked and improved using BioEdit v. 7.0.5.2 [27]. Phylogenetic methods used in this study included maximum-likelihood (ML), maximum parsimony (MP) and Bayesian criteria (BI). Generated phylograms were visualized using the FigTree v1.4.0 program [28], reorganized in Microsoft PowerPoint (2016) and deposited in TreeBASE, submission ID: 26171.
Table 2: The species and strains used in the phylogenetic analyses
For macromorphological observations, colonies were grown on MEA and Czapek yeast extract agar (CYA) media. The cultures were inoculated in the center of each plate and incubated at 28°C for seven days. The culture plates were observed for colony colour, size, texture and production of exudates. Microscopic mounts prepared in lactic acid were used to study the microscopic features. Scanning electron microscopy (SEM, Hitachi S570) was performed as described previously by Khan [15].
The combined alignment (ITS, β- tubulin and CaM) contained 51 strains with 2094 characters representing section Nigri, the new strains proposed in this study, and the outgroup taxon (Aspergillus flavus). The topology of the multi-locus ML, MP phylogenetic trees and Bayesian inference is largely concordant, and the same clades are supported. The overall topology of these yielded trees was similar to previous work [7,14]. The best scoring tree with RAxML analysis gave a final optimization value of −13115.709914 (Fig. 1). The ML model of the concatenated data showed the following parameters: Estimated base frequencies: A = 0.214563, C = 0.284009, G = 0.262361, T = 0.239066; substitution rates: AC = 0.894395, AG = 2.869504, AT = 1.314187, CG = 0.567345, CT = 4.215652, GT = 1.000; proportion of invariable sites: I = 0.397753; gamma distribution shape parameter α = 1.204389. The MP analyses generated the maximum of 26 equally most parsimonious trees, and the most parsimonious is shown in Fig. 1 with the following parameters: Length = 2244, CI = 0.638, RI = 0.869, RC = 0.555 and HI = 0.362. From the analysed characters, 1192 were constant, 180 were variable and parsimony-uninformative and 722 were parsimony-informative. For Bayesian analysis, dirichlet base frequencies and the GTR+I+G model were recommended by MrModelTest. The dataset comprised 1101 unique patterns. Neglecting the initial 20% of trees, 1601 trees remained and were used to calculate Bayesian PP (Fig. 1; third value: PP > 0.95 shown).
In the concatenated sequence data analysis, A. aculeatus, A. carbonarius, A. homomorphus, A. niger and A. tubingensis clades were supported by high bootstrap support values (Fig. 1). But the species in the A. heteromorphus clade are not monophyletic and constitute a basal lineage sister to the A. aculeatus clade. Two of our new isolates, KUMCC 19-0219 and KUMCC 19-0220, clustered in the A. tubingensis clade (Fig. 1). Within the A. tubingensis clade, our new isolates were monophyletic with A. vadensis (CBS 113365 and IHEM 26351), and this relationship was statistically well-supported (ML = 100%, MP = 99% and PP = 1.00, Fig. 1)
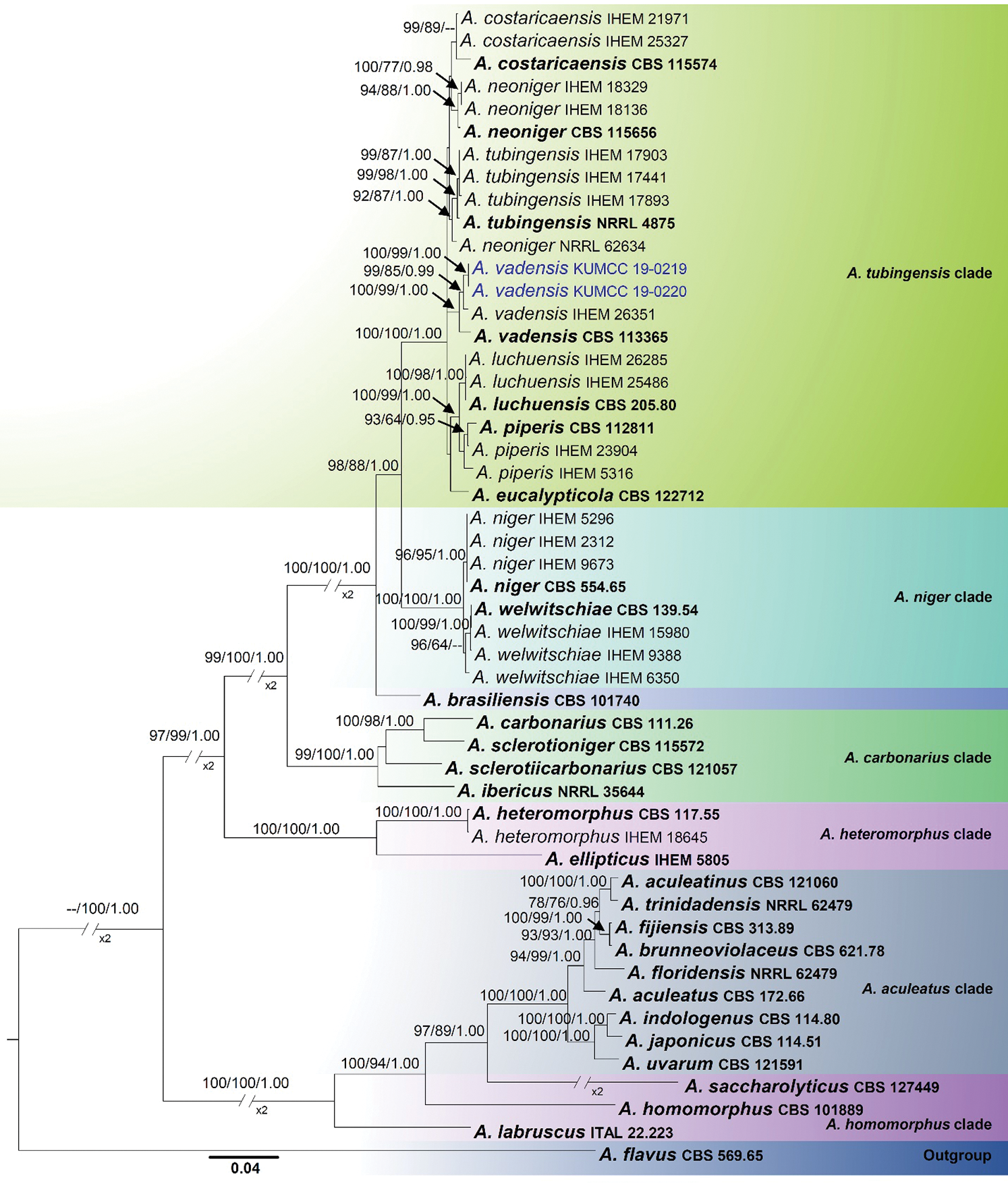
Figure 1: Phylogenetic tree based on analysis of the combined sequence data of ITS, beta tubulin and calmodulin gene region for Aspergillus section Nigri. Bootstrap support values >60% for ML and MP, and Bayesian PP values higher than 0.95% are defined above the internal branches, respectively. The tree is rooted to Aspergillus flavus (CBS 569.65) in the section flavi. Bold = ex-type strains; Blue = new isolate
Aspergillus vadensis Samson, de Vries, Frisvad & Visser, Antonie van Leeuwenhoek 87 (3): 201 (2005)
IndexFungorum number: IF340234
Colony dimensions after 2 days at 25°C were as follows: MEA: 24 mm–25 mm, CYA: 21 mm–22 mm. Colony colours and texture on MEA for 7 days showed seriate margins, upper surface granular and black, flat mycelium white in beginning become black after 10 days, sporulate within 24 hours, densely covered with black conidial heads, conspicuous exudates like big black droplets, and creamy white wrinkled reverse that becomes dull yellow with age. Colony colours and texture on CYA for 7 days showed circular colony with curled edge, mycelium white, flossy, velvety with sporulate after 24 h, densely covered with black conidial heads. Mycelium composed of hyaline, branched, septate, smooth-walled hyphae. Conidial heads radiating. Stipes 4 µm–7.5 µm (n = 15) μm diam., hyaline, smooth to finely rough-walled, variable in length, just below vesicles 5 mm–8 mm. Vesicles 25–35 × 30–35 µm (x− = 31 × 34 µm, n = 15), globose to subglobose, hyaline, entirely covered by phialides, Conidiophores 4–6 × 3–4 µm (x− = 5 × 3.5 µm, n = 15) uniseriate or biseriate, hyaline, cylindrical to ampulliform, smooth-walled, wide at the top, non-septate or with occasional septum, reduced short conidiophores or solitary phialides frequently present on the aerial mycelium. Conidia 3–5 × 3–5 µm (x− = 3.87 × 3.89 µm, n = 30), arranged in long chains, globose to subglobose, echinulate, hyaline to greenish and coarsely roughened.
Material examined: CHINA, Yunnan Province, Kunming, Panlong, 25.135016° N, 102.747458° E, from soil, 22 February 2018, Sadia Nadir, G3, MFLU 20–0141, living culture KUMCC 19–0219; from soil, G7, MFLU 20–0142, living culture KUMCC 19–0220.
Notes: de Vries et al. [20] introduced Aspergillus vadensis from air in Egypt as a taxon in Aspergillus section Nigri. During our investigation on the diversity of soil microfungi in Yunnan Province, two isolates were recovered from soil samples in Kunming. Morphological characters such as conidial heads, vesicles, conidiophores and conidia fit well within the type description of Aspergillus vadensis, which was derived from culture. In the phylogenetic study, our new strains cluster with Aspergillus vadensis as a monophyletic clade with 100% ML, 99% MP and 1.00 PP support (Fig. 1). Comparison of ITS, beta tubulin and calmodulin sequence data revealed there is no significant difference (no differences in ITS, 3 pb differences in beta tubulin and 6 bp differences in calmodulin) between our new isolate and Aspergillus vadensis. Therefore, we would like to keep our new isolates as new records of Aspergillus vadensis from China.
Species within Aspergillus section Nigri require both morphological and molecular analyses for accurate species delamination. For the phylogenetic analyses, ITS alone is not sufficient. Additional gene regions (β-tubulin and calmodulin) are recommended [11,12,29], and our analyses were conducted accordingly. Despite the fact that the new isolates bear close morphological and phylogenetic resemblances to the type species of Aspergillus vadensis, there are some observed differences worth mention. The ex-type culture has brown to olive-green colony on CYA and MEA while we observed white mycelium that became black upon maturation on these media. We observed that the colonies grown on MEA (Fig. 2a) and colonies grown on CYA (Fig. 2b) differed from each other in their appearance (top view), and this was not reported from the ex-type culture. Phialides of the new isolates were uniseriate at the immature stage (3 days) and became biseriate after 7 days. This was also not mentioned in de Vries et al. [20]. Furthermore, the new isolates were found to produce exudates on MEA appearing as big black droplets, which was also not reported from the type description.
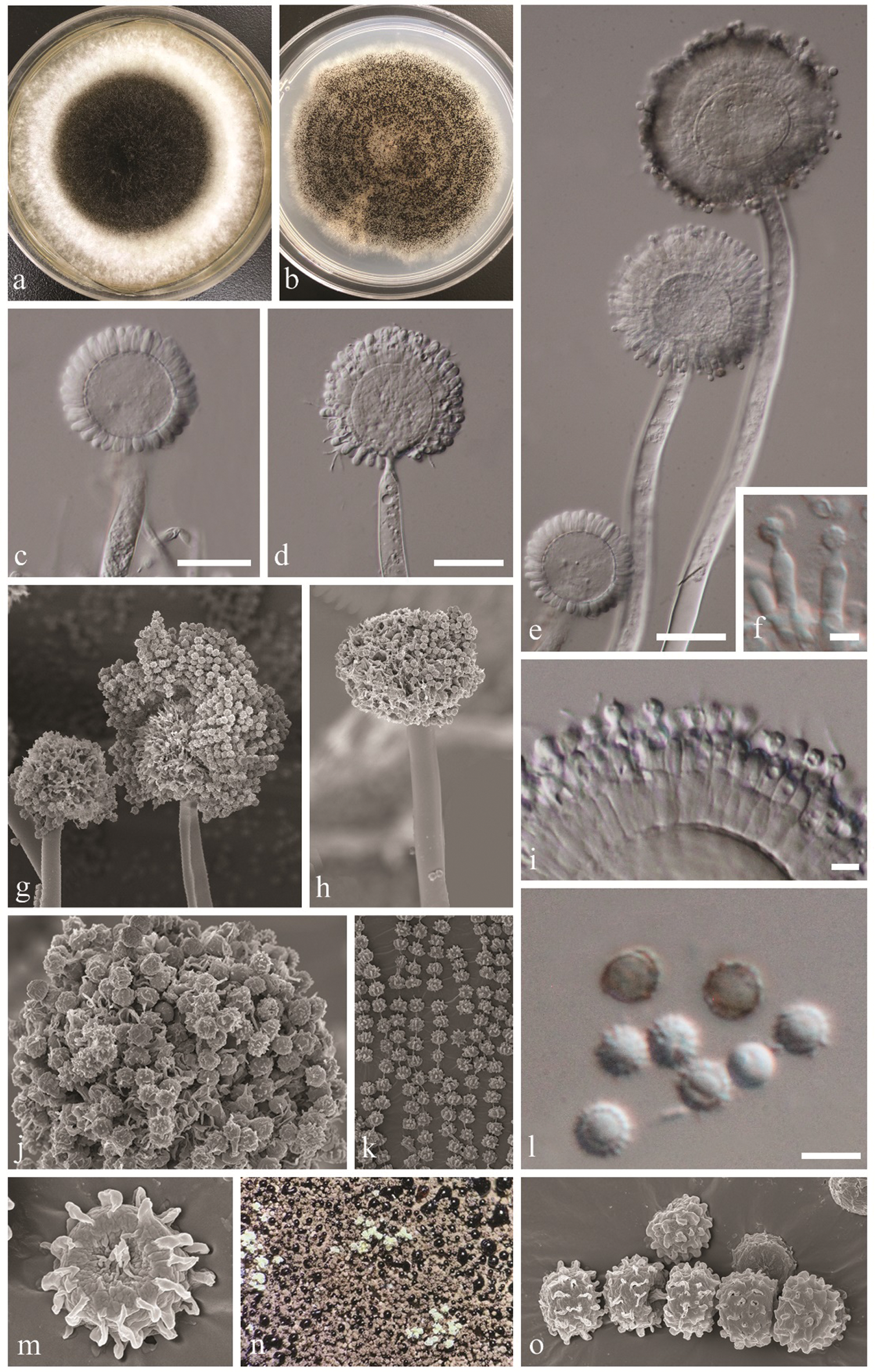
Figure 2: Aspergillus vadensis (KUMCC 19-0219, new record). a: Culture on MEA at 28°C. b: Culture on CYA at 28°C. c, d, e: Conidiophores. f, i: Close up conidia head and conidia (note: phialides attached to metulae in i). g, h: Conidiophores and conidia under SEM. j, k, m, o: Conidia under SEM. l: Close up conidia. n: Culture surface and exudate production. Scale bars: b–e: = 20 µm, f, l = 5 µm, i = 10 µm
Funding Statement: This research was funded by Key Research Project “Agroforestry Systems for Restoration and Bio-industry Technology Development”, Grant No. 2017YFC0505101. S. Khan, S. Tibpromma and D. Wanasinghe are thankful to the CAS President’s International Fellowship Initiative (PIFI Grant Nos. 2019PC0011, 2020PC0009 and 2019PC0008) and S. Tibpromma is thankful to the International Postdoctoral Exchange Fellowship Program (No. Y9180822S1), China Postdoctoral Science Foundation, Yunnan Human Resources and Social Security Department Foundation. D. Wanasinghe is thankful to the 64th batch of China Postdoctoral Science Foundation (Grant No. Y913083271). P. Mortimer is thankful to the National Science Foundation of China (NSFC), Project Codes 41761144055 and 41771063.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Wijayawardene, N. N., Hyde, K. D., Al-Ani, L. K. T. (2020). Outline of Fungi and fungus-like taxa. Mycosphere, 11(1), 1060–1456. DOI 10.5943/mycosphere/11/1/8. [Google Scholar] [CrossRef]
2. Samson, R. A., Visagie, C. M., Houbraken, J. J., Hong, S. B., Frisvad, J. C. (2014). Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology, 78, 141–173. DOI 10.1016/j.simyco.2014.07.004. [Google Scholar] [CrossRef]
3. Punt, P. J., van Biezen, N.,Conesa, A., Albers, A., Mangnus, J. et al. (2002). Filamentous fungi as cell factories for heterologous protein production. Trends in Biotechnology, 20(5), 200–206. DOI 10.1016/S0167-7799(02)01933-9. [Google Scholar] [CrossRef]
4. Wösten, H., Scoltmeijer, K., de Vries, R. (2007). Dijksterhuis, J., Samson, R. (eds.Food mycology: A multifaceted approach to fungi and food. pp. 183–196. Boca Raton, FL, USA: CRC Press. [Google Scholar]
5. Nierman, W. C., Pain, A., Anderson, M. J., Wortman, J. R., Kim, H. S. et al. (2005). Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature, 438(7071), 1151–1156. DOI 10.1038/nature04332. [Google Scholar] [CrossRef]
6. Meijer, M., Houbraken, J. A. M. P., Dalhuijsen, S., Samson, R. A., de Vries, R. P. (2011). Growth and hydrolase profiles can be used as characteristics to distinguish Aspergillus niger and other black aspergilli. Studies in Mycology., 69(Suppl 1), 19–30. DOI 10.3114/sim.2011.69.02. [Google Scholar] [CrossRef]
7. D’hooge, E., Becker, P., Stubbe, D., Normand, A., Piarroux, R. et al. (2019). Black aspergilli: A remaining challenge in fungal taxonomy. Medical Mycology, 57(6), 773–780. DOI 10.1093/mmy/myy124. [Google Scholar] [CrossRef]
8. Vesth, T. C., Nybo, J. L., Theobald, S., Frisvad, J. C., Larsen, T. O. et al. (2018). Investigation of inter- and intraspecies variation through genome sequencing of Aspergillus section Nigri. Nature Genetics, 50(12), 1688–1695. DOI 10.1038/s41588-018-0246-1. [Google Scholar] [CrossRef]
9. Abrunhosa, L., Calado, T., Venancio, A. (2011). Incidence of fumonisin B2 production by Aspergillus niger in Portuguese wine regions. Journal of Agricultural and Food Chemistry, 59(13), 7514–7518. DOI 10.1021/jf202123q. [Google Scholar] [CrossRef]
10. Palumbo, J. D., O’Keefe, T. L., Vasquez, S. J., Mahoney, N. E. (2011). Isolation and identification of ochratoxin A-producing Aspergillus section Nigri strains from California raisins. Letters in Applied Microbiology, 52(4), 330–336. DOI 10.1111/j.1472-765X.2011.03004.x. [Google Scholar] [CrossRef]
11. Perrone, G., de Girolamo, A., Sarigiannis, Y., Haidukowski, M. E., Visconti, A. (2013). Occurrence of ochratoxin A, fumonisin B2 and black aspergilli in raisins from Western Greece regions in relation to environmental and geographical factors. Food Additives & Contaminants Part A Chemistry Analysis, Control, Exposure and Risk Assessment, 30(7), 1339–1347. DOI 10.1080/19440049.2013.796594. [Google Scholar] [CrossRef]
12. Varga, J., Frisvad, J. C., Kocsubé, S., Brankovics, B., Tóth, B. et al. (2011). New and revisited species in Aspergillus section Nigri. Studies in Mycology, 69, 1–17. DOI 10.3114/sim.2011.69.01. [Google Scholar] [CrossRef]
13. Pel, H. J., de Winde, J. H., Archer, D., Dyer, P. S., Hofmann, G. et al. (2007). Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nature Biotechnology, 25(2), 221–231. DOI 10.1038/nbt1282. [Google Scholar] [CrossRef]
14. Fungaro, M. H. P., Ferranti, L. S., Massi, F. P., de Silva, J. J., Sartori, D. et al. (2017). Aspergillus labruscus sp. nov., a new species of Aspergillus section Nigri discovered in Brazil. Scientific Reports, 7(1), 5736. DOI 10.1038/s41598-017-06589-y. [Google Scholar] [CrossRef]
15. Khan, S. (2017). Biodegradation of polyester polyurethane by Aspergillus tubingensis. Environmental Pollution., 225(8), 469–480. DOI 10.1016/j.envpol.2017.03.012. [Google Scholar] [CrossRef]
16. Serra, R., Abrunhosa, L., Kozakiewicz, Z., Venâncio, A. (2003). Black Aspergillus species as ochratoxin A producers in Portuguese wine grapes. International Journal of Food Microbiology, 88(1), 63–68. DOI 10.1016/S0168-1605(03)00085-0. [Google Scholar] [CrossRef]
17. Bui-Klimke, T. R., Wu, F. (2013). Ochratoxin A and human health risk: A review of the evidence. Critical Reviews in Food Science and Nutrition, 55(13), 1860–1869. DOI 10.1080/10408398.2012.724480. [Google Scholar] [CrossRef]
18. Abarca, M. L., Accensi, F., Bragulat, M. R., Cabañes, F. J. (2001). Current Importance of Ochratoxin A–Producing Aspergillus spp. Journal of Food Protection, 64(6), 903–906. DOI 10.4315/0362-028X-64.6.903. [Google Scholar] [CrossRef]
19. Serra, R., Cabanes, F. J., Perrone, G., Castellá, G., Venâncio, A. et al. (2017). Aspergillus ibericus: A new species of section Nigri isolated from grapes. Mycologia, 98(2), 295–306. DOI 10.1080/15572536.2006.11832702. [Google Scholar] [CrossRef]
20. de Vries, R. P., Frisvad, J. C., van de Vondervoort, P. J., Burgers, K. et al. (2005). Aspergillus vadensis, a new species of the group of black Aspergilli. Antonie van Leeuwenhoek, 87(3), 195–203. DOI 10.1007/s10482-004-3194-y. [Google Scholar] [CrossRef]
21. Warcup, J. H. (1950). The soil-plate method for isolation of fungi from soil. Nature, 166(4211), 117–118. DOI 10.1038/166117b0. [Google Scholar] [CrossRef]
22. Taniwaki, M. H., Pitt, J. I., Iamanaka, B. T., Sartori, D., Copetti, M. V. et al. (2012). Aspergillus bertholletius sp. nov. from Brazil Nuts. PLoS One, 7(8), e42480. DOI 10.1371/journal.pone.0042480. [Google Scholar] [CrossRef]
23. White, T. J., Burns, T., Lee, S., Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M. A., Gelfand, D. H., Sninsky, J. J., White, T. J. (eds.PCR Protocols: A guide to methods and applications. pp. 315–322. San Diego: Academic Press. 10.1016/b978-0-12-372180-8.50042-1. [Google Scholar] [CrossRef]
24. Glass, N. L., Donaldson, G. C. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied and Environmental Microbiology, 61(4), 1323–1330. DOI 10.1128/AEM.61.4.1323-1330.1995. [Google Scholar] [CrossRef]
25. Hong, S. B., Cho, H. S., Shin, H. D., Frisvad, J. C., Samson, R. A. (2006). Novel Neosartorya species isolated from soil in Korea. International Journal of Systematic and Evolutionary Microbiology, 56(2), 477–486. DOI 10.1099/ijs.0.63980-0. [Google Scholar] [CrossRef]
26. Katoh, K., Standley, D. M. (2016). A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics, 32(13), 1933–1942. DOI 10.1093/bioinformatics/btw108. [Google Scholar] [CrossRef]
27. Hall, T. (2004). Bioedit version 6.0.7. http://www.mbio.-ncsu.edu/bioedit/bioedit.html. [Google Scholar]
28. Rambaut, A., Drummond, A. (2008). FigureTree: Tree figures drawing tool, version 1.2.2. University of Edinburgh, Institute of Evolutionary Biology. [Google Scholar]
29. Samson, R. A., Houbraken, J. A. M. P., Kuijpers, A. F. A., Frank, J. M., Frisvad, J. C. (2004). New ochratoxin A or sclerotium producing species in Aspergillus section. Studies in Mycology, 50(1), 45–61. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |