
International Journal of
Experimental Botany

 | Phyton- International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.016314
ARTICLE
Overexpression of IbSINA5 Increases Cold Tolerance through a CBF SINA-COR Mediated Module in Sweet Potato
1Shandong Technology and Business University, Yantai, 264005, China
2The Engineering Research Institute of Agriculture and Forestry, Ludong University, Yantai, 264025, China
3School of Life Sciences, Ludong University, Yantai, 264025, China
*Corresponding Authors: Limin Wang. Email: wanglimin9696@163.com; Shenglin Zhou. Email: zhoushenglin@163.com
Received: 20 February 2021; Accepted: 05 March 2021
#These authors contribute equally to this work
Abstract: Seven in absentia (SINA) family proteins play a central role in plant growth, development and resistance to abiotic stress. However, their biological function in plant response to cold stress is still largely unknown. In this work, a seven in absentia gene IbSINA5 was isolated from sweet potato. Quantitative real-time polymerase chain reaction (qRT-PCR) analyses demonstrated that IbSINA5 was ubiquitously expressed in various tissues and organs of sweet potato, with a predominant expression in fibrous roots, and was remarkably induced by cold, drought and salt stresses. Subcellular localization assays revealed that IbSINA5-GFP fusion protein was mainly localized in cytoplasm and nucleus. Overexpression of IbSINA5 in sweet potato led to dramatically improved resistance to cold stress in transgenic plants, which was associated with the up-regulated expression of IbCOR (cold-regulated) genes, increased proline production, and decreased malondialdehyde (MDA) and H2O2 accumulation in the leaves of transgenic plants. Furthermore, transient expression of IbCBF3, a C-repeat binding factor (CBF) gene, in the leaf protoplasts of wild type sweet potato plants up-regulated the expression of both IbSINA5 and IbCOR genes. Our results suggest that IbSINA5 could function as a positive regulator in the cold signaling pathway through a CBF-SINA-COR mediated module in sweet potato, and have a great potential to be used as a candidate gene for the future breeding of new plant species with improved cold resistance.
Keywords: Cold stress; IbSINA5; seven in absentia; sweet potato; transgenic plant
Sweet potato (Ipomoea batatas [L.] Lam) has been grown worldwide as a tuberous crop due to its food and commercial values, and resistance to adverse growth conditions [1–3]. However, owing to its tropical and subtropical origin, abiotic stresses such as low temperature and drought can severely affect the storage root yield of sweet potato, especially during the rapid root expansion period [4,5].
To increase the resistance of sweet potato to abiotic stresses, a number of stress related genes such as IbCBF3, IbMIPS1, IbNAC1, IbOR, IbTPS and IbLEA14 have been isolated, and transgenic sweet potato plants with improved tolerance to biotic and abiotic stresses have been generated [6–9]. Ectopic expression of rice OCI enhanced the resistance to stem nematode in transgenic sweet potato [10]. Constitutive expression of Arabidopsis AtNHX1, HDG11 or spinach BADH in sweet potato increased salt and cold resistance of transgenic plants [4,5,11]. Overexpression of IbMIPS1 or IbCBF3 increased the resistance to stem nematode, salt, drought and cold stress of transgenic sweet potato [12,13].
SINA protein is a subfamily of the tumor necrosis factor receptor-associated factor (TRAF) super family, which function in various developmental processes of animals and plants as ubiquitin ligases. They have been characterized by an N-terminal cysteine-rich really interesting new gene (RING) domain, two zinc-finger motifs and a conserved C-terminal coiled-coil domain responsible for substrate binding and homo- or hetero-dimerization [14–17]. In plants, SINA proteins were found to function in proteasome mediated protein ubiquitination [18]. In Arabidopsis, SINAT2 interacted with RELATED TO APETALA2 (AP2) 2 (AtRAP2.2), whereas SINAT5 worked in the degradation of NAC1 to regulate lateral root growth [19–21]. In rice, the SINA family protein OsDIS1 negatively regulated the drought tolerance in transgenic rice plants [22]. In alfalfa, ectopic expression of the dominant negative form of SINAT5 affected the growth and nodulation of transgenic plant [23]. In tomato, overexpression of SlSINA2 lead to altered chlorophyll level in the leaves of transgenic plants, whereas overexpression of SlSINA5 disturbed flower development [24].
Previously, we investigated the function of Arabidopsis SINA2 (AtSINA2), a SINA protein lacking RING domain, and found that it positively regulated the drought tolerance of Arabidopsis plants [25]. Here, we demonstrate for the first time that IbSINA5, a homolog of SINA2, functions as a positively regulator in the cold signaling pathway in sweet potato.
2.1 Plant Materials and RNA Isolations
Sweet potato cultivars Xushu18 and Taizhong 6 were grown in greenhouse as described previously [13]. For cold, drought and salt stress treatments, three-week-old Taizhong 6 plants were kept at 4°C, or treated with 25% PEG6000 or 250 mM NaCl. The fourth fully expanded leaves counted from the tops of plants were collected at 3 h, 6 h, 12 h, 24 h and 48 h after the treatments for RNA isolation. Ten plants were used for each treatment. For IbSINA5 expression analysis, fibrous roots, storage roots, stems, young leaves, mature leaves and senescent leaves of ten-week-old Taizhong 6 sweet potato plants were used. Total RNA was extracted as described previously [26].
2.2 Quantitative Real-Time Polymerase Chain Reaction Analysis
qRT-PCR was carried out using IbSINA5 primers (forward: 5’-ATGTATAAAATGGAGATTGAAAGC-3’; reverse: 5’-TTAAGTGCTACAAATATTCG-3’) as described previously [26]. The expression level of IbCBF3, IbCOR413, IbCOR314 and IbCOR27 were examined with the house keeping gene IbTubulin as an internal control [13].
The encoding cDNA of IbSINA5 was cloned from sweet potato cultivar Taizhong 6 using its forward and reverse primers based on the gene information from the database [27]. The amplified cDNA fragment was cloned to the pEASY-T5 Zero Cloning Vector (Transgen, China) after sequence confirmation.
2.4 Subcellular Localization of IbSINA5
To detect the subcellular localization of IbSINA5 protein, IbSINA5 was fused in frame to the N-terminal of GFP in the pBI221:GFP vector. The resultant pBI221:IbSINA5-GFP and pBI221:GFP was transiently expressed in onion epidermal cells, respectively, as described previously [28]. The location of GFP and IbSINA5-GFP in the onion cells was observed with a confocal laser scanning microscopy (Leica TCS SP2).
2.5 Plasmid Construction and Sweet Potato Transformation
To construct the plant expression vector pCAMBIA-2301s2:IbSINA5, the full length cDNA of IbSNIA5 was cloned into the modified pCAMBIA-1301s2 vector, driven by two copies of cauliflower mosaic virus (CaMV) 35S promoter. The resultant construct pCAMBIA-2301s2:IbSINA5 was introduced into sweet potato cv. Xushu18 via Agrobacterium-mediated transformation as described previously [29].
2.6 Selection of Transgenic Sweet Potato Plants
For the molecular confirmation of transgenic plants, PCR and qRT-PCR were carried out to verify the integration and expression of IbSNIA5. To assess the expression levels of IbSNIA5, IbCBF3 and IbCOR genes, total RNAs extracted from the mature leaves (the fourth leaves counted from the tops of plants) of four-week-old wild type and transgenic plants before and after cold treatment were used for qRT-PCR analyses. All gene specific primer sequences were the same as used by Jin et al. [13].
For cold resistance analysis, four-week-old wild type and transgenic sweet potato plants grown in soil were kept at 4°C for 48 h and recovery at 25°C for 24 h. The phenotypes of plants before and after the treatments were photographed and the contents of proline, MDA and hydrogen peroxide (H2O2) in the mature leaves were measured as described previously [30,31].
2.8 Transient Transcription Dual-Luciferase Assays
To generate the reporter construct 35S::REN-ProIbSIN5::LUC, the promoter region (2001 bp) of IbSNIA5 was inserted into pGreenII0800. To generate the effecter vector pGreenII62-SK-IbCBF3, IbCBF3 were cloned into pGreenII62-SK. Sweet potato leaf protoplast preparation and transformation were carried out as described by Wang et al. [32]. Transient dual-luciferase assays in sweet potato leaf protoplast were performed as described previously [33,34].
2.9 Transient Expression of IbCBF3 in Sweet Potato Mesophyll Protoplasts
To analyze the transcription function of IbCBF3, the transcriptional factor effector pGreenII62-SK-IbCBF3 was transfected into sweet potato leaf protoplast. The transient expressions of IbCBF3, IbSNIA5 and IbCOR genes in sweet potato leaf mesophyll protoplasts were examine by qRT-PCR as described previously [35].
ANOVA (one-way) was used to generate every P value. The variability was indicated with the standard deviation (SD). *, ** and *** indicate p-value < 0.05, < 0.01 and < 0.001, respectively. Data are shown as mean ± standard deviation (SD) from three biological replicates each.
3.1 IbSNIA5 Encodes a Putative SINA Protein
Ubiquitination mediated protein degradation at different plant growth stages and in response to various abiotic stresses has been well studied [36,37]. The Arabidopsis SINAT5 (AtSINAT5) functioned as a RING type E3 ubiquitin ligase to control lateral root growth by degrading NAC1, whereas SINA2, a TRAF-like SINA family protein lacking RING domain, positively regulated the resistance to drought stress [19,25]. To explore the biological function of SINA family proteins in tuberous crops, we blast searched the sweet potato genome database using Arabidopsis SINA2 (https://www.ipomoea-genome.org/genome_ jbrowse.html). A total number of seven SINA2 homologues were identified. Similar to the previously reported SINA and some other TRAF-like family proteins, they all contain a conserved C-terminal TRAF-like domain [14–16]. However, among them, same as the Arabidopsis SINA2 and animal’s TRAF1 proteins, IbSINA5 also lacks the RING domain (Fig. 1). Therefore, IbSINA5 does not have E3 ligase activity required for its target protein degradation.

Figure 1: Sequence alignment and phylogenetic analyses of SINAs from different plant species. Comparison of Ipomoea batatas IbSINA5 with Arabidopsis thaliana AtSINAT1 (NP_181729.1), AtSINA2 (NP_187978.1), Solanum lycopersicum SlSINA1 (XP_004228679.1), and Populus trichocarpa PtSINA2 (XP_002299212.1), and phylogenetic tree of IbSINA5 homologs in different plants are shown. The gene and amino acid sequences of IbSINA1 (g919), IbSINA2 (g4297), IbSINA3 (g8848), IbSINA4 (g17091), IbSINA5 (g20660), IbSINA6 (g30553) and IbSINA7 (g58368) are extracted from the sweet potato genome database (https://www.ipomoea-genome.org/genome_ jbrowse.html). Based on their conservation levels, residues in the sequence alignment analyses are respectively highlighted in black, dark gray and light gray. The bar in phylogenetic tree represents the number of amino acid substitutions per site. Asterisk and plus signs represent the RING domain and zinc-finger motif, respectively
3.2 IbSNIA5 is Induced by Different Abiotic Stresses in Sweet Potato
To dissect the function of IbSNIA5 in the resistance to abiotic stresses in sweet potato, its expression levels in wild type Taizhong 6 plants were examined by qRT-PCR. IbSNIA5 was ubiquitously expressed in all the tested tissues and organs including fibrous roots, storage roots, stems, young leaves, mature leaves and senescent leaves, with a predominant expression in fibrous roots (Fig. 2A). Previously, we reported that SINA2 was strongly induced by ABA and drought [25]. We examined the expression levels of IbSINA5 in three-week-old Taizhong 6 plants after low temperature (4°C), 25% PEG6000 or 250 mM NaCl treatments by qRT-PCR. We found that IbSNIA5 was remarkably induced by cold, drought and salt stress, indicating its possible roles in plant response to various abiotic stresses (Figs. 2B–2D).

Figure 2: IbSINA5 expression and IbSINA5 subcellular localization. (A) Expression pattern of IbSINA5. The transcription levels of IbSINA5 in fibrous roots, storage roots, stems, young leaves, mature leaves and senescent leaves of ten-week-old sweet potato plants were shown. (B–D) Expression of IbSINA5in response to different abiotic stress treatments. Three-week-old sweet potato plants grown in greenhouse were kept at 4°C, or treated with 25% PEG6000 or 250 mM NaCl. The fourth fully expanded leaves counted from the tops of plants were collected at 3, 6, 12, 24 and 48 h after the treatments for RNA isolation. The transcription levels of IbSINA5 were measured by qRT-PCR and normalized to IbTubulin expression. Values are means and standard deviations of three biological replicates. * and ** indicate p-value < 0.05 and < 0.01, respectively. (E) IbSINA5 is mainly localized in nuclei and cytoplasm. The location of GFP and IbSINA5-GFP in the onion epidermal cells was observed with a confocal laser scanning microscopy (Leica TCS SP2). Scale bars = 100 μm
3.3 IbSINA5 is Mainly Localized in Nuclei and Cytoplasm
The subcellular localization of a certain protein may reflect its possible functions in the relevant biological processes it participates. To determine the subcellular localization of IbSNIA5, pBI221:IbSINA5-GFP and pBI221:GFP was transiently expressed in onion epidermal cells. IbSINA5-GFP fusion protein was mainly localized in nuclei and cytoplasm (Fig. 2E). Similar subcellular localization was also observed with SINA2-YFP fusion protein in Arabidopsis protoplasts [25]. The nuclei and cytoplasm localization of IbSINA5 implies that it may have a regulatory role in the expression of its upstream transcriptional factors in nuclei and its downstream functional genes in cytoplasm.

Figure 3: Molecular confirmation of IbSINA5 transgenic plants. (A) Schematic map of pCAMBIA1301s2-IbSINA5 construct. Expression of IbSINA5 was driven by two copies of the cauliflower mosaic virus 35S promoter. (B) PCR analysis of IbSINA5 integration in different transgenic lines. M, molecular marker; WT, wild type; L1-7, different transgenic lines; P, plasmid. (C) qRT-PCR analyses of IbSINA5 in wild type and different transgenic lines. Gene expression level in WT was set to 1. Values are means and standard deviations of three biological replicates. ** and *** indicate p-value < 0.01 and < 0.001, respectively
3.4 Overexpression of IbSINA5 Improves Cold Tolerance in Sweet Potato
Base on our previous study that overexpression of SINA2 significantly augmented the resistance to drought stress in Arabidopsis plants, and the observation that IbSINA5 was dramatically induced by cold and drought stresses, we speculated that overexpression of IbSINA5 in sweet potato may also promote the cold resistance [25]. To verify this hypothesis, the plant expression construct pCAMBIA-2301s2:IbSINA5 containing the coding sequence of IbSINA5 was introduced into the genome of Xushu18 (Fig. 3A). A total number of 34 independent hygromycin resistant lines were obtained, and seven of them were randomly selected. PCR and qRT-PCR analyses confirmed the integration of IbSINA5 in the sweet potato genome, and the overexpression of IbSINA5 in all the tested transgenic lines (Figs. 3B and 3C).
To determine whether overexpression of IbSINA5 would improve the resistance of transgenic sweet potato plants to cold stress, we compared their growth phenotypes with wild type plants. Three transgenic lines with different IbSINA5 overexpression levels (L2, L5 and L7) were selected. Four-week-old wild type and transgenic plants grown in greenhouse were kept at 4°C for 48 h, then recovered at 25°C for 24 h. All transgenic plants showed less cold damage and successfully recovered after they were transferred back to room temperature, whereas wild type plants exhibited more severe cold damage and failed to recover (Fig. 4A). The damage extents were consistent with IbSINA5 overexpression levels, as indicated by the more severe damage in transgenic line L5, which showed relatively lower IbSINA5 overexpression level, than in transgenic lines L2 and L7, which showed relatively higher IbSINA5 overexpression levels (Fig. 4B).

Figure 4: Cold tolerance and gene expression analyses of wild type and transgenic plants. (A) Phenotypes of sweet potato plants before and after cold stress treatment. Four-week-old wild type and transgenic were treated at 4°C for 48 h and recovered at 25°C for 24 h. Cold damage in the leaves of wild type plants was significantly severe than that of transgenic plants after 48 h cold treatment and 24 h recovery. Ten plants of wild type and each transgenic line were used for the treatment. Photos were the representatives from three replicates. (B) qRT-PCR analyses. The transcription levels of IbSINA5 in the leaves of wild type and transgenic lines L2, L5 and L7 before cold treatments were measured by qRT-PCR. (C) IbCOR gene expression analyses. The transcription levels of IbCOR413, IbCOR314 and IbCOR27 in the leaves of four-week-old wild type and transgenic plants were examined by qRT-PCR. The transcript levels of these genes were normalized to IbTubulin expression. (D–F) Proline, MDA and H2O2 content analyses in the leaves of wild type and transgenic plants before and after the cold treatments. Values are means and standard deviations of three biological replicates. *, ** and *** indicate p-value < 0.05, < 0.01 and < 0.001, respectively
3.5 IbSINA5 Increases IbCOR Gene Expression and Oxidative Stress Tolerance in Transgenic Plants
It is well known that COR genes play a crucial role in plant response to cold stress [38]. We examined the expressions of IbCOR413, IbCOR314 and IbCOR27 in wild type and transgenic plants. As we have expected, the expression levels of IbCOR genes in transgenic plants were significantly higher than in wild type plants (Fig. 4C). Previous study has showed that overexpression of IbCBF3 conferred improved tolerance to low temperature and drought stress on transgenic sweet potato, leading to decreased accumulation of MDA and H2O2 [13]. Similar results were also observed in transgenic sweet potato plants overexpressing IbSINA5. Under normal growth condition, no significant difference was observed. However, under cold stress condition, a higher proline, and lower MDA and H2O2 content was detected in transgenic plants (Figs. 4D–4F). The increased IbCOR gene expression and antioxidant ability might help explain the improved cold resistance in IbSINA5 transgenic plants.
3.6 IbSINA5 is Up-Regulated by IbCBF3 in Sweet Potato
Previous work has demonstrated that IbCOR genes were up-regulated by IbCBF3 [13]. We performed PlantCARE analysis with the promoter sequence of IbSINA5, and a dehydration-responsive element-binding/C-repeat-binding factor (DREB/CBF) binding (DRE core) element was identified (Fig. 5). To explore whether IbSINA5 expression was regulated by CBFs in sweet potato, dual-luciferase assays were carried out. We found that IbCBF3 significantly activated the promoter of IbSINA5, implying that IbSINA5 expression could be regulated by IbCBF3 (Figs. 6A and 6B). We then transiently expressed IbCBF3 in the leaf protoplasts of wild type Xushu 18 plants. We observed that transient expression of IbCBF3 increased the transcription of IbSINA5, IbCOR413, IbCOR314 and IbCOR27 (Figs. 6C and 6D).

Figure 5: Plant CARE analysis. The Cis-acting elements in the promoter region of IbSINA5 (2001 bp) were analyzed. The predicted CBF binding DRE core element was marked in red
Taken together, our results suggests that overexpression of IbSINA5 up-regulates IbCOR gene expression and enhances cold resistance in transgenic sweet potato plants. IbCBF3 could be an efficient activator of IbSINA5 to regulate the expression of IbCOR genes, possible in a CBF-SINA-COR module.
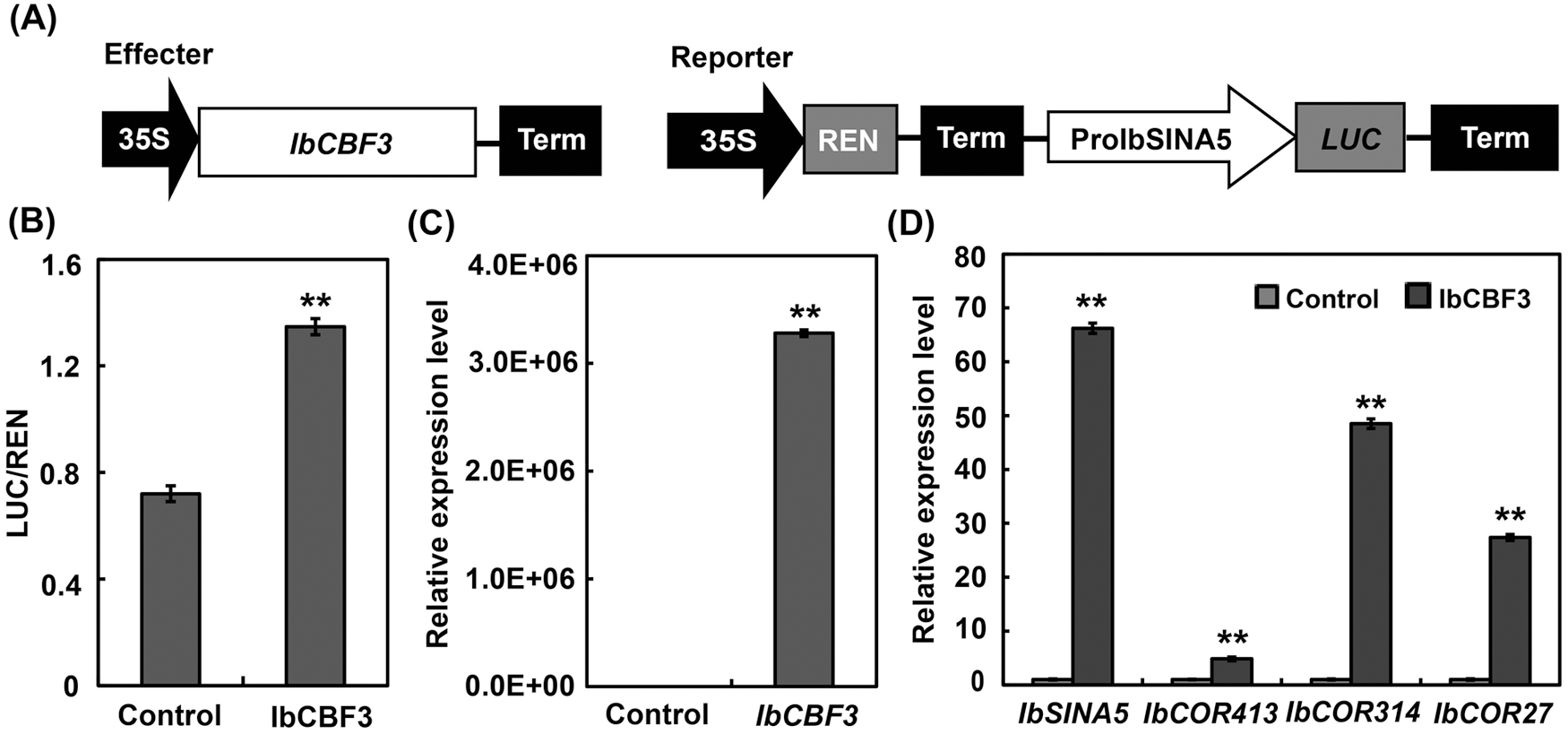
Figure 6: IbCBF3 positively regulates the expression of IbSINA5 and IbCOR genes. (A) Schematic diagrams of the effector and reporter constructs. (B) IbCBF3 activates the promoter of IbSINA5. The control vector or pGreenII62-SK-IbCBF3 were co-expressed with 35S::REN-ProIbSIN5::LUC in the leaf protoplasts of wild type sweet potato plants. LUC/REN ratios indicate the promoter activity of IbSINA5 activated by IbCBF3. Control, protoplasts co-transfected with pGreenII62-SK and 35S::REN-ProIbSIN5::LUC; IbCBF3, protoplasts co-transfected with pGreenII62-SK-IbCBF3 and 35S::REN-ProIbSIN5::LUC. (C) Expression of IbCBF3 in the transfected sweet potato mesophyll protoplasts. (D) Expression levels of IbSINA5 and IbCOR genes in the transfected protoplasts. pGreenII62-SK-IbCBF3 was transfected into the sweet potato mesophyll protoplasts. pGreenII62-SK was used as a negative control. Total RNA was isolated from the transfected protoplasts for qRT-PCR analyses. The expression of gene in control was set to 1. Control, protoplasts transfected with pGreenII62-SK; IbCBF3, protoplasts transfected with pGreenII62-SK-IbCBF3. Values are means and standard deviations of three biological replicates. ** indicates p-value < 0.01
Acknowledgement: We thank Prof. Jun-jie Han (Yantai Academy of Agricultural Sciences, Yantai, Shandong Province, China) for providing the sweet potato cultivars Xushu18 and Taizhong6.
Authors Contribution: LMW, SLZ and HXZ designed the experiments; SYL, XAL, LZZ, HQH and BL performed the experiments; ZZS and LMW analyzed the data; SYL and HXZ wrote the manuscript. All authors have read and approved the final manuscript.
Funding Statement: This work was jointly supported by the following grants: Agricultural Variety Improvement Project of Shandong Province [Grant Nos. 2019LZGC009, 2019LZGC010, 2020LZGC007]; the National Key R&D Program of China [Grant Nos. 2018YFD1000500, 2019YFD1000500]; the National Natural Science Foundation of China [Grant Nos. 31870576, 31901572, 32071733]; the Natural Science Foundation of Shandong Province [Grant Nos. ZR2018PH041, ZR2019PC015, ZR2020MC138]; the Modern Agricultural Industry Technology System Innovation Team of Shandong Province of China [Grant No. SDAIT-02-05]; the Key R&D Program of Shandong Province of China [2019GSF108154]; the Science and Technology Development Project in Yantai [Grant No. 2018XSCC041].
Conflicts of Interest: We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
1. Bovell-Benjamin, A. (2007). Sweet potato: A review of its past, present, and future role in human nutrition. Advances in Food and Nutrition Research, 52(9), 1–59. DOI 10.1016/S1043-4526(06)52001-7. [Google Scholar] [CrossRef]
2. Mukhopadhyay, S. K., Chattopadhyay, A., Chakraborty, I., Bhattacharya, I. (2011). Crops that feed the world 5. Sweetpotato. Sweetpotatoes for income and food security. Food Security, 3(3), 283–305. DOI 10.1007/s12571-011-0134-3. [Google Scholar] [CrossRef]
3. Mohanraj, R., Sivasankar, S. (2014). Sweet potato (Ipomoea batatas [L.] Lam) avaluable medicinal food: A review. Journal of Medicinal Food, 17(7), 733–741. DOI 10.1089/jmf.2013.2818. [Google Scholar] [CrossRef]
4. Fan, W., Zhang, M., Zhang, H., Zhang, P. (2012). Improved tolerance to various abiotic stresses in transgenic sweet potato (Ipomoea batatas) expressing spinach betaine aldehyde dehydrogenase. PLoS One, 7(5), e37344. DOI 10.1371/journal.pone.0037344. [Google Scholar] [CrossRef]
5. Fan, W., Deng, G., Wang, H., Zhang, H., Zhang, P. (2015). Elevated compartmentalization of Na+ into vacuoles improves salt and cold stress tolerance in sweet potato (Ipomoea batatas). Plant Physiology, 154(4), 560–571. DOI 10.1111/ppl.12301. [Google Scholar] [CrossRef]
6. Kim, K. Y., Jeong, J. C., Lee, H. S., Kwak, S. S. (2013). Comparative characterization of sweet potato antioxidant genes from expressed sequence tags of dehydration- treated fibrous roots under different abiotic stress conditions. Molecular Biology Reports, 40(4), 2887–2896. DOI 10.1007/s11033-012-2304-6. [Google Scholar] [CrossRef]
7. Jiang, T., Zhai, H., Wang, F. B., Zhou, H. N., Si, Z. Z. et al. (2014). Cloning and characterization of a salt tolerance-associated gene encoding trehalose-6-phosphate synthase in sweetpotato. Journal of Integrative Agriculture, 13(8), 1651–1661. DOI 10.1016/S2095-3119(13)60534-1. [Google Scholar] [CrossRef]
8. Chen, S. P., Lin, I. W., Chen, X., Huang, Y. H., Chang, S. C. et al. (2016). Sweet potato NAC transcription factor, IbNAC1, upregulates sporamin gene expression by binding the SWRE motif against mechanical wounding and herbivore attack. Plant Journal, 86(3), 234–248. DOI 10.1111/tpj.13171. [Google Scholar] [CrossRef]
9. Park, S., Kim, H. S., Jung, Y. J., Kim, S. H., Ji, C. Y. et al. (2016). Orange protein has a role in phytoene synthase stabilization in sweetpotato. Scientific Reports, 6(1), 111. DOI 10.1038/srep33563. [Google Scholar] [CrossRef]
10. Gao, S., Yu, B., Zhai, H., He, S. Z., Liu, Q. C. (2011). Enhanced stem nematode resistance of transgenic sweetpotato plants expressing oryzacystatin-I gene. Agricultural Sciences in China, 10(4), 519–525. DOI 10.1016/S1671-2927(11)60032-1. [Google Scholar] [CrossRef]
11. Ruan, L., Chen, L., Chen, Y., He, J., Zhang, W. et al. (2012). Expression of Arabidopsis homeodomain glabrous 11 enhances tolerance to drought stress in transgenic sweet potato plants. Journal of Plant Biology, 55(2), 151–158. DOI 10.1007/s12374-011-9198-z. [Google Scholar] [CrossRef]
12. Zhai, H., Wang, F., Si, Z., Huo, J., Xing, L. et al. (2016). A myo-inositol-1-phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotechnology Journal, 14(2), 592–602. DOI 10.1111/pbi.12402. [Google Scholar] [CrossRef]
13. Jin, R., Kim, B. H., Ji, C. Y., Kim, H. S., Min, H. et al. (2017). Overexpressing IbCBF3 increases low temperature and drought stress tolerance in transgenic sweet potato. Plant Physiology and Biochemistry, 118, 45–54. DOI 10.1016/j.plaphy.2017.06.002. [Google Scholar] [CrossRef]
14. Rothe, M., Wong, S. C., Henzel, W. J., Goeddel, D. V. (1994). A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell, 78(4), 681–692. DOI 10.1016/0092-8674(94)90532-0. [Google Scholar] [CrossRef]
15. Pullen, S. S., Miller, H. G., Everdeen, D. S., Dang, T. T. A., Crute, J. J. et al. (1998). CD40-Tumor necrosis factor receptor-associated factor (TRAF) interactions: regulation of CD40 signaling through multiple TRAF binding sites and TRAF hetero-oligomerization. Biochemistry, 37(34), 11836–11845. DOI 10.1021/bi981067q. [Google Scholar] [CrossRef]
16. Leo, E., Welsh, K., Matsuzawa, S., Zapata, J. M., Kitada, S. et al. (1999). Differential requirements for tumor necrosis factor receptor-associated factor family proteins in CD40 mediated induction of NF-kappa B and Jun N-terminal kinase activation. Journal of Biological Chemistry, 274(32), 22414–22422. DOI 10.1074/jbc.274.32.22414. [Google Scholar] [CrossRef]
17. Han, S. Y., Yoon, K., Lee, K., Kim, K., Jang, H. et al. (2003). TNF-related weak inducer of apoptosis receptor, a TNF receptor superfamily member, activates NF-kappa B through TNF receptor-associated factors. Biochemical and Biophysical Research Communications, 305(4), 789–796. DOI 10.1016/S0006-291X(03)00852-0. [Google Scholar] [CrossRef]
18. Wang, M., Jin, Y., Fu, J. J., Zhu, Y., Zheng, J. et al. (2009). Genome-wide analysis of SINA family in plants and their phylogenetic relationships. DNA Sequence, 19(3), 206–216. DOI 10.1080/10425170701517317. [Google Scholar] [CrossRef]
19. Xie, Q., Guo, H. S., Dallman, G., Fang, S. Y., Weissman, A. M. et al. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature, 419(6903), 167–170. DOI 10.1038/nature00998. [Google Scholar] [CrossRef]
20. He, X. J., Mu, R. L., Cao, W. H., Zhang, Z. G., Zhang, J. S. et al. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant Journal, 44(6), 903–916. DOI 10.1111/j.1365-313X.2005.02575.x. [Google Scholar] [CrossRef]
21. Welsch, R., Maass, D., Voegel, T., DellaPenna, D., Beyer, P. (2007). Transcription factor RAP2.2 and its interacting partner SINAT2: Stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiology, 145(3), 1073–1085. DOI 10.1104/pp.107.104828. [Google Scholar] [CrossRef]
22. Ning, Y., Jantasuriyarat, C., Zhao, Q., Zhang, H., Chen, S. et al. (2011). The SINA E3 ligase OsDIS1 negatively regulates drought response in rice. Plant Physiology, 157(1), 242–255. DOI 10.1104/pp.111.180893. [Google Scholar] [CrossRef]
23. Herder, G. D., Keyser, A. D., Rycke, R. D., Rombauts, S., Velde, W. V. et al. (2008). Seven in absentia proteins affect plant growth and nodulation in Medicago truncatula. Plant Physiology, 148(1), 369–382. DOI 10.1104/pp.108.119453. [Google Scholar] [CrossRef]
24. Wang, W., Fan, Y., Niu, X., Miao, M., Kud, J. et al. (2018). Functional analysis of the seven in absentia ubiquitin ligase family in tomato. Plant, Cell & Environment, 41(3), 689–703. DOI 10.1111/pce.13140. [Google Scholar] [CrossRef]
25. Bao, Y., Wang, C. T., Jiang, C. M., Pan, J., Zhang, G. B. et al. (2014). The TRAF-like family protein SINA2 promotes drought tolerance in an ABA-dependent manner in Arabidopsis. New Phytologist, 202(1), 174–187. DOI 10.1111/nph.12644. [Google Scholar] [CrossRef]
26. Li, B., Liu, H., Zhang, Y., Kang, T., Zhang, L. et al. (2013). Constitutive expression of cell wall invertase genes increases grain yield and starch content in maize. Plant Biotechnology Journal, 11(9), 1080–1091. DOI 10.1111/pbi.12102. [Google Scholar] [CrossRef]
27. Yang, J., Moeinzadeh, M. H., Kuhl, H., Helmuth, J., Xiao, P. et al. (2017). Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nature Plants, 3(9), 696–703. DOI 10.1038/s41477-017-0002-z. [Google Scholar] [CrossRef]
28. Liu, T. B., Kim, D. W., Niitsu, M., Berberich, T., Kusano, T. (2014). Oryza sativa polyamine oxidase 1 back-converts tetraamines, spermine and thermospermine, to spermidine. Plant Cell Reports, 33(1), 143–151. DOI 10.1007/s00299-013-1518-y. [Google Scholar] [CrossRef]
29. Park, S. C., Kim, S. H., Park, S., Lee, H. U., Lee, J. S. et al. (2015). Enhanced accumulation of carotenoids in sweetpotato plants overexpressing IbOr-Ins gene in purple-fleshed sweetpotato cultivar. Plant Physiology and Biochemistry, 86, 82–90. DOI 10.1016/j.plaphy.2014.11.017. [Google Scholar] [CrossRef]
30. Wang, W. B., Kim, Y. H., Lee, H. S., Kim, K. Y., Deng, X. P. et al. (2009). Analysis of antioxidant enzyme activity during germination of alfalfa under salt and drought stresses. Plant Physiology and Biochemistry, 47(7), 570–577. DOI 10.1016/j.plaphy.2009.02.009. [Google Scholar] [CrossRef]
31. Bindschedler, L. V., Minibayeva, F., Gardner, S. L., Gerrish, C., Davies, D. R. et al. (2001). Early signalling events in the apoplastic oxidative burst in suspension cultured French bean cells involve cAMP and Ca2+. New Phytologist, 151(1), 185–194. DOI 10.1046/j.1469-8137.2001.00170.x. [Google Scholar] [CrossRef]
32. Wang, H. H., Tang, R. J., Liu, H., Chen, H. Y., Liu, J. Y. et al. (2013). Chimeric repressor of PtSND2 severely affects wood formation in transgenic populus. Tree Physiology, 33(8), 878–886. DOI 10.1093/treephys/tpt058. [Google Scholar] [CrossRef]
33. Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K. et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods, 1(1), 13–26. DOI 10.1186/1746-4811-1-13. [Google Scholar] [CrossRef]
34. Jin, Y. L., Yu, C. Y., Jiang, C. M., Guo, X. T., Li, B. et al. (2020). PtiCYP85A3, a BR C-6 oxidase gene, plays a critical role in brassinosteroid-mediated tension wood formation in poplar. Frontiers in Plant Science, 11, 468–470. DOI 10.3389/fpls.2020.00468. [Google Scholar] [CrossRef]
35. Wang, H. H., Wang, X. Q., Yu, C. Y., Wang, C. T., Jin, Y. L. et al. (2020). MYB transcription factor PdMYB118 directly interacts with bHLH transcription factor PdTT8 to regulate wound-induced anthocyanin biosynthesis in poplar. BMC Plant Biology, 20(1), 485. DOI 10.1186/s12870-020-02389-1. [Google Scholar] [CrossRef]
36. Stone, S. L., Callis, J. (2007). Ubiquitin ligases mediate growth and development by promoting protein death. Current Opinion in Plant Biology, 10(6), 624–632. DOI 10.1016/j.pbi.2007.07.010. [Google Scholar] [CrossRef]
37. Lyzenga, W. J., Stone, S. L. (2012). Abiotic stress tolerance mediated by protein ubiquitination. Journal of Experimental Botany, 63(2), 599–616. DOI 10.1093/jxb/err310. [Google Scholar] [CrossRef]
38. Wang, J. G., Dai, S. Y., Sun, H. W., Liu, E. H., Zhou, M. et al. (2020). The N-terminal and third transmembrane domain of PsCor413im1 are essential for targeting to chloroplast envelope membrane. Biochemical and Biophysical Research Communications, 527(4), 929–934. DOI 10.1016/j.bbrc.2020.05.046. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |