
International Journal of
Experimental Botany

 | Phyton- International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2021.016217
ARTICLE
AFP2 Coordinates the Activity of PIF7 for Thermomorphogenesis in Arabidopsis Seedlings
Shanghai Key Laboratory of Bio-Energy Crops, School of Life Sciences, Shanghai University, Shanghai, 200444, China
*Corresponding Author: Ping Li. Email: liping80@shu.edu.cn
Received: 18 February 2021; Accepted: 09 March 2021
Abstract: Ambient temperature induces the hypocotyl elongation of seedling, called as thermomorphogenesis. It has been reported that the bHLH transcriptional factor PIF7 acts as the critical component to modulate plant thermomorphogenesis, but the underlying mechanism remains elusive. The phytohormone abscisic acid (ABA) suppresses the hypocotyl elongation under high temperature (HT) stress. As the ABI5 binding protein, AFP2 acts as the negative factor to control ABA signaling. In this study, we first identified AFP2 as the interaction protein of PIF7 in vitro and in vivo. Phenotype analysis revealed that overexpressing AFP2 reduced the hypocotyl elongation, while loss-of-function afp2 mutant showed longer hypocotyl under HT. Consistently, overexpressing AFP2 impaired the transactivation effect of PIF7 on auxin biosynthesis related genes YUC8 and IAA19, which possibly resulted into the shorter hypocotyl in the transgenic line overexpressing AFP2 or co-overexpressing AFP2 and PIF7. Thus, these data suggest that AFP2 suppressed PIF7 activity to suppress hypocotyl elongation. Furthermore, we found that HT gradually induced the degradation of AFP2 that possibly released the inhibitory effect of AFP2 on PIF7, thus induced hypocotyl elongation under HT. Taken together, our result reveals the novel function of AFP2 in coordinating thermomorphogenesis through sophistically modulating PIF7 activity.
Keywords: Thermomorphogenesis; AFP2; PIF7; hypocotyl elongation
Plant has evolved an elaborate mechanism to adapt the environment fluctuation. Ambient temperature over the optimal range dramatically affects the plant architecture, for example, longer petioles, up-ward leave and elongated hypocotyl. These architecture changes, also termed as thermomorphogenesis, promote plant to cool down their leaf surface temperature [1–3]. Accumulated evidence demonstrates that plant photoreceptor phytochromes B (phyB) is functional as the thermo-sensor to perceive the environment change and thus coordinate plant thermomorphogenesis [4]. PIF4 belongs to the bHLH transcriptional factor, phyB interacts with PIF4 to repress its activity, even trigger its degradation under light condition [5,6]. However, ambient high temperature inactivates phyB activity, thus release PIF4 activity, which directly activates the expression of several auxin biosynthesis or signaling related genes, subsequently induce cell elongation [1,3]. Besides PIF4 as the key positive thermomorphogenesis, there exist other seven PIFs, such as PIF3 and PIF5 that also modulate hypocotyl elongation for plant response to environmental stress [7–11]. Recently PIF7 is also identified as the necessary factor for thermomorphogenesis under warm cycling daytime temperature, PIF7 directly activates the transcription of key gene such as auxin biosynthesis genes such as YUC8 [12,13]. Warm environment also indued the quick accumulation of PIF7, suggesting the feedback regulation network underlying PIF7 during thermomorphogenesis, though detail mechanism needs more investigated.
Apart from auxin biosynthesis, another phytohormone including ABA also regulates plant thermomorphogenesis, directly treatment with ABA strongly suppresses hypocotyl elongation under high temperature stress, the hypocotyl elongation in ABA biosynthesis deficiency mutant is longer after heat stress, suggesting the negative role of ABA signal during thermomorphogenesis [14–16]. In Arabidopsis, the bZIP transcriptional factor ABI5 acts as the important component for ABA signal transduction [17,18], AFPs as the interaction protein of ABI5 affects the protein level of ABI5, thus control seed germination after ABA treatment [19–21]. There are four members of AFPs in the Arabidopsis genome, including AFP1 to AFP4. AFP1 is reported to negatively regulate ABI5 protein stability, and seed germination of afp1 mutant contains higher ABI5 protein level and show more sensitive to ABA stimulation [19]. Similar to AFP1, AFP2 and AFP3 are also functional epistatically to ABI5, therefore affect the seed germination in response to ABA signal [21]. Seed germination of afp2 or afp3 mutant was lower after saline or drought treatment. Our previous study also showed that AFP2 recruits its corepressor TPR2 to suppress CONSTANS activity to delay flowering time [22]. What is more, we found that overexpressing AFP2 presents high germination percentage under high temperature condition while the seed germination of afp2 was lower, we also found that AFP2 dependents on ABI5 to control seed germination under high temperature [23]. However, whether or how AFP2 control seedling thermomorphogenesis remains elusive so far.
In this study, we first confirm that PIF7 indeed acted as the necessary factor to control seedling hypocotyl elongation in response to ambient high temperature. Further biochemical analysis showed that PIF7 interacts with AFP2 in planta, and AFP2 suppressed PIF7 activity, subsequently suppressing PIF7-mediated thermomorphogenesis. In agreement with it, we found that overexpressing AFP2 suppressed seedling hypocotyl elongation, while afp2 mutant showed longer hypocotyl. Therefore our data reveal the novel function of AFP2 in controlling seedling thermomorphogenesis through PIF7.
2.1 Arabidopsis Materials and Growth
Arabidopsis thaliana ecotype Columbia (Col-0) plants were used as the wild type. The mutant of pif7-1 (Sail_622_G02) were obtained from the Arabidopsis Biological Resource Center. Seeds of each genotype were surface sterilized and sown on Murashige and Skoog medium consisting of 0.8% agar, stratified at 4°C in the dark for 3 days, and transferred to a growth chamber at 23°C under constant light. After 10 days, the seedlings were transferred to soil (3:1 [v/v] potting soil:vermiculite) and grown in a greenhouse under a 16-h white light (100–120 μmol m–2 s–1)/8-h dark cycle. Seeds used for measurement of hypocotyl length analysis were simultaneously harvested from plants grown in the same batches at 22°C under long-day (16 h light/8 h dark) conditions. Dry seeds were stored at room temperature for 2–5 months before germination analysis.
The seed was cold stratified at 4°C for 3 days and then sowed on 1/2MS medium for germination in the grow camber under a 16-h white light (100–120 μmol m–2 s–1)/8-h dark cycle. For hypocotyl elongation measurements, seeds were sown on 1/2MS on the growth media. Seedlings were grown on vertical plates in an incubator for 3 d at 22°C. High temperature treatment (28°C) started on day 4 for 3 days.
To generate transgenic plants overexpressing PIF7, the full-length coding sequence of this gene was amplified using the primers listed in Supplemental Table 1. The amplified fragments were cloned into the Nde/EcoRI (New England Biolabs) site of the pRI101-6Flag vector [6 × Flag tag inserted in the EcoRI/SacI site of the pRI101-AN vector (3262; Takara)] to generate the PIF7-Flash construct, using an In-fusion HD Cloning Kit (638911, Clontech). These constructs were transformed into Arabidopsis plants using the Agrobacterium tumefaciens-mediated floral dip method (Clough and Bent, 1998).
To check the interaction of AFP2 and PIF7 in yeast, sequences encoding AFP2 and PIF7 were amplified using the primers listed in Supplemental Table 1. The PCR products were cloned into the prey vector pGADT7 (630442, Clontech) and bait vector pGBKT7 (630443, Clontech) to generate pGBKT7-AFP2 (BD-AFP2) and pGADT7-PIF7 (AD-PIF7), respectively, using In-fusion Cloning Technology (Clontech). Yeast strains Y187 and AH109 were transformed with the prey and bait vectors, respectively, by polyethylene glycol-mediated yeast transformation as described by the manufacturer. After screening on minus Trp medium (630413, Clontech) or minus Leu medium (630414, Clontech), three independent clones were mated and grown on minus Trp/Leu medium (630417, Clontech) for 3 d to confirm mating success, and the corresponding clones were transferred to minus TPR/Leu/His/Ade medium (630428, Clontech) to measure their growth status.
2.5 Co-immunoprecipitation Analysis
The transgenic plants AFP2-FLAG and PIF7-Flash/AFP2-FLAG were used in Co-IP assays to detect the interaction between AFP2 and PIF7 in planta. Seedlings were grown in a grown cabinet under long-day (16 h light/8 h dark) conditions at 22°C for 7 days. The samples were harvested, ground to a fine powder in liquid nitrogen, and homogenized in IP buffer (50 mM Tris–HCl, 150 mM NaCl, 5 mM EDTA [pH 8.0], 1% Triton X-100, 0.6 mM PMSF, 20 μM MG132, Roche protease inhibitor cocktail). The extracts were centrifuged at 14,000 g for 20 min. The supernatant was mixed with 20 μl anti-MYC resin (ChromoTek) for 4 h at 4°C. The beads were washed three times with washing buffer (50 mM sodium phosphate buffer, pH 7.0, 100 mM NaCl, 5 mM EDTA, 0.1% [v/v] Triton X-100, 20 μM MG132, and a protease inhibitor tablet). After brief centrifugation, the immunoprecipitates were separated on a 12% (w/v) SDS-polyacrylamide gel and detected by immunoblot analysis with anti-Myc (Clontech) or anti-Flag (Sigma-Aldrich) antibodies, and the immunoblot signals were detected using an ECL Kit (Invitrogen).
BiFC experiments were performed using the leaves of Nicotiana benthamiana plants grown in a greenhouse (16 h light/8 h dark) for 2 weeks as described previously [22]. In brief, full-length or truncated AFP2 and PIF7 were recombined into the binary BiFC vector pSPYNE and pSPYCE, respectively. The constructs were transformed into competent A. tumefaciens strain LBA4404 cells (9115, Clontech), and the cultures were incubated in a rotator at 250 rpm for 12 h at 28°C. After culture, Agrobacteria harboring nYFP or cYFP were mixed together and centrifuged at 12000 g for 5 min at 4°C, and the pellets were dissolved in injection solution (10 mM Tri-HCl buffer, 25 mM MgCl2, pH 5.6) to an OD600 of 0.1. The Agrobacterium solution was injected into N. benthamiana leaves, and YFP fluorescence was observed 3 d after injection under a confocal microscope (LSM710; Zeiss).
Total RNA was extracted from the seedling samples using a Plant Total RNA extraction kit (NEB) and reverse-transcribed to cDNA using the reverse transcriptase kit (Takara Bio) according to the manufacturer’s instructions. RT-qPCR was performed using SYBR Premix Ex Taq (Perfect Real Time; Takara Bio) on a LightCycler 96 (Roche). The gene expression results were normalized to the expression of PP2A as the internal control. All experiments were performed three biological replicates and technical repeats [23,24]. Primers are listed in Supplemental Tab. 1.
2.8 Transient Transactivation Assay
The transient transactivation assay was performed in Arabidopsis protoplasts as described previously. For transient transcription activity assays using specific promoters, the YUC8 and IAA19 promoter sequence was amplified from Col-0 genomic DNA and cloned into the pGreenII-0800-LUC vector as a reporter and the REN gene under the control of the cauliflower mosaic virus (CaMV) 35S promoter in the pGreenII 0800-LUC vector as the internal control. The coding sequences of AFP2 and PIF7 were cloned into pGREEN-62-SK under the control of the 35S promoter and used as effectors. Reporters and effectors were transformed into Arabidopsis protoplasts in different combinations and incubated in the dark for 18 h. LUC and REN activities were separately determined at 18 h post-transformation using the Dual-Luciferase Reporter Assay System (Promega, E1910).
2.9 Total Protein Extract and Western Blotting
For immunoblot analysis, freshly harvested or after-ripened seeds were rapidly frozen in liquid nitrogen and ground in extraction buffer (50 mM sodium phosphate, pH 7.0, 100 mM NaCl, 5 mM EDTA, 0.1% [v/v] Triton X-100, 0.1% [w/v] sodium deoxycholate, and protease inhibitor tablet; Roche). Following centrifugation at 14,000 g for 5 min at 4°C, the supernatant was collected. Approximately 10 mg of total protein was separated on a 12% (w/v) SDS–PAGE gel and transferred to a nitrocellulose membrane, which was then probed with the appropriate primary antibody (anti-Myc, anti-GST, 1:3000, Clontech; anti-Flag, 1:3000, Sigma-Aldrich) and horseradish-conjugated goat anti-mouse secondary antibody (1:3000, Promega). Signals were detected using an ECL Kit (Invitrogen).
2.10 Subcellular Localization Analysis
The coding sequence of PIF7 or AFP2 was cloned into pRI101-NGFP, pRI101-m Cherry, or pRI101-CFP. The constructs were transformed into Agrobacterium strain GV3101. The Agrobacterium cultures were infiltrated into N. benthamiana leaves, and fluorescent signals from GFP, CFP, or mCherry fluorescent protein (YFP) were observed at 72 h post-infiltration.
3.1 The Thermomorphogenic Response Requires PIF7 in Arabidopsis
It is known that PIF4 plays the vital role in hypocotyl elongation during thermomorphogenesis [13]. Here we accessed the function of its homology, PIF7, during hypocotyl elongation. At first we generated the transgenic line expressing GUS marker under the control of the native promoter of PIF7 (pPIF7:GUS). GUS staining showed the expression of PIF7 in the cotyledon and the hypocotyl of the young seedling. High temperature treatment partially increased the GUS staining in cotyledon and hypocotyl (Fig. 1A) suggesting the potential function of PIF7 during heat stress. We then obtained the loss-of-function mutant, SAIL_622_G02, from ABRC, in which a T-DNA was inserted into its third and fourth exon through flanking primers analysis, such insertion also abolished the functional transcripts of PIF7 (Fig. 1B), thus such mutant was named as pif7-1. We also generated the transgenic line overexpressing PIF7-Flash fusion under the control of the 35S promoter (35S:PIF7-Flash, abbreviated as PIF7-Flash), the expression of PIF7-Flash was also confirmed by western blotting analysis using anti-MYC antibody (Fig. 1C). Under the normal condition at 22°C, the hypocotyl length of PIF7-Flash was longer than wild-type Col and pif7-1 mutant, ambient HTs treatment promoted the hypocotyl elongation, but HT did not obviously promote the hypocotyl elongation in pif7-1 mutant, and PIF7-Flash seedling still showed longer hypocotyl independent of ambient environment temperature (Figs. 1C and 1D). Thus, these data suggest that PIF7 is associated with the hypocotyl elongation during thermomorphogenesis.
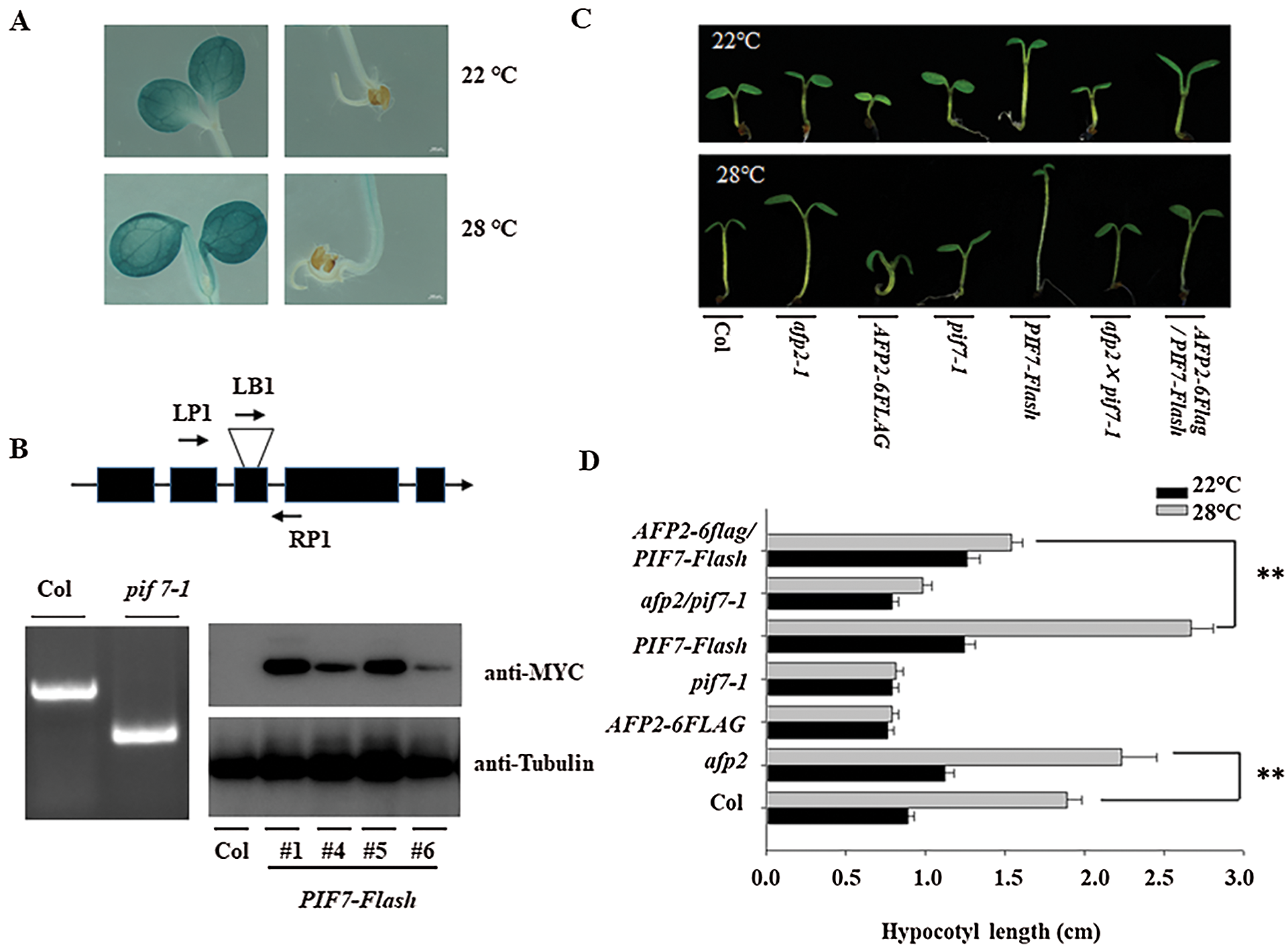
Figure 1: HT induced the hypocotyl length through PIF7. (A) HT slightly induced the expression of GUS in the transgenic pPIF7: GUS. Upper panel: the transgenic line under 22°C; Lower panel: the transgenic line under 28°C. (B) Validating the genotype of pif7-1 and PIF7-Flash overexpression lines by anti-MYC antibody. The T-DNA insertion position was confirmed by corresponding primers in pif7-1 mutant. Upper panel: the genomic structure of PIF7 and the T-DNA insertion position. The exon was shown as a black box, and the T-DNA insertion position was shown as a triangle. Bottom panel (Left): confirming the T-DNA insertion position by corresponding primers. Bottom panel (Right): The strong immunoblotting signal could be detected in the transgenic PIF7-Flash line by anti-MYC antibody. The top diagram indicates the genomic structure of PIF7 and the T-DNA insertion position. (C&D) HT induced the hypocotyl elongation. The seedling of Col, pif7-1, afp2-1, PIF7-Flash, AFP2-FLAG, afp2/pif7 and AFP2-FALG/pif7 seedling was placed under 22°C or 28°C for 3 days, and the hypocotyl length was measured. The values are shown as means SD of triplicate experiments. Asterisks indicate significant difference by Student’s t test (**p < 0.01)
3.2 PIF7 Activates the Expression of YUC8 and IAA19 for Hypocotyl Elongation
The auxin-biosynthesis related genes including YUC8 and IAA19 is dramatically induced by ambient HT, and mediates PIF4-dependent hypocotyl elongation during thermomorphogenesis [1]. Here we also compared their expression in pif7-1 mutant or PIF7-Flash seedlings, and found HT indeed induced the expression of YUC8 and IAA19, such induction was weak in the pfi7-1 seedlings, but stronger in the PIF7-Flash seedlings (Fig. 2). Similarly, PIF7 also regulated the expression of IAA19 and SAUR24, Thus, this genetic analysis suggests that PIF7 promote hypocotyl elongation through auxin-biosynthesis related gene such as YUC8, etc.
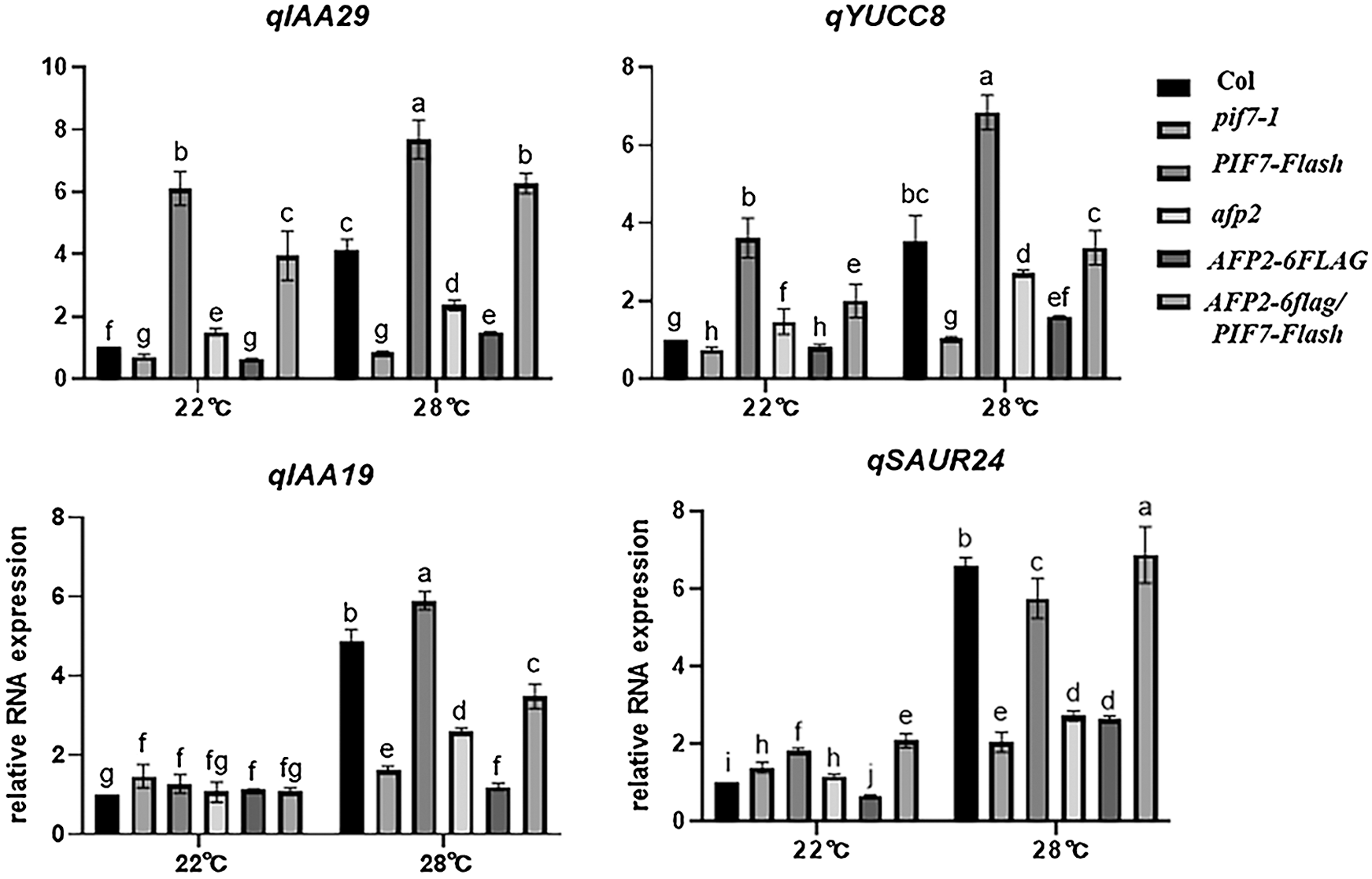
Figure 2: HT induced the expression of auxin-biosynthesis related genes
The wild-type Col seedling was treated with 28°C for indicated time, and the total RNA was extracted for RT-qPCR analysis. The PP2A was used as the internal control. The values are shown as means SD of triplicate experiments. Bars with different letters are significantly different at p < 0.05 (Tukey’s test).
To better understand the mechanism of which PIF7 control hypocotyl elongation during HTs, we used PIF7 as the bait and perform yeast two-hybrid approach to screen the Arabidopsis normalized cDNA library to look for PIF7-interaction protein. After two-round of screening, we obtained several positive clones. One clone encoding APF2 was selected for further study as AFP2 is also reported to particulate plant response to HTs stress. We first confirmed the interaction of PIF7 and AFP2 in yeast cell as the yeast co-expressing BD-PIF7 and AD-AFP2 grow well on the selective medium, but not for the BD-PIF7 or AD-AFP2 with their corresponding empty vector (Fig. 3A). We then generated the construct containing PIF7-Flash fusion under the control of 35S promoter (35S:PIF7-Flash, abbreviated as PIF7-Flash), or the AFP2-FLAG fusion under the control of 35S promoter (35S:AFP2-FLAG, abbreviated as AFP2-FLAG), and co-expressed them in tobacco leaves, and then checked their interaction between AFP2 and PIF7 (Figs. 3C and 3D). As shown in Fig. 3B, we found AFP2-FLAG could be co-immunoreacted by PIF7-Flash, but not by anti-MYC alone, suggesting the interaction between PIF7 and AFP2 in planta.
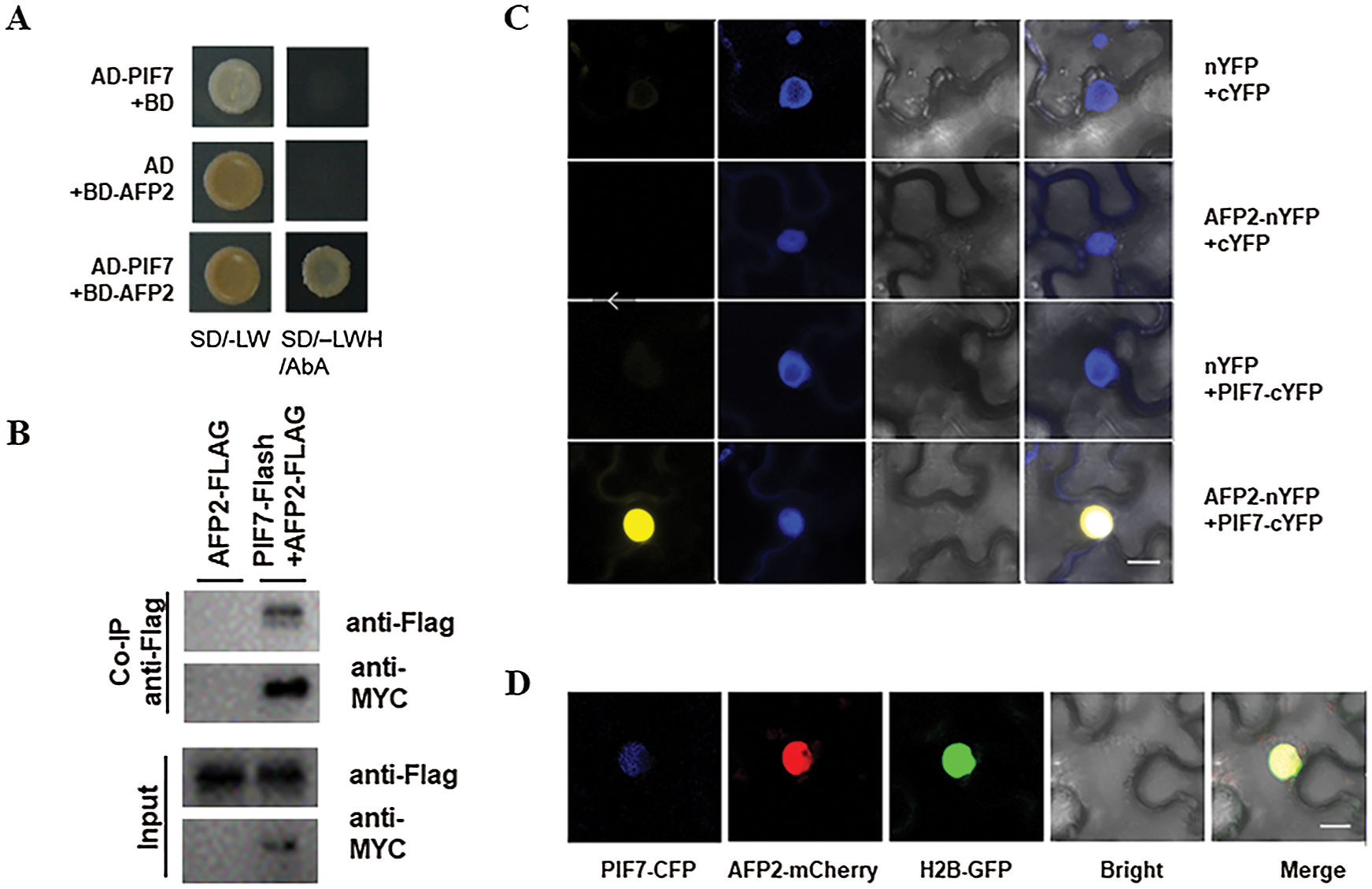
Figure 3: PIF7 interacts with AFP2 in vivo and in vitro (A) The interaction between PIF7 and APF2 by Y2H analysis. Yeast cells co-transformed with the indicated construct combinations were grown on SD medium lacking Trp/Leu (-LW) or Trp/Leu/His (-LWH) with 100 ng mL-1 Aureobasidin A (AbA). AD: DNA-activation domain of GAL4; BD: DNA-binding domain of GAL4. (B) The in-vivo interaction between PIF7 and AFP2 by Co-iP analysis Co-IP showed the interaction of PIF7 with AFP2 in Arabidopsis. Proteins extracted from hydrated seeds from AFP2-FLAG or PIF7-Flash/AFP2-FLAG plants were immunoprecipitated by MYC-Trap beads. Coimmunoprecipitated proteins were detected by anti-FLAG or anti-MYC antibody. Immunoblots show the presence of proteins in total protein extracts from plants (input) and fractions after immunoprecipitation by anti-FLAG or anti-MYC antibodies. (C) The interaction between PIF7 and AFP2 by BiFC analysis BiFC assay showed that PIF7-cYFP interacts with AFP2-nYFP in the nuclei of N. benthamiana epidermal leaf cells. PIF7 was fused to the C-terminal fragment of YFP (cYFP) to form PIF7-cYFP. Full-length AFP2 was fused with the N-terminal fragment of YFP (nYFP) to generate AFP2-nYFP. YFP fluorescence was detected in N. benthamiana leaves co-infiltrated with the indicated constructs. Nuclei were stained with 4, 6-diamidino-2-phenylindole (DAPI). Bar = 10 μm. (D) The co-localization of PIF7 and AFP2 in the Nucleus Analysis of the colocalization of PIF7 and AFP2 in N. benthamiana leaves. PIF7-CFP and AFP2-mCherry colocalize in the nuclei of N. benthamiana epidermal leaf cells. H2B-GFP: a Nuclei marker. Bar = 10 μm.
3.4 AFP2 Influences the Transcriptional Activation of PIF7 to Suppress Hypocotyl Elongation
As we mentioned above, PIF7 modulates the hypocotyl elongation under HTs, we then analyzed the function of AFP2 during thermomorphogenesis. We used the afp2-1 mutant and the transgenic line overexpressing AFP2-FLAG that is used before, to compare the hypocotyl length before or after HT stress. Under normal condition, all of the wild-type Col, afp2-1 and AFP2-FLAG transgenic line presented the similar length, however, HTs treatment increased the hypocotyl length, but HTs-induced hypocotyl elongation was attenuated in the AFP2-FLAG seedlings, but more obvious for afp2-1 mutant seedling (Figs. 1C and 1D), suggesting that AFP2 as the negative regulator controls hypocotyl elongation during seedling thermomorphogenesis process. We then checked the YUC8 and IAA19 expressing among Col, afp2-1 and AFP2-FLAG seedlings. As shown in Fig. 2, expressions of YUC8 and IAA19 were lower in the AFP2-FLAG lines, but higher in the afp2-1 line, compared with Col seedlings. These data suggest that AFP2 control hypocotyl elongation through auxin biosynthesis pathway.
We further dissect the genetic relationship between AFP2 and PIF7 during thermomorphogenesis by reciprocally crossing AFP2-FLAG or afp2 with PIF7-Flash or pif7 mutant, and then checked their hypocotyl elongation after HTs stress. As shown in Figs. 1C and 1D, HTs promoted the hypocotyl elongation for wild-type Col and PIF7-Flash seedling, but such induction was not effective in AFP2-FLAG and pif7-1 mutant seedlings. However, overexpressing AFP2-FALG in PIF7-Flash seedlings (AFP2-FLAG/PIF7-Flash) attenuated the hypocotyl elongation in contrast to PIF7-Flash seedling. In agreement with it, the expressions of YUC8 and IAA19 in AFP2-FLAG/PIF7-Flash was lower than that in PIF7-Flash background alone (Fig. 2). These data suggest that AFP2 antagonizes the function of PIF7 through influencing the auxin biosynthesis.
Furthermore, we checked the effects of AFP2 and PIF7 on the transcriptional level of YUC8 or IAA19 by transient protoplast analysis. As shown in Figs. 4A and 4B, overexpressing PIF7 alone obviously activated the transcriptional level of YUC8 or IAA19, however, coexpressing AFP2 with PIF7 attenuated the activation effect of PIF7 on the transcriptional level of YUC8 or IAA19, confirming the antagonistic effect between AFP2 or PIF7 on YUC8 or IAA19 expressions.
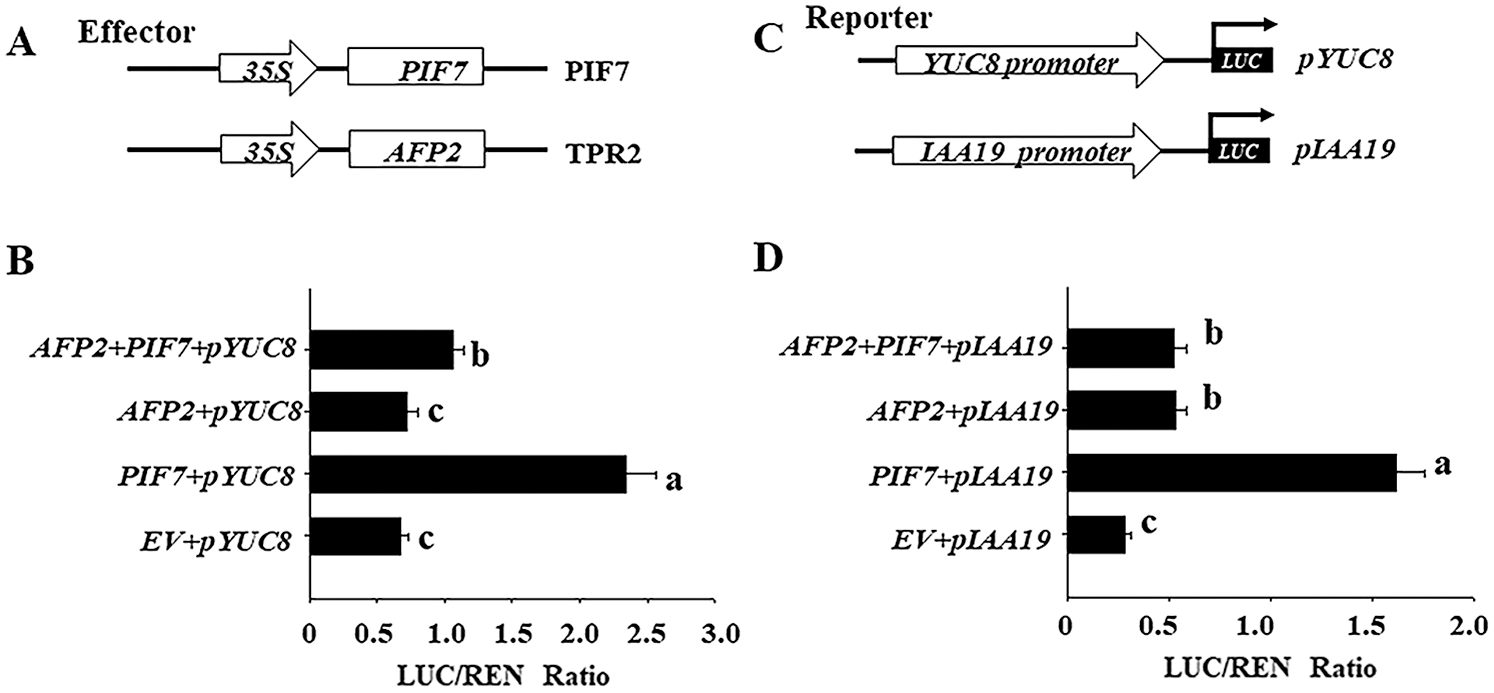
Figure 4: PIF7 and AFP2 antagonistically regulate the expressions of YUC8 and IAA19 by transient protoplast analysis. A) Schematic diagram of the effector (PIF7 or AFP2) and reporter constructs (YUC8 and IAA19) used in the transient transactivation assay. B) AFP2 and PIF7 antagonistically modulate the expressions of YUC8 and IAA19. The YUC8pro:LUC or IAA19pro:LUC reporter were coexpressed with PIF7 or AFP2 effectors for 24 h; the firefly luciferase and Renilla luciferase (LUC/REN) ratio represents YUC8pro:LUC or IAA19pro:LUC activity relative to the internal control (35Spro:REN). Data are means ± SD of three biological replicates. Bars labelled with different letters are significantly different at p < 0.05 (Tukey’s test).
3.5 HT Programmatically Induces the Degradation of AFP2 Protein
We previously study showed that AFP2 present circadian expression during daytime and nighttime. Here we further explore the expressing profile of AFP2 during seedling thermomorphogenesis. HTs treatment did not obviously affect the transcriptional level of AFP2. We then generated the transgenic seedling expressing the AFP2-FLAG fusion under the control of its native promoter (pAFP2:AFP2-FLAG), and checked the endogenous AFP2 protein level after HT treatment. As show in Fig. 5A, HT gradually decreased the protein abundance of AFP2 at the first 24 h treatment and sustained the lower level of AFP2 during the following 48 h treatment, suggesting that HT induced the degradation of AFP2. To further test the effect of HT on the stability of AFP2, we expressed GST-AFP2 in bacteria and used the extracted GST-AFP2 protein to test the effect of HT on AFP2 stability by cell-free analysis. As shown in Fig. 5B upper panel, we incubated GST-AFP2 with the crude protein extraction from the seedling subjected with 48 h of HT treatment, observed the gradually degradation of GST-AFP2. As the control, the crude protein extract from the seedling after HT treatment with additional proteasome inhibitor MG132 did not affect the stability of GST-APF2 (Fig. 5C, bottom panel). These data suggest that HT induced the degradation of AFP2 through ubiquitin-dependent degradation pathway.
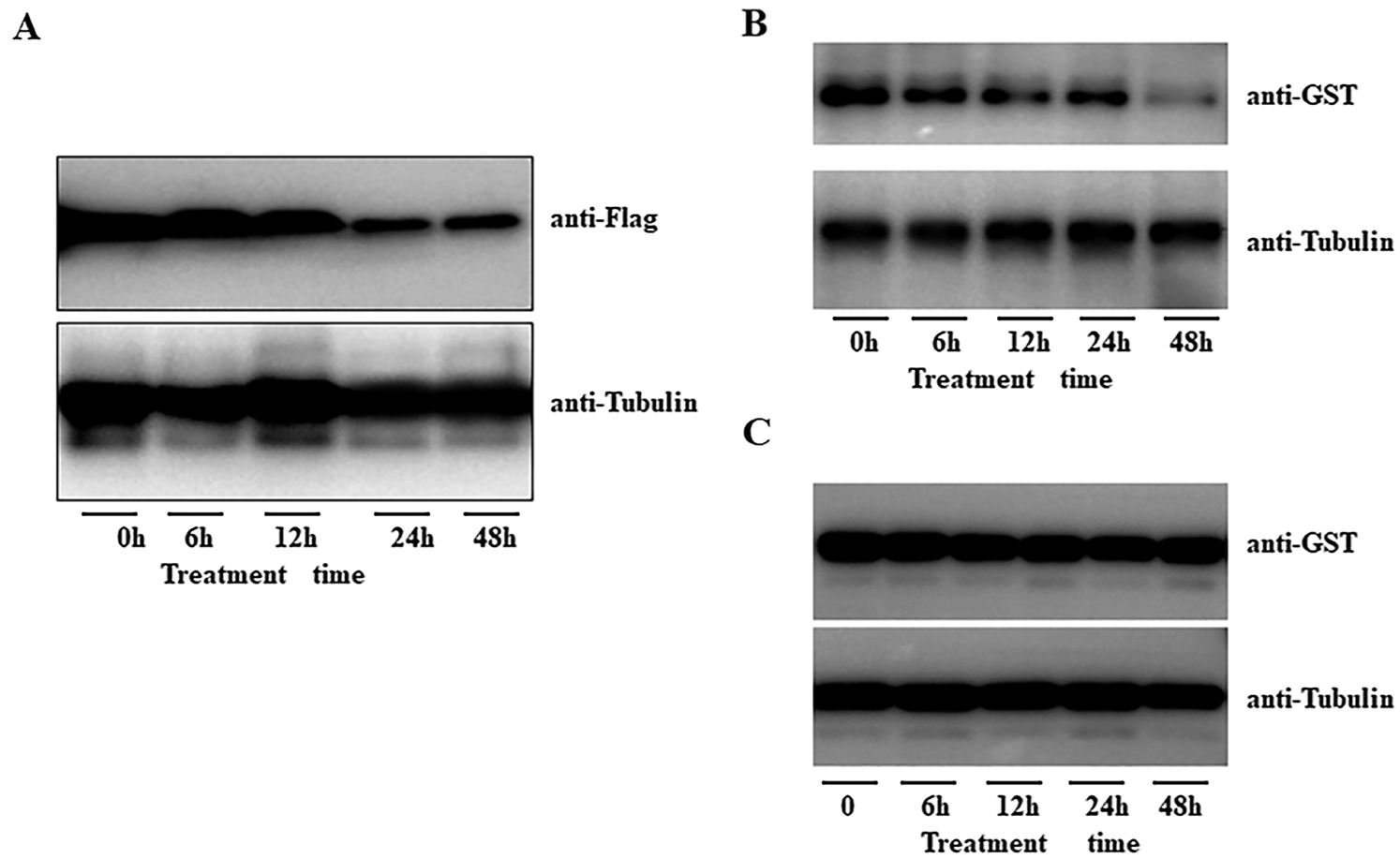
Figure 5: HT induced the degradation of AFP2. A) HT gradually induced the degradation of AFP2-FLAG in the transgenic pAFP2: AFP2-FLAG. One-week-old seedling of pAFP2: AFP2-FLAG was placed under 28°C for indicated time, and the protein abundance of AFP2-FLAG was checked by anti-FLAG. Anti-tubulin was used as the loading control. B, C) Cell-free analysis of the degradation of GST-AFP2 in planta. The purified GST-AFP2 was incubated the total protein extract from the seedling under 28°C for 3 days, and the degradation speed of GST-AFP2 could be detected by anti-GST antibody (upper panel). The anti-tubulin antibody was used as the loading control (lower panel). As for the control, the protein extract for the seedling under normal condition was incubated with GST-AFP2 for 6 h, 12 h or 24 h, and the degradation of GST-AFP2 was detected (B). For inhibitory experiment, the seedling was pretreated with the proteasome inhibitor MG132 at 10 M for 12 h following HT treatment for 3 days of HT at 28°C for cell-free analysis (C).
Previous study demonstrates that PIF7 binds the promoter of CBFs, the key gene family to control plant cold acclimation. To coordinate plant freezing tolerance, different photoperiod length, for example long-day (LD) or short-day (SD) condition affect the expression and protein stability of PIF7; subsequently affect its binding to CBF for freezing tolerance [25]. During this process, PIF4 cooperates with PIF7 to control CBF expressing and freezing tolerance [1,25]. In this study, we first investigated the role of PIF7 during thermomorphogenesis, and found overexpressing PIF7 presented longer hypocotyl, whereas the loss-of-function pif7 mutant showed relative shorter hypocotyl compared with wild-type Col under HT stress, which confirm that PIF7 is necessary for hypocotyl elongation during thermomorphogenesis. RT-qPCR analysis revealed that PIF7 mediated HT-dependent YUC8 and IAA19 expressing, since inactivating PIF7 compromised HT-induced YUC8 and IAA19 expressing, and overexpressing PIF7 continuously activated YUC8 and IAA19 expressing, which can explain the longer hypocotyl of PIF7-Flash before or after HT treatment [13]. In agreement with our result, PIF7 is important for early response to elevated temperature for Arabidopsis seedling. Like cold stress, high temperature also induced the rapid accumulation of PIF7 protein to activate auxin biosynthesis for long hypocotyl elongation, however high temperature represses the transcriptional level of PIF7, suggesting the different mechanism for HT to PIF7 transcription or protein expression. Meanwhile, both of pif4 or pif7 mutant show insensitive to cold stress or HT, suggesting the PIF4 and PIF7 forms heterodimers to regulate plant in response to ambient temperature stress.
Several evidences propose ABA affects thermomorphogenesis process, including auxin biosynthesis or auxin-responsive gene expression [14,19]. AFP2 as the ABI5 binding protein negatively regulates ABA signal [19]. Our previous study also showed that AFP2 enhance seed germination tolerance to HT through weakening ABI5 activity [26]. In this study we identified AFP2 as the interaction of PIF7, such interaction was validated by a series of experiment including yeast two-hybrid experiment, Co-IP experiment, colocalization experiment also revealed both of AFP2 and PIF7 localized in the nucleus (Fig. 3D). Meanwhile, the afp2 mutant displayed longer hypocotyl, while overexpressing AFP2 showed shorter hypocotyl after HT stress, suggesting that AFP2 acts as the negative regulator to control thermomorphogenesis. Furthermore, genetic analysis using different crossed line showed that overexpressing AFP2 in the PIF7-Flash attenuated the longer hypocotyl phenotype in contrast to PIF7-Flash line. Coordinately, the transcriptional level of YUC8 and IAA19 was also lower in the crossed AFP2-FLAG/PIF7-Flash line than that in PIF7-Flash line alone. These data suggest that AFP2 represses the trans-activating of PIF7 to its target YUC8 and IAA19 for hypocotyl elongation (Fig. 2). As AFP2 protein contains the EAR motif, which can recruit TPL/TPR complex and HDAC complex, and HDAC complex can epigenetically erase the histone acetylation modification in the chromatin of target gene locus, as a result, epigenetically repress these target gene expressions. Supporting this possibility, our previous study also showed that CONSNTANS recruits AFP2 and HDACs to coordinates the activation of CONSTANS to FT expression [22]. Besides the acetylation modification, PIF7 is reported to recruits MRG1/MRG2 complex, thus brings histone-acetylases to increase histone acetylation level to promote the expression of YUC8 and IAA19 during shade response [27]. Thus, these evidences, including our data, suggest the complicated regulatory mechanism for PIF7 for plant response to environment stress.
In this study we also noticed that HT gradually increased the protein accumulation of AFP2 in the transgenic pAFP2: AFP2-FLAG lines, but the transcriptional level of AFP2 did not obviously change after HT treatment. Cell-free analysis also showed that HT retarded the degradation of AFP2, suggesting the post-transcriptional regulation mechanism for AFP2 expression, possibly suppressing proteasome-dependent ubiquitin system, as the proteasome inhibitor MG132 suppressed the degradation of AFP2 under normal condition. In consistent with the differential profile of AFP2 transcript and its protein expression, HT also differentially regulated the transcriptional level of PIF7 and its protein expression [13]. We speculate that HT induced the gradually accumulation of AFP2 as the brake to slow down the PIF7 activity during the long-term HT treatment, as AFP2 interacts with PIF7 as the AFP2/PIF7 complex, which possible recruits TPL/TPR complex and HDAC to reduce histone acetylation modification level, ultimately reduce its target gene, such as YUC8 and IAA19 expression, and guarantee the appropriate PIF7 activity to avoid excess elongation of hypocotyl under HT condition (Fig. 6).
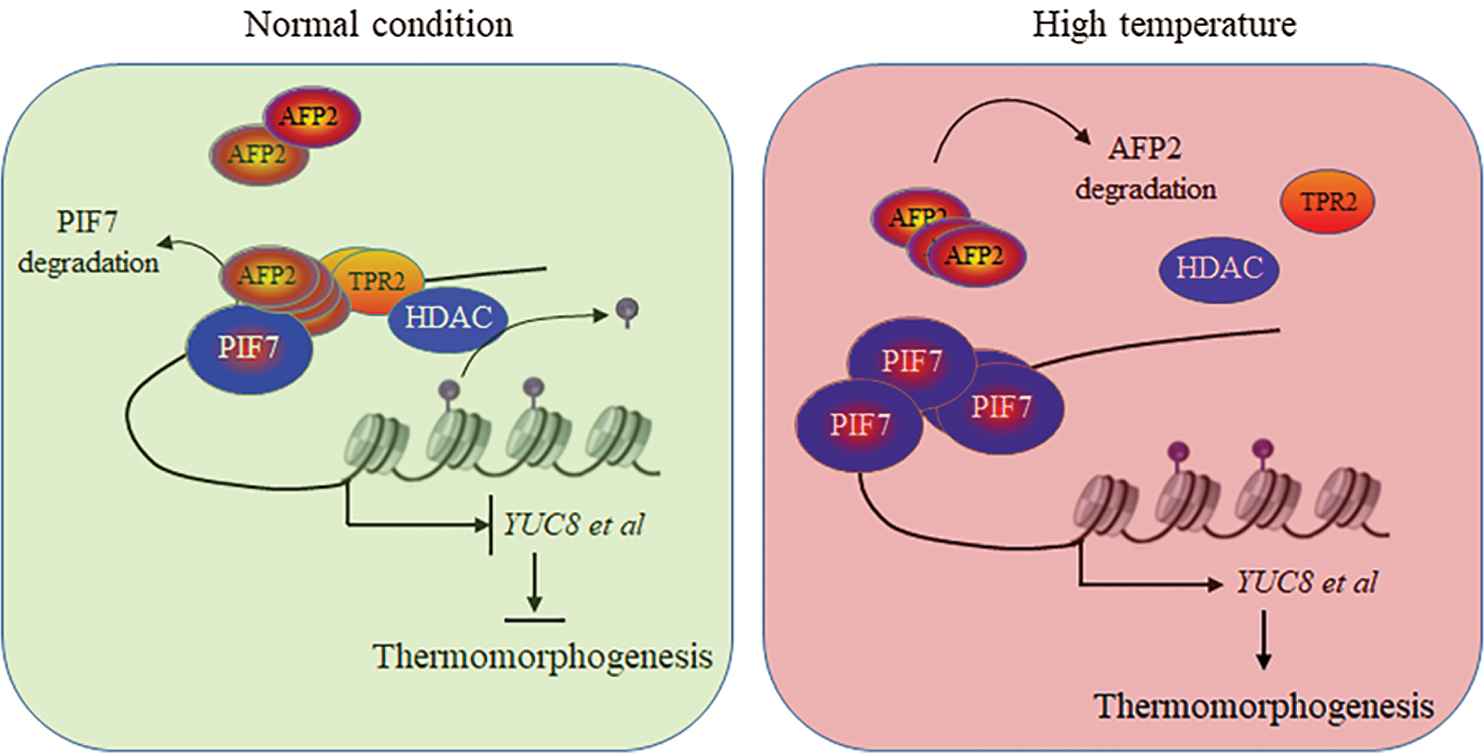
Figure 6: The propose model to illustrate the cooperation mechanism between PIF7 and AFP2 to regulate seedling thermomorphogenesis
After all, in this study we confirmed that PIF7 activated YUC8 and IAA19 expression for longer hypocotyl elongation, or thermomorphogenesis, after HT stress. Meanwhile, we identified AFP2 as the new regulator coordinates PIF7 during thermomorphogenesis. We propose a model to illustrate the possible mechanism by which AFP2 cooperates with PIF7 to modulate thermomorphogenesis. As shown in Fig. 6, HT induced the rapid accumulation of PIF7 to activate YUC8 and IAA19 for longer hypocotyl, meanwhile, long-term HT treatment gradually increased the protein abundance of AFP2, which interacts with PIF7 and recruit HDAC to repress the trans-activation activity of PIF7 on YUC8 and IAA19 to avoid the excessive damage of HT on seedling growth. In a word, AFP2 as the novel regulator interacts with PIF7 as the PIF7/AFP2 complex to sophisticate modulate thermomorphogenesis for the young seedling.
Under normal condition, AFP2 interacts with PIF7, which then recruit the corepressor TPR2 to orchestrate the transactivation activity of PIF7 on auxin-biosynthesis related gene expressions, thus present normal hypocotyl growth. However, HT treatment induces the degradation of AFP2, which could not efficiently recruit TPR2, thus release PIF7 activity to activate the expressions of auxin-biosynthesis genes, ultimately accelerate hypocotyl elongation for thermomorphogenesis.
Funding Statement: This work was funded by the National Natural Science Foundation of China (Grant No. 31970289).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Quint, M., Delker, C., Franklin, K. A., Wigge, P. A., Halliday, K. J. et al. (2016). Molecular and genetic control of plant thermomorphogenesis. Nature Plants, 2, 15190. DOI 10.1038/nplants.2015.190. [Google Scholar] [CrossRef]
2. Park, Y. J., Park, C. M. (2018). External coincidence model for hypocotyl thermomorphogenesis. Plant Signaling & Behavior, 13(4), e1327498. DOI 10.1080/15592324.2017.1327498. [Google Scholar] [CrossRef]
3. Casal, J. J., Balasubramanian, S. (2019). Thermomorphogenesis. Annual Review of Plant Biology, 70, 321–346. DOI 10.1146/annurev-arplant-050718-095919. [Google Scholar] [CrossRef]
4. Lee, C. M., Thomashow, M. F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 109(37), 15054–15059. [Google Scholar]
5. Han, X., Yu, H., Yuan, R., Yang, Y., An, F. et al. (2019). Arabidopsis transcription factor TCP5 controls plant thermomorphogenesis by positively regulating PIF4 activity. iScience, 15, 611–622. DOI 10.1016/j.isci.2019.04.005. [Google Scholar] [CrossRef]
6. Raschke, A., Ibanez, C., Ullrich, K. K., Anwer, M. U., Becker, S. et al. (2015). Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4-dependent auxin response genes. BMC Plant Biology, 15, 197. DOI 10.1186/s12870-015-0566-6. [Google Scholar] [CrossRef]
7. Soy, J., Leivar, P., Monte, E. (2014). PIF1 promotes phytochrome-regulated growth under photoperiodic conditions in arabidopsis together with PIF3, PIF4, and PIF5. Journal of Experimental Botany, 65(11), 2925–2936. DOI 10.1093/jxb/ert465. [Google Scholar] [CrossRef]
8. Leivar, P., Martin, G., Soy, J., Dalton-Roesler, J., Quail, P. H. et al. (2020). Phytochrome-imposed inhibition of PIF7 activity shapes photoperiodic growth in arabidopsis together with PIF1, 3, 4 and 5. Physiologia Plant, 169(3), 452–466. DOI 10.1111/ppl.13123. [Google Scholar] [CrossRef]
9. Job, N., Datta, S. (2020). PIF3/HY5 module regulates BBX11 to suppress protochlorophyllide levels in dark and promote photomorphogenesis in light. New Phytologist. DOI 10.1111/nph.17149. [Google Scholar] [CrossRef]
10. Zhang, X., Huai, J., Shang, F., Xu, G., Tang, W. et al. (2017). A PIF1/PIF3-hY5-bBX23 transcription factor cascade affects photomorphogenesis. Plant Physiology, 174(4), 2487–2500. DOI 10.1104/pp.17.00418. [Google Scholar] [CrossRef]
11. Tavridou, E., Pireyre, M., Ulm, R. (2020). Degradation of the transcription factors PIF4 and PIF5 under UV-b promotes UVR8-mediated inhibition of hypocotyl growth in arabidopsis. Plant Journal, 101(3), 507–517. DOI 10.1111/tpj.14556. [Google Scholar] [CrossRef]
12. Paulisic, S., Qin, W., Veraszto, H. A., Then, C., Alary, B. et al. (2021). Adjustment of the PIF7-hFR1 transcriptional module activity controls plant shade adaptation. EMBO Journal, 40(1), e104273. DOI 10.15252/embj.2019104273. [Google Scholar] [CrossRef]
13. Fiorucci, A. S., Galvao, V. C., Ince, Y. C., Boccaccini, A., Goyal, A. et al. (2020). PHYTOCHROME INTERACTING FACTOR 7 is important for early responses to elevated temperature in arabidopsis seedlings. New Phytologist, 226(1), 50–58. DOI 10.1111/nph.16316. [Google Scholar] [CrossRef]
14. Xu, Y., Zhu, Z. (2020). Abscisic acid suppresses thermomorphogenesis in arabidopsis thaliana. Plant Signaling & Behavior, 15(5), 1746510. DOI 10.1080/15592324.2020.1746510. [Google Scholar] [CrossRef]
15. Lorrai, R., Boccaccini, A., Ruta, V., Possenti, M., Costantino, P. et al. (2018). Abscisic acid inhibits hypocotyl elongation acting on gibberellins, DELLA proteins and auxin. AoB Plants, 10(5). DOI 10.1093/aobpla/ply061. [Google Scholar] [CrossRef]
16. Qi, L., Liu, S., Li, C., Fu, J., Jing, Y. et al. (2020). PHYTOCHROME-INTERACTING FACTORS interact with the ABA receptors PYL8 and PYL9 to orchestrate ABA signaling in darkness. Molecular Plant, 13(3), 414–430. DOI 10.1016/j.molp.2020.02.001. [Google Scholar] [CrossRef]
17. Finkelstein, R. R., Lynch, T. J. (2000). The arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell, 12(4), 599–609. DOI 10.1105/tpc.12.4.599. [Google Scholar] [CrossRef]
18. Yu, F., Wu, Y., Xie, Q. (2015). Precise protein post-translational modifications modulate ABI5 activity. Trends in Plant Science, 20(9), 569–575. DOI 10.1016/j.tplants.2015.05.004. [Google Scholar] [CrossRef]
19. Lopez-Molina, L., Mongrand, S., Kinoshita, N., Chua, N. H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes and Development, 17(3), 410–418. DOI 10.1101/gad.1055803. [Google Scholar] [CrossRef]
20. Lynch, T. J., Erickson, B. J., Miller, D. R., Finkelstein, R. R. (2017). ABI5-binding proteins (AFPs) alter transcription of ABA-induced genes via a variety of interactions with chromatin modifiers. Plant Molecular Biology, 93(4–5), 403–418. DOI 10.1007/s11103-016-0569-1. [Google Scholar] [CrossRef]
21. Garcia, M. E., Lynch, T., Peeters, J., Snowden, C., Finkelstein, R. (2008). A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating arabidopsis seeds and seedlings. Plant Molecular Biology, 67(6), 643–658. DOI 10.1007/s11103-008-9344-2. [Google Scholar] [CrossRef]
22. Chang, G., Yang, W., Zhang, Q., Huang, J., Yang, Y. et al. (2019). ABI5-BINDING PROTEIN2 coordinates CONSTANS to delay flowering by recruiting the transcriptional corepressor TPR2. Plant Physiology, 179(2), 477–490. DOI 10.1104/pp.18.00865. [Google Scholar] [CrossRef]
23. Yu, L., Zhou, W., Zhang, D., Yan, J., Luo, L. (2020). Phytosulfokine-α promotes root growth by repressing expression of pectin methylesterase inhibitor (PMEI) genes in Medicago truncatula. Phyton-International Journal of Experimental Botany, 89(4), 873–881. DOI 10.32604/phyton.2020.011882. [Google Scholar] [CrossRef]
24. Zhang, Q., He, D., Ying, S., Lu, S., Wei, J. et al. (2020). GABA enhances thermotolerance of seeds germination by attenuating the ROS damage in arabidopsis. Phyton–International Journal of Experimental Botany, 89(3), 619–631. DOI 10.32604/phyton.2020.010379. [Google Scholar] [CrossRef]
25. Lee, S., Paik, I., Huq, E. (2020). SPAs promote thermomorphogenesis by regulating the phyB-pIF4 module in arabidopsis. Development, 147(19). DOI 10.1242/dev.189233. [Google Scholar] [CrossRef]
26. Chang, G., Wang, C., Kong, X., Chen, Q., Yang, Y. et al. (2018). AFP2 as the novel regulator breaks high-temperature-induced seeds secondary dormancy through ABI5 and SOM in arabidopsis thaliana. Biochemical and Biophysical Research Communications, 501(1), 232–238. DOI 10.1016/j.bbrc.2018.04.222. [Google Scholar] [CrossRef]
27. Peng, M., Li, Z., Zhou, N., Ma, M., Jiang, Y. et al. (2018). Linking PHYTOCHROME-INTERACTING FACTOR to histone modification in plant shade avoidance. Plant Physiology, 176(2), 1341–1351. DOI 10.1104/17.01189. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |