

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.022421
ARTICLE
Transcriptome Analysis via RNA Sequencing Reveals the Molecular Mechanisms Underlying the Hedera helix Response to High Temperature
1Shanghai Botanical Garden, Shanghai, 200231, China
2Shanghai Engineering Research Center of Sustainable Plant Innovation, Shanghai, 200231, China
3Shanghai Key Laboratory of Bio-Energy Crops, School of Life Sciences, Shanghai University, Shanghai, 200444, China
*Corresponding Authors: Ping Li. Email: liping80@shu.edu.cn; Jiali Wei. Email: jlwei3588@shu.edu.cn
Received: 09 March 2022; Accepted: 23 April 2022
Abstract: Hedera helix is an evergreen ornamental plant that is resistant to cool but not high temperature and deserves to be further researched for improving its adaptability to heat stress. Two Hedera helix cultivars, heat-tolerant (HT) ‘Jessica’ and heat-sensitive (HS) ‘Shamrock’, were used for differences analyses of transcriptome. We detected 6179 differentially expressed genes (DEGs) and 5992 DEGs in ‘Jessica’ and ‘Shamrock’ to heat stress, respectively. Among these, 1983 upregulated DEGs and 1400 downregulated DEGs were shared between both varieties, resulting in enhancement of various pathways such as biosynthesis of secondary metabolites, glyoxylate dicarboxylate metabolism, and protein processing in endoplasmic reticulum (PPER), RNA transport, respectively. Among the common downregulated DEGs, 72 TFs in 25 gene families were found, including members of the MYB and MYB-related families, the bHLH family, etc. In ‘Jessica’ (HT), 634 unique up-regulated DEGs were identified, including genes associated with phenylpropanoid biosynthesis, starch and sucrose metabolism, biosynthesis of amino acids. Most upregulated TFs of HT were upregulated much more rapidly than those of HS in response to high temperature. Eleven TF-encoding genes were selected to verify the RNA sequencing data by qPCR. This study revealed the gene expression patterns of ivy in response to heat stress and the molecular basis of heat tolerance, which provided theoretical references for improving the heat tolerance of ivy.
Keywords: Hedera helix; RNA-sequencing; heat stress; heat tolerance; transcriptome
Sessile plants inevitably face various environmental pressures, such as drought, salt stress, cold stress, and high temperature. Throughout the long-term adaptation process, plants have gradually evolved to cope with abiotic stress caused by environmental changes [1,2]. Plants respond to abiotic stresses through complex molecular regulatory networks that involve a variety of signal transduction pathways and activate the expression of a large number of stress-related genes. At present, global warming is a trend, and plants are experiencing increasingly severe high-temperature stress. High-temperature stress has a severe impact on the normal growth of plants. Although plants have evolved a set of molecular pathways to respond to high-temperature stress, there are still some defects in several species. Therefore, studying the response of plants to high-temperature stress and determining the regulatory factors involved in the high-temperature response can help cultivar improvement.
Heat shock proteins (HSPs) are conserved chaperone family proteins that respond to high-temperature stress and participate in cell homeostasis and protein folding, such as by preventing protein misfolding, denaturation and aggregation caused by heat stress [3,4]. HSPs are divided into several families according to their molecular weight, including small HSPs, Hsp40 s, Hsp70 s, Hsp90 s, Hsp100 s and Hsp110 [5–7]. Heat shock transcription factors (HSFs) are involved in the transcriptional regulation of HSPs. Heat stress (HS) activates HSFs and induces the expression of HSPs [8,9]. The HSF protein family can be divided into three classes, A, B and C. There are 52 genes encoding HSFs in soybean, 25 in rice and 21 in Arabidopsis [10–13]. In Arabidopsis, HSFA1 is constitutively expressed and responsible for the initiation of the heat stress response [14]. In tomato, HSFA1 is considered a master regulator involved in signal perception [15]; HSFA2 is a protein whose structure and function are similar to those of HSFA1, but it is expressed only under stress [16]. HSFA2 forms a heterooligomer with HSFA1 under stress and regulates the expression of HSPs and other downstream genes [17,18]. At the same time, DREB2, a dehydration responsive element binding protein, is involved in the transcriptional regulation of HSFA1 under stress [19]. DREB2B can activate the expression of HSFA3 by binding to the promoter of HSFA3 in the early stage of the heat stress response (HSR) [20], while in the later stage of the HSR, the expression of HSFA3 depends on DREB2C [21].
In addition to HSFs, the members of many transcription factor families are also involved in the regulation of abiotic stress, such as ERFs and WRKYs [22–24]. These transcription factors, through a variety of biological and hormone metabolic pathways, form a complex gene regulatory network and participate in the regulation of the response to abiotic stress, such as heat stress [25]. In Arabidopsis and wheat, HSFA1B regulates plant heat tolerance through the jasmonic acid (JA) pathway-related protein OPR3, and DREB2A expression is also regulated by OPR [26]. ERF family members are downstream targets of EIN3, a key component in the ethylene signaling pathway; these members regulated a variety of stress responses, including response to salt, drought, cold and heat stresses [23]. Further, ERFs are regulated by ABA and JA [23,27]. In Arabidopsis, WRKY39 enhances plant tolerance to high temperature, and WRKY39 is also regulated by the SA and JA hormone pathways [28]. In addition, some biological processes in plant cells are related to the HSR, such as endoplasmic reticulum protein processing and ribosome biosynthesis [29–33].
Ivy is an evergreen plant species widely used in three-dimensional urban landscapes, ground cover greening, and indoor greening. There are more than 600 cultivars of ivy recorded worldwide, most are distributed in Europe, and a small number of cultivars are found in Asia [34]. English ivy (Hedera helix L.), the largest genus of ivy, tolerates cool [35], but not high temperature. In Shanghai, the 24 h mean temperature in the hottest period (July and August) is 27.8°C, and the hot weather with a daily high temperature greater than 35°C lasts about 6–8 days per year. High temperature is a limiting factor affecting the broad application of English ivy cultivars in Shanghai. Thus, it is essential to investigate the molecular mechanisms of the Hedera helix’s response to high temperature for improving its adaptability to heat stress.
Although the adaptation mechanism of ivy to high temperatures is vital to its growth and application in a country with sweltering summers, this topic still lacks research. The relative works about the ivy are carried from other sides. For example, the influence of NaCl on the growth of English ivy (Hedera helix) cuttings and callus tissue is studied in [36]. The study in Varanasi, India, evaluates the air pollution tolerance index of English ivy [37]. The work in [38] document the temporal patterns of photosynthesis vs. freezing resistance during spring in adult and juvenile leaf phenotypes of Hedera helix in Switzerland. In recent years, the development of high-throughput sequencing technology has provided convenience for gene regulation research of many species whose genome has not been completely sequenced. In this paper, we investigated the mechanism of the high-temperature tolerance of ivy by RNA sequencing. We found that members of many transcription factor families, such as HSFs, ERFs, and WRKYs, are involved in the heat stress response of ivy, and their expression profiles differed between the tolerant variety and the sensitive variety.
More than thirty ivy varieties are planted in the Shanghai Botanical Garden in July 2018 (the daily maximum temperature was approximately 35°C under 60% shade for 28 days under naturally high temperatures). Based on the phenotype changes and physiological experiments of those ivy cultivars under the natural high temperature, we select two ivy varieties, Hedera helix ‘Jessica’ and Hedera helix ‘Shamrock’, as samples and materials in this paper research. These plants were two-year-old plants propagated from cuttings generated from branches. Potted seedlings were selected and cultured in an artificial growth incubator (20°C, 16 h of light/8 h of darkness) for two weeks. Afterward, they were subjected to 40°C for 7 days. The conditions of the incubator included a relative humidity of 70% and a light intensity of 8000 lx. During high-temperature treatment, to avoid drought stress, supplemental water was added to maintain moisture.
2.2 Measurement of Chlorophyll Fluorescence
Chlorophyll fluorescence was measured by a laboratory Fluorocam instrument (PSI, Czech Republic), the specific operation was performed according to the user manual [39], and the maximum photochemical efficiency Fv/Fm of PSII was calculated as (Fm-F0)/Fm.
2.3 Library Construction and Sequencing
After total RNA was extracted, mRNA was enriched by oligo (dT) beads. Afterward, the enriched mRNA was fragmented into short fragments using fragmentation buffer and subsequently reverse transcribed into cDNA with random primers. Afterward, second-strand cDNA was synthesized. The cDNA fragments were then ligated to Illumina sequencing adapters after purification with a QiaQuick PCR extraction kit. Sequencing was subsequently conducted on the prepared cDNA library using an Illumina HiSeq 4000 device by Gene Denovo Biotechnology Co. (Guangzhou, China).
The raw data were filtered using in-house Perl scripts. The clean reads were obtained by removing reads containing adapters, more than 10% unknown nucleotides, and more than 40% low-quality (Q-value ≤ 20). Differentially expressed genes were identified and annotated. The gene abundances were calculated and normalized to RPKM (reads per kilobase per million reads) values [40]. Afterward, the calculated gene expression values were used to compare the difference in gene expression among samples. The edgeR package (http://www.r-project.org/) was used to identify differentially expressed genes across samples or groups. Genes with a fold change ≥2 and a false discovery rate (FDR) were identified. The unigenes were aligned to the GO database (http://www.geneontology.org/) via BLASTX.
2.5 KEGG Pathway Enrichment Analysis
Pathway enrichment analysis revealed significantly enriched metabolic pathways or signal transduction pathways of the DEGs compared with those of the whole-genome background. The calculated P-value was subjected to FDR correction, taking FDR ≤ 0.05 as a threshold. Pathways meeting this condition were regarded as significantly enriched pathways of DEGs [41].
The growth and treatment conditions of the plant materials used for qPCR were the same as those used for RNA sequencing. Total RNA was extracted from the leaves using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. cDNA was synthesized by reverse transcription using a PrimeScript™ First Strand cDNA Synthesis Kit (Vazyme, China). QPCR was performed using SYBR Green reagent (Vazyme, China) and an ABI Step One Plus Real-Time PCR Detection System (ABI, United States). Gene expression relative to that of UBI was calculated by the 2-ΔΔCt method. The primers used for qPCR are listed in the supplementary materials [42] (Supplemental data 1).
SOD enzyme activity was measured and total protein was extracted according to [43,44]. 50% of the inhibition of nitroblue tetrazole (NBT) photochemical reduction was considered as one unit of enzyme activity (U). The total SOD activity was expressed as u.g−1 (FW).
3.1 Analysis of Physiological Responses
To study the physiological response of plants under heat stress, two ivy varieties, ‘Jessica’ and ‘Shamrock’, were grown in an incubator at 40°C, and then the phenotypes of plants were observed after heat treatment for 1, 3, 5 and 7 days. There was no visible change in both phenotypes of two varieties on the first day compared with the control plants growing at 40°C (The data is not displayed). The withered leaves began to appear in both ivy varieties after heat treatment for 3 days (Fig. 1A), and more withered leaves occurred for ‘Shamrock’ than in ‘Jessica’. After treatment for 5 days and 7 days, a severe heat-stress phenotype was observed for ‘Shamrock’, while most of the leaves of ‘Jessica’ were still green. This result indicated that, compared with ‘Shamrock’, ‘Jessica’ exhibited higher tolerance to heat stress. Further, we analyzed the photosynthetic capacity of plant leaves (nondried leaves) after heat treatment for 1, 3, 5, and 7 days separately via a chlorophyll fluorescence meter. As shown in Fig. 1B, the maximum quantum yield of photosystem II (Fv/Fm) decreased significantly after treatment, and the decrease in ‘Shamrock’ was more severe than that in Jessica. After treatment for one day, the Fv/Fm value decreased from the control value of 0.8 to approximately 0.7 in ‘Jessica’ and approximately 0.6 in ‘Shamrock’. Chlorophyll fluorescence was hardly detected in most parts of the leaves of ‘Shamrock’ subjected to heat for 5 days, while the Fv/Fm value remained at approximately 0.4 for ‘Jessica’, which further confirmed that ‘Jessica’ is more resistant to heat stress than ‘Shamrock’ (Fig. 1B).
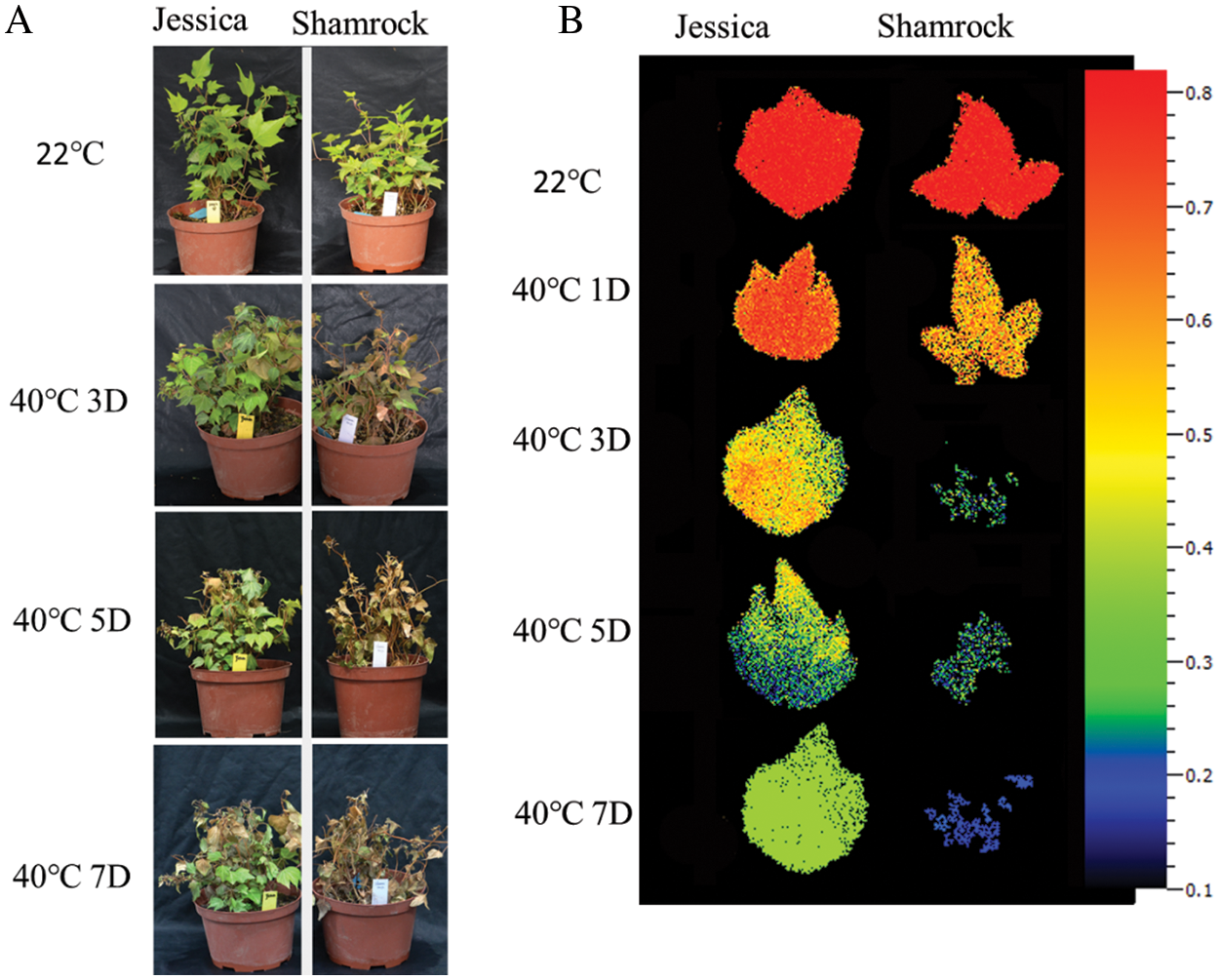
Figure 1: Phenotype of ivy after heat treatment. A. Typical phenotype of ‘Jessica’ and ‘Shamrock’ after 40°C treatment for 3 days, 5 days and 7 days, D: days. B. The maximum quantum yield of photosystem II (Fv/Fm) between ‘Jessica’ and ‘Shamrock’
3.2 Analysis of the Transcriptome
Based on the results above, ‘Jessica’ and ‘Shamrock’ were considered heat-resistant and heat-sensitive varieties, respectively. Further, they were applied to research the heat response mechanism in ivy plants via RNA sequencing. After heat treatment for 6 h and 24 h, six RNA-seq libraries, including those comprising DNA from both treated plants and control plants, were constructed. As shown in the associated table, 3.6 × 107 raw reads were generated, and low-quality reads with Q30 percentages of 0.06%–0.09% were removed. The Q30 percentages of the six samples were 95.6%, 95.4%, 95.4%, 95.2%, 94.9% and 95.4%. Afterward, the high-quality clean reads were mapped to the reference genome, and the mapping percentages were 80.5%, 79.8%, 74.0%, 81.4%, 85.2%, and 78.5%. There were 20,264 genes with 90–100% gene coverage, accounting for 79.0% of the total number of genes. In addition, the number of expressed genes in all the samples was 20, 085, accounting for 67.1% of the reference gene number.
A total of 6178 differentially expressed genes (DEGs) in ‘Jessica’ and 5991 DEGs in ‘Shamrock’ were identified according to their RPKM value (log2 (RPKM ratio GR/CK) ≥ 1) from the heat-treated samples compared with the untreated samples. The temporal characteristics of DEGs were clustered based on their temporal expression patterns by Short Time series Expression Miner software (STEM). STEM analysis revealed eight cluster profiles in both ivy varieties, of which the expression of the DEGs in profile 0, profile 1 and profile 3 was downregulated after the three-day treatment and that in profile 4, profile 6 and profile 7 was upregulated after the three-day treatment (Fig. 2A). DEGs in profile 0, profile 1 and profile 3 were defined as downregulated DEGs, and those in profiles 4, profile 6 and profile 7 were defined as upregulated DEGs (Supplemental data 2). DEGs in the two ivy varieties were submitted to the website http://bioinfogp.cnb.csic.es/tools/venny/index.html for Venn analysis (Fig. 2B). A total of 1983 common upregulated DEGs and 1400 common downregulated DEGs were identified in the two ivy varieties, while 634 upregulated DEGs and 703 downregulated DEGs were identified exclusively in ‘Jessica’ (Supplemental data 3), indicating that plants also employ specific molecular strategies in heat-resistant plants in addition to the molecular pathways common to both heat-resistant plants and heat-sensitive plants.
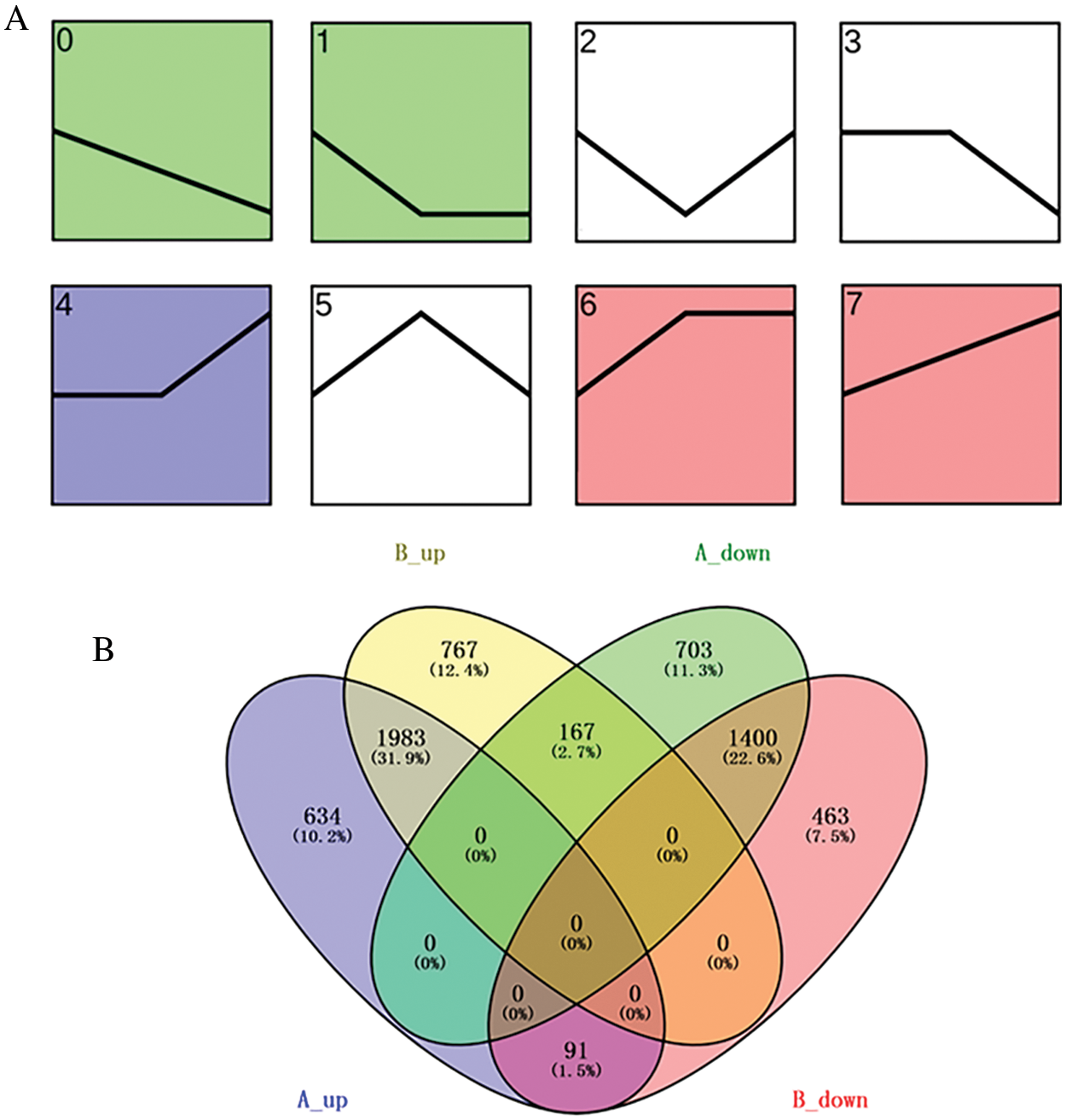
Figure 2: A. Expression profiles of DEGs by trend analysis in ivy. The curse represents the relative expression of DEGs in control sample, and samples heat treated with 6 h and 24 h. B. Venn diagram of DEGs in two ivy varieties, ‘Jessica’ and ‘Shamrock’. A_up and A_down, up and down-regulated DEGs in ‘Jessica’; B_up and B_down, up and down-regulated DEGs in ‘Shamrock’
3.4 KEGG Enrichment Analysis of DEGS
To investigate the common pathways and specific pathways involved in ivy after heat stress, we performed KEGG enrichment analysis with the DEGs common to both varieties and the DEGs only in the heat-tolerant variety (‘Jessica’). Among the most common downregulated DEGs, photosynthesis and other biosynthesis and metabolism-related pathways, such as “biosynthesis of secondary metabolites,” “glyoxylate and dicarboxylate metabolism,” and “fatty acid metabolism,” were enriched, suggesting that the plants needed to decrease their energy consumption in response to high temperature (Fig. 3A, Supplemental data 4a). Among the common upregulated DEGs, six pathways were significantly enriched: “protein processing in endoplasmic reticulum (PPER),” “RNA transport”, “ribosome,” “spliceosome”, “RNA degradation”, and “ribosome biogenesis in eukaryotes” (Fig. 3B, Supplemental data 4b). Fig. 4 shows that both the expression of most upregulated enriched genes in the KEGG pathway of PPER and the expression of the genes involved in protein folding and degradation increased, indicating that the synthesis of the folding proteins increased when degradation of the misfolded proteins was enhanced. Four pathways, “phenylpropanoid biosynthesis”, “starch and sucrose metabolism”, “biosynthesis of amino acids”, and “amino sugar and nucleotide sugar metabolism”, and one pathway, “ribosome”, were specifically enriched in ‘Jessica’ _down and ‘Jessica’ _up individuals (Figs. 3C and 3D, Supplemental data 4b). Interestingly, both “biosynthesis of secondary metabolites” and “ribosome” were enriched not only in the common up-and downregulated DEGs but also in the up-and downregulated DEGs exclusively in ‘Jessica’ (Fig. 3, Supplemental data 4c, 4d), inferring that these pathways are important in regulating plant responses to high temperature and that the expression of the genes related to these pathways was up-and downregulated more drastically in ‘Jessica’. Top 10 significantly enriched pathways of the downregulated DEGs common to two ivy varieties.
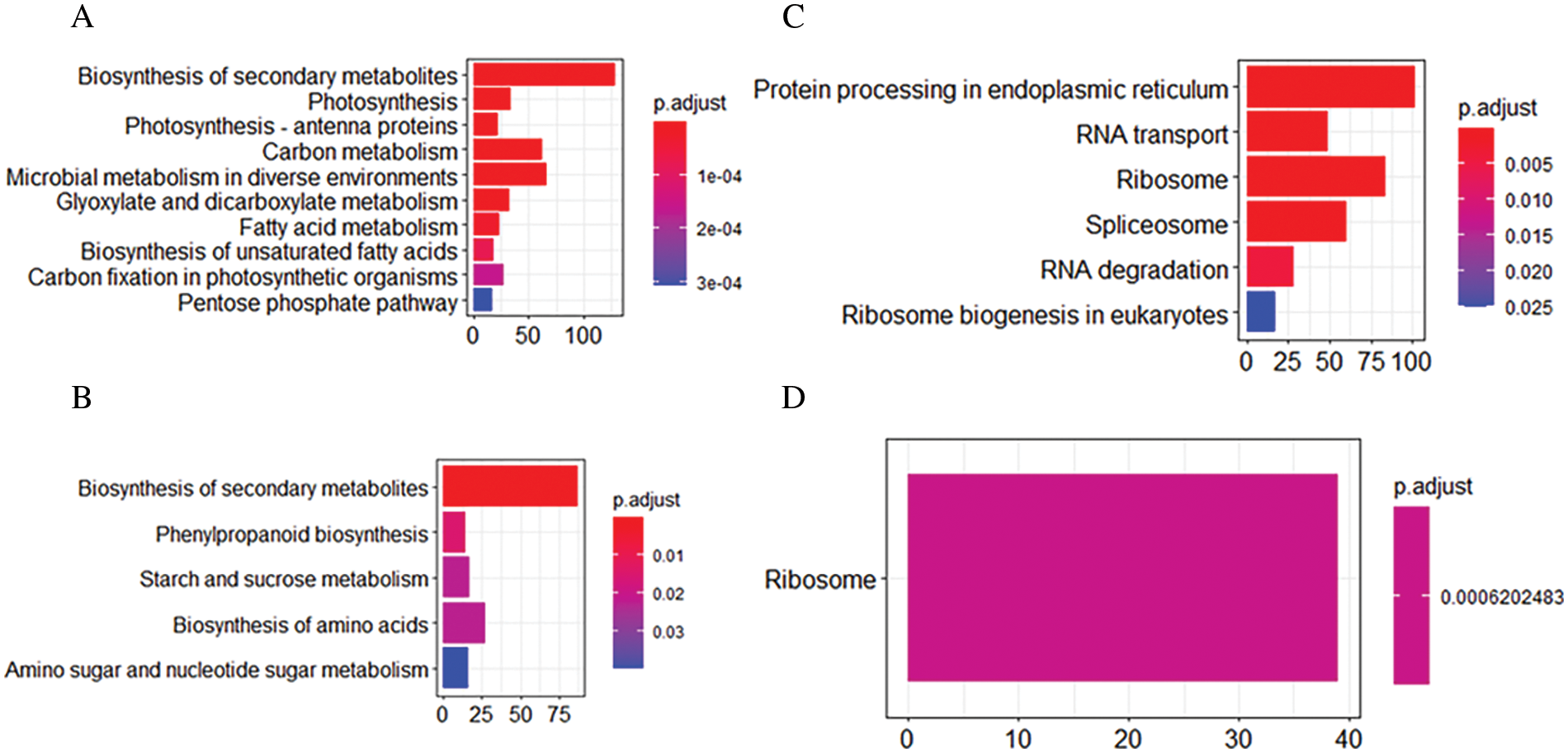
Figure 3: KEGG enrichment analysis in two ivy varieties. A. Top10 of the significant enriched pathway of the common down-regulated DEGs in two ivy varieties. B. The significant enriched pathway in the common up-regulated DEGs in two ivy varieties; C. the significant enriched pathways in the up-regulated DEGs only found in ‘Jessica’; D. the significant enriched pathways in the down-regulated DEGs only found in ‘Jessica’. P. adjust value referred to false discovering rate. Pathways with P. adjust value ≤ 0.05 were significantly enriched in DEGs
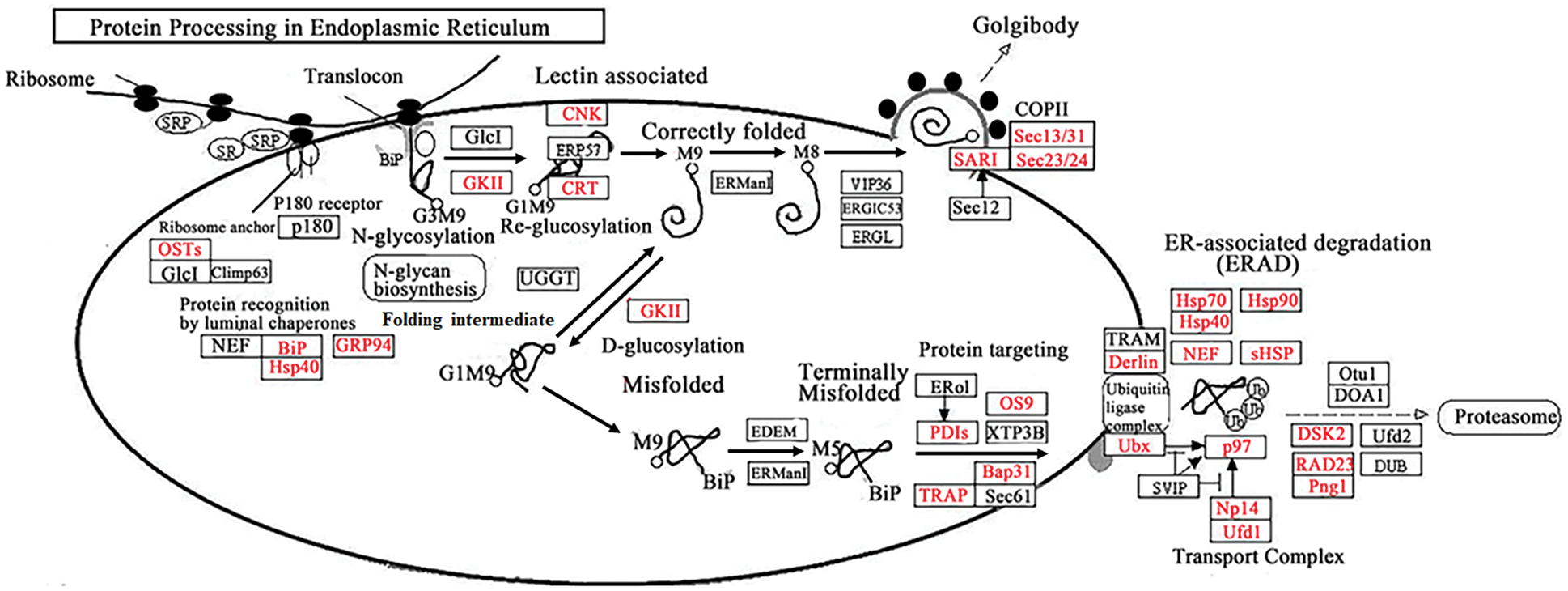
Figure 4: The up-regulated DEGs enriched in protein procession in Endoplasmic Reticulum pathway after heat treatment. The Proteins in red front were encoding by the up-regulated DEGs
3.5 Transcription Factors of DEGS
To further investigate the mechanism of ‘Jessica’ in response to high temperature, we analyzed the transcription factors of DEGs. The full-length cDNAs of plants were obtained by third-generation RNA sequencing technology, and then the sequences of the predicted proteins were compared to the sequences within the Plant TF Database (http://planttfdb.cbi.pku.edu.cn). By integrating the second-generation and third-generation RNA sequencing data, we found 1238 TFs in 53 gene families among the expression data of genes of heat-treated plants (Supplemental data 5a). Among the common downregulated DEGs of the two ivy varieties, 72 TFs in 25 gene families were found, including members of the MYB and MYB-related families, HSF families, the bHLH family, the HD-Zip family, the ERF family, etc (Supplemental data 5b). For 59 transcription factors encoded by the most upregulated DEGs in both varieties, those enriched not only included MYB-and MYB-related family, HSF families, HD-Zip family, and ERF family members but also included WRKY family members, NAC family members, C2H2 family members and so on (Fig. 4, Supplemental data 5c). Although these TFs were also downregulated or upregulated in the two varieties, their expression patterns were significantly different. As shown in Fig. 5, in ‘Jessica’, the expression of most of the genes was greatly reduced after 6 h of heat treatment, while the expression of the genes decreased more moderately in ‘Shamrock’, and the overall gene expression level decreased in ‘Shamrock’ less than that in ‘Jessica’ after 24 h of heat treatment. Similarly, most of the upregulated TFs in ‘Jessica’ were upregulated much more rapidly than those in ‘Shamrock’ were in response to high temperature.
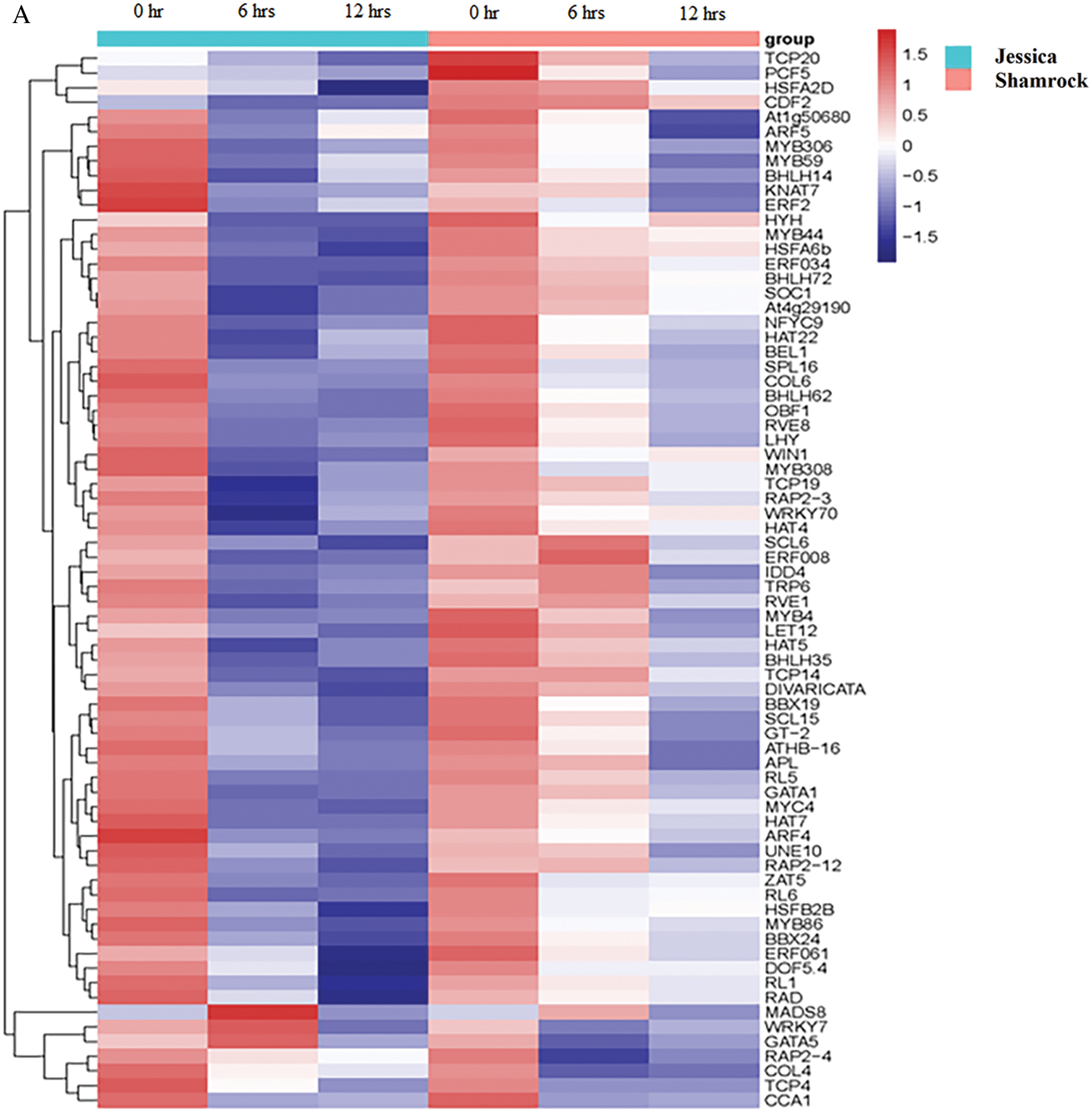

Figure 5: The transcriptional factors in up and down-regulated DEGs of the treatment plants. A, Down-regulated DEGs; B, Up-regulated DEGs
Further, qPCR was performed to verify the RNA sequencing data for 11 TF-encoding genes, including 9 downregulated genes and 2 upregulated genes, in ‘Jessica’ and ‘Shamrock’ (Fig. 6). The qPCR results were consistent with the RNA sequencing data.
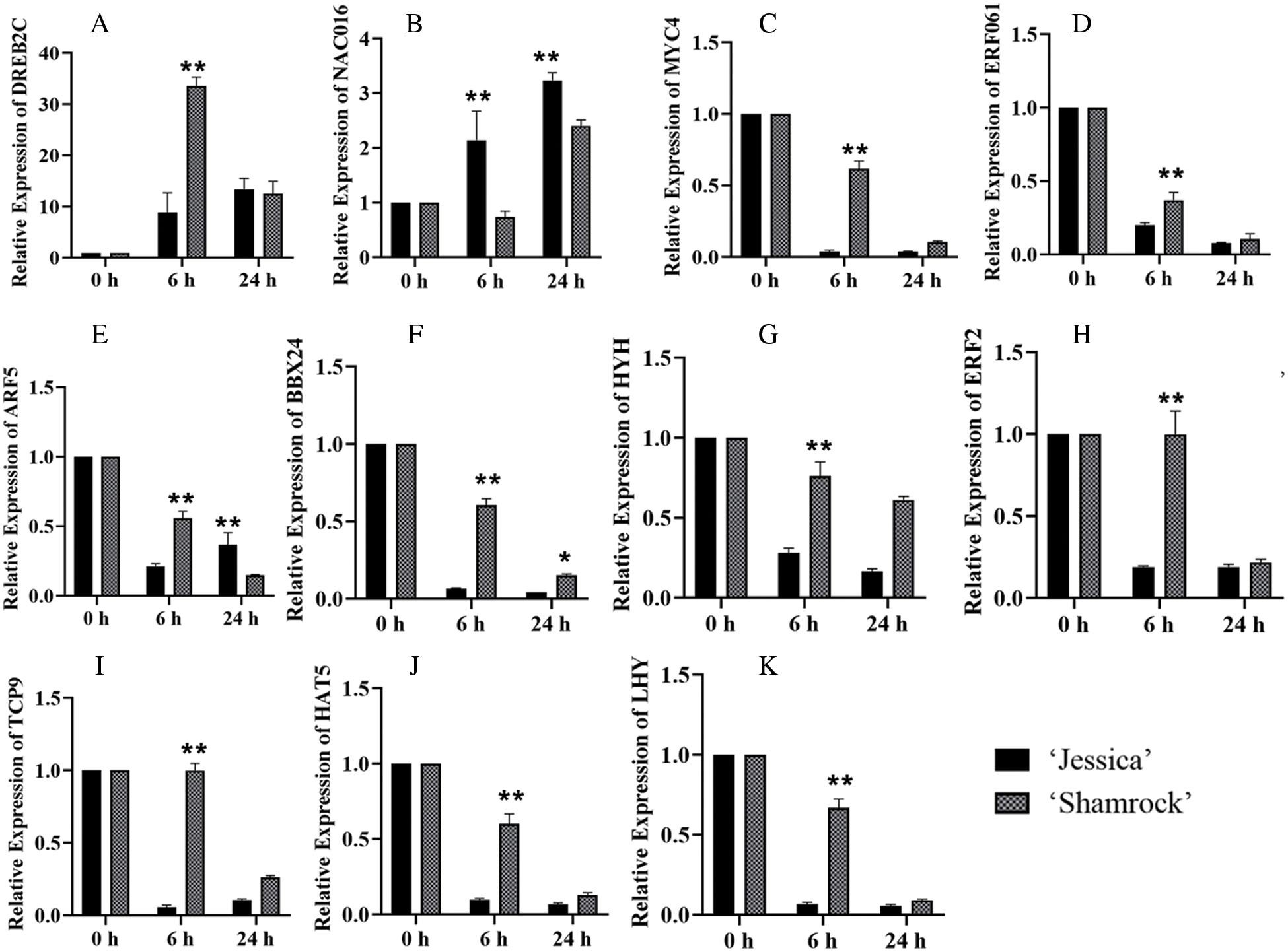
Figure 6: The analysis of relative expressions of 11 TFs in ‘Jessica’ and ‘Shamrock’ under 40°C at 0 h, 6 h and 24 h. The 11 TFs are as follows: A, DREB2C; B, NAC016; C, MYC4; D, ERF061; E, ARF5; F, BBX24; G, HYH; H, ERF2; I, TCP19; J, HAT5; K, LHY. The values are shown as means ± SD of triplicate experiments. Asterisks indicate significant difference between ‘Jessica’ and ‘Shamrock’ under 40°C at the same time, as determined by Student’s t test (*P < 0.05; **P < 0.01)
Further, through analysis of the differential expression of transcription factors between these two ivy strains, it was found that several families of transcription factors, such as HSFs, NACs, WRKYs, and so on, were regulated by the signal molecule H2O2 [45–47]. To investigate whether the expression of these differential genes is caused by the change of the H2O2 content in ivy, we examined the activity of SOD, a major antioxidant enzyme, which can clear active oxygen in plants by converting O2- into H2O2 and O2 [48]. As shown in Fig. 7, the heat tolerant Ivy shows higher SOD enzyme activity compared with sensitive varieties, suggesting that tolerant varieties have a stronger scavenging ability on superoxide anion.
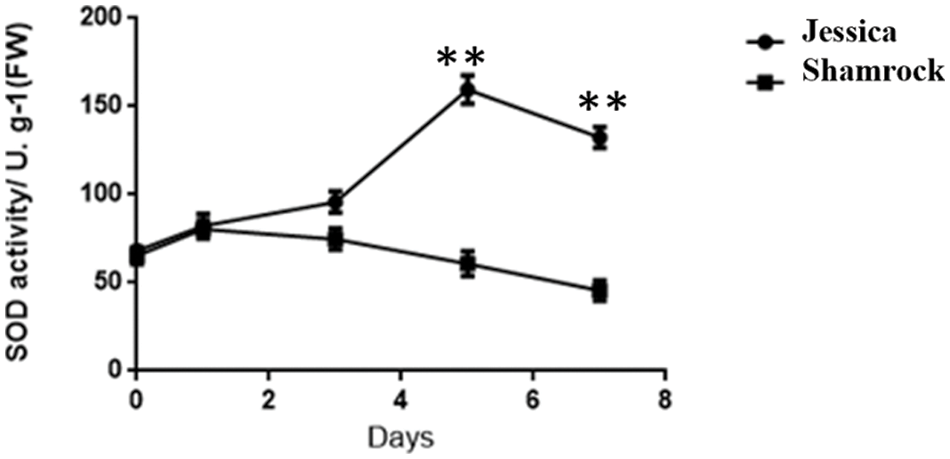
Figure 7: The activity of SOD in ‘Jessica’ and ‘Shamrock’ under 40°C treatment at different days. The values are shown as means ± SD of triplicate experiments. Asterisks indicate significant difference between ‘Jessica’ and ‘Shamrock’ under 40°C at different time, as determined by Student’s t test (**P < 0.01)
In this study, to reveal the mechanism underlying the high-temperature regulatory response of ivy, after high-temperature treatment, heat-resistant and heat-sensitive varieties were subjected to RNA sequencing, and DEGs between the heat-resistant and heat-sensitive varieties were compared and analyzed.
After high-temperature treatment, the two varieties exhibited different degrees of wilting. Compared with that of the sensitive variety, the wilting of the tolerant variety occurred more gradually. Some leaves of the tolerant plants still had photosynthetic capacity after seven days of treatment, while all the leaves of the sensitive varieties wilted and lost their photosynthetic capacity after the third day of treatment. By analyzing differentially expressed genes obtained by sequencing, we found that most of the upregulated and downregulated genes of the two varieties overlapped. The downregulated genes are mainly related to photosynthesis and metabolism, and the upregulated genes are mainly related to the endoplasmic reticulum (ER) protein process pathway and RNA transcription and translation pathway according to KEGG enrichment analysis. The ER is a major organelle in plant cells that is involved in regulating the cell stress response. Stress will cause protein misfolding, which leads to ER stress. Plants adapt to the environment through the ER stress response, such as the unfolded protein response [30,49]. BIP protein is an important regulatory protein of unfolded protein responses in the ER [30]. It has been reported that BIP3 is required when heat activates IRE1 splicing of bZIP60 mRNA [50]. According to our data, BIP protein levels increased significantly (Figs. 5 and 6). In addition, multiple ER protein process-related genes were upregulated. It is suggested that ER stress is also the main pathway through which ivy adapts to heat stress. Furthermore, to reveal the differentially regulated mechanism by analysis of DEGs between tolerant and sensitive varieties, we also performed an enrichment analysis of DEGs whose expression was upregulated and downregulated only in the tolerant individuals. According to the results, the downregulated genes in the tolerant variety were mainly metabolism-and synthesis-related genes, such as those involved in amino acid synthesis and carbohydrate metabolism. Previous studies have also shown that nitrogen metabolism and sugar metabolism are closely related to heat stress, for example on soybean [51]. These changes in the expression of the genes in metabolic pathways may be due to the heat resistance of the tolerant variety. It was found that only “ribosome” was enriched significantly in the upregulated DEGs in the tolerant variety according to the KEGG pathway analysis, and the genes related to this pathway were also significantly enriched among the common downregulated genes.
Our data revealed a large amount of data concerning the expression of transcription factors in high temperature-treated plants through analysis of the sequencing data across three generations. By comparing the second-generation RNA sequencing results with those of the third-generation RNA sequencing, we found differentially expressed transcription factor-encoding genes induced by high temperature. When comparing the expression changes of transcription factors in tolerant plants and sensitive plants at different treatment times, we found that most of the transcription factors in the tolerant plants were upregulated and downregulated faster than those in the heat-sensitive plants, indicating that the gene response of the tolerant plants was more sensitive than that of the sensitive plants. This may be the reason why the tolerant plants are more tolerant to high temperature than the sensitive plants are. However, that why tolerant plants respond more quickly to high temperature than sensitive plants remains to be further studied. Among the upregulated and downregulated transcription factors in the two varieties, the expression of some important heat-responsive gene family members was significantly upregulated (Fig. 5). These included HSFB2A, HSF24 and HSF30 of the HSF gene family; ERF3, ERF5, ERF008 and ERF114 of the ERF family; WRKY1, WRKY6, and WRKY20 of the WRKY family; and NAC002, NAC016, NAC078, and NAC062 of the NAC family. In addition, DREB2C, a transcription factor, that activates the transcription of HSFA3 by binding to its promoter [21], was also upregulated. Both ERFs and EIN3, an important transcription factor in the ethylene signaling pathway [22–23,52], were upregulated, indicating that the ethylene signaling pathway in ivy is involved in the regulation of plants in response to high temperature. Further, the higher enzyme of SOD was found in the tolerant plants Jessica, meaning more H2O2 was accumulated in tolerant plants, and the expression of TFs, such as HSFs, NACs, WRKYs, was induced by H2O2 [43,46,47], and these may explain why the expression of the TFs was more quickly in Jessica.
Overall, we identified differentially expressed genes in the high-temperature response of ivy via RNA sequencing analysis. By comparing the DEGs of the heat-resistant variety and heat-sensitive variety, we revealed the genes and pathways involved in ivy in response to high temperature. This study will provide a convenient way for us to understand the heat stress mechanism of ivy and improve ivy varieties.
Authors Contribution: PL and TZ conceived the study. JL and TZ wrote the manuscript. PL revised the whole manuscript.
Funding Statement: This work was supported by the Shanghai Agriculture Applied Technology Development Program, China (Grant No. C201702-1) and Engineering Research Center Project from Science and Technology Commission of Shanghai Municipality (No. 18DZ2283500).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Deng, G. L., Ying, S. B., Jing, S. S., Zhou, J., Lu, S. Y. et al. (2021). AFP2 coordinates the activity of PIF7 for thermomorphogenesis in Arabidopsis seedlings. Phyton-International Journal of Experimental Botany, 90(4), 1089–1101. DOI 10.32604/phyton.2021.016217. [Google Scholar] [CrossRef]
2. Redha, A., Suleman, P., Al-Hasan, R., Afzal, M. (2012). Responses of conocarpus lancifolius to environmental stress: A case study in the semi-arid land of Kuwait. Phyton-International Journal of Experimental Botany, 81(1), 181–190. DOI 10.32604/phyton.2012.81.181. [Google Scholar] [CrossRef]
3. Hartl, F. U., Bracher, A., Hayer-Hartl, M. (2011). Molecular chaperones in protein folding and proteostasis. Nature, 475(7356), 324–332. DOI 10.1038/nature10317. [Google Scholar] [CrossRef]
4. Balchin, D., Hayer-Hartl, M., Hartl, F. U. (2016). In vivo aspects of protein folding and quality control. Science, 353(6294), aac4354. DOI 10.1126/science.aac4354. [Google Scholar] [CrossRef]
5. Easton, D. P., Kaneko, Y., Subjeck, J. R. (2000). The hsp110 and Grp1 70 stress proteins: Newly recognized relatives of the Hsp70s. Cell Stress Chaperones, 5(4), 276–290. DOI 10.1379/1466-1268(2000)005. [Google Scholar] [CrossRef]
6. Lindquist, S. (1986). The heat-shock response. Annual Review of Biochemistry, 55, 1151–1191. DOI 10.1146/annurev.bi.55.070186.005443. [Google Scholar] [CrossRef]
7. Kampinga, H. H., Hageman, J., Vos, M. J., Kubota, H., Tanguay, R. M. et al. (2009). Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones, 14(1), 105–111. DOI 10.1007/s12192-008-0068-7. [Google Scholar] [CrossRef]
8. Pirkkala, L., Nykänen, P., Sistonen, L. (2001). Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. The FASEB Journal, 15(7), 1118–1131. DOI 10.1096/fj00-0294rev. [Google Scholar] [CrossRef]
9. Akerfelt, M., Morimoto, R. I., Sistonen, L. (2010). Heat shock factors: Integrators of cell stress, development and lifespan. Nature Reviews Molecular Cell Biology, 11(8), 545–555. DOI 10.1038/nrm2938. [Google Scholar] [CrossRef]
10. Nover, L., Bharti, K., Döring, P., Mishra, S. K., Ganguli, A. et al. (2001). Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones, 6(3), 177–189. DOI 10.1379/1466-1268(2001)006. [Google Scholar] [CrossRef]
11. Guo, J. K., Wu, J., Ji, Q., Wang, C., Luo, L. et al. (2008). Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. Journal of Genetics and Genomics, 35(2), 105–118. DOI 10.1016/S1673-8527(08)60016-8. [Google Scholar] [CrossRef]
12. Scharf, K. D., Berberich, T., Ebersberger, I., Nover, L. (2012). The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 2, 104–119. DOI 10.1016/j.bbagrm.2011.10.002. [Google Scholar] [CrossRef]
13. Liu, S., Liu, J., Zhang, Y., Jiang, Y., Hu, S. et al. (2022). Cloning of the soybean sHSP26 gene and analysis of its drought resistance. Phyton-International Journal of Experimental Botany, 91(7), 1465–1482. DOI 10.32604/phyton.2022.018836. [Google Scholar] [CrossRef]
14. Yoshida, T., Ohama, N., Nakajima, J., Kidokoro, J., Mizoi, S. et al. (2011). Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Molecular Genetics and Genomics, 286(5–6), 321–332. DOI 10.1007/s00438-011-0647-7. [Google Scholar] [CrossRef]
15. Mishra, S. K., Tripp, J., Winkelhaus, S., Tschiersch, B., Theres, K. et al. (2002). In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes & Development, 16(12), 1555–1567. DOI 10.1101/gad.228802. [Google Scholar] [CrossRef]
16. Döring, P., Treuter, E., Kistner, C., Lyck, R., Nover, C. L. (2000). The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. The Plant Cell, 12(2), 265–278. DOI 10.1105/tpc.12.2.265. [Google Scholar] [CrossRef]
17. Scharf, K. D., Heider, H., Höhfeld, I., Lych, R., Schmidt, E. et al. (1998). The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Molecular and Cellular Biology, 18(4), 2240–2251. DOI 10.1128/MCB.18.4.2240. [Google Scholar] [CrossRef]
18. Chan-Schaminet, K. Y., Baniwal, S. K., Bublak, D., Nover, L., Scharf, K. (2009). Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. Journal of Biological Chemistry, 284(31), 20848–20857. DOI 10.1074/jbc.M109.007336. [Google Scholar] [CrossRef]
19. Laloum, T., Martín, G., Duque, P. (2018). Alternative splicing control of abiotic stress responses. Trends in Plant Science, 23(2), 140–150. DOI 10.1016/j.tplants.2017.09.019. [Google Scholar] [CrossRef]
20. Sakuma, Y., Maruyama, K., Qin, F., Osakabe, Y., Shinozaki, K. et al. (2006). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proceedings of the National Academy of Sciences, 103(49), 18822–18827. DOI 10.1073/pnas.0605639103. [Google Scholar] [CrossRef]
21. Chen, H., Hwang, J. E., Lim, C. J., Kim, D. Y., Lee, S. Y. et al. (2010). Arabidopsis DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response. Biochemical and Biophysical Research Communications, 401(2), 238–244. DOI 10.1016/j.bbrc.2010.09.038. [Google Scholar] [CrossRef]
22. Mizoi, J., Shinozaki, K., Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta-Gene Regulatory Mechanisms, 1819(2), 86–96. DOI 10.1016/j.bbagrm.2011.08.004. [Google Scholar] [CrossRef]
23. Xie, Z., Nolan, T. M., Jiang, H., Yin, Y. H. (2019). AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Frontiers in Plant Science, 10(228). DOI 10.3389/fpls.2019.00228. [Google Scholar] [CrossRef]
24. Li, W., Pang, S., Lu, Z., Jin, B. (2020). Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants, 9(11), 1515. DOI 10.3390/plants9111515. [Google Scholar] [CrossRef]
25. Tian, X., Wang, F., Zhao, Y., Lan, T., Yu, K. et al. (2020). Heat shock transcription factor A1b regulates heat tolerance in wheat and Arabidopsis through OPR3 and jasmonate signalling pathway. Plant Biotechnology Journal, 18(5),1109–1111. DOI 10.1111/pbi.13268. [Google Scholar] [CrossRef]
26. Huang, Y. C., Niu, C. Y., Yang, C. R., Jinn, T. L. (2016). The heat stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiology, 172(2), 1182–1199. DOI 10.1104/16.00860. [Google Scholar] [CrossRef]
27. Zhu, Z., An, F., Feng, Y., Li, P., Xue, L. et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. The Proceedings of the National Academy of Sciences, 108(30), 12539–12544. DOI 10.1073/pnas.1103959108. [Google Scholar] [CrossRef]
28. Li, S., Zhou, X., Chen, L., Huang, W., Yu, D. (2010). Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Molecules and Cells, 29(5), 475–483. DOI 10.1007/s10059-010-0059-2. [Google Scholar] [CrossRef]
29. Merret, R., Nagarajan, V. K., Carpentier, M. C., Park, S., Favory, J. J. et al. (2015). Heat-induced ribosome pausing triggers mRNA co-translational decay in Arabidopsis thaliana. Nucleic Acids Research, 43(8), 4121–4132. DOI 10.1093/nar/gkv234. [Google Scholar] [CrossRef]
30. Liu, J. X., Howell, S. H. (2016). Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytologist, 211(2), 418–428. DOI 10.1111/nph.13915. [Google Scholar] [CrossRef]
31. Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell, 167(2), 313–324. DOI 10.1016/j.cell.2016.08.029. [Google Scholar] [CrossRef]
32. Park, C. J., Park, J. M. (2019). Endoplasmic reticulum plays a critical role in integrating signals generated by both biotic and abiotic stress in plants. Frontiers in Plant Science, 10(399). DOI 10.3389/fpls.2019.00399. [Google Scholar] [CrossRef]
33. Liu, G., Yan, P., Du, Q., Wang, Y., Guo, Y. et al. (2020). Pre-rRNA processing and its response to temperature stress in maize. Journal of Experimental Botany, 71(4), 1363–1374. DOI 10.1093/jxb/erz488. [Google Scholar] [CrossRef]
34. Hugh, M., Rosalyn, M. (2017). Hedera: The complete. UK: The Royal Horticulture Society Media. [Google Scholar]
35. Metcalfe, D. J. (2005). Hedera helix L. Journal of Ecology, 93, 632–648. DOI 10.1111/j.1365-2745.2005.01021.x. [Google Scholar] [CrossRef]
36. Brawley, J., Mathes, M. C. (1990). The influence of NaCl on the growth of English ivy (Hedera helix) cuttings and callus tissue. Environmental and Experimental Botany, 30(1), 43–47+49–50. DOI 10.1016/0098-8472(90)90007-Q. [Google Scholar] [CrossRef]
37. Pandey, A. K., Pandey, M., Tripathi, B. D. (2015). Air pollution tolerance index of climber plant species to develop vertical greenery systems in a polluted tropical city. Landscape and Urban Planning, 144, 119–127. DOI 10.1016/j.landurbplan.2015.08.014. [Google Scholar] [CrossRef]
38. Rehm, E. M., Lenz, A., Hoch, G., Körner, C. (2014). Spring patterns of freezing resistance and photosynthesis of two leaf phenotypes of Hedera helix. Basic and Applied Ecology, 15(6), 543–550. DOI 10.1016/j.baae.2014.07.009. [Google Scholar] [CrossRef]
39. Rousseau, C., Belin, E., Bove, E., Rousseau, D., Fabre, F. et al. (2013). High throughput quantitative phenotyping of plant resistance using chlorophyll fluorescence image analysis. Plant Methods, 9(1), 17. DOI 10.1186/1746-4811-9-17. [Google Scholar] [CrossRef]
40. Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods, 5(7), 621–628. DOI 10.1038/nmeth.1226. [Google Scholar] [CrossRef]
41. Zhou, J., Sun, H. Q., Wei, J. L., Li, P. (2021). Physiologic and transcriptomic insights into the high alkali response of Dunaliella salina. Phyton-International Journal of Experimental Botany, 90(5), 1401–1414. DOI 10.32604/phyton.2021.016514. [Google Scholar] [CrossRef]
42. Wei, W., Hu, Y., Yang, W., Li, X., Wei, J. et al. (2021). S-Nitrosoglutathion reductase activity modulates the thermotolerance of seeds germination by controlling ABI5 stability under high temperature. Phyton-International Journal of Experimental Botany, 90(4), 1075–1087. DOI 10.32604/phyton.2021.016134. [Google Scholar] [CrossRef]
43. Zhang, J., Kirkham, M. B. (1996). Antioxidant responses to drought in sunflower and sorghum seedlings. New Phytologist, 132(3), 361–373.DOI 10.1111/j.1469-8137.1996.tb01856.x. [Google Scholar] [CrossRef]
44. Hasanuzzaman, M., Fujita, M. (2013). Exogenous sodium nitroprusside alleviates arsenic-induced oxidative stress in wheat (Triticum aestivum L.) seedlings by enhancing antioxidant defense and glyoxalase system. Ecotoxicology, 22(3), 584–596. DOI 10.1007/s10646-013-1050-4. [Google Scholar] [CrossRef]
45. Nishizawa, A., Yabuta, Y., Yoshida, E., Maruta, T., Yoshimura, K. et al. (2006). Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant Journal, 48, 535–547. DOI 10.1111/j.1365-313X.2006.02889.x. [Google Scholar] [CrossRef]
46. Balazadeh, S., Wu, A., Mueller-Roeber, B. (2010). Salt-triggered expression of the ANAC092-dependent senescence regulon in Arabidopsis thaliana. Plant Signal Behavior, 5(6), 733–735. DOI 10.4161/psb.5.6.11694. [Google Scholar] [CrossRef]
47. Besseau, S., Li, J., Palva, E. T. (2012). WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. Journal of Experimental Botany, 63, 2667–2679. DOI 10.1093/jxb/err450. [Google Scholar] [CrossRef]
48. Kaushal, N., Bhandari, K., Siddique, K. H. M., Nayyar, H. (2016). Food crops face rising temperatures: An overview of responses, adaptive mechanisms, and approaches to improve heat tolerance. Cogent Food & Agriculture, 2(1), 1–42. DOI 10.1080/23311932.2015.1134380. [Google Scholar] [CrossRef]
49. Walter, P., Ron, D. (2011). The unfolded protein response: From stress pathway to homeostatic regulation. Science, 334(6059), 1081–1086. DOI 10.1126/science.1209038. [Google Scholar] [CrossRef]
50. Deng, Y., Humbert, S., Liu, J. X., Howell, S. H. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in arabidopsis. Proceedings of the National Academy of Sciences, 108(17), 7247–7252. DOI 10.1073/pnas.1102117108. [Google Scholar] [CrossRef]
51. Das, A., Rushton, P. J., Rohila, J. S. (2017). Metabolomic profiling of soybeans (Glycine max L.) reveals the importance of sugar and nitrogen metabolism under drought and heat stress. Plants, 6(2), 1–21. DOI 10.3390/plants6020021. [Google Scholar] [CrossRef]
52. Chao, Q., Rothenberg, M., Solano, R., Roman, G., Terzaghi, W. et al. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell, 89(7), 1133–1144. DOI 10.1016/S0092-8674(00)80300-1. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |