

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.018284
REVIEW
Chlorogenic Acid Metabolism: The Evolution and Roles in Plant Response to Abiotic Stress
1Engineering Research Centre of Ecology and Agricultural Use of Wetland, Ministry of Education, Hubei Collaborative Innovation Center for Grain Industry, Yangtze University, Jingzhou, 434025, China
2Hubei Sweetpotato Engineering and Technology Research Centre, Hubei Academy of Agricultural Sciences, Wuhan, 430064, China
*Corresponding Authors: Wenying Zhang. Email: wyzhang@yangtzeu.edu.cn; Xinshun Yang. Email: yangxins013@163.com
Received: 13 July 2021; Accepted: 27 August 2021
Abstract: During the evolution, plants acquired the ability to synthesize different phenylpropanoid compounds like chlorogenic acid (CGA), which plays vital roles in resistance mechanisms to abiotic stresses. These environmental factors, including heavy metal, cold, heat, ultraviolet (UV) light, drought, and salinity affect the plant physiological processes, resulting in massive losses of agriculture production. As plants evolve from green algae to bryophytes, ferns, gymnosperms and angiosperms, phenylpropanoids are produced and accumulated in different tissues, giving the plant the capacity to counteract the harmful effects of the adverse environments. Studies have been performed on the metabolic evolution of rosmarinic acid, flavonoids and lignin, showing that the biosynthesis of phenylpropanoids begins in green algae until the emersion of genes found in angiosperms; however, the evolution of the CGA pathway has not yet been reviewed. We hypothesize that CGA could also be synthesized from algae to angiosperms. In the present review, the evolutionary analysis of CGA pathway and the function of this compound in plant tolerance to abiotic stresses are summarized. Bioinformatics analyzes were carried out on CGA-related genes across 37 plant species and revealed that the metabolic pathway starts in algae and gradually increases until it becomes complete in angiosperms. The key genes exhibited different expression patterns in stress and plant tissues. Interestingly, some genes accumulated rapidly during evolution and were more sensitive to environmental stresses, while others appeared only later in angiosperms. Further studies are needed to better understand the evolution of the CGA metabolic pathway in plants under environmentally stressed conditions.
Keywords: Chlorogenic acid; biosynthesis; evolution; plant; abiotic stress
Phenylpropanoids (PPs) are secondary metabolites important for plant development. They play essential roles in different physiological processes including photosynthetic activity, hormonal regulation, nutrient mineralization, metabolism and reproduction [1]. Plant PPs are also vital for the resistance to abiotic stresses [2,3]. In their living habitat, plants are constantly exposed to several constraints such as heavy metals, cold, heat, drought, ultraviolet radiation, salinity, etc., which are detrimental to their productivity [4,5]. To survive, plants develop tolerance to the stresses, and this evolution process results in the accumulation of PPs in different tissues as a response to the harmful conditions [6]. PPs are composed of different compounds including flavonoids, coumarins, monolignols, stilbenes and various phenolic acids [7]. Among the latter, chlorogenic acid (CGA) is the most abundant metabolite found in many plants like Coffea arabica, Solanum lycopersicum, Malus domestica, Solanum tuberosum, etc. [8]. CGA possesses numerous bio-medical, pharmacological and phytoprotective properties such as antioxidant, antiseptic, antibacterial, and anti-inflammatory [9]. These various properties confer CGA the potentiality to be involved in plant tolerance to environmental stresses. Because of its beneficial effects, CGA is receiving more attention. Currently, studies have focused on the biosynthesis of this phenolic acid in plants. However, the mechanism for regulating CGA synthesis under environmental stresses is still not well understood.
Plants emerge from green algae passing through bryophytes (liverworts and mosses), ferns, gymnosperms and end with angiosperms [10]. As they evolve, CGA is synthesized and accumulated, giving them the ability to counter the harmful effects of the unfavorable environments. Previous research has reported that the synthesis of PPs in plants during evolution begins in green algae until the appearance of key enzymatic genes responsible for their accumulation in angiosperms [11]. The metabolic evolution of various phenylpropanoid derived compounds like rosmarinic acid [12], flavonoid [13] and lignin [14] has been studied; however, the evolution of the CGA pathway has not yet been reviewed. Therefore, a thorough understanding of how CGA pathway is genetically controlled during plant evolution under stress conditions is important for agricultural production.
Here, the review summarized recent knowledge regarding the metabolic pathway of CGA in plants. Moreover, the evolutionary analysis of CGA-related genes from algae to angiosperm is surveyed in order to provide a broader view on how this metabolic system originated and developed across evolution. Furthermore, the expression profile of the key enzymatic genes under different abiotic stresses and plant tissues was investigated in an attempt to show their roles in plant resistance mechanisms.
2 Structure, Compositional Diversity and Biosynthetic Pathway of CGA
Classically, CGAs (C16H18O9) are esters produced from cinnamic acid (p-coumaric, caffeic, ferulic acids) and quinic acid (Fig. 1). Due to these various compositions of CGAs, they are also referred to as p-coumaroylquinic acids, caffeoylquinic acids and feruloylquinic acids [15]. CGA exists in different forms of isomers but the most frequent are those derived from caffeoylquinic acids which consist of four different subgroups. The first subgroup, the mono caffeoylquinic acid (monoCQA) is composed of chlorogenic acid, neochlorogenic acid and cryptochlorogenic acid. The second subgroup includes the dicaffeoylquinic acids (diCQA), while the third concerns the tri-caffeic acid (triCQA) and the last one is specific to the tetra-caffeic acid (tetraCQA) [16]. Among these various isomers, chlorogenic acid (5-CQA) is abundantly found in plants. Moreover, a mixture of ferulic and caffeic acids (caffeoyl-feruloylquinic acid) and of sinapic and caffeic acids (caffeoyl-sinapoylquinic acid) have been found in Coffea arabica [17].
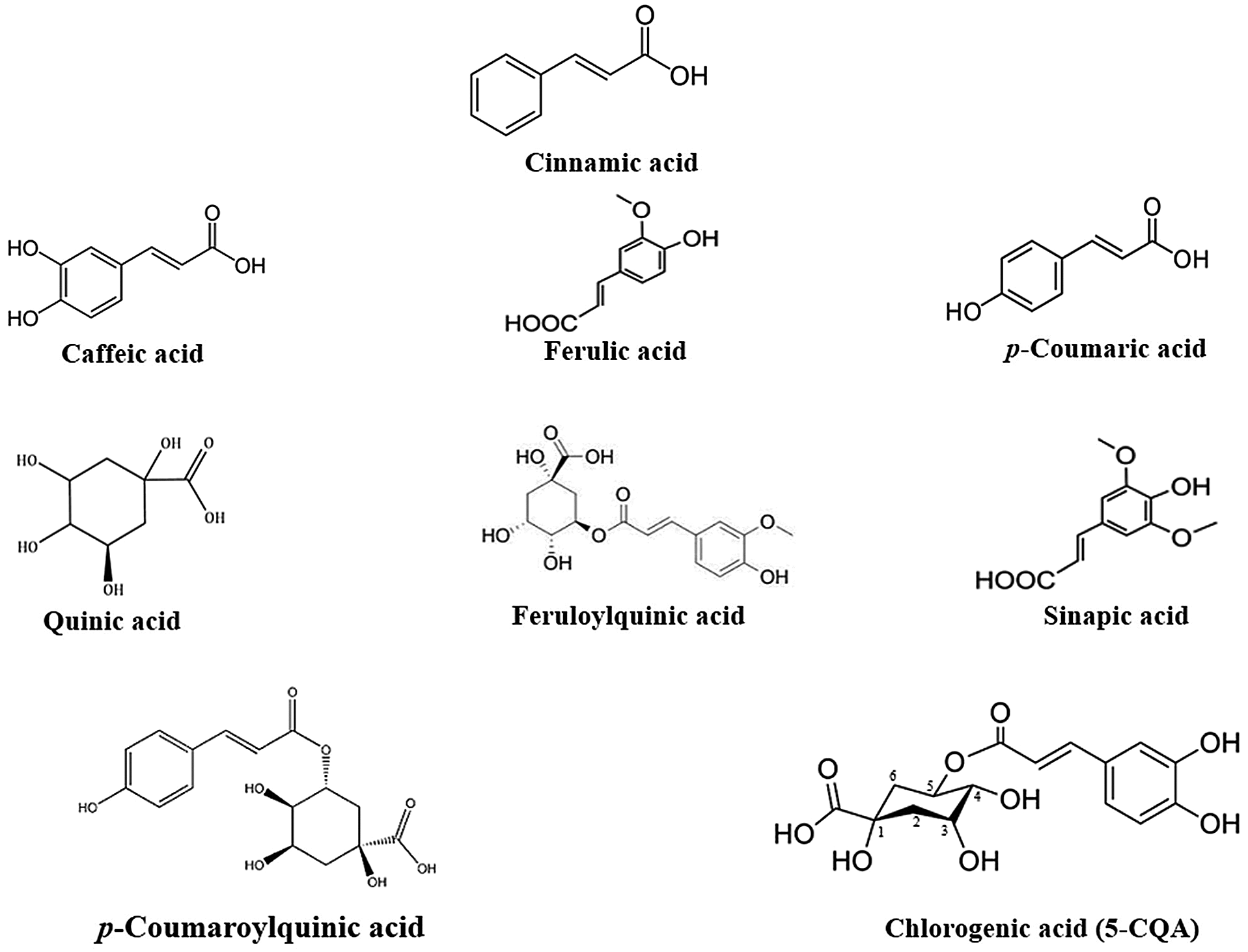
Figure 1: Chemical structures of CGA
CGA is synthesized through the general pathway of PPs under the action of various enzymatic genes. First, trans-cinnamic acid is produced from L-phenylalanine through deamination by phenylalanine ammonia-lyase (PAL) [18]. The isoforms of PAL designated PAL1-PAL4 were found to be responsible for different branches in the pathway [19]. Furthermore, trans-cinnamic acid is transformed into p-Coumaroyl Coenzyme A by cinnamate 4-hydroxylase (C4H) and 4-coumarate CoA ligase (4CL). In the next synthetic step, hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (HCT) translates p-coumaroyl-CoA to p-Coumaroyl quinic acid, followed by hydroxylation via coumarate 3-hydroxylase (C3H) to synthesize CGA [16]. Another proposed route showed that CGA can be generated from caffeoyl-coenzyme A in a process of catalysis by hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase (HQT) [8]. Caffeoyl-coenzyme A also reacts with the enzyme caffeoyl-CoA 3-O methyltransferase (ccoAOMT) to form CGA [20]. These metabolic pathways were identified in different plants such as Cynara cardunculus, Solanum melongena, Prunus domestica, Nicotiana tabacum, and Coffea arabica [21]. Moreover, another route has been described in sweet potato, showing that CGA can originate from caffeoyl-D-glucose catalyzed by hydroxycinnamoyl D-glucose: quinate hydroxycinnamoyl transferase (HCGQT) [22]. So far, several studies have reviewed different routes of PPs pathway that result in CGA synthesis in plants [23,24]. However, among the principal described routes, only two were predominant in most plant species (Fig. 2).
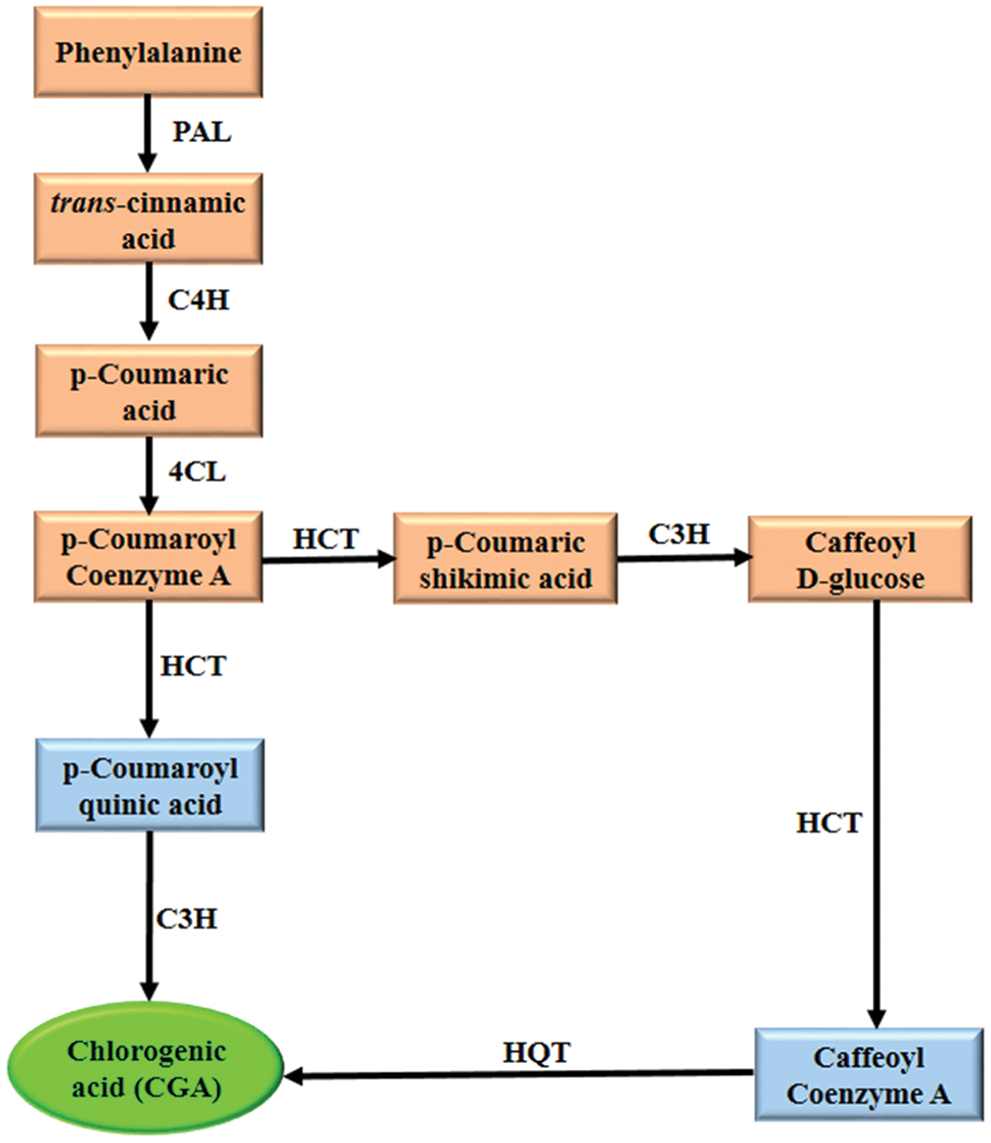
Figure 2: Biosynthetic pathway of CGA. PAL: phenylalanine ammonia-lyase, C4H: cinnamate 4-hydroxylase, 4CL: 4-coumarate CoA ligase, HCT: hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase, C3H: coumarate 3-hydroxylase, HQT: hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase
The biosynthesis of CGA is mostly regulated by DNA-binding proteins transcription factors MYB, ERF, WRKY, etc. [25]. These proteins activate the transcription of the functional key genes PAL, C4H, 4CL, C3H, HCT, HQT and ccoAOMT that are actively involved in the synthesis pathway. MYB has been shown to be the largest family of transcription factors which regulate the function of various genes in plants. It activated the promotor of PAL in Daucus carota [26], HQT in Helianthus annuus [27] and HCT in Solanum lycopersicum [28], resulting in an increased accumulation of CGA in these plants. ERF bounds to the GCC-box homolog and activates the promoter of PAL in Daucus carota [29]. Another study reported that the overexpression of ERF increased phenolic acid content in Salvia miltiorrhiza, but its silencing reduced the level of this acid [30]. Moreover, WRKY transcription factor was found to regulate the synthesis of CGA in Populus trichocarpa by activating the promoter of the HCT gene [31].
3 Evolutionary Analysis of CGA Synthesis Pathway
The origin of PPs metabolism can be traced back to algae with a number of genes in primitive green algae, indicating an early evolution of this metabolic pathway in plants [32]. Since then, algae have evolved into bryophytes, gymnosperms and angiosperms [33]. The evolution of various PPs has been previously shown in each of these different plant clades [12–14]. Here, bioinformatic analyzes were performed to predict the metabolic evolution of key genes essential for CGA synthesis (Fig. 3). Eleven genes including PAL1-PAL4, C4H, 4CL1, 4CL2, C3H, HCT, HQT and ccoAOMT were found related to the pathway across 37 plant species composed of 6 groups, namely algae, liverworts, mosses, ferns, gymnosperms and angiosperms. The protein sequences of these genes were compared with those of Arabidopsis thaliana except HQT, which was compared to Nicotiana tabacum due to the lack of this gene in the genome of Arabidopsis thaliana. The analysis of the Heatmap revealed the presence of most of the predicted genes in all plant groups, with different homology coefficients. Only a few genes have been detected in algae such as Klebsormidium flaccidum and Spirogloea muscicola, suggesting that the ability of plants to produce CGA begins in algae; thus enabling them to cope with the ever-changing environmental constraints during their development. The genes acquired earlier a similarity of protein sequence with their plant counterparts; however, the CGA synthesis pathway in algae remains incomplete. Gene homologies in liverworts, mosses, ferns, gymnosperms and angiosperms were higher than those in algae, indicating various changes in the metabolic pathways during plant development and therefore an evolution of the key genes from algae to angiosperms. For example, the protein sequences of genes in the angiosperms Brassica rapa and Dioscorea esculenta were more similar to those of Arabidopsis thaliana and Nicotiana tabacum compared to the sequences in the algae Mesotaenium endlicherianum and Chara braunii. Most of the CGA-related genes were present in angiosperms; we assumed that as plants evolve from the aquatic environment of algae to terrestrial angiosperm plants, the synthetic pathway of CGA also evolves and becomes complete in angiosperms. This could be explained by the long-term exposure of the angiosperm plants to various environmental stresses, which would stimulate the expression of most of the key genes, resulting in the completion of the synthetic pathway in angiosperms and probably to a high accumulation of CGA. A study reported the ability of angiosperm plants to accumulate CGA, which may be strongly correlated to the expression of the functional genes in their biosynthetic pathways. CGA was abundant in angiosperm species of the orders Apiales, Asterales and Dipsacales [34]. Furthermore, analysis of the evolution of each gene showed that PAL1 and PAL2 were more present in all the plant groups than PAL3 and PAL4, which were mostly observed in angiosperms. Likewise, the C4H and 4CL2 genes were detected in most species, while 4CL1 was just present in angiosperms. This indicates that PAL3, PAL4 and 4CL1 appear later in the process of CGA synthesis, showing their importance for the completion of the metabolic pathway in angiosperms. Moreover, in the downstream of the pathway, C3H, HCT and ccoAOMT appeared in almost all plant groups, but exhibited a low sequence similarity in algae compared to angiosperms, indicating that these genes have undergone evolution during the synthesis of CGA. However, HQT gene has only been detected in angiosperm species. It appeared much later in the metabolic pathway, denoting the significant role of this gene for the synthesis of CGA. Numerous studies have revealed the strong correlation between the HQT gene and CGA metabolism [35,36]. For example, the upregulation of HQT significantly increased the content of CGA in Lonicera macranthoides [21], while its silencing reduced the content in Lonicera japonica [24]. Further studies could help to better understand the metabolic evolution of CGA-related genes in plants.
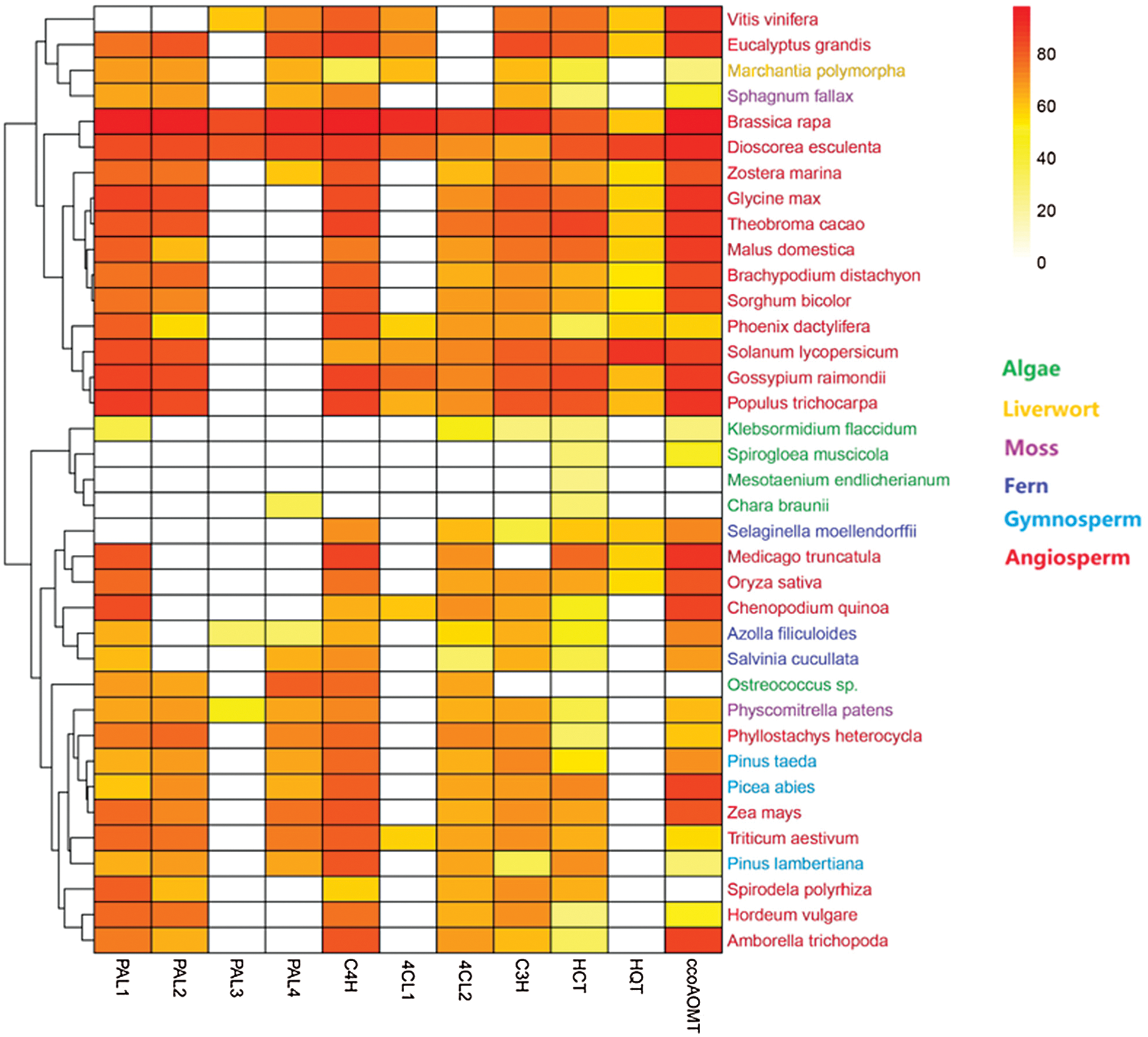
Figure 3: Heatmap describing the evolution of CGA synthesis pathway across different plant species. PAL: Phenylalanine ammonia-lyase, C4H: cinnamate 4-hydroxylase, 4CL: 4-coumarate CoA ligase, C3H: coumarate 3-hydroxylase, HCT: hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase, HQT: hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase, ccoAOMT: caffeoyl-CoA 3-O methytransferase
4 CGA Role in Plant Tolerance to Abiotic Stresses
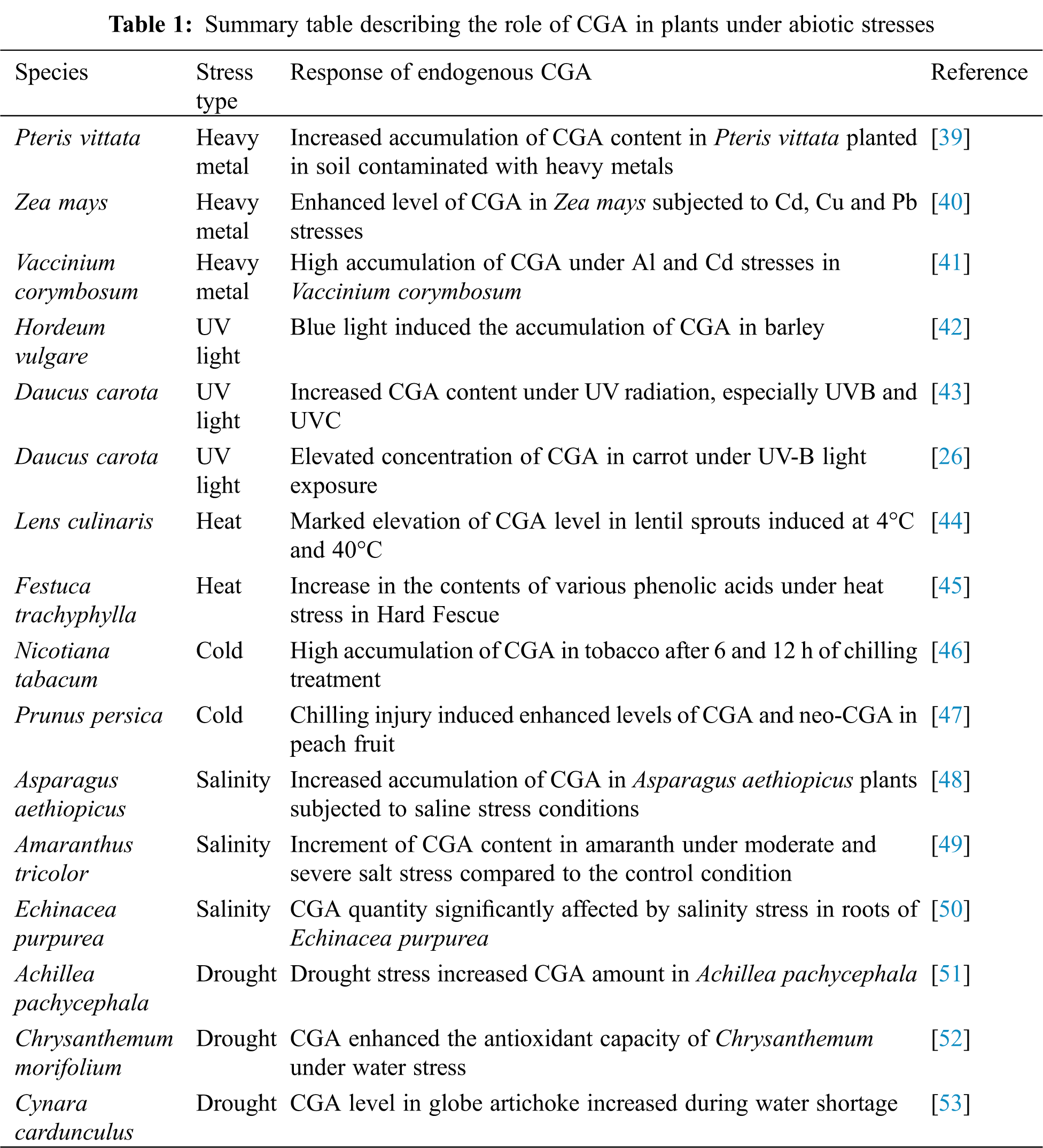
Plant stresses have increased recently due to environmental disturbances caused by climate change [18]. Abiotic stresses including heavy metals, UV radiations, temperature, drought and salinity adversely affect the growth and development of plants [37]. They trigger the generation of reactive oxygen species (ROS), which affect cell membranes, proteins, nucleic acids and lipids leading to cell death [38]. In response to these stresses, the biosynthesis of CGA usually increases in plants. CGA facilitates the plant stress-responsive mechanisms to counteract the effects of challenging environments. It scavenges the overproduced ROS, and reduces cell membrane peroxidation, hence protecting the cells from oxidative stresses. A brief summary of the role of CGA in plants under abiotic stresses is provided in Tab. 1.
Temperature is an environmental factor important for plant growth. However, below or beyond the optimum degree, temperature can trigger a thermal stress, which will affect the plant physiological processes [54,55]. Chilling (0–20°C) directly impacts the plasma membrane, affecting its fluidity and stability. The plasma membrane undergoes a transition from its liquid state to a rigid gel phase resulting in the rigidity of the membrane. This is enhanced at freezing temperatures (<0°C) by the formation of ice crystals in the intracellular and extracellular spaces of plant tissues [56]. In tropical regions, the plasma membrane is also the first organ affected by heat stress. It undergoes a transition from the crystalline liquid phase to the fluid phase [57]. An extremely high temperature can cause instant cell death [58]. Cold and heat stresses negatively impact photosynthesis and electron transport reactions, which affect the cells’ structural components, thus changing different cellular metabolisms and subsequently leading to severe losses of crop yield [59,60]. CGA has been shown to be an excellent antioxidant in cell detoxification under thermal stress. It confers thermotolerance to plants by scavenging the generated ROS due to cold and heat stresses. The accumulation of CGA occurs at a temperature threshold where stress begins to be harmful to plant [18]. For example, significant increases of CGA content were observed at 6 and 12 h after chilling stress (4°C) in Nicotiana tabacum [46], and 2 h after heat stress (50°C) in Nicotiana langsdorffii [61].
4.2 UV-Light and Heavy Metal Stresses
The exposure of plants to UV radiations and heavy metals causes the generation of harmful ROS resulting in retarded growth. Under these stress conditions, plants develop a resistance strategy, consisting in the production of the antioxidant CGA, which reacts in the process of detoxifying cells [62]. It has been reported that the content of CGA in Vaccinium corymbosum significantly increased after UV-B radiation [63]. In addition, Dresler et al. found a high accumulation of CGA in Echium vulgare induced by heavy metal stress [64]. This phenolic compound possesses hydroxyl and carboxyl groups that allow it to bind heavy metals and protect plants from oxidative stresses [40]. It not only acts as a scavenger of ROS but also as a metal chelator and sunscreen in the epidermal layer of tissues to block UV rays, hence protecting plants from the harsh effects of such hostile environments [18,65].
4.3 Drought and Salinity Stresses
As an abiotic factor, drought is the major threat that causes changes in plant biological processes [66]. It occurs when the available water in the soil is reduced to a critical level, affecting the transport of essential mineral nutrients such as potassium, nitrogen, calcium and phosphorus to other parts of the plant. During water deficiency, leaves close their stomata, causing a drop in their conductance, and therefore a decrease of transpiration and photosynthesis activity [2,3]. Salinity stress induces osmotic stress and ion toxicity. Osmotic stress occurs when the level of salt around plant roots is increased to a high level, causing water deficit in the roots and suppression of shoot growth shortly after exposure [67]. Long-term exposure of plants to high salinity triggers ionic stress due to an excessive accumulation of Na+ and Cl− ions in the cell cytoplasm which results in a leaf senescence, decreased crop yield and ultimately plant death [68]. CGA biosynthesis and accumulation under salinity and drought stresses play a detoxifying role, protecting plants from oxidative damages, thus improving antioxidant activities. CGA has been reported to participate in a resistance mechanism of Malus domestica leaves exposed to an oxidative stress [69]. Under drought, CGA has the capacity to reduce the detrimental effects of ROS on plant metabolism by increasing photosynthetic activity [53]. Various studies have also reported the strong antioxidant properties of CGA in removing ROS under high salinity stress in Thymus daenesis [70] and Lonicera japonica [71].
5 Expression Profile of CGA Pathway Genes under Abiotic Stress
Transcriptome data from Arabidopsis thaliana were collected to trace the potential role of CGA-related genes in tolerance to abiotic factors. All the key genes exhibited different expression patterns in a specific manner for UV, drought, salinity, cold and heat stresses (Fig. 4). No expression was observed for these genes under hypoxia stress. CGA might not be involved in the response of Arabidopsis thaliana plants to the stress of hypoxia. To date, no study has reported the involvement of CGA in reducing hypoxia stress in Arabidopsis thaliana. Above, it was noticed that the PAL3 and PAL4 genes appeared later in the process of CGA synthesis, thus showing the importance of these genes for a complete metabolic pathway. However, under stress conditions, PAL3 and PAL4 exhibited lower levels of expression than PAL1 and PAL2. This could be explained by the fact that in hostile living environments, PAL1 and PAL2 genes get activated earlier with the generation of the first ROS and keep expressing throughout the development cycle, thus helping plants to resist altered environments. However, PAL3 and PAL4 were not expressed until later in angiosperms. Hence, there was a higher expression of PAL1 and PAL2 compared to that of PAL3 and PAL4. An example is shown with the Arabidopsis thaliana genes AtPAL1 and AtPAL2, which were more responsive to temperature stress [72]. Concerning the 4CL gene family, it was observed that 4CL1 appeared later in the metabolic evolution of CGA than 4CL2. The transcript abundance of 4CL1 was higher than that of 4CL2 under stress conditions. It could be speculated that 4CL1 accumulates rapidly and might be more sensitive to environmental constraints. This finding is consistent with a previous study [73]. Moreover, the C4H, ccoAOMT and HCT genes exhibited high expression levels compared to C3H, which showed an extremely low accumulation under almost all stress conditions. This suggests that the C3H gene may not function for the tolerance of Arabidopsis thaliana plant to abiotic stresses. This was not the case for C3H in Pyrus betulaefolia and Aegilops tauschii, which were sensitive to salt and drought stresses, respectively [74,75]. Under UV, drought, salt and heat stresses, the ccoAOMT gene displayed the highest level of expression. It was followed by PAL1 and 4CL1 under UV stress and HCT under drought, salt and heat stresses; PAL2 and C4H genes were most induced in cold stress. Further, the downstream genes C3H, HCT and ccoAOMT, responsible for CGA production, were less expressed under cold conditions compared to the other stresses. This indicates that these resistance genes accumulate slowly under low temperature. Various studies reported the down-regulation of the C3H, HCT and ccoAOMT genes during plant adaptation to cold stress [76,77]. We hypothesize that the Arabidopsis thaliana cold resistance genes could also originate from another route in the general phenylpropanoid pathway, parallel to that of the CGA and responsible for the flavonoid synthesis.
Most of the CGA-related genes were relatively highly expressed in almost all tissues other than pollen, where the genes exhibited an extremely low expression level (Fig. 5). The PAL1 and PAL2 genes were mostly expressed in roots, whereas PAL3 and PAL4 showed the highest expression levels in shoots and siliques, respectively. These results do not correspond to those of Tohge et al. who demonstrated that in Arabidopsis thaliana AtPAL1, AtPAL2 and AtPAL3 were expressed in the stems, while AtPAL4 was strongly expressed in the seeds [78]. 4CL1 was high in stems, whereas 4CL2 and C4H transcript accumulated highly in roots, which is consistent with a previous report [79]. The ccoAOMT gene was markedly high in the endosperm and HCT in roots and leaves. In contrast, the C3H gene exhibited a specific expression pattern in flowers. The ability of key genes to catalyze major enzymatic reactions in the CGAmetabolic pathway makes them suitable candidate genes for plant resistance mechanisms.

Figure 4: Expression profiles of CGA-related genes under abiotic stress among 515 transcriptome data of Arabidopsis thaliana. The number in bracket indicates the number of transcriptomes. X axis represents different abiotic stress treatments and Y axis represents gene expression level in FPKM (fragments per kilobase of exon model per million reads mapped). The box plots represent the gene expression level in different transcriptome samples. Horizontal and vertical lines from top to bottom in the plots represent maximum value, upper whisker, upper quartile, median, lower quartile, lower whisker and minimum value, respectively. PAL: Phenylalanine ammonia-lyase, C4H: cinnamate 4-hydroxylase, 4CL: 4-coumarate CoA ligase, C3H: coumarate 3-hydroxylase, HCT: hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase, ccoAOMT: caffeoyl-CoA 3-O methytransferase

Figure 5: Expression profiles of CGA-related genes in different tissues among 1122 transcriptome data of Arabidopsis thaliana. The number in bracket indicates the number of transcriptomes. X-axis represents plant tissues and Y-axis represents gene expression level in FPKM (fragments per kilobase of exon model per million reads mapped). The box plots represent the gene expression level in different transcriptome samples. Horizontal and vertical lines from top to bottom in the plots represent maximum value, upper whisker, upper quartile, median, lower quartile, lower whisker and minimum value, respectively. PAL: Phenylalanine ammonia-lyase, C4H: cinnamate 4-hydroxylase, 4CL: 4-coumarate CoA ligase, C3H: coumarate 3-hydroxylase, HCT: hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase, ccoAOMT: caffeoyl-CoA 3-O methytransferase
Abiotic stresses affect growth and development of plants over evolutionary time. The plant’s resistance mechanisms against these factors induce the production of CGA, which antioxidant properties improve the plant’s ability to overcome the hostile environments (Fig. 6). The overexpression of the CGA-related genes 4CL, PAL, HCT, C3H, C4H and ccoAOMT increases the plant’s resistance to abiotic stresses. The present review showed compelling evidence for the metabolic evolution of the CGA pathway, which evolved from algae until the appearance of the complete pathway observed in angiosperm land plants. The key genes evolved with plant growth and development and some of them were more stress-responsive. The ccoAOMT gene was highly expressed in almost all stresses, while PAL2 and C4H were more specific to cold stress.
In addition to the number of papers on the evolution of different polyphenols, further investigations are needed for understanding the evolution of the CGA metabolic pathway in plants under the ever-changing environmental constraints.

Figure 6: Plant response to abiotic stresses. These stresses (e.g., drought, salinity, ion toxicity, nutrient deficiency, UV radiation, etc.) cause serious damages to plants. As adaptive responses, plants biosynthesize CGA, which help to correct the damages and resist the unfavorable conditions. ROS: reactive oxygen species, PP: phenylpropanoid, PAL: Phenylalanine ammonia-lyase, C4H: cinnamate 4-hydroxylase, 4CL: 4-coumarate CoA ligase, C3H: coumarate 3-hydroxylase, HQT: hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase, HCT: hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase, CGA: chlorogenic acid, ccoAOMT: caffeoyl-CoA 3-O methytransferase The authors thank Abdullah Shalmani for his helpful comments on the manuscript.
Acknowledgement: The authors thank Abdullah Shalmani for his helpful comments on the manuscript.
Funding Statement: This work was supported by the (1) National Key R&D Program of China (2019YFD1001300, 2019YFD1001305), (2) Science and Technology Development Plan Project of Jingzhou City, Hubei Province, China (2018-37), (3) Characteristic Discipline of Hubei Academy of Agricultural Sciences (2015TSXK06) and (4) Science and Technology Innovation Center of Hubei Academy of Agricultural Sciences (2007-620-001-03).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Sharma, A., Shahzad, B., Rehman, A., Bhardwaj, R., Landi, M. et al. (2019). Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules, 24(13), 2452. DOI 10.3390/molecules24132452. [Google Scholar] [CrossRef]
2. Francini, A., Giro, A., Ferrante, A. (2019). Biochemical and Molecular Regulation of Phenylpropanoids Pathway under Abiotic Stresses. Amsterdam, Netherlands: Elsevier. [Google Scholar]
3. Ahmad, B., Raina, A., Khan, S. (2019). Impact of Biotic and Abiotic Stresses on Plants, and Their Responses. Berlin, Germany: Springer. [Google Scholar]
4. Zhang, Y., Lv, Y., Jahan, N., Chen, G., Ren, D. et al. (2018). Sensing of abiotic stress and ionic stress responses in plants. International Journal of Molecular Sciences, 19(11), 3298. DOI 10.3390/ijms19113298. [Google Scholar] [CrossRef]
5. Tohge, T., Perez de Souza, L., Fernie, A. R. (2018). On the natural diversity of phenylacylated-flavonoid and their in planta function under conditions of stress. Phytochemistry Reviews, 17(2), 279–290. DOI 10.1007/s11101-017-9531-3. [Google Scholar] [CrossRef]
6. Naikoo, M. I., Dar, M. I., Raghib, F., Jaleel, H., Ahmad, B. et al. (2019). Role and regulation of plants phenolics in abiotic stress tolerance: An overview. Amsterdam, Netherlands: Elsevier. [Google Scholar]
7. Deng, Y., Lu, S. (2017). Biosynthesis and regulation of phenylpropanoids in plants. Critical Reviews in Plant Sciences, 36(4), 257–290. DOI 10.1080/07352689.2017.1402852. [Google Scholar] [CrossRef]
8. Payyavula, R. S., Shakya, R., Sengoda, V. G., Munyaneza, J. E., Swamy, P. et al. (2015). Synthesis and regulation of chlorogenic acid in potato: Rerouting phenylpropanoid flux in HQT-silenced lines. Plant Biotechnology Journal, 13(4), 551–564. DOI 10.1111/pbi.12280. [Google Scholar] [CrossRef]
9. Naveed, M., Hejazi, V., Abbas, M., Kamboh, A. A., Khan, G. J. et al. (2018). Chlorogenic acid (CGAA pharmacological review and call for further research. Biomedicine & Pharmacotherapy, 97(3), 67–74. DOI 10.1016/j.biopha.2017.10.064. [Google Scholar] [CrossRef]
10. Cannell, N., Emms, D. M., Hetherington, A. J., MacKay, J., Kelly, S. et al. (2020). Multiple metabolic innovations and losses are associated with major transitions in land plant evolution. Current Biology, 30(10), 1783–1800. DOI 10.1016/j.cub.2020.02.086. [Google Scholar] [CrossRef]
11. Yonekura-Sakakibara, K., Higashi, Y., Nakabayashi, R. (2019). The origin and evolution of plant flavonoid metabolism. Frontiers in Plant Science, 10, 943. DOI 10.3389/fpls.2019.00943. [Google Scholar] [CrossRef]
12. Levsh, O., Pluskal, T., Carballo, V., Mitchell, A. J., Weng, J. K. (2019). Independent evolution of rosmarinic acid biosynthesis in two sister families under the Lamiids clade of flowering plants. Journal of Biological Chemistry, 294(42), 15193–15205. DOI 10.1074/jbc.RA119.010454. [Google Scholar] [CrossRef]
13. Wen, W., Alseekh, S., Fernie, A. R. (2020). Conservation and diversification of flavonoid metabolism in the plant kingdom. Current Opinion in Plant Biology, 55, 100–108. DOI 10.1016/j.pbi.2020.04.004. [Google Scholar] [CrossRef]
14. Renault, H., Werck-Reichhart, D., Weng, J.-K. (2019). Harnessing lignin evolution for biotechnological applications. Current Opinion in Biotechnology, 56, 105–111. DOI 10.1016/j.copbio.2018.10.011. [Google Scholar] [CrossRef]
15. Badmos, S., Fu, M., Granato, D., Kuhnert, N. (2020). Classification of Brazilian roasted coffees from different geographical origins and farming practices based on chlorogenic acid profiles. Food Research International, 134, 109218. DOI 10.1016/j.foodres.2020.109218. [Google Scholar] [CrossRef]
16. Clifford, M. N., Jaganath, I. B., Ludwig, I. A., Crozier, A. (2017). Chlorogenic acids and the acyl-quinic acids: Discovery, biosynthesis, bioavailability and bioactivity. Natural Product Reports, 34(12), 1391–1421. DOI 10.1039/c7np00030h. [Google Scholar] [CrossRef]
17. Wei, F., Tanokura, M. (2015). Organic compounds in green coffee beans. Amsterdam, Netherlands: Elsevier. [Google Scholar]
18. Lattanzio, V. (2021). Relationship of phenolic metabolism to growth in plant and cell cultures under stress. Springer, Cham. [Google Scholar]
19. Yuan, Y., Wang, Z., Jiang, C., Wang, X., Huang, L. (2014). Exploiting genes and functional diversity of chlorogenic acid and luteolin biosyntheses in Lonicera japonica and their substitutes. Gene, 534(2), 408–416. DOI 10.1016/j.gene.2012.09.051. [Google Scholar] [CrossRef]
20. Lepelley, M., Cheminade, G., Tremillon, N., Simkin, A., Caillet, V. et al. (2007). Chlorogenic acid synthesis in coffee: An analysis of CGA content and real-time RT-PCR expression of HCT, HQT, C3H1, and CCoAOMT1 genes during grain development in C. canephora. Plant Science, 172(5), 978–996. DOI 10.1016/j.plantsci.2007.02.004. [Google Scholar] [CrossRef]
21. Chen, Z., Liu, G., Liu, Y., Xian, Z., Tang, N. (2017). Overexpression of the LmHQT1 gene increases chlorogenic acid production in Lonicera macranthoides hand-mazz. Acta Physiologiae Plantarum, 39(1), 27. DOI 10.1007/s11738-016-2310-8. [Google Scholar] [CrossRef]
22. Villegas, R., Kojima, M. (1986). Purification and characterization of hydroxycinnamoyl D-glucose. Quinate hydroxycinnamoyl transferase in the root of sweet potato, Ipomoea batatas Lam. Journal of Biological Chemistry, 261(19), 8729–8733. DOI 10.1016/S0021-9258(19)84441-1. [Google Scholar] [CrossRef]
23. Yu, Y., Wang, Y., Yu, Y., Ma, P., Jia, Z. et al. (2021). Overexpression of IbPAL1 promotes chlorogenic acid biosynthesis in sweetpotato. The Crop Journal, 9(1), 204–215. DOI 10.1016/j.cj.2020.06.003. [Google Scholar] [CrossRef]
24. Li, Y., Kong, D., Bai, M., He, H., Wang, H. et al. (2019). Correlation of the temporal and spatial expression patterns of HQT with the biosynthesis and accumulation of chlorogenic acid in Lonicera japonica flowers. Horticulture Research, 6(1), 1–14. DOI 10.1038/s41438-019-0154-2. [Google Scholar] [CrossRef]
25. Ye, J., Han, W., Deng, P., Jiang, Y., Liu, M. et al. (2019). Comparative transcriptome analysis to identify candidate genes related to chlorogenic acid biosynthesis in Eucommia ulmoides Oliv. Trees, 33(5), 1373–1384. DOI 10.1007/s00468-019-01865-y. [Google Scholar] [CrossRef]
26. Bartley, G. E., Avena-Bustillos, R. J., Du, W. X., Hidalgo, M., Cain, B. et al. (2016). Transcriptional regulation of chlorogenic acid biosynthesis in carrot root slices exposed to UV-B light. Plant Gene, 7, 1–10. DOI 10.1016/j.plgene.2016.07.001. [Google Scholar] [CrossRef]
27. Cheevarungnapakul, K., Khaksar, G., Panpetch, P., Boonjing, P., Sirikantaramas, S. (2019). Identification and functional characterization of genes involved in the biosynthesis of caffeoylquinic acids in sunflower (Helianthus annuus L.). Frontiers in Plant Science, 10, 968. DOI 10.3389/fpls.2019.00968. [Google Scholar] [CrossRef]
28. Li, Y., Chen, M., Wang, S., Ning, J., Ding, X. et al. (2015). AtMYB11 regulates caffeoylquinic acid and flavonol synthesis in tomato and tobacco. Plant Cell, Tissue and Organ Culture (PCTOC), 122(2), 309–319. DOI 10.1007/s11240-015-0767-6. [Google Scholar] [CrossRef]
29. Kimura, S., Chikagawa, Y., Kato, M., Maeda, K., Ozeki, Y. (2008). Upregulation of the promoter activity of the carrot (Daucus carota) phenylalanine ammonia-lyase gene (DcPAL3) is caused by new members of the transcriptional regulatory proteins, DcERF1 and DcERF2, which bind to the GCC-box homolog and act as an activator to the DcPAL3 promoter. Journal of Plant Research, 121(5), 499–508. DOI 10.1007/s10265-008-0170-z. [Google Scholar] [CrossRef]
30. Sun, M., Shi, M., Wang, Y., Huang, Q., Yuan, T. et al. (2019). The biosynthesis of phenolic acids is positively regulated by the JA-responsive transcription factor ERF115 in salvia miltiorrhiza. Journal of Experimental Botany, 70(1), 243–254. DOI 10.1093/jxb/ery349. [Google Scholar] [CrossRef]
31. Zhang, J., Yang, Y., Zheng, K., Xie, M., Feng, K. et al. (2018). Genome-wide association studies and expression-based quantitative trait loci analyses reveal roles of HCT2 in caffeoylquinic acid biosynthesis and its regulation by defense-responsive transcription factors in Populus. New Phytologist, 220(2), 502–516. DOI 10.1111/nph.15297. [Google Scholar] [CrossRef]
32. de Vries, J., de Vries, S., Slamovits, C. H., Rose, L. E., Archibald, J. M. (2017). How embryophytic is the biosynthesis of phenylpropanoids and their derivatives in streptophyte algae? Plant and Cell Physiology, 58(5), 934–945. DOI 10.1093/pcp/pcx037. [Google Scholar] [CrossRef]
33. Shabala, S., Chen, G., Chen, Z. H., Pottosin, I. (2020). The energy cost of the tonoplast futile sodium leak. New Phytologist, 225(3), 1105–1110. DOI 10.1111/nph.15758. [Google Scholar] [CrossRef]
34. Petersen, M., Abdullah, Y., Benner, J., Eberle, D., Gehlen, K. et al. (2009). Evolution of rosmarinic acid biosynthesis. Phytochemistry, 70(15–16), 1663–1679. DOI 10.1016/j.phytochem.2009.05.010. [Google Scholar] [CrossRef]
35. Zhang, J., Wu, M., Li, W., Bai, G. (2017). Regulation of chlorogenic acid biosynthesis by hydroxycinnamoyl CoA quinate hydroxycinnamoyl transferase in Lonicera japonica. Plant Physiology and Biochemistry, 121(3), 74–79. DOI 10.1016/j.plaphy.2017.10.017. [Google Scholar] [CrossRef]
36. Liu, Q., Liu, Y., Xu, Y., Yao, L., Liu, Z. et al. (2018). Overexpression of and RNA interference with hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase affect the chlorogenic acid metabolic pathway and enhance salt tolerance in Taraxacum antungense kitag. Phytochemistry Letters, 28, 116–123. DOI 10.1016/j.phytol.2018.10.003. [Google Scholar] [CrossRef]
37. Saleem, S., Mushtaq, N. U., Shah, W. H., Rasool, A., Hakeem, K. R. et al. (2021). Morpho-physiological, biochemical and molecular adaptation of millets to abiotic stresses: A review. Phyton-International Journal of Experimental Botany, 90(5), 1363. DOI 10.32604/phyton.2021.014826. [Google Scholar] [CrossRef]
38. Małkowski, E., Sitko, K., Zieleźnik-Rusinowska, P., Gieroń, Ż., Szopiński, M. (2019). Heavy metal toxicity: Physiological implications of metal toxicity in plants. Berlin, Germany: Springer. [Google Scholar]
39. Pham, H. N., Michalet, S., Bodillis, J., Nguyen, T. D., Nguyen, T. K. O. et al. (2017). Impact of metal stress on the production of secondary metabolites in Pteris vittata L. and associated rhizosphere bacterial communities. Environmental Science and Pollution Research, 24(20), 16735–16750. DOI 10.1007/s11356-017-9167-2. [Google Scholar] [CrossRef]
40. Kısa, D., Elmastaş, M., Öztürk, L., Kayır, Ö. (2016). Responses of the phenolic compounds of Zea mays under heavy metal stress. Applied Biological Chemistry, 59(6), 813–820. DOI 10.1007/s13765-016-0229-9. [Google Scholar] [CrossRef]
41. Manquian-Cerda, K., Cruces, E., Escudey, M., Zuniga, G., Calderon, R. (2018). Interactive effects of aluminum and cadmium on phenolic compounds, antioxidant enzyme activity and oxidative stress in blueberry (Vaccinium corymbosum L.) plantlets cultivated in vitro. Ecotoxicology and Environmental Safety, 150, 320–326. DOI 10.1016/j.ecoenv.2017.12.050. [Google Scholar] [CrossRef]
42. Klem, K., Gargallo-Garriga, A., Rattanapichai, W., Oravec, M., Holub, P. et al. (2019). Distinct morphological, physiological, and biochemical responses to light quality in barley leaves and roots. Frontiers in Plant Science, 10, 1026. DOI 10.3389/fpls.2019.01026. [Google Scholar] [CrossRef]
43. Surjadinata, B. B., Jacobo-Velazquez, D. A., Cisneros-Zevallos, L. (2017). UVA, UVB and UVC light enhances the biosynthesis of phenolic antioxidants in fresh-cut carrot through a synergistic effect with wounding. Molecules, 22(4), 668. DOI 10.3390/molecules22040668. [Google Scholar] [CrossRef]
44. Świeca, M. (2015). Elicitation with abiotic stresses improves pro-health constituents, antioxidant potential and nutritional quality of lentil sprouts. Saudi Journal of Biological Sciences, 22(4), 409–416. DOI 10.1016/j.sjbs.2014.12.007. [Google Scholar] [CrossRef]
45. Wang, J., Yuan, B., Huang, B. (2019). Differential heat-induced changes in phenolic acids associated with genotypic variations in heat tolerance for hard fescue. Crop Science, 59(2), 667–674. DOI 10.2135/cropsci2018.01.0063. [Google Scholar] [CrossRef]
46. Zhou, P., Li, Q., Liu, G., Xu, N., Yang, Y. et al. (2019). Integrated analysis of transcriptomic and metabolomic data reveals critical metabolic pathways involved in polyphenol biosynthesis in Nicotiana tabacum under chilling stress. Functional Plant Biology, 46(1), 30–43. DOI 10.1071/FP18099. [Google Scholar] [CrossRef]
47. Wang, L., Shan, T., Xie, B., Ling, C., Shao, S. et al. (2019). Glycine betaine reduces chilling injury in peach fruit by enhancing phenolic and sugar metabolisms. Food Chemistry, 272, 530–538. DOI 10.1016/j.foodchem.2018.08.085. [Google Scholar] [CrossRef]
48. Al-Ghamdi, A. A., Elansary, H. O. (2018). Synergetic effects of 5-aminolevulinic acid and Ascophyllum nodosum seaweed extracts on Asparagus phenolics and stress related genes under saline irrigation. Plant Physiology and Biochemistry, 129, 273–284. DOI 10.1016/j.plaphy.2018.06.008. [Google Scholar] [CrossRef]
49. Sarker, U., Oba, S. (2018). Augmentation of leaf color parameters, pigments, vitamins, phenolic acids, flavonoids and antioxidant activity in selected Amaranthus tricolor under salinity stress. Scientific Reports, 8(1), 12349. DOI 10.1038/s41598-018-30897-6. [Google Scholar] [CrossRef]
50. Khorasaninejad, S., Zare, F., Hemmati, K. (2020). Effects of silicon on some phytochemical traits of purple coneflower (Echinacea purpurea L.) under salinity. Scientia Horticulturae, 264, 108954. DOI 10.1016/j.scienta.2019.108954. [Google Scholar] [CrossRef]
51. Gharibi, S., Tabatabaei, B. E. S., Saeidi, G., Talebi, M., Matkowski, A. (2019). The effect of drought stress on polyphenolic compounds and expression of flavonoid biosynthesis related genes in Achillea pachycephala Rech. f. Phytochemistry, 162, 90–98. DOI 10.1016/j.phytochem.2019.03.004. [Google Scholar] [CrossRef]
52. Hodaei, M., Rahimmalek, M., Arzani, A., Talebi, M. (2018). The effect of water stress on phytochemical accumulation, bioactive compounds and expression of key genes involved in flavonoid biosynthesis in Chrysanthemum morifolium L. Industrial Crops and Products, 120, 295–304. DOI 10.1016/j.indcrop.2018.04.073. [Google Scholar] [CrossRef]
53. Nouraei, S., Rahimmalek, M., Saeidi, G. (2018). Variation in polyphenolic composition, antioxidants and physiological characteristics of globe artichoke (Cynara cardunculus var. scolymus Hayek L.) as affected by drought stress. Scientia Horticulturae, 233, 378–385. DOI 10.1016/j.scienta.2017.12.060. [Google Scholar] [CrossRef]
54. Ritonga, F. N., Chen, S. (2020). Physiological and molecular mechanism involved in cold stress tolerance in plants. Plants, 9(5), 560. DOI 10.3390/plants9050560. [Google Scholar] [CrossRef]
55. Zou, M., Yuan, L., Zhu, S., Liu, S., Ge, J. et al. (2017). Effects of heat stress on photosynthetic characteristics and chloroplast ultrastructure of a heat-sensitive and heat-tolerant cultivar of wucai (Brassica campestris L.). Acta Physiologiae Plantarum, 39(11–10. DOI 10.1007/s11738-016-2319-z. [Google Scholar] [CrossRef]
56. Chinnusamy, V., Zhu, J., Zhu, J. K. (2007). Cold stress regulation of gene expression in plants. Trends in Plant Science, 12(10), 444–451. DOI 10.1016/j.tplants.2007.07.002. [Google Scholar] [CrossRef]
57. Chinnusamy, V., Zhu, J. K., Sunkar, R. (2010). Gene regulation during cold stress acclimation in plants. Berlin, Germany: Springer. [Google Scholar]
58. Commisso, M., Toffali, K., Strazzer, P., Stocchero, M., Ceoldo, S. et al. (2016). Impact of phenylpropanoid compounds on heat stress tolerance in carrot cell cultures. Frontiers in Plant Science, 7, 1439. DOI 10.3389/fpls.2016.01439. [Google Scholar] [CrossRef]
59. Tsai, W. A., Weng, S. H., Chen, M. C., Lin, J. S., Tsai, W. S. (2019). Priming of plant resistance to heat stress and tomato yellow leaf curl thailand virus with plant-derived materials. Frontiers in Plant Science, 10, 906. DOI 10.3389/fpls.2019.00906. [Google Scholar] [CrossRef]
60. Ghanbari, F., Sayyari, M. (2018). Controlled drought stress affects the chilling-hardening capacity of tomato seedlings as indicated by changes in phenol metabolisms, antioxidant enzymes activity, osmolytes concentration and abscisic acid accumulation. Scientia Horticulturae, 229, 167–174. DOI 10.1016/j.scienta.2017.10.009. [Google Scholar] [CrossRef]
61. Ancillotti, C., Bogani, P., Biricolti, S., Calistri, E., Checchini, L. et al. (2015). Changes in polyphenol and sugar concentrations in wild type and genetically modified Nicotiana langsdorffii Weinmann in response to water and heat stress. Plant Physiology and Biochemistry, 97, 52–61. DOI 10.1016/j.plaphy.2015.09.012. [Google Scholar] [CrossRef]
62. Riaz, U., Kharal, M. A., Murtaza, G., uz Zaman, Q., Javaid, S. et al. (2019). Prospective roles and mechanisms of caffeic acid in counter plant stress: A mini review. Pakistan Journal of Agricultural Research, 32(1), 8. DOI 10.17582/journal.pjar/2019/32.1.8.19. [Google Scholar] [CrossRef]
63. Chen, Z., Ma, Y., Weng, Y., Yang, R., Gu, Z. et al. (2019). Effects of UV-B radiation on phenolic accumulation, antioxidant activity and physiological changes in wheat (Triticum aestivum L.) seedlings. Food Bioscience, 30, 100409. DOI 10.1016/j.fbio.2019.04.010. [Google Scholar] [CrossRef]
64. Dresler, S., Wojciak-Kosior, M., Sowa, I., Stanislawski, G., Bany, I. et al. (2017). Effect of short-term Zn/Pb or long-term multi-metal stress on physiological and morphological parameters of metallicolous and nonmetallicolous Echium vulgare L. populations. Plant Physiology and Biochemistry, 115, 380–389. DOI 10.1016/j.plaphy.2017.04.016. [Google Scholar] [CrossRef]
65. Chen, S., Lin, R., Lu, H., Wang, Q., Yang, J. et al. (2020). Effects of phenolic acids on free radical scavenging and heavy metal bioavailability in Kandelia obovata under cadmium and zinc stress. Chemosphere, 249, 126341. DOI 10.1016/j.chemosphere.2020.126341. [Google Scholar] [CrossRef]
66. Abou-Elwafa, S. F., Shehzad, T. (2021). Genetic diversity, GWAS and prediction for drought and terminal heat stress tolerance in bread wheat (Triticum aestivum L.). Genetic Resources and Crop Evolution, 68(2), 711–728. DOI 10.1007/s10722-020-01018-y. [Google Scholar] [CrossRef]
67. Isayenkov, S. V., Maathuis, F. J. M. (2019). Plant salinity stress: Many unanswered questions remain. Frontiers in Plant Science, 10, 80. DOI 10.3389/fpls.2019.00080. [Google Scholar] [CrossRef]
68. Alqahtani, M., Roy, S. J., Tester, M. A. (2018). Increasing salinity tolerance of crops. New York: Springer. [Google Scholar]
69. Mei, Y., Sun, H., Du, G., Wang, X., Lyu, D. (2020). Exogenous chlorogenic acid alleviates oxidative stress in apple leaves by enhancing antioxidant capacity. Scientia Horticulturae, 274, 109676. DOI 10.1016/j.scienta.2020.109676. [Google Scholar] [CrossRef]
70. Bistgani, Z. E., Hashemi, M., DaCosta, M., Craker, L., Maggi, F. et al. (2019). Effect of salinity stress on the physiological characteristics, phenolic compounds and antioxidant activity of Thymus vulgaris L. and Thymus daenensis Celak. Industrial Crops and Products, 135, 311–320. DOI 10.1016/j.indcrop.2019.04.055. [Google Scholar] [CrossRef]
71. Yan, K., Cui, M., Zhao, S., Chen, X., Tang, X. (2016). Salinity stress is beneficial to the accumulation of chlorogenic acids in honeysuckle (Lonicera japonica Thunb.). Frontiers in Plant Science, 7, 1563. DOI 10.3389/fpls.2016.01563. [Google Scholar] [CrossRef]
72. Olsen, K. M., Lea, U. S., Slimestad, R., Verheul, M., Lillo, C. (2008). Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. Journal of Plant Physiology, 165(14), 1491–1499. DOI 10.1016/j.jplph.2007.11.005. [Google Scholar] [CrossRef]
73. de Santana Costa, M. G., Feltrim, D., Mazzafera, P., Balbuena, T. S. (2020). Revisiting the stem proteome of Eucalyptus grandis and Eucalyptus globulus: Identification of temperature-induced changes. Biochimica et Biophysica Acta (BBA)–Proteins and Proteomics, 1868(12), 140530. DOI 10.1016/j.bbapap.2020.140530. [Google Scholar] [CrossRef]
74. Liu, C., Xu, X., Kan, J., Cheng, Z. M., Chang, Y. et al. (2020). Genome-wide analysis of the C3H zinc finger family reveals its functions in salt stress responses of Pyrus betulaefolia. PeerJ, 8, e9328. DOI 10.7717/peerj.9328. [Google Scholar] [CrossRef]
75. Jiang, A. L., Xu, Z. S., Zhao, G. Y., Cui, X. Y., Chen, M. et al. (2014). Genome-wide analysis of the C3H zinc finger transcription factor family and drought responses of members in Aegilops tauschii. Plant Molecular Biology Reporter, 32(6), 1241–1256. DOI 10.1007/s11105-014-0719-z. [Google Scholar] [CrossRef]
76. Zhang, F., Ji, S., Wei, B., Cheng, S., Wang, Y. et al. (2020). Transcriptome analysis of postharvest blueberries (Vaccinium corymbosum ‘Duke’) in response to cold stress. BMC Plant Biology, 20(1), 1–20. DOI 10.1186/s12870-020-2281-1. [Google Scholar] [CrossRef]
77. Lee, S. C., Lim, M. H., Kim, J. A., Lee, S. I., Kim, J. S. et al. (2008). Transcriptome analysis in Brassica rapa under the abiotic stresses using Brassica 24K oligo microarray. Molecules & Cells, 26(6595–605. [Google Scholar]
78. Tohge, T., Watanabe, M., Hoefgen, R., Fernie, A. R. (2013). The evolution of phenylpropanoid metabolism in the green lineage. Critical Reviews in Biochemistry and Molecular Biology, 48(2), 123–152. DOI 10.3109/10409238.2012.758083. [Google Scholar] [CrossRef]
79. Chen, X., Fang, X., Zhang, Y., Wang, X., Zhang, C. et al. (2019). Overexpression of a soybean 4-coumaric acid: Coenzyme A ligase (GmPI4L) enhances resistance to Phytophthora sojae in soybean. Functional Plant Biology, 46(4), 304–313. DOI 10.1071/FP18111. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |