

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.017811
ARTICLE
OsMAPK6 Affects Male Fertility by Reducing Microspore Number and Delaying Tapetum Degradation in Oryza sativa L.
Key Laboratory of Application and Safety Control of Genetically Modified Crops, College of Agronomy and Biotechnology, Southwest University, Chongqing, 100715, China
*Corresponding Authors: Guanghua He. Email: heghswu@163.com; Nan Wang. Email: wangnan_xndx@126.com
#Wuzhong Yin & Hongxia Yang contributed equally to this work
Received: 08 June 2020; Accepted: 05 August 2021
Abstract: The mitogen-activated protein kinase (MAPK) cascade is important in stress signal transduction and plant development. In the present study, we identified a rice (Oryza sativa L.) mutant with reduced fertility, Oryza sativa mitogen-activated protein kinase 6 (osmapk6), which harbored a mutated MAPK gene. Scanning and transmission electron microscopy, quantitative RT-PCR analysis, TUNEL assays, RNA in situ hybridization, longitudinal and transverse histological sectioning, and map-based cloning were performed to characterize the osmapk6 mutant. The gene OsMAPK6 was expressed throughout the plant but predominantly in the microspore mother cells, tapetal cells, and microspores in the anther sac. Compared with the wild type, the total number of microspores was reduced in the osmapk6 mutant. The formation of microspore mother cells was reduced in the osmapk6 anther sac at an early stage of anther development, which was the primary reason for the decrease in the total number of microspores. Programmed cell death of some tapetal cells was delayed in osmapk6 anthers and affected exine formation in neighboring microspores. These results suggest that OsMAPK6 plays pivotal roles in microspore mother cell formation and tapetal cell degradation.
Keywords: Mitogen-activated protein kinase; tapetal programmed cell death; microspore mother cell formation; male sterility; rice
Rice is both a model plant for crop research and a globally important food crop. The manipulation of microspore fertility is important for rice grain yield [1]. Microspores are generated by the meiotic division of microspore mother cells in the anther sac, and the peripheral tapetum (the inner cell layer of the anther wall) plays a pivotal role in microspore maturation, especially exine formation and assimilate accumulation of microspores [1]. The exine is the outer layer of the microspore wall and plays a pivotal role in microspore surviving, and it is mainly composed of sporopollenin, which is secreted by the tapetal cells [2]. At the later stage of anther development, the microspores are filled with nutrients such as sugars and lipids, which are also secreted by the tapetal cells [3,4]. The secretion of tapetum is affected by the programmed cell death (PCD) of it [1], and a suite of genes involved in tapetal PCD have been identified in rice. TDR INTERACTING PROTEIN2 interact with TAPETUM DEGENERATION RETARDATION and cooperatively regulate the expression of ETERNAL TAPETUM1, a basic helix-loop-helix transcription factor and positively regulates PCD in tapetal cells [5–8]. The mutation of MICROSPORE AND TAPETUM REGULATOR1, a fasciclin glycoprotein gene that specifically expressed in the male reproductive cells, retards the PCD of tapetum [9]. The mutation of PERSISTENT TAPETAL CELL1, the homologous gene of Arabidopsis thaliana MALE STERILITY1, also delays the PCD of tapetum [10]. APOPTOSIS INHIBITOR5, a nuclear protein that interacts with API5-INTERACTING PROTEIN1 and API5-INTERACTING PROTEIN2, affects the tapetum PCD through the regulation of an Oryza sativa CYSTEINE PROTEASE1 expression [11]. OsMADS3, a Oryza sativa MADS-box gene, influences the tapetum PCD through reactive oxygen species homeostasis [12]. A tapetum-specific gene in Oryza sativa, RTS, affects the development of tapetum [13]. GAMYB, a MYB transcriptional factor involved in gibberellins signaling regulation, affects the tapetum PCD through gibberellins regulation [14]. Compared with genes involved in tapetal PCD, genes involved in microspore mother cells formation are few reported in rice. MULTIPLE SPOROCYTE1 (MSP1), a Leu-rich repeat receptor-like protein kinase, controls the early sporogenic development [15]. MICROSPORELESS2, the homologous protein of Arabidopsis thaliana TAPETUM DETERMINANT1, binds MSP1 and limits sporocyte numbers [16,17]. The importance of manipulation of microspore fertility motivates the need for research on other genes that regulate pollen development [18].
The mitogen-activated protein kinase (MAPK) cascade is involved in biotic and abiotic stress responses, and participates in the regulation of plant development [19–21]. As an important member of the MAPK cascade in Arabidopsis thaliana L., AtMAPK6 participates in microspore mother-cell formation through phosphorylation of SPOROCYTELESS (SPL), which is an essential regulator of diploid germ cell differentiation in the stamens and ovules [22,23]. The atmapk6 mutant develops abnormal anthers and pollen grains, which results in decreased male fertility [21]. In rice, OsMAPK6 is a vital component of the MAPK cascade and shows 84.5% identity with AtMAPK6 [24]. The OsMKK4-OsMAPK6 cascade plays a crucial role in defense responses triggered by microbe-associated molecular patterns [20]. OsMAPK6 is activated in response to a moderately low temperature and is phosphorylated directly by OsMKK6 in vitro [19]. The bHLH transcription factor RAC IMMUNITY 1 is activated by OsRac1 via the OsMKK4-OsMAPK3/6 cascade in rice immunity [25]. The OsMKK4-OsMAPK6-OsWRKY53 cascade is important in the response to wounding [26]. OsMAPK6 regulates grain size in rice by means of cell proliferation and the brassinolide pathway [24], and is essential for cell differentiation during early embryogenesis in rice [27]. The OsMKKK10-OsMKK4-OsMAPK6 signaling pathway positively regulates grain size and weight in rice [28]. The GSN1-OsMKKK10-OsMKK4-OsMAPK6 module coordinates the trade-off between grain number and grain size by integration of localized cell differentiation and proliferation [29], and the OsER1-OsMKKK10-OsMKK4-OsMAPK6 pathway is required to maintain cytokinin homeostasis [30]. However, no previous study has focused on the effect on the male fertility of OsMAPK6.
In this study, we identified a mutant with reduced male fertility, osmapk6, which exhibited a decreased total number of microspores in the anther. Longitudinal sections of anthers indicated that the number of microspore mother cells was reduced in osmapk6 anthers, which was the primary reason for the decrease in the number of microspores. Further investigations showed that degradation of some tapetal cells was delayed and resulted in the abortion of adjacent microspores.
The rice osmapk6 mutant was isolated from an ethyl methanesulfonate (EMS)-induced mutant library of ‘Xilong1B’ (O. sativa L. indica). Plants of the mutant were grown in the experimental paddy fields (latitude 29.59° N, longitude 106.54° E) of Southwest University, Chongqing, China.
2.2 Phenotypic Analysis of the Osmapk6 Mutant
An Olympus MVX10 stereomicroscope and a Canon EOS 5D digital camera were used to examine and photograph the plant and floral morphology. Mature pollen grains from six individual anthers were observed and photographed using an Olympus BX53 microscope after staining with 1% iodine–potassium iodide (I2-KI) solution.
2.3 Preparation of Histological Sections
Anthers at developmental stages were selected based on the length and color of the spikelet, and fixed in 4% (w/v) paraformaldehyde and 0.25% (v/v) glutaraldehyde in 0.1 M sodium phosphate buffer (PBS; pH 7.2) for 16 h at 4°C, before being washed and post-fixed with 1% osmic acid for 2 h. After dehydration through a graded ethanol series and acetone replacement, the anthers were embedded in Epon 812 resin and polymerized at 60°C. Longitudinal or transverse sections of 2 μm thickness were cut using a Leica RM2265 microtome, stained with 1% toluidine blue O, then observed and photographed with an Olympus BX53 microscope.
2.4 Statistical Analysis of Microspore Number
Anthers at developmental Stages 9–14 were selected based on the length and color of the spikelet, and the stage was confirmed by observation of transverse sections. Eighteen anthers were crushed in fine quartz sand in 300 μl PBS, and the released microspores were counted using a hemocytometer. The microspore mother cells were counted by observation of longitudinal sections of anthers. Student’s t-test was performed using SPSS 26.0 statistical software (IBM Corporation, Armonk, NY, USA).
2.5 Electron Microscopic Examination
For scanning electron microscopy (SEM), mature anthers and pollen grains were examined using a Hitachi SU3500 scanning electron microscope. The Hitachi SU3500 scanning electron microscope is suitable for direct detecting. The anther samples were selected and adhered on a strip, then transported into the scanning electron microscope and frozen to −20°C immediately, and detected at 5 kV. For transmission electron microscopy (TEM), the samples were prepared as described to examine the histological sections. Ultra-thin sections of 50 nm were cut using a Leica UC6 microtome, and picked up by a mesh copper, then double-stained with 2% uranyl acetate (w/v) and 2.6% lead citrate aqueous (w/v) solution, and examined with a Hitachi H-7500 transmission electron microscope at 80 kV.
Spikelets at different developmental stages were selected from wild-type and osmapk6 mutant plants, and fixed in 1.9% formaldehyde, 5% acetic acid, and 50% ethanol in distilled water for 48 h at 4°C. After dehydration through a graded ethanol series, the samples were embedded in Paraplast Plus® (Sigma-Aldrich), and sections of 5 μm thickness were cut using a Leica RM2245 microtome. The sections were treated using the DeadEnd™ Fluorometric TUNEL System (Promega) in accordance with the supplier’s instructions. After deparaffinization with xylene and rehydration through a graded ethanol series, the sections were incubated with a terminal deoxynucleotidyl transferase and fluorescein isothiocyanate (FITC)-labeled nucleotide mix at 37°C for 1 h, then washed, and incubated with propidium iodide (PI). The samples were observed and photographed using a Zeiss LSM800 confocal microscope in the FITC (excitation 488 nm, emission 520 ± 20 nm) and PI (excitation 488 nm, emission 640 ± 40 nm) channels.
2.7 Quantitative RT-PCR Analysis
Total RNA was isolated from the root, stem, leaf, sheath, and spikelet of rice at the flowering stage, and from spikelets at various developmental stages using the Eastep® Super Total RNA Extraction Kit (Promega). Spikelets were selected at different developmental stages based on length and color. The purified total RNA was used to synthesize the oligo(dT)-primed first-strand cDNA using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Promega). The first-strand cDNA was used for quantitative RT-PCR (qRT-PCR) analysis using TB Green™ Premix Ex Taq™ II (Takara) and the CFX Connect Real-Time PCR Detection System (Bio-Rad) following the suppliers’ instructions.
The procedures of qRT-PCR:
Step 1 95°C 30 s, Plate read
Step 2 95°C 5 s
Step 3 60°C 20 s, Plate read, Go to step 2 with 40 cycle
Step 4 Melt curve, 65°C to 95°C, Increment 0.5°C for 5 s, Plate read, End
Statistical analysis was performed using SPSS 26.0. The primers of OsMAPK6, OsMADS3, RTS and Actin, an internal reference gene, are listed in Supplemental Table 1.
Wild-type spikelets at different developmental stages were selected and fixed in 3.8% formaldehyde, 5% acetic acid, and 50% ethanol for 16 h at 4°C. After dehydration through a graded ethanol series, the samples were embedded in Paraplast Plus® (Sigma-Aldrich), and transverse sections of 8 μm thickness were cut using a Leica RM2245 microtome in preparation for RNA in situ hybridization. A 320 bp gene-specific cDNA amplified from the full-length OsMAPK6 cDNA was transcribed and labeled in vitro with the DIG RNA Labeling Kit (Roche) using a general method for the preparation of probes for floral organ genes. Pretreatment of sections, hybridization, and immunological detection were performed as described previously [31]. The primers used are listed in Supplemental Table 1.
2.9 Identification of the Mutated Gene
The F2 mapping population was raised from a cross between ‘Jinhui 10’ and the osmapk6 mutant. Individuals in the F2 population that exhibited reduced male fertility were selected. A suite of insertion/deletion (InDel) molecular markers that were polymorphic between ‘Jinhui 10’ and ‘Xilong 1B’ were used for mapping. Statistical analysis was performed using SPSS 26.0. For functional complementation, the coding sequence of OsMAPK6 was cloned into the binary vector PTCK303 driven by the Ubiquitin promoter. Calli induced from osmapk6 mutant seeds were used for transformation with Agrobacterium tumefaciens (strain EHA105) carrying PTCK303-OsMAPK6. Transgenic plants were then confirmed by sequencing and subject to analysis. The primers used are listed in Supplemental Table 1.
3.1 The Osmapk6 Mutant Exhibited Reduced Male Fertility
We isolated the osmapk6 mutant from an EMS-induced rice mutant library on account of its reduced male fertility and dwarf phenotypes. The osmapk6 mutants do not exhibited phenotypic segregation. Compared with the wild type, the mutant exhibited a reduced plant height and produced few filled grains at the maturity stage (Figs. 1A and 1B). The spikelets and anthers of the osmapk6 mutant were smaller than those of the wild type (Figs. 1C and 1D; Supplemental Figs. 1A–1D). Staining of pollen grains in the anthers with I2-KI solution revealed a reduced number of pollen grains, and some collapsed pollen grains were observed (Figs. 1E and 1F). We found that the pollen grains in the wild type anthers showed 89.8% fertility while those in osmapk6 anthers had 48.3% fertility (Fig. 1G).
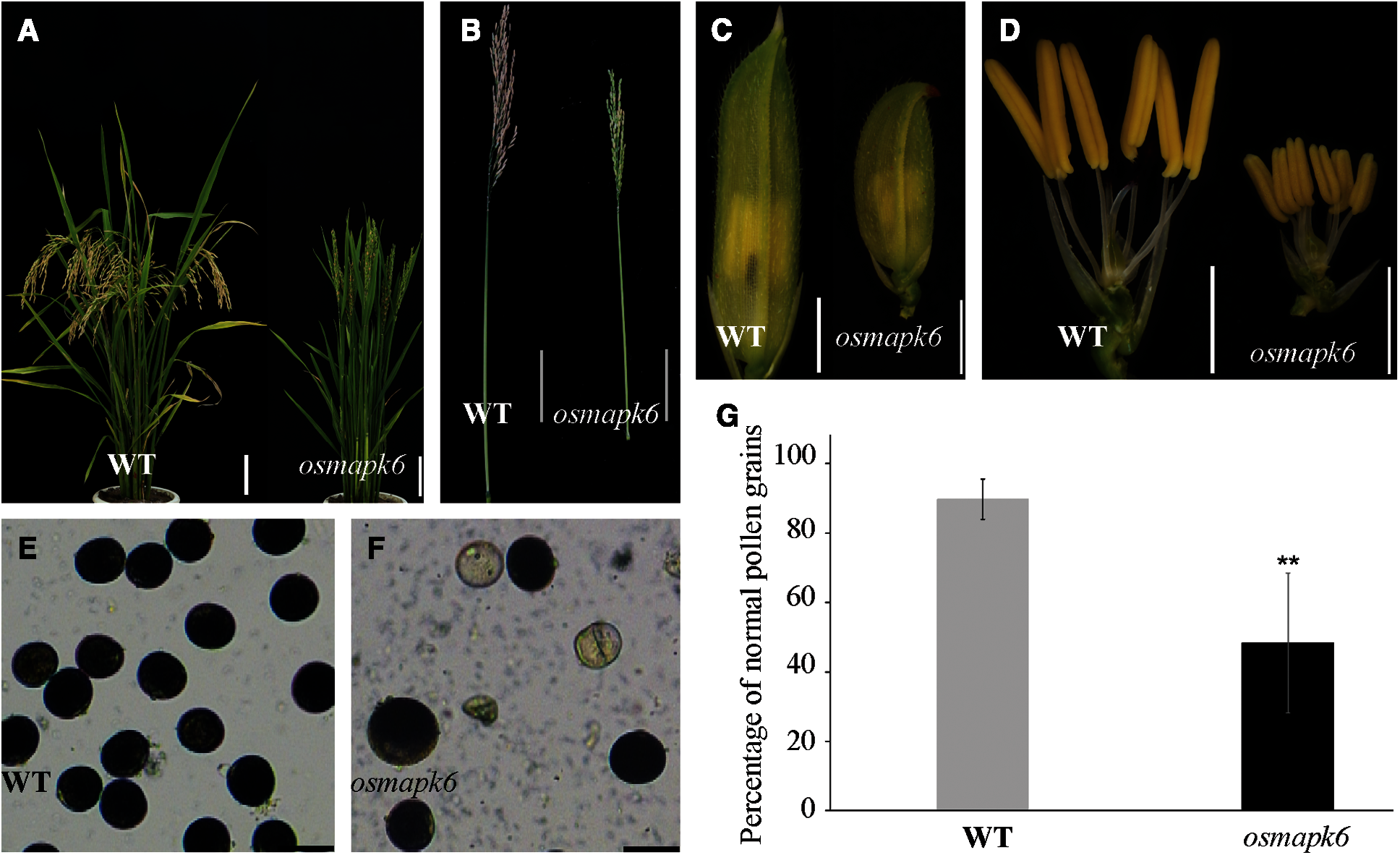
Figure 1: Male fertility of the osmapk6 mutant. (A) plant phenotype at the seed maturation stage. (B) Panicle at the seed maturation stage. (C) spikelet before anthesis. (D) spikelet before anthesis with palea and lemma removed. (E and F) Pollen grains at Stage 12 stained with I2-KI. (G) Fertility of pollen gains detected by I2-KI. Error bars represent the SD (n = 6). * p < 0.01. WT, wild type. Bars = 10 cm (A and B), 2 mm (C and D), 50 μm (E and F)
3.2 Anthers of the Osmapk6 Mutant Contained Fewer Microspores
The number of pollen grains in the osmapk6 anther was counted using a hemocytometer. Based on a recent classification of anther developmental stages, the anthers were classified into 14 developmental stages in this study [1]. The total number of microspores per anther in the wild type was 2048, 2104, 2056, 2354, and 1992 at the Stages 9–10, 10–11, 11–12, 12–13, and 13–14, respectively. Compared with the wild type, the total number of microspores per anther in the osmapk6 mutant was reduced by as much as 70.2%, 60.4%, 50.5%, 63.0%, and 63.7% at the Stages 9–10, 10–11, 11–12, 12–13, and 13–14, respectively (Fig. 2A, Supplemental Tab. 2). Before their release from tetrads at Stage 9, the microspores were weak and difficult to isolate due to the lack of a pollen wall [2]. Therefore, we prepared longitudinal and transverse sections of anthers to count the number of microspore mother cells. At Stage 7, the transverse sections did not reveal a distinct difference in the number of microspore mother cells between the wild type and the osmapk6 mutant (Figs. 3A and 3E), whereas longitudinal sections did reveal significant differences (Figs. 2B and 2C). In the longitudinal sections of wild-type anther sac, almost 39 microspore mother cells were counted (Supplemental Tab. 3), whereas the number was reduced by as much as 47.8% in the osmapk6 anther sac (Fig. 2D).
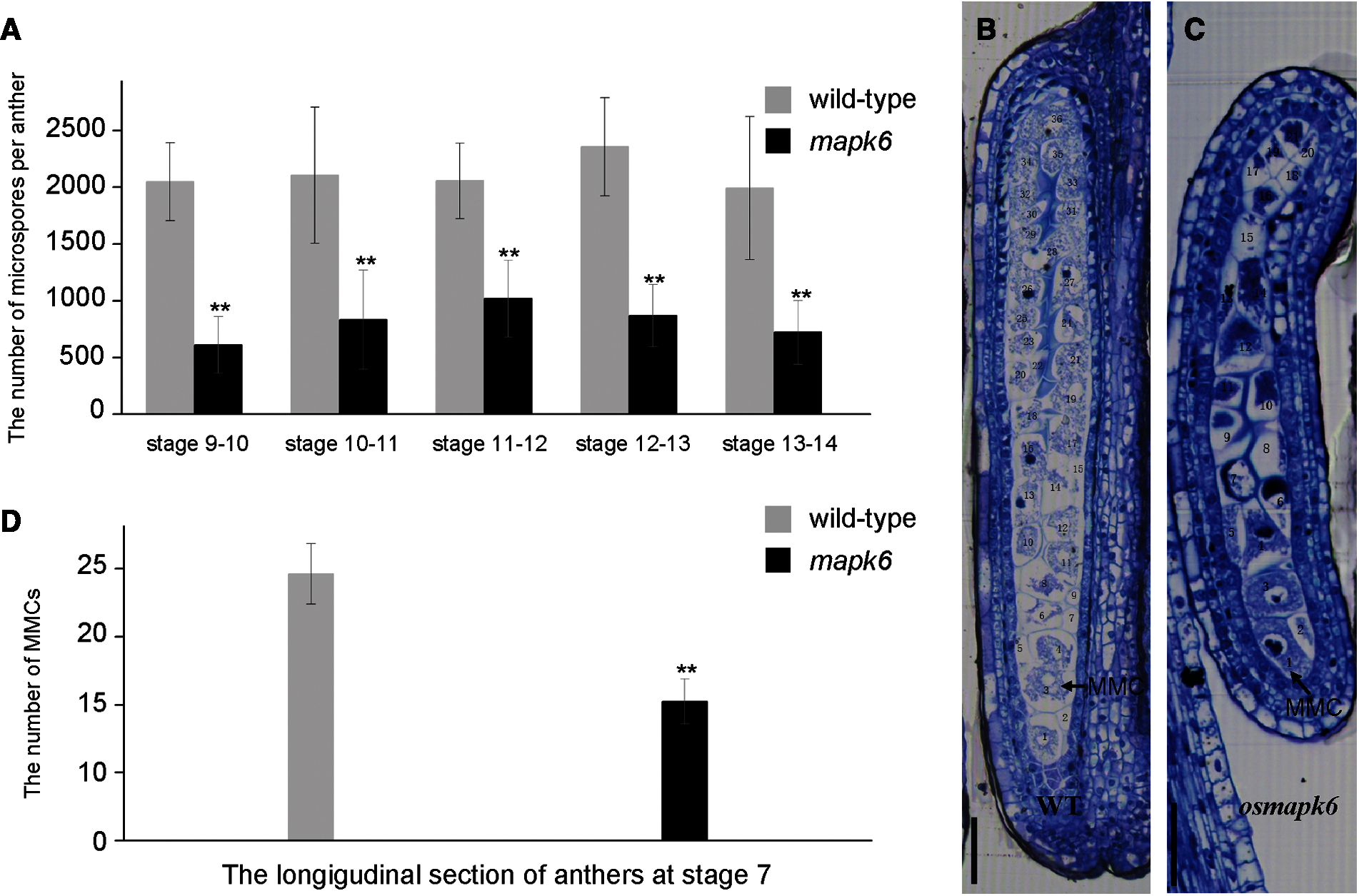
Figure 2: Total number of microspores per anther in the osmapk6 mutant. (A) total number of microspores per anther. Error bars represent the SD (n = 8). The experiment was repeated three times with similar results. * p < 0.01. (B and C) longitudinal sections of wild-type and osmapk6 anthers, respectively. (D) Number of microspore mother cells in longitudinal sections of wild-type and osmapk6 anthers. Error bars represent the SD (n = 4). * p < 0.01. MMC, microspore mother cell. Bars = 50 μm (B and C)
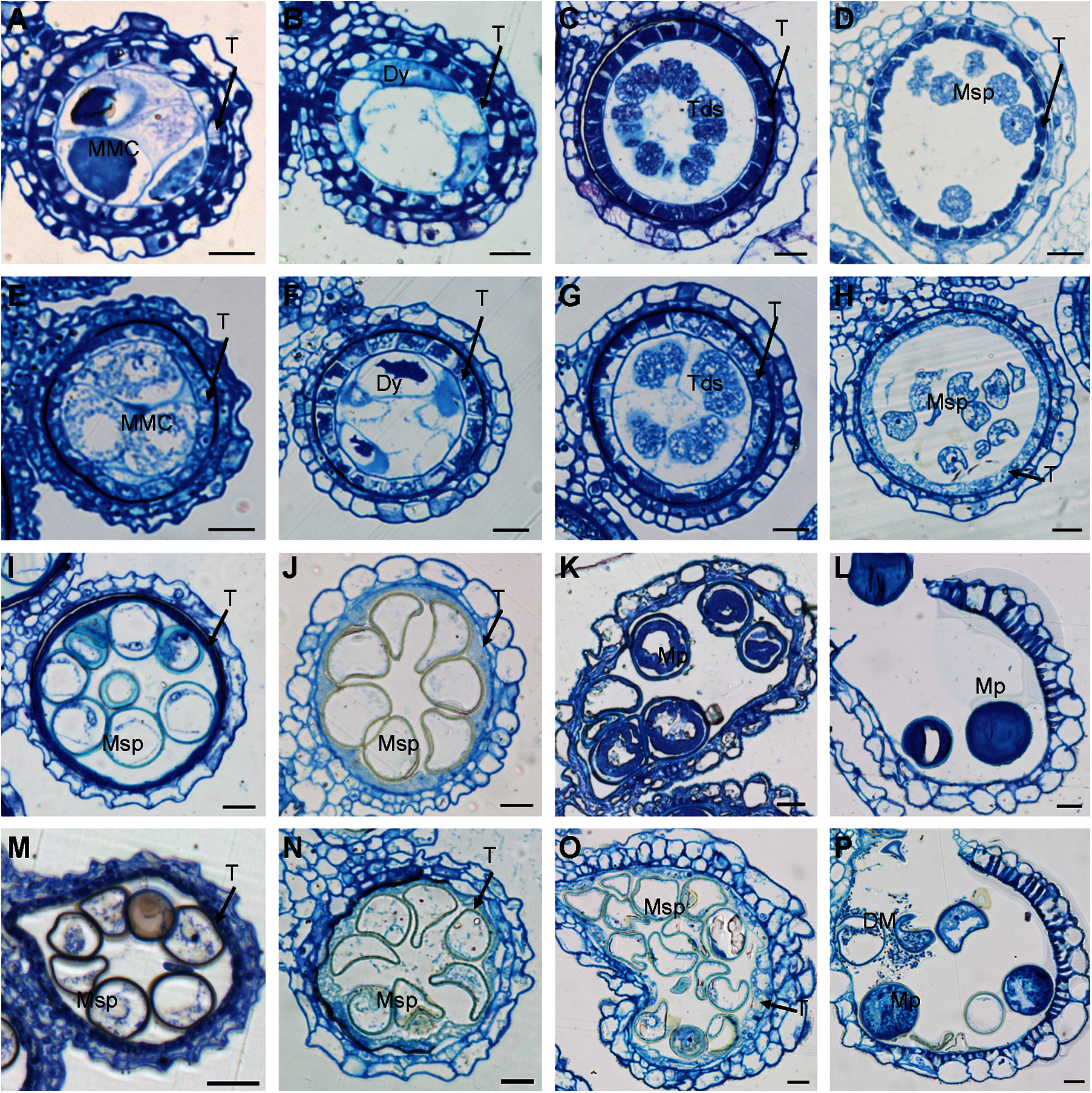
Figure 3: Transverse sections of osmapk6 anthers. (A to D, I to L) Wild-type anthers. (E to H, M to P) osmapk6 anthers. Anthers were sampled at Stages 7 (A and E), 8a (B and F), 8b (C and G), 9 (D and H), 10 (I and M), 11 (J and N), 12 (K and O), and 14 (L and P). DM, degraded microspore; Dy, dyad cell; MMC, microspore mother cell; Msp, microspore; Mp, mature pollen; T, tapetum; Tds, tetrads. Bars = 10 μm
3.3 Anthers of the Osmapk6 Mutant Displayed Abnormal Tapetal Cell Degradation
Transverse sections were observed to determine the morphological defects in osmapk6 anthers. No difference between the wild type and osmapk6 anthers was detected at Stages 7 to 11 (Figs. 3A–3J, 3M, and 3N). At Stage 7, the microspore mother cells were formed and initiated the meiotic division (Figs. 3A and 3E). At Stage 8a, the microspore mother cells continued meiosis I and formed dyads, and PCD of the tapetal cells was initiated (Figs. 3B and 3F). At Stage 8b, the microspore mother cells continued meiosis II and formed tetrads (Figs. 3C and 3G). At Stage 9, the microspores were released from the tetrads, and the tapetal cells became condensed and darkly stained (Figs. 3D and 3H). At Stage 10, the microspores were vacuolated and spherical, and the tapetal cells were more condensed and darkly stained (Figs. 3I and 3M). At Stage 11, the vacuolated microspores underwent the asymmetric first mitotic cell division and were falcate-shaped, and the tapetum degenerated into cellular debris on the internal surface of the anther wall (Figs. 3J and 3N). At Stage 12, the wild-type pollen grains underwent the second mitotic division and were nutrient-rich, and the tapetum had completely degenerated (Fig. 3K). In contrast, the osmapk6 anther retained a highly vacuolated tapetum and the pollen grains showed delayed filling (Fig. 3O). At stage 14, the wild-type anthers dehisced and released mature pollen grains (Fig. 3L), whereas the osmapk6 anthers released viable and aborted pollen grains (Fig. 3P).
3.4 The Abnormal Tapetum of the Osmapk6 Mutant Affected Exine Formation of Neighboring Microspores
We used TEM to explore further the anther defects in the osmapk6 mutant revealed by the aforementioned phenotypic analysis. At Stage 10, in the wild-type anthers, the highly vacuolated microspores were in contact with the tapetum, and the microspore exine was composed of the tectum, bacula, and nexine. In addition, the tapetum was condensed and the Ubisch bodies were darkly stained and dispersed on the inner surface of tapetal cells (Figs. 4A and 4B). In osmapk6 anthers, some tapetal cells and neighboring microspores were similar to those of the wild type (Figs. 4C and 4D); however, some tapetal cells were highly vacuolated and detached from the inner surface of the anther wall, and exine formation of adjacent microspores was retarded (Figs. 4E and 4F). Then, we used SEM to explore the inner surface of anthers. The Ubisch bodies were uniformly dispersed on the inner surface of the tapetum of the wild-type anthers, whereas the inner surface of the tapetum in the osmapk6 anthers displayed abnormal humps, which indicated abnormal tapetum degeneration in the osmapk6 anthers (Figs. 4G and 4H).
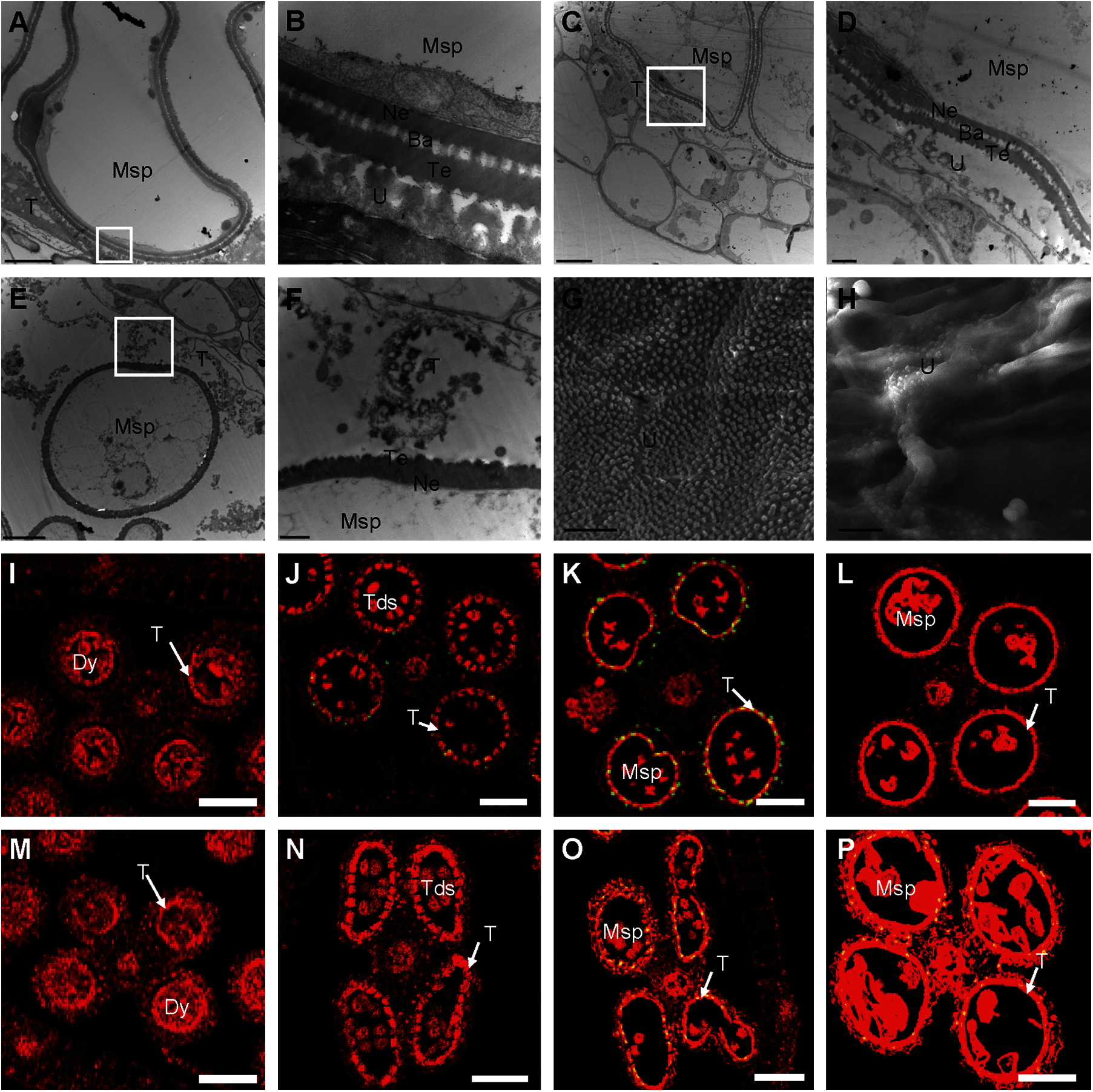
Figure 4: Electron microscopic examination and TUNEL assay of osmapk6 anthers. (A) transmission electron micrograph of wild-type anther at Stage 10. (B) Higher-magnification image of the region enclosed by the white box in (A). (C and E) Transmission electron micrographs of osmapk6 anthers at Stage 10. (D and F) Higher-magnification images of the region enclosed by the white box in (C) and (E), respectively. (G and H) Scanning electron micrographs of the inner surface of the tapetum of wild-type and osmapk6 anthers, respectively. (I to L) TUNEL assay of wild-type anthers. (M to P) TUNEL assay of osmapk6 anthers. The yellow points represent positive signals. Anthers were sampled at Stages 8a (I and M), 8b (J and N), 9 (K and O), and 10 (L and P). Ba, bacula; Dy, dyad cell; Msp, microspore; Ne, nexine; T, tapetum; Tds, tetrads; Te, tectum; U, Ubisch body. Bars = 5 μm (A, C, E, G, and H), 1 μm (B, D, and F), 50 μm (I to P)
3.5 The Osmapk6 Tapetum Exhibited Delayed PCD
Tapetum degeneration is considered to be the result of tapetal PCD, which is characterized by genomic DNA cleavage [8]. To examine whether tapetal cell PCD was abnormal in osmapk6 anthers, we performed a terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay on the wild-type and osmapk6 anthers. In the wild-type anther, positive signals commenced at Stage 8b and became intense at Stage 9, whereas no positive signals were detected at Stages 8a and 10 (Figs. 4I to 4L). By contrast, in the osmapk6 anther, positive signals were detected at Stages 9 and 10, and no positive signals were detected at Stages 8a and 8b (Figs. 4M–4P). These results suggest that PCD of the tapetal cells in the osmapk6 anther was slightly delayed.
3.6 Identification of the Mutated Gene
Map-based cloning was conducted to identify the mutated gene in the osmapk6 mutant. All F1 individuals from a cross between the osmapk6 mutant and rice ‘Jinhui 10’ showed a normal phenotype. In the F2 population, the phenotypic segregation was approximately 3:1 (normal phenotypes:reduced fertility and dwarfing = 729: 207, χ2 = 0.019, p > 0.05), which suggests that the mutant phenotype was caused by a single recessive mutation. The F2 mapping population of 207 individuals exhibiting the mutant phenotype was used for map-based cloning. We found the linkage relationship between the target gene and the molecular marker RM19552 on chromosome 6. Further linkage analysis was performed and we found that the recombinants decreased at the RM19549 (38 recombinants), ZTQ52 (23 recombinants) and In6-24 (4 recombinants) loci. In addition, we identified another 7 recombinants at the Chr6-1 loci, and the recombinants decreased at the Chr6-2 (5 recombinants) and Ind6-17 (1 recombinant) loci. These results suggest that the mutant gene was located in the 188 kb region between Ind6-17 and In6-24 (Fig. 5A). By sequencing candidate genes within this region, we detected a transition of “G” to “A” in the fourth exon of LOC_Os06g06090, which resulted in an amino acid change of “Gly” to “Glu” (Fig. 5A). Then, we performed functional complementation assay, and the complemented lines developed normal spikelets and anthers (Figs. 5B–5D), and the male fertility was partially rescued (Fig. 5E). Therefore, we suggested that LOC_Os06g06090 was OsMAPK6.
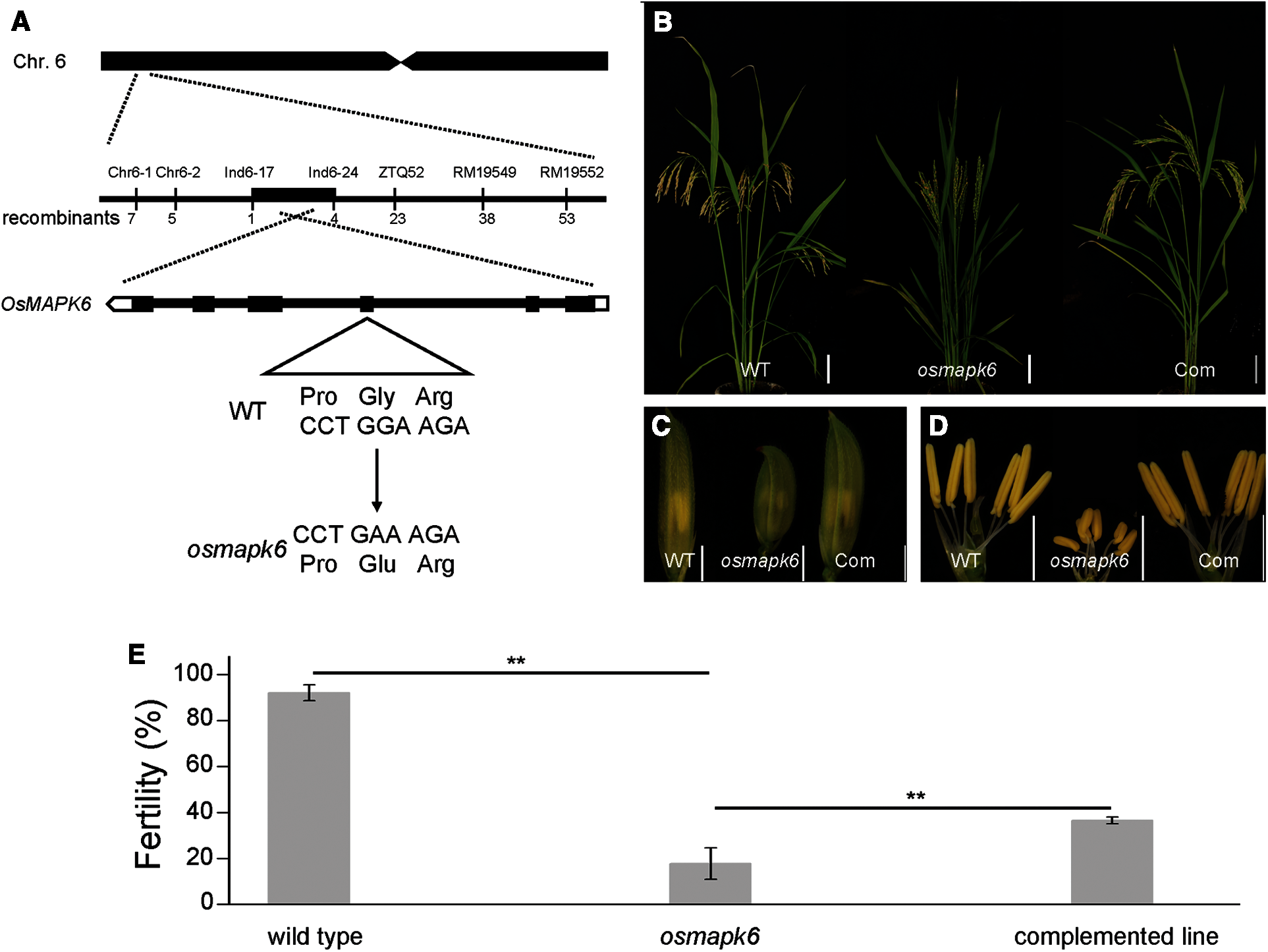
Figure 5: Identification of the mutated gene in the osmapk6 mutant. (A) Fine-mapping of the mutated gene to a 188 kb region on chromosome 6 of the osmapk6 mutant. The mutated sequence shows a transition of “G” to “A” in the fourth exon and results in an amino acid change of “Gly” to “Glu”. (B) Plant phenotypes of the wild type, osmapk6 and complemented line at the seed maturation stage, respectively. (C) Spikelets of the wild type, osmapk6 mutant, and complemented line, respectively. (D) Anthers of the wild type, osmapk6 mutant, and complemented line, respectively. (E) Number of filled grains in the wild type, osmapk6 mutant, and complemented line, respectively. Error bars represent the SD (n = 4). * P < 0.01. Com, complemented line. Bars = 10 cm in (B), 2 mm (C and D)
To investigate the spatiotemporal expression pattern of OsMAPK6 in rice, qRT-PCR and RNA in situ hybridization analyses were performed. The qRT-PCR results indicated that OsMAPK6 was expressed in the root, stem, leaf, sheath, and spikelet with a small range of variation (Fig. 6A). RNA in situ hybridization revealed more detailed information on OsMAPK6 expression in the anther sac. At Stages 6 to 8a, OsMAPK6 was expressed in the male reproductive cells and tapetal cells (Figs. 6B–6G). At Stage 8b, OsMAPK6 was predominantly expressed in the tetrads (Figs. 6H and 6K). At Stage 9, OsMAPK6 was expressed in the microspores and tapetum (Figs. 6I and 6L). At Stage 10, OsMAPK6 was weakly expressed in the anther sac (Figs. 6J and 6M). The sense probe for OsMAPK6 was used as a negative control.

Figure 6: Expression analysis. (A) Relative expression level of OsMAPK6 in the root, stem, leaf, sheath, and spikelet of 90-day-old wild-type plants. Error bars represent the SD (n = 3). The experiment was repeated three times with similar results. (B to D, H to J) Wild-type anthers hybridized with antisense DIG-labeled probe of OsMAPK6. (E to G, K to M) Wild-type anthers hybridized with sense DIG-labeled probe of OsMAPK6 as a negative control. Anthers were sampled at Stages 6 (B and E), 7 (C and F), 8a (D and G), 8b (H and K), 9 (I and L), and 10 (J and M). (N) and (O) Expression of OsMADS3 and RTS at Stages 8b–9, 9–10, 10–11, and 11–12, respectively. Error bars represent the SD (n = 3). The experiment was repeated three times with similar results. * P < 0.01. Dy, dyad cell; MMC, microspore mother cell; Msp, microspore; T, tapetum; Tds, tetrads. Bars = 50 μm
We also investigated the expression of genes associated with advanced stages of tapetum development by means of qRT-PCR analysis. OsMADS3 participates in reactive oxygen species homeostasis at advanced stages of tapetum development [12], and RTS, a tapetum-specific gene in rice, is essential for tapetum development at advanced stages [13]. In our results, the expression of OsMADS3 was dramatically reduced at Stages 8b–9, 10–11, and 11–12 in the osmapk6 spikelet (Fig. 6N), and the expression of RTS was dramatically reduced at Stages 10–11 and 11–12 (Fig. 6O). These results also indicate that osmapk6 was defective in tapetum degradation.
OsMAPK6 was expressed in the root, stem, leaf, sheath, and spikelet of rice, but the expression was predominantly in the male reproductive cells and tapetal cells in the anther sac (Figs. 6A–6M). In previous study, the mutant of OsMAPK6 affected the homodimerization and kinase activity of it, which in turn influenced the plant height and grain size during rice development [24]. In our results, the mutant of OsMAPK6 also influenced the plant height and grain size (Supplemental Figs. 1E–1I). In addition, the osmapk6 mutant exhibited small anthers and reduced fertility phenotypes (Figs. 1B and 1D). Further studies suggested that the microspore mother cells formation and tapetal PCD were affected in osmapk6 mutant (Figs. 2D, 4K and 4O). We examined pollen grain development in the osmapk6 mutant by staining with I2-KI and observed that the total number of pollen grains was decreased in the mutant (Figs. 1E and 1F). Further investigation revealed that the total number of microspores decreased at Stages 9 to 14 with a small range in variation, which indicates that reduction in the number of microspores occurred before Stage 9. Longitudinal and transverse histological sections revealed that the reduction in the numbers of microspore mother cells and microspores was comparable, which suggests that the reduction in the number of microspore mother cells was responsible for the decrease in the number of microspores (Figs. 2A and 2D). In Arabidopsis thaliana, AtMAPK6 participated in microspore mother cell formation through phosphorylation of SPL, which was reported to affect diploid germ cell differentiation [22,23]. On the other hand, no associated genes of SPL were reported in rice. Therefore, we suggest that OsMAPK6 plays an important role in microspore mother cell formation, however, the relevant underlying mechanism remains unclear.
The I2-KI staining also revealed aborted pollen grains in the osmapk6 mutant (Figs. 1E and 1F). Transverse sections indicated no detectable difference between the wild-type and osmapk6 anthers at Stages 7 to 11, but the osmapk6 anthers displayed highly vacuolated tapetal cells and pollen grains with delayed filling at Stage 12 (Figs. 3K and 3O). Nutrient accumulation and exine formation of microspores is mainly affected by the tapetum [1]. Therefore, we considered that the abnormal tapetal degeneration might affect the filling of microspores, and focused our attention on the abnormal tapetal degeneration. The TUNEL assay showed that PCD of tapetal cells was slightly delayed in osmapk6 anthers (Figs. 4I–4P). In previous studies, the seriously abnormal PCD of tapetal cells always caused complete male sterility [1,5–7]. However, in osmapk6 anthers, the slightly delayed PCD of tapetal cells caused semi-sterility, which exhibited viable and aborted pollen grains at the same time (Figs. 1F and 3P).
The results of TEM revealed more detailed information on the tapetum degradation of osmapk6. In our results, at Stage 10, condensed and abnormal tapetal cells appeared in the osmapk6 anthers at the same time. The condensed tapetal cells were similar to those of the wild type, and the neighboring microspores exhibited complete pollen wall (Figs. 4C and 4D). By contrast, the abnormal tapetal cells were highly vacuolated and became detached from the inner surface of the anther wall in the osmapk6 mutant, and exine formation in neighboring microspores was abnormal (Figs. 4E and 4F). The exine is mainly composed of sporopollenin, which is secreted by tapetal cells [2]. In rice, OsC6 participates in the transportation of sporopollenin from the tapetum to the exine, which is specific and efficient [2,32]. Therefore, we considered that abnormal degradation of the tapetum was responsible for abnormal exine formation in adjacent microspores.
In a previous study, tapetum degeneration is considered to be the result of tapetal PCD [8]. We consider that the slightly delayed PCD in osmapk6 anthers caused abnormal degeneration of some tapetal cells, which prevented the transportation of sporopollenin from the tapetum to the exine of neighboring microspores and affected the formation of it. As exine is the outer layer of microspores and necessary for pollen survival [2], we suggest that the slightly delayed PCD in the tapetal cells was responsible for the aborted pollen grains. By contrast, the viable pollen grains were due to the other tapetal cells which exhibited normal degradation. We found that the amino acid change from “Gly” to “Glu” might appear at the surface of OsMAPK6, which is a conserved domain in various plants (Supplemental Fig. 2). We thought that the amino acid change from neutral amino acids (Gly) to acidic amino acids (Glu) may influence the charge on the surface of OsMAPK6 , which could affect the function of OsMAPK6 [33]. However, the mechanism by which only some tapetal cells underwent PCD at Stage 9 in the osmapk6 mutant needs further exploration.
Author Contributions: W.Y., G.H. and N.W. conceived and designed the experiments. W.Y. and Y.W. conducted experiments on TUNEL and RNA in situ hybridization assays. W.Y. and H.Y. performed qRT-PCR analysis, scanning and transmission electron microscopy. W.Y. and P.F. performed on the transgenic lines. W.Y. and Y.D. performed the genetic analysis and map-cloning. W.Y., L.Y., C.D., B.Y. and L.W. performed the longitudinal and transverse histological sectioning, and hemocytometer assay. W.Y., G.H. and N.W. collected and analyzed the data. W.Y. wrote the manuscript.
Funding Statement: This work was supported by the National Natural Science Foundation of China (31771750, 31730063), National Key Research and Development Project (2017YFD0100201, 2017YFD0100202), Natural Science Foundation of Chongqing, China (cstc2018jcyjAX0424).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Zhang, D. B., Luo, X., Zhu, L. (2011). Cytological analysis and genetic control of rice anther development. Journal of Genetics and Genomics, 38(9), 379–390. DOI 10.1016/j.jgg.2011.08.001. [Google Scholar] [CrossRef]
2. Shi, J. X., Cui, M. H., Yang, L., Kim, Y. J., Zhang, D. B. (2015). Genetic and biochemical mechanisms of pollen wall development. Trends in Plant Science, 20(11), 741–753. DOI 10.1016/j.tplants.2015.07.010. [Google Scholar] [CrossRef]
3. Zhang, H., Liang, W. Q., Yang, X. J., Luo, X., Jiang, N. et al. (2010). Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell, 22(3), 672–689. DOI 10.1105/tpc.109.073668. [Google Scholar] [CrossRef]
4. Pacini, E., Guarnieri, M., Nepi, M. (2006). Pollen carbohydrates and water content during development, presentation, and dispersal: A short review. Protoplasma, 228(1–3), 73–77. DOI 10.1007/s00709-006-0169-z. [Google Scholar] [CrossRef]
5. Ko, S. S., Li, M. J., Sun-Ben Ku, M., Ho, Y. C., Lin, Y. J. et al. (2014). The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell, 26(6), 2486–2504. DOI 10.1105/tpc.114.126292. [Google Scholar] [CrossRef]
6. Fu, Z., Yu, J., Cheng, X., Zong, X., Xu, J. et al. (2014). The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell, 26(4), 1512–1524. DOI 10.1105/tpc.114.123745. [Google Scholar] [CrossRef]
7. Niu, N., Liang, W., Yang, X., Jin, W., Wilson, Z. A. et al. (2013). EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nature Communications, 4, 1445. DOI 10.1038/ncomms2396. [Google Scholar] [CrossRef]
8. Li, N., Zhang, D. S., Liu, H. S., Yin, C. S., Li, X. X. et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell, 18(11), 2999–3014. DOI 10.1105/tpc.106.044107. [Google Scholar] [CrossRef]
9. Tan, H. X., Liang, W. Q., Hu, J. P., Zhang, D. B. (2012). MTR1 encodes a secretory fasciclin glycoprotein required for male reproductive development in rice. Developmental Cell, 22(6), 1127–1137. DOI 10.1016/j.devcel.2012.04.011. [Google Scholar] [CrossRef]
10. Li, H., Yuan, Z., Vizcay-Barrena, G., Yang, C. Y., Liang, W. Q. et al. (2011). PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiology, 156(2), 615–630. DOI 10.1104/pp.111.175760. [Google Scholar] [CrossRef]
11. Li, X. W., Gao, X. Q., Wei, Y., Deng, L., Ouyang, Y. D. et al. (2011). Rice APOPTOSIS INHIBITOR5 coupled with Two DEAD-box adenosine 5 ‘-Triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell, 23(4), 1416–1434. DOI 10.1105/tpc.110.082636. [Google Scholar] [CrossRef]
12. Hu, L. F., Liang, W. Q., Yin, C. S., Cui, X. A., Zong, J. et al. (2011). Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell, 23(2), 515–533. DOI 10.1105/tpc.110.074369. [Google Scholar] [CrossRef]
13. Luo, H., Lee, J. Y., Hu, Q., Nelson-Vasilchik, K., Eitas, T. K. et al. (2006). RTS, a rice anther-specific gene is required for male fertility and its promoter sequence directs tissue-specific gene expression in different plant species. Plant Molecular Biology, 62(3), 397–408. DOI 10.1007/s11103-006-9031-0. [Google Scholar] [CrossRef]
14. Aya, K., Ueguchi-Tanaka, M., Kondo, M., Hamada, K., Yano, K. et al. (2009). Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell, 21(5), 1453–1472. DOI 10.1105/tpc.108.062935. [Google Scholar] [CrossRef]
15. Nonomura, K. I., Miyoshi, K., Eiguchi, M., Suzuki, T., Miyao, A. et al. (2003). The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell, 15(8), 1728–1739. DOI 10.1105/tpc.012401. [Google Scholar] [CrossRef]
16. Hong, L. L., Tang, D., Shen, Y., Hu, Q., Wang, K. J. et al. (2012). MIL2 (MICROSPORELESS2) regulates early cell differentiation in the rice anther. New Phytologist, 196(2), 402–413. DOI 10.1111/j.1469-8137.2012.04270.x. [Google Scholar] [CrossRef]
17. Zhao, X. A., de Palma, J., Oane, R., Gamuyao, R., Luo, M. et al. (2008). OsTDL1A binds to the LRR domain of rice receptor kinase MSP1, and is required to limit sporocyte numbers. Plant Journal, 54(3), 375–387. DOI 10.1111/j.1365-313X.2008.03426.x. [Google Scholar] [CrossRef]
18. Chang, Z. Y., Chen, Z. F., Wang, N., Xie, G., Lu, J. W. et al. (2016). Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proceedings of the National Academy of Sciences of the United States of America, 113(49), 14145–14150. DOI 10.1073/pnas.1613792113. [Google Scholar] [CrossRef]
19. Xie, G. S., Kato, F., Imai, R. (2012). Biochemical identification of the OsMKK6-osMPK3 signalling pathway for chilling stress tolerance in rice. Biochemical Journal, 443, 95–102. DOI 10.1042/BJ20111792. [Google Scholar] [CrossRef]
20. Kishi-Kaboshi, M., Okada, K., Kurimoto, L., Murakami, S., Umezawa, T. et al. (2010). A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant Journal, 63(4), 599–612. DOI 10.1111/j.1365-313X.2010.04264.x. [Google Scholar] [CrossRef]
21. Bush, S. M., Krysan, P. J., (2007). Mutational evidence that the arabidopsis MAP kinase MPK6 is involved in anther, inflorescence, and embryo development. Journal of Experimental Botany, 58(8), 2181–2191. DOI 10.1093/jxb/erm092. [Google Scholar] [CrossRef]
22. Zhao, F., Zheng, Y. F., Zeng, T., Sun, R., Yang, J. Y. et al. (2017). Phosphorylation of SPOROCYTELESS/NOZZLE by the MPK3/6 kinase is required for anther development. Plant Physiology, 173(4), 2265–2277. DOI 10.1104/pp.16.01765. [Google Scholar] [CrossRef]
23. Liu, X. D., Huang, J., Parameswaran, S., Ito, T., Seubert, B. et al. (2009). The SPOROCYTELESS/NOZZLE gene is involved in controlling stamen identity in arabidopsis. Plant Physiology, 151(3), 1401–1411. DOI 10.1104/pp.109.145896. [Google Scholar] [CrossRef]
24. Liu, S. Y., Hua, L., Dong, S. J., Chen, H. Q., Zhu, X. D. et al. (2015). OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant Journal, 84(4), 672–681. DOI 10.1111/tpj.13025. [Google Scholar] [CrossRef]
25. Kim, S. H., Oikawa, T., Kyozuka, J., Wong, H. L., Umemura, K. et al. (2012). The bHLH Rac immunity1 (RAI1) is activated by osrac1 via OsMAPK3 and OsMAPK6 in rice immunity. Plant and Cell Physiology, 53(4), 740–754. DOI 10.1093/pcp/pcs033. [Google Scholar] [CrossRef]
26. Hu, L. F., Ye, M., Li, R., Zhang, T. F., Zhou, G. X. et al. (2015). The rice transcription factor WRKY53 suppresses herbivore-induced defenses by acting as a negative feedback modulator of mitogen-activated protein kinase activity. Plant Physiology, 169(4), 2907–2921. DOI 10.1104/pp.15.01090. [Google Scholar] [CrossRef]
27. Yi, J., Lee, Y. S., Lee, D. Y., Cho, M. H., Jeon, J. S. et al. (2016). OsMPK6 plays a critical role in cell differentiation during early embryogenesis in Oryza sativa. Journal of Experimental Botany, 67(8), 2425–2437. DOI 10.1093/jxb/erw052. [Google Scholar] [CrossRef]
28. Xu, R., Duan, P. G., Yu, H. Y., Zhou, Z. K., Zhang, B. L. et al. (2018). Control of grain size and weight by the OsMKKK10-osMKK4-osMAPK6 signaling pathway in rice. Molecular Plant, 11(6), 860–873. DOI 10.1016/j.molp.2018.04.004. [Google Scholar] [CrossRef]
29. Guo, T., Chen, K., Dong, N. Q., Shi, C. L., Ye, W. W. et al. (2018). GRAIN SIZE and NUMBER1 negatively regulates the OsMKKK10-osMKK4-osMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell, 30(4), 871–888. DOI 10.1105/tpc.17.00959. [Google Scholar] [CrossRef]
30. Guo, T., Lu, Z. Q., Shan, J. X., Ye, W. W., Dong, N. Q. et al. (2020). ERECTA1 Acts upstream of the OsMKKK10-osMKK4-osMPK6 cascade to control spikelet number by regulating cytokinin metabolism in rice. Plant Cell, 32(9), 2763–2779. DOI 10.1105/tpc.20.00351. [Google Scholar] [CrossRef]
31. Zhang, T., Li, Y., Ma, L., Sang, X., Ling, Y. et al. (2017). LATERAL FLORET 1 induced the three-florets spikelet in rice. Proceedings of National Academy of Sciences of the United States of America, 114(37), 9984–9989. DOI 10.1073/pnas.1700504114. [Google Scholar] [CrossRef]
32. Zhang, D. S., Liang, W. Q., Yin, C. S., Zong, J., Gu, F. W. et al. (2010). OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiology, 154(1), 149–162. DOI 10.1104/pp.110.158865. [Google Scholar] [CrossRef]
33. Wang, B., Qin, X. H., Wu, J., Deng, H. Y., Li, Y. et al. (2016). Analysis of crystal structure of arabidopsis MPK6 and generation of its mutants with higher activity. Scientific Reports, 6, 25646. DOI 10.1038/srep25646. [Google Scholar] [CrossRef]
Supplemental Material
The following supplemental materials are available:
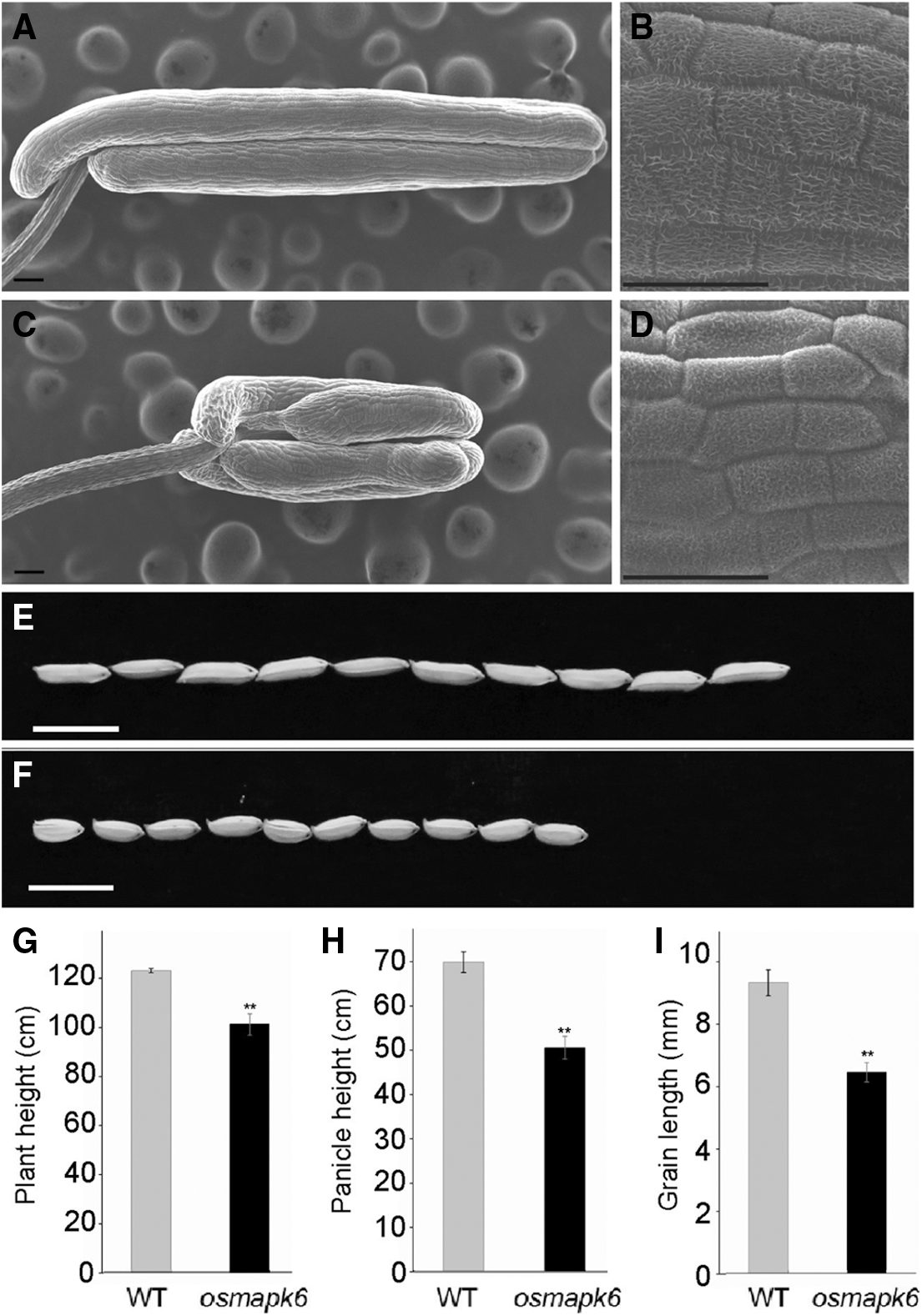
Figure S1: The phenotypic analysis of wild type and osmapk6. (A) and (C) are the scanning electron micrographs of wild type and osmapk6 anthers, respectively. (B) and (D) are the higher magnification of the surface of wild type and osmapk6 anthers, respectively. (E) and (F) are gains of wild type and osmapk6, respectively. (G), (H) and (I) are plant height, panicle height and grain length analysis, respectively. Error bars represent the SD (n = 3). ** P < 0.01. Bars = 100 μm in (A) and (C), 50 μm in (B) and (D), 10 mm in (E) and (F)

Figure S2: The predecrted model of OsMAPK6. (A) Multiple amino acid sequence alignment of LOC_Os06g06090 (Oryza sativa L.), AT2G43790 (Arabidopsis thaliana), Bradi1g49100 (Brachypodium distachyon), GRMZM2G002100 (Zea mays), Gorai.005G011100 (Gossypium raimondii) and TraesCS7A02G111300 (Triticum aestivum). The red box present the mutant in osmapk6. (B) The 3D model of OsMAPK6 was built based on 6dtl.2.A(Mitogen-activated protein kinase 6) in Swiss-model (https://swissmodel.expasy.org/). The red circle present the mutant in osmapk6.



 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |