

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.018853
ARTICLE
Cloning and Drought Resistance Analysis of Soybean GmHsps_p23-like Gene
1College of Life Sciences, Jilin Agricultural University, Changchun, 130118, China
2College of Agronomy, Jilin Agricultural University, Changchun, 130118, China
*Corresponding Authors: Siyan Liu. Email: siyan_2001@163.com; Shuyan Guan. Email: guanshuyan@jlau.edu.cn
#Yuzhe Zhang, Guanglong Li contributed equally to this work
Received: 21 August 2021; Accepted: 03 November 2021
Abstract: Soybean (Glycine max (L.) Merr.) is an important cultivated crop, which requires much water during its growth, and drought seriously affects soybean yields. Studies have shown that the expression of small heat shock proteins can enhance drought resistance, cold resistance and salt resistance of plants. In this experiment, soybean GmHsps_p23-like gene was successfully cloned by RT-PCR, the protein encoded by the GmHsps_p23-like gene was subjected to bioinformatics analysis, and the pCAMBIA3301-GmHsps_p23-like overexpression vector and pCBSG015-GmHsps_p23-like gene editing vector were constructed. Agrobacterium-mediated method was used to transform soybeans to obtain positive plants. RT-PCR detection, rehydration experiment and drought resistance physiological and biochemical index detection were performed on the T2 generation positive transgenic soybean plants identified by PCR and Southern hybridization. The results showed that the overexpression vector plant GmHsps_p23-like gene expression increased. After rehydration, the transgenic overexpression plants returned to normal growth, and the damage to the plants was low. After drought stress, the SOD and POD activities and the PRO content of the transgenic overexpression plants increased, while the MDA content decreased. The reverse was true for soybean plants with genetically modified editing vectors. The drought resistance of the overexpressed soybeans under drought stress was higher than that of the control group, and had a stronger drought resistance. It showed that the expression of soybean GmHsps_p23-like gene can improve the drought resistance of soybean. The cloning and functional verification of soybean GmHsps_p23-like gene had not been reported yet. This is the first time that PCR technology has been used to amplify the soybean GmHsps_p23-like gene and construct an expression vector for this gene. This research has laid the foundation for transgenic technology to improve plant drought resistance and cultivate new drought-resistant transgenic soybean varieties.
Keywords: Soybean; small heat shock protein; CRISPR/Cas9; drought resistance
During growth of soybean, the demand for water is high, and drought seriously affects soybean yield. The annual loss of soybean yield and quality in China is immeasurable [1,2]. As a major grain producing area, the cultivation and production of soybean in Northeast China is greatly affected by extreme weather events, such as high temperatures and drought [3]. Improving plant drought resistance through transgenic technology has shown great application and economic value. Cloning of genes related to drought resistance and transferring them into the genome is one of the most effective methods to cultivate drought-tolerant soybean varieties.
sHsps are a class of low molecular weight heat shock proteins (12–40 kDa). In plants, sHsps are more diverse in sequence similarity, cell localization and biological function than other Hsps families. However, under unfavorable conditions of the external environment, especially when stimulated by heat stress, the structure is conservative. sHsps have a molecular chaperone function and play a vital role in protecting plants from stress and rebuilding cell homeostasis [4–6]. Plant sHsps can deal with various environmental stresses, including heat, cold, drought, salinity and oxidative stress. sHsps also interact with thylakoid membranes and participate in plant resistance [7]. Maize (Zea mays) mitochondrial sHsps showed protection against NADH and improved mitochondrial electron b transport during salt stress [8]. The accumulation of SlHSP21.5A can increase the tolerance of rice (Oryza sativa) to salt stress [9]; overexpression of CaHSP25.9 can enhance pepper (Piper nigrum) tolerance to heat, salt and drought stress; CaHSP16.4 is related to pepper (Piper nigrum) heat and drought tolerance [10,11]; overexpression of KSlHSP21.5A can enhance tomato (Solanum lycopersicum) tolerance to heat stress [12]; the expression of CvHSF30-2 can improve the heat resistance of clematis (Clematis tibetana) [13]. More and more studies have shown that there is a strong correlation between the accumulation of sHsps and the stress tolerance of plants.
CRISPR/Cas9 is an acquired immune defense mechanism evolved from bacteria and archaea, which can cope with the constant attack of viruses and plasmids [14]. In recent years, CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats)-mediated gene editing technology has become more and more mature, and has become one of the more popular technologies in the field of life sciences. Knocking out the ZmCEP1 gene through CRISPR/Cas9 significantly increased the ear height, grain size and yield of corn plants [15]. CRISPR/Cas9-mediated genome editing improved wheat grain quality, successfully transformed soft wheat into durum wheat, and increased the genetic resources for wheat (Triticum aestivum) grain hardness improvement [16]. CRISPR/Cas9 technology is used in soybean (Glycine max) to specifically design double sgRNA/Cas9 to induce targeted deletion of the soybean GmFATB1 gene. This system is conducive to gene function research and improves soybean molecular breeding [17].
In this experiment, the soybean GmHsps_p23-like gene was successfully cloned by RT-PCR technology, and the function of the gene was analyzed by molecular biology detection and drought resistance analysis of the positively transformed soybean plants. This research lays the foundation for the use of genetically modified technology to improve the drought resistance of soybeans and cultivate new lines of drought-resistant genetically-modified soybean.
We worked with the Ji Nong 18 Soybean (‘JN 18’), Escherichia coli strain DH5α, Agrobacterium strain EHA105 and plant expression vector pCAMBIA3301 from Jilin Agricultural University Corn and Oil Crop Germplasm Resources Innovation and Molecular Breeding Innovation Team Laboratory.
2.2 Cloning of Soybean GmHsps_p23-like Gene
Refer to the recommended procedure of RNAiso plus (Bao Bioengineering (Dalian) Co., Ltd., China) for total RNA extraction from soybean ‘JN18’ seven-leaf stage. Soybean cDNA was obtained using All-in-Onetm First-Strand cDNA Synthesis (Gene Copoeia), and primer p23s/p23As was designed using Primer Premier 5.0 software (sequences are shown in Table 1, synthesized by Jilin Changchun Kumei Biotechnology Co., Ltd., China). The cDNA was used as a template for PCR amplification. The amplification procedure was pre-denaturation at 94°C for 5 min, denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 30 s during 32 cycles, extension at 72°C for 8 min, and storage at 4°C. The fragment of the gel-removing recovery was sent to Changchun Kumei Biotechnology Co., Ltd. of Jilin Province for sequencing, and the sequencing results were compared. The correct target fragment was ligated with the cloning vector pMD-18T to transform competent cells of E. coli DH5α.
2.3 Bioinformatics Analysis of Protein Encoded by GmHsps_p23-like Gene
A different software was used to analyze the protein encoded by GmHsps_p23-like gene, such as SignalP analysis of protein signal peptide and Softberry analysis of protein subcellular localization. In addition, TMHMM server 2.0, ExPASy-protSCale, and SOPMA models analyzed protein transmembrane, protein hydrophobicity and hydrophilicity, and protein secondary structure, respectively.
2.4 Construction of Plant Overexpression Vector pCAMBIA3301-GmHsps_p23-like
The plant expression vector pCAMBIA3301 plasmid was double-digested with BGL II and BstE II endonuclease according to the double digestion system recommended by the Double Digest Calculator of Thermo Scientific. The incubation was carried out at 37°C for 4.5 h, the digestion reaction was terminated by 80°C for 5 min. The double-digested large vector fragment and the target gene fragment were ligated using T4 DNA Linking Enzyme, and then transformed into E. coli DH5α competent cells. After the extracted monoclonal colony plasmid was subjected to PCR and double enzyme digestion verification, sequencing alignment was performed. The T-region structure of the plant overexpression vector is shown in Fig. 1.
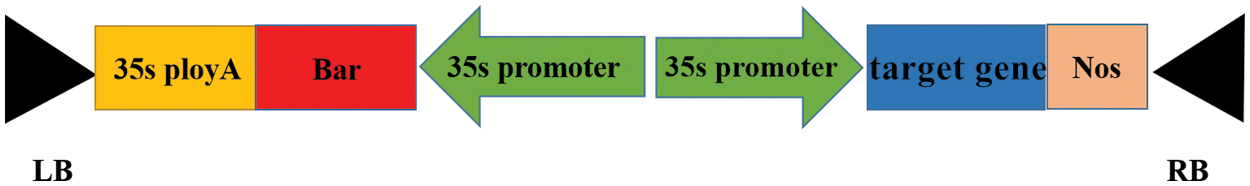
Figure 1: T-DNA structure of plant overexpression vector
2.5 Construction of Plant Gene Editing Vector pCBSG015-GmHsps_p23-like
The construction was carried out using the CRISPR/Cas9 vector construction kit of Baige Biotechnology, and the construction process was carried out in strict accordance with the instructions. The construction process consisted of designing Oligo primers, preparation of Oligo dimers, linkage of the CRISPR/Cas9 vector backbone and transformation of E. coli. The plasmid was extracted, and the constructed CRISPR/Cas9 vector was subjected to PCR detection using the vector-labeled marker Bar gene primer and the promoter 35S primer, and then sequenced (see Table 1 for the sequence).
2.6 Genetic Transformation of Soybean
In this experiment, Agrobacterium tumefaciens was used to infect the soybean cotyledonary node method, and plant expression vectors were genetically transformed into the recipient soybean variety (JN18) to finally obtain transformed plants.
2.7 PCR Detection of Transgenic Plants
The genomic DNA of the transformed plant leaves was extracted. Because GmHsps_p23-like gene is derived from the receptor soybeans, so the bar, 35 s sequence (Table 1) was subjected to PCR detection. The PCR reaction procedure was: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 30 s, annealing at 58°C for 30 s, extension at 72°C during 30 s for 32 cycles; extension at 72°C during 8 min; storage at 4°C. The bar gene amplification product had a size of 552 bp, and the 35 s gene amplification product had a size of 500 bp.
2.8 T2 Generation Plant Southern Bloting Detection
For soybean plants that were tested positive by PCR in T2 generation, the leaf genomic DNA was extracted by the SDS method, and the genomic DNA of control soybean plants was extracted. The extracted DNA was digested with Hind III restriction endonuclease overnight. Probes were prepared with the bar gene as the target. Southern blot detection was performed using DIG DNA Labeling and Detection Kit I.
2.9 T2 Generation Transformed Plant Gene Editing Test
In order to determine whether the CRISPR/Cas9 gene editing vector can effectively edit the target site of soybean Gm_Hsps23-like gene, the transformed plant DNA was used as a template to sequence the PCR products, and the DNAMAN software was used for sequence comparison.
2.10 T2 Generation Plant Fluorescence Quantitative PCR Detection
RNA from T2-transformed positive soybean plants and untransformed plants was separately extracted for reverse transcription. Using cDNA as a template, soybean actin (designed according to β-soybean actin gene sequence, Gen Bank ID NM_001252731.2) was used as the internal reference gene, and Yp23S/Yp23AS was used to quantify the target gene primer for qRT-PCR. According to the qPCR Mix manual produced by Kangwei Company, in the Mx 3000P real-time PCR, the two-step method was used, and each sample was repeated three times. When the program was finished, the result was output and saved. Relative expression levels were calculated and analyzed according to the 2−△△Ct method.
2.11 Identification of Drought Resistance of Genetically Modified Soybean
T2 transgenic soybean and receptor ‘JN18’ soybean were planted in the same potted nutrient soil in the artificial climate chamber, and the water was cultured normally. When the soybean plant grew to the three-leaf-leaf stage, the water supply was stopped to simulate natural drought stress, and the growth of the transgenic material and the control were observed. Water supply was restored after 10 days of the drought stress treatment. Growth recovery status was recorded on the genetically modified material and the control soybean.
2.11.2 Determination of Physiological and Biochemical Indicators Related to Drought Resistance
For the soybean leaves before and after simulated drought stress, the kits of Suzhou Keming Biotechnology Co., Limited Company were used to detect the drought-related physiological and biochemical indicators, including superoxide dismutase (SOD), peroxidase (POD), malondialdehyde (MDA) and proline Acid (PRO).
3.1 Cloning and Identification of Soybean GmHsps_p23-Like Gene
The extracted total RNA of soybeans was detected by 2% agarose gel electrophoresis (Fig. 2), and the good quality total RNA was reverse transcribed to generate cDNA. The specific band obtained after PCR amplification using cDNA as a template is shown in Fig. 3. The band size was 480 bp, which is consistent with the size of soybean GmHsps_p23-like gene. PCR (Fig. 4) and double digestion identification (Fig. 5) of the recombinant cloning vector proved that the GmHsps_p23-like gene was successfully cloned, and the cloning vector of the gene was recombined.
Figure 2: Electrophoresis of total soybean RNA extraction. 1–4: Total RNA of soybean leaves
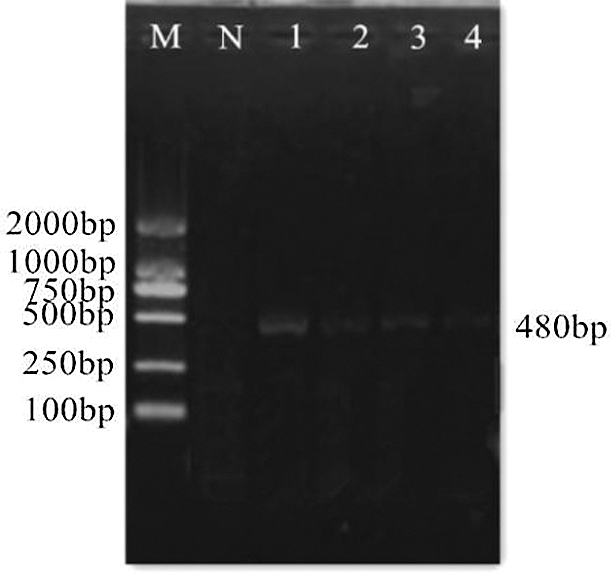
Figure 3: Cloning of the GmHsps_p23-like gene. M: DL2000 Marker; N: Negative control; 1–4: RT-PCR product
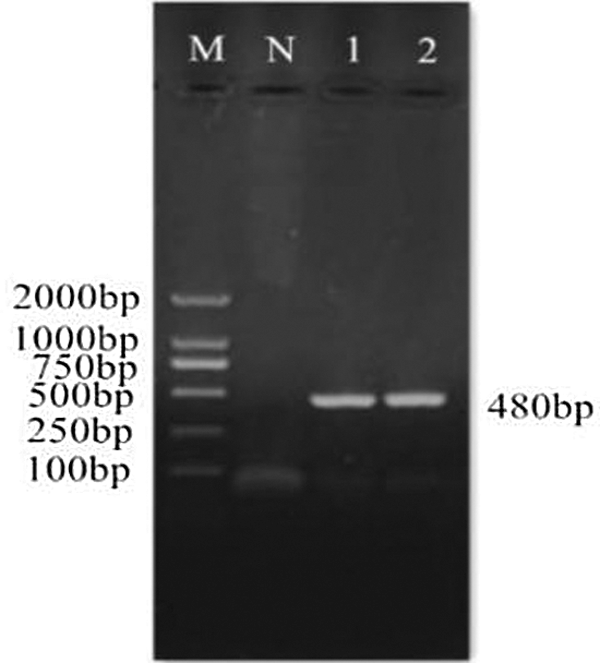
Figure 4: PCR identification of cloning vector. M: DL2000 Marker; N: Negative control; 1–2: PCR product
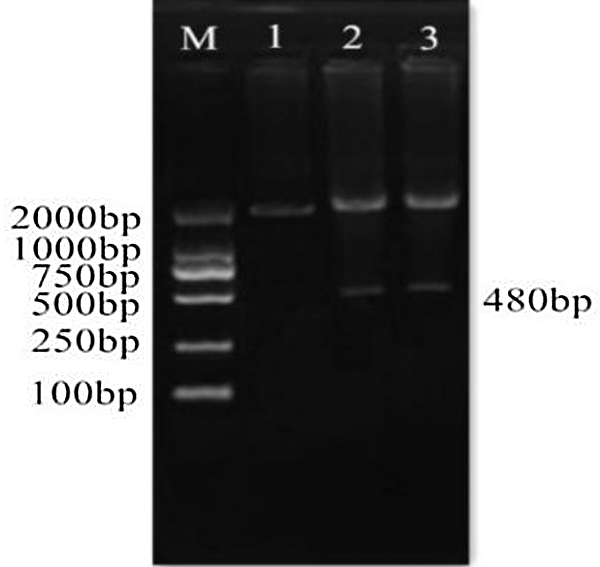
Figure 5: Cloning vector verification by double digestion. M: DL2000 Marker; 1: Cloning vector plasmid; 2–3: Double digested product
3.2 Bioinformatics Analysis of Protein Encoded by GmHsps_p23-like Gene
Using ExPASy-ProtScale software to analyze the hydrophobicity and hydrophilicity of the protein, the protein encoded by the GmHsps_p23-like gene is hydrophobin (Fig. 6). Online prediction of the protein interacting with this protein through STRING (Fig. 7). According to the online prediction of the protein transmembrane region of the TMHMM server 2, the protein does not have a transmembrane structure and is a non-transmembrane protein (Fig. 8). Through online analysis of Signalp to predict the protein signal peptide, the protein is a non-secreted protein (Fig. 9). Use the online website Softberry to predict the subcellular location of the protein, which may play a major role in the cytoplasm (Fig. 10). Using SOPMA to analyze the secondary structure of the protein online, the secondary structure of the protein accounted forα helix (31.45%), extended chain (21.16%), β angle (6.29%) and random coil (37.11%) (Fig. 11).
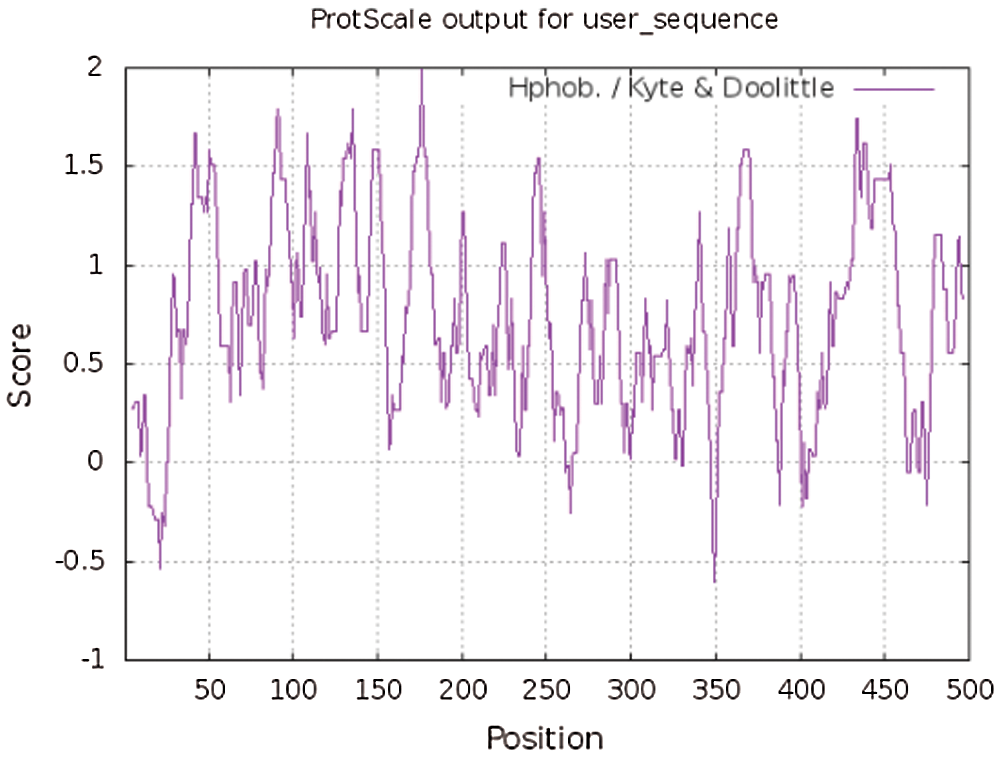
Figure 6: Hydrophobic and hydrophilic analysis of GmHsps_p23-like protein
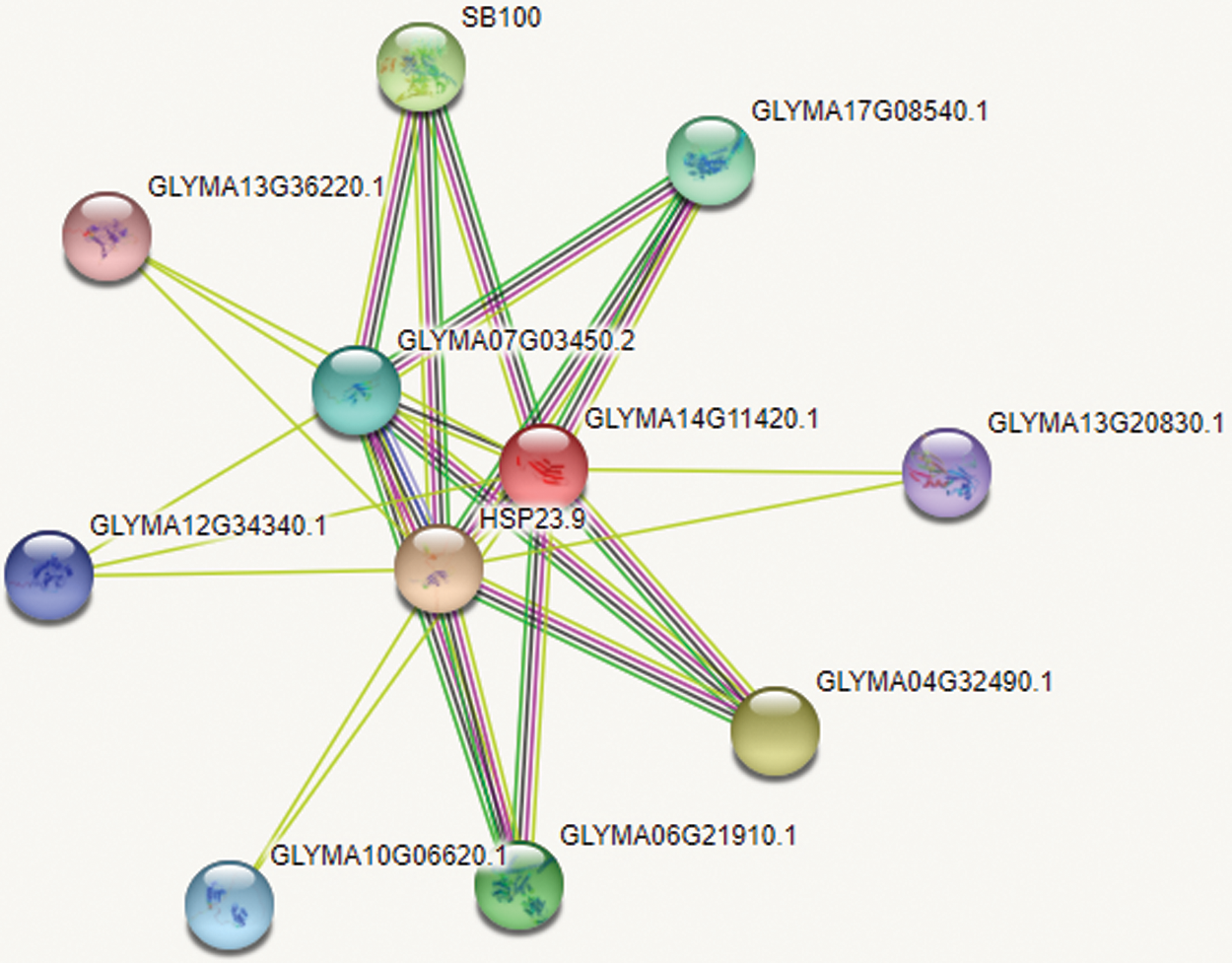
Figure 7: Interacting protein prediction of GmHsps_p23-like protein
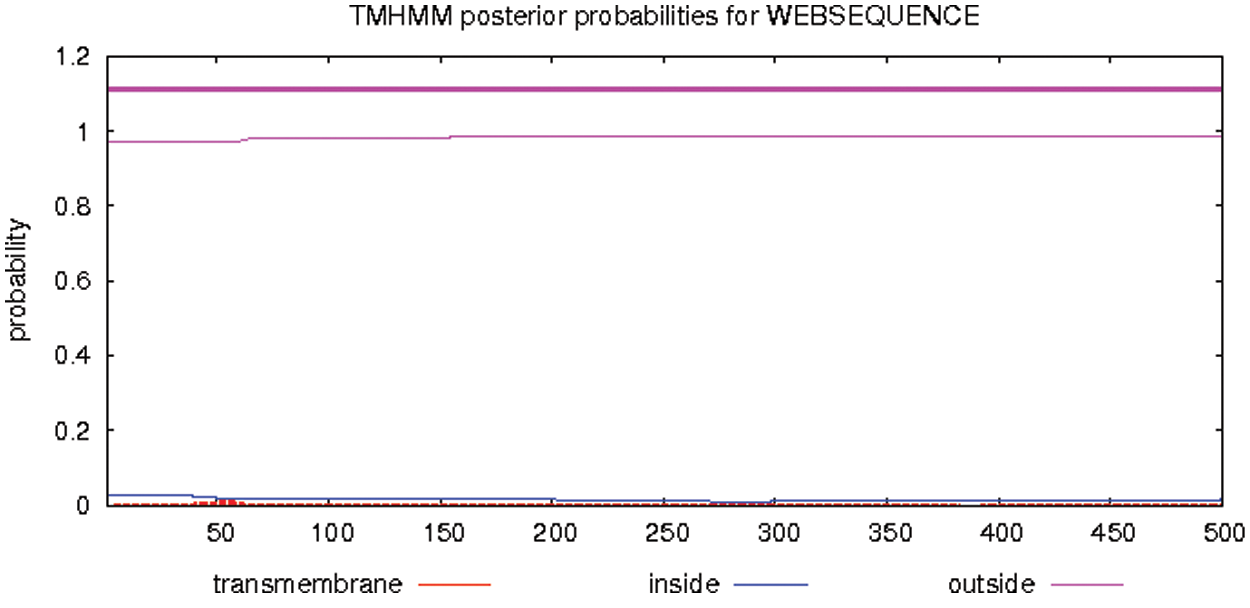
Figure 8: Transmembrane region prediction of GmHsps_p23-like protein
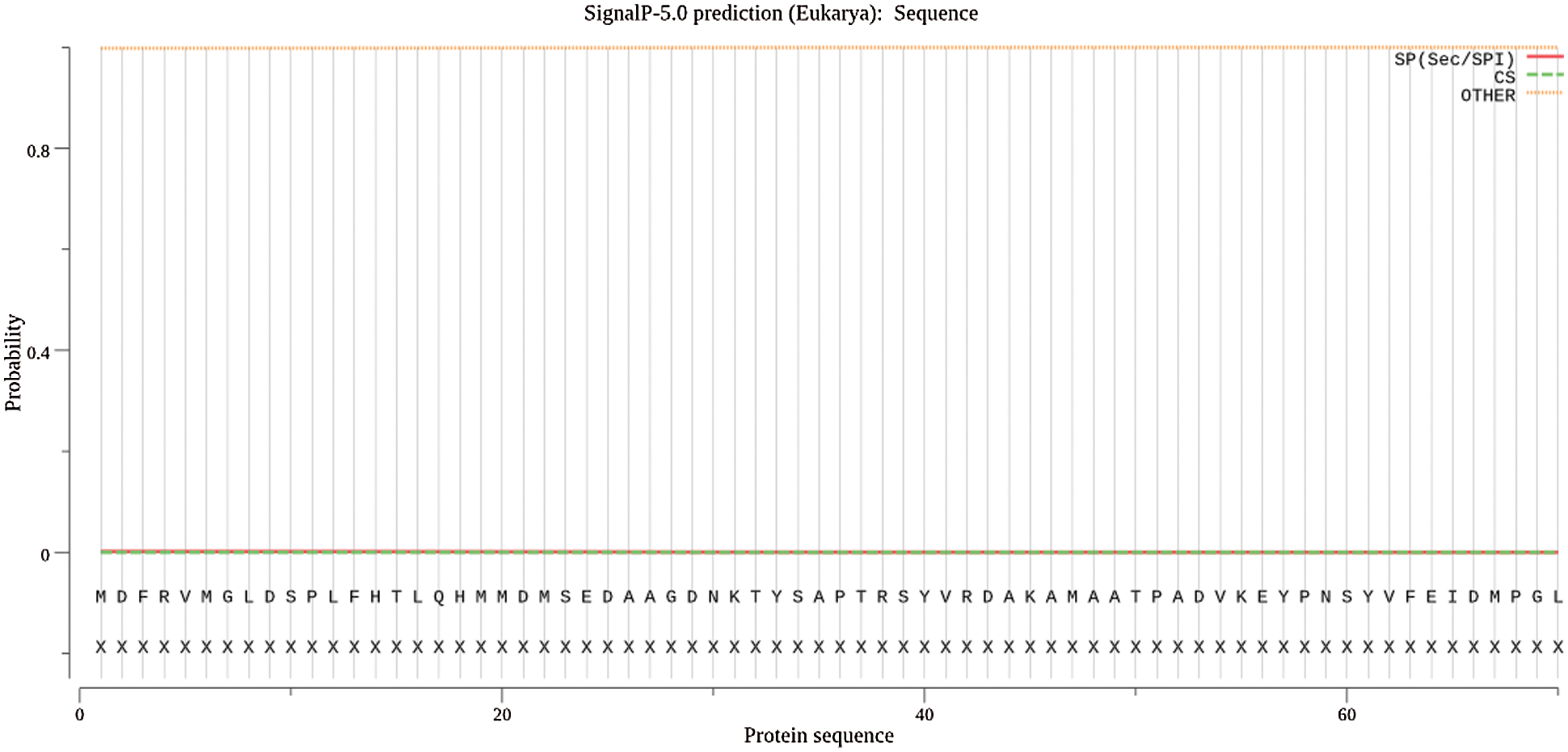
Figure 9: Signal peptide prediction of GmHsps_p23-like protein
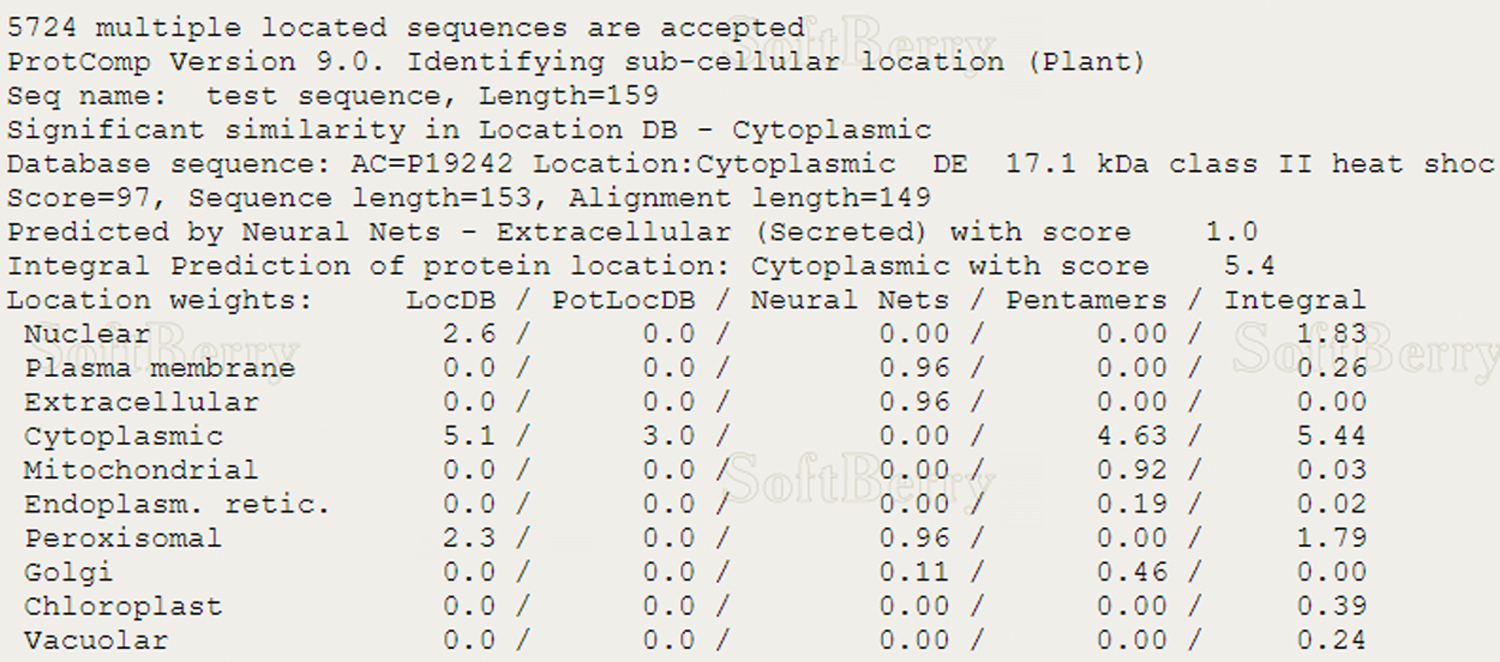
Figure 10: Subcellular localization prediction of GmHsps_p23-like protein
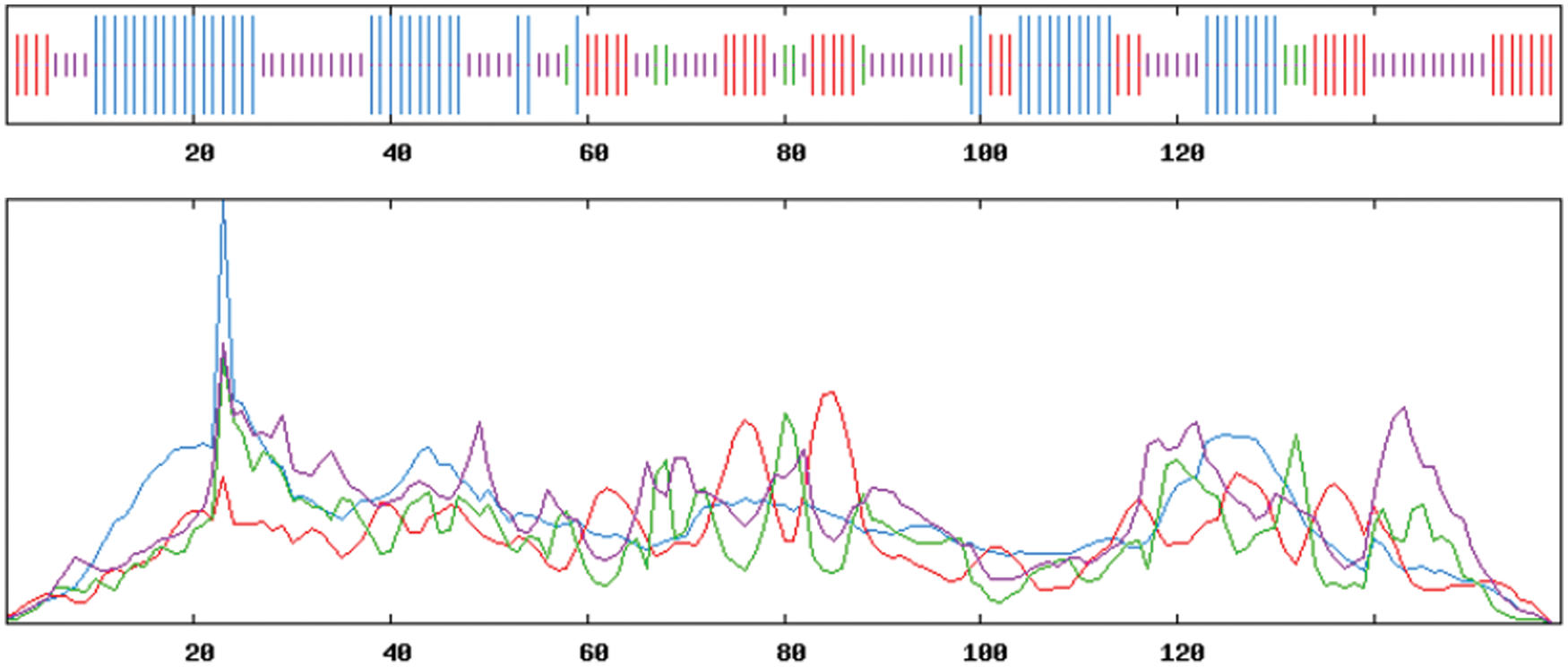
Figure 11: Secondary structure for protein encoded of GmHsps_p23-like protein
3.3 Construction and Identification of Plant Overexpression Vector pCAMBIA3301-GmHsps_p23-like
The double-digested product of pCAMBIA3301 plasmid was separated by 1% agarose gel electrophoresis (Fig. 12). The large vector fragment was recovered by gel, and the target fragment was ligated with the large vector fragment by T4 DNA ligase, furthermore the ligation product was transformed into E. coli. The plasmid was subjected to PCR (Fig. 13) as well as double restriction enzyme identification (Fig. 14) with reference to the test 2.1, and the obtained band size was consistent with the expectation. Moreover, the sequence alignment was 100% consistent, which proved that the plant overexpression vector was successfully constructed.
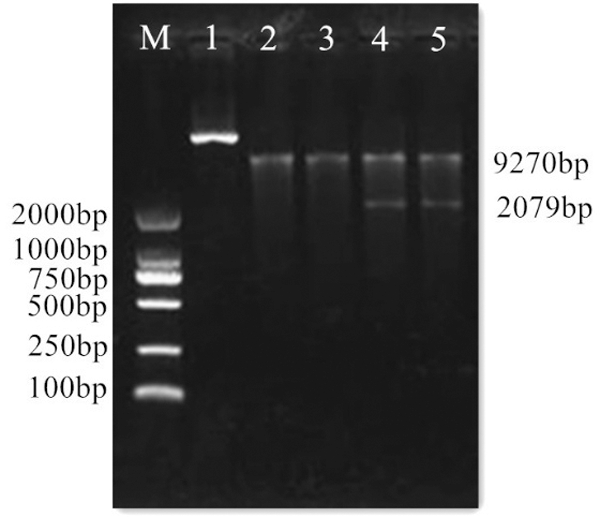
Figure 12: Digestion of vector pCAMBIA3301. M: DL2000 Marker; 1: Vector plasmid; 2–5: Double digested product
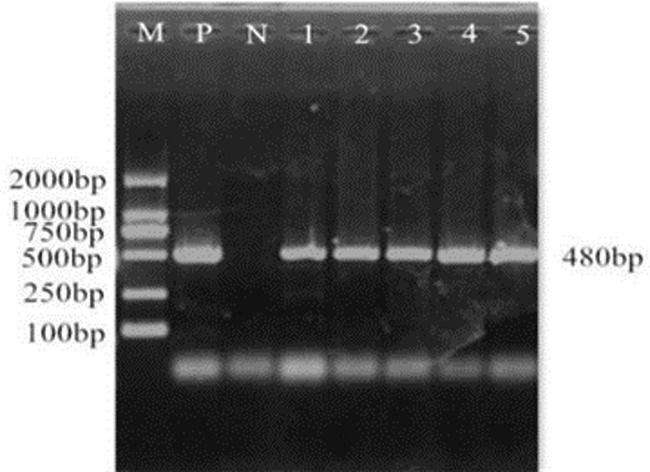
Figure 13: PCR identification of overexpression vector. M: DL2000 Marker; P: Positive control; N: Negative control; 1–5: PCR product
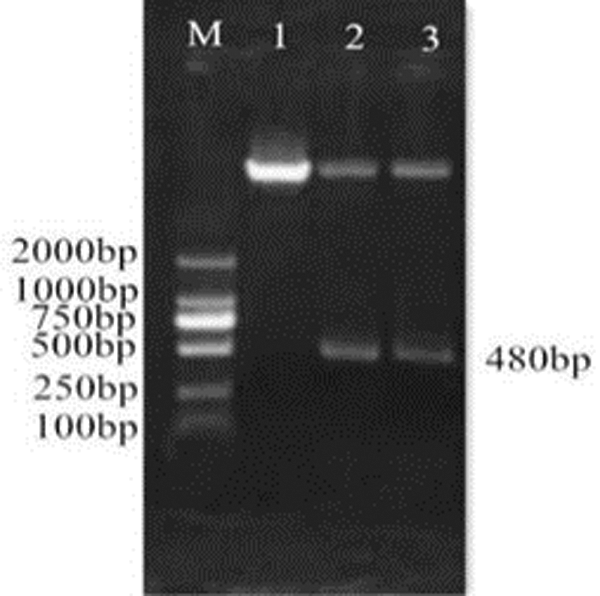
Figure 14: Double enzyme digestion identification of the overexpression vector pCAMBIA3301-GmHsps_p23-like. M: DL2000 Marker; 1: Overexpression vector plasmid; 2–3: Double digested production
3.4 Construction and Identification of Plant Gene Editing Vector pCBSG015-GmHsps_p23-like
The transformed monoclonal colony plasmid was extracted, and the plasmid was identified by PCR using the Bar gene primer and the 35S promoter primer (Fig. 15). It can be seen from the figure that specific bands of 552 bp and 500 bp were obtained. It was a preliminary proof that the vector was successfully constructed. Using sequencing primers (CRI), the constructed CRISPR/Cas9 vector plasmid was sequenced and identified. Using DNAMAN for comparison, the sequencing results were 100% identical to the target fragment (Fig. 16), further proving that the CRISPR/Cas9 gene editing vector for the GmHsps_p23-like gene was successfully constructed.
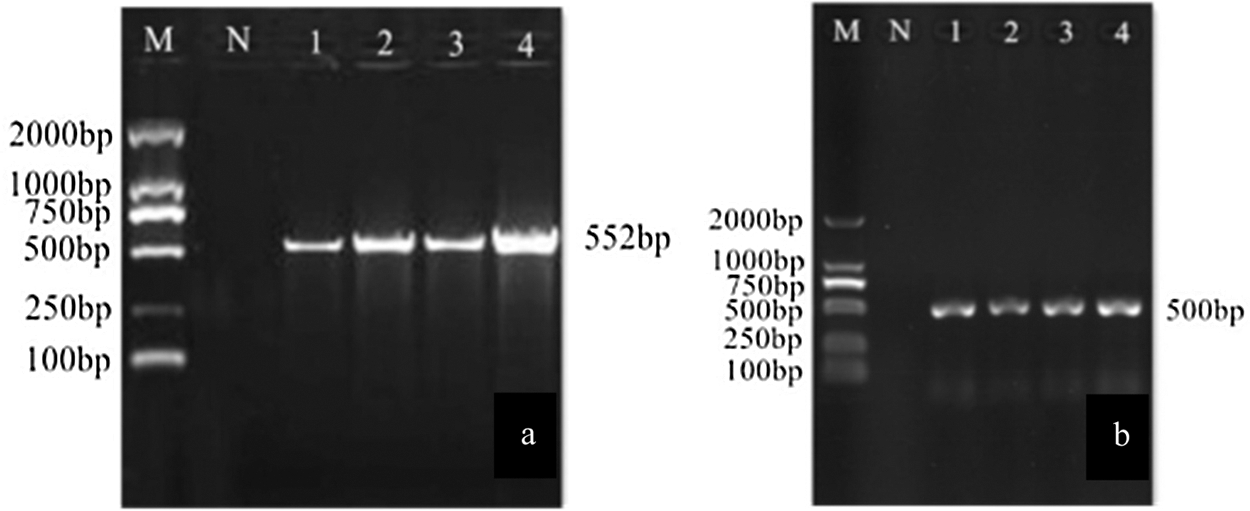
Figure 15: PCR identification of overexpression vectors. M: DL2000 Marker; N: Negative control; 1–4: PCR product. (a) Bacteria solution PCR; (b) Plasmid PCR
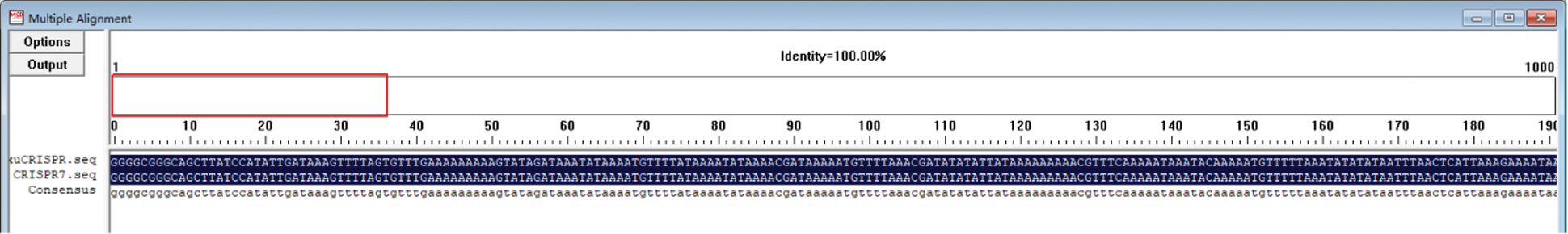
Figure 16: Sequencing alignment results of the CRISPR-Cas9 vector
3.5 Genetic Transformation of Soybean and PCR Detection of Transgenic Plants
Two sets of plant expression vectors were genetically transformed into soybean (JN18) by Agrobacterium-mediated soybean cotyledon node method (Fig. 17), and the obtained transformed plants were subjected to PCR detection. The results showed that 3 strains of the overexpression vector T0 generation soybeans were positive. The transformation rate of the plants was 1.24%; 5 positive soybean plants of the T0 generation of transgenic editing vector were obtained, and the positive rate was 2.11%. Subculture, the PCR detection result was 14 plants, and 21 plants of T1 generation were obtained. In the artificial climate room, 21 and 13 T2 generation plants were planted in flower pots, and the PCR detection results obtained 17 and 11 T2 generation positive plants, respectively. The PCR detection results of each generation are shown in Fig. 18. The amplified fragment showed the same size as the 552 bp of the bar gene.
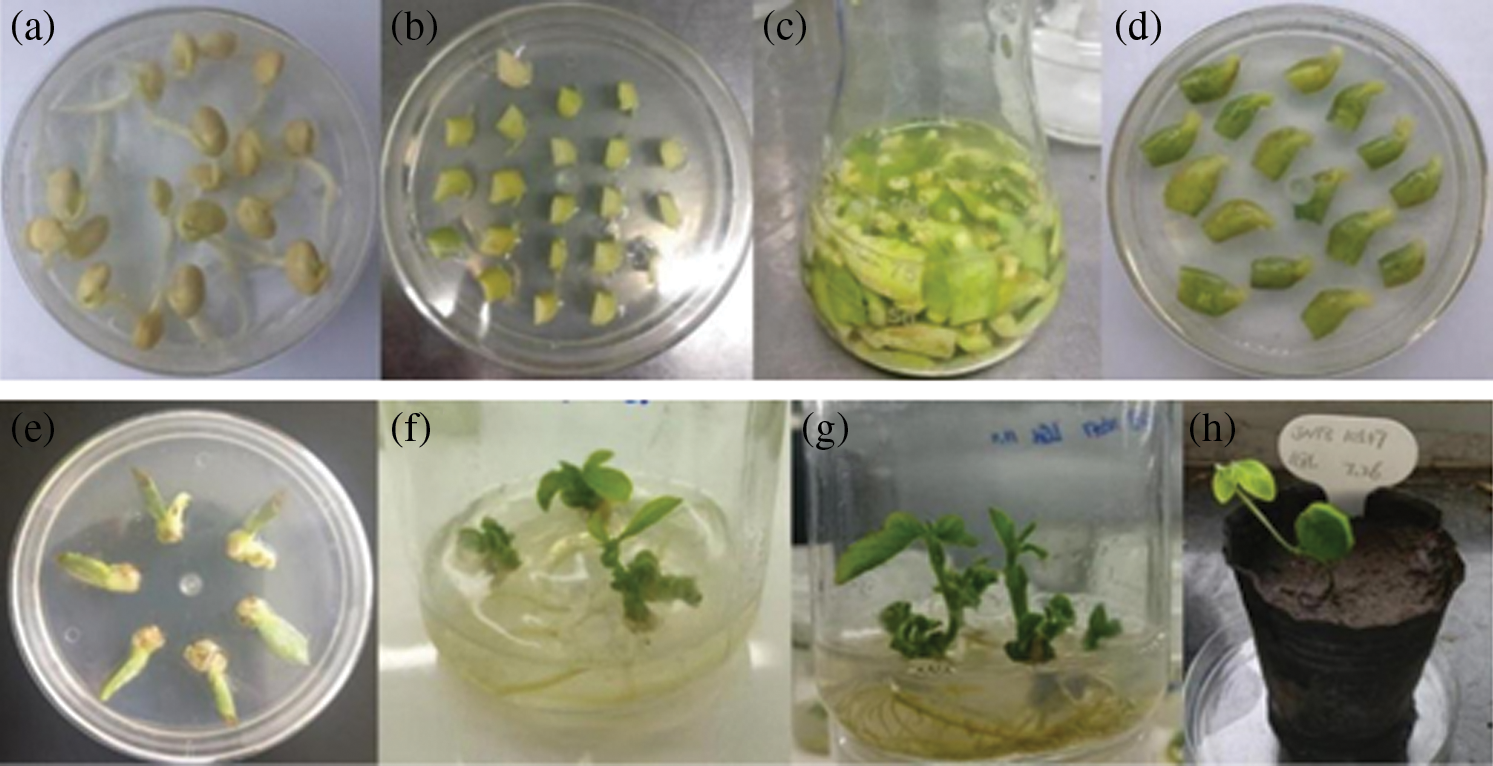
Figure 17: Soybean cotyledons transformation processes by Agrobacterium. (a) Seed germination; (b) Pre-culture; (c) Infection; (d) Co-culture; (e) Selective culture; (f) Elongation culture; (g) Rooting culture; (h) Seedling transplanting picture
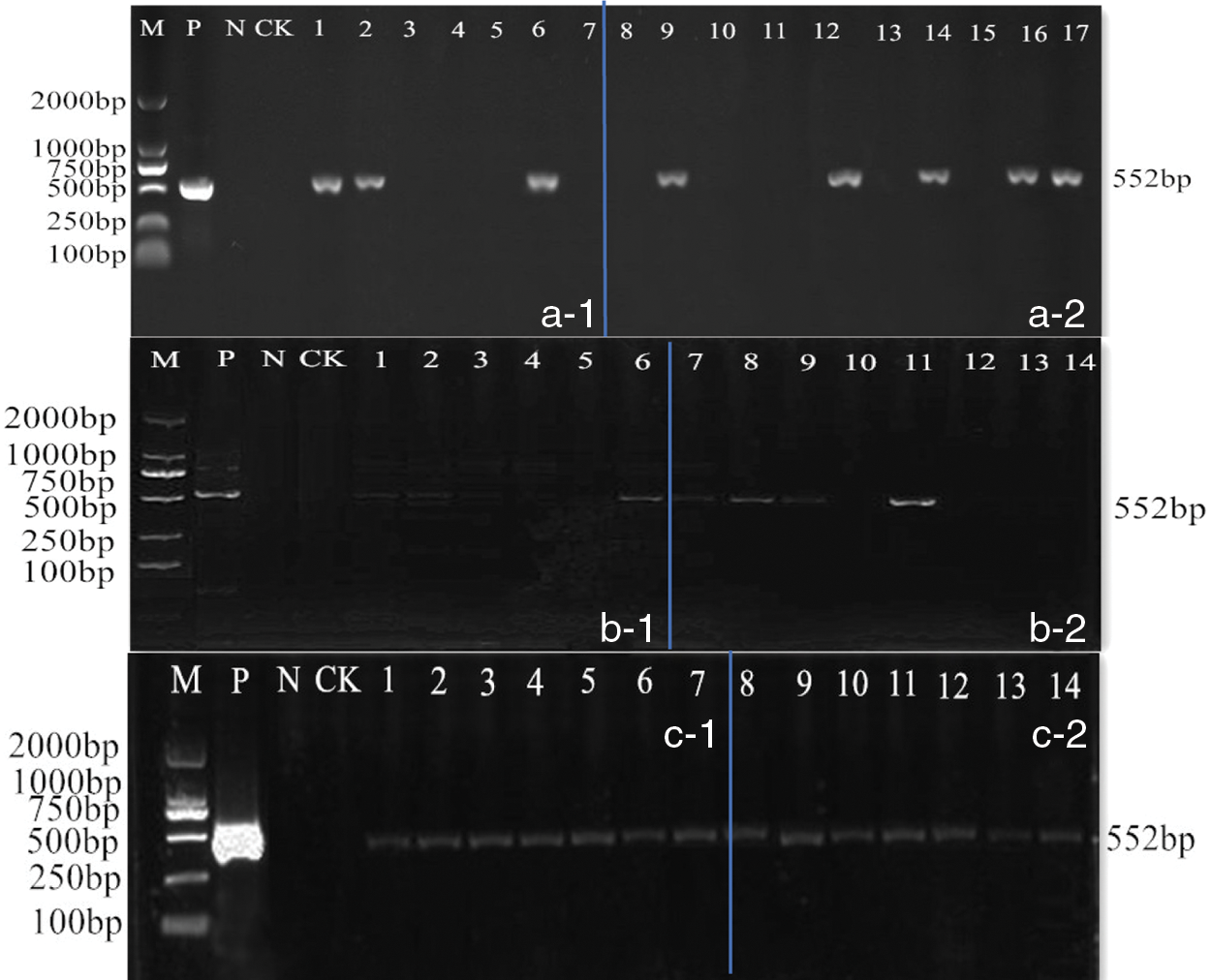
Figure 18: Detection of generation transgenic plants with Bar. M: DL2000 Marker; P: Positive control; N: Negative control; 1–17: PCR product. (a-1) Detection of T0 generation transgenic overexpression vector by PCR; (a-2) Detection of T0 generation transgenic CRISPR/Cas9 vector by PCR; (b-1) Detection of T1 generation transgenic overexpression vector by PCR; (b-2) Detection of T1 generation transgenic CRISPR/Cas9 vector by PCR; (c-1) Detection of T2 generation transgenic overexpression vector by PCR; (c-2) Detection of T2 generation transgenic
3.6 T2 Generation Plant Southern Bloting Detection
Extract the genomic DNA from the leaves of the receptor material ‘JN18’ and T2 generations transfected with overexpression vector PCR-positive soybean plants, digest them overnight with Hind III restriction enzymes, select the Bar gene to prepare probes, and perform Southern Bloting detection. The results showed that among the tested T2 generation transformed plants, three plants showed hybridization signals, and their integration sites in the genome were different. (Fig. 19).
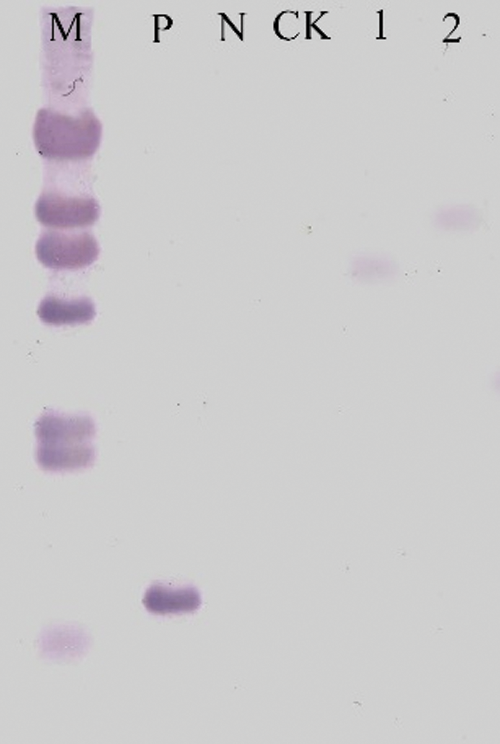
Figure 19: Southern Bloting detection of transgenic plants. M: Southern marker; P: Positive control; N: Negative control; 1–2: T2 transformed plants
3.7 T2 Generation Transformed Plant Gene Editing Test
PCR (p23 S/p23 AS) primers were used to carry out PCR on the genomic DNA of CRISPR/Cas9 vector-positive plants. The sequencing results of CR1-1 plants are shown in Fig. 20a. The mismatch mutation occurred in the base of this locus. The sequencing results of CR1-2 plants are shown in Fig. 20b. It can be seen that the bases at 119 bp and 130 bp were mismatched, and the target genes of the two transgenic plants were edited by CRISPR/Cas9 vector.
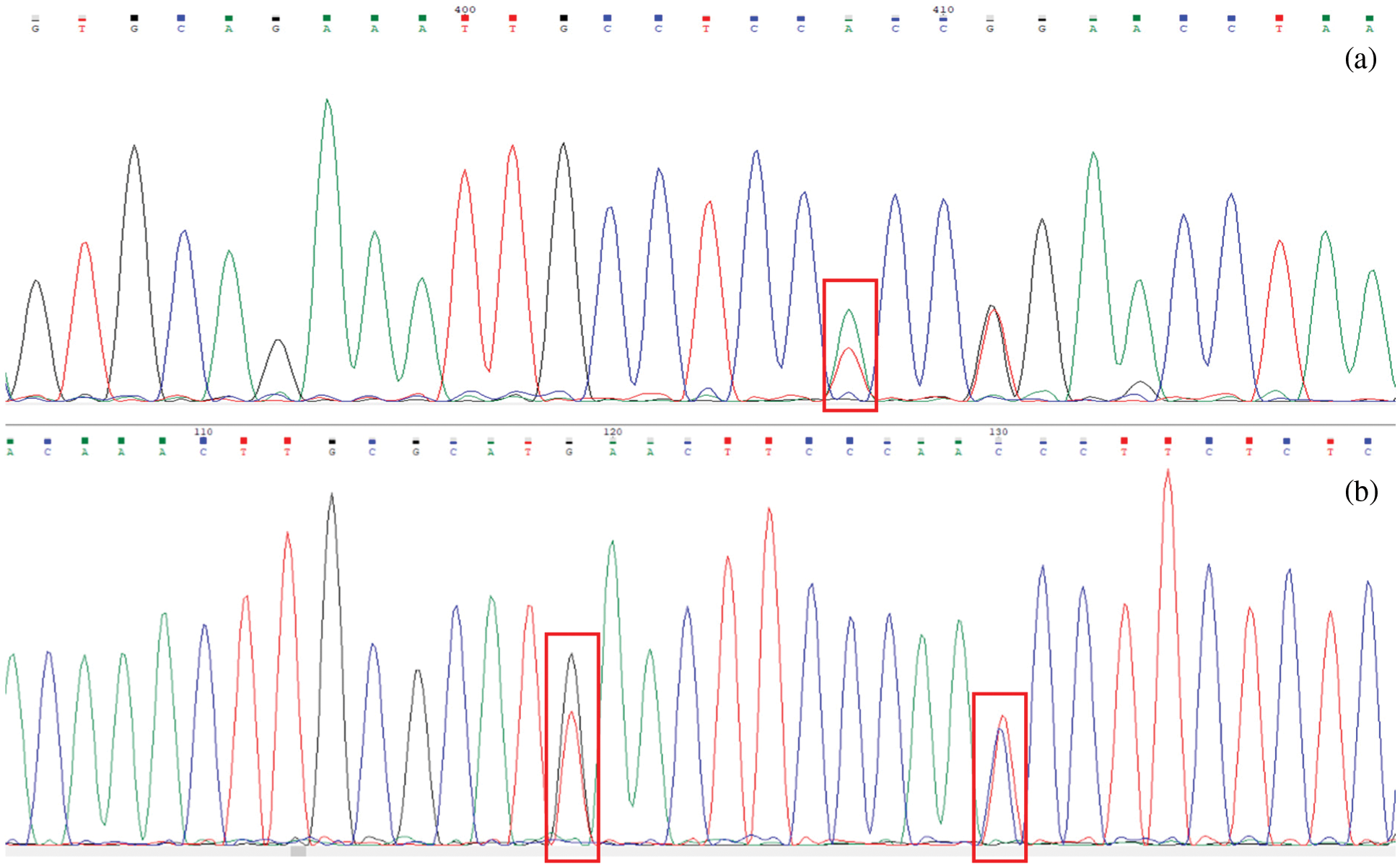
Figure 20: Sequencing peak map analysis. (a) CR1-1 sequencing results; (b) CR1-2 sequencing results
3.8 T2 Fluorescence Quantitative PCR Detection of Plants
SYBR green was selected as the dye, and the expression level of GmHsps_p23-like gene in the leaves of transgenic plants was detected by real-time PCR; the data were sorted into a column chart, as shown in Fig. 21. As can be seen from the figure, the relative expression level of the trans-overexpression vector plants was 343% and 188% of the control, which proved that the gene was over-expressed; while the gene-edited target gene expression amount was 74.65% and 86.91% of the control. The expression level was affected by a decrease.
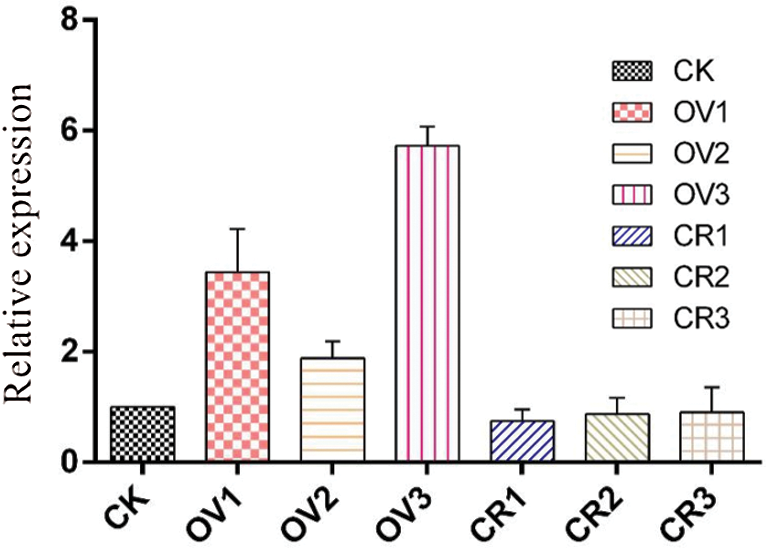
Figure 21: Relative expression level of GmHsps_p23-like gene
3.9 Identification of Drought Resistance of Genetically Modified Soybean
Watering was resumed after 10 days of drought stress, and the growth of soybean plants before and after observation was determined (Fig. 22). Transgenic plants (OV) were rapidly diastolic, fresh green leaves, and drought stress had little effect on their growth status. Compared with the control group, the transfected CRISPR/Cas9 vector (CR) showed no significant improvement in leaf curl, the plant was short, and drought stress had a significant effect on its growth status. It indicated that the plants with transgenic overexpression after rehydration resumed normal growth, and the degree of damage to plants was low. However, the plants transfected with CRISPR/Cas9 gene editing vector returned to normal growth slow, and drought stress had a high degree of damage to plants. It preliminarily proved that the expression of the GmHsps_p23-like gene can improve the drought resistance of soybean.

Figure 22: Rehydration test. JN18: acceptor soybean plants; OV: Trans-overexpression vector plants; CR: Trans-CRISPR/Cas9 vector plants
3.9.2 Detection of Physiological and Biochemical Indicators Related to Drought Resistance
The physiological and biochemical indicators related to drought resistance of superoxide dismutase (SOD), peroxidase (POD), malondialdehyde (MDA)and proline Acid (PRO) were detected by kit method (Suzhou Keying Biotechnology Co., Ltd., China). The superoxide dismutase (SOD) and peroxidase (POD) activities and the proline Acid (PRO) content of the transgenic plants were significantly increased after drought stress (Fig. 23). However, the increase of malondialdehyde (MDA) content under drought stress was not significant. Actively coping with alleviating the adverse oxidation reaction caused by drought stress, and having a strong drought resistance; the physiological and biochemical indicators of soybean plants transgenic editing vector showed that the plants were affected by drought; the degree of cell damage was obvious, and the plants were highly affected by drought and plant drought resistance decreased.
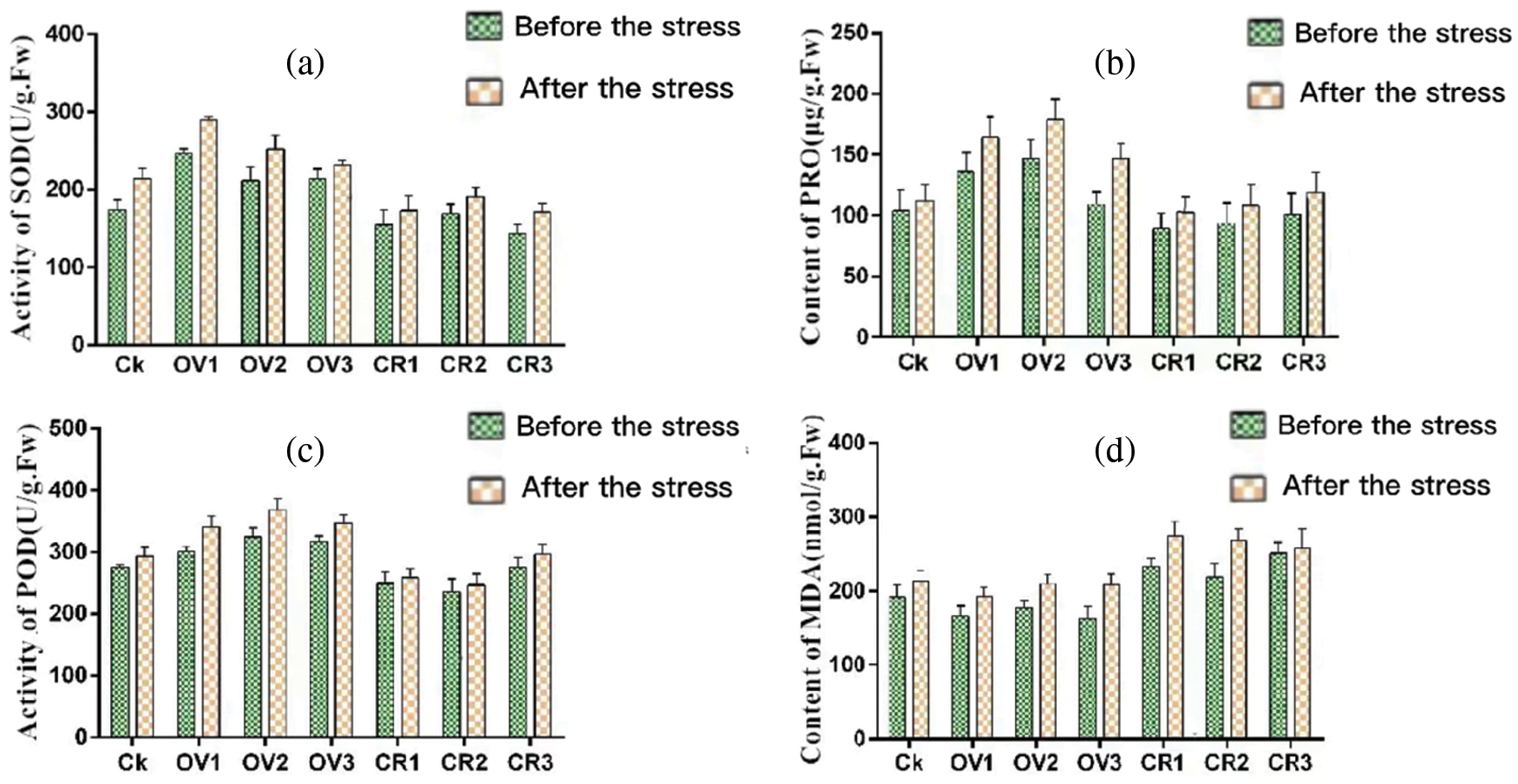
Figure 23: Detection of physiological and biochemical indicators related to drought resistance at different times (a) SOD activity (b) PRO content (c) POD activity (d) MDA content
Drought has been considered as the most important factor limiting soybean yield among various abiotic stresses such as high temperature, salinity, heavy metals, and freezing damage. The study of Hsps in soybeans have been carried out for many years. Different types of Hsps have been cloned in soybeans and their functions have been studied. Overexpression of the GmHsf-34 gene enhanced drought and heat tolerance in Arabidopsis (Arabidopsis thaliana) plants [18]. By constructing the Hsp17.4 gene overexpression vector and RNAi interference vector were constructed respectively, and the relevant indexes were measured after genetic transformation of soybean. It was concluded that soybean Hsp17.4 gene was associated with high temperature resistance of soybean and could improve heat tolerance [19]. Studies on the GmHsp90A2 gene showed that the heat tolerance of the transgenic soybean was higher than that of the receptor control. The transgenic soybean materials with higher chlorophyll content and lower Malondialdehyde (MDA) content showed an increased tolerance to heat stress [20]. More and more studies have shown that the expression of heat shock protein family genes can improve the ability of plants to resist drought and heat.
CRISPR/Cas9 gene editing technology has been applied in many fields. Compared with other gene editing technologies, this technology has many advantages such as simple and feasible experiments, precise editing of target sites, and the ability to edit multiple target sites. For example, by editing the OsSWEET14 promoter improved the resistance of rice to bacterial leaf blight [21]. Using CRISPR/Cas9 technology, editing soybean GmFAD2-1A to obtain pure mutants proved that the GmFAD2-1A gene is a key gene in oleic acid metabolism [22]. In this paper, aiming at the soybean GmHsps_p23-like gene, a CRISPR/Cas9 plant expression vector was constructed, and U6 dicot plant promoter was used to guide the expression of gRNA sequence. The cloning of soybean GmHsps_p23-like gene and the analysis of drought resistance function had not yet been reported. Soybean GmHsps_p23-like gene has three conserved domains, ACD-ScHsp26-like, HSP20, and IbpA. It belongs to a typical sHsps and has the related function of sHsps. It is speculated that the expression of this gene is related to plant drought resistance. In-depth research on it can be used as a candidate gene for soybean drought and stress resistance molecular breeding. In this experiment, the soybean GmHsps_p23-like gene was successfully cloned, and an overexpression vector and a gene editing vector were constructed. Agrobacterium-mediated genetic transformation of soybeans was used to obtain transgenic soybean plants, and PCR and Southern hybridization techniques were used for plant identification. The T2 generation positive transgenic soybean plants identified by PCR and Southern hybridization were tested by RT-PCR, a rehydration experiment, and drought resistance physiological and biochemical indicators. The results showed that the overexpression vector plant GmHsps_p23-like gene expression increased significantly. After rehydration, the transgenic overexpression plants returned to normal growth, and the damage to the plants was low. After drought stress, the SOD, POD activity and PRO content of the transgenic overexpression plants increased, while the MDA content decreased. The reverse was true for soybean plants with genetically modified editing vectors. Under drought stress, the drought resistance of overexpression soybeans was higher than that of the control group. It shows that the transgenic plants have strong drought resistance. The results showed that the overexpression vector plant in soybeans verified that the expression of this gene can improve soybean drought resistance, which is consistent with previous research results on heat shock protein family genes. It shows that this gene can be used as a candidate gene for soybean drought resistance breeding. In this experiment, there were differences in drought resistance between different transgenic strains. It may be due to the different insertion sites and post-transcriptional regulation of the target gene in the recipient genome. Therefore, it is necessary to continue screening and exploring high-generation materials, and the protein expression level needs further study.
Funding Statement: The research was jointly funded by Jilin Province Education Department Science and Technology Research Project [JJKH20210350KJ], Jilin Province Science and Technology Guidance Program Project [20200402023NC], Jilin Provincial Natural Science Foundation Project [20200201027JC] and Innovation and Entrepreneurship Training Program for College Students in Jilin Province [2021].
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Wang, C., Linderholm, H. W., Song, Y., Wang, F., Liu, Y. et al. (2020). Impacts of drought on maize and soybean production in Northeast China during the past five decades. International Journal of Environmental Research, 17(7), 2459. DOI 10.3390/ijerph17072459. [Google Scholar] [CrossRef]
2. Imran, M., Latif Khan, A., Shahzad, R., Aaqil Khan, M., Bilal, S. et al. (2021). Exogenous melatonin induces drought stress tolerance by promoting plant growth and antioxidant defence system of soybean plants. AOB Plants, 13(4), plab026. DOI 10.1093/aobpla/plab026. [Google Scholar] [CrossRef]
3. Li, X. F., Guo, Z. B., Zhu, H. X., Wang, P., Gong, L. J. et al. (2020). Time-series characteristics of drought and flood in spring soybean growing season and its effect on soybean yield in Heilongjiang Province, China. Ying Yong Sheng Tai Xue Bao, 31(4), 1223–1232 (in Chinese). DOI 10.13287/j.1001-9332.202004.016. [Google Scholar] [CrossRef]
4. Shan, Q., Ma, F., Wei, J., Li, H., Ma, H. et al. (2020). Physiological functions of heat shock proteins. Current Protein & Peptide Science, 21(8), 751–760. DOI 10.2174/1389203720666191111113726. [Google Scholar] [CrossRef]
5. Li, Z., Long, R., Zhang, T., Wang, Z., Zhang, F. et al. (2007). Molecular cloning and functional analysis of the drought tolerance gene MsHSP70 from alfalfa (Medicago sativa L.). Journal of Plant Research, 130(2), 387–396. DOI 10.1007/s10265-017-0905-9. [Google Scholar] [CrossRef]
6. Ul Haq, S., Khan, A., Ali, M., Khattak, A. M., Gai, W. X. et al. (2019). Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. International Journal of Molecular Sciences, 20(21), 5321. DOI 10.3390/ijms20215321. [Google Scholar] [CrossRef]
7. Bernfur, K., Rutsdottir, G., Emanuelsson, C. (2017). The chloroplast-localized small heat shock protein Hsp21 associates with the thylakoid membranes in heat-stressed plants. Protein Science, 26(9), 1773–1784. DOI 10.1002/pro.3213. [Google Scholar] [CrossRef]
8. Hamilton, E. W.3rd., Heckathorn, S. A. (2001). Mitochondrial adaptations to NaCl. Complex I is protected by anti-oxidants and small heat shock proteins, whereas complex II is protected by proline and betaine. Plant Physiology, 126(3), 1266–1274. DOI 10.1104/pp.126.3.1266. [Google Scholar] [CrossRef]
9. Wang, X., Zhang, H., Shao, L. Y., Yan, X., Peng, H. et al. (2019). Expression and function analysis of a rice OsHSP40 gene under salt stress. Genes Genomics, 41(2), 175–182. DOI 10.1007/s13258-018-0749-2. [Google Scholar] [CrossRef]
10. Feng, X. H., Zhang, H. X., Ali, M., Gai, W. X., Cheng, G. X. et al. (2019). A small heat shock protein CaHsp25.9 positively regulates heat, salt, and drought stress tolerance in pepper (Capsicum annuum L.). Plant Physiology and Biochemistry, 142, 151–162. DOI 10.1016/j.plaphy.2019.07.001. [Google Scholar] [CrossRef]
11. Huang, L. J., Cheng, G. X., Khan, A., Wei, A. M., Yu, Q. H. et al. (2019). CaHSP16.4, a small heat shock protein gene in pepper, is involved in heat and drought tolerance. An International Journal of Animal, Fungal and Plant Cell Biology, 256(1), 39–51. DOI 10.1007/s00709-018-1280-7. [Google Scholar] [CrossRef]
12. Zhuang, K., Gao, Y., Liu, Z., Diao, P., Sui, N. et al. (2020). WHIRLY1 regulates HSP21.5A expression to promote thermotolerance in tomato. Plant and Cell Physiology, 61(1), 169–177. DOI 10.1093/pcp/pcz189. [Google Scholar] [CrossRef]
13. Wang, R., Mao, C., Jiang, C., Zhang, L., Peng, S. et al. (2021). One heat shock transcription factor confers high thermal tolerance in clematis plants. International Journal of Molecular Sciences, 22(6), 2900. DOI 10.3390/ijms22062900. [Google Scholar] [CrossRef]
14. Hsu, P. D., Lander, E. S., Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell, 157(6), 1262–1278. DOI 10.1016/j.cell.2014.05.010. [Google Scholar] [CrossRef]
15. Xu, R., Li, Y., Sui, Z., Lan, T., Song, W. et al. (2021). A C-terminal encoded peptide, ZmCEP1, is essential for kernel development in maize. Journal of Experimental Botany, 72(15), 5390–5406. DOI 10.1093/jxb/erab224. [Google Scholar] [CrossRef]
16. Zhang, S., Zhang, R., Gao, J., Song, G., Li, J. et al. (2021). CRISPR/Cas9-mediated genome editing for wheat grain quality improvement. Plant Biotechnology Journal, 19(9), 1684–1686. DOI 10.1111/pbi.13647. [Google Scholar] [CrossRef]
17. Ma, J., Sun, S., Whelan, J., Shou, H. (2021). CRISPR/Cas9-mediated knockout of GmFATB1 significantly reduced the amount of saturated fatty acids in soybean seeds. International Journal of Molecular Sciences, 22(8), 3877. DOI 10.3390/ijms22083877. [Google Scholar] [CrossRef]
18. Li, P. S., Yu, T. F., He, G. H., Chen, M., Zhou, Y. B. et al. (2014). Genome-wide analysis of the HSF family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genomics, 15(1), 1009. DOI 10.1186/1471-2164-15-1009. [Google Scholar] [CrossRef]
19. Rong, J., Wang, P. W., Wu, N., Qu, J., Yu, M. et al. (2018). Identification of heat resistance function of soybean heat shock protein gene HSP17.4. Journal of Jilin Agricultural University, 40(5), 568–576. DOI 10.13327/j.jjlau.2018.3698. [Google Scholar] [CrossRef]
20. Huang, Y., Xuan, H., Yang, C., Guo, N., Wang, H. et al. (2019). GmHsp90A2 is involved in soybean heat stress as a positive regulator. Plant Science, 285(7), 26–33. DOI 10.1016/j.plantsci.2019.04.016. [Google Scholar] [CrossRef]
21. Duy, P. N., Lan, D. T., Thu, H. P. (2019). Improved bacterial leaf blight disease resistance in the major elite Vietnamese rice cultivar TBR225 via editing of the OsSWEET14promoter. PLoS One, 16(9), e0255470. DOI 10.1371/journal.pone. [Google Scholar] [CrossRef]
22. Do, P. T., Nguyen, C. X., Bui, H. T., Tran, L. T. N., Stacey, G. et al. (2019). Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC Plant Biology, 15, 19(1), 311. DOI 10.1186/s12870-019-1906-8. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |