

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.019912
ARTICLE
Phytohormones Accumulation and Distribution in Shoots and Roots of Haploid, Diploid and Tetraploid Barley Seedlings Derived from Microspore Culture
1Biotech Research Institute, Shanghai Academy of Agricultural Sciences, Shanghai, 200000, China
2Biotechnology Research Institute, Shanghai Key Laboratory of Agricultural Genetics and Breeding, Shanghai, 200000, China
3Shanghai Baixin Biotechnology Company Limited, Shanghai, 200000, China
*Corresponding Authors: Zhiwei Chen. Email: chenzhiwei@saas.sh.cn; Chenghong Liu. Email: liuchenghong@saas.sh.cn
#These two authors contributed equally to this work
Received: 23 October 2021; Accepted: 22 December 2021
Abstract: Phytohormones play important roles in plant growth and development, and polyploids are thought to be an important method for plant breeding. However, the relationship between ploidy and phytohormone is still unclear. In this study, barley at three ploidy levels were produced by microspore culture. Therefore, we further analyzed the phytohormone content in the shoots and roots of the three kinds of barley materials to study the effect of ploidy on phytohormones accumulation and distribution. The results showed that Abscisic acid (ABA), gibberellin (GA), jasmonic acid (JA), auxin (IAA), salicylic acid (SA) and cytokinin (CTK) were successfully determined in shoots and roots using LC-MS (liquid chromatography mass spectrometry). By comparing the shoots of the haploid and diploid plants, it was found that the distribution trend of the six phytohormones was consistent, and another consistent trend was found in the roots of the diploid and tetraploid plants. In addition, we further analyzed the shoot/root ratio of the different phytohormones to identify the potential differences for haploid, diploid and tetraploid. Here, the relationship between ploidy and phytohormone we provided would provide new insights into understanding the new phenotypes that occur in polyploid species.
Keywords: Barley; polyploidy; microspore culture; phytohormone; shoot; root
The new phenotype normally occurs in natural polyploidy and has certain advantages in agriculture applications. Polyploidy is considered to be an important force influencing the evolution of eukaryotes, especially flowering plants [1]. Many important crops, such as wheat, soybean, cotton or rapeseed, are almost the selective result of spontaneous interspecific hybridization and then undergo a process of polyploidization. In most cases, the percentage of polyploids in angiosperms is estimated to be about 70% [1].
Barley is the fourth largest cereal crop produced in the world. Interestingly, barley is also suitable for polyploidy research. One reason for this characteristic is that barley has a high self-breeding rate and its genetic background is not easy to cross. Moreover, barley is one of the cereal crops that have a high response to embryogenesis and can effectively regenerate homozygous plants [2,3]. In the process of embryogenesis, the genome copies of some special plant cells can automatically replicate themselves, eventually forming aneuploidy plants [4]. Thus, these crucial points give barley advantages for use as a potential crop in producing the polyploid materials.
Phytohormones are crucial to the development of plants. In view of the morphology form of plant, the vegetative phase of plant development can be divided into an overground part and an underground part. The leaves and stems make up the overground part (also called shoot), and the underground part is mainly composed of roots. Many factors, including soil nutrition, humidity, temperature, light and plant growth regulators, could affect the physiological growth activities of the plant overground and underground part. Among these factors, the plant growth regulators phytohormones could regulate various biological processes for the plants overground and underground parts. For example, auxin is the first plant growth phytohormone discovered and has been proven to be the central regulator for root growth [5]. Cytokinin (CTK) stimulates root cell division [6]. Abscisic acid (ABA) and enthylene (ETH) are involved in OsNAC2-regulated pathway to regulate plant seedling morphogenesis [7]. Gibberellin (GA) controls seed germination and plays an important role in the ‘green revolution’ [8]. Salicylic acid (SA) and jasmonic acid (JA) act as the important signal molecules during the immune and defense responses of plants [9,10].
A convenient way to study the effects of polyploid on phenotypes is to use a series of plants with different ploidy levels. Dana et al. [11] simultaneously quantified the cell and organ sizes of the sepals of Arabidopsis diploid, tetraploid, and octoploid plants and examined the proportional relationship between ploidy and size. Xiang et al. [12] found that a significant difference of morphology, physiology and tolerance between autotetraploid (4x = 44) Stevia rebaudiana (bertoni) and its diploids (2x = 22). Wang et al. [13] also found that autotetraploid Paulownia australis plants exhibited superior traits compared with their diploid progenitors. In addition, the autopolyploid cabbage and maize had also been reported to affect the ultimate phenotypes [14,15].
As discussed above, the phytohormones could regulate phenotype, and the different ploidy levels could also influence phenotype. However, to date, the relationship between the phytohormones and the different ploidy levels of plants is unclear. Here, we chose to use barley to solve this problem, as barley is a good model for genetic and physiology research [16,17]. The haploid, diploid and tetraploid barley plants were successfully obtained through microspore in vitro culture by barley cultivar Hua-30. Finally, 6 kinds of phytohormones were successfully detected by using liquid chromatography-mass spectrometry (LC-MS) technology, and phytohormones distribution characteristics (root/shoot ratio) of haploid, diploid and tetraploid were analyzed.
2.1 Plant Materials Construction
The barley cultivar “Hua-30”, two-row spring barley, was applied in isolated microspore culture.
2.1.1 Extraction Solution (ES), Pretreatment Solution (PS) and Medium Preparation
First, 27.2 mg KH2PO4, 246.0 mg MgSO4·2H2O, 101.0 mg KNO3 and 1480.0 mg CaC12·2H20 dissolved into 1000 ml sterilized water to achieve solution CPW. Then, 0.3 mol mannitol dissolved into 1000 ml CPW solution to obtain extraction solution ES. And 10 mg colchicine dissolved into ES solution to obtain pretreatment solution PS. The induction medium was prepared by adding 0.5 mg KT, 0.5 mg 2,4-D and 90 g maltose into 1000 ml N6 medium. For differentiation medium preparation, 0.5 mg 6-BA, 1.5 mg KT, 0.05 mg NAA and 30 g maltose were added into 1000 ml MS medium. The treatment solution and culture medium were used after filter-sterilization.
Panicles from main stems containing early- and middle-stage uninucleate microspores were collected and put at a 4°C refrigerator before this experiment. During the inoculation period, panicles were disinfected in saturated bleach solution for 15 min and were washed 3 times with sterile water. Then, ten panicles were transferred into a test tube including 15 ml extract. Mixed liquids were cut using a high-speed rotary disperser and filtered through a 150-mesh screen to produce filtrate. The filtrate was centrifuged for 5 min at 100 g at room temperature, and the microspores were gently resuspended in pretreatment liquid and placed for 2 days at 25°C in the dark. Microspores were washed once with induction medium before being cultured, and then the induction medium was used to adjust microspore density to 2.0 × 105~3.0 × 105/ml. The 4 ml microspore suspension was inoculated into a 60-mm × 15-mm petri dish and tightly sealed with parafilm and cultured at 25°C in dark.
2.1.3 Embryoid Body Regeneration
The embryoid body formation on the 21st day in induction medium was transferred to the differentiation medium, and the seedlings with roots and green leaves were marked for further ploidy identification.
2.1.4 Flow Cytometry Analysis of Ploidy
The fine suspension was achieved by gently grinding the 1.5 cm barley leaves into 1.5 mL EP tubes using liquid nitrogen. 0.5 mL LB-01 buffer (Tris 15 mmol · L−1, Na2EDTA·2H2O 2 mmol · L−1, Spermine 0.5 mmol · L−1, KCl 80 mmol · L−1, NaCl 20 mmol · L−1, Trixon X-100 0.1%) and 0.2 mL PI/RNase staining buffer (BD#3179921) were added and pipetted up and down quickly five times. The suspension was placed on an ice-water mixture and protected from light for 20 min, and then was filtered through nylon mesh (pore size, 37.5 μm; Tokyo Screen Co., Japan), the harvested filtrate was transferred to a new EP tube. The cellular ploidy was detected by flow cytometry (BD Accrui C6) and analyzed via BD Accuri C6 Software.
2.1.5 Checking the Chromosome Number in Root-Tip Cells by Microscopy
First, 2 cm roots were cut at a fixed distance from the root tip at 8:30~9:00, next, the roots were transferred into 1.5 ml centrifuge tubes containing 1 ml of colchicine (0.1%), the tubes were treated in the cell culture incubator at 13°C for 3 h. Subsequently, roots in the tubes were washed three times in distilled water (5 min/wash), excess water removed with absorbent tissue paper and transferred into new EP tubes, immediately 1 ml of Carnoy’s fixative was added for 24 h to make slices. Using acetic acid magenta solution to stain for roots and observing the chromosome number from slides pre-soaked in 45% glacial acetic acid.
2.2 Planting Plants in a Climate-Controlled Room and Plants Conservation
After the above series of operations, haploid, diploid and tetraploid seedlings were selected, followed by growing in a climate-controlled room. Briefly, seedlings were grown in half-strength Hoagland nutrient solution for 4 to 6 weeks for the production of new fine roots, with a nutrient solution change every three days. Then, uniform haploid, diploid and tetraploid seedlings were fixed on a plastic sheet and grown in a plastic box filled to the half with tap water, after 3 days, we used Hoagland nutrient solution to replace water, with a change every three days. After 2 weeks, shoots and roots derived from the haploid, diploid and tetraploid were separately collected as one biological replicate from three plates.
Seeds were harvested from each independent plant, the ploidy levels of the haploid, diploid and tetraploid were further confirmed according to its pollen fertility. Seeds of diploid and tetraploid were harvested. The brief method of haploid plants subculture is described as follows. First, the plants identified as haploids were transferred from strong seedling rooting medium (1/2 MS + NAA 0.05 mg · L−1 + chlormequat chloride 3.0 mg · L−1 + sucrose 30 g · L−1 + agar powder 6.0 g · L−1) to basic medium (1/2 MS + TDZ 0.5 mg · L−1 + chlormequat chloride 1.0 mg · L−1 + sucrose 30 g · L−1 + agar powder 6.0 g · L−1). The stages of proliferation and subculture were implemented in the basic medium and the frequency of subculture was planned for once every 4–6 weeks. After 5 subculture periods, the haploid plants were transferred to the strong seedling rooting medium, and the ploidy of each line was checked again by the above two methods.
2.3 Phytohormones Extraction and Ultra-High-Performance Liquid Chromatography
The phytohormones determination was performed by Shanghai Luming Biotechnology Co., Ltd. (China). Chloroform and ultrasound disruption were used to extract phytohormones from leaf samples. After evaporation, reconstitute the samples with methanol: water: formic acid (7.9:2:0.1) in an ice bath. The MS system included of an API5500 triple quadrupole (AB Sciex), an ESI source, and an Analyst 1.6.2 software workstation. Use the ESI source to perform a Multiple Reaction Detection Mode (MRM) analysis with positive and negative ion scanning. The MS conditions were as follows: CUR (Curtain Gas): 30 Psi; CAD (Collision Gas): 8 Psi; IS (IonSpray Voltage): 3000 V; TEM (Temperature): 500°C; GS1 (Ion Source Gas1): 35 Psi; GS2 (Ion Source Gas2): 45 Psi. The chromatographic system uses Waters high-performance liquid chromatograph and ACQUITY UPLC BEH C18 liquid chromatography column (2.1 × 100 mm, 1.7 μm). The column temperature is 40°C, and the injection volume is 5 μL. The gradient elution conditions of mobile phase A is 0.1% formic acid in water and B is 0.1% formic acid in acetonitrile.
The values for shoot length, root length, the number of tillers and the phytohormones content are mean values with corresponding standard error (i.e., mean ± SE). Each phenotypic parameter includes three biological repetitions. In analyzing the content of phytohormones in shoots and roots of haploid, diploid and tetraploid barley plants, one-way analysis of variance (ANOVA) followed by Fisher’s LSD multiple comparison test was carried out by using SPSS 24.0. In analyzing the shoot/root ratio of the phytohormones, statistical analysis was performed using the two tailed unpaired Student’s t-test. * represents the P value < 0.05, ** represents the P value < 0.01, ns indicates not significant difference.
We grew the barley cultivar (Hordeum vulgare L. cv. Hua-30) at China Shanghai Academy of Agriculture Sciences (31°22′N, 121°33′E) in November 2017. Panicles from cv. Hua-30 were collected in March 2018, followed by the isolation and cultivation of microspores, and further “embryoid body regeneration” and “ploidy identification” stages were the following steps (Fig. 2A). For the confirmation of haploid, diploid and tetraploid (Fig. 2B), a preliminary identification was carried out using flow cytometry technology. The peaks of flow cytometry that located in ‘1C’ were from haploid (Fig. 1A), while the peaks that located in ‘2C’ and ‘4C’ respectively were from diploid (Fig. 1B) and tetraploid (Fig. 1C). In addition, the chromosome number was measured by a Zeiss Axioskop microscope at 100x magnification, results showed that the root cell of haploid had 8 chromosomes (Fig. 1D), the diploid had 16 chromosomes (Fig. 1E) and the tetraploid had 24 chromosomes (Fig. 1F).
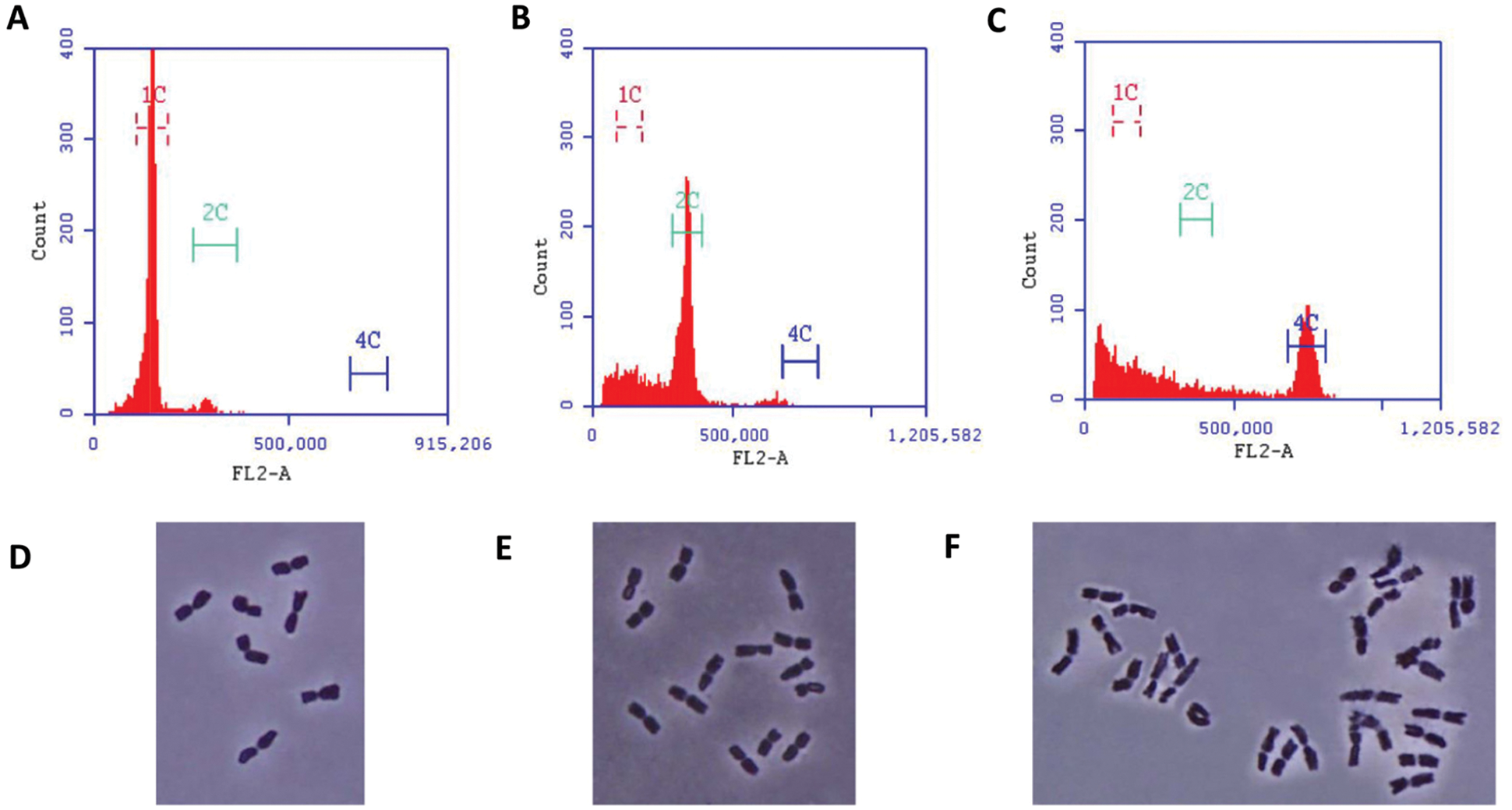
Figure 1: Flow cytometric identification of haploid (A), diploid (B) and tetraploid (C). Fluorescence microscope identification of chromosome numbers in root tips cells separately for haploid (D), diploid (E) and tetraploid (F)
3.2 Phenotypic Observation of Haploid, Diploid and Tetraploid
Observation of the seedling morphology at the sampling stage indicated that the haploid plant showed a highly significant lower shoot height than the diploid plant (P < 0.01), and the shoot length of the tetraploid plant had no significant difference with haploid, or diploid (Fig. 2C). For analysis of root length, the difference between the haploid, diploid or tetraploid plants reached a statistically significant level (P < 0.05) (Fig. 2D). Finally, we found that the haploid plant had 5∼6 tillers, diploid had 4∼5 tillers, while tetraploid had only 1 tiller (Fig. 2E).
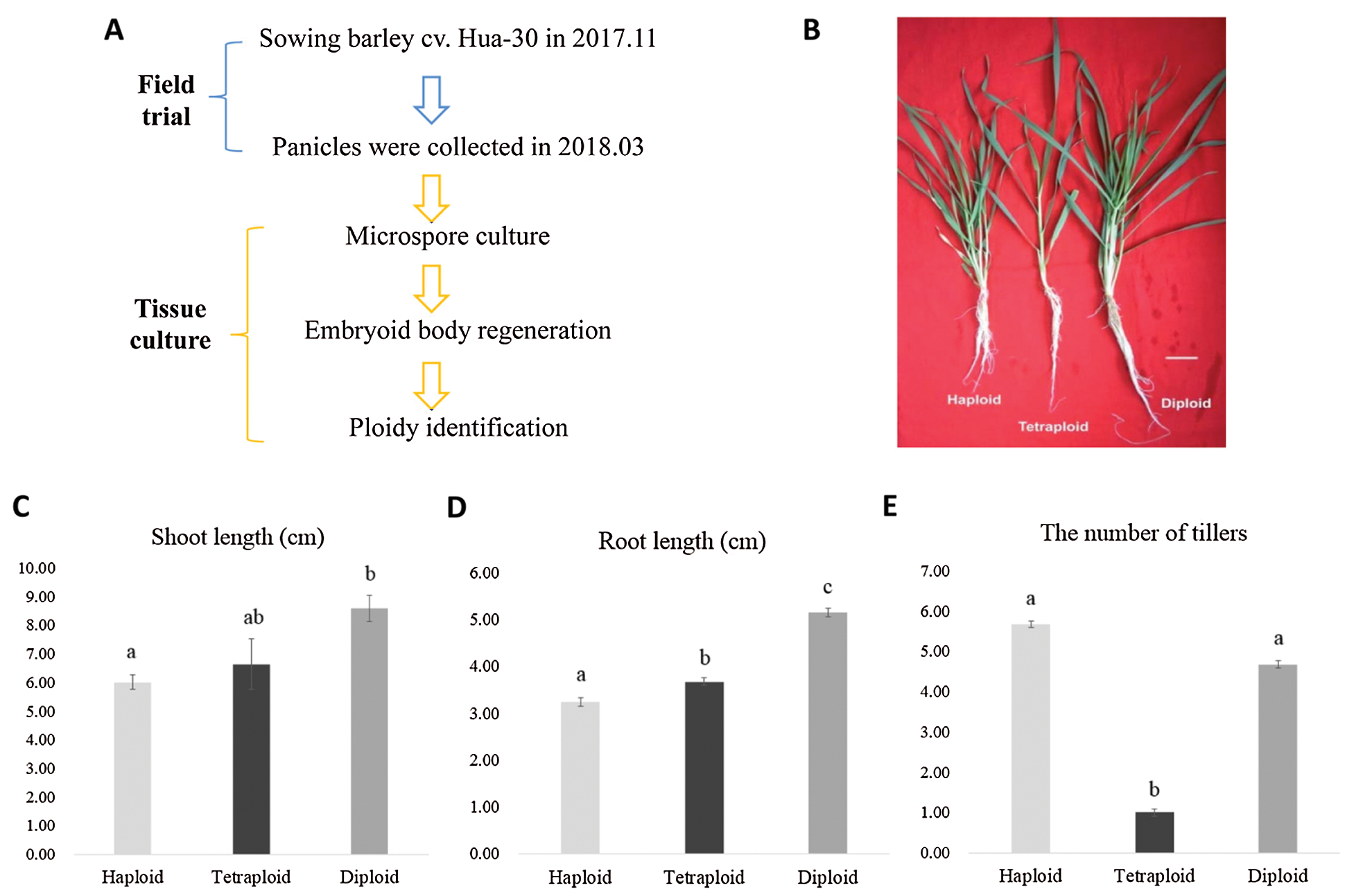
Figure 2: (A) Schematic drawings of material creation. (B) Seedling morphological observation of the materials at three different ploidy levels, bar = 1 cm. Statistical analysis of shoot length (cm) (C), root length (cm) (D) and the number of tillers (E) for haploid, tetraploid and diploid. Note, significant level was P < 0.05
3.3 Detection and Analysis of Phytohormones in Haploid, Diploid and Tetraploid
Totally, six kinds of phytohormones were detected in shoots and roots of haploid, diploid and tetraploid. Among the haploid, diploid and tetraploid plants, JA displayed the highest content in shoots and roots. Interestingly, by comparing the shoots of the haploid and diploid plants, it was found that the distribution trend of the six phytohormones was consistent (Fig. 3 red frame), while another consistent trend was found in the roots of the diploid and tetraploid plants (Fig. 3 purple frame). More, for comparison of the distribution trend between shoot and root of the haploid plant, we found that the distribution trend was not consistent (Fig. 3), this is the same as the diploid and tetraploid plant.
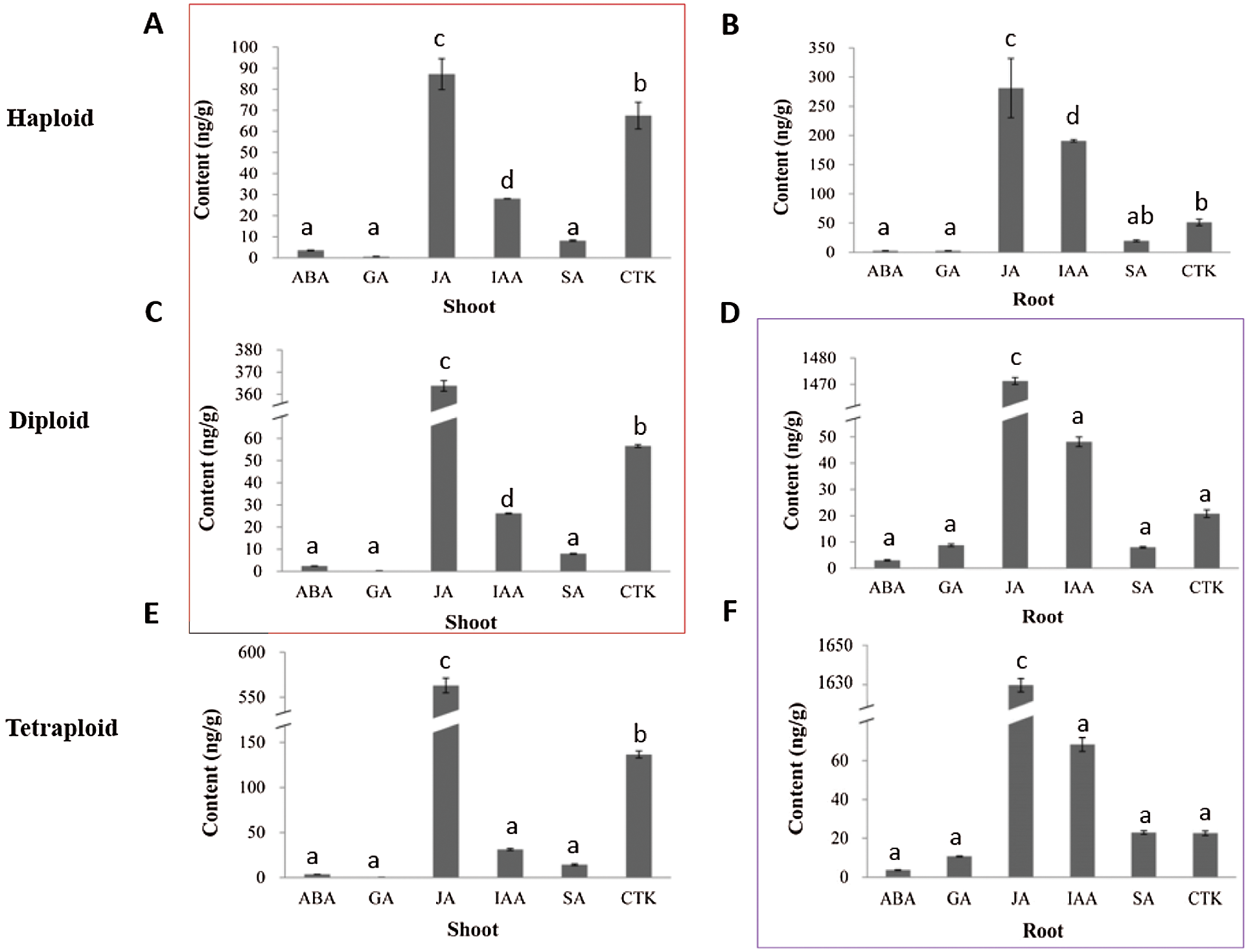
Figure 3: Analysis of the content of the six kinds of phytohormones in shoots and roots of the haploid, diploid and tetraploid plants. (A, B) represent the haploid plant, (C, D) represent the diploid plant, (E, F) represent the tetraploid plant. Note, significant level was P < 0.05
3.4 Analysis of Phytohormones Ratio (Shoot/Root) in Haploid, Diploid and Tetraploid
Considering that phytohormones have a great influence on the overground and underground parts of plants (see introduction), therefore, mining which hormones among the haploid, diploid and tetraploid plants tend to be enriched in shoot, and which hormones tend to be enriched in root, has certain reference significance in analyzing the phenotypic differences between the haploid, diploid, and tetraploid plants. Based on the significant results, in haploid plants, we found that the ABA content in the shoot was higher than the root (P < 0.05), while the haploid root contained higher JA (P < 0.05), SA (P < 0.01), GA (P < 0.01) and IAA (P < 0.01) content. In diploid plants, comparing to the root, the shoot had higher CTK content (P < 0.01), while the root had higher JA (P < 0.01), GA (P < 0.01), IAA (P < 0.01) content. In tetraploid plants, the CTK content in the shoot was higher (P < 0.01) than the root, but the contents of JA (P < 0.01), SA (P < 0.05), GA (P < 0.01) and IAA (P < 0.01) in the root were higher (Figs. 4 and 5).
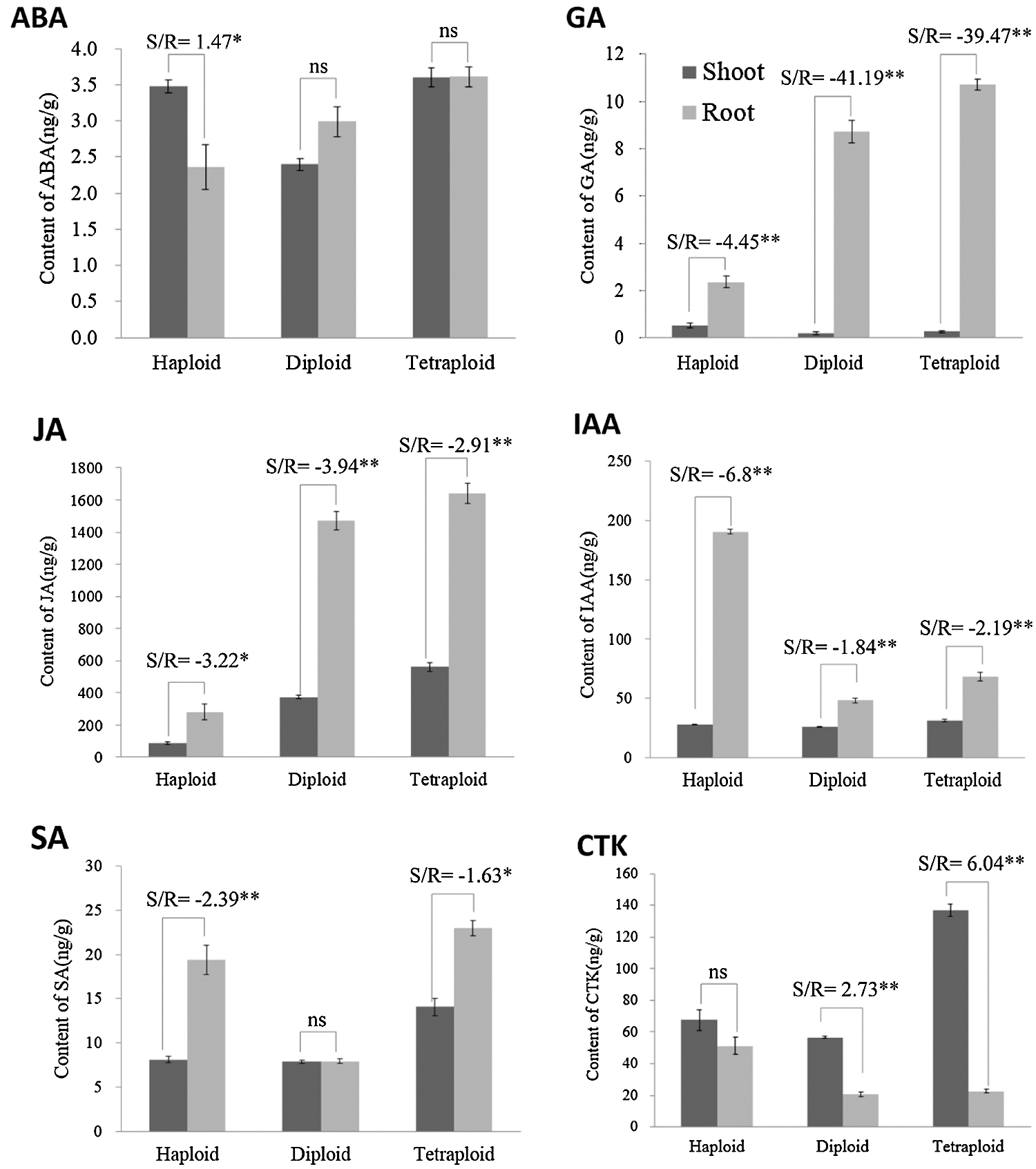
Figure 4: Statistical analysis of the shoot/root ratio for ABA, JA, SA, GA, IAA and CTK in haploid, diploid and tetraploid. * represents P < 0.05, ** represents P < 0.01
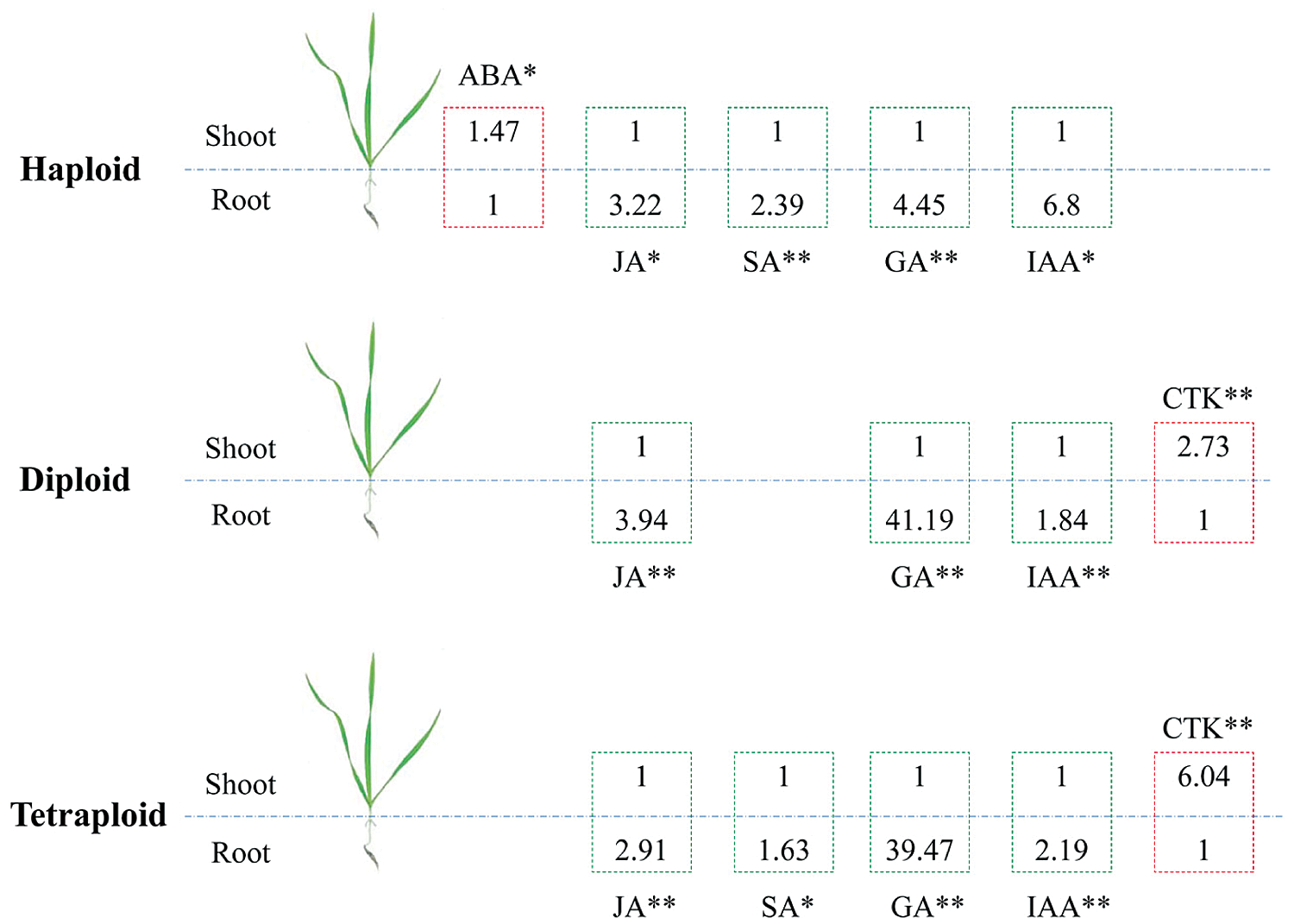
Figure 5: Summary of those phytohormones with significant difference between shoots and roots in haploid, diploid and tetraploid. * represents P < 0.05, ** represents P < 0.01
Our results showed that the distribution trend of six phytohormones (i.e., ABA, GA, JA, IAA, SA and CTK) in diploid root was consistent with tetraploid root. From diploid to tetraploid, it is a process of genome duplication. Genome duplication leads to a good interaction between individual copies of genome in the offspring. Compared to their parent species, this characteristic can permanently change their internal genetic mechanisms and produce new phenotypes [1,18]. Our results are beneficial for the mechanisms study of the new phenotypes that occur in polyploidy. More, the accumulation characteristics of the different phytohormones we provided could be expanded to analyze the potential differences between haploid, diploid and tetraploid.
In recent years, the roles of polyploidy in regulating plant metabolism and physiology had been studied widely [19–22], however, those conclusions were miscellaneous [23], one major reason we drew was that the genome copies of the polyploid were usually from different parental species (allopolyploidy), rather than multiple copies of the same genome (autopolyploidy), the pure genome duplication effect could be easily mixed by other genetic factors, such as the allele effect [24].
To overcome this defect and draw a clear conclusion on the pure genome duplication effects, this article used an efficient microspore regeneration experimental system based on barley cv. Hua-30, successfully produced the haploid, diploid and tetraploid homozygous lines. And, an earlier study also supported that it was possible to induce uninucleate microspores to obtain fully homozygous diploid plants [3]. In addition, the effects we investigated by using the haploid, diploid and tetraploid homozygous lines we created could also be called “fixed heterosis” [25,26], the fact was that the “fixed heterosis” would not be lost during inbreeding.
Last, the reason for we selected phytohormone was that one study had showed that in the comparative analysis of proteomics, autotetraploids and diploids Brassica napus were no significant difference [27]. Thus, we concluded that selection of a suitable sensitive phenotypic parameter for investigating the effects of genome duplication was important.
In conclusion, through providing a first work of describing the effects of doubling the barley genome copies on phytohormones accumulation and distribution, we provided a certain meaningful way to help to understand the mechanisms of new phenomenon produced in polyploidy crops.
Funding Statement: This work was financially supported by the project from the Key Technology R&D Project of Shanghai Agriculture-developed with Science & Technology Program, China (Grant No. 2018(1–2)); the Natural Science Foundation of Shanghai, China (Grant No. 19ZR1417000); the National Key Research and Development Program of China (2018YFD1000702-5); the Climbing Plan of Shanghai Academy of Agricultural Sciences (2022); the earmarked fund for China Agriculture Research System of MOF and MARA (CARS-05-01A-02).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Oleszczuk, S., Lukaszewski, A. J. (2014). The origin of unusual chromosome constitutions among newly formed allopolyploids. American Journal of Botany, 101(2), 318–326. DOI 10.3732/ajb.1300286. [Google Scholar] [CrossRef]
2. Soriano, M., Li, H., Boutilier, K. (2013). Microspore embryogenesis: Establishment of embryo identity and pattern in culture. Plant Reproduction, 26(3), 181–196. DOI 10.1007/s00497-013-0226-7. [Google Scholar] [CrossRef]
3. Belanger, S., Marchand, S., Jacques, P. E., Meyers, B., Belzile, F. (2018). Differential expression profiling of microspores during the early stages of isolated microspore culture using the responsive barley cultivar Gobernadora. G3-Genes Genomes Genetics, 8(5), 1603–1614. DOI 10.1534/g3.118.200208. [Google Scholar] [CrossRef]
4. Lu, R. J., Wang, Y. F., Sun, Y. F., Shan, L. L., Chen, P. D. et al. (2008). Improvement of isolated microspore culture of barley (Hordeum vulgare L.The effect of floret co-culture. Plant Cell Tissue and Organ Culture, 93(1), 21–27. DOI 10.1007/s11240-008-9338-4. [Google Scholar] [CrossRef]
5. Gelova, Z., Gallei, M., Pernisova, M., Brunoud, G., Zhang, X. et al. (2021). Developmental roles of Auxin Binding Protein 1 in Arabidopsis thaliana. Plant Science, 303, 110750. DOI 10.1016/j.plantsci.2020.110750. [Google Scholar] [CrossRef]
6. Ohashi-Ito, K., Saegusa, M., Iwamoto, K., Oda, Y., Katayama, H. et al. (2014). A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Current Biology, 24(17), 2053–2058. DOI 10.1016/j.cub.2014.07.050. [Google Scholar] [CrossRef]
7. Yu, J. T., Mao, C. J., Zhong, Q., Yao, X. F., Li, P. et al. (2021). OsNAC2 is involved in multiple hormonal pathways to mediate germination of rice seeds and establishment of seedling. Frontiers in Plant Science, 12, 1664–462X. DOI 10.3389/Fpls.2021.699303. [Google Scholar] [CrossRef]
8. Tzatzani, T. T., Basdeki, E., Ladikou, E. V., Sotiras, M. I. N., Panagiotakis, G. et al. (2020). Seed germination traits of loquat (Eriobotrya japonica Lindl.) as affected by various pre-sowing treatments (cutting of cotyledons, removal of perisperm, moist chilling and/or exogenous application of gibberellin). Phyton-International Journal of Experimental Botany, 89(3), 645–656. DOI 10.32604/phyton.2020.010532. [Google Scholar] [CrossRef]
9. Mollah, M. M. I., Choi, H. W., Yeam, I., Lee, J. M., Kim, Y. (2021). Salicylic acid, a plant hormone, suppresses phytophagous insect immune response by interrupting HMG-Like DSP1. Frontiers in Physiology, 12, 744272. DOI 10.3389/fphys.2021.744272. [Google Scholar] [CrossRef]
10. Smirnova, E., Marquis, V., Poirier, L., Aubert, Y., Zumsteg, J. et al. (2017). Jasmonic Acid Oxidase 2 hydroxylates jasmonic acid and represses basal defense and resistance responses against botrytis cinerea infection. Molecular Plant, 10(9), 1159–1173. DOI 10.1016/j.molp.2017.07.010. [Google Scholar] [CrossRef]
11. Robinson, D. O., Coate, J. E., Singh, A., Hong, L., Bush, M. et al. (2018). Ploidy and size at multiple scales in the arabidopsis sepal. Plant Cell, 30(10), 2308–2329. DOI 10.1105/tpc.18.00344. [Google Scholar] [CrossRef]
12. Xiang, Z. X., Tang, X. L., Liu, W. H., Song, C. N. (2019). A comparative morphological and transcriptomic study on autotetraploid Stevia rebaudiana (bertoni) and its diploid. Plant Physiology and Biochemistry, 143(2), 154–164. DOI 10.1016/j.plaphy.2019.09.003. [Google Scholar] [CrossRef]
13. Wang, Z., Fan, G. Q., Dong, Y. P., Zhai, X. Q., Deng, M. J. et al. (2017). Implications of polyploidy events on the phenotype, microstructure, and proteome of Paulownia australis. PLoS One, 12(3), 1932–6203. DOI 10.1371/journal.pone.0172633. [Google Scholar] [CrossRef]
14. Gu, A. X., Zhao, J. J., Li, L. M., Wang, Y. H., Zhao, Y. J. et al. (2016). Analyses of phenotype and ARGOS and ASY1 expression in a ploidy Chinese cabbage series derived from one haploid. Breeding Science, 66(2), 161–168. DOI 10.1270/jsbbs.66.161. [Google Scholar] [CrossRef]
15. Riddle, N. C., Kato, A., Birchler, J. A. (2006). Genetic variation for the response to ploidy change in Zea mays L. Theoretical and Applied Genetics, 114(1), 101–111. DOI 10.1007/s00122-006-0414-z. [Google Scholar] [CrossRef]
16. Purugganan, M. D., Fuller, D. Q. (2009). The nature of selection during plant domestication. Nature, 457(7231), 843–848. DOI 10.1038/nature07895. [Google Scholar] [CrossRef]
17. Russell, J., Dawson, I. K., Flavell, A. J., Steffenson, B., Weltzien, E. et al. (2011). Analysis of >1000 single nucleotide polymorphisms in geographically matched samples of landrace and wild barley indicates secondary contact and chromosome-level differences in diversity around domestication genes. New Phytologist, 191(2), 564–578. DOI 10.1111/j.1469-8137.2011.03704.x. [Google Scholar] [CrossRef]
18. Koh, O., Shigehiro, K. (2018). Inference of the ancestral vertebrate phenotype through vestiges of the whole-genome duplications. Briefings in Functional Genomics, 17(5), 352–361. DOI 10.1093/bfgp/ely008. [Google Scholar] [CrossRef]
19. Nicole, C. R., James, A. B. (2008). Comparative analysis of inbred and hybrid maize at the diploid and tetraploid levels Theoretical and Applied Genetics, 116(4), 563–576. DOI 10.1007/s00122-007-0691-1. [Google Scholar] [CrossRef]
20. Pumphrey, M., Bai, J., Laudencia, C., Anderson, D., Gill, O. et al. (2009). Nonadditive expression of homoeologous genes is established upon polyploidization in hexaploid wheat. Genetics, 181(3), 1147–1157. DOI 10.1534/genetics.108.096941. [Google Scholar] [CrossRef]
21. Tan, F. Q., Tu, H., Wang, R., Wu, X. M., Xie, K. D. et al. (2017). Metabolic adaptation following genome doubling in citrus doubled diploids revealed by non-targeted metabolomics. Metabolomics, 13(11), 284. DOI 10.1007/s11306-017-1276-x. [Google Scholar] [CrossRef]
22. Yang, X. H., Cheng, J. L., Yao, B. B., Lu, H., Zhang, Y. et al. (2021). Polyploidy-promoted phenolic metabolism confers the increased competitive ability of Solidago canadensis. Oikos, 130(6), 1014–1025. DOI 10.1111/oik.08280. [Google Scholar] [CrossRef]
23. Veitia, R. A., Bottani, S., Birchler, J. A. (2008). Cellular reactions to gene dosage imbalance: Genomic, transcriptomic and proteomic effects. Trends in Genetics, 24(8), 390–397. DOI 10.1016/j.tig.2008.05.005. [Google Scholar] [CrossRef]
24. Ryder, P., McHale, M., Fort, A., Spillane, C. (2017). Generation of stable nulliplex autopolyploid lines of Arabidopsis thaliana using CRISPR/Cas9 genome editing. Plant Cell Reports, 36(6), 1005–1008. DOI 10.1007/s00299-017-2125-0. [Google Scholar] [CrossRef]
25. Abel, S., Mollers, C., Becker, H. C. (2005). Development of synthetic Brassica napus lines for the analysis of “fixed heterosis” in allopolyploid plants. Euphytica, 146(1–2), 157–163. DOI 10.1007/s10681-005-3364-7. [Google Scholar] [CrossRef]
26. Saminathan, T., Nimmakayala, P., Manohar, S., Malkaram, S., Almeida, A. et al. (2015). Differential gene expression and alternative splicing between diploid and tetraploid watermelon. Journal of Experimental Botany, 66(5), 1369–1385. DOI 10.1093/jxb/eru486. [Google Scholar] [CrossRef]
27. Albertin, W., Brabant, P., Catrice, O., Eber, F., Jenczewski, E. et al. (2005). Autopolyploidy in cabbage (Brassica oleracea L.) does not alter significantly the proteomes of green tissues. Proteomics, 5(8), 2131–2139. DOI 10.1002/pmic.200401092. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |