

 | Phyton-International Journal of Experimental Botany |  |
DOI: 10.32604/phyton.2022.018854
ARTICLE
Development of a New Cold-Tolerant Maize (Zea mays L.) Germplasm Using the ICE1 Gene from Arabidopsis thaliana
1College of Agronomy, Jilin Agricultural University, Changchun, 130118, China
2College of Life Sciences, Jilin Agricultural University, Changchun, 130118, China
*Corresponding Authors: Shuyan Guan. Email: guanshuyan@jlau.edu.cn; Yiyong Ma. Email: m18404319202_1@126.com
Received: 21 August 2021; Accepted: 14 December 2021
Abstract: To develop cold-tolerant maize germplasms and identify the activation of INDUCER OF CRT/DRE-BINDING FACTOR EXPRESSION (ICE1) expression in response to cold stress, RT-PCR was used to amplify the complete open reading frame sequence of the ICE1 gene and construct the plant expression vector pCAMBIA3301-ICE1-Bar. Immature maize embryos and calli were transformed with the recombinant vector using Agrobacterium tumefaciens-mediated transformations. From the regenerated plantlets, three T1 lines were screened and identified by PCR. A Southern blot analysis showed that a single copy of the ICE1 gene was integrated into the maize (Zea mays L.) genomes of the three T1 generations. Under low temperature-stress conditions (4°C), the relative conductivity levels decreased by 27.51%–31.44%, the proline concentrations increased by 12.50%–17.50%, the malondialdehyde concentrations decreased by 16.78%–18.37%, and the peroxidase activities increased by 19.60%–22.89% in the T1 lines compared with those of the control. A real-time quantitative PCR analysis showed that the ICE1 gene was ectopically expressed in the roots, stems, and leaves of the T1 lines. ICE1 positively regulates the expression of the CBF genes in response to cold stress. Thus, this study showed the successful transformation of maize with the ICE1 gene, resulting in the generation of a new maize germplasm that had increased tolerance to cold stress.
Keywords: Maize; ICE1; cold tolerance; inducer; maize; Arabidopsis thaliana
Maize (Zea mays L.) is an important crop worldwide [1]. Frost in late spring severely impacts maize production in Northeastern China [2]. The germination of maize seeds has been examined using a series of enzymatic reactions to identify tolerance to this stress. The catalytic activities of enzymes are closely correlated with changes in temperature. Low temperatures during the sowing period delay emergence, with an average low temperature of 10°C, resulting in a reduction of the seed germination potential to 9.4% and the germination rate to 6.7%, compared with those of the control, thereby seriously affecting maize yield [3].
In 1998, Gilmour et al. [4] proposed that the presence of an inducer of the CRT/DRE-BINDING FACTOR (CBF/DREB) gene (INDUCER OF CBF EXPRESSION, ICE) plays an important role in the regulation of CBF/DREB1 gene expression. The ICE1 gene encodes a bHLH transcription factor similar to the transcriptional activator MYC. At normal temperatures, ICE1 exists in an inactive form that is unable to activate transcription. However, when plants suffer low-temperature stress, ICE1 is activated and regulates CBF/DREB1 expression by binding to a DRE/CRT element in the gene’s promoter, initiating the ICE1–CBF/DREB1–COR cascade, and improving plant cold resistance. Post-transcriptional modifications of the ICE1 protein, such as phosphorylation, dephosphorylation, ubiquitination, and a small ubiquitin-like modifier, are important for its activity. Low temperature activates MPK3/MPK6 activity, and activated MPK3/MPK6 inhibits the stability and transcriptional activity of the ICE1 protein through the phosphorylation of its six conserved phosphorylation sites at low temperatures, thereby inhibiting the expression of the CBF gene and negatively regulating the tolerance of Arabidopsis thaliana to low-temperature stress [5]. The ICE1 activity is negatively regulated by protease degradation through HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 ligase of the ubiquitin E3 pathway [6,7]. The expression levels of a series of anti-stress-related genes may be regulated by the addition of an anti-stress-related transcription factor in plants that would be more effective than the insertion of a single anti-stress-related gene. In this study, the plant expression vector pCAMBIA3301-ICE1 was constructed to include the Bar gene as the selective marker. Maize was transformed with this vector using Agrobacterium tumefaciens-mediated transformation. This work is important for the improvement of cold resistance in maize, and it lays a theoretical foundation for further studies of anti-stress mechanisms in maize.
Immature embryos and embryogenic calli of the maize inbred line GSH9901 (a homozygous and stable inbred line covered by independent intellectual property rights bred by the research group), Escherichia coli (DH5α), Agrobacterium (EHA105), the recombinant cloning vector plasmid pMD18T-ICE1, and the plant expression vector pCAMBIA3301 were provided by the Center for Plant Biotechnology of Jilin Agricultural University. PCR amplification reagents, DNA markers, and restriction enzymes were purchased from MBI. Other reagents were of domestic analytical grade.
2.2.1 Construction of the Expression Vector pCAMBIA3301-ICE1
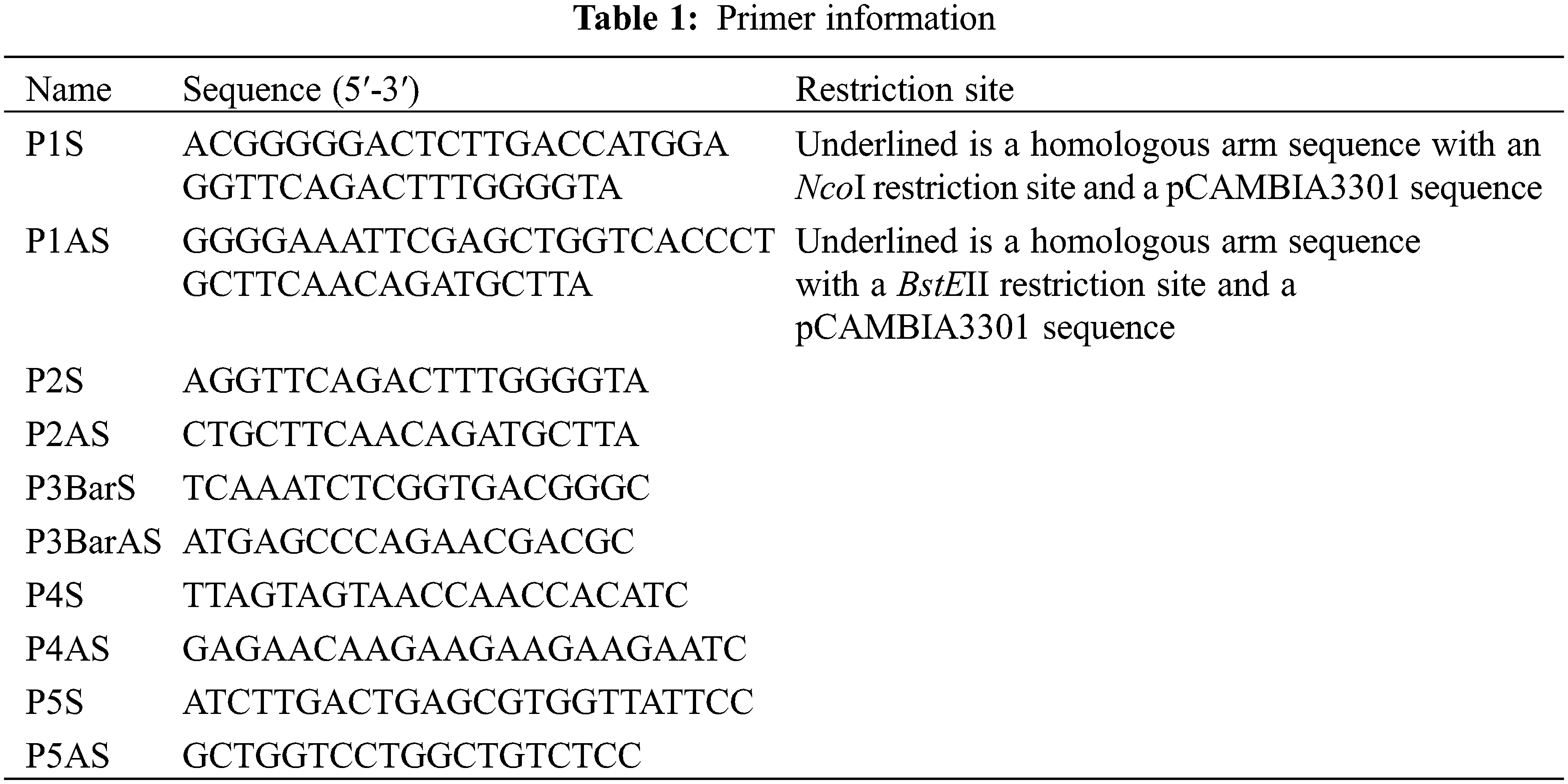
The complete open reading frame sequence of the ICE1 gene (GenBank accession number: NM_113586.3) was cloned from wild-type Arabidopsis. Using seamless cloning technology, pMD18T-ICE1 was used as the template to design the specific primers (P1S/P1AS) to amplify ICE1 (Table 1). The 20-μL PCR reaction comprised 10 μL 2× Es Taq MasterMix, 1 μL P1S, 1 μL P1AS, 1 μL ICE1 plasmid, and 7 μL dd water. The PCR conditions for ICE1 amplification were as follows: 94°C for 5 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min, followed by a final extension at 72°C for 10 min. The expression vector pCAMBIA3301 was double digested with NcoI and BstEII and then incubated at 37°C for 3 h, followed by 65°C for 20 min. The PCR products and the enzyme digestion products were purified and recycled, respectively, and then combined using a seamless connection kit (CE Design Company) in accordance with the manufacturer’s instructions. The reaction mixture comprised 50–200 ng vector fragment, 10–50 ng target fragment, 4 μL 5× buffer, 2 μL ligase, and dd water up to 20 μL. The reaction was carried out at 37°C for 30 min, followed by 0°C for 5 min. The ligation products were transformed into E. coli DH5α competent cells, and the positive clones were identified.
2.2.2 Agrobacterium-Mediated Transformation of Maize
The recombinant plasmid was transformed into Agrobacterium, and the colonies containing the recombinant plasmid pCAMBIA3301-ICE1 were selected and inoculated into YEP liquid medium containing Kan following the method of Jiao et al. [8] with minor modifications. The vials containing the infection broth and calli were placed at 140 rpm on a shaker at 28°C during the infection process to allow greater contact between the infection fluid and calli, resulting in a greater infection rate.
2.2.3 Detection of Transgenic Plants
Genomic DNAs of the T1 lines were extracted using the CTAB method [9] and used as the templates for PCR detection of the ICE1 and Bar genes. Transformed plants tested positive for the ICE1 and Bar genes, whereas untransformed plants tested negative for these genes. The recombinant plasmid pCAMBIA3301-ICE1 was used as the positive control, and untransformed recipient maize plants were used as negative controls. The primers P2S, P2AS, P3BarS, and P3BarAS (Table 1) were designed on the basis of known sequences using Primer 5.0 software. The PCR reaction system and conditions for amplification of ICE1 were as described earlier. PCR conditions for Bar gene amplification were as follows: 94°C for 5 min, 32 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s, followed by a final extension at 72°C for 10 s.
Southern Blot Analysis of Transgenic Plants
DNA was extracted from the leaves of T1 PCR-positive transgenic plants and digested with BamHI. ICE1 was used as the probe, and the biotin and DIG-labeled λ HindIII DNA (Beijing Bioco Laibo Technology Co., Ltd. (Beijing, China)) was used as the marker. Southern blotting was performed using a DIG DNA Labeling and Detection Kit I kit (Roche) in accordance with the manufacturer’s instructions.
Quantitative Real-Time PCR Detection of Transgenic Lines
Transgenic plants determined to be positive for the transgene by Southern blotting were maintained at 4°C for 24 h. Total RNA was then extracted from the roots, stems, and leaves of these plants, reverse transcribed into cDNA, and diluted four-fold. Fluorescent quantitative real-time PCR (qRT-PCR) was carried out using the P4S, P4AS, P5S, and P5AS primers (Table 1). The maize β-actin gene (GenBank accession number: NM_001252731.2) was used as a reference gene. The cDNAs of the cold-treated maize roots, stems, and leaves were analyzed using an Mx 3000 P FQ-PCR System (Jitai Biotech). A TaKaRa SYBR Premix Ex TaqTM kit was used in accordance with the manufacturer’s instructions. Each 20-μL PCR reaction comprised 10 μL 2× SYBR Premix Ex Taq polymerase, 2 μL primer P4S, 2 μL primer P4AS, 2 μL template, and dd water up to 20 μL. Rapid two-step PCR conditions were used with an initial pre-denaturation at 95°C for 3 min followed by 40 cycles of 95°C for 10 s for denaturation and 60°C for 30 s for the reaction. The relative expression of the target gene was analyzed using the 2−ΔΔCT method [10,11].
Determination of Physiological and Biochemical Indicators
The positive and untransformed plants were maintained at 4°C for 0, 24, or 72 h before changes in leaf relative conductivity, proline content, malondialdehyde (MDA) content, and peroxidase (POD) activity were measured. Each experiment was repeated three times to obtain an average. Relative conductivity was measured using the method of Petrov et al. [12]. The proline content was determined using the sulfosalicylic acid method, the MDA content by the thiobarbituric acid-spectrophotometric method, and POD activity using the guaiacol method [13].
All results in this study were performed in more than three replicates. SPSS 19.0 software was used for statistical analysis of experimental measurement data. One way ANOVA were used to confirm the variability of results between treatments, respectively, P < 0.05 (*), P < 0.01 (**).
3.1 Construction and Identification of the pCAMBIA3301-ICE1 Plant Expression Vector
The plant expression vector pCAMBIA3301-ICE1 was digested with NcoI and BstEII. The inducer sequence was amplified with its specific primer (Table 1). The products were separated by gel electrophoresis to identify the successful integration of the inducer sequence of the ICE1 gene into the pCAMBIA3301 vector (Fig. 1).

Figure 1: PCR and double digested verification recombinant plant expression vector. pCAMBIA3301-ICE1. (A): PCR to confirm the recombinant vector pCAMBIA3301-ICE1; M: DNA Marker DL 2000; 1–3: Recombinant plasmid; (B): double digested verification recombinant plant expression vector pCAMBIA3301-ICE1; M: DNA Marker DL 2000; N: water; 1–2: Recombinant plasmid
3.2 Production and Detection of Transgenic Plants on T1 Generation
3.2.1 PCR Detection of T1 Generation Plants
The plant expression vector pCAMBIA3301-ICE1 was transformed into the maize variety GSH9901, and three T0-positive plants were detected by PCR. In total, 20 seeds of each of these T0 plants were harvested and planted indoors to obtain T1 plants. Genomic DNA was extracted from these plants and screened using PCR to determine the presence or absence of the ICE1 gene (1,641 bp) and the herbicide-resistant Bar gene (552 bp). The recombinant plasmid pCAMBIA3301-ICE1 was used as the positive control, and untransformed recipient maize plants were used as negative controls. The results are shown in Fig. 2. Three transgenic positive maize plants GSH9901-1, -5, and -7 were successfully obtained.

Figure 2: PCR detection of ICE1 (A) and Bar (B) on T1 generation plants. M: DNA Marker DL 2000; P: Positive control; N: water ; CK: Non-transformed plant genome; 1–3: T1 transgenic plants
3.2.2 Southern Blot Analysis of T1 Plants
Genomic DNAs were extracted from PCR positive plants and digested with BamHI for 20 h before Southern blotting was performed using ICE1 as the probe. As shown in Fig. 3, the untransformed plants did not show hybridization signals, whereas the three transgenic plants showed significant hybridization signals. This indicated that the ICE1 gene was integrated into the maize genome as a single copy at different insertion sites.

Figure 3: Southern blotting detection on T1 generation of positive plants. M: λ HindIII DNA Marker; P: Positive control; N: Non-transformed plant genome; 1–3: T1 transgenic plants
3.2.3 Detection of Physiological and Biochemical Indicators Related to Cold Tolerance in Transgenic Maize
First, the three-leaf stage overexpression (GSH9901-1, -5, and -7) and wild type (WT) plants were treated at 4°C for 0, 24, and 72 h (Fig. 4), and then, phenotypes were observed. After a 24-h cold treatment, less wilting and lodging were observed in the transformed than in the control plants. The relative electrical conductivity, proline content, MDA content, and POD activity of the overexpressing transgenic and control plants were measured.

Figure 4: Phenotypic changes of maize under 4°C treatment
3.2.4 Determination of Relative Conductivity
Changes in the conductivity of plant tissue extravasation fluid reflect cell membrane damage and plant stress resistance. The greater the injury, the more likely the conductivity increases. As shown in Fig. 5, the relative electrical conductivities of ICE1 transgenic plants at ambient temperatures did not differ significantly from the non-transformed plants. The relative electric conductivities of the leaves of transformed strains GSH9901-1, -5, and -7 increased by 21.04%, 35.59%, and 24.74%, respectively, after 24 h at 4°C and by 49.64%, 62.86%, and 48.16%, respectively, after 72 h at 4°C. The relative electrical conductivities of the WT leaves increased by 84.50% and 110.84% after 24 h and 72 h, respectively, at 4°C, indicating that the cell membranes of the transgenic lines were less damaged by the cold treatment.

Figure 5: Relative conductivity changes on plants exposed to low temperature at different times (Duncan’s method, P < 0.05)
3.2.5 Determination of Proline Concentrations
Proline is an important organic osmotic regulator in plant cells that accumulates under stress conditions, proline levels can, therefore, reflect the stress resistance of cells and the degree of cell damage. At room temperature, the proline concentrations of transformant leaves did not differ significantly from those of untransformed leaves (Fig. 6). After 24 h and 72 h at 4°C, the proline concentrations of the leaves of the transformed lines GSH9901-1 and -7 averaged 18.13 µg·g−1 and 20.96 µg·g−1, respectively, which were significantly higher than those of the untransformed WT line. The difference between 15.43 µg·g−1 and 18.63 µg·g−1 was significant at P < 0.05.

Figure 6: Proline concentrations (μg·g−1) of plants exposed to low temperature at different times (Duncan’s method, P < 0.05)
3.2.6 Determination of Malondialdehyde Content
The MDA level reflects the degree of cell membrane peroxidation and a response to stress. After 24 h and 72 h at 4°C, the average MDA concentrations in the leaves of the transformed lines were 0.0500 µmol·g−1 and 0.0513 µmol·g−1 (Fig. 7), respectively, which were significantly lower than the 0.0598 µmol·g−1 and 0.0628 µmol·g−1, respectively, on the untransformed WT line. These differences were significant, but there were no significant differences at room temperature. Thus, the leaves of the transgenic lines exhibited reduced stress responses when compared with the leaves of the untransformed lines. The expression of ICE1 in the leaves of the transgenic maize may, therefore, enhance membrane oxidization.

Figure 7: MDA concentration changes (μmol·g−1) on plants exposed to low temperature at different times (Duncan’s method, P < 0.05)
3.2.7 Determination of Peroxidase Activity
In plants, POD is a key enzyme responsible for scavenging active oxygen free radicals. POD activity can, therefore, be used to measure the level of stress resistance in plants. At room temperature, there was no significant difference in the leaf POD activities of transformed and untransformed plants (Fig. 8). After treatments at 4°C for 24 h and 72 h, the POD activity showed a trend of first increasing and then decreasing. The average POD activities of transformed plants increased by 19.60% and 22.89% at 24 h and 72 h, respectively, compared with the activities of untransformed plants, and the differences were significant. This indicates that the antioxidant capacity of the transgenic plants was greater than that of the non-transgenic plants.

Figure 8: Peroxidase activity changes of plants exposed to low temperature at different times (U·g−1·min−1) (Duncan’s method, P < 0.05)
3.2.8 Detection of Transgenic Plants by qPCR
qRT-PCR with SYBR Green I was used to confirm that the Southern blot-positive transgenic plants contained the transgene. The relative expression levels of ICE1 in the roots of transgenic maize plants GSHH9901-1, -5, and -7 were 5.19, 5.01, and 4.27, respectively, whereas the relative expression levels of ICE1 in the stem tissue were 8.96, 9.09, and 7.62, respectively (Fig. 9). The relative expression levels of ICE1 were the highest (P < 0.05) in the GSHH9901-1, -5, and -7 leaves, at 10.82, 10.42, and 11.26, respectively.

Figure 9: The relative expression of ICE1 gene (Duncan’s method, P < 0.05)
3.2.9 ICE1 Positively Regulates CBFs Gene Expression under Cold Stress
To identify the relationships between AtICE1 and the molecular components of the ICE–CBF pathway, we analyzed the CBF gene’s expression pattern under cold conditions. Under normal conditions, the expression levels of the identified genes were relatively low in both WT and transgenic plants (GSH9901-1, -5, and -7). However, compared with those in the control group, all the family genes, such as CBF1, CBF2, and CBF3, were significantly (P < 0.01) up-regulated in the transgenic plants after 24 h of cold treatment. After 72 h of cold treatment, the expression levels of these three genes decreased (Fig. 10). Thus, ICE1 may positively regulate the expression of the CBF genes in response to cold stress and may, thereby, improve cold tolerance.

Figure 10: The effect of transgenic corn with ICE1 on the transcription level of CBFs genes in the cold stress pathway (Duncan’s method, * P < 0.05, ** P < 0.01)
The generation of stress-resistant plants using genetic engineering generally involves the introduction of target genes, or the overexpression of existing genes of interest, in the plant species. The overexpression of ICE1 in plants under normal growth conditions has a small effect on plant growth. Transgenic ICE1-expressing rice [14], tomato [15], Arabidopsis [16,17], tobacco [18], and lemon [7] plants show normal growth, with very few instances of mutation or dwarfing, indicating that the introduction of ICE1 has fewer adverse effects on plant growth than the introduction of CBF/DREB1. When CBF/DREB expression increases in a plant, COR expression also increases, which regulates the expression of a series of genes and changes the physiological and biochemical indicators of the plant itself, resulting in the plant having a certain cold tolerance. Therefore, this study showed the successful transformation of maize with AtICE1 resulting in the generation of a new maize germplasm that had increased tolerance to cold stress. However, Lu et al. [19] indicated that ZmICE1 is the homolog of Arabidopsis AtICE1/2 and plays an important role in the regulation of freezing stress responses. A comparative analysis indicated that the expression of the ICE1 gene in A. thaliana changed more rapidly during the cold-response process. Therefore, compared with the transfer of the ZmICE1 gene into maize, the transfer of the AtICE1 gene into maize has a greater application value.
In this study, the pCAMBIA3301-ICE1 plant expression vector was constructed using a rapid and seamless cloning technique and was transformed into maize using Agrobacterium-mediated transformation. Three T1-positive plants were obtained, with the Southern blotting indicating that the target gene was integrated into the maize genome. Under low-temperature stress conditions, the expression of ICE1 decreased the relative conductivity of plant tissue extravasation fluid and the MDA concentrations, whereas the proline concentrations and POD activity increased. These results are consistent with those of Zhou et al. [20] and Feng et al. [21]. Using qRT-PCR, the ICE1 gene was determined to be expressed in the roots, stems, and leaves of transgenic maize plants under low-temperature conditions, indicating that low temperature induced ICE1 expression and increased the cold tolerance of plants. At normal temperatures, the physiological indicators of transformed and untransformed plants were almost the same, indicating that ICE1 was inactive under normal temperature conditions. This is consistent with the findings of Thomashow et al. [22]. Thus, further studies are required to confirm that ICE1 transgenic maize plants can tolerate low-temperature stress and that the inserted ICE1 is stable in the offspring.
Funding Statement: This work was supported by the Sub-Project of National Key R&D Plan [2019YFD1002603-1], Science and Technology Project of Jilin Provincial Department of Education [JJKH20200341KJ, JJKH20210351KJ, JJKH20210346KJ], Jilin Province Science and Technology Development Plan Project [20200402023NC].
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Jiao, P., Ma, R., Qi, Z., Jiang, Z., Ma, Y. (2019). Optimization of callus induction conditions from immature embryos in maize and plant regeneration. Phyton-International Journal of Experimental Botany, 88(3), 121–130. DOI 10.32604/phyton.2020.07980. [Google Scholar] [CrossRef]
2. Li, Z., Hu, G., Liu, X., Yao, Z., Li, Y. et al. (2016). Transcriptome sequencing identified genes and gene ontologies associated with early freezing tolerance in maize. Frontiers in Plant Science, 7(106), 1477. DOI 10.3389/fpls.2016.01477. [Google Scholar] [CrossRef]
3. Rxs, A., Kky, A., Hwl, A., Sjj, A., Qhy, A. et al. (2021). The effect of elevating temperature on the growth and development of reproductive organs and yield of summer maize. Journal of Integrative Agriculture, 20(7), 1783–1795. DOI 10.1016/S2095-3119(20)63304-4. [Google Scholar] [CrossRef]
4. Gilmour, S. J., Zarka, D. G., Stockinger, E. J. (2010). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant Journal, 16(4), 433–442. DOI 10.1046/j.1365-313x.1998.00310.x. [Google Scholar] [CrossRef]
5. Hui, L., Ding, Y., Shi, Y., Zhang, X., Yang, S. (2017). MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Developmental Cell, 43(5), 1–13. DOI 10.1016/j.devcel.2017.09.025. [Google Scholar] [CrossRef]
6. Zhan, X., Zhu, J. K., Lang, Z. (2015). Increasing freezing tolerance: Kinase regulation of ICE1. Developmental Cell, 32(3), 257–258. DOI 10.1016/j.devcel.2015.01.004. [Google Scholar] [CrossRef]
7. Huang, X. S., Zhang, Q. H., Zhu, D., Fu, X. Z., Wang, M. et al. (2015). ICE1 of Poncirus trifoliata functions in cold tolerance by modulating polyamine levels through interacting with arginine decarboxylase. Journal of Experimental Botany, 66(11), 3259–3274. DOI 10.1093/jxb/erv138. [Google Scholar] [CrossRef]
8. Jiao, P., Yuan, W. Y., Zhao, H. D., Qu, J., Wang, P. W. et al. (2021). Construction of a new plant expression vector and the development of maize germplasm expressing the Aspergillus ficuum phytase gene PhyA2. Genetic Resources and Crop Evolution, 68(3), 1103–1115. DOI 10.1007/s10722-020-01052-w. [Google Scholar] [CrossRef]
9. Ling, G. (2005). Mapping of rice long-protective mutant gene map. Xianyang: Northwest A&F University. [Google Scholar]
10. Livak, K. L., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the −ΔΔCT method. Methods, 25(4), 402–408. DOI 10.1006/meth.2001.1262. [Google Scholar] [CrossRef]
11. Schmittgen, T. D. (2008). Analyzing real-time PCR data by the comparative CT method. Nature Protocols, 6(3), 1101–1108. DOI 10.1038/nprot.2008.73. [Google Scholar] [CrossRef]
12. Petrov, N., Biryulina, N., Sidorova, Y., Mazo, V. (2021). A food ingredient containing phytoecdysteroids and polyphenols from quinoa grain: Technology and physiological and biochemical evaluation in vivo. E3S Web of Conferences, 285, 05014. DOI 10.1051/e3sconf/202128505014. [Google Scholar] [CrossRef]
13. Doru, A. (2021). Effects of heat stress on photosystem II activity and antioxidant enzymes in two maize cultivars. Planta, 253(4), 1–15. DOI 10.1007/s00425-021-03611-6. [Google Scholar] [CrossRef]
14. Xiang, D., Man, L., Yin, K., Song, Q., Xu, Z. (2013). Overexpression of an ItICE1 gene from Isatis tinctoria enhances cold tolerance in rice. Molecular Breeding, 32(3), 617–628. DOI 10.1007/s11032-013-9894-0. [Google Scholar] [CrossRef]
15. Juan, J. X., Yu, X. H., Jiang, X. M., Gao, Z., Zhang, Y. et al. (2015). Agrobacterium-mediated transformation of tomato with the ICE1 transcription factor gene. Genetics and Molecular Research, 14(1), 597–608. DOI 10.4238/2015.January.30.1. [Google Scholar] [CrossRef]
16. Bredow, M., Vanderbeld, B., Walker, V. K. (2017). Ice-binding proteins confer freezing tolerance in transgenic Arabidopsis thaliana. Plant Biotechnology Journal, 15(1), 68–81. DOI 10.1111/pbi.12592. [Google Scholar] [CrossRef]
17. Kidokoro, S., Kim, J. S., Ishikawa, T., Suzuki, T., Yamaguchi-Shinozaki, K. (2020). DREB1A/CBF3 is repressed by transgene-induced dna methylation in the Arabidopsis ICE1-1 mutant. The Plant Cell, 32(4), 1035–1048. DOI 10.1105/tpc.19.00532. [Google Scholar] [CrossRef]
18. Zou, C., Yu, D. (2010). Analysis of the cold-responsive transcriptome in the mature pollen of Arabidopsis. Journal of Plant Biology, 53(6), 400–416. DOI 10.1007/s12374-010-9129-4. [Google Scholar] [CrossRef]
19. Lu, X., Yang, L., Yu, M., Lai, J., Wang, C. et al. (2017). A novel Zea mays ssp. mexicana L. MYC-type ICE-like transcription factor gene ZmmICE1, enhances freezing tolerance in transgenic Arabidopsis thaliana. Plant Physiology and Biochemistry, 113, 78–88. DOI 10.1016/j.plaphy.2017.02.002. [Google Scholar] [CrossRef]
20. Zhou, L., He, Y. J., Li, J., Li, L. Z., Liu, Y. et al. (2020). An eggplant SmICE1a gene encoding MYC-type ICE1-like transcription factor enhances freezing tolerance in transgenic Arabidopsis thaliana. Plant Biology, 22(3), 450–458. DOI 10.1111/plb.13095. [Google Scholar] [CrossRef]
21. Feng, H. L., Ma, N. N., Meng, X., Zhang, S., Wang, J. R. et al. (2013). A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiology Biochemistry, 73, 309–320. DOI 10.1016/j.plaphy.2013.09.014. [Google Scholar] [CrossRef]
22. Thomashow, M. F., Torii, K. U. (2020). Screaming twist on the role of ICE1 in freezing tolerance. The Plant Cell, 32(4), 816–819. DOI 10.1105/tpc.20.00124. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |