 Open Access
Open Access
ARTICLE
A Hybrid Attention-Based Residual Unet for Semantic Segmentation of Brain Tumor
1 Department of Computer Science, COMSATS University Islamabad, Islamabad, 45550, Pakistan
2 Department of Electrical and Computer Engineering, COMSATS University Islamabad, Wah Campus, Pakistan
3 Department of Computer Science, HITEC University, Taxila, Pakistan
4 Department of Computer Science, College of Computer and Information Sciences, Princess Nourah bint Abdulrahman University, Riyadh, 11671, Saudi Arabia
5 Department of Management Information Systems, CoBA, Prince Sattam Bin Abdulaziz University, Al-Kharj, 16273, Saudi Arabia
6 Department of Computer Science, Hanyang University, Seoul, 04763, Korea
* Corresponding Author: Jae-Hyuk Cha. Email:
Computers, Materials & Continua 2023, 76(1), 647-664. https://doi.org/10.32604/cmc.2023.039188
Received 13 January 2023; Accepted 10 March 2023; Issue published 08 June 2023
Abstract
Segmenting brain tumors in Magnetic Resonance Imaging (MRI) volumes is challenging due to their diffuse and irregular shapes. Recently, 2D and 3D deep neural networks have become famous for medical image segmentation because of the availability of labelled datasets. However, 3D networks can be computationally expensive and require significant training resources. This research proposes a 3D deep learning model for brain tumor segmentation that uses lightweight feature extraction modules to improve performance without compromising contextual information or accuracy. The proposed model, called Hybrid Attention-Based Residual Unet (HA-RUnet), is based on the Unet architecture and utilizes residual blocks to extract low- and high-level features from MRI volumes. Attention and Squeeze-Excitation (SE) modules are also integrated at different levels to learn attention-aware features adaptively within local and global receptive fields. The proposed model was trained on the BraTS-2020 dataset and achieved a dice score of 0.867, 0.813, and 0.787, as well as a sensitivity of 0.93, 0.88, and 0.83 for Whole Tumor, Tumor Core, and Enhancing Tumor, on test dataset respectively. Experimental results show that the proposed HA-RUnet model outperforms the ResUnet and AResUnet base models while having a smaller number of parameters than other state-of-the-art models. Overall, the proposed HA-RUnet model can improve brain tumor segmentation accuracy and facilitate appropriate diagnosis and treatment planning for medical practitioners.Keywords
A tumor is the abnormal growth of cells within the brain tissues. These unusual cells can be fatal if not diagnosed early [1]. In brain tumors, the most typical type of malignancies reported in adults are gliomas, and they are graded as High-Grade Gliomas (HGG) that increase rapidly and Low-Grade Gliomas (LGG) that grow slowly [2]. Meningiomas are more accessible to segment. However, glioblastomas are challenging to identify because of their high contrast and diffusion [3]. Its appearance varies in size, shape, and form, making it more challenging. Most widely used medical imaging modalities, i.e., Computed Tomography (CT) scans, X-rays, and Magnetic Resonance Imaging (MRI), are commonly used to diagnose diseases. High-resolution MRI is commonly used for brain tumor diagnosis and treatment planning [4]. In 2015, [5] suggested a unique, fully convolutional-based network (FCN) end-to-end model for natural image segmentation that helped transform the entire image segmentation field.
Similarly, [6] Introduced an FCN termed Unet for segmentation in medical images, inspired by the FCN architecture. Unet has two parts: One is an encoder, and the other is the decoder. It also consists of several skip connections between encoder-decoder parts to merge cropped feature maps, significantly improving medical image segmentation performance. Unet has now established itself as a benchmark for tackling the brain tumor segmentation problem. Meanwhile, many enhanced Unet models, such as Residual Unet [7] and Ensemble Unet [8], have also been proposed to enhance the performance of tumor segmentation tasks.
Brain tumor segmentation is a challenging task that has been the focus of extensive research in recent years due to its potential for improving patient diagnosis, treatment, and prognosis. The Brain Tumor Segmentation (BraTS) dataset has emerged as a popular benchmark for evaluating the performance of different segmentation models, providing a standardized platform for comparing results across studies.
However, the BraTS dataset poses several challenges that make accurate segmentation difficult. For instance, the images in the dataset exhibit significant variability in terms of tumor size, shape, location, and appearance, as well as in the presence of confounding factors such as edema and necrosis. Additionally, the dataset is highly imbalanced, with a relatively small number of tumor regions compared to normal brain tissue regions, which can affect the performance of segmentation models.
To address these challenges, researchers have developed various segmentation techniques [9–11] that incorporate advanced machine learning algorithms, such as convolutional neural networks (CNNs), and feature engineering methods. Some of these techniques use sophisticated pre-processing methods, such as intensity normalization, bias correction, and skull stripping, to improve the quality of the input data and enhance the performance of segmentation models.
Several studies have explored the potential of these techniques in the context of the BraTS dataset, showing promising results regarding segmentation accuracy and efficiency. However, further research is still needed to develop robust and reliable segmentation models that can address the challenges of brain tumor segmentation in the BraTS dataset.
As Unet must progressively recover down-sampled images due to intrinsic stridden convolution and pooling layers, attention methods can bridge the information flow between shallow and deep layers to facilitate the up-sampling. As a result, this improved the accuracy of the pre-existing Unet models for the MRI segmentation tasks by increasing its capacity to capture multiple local and global responses. Inspired by the performance of Unet, this research integrates the residual attention and Squeeze-Excitation (SE) modules in Residual Unet to explore further the impacts of both local and global features mapping for the segmentation of brain tumors in MRIs. Notably, a 3D deep model for brain tumour segmentation is proposed by stacking attention and Squeeze-Excitation modules at different levels into the Residual Unet for performance enhancement. The following are the contributions of our work:
• This research proposes a 3D deep and light-weighted, end-to-end model to address the challenge of accurately segmenting brain tumors in MR scans by successfully embedding the residual blocks, attention, and Squeeze-Excitation (SE) modules into the basic Unet model.
• The proposed 3D Residual Unet model integrates low- and high-level features using residual blocks. A series of Attention modules are added on Skip-Connections between corresponding contracting and expanding parts to adaptively enhance local responses of different down-sampled feature maps used for feature recovery in the subsequent up-sampled phase to reduce the semantic gap between both parts. Finally, Squeeze-Excitation (SE) modules are added before up-sampling in the expanding part, recalibrating the channels to suppress the less relevant features and learn the inter-channel relationships.
• Experiments on the publicly available dataset, i.e., BraTs-2020, show that the proposed 3D deep model with attention and Squeeze-Excitation modules outperforms its baseline model and shows competitive performance with a smaller number of parameters for segmenting the brain tumors.
The rest of the sections in this paper are organized as Section 2, which contains the state-of-the-art methods for semantic segmentation of tumors. Section 3 gives a detailed overview of our proposed methodology, including the dataset, pre-processing strategy, and deep Unet model. Section 4 evaluates the proposed model, presents experimental findings, and compares it to existing models. Section 5 contains the conclusion.
Healthcare professionals consider medical image segmentation as a crucial task, which helps them to pinpoint a patient’s area of interest and perform a diagnosis. Traditional approaches such as the watershed transform, graph cut, and other methods required handcrafted features and prior information on the input data to obtain impressive performance. However, this still does not fill the gap between computational cost and domain specialists in healthcare. Introducing deep neural networks (DNNs) has been a significant breakthrough in medical image processing. Unlike previous approaches, DNNs can automatically acquire discriminative features, making them capable of segmenting images effectively. Convolutional Neural Networks (CNNs) are the most common deep network for segmenting videos and images. In 2014, a novel fully convolutional-based neural network (FCN) was proposed to enhance the effectiveness of CNN for segmentation in medical images. In 2015, a deep CNN method was proposed for segmenting tissues in the brain using multiple modalities of magnetic resonance images. CNNs are not fully capable of separating tumors with uneven densities and indistinct boundaries. Unet was designed to have a structure comparable to a fully convolutional network to address the volumetric aspect of medical imaging. The attention mechanism has been widely employed in computer vision for various tasks, and its use in medical imaging tasks is minimal. In 2018, a novel attention gate model for diagnosis in medical images called attention Unet was proposed, which could learn to focus on target structures of diverse sizes and shapes. Another study created a hybrid attention-aware deep model to identify liver and extract malignancies from liver CT images in the same year. Customized U-Net architectures have been utilized for effective medical image segmentation.
Table 1 presents a summary of the methods and their respective applications in medical image segmentation from the year 2007 to 2022. The methods included traditional approaches such as the watershed transform, graph cut, and other methods, deep neural networks (DNNs), and customized U-Net architectures. The attention mechanism has also been introduced for medical image segmentation to identify target structures of diverse sizes and shapes. These methods have demonstrated impressive performance in various medical imaging applications, including tissue segmentation in the brain, brain tumor segmentation, and malignancy extraction from liver CT images.
This research proposes a new hybrid attention-based deep model for the semantic segmentation of brain tumors. The proposed deep learning model comprises three core blocks that segment the brain’s tumorous cells. Initially, a 3D residual Unet is developed. Many residual blocks are layered together to acquire multi-scale information by merging low-level features with high-level ones in brain MRIs. Multiple attention modules are embedded at the skip connections between the encoder and decoder to learn multiple attention-aware features of different tumor parts. Finally, squeeze and excitation blocks enhance essential features by recalibrating the feature maps across various channels.
3.1 The Proposed Deep Learning Model
Our proposed deep-learning model is depicted in Fig. 1. It is relatively like a regular Unet, simultaneously embedded with residual, attention, and squeeze-excitation mechanisms better suited for segmentation tasks in the medical image when the availability of correctly annotated data is limited. The contracting path of the model propagates the contextual information through residual, allowing for a more complex extraction of hierarchical feature maps. The expanding path takes these hierarchical features of varying complexity and reconstructs them from coarse to fine. The Unet, like deep models, benefits from introducing long-range connections across the contracting and expanding parts, allowing distinct hierarchical feature maps to be combined and making the model more robust.
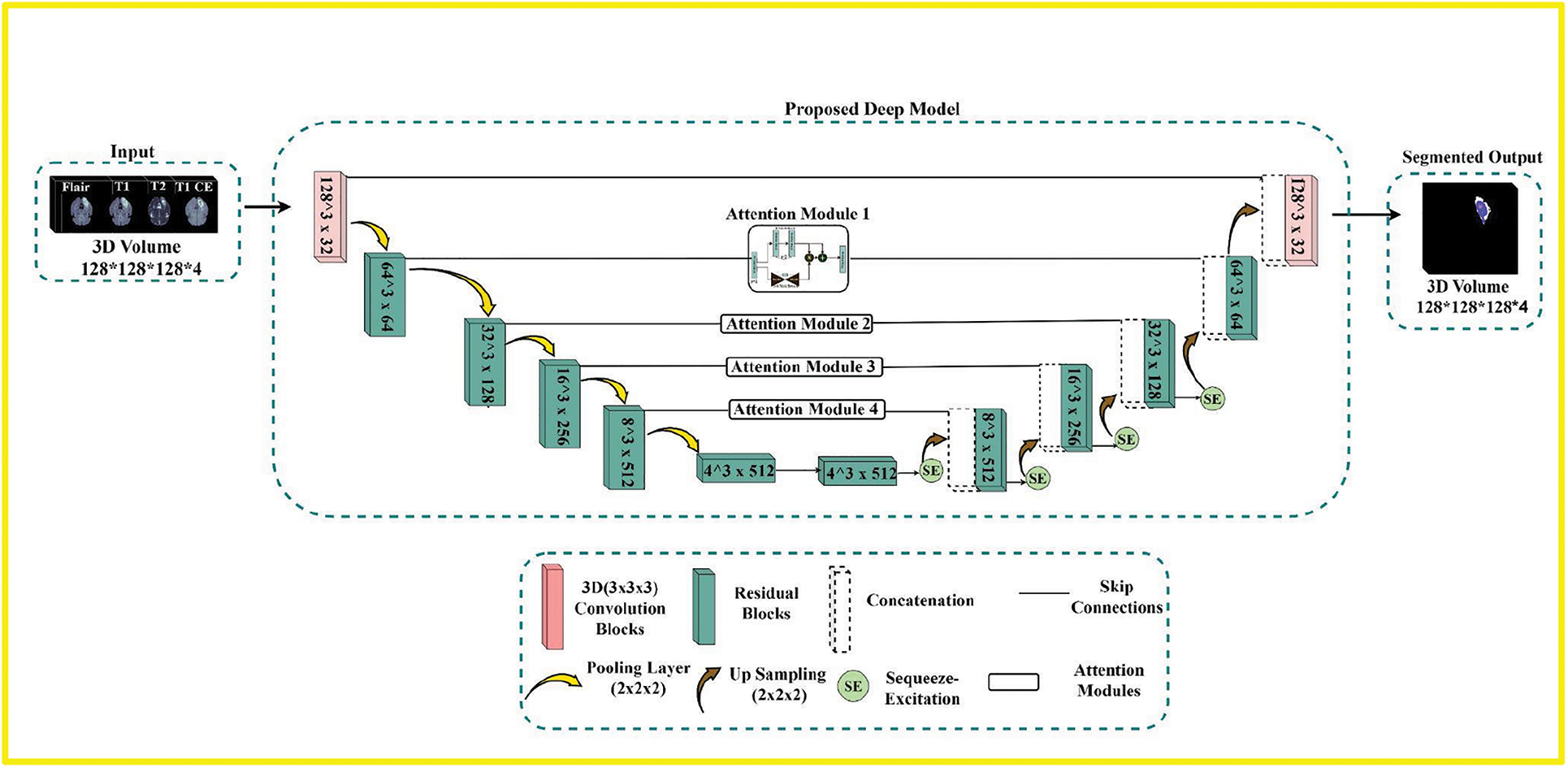
Figure 1: The overview of the proposed pipeline. First is the input module that comprises four modalities, i.e., Flair, T1, T2, and T1ce, which are stacked together to form 3D volumes. The second is the proposed deep Residual-based Unet model with attention and Squeeze-Excitations blocks to process the input volumes and segment the Tumor. The third is the output module with three labels
3.2 Residual Learning Mechanism
The proposed model consists of Residual blocks as basic building blocks, with basic architecture illustrated in Fig. 2. The deep models’ depth is critical in all computer vision tasks. However, the vanishing of the gradient is a typical problem during the backpropagation in a deep neural network, resulting in poor training outcomes. Reference [23], presented a residual learning network to learn the identity maps. Residual blocks are layered except for the first and final layers to unlock the learning capacity of our proposed deep model. A shortcut identity mapping achieves the residual learning mechanism as fast connections through element-wise addition operation, significantly improving accuracy and training speed without extra parameters. The residual learning mechanism is presented in Eq. (1):
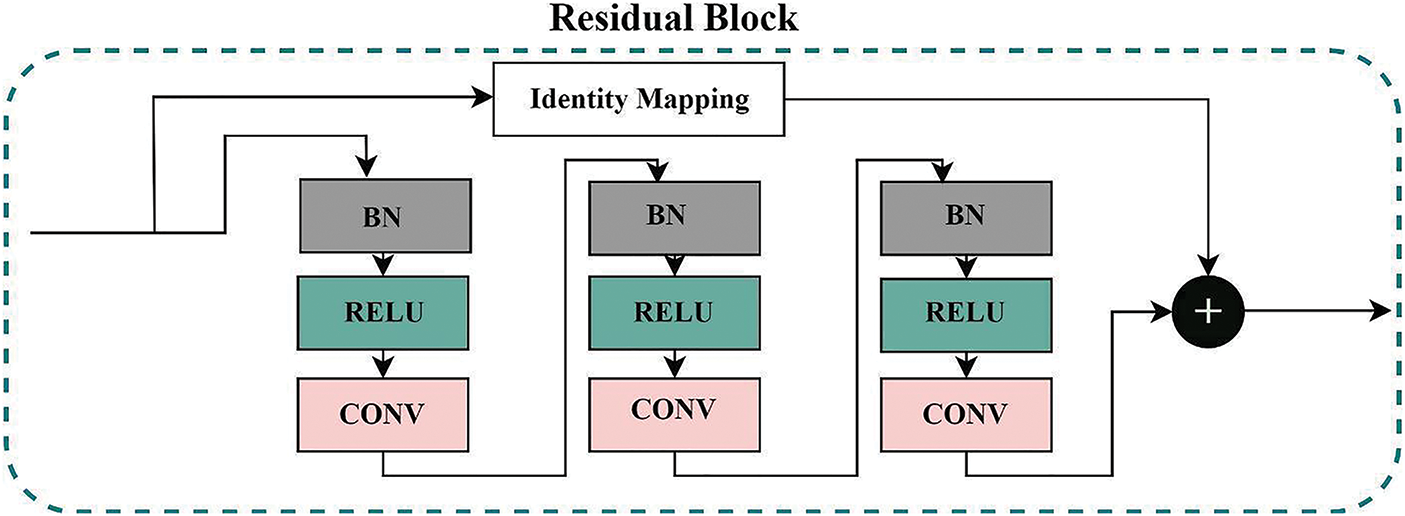
Figure 2: The residual-block used in the proposed deep model. The final output is the sum of Identity mapping and features from three sub blocks containing batch normalization, relu activation, and convolution blocks
Here,
3.3 Residual-Attention Learning Mechanism
If attention modules are naively stacked in any deep model, the performance of that model will surely suffer. A study [24] presented a novel attention-learning mechanism to solve this problem. This Residual-attention learning mechanism is based on Residua-Bocks and splits into two branches: Trunk-Branch and Soft-Mask-Branch; original features are processed in the Trunk-Branch with t number of residual blocks. The Soft-Mask-Branch constructs the identity mapping using the r number of residual blocks in encoder-decoder type structures. (2) depicts the final output of the residual-attention module for the Residual-Attention learning mechanism
Here, S(I) is the Output of Soft-Mask-Branch and has values between 0 and 1; if S(I) has values near 0, then
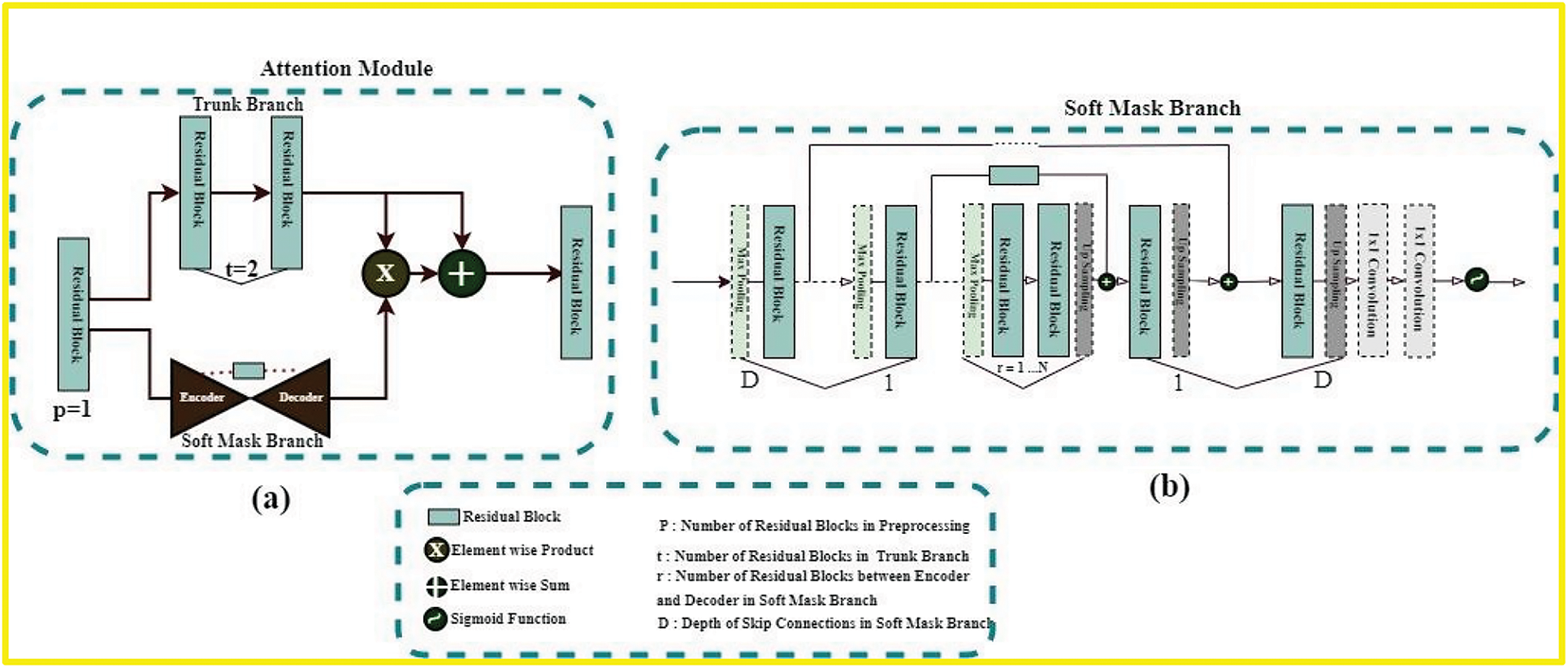
Figure 3: (a) Is the residual-based attention module’s architecture used in our proposed model. The architecture of soft-mask-branch is depicted in (b). Here p, t, and r represent the hyperparameters. Soft-mask branch has an encoder-decoder structure. D denotes the depth of these blocks. In our model, D is equal to {1, 2, 3, 4} according to the skip-connections of the proposed model
In general, the original feature maps are conserved by the trunk branch in the proposed attention mechanism, and it pays attention to the features of the brain tumor through the soft mask branch.
3.4 Squeeze-Excitation Learning Mechanism
Every deep model creates valuable feature maps by combining H × W (spatial) and C (Channel) information inside local-receptive fields at different stages to perform correctly. Soft-Mask-Branch’s attention varies adaptably based on the trunk-branch’s features in the attention mechanism mentioned above. However, the “Squeeze-Excitation” (SE) module in the expanding part of the proposed model addresses this limitation of the Residual-attention mechanism. This SE module [26] finds the interdependencies across distinct channels to recalibrate channel-wise feature maps. Fig. 4 depicts the architecture of the SE module. As a result of this channel-wise feature extraction mechanism, the model can concentrate on the channel dependencies that the model ignores as it recreates the feature maps. The SE module works the following way: First, each channel’s features (H × W) are compressed into an actual number using global average pooling. Then, a nonlinear transformation is performed using a fully connected neural network to obtain each feature channel’s weight. Finally, the normalized weights obtained above are weighted to the Residual-based channels. These weighted features of respective channels are then concatenated with the up-sampled features maps of the residual blocks, which are then fed into subsequent layers of the corresponding expanding part.
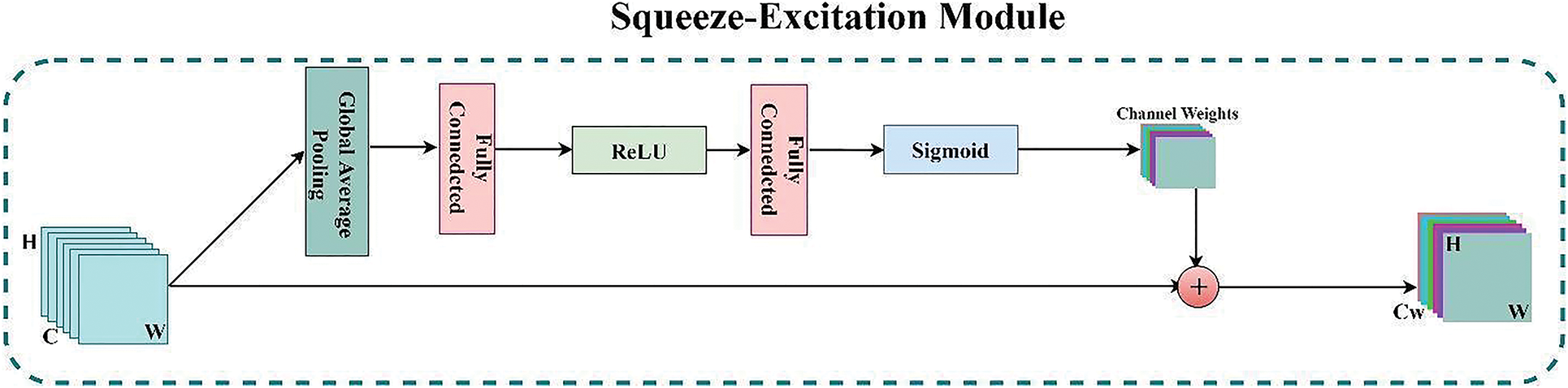
Figure 4: The architecture of the SE (Squeeze-Excitation) module
The SE normalization technique has shown the potential to enhance the performance of CNNs in brain tumor segmentation. By weighting the feature maps adaptively, SE normalization allows the model to focus on the most significant features that distinguish between different brain tissue types and tumors. At the same time, it can down-weight features prone to noise or artefacts, resulting in improved accuracy and robustness of CNN-based segmentation models for brain tumor analysis.
Generally, a SE block is a structural unit that allows the proposed model to execute dynamic channel-wise feature recalibration to increase its segmentation power. Furthermore, SE blocks shed light on the residual attention mechanism’s incapacity to appropriately describe channel-wise feature relationships.
Extensive experimentation is performed in this study to evaluate the effectiveness of the proposed tumor segmentation model. All the experiments were performed using python language using the Keras and Tensor Flow frameworks on NVIDIA Titan 1080 TI GPU with a 12 GB GPU workstation. The rest of this section provides the details of data pre-processing, loss function, evaluation matrices, results and finally, the comparison with baseline models.
This study used the publicly available BraTs-2020 dataset [11] to evaluate the proposed model. BraTs-2020 contains the pre-operative MRI scans collected from multiple institutes to segment inherently diverse and complex brain tumors with four commonly used modalities. These are T1 weighted (T1), T1 weighted and contrast-enhanced (T1Gd), T2 weighted (T2), and fluid-attenuated inversion recovery (FLAIR) for each case, as shown in Fig. 5. All the MRI scans are of 240 × 240 resolution, and 155 slices for each modality with manually annotated ground-truth labels for every scan. The BraTs-2020 training set comprises 369 patients. The hyperparameters were defined as a learning rate of 0.001, a dropout rate of 0.2 the number of epochs was 100 with a batch size of 1.
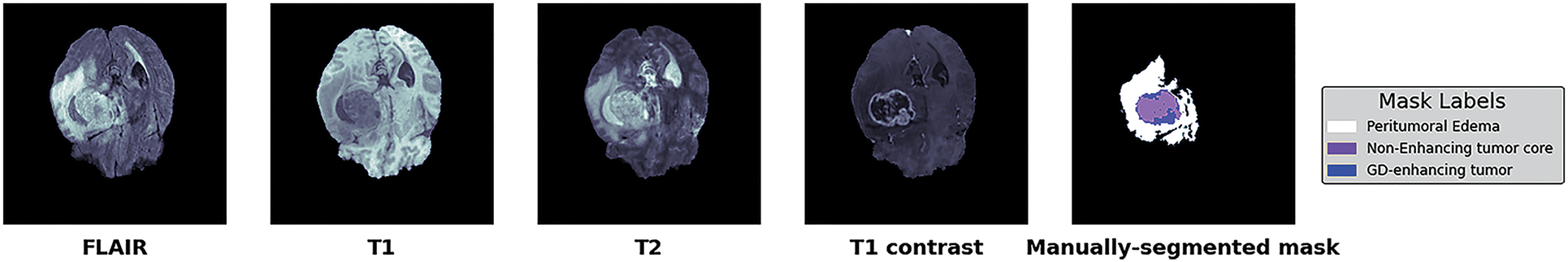
Figure 5: BraTs-2020 dataset with four different modalities with segmented masks and labels
Similarly, the validation set includes 125 patients. However, the ground-truth labels are made public for the training set. In contrast, withheld labels for validation sets validate the proposed model’s generalization and scalability using the online validation approach.
Segmenting brain tumors in MRIs is a difficult challenge because of the brain’s and biological tissue’s complex structure and the medical imaging quality. Due to the restricted processing resources, this model downsized each image from 240 × 240 × 155 voxels to 128 × 128 × 128 voxels. Input MRI scans must be normalized because pixel intensities vary due to manufacturers, acquisition conditions, and sequences. Each MRI sequence’s non-zero voxels are normalized independently through zero-score normalization and shrink input data. (3) illustrates Z-Score normalization:
Here, Z is the output image obtained after normalization on the input image I, μ is the mean, and δ is the standard deviation. There is no additional image processing. All MR images fed into our model have a final input size of 128 × 128 × 128 × 4, where 128 × 128 indicates the spatial dimensions of the input image (i.e., width and height), 128 represents the number of slices, and 4 represents the four MRI modalities. This research stacked all the modalities together to generate four channels to feed into the model. All the annotated training sets’ masks are handled the same way. The ultimate size of the segmented masks input into the model is 128 × 128 × 128 × 4. The first three numbers reflect the resolution and the number of scan slices. 4 represents the number of classes provided, namely: label 0 (Background), label 4 (ET (GD Enhancing-Tumor)), label 2 (ED (Peritumoral-Edema)), and label 1 (NCR/NET (Necrotic and Non-Enhancing Tumor-Core)). Even though deep models are mainly resistant to noise, data processing is still necessary for improving brain tumor segmentation accuracy. In this research, we performed data processing to make the available brain MRI scans better suited to train the deep model according to the available resources, as shown in Fig. 6.
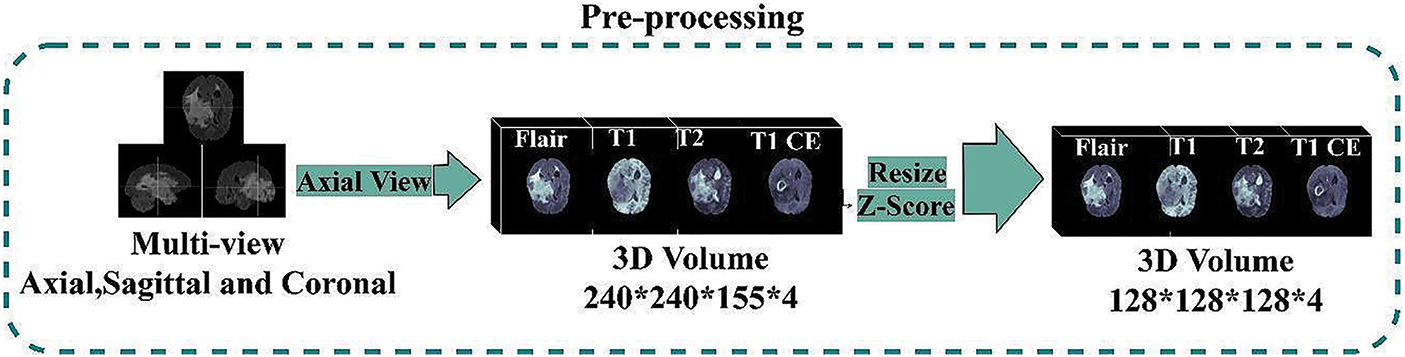
Figure 6: Dataset pre-processing steps
The proposed model determines if a pixel represents a specific brain tumor tissue or a regular pixel. However, severe class imbalance problems are usually exhibited using the BraTs dataset for segmentation. Table 2 depicts the distribution of tumor subclasses and healthy tissues in the Brats-2020 training set.

As brain tumors, mainly the TC (tumor core), are often tiny in the overall MRI Scans, CE (Cross-Entropy) loss is ineffective in solving this class imbalance problem in the segmentation task. The well-known, class-wise Dice overlap coefficient is adopted as a loss function in such tasks, outperforming the CE. For multi-class segmentation problems, the Dice-Coefficient for each respective class can be determined using Eq. (4):
Here N represents the number of volumes (subjects) that, in our case, is 365, s (c, i) ∈ [0,1,2,3] represents the predicted output (softmax output) of the model, and gt (c, i) ∈ [0,1,2,3] is the given mask with four labels for tumor sub-classes, respectively. This loss function is used to check the similarity index of multi-class samples directly.
An overlap and a distance metric are two primary evaluation metrics for the deep model's evaluation. The Dice Similarity Coefficient (DSC) is a regularly used metric for determining overlap between two sets. It compares two groups, S1 and S2, by normalizing their intersection sizes over the average of their sizes. It is defined as follows in the context of ground truth comparison in Eq. (5):
Sensitivity, specificity, and Hausdorff distance are statistical decision theory metrics determined using Eqs. (6) to (8), respectively.
Hausdorff-Distance is complementary with the Dice coefficient metric to quantify the maximum distance between the margins of original and predicted volumes. In (8), A (Actual Mask) and B (Predicted Mask) are the two sets of points being compared, and H (A,B) is the directed Hausdorff distances between them. Here a is a point in set A, b is a point in set B, and the function max and min (a,b) calculate the maximum and minimum Euclidean distance between a point in set A and a point in set B. It penalizes outliers heavily, as a model prediction might have voxel-perfect overlap. However, its value will be significant if the predicted voxel is far from the actual segmentation mask. This metric may appear noisier than the Dice-Coefficient, but it helps determine the segmentation’s clinical utility. For example, suppose the predicted segmented mask includes surrounding healthy brain tissues. It will require manual adjustment from the radiation oncologist, even if the overlap of predicted and actual masks assessed through the Dice metric is good enough to avoid fatal effects on the patient.
4.4 Comparison with SOTA Methods
In this study, a new method for brain tumor segmentation was proposed that achieves state-of-the-art performance on the BraTS 2020 challenge dataset. Our method leverages the power of deep learning. It combines several innovative components, such as a dual-attention mechanism, a residual-attention mechanism, and a squeeze and excitation attention mechanism, to improve the segmentation's accuracy and robustness. We extensively evaluate our approach using standard evaluation metrics and statistical tests and show that it outperforms previous state-of-the-art methods on BraTS 2020. In particular, our method achieves an average dice score of 0.87 on the Training set and 0.86 on the test set, with less number of parameters as shown in Table 3. Furthermore, in Tables 4–7, we performed an ablation study to investigate the contribution of each component to the overall performance.




The proposed 3D deep model completely utilizes volume information while capturing three-dimensional data. The proposed 3D lightweight model integrates different modules to create a precise segmentation mask. Meanwhile, training with large MRI volumes of 128 × 128 × 128 × 4 gives significantly more contextual-based information than training with small patches. The brain volumes received by the model are passed from top to bottom during the contracting phase. Lower features were then passed top-down in the contracting path through different pooling layers and reversed during the expanding phase, with doubled resolution thanks to up-sampling. The attention blocks are used to realize the links between both parts. The feature maps from these attention blocks were concatenated with corresponding up-sampling and SE level of expanding part. The final brain tumor segmentation probability map was generated using an activation layer (i.e., Softmax). Table 4 presents our model's training and validation results (Mean, Standard Deviation, Median and 25 & 75 Quantile) obtained through the online evaluation system for Brats-2020.
Our proposed deep learning model relies on three primary modules. 1st: a simple 3D basic Unet model that uses residual blocks rather than convolutional blocks like traditional Unet. 2nd: Residual Attention module added to the base model on skip Connections. 3rd: SE (Squeeze and excitation) module to derive information from the feature maps; SE module is utilized with upsampling layer after each residual unit in the expanding part. In this research, we trained the proposed deep model in this way; first, our base residual Unet model was trained, then added the attention modules, and finally added SE modules in the proposed model. For comparison, we acquired quantitative evaluations of each tumor class with baseline models regarding Dice-Coefficient and Hausdorff-Distance. This research trained all three models without weight initialization for the convolutional kernel parameters with Adam optimizer and Dice Loss. LR = 0.001 was the initial learning rate. The time cost and the total number of trainable parameters for each model are depicted in Table 5.
In this research, we evaluate the performance after adding each corresponding module in the baseline model through BraTs-Online Evaluation-System for training and validation sets. Fig. 7 gives a graphical representation of evaluation metrics for all models, while Tables 6 and 7 depict the numeric comparison of Evaluation metrics of the proposed model with base models.
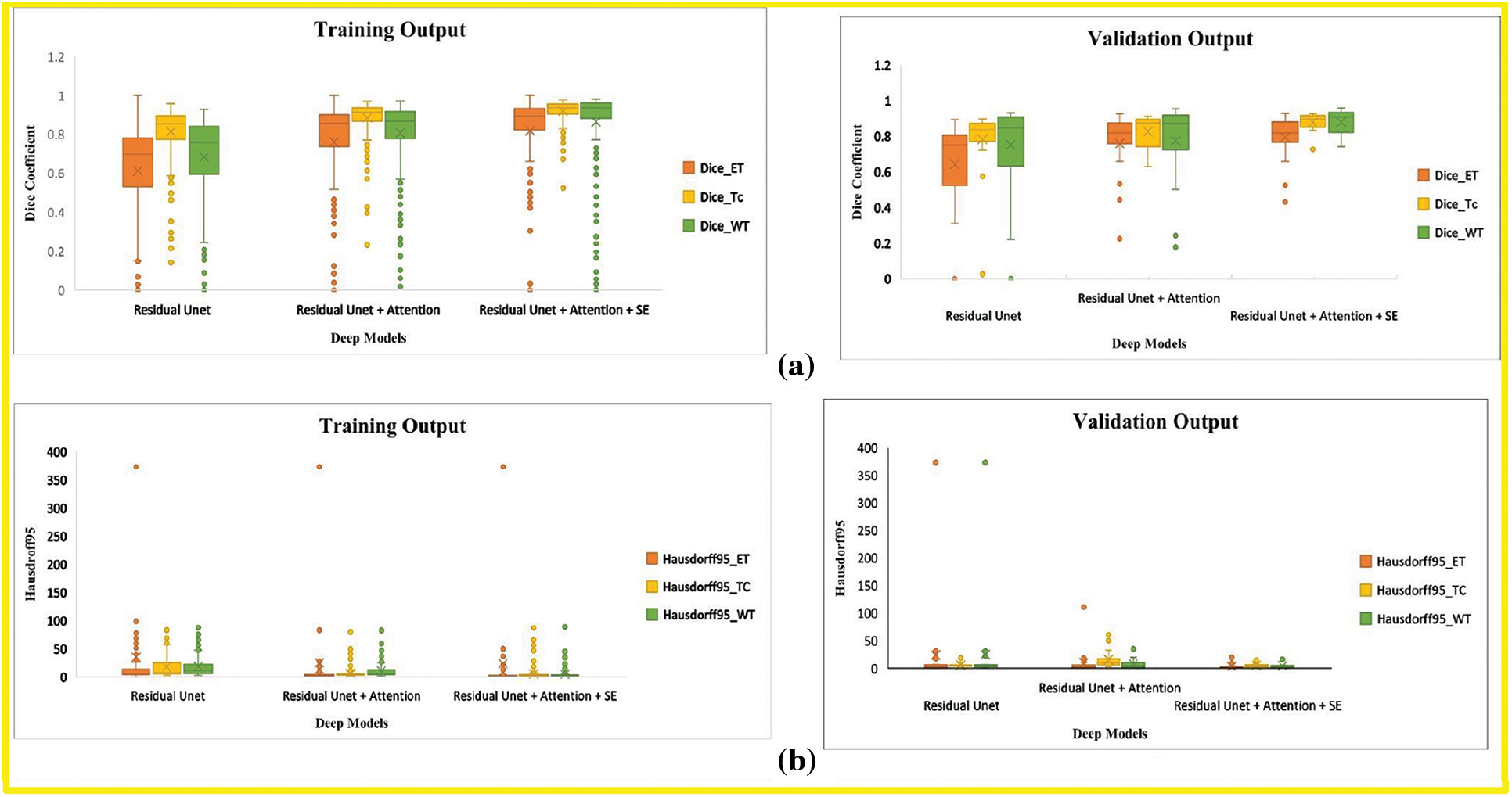
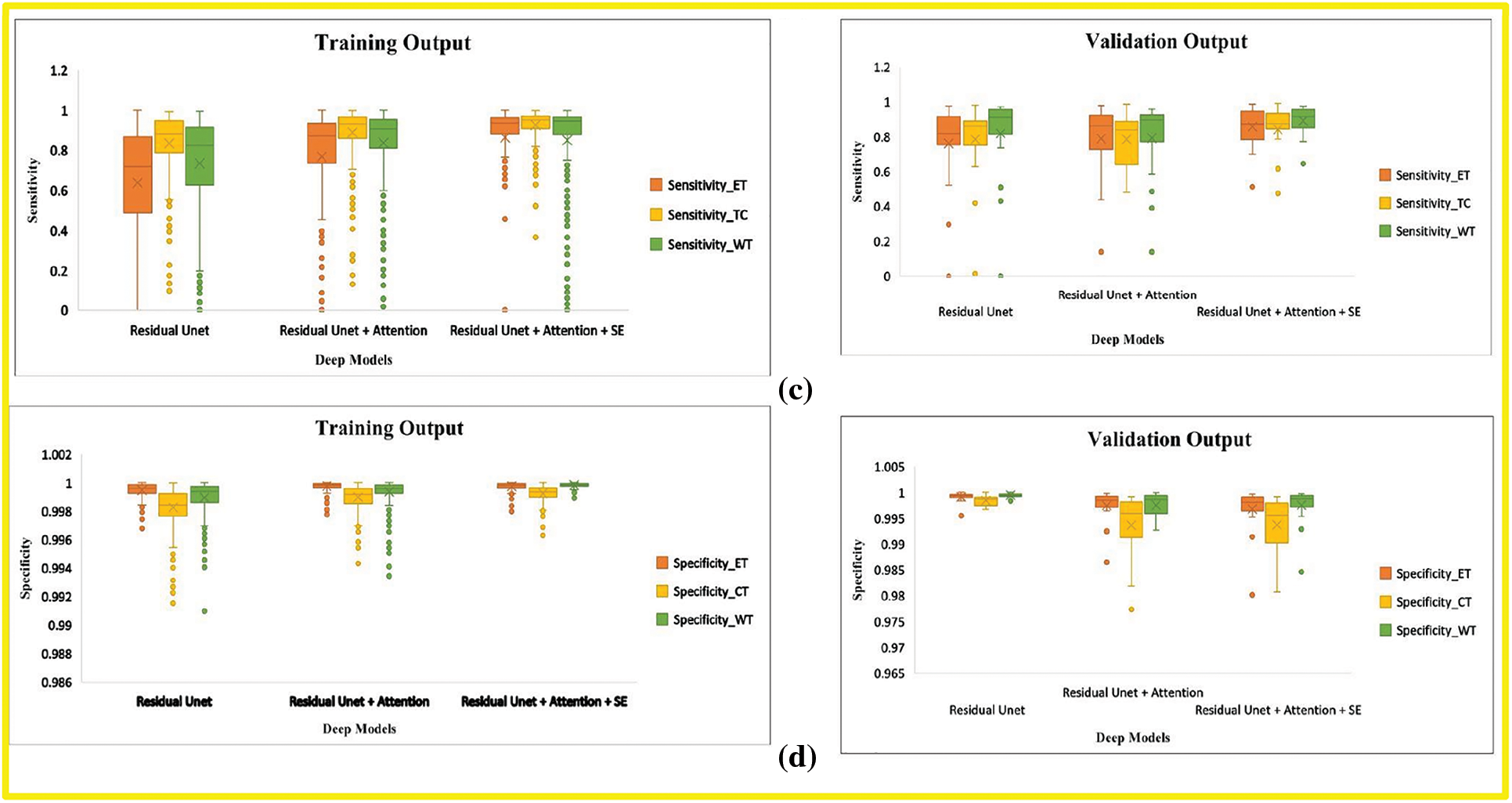
Figure 7: Boxplot demonstrates the evaluation metric for three deep models, including the base model, base model with attention, and proposed model. (a) Dice-coefficient for both training and validation sets, (b) Hausdroff95 for training and validation sets, (c) Sensitivity for training and validation sets (d) Specificity for training and validation sets. The different tumor types explained in Section 4.1 represent the abbreviation in the legend. (orange) ET is an acronym for Enhancing-Tumor (yellow) TC stands for Tumor-Core, and (green) WT is for Whole-Tumor
By comparing the (1) and (3) rows of Table 6, this research finds that our final model’s Dice Score of WT, TC, and ET of base model Residual Unet increased for training and validation sets. The Hausdorff-Distance of all tumors was also reduced by adding deep supervision through two additional modules in the base model. These two modules’ inclusion helped extract other discriminative tumor features, as seen by the improvement in most measures. By comparing the results of the last two rows in Table 5, the Dice of all tumor classes in the base model with both attention and SE modules were better than those of the base model with only the attention module.
The proposed 3D deep model has fewer parameters than the traditional Unet. Adding Attention and SE modules to the base model increases the trainable parameters. However, the above-mentioned results demonstrated the effectiveness of each module individually that is added base residual Unet model along with model’s depth. A few segmented examples of our proposed model and baseline models are shown in Fig. 8.
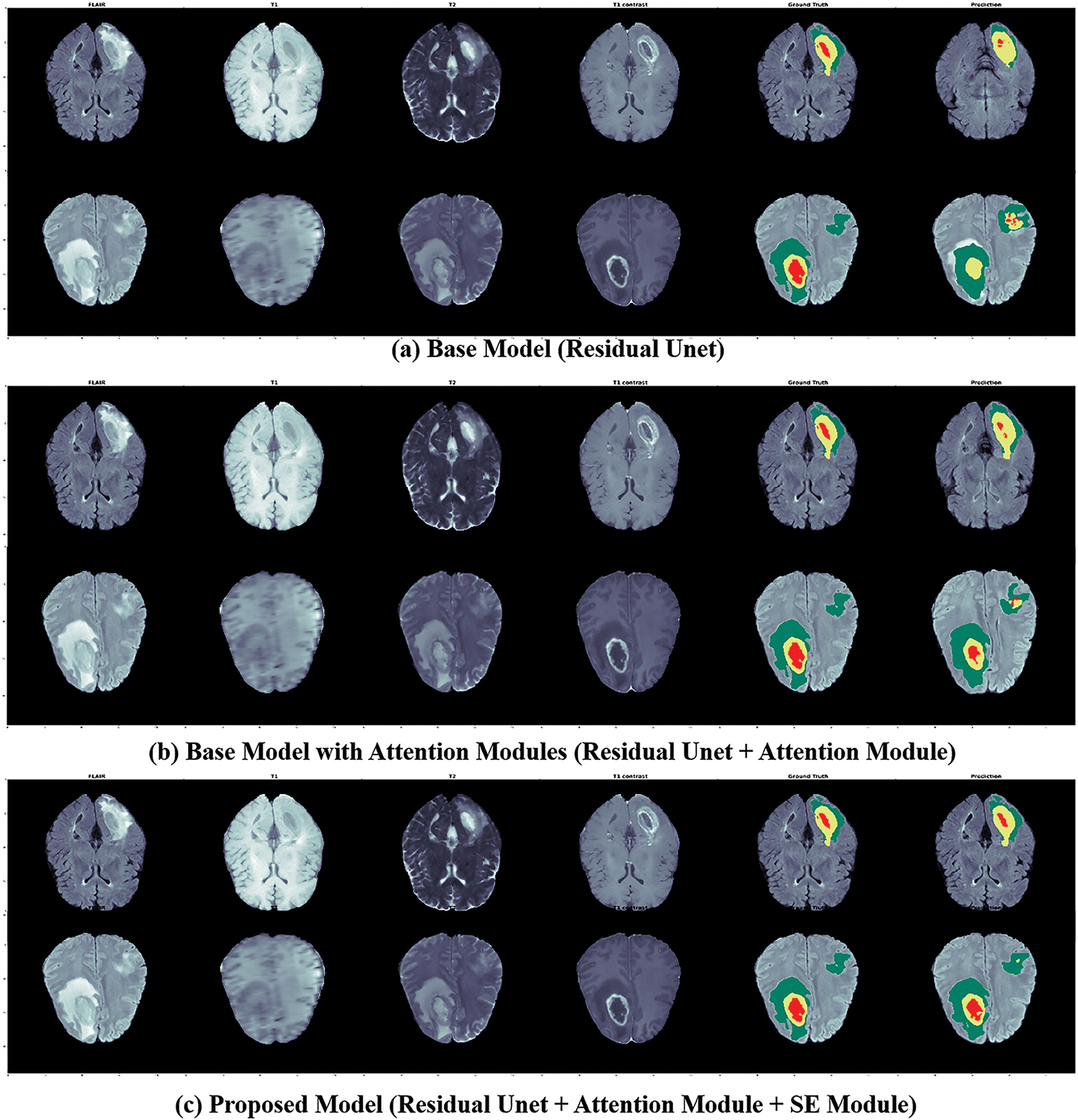
Figure 8: The segmented outputs for unseen MRIs from the BraTs training set, from left to right: four modalities (FLAIR, T1, T1CE, T2), ground truth, and prediction of our models; for this instance, 1st Row: Patient ID 366, slice 90; 2nd Row: Patient ID 369, slice 100; Colors: Red is (Tumor-Core), Yellow is (GD-Enhancing-Tumor), and Green is (Peritumoral-Edema)
This research presented a light-weighted 3D deep model for the segmentation task of tumors using brain MRIs by embedding attention, Squeeze-Excitation mechanisms, and residual blocks into Unet to perform pixel-to-pixel segmentation. By including a series of attention modules on the skip connections and Squeeze-Excitation modules on the corresponding expanding path, the proposed model adaptively enhances local responses of residual features maps by reducing the noise and rescaling these features maps by assigning channel weights in the recovery process of the expanding part. The effectiveness of the proposed model is demonstrated through experimental results. Our model outperformed the baseline models, i.e., Residual Unet and Residual-attention Unet, and showed comparable performance with other state-of-the-art brain tumor segmentation methods with the fewest parameters on the benchmark of the BraTs database. The proposed model expands the Unet family and can be extended to different medical imaging domains critical for real-world applications. However, there is a drawback in using resized MRI volumes as inputs to train the segmentation model, which leads to the potential loss of information on brain tumors due to the limitation of available computational resources. Therefore, in the future, some post-processing methods will be explored to improve the segmentation results further. In addition, to make the architecture more general for various datasets and more flexible for everyday medical imaging tasks, this research will further explore other powerful feature extraction modules to improve extra performance.
Funding Statement: This work was supported by “Human Resources Program in Energy Technology” of the Korea Institute of Energy Technology Evaluation and Planning (KETEP), granted financial resources from the Ministry of Trade, Industry & Energy, Republic of Korea. (No. 20204010600090). This study was conducted at the Medical Imaging and Diagnostics Lab at COMSATS University Islamabad and was funded by Pakistan's National Center of Artificial Intelligence.
Availability of Data and Materials: A publicly available BRATS 2020 dataset was used for analyzing our model. This dataset can be found at http://ipp.cbica.upenn.edu.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. N. Gordillo, E. Montseny and P. Sobrevilla, “State of the art survey on MRI brain tumor segmentation,” Magnetic Resonance Imaging, vol. 31, no. 2, pp. 1426–1438, 2013. [Google Scholar] [PubMed]
2. F. Demir, Y. Akbulut, B. Taşcı and K. Demir, “Improving brain tumor classification performance with an effective approach based on new deep learning model named 3ACL from 3D MRI data,” Biomedical Signal Processing and Control, vol. 81, no. 11, pp. 104424, 2023. [Google Scholar]
3. X. Du, Q. He, B. Zhang, N. Li and W. Li, “Diagnostic accuracy of diffusion-weighted imaging in differentiating glioma recurrence from posttreatment-related changes: A meta-analysis,” Expert Review of Anticancer Therapy, vol. 22, no. 4, pp. 123–130, 2022. [Google Scholar] [PubMed]
4. A. Chattopadhyay and M. Maitra, “MRI-based brain tumor image detection using CNN based deep learning method,” Neuroscience Informatics, vol. 21, no. 2, pp. 100060, 2022. [Google Scholar]
5. R. Mehta, A. Filos, U. Baid, C. Sako and R. McKinley, “QU-BraTS: MICCAI BraTS, 2020 challenge on quantifying uncertainty in brain tumor segmentation-analysis of ranking scores and benchmarking results,” Journal of Machine Learning for Biomedical Imaging, vol. 1, no. 2, pp. 1–21, 2022. [Google Scholar]
6. J. Long, E. Shelhamer and T. Darrell, “Fully convolutional networks for semantic segmentation,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, NY, USA, pp. 3431–3440, 2015. [Google Scholar]
7. H. Dong, G. Yang, F. Liu and Y. Guo, “Automatic brain tumor detection and segmentation using U-Net based fully convolutional networks,” in Annual Conf. on Medical Image Understanding and Analysis, NY, USA, pp. 506–517, 2017. [Google Scholar]
8. K. V. Kumari and S. S. Barpanda, “Residual UNet with dual attention—An ensemble residual UNet with dual attention for multi-modal and multi-class brain MRI segmentation,” International Journal of Imaging Systems and Technology, vol. 4, no. 5, pp. 1–23, 2022. [Google Scholar]
9. J. Hu, X. Gu and X. Gu, “Mutual ensemble learning for brain tumor segmentation,” Neurocomputing, vol. 504, no. 5, pp. 68–81, 2022. [Google Scholar]
10. H. Ng, S. Huang, S. Ong, K. Foong and W. Nowinski, “Medical image segmentation using watershed segmentation with texture-based region merging,” in 2008 30th Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society, NY, USA, pp. 4039–4042, 2008. [Google Scholar]
11. A. Hamza, M. Alhaisoni, A. Al Hejaili, K. A. Shaban and S. Alsubai, “D2BOF-COVIDNet: A framework of deep bayesian optimization and fusion-assisted optimal deep features for COVID-19 classification using chest x-ray and MRI scans,” Diagnostics, vol. 13, no. 2, pp. 101, 2023. [Google Scholar]
12. M. Ajmal, T. Akram, A. Alqahtani, M. Alhaisoni and A. Armghan, “BF2SkNet: Best deep learning features fusion-assisted framework for multiclass skin lesion classification,” Neural Computing and Applications, vol. 5, no. 2, pp. 1–17, 2022. [Google Scholar]
13. A. Khan, M. Alhaisoni, A. Alqahtani, S. Alsubai and M. Alharbi, “Multimodal brain tumor detection and classification using deep saliency map and improved dragonfly optimization algorithm,” International Journal of Imaging Systems and Technology, vol. 21, no. 4, pp. 1–21, 2022. [Google Scholar]
14. M. Alhaisoni, M. Nazir, A. Alqahtani, A. Binbusayyis and S. Alsubai, “A healthcare system for COVID19 classification using multi-type classical features selection,” Computers, Material and Continua, vol. 73, no. 2, pp. 1–16, 2023. [Google Scholar]
15. M. Masood, R. Maham, A. Javed, U. Tariq and S. Kadry, “Brain MRI analysis using deep neural network for medical of internet things applications,” Computers and Electrical Engineering, vol. 103, no. 4, pp. 108386, 2022. [Google Scholar]
16. A. Hamza, S. H. Wang, A. Alqahtani, S. Alsubai and A. Binbusayyis, “COVID-19 classification using chest X-ray images: A framework of CNN-LSTM and improved max value moth flame optimization,” Frontiers in Public Health, vol. 10, no. 2, pp. 1–24, 2022. [Google Scholar]
17. N. Zhang, S. Ruan, S. Lebonvallet, Q. Liao and Y. Zhu, “Kernel feature selection to fuse multi-spectral MRI images for brain tumor segmentation,” Computer Vision and Image Understanding, vol. 115, no. 6, pp. 256–269, 2011. [Google Scholar]
18. C. Botoca, “Cellular neural networks assisted automatic detection of elements in microscopic medical images. A preliminary study,” in 2014 11th Int. Symp. on Electronics and Telecommunications (ISETC), NY, USA, pp. 1–4, 2014. [Google Scholar]
19. W. Zhang, R. Li, H. Deng, L. Wang and W. Lin, “Deep convolutional neural networks for multi-modality isointense infant brain image segmentation,” NeuroImage, vol. 108, no. 6, pp. 214–224, 2015. [Google Scholar] [PubMed]
20. K. L. Hua, C. H. Hsu, S. C. Hidayati and Y. J. Chen, “Computer-aided classification of lung nodules on computed tomography images via deep learning technique,” OncoTargets and Therapy, vol. 8, no. 6, pp. 1–21, 2015. [Google Scholar]
21. S. Pereira, A. Pinto, V. Alves and C. A. Silva, “Brain tumor segmentation using convolutional neural networks in MRI images,” IEEE Transactions on Medical Imaging, vol. 35, no. 4, pp. 1240–1251, 2016. [Google Scholar] [PubMed]
22. O. Ronneberger, P. Fischer and T. Brox, “U-net: Convolutional networks for biomedical image segmentation,” in Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, NY, USA, pp. 234–241, 2015. [Google Scholar]
23. F. Milletari, N. Navab and S. Ahmadi, “V-Net: Fully convolutional neural networks for volumetric medical image segmentation,” in Proc. 2016 4th Int. Conf. 3D, Vision, NY, USA, pp. 565–571, 2016. [Google Scholar]
24. Z. Liu, L. Tong, L. Chen, Z. Jiang and F. Zhou, “Deep learning based brain tumor segmentation: A survey,” Complex & Intelligent Systems, vol. 6, no. 2, pp. 1–26, 2022. [Google Scholar]
25. F. Wang, M. Jiang, C. Qian, S. Yang and C. Li, “Residual attention network for image classification,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, NY, USA, pp. 3156–3164, 2017. [Google Scholar]
26. Y. Chen, Y. Kalantidis, J. Li and J. Feng, “A 2-nets: Double attention networks,” Advances in Neural Information Processing Systems, vol. 31, no. 5, pp. 1–22, 2018. [Google Scholar]
27. H. Nam, J. W. Ha and J. Kim, “Dual attention networks for multimodal reasoning and matching,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, NY, USA, pp. 299–307, 2017. [Google Scholar]
28. C. Yu, J. Wang, C. Peng, C. Gao, G. Yu et al., “Learning a discriminative feature network for semantic segmentation,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, NY, USA, pp. 1857–1866, 2018. [Google Scholar]
29. O. Oktay, J. Schlemper, L. L. Folgoc and M. Heinrich, “Attention u-net: Learning where to look for the pancreas,” ArXiv Preprint, vol. 4, no. 2, pp. 1–11, 2018. [Google Scholar]
30. Q. Jin, Z. Meng, C. Sun, H. Cui and R. Su, “RA-UNet: A hybrid deep attention-aware network to extract liver and tumor in CT scans,” Frontiers in Bioengineering and Biotechnology, vol. 5, no. 2, pp. 1471, 2020. https://doi.org/10.3389/fbioe.2020.605132 [Google Scholar] [PubMed] [CrossRef]
31. Y. Li, J. Yang, J. Ni, A. Elazab and J. Wu, “TA-Net: Triple attention network for medical image segmentation,” Computers in Biology and Medicine, vol. 137, no. 2, pp. 104836, 2021. https://doi.org/10.1016/j.compbiomed.2021.104836 [Google Scholar] [PubMed] [CrossRef]
32. Y. Gao, Y. Zhao, X. Luo and C. Liang, “Dense encoder-decoder network based on two-level context enhanced residual attention mechanism for segmentation of breast tumors in magnetic resonance imaging,” in 2019 IEEE Int. Conf. on Bioinformatics and Biomedicine (BIBM), NY, USA, pp. 1123–1129, 2019. [Google Scholar]
33. Z. Wang, Y. Zou and P. X. Liu, “Hybrid dilation and attention residual U-Net for medical image segmentation,” Computers in Biology and Medicine, vol. 134, no. 13, pp. 104449, 2021. [Google Scholar] [PubMed]
34. J. Hu, L. Shen and G. Sun, “Squeeze-and-excitation networks,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, NY, USA, pp. 7132–7141, 2018. [Google Scholar]
35. B. Menze, A. Jakab, S. Bauer, K. Farahani and J. Kirby, “The multimodal brain tumor image segmentation benchmark (brats) medical imaging,” IEEE Transactions on Medical Imaging, vol. 24, no. 1, pp. 1–32, 2014. [Google Scholar]
36. M. Shah, Y. Xiao, N. Subbanna, S. Francis and D. L. Arnold, “Evaluating intensity normalization on MRIs of human brain with multiple sclerosis,” Medical Image Analysis, vol. 15, no. 7, pp. 267–282, 2011. [Google Scholar] [PubMed]
37. Z. Liu, “Deep learning based brain tumor segmentation: A survey,” Sensors, vol. 2, no. 4, pp. 1–21, 2020. [Google Scholar]
38. D. Ni, “A novel multi-modal MRI synthesis network with enhanced quality and controllability for brain tumor segmentation,” Medical Image Analysis, vol. 66, no. 2, pp. 101770, 2020. [Google Scholar]
39. Y. Pan, Z. Chen and Y. Yin, “BRAINS: Boundary-aware attentive network for brain tumor segmentation,” Medical Image Analysis, vol. 61, no. 2, pp. 101670, 2020. [Google Scholar]
40. Y. Li, J. Yang, J. Ni and A. Elazab, “TA-Net: Triple attention network for medical image segmentation,” Computer Biology in Medicine, vol. 137, no. 3, pp. 1–21, 2021. [Google Scholar]
41. Z. Wang, Y. Zou and P. X. Liu, “Hybrid dilation and attention residual U-Net for medical image segmentation,” Computer Biology in Medicine, vol. 134, no. 12, pp. 104449, 2021. [Google Scholar]
42. Y. Lu, “Multilevel volumetric attention network for brain tumor segmentation,” IEEE Transactions on Neural Networks and Learning Systems, vol. 32, no. 8, pp. 3295–3307, 2021. [Google Scholar]
43. C. Zhang, “CBAM-Unet: A novel convolutional block attention module based Unet for accurate brain tumor segmentation,” IEEE Access, vol. 9, no. 3, pp. 115399–115409, 2021. [Google Scholar]
44. X. Li, “FCGAN: Fully convolutional generative adversarial network for brain tumor segmentation,” Frontiers in Neuroscience, vol. 15, no. 3, pp. 602391, 2021. [Google Scholar]
45. Y. Yuan, “Automatic brain tumor segmentation with scale attention network,” in BrainLes@MICCAI Conf., NY, USA, pp. 1–7, 2020. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF

 Downloads
Downloads
 Citation Tools
Citation Tools