 Open Access
Open Access
ARTICLE
Topological Aspects of Dendrimers via Connection-Based Descriptors
1 Department of Mathematics, School of Science, University of Management and Technology, Lahore, 54770, Pakistan
2 Department of Mathematics, College of Science, University of Tabuk, Tabuk, 7149, Saudi Arabia
* Corresponding Author: Aqsa Sattar. Email:
(This article belongs to the Special Issue: Resolvability Parameters and their Applications)
Computer Modeling in Engineering & Sciences 2023, 135(2), 1649-1667. https://doi.org/10.32604/cmes.2022.022832
Received 28 March 2022; Accepted 06 June 2022; Issue published 27 October 2022
Abstract
Topological indices (TIs) have been practiced for distinct wide-ranging physicochemical applications, especially used to characterize and model the chemical structures of various molecular compounds such as dendrimers, nanotubes and neural networks with respect to their certain properties such as solubility, chemical stability and low cytotoxicity. Dendrimers are prolonged artificially synthesized or amalgamated natural macromolecules with a sequential layer of branches enclosing a central core. A present-day trend in mathematical and computational chemistry is the characterization of molecular structure by applying topological approaches, including numerical graph invariants. Among topological descriptors, Zagreb connection indices (ZCIs) have much importance. This manuscript involves the establishment of general results to calculate ZCIs, namely first ZCI (FZCI), second ZCI (SZCI), third ZCI (TZCI), modified FZCI, modified SZCI and modified TZCI of two special types of dendrimers nanostars, namely, poly propylene imine octamin (PPIO) dendrimer and poly (propyl) ether imine (PPEtIm) dendrimer. Furthermore, we provide the numerical and graphical comparative analysis of our calculated results for both types of dendrimers with each other.Keywords
Nanobiotechnology is a swiftly growing field of technological and analytical opportunity that applies the mechanism of nanofabrication to fabricate devices for exploring biosystems. One of the main elements of this field is a dendrimer. Dendrimers are prolonged artificially synthesized or amalgamated natural macromolecules with a sequential layer of branches enclosing a central core. Dendrimers are considered to be a primary element of nanobiotechnology. The cores, end groups and branches are the main three parts of dendrimers. Nowadays, dendrimers are getting much more consideration from the researchers due to their exceptional attributes and a large span of utilization in various areas of bioscience, involving immunology, drug delivery and the development of vaccines and antivirals, for more details see [1–3].
At present, in computational and mathematical chemistry, researchers and scientists are taking heed to characterize the molecular structures by utilizing different topological perspectives incorporating graph invariants. These topological descriptors are broadly utilized in the study of quantitative structure-activity relationships (QSAR) and quantitative structure-property relationships (QSPR) [4]. Graph invariants are essential to achieve the mathematical characterization of molecular structures successfully. The field of network theory (NT) has played an indispensable role in distinct areas of life. During the last decade, NT has found a remarkable use in the field of nanobiotechnology. Topological indices (TIs), the numerical parameters which link a number with a molecular network, are widely used in chemical network theory and mathematical chemistry to characterize the topology of a molecular network. TIs can predict many psychochemical properties of molecular structures in theocratical chemistry. TIs enable researchers to find the chemical reactivity, physical attributes and biological actions of molecular compounds. A molecular network in terms of a graph is a portrayal of structural information of a chemical compound in which the atoms are displayed by vertices while the bonds are represented by edges between the vertices. The applications of TIs in various fields of science are boundless, as one can see in [5–7]. According to the reported literature, TIs have great importance in nanotechnology and theoretical chemistry. Some important categories of TIs are degree-based, distance-based and connection-based TIs. In 1972, Wiener [8] introduced the concept of a distance-based TI, which is known as the Wiener index. By theoretical and conceptual framework, the Wiener index was the most studied TI. Moreover, in 2019, Gao et al. [9] utilized distance-based indices to study the topological aspects of dendrimers.
The idea of the first ZI (FZI) was invented by Gutman et al. [10]. The second and third ZIs were discovered by Gutman et al. [11] and Furtula et al. [12]. All of these degree-based TIs have a variety of applications in the field of cheminformatics which is a combination of three major fields, namely, chemistry, mathematics and information technology [13–15]. These TIs have been utilized to distinct wide-ranging physicochemical applications, especially to characterize the different chemical structures such as dendrimers nanostars, for details see [16–18]. Among these defined TIs, the connection-based TIs have much importance because of their utilizations in characterizing the molecular chemical compounds and their ability to predict various physical and chemical properties in a comprehensive and logical way. A number of those vertices at a distance of two from a vertex v is called CN of a vertex v. According to researchers, connection-based TIs as compared to the other classical TIs provide a better absolute value of the correlation coefficient. Ali et al. [19] initiated Zagreb connection indices (ZCIs) and used octane isomers to examine their applicability. For a detailed review of some connection-based TIs, the readers are referred to [20–22]. Haoer et al. [23] computed multiplicative ZIs of some T-thorny graphs. Moreover, Javaid et al. [24] found the topological aspects of distinct wheel graphs. Further, Liu et al. [25] found the topological properties of neural networks. Recently, Sattar et al. [26–28] worked on computing the general expression of ZIs for two types of dendrimers. For the other terminologies not discussed in this paper, the readers are referred to [29–31]. The motivation for this article is as follows:
1. TIs, the numerical descriptors, are efficient enough to characterize the topology of molecular structures and assist in correlating their distinct psychochemical properties.
2. Dendrimers are symmetric, versatile and well defined chemical polymers forming a tree like structure. These nanoparticles are signalized by a numerous attributes which make them advantageous for wide ranging utilizations in various fields of science.
3. The connection-based ZIs have better applicability to predict the various psychochemical properties of distinct molecular structures in chemistry rather than the other classical ZIs present in the literature.
In this paper, first, we define the third Zagreb connection index. Further, we calculate ZCIs of two special types of dendrimer nanostars, namely, PPEtIm dendrimer and PPIO dendrimer. Moreover, we compare the results of both types of dendrimers.
This research article is structured as: in Section 1, we stated preliminaries which help the readers to understand the idea of this article. In Section 2, we computed ZCIs for PPEtIm dendrimer. In Section 3, we calculated ZCIs for the other type of dendrimer, namely, PPIO dendrimer in a comprehensive way. In Section 4, we compared the computed values of both types of dendrimers with each other. Section 5 holds the conclusions. The acronyms used in this paper are given in Table 1.
This section gives some primary definitions which are helpful for the good understanding of this manuscript.
Definition 2.1. In [10] let
which can also be rewritten in the given form
where
Definition 2.2. In [11] for network
where
Definition 2.3. In [12] for network
Third Zagreb index is also called forgotten index.
Definition 2.4. In [19] for a network
1.
2.
where
Definition 2.5. In [19] for a network
Definition 2.6. In [20] for a network
1.
2.
Now, we rewrite the above defined ZCIs. Let A be the set of all CNs and B be the set of all degrees of the vertices in
Definition 2.7. For a network
where
The SZCI rewritten as
Similarly, the modified FZCI can be rewritten as
where
Similarly, for the modified SZCI, we have
The modified TZCI can be rewritten as
where
3 ZCIs of Poly (propyl) Ether Imine Dendrimer
In this section, we calculate the ZCIs, namely, FZCI, SZCI, TZCI, modified FZCI, modified SZCI and modified TZCI, of PPEtIm dendrimer. PPEtIm dendrimer is a special type of dendrimer constructed by including an ether component as the linker and an imine component as the branching unit. It grows three-dimensionally in which the core is the oxygen element and tertiary nitrogen atoms are at the branches which are separated by eight bonds for each growth of dendrimer. Let
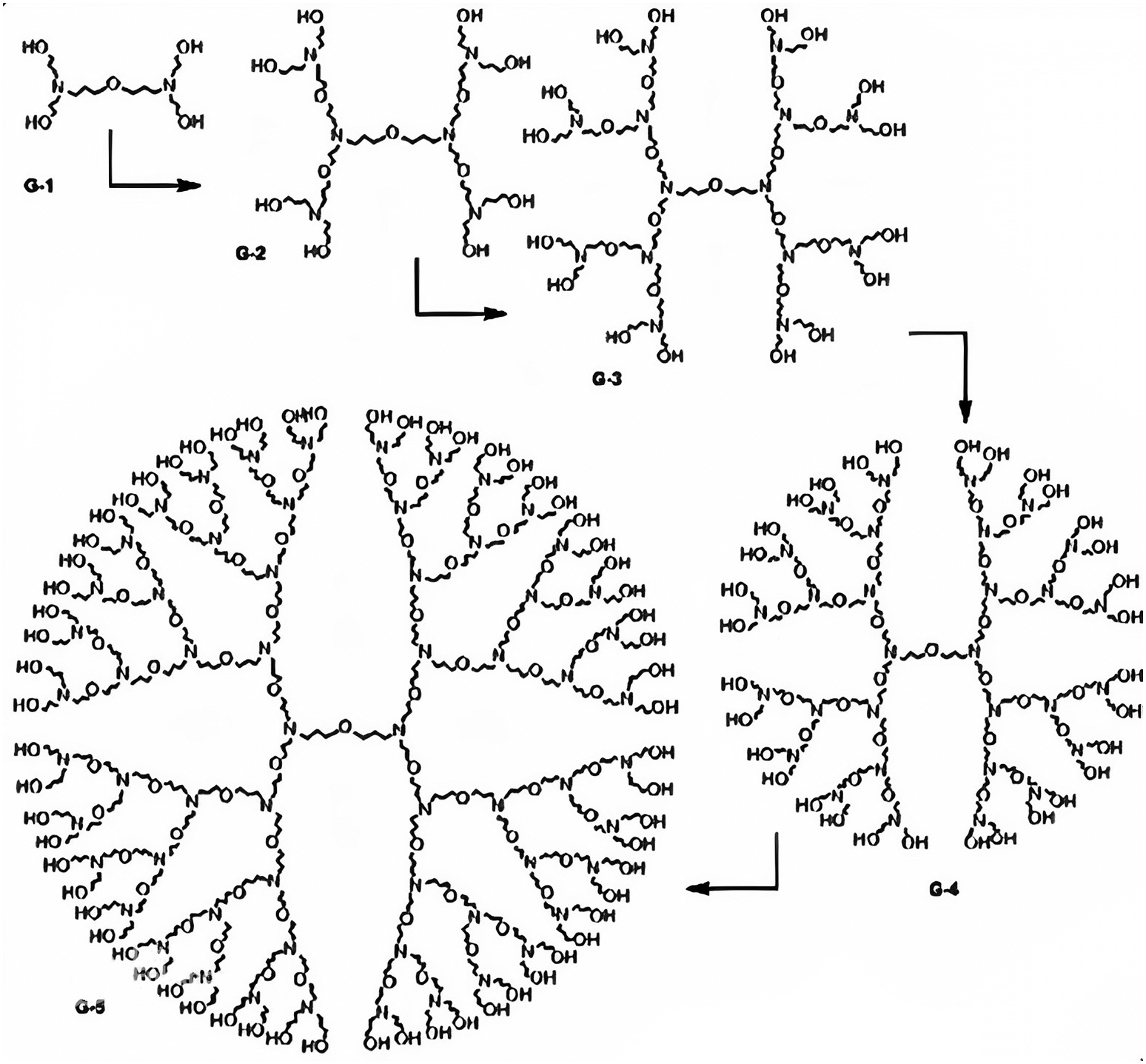
Figure 1: Chemical structural formula of PPEtIm dendrimer
Definition 3.1. Let
where
This can also be rewritten as
where
Theorem 1. Let
1.
2.
3.
Proof.
1. Firstly, we find the cardinality of vertices and edges of
As
Next, we find cardinality of those vertices which have CN 1, 2 and 3 in
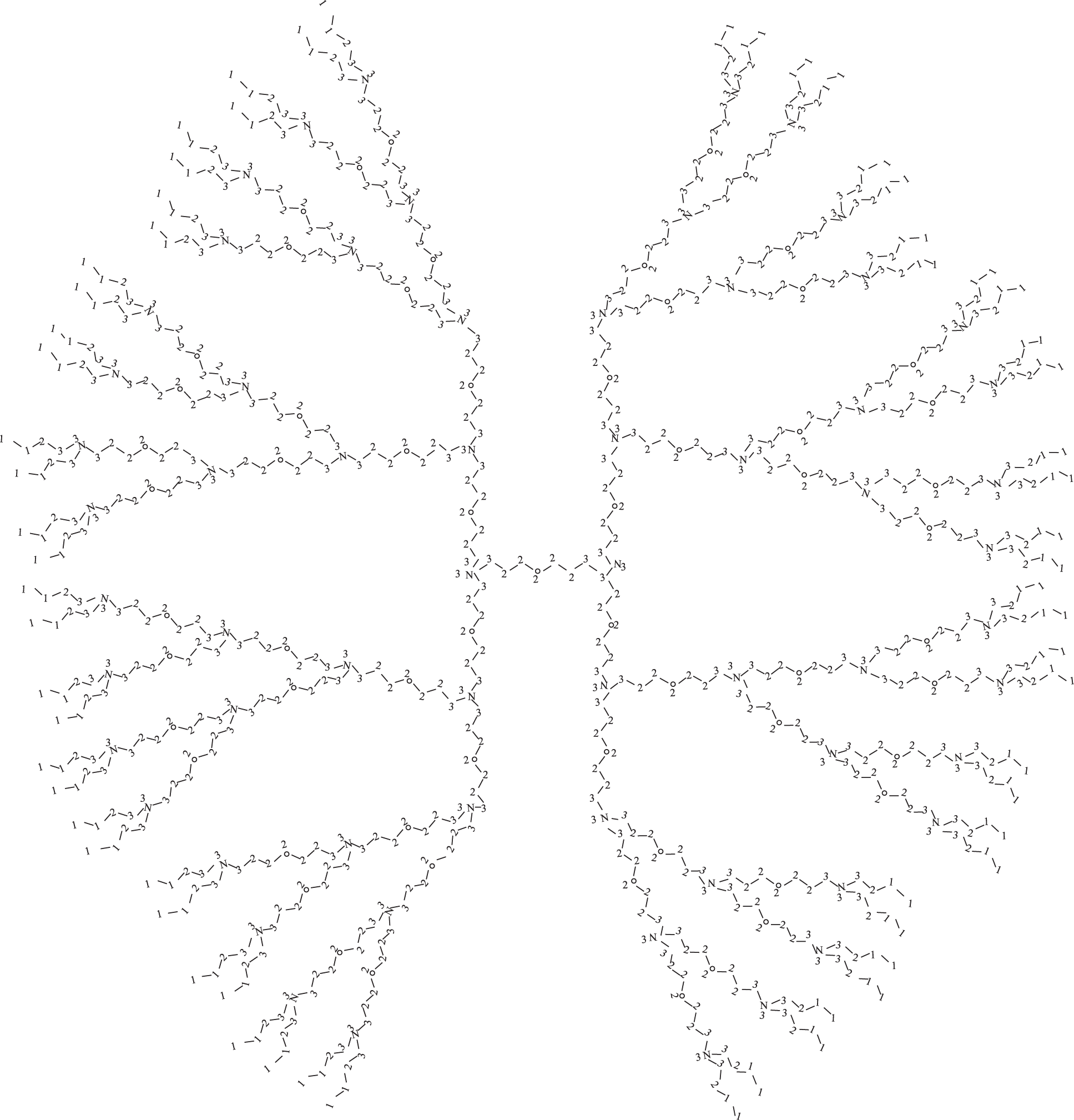
Figure 2: Structural formula of PPEtIm dendrimer for
Thus, number of vertices with connection numbers 1, 2 and 3 are given as;
Now, after placing the values of
2. Now, we classify the edge set of
Now,
Here, we have used the following formula to find the sum of the series [32].
where a is the first term and r is the common difference between two terms of the series. By placing the above calculated values in Eq. (2), we have
3. By placing the values of
This proves the theorem.
Theorem 2. Let
(1)
(2)
(3)
Proof.
1. For the proof, the readers are referred to see [33].
2. Now, we classify the edges on the basis of their degrees of incident vertices. It can be seen that
Row 1 of Table 2 shows that there are total
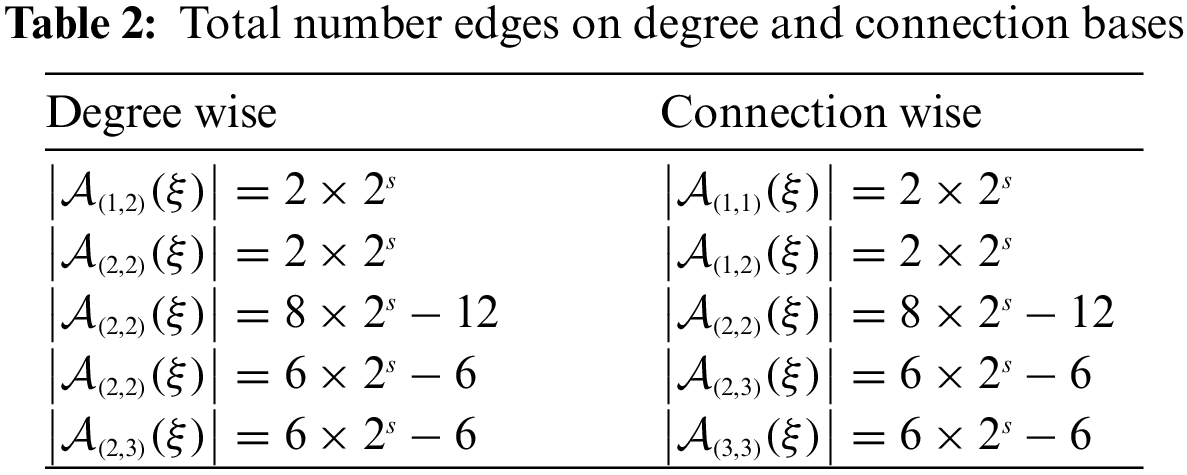
After placing the calculated values of
3. By placing the
This proves the theorem.
4 ZCIs of Poly Propylene Imine Octamin Dendrimer
In this section, we find the general expressions to compute the connection-based Zagreb indices, namely, FZCI, SZCI, TZCI, modified FZCI, modified SZCI and modified TZCI of another special type of dendrimer nanostar named as, PPIO dendrimer. PPIO dendrimer grows in three dimensions and it has five bonds in the core. The structural formula of PPIO dendrimer is shown in Fig. 3 for five generations.
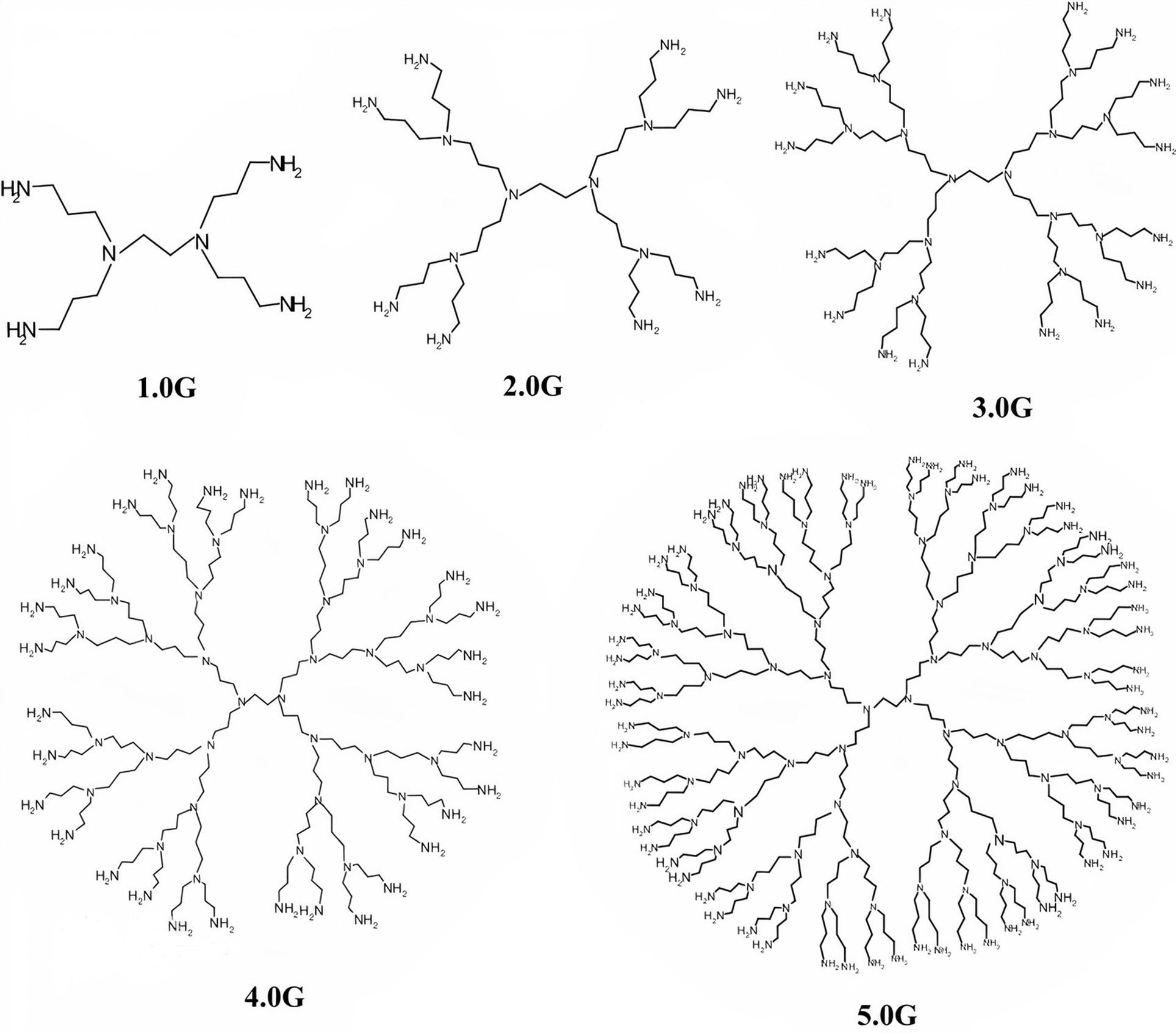
Figure 3: Structural formula of PPIO dendrimer
Theorem 3. Let
1.
2.
3.
Proof.
1. Initially, we find cardinality of edges of
As
Furthermore, we find the cardinality of those vertices which have CN 1, 2 and 3 in
In Fig. 4, we have labeled the vertices of PPIO dendrimer for
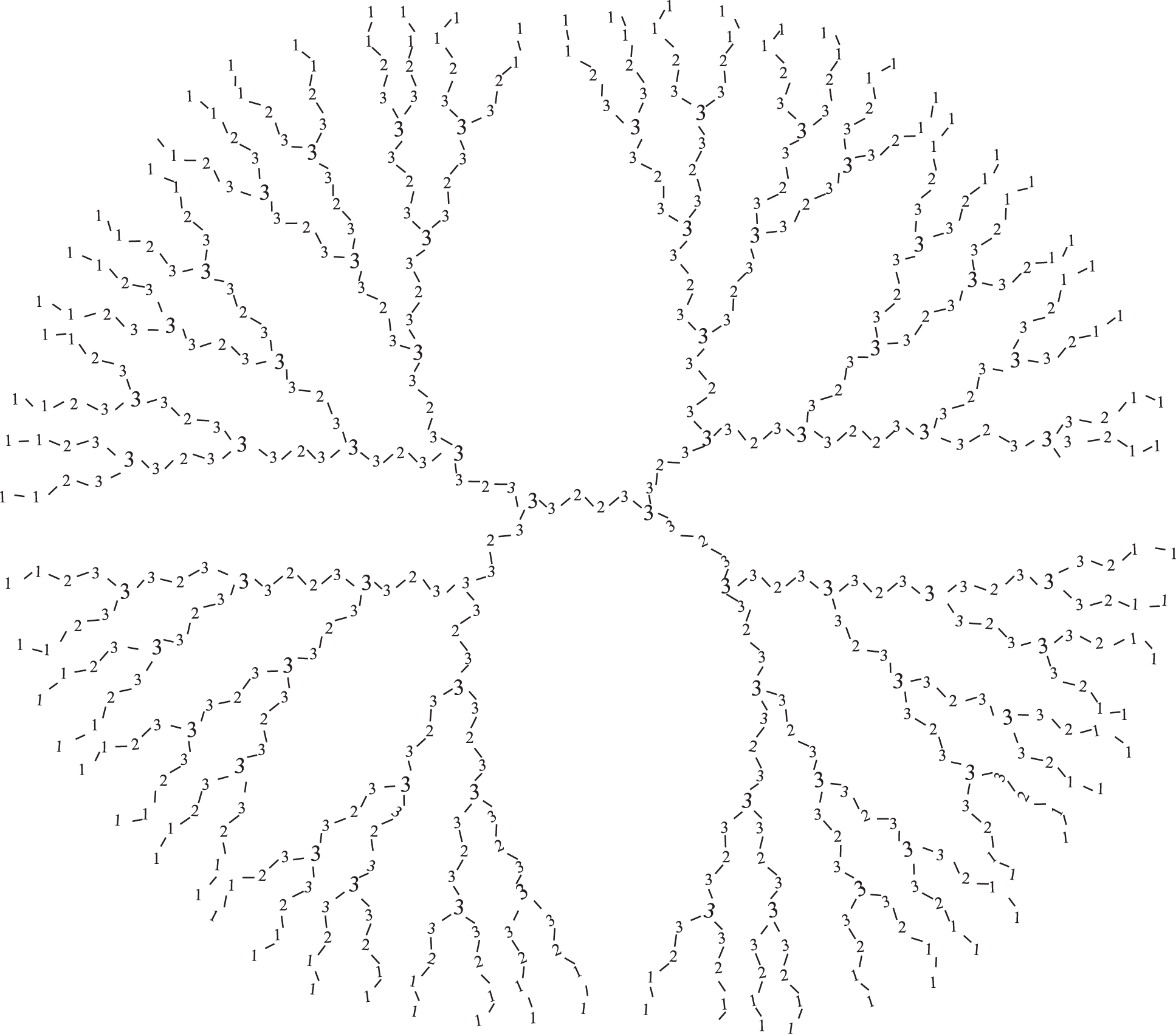
Figure 4: Structural formula of PPIO dendrimer for
2. Now, we calculate
After placing the above calculated values in Eq. (2), we have
3. After placing the values of
This proves the theorem.
Theorem 4. Let
1.
2.
3.
Proof.
1. For the proof, the readers are referred to see [32].
2. Now we make the partition of the edges based on their degrees of incident vertices. It can be easily seen that
Thus, we have
By putting the
3. By putting the
This proves the theorem.
5 Comparative Analysis and Concluding Remarks
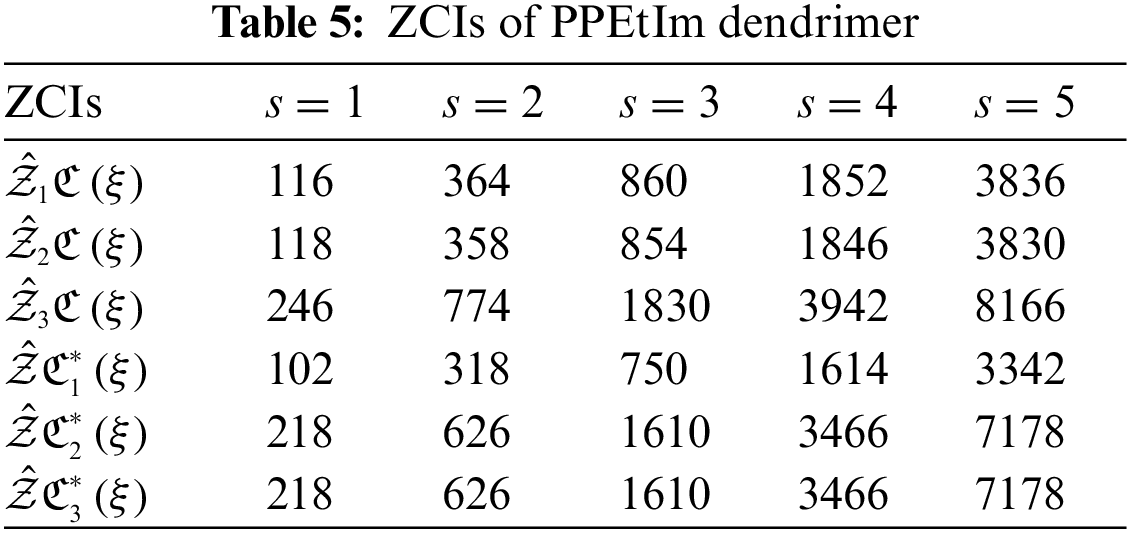
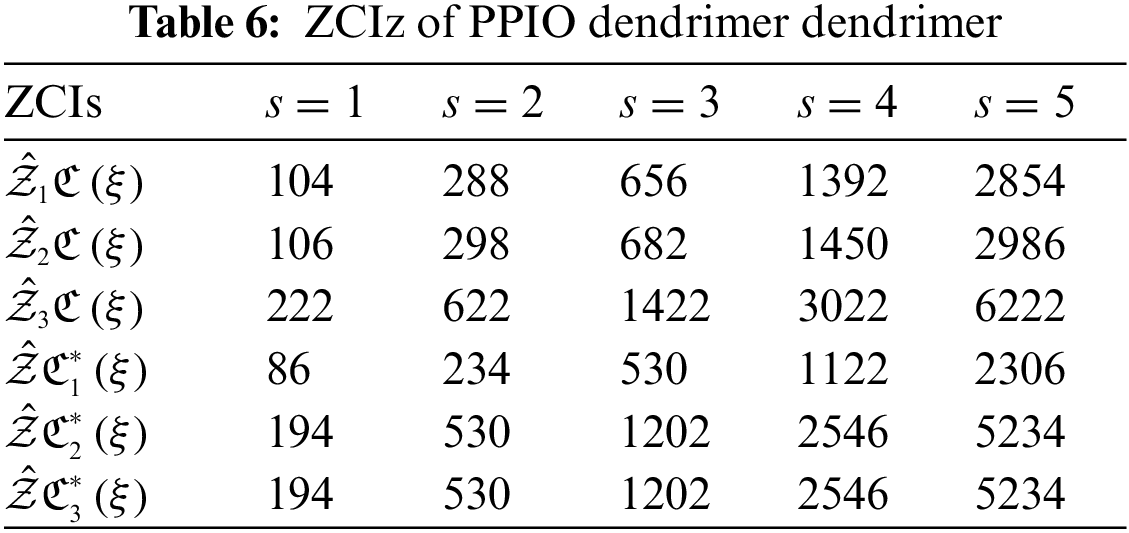
The analysis of networks plays a remarkable role to conclude their fundamental topologies. TIs specified on chemical structures can assist the researchers to recognize the biological activity, chemical reactivity and physical features. Through TIs, researchers can easily predict the distinct psychochemical properties of the molecular structures. To check the superiority of consequences of this research article, we compare our computed values for two nanostars. Table 4 displays the comparison between the calculated results of PPEtIm and PPIO dendrimers.
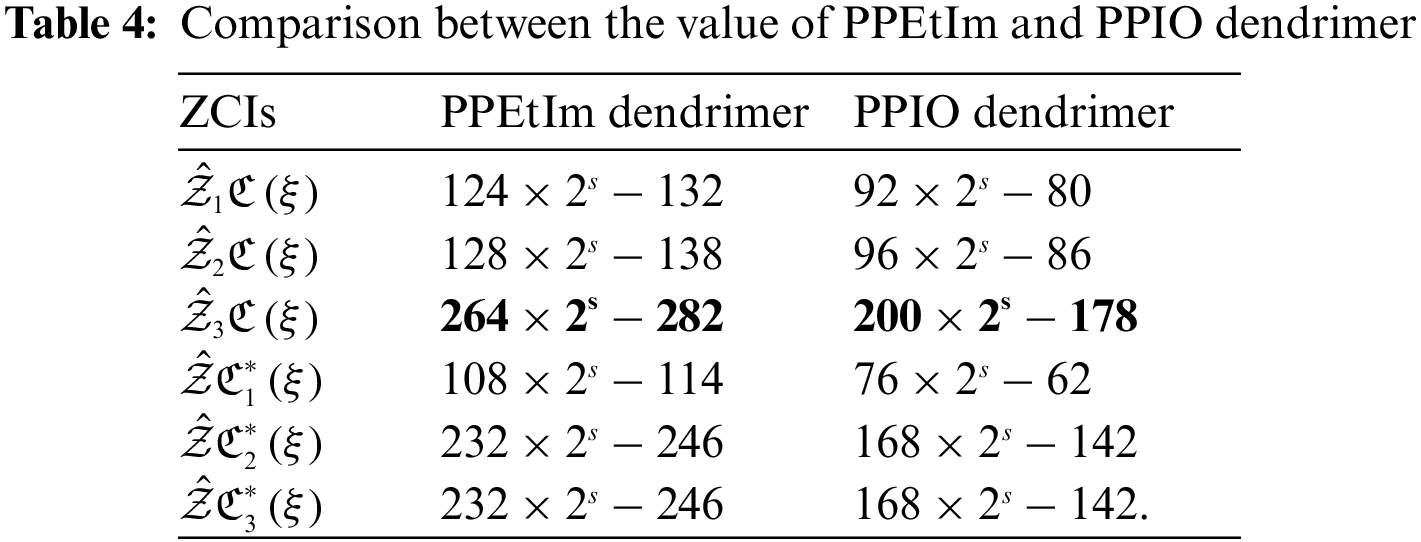
From Table 4, it can be easily seen that PPEtIm dendrimer and PPIO dendrimer get the greatest value of TZCI
The graphical comparison of
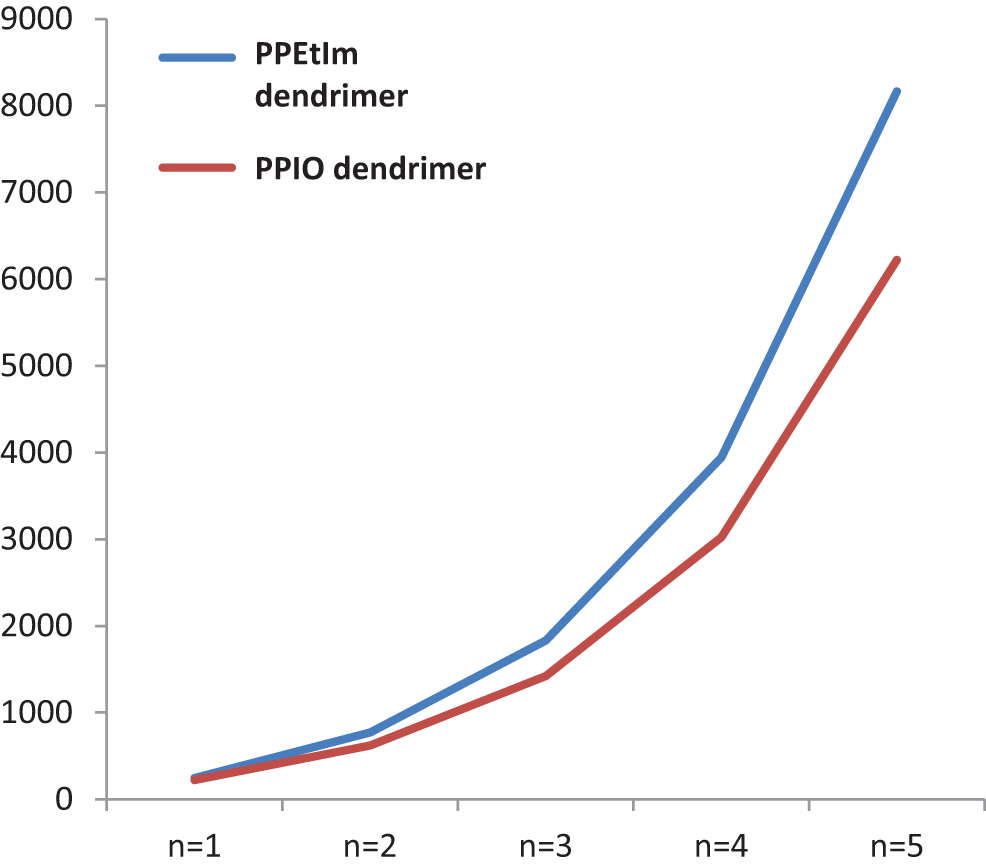
Figure 5: Graphical display of TZCI for PPEtIm and PPIO dendrimers
Now, we conclude our discussion with the following lines. In this manuscript, we have established the general results to compute ZCIs namely, FZCI, SZCI and TZCI of two special types of dendrimer nanostars, namely, PPEtIm dendrimer and PPIO dendrimer which will be helpful in computational chemistry to predict many physical and chemical attributes of the chemical substances. Moreover, we have computed modified FZCI, modified SZCI and modified TZCI for these dendrimers. The computed results just depend upon the value of
Data Availability: The data used to support the findings of this study are included within this article. However, the reader may contact the corresponding author for more details on the data.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Kesharwani, P., Gothwal, A., Iyer, A. K., Jain, K., Chourasia, M. K. et al. (2018). Dendrimer nanohybrid carrier systems: An expanding horizon for targeted drug and gene delivery. Drug Discovery Today, 23(2), 300–314. [Google Scholar]
2. Klajnert, B., Peng, L., Ceoa, V. (2013). Dendrimers in biomedical applications. RSC (Royal Society of ChemistryThomas Graham House, Science Park, Milton Road, Cambridge, UK. [Google Scholar]
3. Kurczewska, J., Ceglowski, M., Messyasz, B., Schroeder, G., (2018). Dendrimer-functionalized halloysite nanotubes for effective drug delivery. Applied Clay Science, 153, 134–143. [Google Scholar]
4. Todeschini, R., Consonni, V. (2009). Molecular descriptors for chemoinformatics: Volume I: Alphabetical listing/volume II: Appendices, references. 2nd edition. John Wiley and Sons, New Jersey, USA. [Google Scholar]
5. Afzal, H. U., Fatima, T. (2019). On topological indices of OT [m, n] octagonal tillings and TiO2 nanotubes. Acta Chimica Slovenica, 66(2), 435–442. [Google Scholar]
6. Gao, W., Younas, M., Farooq, A., Mahboob, A., Nazeer, W. (2018). M-polynomials and degree-based topological indices of the crystallographic structure of molecules. Biomolecules, 8(4), 107. [Google Scholar]
7. Imran, M., Baig, A. Q., Ali, H. (2016). On molecular topological properties of hex-derived networks. Journal of Chemometrics, 30(3), 121–129. [Google Scholar]
8. Wiener, H. (1947). Structural determination of paraffin boiling points. Journal of the American Chemical Society, 69(1), 17–20. [Google Scholar]
9. Gao, W., Iqbal, Z., Ishaq, M., Aslam, A., Sarfraz, R. (2019). Topological aspects of dendrimers via distance based descriptors. IEEE Access, 7, 35619–35630. [Google Scholar]
10. Gutman, I., Trinajstic, N. (1972). Graph theory and molecular orbitals. Total
11. Gutman, I., Ruscic, B., Trinajstic, N., Wilcox Jr, C. F. (1975). Graph theory and molecular orbitals. XII. Acyclic polyenes. The Journal of Chemical Physics, 62(9), 3399–3405. DOI 10.1063/1.430994. [Google Scholar] [CrossRef]
12. Furtula, B., Gutman, I. (2015). A forgotten topological index. Journal of Mathematical Chemistry, 53(4), 1184–1190. DOI 10.1007/s10910-015-0480-z. [Google Scholar] [CrossRef]
13. Ali, A., Gutman, I., Milovanovic, E., Milovanovic, I. (2018). Sum of powers of the degrees of graphs extremal results and bounds. MATCH Communications in Mathematical and in Computer Chemistry, 80(1), 5–84. [Google Scholar]
14. Borovicanin, B., Das, K. C., Furtula, B., Gutman, I. (2017). Bounds for Zagreb indices. MATCH Communications in Mathematical and in Computer Chemistry, 78(1), 17–100. [Google Scholar]
15. Javaid, M., Ali, U., Liu, J. B. (2021). Computing analysis for first Zagreb connection index and coindex of resultant graphs. Mathematical Problems in Engineering, 2021, 1–19. [Google Scholar]
16. Bashir, Y., Aslam, A., Kamran, M., Qureshi, M. I., Jahangir, A. et al. (2017). On forgotten topological indices of some dendrimers structure. Molecules, 22(6), 867. DOI 10.3390/molecules22060867. [Google Scholar] [CrossRef]
17. Bokhary, S. A. U. H., Imran, M., Manzoor, S. (2016). On molecular topological properties of dendrimers. Canadian Journal of Chemistry, 94(2), 120–125. DOI 10.1139/cjc-2015-0466. [Google Scholar] [CrossRef]
18. Dorosti, N., Iranmanesh, A., Diudea, M. V. (2010). Computing the cluj index of the first type dendrimer nanostar. Optoelectronics and Advanced Materials-Rapid Communications, 4, 381–384. [Google Scholar]
19. Ali, A., Trinajstic, N. (2018). A novel/old modification of the first Zagreb index. Molecular Informatics, 37(6–7), 1800008. DOI 10.1002/minf.201800008. [Google Scholar] [CrossRef]
20. Ali, U., Javaid, M., Kashif, A. (2020). Modified Zagreb connection indices of the T-sum graphs. Main Group Metal Chemistry, 43(1), 43–55. DOI 10.1515/mgmc-2020-0005. [Google Scholar] [CrossRef]
21. Cao, J., Ali, U., Javaid, M., Huang, C. (2020). Zagreb connection indices of molecular graphs based on operations. Complexity, 2020, 7385682. DOI 10.1155/2020/7385682. [Google Scholar] [CrossRef]
22. Du, Z., Ali, A., Trinajstic, N. (2019). Alkanes with the first three maximal/minimal modified first Zagreb connection indices. Molecular Informatics, 38(4), 1800116. DOI 10.1002/minf.201800116. [Google Scholar] [CrossRef]
23. Haoer, R. S., Mohammed, M. A., Selvarasan, T., Chidambaram, N., Devadoss, N. (2020). Multiplicative leap Zagreb indices of T-thorny graphs. Eurasian Chemical Communications, 2(8), 841–846. [Google Scholar]
24. Javaid, M., Ali, U., Siddiqui, K. (2021). Novel connection-based Zagreb indices of several wheel-related graphs. Computational Journal of Combinatorial Mathematics, 1, 1–28. [Google Scholar]
25. Liu, J. B., Raza, Z., Javaid, M. (2020). Zagreb connection numbers for cellular neural networks. Discrete Dynamics in Nature and Society, 2020, 8038304. DOI 10.1155/2020/8038304. [Google Scholar] [CrossRef]
26. Sattar, A., Javaid, M., Bonyah, E. (2021). Connection-based multiplicative Zagreb indices of dendrimer nanostars. Journal of Mathematics, 2021, 2107623. DOI 10.1155/2021/2107623. [Google Scholar] [CrossRef]
27. Sattar, A., Javaid, M., Bonyah, E. (2022). On the studies of dendrimers via connection-based molecular descriptors. Mathematical Problems in Engineering, 2022, 1–13. DOI 10.1155/2022/1053484. [Google Scholar] [CrossRef]
28. Sattar, A., Javaid, M., Bonyah, E. (2022). Computing connection-based topological indices of dendrimers. Journal of Chemistry, 2022, 1–15. DOI 10.1155/2022/7204641. [Google Scholar] [CrossRef]
29. Julietraja, K., Venugopal, P., Prabhu, S., Liu, J. B. (2020). M-polynomial and degree-based molecular descriptors of certain classes of benzenoid systems. Polycyclic Aromatic Compounds, 1–30. DOI 10.1080/10406638.2020.1867205. [Google Scholar] [CrossRef]
30. Liu, J. B., Arockiaraj, M., Arulperumjothi, M., Prabhu, S. (2021). Distance based and bond additive topological indices of certain repurposed antiviral drug compounds tested for treating COVID-19. International Journal of Quantum Chemistry, 121(10), 26617. DOI 10.1002/qua.26617. [Google Scholar] [CrossRef]
31. Yang, Z., Arockiaraj, Z., Prabhu, S., Arulperumjothi, M., Liu, J. B. (2021). Second Zagreb and sigma indices of semi and total transformations of graphs. Complexity, 2021, 1–15. DOI 10.1155/2021/6828424. [Google Scholar] [CrossRef]
32. James, S. (2002). Calculus. Thomson brooks cole, 5th edition. Canada. [Google Scholar]
33. Noureen, S., Bhatti, A. A., Ali, A. (2020). Extremal trees for the modified first Zagreb connection index with fixed number of segments or vertices of degree 2. Journal of Taibah University for Science, 14(1), 31–37. DOI 10.1080/16583655.2019.1699227. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools