 Open Access
Open Access
ARTICLE
BioSkinNet: A Bio-Inspired Feature-Selection Framework for Skin Lesion Classification
1 Department of Information Systems, College of Computer Engineering and Sciences, Prince Sattam bin Abdulaziz University, Al-Kharj, 11942, Saudi Arabia
2 Department of Electrical and Computer Engineering, COMSATS Univeristy Islamabad, Wah Campus, WahCantt, 47010, Pakistan
3 Department of Computer Science, Tulane University, New Orleans, LA 70118, USA
* Corresponding Author: Tallha Akram. Email:
(This article belongs to the Special Issue: Machine Learning and Deep Learning-Based Pattern Recognition)
Computer Modeling in Engineering & Sciences 2025, 143(2), 2333-2359. https://doi.org/10.32604/cmes.2025.064079
Received 04 February 2025; Accepted 08 April 2025; Issue published 30 May 2025
Abstract
Melanoma is the deadliest form of skin cancer, with an increasing incidence over recent years. Over the past decade, researchers have recognized the potential of computer vision algorithms to aid in the early diagnosis of melanoma. As a result, a number of works have been dedicated to developing efficient machine learning models for its accurate classification; still, there remains a large window for improvement necessitating further research efforts. Limitations of the existing methods include lower accuracy and high computational complexity, which may be addressed by identifying and selecting the most discriminative features to improve classification accuracy. In this work, we apply transfer learning to a Nasnet-Mobile CNN model to extract deep features and augment it with a novel nature-inspired feature selection algorithm called Mutated Binary Artificial Bee Colony. The selected features are fed to multiple classifiers for final classification. We use , ISIC-2016, and HAM datasets for experimentation, supported by Monte Carlo simulations for thoroughly evaluating the proposed feature selection mechanism. We carry out a detailed comparison with various benchmark works in terms of convergence rate, accuracy histogram, and reduction percentage histogram, where our method reports 99.15% (2-class) and 97.5% (3-class) accuracy on the dataset, while 96.12% and 94.1% accuracy for the other two datasets, respectively, against minimal features.Keywords
Cancer is a pathological condition characterized by aberrant proliferation of cells in many anatomical locations within the human body [1]. Skin is a vital organ in the human body that includes all the muscular, skeletal, and visceral components. The fundamental role of the skin is to protect the body from external factors, including chemicals, pathogens, and temperature variations [2]. Naturally, a skin disease, such as skin cancer, will have a consequential impact on all other bodily organs. Several prominent risk factors for skin cancer include genetic susceptibility, fair skin, repeated sunburns, and other medical conditions [3]. Although skin cancer continues to increase worldwide as well, studies suggest that it is the most prevalent kind of cancer in the US [4]. Recent studies [5] suggest that around 9500 people are diagnosed with general skin cancers every day, where more than 2 die every hour only in the US. Similarly, another study by the American Cancer Society [6] reveals that 104,960 people are projected to be diagnosed only with melanoma in 2025, leading to an alarming number of 8430 deaths. The situation, unfortunately, isn’t encouraging in the rest of the world either, especially in Australia and Scandinavia [7]. Due to its rapid growth and metastasis, melanoma is the most severe form of cancer [8]; however, it is curable with early detection and treatment [9]. Malignant and benign examples from various melanoma datasets are shown in Fig. 1.
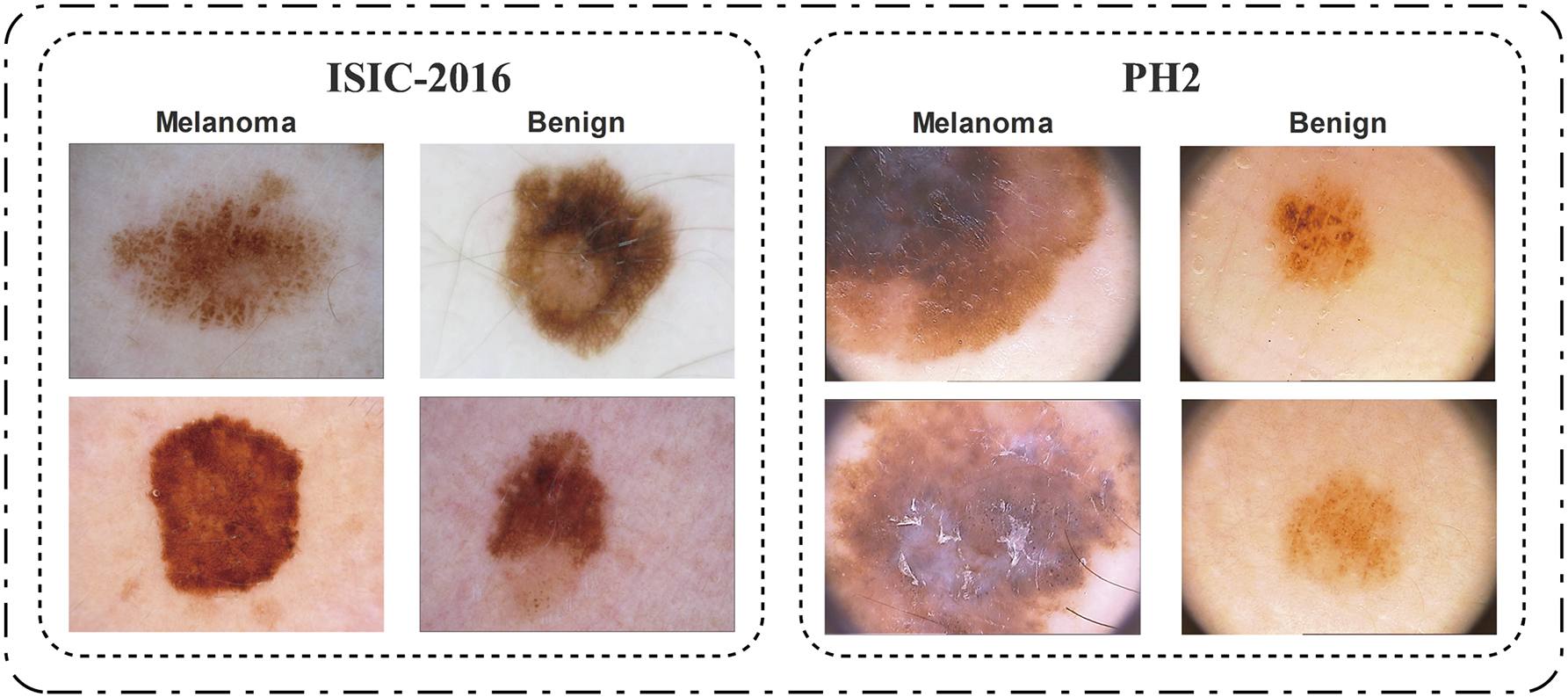
Figure 1: A few image samples from the selected datasets of ISIC-2016 and
Considerable progress has been made by numerous researchers in the field of computer vision for skin cancer detection [4,10–13].
One significant challenge associated with this approach is the substantial data requirement for model training. The problem has been successfully resolved by utilizing transfer learning (TL) [14]. Several researchers utilized multiple pre-trained models to categorize computer vision and medical imaging tasks. These models include VGG [15], GoogleNet [16], ResNet [17], and Densnet201 [18].
Despite the great strides taken thus far, a lot more work is needed to fix the problem with the detection and classification phases. Given the significance of feature selection for improving the model’s overall accuracy and decreasing computation time, this research focuses primarily on the feature selection process as a strategy to alleviate the challenge referred to as the curse of dimensionality. Given the increasing recognition of bio-inspired techniques in the field of feature selection, we present a unique approach known as the mutated binary artificial bee colony (MBABC) algorithm. The primary advantage of employing MBABC compared to traditional methods, such as particle swarm optimization (PSO), genetic algorithm (GA), grey wolf optimization (GWO), and BMNABC, resides in its improved exploration and exploitation balance, broader search space, and its ability to adjust the selection strategy. Moreover, the implementation of the entropy function for fitness evaluation has necessitated adaptations in the employed bee phase. This extension not only enhances diversity but also facilitates fast convergence and reduces the likelihood of being stuck in local minima.
The following is the logical progression of this article: The current approaches are discussed in Section 2, and then the problem statement and contributions are presented in Section 3. Section 4 will focus on the datasets and models used in the study, followed by the proposed section. Section 5 discusses the results, followed by Section 6, which concludes our work.
Skin cancer classification is a well-researched field, with numerous contributions, directly towards deep learning models and feature selection methods, proposed in the past few years. Bassel et al. [4] introduced an automated technique for classifying benign and malignant skin cancer by utilizing a hybrid deep learning approach. They utilized several pre-trained deep models, such as Xception, ResNet50, and VGG16, to perform feature extraction on the ISIC-2019 dataset. Afterwards, the deep features wee passed to the classifiers, that included support vector machines (SVM), random forest (RF), neural networks (NN), and K-nearest neighbors (KNN). To evaluate the effectiveness of the proposed framework, various performance metrics such as F1 score, accuracy, AUC, and sensitivity were employed. The proposed method achieved an accuracy of around 90.9% when applied to the extracted features using the Xception model.
Ali et al. presented a multi-class classification of skin cancer by applying pre-processing and transfer learning techniques [11]. During the preprocessing stage, the images from HAM10000 dataset were scaled and augmented, and the hair were removed using existing techniques. They evaluated the effectiveness of different variants of EfficientNet. To accomplish this, they applied transfer learning to Efficientnet B0-B7 nets. The evaluation criteria used in this article were recall, accuracy, and F1 score. Among all, EfficientNet models, B4 performed well with an accuracy of
In their study, Bechelli et al. [12] employed a combination of conventional and deep learning approaches to classify skin cancer from dermoscopic images. They used multiple machine learning algorithms, including logistic regression (LR), linear discriminant analysis (LDA), decision tree classifiers (DTC), k-nearest neighbors classifiers (KNN), and Gaussian Naive bayes (GNB). Additionally, they utilized their own 11-layered custom convolutional neural network (CNN) model, as well as pre- models such as ResNet50, Xception, and VGG16. Their model achieved an accuracy rate of 84%, while among the conventional machine learning models, LR exhibited the highest performance with an accuracy rate of 72%. On the other hand, among the deep learning models, VGG16 outperformed the rest with an accuracy of 88%.
Chen et al. [19] proposed a multimodal data fusion diagnosis network (MDFNet) for the purpose of classifying skin cancer. The proposed approach extensively integrated feature fusion in conjunction with the attention mechanism. The proposed method consisted of three main steps: 1. Construction of a feature extraction mechanism based on two modes; 2. Implementation of attention mechanisms to effectively handle multimodal features; and 3. Integration of multimodal characteristics through fusion. Various models, including ResNet50, VGGNet19, Inception-V3, and DenseNet121, were employed for feature extraction. The developed approach achieved an accuracy of 80.42%, exhibiting a 9% improvement in comparison to models solely utilizing medical images.
Hosny et al. [20] proposed a refined residual deep convolutional neural network for skin cancer classification. This research work utilized six skin cancer datasets: ISIC (2016, 2017, 2018), MED-NODE, DermIS, Quest, and PH2 and considered a set of experiments for evaluating the proposed model. Initially, the proposed residual deep convolutional neural network (RDCNN) was trained and tested on the dataset without undergoing any preprocessing. Subsequently, during the second experimentation, the model underwent testing on the segmented images. Finally, the model obtained from the second experimentation was employed as a pre-trained model for final classification. The results demonstrated that their proposed RDCNN model surpassed other models and achieved the best accuracy, around 96.29%, on the ISIC 2017 dataset.
Tembhurne et al. [21] proposed a method for detecting skin cancer using a combination of traditional and modern machine learning approaches. The dataset used in this study was sourced from Kaggle, a publicly accessible platform, and consisted of processed images from the ISIC dataset. The developed technique relied on four key steps: 1. Feature extraction; 2. Concatenation; 3. Dimensionality reduction; and 4. Classification. The features were initially retrieved using the contourlet transform (CT) and local binary pattern histogram (LBP). Subsequently, the extracted features from both algorithms were combined before applying the principal component analysis (PCA) for dimensionality reduction. The concatenated features were later classified using two classifiers: logistic regression (LR) and linear support vector machines (SVM). The proposed technique achieved a precision of 93%. Similarly, other researchers have also explored several different pre-trained CNN models, either solely or in conjunction with the other pre-trained models [22–25]. In Table 1, the existing techniques are summarized briefly on the basis of the methodologies proposed, the precision, and the datasets utilized.
Despite extensive work on skin lesion classification, several gaps remain. First, many methods strive to boost accuracy but pay limited attention to dimensionality reduction—leading to high feature redundancy and potential overfitting. Second, computational complexity is seldom discussed in detail, making it difficult to assess a model’s practical feasibility. Third, bio-inspired optimization techniques, such as swarm and evolutionary strategies, are underutilized despite their potential to mitigate the “curse of dimensionality”. Especially, the advanced metaheuristics remain underexplored for class-imbalanced datasets like HAM10000. Fourth, many approaches evaluate performance on a single or closely related dataset, leaving questions about cross-dataset generalization. Finally, although some studies handle multi-class problems, many focus primarily on binary classification, inhibiting broader clinical adoption.
3 Problem Statement & Contributions
Existing methods in automated skin lesion classification are limited in their practical efficacy due to numerous significant challenges that have not been responded: despite substantial research in the field. Mostly, several presented methods emphasize accuracy improvement without sufficient fixing of dimensionality reduction, leading to duplicated features and elevated overfitting risk. Additionally, computational complexity and efficiency are often overlooked, posing difficulties in real-world implementation, particularly in resource-constrained clinical environments. Although bio-inspired optimization methods, especially advanced metaheuristics, have shown the potential to resolve these challenges, their use remains constrained, mostly in class-imbalanced datasets. Ultimately, the majority of research primarily focuses on binary classification, overlooking the complex challenges present in multi-class clinical scenarios. Therefore, it is evident that a strong and computationally efficient feature selection method is necessary to overcome these limitations, thereby improving the reliability, scalability, and practical applicability of skin lesion classification systems.
In this study, we consider a labeled dataset
To balance accuracy with feature reduction, we solve
where
The significant contributions of this work are as follows:
1. A novel bio-inspired metaheuristic algorithm, mutated binary artificial bee colony (MBABC), for feature selection is proposed to address the challenges posed by over-fitting, “curse of dimensionality”, and computational cost. The core objective is to identify and ultimately choose the most discriminatory feature information for the final classification.
2. A thorough evaluation framework is developed that not only assesses robustness and generalization of the proposed feature selection method using multiple datasets, but also compares it with the baseline models using various metrics The results are compiled by conducting Monte-Carlo simulations for stability analysis.
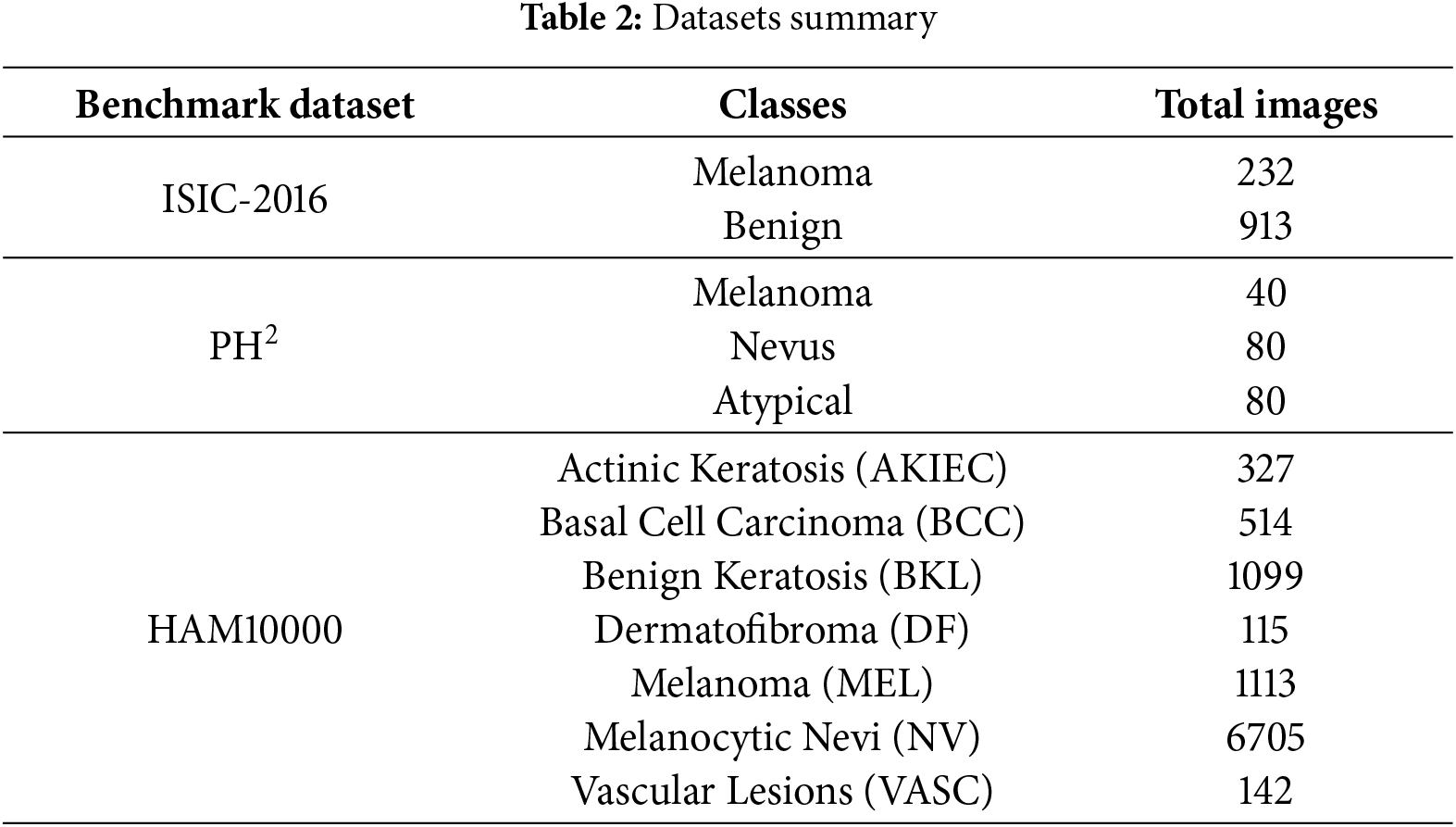
Our proposed methodology in this research is assessed using three publicly accessible benchmark datasets: ISIC-2016 [34],
To evaluate the performance of the proposed method comprehensively, several standard evaluation metrics were utilized. Initially, a confusion matrix was constructed to visualize the performance through the classification of predictions into four distinct categories: True Positive (TP), True Negative (TN), False Positive (FP), and False Negative (FN) [37]. Based on these categories, the following performance measures are calculated:
1. Accuracy calculates the ratio of accurate predictions relative to the total number of predictions:
2. Precision quantifies how precise the positive predictions are, defined as the ratio of correctly identified positive cases to all predicted positive cases:
3. Recall (Sensitivity) evaluates the method’s ability to identify positive instances correctly. High recall is especially critical in medical diagnosis to minimize the risk of missing potentially malignant lesions:
4. Specificity measures the proportion of correctly identified negative instances:
5. F1-score is the harmonic mean of precision and recall, providing a balanced measure especially useful when dealing with imbalanced datasets:
6. Matthews Correlation Coefficient (MCC) evaluates the quality of binary classifications, considering all four confusion matrix categories (TP, TN, FP, FN):
Convolutional Neural Network (CNN) is a specialized architecture that has been specifically designed for computer vision applications. These architectures have significantly transformed the domain of computer vision by facilitating the learning of complex visual patterns and automatic recognition, thus enabling the implementation of image segmentation, object detection, and classification, among other applications.
Nasnet-mobile
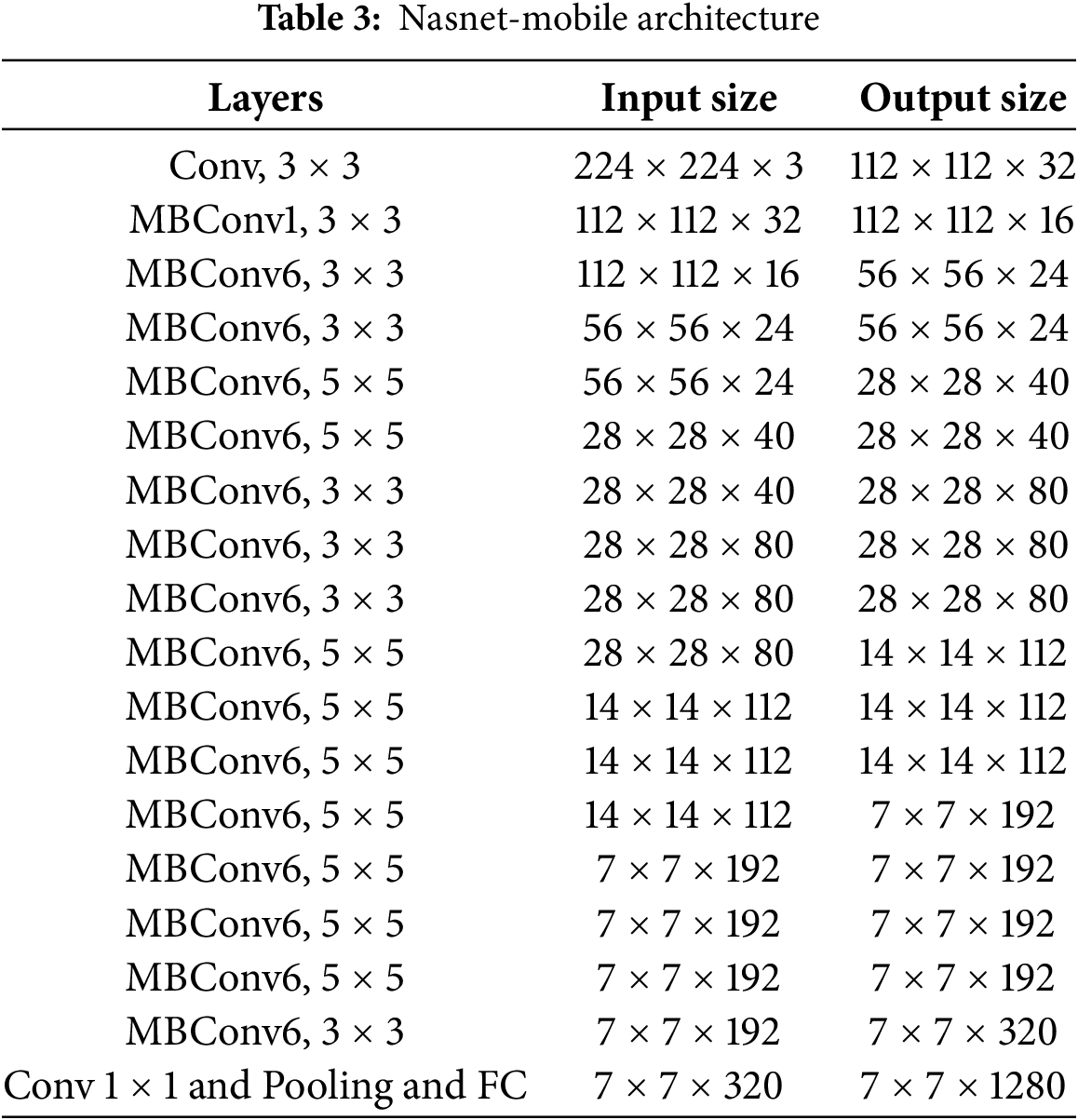
Construction of the Nasnet-Mobile convolutional neural network model utilizes the neural architecture search (NAS) technique. Neural architecture search is a procedure that autonomously looks for the optimal network architecture for a specified task. Designed specifically for mobile devices, this version of the NasNet architecture is more compact and efficient.
Compared to deep models such as ResNet and InceptionV3, NasNet-mobile is an exceptionally quick, shallow, and compact convolutional neural network model with 70 layers and 4.2 million parameters. The Nasnet-mobile model utilizes the Adam Optimizer in combination with the Rectified Linear Unit (ReLU) activation function. The comprehensive Nasnet-mobile architecture is illustrated in Table 3.
A critical concern arises when confronted with the substantial volume of data needed for the CNN framework. Deep learning models demand a significantly greater volume of data for training purposes when compared to conventional machine learning models. Nevertheless, this issue has been resolved since the inception of transfer learning [38]. By utilizing transfer learning, it is feasible to train a deep learning model despite the limited availability of data. Fig. 2 demonstrates a visual illustration of transfer learning. In this process, the final layers of the model are replaced with the new layers, and the weights in the previous layers remain frozen.
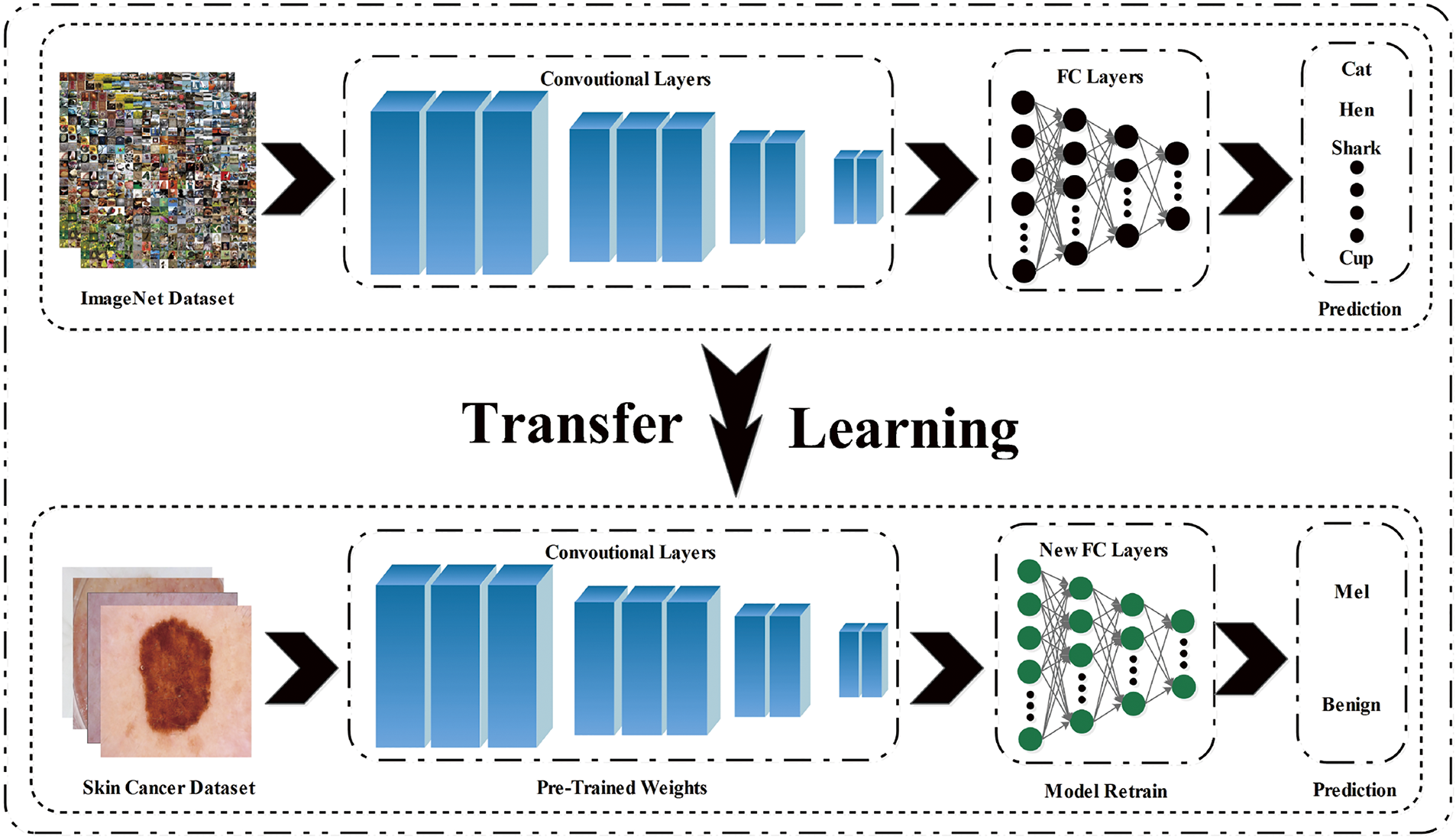
Figure 2: Transfer Learning: A CNN model trained on ImageNet is adapted for skin lesion classification by substituting and fine-tuning its final layers
The steps involved in transfer learning are listed below:
1. Selection of a pre-trained model
2. Change the fully-connected (FC) layer of the
3. The output model is fine-tuned on new data
The proposed framework in Fig. 3 encompasses a comprehensive pipeline that spans from the initial acquisition of images to the last stage of classification. The images obtained from the chosen databases are initially fed into the selected pre-trained model, followed by transfer learning application. This process involves substituting the last three layers with the newly initialized layers. By maintaining the frozen state of the existing weights in the previous layers, the model undergoes retraining on the chosen datasets, which then extracts the resulting features. It is crucial to acknowledge that the last layers possess the potential to provide redundant information. Consequently, it becomes necessary to employ a feature selection strategy to improve efficiency.
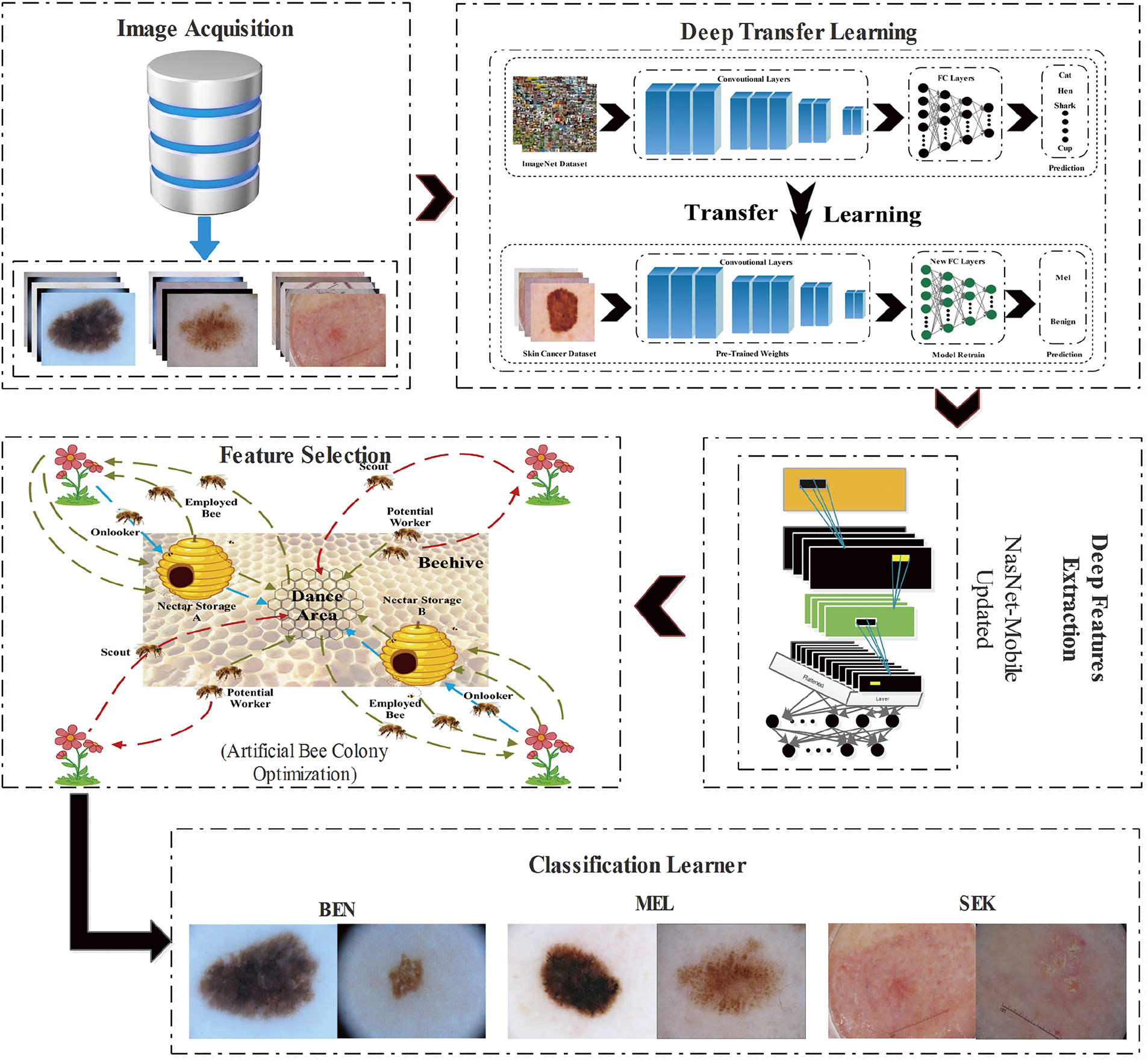
Figure 3: Schematic of the proposed BioSkinNet framework: From the image acquisition block to the final classification block
In the field of machine learning, and feature selection in particular, bio-inspired algorithms have recently emerged as key players. These algorithms draw inspiration from the principles of evolution and biological systems, and encompass a category of optimization methodologies that have been effectively employed in diverse sectors for problem-solving purposes [18]. In this research, we focus on the artificial bee colony (ABC) algorithm, which emulates the foraging behavior of honeybees to explore and improve the solutions. The ABC algorithm employs two distinct categories of bees: employed bees and observer bees. The foraging behavior involves employed bees actively exploring the solution space surrounding their present food source. On the other hand, the observer bees make decisions regarding food sources by relying on the information acquired from the employed bees. Bees engage in a dance-like technique to transmit the quality of their food sources to one another. As the algorithm advances, an increasing number of spectator bees opt for the most optimal food sources, thereby facilitating the exploration of novel and superior problem-solving strategies. The proposed method integrates an entropy-based fitness function with a balanced exploration-exploitation mechanism. Moreover, with mutation-driven diversity, it often surpasses other feature selection methods. While many traditional algorithms (e.g., PSO, GA, GWO) risk early convergence or poor local minima in high-dimensional spaces, MBABC’s added mutation phase that flips feature bits stochastically - allowing swarm to escape suboptimal regions. Simultaneously, its entropy-based fitness penalizes redundancy more precisely than just accuracy-based approaches by rewarding the most discriminative feature subsets. This combination consistently yields higher classification performance, more robust convergence, and better feature reduction than competing approaches, making MBABC well-suited to real-world tasks that demand both accuracy and efficiency.
4.5.1 Mutated Binary Artificial Bee Colony Algorithm (MBABC)
In order to optimize the mathematical operations, Karaboga [39] presented the ABC algorithm as a swarm intelligence method. It is based on how honey bee colonies hunt for food. The bee colony consists of three distinct types of bees, namely employed bees, onlooker bees, and scout bees.
In the initial phase, the employed bees engage in foraging activities to locate food sources. They communicate their findings to onlookers, who then evaluate the information provided by the employed bees to determine which food sources to exploit. The decision-making process is predicated on the employed bees’ perception of whether a specific food source constitutes a geographically optimal solution. When a food source does not experience any advancements within a designated period of time, referred to as the abandonment counter (AC), the bee linked to that food source transitions into a scout bee. During the scout bees’ phase, the bees look for more food sources using an algorithm called AC that operates with a threshold value set by the user.
The variables
for the given objective function
During this phase, the value of AC is set to zero and a predetermined limit value is established. Moreover, within the predetermined boundaries, the food sources are designated at random.
The variable
where
The new solution is produced as follows with the improved solution at this phase:
where
As the employed bees return to the hive, they impart knowledge to the observer bees concerning the food sources they have utilized. The observer bee selects its food source through the utilization of probabilistic information at its disposal, which leads to the discovery of the new solution denoted as
In this phase, each food source’s AC value is explored. If the count goes above the maximum allowed, the AC is set back to zero. Eq. (5) is used to generate a new supply of food, and the current solution is the best solution, that is, (
Algorithm 1 explains the detailed flow of our proposed MBABC technique. The process initiates with a random initialization of a population of candidate solutions, where each solution is represented as a binary vector, indicating features that are either selected or discarded. Each iteration cycle employs three distinct bee phases: Employed bees perturb existing solutions to explore neighboring subsets, guided by a mutation operator to improve solution diversity. Subsequently, based on an entropy-controlled fitness criterion, onlooker bees probabilistically select the best possible solutions, ensuring a balanced exploration and exploitation of the search space. Scouts replace solutions stalling beyond a threshold level, therefore fostering diversity and lowering the chance of early convergence. The method balances exploration and exploitation until a stopping criterion—such as a convergence threshold or a maximum iteration limit—is fulfilled, so iteratively updating a global best solution.

The simulations are performed using three publicly available benchmark datasets, including PH
For the selected pre-trained deep model of Nasnet-mobile, the resultant feature vector is of dimension
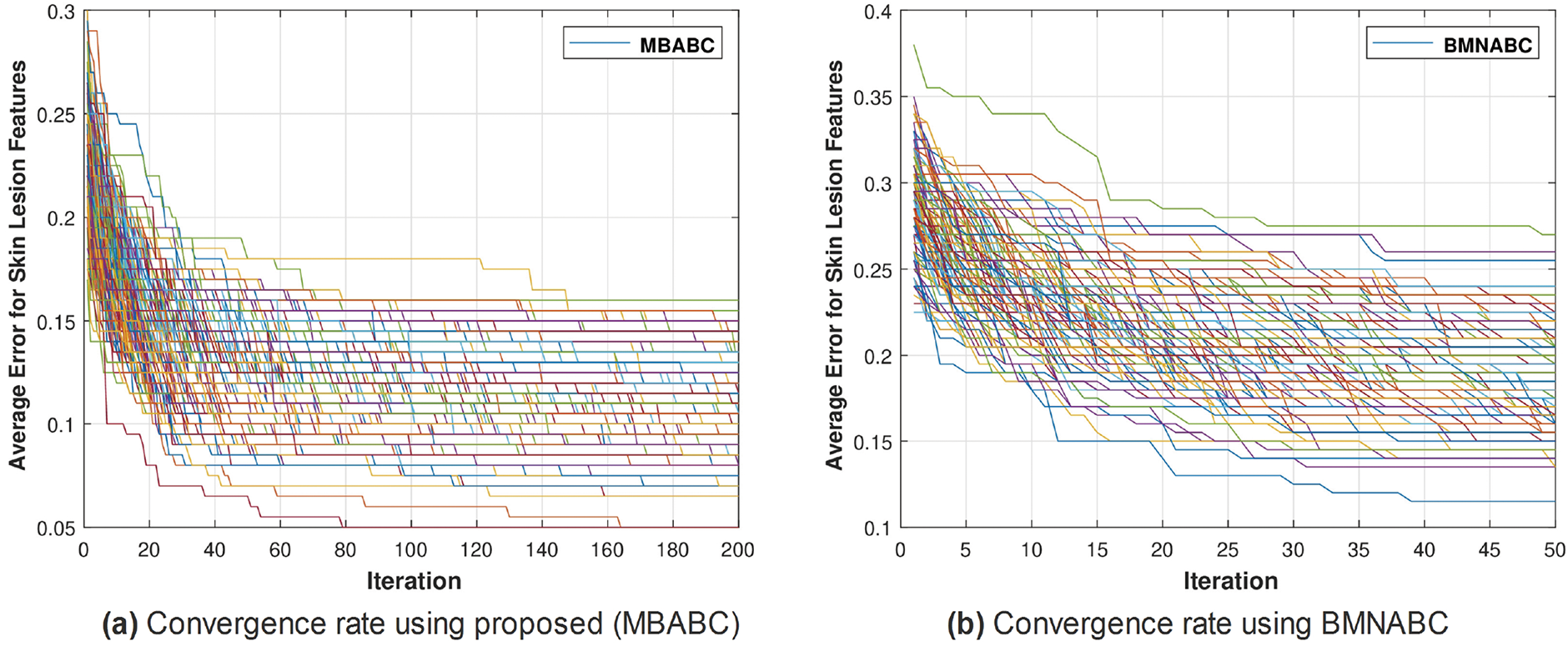
Figure 4: Comparison of MBABC vs. BMNABC convergence on ISIC-2016. Plot (a) illustrates faster convergence rate to a lower average error using proposed method, while plot (b) depicts the slower convergence rate of BMNABC
One could observe two main events: 1) a sudden drop in average error; and 2) a minimum average error. It is clear from the comparison that the proposed method has shown a sudden drop in average error compared to the existing method, as well as a greater number of times the algorithm touches the lowest value of less than 0.1. On the contrary, the existing method has not shown a sharp trend or a minimum average error. Similar trends have been observed in Fig. 5, where with the proposed feature selection algorithm, the classification framework performs better compared to the existing one. Fig. 5a has an accuracy range of (84%–96%) compared to (72–90) for the existing Fig. 5b.
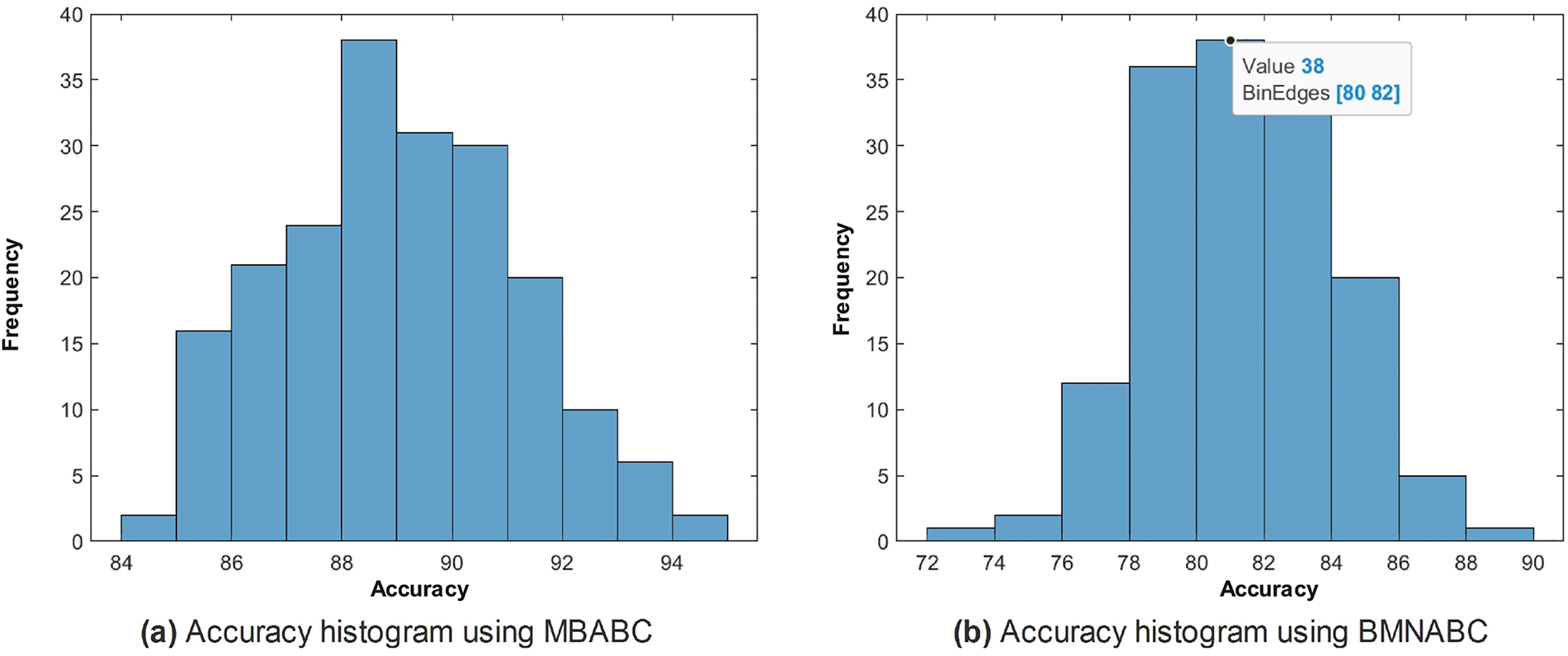
Figure 5: Comparison of accuracy histogram (%) on ISIC-2016 after Monte-Carlo simulations
Utilizing a combination of parameters to obtain a more comprehensive understanding of the model’s performance is often recommended. As a result, we opted for the collection of performance parameters mentioned earlier. The majority of these parameters exhibit interdependence and collectively exert an impact on the average accuracy of the model. Accuracy is a measure that tests the overall correctness of a system or model. Sensitivity, on the other hand, quantifies the ability of the system or model to correctly identify real positives. Specificity, in contrast, evaluates the system’s or model’s ability to avoid false positives. Precision is a metric that specifically reviews the accuracy of positive predictions made by a system or model. Lastly, NPV is a measure that assesses the accuracy of negative predictions made by the system or model. FDR is used to assess the fraction of inaccurate positive predictions, while FNR quantifies the rate of false negatives. The F1-Score is a metric that strikes a compromise between precision and sensitivity, giving preference to a model that exhibits both high precision and recall. In conclusion, MCC provides a comprehensive evaluation of the model’s performance by taking into account both true and erroneous positives and negatives. A thorough examination of these metrics is crucial to improve a classification model for particular applications since they jointly provide insights into its accuracy and efficacy.
In the case of the proposed, the frequency is maximum for an accuracy of (88%–90%), whereas in the case of the existing, the frequency is maximum within the range of (78%–84%). This is clear evidence of the performance of the proposed feature selection method.
Similar trends have been observed in the reduction plot in Fig. 6, where, after applying Monte-Carlo simulations, the histogram is plotted. It could be observed that in Fig. 6a, with the proposed reduction method, maximum features are reduced within the range of (70%–75%) percent, whereas on the other hand with the existing method, mostly the features are reduced within the range of (60%–75%). For a fair comparison, a confidence interval is plotted in terms of a box plot, where it could be seen that the proposed framework outperforms the existing method with a greater margin, whereas in terms of a reduction percentage, the difference is somewhat comparable, see Fig. 7a,b. For better understanding and to make well-informed decisions, the confusion matrix and receiver-operating characteristic curve (ROC) plots have been included in the analysis. These visualizations, depicted in Fig. 8, help to distinguish between positive and negative examples. The ISIC-2016 dataset presents TPR and FNR values that are inversely related: when TPR increases, FNR drops, and vice versa. The elevated true positive rate (TPR) values demonstrate the effectiveness of the proposed approach in accurately identifying positive events. Similarly, the low FNR value plainly implies that the model is successfully minimizing the amount of false negatives, which reveals that the model is good at finding positive cases. Similarly, the Area under the curve-Receiver-operating characteristic curve (AUC-ROC) score demonstrates superior discriminatory capability, as observed in our ISIC-2016 case.
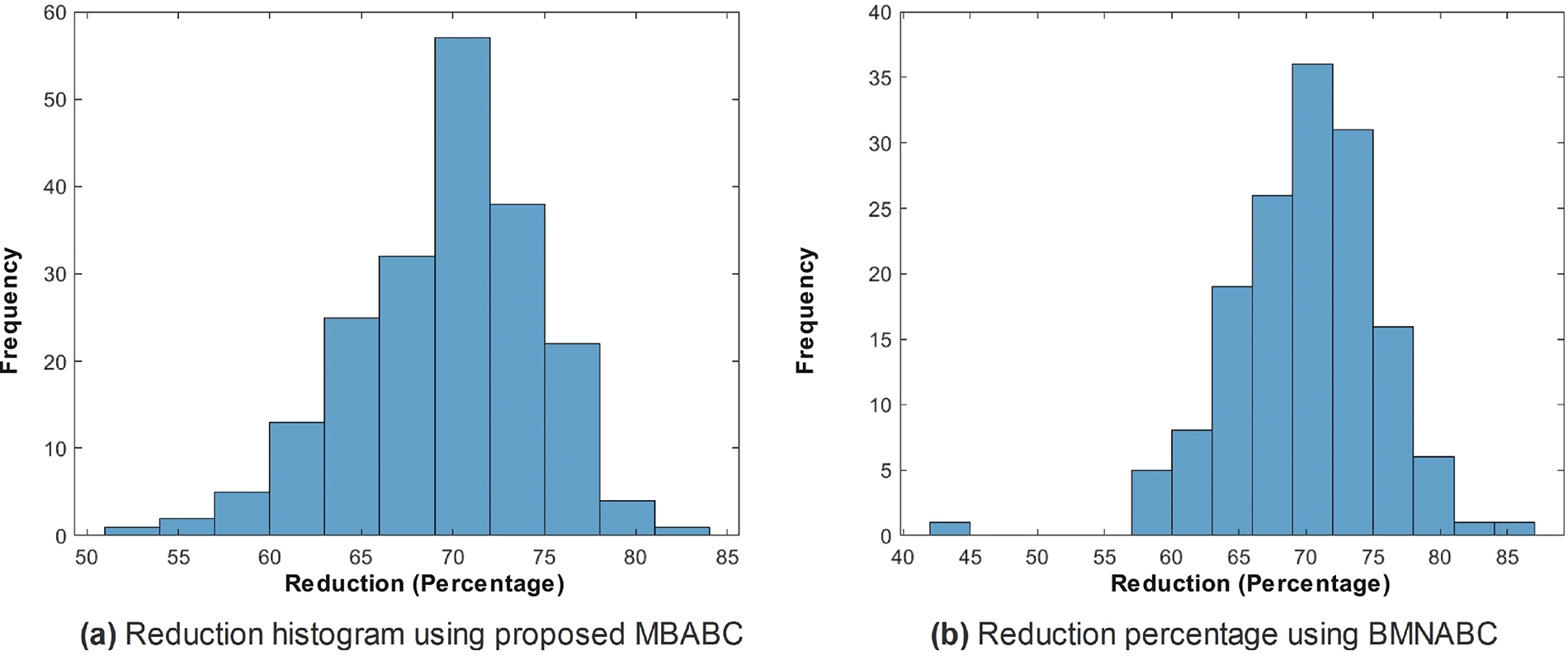
Figure 6: Comparison of feature reduction percentage histograms using proposed MBABC (a) vs. BMNABC (b) on ISIC-2016 dataset
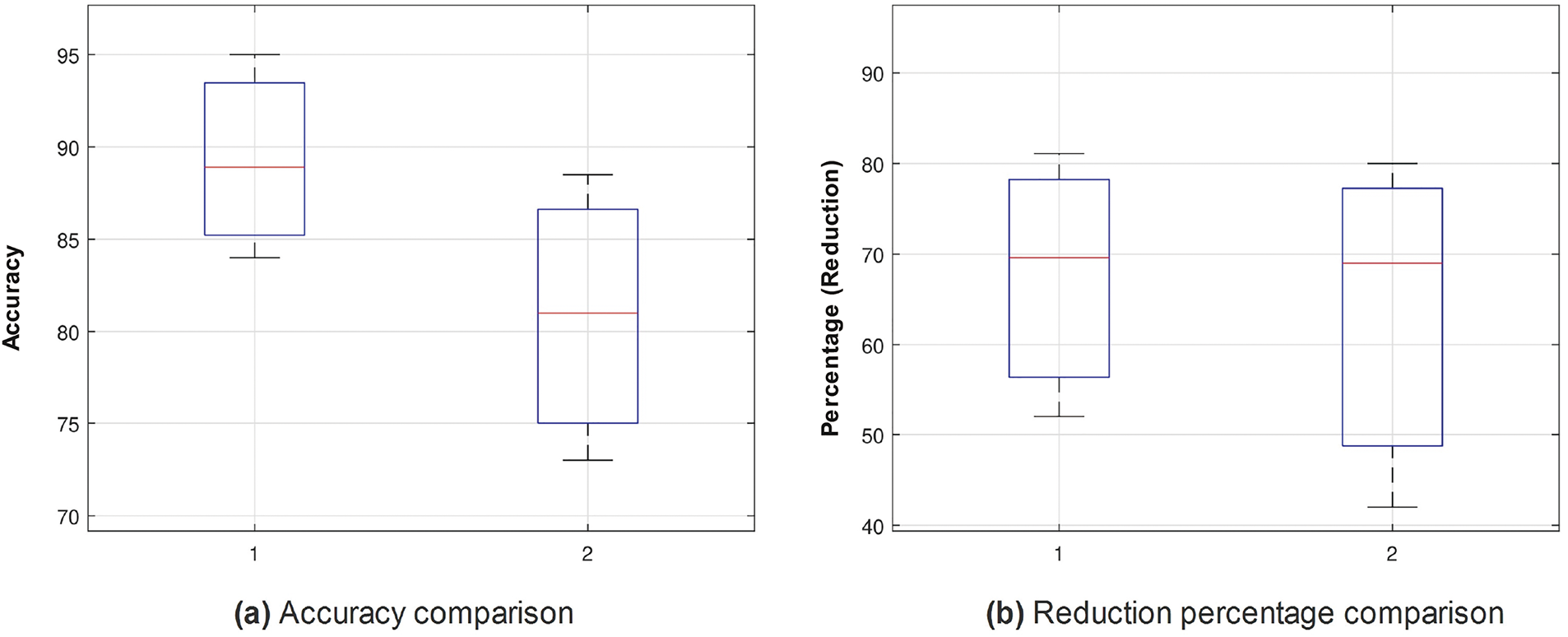
Figure 7: Box plots for accuracy (%) (a) and feature reduction percentage (b): Comparing MBABC (1) vs. BMNABC (2) on ISIC-2016 dataset
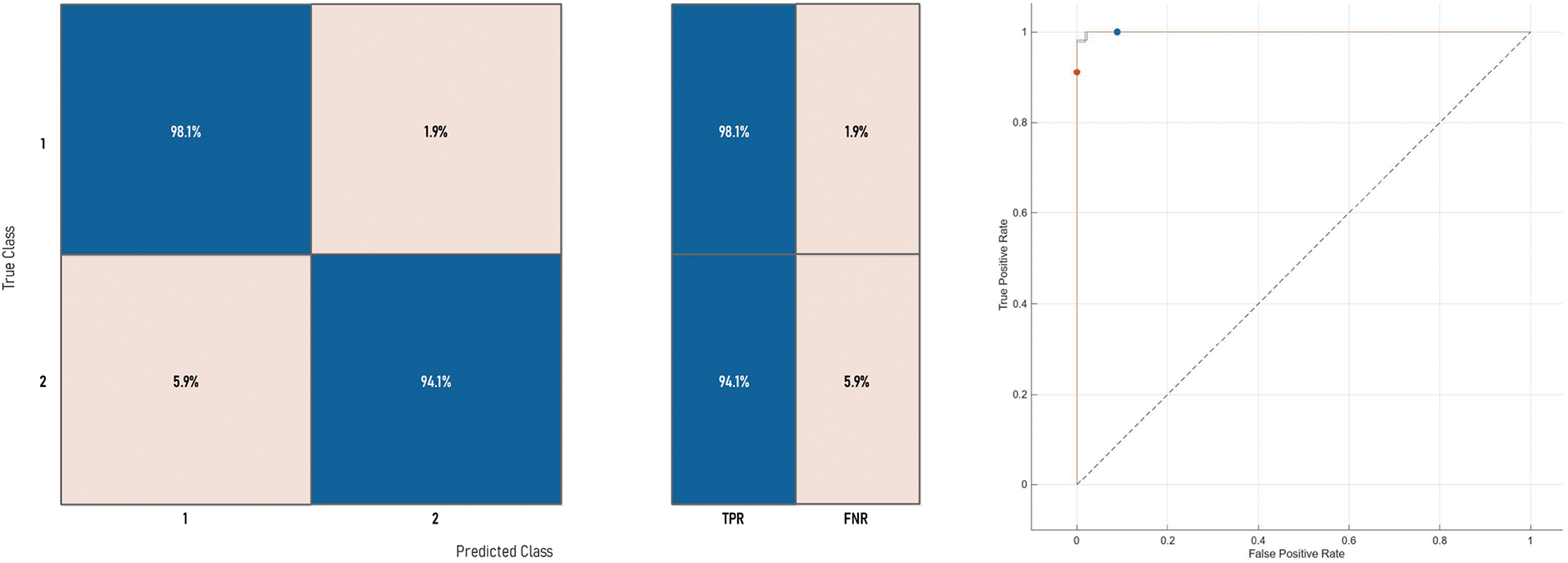
Figure 8: Confusion matrix and ROC curve of Medium-NN on ISIC-2016 dataset
In the case of
Fig. 9 demonstrates the convergence of the proposed framework to that of the existing one. The convergence rate of the proposed framework clearly shows a sharp drop as well as a low average error in most of the cases. On the contrary, the existing method shows a few cases of zero average error. Considering the accuracy histogram, Fig. 10a,b, for the
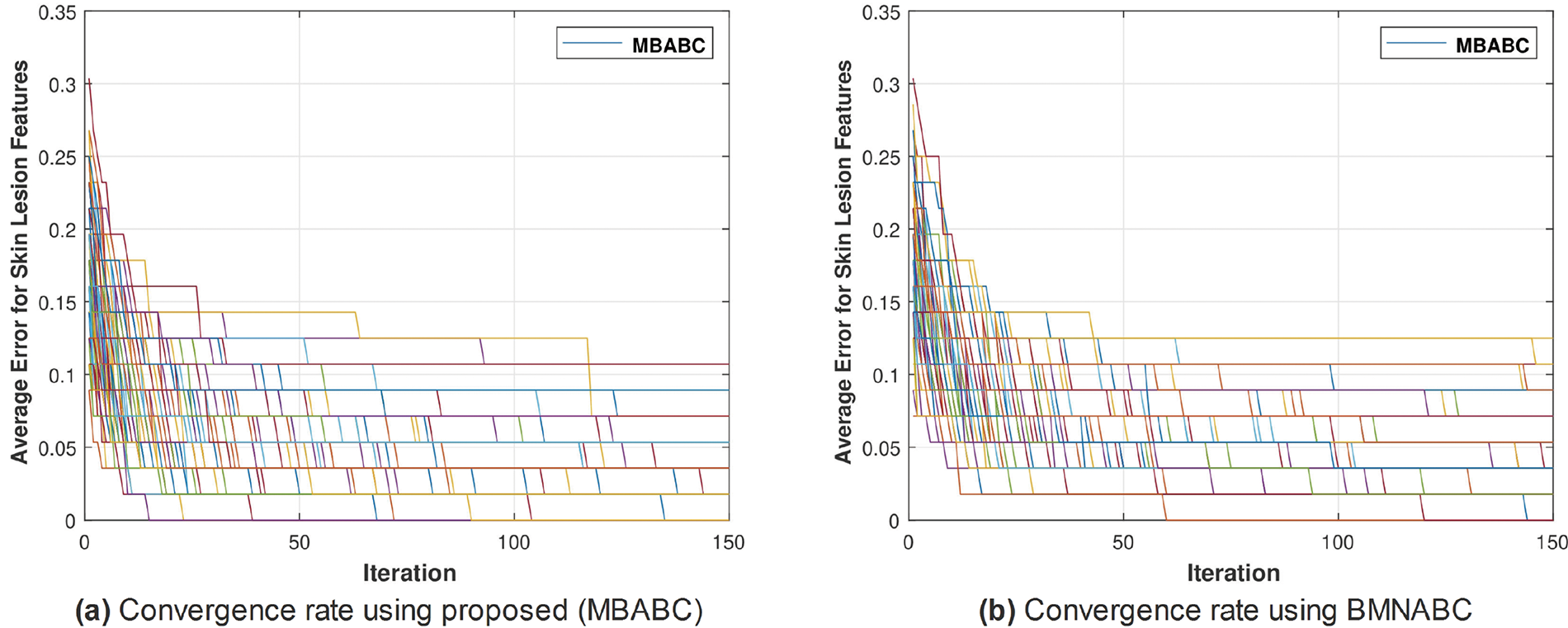
Figure 9: Comparison of MBABC vs. BMNABC convergence on
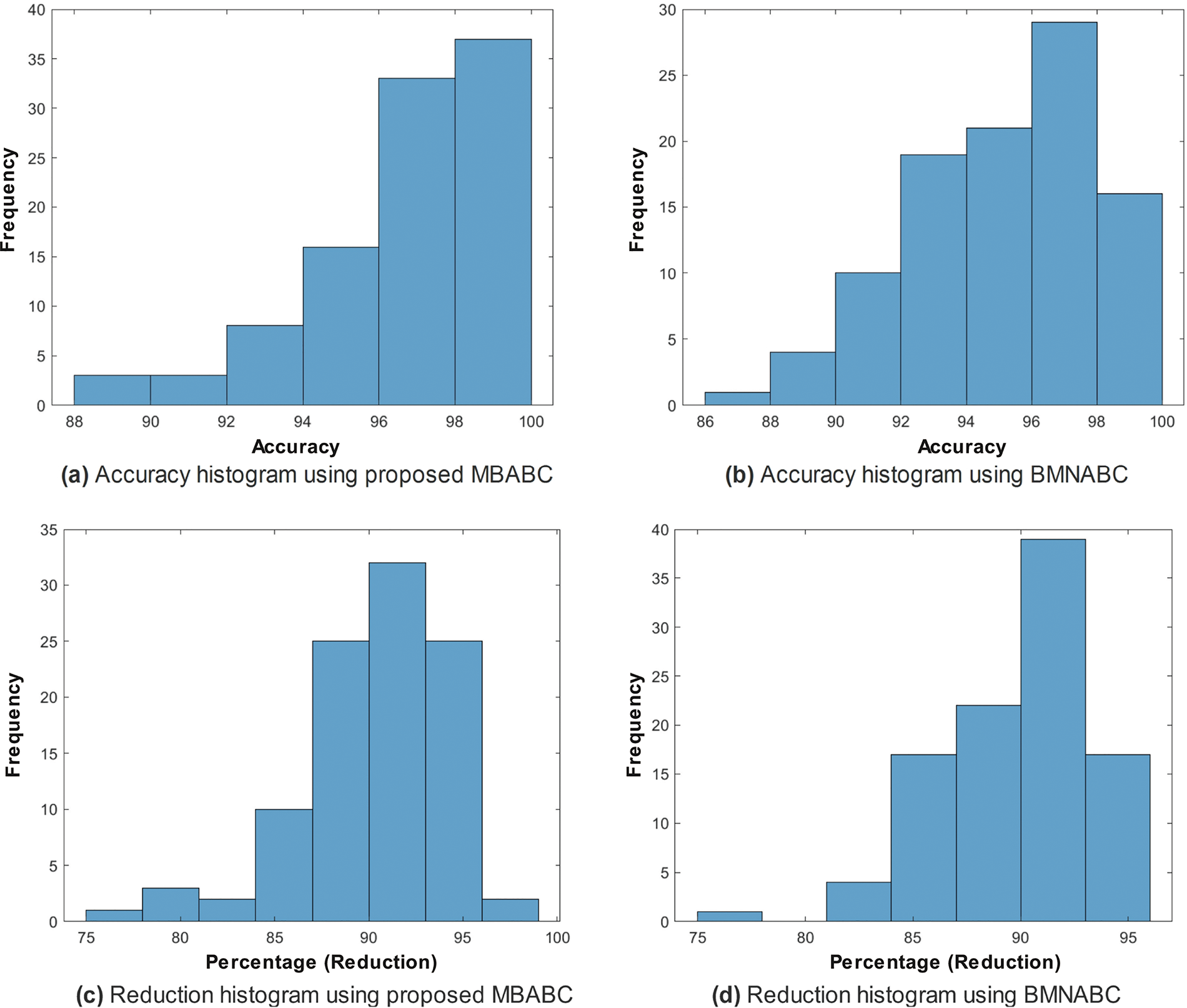
Figure 10: Top: comparison of accuracy (%) histograms on
Similarly, Fig. 10c demonstrates a maximum reduction trend, as it reaches the limit of 98%. This shows, that even with only
The box plot in Fig. 11 shows the confidence interval for both accuracy and reduction percentages. From both plots, one could easily observe the difference in accuracy as well as the reduction percentage.
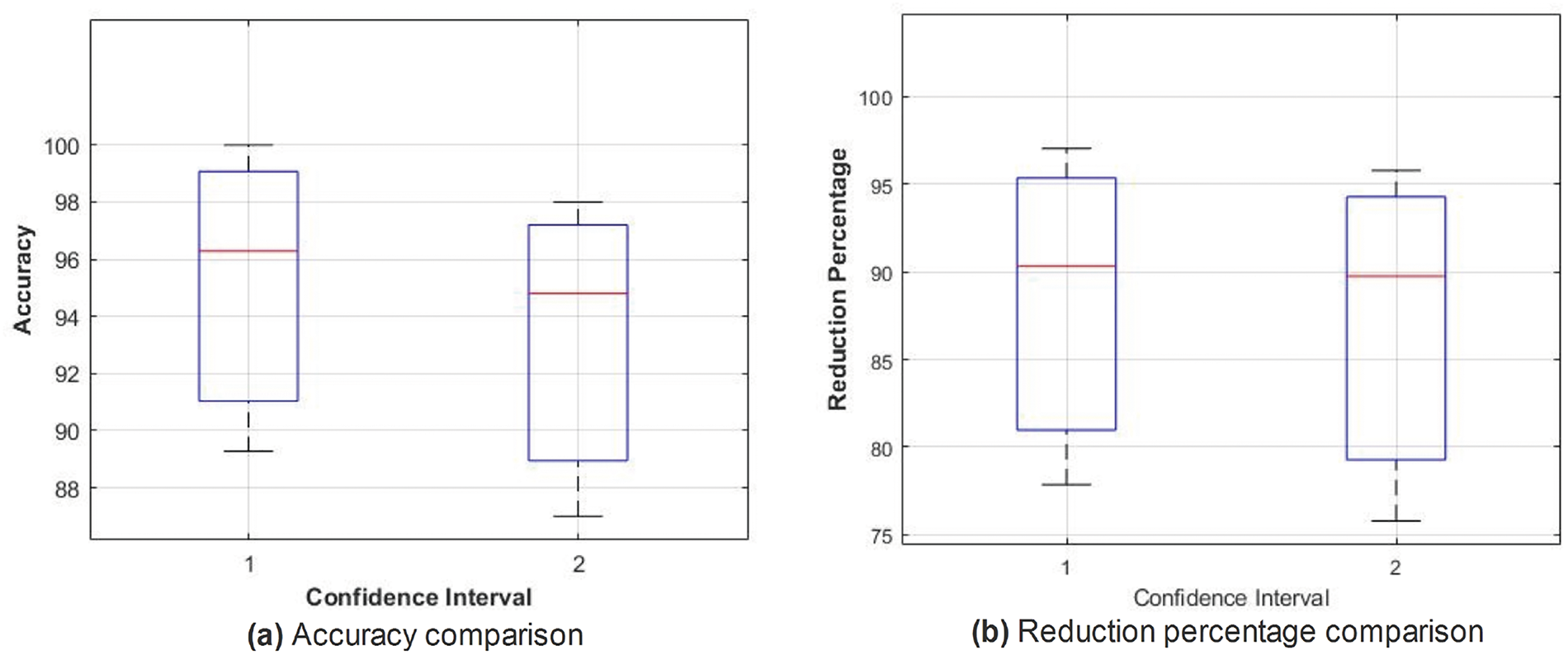
Figure 11: Box plots for accuracy (%) (a) and feature reduction percentage (b): Comparing MBABC (1) vs. BMNABC (2) on
Similarly, as in ISIC-2016, the confusion matrix and AUC-ROC for the
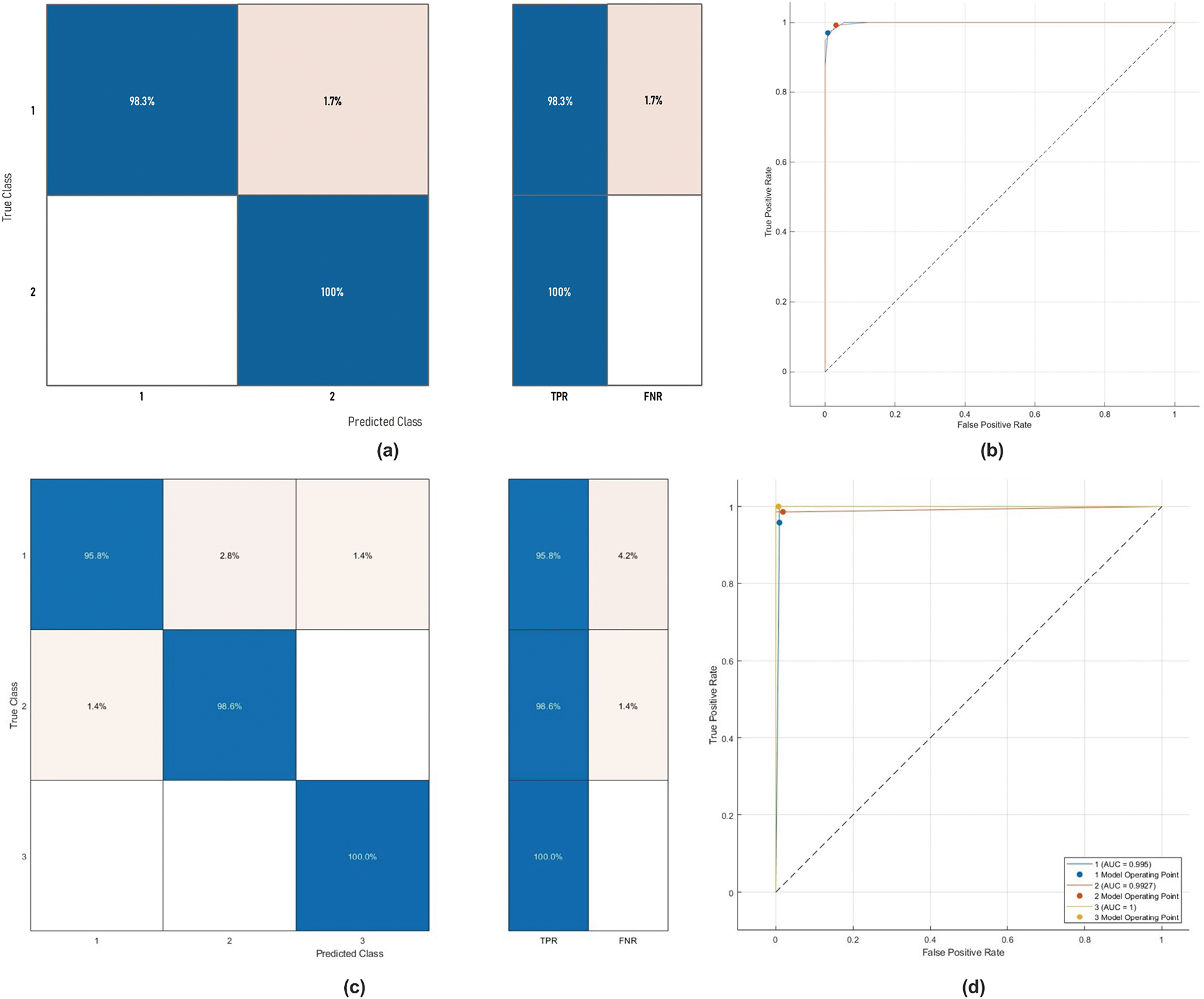
Figure 12: The ROC curve and confusion matrix of
In the case of HAM10000, after applying Monte-Carlo simulations, we generated convergence and accuracy plots. The proposed feature selection strategy exhibits distinct behavior when applied to the HAM10000 dataset compared to the ISIC-2016 and
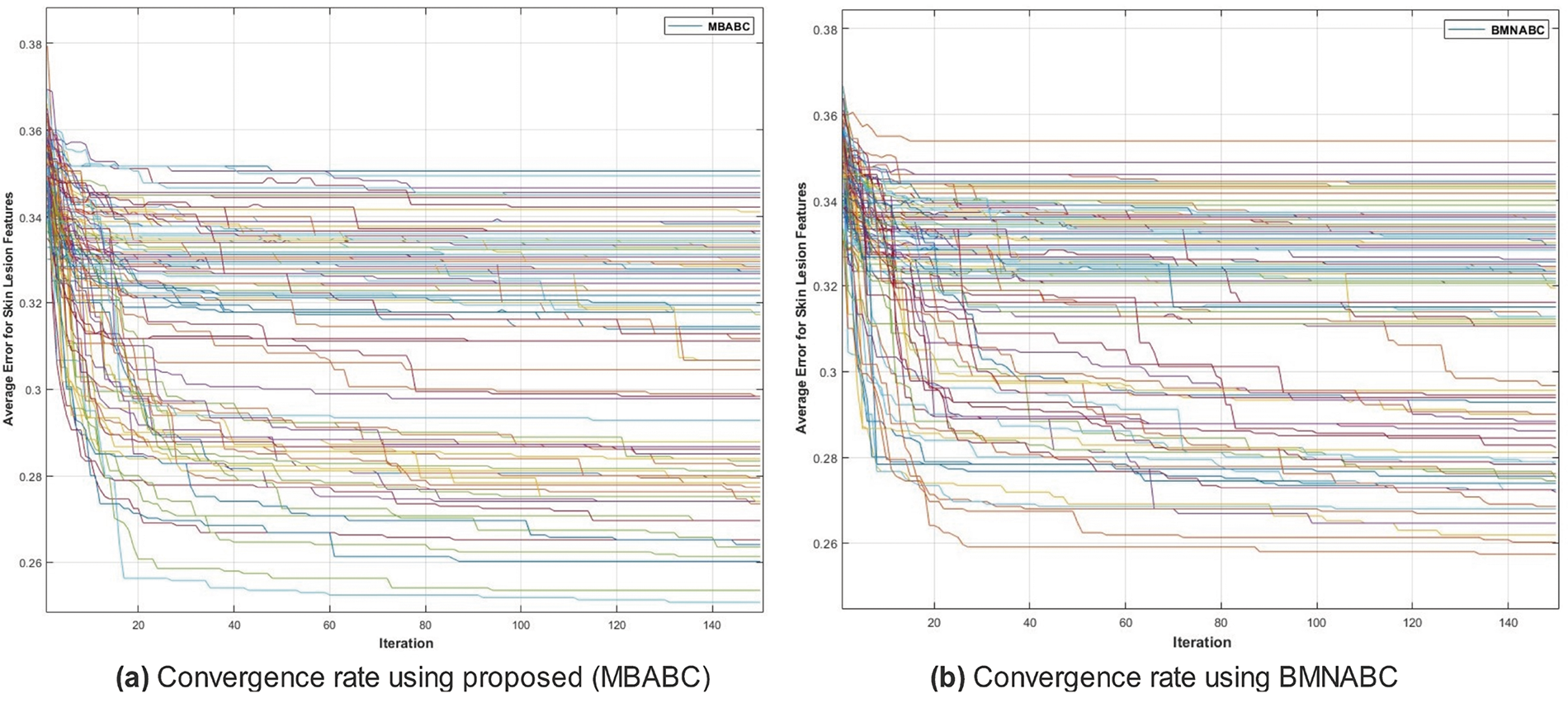
Figure 13: Comparison of MBABC vs. BMNABC convergence on HAM10000 dataset. Plot (a) illustrates faster convergence rate to a lower average error using the proposed method, while plot (b) depicts the slower convergence rate of BMNABC
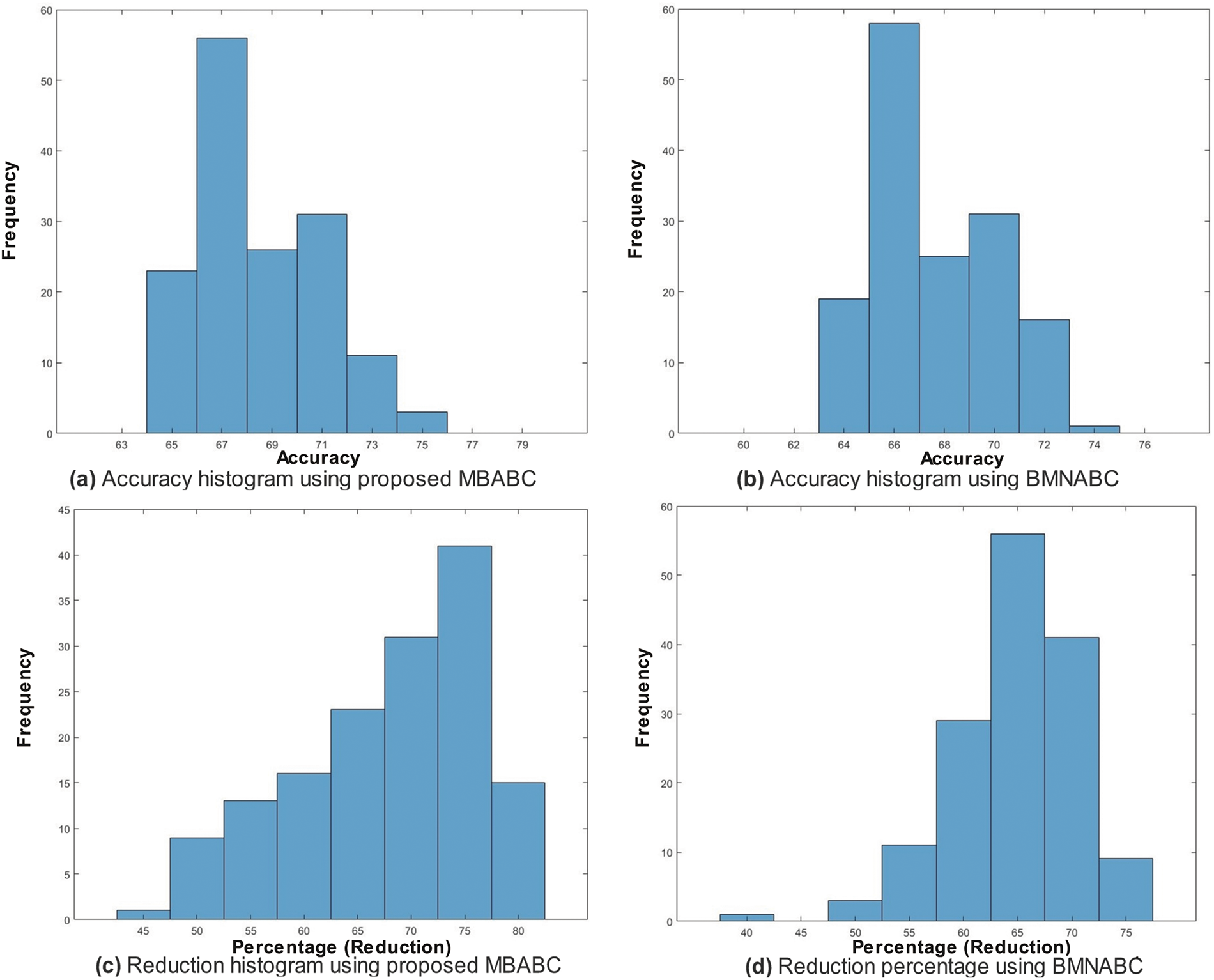
Figure 14: Top: comparison of accuracy (%) histograms on HAM10000 dataset using the proposed BMABC (a) and BMNABC algorithms (b); Bottom: comparison of feature reduction percentage histograms using the proposed MBABC (c) vs. BMNABC algorithms (d) on HAM10000 dataset
Furthermore, Fig. 15a presents the confusion matrix for HAM10000. The high TPR values of
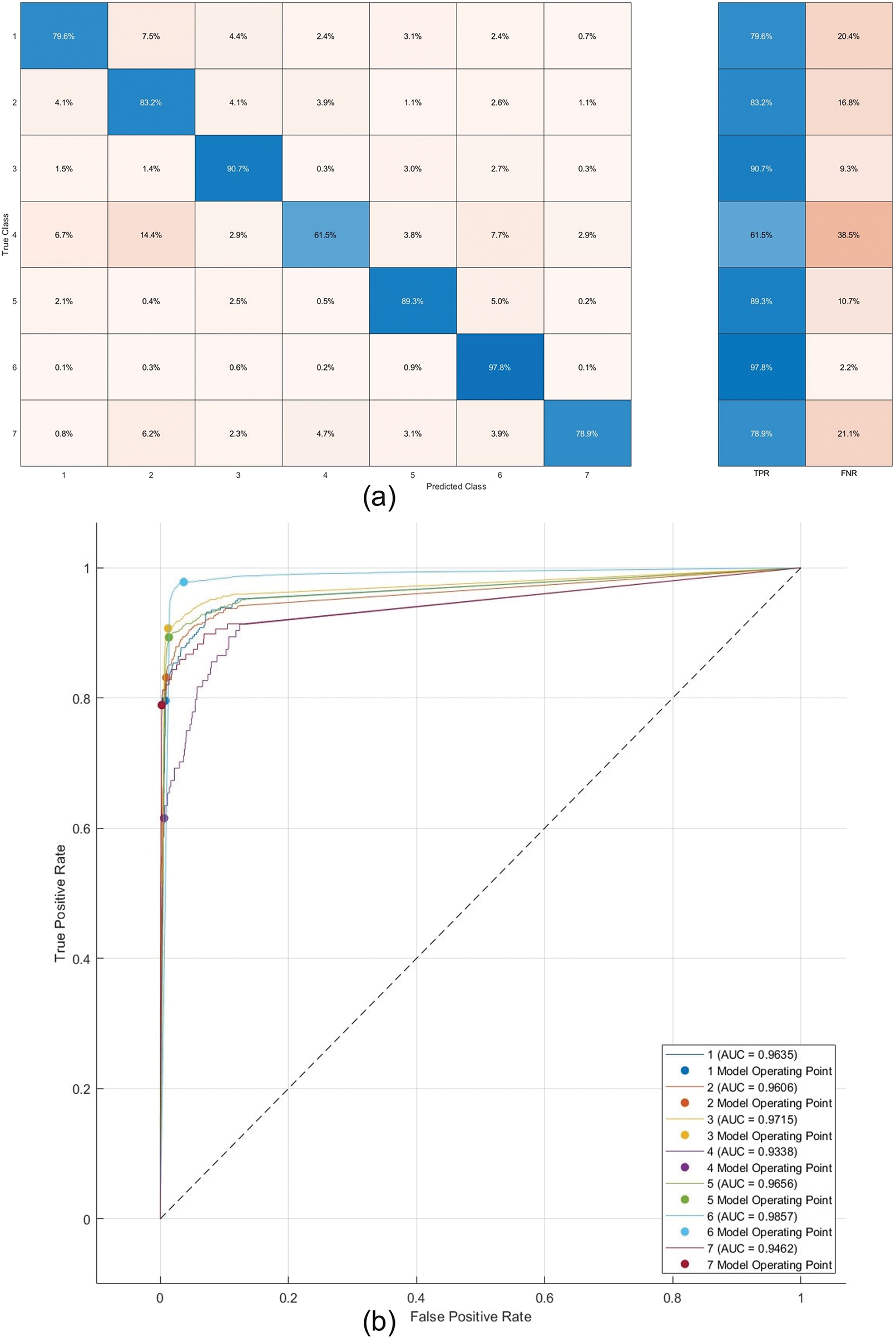
Figure 15: Confusion matrix and ROC of Trilayered-NN on HAM10000 dataset
Table 4 shows the list of classifiers in a given order from highest accuracy to the lowest. Similarly, other parameters also show the variations as per the generated confusion matrix. It is quite clear that for both datasets, different classifiers have achieved maximum and minimum accuracies. In the case of the
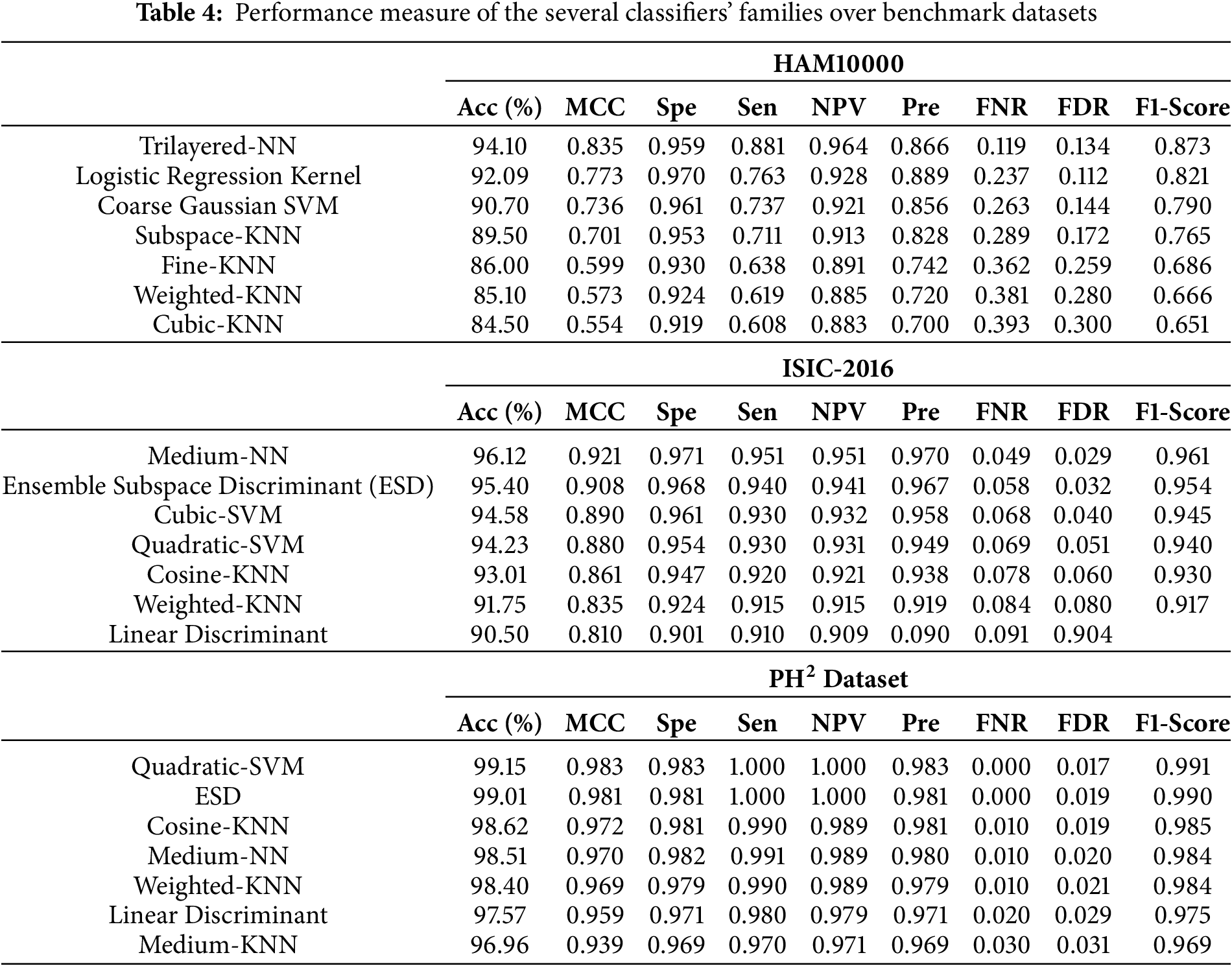
In this table, seven selected classifiers with improved classification accuracy are provided. The minimum accuracy achieved is
Regarding ISIC-2016, both the SVM and KNN families have exhibited excellent performance in comparison to other families, such as linear discriminant and ensemble. In the concluding instance of HAM10000, the ensemble of neural networks has exhibited excellent performance in comparison to alternative groups of classifiers. The results clearly demonstrate that the neural network family possesses the capability to achieve outstanding performance across several classes.
To ensure a fair evaluation of feature selection strategies in optimizing classification accuracy for this application, a comparison table is also included (Table 5). In contrast to established feature selection methods such as genetic algorithm (GA), particle swarm optimization (PSO), grey wolf optimization (GW), and BMNABC, the proposed method demonstrated remarkably high performance, as evidenced by its classification accuracy. As compared to alternative methodologies, GA achieves a higher average classification accuracy across all datasets. Undoubtedly, the genetic algorithm stands out as the optimal solution for feature selection in this specific application, surpassing other alternatives. Additionally, F1-score has been incorporated alongside accuracy metrics for skin lesion classification. The rationale for adding the F1-score is twofold: 1) HAM10000 skin lesion dataset exhibits significant class imbalance, where accuracy alone can mask poor performance on minority classes. The F1-score, which balances precision and recall, provides a more robust evaluation of model effectiveness in such scenarios; and 2) by reporting both metrics, we demonstrate that MBABC not only achieves higher accuracy but also maintains superior F1-scores compared to PSO and GWO. This emphasizes its ability to optimize feature selection for both majority and minority classes, a critical requirement in medical diagnostics. Table 6 presents a comparison of classification accuracy between a baseline method (without feature selection), and a proposed method for all selected datasets. The results clearly demonstrate a consistent improvement in accuracy when the proposed feature selection method is applied. The baseline model attained an accuracy of 96.12% on the Ph2 dataset with quadratic SVM classifiers, whereas the proposed method achieved an accuracy of 99.15%. Moreover, employing ISIC-2016, the achieved accuracy is 92.68% with a medium-NN classifier, in contrast to 96.64% with the proposed method. Ultimately, the baseline model attained an accuracy of 87.41% on the HAM10000 dataset with a quadratic SVM, whereas the proposed technique achieved an accuracy of 94.10%. These findings underscore the effectiveness of the proposed feature selection technique in improving model’s performance across diverse datasets and classifiers, highlighting its potential to optimize classification tasks in medical imaging and related domains.
Melanoma is the form of skin cancer that has the highest mortality rate, and its incidence has been rapidly rising over the past several years. The main objective of this study is to prioritize the identification and selection of feature information that exhibits the highest level of discrimination, ultimately resulting in improved accuracy in classification. Three benchmark datasets, namely
In subsequent research, the number of image samples in specific categories will be augmented through the utilization of either data augmentation techniques or generative adversarial networks. Additionally, exploring the use of additional pre-trained models and developing a customized CNN model could be a promising avenue to pursue. When using a feature selection method, the effectiveness of the proposed algorithm relies heavily on the fitness function. Therefore, it is important to build a suitable fitness function that can efficiently handle a large feature set with more number of classes.
Acknowledgement: The authors extend their appreciation to Prince Sattam bin Abdulaziz University for funding this research work.
Funding Statement: The authors extend their appreciation to Prince Sattam bin Abdulaziz University for funding this research work through the project number (PSAU/2024/03/31540).
Author Contributions: Tallha Akram: Conceptualization, methodology, original draft, simulation, funding; Fahdah Almarshad: Methodology, review & editing; Anas Alsuhaibani: Methodology, original draft, review & editing; Syed Rameez Naqvi: Conceptualization, methodology, original draft, review & editing. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data used in this study were obtained from Kaggle.com. The dataset is publicly available and open to research community.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present research.
References
1. Malik S, Akram T, Awais M, Khan MA, Hadjouni M, Elmannai H, et al. An improved skin lesion boundary estimation for enhanced-intensity images using hybrid metaheuristics. Diagnostics. 2023;13(7):1285. doi:10.3390/diagnostics13071285. [Google Scholar] [PubMed] [CrossRef]
2. Peate I. The skin: largest organ of the body. British J Healthcare Assist. 2021;15(9):446–51. doi:10.12968/bjha.2021.15.9.446. [Google Scholar] [CrossRef]
3. Calabrò F, Sternberg C. Cancer and its management. BJU Int. 2006;97(3):651–1. doi:10.1111/j.1464-410X.2006.06115_2.x. [Google Scholar] [CrossRef]
4. Bassel A, Abdulkareem AB, Alyasseri ZAA, Sani NS, Mohammed HJ. Automatic malignant and benign skin cancer classification using a hybrid deep learning approach. Diagnostics. 2022;12(10):2472. doi:10.3390/diagnostics12102472. [Google Scholar] [PubMed] [CrossRef]
5. Foundation SC. Nonmelanoma Skin Cancer Statistics. Skin Cancer Facts, Estimated cases of basal cell carcinoma and squamous cell carcinoma in the United States; 2025. [cited 2025 Apr 7]. Available from: https://www.skincancer.org/skin-cancer-information/skin-cancer-facts/. [Google Scholar]
6. Society AC. Melanoma Statistics 2025. Cancer Facts & Figures. 2025. Estimated new cases and deaths for melanoma in the United States. [cited 2025 Apr 7]. Available from: https://www.cancer.org/cancer/types/melanoma-skin-cancer/about/key-statistics.html. [Google Scholar]
7. World Cancer Research Fund. Skin Cancer Statistics; 2025. [cited 2025 Mar 6]. Available from: https://www.wcrf.org/preventing-cancer/cancer-statistics/skin-cancer-statistics/. [Google Scholar]
8. Khayyati Kohnehshahri M, Sarkesh A, Mohamed Khosroshahi L, HajiEsmailPoor Z, Aghebati-Maleki A, Yousefi M, et al. Current status of skin cancers with a focus on immunology and immunotherapy. Cancer Cell Int. 2023;23(1):174. doi:10.1186/s12935-023-03012-7. [Google Scholar] [PubMed] [CrossRef]
9. Bhatt H, Shah V, Shah K, Shah R, Shah M. State-of-the-art machine learning techniques for melanoma skin cancer detection and classification: a comprehensive review. Intell Medi. 2023;03(3):180–90. doi:10.1016/j.imed.2022.08.004. [Google Scholar] [CrossRef]
10. Atta A, Khan MA, Asif M, Issa GF, Said RA, Faiz T. Classification of Skin Cancer empowered with convolutional neural network. In: 2022 International Conference on Cyber Resilience (ICCR). Dubai, United Arab Emirates; 2022. p. 1–6. [Google Scholar]
11. Ali K, Shaikh ZA, Khan AA, Laghari AA. Multiclass skin cancer classification using EfficientNets—a first step towards preventing skin cancer. Neurosci Inform. 2022;2(4):100034. doi:10.1016/j.neuri.2021.100034. [Google Scholar] [CrossRef]
12. Bechelli S, Delhommelle J. Machine learning and deep learning algorithms for skin cancer classification from dermoscopic images. Bioengineering. 2022;9(3):97. doi:10.3390/bioengineering9030097. [Google Scholar] [PubMed] [CrossRef]
13. Malik S, Akram T, Ashraf I, Rafiullah M, Ullah M, Tanveer J. A hybrid preprocessor DE-ABC for efficient skin-lesion segmentation with improved contrast. Diagnostics. 2022;12(11):2625. doi:10.3390/diagnostics12112625. [Google Scholar] [PubMed] [CrossRef]
14. Ribani R, Marengoni M. A survey of transfer learning for convolutional neural networks. In: 2019 32nd SIBGRAPI Conference on Graphics, Patterns and Images Tutorials (SIBGRAPI-T). Rio de Janeiro, Brazil: IEEE; 2019. p. 47–57. [Google Scholar]
15. Devi GM, Neelambary V. Computer-aided diagnosis of white blood cell leukemia using VGG16 convolution neural network. In: 2022 4th International Conference on Inventive Research in Computing Applications (ICIRCA). Tamil Nadu, India: IEEE; 2022. p. 1064–8. [Google Scholar]
16. Bhatt B, Iyer A, Writer D, Schau G. A comparative study of transfer learning networks and siamese networks for acute lymphoblastic leukemia (ALL) diagnosis. J Student Res. 2022;11(4):1. doi:10.47611/jsrhs.v11i4.3489. [Google Scholar] [CrossRef]
17. Ain QU, Akbar S, Hassan SA, Naaqvi Z. Diagnosis of leukemia disease through deep learning using microscopic images. In: 2022 2nd International Conference on Digital Futures and Transformative Technologies (ICoDT2). Rawalpindi, Pakistan: IEEE; 2022. p. 1–6. [Google Scholar]
18. Ahmad R, Awais M, Kausar N, Akram T. White blood cells classification using entropy-controlled deep features optimization. Diagnostics. 2023;13(3):352. doi:10.3390/diagnostics13030352. [Google Scholar] [PubMed] [CrossRef]
19. Chen Q, Li M, Chen C, Zhou P, Lv X, Chen C. MDFNet: application of multimodal fusion method based on skin image and clinical data to skin cancer classification. J Cancer Res Clinic Oncol. 2023;149(7):3287–99. doi:10.1007/s00432-022-04180-1. [Google Scholar] [PubMed] [CrossRef]
20. Hosny KM, Kassem MA. Refined residual deep convolutional network for skin lesion classification. J Digit Imaging. 2022;35(2):258–80. doi:10.1007/s10278-021-00552-0. [Google Scholar] [PubMed] [CrossRef]
21. Tembhurne JV, Hebbar N, Patil HY, Diwan T. Skin cancer detection using ensemble of machine learning and deep learning techniques. Multimed Tools Appl. 2023;82(18):27501–24. doi:10.1007/s11042-023-14697-3. [Google Scholar] [CrossRef]
22. Padhy S, Dash S, Kumar N, Singh SP, Kumar G, Moral P. Temporal integration of ResNet features with LSTM for enhanced skin lesion classification. Res Eng. 2025;25(1):104201. doi:10.1016/j.rineng.2025.104201. [Google Scholar] [CrossRef]
23. Srinivasu PN, SivaSai JG, Ijaz MF, Bhoi AK, Kim W, Kang JJ. Classification of skin disease using deep learning neural networks with MobileNet V2 and LSTM. Sensors. 2021;21(8):2852. doi:10.3390/s21082852. [Google Scholar] [PubMed] [CrossRef]
24. Zhong L, Li T, Cui M, Cui S, Wang H, Yu L. DSU-Net: dual-stage U-Net based on CNN and transformer for skin lesion segmentation. Biomed Signal Process Cont. 2025;100(1):107090. doi:10.1016/j.bspc.2024.107090. [Google Scholar] [CrossRef]
25. Tan L, Wu H, Zhu J, Liang Y, Xia J. Clinical-inspired skin lesions recognition based on deep hair removal with multi-level feature fusion. Pattern Recognit. 2025;161(1):111325. doi:10.1016/j.patcog.2024.111325. [Google Scholar] [CrossRef]
26. Bibi S, Khan MA, Shah JH, Damaševičius R, Alasiry A, Marzougui M, et al. MSRNet: multiclass skin lesion recognition using additional residual block based fine-tuned deep models information fusion and best feature selection. Diagnostics. 2023;13(19):3063. doi:10.3390/diagnostics13193063. [Google Scholar] [PubMed] [CrossRef]
27. Tahir M, Naeem A, Malik H, Tanveer J, Naqvi RA, Lee SW. DSCC_Net: multi-classification deep learning models for diagnosing of skin cancer using dermoscopic images. Cancers. 2023;15(7):2179. doi:10.3390/cancers15072179. [Google Scholar] [PubMed] [CrossRef]
28. Gilani SQ, Syed T, Umair M, Marques O. Skin cancer classification using deep spiking neural network. J Digit Imaging. 2023;36(3):1137–47. doi:10.1007/s10278-023-00776-2. [Google Scholar] [PubMed] [CrossRef]
29. Kibriya H, Abdullah I, Kousar F. Melanoma lesion segmentation and classification using SegNet; 2023:1–6. [Google Scholar]
30. Khan MA, Sharif MI, Raza M, Anjum A, Saba T, Shad SA. Skin lesion segmentation and classification: a unified framework of deep neural network features fusion and selection. Expert Syst. 2022;39(7):e12497. doi:10.1111/exsy.12497. [Google Scholar] [CrossRef]
31. Kaur R, GholamHosseini H, Sinha R, Lindén M. Melanoma classification using a novel deep convolutional neural network with dermoscopic images. Sensors. 2022;22(3):1134. doi:10.3390/s22031134. [Google Scholar] [PubMed] [CrossRef]
32. Saarela M, Geogieva L. Robustness, stability, and fidelity of explanations for a deep skin cancer classification model. Appl Sci. 2022;12(19):9545. doi:10.3390/app12199545. [Google Scholar] [CrossRef]
33. Khan MA, Sharif M, Akram T, Bukhari SAC, Nayak RS. Developed Newton-Raphson based deep features selection framework for skin lesion recognition. Pattern Recognit Letters. 2020;129(4/5):293–303. doi:10.1016/j.patrec.2019.11.034. [Google Scholar] [CrossRef]
34. Mahbod A, Schaefer G, Wang C, Ecker R, Ellinge I. Skin lesion classification using hybrid deep neural networks. In: ICASSP, 2019-2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); Brighton, UK; 2019. p. 1229–33. [Google Scholar]
35. Akram T, Junejo R, Alsuhaibani A, Rafiullah M, Akram A, Almujally NA. Precision in dermatology: developing an optimal feature selection framework for skin lesion classification. Diagnostics. 2023;13(17):2848. doi:10.3390/diagnostics13172848. [Google Scholar] [PubMed] [CrossRef]
36. Akram T, Alsuhaibani A, Khan MA, Khan SU, Naqvi SR, Bilal M. Dermo-optimizer: skin lesion classification using information-theoretic deep feature fusion and entropy-controlled binary bat optimization. Int J Imaging Syst Technol. 2024;34(5):e23172. doi:10.1002/ima.23172. [Google Scholar] [CrossRef]
37. Vanacore A, Pellegrino MS, Ciardiello A. Fair evaluation of classifier predictive performance based on binary confusion matrix. Computat Statist. 2024;39(1):363–83. doi:10.1007/s00180-022-01301-9. [Google Scholar] [CrossRef]
38. Torrey L, Shavlik J. Transfer learning. Vol. 2, In: Handbook of research on machine learning applications and trends: algorithms, methods, and techniques. IGI Global. 2010. p. 242–64. [Google Scholar]
39. Karaboga D. Artificial bee colony algorithm. Scholarpedia. 2010;5(3):6915. [Google Scholar]
40. Beheshti Z. BMNABC: binary multi-neighborhood artificial bee colony for high-dimensional discrete optimization problems. Cybernet Syst. 2018;49(7–8):452–74. doi:10.1080/01969722.2018.1541597. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF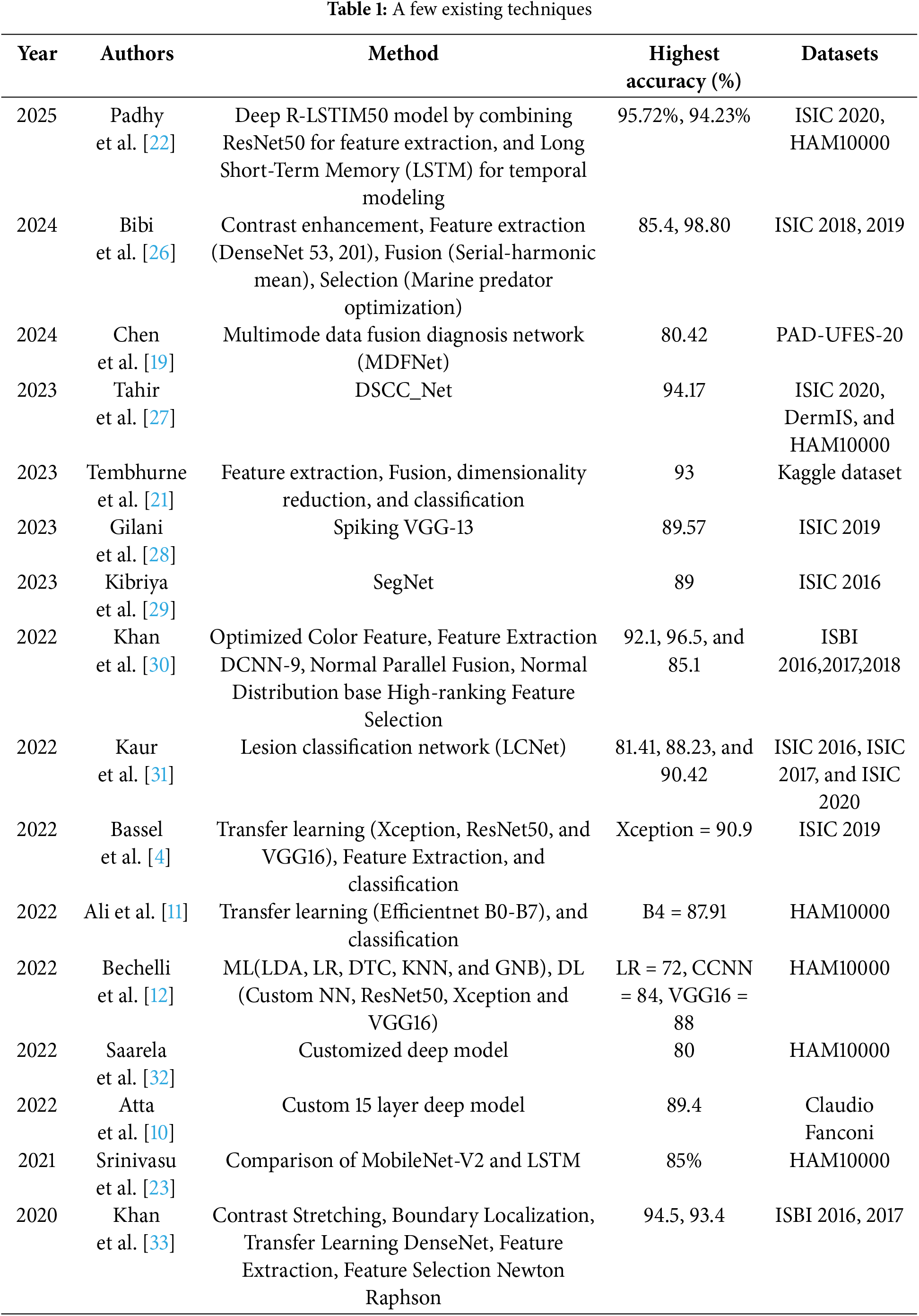
 Downloads
Downloads
 Citation Tools
Citation Tools