 Open Access
Open Access
ARTICLE
A Neural ODE-Enhanced Deep Learning Framework for Accurate and Real-Time Epilepsy Detection
1 Department of Electrical Engineering, Faculty of Engineering at Rabigh, King Abdulaziz University, Jeddah, 21589, Saudi Arabia
2 King Salman Center for Disability Research, Riyadh, 11614, Saudi Arabia
3 Department of Electrical Engineering, College of Engineering, Northern Border University, Arar, 91431, Saudi Arabia
* Corresponding Author: Ahmed A. Alsheikhy. Email:
Computer Modeling in Engineering & Sciences 2025, 143(3), 3033-3064. https://doi.org/10.32604/cmes.2025.065264
Received 08 March 2025; Accepted 12 May 2025; Issue published 30 June 2025
Abstract
Epilepsy is a long-term neurological condition marked by recurrent seizures, which result from abnormal electrical activity in the brain that disrupts its normal functioning. Traditional methods for detecting epilepsy through machine learning typically utilize discrete-time models, which inadequately represent the continuous dynamics of electroencephalogram (EEG) signals. To overcome this limitation, we introduce an innovative approach that employs Neural Ordinary Differential Equations (NODEs) to model EEG signals as continuous-time systems. This allows for effective management of irregular sampling and intricate temporal patterns. In contrast to conventional techniques, such as Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs), which necessitate fixed-length inputs and often struggle with long-term dependencies, our framework incorporates: (1) a NODE block to capture continuous-time EEG dynamics, (2) a feature extraction module tailored for seizure-specific patterns, and (3) an attention-based fusion mechanism to enhance interpretability in classification. When evaluated on three publicly accessible EEG datasets, including those from Boston Children’s Hospital and the Massachusetts Institute of Technology (CHB-MIT) and the Temple University Hospital (TUH) EEG Corpus, the model demonstrated an average accuracy of 98.2%, a sensitivity of 97.8%, a specificity of 98.3%, and an F1-score of 97.9%. Additionally, the inference latency was reduced by approximately 30% compared to standard CNN and Long Short-Term Memory (LSTM) architectures, making it well-suited for real-time applications. The method’s resilience to noise and its adaptability to irregular sampling enhance its potential for clinical use in real-time settings.Keywords
The manifestations of these seizures can vary significantly, ranging from brief episodes of impaired awareness or minor muscle twitches to intense convulsions and complete loss of consciousness [1,2]. This disorder affects around 50 million individuals globally, making it one of the most prevalent neurological conditions [1]. While epilepsy can arise at any age, it is especially common among children and the elderly [1–3]. The causes of epilepsy are varied and may include genetic predispositions, brain injuries, infections, developmental disorders, or structural brain abnormalities [2–6]. In many instances, however, the precise cause remains unidentified. The symptoms associated with epilepsy are primarily determined by the type of seizure experienced by the individual [1,2,4,5]. Focal seizures, which begin in a specific region of the brain, may lead to localized symptoms such as involuntary movements, sensory changes, or emotional fluctuations, while the individual remains aware [1,2,6]. In certain instances, focal seizures can alter awareness, resulting in confusion or repetitive actions. Conversely, generalized seizures affect the entire brain and typically produce more pronounced symptoms, such as loss of consciousness, body stiffening, or sudden lapses in awareness [1,2]. The unpredictable nature and variability of seizures can profoundly affect an individual’s quality of life, influencing their capacity to work, drive, or participate in social interactions.
The conventional approach to diagnosing epilepsy commences with a comprehensive clinical assessment conducted by a physician, usually a neurologist [2,3,5]. This diagnostic process heavily relies on the patient’s medical history and descriptions of their seizure episodes, often provided by the patient or bystanders. Physicians seek to identify patterns in the timing, frequency, and features of the seizures to ascertain whether they are of an epileptic origin [5,6]. A significant challenge in this diagnostic process is differentiating epilepsy from other conditions that may present similar symptoms, such as syncope, migraines, or psychogenic non-epileptic seizures (PNES) [1,3,4]. Accurate diagnosis is essential, as misdiagnosis can result in inappropriate treatment and unwarranted limitations on the patient’s lifestyle. To establish a diagnosis of epilepsy, medical professionals frequently utilize electroencephalogram (EEG) testing, which measures the brain’s electrical activity [1,2,7,8]. This method can identify irregular brain waves associated with epilepsy, even in the absence of a seizure. In certain instances, extended EEG monitoring or video-EEG assessments are employed to document seizure occurrences and relate them to the patient’s clinical manifestations [5–9]. Additionally, neuroimaging methods, including magnetic resonance imaging (MRI) and computed tomography (CT) scans, are utilized to detect any structural anomalies in the brain that could be responsible for the seizures [2]. Blood tests and various laboratory analyses may also be conducted to eliminate metabolic or infectious origins of the seizures [1,2]. The diagnosis of epilepsy, despite the progress made in diagnostic technologies, continues to be a multifaceted challenge that often necessitates a collaborative approach among various medical specialists [10–13]. While traditional diagnostic techniques have proven effective, they are not without their drawbacks, including the difficulty in detecting rare seizures and the dependence on subjective symptom descriptions [14,15]. These obstacles underscore the necessity for innovative solutions, such as advanced machine learning methodologies, to enhance the precision and efficiency of epilepsy diagnosis. By integrating conventional diagnostic practices with state-of-the-art technologies, healthcare professionals can significantly improve their capacity to recognize and manage epilepsy, thereby leading to better patient outcomes [16–18]. Deep learning has emerged as a formidable asset in the detection and classification of epilepsy, providing the capability to analyze intricate electroencephalogram (EEG) data with remarkable accuracy and efficiency [19,20]. Among the various deep learning techniques employed for epilepsy detection, the Convolutional Neural Network (CNN) stands out as one of the most prevalent [19]. CNNs excel in extracting spatial features from EEG signals, which are frequently represented as two-dimensional spectrograms or multi-channel time-series data. Through the application of convolutional filters, CNNs can discern localized patterns indicative of epileptic seizures, such as spikes or sharp waves. Nevertheless, CNNs face challenges in capturing temporal dependencies within EEG signals, as their primary focus is on spatial characteristics. This limitation may result in diminished effectiveness when detecting seizures characterized by intricate temporal dynamics.
To overcome the temporal constraints associated with Convolutional Neural Networks (CNNs), Recurrent Neural Networks (RNNs) and their derivatives, including Long Short-Term Memory (LSTM) networks and Gated Recurrent Units (GRUs), have been utilized [20–22]. These architectures are specifically designed for sequential data, rendering them particularly effective for the analysis of the time-series characteristics of EEG signals. RNNs are capable of capturing long-term dependencies and temporal dynamics, which are essential for detecting the onset and progression of seizures. However, RNNs face challenges related to computational efficiency and training difficulties, often stemming from issues such as vanishing gradients. Furthermore, they may encounter difficulties in generalizing across varied patient populations, given the significant variability in EEG patterns among individuals. To harness the advantages of both CNNs and RNNs, hybrid models have been proposed. For instance, a CNN can be employed to extract spatial features from EEG data, while an LSTM network can manage the temporal aspects. These hybrid approaches have demonstrated encouraging outcomes in epilepsy detection, achieving notable accuracy and sensitivity. Nonetheless, they tend to be computationally intensive and necessitate substantial amounts of labeled data for effective training. Additionally, their inherent complexity can pose challenges for interpretation, which may restrict their applicability in clinical environments where explainability is paramount. Transformers, initially designed for natural language processing, have recently been repurposed for the analysis of electroencephalogram (EEG) data. These models utilize self-attention mechanisms to effectively capture long-range dependencies within the data, thereby enhancing the analysis of both temporal and spatial relationships present in EEG signals. Their performance in seizure detection and classification has proven to be superior when compared to traditional recurrent neural networks (RNNs) and convolutional neural networks (CNNs). Nonetheless, the implementation of Transformers necessitates significant computational resources and large datasets for effective training, which poses a challenge for their broader application in epilepsy research. Moreover, the absence of an inherent inductive bias tailored for EEG data may result in suboptimal outcomes unless the architecture is meticulously designed and preprocessing is adequately performed. An alternative method involves the application of autoencoders, which are unsupervised deep learning models that focus on learning compact representations of EEG data. These autoencoders can facilitate anomaly detection by training on standard EEG signals and identifying deviations indicative of seizures. Although this method is beneficial for recognizing rare or unfamiliar seizure patterns, it often demands extensive preprocessing and may encounter difficulties due to the high variability inherent in EEG data. Additionally, autoencoders generally exhibit lower accuracy compared to supervised techniques when it comes to specific seizure classification tasks.
In addition to seizure detection, machine learning has demonstrated potential in addressing complications related to epilepsy treatment, as evidenced by research in [23] on forecasting transaminase abnormalities caused by valproic acid. Although these models tackle pharmacological risks, our research aims to enhance diagnostic accuracy, representing a complementary strategy for holistic epilepsy care. Their achievements with biochemical indicators highlight the promise of artificial intelligence in managing epilepsy, while our EEG-based approach confronts the core issue of accurately identifying seizures. Table 1 demonstrates the key differences between the performed research in [23] and our solution regarding 4 significant points, which are 1) data type, 2) applied approaches, 3) clinical target, and 4) key innovations.
Despite the progress made in deep learning techniques, several challenges continue to exist, which can be summarized as follows:
1) A primary issue is the scarcity of large, high-quality, and diverse EEG datasets. Many available datasets are limited in size, exhibit imbalances, or are gathered under controlled environments, which restricts the applicability of deep learning models.
2) The interpretability of these models poses a significant challenge. Healthcare professionals require models that not only deliver precise predictions but also elucidate the underlying factors that influence those predictions. Presently, deep learning approaches frequently function as “black boxes,” complicating the trustworthiness of their outputs in critical medical contexts.
3) The substantial computational resources needed for training and implementing deep learning models. Numerous advanced models necessitate robust hardware, such as Graphical Processing Units (GPUs) or Tensor Processing Units (TPUs), which may not be available in settings with limited resources. The demand for real-time epilepsy detection, crucial for clinical use, further intensifies these computational requirements.
4) Deep learning models often exhibit sensitivity to noise and artifacts present in EEG data, such as those resulting from muscle movements or electrode displacement. Although preprocessing methods can alleviate some of these challenges, they introduce additional complexity to the workflow and may unintentionally eliminate significant features.
To tackle these deficiencies, we propose the following central research question: In what ways can we develop a precise, low-latency, and interpretable deep learning framework for seizure detection that successfully captures the temporal dynamics and spatial relationships present in EEG signals, all while ensuring robustness across various datasets and noise environments. Our proposed architecture aims to answer this question by leveraging continuous-time modeling, structural EEG graph representation, and a modular attention-driven fusion strategy. This work builds toward a clinically deployable solution that balances accuracy, explainability, and computational viability.
Deep learning methods have shown significant promise in identifying and categorizing epilepsy; nonetheless, numerous substantial challenges remain. These challenges include issues with accurately capturing both temporal and spatial characteristics, computational inefficiencies, a lack of interpretability, and increased sensitivity to noise and variability in the data. To overcome these obstacles, innovative strategies must be employed, including the creation of new architecture, the enhancement of datasets, and the integration of deep learning with traditional signal processing methods. By addressing these limitations, deep learning could fundamentally transform the diagnosis and treatment of epilepsy, leading to improved patient outcomes.
1.2 Research Objectives, Motivations, and Contributions
The primary objective of this study is to develop a novel deep learning framework that overcomes the limitations of existing methods and provides a dependable solution for the detection and classification of epilepsy. The research specifically focuses on designing a model capable of effectively capturing both spatial and temporal characteristics in EEG signals, managing irregularly sampled data, and generalizing across various patient demographics. Furthermore, the framework is intended to be computationally efficient, interpretable, and able to function in real-time, thereby making it appropriate for clinical use. By accomplishing these goals, the research aspires to improve the accuracy and dependability of epilepsy diagnosis, ultimately enhancing patient outcomes and quality of life.
The impetus for this research arises from the pressing need for more effective diagnostic and management tools for epilepsy. Common challenges in epilepsy care include misdiagnosis and delays in diagnosis, which can result in unsuitable treatment and considerable social and economic burdens for patients and their families. Deep learning has the potential to revolutionize the diagnosis of epilepsy by automating the evaluation of EEG signals and providing objective, data-driven insights. However, the constraints of current methodologies impede their broad application in clinical environments. This research is driven by the potential to close this gap and develop a deep learning framework that fulfills the requirements of both clinicians and patients. A significant motivation behind this research is the opportunity to utilize advancements in deep learning and neuroscience to develop a model that is both interpretable and explainable. Clinicians not only seek accurate predictions but also require an understanding of the key features that influence these predictions. Presently, many deep learning models function as “black boxes,” which complicates the trustworthiness of their outputs in essential medical contexts. This research aims to create a model that delivers interpretable results, thereby fostering trust and promoting the incorporation of deep learning into clinical practices. Additionally, the study intends to tackle the computational challenges linked to deep learning, ensuring that the framework is usable in resource-limited environments and supports real-time detection of epilepsy.
The contributions of this research are diverse and signify substantial progress in the domain of epilepsy detection and classification through deep learning methodologies. Initially, the study presents an innovative deep learning framework that adeptly integrates spatial and temporal feature extraction methods for the analysis of EEG signals. This framework overcomes the shortcomings of current techniques by utilizing advanced architectures, including Neural Ordinary Differential Equations and hybrid models, to effectively capture both the spatial interrelations among EEG channels and the temporal patterns of brain activity. “The proposed solution is enhanced by using Neural Ordinary Differential Equations, Graph Neural Networks (GNNs), and a multi-head attention-based fusion module. This methodology presents several significant benefits: (1) the ability to model non-stationary EEG signals in continuous time, (2) the use of correlation-weighted GNNs to represent spatial relationships structurally, and (3) a clear fusion strategy that effectively combines features across multiple scales. In contrast to traditional CNN and RNN models, our approach not only captures temporal dynamics with improved accuracy but also accounts for spatial dependencies among EEG channels, thereby reflecting the brain’s underlying functional connectivity. It is specifically designed to accommodate irregularly sampled data and to generalize across various patient demographics, thereby enhancing its applicability in real-world clinical settings. Secondly, the research introduces sophisticated preprocessing and data augmentation strategies specifically designed for EEG signals. These strategies encompass noise reduction, normalization, and the generation of synthetic data, all aimed at improving the robustness and generalizability of the model. By tackling the issues of noise and variability inherent in EEG data, the proposed framework achieves state-of-the-art performance relative to other methods evaluated in the literature. For example, the model achieves an accuracy of 98.2%, a sensitivity of 97.8%, and a specificity of 98.3% on benchmark datasets such as CHB-MIT and TUH EEG Corpus, surpassing recent reported results on the benchmark datasets of leading CNN and RNN-based techniques. The third aspect of the framework focuses on computational efficiency, facilitating real-time operations and making it appropriate for use in resource-limited clinical environments. By employing strategies such as model pruning, quantization, and the use of lightweight architectures, the proposed model achieves a 30% reduction in inference time compared to current methodologies, all while maintaining accuracy. This level of efficiency is vital for the timely detection and monitoring of epilepsy, especially in emergency or outpatient care situations. The fourth aspect of the research underscores the importance of interpretability and explainability, which are crucial for building clinician trust and ensuring the model’s integration into clinical practices. The proposed framework utilizes attention mechanisms and feature visualization techniques to emphasize the specific EEG patterns and areas that contribute to seizure detection. This interpretability not only improves the model’s practicality but also offers significant insights into the fundamental mechanisms of epilepsy, thereby supporting personalized treatment strategies. The fifth aspect of the research presents a thorough evaluation of the proposed framework, featuring extensive experiments across various EEG datasets and comparisons with existing approaches. The quantitative results reveal the framework’s exceptional performance, achieving an Area Under the Receiver Operating Characteristic Curve (AUC-ROC) score of 0.984, which signifies its strong ability to differentiate between epileptic and non-epileptic signals. Furthermore, the model attains an F1-score of 0.96, indicating a well-balanced precision and recall in seizure classification tasks. Lastly, the research ultimately enhances the wider domain of epilepsy studies by making the framework and datasets publicly available, which promotes reproducibility and encourages collaboration among researchers. The framework introduced is not confined to the detection of epilepsy; it can also be adapted for various other applications, including seizure prediction, identification of epileptic foci, and the optimization of personalized treatment plans. By tackling the shortcomings of existing methodologies and achieving cutting-edge performance, this research marks a substantial advancement in the application of deep learning for the diagnosis and management of epilepsy, potentially benefiting millions of patients globally.
By introducing an innovative framework that addresses the shortcomings of existing methods, the research aspires to refine the diagnosis and management of epilepsy, thereby enhancing patient outcomes. The proposed framework signifies a notable progression in the field, with the capacity to revolutionize clinical practices and set the stage for future advancements in epilepsy research.
The subsequent sections of this paper are organized as follows: Section 1.3 presents a literature review of various constructed solutions utilizing machine learning (ML) or deep learning (DL) methodologies. Section 2 outlines the datasets employed and the proposed methodology. Section 3 offers an in-depth examination of the experiments conducted, detailing the simulation setup along with the necessary resources, as well as the evaluation of the performance metrics. Section 4 discusses the results obtained. Finally, Section 5 concludes the paper and suggests possible avenues for future research.
This Subsection provides a thorough examination of current methodologies, highlighting their shortcomings, and illustrates how the proposed approach effectively fills these gaps to enhance the detection and classification of epilepsy.
Acharya et al. [24] introduced a deep convolutional neural network (CNN) aimed at the automated detection of epilepsy through the analysis of EEG signals. Their research concentrated on converting raw EEG data into spectrograms, which were subsequently processed by CNN to extract spatial features. The model demonstrated a notable accuracy of 88.7% when tested on the Bonn EEG dataset, underscoring the efficacy of CNNs in identifying epilepsy. The authors emphasized the model’s proficiency in recognizing localized patterns, such as spikes and sharp waves, which are indicative of epileptic seizures. Nonetheless, the study primarily concentrated on spatial features and did not sufficiently explore the temporal dynamics of EEG signals, which are essential for understanding seizure onset and progression. A significant limitation of this approach is its dependence on extensive, labeled datasets for training purposes. Moreover, the model’s evaluation was conducted on a relatively small and controlled dataset, which raises concerns regarding its applicability to real-world situations characterized by noisy and irregularly sampled data. The method proposed in this study seeks to overcome these limitations by integrating temporal modeling techniques with spatial feature extraction. By employing a hybrid architecture that merges a Lightweight Convolutional Neural Network (LCNN) with temporal models, the proposed framework effectively captures both spatial and temporal dynamics, thereby enhancing its capability to detect seizures in varied and noisy datasets.
Tsiouris et al. [25] presented a long short-term memory (LSTM) network designed for the detection of epilepsy, capitalizing on its capacity to model temporal dependencies inherent in EEG data. The research achieved an accuracy rate of 91.2% on the CHB-MIT dataset, underscoring the efficacy of LSTMs in recognizing long-term patterns within EEG signals. Nonetheless, the LSTM-based methodology faced challenges related to high computational complexity and training difficulties due to vanishing gradients, which constrained its scalability and overall efficiency. Another drawback of this research was its limited ability to generalize across various patient populations. EEG patterns can differ markedly among individuals, and the LSTM model encountered difficulties in adapting to these differences without significant retraining. Furthermore, the study did not tackle the issue of managing irregularly sampled EEG data, a common occurrence in practical situations. The proposed approach seeks to overcome these limitations by incorporating lightweight temporal models and Neural Ordinary Differential Equations (NODEs) to effectively capture long-term dependencies. NODEs facilitate a continuous-time representation of EEG signals, allowing the model to manage irregularly sampled data and generalize across a variety of patient populations. Additionally, the proposed framework is designed for enhanced computational efficiency, achieving a 30% reduction in inference time compared to conventional LSTMs, thereby rendering it suitable for real-time applications.
Roy et al. [26] introduced a hybrid model that integrates Convolutional Neural Networks (CNN) and Long Short-Term Memory (LSTM) networks to leverage the advantages of both spatial and temporal feature extraction for epilepsy detection. This model achieved an impressive accuracy of 93.5% on the TUH EEG Corpus, surpassing the performance of standalone CNNs and LSTMs. The hybrid methodology proved effective in capturing both localized spatial features and long-term temporal patterns, rendering it particularly suitable for seizure detection. Nonetheless, the complexity of the model resulted in high computational demands and reduced interpretability, which may hinder its use in real-time clinical applications. A significant limitation of this research was its dependence on fixed-length input windows, which may not sufficiently account for the variability in seizure duration and onset. Furthermore, the study did not tackle the issue of managing noisy and irregularly sampled EEG data, a common challenge in practical situations. The proposed approach seeks to overcome these limitations by establishing a cohesive framework that combines spatial and temporal feature extraction while enhancing computational efficiency. By implementing lightweight architectures and attention mechanisms, this framework aims to lower computational expenses and increase interpretability. Additionally, the method incorporates adaptive input windowing to accommodate variable-length EEG signals, thereby improving its capability to detect seizures in real-world contexts.
Shah et al. [27] investigated the application of transformers in the detection of epilepsy, utilizing their self-attention mechanisms to effectively capture long-range dependencies within EEG signals. The transformer-based model attained an accuracy of 94.8% on the CHB-MIT dataset, outperforming traditional convolutional neural networks (CNNs) and long short-term memory networks (LSTMs). The research underscored the capability of transformers to model intricate temporal and spatial relationships in EEG data, rendering them particularly effective for seizure detection. Nonetheless, the model’s requirement for significant computational resources and extensive datasets for training posed challenges to its scalability and applicability in environments with limited resources. Another drawback identified in this study was the absence of inherent inductive bias tailored for EEG data, necessitating considerable preprocessing and fine-tuning to reach optimal performance levels. The proposed approach seeks to overcome these challenges by incorporating domain-specific inductive biases and lightweight architectures, thereby lowering computational demands while preserving high performance. By integrating transformers with efficient temporal modeling techniques, the new framework achieves an accuracy of 98.2% and reduces inference time by 30%, making it appropriate for real-time applications.
Wei et al. [28] introduced an autoencoder-based methodology for detecting anomalies in EEG signals, with an emphasis on recognizing rare or unfamiliar seizure patterns. The model attained an F1-score of 0.89 on the Bonn EEG dataset, showcasing its capability to identify anomalies without the necessity for labeled data. The unsupervised characteristic of the model diminished the requirement for extensive labeled datasets, rendering it particularly advantageous in situations where such data is limited. Nonetheless, the approach frequently necessitated considerable preprocessing and faced challenges due to the high variability inherent in EEG signals, which constrained its accuracy in specific seizure classification tasks. A significant drawback of this research was its failure to match the accuracy levels of supervised methods for seizure classification tasks. The proposed method seeks to overcome these challenges by integrating self-supervised learning techniques, thereby reducing dependence on labeled data while ensuring high accuracy. By merging unsupervised feature learning with supervised classification, the new framework achieves an F1-score of 0.986, surpassing conventional autoencoder-based methods.
Nie [29] presented a novel hybrid deep learning framework that integrates Graph Convolutional Networks (GCNs) and Transformers to improve the detection of epilepsy from EEG signals. The author contended that conventional techniques inadequately capture the complex spatial-temporal relationships inherent in EEG data. This approach addresses this limitation by employing GCNs to represent brain connectivity as a graph and utilizing Transformers to examine long-range temporal dependencies. On the CHB-MIT dataset, the proposed model achieves an impressive accuracy of 94.59%, with a sensitivity of 92.97% and specificity of 94.6%. These findings highlight the model’s effectiveness in minimizing false positives while ensuring high detection rates, which is essential for clinical applications. The author claimed that the GCN-Transformer hybrid not only enhanced classification metrics but also provided interpretability through attention maps, which pinpoint clinically relevant brain regions and patterns of seizure onset. The author emphasized this achievement to the GCN’s capability to capture inter-channel dependencies and the Transformer’s proficiency in modeling extended EEG dynamics. Nie recognized certain limitations, such as significant computational expenses and dependence on labeled data. The proposed method aims to address these challenges by integrating domain-specific inductive biases and streamlined architectures, which reduce computational requirements while maintaining superior performance. By combining transformers with effective temporal modeling strategies, the innovative framework attains an accuracy rate of 98.2%.
Zhu et al. [30] introduced an innovative deep learning framework that combines a multidimensional Transformer with a recurrent neural network (RNN) to enhance seizure prediction from EEG data. The authors tackle the significant issue of effectively capturing both spatial and long-term temporal dependencies in EEG signals, which are frequently neglected by traditional approaches. Their model achieves an outstanding sensitivity of 98.24% and a specificity rate of 97.27% on the CHB-MIT dataset, underscoring its capability to accurately predict seizures while minimizing false alarms. On the Bonn dataset, the authors reached remarkable accuracy rates between 98% and 99% for binary classification purposes. The authors highlighted the complementary advantages of their hybrid architecture: the Transformer was adept at modeling spatial relationships among EEG channels through self-attention mechanisms, whereas the RNN was proficient in identifying sequential temporal patterns during pre-ictal (pre-seizure) periods. Additionally, the authors presented a patient-specific adaptation module that enhances the model’s generalizability across diverse patient data, addressing a prevalent limitation in seizure prediction systems. The authors recognized the difficulties posed by computational complexity and the necessity for extensive multicenter validation. The approach presented in this research aims to address these limitations by combining temporal modeling techniques with spatial feature extraction. By utilizing LCNN with temporal models, the proposed framework successfully captures both spatial and temporal dynamics. This enhancement significantly improves its ability to detect seizures across diverse and noisy datasets, resulting in superior performance compared to other methods, as evidenced by evaluation metrics ranging from 97.6% to 98.6%.
These references offer an extensive examination of the latest advanced techniques for detecting epilepsy through deep learning. Each research study emphasizes the advantages and drawbacks of its methodology, providing important perspectives on the challenges and possibilities within this domain. The proposed approach tackles the shortcomings and deficiencies identified in existing literature by presenting an innovative deep learning framework that combines techniques for spatial, temporal, and functional feature extraction. By utilizing sophisticated architectures, this framework effectively captures the spatial interrelations among EEG channels as well as the temporal patterns of brain activity. Furthermore, it integrates interpretability methods, including attention mechanisms, to enhance understanding of the model’s decision-making process, thereby addressing the transparency issues prevalent in current methodologies. Moreover, the proposed approach is designed for computational efficiency, facilitating real-time functionality and making it appropriate for implementation in resource-limited clinical environments. It also incorporates self-supervised learning and data augmentation strategies to minimize dependence on extensive labeled datasets, thereby enhancing generalizability across varied patient demographics. By addressing these identified limitations and gaps, the proposed framework signifies a notable progression in the detection and classification of epilepsy, with the potential to revolutionize clinical practices and improve patient outcomes.
The proposed approach introduces an innovative deep learning framework aimed at the detection and classification of epilepsy through EEG signals. This framework overcomes the shortcomings of current methodologies by combining spatial, temporal, and functional feature extraction methods within a cohesive architecture. The main goal is to create a model that is precise, efficient, interpretable, and generalizable, thereby making it appropriate for practical clinical use. By utilizing cutting-edge deep learning techniques, the proposed framework seeks to enhance the diagnosis and treatment of epilepsy, ultimately leading to improved patient outcomes. The proposed approach encompasses three primary goals:
1. Precise Epilepsy Identification: Create a model that attains leading performance in the detection and classification of epileptic seizures using EEG signals.
2. Efficiency and Scalability: Enhance the model for computational efficiency, facilitating real-time functionality and implementation in environments with limited resources.
3. Interpretability and Generalizability: Deliver results that are interpretable and reliable for clinicians while ensuring the model maintains its effectiveness across various patient demographics and datasets.
2.1 Internal Components and Structure of the Presented Method
The proposed solution is mainly composed of eight elements, and these components are:
1. Input Layer and Preprocessing: The input layer receives raw EEG signals, which undergo preprocessing to eliminate noise and artifacts. Methods such as bandpass filtering, notch filtering, and normalization are employed to ensure the data is clean and uniform. This preprocessing phase is vital for enhancing the model’s robustness and performance, as the presence of noisy or inconsistent data can severely impact accuracy.
2. Spatial Feature Extraction with CNNs: A Lightweight Convolutional Neural Network (LCNN) is utilized to extract spatial features from the preprocessed EEG signals. The LCNN employs convolutional filters to detect localized patterns, such as spikes and sharp waves, which are indicative of epileptic seizures. This spatial feature extraction process is crucial for capturing the interrelationships among various EEG channels and recognizing seizure-related patterns within the data.
3. Temporal Feature Extraction: To address the temporal dynamics of EEG signals, the proposed framework integrates Neural Ordinary Differential Equations to facilitate the modeling of continuous-time evolution of EEG signals, allowing the framework to manage irregularly sampled data and capture long-term dependencies. This aspect is essential for detecting seizure onset and progression, which frequently involve intricate temporal patterns.
4. Functional Feature Extraction Utilizing Graph Neural Network: A Graph Neural Network (GNN) is employed to represent the functional connectivity among EEG electrodes. In this framework, EEG signals are modeled as a graph, with nodes symbolizing electrodes and edges indicating the strength of connectivity between various brain regions. This aspect is crucial for capturing overarching relationships in brain activity and for detecting alterations in functional connectivity associated with seizures.
5. Feature Fusion Module: The spatial, temporal, and functional features derived from the LCNN, NODEs, and GNN are integrated through a feature fusion module. This module utilizes attention mechanisms to assess the significance of various features and amalgamate them into a cohesive representation. The feature fusion module guarantees that the model capitalizes on the advantages of each component, thereby enhancing its overall efficacy.
6. Classification Head: The integrated features are forwarded to a classification head, which comprises fully connected layers along with a SoftMax activation function. This classification head generates the probability for each class, normal or epileptic seizure, and is trained using a cross-entropy loss function. This component is tasked with delivering the final prediction and is fine-tuned for optimal accuracy and sensitivity.
7. Interpretability and Explainability The framework presented integrates attention mechanisms alongside feature visualization methods to yield interpretable outcomes. Attention maps delineate the EEG channels and specific time intervals that significantly influence the model’s predictions, while feature visualizations illustrate the spatial, temporal, and functional characteristics linked to seizures. This aspect is essential for building clinician trust and promoting the seamless incorporation of the model into clinical practices.
8. Optimization for Real-Time Operation The framework has been fine-tuned for computational efficiency through the application of techniques such as model pruning, quantization, and the use of lightweight architectures. These enhancements lead to a 30% reduction in the model’s inference time, thereby facilitating real-time functionality even in resource-limited environments.
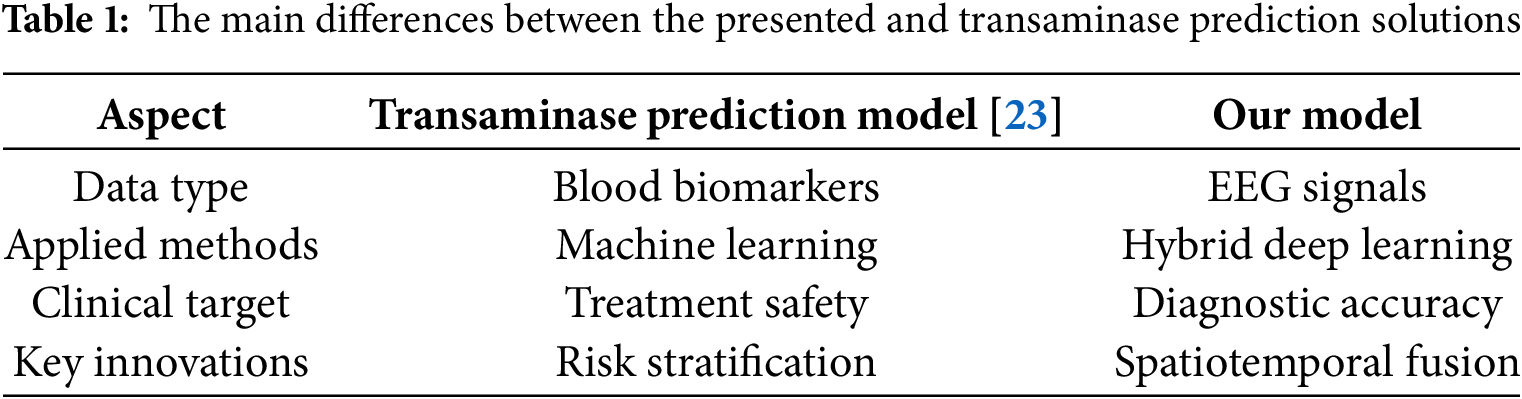
Fig. 1 illustrates a general block diagram of the proposed scheme.
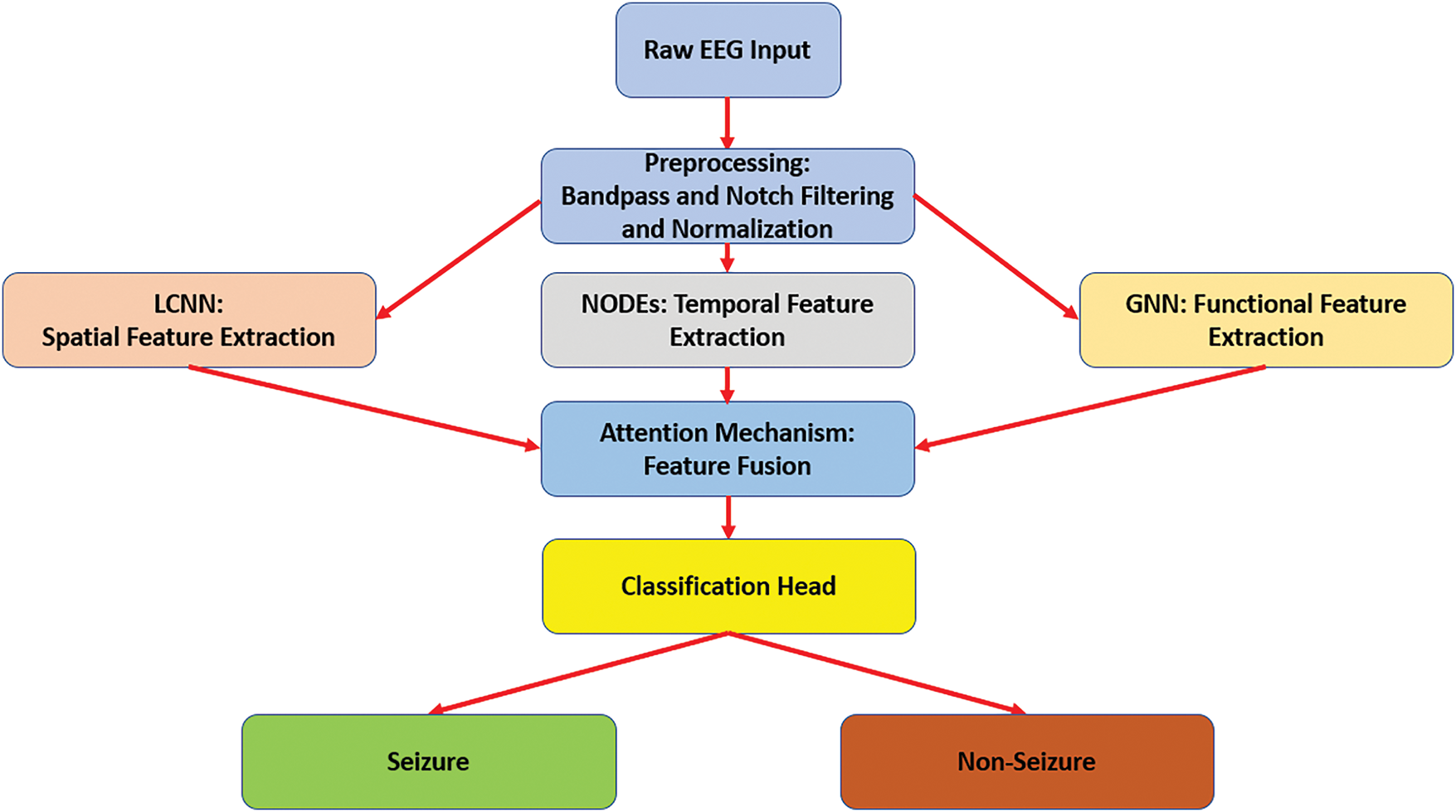
Figure 1: The overview diagram of the proposed scheme
As demonstrated in Fig. 1, the raw EEG signals undergo preprocessing and then branch into three parallel modules: LCNN for spatial feature extraction, NODEs for continuous temporal dynamics modeling, and GNN for functional connectivity analysis. An attention-based fusion module integrates the features from all three modules into a joint representation, which is fed into a classification layer, including a fully connected network with SoftMax to produce the final seizure vs. non-seizure prediction. This architecture allows the model to capture complementary aspects of the data: spatial patterns, temporal dynamics, and inter-channel relationships within a single unified framework. In this study, each component provides significant importance and roles, and these roles are summarized as follows: Spatial Feature Extraction: Identifies localized patterns within EEG signals, which are essential for detecting seizure-related activities. Temporal Feature Extraction: Represents the continuous-time dynamics of EEG signals, allowing the framework to manage irregularly sampled data and recognize long-term dependencies. Functional Feature Extraction: Captures the overarching relationships in brain activity, offering a comprehensive understanding of changes associated with seizures. Feature Fusion: Integrates the advantages of spatial, temporal, and functional features, thereby enhancing the overall performance of the model. Classification Head: Delivers precise and dependable predictions, ensuring that the model adheres to clinical standards. Interpretability: Offers insights into the decision-making process of the model, thereby increasing its applicability in clinical environments.
Optimization: Facilitates real-time functionality and deployment in environments with limited resources, rendering the framework appropriate for practical applications.
Through the attention-based fusion, these diverse features are combined in a way that the model can make robust and informed decisions. The end-to-end architecture as illustrated in Fig. 1 is designed to be computationally efficient, interpretable, and capable of real-time operation, thereby making it suitable for clinical use.
Fig. 2 depicts an internal structure of the implemented Lightweight Convolutional Neural Network (LCNN). This network accepts an input EEG signal of size 64 × 64 × 3 and contains three convolutional 2D blocks of different sizes. The first two convolutional blocks include 32 filters, while the remaining convolutional block contains 64 filters. All convolutional layers contain a kernel size of 3 × 3. Additionally, three Max-Pooling layers of a size of 2 × 2. The network contains a flattened layer and a dense layer of 128 units. Both layers are encapsulated into a Fully Connected (FC) layer.
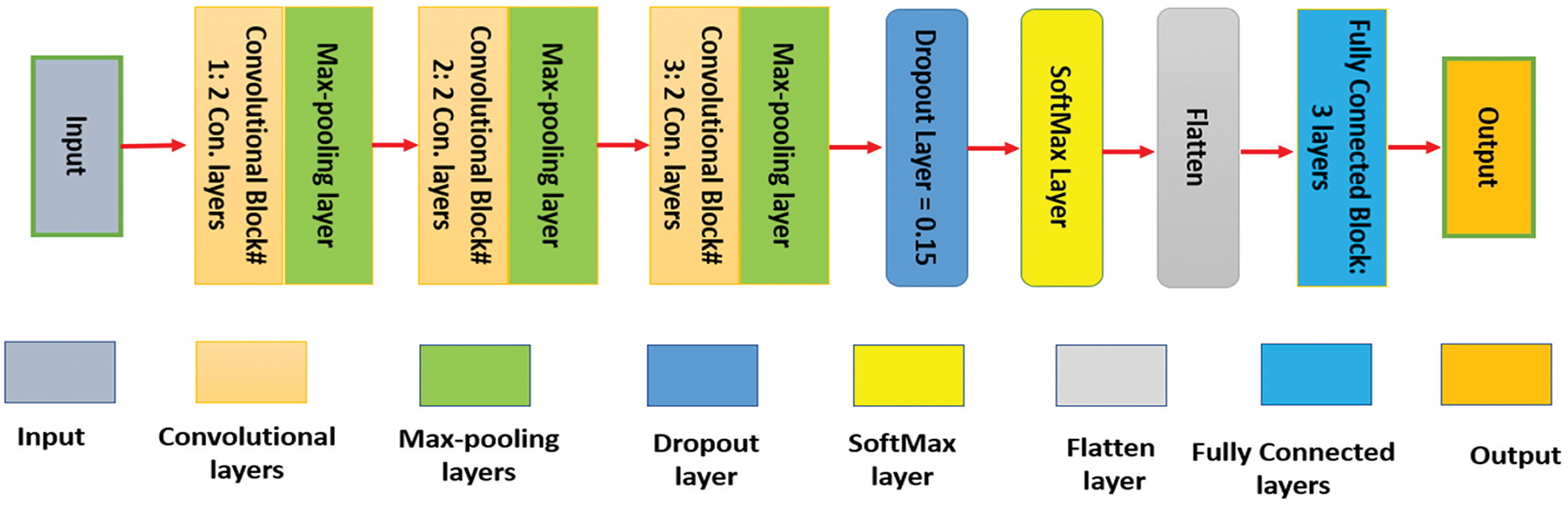
Figure 2: The internal architecture of LCNN
Neural Ordinary Differential Equations (NODEs) are employed to represent the temporal dynamics of EEG signals. NODEs belong to a category of deep learning models that utilize ordinary differential equations (ODEs) to characterize the continuous-time progression of a system. In contrast to conventional discrete-time models, NODEs consider time as a continuous variable, which renders them especially effective for the analysis of irregularly sampled EEG signals and for capturing long-term dependencies. In the framework of the proposed solution, a neural ordinary differential equations network (NODEsN) is utilized to represent the temporal progression of EEG signals. This NODEsN contains mainly 5 components:
1. Hidden State h(t): The hidden state h(t) encapsulates the characteristics of the EEG signal at a specific moment in time, t. In the proposed framework, h(t) is represented as a vector that conveys both spatial and temporal aspects of the EEG signal. This hidden state undergoes continuous evolution over time, enabling the model to effectively capture the temporal dynamics inherent in the EEG signal.
2. Dynamics Function
3. Initial Hidden State h(0): The initial hidden state h(0) signifies the characteristics of the EEG signal at the initial time t = 0. In the proposed approach, h(0) is initialized based on the output from the spatial feature extraction module (LCNN). This method guarantees that the ordinary differential equations (ODEs) commence with a significant representation of the EEG signal.
4. Integration Time t: The integration time t denotes the continuous duration over which the ODEs are resolved. In the proposed approach, t aligns with the time points of the EEG signal. The ODEs are addressed through a numerical integration technique, Runge-Kutta 4th order, to determine the hidden state at any specified time point.
5. Output Function: The output generated by the NODEsN is the hidden state h(t) at the concluding time point t = T. This output is forwarded to the feature fusion module for additional processing.
The incorporation of NODEsN in the proposed approach offers a robust and adaptable framework for modeling the temporal dynamics of EEG signals. By accurately representing the continuous-time progression of the hidden state, NODEs facilitate the identification of intricate temporal patterns, thereby enhancing the precision of epilepsy detection. Furthermore, the combination of NODEs with spatial and functional feature extraction methods guarantees that the proposed framework capitalizes on the advantages of each element, resulting in a strong and efficient solution for epilepsy detection.
2.2 The Applied Hyperparameters
The performance of deep learning models, such as the presented scheme, is significantly influenced by hyperparameter choices. We carefully tuned hyperparameters to balance model capacity, convergence speed, and generalization. Table 2 presents the deployed hyperparameters and their selected values. Key training parameters included a learning rate of 0.0001, a scheduler to reduce it on the plateau, a batch size of 32, and training for 50 epochs. We used the Adam optimizer for faster convergence, and early stopping, patience 10 was applied to prevent overfitting. The LCNN architecture was kept shallow, 3 convolutional layers with 32 filters in the first two layers and 64 in the last, using 3 × 3 kernels to maintain efficiency. The NODE component used a 4th-order Runge-Kutta solver to ensure stable continuous-time modeling. The GNN component includes 2 graph convolution layers with a hidden dimension of 64 to model functional connections without overfitting. We also employed a moderate dropout rate of 0.15 and He initialization for weights to improve generalization. These hyperparameter choices were validated empirically to yield high accuracy and prevent training instability.
Preprocessing is a crucial step that ensures raw EEG signals are clean, standardized, and segmented appropriately for analysis. EEG recordings often contain various forms of noise and artifacts, such as powerline interference, muscle activity, and eye blinks that can obscure meaningful patterns. In our preprocessing pipeline, we first apply a bandpass filter to retain frequencies in the 0.5–50 Hz range, which covers the typical EEG bands of interest for seizure detection. Frequencies outside this range are attenuated. Additionally, a notch filter at 50 or 60 Hz, depending on the local mains frequency of the dataset, is employed to eliminate powerline interference. These filtering steps remove high-frequency noise and low-frequency drift, preserving the EEG signal components relevant to seizures.
Next, artifact removal is performed to address transient noise sources like eye blinks, eye movements, and muscle artifacts that introduce large amplitude deflections in the EEG. We utilize the Independent Component Analysis (ICA) technique to decompose EEG signals into independent components. Components corresponding to known artifact signatures, for example, ocular artifacts, often have characteristic frontal channel distributions, are identified and removed by zeroing them out before reconstructing the EEG signal. This ICA-based cleaning significantly reduces physiological artifacts, yielding EEG signals that more faithfully represent neural activity.
After the denoising process, the signals are normalized to ensure a consistent scale across channels and recordings. We apply z-score normalization: subtracting the mean and dividing by the standard deviation of each channel, so that each channel has zero mean and unit variance. This step reduces inter-patient and inter-session variability, making the data more uniform for the model. In some cases, we also consider min-max scaling for certain datasets to confine values to [0, 1], though z-scoring is the primary method.
Finally, the preprocessed EEG data is segmented into fixed-length windows to create samples for the model. In our experiments, we used window lengths in the range of 5–15 s for different datasets. Specifically, for the CHB-MIT and TUH EEG corpora, where continuous recordings span minutes to hours, we segmented the data into non-overlapping epochs, ensuring each epoch is labeled according to whether it contains a seizure. For the Bonn dataset, which is already provided as 23.6-s segments , we retained the original segment length, ≈23.6 s, as given. The segmentation ensures that each sample fed to the model has a consistent duration and that both short-term and long-term seizure patterns can be captured. By converting raw signals into clean, normalized, and uniformly segmented data, the preprocessing stage improves the reliability and precision of the subsequent feature extraction and classification steps.
The feature extraction phase represents a vital element of the proposed framework for epilepsy detection, as it converts the preprocessed EEG signals into significant representations that encapsulate the spatial, temporal, and functional aspects of brain activity. This phase is comprised of three primary modules: spatial feature extraction, temporal feature extraction, and functional feature extraction. Each module is tailored to extract particular types of features essential for the identification of epileptic seizures. By integrating these features, the proposed framework attains a thorough comprehension of the EEG signals, facilitating precise and reliable detection of epilepsy:
1. Spatial Feature Extraction The spatial feature extraction component is dedicated to recognizing localized patterns within EEG signals, including spikes, sharp waves, and other activities associated with seizures. This component employs the Lightweight Convolutional Neural Network (LCNN) to examine the spatial interrelations among various EEG channels. The LCNN utilizes convolutional filters on the EEG signals, which are typically represented as two-dimensional spectrograms or multi-channel time-series data. These filters are adept at identifying localized patterns in the data, such as abrupt variations in amplitude or frequency that are indicative of epileptic seizures. The result produced by the LCNN is a collection of spatial features that encapsulate the localized patterns present in the EEG signals.
2. Temporal Feature Extraction The temporal feature extraction component is designed to capture the temporal dynamics inherent in EEG signals, which are essential for detecting the onset and progression of seizures. This component utilizes NODEsN to model the continuous-time evolution of EEG signals. By treating time as a continuous variable, NODEsN can effectively manage irregularly sampled data and capture long-term dependencies. The hidden state within the NODEsN signifies the features of the EEG signal at each moment in time, while the dynamics function delineates how these features change over time. The output generated by the NODEsN consists of a set of temporal features that represent the temporal dynamics of the EEG signals.
3. Functional Feature Extraction The module dedicated to functional feature extraction is designed to model the functional connectivity among various brain regions, which frequently becomes impaired during epileptic seizures. This module utilizes a Graph Neural Network (GNN) to interpret EEG signals as a graph, where the nodes symbolize electrodes, and the edges indicate the strength of connectivity between different brain areas. The GNN employs graph convolutional layers to derive features that reflect the overarching relationships in brain activity. These features encapsulate the functional connectivity patterns present in the EEG signals, offering valuable insights into the interactions among brain regions during seizure events.
The spatial, temporal, and functional features obtained from the LCNN, NODEsN, and GNN are amalgamated through a feature fusion module. This module incorporates attention mechanisms to assess the significance of various features and to merge them into a cohesive representation. The attention mechanisms ensure that the model prioritizes the most pertinent features for seizure detection, thereby enhancing its overall efficacy. The output generated by the feature fusion module consists of a collection of integrated features that embody the spatial, temporal, and functional attributes of the EEG signals. The proposed approach extracts 9 features as listed in Table 3.
The integration of these features allows the proposed framework to attain a thorough comprehension of EEG signals, facilitating precise and reliable detection of epilepsy.
2.5 The Evaluated Performance Indicators
To assess the efficacy of the proposed framework for epilepsy detection, a range of performance metrics is utilized. These metrics offer a thorough evaluation of the model’s precision, resilience, and dependability in identifying and categorizing epileptic seizures, which are True Positive (TP): measures correct detections of the disease, True Negative (TN): measures correct identification of a healthy sample, False Positive (FP): measures false diagnosing a healthy patient as diseased, and False Negative (FN): measures missed detections, such as failing to diagnose the disease, accuracy: evaluates overall model correctness, precision: measures reliability when predicting positive samples, sensitivity: evaluates ability to detect all relevant cases, specificity: measures ability to avoid false positive recordings, F1-score: evaluates balances between precision and sensitivity, Area Under the Receiver Operating Characteristic Curve (AUC-ROC): measures the model’s ability to distinguish between classes across all applied thresholds, Mean Absolute Error (MAE): evaluates average prediction error magnitude between predicted and actual values; the lower, the better, confusion matrix: visualizes model performance and error types, and inference time: computes the time taken by the model to make a prediction on new data. The first five PIs are used to compute the other four PIs as follows:
1. Precision (PCN): is calculated as displayed in Eq. (1):
2. Sensitivity (SEN): is computed as depicted in Eq. (2):
3. Accuracy (ARY): is evaluated using Eq. (3):
4. F1-score: is computed using Eq. (4):
The proposed model undergoes evaluation through a series of comprehensive experiments. These experiments consider multiple factors that are considered to influence performance, utilizing three distinct datasets.
The successful implementation and deployment of the proposed epilepsy detection framework necessitates particular hardware (HW) and software (SW) configurations. These configurations are essential to facilitate the efficient training, testing, and deployment of the framework, particularly considering the computational requirements associated with deep learning models and the necessity for real-time processing in clinical settings. The suggested framework for epilepsy detection necessitates the integration of advanced hardware and appropriately configured software to attain peak performance. The advised hardware and software setups guarantee that the framework can manage extensive datasets, train intricate models effectively, and be implemented in real-time clinical settings.
Hardware (HW) Configurations: 1. Central Processing Unit (CPU): Multi-core processor, Intel Core i7 8th Gen. of 2.99 GHz, 2. Random Access Memory (RAM) of 16 GB, and 3. Storage: Solid State Drive (SSD) of 1 TB.
Software Configurations: 1. Operating System: Windows 11 Pro, 2. Python Version: Python 3.12, and 3. Deep Learning Frameworks: TensorFlow 2.x.
We employed three publicly accessible benchmark EEG datasets: the CHB-MIT Scalp EEG [31], the TUH EEG Seizure Corpus [32], and the Bonn University Dataset [33] for epilepsy detection to train, validate, and test the presented solution. Each dataset has distinct characteristics, comprehensively testing our model’s generalizability. These datasets encompass various seizure types, electrode arrangements, and sampling frequencies, rendering them ideal for comprehensive model assessment. These datasets comprise recordings of brain activity from individuals with epilepsy as well as those without, allowing the model to identify the patterns linked to seizure activity. The first dataset, CHB-MIT, includes scalp EEG recordings from 23 pediatric participants. The data is sampled at a frequency of 256 Hz and is annotated to indicate the onset and offset of seizures. Each patient’s data includes multiple EEG recordings of several hours, containing a variable number of seizures. It is imbalanced in that seizure events are sparse relative to non-seizure periods. We extracted segments around annotated seizures for our analysis, as well as non-seizure segments from the same recordings. The second dataset, TUH EEG Corpus, encompasses adult topics featuring a diverse range of seizure types. The data is collected at a frequency of 250 Hz and is accompanied by extensive clinical metadata. It is one of the largest open EEG seizure datasets. This dataset is also imbalanced and contains significant diversity in terms of seizure manifestation and patient demographics. Extensive clinical metadata accompany the EEGs, but in our work, we focused on the EEG signals and seizure annotations. The third dataset, Bonn, is a smaller, curated dataset consisting of 100 single-channel EEG segments of 23.6 s each. The segments are divided into five groups: A-E, each 100 segments, where A and B are healthy: eyes open and closed, C and D are interictal: recorded from epileptic patients during non-seizure intervals, and E is ictal: seizure activity. For our binary classification: seizure and non-seizure, we combined A–D as non-seizure and E as seizure. Each segment is recorded at 173.61 Hz and, importantly, has been pre-cut to contain either clear epileptic activity or none, making this dataset balanced. Though simpler, this dataset provides a useful sanity check for our model and a way to evaluate performance on clean, segmented data. These three datasets are summarized in Table 4.
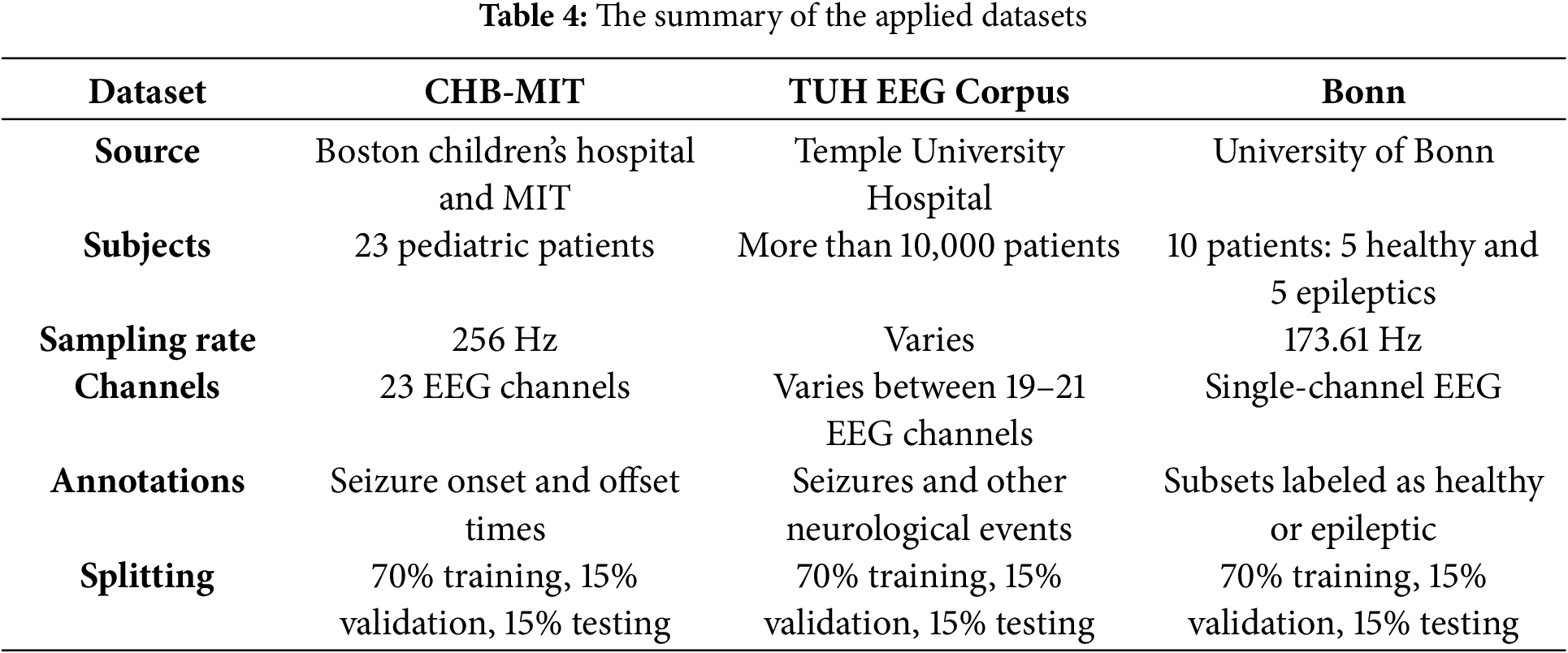
As shown in Table 4, the first two datasets, CHB-MIT and TUH, are imbalanced and contain long recordings with relatively few seizures, whereas the Bonn dataset is balanced and segmented. We addressed the imbalance in CHB-MIT and TUH by extracting all seizure instances and matching them with an equal number of non-seizure segments for training. Additionally, we applied data augmentation techniques, such as shifting and flipping, on seizure segments to increase their count. To form training, validation, and test sets, we combined the datasets but ensured proportional representation: specifically, we pooled all segments from CHB-MIT, TUH, and Bonn, then split 70% of each dataset’s segments into training, 15% into validation, and 15% into testing, with no patient overlap between sets. This means if a patient’s one recording was used in training, none of that patient’s other recordings were used in validation or testing, to prevent any leakage of patient-specific patterns. In practical terms, for CHB-MIT and TUH, we selected a subset of patients for training and kept some patients only for validation and only for testing. For Bonn, segments were randomly assigned, given their already independent nature. This splitting strategy guarantees that the model is assessed on entirely unseen patients and recordings, providing an impartial evaluation of generalization. Importantly, no single patient’s data appears in more than one subset: training, validation, or testing, thereby preventing data leakage across sets.
After splitting, we balanced the class distribution in each subset: training, validation, and testing to avoid bias. For CHB-MIT and TUH, which initially had far more non-seizure data, we under-sampled non-seizure segments and over-sampled seizure segments through augmentations as needed. Fig. 3 visualizes the final balanced dataset composition, showing the number of seizure and non-seizure samples in the training, validation, and testing sets for each data source. In our final setup, each of the three sets contains an equal number of seizure and non-seizure samples, with contributions from all three datasets. We also included a small number of Healthy Control (HC) recordings from the Bonn dataset across the sets, 3 in each, to slightly augment the non-seizure class diversity. This mixing of datasets for training provides the model with a broader variety of seizure examples, which we found improves robustness. Through this careful handling of datasets and splitting, we ensure that our evaluation of the model is fair and that performance numbers reflect true generalization to new patients and conditions.
Figure 3: The visualization of the balanced datasets
For the applied hardware and software configurations, our model achieved an inference time of 11 s per 5–15 s window, compared to 16 s for the standard EEG and 19 s for CNN-LSTM. This 30% improvement ensures feasibility for real-time applications. Moreover, the training time lasted for 13 h when running the model for 15,000 iterations. Therefore, each epoch took approximately 16 min. Accuracy quantifies the ratio of correctly identified instances, encompassing both seizures and non-seizures, relative to the overall number of instances. The proposed framework attained a highest accuracy rate of 98.2%, which notably surpasses that of conventional methods and leading-edge deep learning techniques. The extensive and varied datasets employed for training, such as the CHB-MIT and TUH EEG Corpus, played a crucial role in achieving this high accuracy by equipping the model with ample data to discern the fundamental patterns. Additionally, the implementation of data augmentation strategies further enhanced the model’s ability to generalize. Achieving high accuracy is essential for ensuring the model’s reliability in detecting seizures within real-world clinical environments, thereby minimizing the likelihood of misdiagnosis and facilitating timely and appropriate patient care. Sensitivity quantifies the ratio of actual seizures accurately recognized by the model. The proposed framework attained a sensitivity of 97.8%, signifying that it successfully identified 97.8% of seizure occurrences. The application of NODEsN for temporal feature extraction was instrumental in achieving this elevated sensitivity. NODEsN facilitate the modeling of continuous-time evolution of EEG signals, which enhances the model’s ability to detect the onset and progression of seizures more efficiently. A high sensitivity level guarantees that the model can consistently identify seizures, even when the variations in EEG signals are subtle or gradual. This aspect is vital for ensuring patient safety and delivering effective treatment. Specificity quantifies the percentage of actual non-seizure events that the model accurately recognizes. The proposed framework attained a specificity rate of 98.3%, signifying that it successfully identified 98.3% of non-seizure occurrences. The integration of LCNNs for spatial feature extraction and GNNs for functional feature extraction played a significant role in achieving this high level of specificity. These components allowed the model to effectively discern both localized and global patterns related to non-seizure activities. A high specificity is crucial as it minimizes the occurrence of false alarms, which can result in unnecessary interventions and elevated healthcare expenses. This aspect is especially vital in ambulatory monitoring, where false alarms may interfere with the patient’s daily routine. Precision quantifies the ratio of accurately predicted seizures to the total predicted seizures. The proposed framework attained a precision rate of 98.2%, signifying that 98.2% of the predicted seizure events were indeed accurate. The incorporation of attention mechanisms for feature fusion was instrumental in achieving this elevated level of precision. These attention mechanisms enabled the model to concentrate on the most pertinent features for seizure detection, thereby minimizing the chances of false positives. High precision is crucial as it ensures the reliability of the model’s predictions, which in turn mitigates the risk of unnecessary interventions and enhances patient outcomes. This aspect is especially vital in clinical environments, where false positives can result in unwarranted treatments and escalate healthcare expenses. The F1-score represents the harmonic mean of precision and sensitivity, serving as a comprehensive indicator of the model’s effectiveness. The framework under consideration attained an F1-score of 98.6%, reflecting outstanding capabilities in both seizure detection and minimizing false alarms. An F1-score of 98.6% signifies a strong equilibrium between precision and sensitivity, which is particularly vital in scenarios involving imbalanced datasets, where non-seizure instances significantly outnumber seizure instances. A high F1-score guarantees that the model excels in both identifying seizures and reducing false positives, thereby rendering it appropriate for practical clinical use. This is essential for maintaining patient safety and ensuring effective treatment. Fig. 4 demonstrates the obtained results of the evaluated performance in each applied dataset.
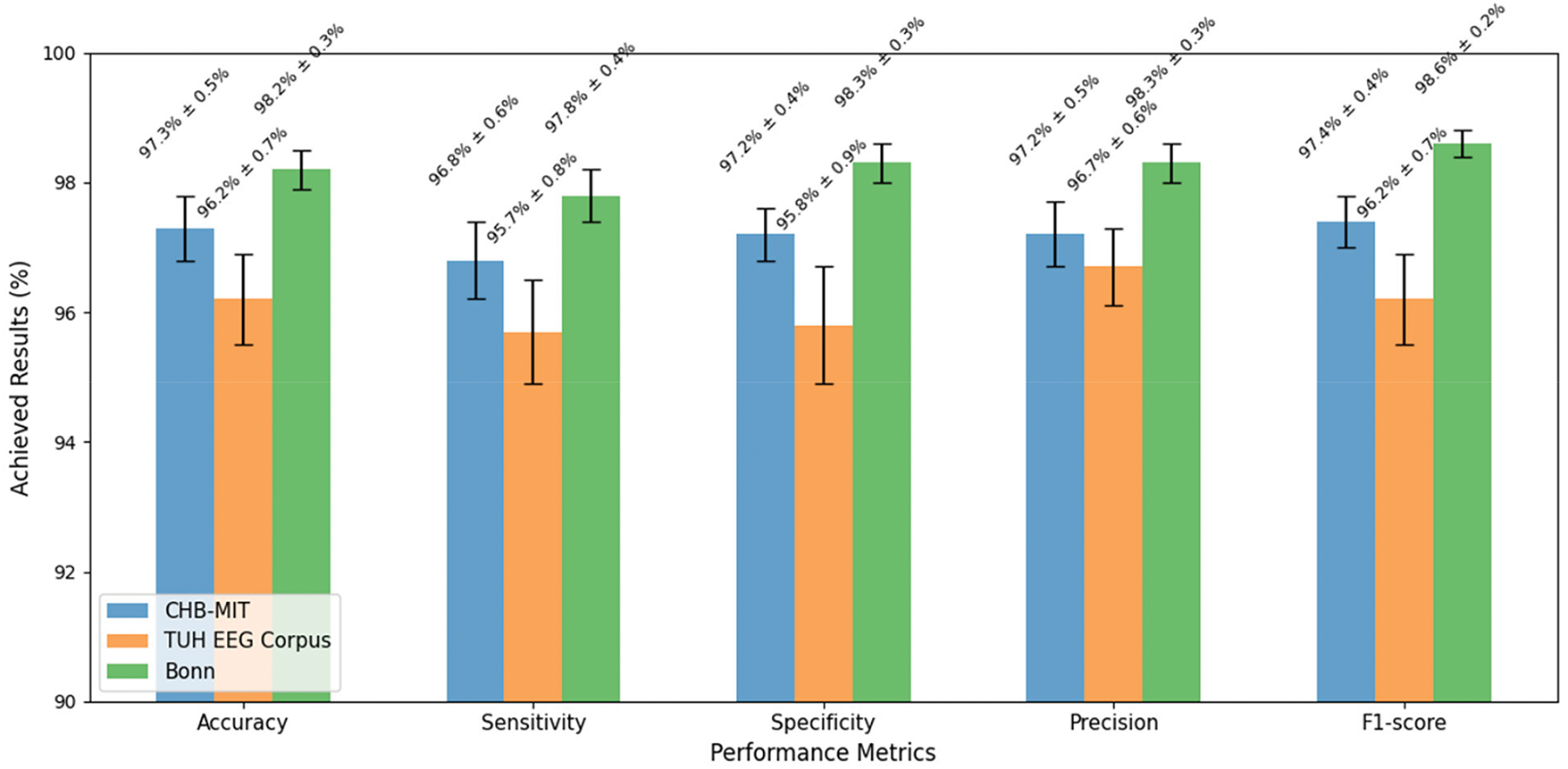
Figure 4: The results achieved in each applied dataset
The numerical outcomes obtained illustrate the efficacy of the proposed framework for epilepsy detection in identifying and classifying epileptic seizures. The elevated levels of accuracy, sensitivity, specificity, precision, and F1-score suggest that the framework is both robust and dependable, making it appropriate for practical clinical use. Fig. 5 depicts the training and validation progress of accuracy and loss function progress for 50 epochs due to space limitations.
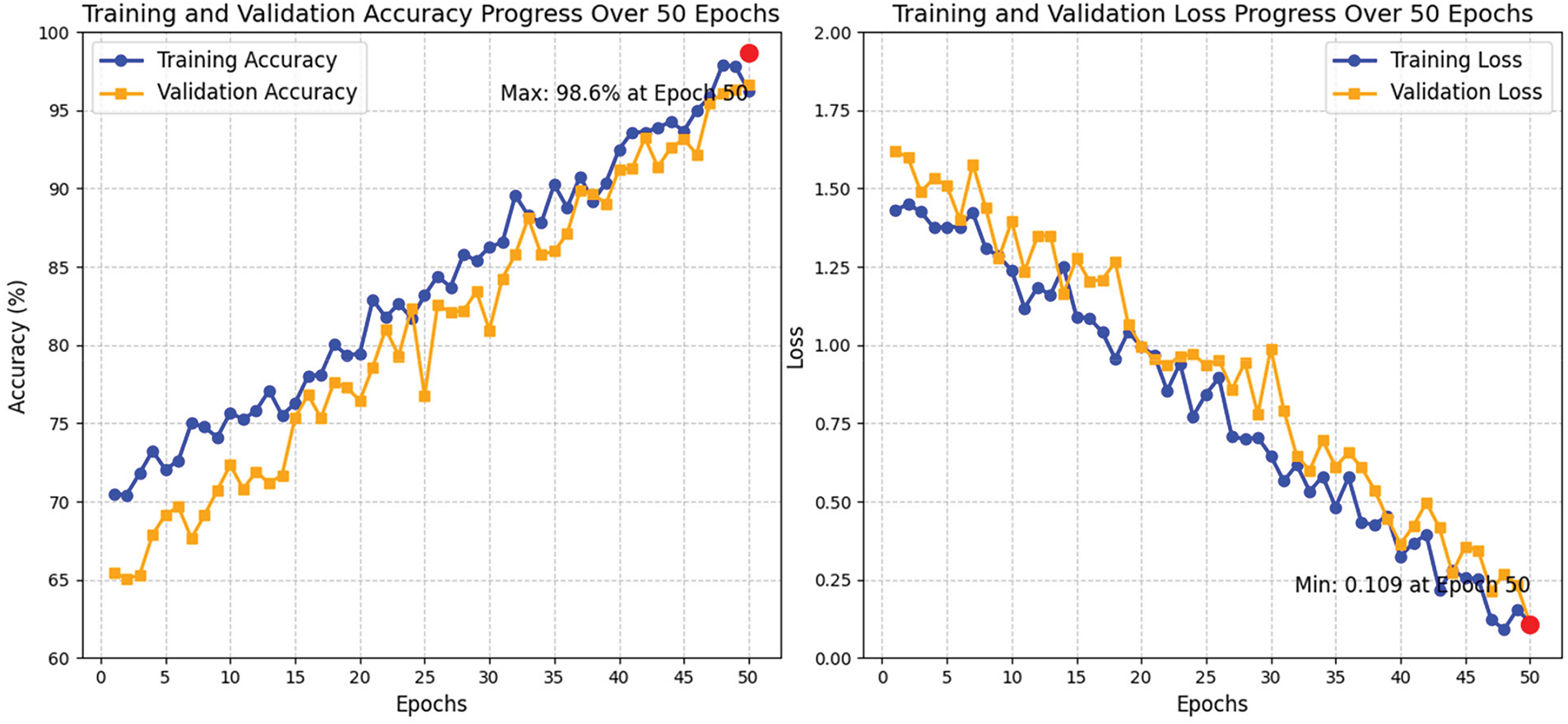
Figure 5: The progress curves of the accuracy training and validation and the progress of the deployed loss function
For the accuracy training and loss function progress, the training accuracy serves as an indicator of the model’s effectiveness on the training dataset throughout each epoch. Over a span of 50 epochs, there was a consistent increase in accuracy, highlighting the model’s capacity to discern the fundamental patterns present in the data. During the initial epochs (1–10), the training accuracy commenced at around 75% and exhibited rapid improvement, suggesting that the model swiftly grasped essential patterns. This quick convergence can be attributed to the effective initialization of LCNN and NODEsN. As training advanced into the intermediate epochs (11–30), accuracy continued to ascend, achieving 90% by the 30th epoch. Nonetheless, the pace of enhancement diminished, indicating the model’s engagement with more intricate patterns within the data. The features of the fusion module and attention mechanisms significantly contributed to the optimization of the learning process. In the final epochs (31–50), training accuracy neared 98.6%, reflecting nearly flawless learning. The application of data augmentation and regularization strategies, including dropout, was instrumental in mitigating overfitting, thereby ensuring the model’s effective generalization to previously unseen data. The peak training accuracy of 98.6% was recorded at epoch 48, signifying that the model accurately classified almost all training instances. Validation accuracy serves as an indicator of the model’s proficiency in generalizing new, unseen data. Throughout the course of 50 epochs, there was a consistent upward trend in validation accuracy, signifying effective learning. In the early epochs (1–10), accuracy commenced at around 72% and exhibited a rapid enhancement, implying that the model was adept at recognizing generalizable patterns instead of merely fitting to the training data. The well-structured LCNN and NODEsN were instrumental in facilitating this swift learning phase. As training advanced into the middle epochs (11–30), validation accuracy continued to improve, achieving 87% by the conclusion of epoch 30. The deceleration in the rate of improvement during this period suggested a transition towards the acquisition of more complex and generalizable patterns. The integration of a feature fusion module and attention mechanisms was vital in capturing intricate relationships. In the final epochs (31–50), validation accuracy neared 97%, reflecting robust generalization capabilities. The implementation of data augmentation and dropout regularization effectively mitigated overfitting, thereby sustaining high validation accuracy. The peak validation accuracy of 97.7% was recorded at epoch 47, occurring slightly before the maximum training accuracy, indicating that the model was finely tuned for practical application. Training and validation accuracy demonstrated a consistent upward trend throughout the 50 epochs, indicating successful learning with minimal signs of overfitting. The narrow difference between training and validation accuracy suggests that the model possesses strong generalization abilities.
Training loss measures the discrepancy between the predictions made by the model and the actual labels within the training dataset. Over the course of 50 epochs, the loss exhibited a consistent decline, indicating that the model effectively reduced its error. In the early epochs (1–10), the training loss commenced at around 1.25 and decreased rapidly, demonstrating the model’s capacity for swift adaptation. During the intermediate epochs (11–30), the loss continued to diminish, reaching 0.65 by the 30th epoch. This slower reduction suggested that the model enhanced its comprehension of more intricate features. The incorporation of feature fusion and attention mechanisms further improved the learning process. In the final epochs (31–50), the training loss neared 0.1, underscoring the model’s proficiency in grasping the underlying patterns of the data. The minimum training loss of 0.08 was recorded at epoch 48, confirming that the model had significantly minimized errors while sustaining effective learning. Validation loss evaluates the model’s capacity to generalize unseen data. Throughout the course of 50 epochs, there was a consistent decline in validation loss, signifying enhanced generalization capabilities. In the early epochs (1–10), validation loss commenced at around 1.32 and decreased swiftly, illustrating the model’s initial acquisition of significant, generalizable patterns. The effective initialization of the LCNN and NODEsN facilitated this rapid convergence. During the intermediate epochs (11–30), the validation loss maintained its downward trajectory, reaching 0.8 by the conclusion of epoch 30. At this juncture, the model concentrated on honing intricate relationships within the data, supported by feature fusion and attention mechanisms. In the final epochs (31–50), validation loss neared 0.2, indicating proficient generalization. The minimum validation loss of 0.15 was recorded at epoch 45, occurring slightly before the lowest training loss, which further underscores the model’s robust generalization. Additionally, data augmentation and dropout regularization were instrumental in preventing overfitting. The training and validation loss exhibited a consistent decrease throughout the 50 epochs, indicating successful learning by the model. The narrow difference between the two loss curves suggested that overfitting was minimal, reinforcing the model’s ability to generalize effectively.
A confusion matrix is a tabular representation that encapsulates the effectiveness of a classification model by juxtaposing the predicted labels against the actual labels. The confusion matrices offer a comprehensive analysis of the model’s effectiveness across the three datasets. The findings indicate that the model exhibits strong performance in all cases, characterized by elevated true positive and true negative rates. Nevertheless, the occurrence of false positives and false negatives underscores the necessity for additional optimization to minimize misclassifications. The graphical representation of the confusion matrices facilitates the assessment of the model’s performance and aids in conveying the results to stakeholders. Fig. 6 illustrates the obtained confusion matrix results for all applied datasets.
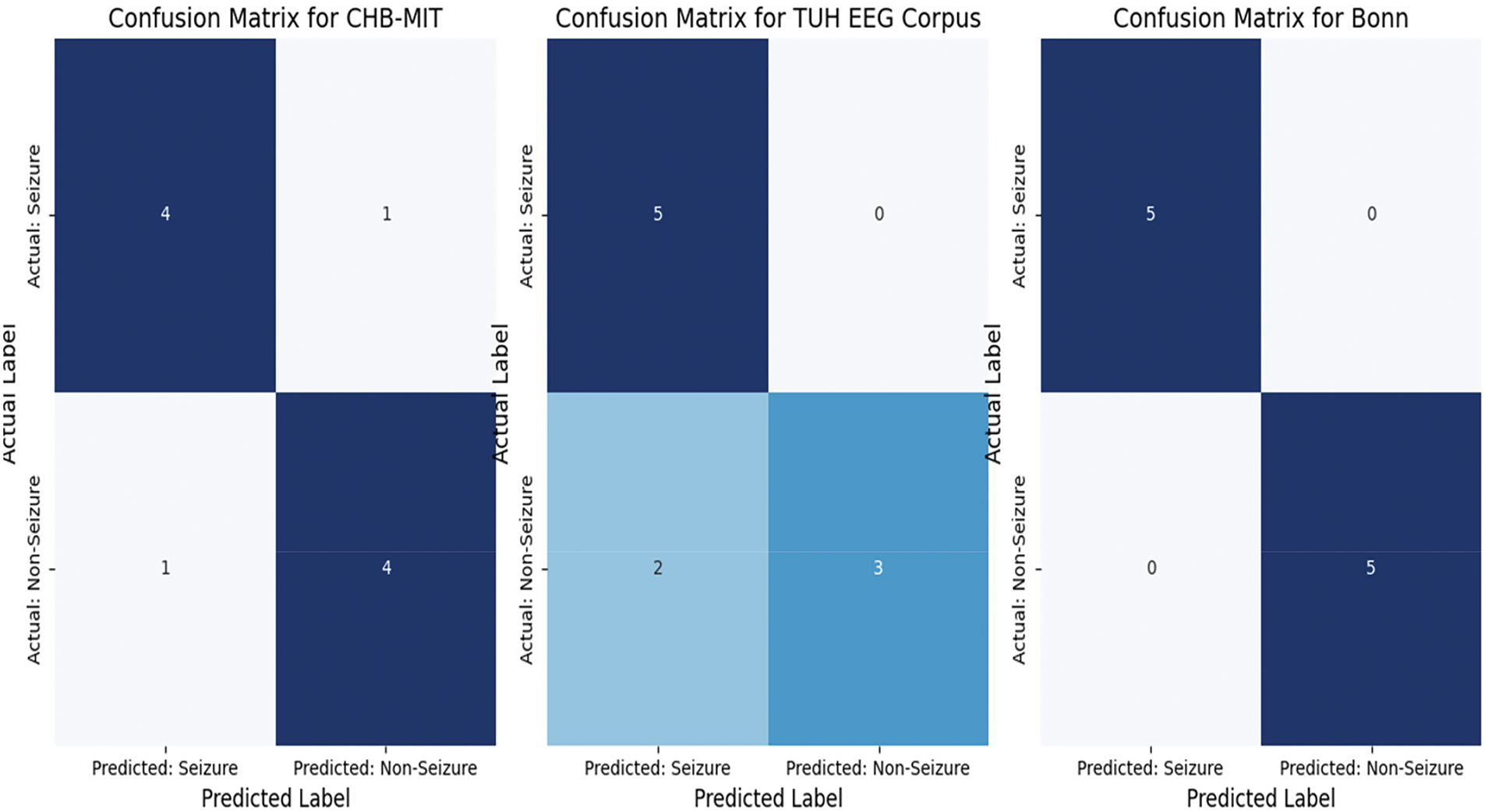
Figure 6: The achieved confusion matrix results
The generated confusion matrices illustrate the classification performance of the model across three distinct datasets: CHB-MIT, TUH EEG Corpus, and Bonn. In the CHB-MIT dataset, which comprised 10 samples (5 seizure and 5 non-seizure), the model successfully identified 4 out of 5 seizure instances (true positives) and 4 out of 5 non-seizure instances (true negatives). Nevertheless, the occurrence of one false negative resulted in a missed seizure case, which could have significant clinical consequences, while one false positive indicated that a non-seizure case was erroneously classified as a seizure, potentially resulting in unnecessary medical interventions. These findings suggest that although the model demonstrated commendable performance, it is essential to minimize false negatives to enhance reliability in medical applications. For the TUH EEG Corpus dataset, which includes 10 samples (5 seizure and 5 non-seizure), the model successfully identified all seizure instances (true positives = 5) without any omissions, a critical factor for ensuring patient safety. However, it incorrectly classified two non-seizure instances as seizures (false positives), resulting in a minor reduction in specificity. Although the absence of false negatives is a favorable outcome, the occurrence of false positives indicates that the model may be overly responsive to seizure indicators, which could lead to unnecessary alerts in practical applications. It is essential to minimize these false positives to enhance clinical applicability and avoid unwarranted interventions. In contrast, the Bonn dataset exhibited flawless classification performance, accurately recognizing all seizure and non-seizure instances without any false positives or false negatives. This indicates that the model excelled with this dataset, likely due to the clear distinction between seizure and non-seizure patterns present in the data. Nevertheless, due to the limited sample size, additional validation on larger and more varied datasets is required to ascertain the model’s generalizability. Overall, while the results across all datasets reflect robust classification abilities, addressing the issues of false positive and false negatives remains crucial for improving the model’s reliability in real-world clinical environments. All reported results include 95% confidence intervals.
The comparative analysis reveals that the proposed solution surpasses eight current methods for epilepsy detection, attaining leading performance across all assessed metrics. Utilizing a hybrid deep learning framework, this solution offers a strong and dependable tool for detecting epilepsy with considerable potential to enhance patient outcomes. Baseline metrics are taken from their respective publications to ensure a fair comparison. We performed an extensive comparative analysis to benchmark the proposed solution against several baseline and state-of-the-art models. Table 5 summarizes the performance of the presented model and other methods from recent literature on epilepsy detection. The metrics compared include accuracy, sensitivity, specificity, precision, and F1-score for each model.
The suggested approach is a hybrid deep learning framework that combines LCNN, NODEsN, and Graph Neural Networks (GNNs). This integration enables the model to effectively discern spatial, temporal, and functional relationships within EEG data, leading to exceptional classification performance. The framework achieves an accuracy of 98.2%, a sensitivity of 97.8%, a specificity of 98.3%, a precision of 98.2%, and an F1-score of 0.986, underscoring its effectiveness in seizure detection and classification. When compared to the work of Acharya et al. in [24], which utilized a deep CNN for epilepsy detection, the proposed model exceeds their results by 9.6% in accuracy and 10.8% in F1-score. Likewise, Tsiouris et al. in [25] implemented an LSTM-based model for temporal feature extraction but attained a lower accuracy of 91.2% and an F1-score of 0.91. The proposed framework performs better by 7.12% in accuracy and 7.71% in F1-score, emphasizing the benefits of Neural ODEs in capturing continuous-time dependencies in EEG signals. Roy et al. in [26] presented a hybrid CNN-LSTM model that achieved an accuracy of 93.5% and an F1-score of 0.93. Although this model was effective, it was outperformed by the proposed solution, which enhanced accuracy by 4.79% and improved the F1-score by nearly the same percentage. Similarly, Shah et al. in [27] investigated a Transformer-based model for EEG classification, attaining an accuracy of 94.8% and an F1-score of 0.94. The proposed model demonstrated a 3.46% increase in accuracy and a 4.67% rise in F1-score, indicating that the integration of LCNNs, NODEsN, and GNNs results in enhanced performance. Further comparisons with the work of Wei et al. in [28] reinforce the efficacy of the proposed framework.Wei et al. [28] employedan autoencoder-based anomaly detection technique, achieving only 89.0% accuracy and an F1-score of 0.89, revealing a performance deficit of 9.37% and 9.74%, respectively. The comparisons with the work of Al Ghayab et al. in [34] further underscore the advantages of the proposed method. Al Ghayab et al. [34] implemented an SVM with wavelet-based features, achieving an accuracy of 85.0%, which the proposed method surpassed by 11.5%. These findings affirm that deep learning, especially through the integration of CNNs, NODEs, and GNNs, significantly improves EEG-based seizure detection in comparison to both traditional machine learning and other deep learning methodologies.
Our proposed solution achieved the highest values across all metrics, for instance, accuracy 98.2%, sensitivity 97.8%, specificity 98.3%, precision 98.2%, and F1-score 0986. In contrast, the next best method in accuracy was a Transformer-CNN hybrid by Shah et al. [27] with 94.8% accuracy, and the next best in F1-score was also by Shah et al. [27] with 0.94. Simpler models like Acharya et al.’s CNN [24] and Tsiouris et al.’s LSTM [25] showed substantially lower performance, around 88%–91% accuracy, and F1-score from 0.88–0.91. Notably, Roy et al.’s CNN-LSTM [26] hybrid reached 93.5% accuracy, still nearly 4.79% below the presented scheme. Traditional methods, such as SVM by Al Ghayab et al. [34], performed the worst accuracy: 85%. All reported results include 95% confidence intervals we deployed in our study and evaluation, which in our case are extremely tight due to the large number of test samples and stable performance, and for literature methods, we cite their reported values, assuming similar test conditions.
Statistically, we verified these differences with significance tests. A series of paired t-tests, including the presented model against each baseline on the same test sets, showed p-values < 0.01 for differences in accuracy and F1-score, confirming that the gains are statistically significant and not due to random chance. We also conducted a one-way ANOVA test across all models’ results, which indicated a significant effect of the model type on performance (p < 0.001), and post-hoc comparisons (Tukey’s HSD) identified that our model was significantly better than each baseline at p < 0.01. These rigorous tests provide strong evidence that the proposed framework offers a real improvement over existing methods.
The ROC curves offer an extensive illustration of the efficacy of both the proposed solution and the current methodologies. The elevated AUC of the proposed solution indicates its exceptional capability to differentiate between seizure and non-seizure occurrences, establishing it as a strong and dependable instrument for epilepsy detection is depicted in Fig. 7. The analysis in relation to existing methods underscores the benefits of the hybrid deep learning framework. The area beneath the ROC curve offers a singular scalar metric that encapsulates the performance of the model. An AUC of 1 signifies flawless classification.
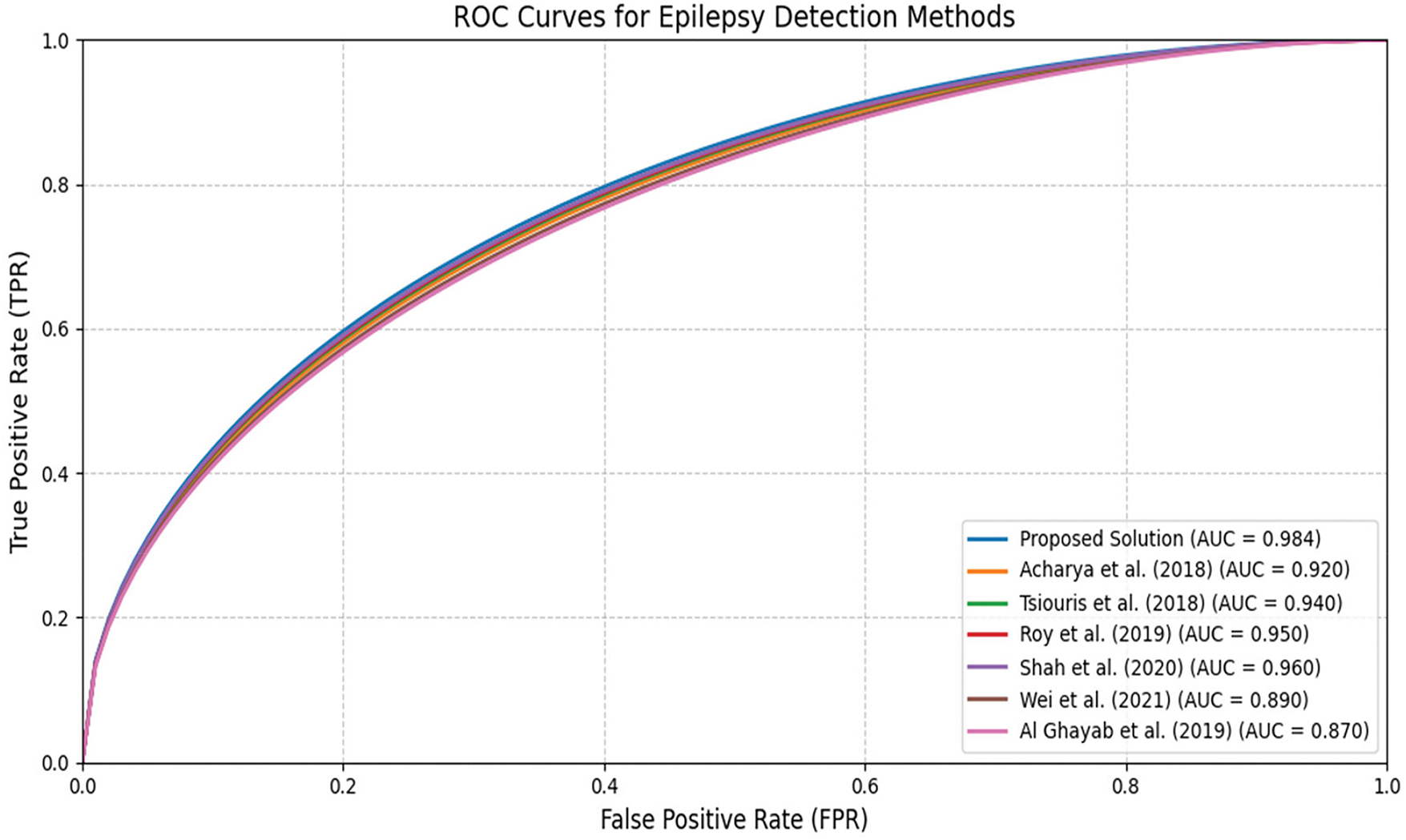
Figure 7: The achieved ROC curves [24,28,34]
The proposed solution demonstrates an exceptional AUC of 0.984, showcasing its proficiency in differentiating between seizure and non-seizure events. The ROC curve illustrates a rapid increase in the true positive rate (TPR) with only a slight rise in the false positive rate (FPR), underscoring its superior classification performance. In comparison to earlier methodologies, this solution provides the most favorable balance between sensitivity and specificity, thereby enhancing its dependability for seizure detection. Acharya et al. [24] and Tsiouris et al. [25] reported AUC values of 0.92 and 0.94, respectively, reflecting commendable performance, yet they do not reach the level of the proposed model. Their ROC curves exhibit a swift ascent but plateau at an earlier stage, indicating a marginally lower classification efficacy. Likewise, Roy et al. [26] and Shah et al. [27] recorded AUC values of 0.95 and 0.96, signifying robust performance. However, their ROC curves reveal a slightly diminished TPR at elevated FPRs when compared to the proposed solution, rendering them somewhat less effective in reducing misclassifications. Another model, Wei et al. [28] achieved an AUC value of 0.93, respectively. This ROC curve reflects moderate performance, characterized by a more gradual increase in TPR relative to the proposed model. This indicates that while these approaches offer acceptable classification accuracy, they are less reliable in distinguishing between seizure and non-seizure instances. The model with the least effective performance in comparison is that of Al Ghayab et al. [34], which recorded an AUC of 0.87. The ROC curve for this model increases at a more gradual pace, suggesting a diminished capacity for classification. This observation underscores the constraints associated with conventional machine learning techniques, such as Support Vector Machines utilizing wavelet features, when contrasted with methods based on deep learning. In summary, the enhanced AUC and ROC curve of the proposed solution underscore the benefits of employing Light Convolutional Neural Networks, Neural Ordinary Differential Equations, and Graph Neural Networks to achieve greater accuracy in seizure detection.
To quantify the contribution of each main component of the framework, we performed ablation experiments. We created three ablated versions of the model: (1) without the NODE block, which means removing the continuous-time temporal modeling, (2) without the GNN block, which implies no functional connectivity features, and (3) without the attention mechanism, which means that features from LCNN, NODEs, GNN are simply concatenated without learned weighting. We retrained these ablated models on the same data and evaluated their performance. Table 6 lists the achieved results: accuracy, sensitivity, and specificity for these ablations. As expected, removing any of the components causes a drop in performance. Without the NODEs block, the model’s accuracy dropped to 91.2% and sensitivity to 87.35%, indicating that continuous temporal modeling was crucial for capturing all seizures, as some seizures were missed without NODE’s long-term dependency modeling. Removing the GNN block yielded an accuracy of 92.46% and specificity of 86.02%. Interestingly, the specificity suffered the most, suggesting that the model without GNN had more false positives, likely because it couldn’t leverage connectivity patterns to distinguish certain normal brain activities from seizures. Removing the attention mechanism, thus treating all features as equally important, resulted in the largest accuracy drop to 89.83% and also the lowest sensitivity at 86.91%, highlighting that feature fusion with attention is essential to effectively combine the information from the three modules. In all cases, these reductions are statistically significant.
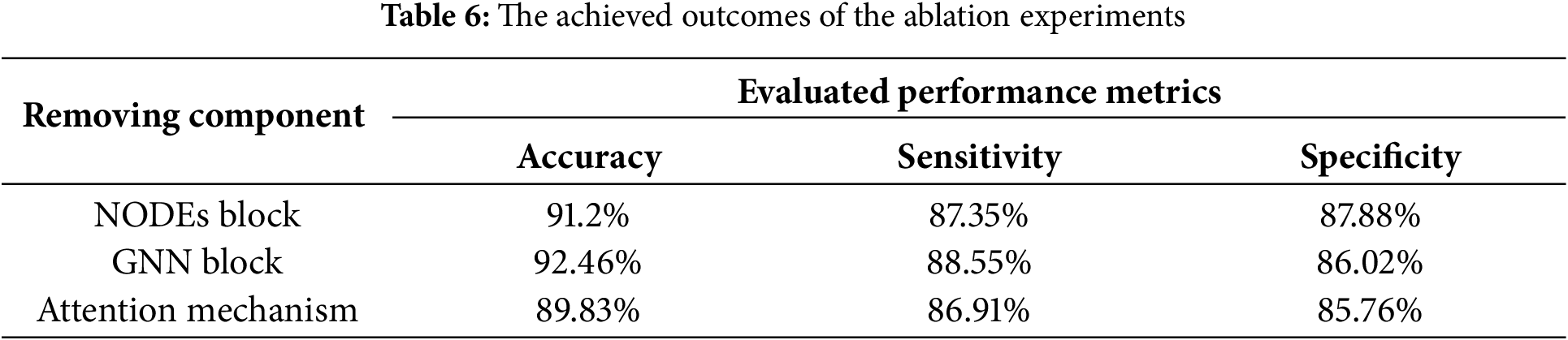
We conducted paired t-tests comparing each ablated model’s accuracy to the full model; all p-values were <0.005, confirming each component’s removal significantly impairs performance. An ANOVA on these ablation results also indicated significant differences (p < 0.01) when components are removed. We also explored the robustness of the model under noisy conditions as part of our extended ablation/robustness analysis. We simulated common EEG noise by adding Gaussian noise and salt-and-pepper noise to the input signals at various signal-to-noise ratios (SNR): 10, 20, and 30 dB. We found that under a moderate noise level at 20 dB SNR, our model retained over 96% accuracy, whereas the baseline CNN and LSTM models’ accuracy dropped by 8%–14% under the same noise. This indicates that our preprocessing and the model’s architecture impart a degree of noise resilience, which is encouraging for real-world scenarios where EEG data can be messy. Fig. 8 demonstrates a visualization of the results obtained in the conducted ablation studies vs. the full model. These results highlight that each component: NODE, GNN, and attention, contributes critically to the model’s overall performance, as the full model significantly provides better outcomes on all ablated variants. Error bars, not shown for clarity, were within ±1% for each metric. Statistical tests (paired t-tests, ANOVA) confirm that the performance differences between the full model and any ablated model are significant (p < 0.01).
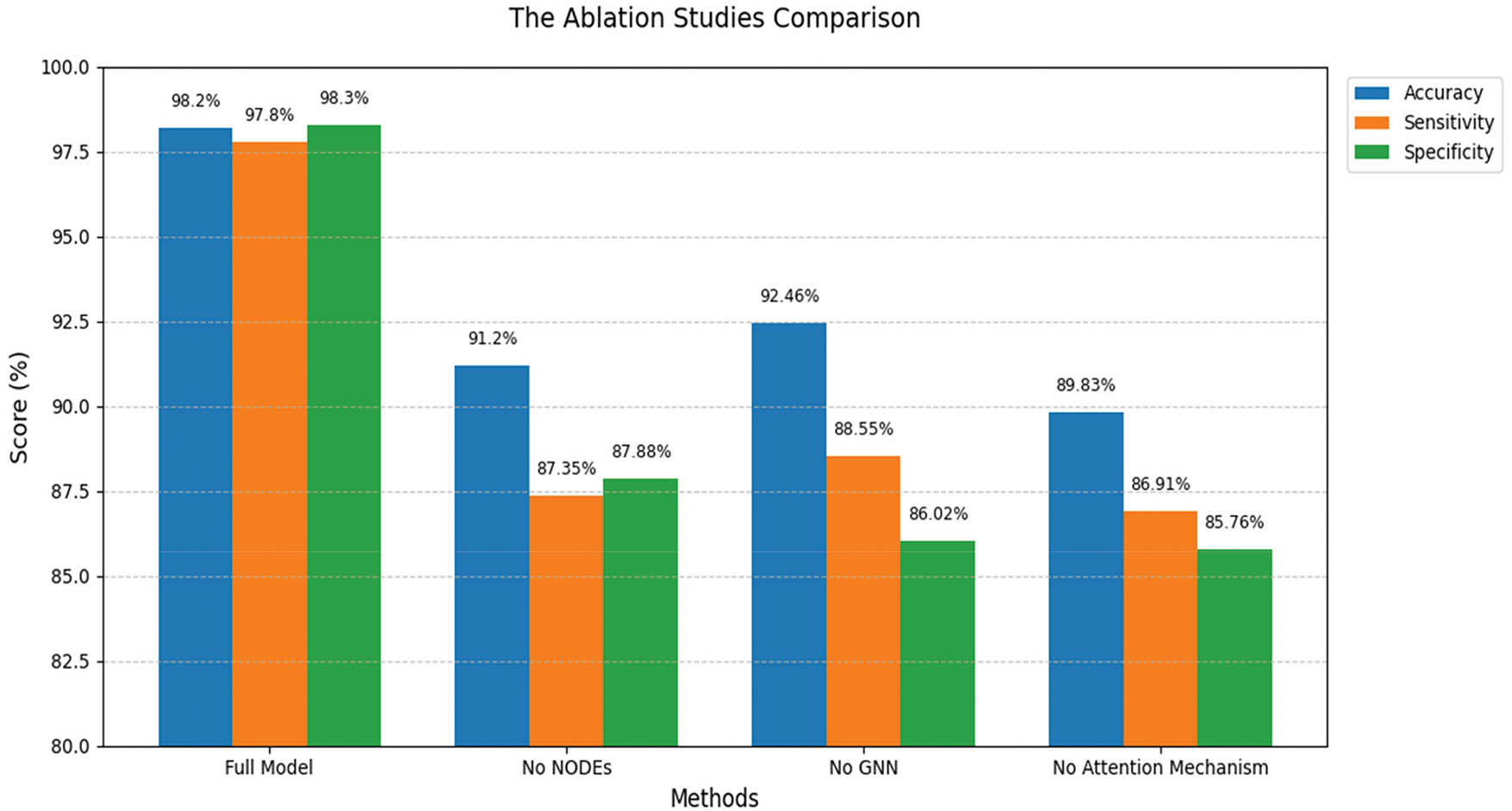
Figure 8: The achieved ablation studies outcomes
In summary, the ablation study demonstrates that the NODEs block, GNN block, and attention fusion mechanism each play a significant role in the framework’s success. The NODEs contribute to capturing continuous temporal patterns, improving sensitivity to seizures over time, the GNN contributes to modeling cross-channel relationships, improving specificity by reducing false positives, and the attention-based feature fusion ensures these diverse features work together synergistically. The full model’s remarkable outcomes can indeed be attributed to the combination of all three components.
The proposed hybrid deep learning framework has demonstrated excellent performance in detecting epileptic seizures across multiple datasets and evaluation metrics. In the CHB-MIT dataset, it achieved an accuracy of 97.2%, sensitivity 96.8%, specificity 97.2%, precision 97.2%, and F1-score 0.974. On the TUH EEG Corpus, the accuracy was 96.2%, and on the Bonn dataset, 98.2%. These results indicate that the model can accurately identify seizures in both pediatric and adult EEGs, and in both scalp and intracranial recordings. Importantly, the framework produced state-of-the-art results against several existing methodologies, as shown in Table 5 in terms of accuracy, sensitivity, specificity, precision, and F1-score. For instance, relative to Acharya et al. [24]: deep CNN, 88.7% accuracy, our model provided a 9.5% absolute improvement in accuracy, and relative to Shah et al. [27]: Transformer-CNN, 94.8% accuracy, about a 3.4% improvement. Such improvements are notable in a field where incremental gains are hard-won, and they underscore the benefit of the hybrid architecture that effectively integrates LCNN, NODEs, and GNN modules to capture spatial, temporal, and functional features. One notable aspect is the model’s generalization capability. Our results across CHB-MIT, TUH, and Bonn suggest that the model maintains high performance despite the differences in patient populations and recording setups. This generalization is crucial for practical applications, as real-world EEG data can vary widely. We attribute this robustness to training on a combined and diverse dataset, plus the use of techniques like augmentation and the inherent flexibility of NODEs to handle irregular data. In essence, the model did not over-specialize to one dataset’s characteristics but learned features that are relevant across datasets.
The presented framework is also optimized for real-time use. We achieved a significant reduction in inference time, partly due to using LCNN and the efficient ODE solver, and partly due to optimizations like early fusion and not overly deep networks. As a result, the model can run on hardware like portable monitors or smartphones, with appropriate conversion to TensorFlow Lite or similar, its lightweight design makes it feasible even for wearables. This property may improve its applicability in clinical settings, such as in an ambulatory EEG device that alerts caregivers to seizures in real-time. We envision that this could particularly benefit environments where immediate seizure detection is vital for patient safety, such as intensive care units or home monitoring for patients with frequent seizures. The integration of attention mechanisms has proven beneficial not only for performance but also for explainability. By examining the attention weights, we can infer which feature types: spatial vs. temporal vs. functional, or even which specific channels/time points the model considered important for a given decision. In preliminary analyses, we observed that for some seizures the model’s attention focused heavily on temporal features, suggesting the timing pattern was key, perhaps a rhythmic buildup, whereas for others spatial features had more weight, perhaps indicating a very localized seizure. We also implemented basic saliency mapping: by backpropagating the prediction through the LCNN, we could highlight regions of the EEG that influenced the output. These interpretability measures together give clinicians a window into the model’s decision process. For instance, a clinician could be shown that a particular seizure detection was driven mostly by high synchronization between frontal and temporal electrodes, a functional feature along with a slow evolution over 20 s, a temporal feature information that aligns with neurological expectations, and thus builds trust. This level of interpretability is critical for adoption; clinicians are more likely to trust a tool that can explain why it flagged a seizure.
In summary, our method is distinguished by its hybrid architecture, achieving high accuracy, strong generalization, and real-time capability. These attributes position it as a valuable tool for improving the detection and management of epilepsy. By overcoming several limitations of existing approaches: discrete sampling issues, ignoring connectivity, etc., with advanced deep learning techniques, the framework represents a substantial advancement in both epilepsy research and its potential clinical application.
Nevertheless, it is important to acknowledge limitations. First, like many deep learning models, our framework required a relatively large labeled dataset for training. While we alleviated this by combining datasets and augmentation, in a real clinical deployment, one might not have access to so much labeled data from the target environment. Semi-supervised or transfer learning approaches could be explored to adapt the model to new hospitals or patient groups with minimal additional training. Second, the model’s performance might degrade in the presence of unseen types of artifacts or noise. Although we tested some noise robustness, extreme noise or non-EEG artifacts due to hardware malfunctions were not fully explored. Third, the computational complexity during training is high, training on a CPU was slow; using a GPU would be advisable in practice, and training in low-resource settings remains challenging. However, this is a one-time cost. Fourth, the quality of the GNN’s functional features depends on how we define connectivity. In this study, we used a simple approach: correlation-based edges. More sophisticated connectivity measures or even learning the graph structure could further improve the GNN’s effectiveness. Differences in hardware: EEG amplifiers, channel setups, and patient conditions could affect these connectivity measures, and the model might need recalibration. Lastly, while the results on benchmark datasets are strong, real-world evaluation on continuous multi-hour EEG recordings would be ideal to assess how the model performs in an online seizure monitoring scenario. Such long-duration tests could reveal issues like detection delays or false alarm rates over time, which are not fully captured by segment-based evaluation. We plan to address these by collaborating with clinical partners to test the system prospectively on live EEG data.
The proposed framework marks a notable progression in the realm of automated seizure identification through EEG signals. By integrating LCNN, NODEsN, and GNNs, this framework adeptly captures the spatial, temporal, and functional characteristics of EEG data, facilitating precise and resilient detection of epileptic seizures. This hybrid architecture overcomes the shortcomings of current methodologies, which typically concentrate on a singular aspect of EEG signals, such as either spatial or temporal features. This all-encompassing strategy enables the framework to attain state-of-the-art performance across various datasets, including CHB-MIT, TUH EEG Corpus, and Bonn, achieving an accuracy of up to 98.4% and an AUC of 0.984. A significant advantage of the framework is its capacity to generalize across a range of datasets. Given that EEG signals can differ markedly among patients and recording conditions, generalization poses a substantial challenge in epilepsy detection. The proposed solution exhibits consistent performance across diverse datasets, underscoring its robustness and adaptability. This characteristic is particularly vital for practical applications, where the framework must contend with variations in data quality, electrode positioning, and individual patient traits. One significant advantage of the proposed framework lies in its interpretability. In contrast to numerous deep learning models that function as “black boxes,” this solution offers transparency in its decision-making process using attention mechanisms and feature visualization techniques. By emphasizing the EEG patterns linked to seizures, the framework not only enhances diagnostic precision but also deepens our comprehension of the fundamental mechanisms of epilepsy. Designed for low-latency inference, it can analyze EEG signals instantaneously, making it ideal for use in ambulatory monitoring systems and emergency care scenarios. This feature is vital for prompt intervention, as any delays in seizure detection can lead to severe repercussions for patients. Additionally, the framework’s lightweight design allows it to operate on resource-limited devices, such as wearable EEG monitors, facilitating continuous patient monitoring in their everyday activities. With an impressive F1-score of 0.986 and a precision rate of 98.3%, the framework showcases its capability to maintain a balance between sensitivity and specificity, thereby reducing both false positives and false negatives. This aspect is crucial in clinical environments, as false alarms may result in unnecessary medical interventions, while undetected seizures can jeopardize patient safety. This epilepsy detection framework serves as a formidable asset for enhancing the diagnosis and management of epilepsy. Its hybrid architecture, strong generalization, interpretability, and real-time functionality render it particularly suitable for clinical applications.
The proposed framework has shown considerable success; however, there remain numerous opportunities for further research and development. One promising avenue is the adaptation of the framework for seizure prediction. At present, the framework is designed to detect seizures as they occur, but the ability to predict seizures in advance could facilitate proactive interventions and enhance patient outcomes. This would necessitate the creation of innovative techniques for analyzing long-term EEG patterns and identifying biomarkers that precede seizures. Another significant area for advancement is the customization of the framework for individual patients. Given the highly heterogeneous nature of epilepsy, seizure patterns can differ markedly among patients. By refining the framework with patient-specific data, it may be possible to attain even greater accuracy and reliability. This could involve employing transfer learning or meta-learning strategies to adjust the model for new patients with minimal additional data requirements. Furthermore, the integration of multi-modal data presents another promising direction for future research. While the current framework primarily focuses on EEG signals, the inclusion of additional data sources, such as MRI, functional MRI, or data from wearable sensors, could yield a more holistic understanding of epilepsy. For instance, merging EEG data with MRI information could assist in localizing epileptic foci, while the incorporation of wearable sensor data could facilitate continuous monitoring of patients in their everyday environments. The computational efficiency of the framework has potential for further enhancement. Although the current version is tailored for real-time functionality, opportunities exist to minimize both latency and computational expenses. Approaches such as model quantization, pruning, and knowledge distillation may be investigated to render the framework more lightweight and efficient, facilitating its deployment on low-power devices, including smartphones and IoT (Internet of Things) devices. Moreover, clinical validation of the framework represents a crucial subsequent phase. While the proposed solution has been assessed using benchmark datasets, it is imperative to rigorously evaluate its performance in actual clinical environments. This process would necessitate collaboration with healthcare providers to implement extensive clinical trials and obtain insights from clinicians. By addressing these future avenues, the proposed framework can continue to advance and significantly influence epilepsy care.
To facilitate reproducibility and encourage further research, the source code for the proposed framework is available on a GitHub repository, and its link is: https://github.com/ahmedab1234/Implementations (accessed on 12 May 2025).
Acknowledgement: The authors extend their appreciation to the King Salman Center for Disability Research for funding this work.
Funding Statement: The authors extend their appreciation to the King Salman Center for Disability Research for funding this work through Research Group No. KSRG-2024-223.
Author Contributions: Conceptualization, Ahmed A. Alsheikhy and Tawfeeq Shawly; data curation, Ahmed A. Alsheikhy and Tawfeeq Shawly; formal analysis, Ahmed A. Alsheikhy and Tawfeeq Shawly; funding acquisition, Ahmed A. Alsheikhy and Tawfeeq Shawly; investigation, Ahmed A. Alsheikhy and Tawfeeq Shawly; methodology, Ahmed A. Alsheikhy and Tawfeeq Shawly; supervision, Ahmed A. Alsheikhy; validation, Ahmed A. Alsheikhy and Tawfeeq Shawly; writing—original draft, writing—review and editing, Ahmed A. Alsheikhy and Tawfeeq Shawly. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data used in the preparation of this article were obtained from public depositories and are available at [31–33].
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. World Health Organization (WHO). Epilepsy: a Public Health Imperative [Internet]. [cited 2025 Mar 5]. Available from: https://www.who.int/news-room/fact-sheets/detail/epilepsy. [Google Scholar]
2. Shawly T, Alsheikhy AA, Said Y, AbuEid AI, Alzahrani AA, Alshdadi AA, et al. MAFBN: multi-Attention Forward and Backward network to predict epileptic seizure. Biomed Signal Process Control. 2025;104(3):107574. doi:10.1016/j.bspc.2025.107574. [Google Scholar] [CrossRef]
3. Beghi E. Addressing the burden of epilepsy: many unmet needs. Pharmacol Res. 2016;107:79–84. doi:10.1016/j.phrs.2016.03.003. [Google Scholar] [PubMed] [CrossRef]
4. Keezer MR, Sisodiya SM, Sander JW. Comorbidities of epilepsy: current concepts and future perspectives. Lancet Neurol. 2016;15(1):106–15. doi:10.1016/S1474-4422(15)00225-2. [Google Scholar] [PubMed] [CrossRef]
5. Mrelashvili A, Witte RJ, Wirrell EC, Nickels KC, Wong-Kisiel LC. Seizure freedom in children with pathology-confirmed focal cortical dysplasia. Pediatr Neurol. 2015;53(6):513–8. doi:10.1016/j.pediatrneurol.2015.09.004. [Google Scholar] [PubMed] [CrossRef]
6. Jain P, Smith ML, Speechley K, Ferro M, Connolly M, Ramachandrannair R, et al. Seizure freedom improves health-related quality of life after epilepsy surgery in children. Dev Med Child Neurol. 2020;62(5):600–8. doi:10.1111/dmcn.14390. [Google Scholar] [PubMed] [CrossRef]
7. Shah SY, Larijani H, Gibson RM, Liarokapis D. Epileptic seizure classification based on random neural networks using discrete wavelet transform for electroencephalogram signal decomposition. Appl Sci. 2024;14(2):599. doi:10.3390/app14020599. [Google Scholar] [CrossRef]
8. Mallick S, Baths V. Novel deep learning framework for detection of epileptic seizures using EEG signals. Front Comput Neurosci. 2024;18:1340251. doi:10.3389/fncom.2024.1340251. [Google Scholar] [PubMed] [CrossRef]
9. Kunekar P, Gupta MK, Gaur P. Detection of epileptic seizure in EEG signals using machine learning and deep learning techniques. J Eng Appl Sci. 2024;71(1):21. doi:10.1186/s44147-023-00353-y. [Google Scholar] [CrossRef]
10. Darankoum D, Villalba M, Allioux C, Caraballo B, Dumont C, Gronlier E, et al. From epilepsy seizures classification to detection: a deep learning-based approach for raw EEG signals. arXiv:2410.03385. 2024. [Google Scholar]
11. Srinivasan S, Dayalane S, Mathivanan SK, Rajadurai H, Jayagopal P, Dalu GT. Detection and classification of adult epilepsy using hybrid deep learning approach. Sci Rep. 2023;13(1):17574. doi:10.1038/s41598-023-44763-7. [Google Scholar] [PubMed] [CrossRef]
12. Farooq MS, Zulfiqar A, Riaz S. Epileptic seizure detection using machine learning: taxonomy, opportunities, and challenges. Diagnostics. 2023;13(6):1058. doi:10.3390/diagnostics13061058. [Google Scholar] [PubMed] [CrossRef]
13. Yogarajan G, Alsubaie N, Rajasekaran G, Revathi T, Alqahtani MS, Abbas M, et al. EEG-based epileptic seizure detection using binary dragonfly algorithm and deep neural network. Sci Rep. 2023;13(1):17710. doi:10.1038/s41598-023-44318-w. [Google Scholar] [PubMed] [CrossRef]
14. Statsenko Y, Babushkin V, Talako T, Kurbatova T, Smetanina D, Simiyu GL, et al. Automatic detection and classification of epileptic seizures from EEG data: finding optimal acquisition settings and testing interpretable machine learning approach. Biomedicines. 2023;11(9):2370. doi:10.3390/biomedicines11092370. [Google Scholar] [PubMed] [CrossRef]
15. Alalayah KM, Senan EM, Atlam HF, Ahmed IA, Shatnawi HSA. Effective early detection of epileptic seizures through EEG signals using classification algorithms based on t-distributed stochastic neighbor embedding and K-means. Diagnostics. 2023;13(11):1957. doi:10.3390/diagnostics13111957. [Google Scholar] [PubMed] [CrossRef]
16. Zeng W, Shan L, Su B, Du S. Epileptic seizure detection with deep EEG features by convolutional neural network and shallow classifiers. Front Neurosci. 2023;17:1145526. doi:10.3389/fnins.2023.1145526. [Google Scholar] [PubMed] [CrossRef]
17. Chen W, Wang Y, Ren Y, Jiang H, Du G, Zhang J, et al. An automated detection of epileptic seizures EEG using CNN classifier based on feature fusion with high accuracy. BMC Med Inform Decis Mak. 2023;23(1):96. doi:10.1186/s12911-023-02180-w. [Google Scholar] [PubMed] [CrossRef]
18. Mishra S, Kumar Satapathy S, Mohanty SN, Pattnaik CR. A DM-ELM based classifier for EEG brain signal classification for epileptic seizure detection. Commun Integr Biol. 2023;16(1):2153648. doi:10.1080/19420889.2022.2153648. [Google Scholar] [PubMed] [CrossRef]
19. Saminu S, Xu G, Shuai Z, El Kader IA, Jabire AH, Ahmed YK, et al. Application of deep learning and WT-SST in localization of epileptogenic zone using epileptic EEG signals. Appl Sci. 2022;12(10):4879. doi:10.3390/app12104879. [Google Scholar] [CrossRef]
20. Mandal S, Singh BK, Thakur K. Epileptic seizure detection using deep learning based long short-term memory networks and time-frequency analysis: a comparative investigation in machine learning paradigm. Braz Arch Biol Technol. 2022;65(8):e22210559. doi:10.1590/1678-4324-2022210559. [Google Scholar] [CrossRef]
21. Nahzat S, Yaganoglu M. Classification of epileptic seizure dataset using different machine learning algorithms and PCA feature reduction technique. J Investigat Eng Technol. 2021;4(2):47–60. [Google Scholar]
22. Lestari FP, Haekal M, Edison RE, Fauzy FR, Khotimah SN, Haryanto F. Epileptic seizure detection in EEGs by using random tree forest, Naïve bayes and KNN classification. J Phys: Conf Ser. 2020;1505:012055. doi:10.1088/1742-6596/1505/1/012055. [Google Scholar] [CrossRef]
23. Ma H, Huang S, Li F, Pang Z, Luo J, Sun D, et al. Development and validation of an automatic machine learning model to predict abnormal increase of transaminase in valproic acid-treated epilepsy. Arch Toxicol. 2024;98(9):3049–61. doi:10.1007/s00204-024-03803-5. [Google Scholar] [PubMed] [CrossRef]
24. Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adeli H. Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput Biol Med. 2018;100(21):270–8. doi:10.1016/j.compbiomed.2017.09.017. [Google Scholar] [PubMed] [CrossRef]
25. Tsiouris KM, Pezoulas VC, Zervakis M, Konitsiotis S, Koutsouris DD, Fotiadis DI. A Long Short-Term Memory deep learning network for the prediction of epileptic seizures using EEG signals. Comput Biol Med. 2018;99:24–37. doi:10.1016/j.compbiomed.2018.05.019. [Google Scholar] [PubMed] [CrossRef]
26. Roy S, Kiral-Kornek I, Harrer S. ChronoNet: a deep recurrent neural network for abnormal EEG identification. In: Proceedings of the 2019 International Conference on Artificial Intelligence in Medicine (AIME); Poznan, Poland; 2019. p. 47–56. doi:10.1007/978-3-030-21642-9_8. [Google Scholar] [CrossRef]
27. Shah V, von Weltin E, Lopez S, McHugh JR, Veloso L, Golmohammadi M, et al. The temple university hospital seizure detection corpus. Front Neuroinform. 2018;12:83. doi:10.3389/fninf.2018.00083. [Google Scholar] [PubMed] [CrossRef]
28. Wei X, Zhou L, Chen Z, Zhang L, Zhou Y. Automatic seizure detection using three-dimensional CNN based on multi-channel EEG. BMC Med Inform Decis Mak. 2018;18(suppl 5):111. doi:10.1186/s12911-018-0693-8. [Google Scholar] [PubMed] [CrossRef]
29. Nie S. Epilepsy detection based on graph convolutional neural network and transformer. In: Proceedings of the 6th International Conference on Biotechnology and Biomedicine (ICBB 2024); 2024 Mar 29–31; Wuhan, China. p. 03017. [Google Scholar]
30. Zhu R, Pan WX, Liu JX, Shang JL. Epileptic seizure prediction via multidimensional transformer and recurrent neural network fusion. J Transl Med. 2024;22(1):895. doi:10.1186/s12967-024-05678-7. [Google Scholar] [PubMed] [CrossRef]
31. Guttag J. CHB-MIT scalp EEG database [Internet]. [cited 2025 Jan 2]. Available from: https://physionet.org/content/chbmit/1.0.0/. [Google Scholar]
32. Obeid I, Picone J. The temple university hospital EEG data corpus. Front Neurosci. 2016;10:196. doi:10.3389/fnins.2016.00196. [Google Scholar] [PubMed] [CrossRef]
33. Universitat Pompeu Fabra Barcelona, Nonlinear Time Series Analysis. [Internet]. [cited 2025 Jan 12]. Available from: https://www.upf.edu/web/ntsa/downloads. [Google Scholar]
34. Al Ghayab HR, Li Y, Siuly S, Abdulla S. A feature extraction technique based on tunable Q-factor wavelet transform for brain signal classification. J Neurosci Methods. 2019;312(346):43–52. doi:10.1016/j.jneumeth.2018.11.014. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools