 Open Access
Open Access
ARTICLE
Early-Stage Cervical Cancerous Cell Detection from Cervix Images Using YOLOv5
1 Department of Software Engineering (SWE), Daffodil International University (DIU), Sukrabad, Dhaka, 1207, Bangladesh
2 Group of Biophotomatiχ, Department of Information and Communication Technology, Mawlana Bhashani Science and Technology University, Santosh, Tangail, 1902, Bangladesh
3 Department of Electrical and Computer Engineering, University of Saskatchewan, 57 Campus Drive, Saskatoon, S7N5A9, SK, Canada
4 Computer Engineering Department, Umm Al-Qura University, Mecca, 24381, Saudi Arabia
5 Department of Computer Science, American International University-Bangladesh (AIUB), Kuratoli, Dhaka, 1229, Bangladesh
6 College of Engineering, IT and Environment, Charles Darwin University, Casuarina, 0909, NT, Australia
* Corresponding Authors: Kawsar Ahmed. Email: ,
Computers, Materials & Continua 2023, 74(2), 3727-3741. https://doi.org/10.32604/cmc.2023.032794
Received 30 May 2022; Accepted 26 August 2022; Issue published 31 October 2022
Abstract
Cervical Cancer (CC) is a rapidly growing disease among women throughout the world, especially in developed and developing countries. For this many women have died. Fortunately, it is curable if it can be diagnosed and detected at an early stage and taken proper treatment. But the high cost, awareness, highly equipped diagnosis environment, and availability of screening tests is a major barrier to participating in screening or clinical test diagnoses to detect CC at an early stage. To solve this issue, the study focuses on building a deep learning-based automated system to diagnose CC in the early stage using cervix cell images. The system is designed using the YOLOv5 (You Only Look Once Version 5) model, which is a deep learning method. To build the model, cervical cancer pap-smear test image datasets were collected from an open-source repository and these were labeled and preprocessed. Then the YOLOv5 models were applied to the labeled dataset to train the model. Four versions of the YOLOv5 model were applied in this study to find the best fit model for building the automated system to diagnose CC at an early stage. All of the model’s variations performed admirably. The model can effectively detect cervical cancerous cell, according to the findings of the experiments. In the medical field, our study will be quite useful. It can be a good option for radiologists and help them make the best selections possible.Keywords
Cancer is a condition when aberrant cells divide uncontrollably and infect neighboring tissues [1,2]. Cervical cancer (CC) is not different from the legislation of cancer. It originates in the woman’s cervix. At first, it attacks any cell of the cervix then it spreads to other cells. It is one of the most malignant and critical cancers throughout the world. Many lives have been falling for this deadly disease. Human papillomaviruses (HPV) are responsible for developing this disease. According to the National Cancer Institute, there are about 14 high-risk HPV types [3]. It ranks fourth on the list of deadly diseases among women. In the year 2018, 570 000 women were diagnosed all over the world where 311 000 women died from cancer [4]. CC is distributed differently over the world, with more than 85 percent of deaths taking place in underdeveloped countries [5]. CC is the most common in Sub-Saharan Africa, accounting for more than 90% of all cases and 70% of cervical cancers occur in developing countries [6]. HIV (Human Immunodeficiency Virus) affected women must be aware of this cancer because these patients are significantly vulnerable to getting infected by cancer [7].
Since the affected women belong to lower or middle-class families for this reason they are not properly concerned about the disease. Even they are not financially capable enough for the treatment of cancer though the disease is very malignant. CC begins in the cervix and has no symptoms until it is accelerated [8]. When it is examined under a microscope, it has two forms [9]. Squamous cell carcinoma is the first form, and it begins in the cells that line the bottom of the cervix. Adenocarcinoma is another type of cancer that originates in gland-related cells in the upper section of the cervix [10]. According to the Cancer.Net Editorial Board, there are some types of tests to observe cervical cancer, such as Pap test, HPV typing test, Biopsy, Molecular testing of the tumor, Cystoscopy, and Sigmoidoscopy (also called a proctoscopy), etc. [11]. All of the treatments are very costly and time-consuming for this reason lower-class and middle-class families are unable to bear the cost of treatments.
Despite being a deadly disease, CC can be cured if it can be detected in the early stages [12,13]. For clinical initiative, it is a very crucial task to identify the cancerous cell which is known as a cyst [14]. Detecting cyst from cervix images ensures clinicians for patients regarding CC. However, it is a challenging task to identify the infected cells. Machine learning (ML) is developing as a potential method for the development of complicated multi-parametric decision algorithms in a variety of domains of science [15–17]. To address the issue, the study is designed to detect the cyst to ensure CC. To fulfill the objective, YOLO (You Only Look Once) architecture was employed in this study to build the expected model. To get the desired model, we applied YOLOv5 models. At first, we collected cervix images and applied preprocessing techniques to make the dataset fit for the YOLOv5 models. Then we applied four versions of YOLOv5 model and compared them to find the best model as our desired approach to detect cancerous cell in initial phase.
In recent years, a variety of studies have been conducted by researchers throughout the world to detect and predict cervical cancers using different approaches. William et al. [18] applied some machine learning classification techniques to the Trainable Weka Segmentation toolkit, such as feature extraction, feature selection, and defuzzification. Park et al. 2021 employed cervicography images to compare deep learning and machine learning models to detect cervical cancer and found that the deep learning approach, ResNet-50, generated significantly greater performance compared to other machine learning models [19]. The form and texture features of the segmentation and classification approach, as well as Gabor properties, are used to classify cervical cancer cells. Both normal and cancer cell classification were shown to have a higher level of accuracy [20]. To extract deep learning features from cervical pictures, a method based on CNN (Convolutional Neural Network) was devised by Goncalves et al. [21] to identify and classify early cervical cancer cells. The input photos were categorized using the extreme learning machine (ELM). They considered the CNN paradigm for fine-tuning and transfer learning to detect CC. Sharma et al. in 2016 [22] tested the KNN (K-Nearest Neighbor) approach for cervical cell categorization. These approaches rely largely on cytopathologists’ expertise in defining handcraft characteristics, as well as correct cell and nuclei segmentation. However, their obtained accuracy is not satisfactory at all to detect a hazardous disease like cervical cancer.
Ali et al. 2021 proposed a machine learning model to predict cervical cancer using four class attributes Biopsy, Schiller, Cytology, and Hinselmann [23]. They applied three feature transformation (FT) methods and five classifiers. It is found from the study that for Biopsy prediction logarithmic FT methods and Random Tree (RT) outperformed, where sine function FT methods and RT for Cytology, sine function FT methods and RF for Hinselmann, and ZScore FT methods and IBk classifier for Schiller prediction. Kuko et al. [24] compared machine vision, ensemble learning, and deep learning methods and showed that the ensemble outperformed with 91.6% accuracy. Bethesda system was used for classification and cell clusters for reporting cervical cytology. Image regeneration and data classification using a variational convolutional autoencoder network was proposed by Khamparia et al. in 2021 [25]. The authors proposed hybrid convolutional and variational encoder methods. Alkhawaldeh et al. [26] expressed some machine learning and deep learning algorithms and approaches. The DNN (Deep Neural Network) model was designed for providing risk scores for cervical cancer.
To growth of cervical cancer cells and controlling angiogenic factors Jianfang et al. [27] proposed Chaihu-shugan-san and paclitaxel+cisplatin method. Chitra et al. [28] applied different kinds of augmentation techniques named MASO-optimized DenseNet 121 architecture for cervical cancer detection and this was performed well. Elakkiya et al. [29] Used Small-Object Detection (SOD), Generative Adversarial Networks (GAN) with Fine-tuned Stacked Autoencoder (F-SAE). SOD-GAN obtained good accuracy and less loss. Arezzo et al. [30] applied some machine learning algorithms, Logistic Regression (LR), Random Forest (RFF), and K-nearest neighbors (KNN). Among these methods, RFF performed the best accuracy, precision, and recall. Here also used feature selection to ordain quality core set of CC.
From the above-mentioned discussion, it can be said that more efficient and advanced technology should be introduced to detect cancerous cells and detect cervical cancer using cervix images. From this perspective, the study aims to find an efficient version of YOLOv5 architecture and build an effective model to detect cysts and CC.
In this experiment, Python (version 3.8.5) was applied to conduct all the analyses. The flow chart of the research methodology is illustrated in Fig. 1. In addition to that, all the methods are described briefly in this section.
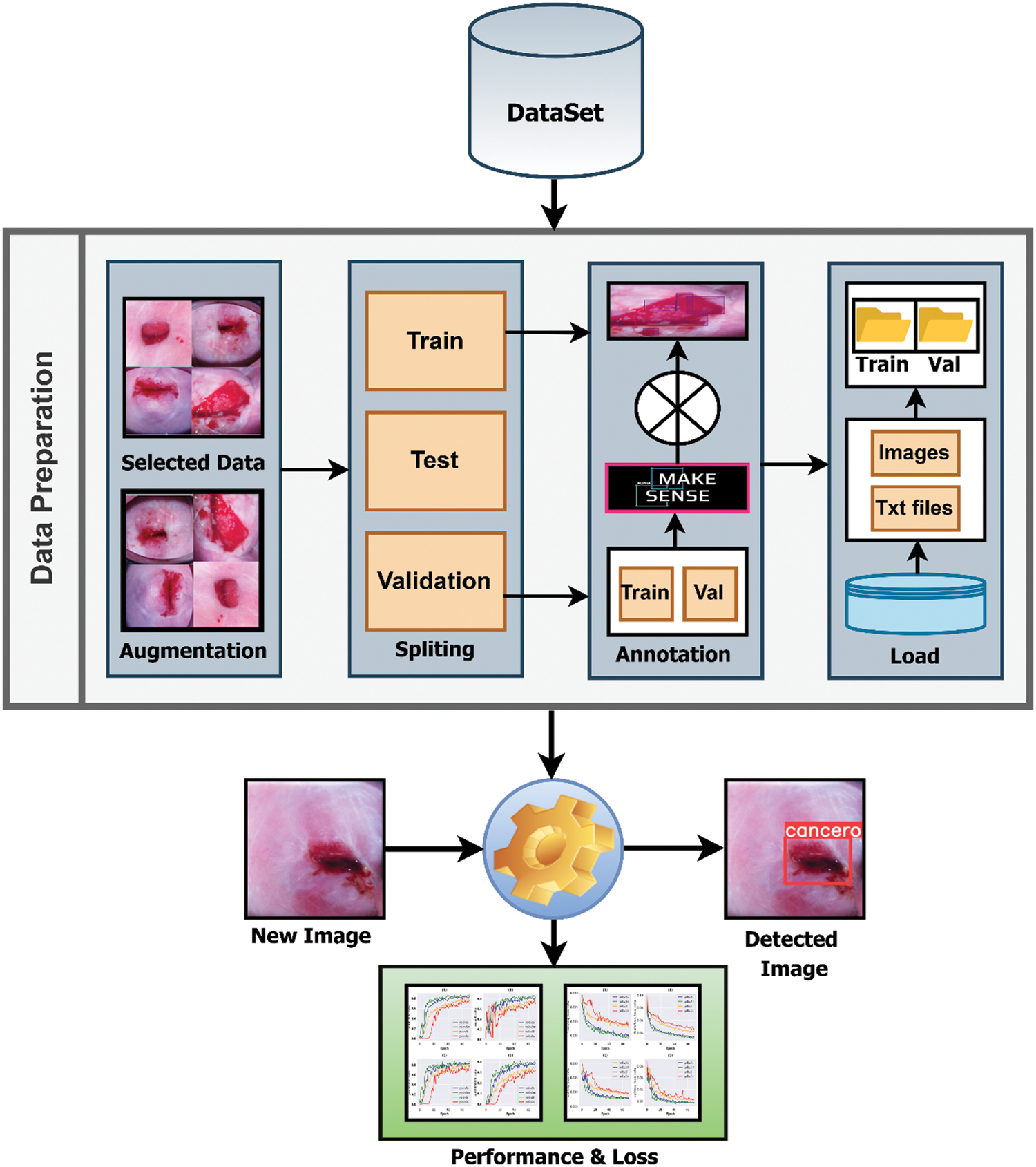
Figure 1: Pipeline of the research methodology
3.1 Data Collection & Preprocessing
A cervix image dataset was collected from an online repository called Kaggle [31]. The dataset contains pap-smear test image data. Pap (cervical) smear testing is an excellent tool for identifying, preventing and delaying cervical cancer progression [32]. The Papanicolaou test is a cervix and colon screening treatment that can detect precancerous and cancerous processes. If abnormal results are found, more sensitive diagnostic procedures are used, as well as, taking relevant treatments to prevent developing CC to its afflux. There were 781 in the dataset, from there 395 clean and fresh images were collected for further study based on the quality of the images. These images are suitable for performing any deep learning task.
After collecting the dataset, different augmentation techniques were applied to increase the number of images and add images from different dimensions. The applied augmentation techniques are random rotation, height and width shift, shearing transformations, random zoom and horizontal shift. After augmentation, the number of images was 1185. Then these images were split into 3 parts such as 638 for training, 274 for validation, and 273 for test data. The validation dataset was used to illustrate how models are performing after hyper parameter tuning and data preparation. On the other hand, the test dataset was used to describe a final tuned model’s evaluation when compared to other final models. After splitting the three parts, the images were labeled and annotated to prepare for YOLOv5 architecture. Data labeling is the process of identifying raw data and providing context by adding one or more relevant and useful labels. Data annotation is the process of labeling data so that machines may use it. It’s particularly beneficial for supervised learning, in which the system uses labeled datasets to interpret, understand, and learn from input patterns to produce desired outputs. Without data labeling and annotation models cannot identify objects properly so this step is very important. All of the train and validation data were labeled and annotated using an online tool called makesense.ai [33]. Fig. 2 shows the data annotation process. First, upload the raw images and complete the process of labeling. After that exported these labeled images as txt files belonging to the same name of those images.
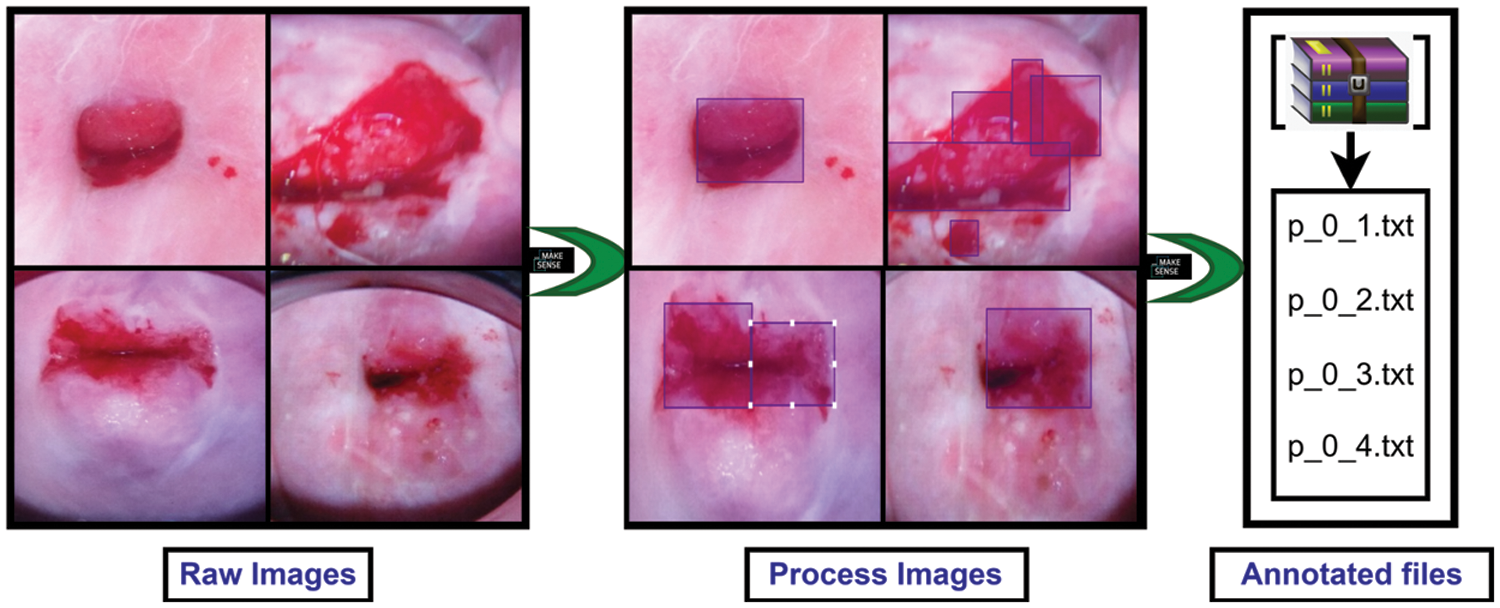
Figure 2: Preprocessing steps for preparing data for the applied models
YOLOv5 models were imported from the GitHub repository. Then set YAML file in the data folder that belongs to the YOLOv5 model folder, which gives the path of training and testing data with its respective images and labels. Google Colab open-source GPUs (Graphics processing units) are employed to train and test the model. Input image size was fixed 640, and all the versions of YOLOv5 models such as YOLOv5s (You Only Look Once Version 5 s), YOLOv5m (You Only Look Once Version 5 m), YOLOv5l (You Only Look Once Version 5 l), and YOLOv5x (You Only Look Once Version 5x) were employed to build the desired model and compared their performance. For training the model, the number of epochs was set to 70 along with 16 batch sizes. Then the models are trained and validated using a validation dataset. To compare their performance, a test dataset was applied and performance evaluation results were generated to find the best fit and most efficient versions of YOLOv5 architectures. The parameters used in this study for building YOLOv5 is mentioned in Table 1.
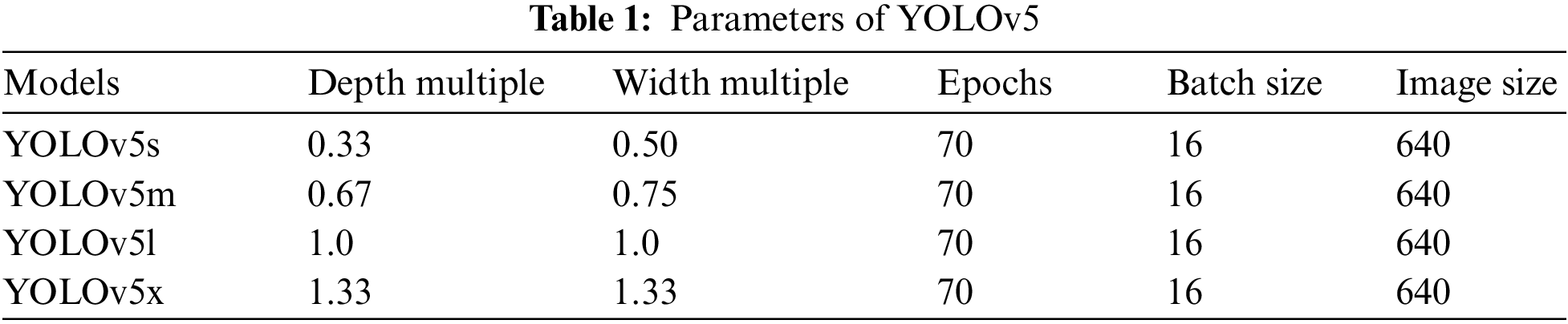
Table 1 shows all the parameters of the applied models of YOLOv5. From the above table, it is found that same number image_size, batch_size, and epochs are employed for all of the versions. However, depth_multiple and width_multiple is changed accordingly, which are represented in Table 1.
3.3 YOLOv5 Model & Architecture
In recent times, object detection is an important task in deep learning. Object detection, a subset of computer vision, is a method for automatically detecting interesting things in an image about the background. YOLO is a popular algorithm of deep learning to detect real-time objects. It is a One-Stage Method algorithm. The latest version of YOLO is YOLOv5. It’s a collection of compound-scaled object identification models trained on the COCO (Common Objects in Context) dataset, with easy capabilities for TTA (Test Time Augmentation), model assembly, hyperparameter development, and export to ONNX (Open Neural Network Exchange), CoreML (Core Machine Learning), and TFLite (TensorFlow Lite) Model. It was also the first time a YOLO model was constructed entirely within PyTorch, allowing for quicker FP16 (Floating Point 16) training and quantization-aware training (QAT). The new YOLOv5 features resulted in quicker and more accurate models on GPUs, but they also increased the complexity of CPU (Central Processing Unit) deployments. It outperforms other YOLO models, which achieve 90.8 percent recall and 7.8 percent FPR when paired with unique pre-and post-processing processes, and 9.1 percent recall and 10.0 percent FPR when not completed. It is the Leader in Real-time Object Detection. YOLO is an object detection model that integrates a bounding box to predict and classify an object into a single end-to-end differentiable network. Darknet was used to write it and is still used to maintain it. YOLOv5 is initially implemented in PyTorch, and it benefits from the PyTorch ecosystem established to support and deploy, it is much more lightweight and easier to use. Iterating on YOLOv5 may also be easier for the broader research community, given it is a more well-known study paradigm [33]. It can detect any object very fast and easily. This model has 4 versions: YOLOv5-s, YOLOv5-m, YOLOv5-l, and YOLOv5-x, and these are applied and compared in this experiment. It has three main architectural blocks: Backbone, Neck, and Head. Fig. 3 shows the full architecture form of the YOLOv5 model.
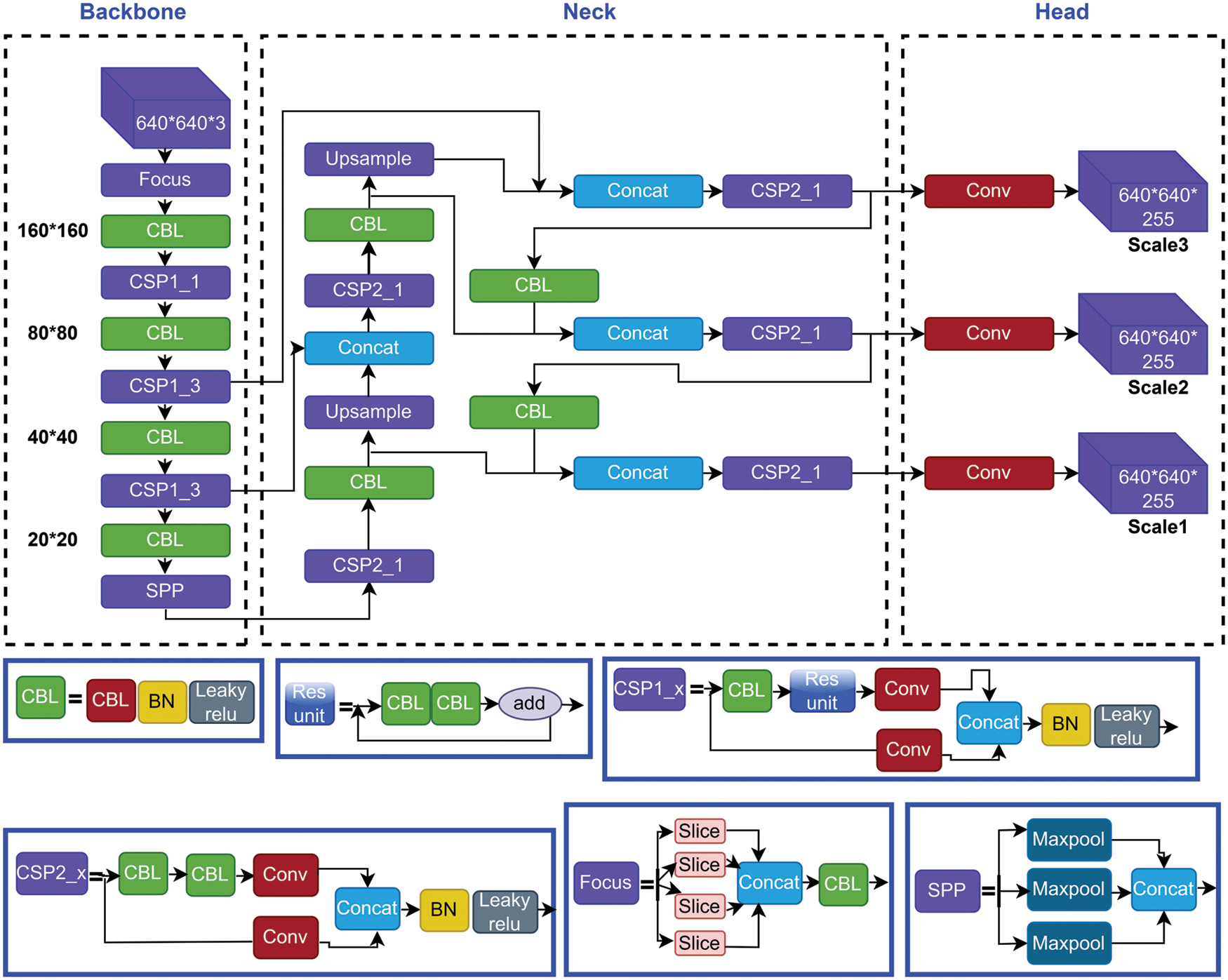
Figure 3: Structural architecture network of yolov5 models
The basic purpose of Backbone is to extract key features from the input image [21]. Cross Stage Partial Networks (CSPNet) and Focus are employed as a backbone in YOLOv5 to select effective prominence from an input image. CSPNet overcomes the problem of repeating network optimization gradient information in the backbone network and reduce the gradient information and develops CNN learning ability [34,35]. The base layer’s feature map is replicated by the backbone network, which then employs a dense block to send the copied feature map to the next level, separating the base layer’s feature map.
To improve the ability of network characteristic fusion, CSP2 structure is adopted here. The most common application of Neck is used to build feature pyramids [33]. Feature pyramids support models in achieving good object scaling generalization. It makes it easier to recognize the same item in different sizes and scales. The neck was created to optimize the benefits of the backbone’s characteristics. It is frequently made up of several bottom-up and top-down routes. The usage of an up and down sampling block is the earliest neck. It reprocesses and uses the feature maps extracted by Backbone at various stages in a reasonable manner. The usage of an up and down sampling block is the earliest neck. The target detection architecture relies heavily on the neck. This approach differs from SSD (single-shot detector) [36] in that it does not use a feature layer aggregation process.
The Head is basically in charge of the last detection procedure. After anchor boxes are applied to features, it generates final output vectors with class probabilities and bounding boxes [37]. The head is responsible for determining the object’s location and category using characteristics maps collected from the backbone. There are two types of heads: one-stage object detectors and two-stage object detectors. The RCNN (Region-based Convolutional Neural Network) series is the most representative of two-stage detectors, which have long been the dominating method in the field of object detection. The head model in the YOLOv5 model is the same as in the Yolov3 and Yolov4 models, and it is mostly utilized in the final inspection stage [38]. After applying anchor boxes to the feature map, it generates a final output vector comprising class probabilities, object scores, and bounding boxes. Choosing an activation function for deep learning networks is crucial, according to YOLOv5.
An artificial neural network’s activation function calculates a weighted sum and then adds bias to it to determine whether a neuron should be activated or not. The goal of this function is to add non-linearity to the output of a neuron. There are many kinds of activation functions. Activation functions are the most crucial part of any deep neural network. Activation features such as Leaky ReLU (Rectified Linear Unit), Sigmoid, mish, swish, and others have recently been implemented. YOLOv5 uses Leaky ReLu and Sigmoid activation functions. Leaky ReLu used in applications involving sparse gradients, such as training generative adversarial networks. It is employed in the middle/hidden layers of YOLOv5. On the other hand, the sigmoid activation function is used in the final detection layer of YOLOv5.
It is expressed as a distance metric on the space of forms, rather than on the space of regions. In severely uneven settings, it may be simply paired with typical regional losses to solve the segmentation job. Furthermore, the suggested term may be used to solve any N-D segmentation problem in any current deep network architecture. The IoU (Intersection over Union) [36] is a typical target detection indicator whose major role is to determine the positive and negative samples as well as to calculate the distance between the output box and the proper label. IoU is scale-invariant, which means it is unaffected by changes in scale. Because it employs integrals across the border (interface) between regions instead of unbalanced integrals over regions, it helps alleviate the issues of regional losses in the setting of extremely unbalanced segmentation problems. The loss function of the Bounding box in YOLOv5 is GIOU Loss, which is as follows.
Optimizers are the way for reducing losses by adjusting the features of neural networks, like weights and learning rate. The purpose of optimization is to create the best and most feasible design based on a set of prioritized criteria or restrictions. These include increased production, strength, reliability, lifespan, efficiency, and usage, among other things. YOLOv5 has two optimization functions and these are Stochastic Gradient Descent (SGD) & Adaptive Moment Estimation (Adam). The weights were adjusted during the backpropagation phase to minimize the error between the actual output and the expected output using the delta rule. The backpropagation is repeated until the output error is as low as possible. Optimization methods are used in a variety of machine learning applications [37]. In YOLOv5, SGD is the default optimization function for training. Adam works with first and second-order momentums, as well as an exponentially decaying average of previous gradients [38]. It uses less memory and is better suited to issues with a lot of noise or sparse gradients.
3.7 Model Evaluation & Exhibitor
Evaluation metrics were used to assess the quality of a statistical or machine learning model [39]. Any endeavor will necessitate a review of machine learning models or algorithms. Several assessment measures are employed to put a model to the test. To ensure that a model is performing efficiently and optimally, evaluation metrics are considered. A confusion matrix is derived from the testing result and notation as True Positive (TP), True Negative (TN), False Positive (FP), and False Negative (FN). Based on these notations, precision and recall values are calculated to evaluate a model. A good precision measures the good number of results because it is the attribute of being precise, and it refers to the distance between two or more measures, whether or not they are correct as well as denotes the proportion of Predicted Positive cases that are correctly Real Positives and reflects how repeatable measurements are, even if they are far from the accepted value. Recall indicates how well the projected positive rule covers the Real Positive situations. It has the advantage of reflecting how many relevant situations the expected positive rule picks up. It refers to the percentage of total relevant results that our system accurately classifies. The precision and recall are calculated using the following Eqs. (4) and (5) respectively [40,41].
For further evaluation of a deep learning model, average precision (AP) and mean average precision (mAP) play a vital role. The (AP) shows whether the model can accurately identify all positive cases without mistakenly labeling too many negative ones as positive. It was used as a metric for determining object detector accuracy, and it computed the average precision value for recall values ranging from 0 to 1. mAP is used as an object detection assessment metric. The mAP is used to evaluate the object detection models. Depending on the multiple detecting challenges that exist, it is determined by calculating the mean AP across all courses and/or overall IoU thresholds. AP and mAP are calculated using the following Eqs. (6) and (7) respectively [42–45].
Here, APk refers the average precision of the class k and n refers to the number of classes.
In this exploration, all the versions of this model are applied to train and test to compare their performances to find the best-fit version. This study considered precision and recalls to assess the algorithm’s efficacy in detecting cancer cells. mAP, box loss, object loss, and cls loss also were considered to evaluate the applied models. PyTorch framework on GPU with CUDA (Compute Unified Device Architecture) environment from google collaboratory was used for the experiment.
Table 2 demonstrates the performance generated by the applied models. The table demonstrates that YOLOv5x and YOLOv5l gained lower precision and recall compared to YOLOv5s and YOLOv5m, though the recall value of YOLOv5l and YOLOv5x is equal to each other. Overall, YOLOv5m outperformed compared to all the applied YOLO models with 88.4% precision and 86.4% recall value for the applied dataset.
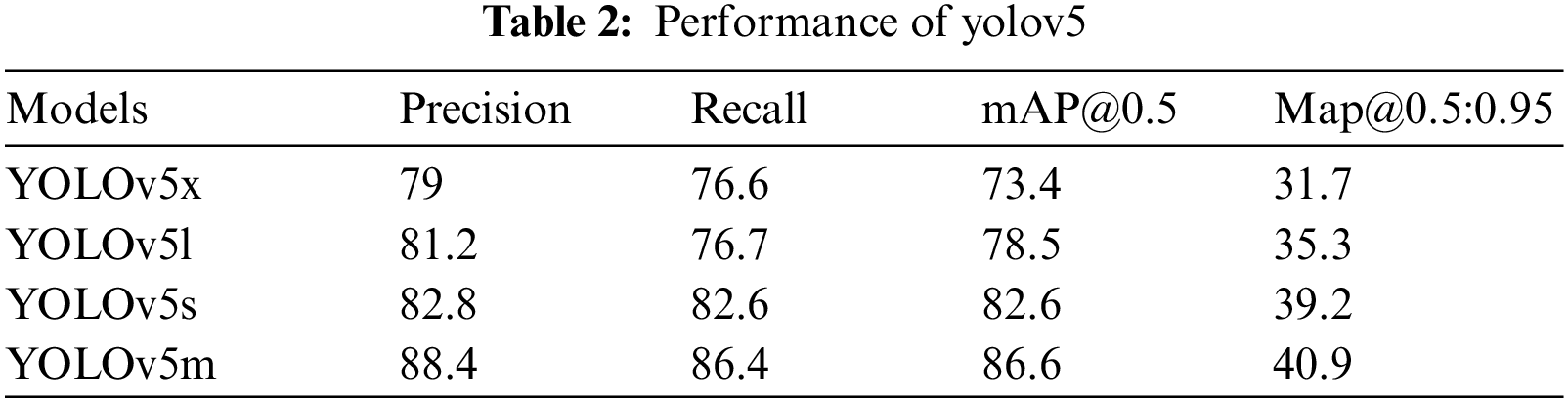
Fig. 4 depicts the performance results of the applied models. This figure represents precision, recall, and mean average precision (mAP_0.5 and mAP0.5:0.95). According to the figure, YOLOv5l performed better than YOLOv5x, and YOLOv5s was more efficient than YOLOv5x in every aspect of the applied parameters. After all, YOLOv5m outperformed all the applied YOLO models.
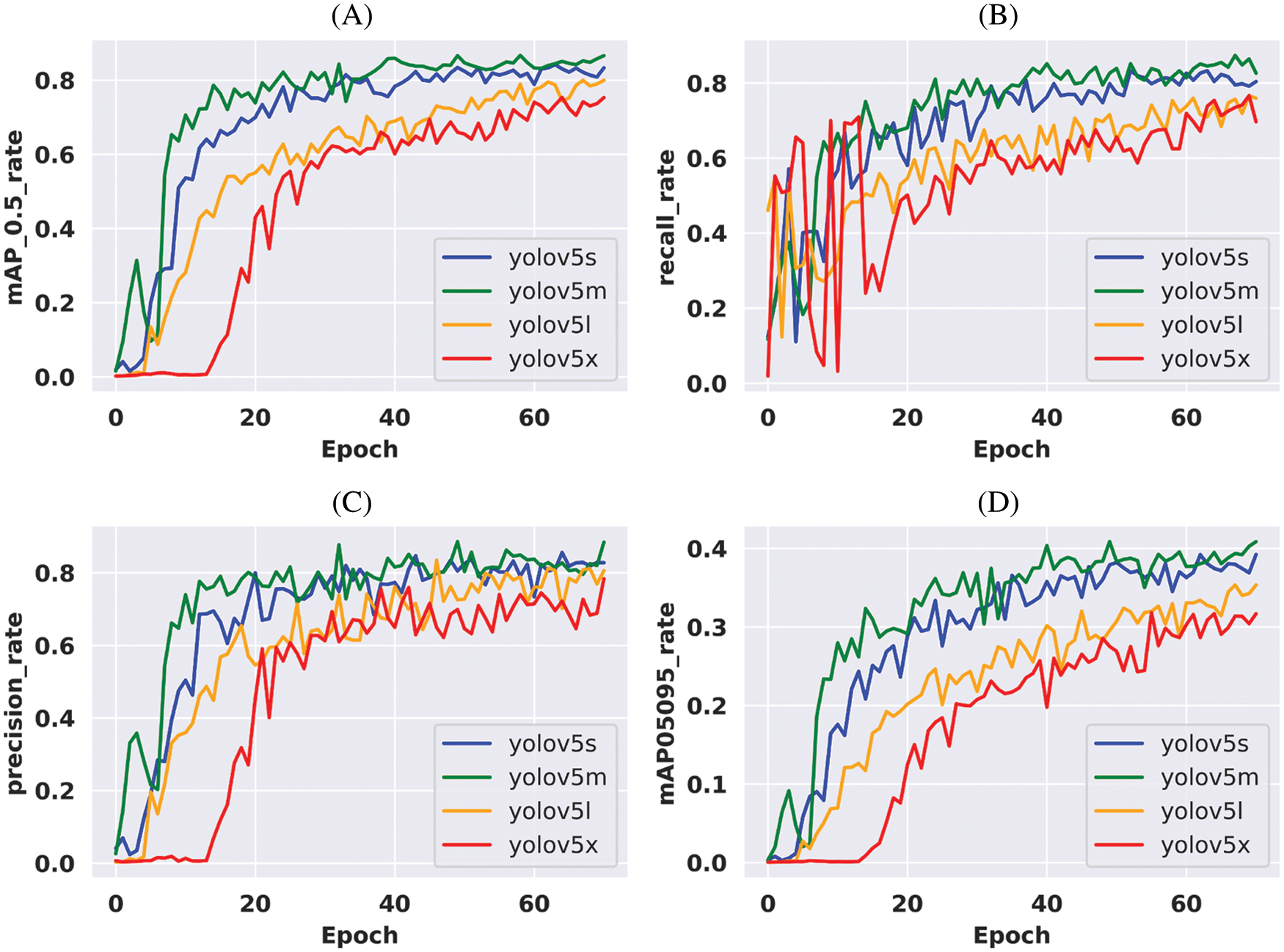
Figure 4: Performance comparison of the applied models according to the corresponding epoch values. (A) Mean average precision (mAP 0.5) values for all the variants of YOLOv5, (B) Recall values for all the variants of YOLOv5, (C) Precision values for all the variants of YOLOv5 & (D) Mean average precision (mAP 0.5:0.95) values for all the variants of YOLOv5
The loss function is a crucial evaluation metric for determining a model’s generalization capacity. The final objective of the model optimization was to lower the loss value as much as feasible while avoiding fitting. One of the most significant aspects of Neural Networks is the loss function. Loss is nothing but a prediction mistake in a Neural Network. The Loss Function is a method of calculating the loss of an applied model. In YOLOv5, the Bounding box’s loss function is GIOU (Generalized Intersection over Union) Loss.
The loss value of all the applied models is illustrated in Fig. 5. Train abject loss, train box loss, validation object loss, and validation box loss are all shown in the figure. According to the plot, it is found that YOLOv5x and YOLOv5l are gaining more loss than other applied models. Though YOLOv5s and YOLOv5m are performing close to each other, YOLOv5 m gained the least loss and outperformed in every aspect.
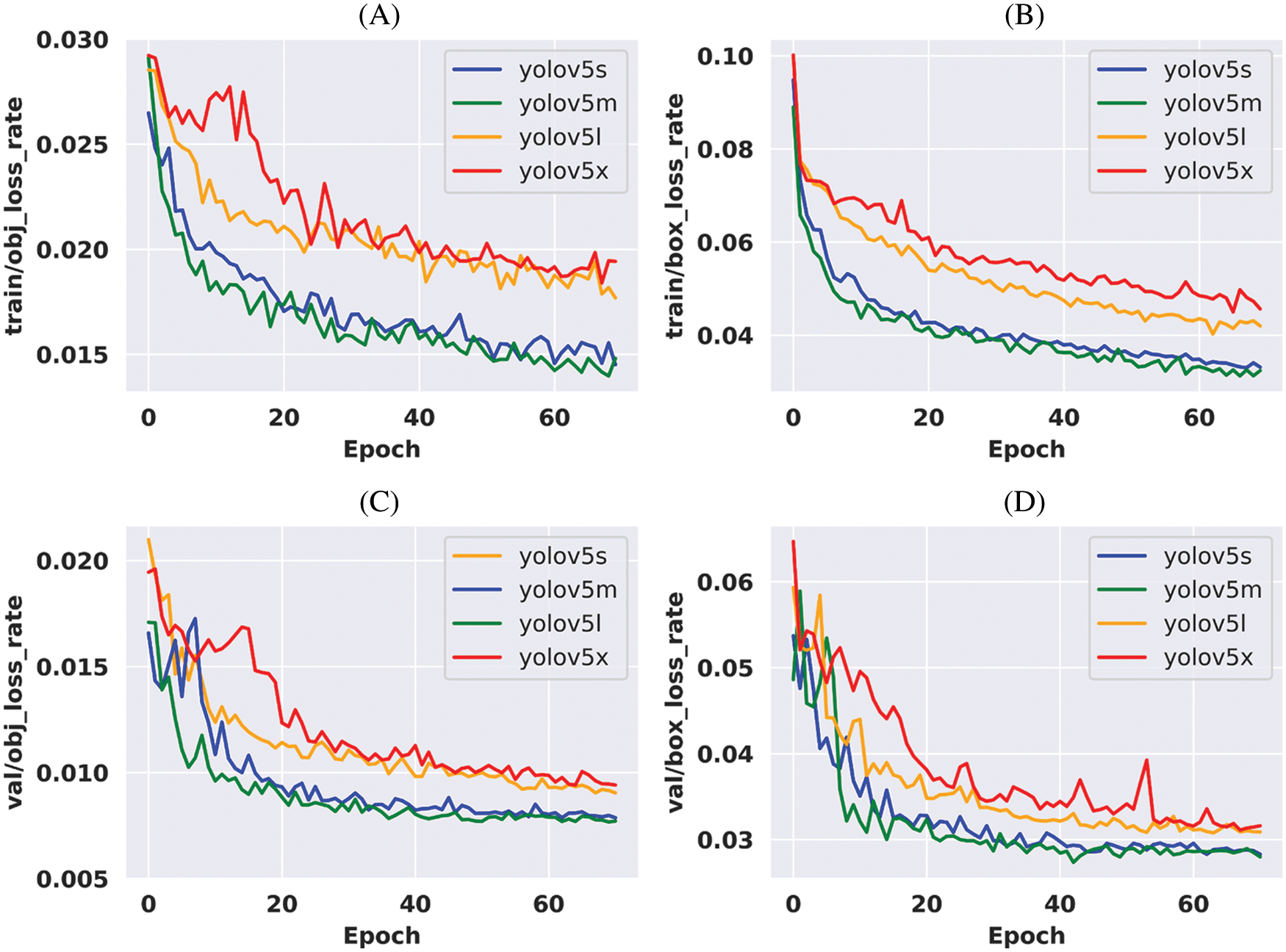
Figure 5: Loss comparison of the applied models during training according to the corresponding epoch values. (A) Object loss values during training for all the variants of YOLOv5, (B) Box loss values during training for all the variants of YOLOv5, (C) Validation object loss values during training for all the variants of YOLOv5 & (D) Validation box loss values during training for all the variants of YOLOv5
Fig. 6 shows the detection process of our applied models. To detect cysts, an image is imputed some images from the split test dataset. When those images pass to enter the model then the model identifies whether they are cancerous or not. The left side of the above figure contains imputed images, the middle is a model and the right side contains detected images.
Figure 6: Cervical cancerous cell detection methods and detected images of the proposed model
In brief, an open-source cervix image dataset was collected from a repository known as Kaggle. From all the images, 395 images were selected based on image quality to train all the applied models. Then feature enhancement technique is applied to the selected images to enhance the number of images and train the model in different dimensions. These images were annotated for fitting the YOLOv5 architecture. Four different versions of the YOLOv5 model such as YOLOv5s, YOLOv5m, YOLOv5l, and YOLOv5x were trained in this study and compared their performance to find the best fit version for the designed model. It is found that YOLOv5x performed worst compare to other versions where YOLOv5m outperformed with 88.4% and 86.4% precision and recall value among all the versions of YOLOv5. YOLOv5m also outperformed in terms of the loss function. Above all, the study indicates the CC risk factors are really invasive for women who are from developing and underdeveloped countries [36]. In addition to that the performance of all the applied models indicates that all the applied models are capable to identify cancerous cell from cervix images. However, the YOLOv5m is outperforming compare to other applied models. So, it can be decided that YOLOv5m is highly potential to identify cancerous cell from cervix images in early stages. The proposed model, YOLOv5m, will enable all the stakeholders to detect CC in early stage and it will reduce the mortality rate of women due to CC in developing and underdeveloped countries.
Cervical cancer is a rapidly growing deadly cancer among women, which is responsible for a large number of deaths. The mortality rate can be reduced and the patients can be cured if the cancerous cells are detected at an early stage and taken proper treatment. For detecting cancerous cells with low-cost diagnosis, YOLOv5 models are applied and compared their performances. It is found that YOLOv5x performed the worst compared to other variants of the YOLOv5 architecture. Though YOLOv5s also performed close to YOLOv5m, YOLOv5m outperformed all the variants of YOLOv5 models. Overall, the performances of the applied models indicate that the introduced patterns are highly susceptible for detecting cancerous cells from cervix images. It is an automatic and low-cost approach, which will enable doctors and patients to detect cervical cancerous cells using cervix images. In the future, more advanced technology will be applied to upgrade the proposed system with more images to increase the efficiency of the model.
Acknowledgement: The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for supporting this work by Grant Code: (22UQU4170008DSR07).
Funding Statement: The project funding number is 22UQU4170008DSR07. This work was supported in part by funding from the Natural Sciences and Engineering Research Council of Canada (NSERC).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. M. Brisson, J. Kim, K. Canfell, M. Drolet, G. Gingras et al., “Impact of HPV vaccination and cervical screening on cervical cancer elimination: A comparative modelling analysis in 78 low-income and lower-middle-income countries,” The Lancet, vol. 395, no. 10224, pp. 575–590, 2020. [Google Scholar]
2. S. Yu, N. Zhao, M. He, K. Zhang and X. Bi, “MiRNA-214 promotes the pyroptosis and inhibits the proliferation of cervical cancer cells via regulating the expression of NLRP3,” Cellular and Molecular Biology, vol. 66, no. 6, pp. 59–64, 2020. [Google Scholar]
3. Cervical Cancer, “Cancers For Disease Control and Prevention (CDC),” 2022. [Online]. Available: https://www.cdc.gov/cancer/cervical/basic_info/symptoms.html. [Google Scholar]
4. Cervical Cancer: Diagnosis, “Cancer.Net Editorial Board,” 2022. [Online]. Available: https://www.cancer.net/cancer-types/cervical-cancer/diagnosis. [Google Scholar]
5. Cervical Cancer, “World Health Organization,” 2022. [Online]. Available: https://www.who.int/health-topics/cervical-cancer#tab=tab_1. [Google Scholar]
6. S. K. Kjaer, C. Dehlendorff, F. Belmonte and L. Baandrup, “Real-world effectiveness of human papillomavirus vaccination against cervical cancer,” Journal of the National Cancer Institute, vol. 113, no. 10, pp. 1329–1335, 2021. [Google Scholar]
7. Y. Qiu, Q. Han, H. Lu and C. Shi, “MiR-196a targeting LRIG3 promotes the proliferation and migration of cervical cancer cells,” Cellular and Molecular Biology, vol. 66, no. 7, pp. 180–185, 2020. [Google Scholar]
8. Y. Gao, H. Wang, A. Zhong and T. Yu, “Expression and prognosis of CyclinA and CDK2 in patients with advanced cervical cancer after chemotherapy,” Cellular and Molecular Biology, vol. 66, no. 3, pp. 85–91, 2020. [Google Scholar]
9. What Is Cervical Cancer? “American Cancer Society,” 2022. [Online]. Available: https://www.cancer.org/cancer/cervical-cancer/about/what-is-cervical-cancer.html. [Google Scholar]
10. R. A. MD, D. AJaffray, M. Barton, F. Bray, M. B. MD et al., “Expanding global access to radiotherapy,” The Lancet Oncology, vol. 16, no. 10, pp. 1153–1186, 2015. [Google Scholar]
11. HPV and Cancer, “National Cancer Institute at the National Institutes of health,” 2021. [Online]. Available: https://www.cancer.gov/about-cancer/causes-prevention/risk/infectious-agents/hpv-and-cancer. [Google Scholar]
12. T. M. Zhang and W. Wang, “Development of poly ADP-ribose polymerase-1 inhibitor with anti-cervical carcinoma activity,” Cellular and Molecular Biology, vol. 66, no. 7, pp. 31–34, 2020. [Google Scholar]
13. H. Li, X. An and Q. Fu, “miR-218 affects the invasion and metastasis of cervical cancer cells by inhibiting the expression of SFMBT1 and DCUNIDI,” Cellular and Molecular Biology, vol. 68, no. 2, pp. 81–86, 2022. [Google Scholar]
14. T. Yu and C. Wang, “Clinical significance of detection of human papilloma virus DNA and E6/E7 mRNA for cervical cancer patients,” Cellular and Molecular Biology, vol. 67, no. 6, pp. 155–159, 2021. [Google Scholar]
15. Z. Zheng, X. Yang, Q. Yu, L. Li and L. Qiao, “The regulating role of miR-494 on HCCR1 in cervical cancer cells,” Cellular and Molecular Biology, vol. 67, no. 5, pp. 131–137, 2021. [Google Scholar]
16. M. M. Ali, B. K. Paul, K. Ahmed, F. M. Bui, J. M. Quinn et al., “Heart disease prediction using supervised machine learning algorithms: Performance analysis and comparison,” Computers in Biology and Medicine, vol. 136, pp. 104–672, 2021. [Google Scholar]
17. V. Venerito, O. Angelini, G. Cazzato, G. Lopalco, E. Maiorano et al., “A convolutional neural network with transfer learning for automatic discrimination between low and high-grade synovitis: A pilot study,” Internal and Emergency Medicine, vol. 16, pp. 1457–1465, 2021. [Google Scholar]
18. W. William, A. Ware, A. H. B. Ejiri and J. Obungoloch, “Cervical cancer classification from pap-smears using an enhanced fuzzy c-means algorithm,” Informatics in Medicine Unlocked, vol. 14, pp. 23–33, 2019. [Google Scholar]
19. Y. R. Park, Y. J. Kim, W. Ju, K. Nam, S. Kim et al., “Comparison of machine and deep learning for the classification of cervical cancer based on cervicography images,” Scientific Reports, vol. 11, no. 16143, pp. 1–11, 2021. [Google Scholar]
20. M. F. Ijaz, M. Attique and Y. Son, “Data-driven cervical cancer prediction model with outlier detection and over-sampling methods,” Sensors, vol. 20, no. 10, pp. 2809, 2020. [Google Scholar]
21. A. Goncalves, P. Ray, B. Soper, D. Widemann and M. Nygard, “Bayesian multitask learning regression for heterogeneous patient cohorts,” Journal of Biomedical Informatics, vol. 100, pp. 100059, 2019. [Google Scholar]
22. M. Sharma, S. K. Singh, P. Agrawal and V. Madaan, “Classification of clinical dataset of cervical cancer using KNN,” Indian Journal of Science and Technology, vol. 9, no. 28, pp. 1–5, 2016. [Google Scholar]
23. M. M. Ali, K. Ahmed, F. M. Bui, B. K. Paul, S. M. Ibrahim et al., “Machine learning-based statistical analysis for early stage detection of cervical cancer,” Computers in Biology and Medicine, vol. 139, pp. 104985, 2021. [Google Scholar]
24. M. Kuko and M. Pourhomayoun, “Single and clustered cervical cell classification with ensemble and deep learning methods,” Information Systems Frontiers, vol. 22, no. 5, pp. 1039–1051, 2020. [Google Scholar]
25. A. Khamparia, D. Gupta, J. J. P. C. Rodrigues and V. H. C. D. Albuquerque, “DCAVN: Cervical cancer prediction and classification using deep convolutional and variational autoencoder network,” Multimedia Tools and Applications, vol. 80, no. 20, pp. 30399–30415, 2021. [Google Scholar]
26. R. Alkhawaldeh, S. Mansi and S. Lu, “Prediction of cervical cancer diagnosis using deep neural networks,” in IIE Annual Conference Proceedings, New York, pp. 438–443, 2019. [Google Scholar]
27. X. Jianfang, Z. Ling, J. Yanan and G. Yanliang, “The effect of Chaihu-shugan-san on cytotoxicity induction and PDGF gene expression in cervical cancer cell line HeLa in the presence of paclitaxel+cisplatin,” Cellular and Molecular Biology, vol. 67, no. 3, pp. 143–147, 2021. [Google Scholar]
28. B. Chitra and S. S. Kumar, “An optimized deep learning model using mutation-based atom search optimization algorithm for cervical cancer detection,” Soft Computing, vol. 25, no. 24, pp. 15363–15376, 2021. [Google Scholar]
29. R. Elakkiya, K. S. S. Teja, L. J. Deborah, C. Bisogni and C. Medaglia, “Imaging based cervical cancer diagnostics using small object detection-generative adversarial networks,” Multimedia Tools and Applications, vol. 81, pp. 1–17, 2021. [Google Scholar]
30. F. Arezzo, D. L. Forgia, V. Venerito, M. Moschetta, A. S. Tagliafico et al., “A machine learning tool to predict the response to neoadjuvant chemotherapy in patients with locally advanced cervical cancer,” Applied Sciences, vol. 11, no. 2, pp. 823, 2021. [Google Scholar]
31. Cervical cancer screening, 2020. [Online]. Available: https://www.kaggle.com/datasets/ofriharel/224-224-cervical-cancer-screening. [Google Scholar]
32. M. Ai, 2022. [Online]. Available: https://www.makesense.ai/ [Accessed on 28–03–2022]. [Google Scholar]
33. W. Liu, D. Anguelov, D. Erhan, C. Szegedy, S. Reed et al.,, October. “Ssd: Single shot multibox detector,” in European Conf. on Computer Vision, Cham, Springer, vol. 9905, pp. 21–37, 2016. [Google Scholar]
34. X. R. Zhang, J. Zhou, W. Sun and S. K. Jha, “A lightweight CNN based on transfer learning for COVID-19 diagnosis,” Computers, Materials & Continua, vol. 72, no. 1, pp. 1123–1137, 2022. [Google Scholar]
35. W. Sun, G. C. Zhang, X. R. Zhang, X. Zhang and N. N. Ge, “Fine-grained vehicle type classification using lightweight convolutional neural network with feature optimization and joint learning strategy,” Multimedia Tools and Applications, vol. 80, no. 20, pp. 30803–30816, 2021. [Google Scholar]
36. H. K. Jung and G. S. Choi, “Improved YOLOv5: Efficient object detection using drone images under various conditions,” Applied Sciences, vol. 12, no. 14, pp. 7255, 2022. [Google Scholar]
37. M. A. Rahaman, M. M. Ali, K. Ahmed, F. M. Bui and S. H. Mahmud, “Performance analysis between YOLOv5s and YOLOv5m model to detect and count blood cells: Deep learning approach,” in Proc. of the 2nd Int. Conf. on Computing Advancements, Dhaka, pp. 316–322, 2022. [Google Scholar]
38. B. Jiang, R. Luo, J. Mao, T. Xiao and Y. Jiang, “Acquisition of localization confidence for accurate object detection,” in European Conf. on Computer Vision (ECCV), Munich, pp. 784–799, 2018. [Google Scholar]
39. J. Wan, B. Chen and Y. Yu, “Polyp detection from colorectum images by using attentive YOLOv5,” Diagnostics, vol. 11, no. 12, pp. 2264, 2021. [Google Scholar]
40. H. Sun and R. Grishman, “Lexicalized dependency paths based supervised learning for relation extraction,” Computer Systems Science and Engineering, vol. 43, no. 3, pp. 861–870, 2022. [Google Scholar]
41. H. Sun and R. Grishman, “Employing lexicalized dependency paths for active learning of relation extraction,” Intelligent Automation & Soft Computing, vol. 34, no. 3, pp. 1415–1423, 2022. [Google Scholar]
42. M. Ragab, H. A. Abdushkour, A. F. Nahhas and W. H. Aljedaibi, “Deer hunting optimization with deep learning model for lung cancer classification,” Computers, Materials & Continua, vol. 73, no. 1, pp. 533–546, 2022. [Google Scholar]
43. Y. Y. Ghadi, I. Akhter, S. A. Alsuhibany, T. A. Shloul, A. Jalal et al., “Multiple events detection using context-intelligence features,” Intelligent Automation & Soft Computing, vol. 34, no. 3, pp. 1455–1471, 2022. [Google Scholar]
44. M. M. El Khair, R. A. Mhand, M. E. Mzibri and M. M. Ennaji, “Risk factors of invasive cervical cancer in Morocco,” Cellular and Molecular Biology, vol. 55, no. 4, pp. 1175–85, 2009. [Google Scholar]
45. M. Z. H. Ontor, M. M. Ali, S. S. Hossain, M. Nayer, K. Ahmed et al., “YOLO_CC: Deep learning based approach for early stage detection of cervical cancer from cervix images using YOLOv5s model,” in 2022 Second Int. Conf. on Advances in Electrical, Computing, Communication and Sustainable Technologies (ICAECT), Bhilai, pp. 1–5, 2022. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools