 Open Access
Open Access
ARTICLE
Crop Disease Recognition Based on Improved Model-Agnostic Meta-Learning
1 College of Information Technology, Jilin Agricultural University, Changchun, 130118, China
2 College of Engineering and Technology, Jilin Agricultural University, Changchun, 130118, China
* Corresponding Author: Lidong Chu. Email:
Computers, Materials & Continua 2023, 75(3), 6101-6118. https://doi.org/10.32604/cmc.2023.036829
Received 13 October 2022; Accepted 08 February 2023; Issue published 29 April 2023
Abstract
Currently, one of the most severe problems in the agricultural industry is the effect of diseases and pests on global crop production and economic development. Therefore, further research in the field of crop disease and pest detection is necessary to address the mentioned problem. Aiming to identify the diseased crops and insect pests timely and accurately and perform appropriate prevention measures to reduce the associated losses, this article proposes a Model-Agnostic Meta-Learning (MAML) attention model based on the meta-learning paradigm. The proposed model combines meta-learning with basic learning and adopts an Efficient Channel Attention (ECA) module. The module follows the local cross-channel interactive strategy of non-dimensional reduction to strengthen the weight parameters corresponding to certain disease characteristics. The proposed meta-learning-based algorithm has the advantage of strong generalization capability and, by integrating the ECA module in the original model, can achieve more efficient detection in new tasks. The proposed model is verified by experiments, and the experimental results show that compared with the original MAML model, the proposed improved MAML-Attention model has a better performance by 1.8–9.31 percentage points in different classification tasks; the maximum accuracy is increased by 1.15–8.2 percentage points. The experimental results verify the strong generalization ability and good robustness of the proposed MAML-Attention model. Compared to the other few-shot methods, the proposed MAML-Attention performs better.Keywords
Agriculture represents one of the basics of the national economy, so crop yields directly affect the economic and social welfare of countries. Diseases and pests have been the most common and severe threats to agriculture, and the effectiveness of their prevention and control directly affects the agricultural sector’s productivity and efficiency. According to the forecast of the United Nations’ Food and Agriculture Organization, by 2050, the global population will reach about 10 billion, so the world’s grain output has to increase by 60% compared to the current levels to meet human needs in the future [1,2]. However, in recent years, grain supply has faced significant challenges, including supply disruptions and labor shortages caused by the new crown rot epidemic. With the increase in population associated with large-scale urbanization, land use, soil, and water are no longer inexhaustible resources. This leads to a reduction in arable land and pressure on agriculture increase over time [3]. In addition, global warming affects the management of virus-affected crops, increasing crop diseases and thus causing significant losses of crop yields [4,5].
There have been various ways to detect diseased crops and pest infestations, including image processing and feature extraction methods [6]. Since 2012, deep learning has been developing rapidly, achieving great performance in feature extraction using multi-layer convolution-based models. However, these models require a large number of training samples but can achieve higher classification and prediction accuracy compared to the traditional techniques. In addition, these methods have been applied to many agricultural tasks, and they have replaced the time-consuming tasks requiring human presence and expertise to a certain extent. For instance, Zhou et al. [7] proposed a reconstructed residual-dense network, which was trained on a dataset consisting of 50,000 samples and achieved a 95% accuracy in tomato disease leave recognition. Brahimi et al. [8] employed the AlexNet and GoogLet models trained on a set of 14,828 samples and achieved the final classification accuracy of 98.66% and 99.18%, respectively. In addition, Zhang et al. [9] used the PlantVillage dataset consisting of 54,305 samples and a self-built dataset of 9,645 samples to train the improved GoogLeNet and Cifar10 models for the classification and detection of nine types of maize leaves. The recognition accuracies of the improved GoogLeNet and Cifar10 models were 98.9% and 98.8%, respectively. Dyrmann et al. [10] used a convolutional neural network to train and test the model with 10,413 samples, and the final average accuracy reached 89.51%. In 2020, Zhang et al. [11] proposed an improved Faster Region-based Convolution Neural Network (Faster RCNN [12]) feature classification model for tomato diseases, including pink mildew, powdery mildew, wilting, chlorosis, and spotted wilt virus. This model adopts a deep residual network instead of the Visual Geometry Group (VGG-16) network [13] and uses the K-means algorithm to cluster candidate boxes. The identification accuracy of this method can reach 97.18%, showing an increase of 2.71 percentage points (pp) compared to the traditional Faster RCNN. In 2021, Saeed et al. [14] proposed a CNN-based automated crop disease recognition model based on the partial least squares regression for feature selection from an extracted deep feature set and achieved average recognition accuracy of 90.1%. In 2021, Hu et al. [15] improved the Residual Network (ResNet) model [16] and developed a multi-dimensional feature compensation residual neural network model. Compared with the VGG and ResNet-50 [16] models, the accuracy of their model was improved by 8.40 and 5.31 pp, respectively. However, although the availability of large datasets and sufficient model complexity allow deep learning-based models to fit data well and achieve high recognition accuracy, there have been many challenges in the deep learning field. For instance, model generalization ability is often low, and there is also a problem of model overfitting. Moreover, models often perform well only in specific tasks they are trained for but require model parameter adjustment and model re-training when used in tasks unseen during the training phase. In addition, acquiring a sufficient amount of training data in agricultural applications has been challenging and labor-intensive, but using insufficient data limits the classification effect of a model.
Judging from the development of agriculture in recent years, agricultural digital transformation has evolved into artificial intelligence system in various aspects [17]. Meta-learning [18], which is also referred to as “learning to learn” [19], has been recognized as an alternative solution that can potentially handle the above-mentioned shortcomings and limitations. The implementation of meta-learning includes two steps. The first step is to let the model learn specific data so that it can complete certain tasks. The second step is to allow the model to generalize to new data and integrate new knowledge to be able to accomplish new tasks. As a mainstream meta-learning model, the MAML model [20] is suitable for learning tasks that are optimized using the gradient descent algorithm. This model is applicable to a wide range of use cases and easy to understand and implement. Therefore, in most deep learning-based models, the MAML model has been used to strengthen their generalization ability. Under the conditions of a relatively small dataset, large environmental changes, and a requirement for fast and highly-accurate prediction, the MAML model can represent an efficient task generalization model. Therefore, the MAML model is very suitable for application scenarios with fewer disease samples, complex diseases, and numerous categories.
In this paper, prediction model accuracy is improved through the adaptation of the network structure based on the MAML model while retaining the MAML model’s advantage of strong generalization. The main aim of this study is to apply an improved meta-learning-based model and use common crop disease samples to train the model to avoid the need for large training datasets of unknown diseases.
The rest of this paper is organized as follows. Section 2 describes the dataset partitioning, the training method, and the base and meta-learners. Section 3 introduces the proposed model. Section 4 evaluates the proposed model, compares it with several common models, and analyzes the comparison results. Finally, Section 5 concludes the paper and presents future work directions.
Compared with traditional deep learning, the proposed model requires less data. Namely, the proposed model can achieve good classification performance using a small amount of sample data. Through internal and external cycle training, the proposed improved MAML model can rapidly apply the knowledge acquired during the training phase to new tasks unseen during the model training phase.
In this study, the research data included the Plant-Village dataset (available at https://plantvillage.psu.edu/) and the Artificial Intelligence (AI) Challenger 2018 crop disease detection competition dataset (available at https://challenger.ai/competition/pdr2018). The datasets consisted of images of apples, cherries, corn, peach, potato, tomato, grapes, peppers, strawberries, and citrus, having a total of 10 crop leaf types and 58 types of disease sample data. There were 50 photos for each condition, including images with the mere presence of a specific condition and more severe infections.
The machine learning-based model used in this study was trained using data on 51 categories, such as healthy apple leaf blades, and its generalization ability was tested on the dataset consisting of seven new categories (i.e., meta-test), including apple mosaic, tomato spot blight, healthy strawberry leaves, tomato late blight fungus, grape brown spot, cherry powdery mildew, and potato late blight. The purpose of the trained model was to discover the universal features of the 51 categories, extract their common characteristics, and identify new diseases based on common associations. Through meta-training, the common characteristics of diseases are obtained, and through meta-testing, the trained model is tested and verified for specific disease types.
In the experiment conducted in this study, each task was considered a basic unit, involving a random combination of N categories selected from the 51 categories in the meta-training set, where each category had K samples, and an N-way K-shot training task was defined. The seven categories were extracted in the same way and combined into the test learning tasks. In contrast to traditional machine learning, in meta-learning training, a large number of tasks are used with the aim of improving the generalization ability of a model so that the trained model can provide better solutions when dealing with similar problems. Fig. 1 shows the dataset division of a three-way one-shot classification task. In this example, the characteristics of healthy corn leaves, grape black rot leaves, and tomato powdery mildew are learned, and the trained model can generalize its identification capability to tomato leaf blight, healthy strawberry leaves, and cherry powdery mildew. Compared with tens of thousands or even millions of samples required for the training of traditional deep learning-based models, the proposed model is trained with only 2,900 samples.
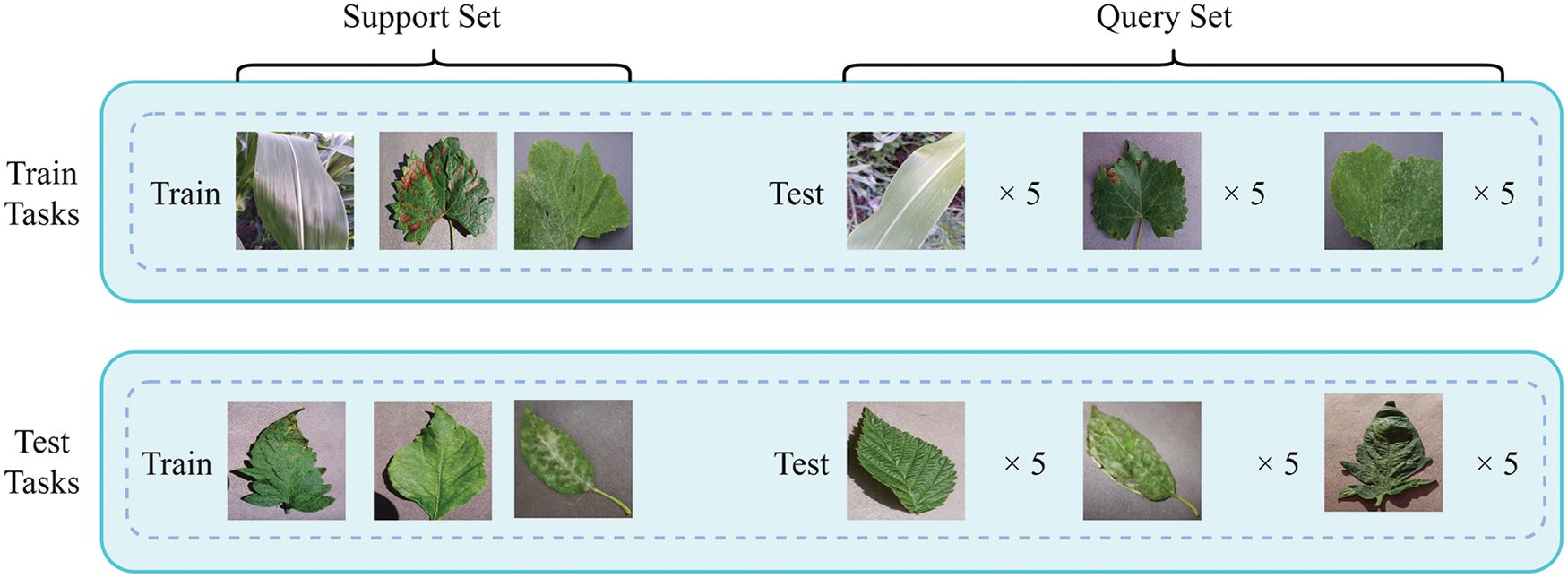
Figure 1: The illustration of a three-way one-shot task
In the proposed model, the MAML model was used as a backbone network, and Convolutional Neural Networks (CNNs) were used as a learning module. The gradient descent method was used to optimize the basic learner and find a global optimal solution. In the experiment, a combination of a basic learner and a meta-learner was used to obtain optimal model parameters by minimizing the loss function and model the characteristics and commonalities of the classification tasks. The collected data on 51 categories were divided into several meta-training sets, and data on the seven new categories were used as a meta-test set. The meta-training and meta-test sets included supporting and query sets, respectively. The overall experimental process is shown in Fig. 2.
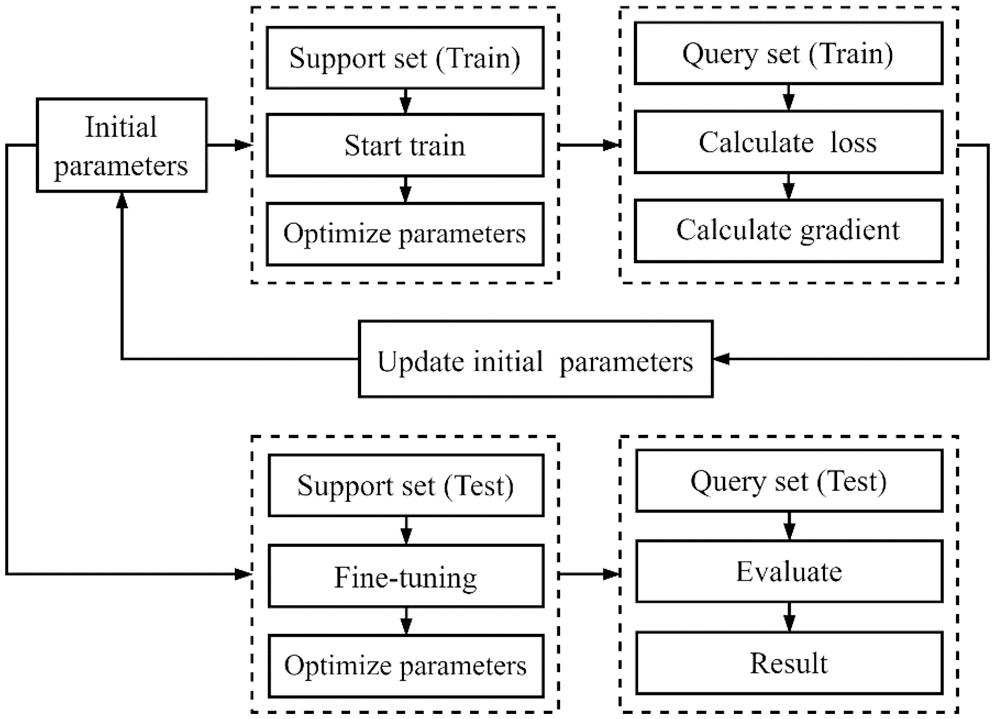
Figure 2: The flowchart of the experimental process
As shown in Fig. 2, in the experiment, the initial parameter values were obtained through Kaiming initialization, and the basis was formed for subsequent training. Then, the support set (for training) was used for training and optimization of the weight parameters for each task, while the query set (again for training) was used for verify the model’s performance. Further, the initial model parameters were updated according to the gradient value, and finally, the support set (for testing) was used to fine-tune the model on the test set, while the query set (for testing) was used to evaluate the optimized model. After the model was trained on the training set, the weight parameters were adjusted to make the model fit the commonality of the tasks better and strengthen its generalization ability. The model focused on learning characteristics of specific categories in the support set in the test task. Because of the generalization process, the model inherited common parameters in the training set in the same manner as the initial parameter optimization shown in Fig. 2. For new tasks, the model could be tuned in a few simple steps using the support set (meta-test) to achieve a good recognition effect. In general, in the meta-training phase, the common characteristics of all classes were modeled through a specific task combination in the meta-training set, with the aim to determine weight parameters that allow fast adaptation to new tasks. During the meta-test phase, the model was generalized and tuned for tasks using the support test (meta-test).
The MAML model is a meta-learning model with excellent performance. In view of this, this paper improves the MAML model and proposes the improved Maml-Attention model. While improving the recognition accuracy, the MAML-Attention model also maintains the advantage of strong meta-learning generalization.
During the training process, the basic learner considers a dataset for a specific classification task. The main function of the basic learner is fourfold. First, for a specific task, the characteristics of the task are learned. Second, the model and initial parameters of meta-learning are inherited. Third, a suitable objective function is formulated according to the problem for which the optimization is required. Fourth, the trained model and parameters are fed back to the meta-learner. On new tasks, the basic learner is iteratively updated using the gradient descent method, and the learned results are passed to the meta-learner.
The network structure of the basic learner is shown in Fig. 3. The overall network structure includes four convolutional layers with the ReLU activation function, reduces the data dimension using maximum pooling, and adopts a fully-connected layer to obtain specific classification results. The basic learner shown above has two main functions: to analyze specific characteristics of each disease in input data and to provide feedback on the meta-learner these characteristics to improve the generalization ability of the meta-learner.
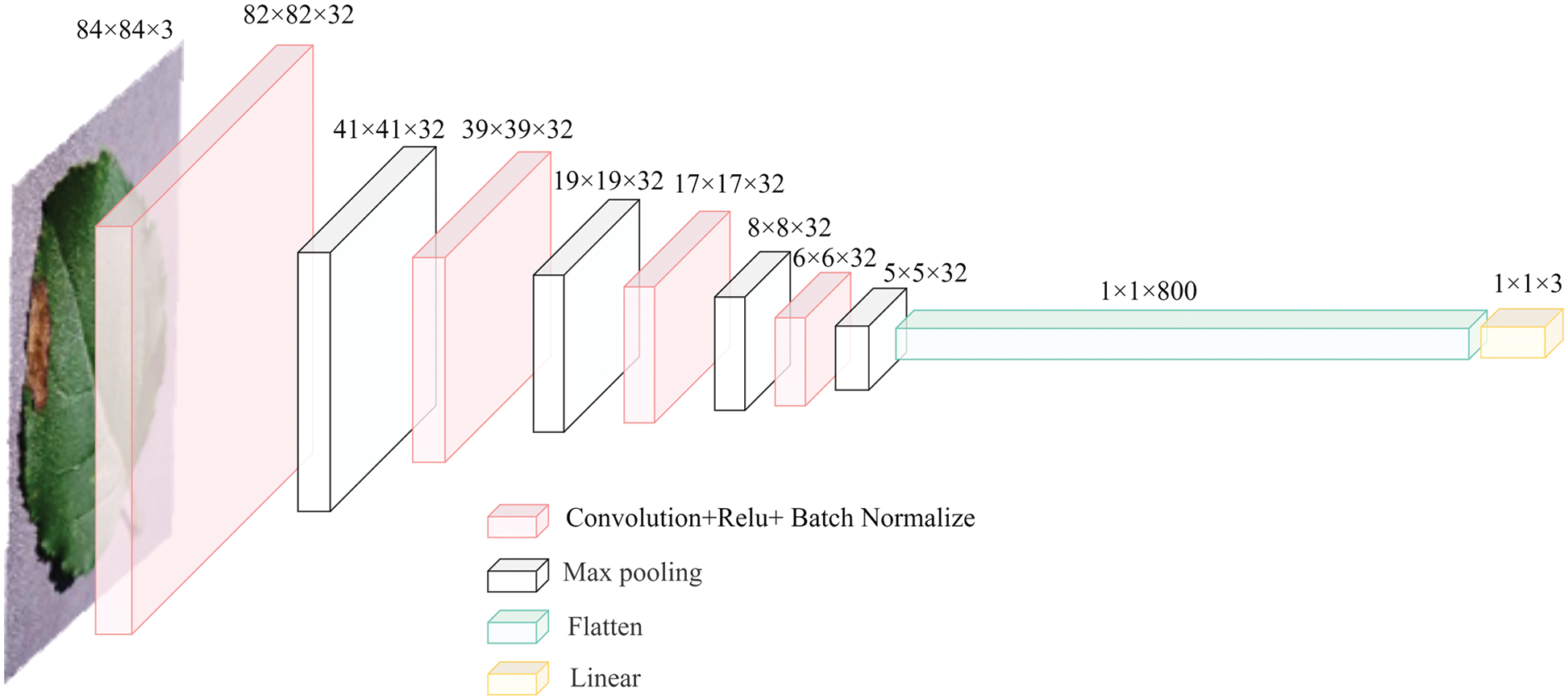
Figure 3: The network structure of the basic learner (three-way classification)
The MAML model consists of a basic learner and a meta-learner. The flowchart of the model is shown in Fig. 4. The meta-learner consists of two parts: the initial values of model parameters and the gradient value of the loss function. For Task 1, when training the basic learner, the initial values of model parameters are obtained by the meta-learning memory module and passed to the basic learner. During the training process of Task 1, the gradient value of the basic learner’s loss function is sent to the meta-learner. This process is repeated for all tasks, namely Task 2, Task 3, ... , Task N. The meta-learner models the common characteristics of the tasks, synthesizes the disease features obtained by the basic learner, and obtains the common parameters allowing it to perform similar tasks with minimal training.
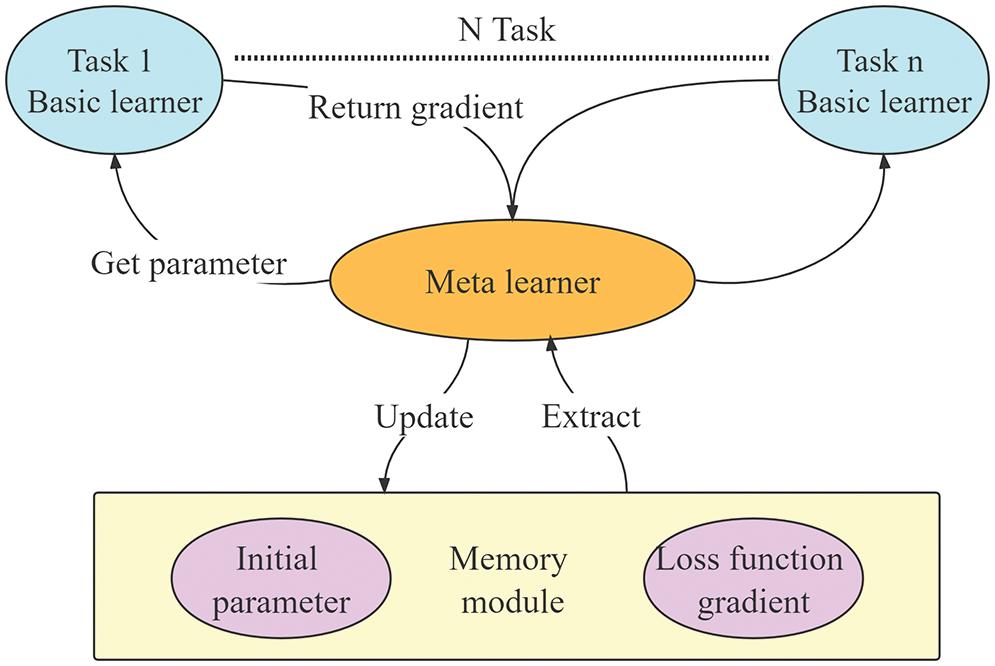
Figure 4: Basic learner and meta-learner training update process
Assume the training task of a basic learner during an experiment is denoted as
Due to the large number of sample categories in the experimental dataset, increasing the number of iterations (basic leaner) can help to improve the accuracy of the basic learner during training, so multiple iterations are performed, as shown in Eq. (2).
After each iteration, the basic learner passes the function loss’ gradient values to the meta-learner, and the meta-learner uses these values to update the gradient values in the memory module, as shown in Eq. (3), where
In the experiment, a meta-training task was a batch of training tasks. After the model was trained on the batch tasks, the common characteristics of different disease samples contained in the corresponding training datasets were obtained. As shown in Fig. 5, after each gradient descent calculation, a model evaluation was performed to obtain the loss function value, and the initial parameter values of the meta-learner were updated after P tasks were completed.
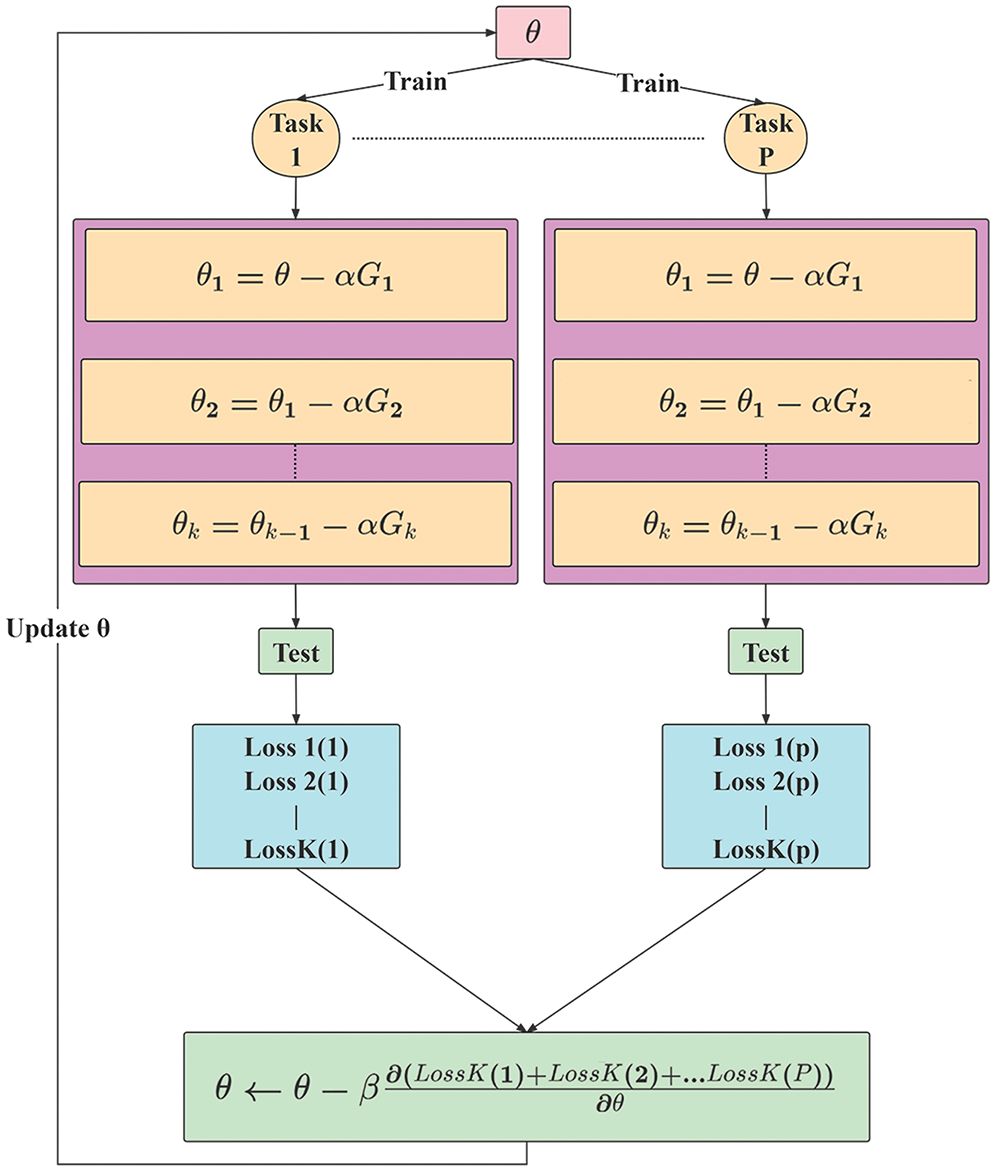
Figure 5: Calculation process flowchart
Based on the interaction process between the meta-learner and the basic learner, the meta-learner mainly integrates the results of the basic learner, summarizes the commonalities of multiple tasks, and obtains accurate results on new tasks rapidly. As shown in Fig. 5, when the network model processes the first task, the meta-learner passes the initial parameter values to the basic learner; the basic learner is trained on the training set, and its weight parameters are updated. These weight parameters denote the specific characteristics corresponding to the current task, for example, the parameters corresponding to diseased leave recognition. As shown in Fig. 5, after k updates, the basic learner obtains parameter
The attention mechanism aims to simulate human cognition processes, particularly the process of extracting key information about external stimuli without the need to understand them as entities. Similarly, for deep learning-based models, the attention mechanism gives different weights to different parts of the model so that the model can extract more valuable information without generating too much computational overhead. Since the attention mechanism was proposed, its development can be roughly divided into two directions, the enhancement of feature aggregation and the combination of channel and spatial attention. In 2017, Hu et al. [21] proposed the Squeeze-and-Excitation Networks (SENet) structure, whose main idea was to establish the interdependence between channels and continuously optimize the correlation and dependencies between channels using a global loss function while reducing the weight of redundant features. In 2019, Hu et al. [22] proposed a simple, lightweight method to use contextual information in CNN algorithms better. This method adopted Gather (G) operator to aggregate contextual information within a given spatial range and Excite (E) operator to accept aggregate and feature inputs to generate new tensors. In applications where the Faster R-CNN model is a basic framework, and the ResNet-50 model is a backbone network, this method can improve the mAP value by 1.3% after the G-E operator optimization. In recent years, the attention mechanism has been proposed to improve the performance of networks. Nie et al. [23] introduced the attention mechanism into the field of disease detection. Compared to the traditional disease detection networks, network structures with an additional attention module had better performance, and the Mean Average Precision (mAP) values on the four categories of Healthy_leaf, Healthy_petiole, Verticillium_leaf, and Verticillium_petiole reached 77.54%, while the accuracy of strawberry verticillium wilt detection reached 99.95%. In 2018, Woo et al. [24] proposed a simple but effective convolutional block attention module, which can perform attention tasks in both channel and spatial dimensions. Compared to the SENet model, which pays attention only to the channel, the Convolutional Block Attention Module (CBAM) model has an additional spatial attention channel processing spatial location information, so it is suitable for plant disease images that require focusing on specific local features of diseases.
Although the above-mentioned attention mechanisms can achieve good performance on most networks, they often increase model complexity to a certain extent. In 2020, Wang et al. [25] proposed the ECA-Net network model, which is shown in Fig. 6, to generate channel weights by performing a fast one-dimensional convolution with a size K, where K (KernelSize) was automatically passed through the channel dimension function. Due to the adaptive properties and the fact that no dimensionality reduction occurred, the ECA module could capture local cross-channel interactions by considering each channel and its K neighbors. In 2022, Yu et al. [26] proposed an ECA-Net to suppress the noise from a complex background; the LW-ResNet model achieved a 97.80% accuracy in the apple disease recognition task. Further, in 2022, Li et al. [27] applied the attention mechanism (ECA-Net) to the spectral classification task to identify the most informative spectral context features; the experiments showed that this method was superior to the state-of-the-art hyperspectral image classification methods. Compared with the aforementioned attention modules, the ECA module learns channel attention more effectively, avoids dimensionality reduction, and captures cross-channel interaction information more effectively without increasing the computational cost significantly. The number of parameters introduced by the ECA module is almost negligible, but it can provide significant performance gains to the network model. Namely, because the disease location is given more weight, the disease features can be obtained more rapidly and accurately.
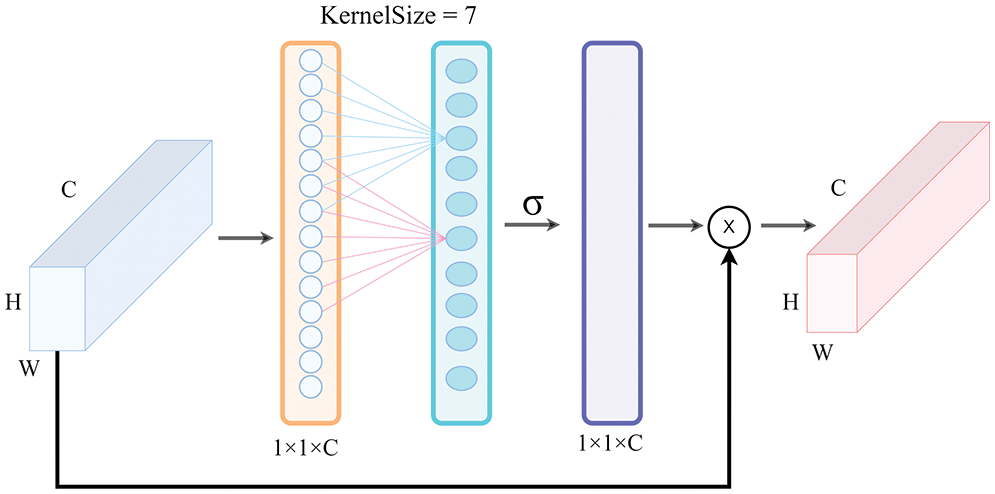
Figure 6: The ECA attention module
In this experiment, four ECA-Net layers were added to the original basic learner network, and each ECA-Net layer was added after the convolution layer. As shown in Fig. 7, the KernelSize of the ECA-Net model was seven. At the model end, the feature space of diseased leaves was mapped to the sample label space through a fully-connected layer.
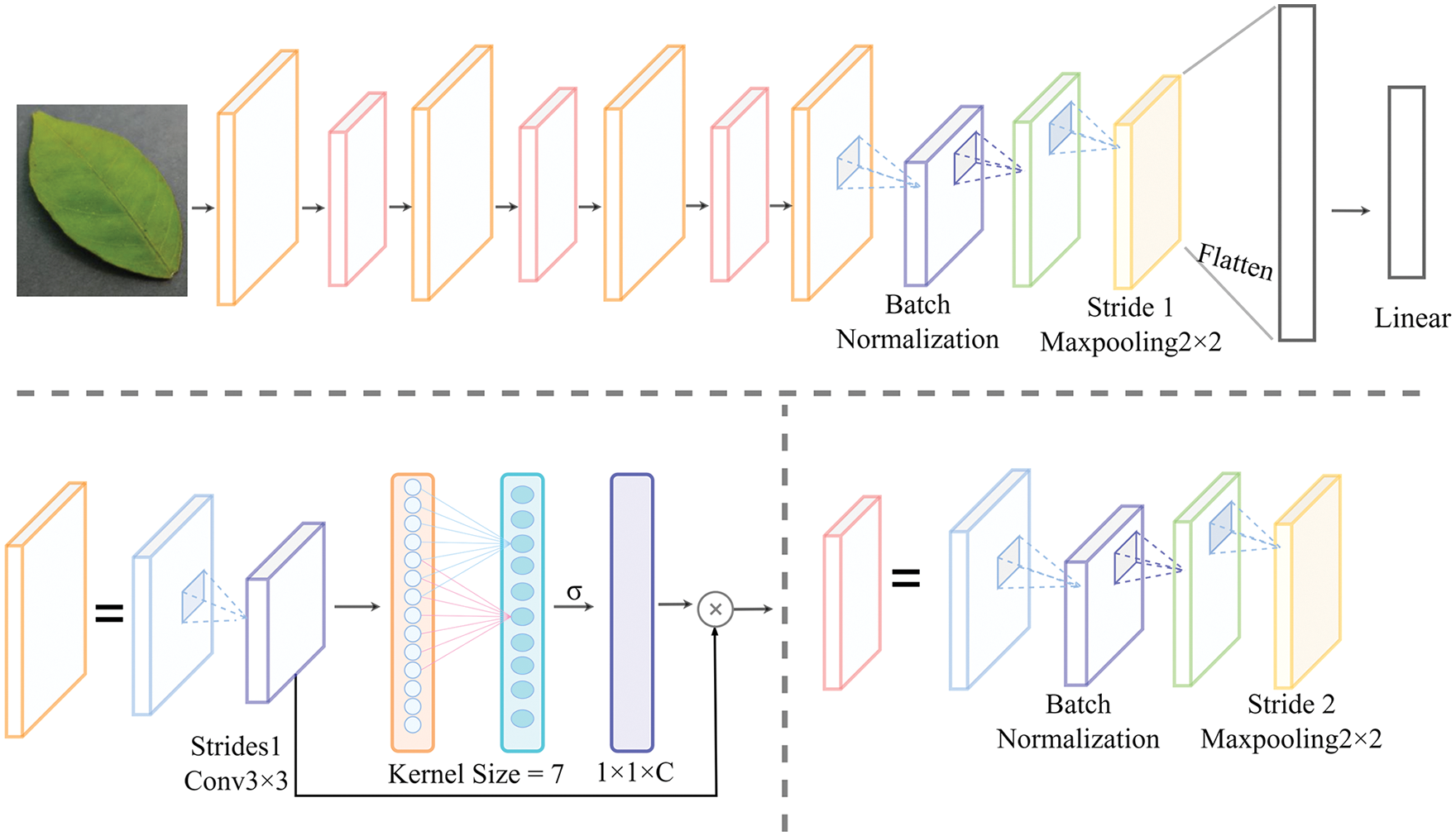
Figure 7: Improved MAML network structure
In the network model presented in Fig. 7, the input denotes the Red, Green, Blue (RGB) gan 3-channel image data, the convolution kernel’s size is 3 × 3, the stride is one, and the padding is zero. The ECA-Net module is added before the corresponding convolutional layer inputs 32-dimensional feature data. After average pooling, a one-dimensional convolution is performed on the feature data with a 7 × 7 convolution kernel. The kernel size parameter, which is seven in this case, represents the coverage of local cross-channel interaction, indicating how many neighbors of the channel participate in the prediction of the current channel. Then, the weight of each channel is obtained using the sigmoid function (σ). Finally, the weight of each channel is assigned to the data before convolution to complete the operation of the attention module. After the attention layer, the Rectified Linear Unit (ReLU) activation function is added, and normalization (BN) is performed, followed by maximum pooling with a sliding window of 2 × 2 and a padding of 0. The above operation is repeated four times, once for each block in the network. In the last layer, the maximum pooling step size is set to one, while the other values are kept unchanged. The classification of diseased leaves is achieved by the fully-connected layer. During the training process, the basic learner continuously optimizes its parameters. After completing the batch of tasks, all gradient values of the loss function are fed back to the meta-learner and used to adjust its parameters accordingly.
As shown in Fig. 8, an efficient channel attention module is added after each convolutional layer, and the local features of each channel are given different weights through one-dimensional convolution. The proposed model can perform subsequent analysis and processing on specified key features according to different weight information. Since the proposed module is more lightweight than the existing modules, it is convenient and easy to integrate while avoiding the addition of unnecessary dimensions. Therefore, it can be easily added to the conventional CNN models to improve their performance.
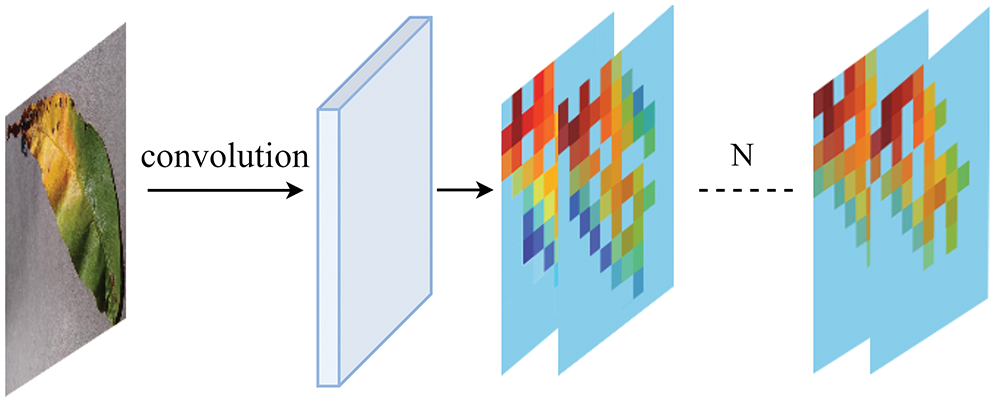
Figure 8: Illustration of the attention layer
4 Experimental Result and Analysis
To develop a model with fast adaptability, meta-learning training has been generally conducted in the form of few-shot learning, which represents learning with fewer samples. A small amount of sample data was used in a task, and the meta-learning model was trained on many tasks created in this way. Therefore, the trained meta-learning model could rapidly adapt to a new task with a small amount of data after several training iterations. The proposed model was compared with other small sample learning-based models. The advantages of the MAML-Attention were reflected in different aspects. A detailed description of the experiments of the proposed MAML-Attention model is given in the following, and the experimental results are analyzed.
The meta-training dataset included data on 51 diseases, and the meta-test set included data on seven diseases. In the training set, the batch size was set to 800, the input size was set to 84 × 84, the initial learning rate of the meta-learner was set to 0.001, and the learning rate of the basic learner was set to 0.01. Both learners were optimized using the gradient descent algorithm. In the test set, the parameters were set the same as in the training set, but the batch size was set to 200. A total of 60 training epochs were performed, each of which included 200 training steps and one optimization round, as shown in the “Fine-tuning” step in Fig. 2; there were five iterations per optimization round. Further, N types were randomly selected from the 51 disease types, and K samples were randomly selected for each disease type to form an N-way K-shot training task. Similarly, seven diseases were selected to obtain the test set. It should be noted that 12 groups of the N-way K-shot training tasks before model improvement in this experiment included: three-way one-shot, three-way two-shot, three-way three-shot, four-way one-shot, four-way two-shot, four-way three-shot, four-way four-shot, five-way one-shot, five-way two-shot, five-way three-shot, five-way four-shot, and five-way five-shot tasks. Similarly, the test set included N-way K-shot sets. Finally, 12 sets of comparative experiments were performed using the same N-way K-shot set, and the improved network model was used for comparison.
4.2 Experiment Results Analysis
After completing 24 groups of experiments, the obtained results were analyzed, and the model’s performances before and after the improvement were compared from four aspects: the number of iterations, data sample size, overall accuracy, and model structure.
The number of iterations. The three-way one-shot training task was used as an example, as shown in Fig. 9. Since the model’s weight parameters were adjusted according to the common features of the disease after training, after one round of fine-tuning, the accuracy of disease identification was improved significantly compared to the results before fine-tuning. After five fine-tuning iterations, the overall model accuracy was basically stable. As shown in Fig. 10, the disease identification accuracy improved rapidly after only one round of fine-tuning iterations. At the same time, compared with the model before the improvement, the accuracy was higher, and the stability of the model was better. The accuracy has greatly increased after one iteration, as seen in the next two figures, but has not significantly increased with the successive increase in iteration times, proving that model independent meta-learning may swiftly generalize to specific characteristics from generalities. The recognition accuracy is observed to fluctuate around the average accuracy, and the model has essentially been steady, according to the statistical average accuracy of the model in the later time.
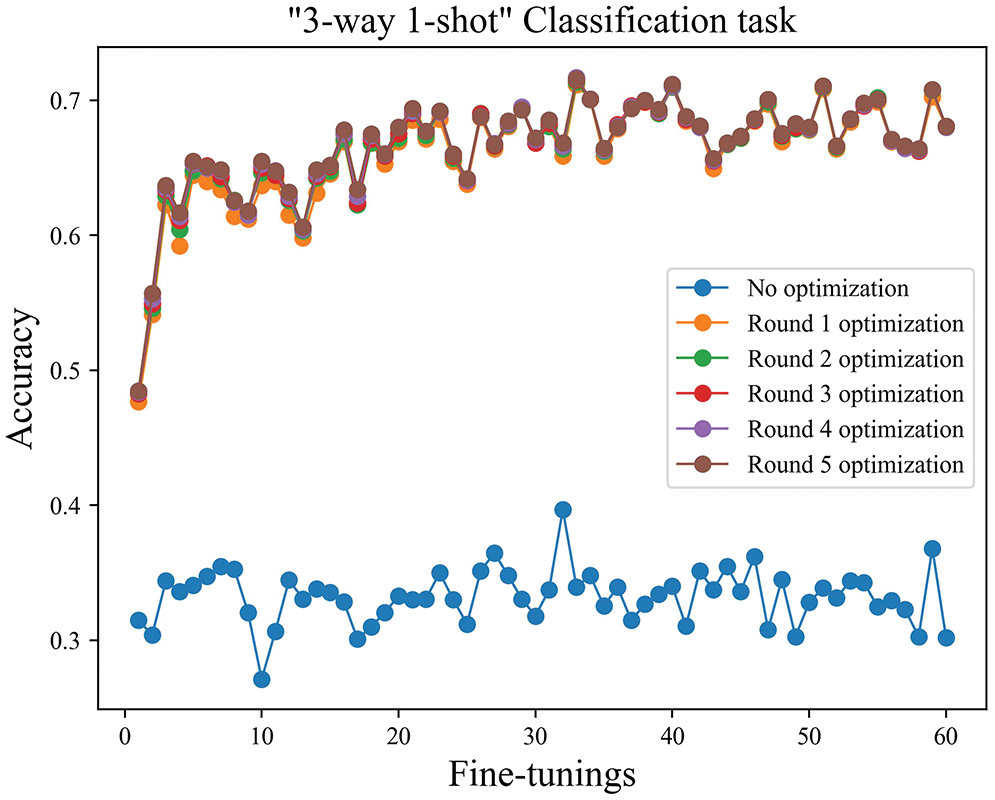
Figure 9: The three-way one-shot iteration diagram before improvement
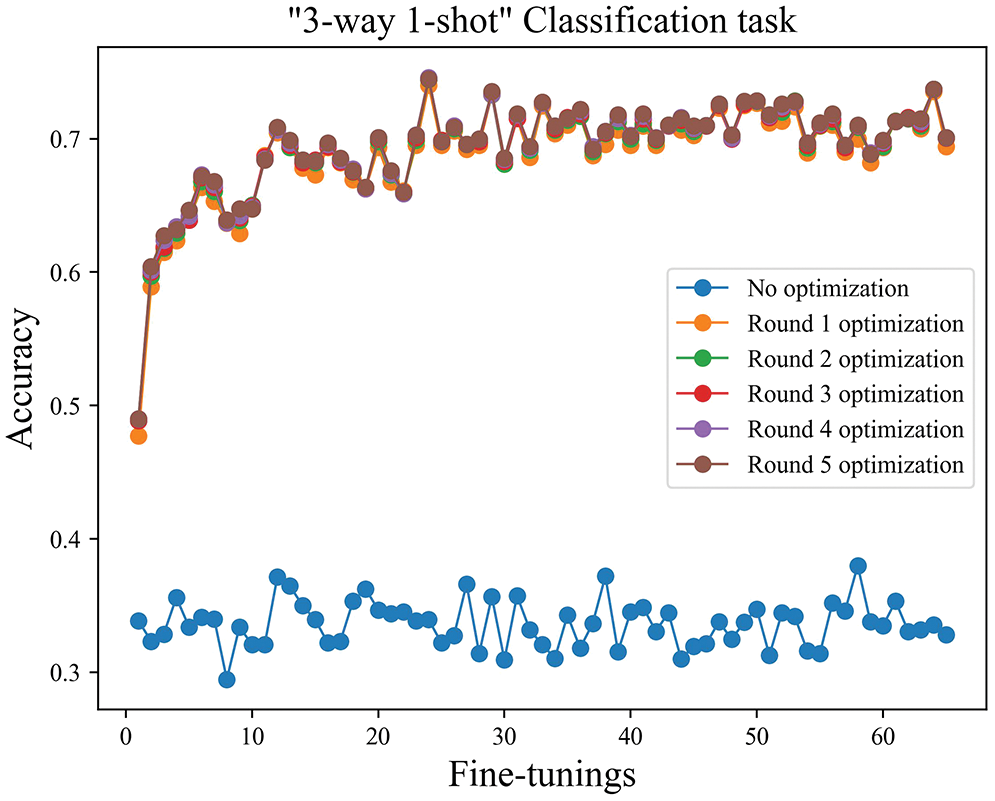
Figure 10: The three-way one-shot iteration diagram after improvement
Sample size analysis. For this analysis, the three-way tasks were selected as an example, and the test results of the one-, two-, and three-shot tasks were compared in terms of the model accuracy after five optimization rounds. As shown in Fig. 11, although model accuracy before improvement was increased with the sample size, this effect was not obvious between two- and three-shot tasks, and the effect of the two-shot task was even better than that of the three-shot in the later stages. When the number of samples increased, the model learned more about the specific disease features, so the overall accuracy was improved. As shown in Fig. 12, the improved model’s performance on the one-, two-, and three-shot tasks showed clearer and more reasonable layering and improved accuracy in disease identification as the number of samples increased.
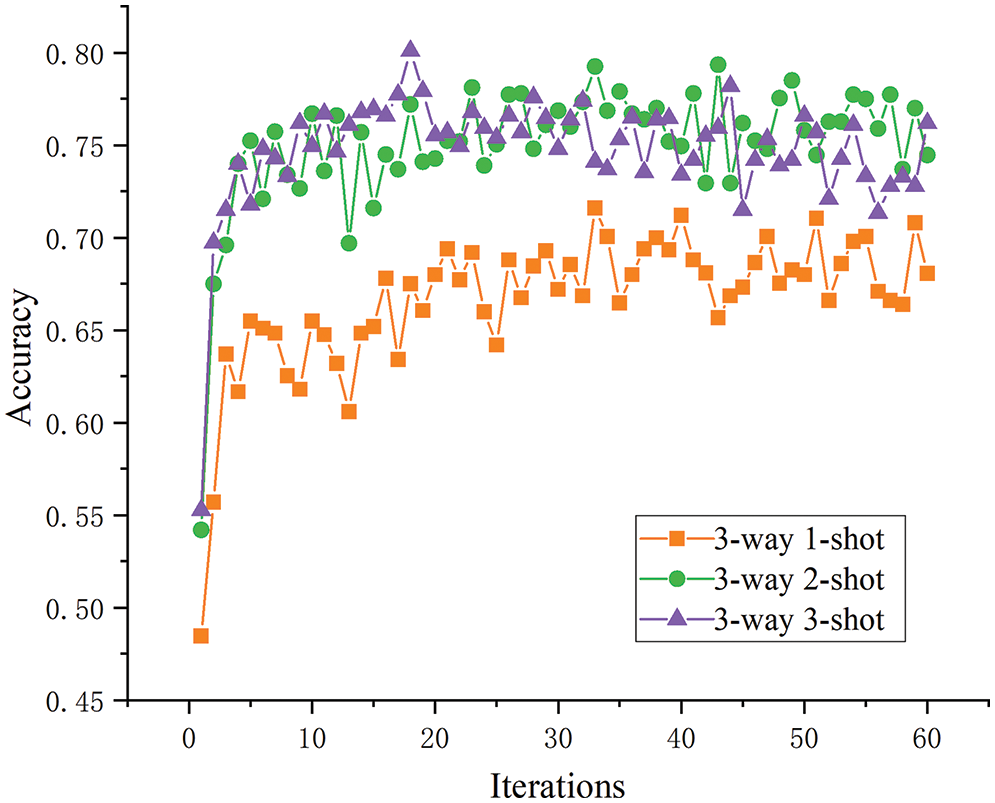
Figure 11: The three-way K-shot test accuracy before improvement
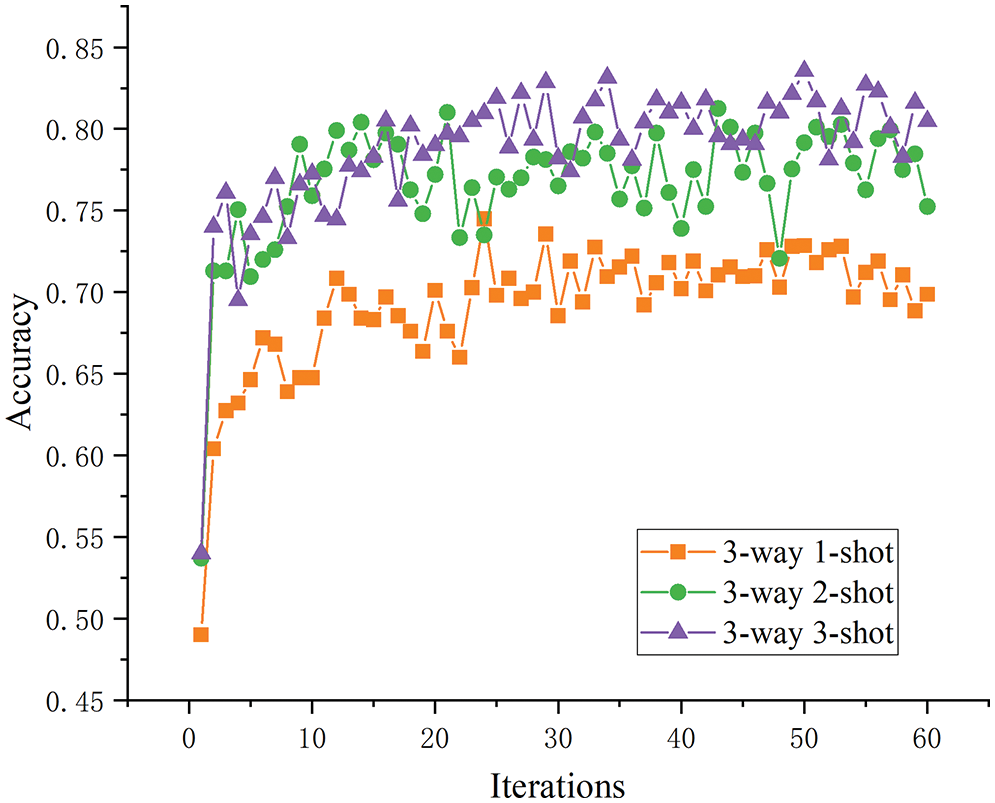
Figure 12: The three-way K-shot test accuracy after improvement
Overall accuracy analysis. The abscissa in Fig. 13 indicates the index of the 12 sets of N-way K-shot tasks. In Fig. 13, it can be observed that the overall recognition accuracy of the model on the training set was higher after improvement compared to that before the improvement, but the performance on the test set was consistent. The accuracy of the three-way three-shot task on the test set before the model improvement decreased by 2.31 percentage points (pp) compared to the three-way two-shot task. Theoretically, by increasing the number of samples, the recognition accuracy should not decrease significantly. However, this anomaly indicated that the network had poor robustness before improvement but performed better after the proposed improvement. In other cases, the average recognition accuracy increased with the number of samples, indicating that the improved model was more robust.
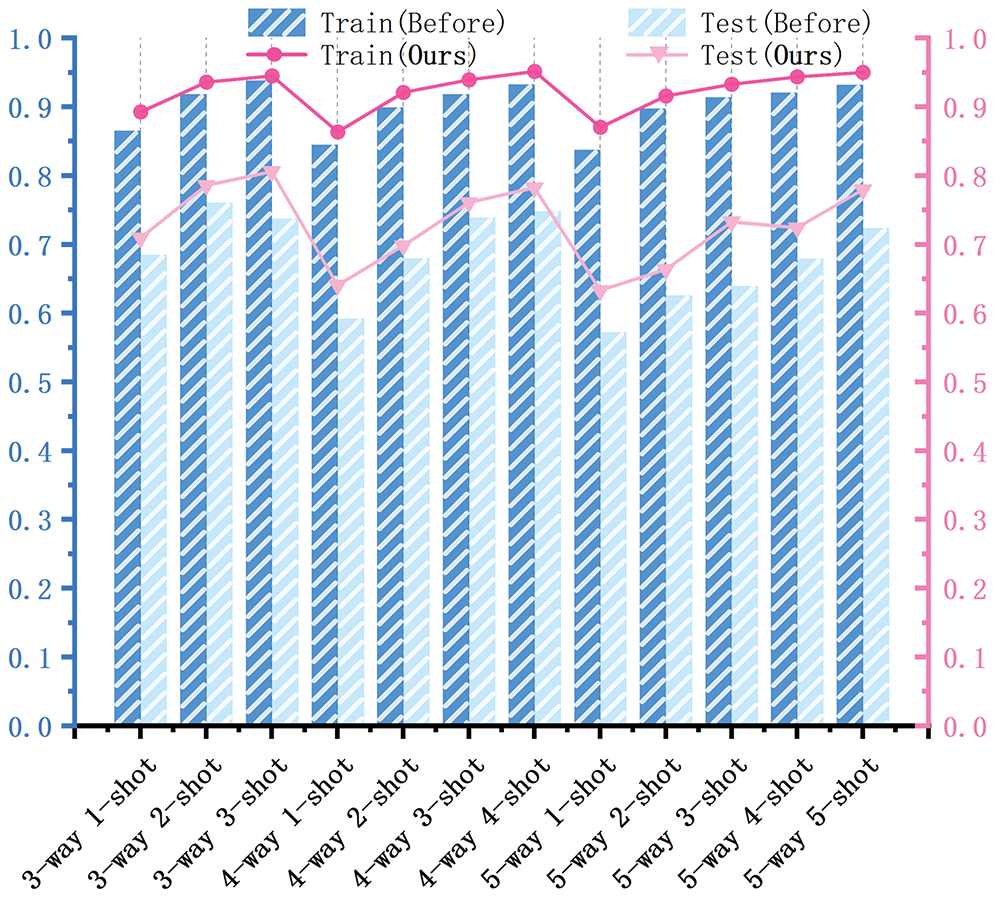
Figure 13: Comparison of the recognition accuracy of the model on the training and test sets before and after the improvement
Model structure analysis. Since the proposed model was based on the MAML model, it was suitable for small datasets and could rapidly update the parameters of a basic learner. The improved model achieved significantly higher accuracy using a local cross-channel interaction strategy without dimensionality reduction by adding only fewer parameters. In the improved model, different weights were assigned to the local features on channels after the convolutional layers to strengthen important information while suppressing redundant information, thus improving both the performance and the robustness of the model, as given in Eq. (4). Therefore, the improved model could focus on the disease characteristics while ignoring the background data of samples.
In Eq. (4),
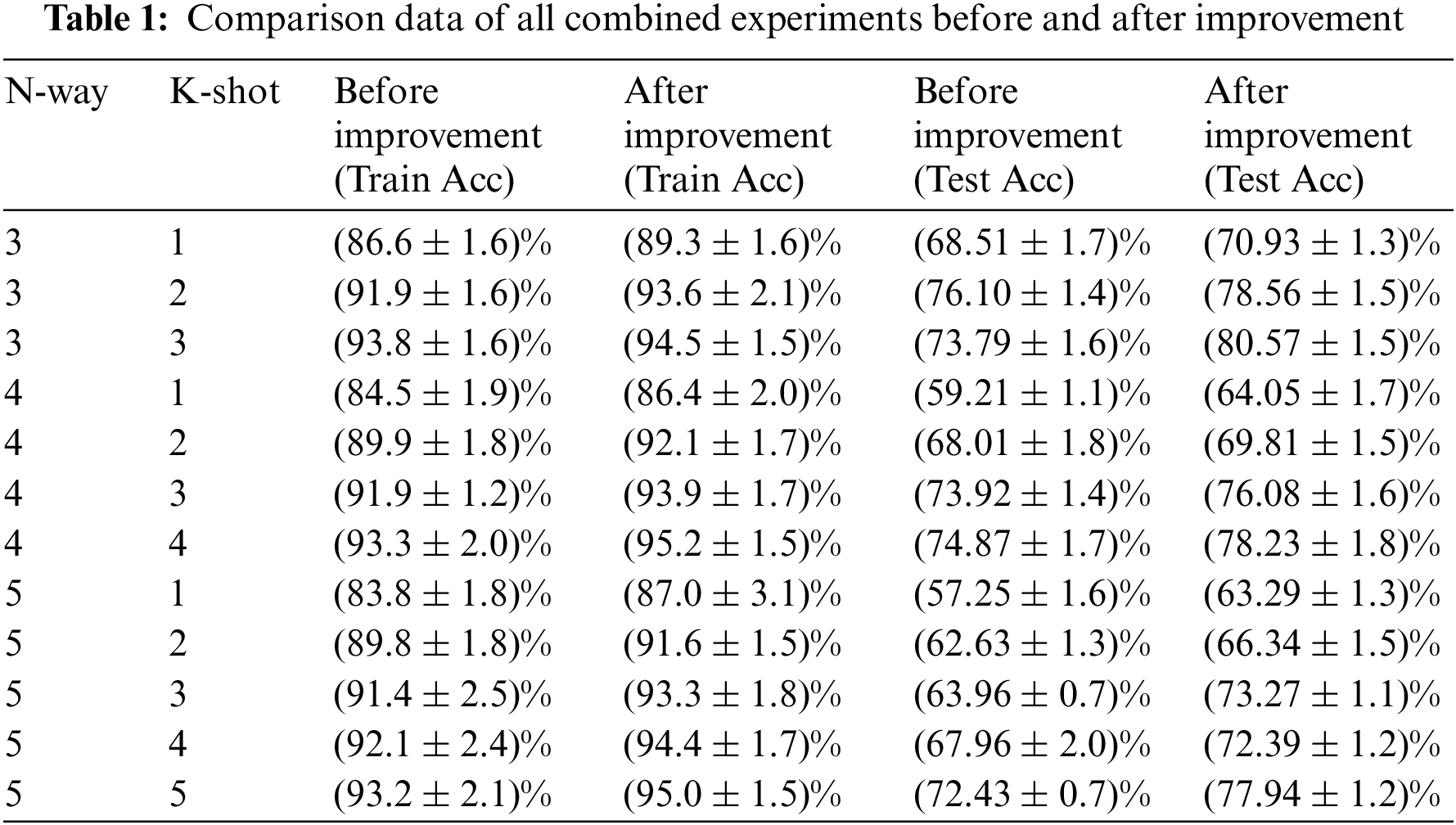
The ablation experiment was also conducted in this study based on the comparison experiment mentioned before. By altering the kernel size and the attention module, as indicated in Table 3, and using the 3 way 1 shot as an example, the superiority of the modified model is demonstrated. As seen in the table, the model performs better when the kernel size is 7 and ECA modules are introduced to the four convolutional layers. It is confirmed that the upgraded model once more has a greater recognition effect.
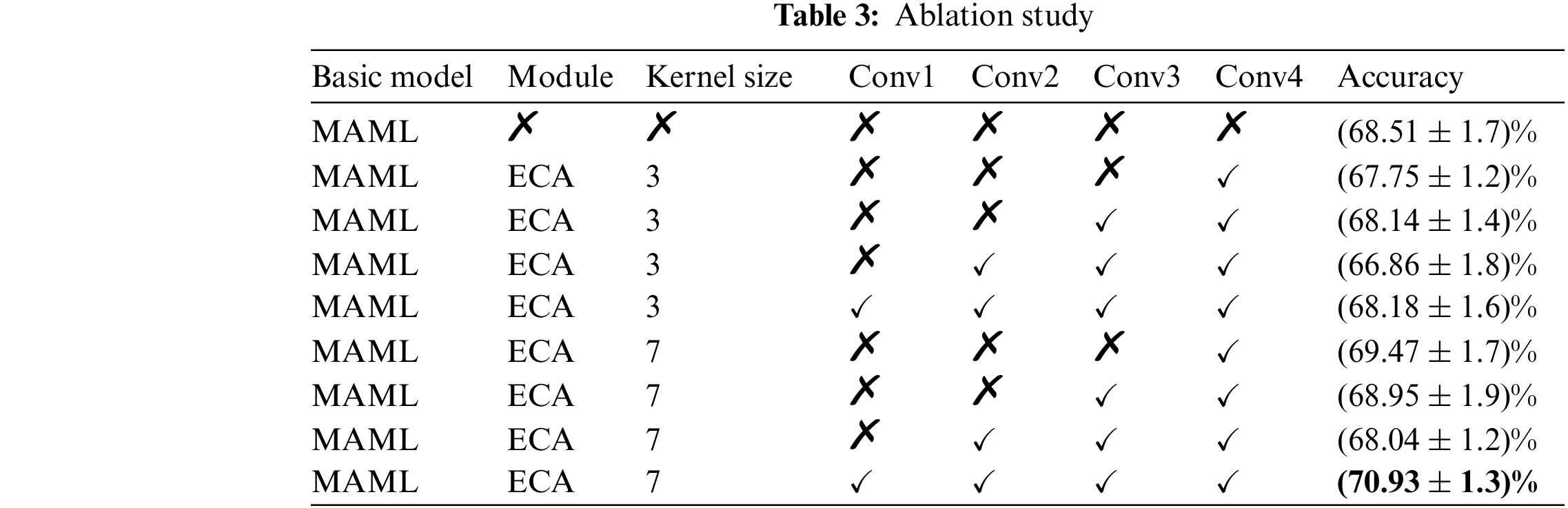
Traditional deep learning-based models require numerous data samples to train complex models, but in practical applications, data have been limited. To overcome this limitation, this paper proposes the MAML-attention model based on the MAML model to improve recognition accuracy under the condition of a limited number of data samples. The improved MAML-Attention model based on the MAML meta-learning algorithm has all advantages of the MAML model but strengthens its performance in classification tasks. The improved MAML-Attention model adds an attention mechanism after the convolution layer, so the specific features of diseased leaves can be extracted better, and their common features can be more accurately summarized. Namely, it can extract the same rules between diseases more efficiently than MAML, demonstrating a stronger generalization effect. In agricultural disease detection and classification applications, the MAML-Attention can identify diseases more efficiently and conveniently. Compared with the traditional deep learning-based approaches, which require large data sets, the MAML-Attention can achieve a good recognition effect using a small number of samples, so it has a good application prospect in the field of disease recognition. Compared with the other methods, the MAML-Attention has a better generalization accuracy in the case of small datasets.
Acknowledgement: We would like to thank MogoEdit (https://www.mogoedit.com) for its English editing during the preparation of this manuscript.
Funding Statement: This work was supported by the Science and Technology Project of Jilin Provincial Department of Education [JJKH20210346KJ], the Capital Construction funds within the budget of Jilin Province in 2021 (Innovation capacity construction) [2021C044-4], and the Jilin Province Science and Technology Development Plan Project [20210101185JC].
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Food and Agriculture Organization of the United Nations, “Moving forward on food loss and waste reduction,” in Proc. the State of Food and Agriculture, Rome, Italy, pp. 156, 2019. [Google Scholar]
2. N. V. Fedoroff, “Food in a future of 10 billion,” Food and Agriculture Organization of the United Nations, vol. 4, no. 1, pp. 1–10, 2015. [Google Scholar]
3. X. J. Tan, W. L. Cheor, K. S. Yeo and W. Z. Leow, “Expert systems in oil palm precision agriculture: A decade systematic review,” Journal of King Saud University—Computer and Information Sciences, vol. 34, no. 4, pp. 1569–1594, 2022. [Google Scholar]
4. R. A. C. Jones, “Global plant virus disease pandemics and epidemics,” Plants, vol. 10, no. 2, pp. 233, 2021. [Google Scholar] [PubMed]
5. D. Laborde, W. Martin, J. Swinnen and R. Vos, “COVID-19 risks to global food security,” Science, vol. 369, no. 6503, pp. 500–502, 2020. [Google Scholar] [PubMed]
6. S. A. Tsaftaris, M. Minervini and H. Scharr, “Machine learning for plant phenotyping needs image processing,” Trends in Plant Science, vol. 21, no. 12, pp. 989–991, 2016. [Google Scholar] [PubMed]
7. C. Zhou, S. Zhou, J. Xing and J. Song, “Tomato leaf disease identification by restructured deep residual dense network,” IEEE Access, vol. 9, pp. 28822–28831, 2021. [Google Scholar]
8. M. Brahimi, K. Boukhalfa and A. Moussaoui, “Deep learning for tomato disease: Classification and symptoms visualization,” Applied Artificial Intelligence, vol. 31, no. 4, pp. 299–315, 2017. [Google Scholar]
9. X. Zhang, Y. Qiao, F. Meng, C. Fan and M. Zhang, “Identification of maize leaf diseases using improved deep convolutional neural networks,” IEEE Access, vol. 6, pp. 30370–30377, 2018. [Google Scholar]
10. M. Dyrmann, H. Karstoft and H. S. Midtiby, “Plant species classification using deep convolutional neural network,” Biosystems Engineering, vol. 151, no. 1, pp. 72–80, 2016. [Google Scholar]
11. Y. Zhang, C. Song and D. Zhang, “Deep learning-based object detection improvement for tomato disease,” IEEE Access, vol. 8, pp. 56607–56614, 2020. [Google Scholar]
12. S. Ren, K. He, R. Girshick and J. Sun, “Faster R-CNN: Towards real-time object detection with region proposal networks,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 39, no. 6, pp. 1137–1149, 2017. [Google Scholar] [PubMed]
13. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” in Proc. Int. Conf. on Learning Representations, San Diego, CA, USA, pp. 1–14, 2015. [Google Scholar]
14. F. Saeed, M. A. Khan, M. Sharif, M. Mittal, L. M. Goyal et al., “Deep neural network features fusion and selection based on PLS regression with an application for crops diseases classification,” Applied Soft Computing, vol. 103, no. 2, pp. 107164, 2021. [Google Scholar]
15. W. -J. Hu, J. Fan, Y. -X. Du, B. -S. Li, N. Xiong et al., “MDFC-ResNet: An agricultural IoT system to accurately recognize crop diseases,” IEEE Access, vol. 8, pp. 115287–115298, 2020. [Google Scholar]
16. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in Proc. IEEE Conf. on Computer Vision and Pattern Recognition, Las Vegas, Nevada, USA, pp. 770–778, 2016. [Google Scholar]
17. L. Benos, A. C. Tagarakis, G. Dolias, R. Berruto, D. Kateris et al., “Machine learning in agriculture: A comprehensive updated review,” Sensors, vol. 21, no. 11, pp. 3758, 2021. [Google Scholar] [PubMed]
18. J. Schmidhuber, “Evolutionary principles in self-referential learning, or on learning how to learn: The meta-meta-.. hook,” Ph.D. Dissertation, Technische Universität München, 1987. [Google Scholar]
19. T. Hospedales, A. Antoniou, P. Micaelli and A. Storkey, “Meta-learning in neural networks: A survey,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 44, no. 9, pp. 5149–5169, 2022. [Google Scholar] [PubMed]
20. C. Finn, P. Abbeel and S. Levine, “Model-agnostic meta-learning for fast adaptation of deep networks,” in Proc. Int. Conf. on Machine Learning, Sydney, NSW, Australia, vol. 70, pp. 1126–1135, 2017. [Google Scholar]
21. J. Hu, L. Shen, S. Albanie, G. Sun and E. Wu, “Squeeze-and-excitation networks,” in Proc. Computer Vision and Pattern Recognition, SLC, Utah, USA, pp. 7132–7141, 2018. [Google Scholar]
22. J. Hu, L. Shen, S. Albanie, G. Sun and A. Vedaldi, “Gather-excite: Exploiting feature context in convolutional neural networks,” in Proc. Advances in Neural Information Processing Systems, Montreal, Canada, vol. 31, pp. 9423–9433, 2018. [Google Scholar]
23. X. Nie, L. Wang, H. Ding and M. Xu, “Strawberry verticillium wilt detection network based on multi-task learning and attention,” IEEE Access, vol. 7, pp. 170003–170011, 2019. [Google Scholar]
24. S. Woo, J. Park, J. Lee and I. S. Kweon, “CBAM: Convolutional block attention module,” in Proc. European Conf. on Computer Vision, Munich, Bayern, Germany, pp. 3–19, 2018. [Google Scholar]
25. Q. Wang, B. Wu, P. Zhu, P. Li, W. Zuo et al., “ECA-Net: Efficient channel attention for deep convolutional neural networks,” in Proc. Computer Vision and Pattern Recognition, Seattle, WA, USA, pp. 11531–11539, 2020. [Google Scholar]
26. H. Yu, X. Cheng, Z. Li, Q. Cai, C. Bi et al., “Disease recognition of apple leaf using lightweight multi-scale network with ECANet,” Computer Modeling in Engineering & Sciences, vol. 132, no. 3, pp. 711–738, 2022. [Google Scholar]
27. X. Li, Y. Gu and A. Pižurica, “A unified multiview spectral feature learning framework for hyperspectral image classification,” IEEE Transactions on Geoscience and Remote Sensing, vol. 60, pp. 1–14, 2022. [Google Scholar]
28. J. Snell, K. Swersky and R. S. Zemel, “Prototypical networks for few-shot learning,” in Proc. Neural Information Processing Systems, Long Beach, California, USA, pp. 4077–4087, 2017. [Google Scholar]
29. J. Oh, H. Yoo, C. Kim and S. Yun, “BOIL: Towards representation change for few-shot learning,” in Proc. Int. Conf. on Learning Representations, Vienna, Austria, 2021. [Google Scholar]
30. A. Raghu, M. Raghu, S. Bengio and O. Vinyals, “Rapid learning or feature reuse? Towards understanding the effectiveness of MAML,” in Proc. Int. Conf. on Learning Representations, Addis Ababa, Ethiopia, 2020. [Google Scholar]
31. A. Antoniou, H. Edwards and A. J. Storkey, “How to train your MAML,” in Proc. Int. Conf. on Learning Representations, New Orleans, Louisiana, USA, 2019. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools