 Open Access
Open Access
ARTICLE
A Modified Deep Residual-Convolutional Neural Network for Accurate Imputation of Missing Data
Intelligent System Research Group, Faculty of Computer Science, Universitas Sriwijaya, Palembang, 30139, Indonesia
* Corresponding Author: Siti Nurmaini. Email:
Computers, Materials & Continua 2025, 82(2), 3419-3441. https://doi.org/10.32604/cmc.2024.055906
Received 10 July 2024; Accepted 21 October 2024; Issue published 17 February 2025
Abstract
Handling missing data accurately is critical in clinical research, where data quality directly impacts decision-making and patient outcomes. While deep learning (DL) techniques for data imputation have gained attention, challenges remain, especially when dealing with diverse data types. In this study, we introduce a novel data imputation method based on a modified convolutional neural network, specifically, a Deep Residual-Convolutional Neural Network (DRes-CNN) architecture designed to handle missing values across various datasets. Our approach demonstrates substantial improvements over existing imputation techniques by leveraging residual connections and optimized convolutional layers to capture complex data patterns. We evaluated the model on publicly available datasets, including Medical Information Mart for Intensive Care (MIMIC-III and MIMIC-IV), which contain critical care patient data, and the Beijing Multi-Site Air Quality dataset, which measures environmental air quality. The proposed DRes-CNN method achieved a root mean square error (RMSE) of 0.00006, highlighting its high accuracy and robustness. We also compared with Low Light-Convolutional Neural Network (LL-CNN) and U-Net methods, which had RMSE values of 0.00075 and 0.00073, respectively. This represented an improvement of approximately 92% over LL-CNN and 91% over U-Net. The results showed that this DRes-CNN-based imputation method outperforms current state-of-the-art models. These results established DRes-CNN as a reliable solution for addressing missing data.Keywords
An electronic health record (EHR) is a digital file containing medical details, such as lab results, of an individual, and it’s stored online [1,2]. Primarily, EHRs serve the purpose of establishing goals, devising patient care plans, recording care delivery, and evaluating the resulting outcomes [2]. Thus, it can be shared among various facilities and accessed swiftly by either patients or medical personnel. These records offer opportunities to improve patient care, integrate performance metrics into clinical practice, and streamline clinical research [2,3]. For instance, one research utilizing EHRs involves using machine learning regression techniques to forecast cardiovascular risk [4]. Such predictive models can function as decision-support tools aiding doctors and healthcare providers in patient management and proactive interventions [5]. However, EHR datasets often exhibit a certain degree of incompleteness, with missing data affecting the quality of results and potentially introducing bias [6]. Missing data occurs when values for variables of interest are not recorded or measured for all subjects in the sample. There are several reasons for missing data, including patient refusal to complete certain questions, data acquisition equipment errors, and measurements with different sampling periods [7].
The most commonly used methods for estimating missing data generally fall into three categories [3]: deletion methods, methods for linear imputation based on statistics, and methods for non-linear imputation based on machine learning. Deletion approaches entail the direct removal of missing data, which, as prior research has demonstrated, can cause considerable changes in the analysis results due to the absence of critical information [8]. In contrast, statistically-based linear imputation methods involve replacing missing values with statistical measures such as the mean [9], or the most common value within a global or local range of the time series data [10]. While they use correlations between measurements across multiple data streams simultaneously, they ignore correlations within individual data streams [11]. As a result, these approaches may overlook important information. Furthermore, the advantages of these methods are the ease of implementation and interpretation, and the availability of various tools and techniques to support their use [12]. However, they often do not consider complex structures in the data and may produce biased or inaccurate results depending on the case at hand [13]. Alternatively, machine learning-based non-linear imputation methods can handle more complex correlations in the data, but they require greater computational resources and more difficult interpretation [14]. In practical use, method selection should consider the balance between practicality, accuracy, and interpretation of results. In the field of imputation missing data analysis, different strategies, such as deep learning (DL) algorithms have become more popular recently [15,16].
Alternative approaches such as deep learning (DL) methods are increasingly attracting attention in missing data imputation analysis [16]. This is because DL methods have proven to be able to take advantage of and manage large data sets with remarkable capabilities [17]. The Convolutional Neural Network (CNN) excels in capturing complex patterns and relationships within structured data [18]. In [19–21], several works have demonstrated the ability of CNNs to effectively process structured data, such as high-dimensional data, through the application of convolutional layers that focus on local features and hierarchical pattern recognition. However, CNN may struggle with imputing missing values if the data lacks spatial relationships, as the learning of kernel weights relies heavily on identifying spatial features. On the other hand, U-Net is specifically designed for spatial relationships that are essential for effective imputation [22]. However, if there isn’t any discernible spatial relationship in the data, the kernel’s learning process can go off course and provide inefficient imputation. The U-Net architecture was found to be less effective in managing missing data when the complex data structure did not align with the patterns that the U-Net is designed to recognize [22].
Current imputation methods, including traditional statistical methods and machine learning approaches like CNN and U-Net, have limitations in handling complex data structures and correlations within individual data streams. These methods either oversimplify the imputation process or require substantial computational resources without guaranteeing accurate results [18]. CNN and U-Net struggle to handle complex data structures and correlations within individual data streams effectively [18,22]. This gap underscores the need for a more advanced imputation method that can address these challenges by maintaining computational efficiency while improving accuracy, particularly in the context of medical datasets.
To address these limitations, we propose a novel neural network architecture called Deep Residual-Convolutional Neural Network (DRes-CNN). The DRes-CNN model modifies the foundation of the original Residual Neural Network (ResNet) architecture [22]. DRes-CNN integrates advanced techniques to enhance both feature extraction and learning capabilities. One key aspect of DRes-CNN is the incorporation of residual connections not only within individual layers but also across different network modules. This facilitates the flow of gradients during training, alleviating the vanishing gradient problem and enabling more efficient optimization. Additionally, by promoting information flow across the network, these connections enable better preservation of useful information throughout the deep layers, ultimately improving the network’s ability to learn intricate patterns [23]. We also explore two different neural network architectures, including Low Light-Convolutional Neural Network (LL-CNN) and U-Net to illustrate how the imputation process’s use of time information inclusion has improved.
The purpose of this study is to evaluate the robustness of our imputation method and fill in the missing information. The main conclusions of our research demonstrate how well DRes-CNN imputes missing values in medical information datasets with remarkable accuracy. The research presents novel findings in four areas:
• Development of a novel DRes-CNN architecture: We propose an advanced Deep Residual-Convolutional Neural Network (DRes-CNN) specifically designed to improve data imputation accuracy, addressing the limitations of existing methods.
• Comprehensive evaluation of diverse datasets: Our approach is tested on three publicly available datasets (MIMIC-III, MIMIC-IV, and the Beijing Multi-Site Air Quality dataset) with various types of missingness, ensuring robustness across different scenarios.
• Application to real-world healthcare data: The proposed DRes-CNN model is implemented for multivariate data imputation in medical applications, demonstrating its practical relevance in clinical settings.
• Superior performance in imputation and prediction tasks: Quantitative comparisons show significant improvements in RMSE over existing state-of-the-art methods, confirming the effectiveness of our approach in both imputation and downstream prediction tasks
Missing data imputation has been a longstanding challenge in various domains. Traditional approaches often rely on statistical methods or simple imputation techniques, which can introduce bias if assumptions about the data distribution are incorrect. In recent years, deep learning-based methods have emerged as promising alternatives, offering more flexible and powerful approaches.
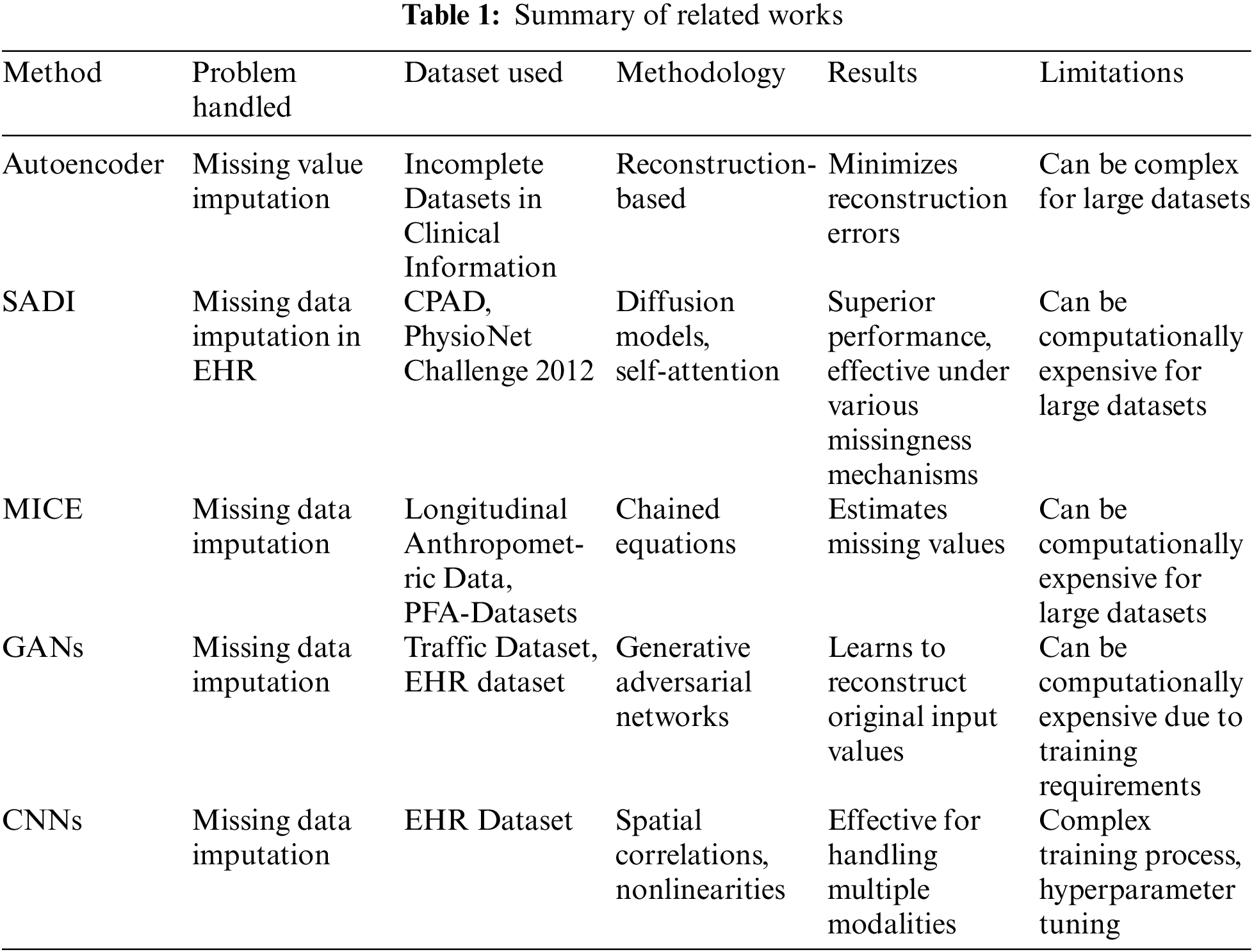
Previous studies have explored a range of techniques for handling missing data. For instance, autoencoder models [24] aim to learn representations that can reconstruct the original data, minimizing reconstruction errors. This study used clinical data to train the power of the model. However, for large and complex datasets this model cannot yield promising effectiveness. Similarity-aware diffusion Model-Based Imputation (SADI) [25] leverages diffusion models and self-attention to effectively impute missing values in EHRs. Current models typically rely on correlations between time points and features, which is effective for data with strong correlations, such as Intensive Care Unit (ICU) data. However, this method is less effective for non-ICU data with weak correlations. This study uses the Critical Path for Alzheimer’s Disease (CPAD) and PhysioNet Challenge 2012 datasets. The Similarity-Aware Diffusion Model-Based Imputation (SADI) method demonstrates superior performance compared to current state-of-the-art models. SADI, which leverages diffusion models and a self-attention mechanism, is effective in imputing missing data under various missingness mechanisms, including Missing Completely At Random (MCAR), Missing At Random (MAR), and Missing Not At Random (MNAR), and is better at improving accuracy and reducing bias Applying the clustering process to big datasets with many of patients can be computationally expensive, which is one of SADI’s limitations. In [26,27], using the method of multiple imputation by chained equations (MICE), a chained approach is used to mine all relevant evidence of missing values and to further estimate those missing values. The datasets used in this study include the Longitudinal Anthropometric Data [26] and the PFA-Datasets in Clinic from [27]. However, in the case of very large datasets, machine learning-based imputation approaches require significant computation time, making these approaches less practical for the task of imputing a large amount of missing data.
In Generative Adversarial Networks (GANs) [28,29] have also been applied to missing data imputation, generating realistic samples to fill in missing values. GANs can suffer from mode collapse, where the generator produces limited diversity in the imputed data. In clinical settings, where variability and accuracy are crucial, mode collapse can lead to biased or incomplete imputations that do not adequately represent the range of possible values These weaknesses make GANs less suitable for clinical data imputation compared to other methods that might offer more stability, transparency, and accuracy. CNNs have also been studied in [30] for the imputation of missing data, utilizing their ability to find spatial correlations and nonlinearities within the data. CNNs have the ability to use techniques such as fuzzy c-means clustering to group data into clusters according to membership values. As a result, it is possible to arrange the data in a way that captures robust attribute relationships. Multiple modalities’ worth of data can be handled and imputed simultaneously by CNNs. However, because the training process is complex and precise hyperparameter tweaking is necessary, imputation tasks may be challenging to comprehend. Due to its complexity, the model may be less useful for real-world applications, particularly in situations where computing resources are scarce or when access to professionals with the necessary experience to fine-tune such models is difficult.
In contrast to existing methods, our proposed approach, DRes-CNN, leverages deep residual-based CNNs to extract deep features from structured data and impute missing values using a learned kernel. This novel approach offers promising results, outperforming existing methods in key performance metrics. This novel approach offers promising results, outperforming existing methods in key performance metrics. Table 1 for details of the conclusions of several approaches to methods used for handling missing data.
We provide a brief overview of the datasets utilized in the study in this section, the benchmarking exercise, and the method for assessing the quality of imputation illustrated in Fig. 1.
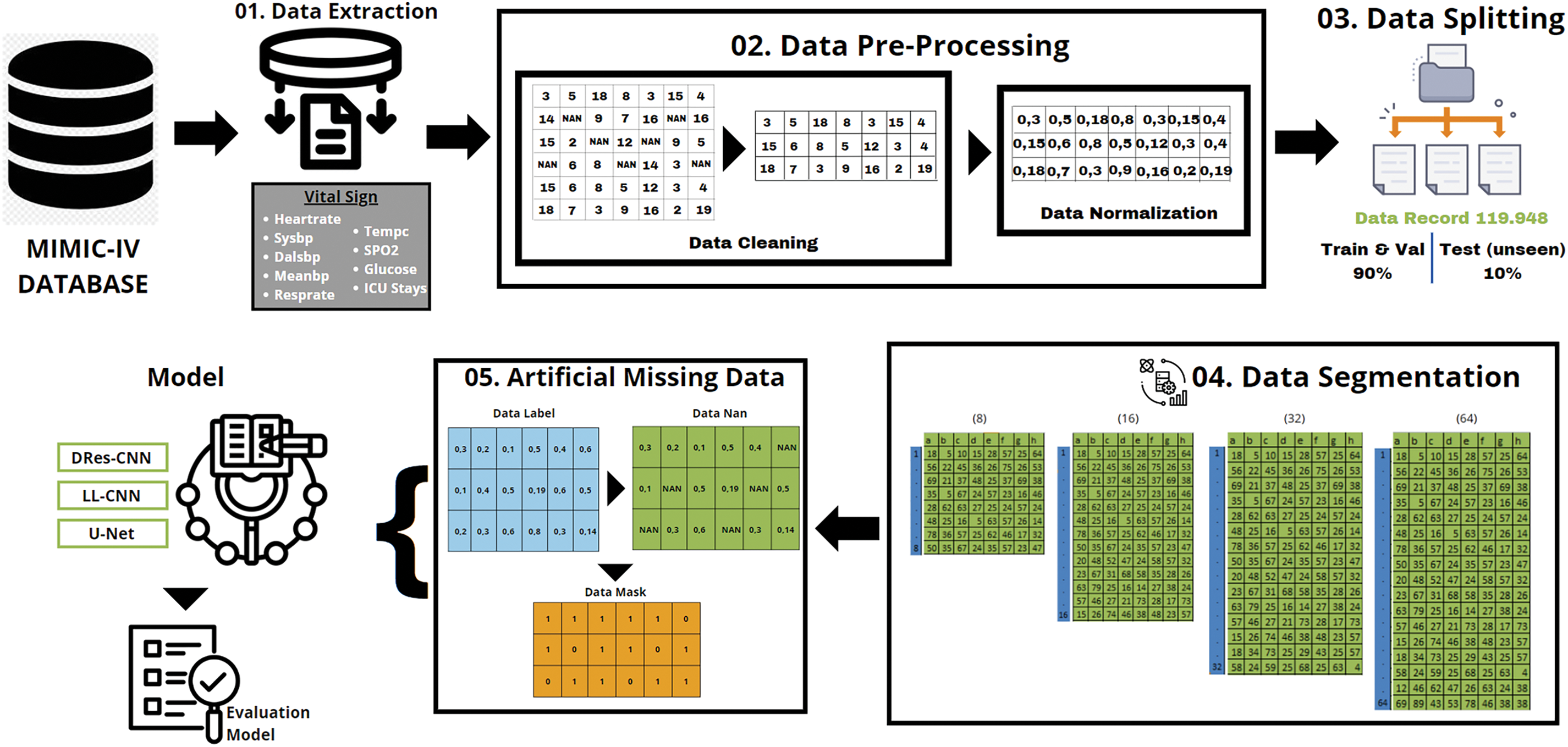
Figure 1: The research methodology
Vital signs and pertinent data taken from the MIMIC-IV database were used in our investigation [31,32]. We received permission to extract data from the MIMIC-IV for research purposes (Certification Number: 50640755) and completed the web-based training course offered by the National Institutes of Health to ensure the safety of human study participants before gaining access to the database. High-quality data from 2008 to 2019 is included in the publicly accessible MIMIC-IV database, which was created by the review boards of Beth Israel Deaconess Medical Center and Massachusetts Institute of Technology (MIT), both located in Cambridge, Massachusetts. MIMIC-IV contains extensive patient data, including demographics, hospitalization records, diagnoses made under the International Classification of Diseases, Ninth Revision (ICD-9), laboratory tests, prescription drugs, procedures, fluid balance, discharge summaries, bedside vital sign measurements, treatment notes, radiology reports, and survival data. The MIMIC-IV database consists of numerous linked tables that use standardized ICD-9 numbers to describe the data of each ICU patient.
Data preprocessing in this study entails some procedures and techniques meant to handle missing or insufficient data, as depicted in Fig. 1. The primary procedures for Preprocessing data for imputation usually entail: Missing Data Detecting: Identifying that attributes or variables within the dataset contain missing values. To approximate missing values, one can do the following: remove rows or columns containing missing values; replace missing values with the mean of the available values within the corresponding attribute; create predictive models to estimate missing values based on relationships with other variables, and utilize statistical distributions to create random values that roughly match the distribution of the available data. Use a large query based on a specific value from the chart events table of the MIMIC-IV database in the itemid column to obtain the vital signs dataset of ICU patients.
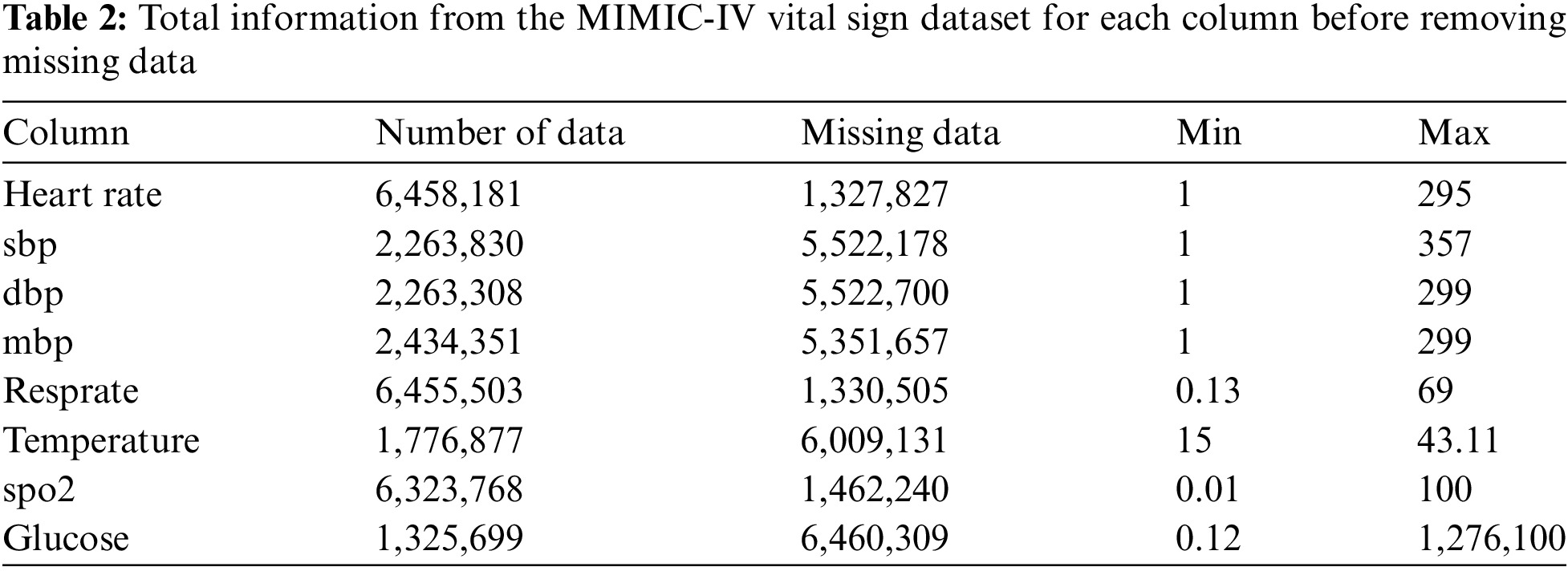
• The total raw data from the ICU patient vital signs dataset is 7.786.008 with missing data (Table 2), has a unique id or stay_id for each patient from 38675535 to 37145532, and the duration of each patient’s stay in the ICU from 2182-01-25 07:00:00+00:00 to 2146-04-27 09:33:00+00:00.
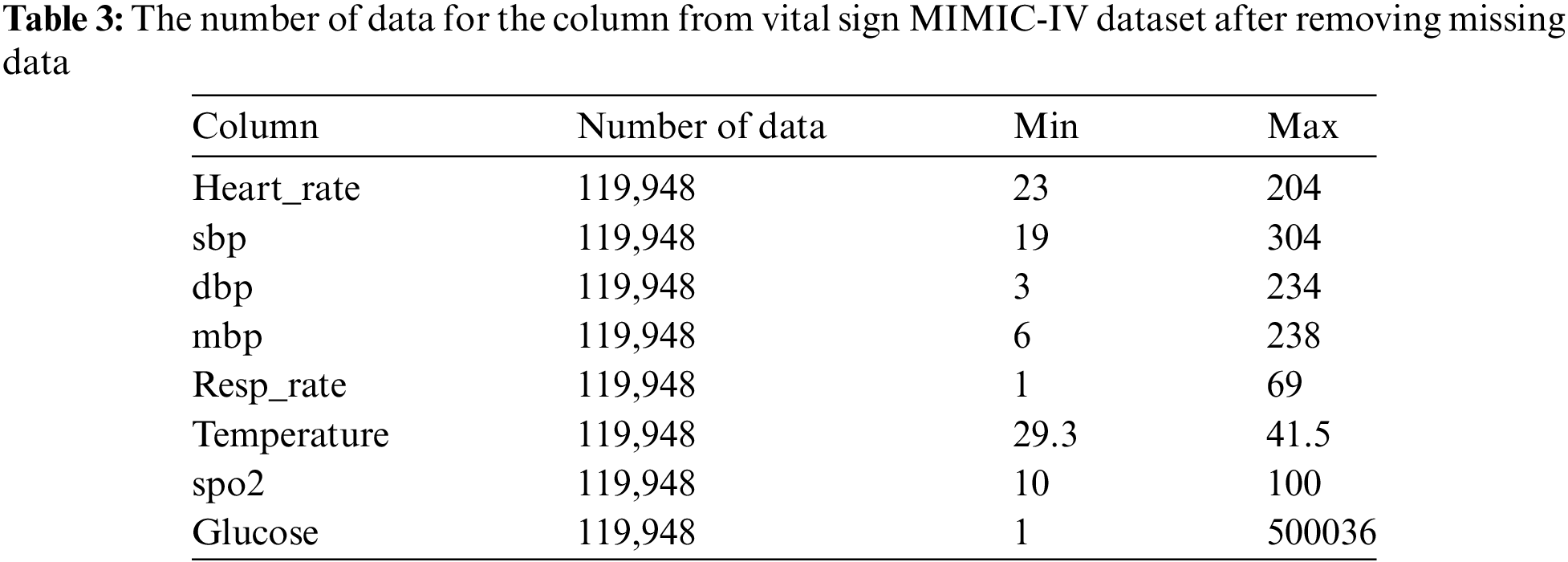
Our proposed model analyzes eight vital sign features: heart rate, systolic blood pressure, diastolic blood pressure, mean blood pressure, respiratory rate, body temperature, oxygen saturation, and glucose level. We used an inner join to get a more extensive dataset. Consequently, our final dataset contained 119,948 unique ICU patients (vital signs) as a result (Table 3). This entire dataset includes the min and max data of vital signs recorded during a patient’s ICU stay.
• Standardization, after imputation of the missing data the dataset was normalized using the Min-Max Scaler [33,34], this procedure is vital to guarantee uniformity or standardization among the features. The following Eq. (1) illustrates the data normalization:
where
In this step, concerning MIMIC-IV datasets, a data adjustment process ensues, aiming to extract the minimum volume of data from each icustay_id. The meticulous step serves as the foundation for generating multiple data segments from every icustay_id, which is pivotal for subsequent analysis or modeling endeavors. The segmentation strategy employed entails categorizing the data into various subsets, comprising 8, 16, 32, and 64 groups based on the Eq. (2) as follows:
3.3 Create Artificial Missing Data
We have a dataset structured as
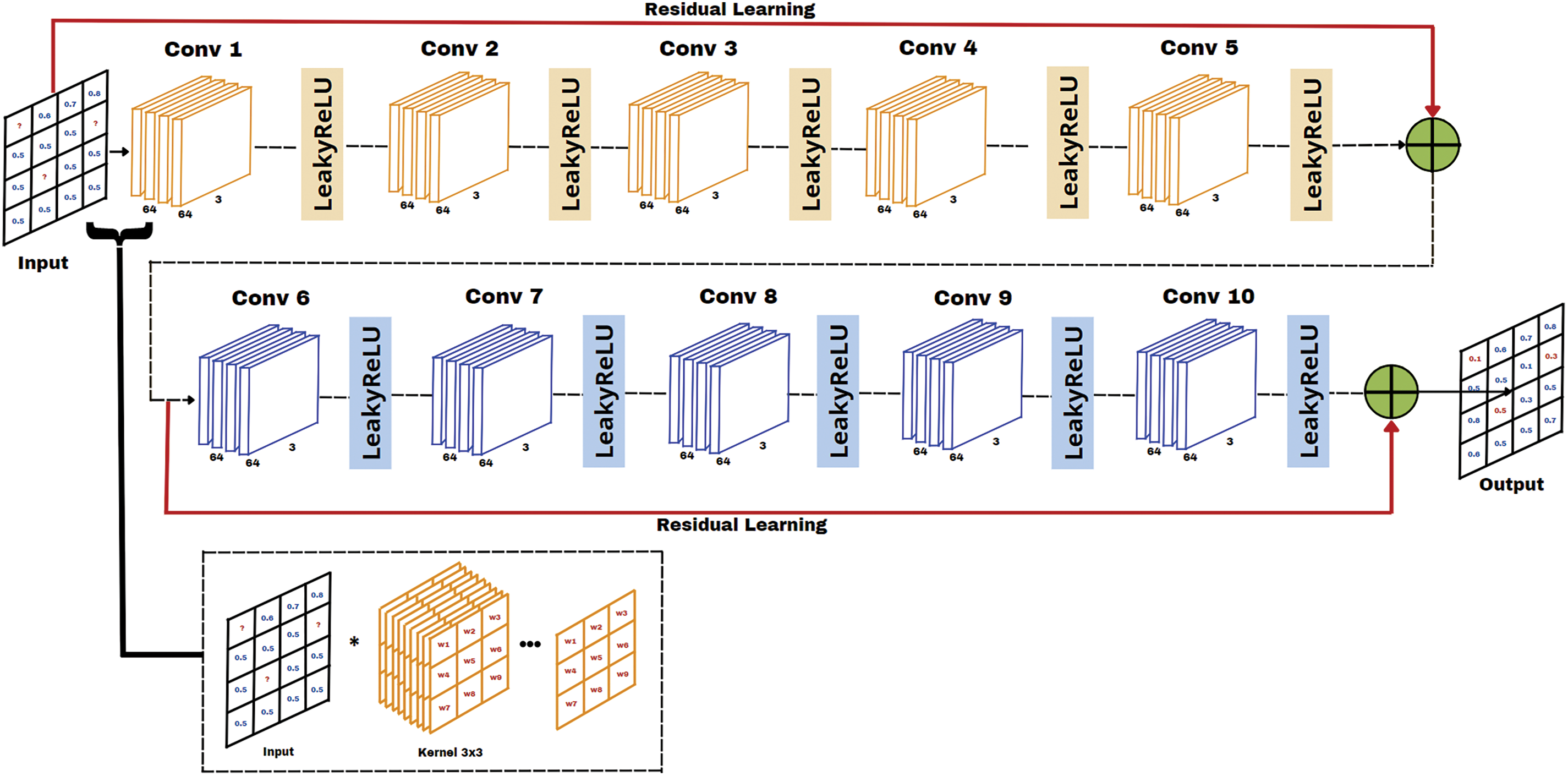
Figure 2: Proposed DRes-CNN architecture

3.4 The Proposed Model DRes-CNN
The proposed method utilizes a specific type of neural network architecture known as Convolutional Neural Networks (CNN). Our network architecture, which is a dense convolutional neural network with residual connections (DRes-CNN), is depicted in Fig. 2. This DRes-CNN network structure resembles the original CNN architecture for image super-resolution [20], but is modified to fit tabular or time-series data in capturing complex patterns and relationships within structured data [18]. This architecture is made to maximize the information flow between network levels using a straightforward connecting scheme. All other levels are directly connected. Every layer passes its feature map to all the following levels and receives extra input from all prior layers to preserve the feed-forward character of the system. Dense networks are one type of CNN that connects each layer layer in a feed-forward way. These dense networks increase feature propagation, stimulate feature reuse, and solve the vanishing-gradient problem in contrast to typical convolutional networks, where each layer is merely connected to the next layer.
The convolutional layers in our proposed DRes-CNN architecture are specifically designed to detect dependencies between features in the EHR dataset, both temporally and contextually. By employing dense and residual connections, the model enhances feature propagation, reuses information from earlier layers, and improves the learning process for long-range feature dependencies. Dense and residual connections function differently. While feature maps in residual connections are combined by summation before being transmitted to a specific layer, feature maps in dense connections are merged based on all prior feature maps. The benefit of having both kinds of connections is that, in a dense connection, all of the features from the prior convolution block may be used, whereas, in a residual connection, we are limited to using one historical feature map.
As can be seen in Fig. 2, the suggested DRes-CNN architecture consists of two residual connections and ten convolutional layers with progressively more filters. The residual connection in Fig. 2 is the solid line that carries the input layer to the summing operator. In ideal circumstances, a deep neural network with more layers can outperform shallower networks and drastically minimize error rates. To outperform a straightforward deep neural network, we incorporate residual blocks.
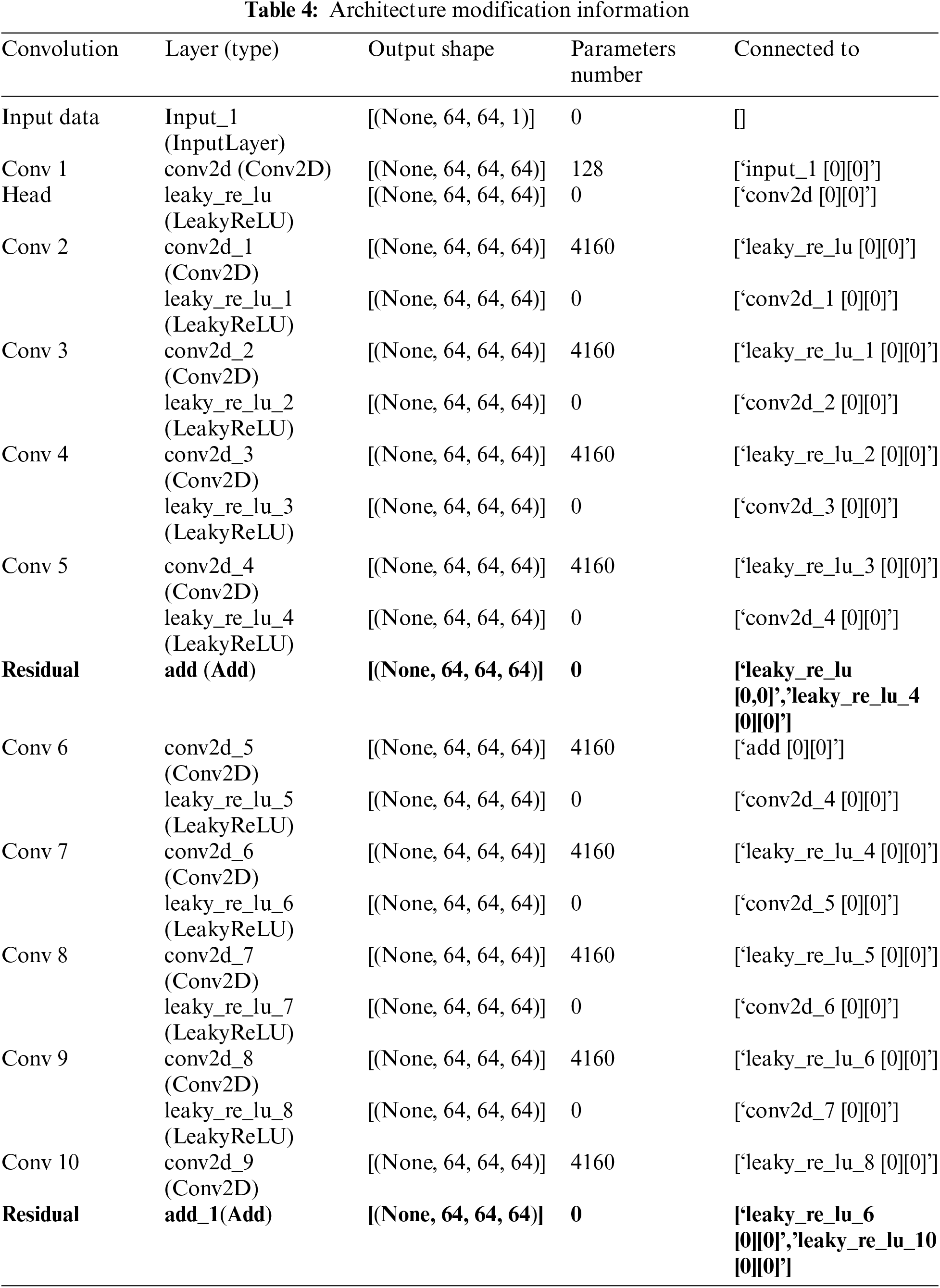
The architecture in our model has 64 filters with multiple kernel sizes of 3 × 3, followed by the LeakyReLU activation function. After multiple convolution layers, there is residual linking which is done by adding the output of the first layer to the output of the last layer before the next convolution layer. This architectural design helps in learning more detailed features and retains the original information of the input. This study, provided detailed descriptions of our architecture, specifying that it consists of ten convolutional layers with both dense and 2 residual connections. We explained the advantages of using both connection types, as dense connections facilitate the utilization of features from previous convolutional blocks, while residual connections enhance the flow of information by summing the outputs of earlier layers with later ones. We also justified the choice of architecture by noting that deeper networks tend to outperform shallower counterparts, especially when incorporating residual blocks that minimize error rates. The total parameters in the model amount to 8.545, which also includes the weights and biases in each convolution layer. To offer transparency, we included Table 3, which outlines the architecture modification information, detailing output shapes and the number of parameters for each layer. The architecture in our model has modification information shown in Table 4.
Residual connections are introduced after Conv 5 and Conv 10, which are created by adding a convolutional layer’s output to a preceding layer’s output before the layer’s activation function is sent through (Table 3). A situation with randomly generated missing data in the input data matrix and the same whole data matrix produced at the output is shown in Fig. 3. Assume that we have a kernel with dimensions M × N (where M represents the number of rows and N denotes the number of columns), and there’s a missing value for a unit
where
where

Figure 3: The performance analysis of RMSE (MIMIC-IV) with U-Net, LL-CNN and DRes-CNN. The x-axis shows the missing ratio datasets and the y-axis shows the RMSE
The evaluation process entails using the available data to train the network to predict missing data, with the model being adaptable for tabular or time series data. Important features are preserved across levels by using jump connections. Several hyperparameters, such as the Adam optimizer, 200 epochs, batch sizes of 16 and 32, and a learning rate of 0.001, are used to maximize the model’s performance. The model’s activation function is linear, and the loss function is a mean squared error. By partitioning the dataset into multiple subsets, we were able to validate the model’s performance across different data segments, thereby enhancing the generalizability of the results. Additionally, a learning rate schedule based on exponential decay is employed to enhance the training. Algorithm 2 presents the pseudocode for the DRes-CNN algorithm.

During training, the learning rate of the model can be adjusted to facilitate a more efficient convergence to the optimal solution. Combining optimization methods with hyperparameters improves the performance of the DRes-CNN algorithm. The Intel® CoreTM i9-9920X CPU running at 3.50 GHz, 490,191 MB of RAM, and GeForce 2080 RTX Ti from NVIDIA Corporation GV102 (rev a1) were the characteristics of the personal computer (PC) used to train the network. Pytorch 1.7.1 package together with Python were used to create the network.
Model evaluation in filling data with DRes-CNN involves evaluating how the trained deep learning model is used to predict missing values in the dataset. Model performance evaluation uses the root mean squared error (RMSE) metric to assess the average error produced by the model [9]. The smaller the value of mean absolute error (MAE) and RMSE, the better the performance of our imputation method. MAE and RMSE are used as model evaluation metrics [9] and are described in Eqs. (5) and (6).
where
4 Experimental Results and Discussions
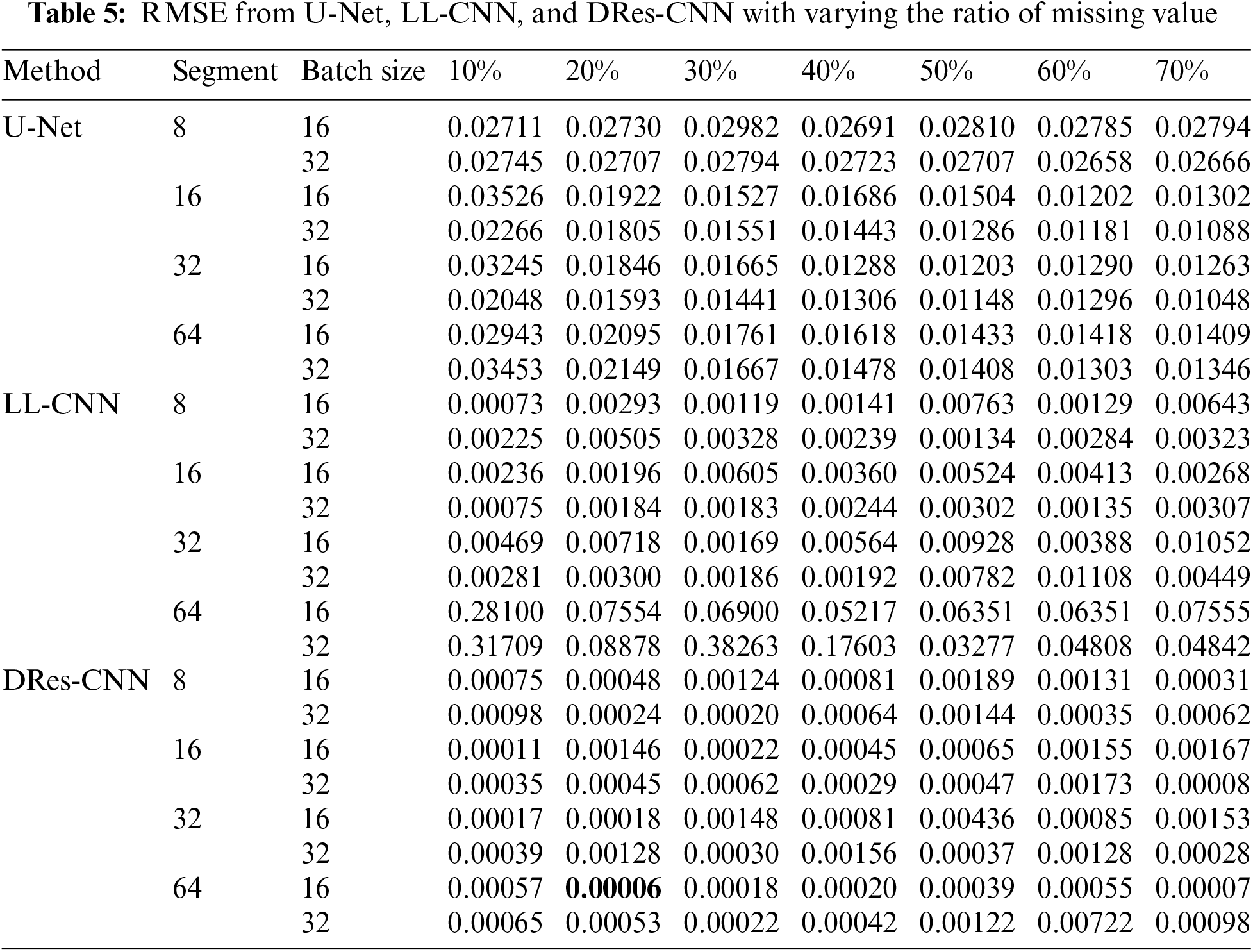
This section presents the study results of our suggested approach and evaluates the significant improvement in handling missing data performance compared to other methods. We use the MIMIC-IV dataset described in Section 3.2. At first, data cleaning for missing data are randomly added, replacing the missing values with zero. Next, our proposed DRes-CNN approach is used to impute those missing values, and other imputation techniques, such as U-Net and LL-CNN, are selected for comparison purposes. Moreover, the robustness of the DRes-CNN approach is tested with various missing ratios (10%, 20%, 30%, 40%, 50%, 60%, and 70%) on the MIMIC-IV dataset. We also evaluated the imputation performance of DRes-CNN by using three supervised learning algorithms to train classifiers on the imputed data. Experiments were conducted three times with random data partitioning (90% for training and 10% for testing), and the average classification accuracy along with the standard deviation was calculated as the final performance score. Initially, 10% of attribute values from all clean data were artificially removed to generate a dataset with missing values.
This section provides a thorough examination of comparing the proposed technique with alternative imputation methods. Table 4 illustrates the assessment outcomes (RMSE) acquired through various approaches when dealing with randomly generated missing values, alongside other advanced imputation methods like U-Net, LL-CNN, and DRes-CNN. Experimental results were obtained through various missing ratios (i.e., 10%–70%). It can be seen in Table 5 that the a substantial difference in performance between the proposed method and the others. Notably, regardless of the missing ratio, the RMSE results remain relatively stable, indicating that the proposed approach maintains its efficacy even when the level of missing data increases. Fig. 3, a graphical depiction of Table 5, makes this distinction easy to see. The best value for each dataset is displayed in a column of Table 4 in bold type. Particularly, the superiority of the DRes-CNN method is underscored, showcasing its adeptness in accurately imputing missing values within vital signs data, even under high missing data scenarios. Our results demonstrated that the Dres-CNN model consistently outperformed the other models, particularly when handling higher ratios of missing data. This performance can be attributed to key architectural enhancements, such as the use of skip and residual connections, which allow the model to retain both high-level and low-level features across layers. This retention is critical for capturing complex spatial relationships and non-linear dependencies, which are essential for accurate imputation.
The experimental findings showcased that the several models, trained used batch size of 16, 64 segments, a 20% missing ratio, and undergoing 200 epochs, attained an impressive on the MIMIC-IV dataset (as depicted in Table 6). Through scrutinizing the model’s efficacy across varied segment sizes and missing rates, we discern its capability to address an array of data patterns and handle missing data occurrences that mirror real-world scenarios, wherein data may manifest varying levels of incompleteness.
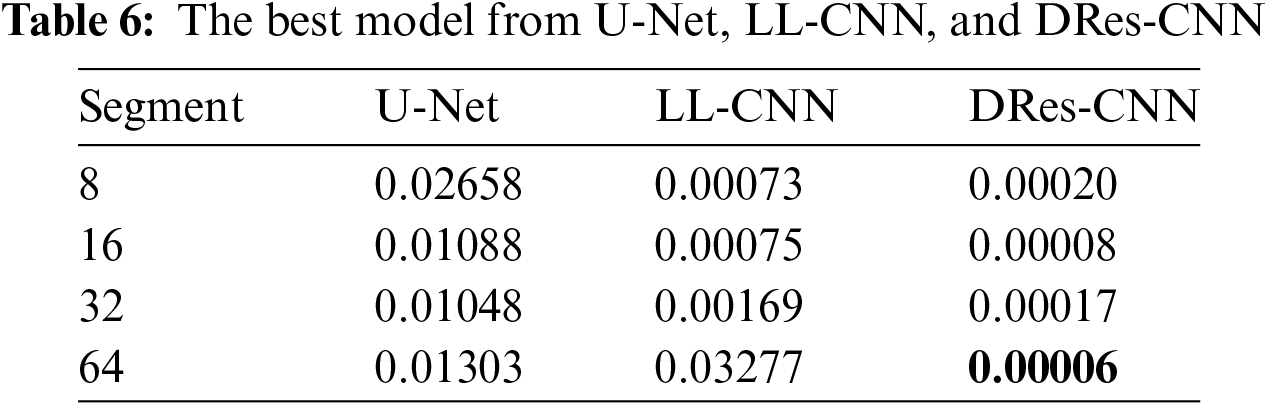
From the data presented in Tables 5 and 6, it’s evident that DRes-CNN surpasses both U-Net and LL-CNN in performance. The model’s ability to generate low RMSE suggests that the imputed data closely aligns with the true values missing from the dataset. A primary factor contributing to DRes-CNN superiority in data imputation is its tailored design for data processing tasks, where missing or corrupted data is common. Also, we conduct a comparison of each attribute from U-Net, LL-CNN, and DRes-CNN (Table 7).
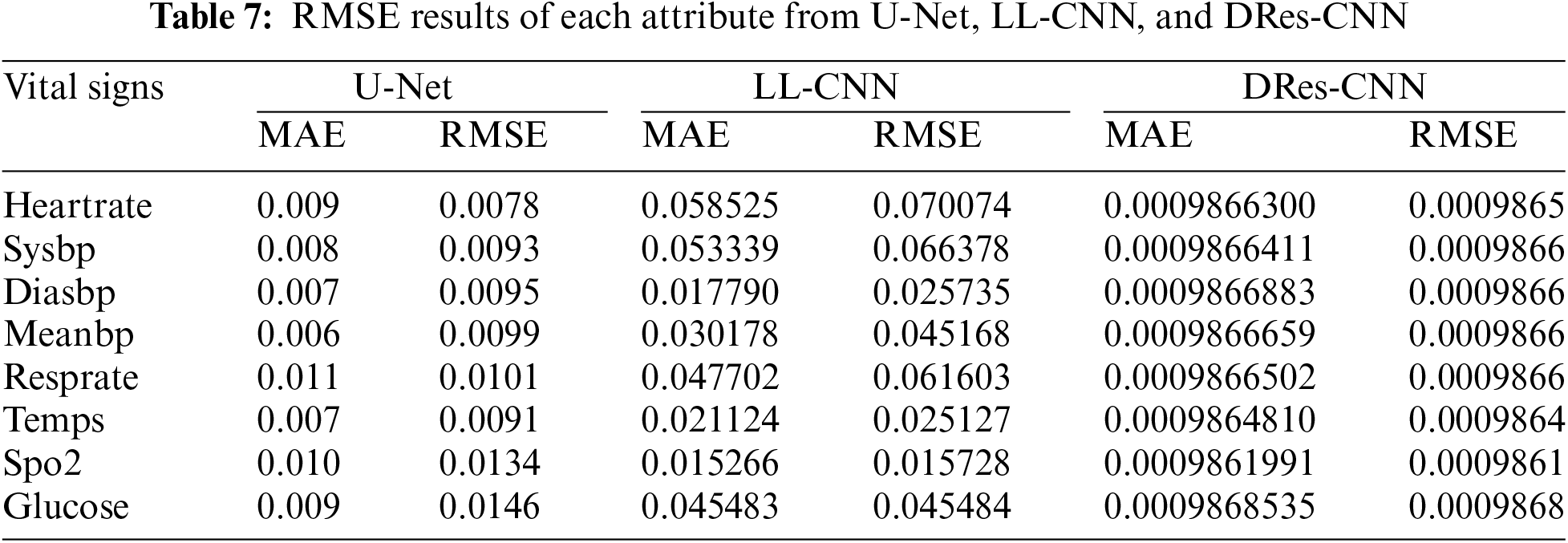
Table 6 presents a comparison of the RMSE values of three different models: U-Net, LL-CNN, and DRes-CNN, in predicting various vital signs attributes. The analysis results show that DRes-CNN consistently has a lower RMSE compared to U-Net and LL-CNN for each attribute. This indicates that DRes-CNN tends to perform better in predicting vital signs values. However, there were variations in model performance between attributes, where some attributes had smaller RMSE than others, such as “spo2” which had relatively low RMSE for all models. The lower the RMSE value, the closer the model prediction is to the true value, so models with lower RMSE tend to have more accurate predictions. A direct comparison between the models can provide insight Into the relative merits of each in predicting certain attributes, although overall DRes-CNN dominates. The best model to use for the vital sign prediction challenge can be chosen with the help of this analysis, as well as highlighting areas where improvements or adjustments may be needed.
The Ablation Study
In this study, we conducted an ablation study to evaluate the performance of our proposed Dres-CNN model for handling missing data in structured datasets. The study focused on assessing the impact of different hyperparameters, and missing data ratios. We tested the model with varying missing data percentages (10% to 70%) and systematically tuned hyperparameters such as segment size (8, 16, 32, 64), batch size (16, 32), and learning rate (0.001), as well as the optimizer (Adam) and the number of epochs (200). to determine their influence on imputation accuracy. A comprehensive experiment and a detailed comparison of the results are provided in the subsequent section with an in-depth analysis shown in Fig. 4.
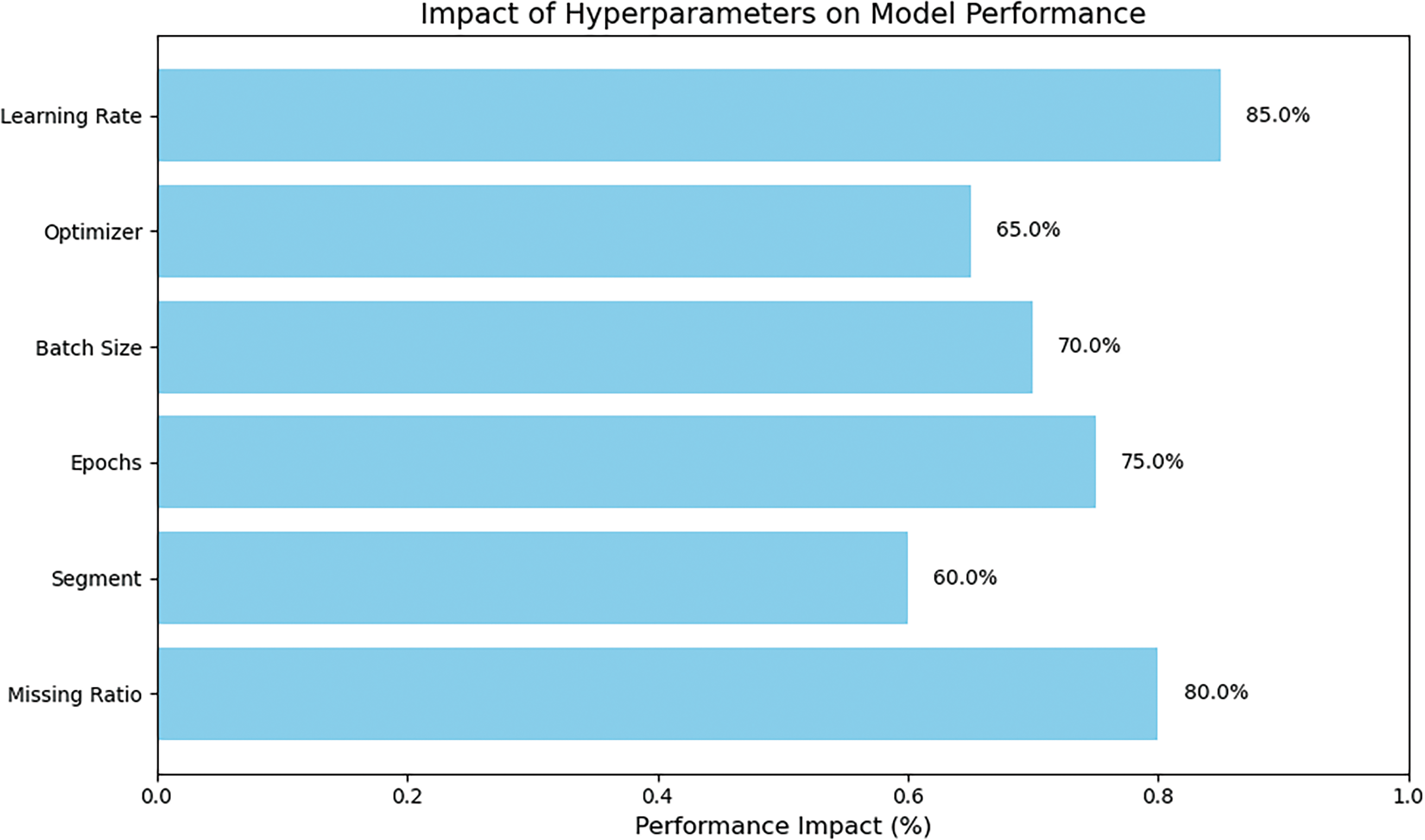
Figure 4: The performance of Dres-CNN model based on hyperparameters
The performance of each model was assessed using the parameter RMSE. Our results demonstrate that the Dres-CNN model outperforms traditional methods, such as LL-CNN and U-Net, especially when dealing with higher missing data ratios. First, we evaluated the effect of different missing ratios, ranging from 10% to 70%. The results showed that the model performed best at lower missing ratios (around 10%), with performance gradually decreasing as the missing ratio increased. This indicates that the model is more efficient at handling datasets with fewer missing entries. Next, we examined segment sizes (8, 16, 32, 64) and found that larger segment sizes (32 and 64) provided better accuracy, as they allowed the model to capture a broader context, though they required more computational resources. We also set the number of training epochs to 200, which appeared to strike a balance between sufficient training time and avoiding overfitting. In terms of batch size, we tested sizes 16 and 32, with the larger batch size proving to be more effective due to its more stable gradient updates during training. The Adam optimizer was chosen for its adaptive learning rate, which allowed the model to converge efficiently. Finally, we found that a learning rate of 0.001 provided the best balance between speed and accuracy, ensuring the model did not converge too quickly or slowly.
Our proposed DRes-CNN approach shows superior performance compared to other state-of-the-art imputation methods, such as U-Net and LL-CNN. Our proposed validation technique involves splitting the data into 90% for training and validation and 10% for testing. This split allows the model to train on a significant portion of the data while using the separate test set to rigorously evaluate the model’s performance on unseen data. Performance evaluation based on the RMSE value shows that the DRes-CNN architecture is more effective in learning features and estimating missing values accurately, even with a significant amount of missing data. Fig. 3, a graphical depiction of Table 4, makes this distinction easy to see. The best value for each dataset is displayed in a column of Table 4 in bold type. As a result, the DRes-CNN architecture produces lower RMSE values compared to the U-Net and LL-CNN architectures. The experimental results also show that in most cases, the DRes-CNN approach outperforms other methods in various data imputations. Although the imputation of missing data with LL-CNN architecture gives better results on the original (clean) version compared to U-Net architecture, DRes-CNN architecture shows better performance on the MIMIC-IV dataset. The superior performance of these imputation methods is most likely due to the suitability of the particular learning algorithm to the data used. Residual connections facilitate deeper network training and utilize convolutional layers with the LeakyReLU activation function to extract features from the input data.
The architecture of DRes-CNN incorporates specialized features like convolutional layers with LeakyReLU activation functions to extract features from input data and have residual connections to facilitate the training of deeper networks. These characteristics render it well-equipped to handle missing or corrupted data within datasets. The ability of DRes-CNN to handle temporal and spatial dependencies within data is very useful in situations when missing data appears sequentially, like in time series data. By leveraging convolutional and pooling layers, DRes-CNN effectively captures these dependencies, enhancing predictive accuracy and enabling more effective imputation of missing data values.
Additionally, DRes-CNN considers spatial relationships among features by reorganizing data or applying convolution operations with larger windows to capture broader contexts, facilitating better data imputation. Non-linear information is utilized through non-linear activation functions and optimized training methods, allowing the network to adjust its weights and biases according to the complex data structure. Through optimized training and testing phases, the network is trained on incomplete data to optimize imputation performance and then tested on incomplete data to evaluate its ability to accurately impute missing values.
To see the robustness of the DRes-CNN model, we use box plots to illustrate the impact of handling data. Box plots show the median value, interquartile range, and outliers, giving a clear view of the distribution of the data. Initially, the box plot depicts the data distribution before imputation (Fig. 5a), followed by a representation of the post-imputation process (Fig. 5b to d). The noticeable trend is that the proposed imputation method consistently handles missing data with values to the data mean. Comparing the box plots of the imputed data with the raw data shows a narrower interquartile range and a reduction in data dissemination (Fig. 5d). These observations suggest that the imputation method, rooted in DRes-CNN, effectively minimizes data variability without introducing bias, confirming its suitability for the task at hand.
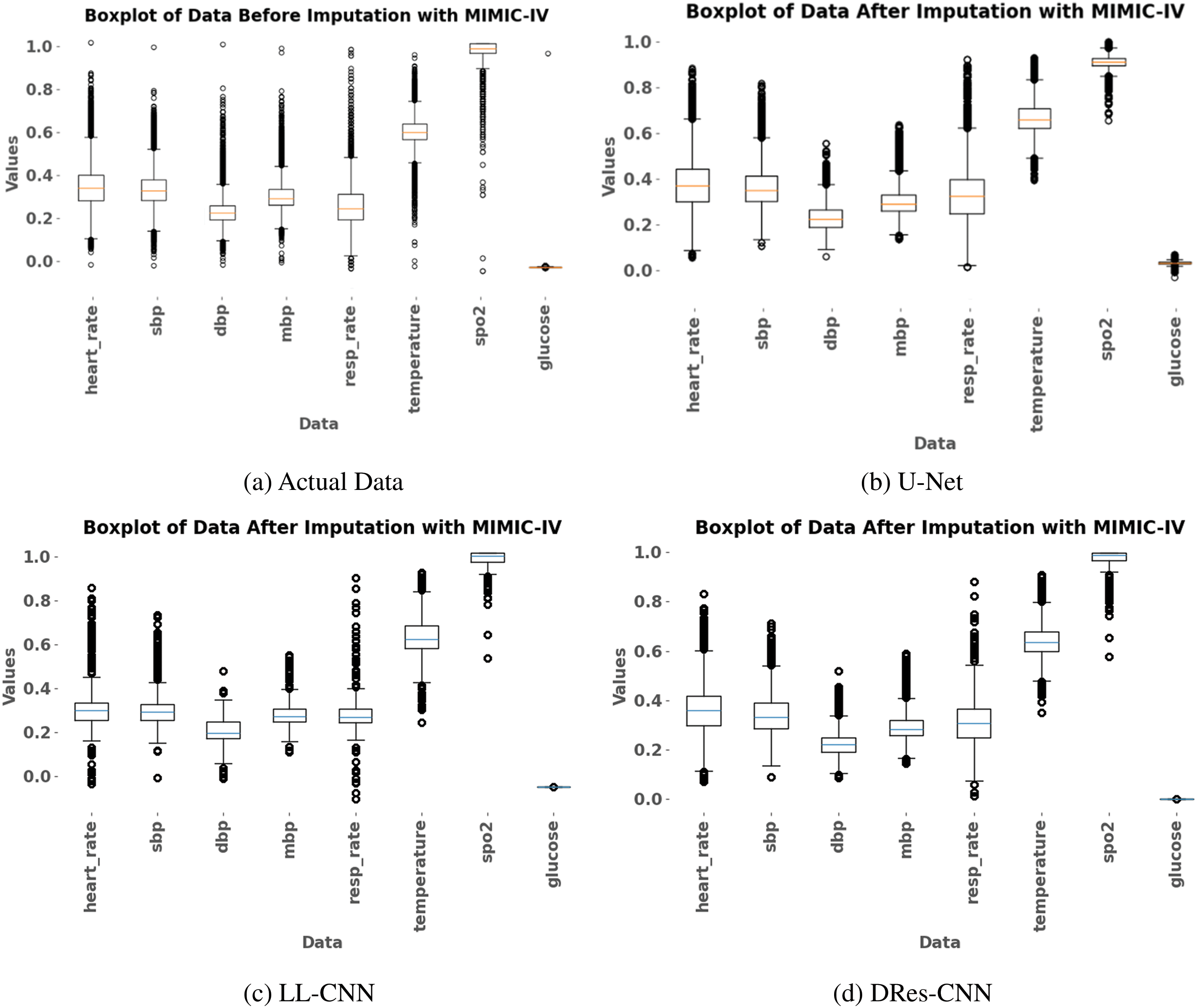
Figure 5: The box plot is utilized for visualizing data representation both pre- and post-imputation using three distinct methods: U-Net, LL-CNN, DRes-CNN
To Validate the robustness of our proposed data imputation model, we conducted a comparative analysis against recent studies that use the existing architecture of the CNN method. However, the use of DRes-CNN architecture for data imputation is still not common. We also use some non-health datasets to test the robustness of our architecture (Table 8). To the best of our knowledge, we are the pioneer in applying the DRes-CNN architecture for data imputation in medical records or other datasets. Our simple DRes-CNN model shows outstanding performance, as evidenced by Table 7, in terms of RMSE beyond previous studies. In the research conducted by Yoon et al. [35], the imputation method used was multi-directional recurrent neural network (M-RNN), which was evaluated on the MIMIC-III database. The evaluation results showed that M-RNN had an RMSE of 0.0295, indicating a fairly good level of imputation accuracy in the context of such medical data.
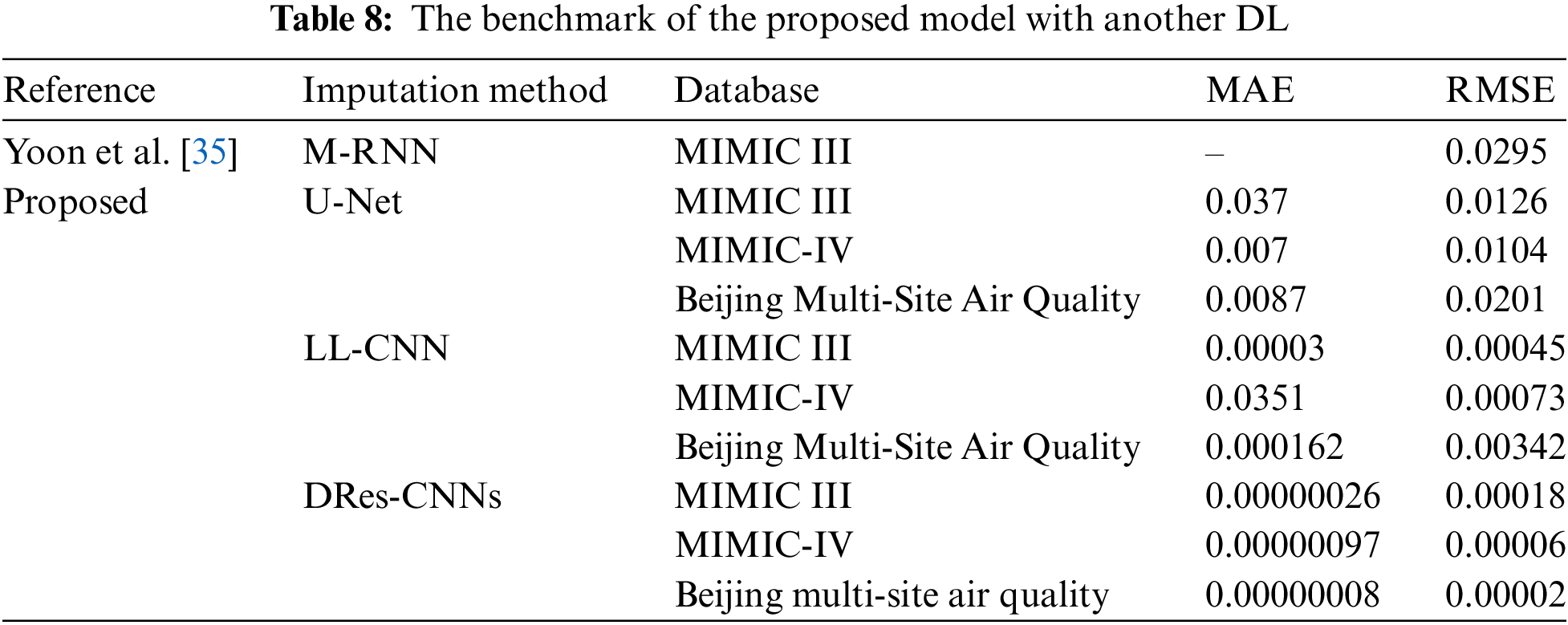
When it comes to imputing missing data across different datasets, the DRes-CNNs model performs exceptionally well, beating other current techniques such as M-RNN and U-Net50. The DRes-CNNs continue to demonstrate greater accuracy in other databases, including Beijing Multi-Site Air Quality, MIMIC III, and IV, where this trend is also evident. The DRes-CNNs routinely yield the lowest errors even among the suite of suggested models, which includes U-Net and LL-CNN, demonstrating its efficacy in managing challenging data imputation jobs. The DRes-CNNs model’s precision and resilience are highlighted by this comparative analysis, which makes it a highly useful tool for situations where exact imputation is crucial.
Despite the proposed DRes-CNN has a good performance. However, to capture the non-linear correlations in the data, for example, the DRes-CNN technique employs learnable weights that are updated iteratively. However, if there are no geographical links in the data, the learning process can proceed incorrectly and produce poor imputation. As is common with most healthcare datasets, our datasets comprised both numerical and categorical factors. It’s important to keep in mind, though, that the results of our method could not be the same for data that just includes categorical variables. Imputation may be difficult with this kind of data since there may not be many learnable inter-variable correlations. But solving this problem was outside the purview of our work.
In this research, we reviewed and evaluated deep learning (DL) methods for missing value imputation, particularly in healthcare datasets. Our proposed Deep Residual-Convolutional Neural Networks (DRes-CNN) architecture demonstrated significant improvements in imputing missing values over existing methods. The model’s performance was assessed using multiple datasets (MIMIC-III, MIMIC-IV, and the Beijing Multi-Site Air Quality), and the results showed that DRes-CNN achieved superior RMSE values compared to other techniques. Despite the proposed DRes-CNN has a good performance. However, to capture the non-linear correlations in the data, for example, the DRes-CNN technique employs learnable weights that are updated iteratively. However, if there are no geographical links in the data, the learning process can proceed incorrectly and produce poor imputation. As is common with most healthcare datasets, our datasets comprised both numerical and categorical factors. It’s important to keep in mind, though, that the results of our method could not be the same for data that just includes categorical variables. Imputation may be difficult with this kind of data since there may not be many learnable inter-variable correlations. However, solving this problem was outside the purview of our work. Future work could focus on addressing these limitations by optimizing the architecture for better scalability and reducing computational cost, while also exploring methods to improve the generalization of the DRes-CNN approach to a broader range of datasets, including those dominated by categorical variables. Additionally, expanding the method to handle multimodal data in more complex healthcare scenarios would be another valuable direction for further research.
Acknowledgement: The research infrastructure and financial support for this work was provided by the Intelligent System Research Group (ISysRG), Universitas Sriwijaya, Indonesia.
Funding Statement: This work was supported by the Intelligent System Research Group (ISysRG). This work was also supported by Universitas Sriwijaya funded by the Competitive Research 2024.
Author Contributions: Firdaus Firdaus and Siti Nurmaini: wrote the manuscript and funding acquisition. Anggun Islami and Annisa Darmawahyuni: formal analysis, methodology. Ade Iriani Sapitri and Muhammad Naufal Rachmatullah: designed computer programs, and curated the data. Bambang Tutuko and Akhiar Wista Arum: formal analysis and methodology. Muhammad Irfan Karim and Yultrien Yultrien: resources, analysis, formal analysis, and methodology. Ramadhana Noor Salassa Wandya: contributed data or analysis tools and formal analysis. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated and analyzed in this study are available at https://physionet.org/content/mimiciv/2.2/ (accessed on 23 January 2023).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. K. Psychogyios, L. Ilias, C. Ntanos, and D. Askounis, “Missing value imputation methods for electronic health records,” IEEE Access, vol. 11, no. 2, pp. 21562–21574, 2023. doi: 10.1109/ACCESS.2023.3251919. [Google Scholar] [CrossRef]
2. M. Y. Landolsi, L. Hlaoua, and L. Ben Romdhane, “Information extraction from electronic medical documents: State of the art and future research directions,” Knowl. Inf. Syst., vol. 65, no. 2, pp. 463–516, 2023. doi: 10.1007/s10115-022-01779-1. [Google Scholar] [PubMed] [CrossRef]
3. M. A. U. Zaman and D. Du, “A stochastic multivariate irregularly sampled time series imputation method for electronic health records,” BioMedInformatics, vol. 1, no. 3, pp. 166–181, 2021. doi: 10.3390/biomedinformatics1030011. [Google Scholar] [CrossRef]
4. C. Li et al., “Improving cardiovascular risk prediction through machine learning modelling of irregularly repeated electronic health records,” Eur. Hear. J.-Digit. Heal., vol. 5, no. 1, pp. 30–40, 2024. doi: 10.1093/ehjdh/ztad058. [Google Scholar] [PubMed] [CrossRef]
5. R. H. H. Groenwold, “Informative missingness in electronic health record systems: The curse of knowing,” Diagn. Progn. Res., vol. 4, no. 1, pp. 4–9, 2020. doi: 10.1186/s41512-020-00077-0. [Google Scholar] [PubMed] [CrossRef]
6. K. Yin, D. Qian, and W. K. Cheung, “PATNet: Propensity-adjusted temporal network for joint imputation and prediction using binary EHRs with observation bias,” IEEE Trans. Knowl. Data Eng, vol. 36, no. 6, pp. 2600–2613, 2024. doi: 10.1109/TKDE.2023.3321738. [Google Scholar] [CrossRef]
7. H. Khan, X. Wang, and H. Liu, “Missing value imputation through shorter interval selection driven by Fuzzy C-Means clustering,” Comput. Electr. Eng., vol. 93, no. 2, pp. 1–16, 2021. doi: 10.1016/j.compeleceng.2021.107230. [Google Scholar] [CrossRef]
8. A. D. Woods et al., “Best practices for addressing missing data through multiple imputation,” Infant. Child Dev., vol. 33, no. 1, pp. 1–37, 2024. doi: 10.1002/icd.2407. [Google Scholar] [CrossRef]
9. L. O. Joel, W. Doorsamy, and B. S. Paul, “On the performance of imputation techniques for missing values on healthcare datasets,” 2024, arXiv:2403.14687. [Google Scholar]
10. Q. Suo, W. Zhong, G. Xun, J. Sun, C. Chen and A. Zhang, “GLIMA: Global and local time series imputation with multi-directional attention learning,” in Proc. 2020 IEEE Int. Conf. Big Data (Big Data), Atlanta, GA, USA, 2020, pp. 798–807. doi: 10.1109/BigData50022.2020.9378408. [Google Scholar] [CrossRef]
11. P. C. Austin, I. R. White, D. S. Lee, and S. van Buuren, “Missing data in clinical research: A tutorial on multiple imputation,” Can J. Cardiol., vol. 37, no. 9, pp. 1322–1331, 2021. doi: 10.1016/j.cjca.2020.11.010. [Google Scholar] [PubMed] [CrossRef]
12. X. Weng et al., “A joint learning method for incomplete and imbalanced data in electronic health record based on generative adversarial networks,” Comput. Biol. Med., vol. 168, 2024, Art. no. 107687. doi: 10.1016/j.compbiomed.2023.107687. [Google Scholar] [PubMed] [CrossRef]
13. J. Wang et al., “Deep learning for multivariate time series imputation: A survey,” 2024, arXiv:2402.04059. [Google Scholar]
14. S. Hong and H. S. Lynn, “Accuracy of random-forest-based imputation of missing data in the presence of non-normality, non-linearity, and interaction,” BMC Med. Res. Methodol., vol. 20, no. 1, pp. 1–13, 2020. doi: 10.1186/s12874-020-01080-1. [Google Scholar] [PubMed] [CrossRef]
15. L. Cappelletti, T. Fontana, G. W. Di Donato, L. Di Tucci, E. Casiraghi and G. Valentini, “Complex data imputation by auto-encoders and convolutional neural networks—A case study on genome gap-filling,” Computers, vol. 9, no. 2, pp. 37, 2020. doi: 10.3390/computers9020037. [Google Scholar] [CrossRef]
16. E. Macias Toro, G. Boquet, J. Serrano, J. Lopez Vicario, J. Ibeas and A. Morell, “Novel imputing method and deep learning techniques for early prediction of sepsis in intensive care units,” in 2019 Comput. Cardiol. Conf. (CinC). , Singapore, 2019, vol. 45, pp. 1–4. doi: 10.22489/cinc.2019.038. [Google Scholar] [CrossRef]
17. D. A. Neu, J. Lahann, and P. Fettke, “A systematic literature review on state-of-the-art deep learning methods for process prediction,” Artif. Intell. Rev., vol. 55, no. 2, pp. 801–827, 2022. doi: 10.1007/s10462-021-09960-8. [Google Scholar] [CrossRef]
18. H. Khan, X. Wang, and H. Liu, “Handling missing data through deep convolutional neural network,” Inf. Sci., vol. 595, no. 5–6, pp. 278–293, 2022. doi: 10.1016/j.ins.2022.02.051. [Google Scholar] [CrossRef]
19. A. D. Kulkarni, “Fuzzy convolution neural networks for tabular data classification,” IEEE Access, vol. 12, pp. 151846–151855, 2024. doi: 10.1109/ACCESS.2024.3479882. [Google Scholar] [CrossRef]
20. M. I. Iqbal, M. S. H. Mukta, A. R. Hasan, and S. Islam, “A dynamic weighted tabular method for convolutional neural networks,” IEEE Access, vol. 10, no. 1, pp. 134183–134198, 2022. doi: 10.1109/ACCESS.2022.3231102. [Google Scholar] [CrossRef]
21. Z. Li, F. Liu, W. Yang, S. Peng, and J. Zhou, “A survey of convolutional neural networks: Analysis, applications, and prospects,” IEEE Trans. Neural Netw. Learn. Syst., vol. 33, no. 12, pp. 6999–7019, 2021. doi: 10.1109/TNNLS.2021.3084827. [Google Scholar] [PubMed] [CrossRef]
22. Y. Wen, K. Yi, J. Ke, and Y. Shen, “DiffImpute: Tabular data imputation with denoising diffusion probabilistic model,” 2024, arXiv:2403.13863. [Google Scholar]
23. C. C. Nguyen, “Deep learning for simultaneous imputation and classification of time series incomplete data,” J. Sci. Tech., vol. 12, no. 1, pp. 110–125, 2023. doi: 10.56651/lqdtu.jst.v12.n1.661.ict. [Google Scholar] [CrossRef]
24. X. Lai, X. Wu, and L. Zhang, “Autoencoder-based multi-task learning for imputation and classification of incomplete data,” Appl. Soft Comput., vol. 98, 2021, Art. no. 106838. doi: 10.1016/j.asoc.2020.106838. [Google Scholar] [CrossRef]
25. Z. Dai, E. Getzen, and Q. Long, “SADI: Similarity-aware diffusion model-based imputation for incomplete temporal EHR data,” in Proc. 27th Int. Conf. Artif. Intell. Statist., València, Spain, 2024, pp. 4195–4203. [Google Scholar]
26. D. Varma, C. S. Yainik, A. Thorave, and N. Sharma, “Handling missing data in longitudinal anthropometric data using multiple imputation method,” in Int. Conf. Data Manag., Analyt. Innov. (ICDMAI 2024). Lect. Not. Netw. Syst., Vellore, India, 2024, vol. 997, pp. 273–287. [Google Scholar]
27. J. Li et al., “Method for incomplete and imbalanced data based on multivariate imputation by chained equations and ensemble learning,” IEEE J. Biomed. Health Inform., vol. 28, no. 5, pp. 3102–3113, 2024. doi: 10.1109/JBHI.2024.3376428. [Google Scholar] [PubMed] [CrossRef]
28. P. Brimos, P. Seregkos, A. Karamanou, E. Kalampokis, and K. Tarabanis, “Deep learning missing value imputation on traffic data using self-attention and GAN-based methods,” in 2024 Panhellenic Conf. Elect. Telecommun. (PACET), 2024, pp. 1–4. [Google Scholar]
29. Y. Yin, Z. Yuan, I. M. Tanvir, and X. Bao, “Electronic medical records imputation by temporal generative adversarial network,” BioData Min., vol. 17, no. 1, p. 19, 2024. doi: 10.1186/s13040-024-00372-2. [Google Scholar] [PubMed] [CrossRef]
30. H. Khan, M. T. Rasheed, H. Liu, and S. Zhang, “High-order polynomial interpolation with CNN: A robust approach for missing data imputation,” Comput. Electr. Eng., vol. 119, 2024, Art. no. 109524. doi: 10.1016/j.compeleceng.2024.109524. [Google Scholar] [CrossRef]
31. A. Johnson, L. Bulgarelli, T. Pollard, S. Horng, L. A. Celi and R. Mark, “MIMIC-IV,” PhysioNet. 2020. Accessed: Jan. 23, 2023. [Online]. Available: https://physionet.org/content/mimiciv/1.0/ [Google Scholar]
32. A. E. W. Johnson et al., “MIMIC-IV, a freely accessible electronic health record dataset,” Sci. Data., vol. 10, no. 1, pp. 1–10, 2023. doi: 10.1038/s41597-022-01899-x. [Google Scholar] [PubMed] [CrossRef]
33. C. Ghosh, “Data pre-processing,” in Data Analysis with Machine Learning for Psychologists: Crash Course to Learn Python 3 and Machine Learning in 10 Hours. Berlin, Heidelberg, Germany: Springer, 2022, pp. 55–85. [Google Scholar]
34. I. Izonin, R. Tkachenko, N. Shakhovska, B. Ilchyshyn, and K. K. Singh, “A two-step data normalization approach for improving classification accuracy in the medical diagnosis domain,” Mathematics, vol. 10, no. 11, pp. 1–18, 2022. doi: 10.3390/math10111942. [Google Scholar] [CrossRef]
35. J. Yoon, W. R. Zame, and M. van der Schaar, “Estimating missing data in temporal data streams using multi-directional recurrent neural networks,” IEEE Trans. Biomed. Eng., vol. 66, no. 5, pp. 1477–1490, 2019. doi: 10.1109/TBME.2018.2874712. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools