 Open Access
Open Access
ARTICLE
DMHFR: Decoder with Multi-Head Feature Receptors for Tract Image Segmentation
1 Department of Endoscopy Center, Zhongshan Hospital (Xiamen), Fudan University, Xiamen, 361015, China
2 School of Computing and Data Science, Xiamen University Malaysia, Sepang, 43900, Malaysia
3 School of Computer Science and Techonology, Tongji University, Shanghai, 200092, China
4 Guangxi Key Laboratory of Machine Vision and Intelligent Control, Wuzhou University, Wuzhou, 543002, China
5 Xiamen Clinical Research Center for Cancer Therapy, Xiamen, 361015, China
* Corresponding Author: Jie He. Email:
Computers, Materials & Continua 2025, 82(3), 4841-4862. https://doi.org/10.32604/cmc.2025.059733
Received 15 October 2024; Accepted 18 December 2024; Issue published 06 March 2025
Abstract
The self-attention mechanism of Transformers, which captures long-range contextual information, has demonstrated significant potential in image segmentation. However, their ability to learn local, contextual relationships between pixels requires further improvement. Previous methods face challenges in efficiently managing multi-scale features of different granularities from the encoder backbone, leaving room for improvement in their global representation and feature extraction capabilities. To address these challenges, we propose a novel Decoder with Multi-Head Feature Receptors (DMHFR), which receives multi-scale features from the encoder backbone and organizes them into three feature groups with different granularities: coarse, fine-grained, and full set. These groups are subsequently processed by Multi-Head Feature Receptors (MHFRs) after feature capture and modeling operations. MHFRs include two Three-Head Feature Receptors (THFRs) and one Four-Head Feature Receptor (FHFR). Each group of features is passed through these MHFRs and then fed into axial transformers, which help the model capture long-range dependencies within the features. The three MHFRs produce three distinct feature outputs. The output from the FHFR serves as auxiliary auxiliary features in the prediction head, and the prediction output and their losses will eventually be aggregated. Experimental results show that the Transformer using DMHFR outperforms 15 state of the arts (SOTA) methods on five public datasets. Specifically, it achieved significant improvements in mean DICE scores over the classic Parallel Reverse Attention Network (PraNet) method, with gains of 4.1%, 2.2%, 1.4%, 8.9%, and 16.3% on the CVC-ClinicDB, Kvasir-SEG, CVC-T, CVC-ColonDB, and ETIS-LaribPolypDB datasets, respectively.Keywords
Colorectal cancer (CRC) is one of the leading causes of mortality worldwide, with early detection and removal of precursors, such as polyps, being crucial for improving survival rates. Timely and accurate localization of polyps during colonoscopy can significantly reduce the incidence of CRC. However, manual inspection of colonoscopy images is often subjective, tedious, time-consuming, and prone to errors. Therefore, developing automatic and accurate polyp detection systems is urgent to assist physicians and reduce diagnostic mistakes. This problem is typically framed as a dense prediction task, which create segmentation maps of the organs or lesions by performing classification of pixel-wise. Methods based on convolutional neural networks (CNNs) have seen significant success in computer vision tasks [1–5], largely due to their inductive bias and ability to maintain translation invariance. One of the most notable methods is U-Net [1], which has made significant contributions to image detection and segmentation tasks through its encoder-decoder architecture. However, the receptive field of a CNN may restrict the model’s focus to a localized area [4]. To address this issue, some works integrate attention mechanisms into their model [2,6,7] to enhance pixel-level features for better classification. While these methods have led to performance improvements, their ability to capture long-range dependencies can still be insufficient.
In addition to the aforementioned studies, the vision transformer [8], a novel transformer model, has made significant breakthroughs in medical image segmentation. By leveraging self-attention mechanisms to learn relationships among all tokens, it effectively addresses previous challenges in capturing long-range dependencies in medical images by its unique multi-head self-attention and multi-head perception fusion modules. Numerous vision-related tasks have adopted transformers in place of CNNs for feature extraction, achieving impressive performance [9–11]. MedCLIP [12] demonstrates impressive performance on small-scale pre-training data by separating text and images for multimodal contrastive learning. Moreover, recent advancements in hierarchical vision transformers have helped reduce computational costs. Notable examples include the Swin transformer [9], which employs window attention mechanisms, and the pyramid vision transformer [13], which utilizes spatial reduction attention mechanisms. However, transformers may lose some fine-grained details and local pixel relationships during the encoding process due to downsampling and lack the ability to model these local details as effectively as CNNs, leading to suboptimal performance in some specific segmentation tasks. Although models like PVTv2 [11] and SegFormer [14] have embedded convolutional layers to attempt to overcome this limitation, the discriminative capability of these methods is restricted by the positioning of convolutional layers, which may limit their ability to effectively model features. Additionally, there is still room for improvement in processing multi-scale features from the encoder.
To address these challenges, we propose a novel Decoder with Multi-Head Feature Receptors (DMHFR), which receives pyramid features from the encoder backbone. We integrate the Frequency Channel Attention Network (FcaNet) [15] into the DMHFR. Before passing through the MHFRs, FcaNet models the features in the frequency domain, emphasizing and learning the most important frequency components through a frequency channel attention module. This enables the network to better capture image details and texture information. The Multi-Head Feature Receptors (MHFRs) consist of two Three-Head Feature Receptors (THFRs) and one Four-Head Feature Receptor (FHFR). THFRs process finer and coarser granularity features separately, while FHFR receives all the features from the encoder backbone. These receptors perceive and fuse multiple sets of features of varying granularity in parallel. Each MHFR produces an output, and the features passing through the FHFR are ultimately used to generate an auxiliary prediction map in the prediction head. Axial transformers are integrated after the MHFRs to capture long-range dependencies in the feature map. This enhances overall feature representation, helping the model to focus on both the boundaries and internal textures of polyps. This capability is crucial for detecting polyps of various shapes and sizes and significantly contributes to the generalization ability of DMHFR. The superiority and effectiveness of DMHFR were validated through experiments on five public colorectal polyp datasets, where it achieved SOTA results in polyp segmentation. The proposed method contributing to the development of the field of medical image segmentation, and early diagnosis and treatment of colorectal cancer. Our contributions can be summarized as follows:
• A novel network architecture, we propose a novel Decoder with Multi-Head Feature Receptors (DMHFR) for 2D medical image segmentation, offering high accuracy and robustness, and can be integrated with other hierarchical visual encoders to enhance network performance.
• A novel method to process multi-scale features, our DMHFR groups four multi-scale features into three feature groups with different granularities: coarse, fine-grained, and full set. This way of handling multi-scale features can help global and local features to be better perceived and integrated.
• Two novel modules, The THFR and FHFR modules, collectively referred to as MHFRs, enhance feature representation by perceiving and aggregating feature maps at multiple resolutions, which offers great potential for improving deep learning in medical image segmentation.
• Experimental results show that the proposed method exhibits outstanding learning ability and generalization ability compared with the SOTA methods on five public polyp datasets.
2.1 Traditional Methods for Polyp Segmentation
Early polyp segmentation techniques primarily relied on low-level feature processing, such as texture, color, and geometric characteristics [1]. Methods to focus on these features include region growing, watershed, and active contour analysis. The method of Sasmal et al. [16] consists of principal component pursuit and active contour model. The region-based method [17] divides the image into multiple regions to judge the features within regions respectively. A commonly used morphology-based method [17] is to perform preprocessing and subsequent processing on images to enhance edge features. The polyp segmentation method proposed by Gross et al. [18] combines Canny operator, nonlinear diffusion filtering, and other methods. However, due to the high similarity between polyps and the surrounding tissue, these traditional methods often struggle with accuracy, leading to an increased likelihood of missed or incorrect detections.
2.2 CNNs for Polyp Segmentation
CNN-based methods [19–23] have made significant contributions to the development of polyp segmentation and have outperformed traditional methods in feature modeling, noise suppression, inference speed, and generalization ability. Akbari et al. [24] proposed a polyp segmentation model using a fully convolutional neural network, which used a new image patch selection method in the training phase, and the probability map generated by the network was effectively post-processed in the test phase. Brandao et al. [25] obtained the shape through a shading strategy and used it to recover the depth, and used the RGB model to receive the result to enrich the feature representation. Encoder-decoder-based models have demonstrated impressive performance in image segmentation. UNet [1] aggregates the encoder features with the upsampled features of the decoder through skip connections to generate high-resolution segmentation maps. UNet++ [3] links the encoder and decoder through nested and dense skip connections. The skip connections of UNet 3+ [5] include full-size internal connections between decoder blocks. Dilated convolutions extract and aggregate high-level semantic features with resolution preservation to achieve improvements in the encoder network. With the advancement of computer vision, ResNet [4] has become a widely adopted backbone in medical image segmentation methods. In our proposed PVT-DMHFR, the PreActBottleneck of ResNet is employed to optimize feature perception in MHFRs. Mask R-CNN [26] was adapted with a deeper feature extractor [27] for polyp segmentation. PraNet [2] generated a global attention feature map by inverting attention and used it to derive boundary information. PNS-Net [28] incorporated temporal and spatial cues based on self-attention for video polyp segmentation, while Spatial-Temporal Feature Transformation [29] aggregated features from adjacent frames to achieve notable performance in video polyp segmentation. U-KAN [30] redesigned the UNet by integrating the dedicated Kolmogorov-Arnold Networks (KAN) layers on the tokenized intermediate representation to improve segmentation performance. GCN-DE [31] projects both support and query images into a feature space, computes long-range and short-range dependencies within a global correlation module that processes the embeddings to reduce complexity and applies discriminative regularization to draw features of the same foreground class closer together, enhancing the accuracy of few-shot medical image segmentation. Polyp-Net [32] utilized a local gradient weighting-embedded level-set method, effectively reducing false-positive instances caused by high-intensity regions during prediction. UACANet [33] modified the U-Net shape network, and in its prediction module, the foreground, background and uncertainty region maps are aggregated with features, and the saliency map that provides this calculation assistance is calculated and propagated to the next prediction module. SANet [34] minimized the impact of irrelevant features on predictions through color transformation. MSNet [35] reduced complementary and redundant information across multi-scale features via a multi-scale subtraction network. While these CNN-based networks have achieved good results, they share a common limitation: an inability to efficiently capture both global information and fine details simultaneously. The decoder struggles to effectively aggregate global features for enhanced information supplementation, the overall generalization performance is not satisfactory, and the results in polyp segmentation also require further improvement.
2.3 Transformer for Polyp Segmentation
Transformer-based methods outperform CNN-based approaches due to their advantages in integrating global context, capturing long-range dependencies, and modeling features more effectively. DC-Net [36] is a dual context network that enhances segmentation performance by reshaping the original image and multiscale feature maps and integrating global and multiscale contextual information. Transfuse [10] combined CNN and transformer architectures through sequential and parallel connections in the encoder, performing well but limited in scalability due to the resulting increase in network size. Many methods [37–40] use PVTv2 as the encoder backbone. SSFormer [37] sought to simplify the structure of the transformer encoder’s backend, aiming to reduce model parameters while enhancing local information. However, this simplification led to a decline in performance. HSNet [38] combined CNN and Transformer in parallel within the decoder, though it did not fully address the differences in representation between the two architectures. Polyp-PVT [39] utilized the Cascaded Fusion Module (CFM) and Camouflage Identification Module (CIM) to merging features and directly extract intricate details from low-level features, respectively. Additionally, the Similarity Aggregation Module (SAM) was implemented to investigate higher-order relationships between the local features of low-level from CIM and the cues of high-level from CFM. Nevertheless, Polyp-PVT did not thoroughly explore and augment the encoder’s output information, resulting in suboptimal feature aggregation by CFM. Furthermore, CIM’s direct extraction of detailed information from low-level features introduced noise. PVT-CASCADE [40] accurately identified the most critical local features through its CASCADE module, which included an Attention Gate (AG), a multi-stage loss and feature aggregation component, as well as an upconv module. However, the upconv module risked losing fine-grained details, and the CASCADE module could result in redundant features, especially when dealing with objects with indistinct edges.
In conclusion, although previous methods have yielded impressive results in addressing the polyp segmentation challenge, they often fall short of effectively bridging the semantic gap between Transformer and CNN architectures. This unresolved gap can negatively affect network performance. Additionally, many Transformer-based polyp segmentation models struggle to adequately process the four pyramid features of varying granularities generated by the encoder backbone, which encapsulates rich spatial details and semantic information. To address these issues, we propose a novel decoder with multi-headed feature receptors.
This section first presents the overall architecture of the PVT-DMHFR. Then the proposed method MHFRs (THFR and FHFR) is described in detail.
The network architecture of PVT-DMHFR is illustrated in Fig. 1. It consists primarily of a Transformer encoder and DMHFR. DMHFR mainly consists of MHFRs, FcaNet, and axial transformers. PVTv2 [11] functions as the Transformer encoder to capture features that represent long-range dependencies across multiple scales from the input image. As shown in Fig. 1a,b, DMHFR receives four pyramid features from the PVTv2 encoder (
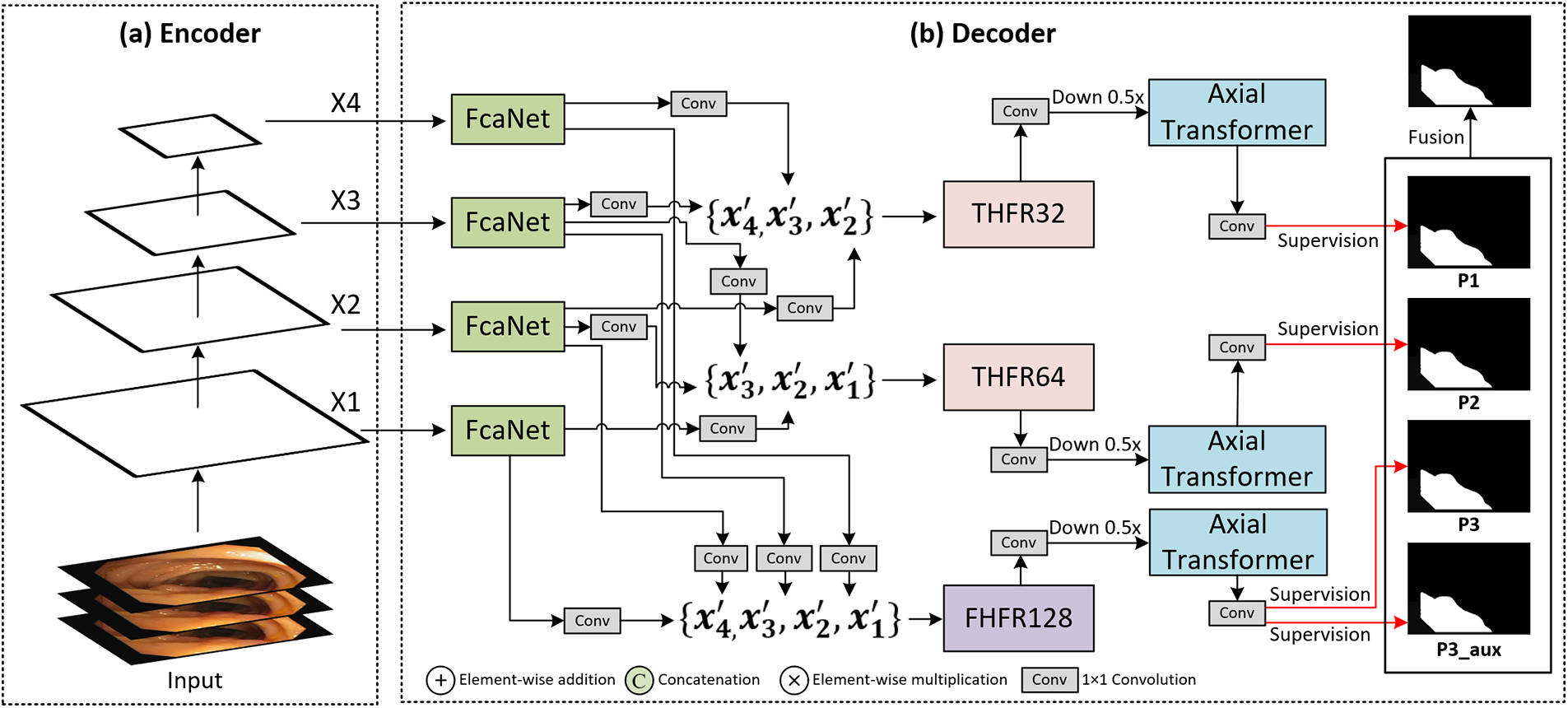
Figure 1: Architecture of PVT-DMHFR network. (a) Backbone: PVTv2-b2 Encoder; (b) DMHFR decoder
DMHFR allows features of multiple different dimensional combinations to be perceived in parallel and be fused, retaining the features in the input image to a high degree, and most of the context output by MHFRs can be calculated in parallel by the axial transformer to express the global dependencies of features. This architecture achieves SOTA performance on several polyp segmentation benchmarks. Details are presented in the experimental section.
Recent studies [8,13,14,42] on vision tasks have demonstrated that transformer-based pyramid structures are better than CNNs in terms of generalization, robustness, and capturing multi-scale and multi-level features. In this proposed method, PVTv2 [11] is employed to extract multi-scale features. Unlike traditional Transformers, PVTv2 does not use a patch embedding module but uses convolution operations to consistently capture spatial information, delivering state-of-the-art performance across various dense prediction applications. PVTv2 generates pyramid features
3.3 Decoder with Multi-Head Feature Receptors (DMHFR)
Due to the high similarity between polyps and backgrounds and the limited ability of existing Transformer-based models to process (local) contextual information among pixels, the localization of local features with higher discrimination in the segmentation task is challenging. To address this challenge, we propose a novel Decoder with Multi-Head Feature Receptors (DHMFR) for pyramid features.
As Fig. 1b shows, DHMFR includes FcaNet [15] to model features in the frequency domain, perceive details and texture information of images, our proposed MHFRs (two THFRs and one FHFR) to perceive and fuse pyramid features in parallel, and axial transformer [41] to keep the full expressiveness of joint distribution over features. The features of four different dimensions from the encoder backbone are passed through four FcaNet blocks, and then the four features are grouped into two three-element feature groups and one four-element feature group, these three feature groups are coarse-grained feature group, a fine-grained feature group, and full-set feature group. The channel of each feature in feature groups:
3.3.1 Integration of Frequency Channel Attention Network (FcaNet)
In our proposed method, we integrate FcaNet [15] into our network architecture by feeding the features from the PVTv2 encoder into FcaNet, as shown in Fig. 1, this integration enhances the model’s feature extraction capabilities, by leveraging frequency domain attention, the network becomes more sensitive to subtle texture and boundary details of polyps. Despite the additional frequency processing, the integration of FcaNet ensures that the network remains computationally efficient. It selectively applies attention to only the most important frequency components, minimizing overhead while boosting performance. The flexibility of FcaNet’s frequency-based attention mechanism also improves the model’s generalization ability across diverse polyp datasets and imaging conditions. The incorporation of FcaNet into our segmentation network plays a crucial role in enhancing the accuracy and robustness of polyp segmentation tasks by providing a more detailed and contextually aware feature extraction process. This process is formulated as Eq. (1):
3.3.2 Multi-Head Feature Receptor (MHFR)
To effectively perceive and fuse multi-scale features, we propose the three-head feature receptor (THFR) and the four-head feature receptor (FHFR), collectively referred to as Multi-Head Feature Receptors (MHFRs). MHFRs are primarily composed of PreActBottleneck blocks [43], which is an enhanced version of the bottleneck structure originally developed in ResNet [4]. These blocks combine element-wise operations such as addition, multiplication, and concatenation. Due to the simplicity of these operations, this approach only results in a slight increase in resource demands and significantly enhances segmentation accuracy. This design effectively bridges spatial and semantic information, balancing the increased resource requirements with notable improvements in segmentation quality. The PreActBottleneck blocks help improve representation capabilities and allow efficient training through better gradient flow. The collaboration of these components allows the model to perceive and aggregate feature maps at multiple resolutions, thereby enhancing feature representation. Additionally, MHFRs are versatile and can be easily adapted to process pyramid features in other models by adjusting the channel setting of MHFRs, offering significant potential to enhance deep learning performance in various medical image segmentation tasks.
Three-Head Feature Receptor (THFR)
In PVT-DMHFR, we assume that adjacent features within the pyramid structure exhibit higher correlation, therefore, THFR receives two sets of features: three adjacent finer-grained features
As shown in Fig. 2, THFR receives three features of different sizes, with the height and width of each feature being double that of the next. The input channel and channel of the features need to be uniformly set to multiples of 32. The finest-grained feature,
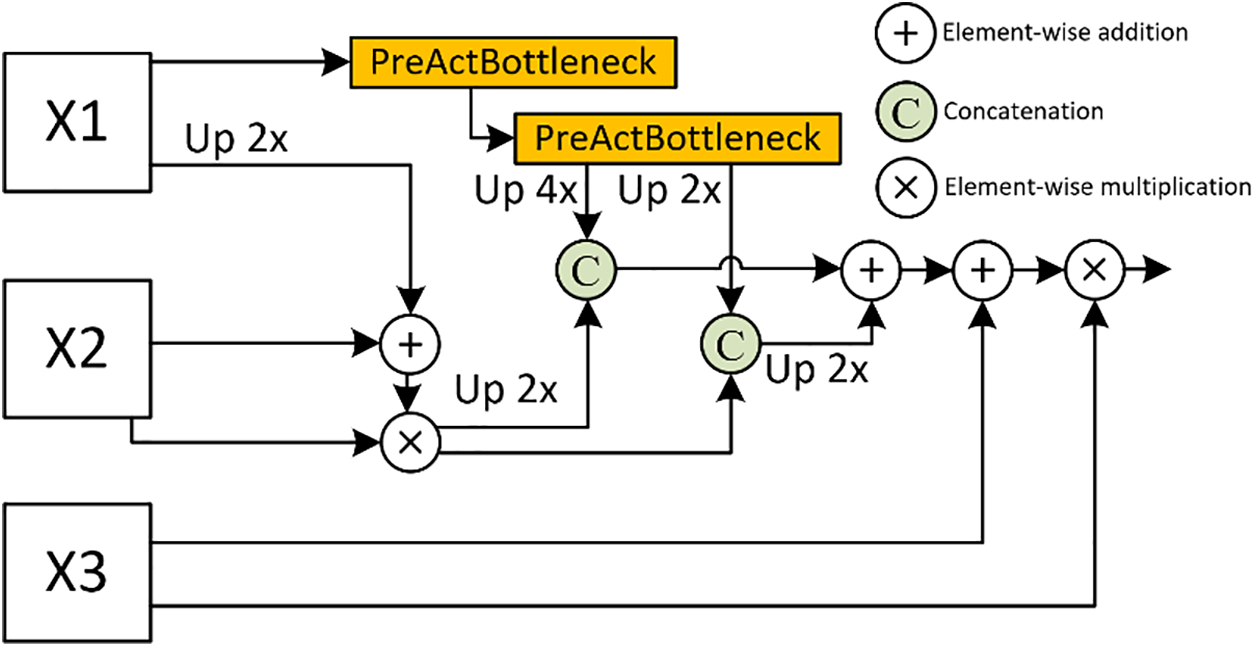
Figure 2: Details of the introduced THFR, X1, X2, and X3 are multi-scale features, each with spatial dimensions that are double those of the preceding feature
where
Four-Head Feature Receptor (FHFR)
As shown in Fig. 3, FHFR etends this approach to four encoder layers to receive four features of different sizes, with all pyramid features being perceived and fused within it. Therefore, we assume that the FHFR output holds a higher priority and carries more weight in the prediction output, to achieve this, we duplicate the output prediction to create an auxiliary prediction, which is then fused into the final prediction. Same as THFR, the height, and width of each feature are double that of the next. The input channel and channel of the features need to be uniformly set to multiples of 32. The finest-grained feature,
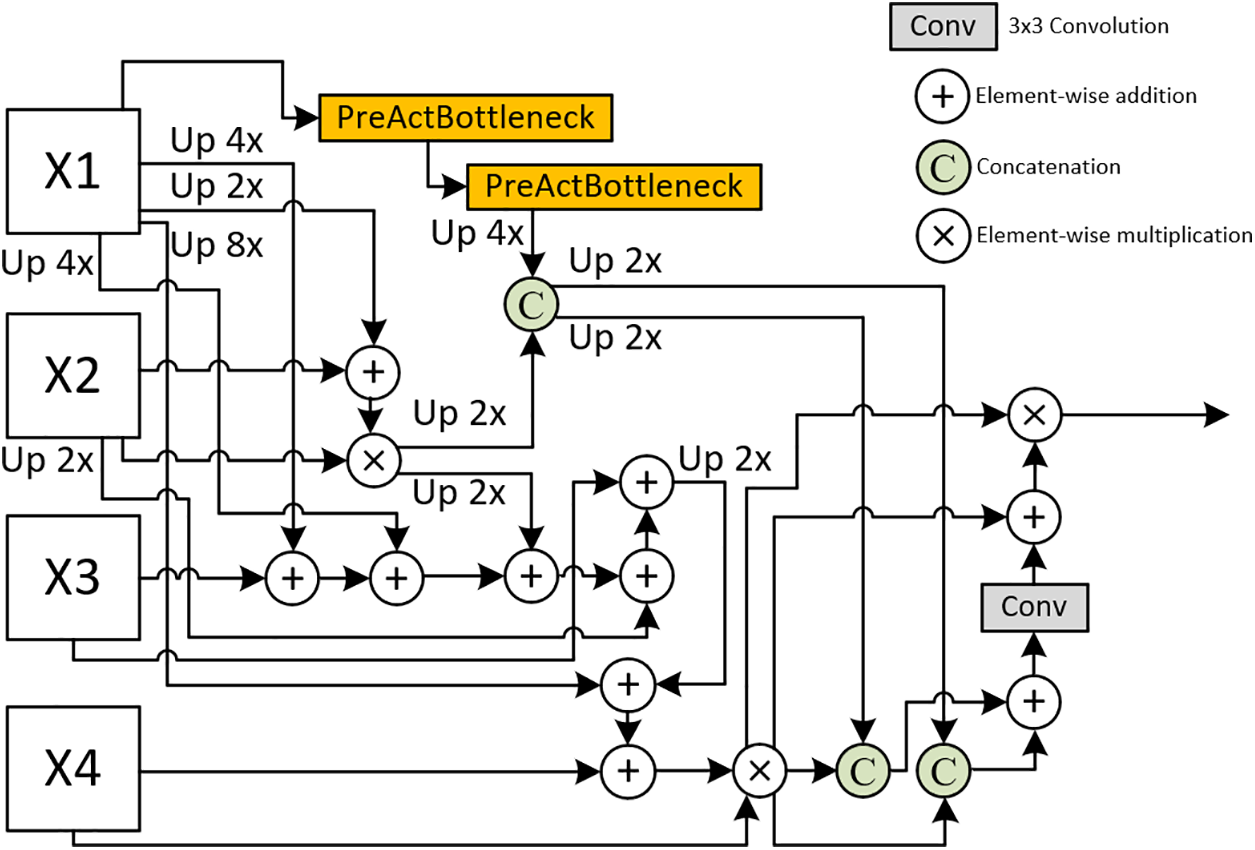
Figure 3: Details of the introduced FHFR, X1, X2, X3, and X4 are multi-scale features, each with spatial dimensions that are double those of the preceding feature
where
3.3.3 Integration of Axial Transformer
In our proposed method, we integrate axial transformers into our network architecture by feeding the features processed by MHFRs into the axial transformers to capture long-range dependencies of the feature maps, as shown in Fig. 1, thereby enhancing the overall feature representation. This integration allows the model to focus on both the boundaries and internal textures of polyps. By applying axial attention across the feature maps, the model efficiently captures the long-range dependencies crucial for detecting polyps in various shapes and sizes, even in cases where polyps are small or occluded by other tissues. This process can be written as Eq. (4):
where
3.4 Loss function and feature fusion
We utilize additive aggregation with four prediction heads to compute the final prediction map, as expressed in Eq. (5):
where
The loss function for feature maps of each prediction head can be expressed as Eq. (6):
where
where
In this section, we first experimentally evaluate the performance of our proposed DMHFR decoder by comparing its results with those of state-of-the-art methods. Additionally, we conduct ablation studies to assess the effectiveness of the DMHFR decoder.
We validate the performance of the proposed method on five public polyp datasets: CVC-ClinicDB [44] has 612 polyp images extracted from 31 colonoscopy videos. Kvasir-SEG [45] has 1000 polyp images collected from the Kvasir dataset’s polyp class. CVC-T [46], a subset of the EndoScene dataset, has 60 polyp images. CVC-ColonDB [47] has 380 polyp images. ETIS-LaribPolypDB [48] has 196 polyp images.
We utilize several key metrics as PraNet [2] used to assess the performance of our proposed method comprehensively. Mean Dice (mDic) [49] quantifies the overlap between predicted and ground truth segmentations, producing a score between 0 and 1, with higher values indicating better accuracy. Mean intersection over union (mIoU) also measures overlap but is more stringent, providing a ratio of the intersection to the union of the predicted and actual regions. Mean absolute error (MAE) calculates the average absolute differences between predictions and ground truth, providing insight into overall accuracy. Weighted F-measure (
where
where
where
where
where
Our proposed method is implemented using the PyTorch 2.0.0 framework. The model is trained using an NVIDIA RTX 3090 GPU with 24 GB of memory. We use the Adam optimizer [54] and set the learning rate to 5 × 10−5 without decay. Following Polyp-PVT [39], we use a multi-scale {0.75, 1.0, 1.25} training strategy with gradient clipping set to 0.5, configure the batch size to 16, set the maximum number of epochs to 100, and resize input images to 352 × 352 pixels. To ensure fairness in our comparative experiments, we adopt the same data division method as used in PraNet. A total of 900 images from Kvasir-SEG and 548 images from CVC-ClinicDB are used as training sets, while the remaining 100 images from Kvasir-SEG and 64 images from CVC-ClinicDB are reserved as test sets for evaluating the model’s learning ability. We further assess the model’s generalization ability on three additional datasets: CVC-ColonDB, CVC-T, and ETIS-Larib. In this paper, we compare the proposed method with 15 state-of-the-art image segmentation models, including UNet [1], UNet++ [3], PraNet [2], MSNet [35], SANet [34], Transfuse [10], UACANet [33], TMF-Net [55], C2F-Net [21], Polyp-PVT [39], SSFormer [37], ColonFormer [56], ESFPNet [57], FCBFormer [58], and PVT-CASCADE [40].
4.4 Quantitative Analysis of Learning Ability
Table 1 presents the quantitative results of the feature modeling capabilities comparison between PVT-DMHFR and 15 different SOTA methods trained on the ClinicDB and Kvasir-SEG datasets. The PVT-DMHFR exhibits superior feature modeling performance compared to other methods. Compared to the transformer-based method PVT-Cascade, our approach improves the mDic and mIoU on the CVC-ClinicDB dataset by 1.2% and 2.1%, respectively. Additionally, it increases the
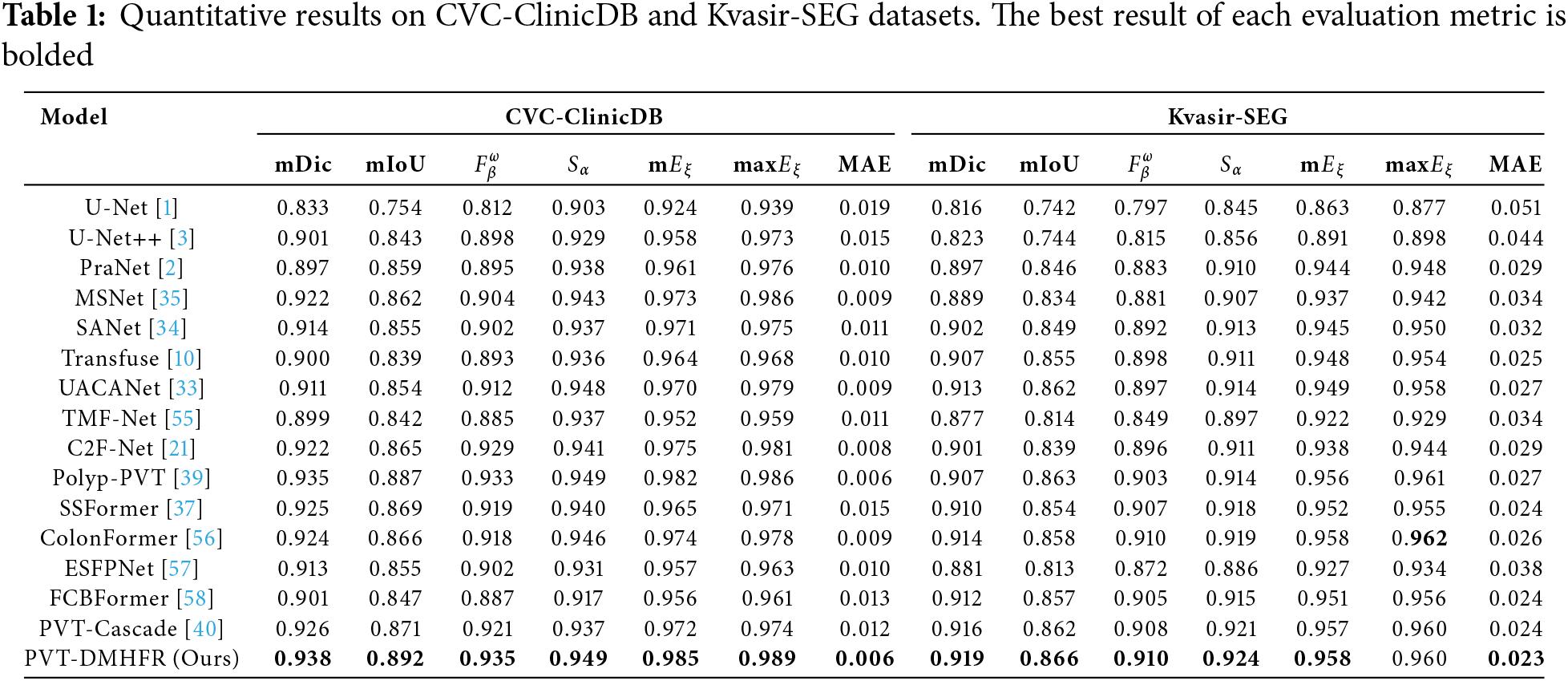
On the Kvasir-SEG dataset, compared to the best CNN-based method, UACANet, our method achieves a 0.6% improvement in the mDic, a 0.4% increase in mIoU, a 1.3% rise in the
4.5 Quantitative Analysis of Generalization Ability
Tables 2 and 3 present the results of performance comparison between our PVT-DMHFR and 15 methods on three unseen datasets: CVC-T, CVC-ColonDB, and ETIS-Larib. On the CVC-T dataset, our method achieves an mDic score of 0.8% higher than the best CNN-based method, SANet, and 0.7% higher than the best transformer-based method, PVT-Cascade. Our method performs best on all metrics, except for max
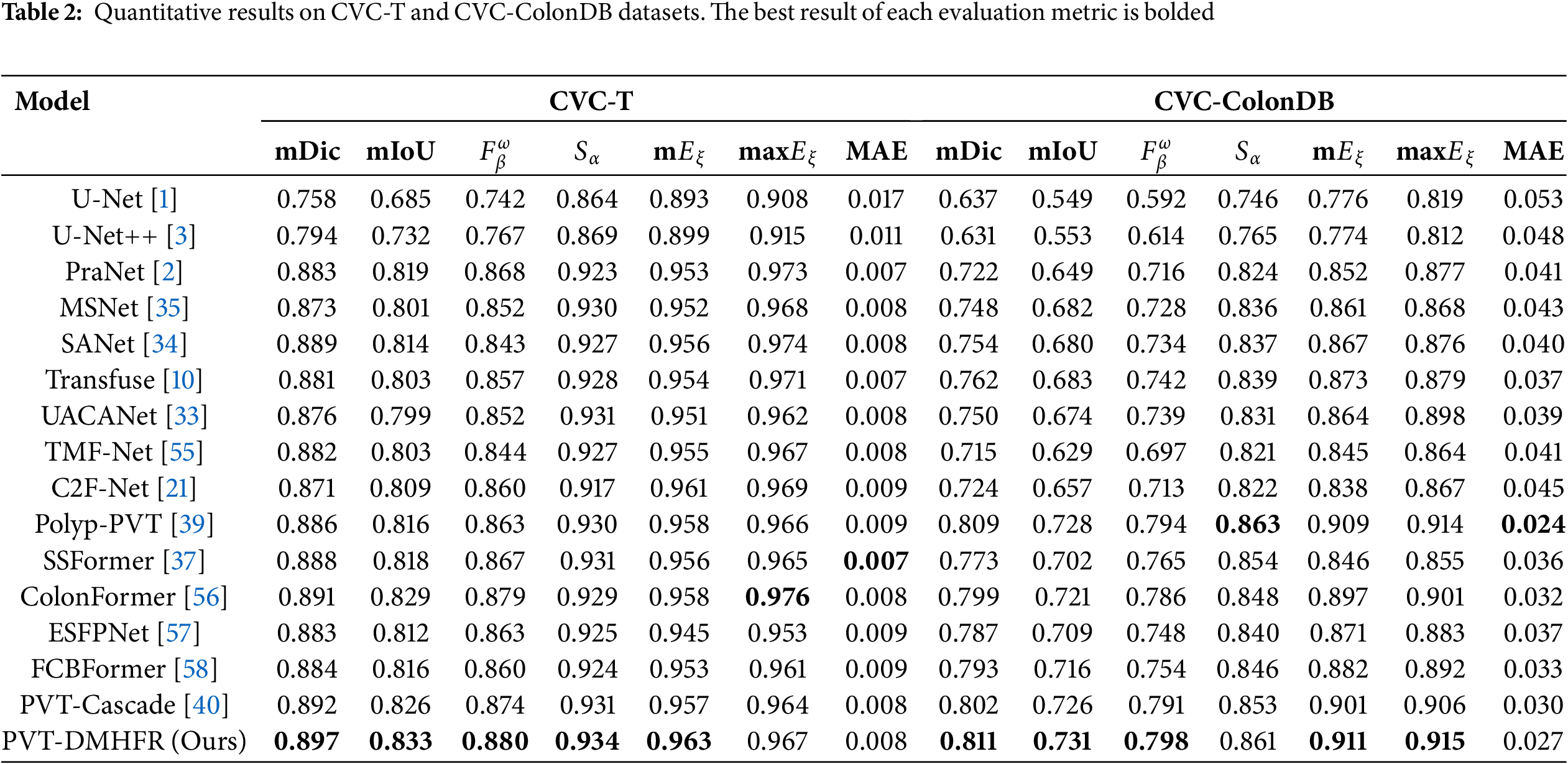
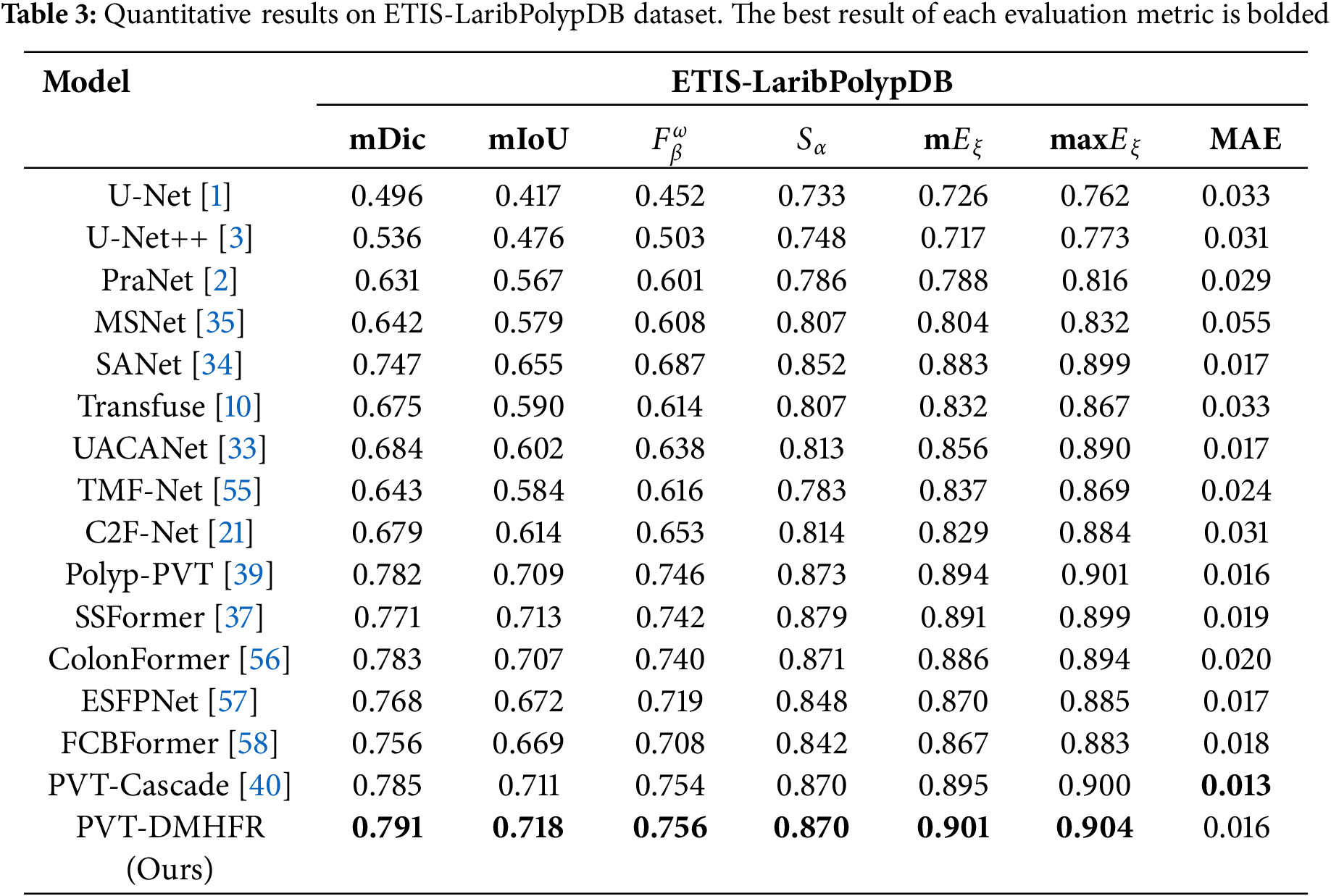
Table 4 presents the results of performance of combination of DMHFR and different backbones, including hierarchical transformer-based backbones (PVTv2b1 [11], PVTv2b2 [11], PVTv2b3 [11], PVTv2b4 [11], PVTv2b5 [11], mitb5 [14]), transformer-based backbone (R50+ViT-B_16 [8]), and CNN-based backbone (ResNetV2 [43]). The results demonstrate that hierarchical transformer-based backbones, particularly PVTv2b3 and PVTv2b2, consistently outperform both transformer-based (R50+ViT-B_16) and CNN-based (ResNetV2) backbones across all datasets and metrics.
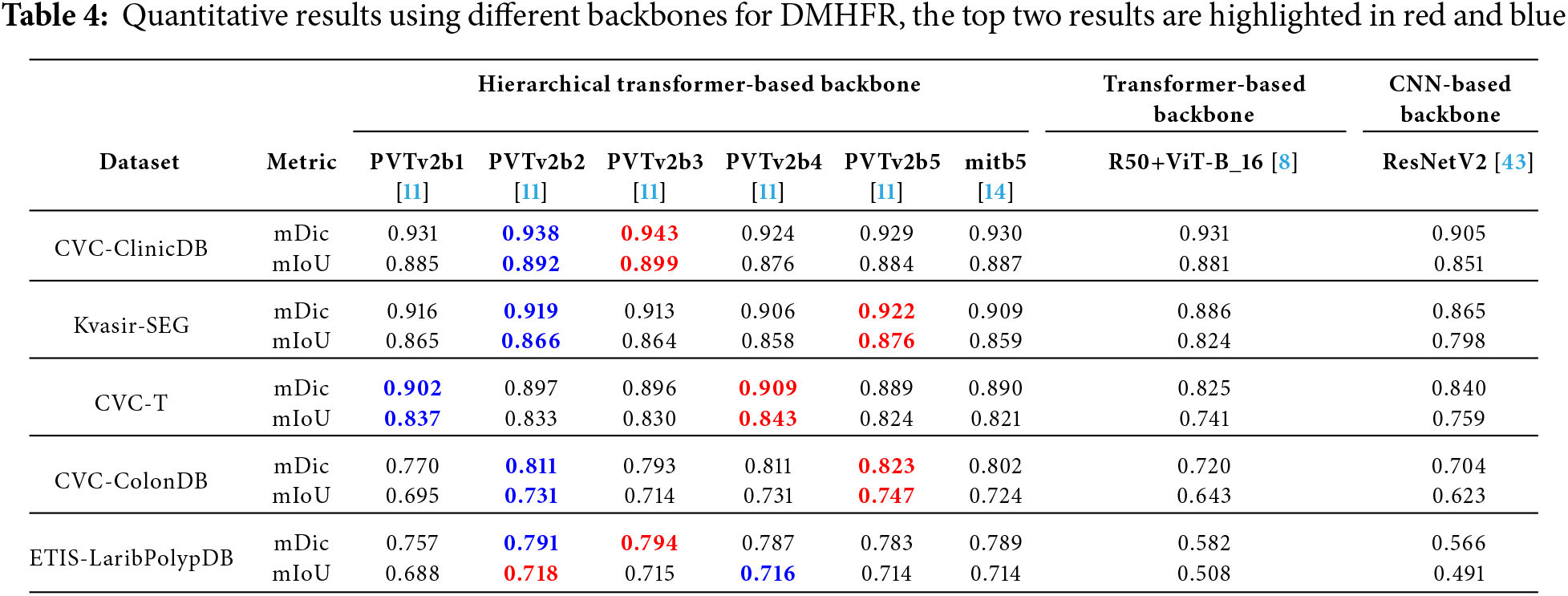
For example, PVTv2b3 achieves the highest mDic (0.943) and mIoU (0.899) on the CVC-ClinicDB dataset, while PVTv2b5 excels on Kvasir-SEG with mDic of 0.922 and mIoU of 0.876. Additionally, mitb5 demonstrates well-balanced and strong performance across all five datasets. In contrast, the transformer-based backbone typically performs significantly worse than its hierarchical counterparts, achieving top scores only in one case (an mDic of 0.931 on CVC-ClinicDB). Meanwhile, the CNN-based backbone consistently underperforms, underscoring their limited effectiveness in these tasks when working in conjunction with DMHFR. This performance disparity can be attributed to the alignment between the hierarchical transformer-based backbones and DMHFR’s input requirements. The hierarchical transformer-based backbones naturally produce outputs with dimensions that align with DMHFR’s architecture, minimizing the need for extensive preprocessing or additional layers. Conversely, both transformer-based and CNN-based backbones require supplementary adjustments, such as additional transformations or layers, to ensure compatibility with DMHFR. These modifications not only introduce computational overhead but may also disrupt the original feature distributions, leading to reduced performance.
By contrast, the seamless integration of hierarchical transformer-based backbones with DMHFR enhances efficiency and preserves the integrity of feature representations. This synergy allows for streamlined processing and optimized information flow, resulting in superior performance across diverse datasets.
Overall, the combination of DMHFR and hierarchical transformer-based backbones demonstrates remarkable adaptability and effectiveness across diverse datasets.
Based on the above analysis, our PVT-DMHFR shows impressive learning and generalization capabilities on the challenging task of polyp segmentation, as well as performance that is superior to other SOTA methods.
4.6 Analysis of Visual Results
To thoroughly evaluate the performance of our proposed method, we compared our PVT-DMHFR with SOTA methods in terms of visual results. As shown in Fig. 4, our PVT-DMHFR demonstrates several advantages over these SOTA methods: First, the transformer-based encoder backbone we employed, PVTv2, enhances polyp localization accuracy. Second, PVT-DMHFR consistently produces segmentation results with high accuracy for polyps of various sizes and shapes. This stability and accuracy are largely attributed to the proposed MHFRs, which effectively capture and fuse multiple groups of multi-scale information. Additionally, the integration of FcaNet and the axial transformer, applied before and after the MHFRs, strengthens the model’s ability to extract features from the encoder backbone and capture long-range dependencies within the feature map, significantly improving overall feature representation.
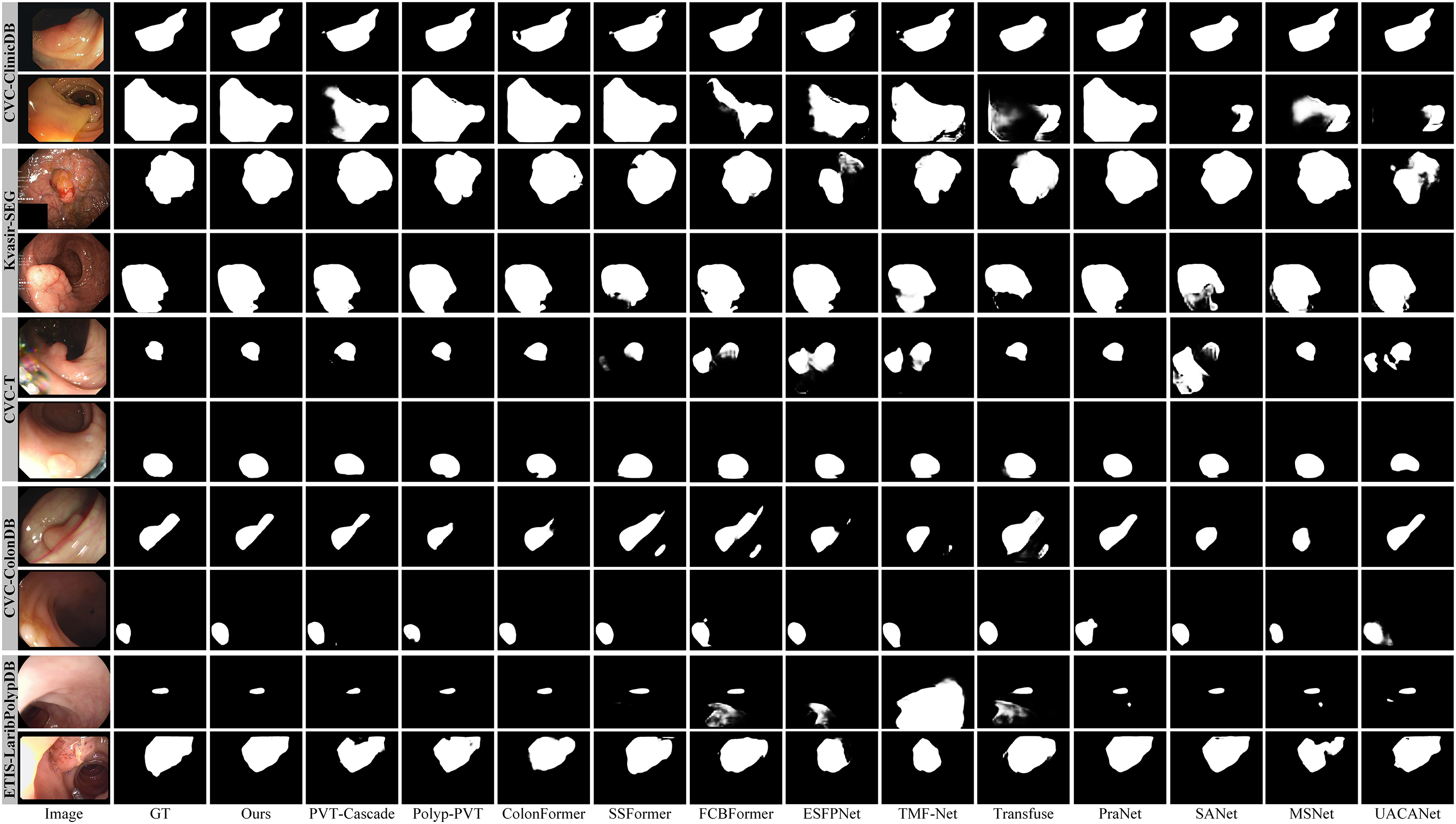
Figure 4: Visualization results of SOTA methods on five datasets
4.7 Analysis of Computational Efficiency
We comprehensively evaluate PVT-DMHFR and SOTA methods in terms of floating-point operations (FLOPs) and the number of parameters (Params). As shown in Table 5, the proposed PVT-DMHFR demonstrates an advantage over several strong competitors (e.g., PVT-Cascade, ColonFormer, SSFormer, FCBFormer, and ESFPNet) in terms of Params. However, it is slightly less efficient in FLOPs, surpassing only FCBFormer in this aspect. Overall, PVT-DMHFR achieves a well-balanced trade-off between computational efficiency and segmentation accuracy.
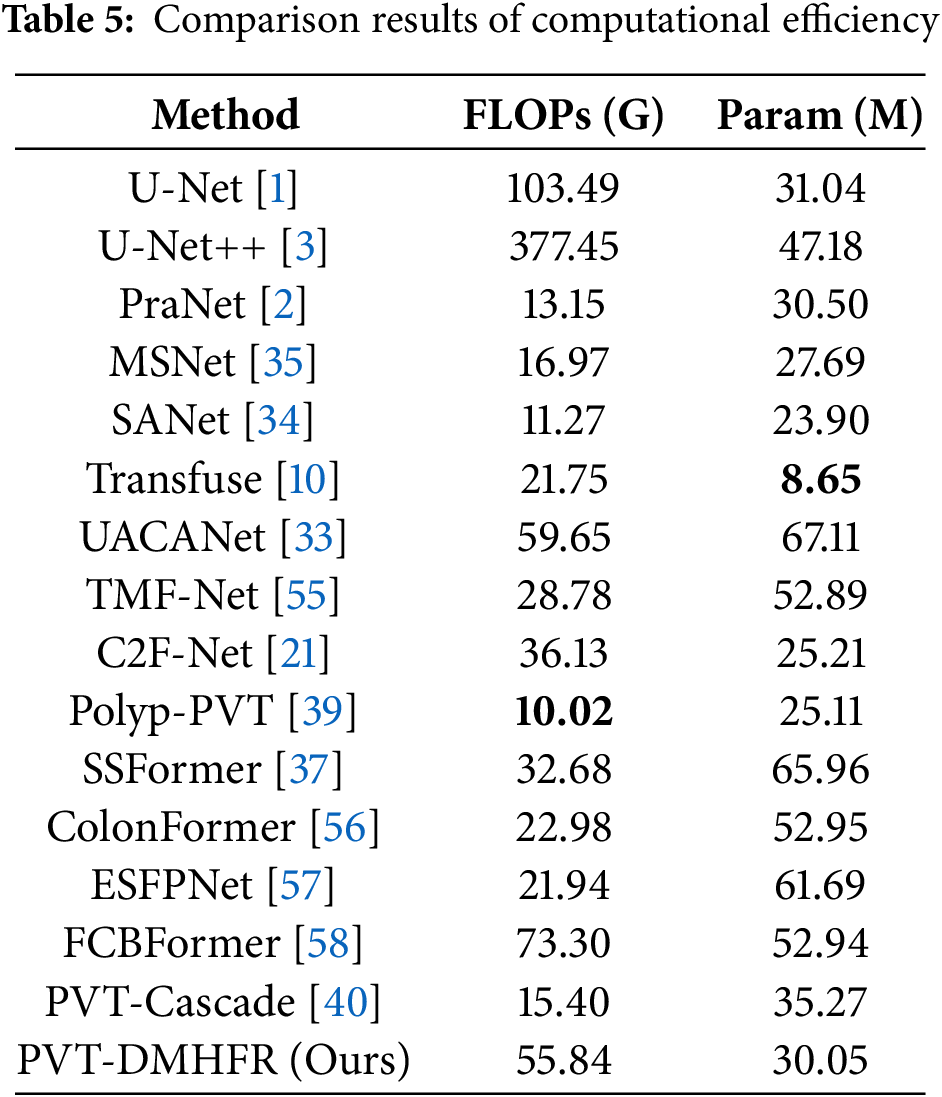
We performed ablation experiments to assess the contribution of each component in our method. The mDic and mIoU metrics were selected to represent network performance, and the results are summarized in Table 6. When the FcaNet modules were removed, the mDic and mIoU scores on the CVC-ClinicDB dataset dropped by 0.4% and 0.5%, respectively. This indicates that passing features through the FcaNet module before processing them with the MHFRs helps the model learn and represent features more effectively. On the ETIS-LaribPolypDB dataset, the mDic and mIoU scores decreased by 0.3% and 1.1%, respectively, suggesting that FcaNet improves the model’s generalization ability through superior feature capture. After removing the MHFRs, the mDic and mIoU scores fell by 1.1% and 0.8%, respectively, on the CVC-ClinicDB dataset, and by 1.4% and 1.6% on the CVC-ColonDB dataset. These results demonstrate that MHFRs play a crucial role in fusing multiple feature sets at various resolutions, leading to a significant improvement in segmentation accuracy. When the axial transformers were excluded, the mDic and mIoU scores on the CVC-ClinicDB dataset dropped by 0.5% and 1.6%, respectively. The mDic and mIoU scores on the CVC-ColonDB dataset dropped by 0.8% and 0.7%. This underscores the importance of axial transformers in capturing long-range dependencies within feature maps post-MHFR processing, further enhancing feature representation and improving both segmentation accuracy and generalization.
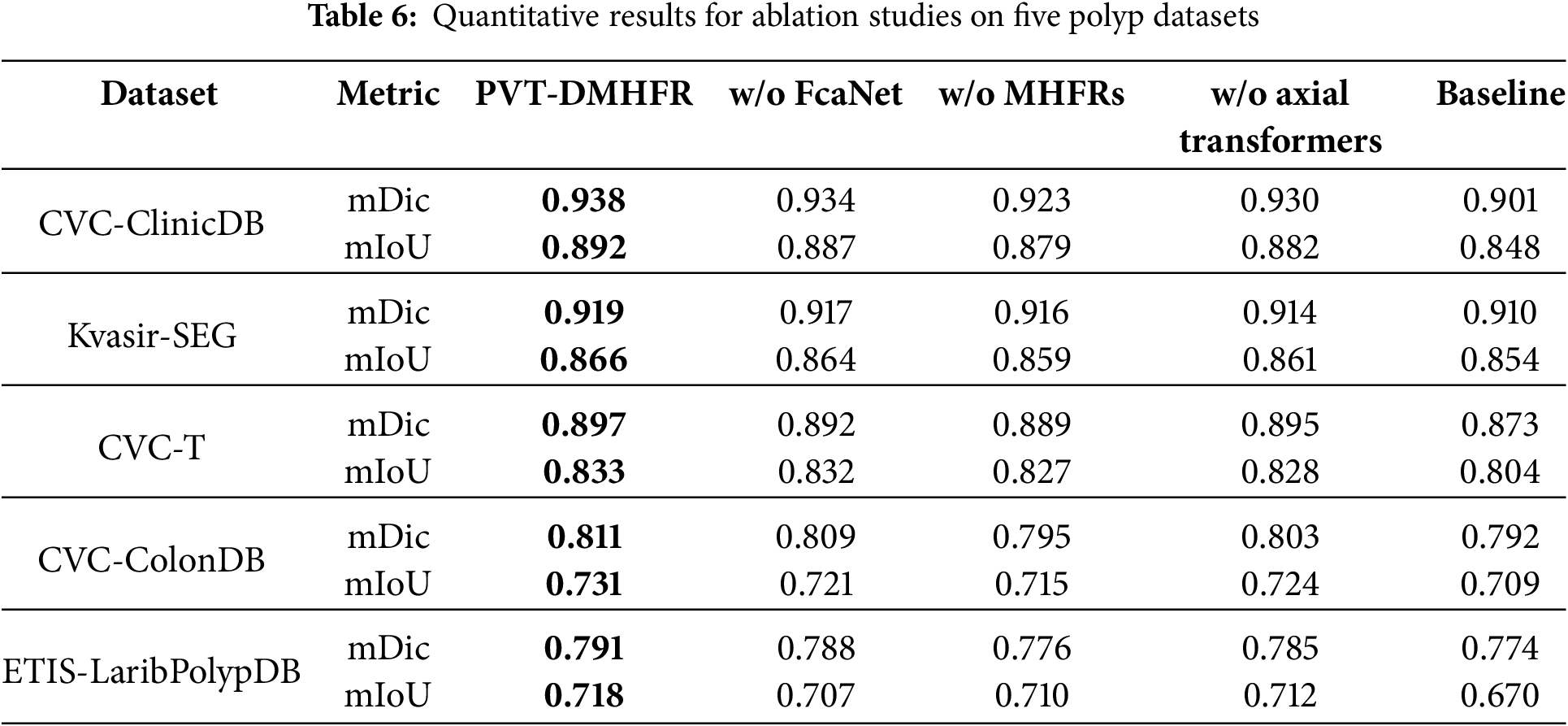
We present visualization results to better demonstrate the impact of our proposed MHFRs and integration of FcaNet modules and axial transformers in PVT-DMHFR. As illustrated in Fig. 5, the removal of any module from PVT-DMHFR results in a noticeable decline in segmentation accuracy. This performance degradation may stem from several factors: excluding FcaNet reduces the model’s capacity to capture detailed features, removing the axial transformers diminishes its ability to account for long-range feature dependencies, and omitting MHFRs impairs the fusion of multi-level features from the encoder backbone, leading to the loss of crucial semantic information.


Figure 5: Segmentation results under different configurations of PVT-DMHFR
Additionally, we explore the impact of replacing the Axial-Transformer with four alternative widely-used attention mechanisms: SE-Attention [62], CoT-Attention [59], EMA [60], and PSA [61], to evaluate their performance. As shown in Table 7, none of these attention mechanisms outperforms the Axial-Transformer. For instance, on CVC-ClinicDB, Axial-Transformer achieves the highest mDic (0.938) and mIoU (0.892), while PSA and CoT-Attention fall slightly short with mDic values of 0.936 and 0.932, and mIoU values of 0.889 and 0.887, respectively. Similarly, on the CVC-T dataset, the Axial-Transformer maintains its lead with mDic and mIoU scores of 0.897 and 0.833, respectively. While EMA and PSA achieve marginal improvements in mIoU on Kvasir-SEG and ETIS-LaribPolypDB, these gains are isolated and do not match the overall performance of the Axial-Transformer.
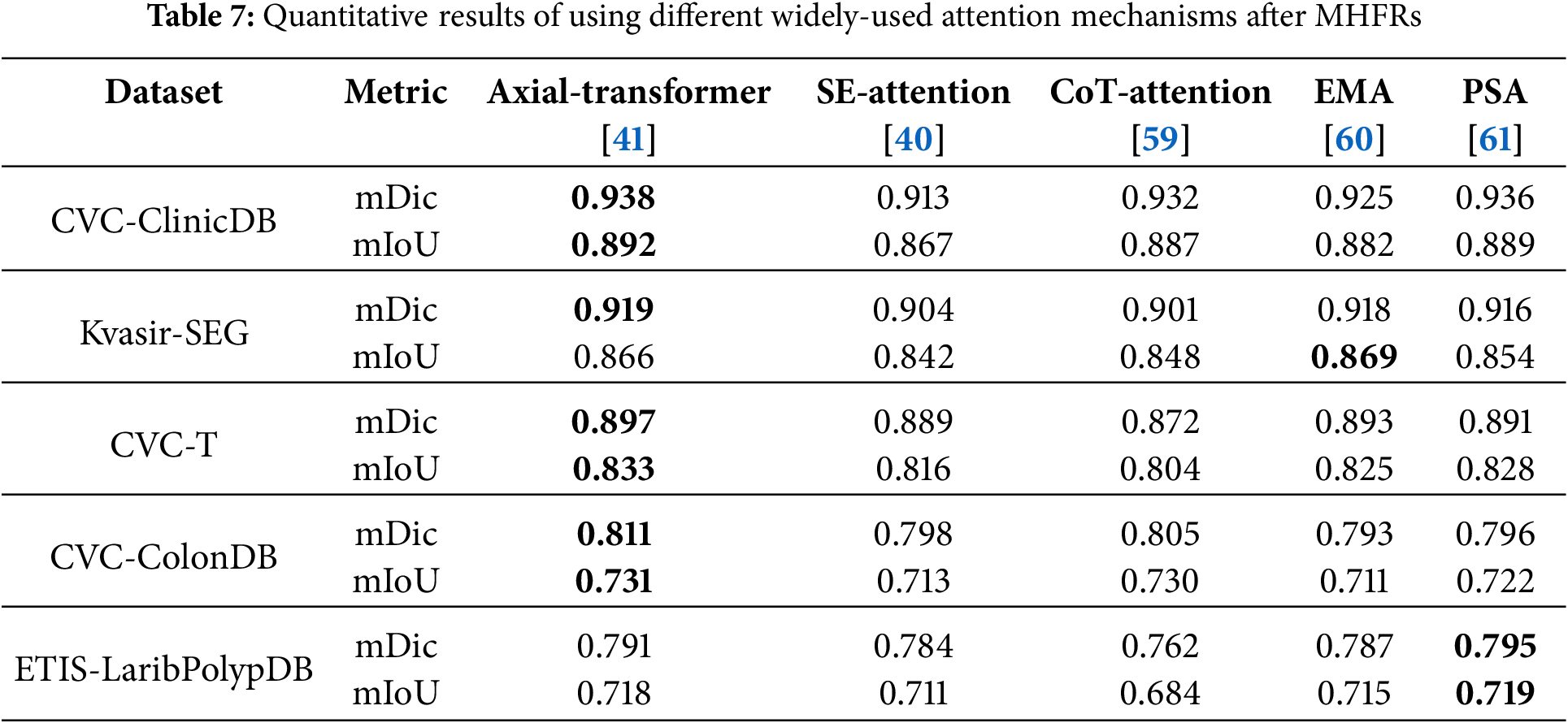
The consistent underperformance of SE-Attention and CoT-Attention reflects their limited ability to model global dependencies. In addition, while EMA and PSA perform better, their results lack consistency across datasets. Axial-Transformer’s superior ability to model long-range dependencies and spatial features explains its dominance, making it the most effective attention mechanism in this study.
In this paper, we propose the DMHFR for the aggregation of pyramid features. The MHFRs (THFR and FHFR), perceive and fuse multiple sets of pyramid features from fine to coarse granularity, as well as the full set. Before these features are processed by the MHFRs, they pass through FcaNet to achieve better feature modeling. After the features are processed MHFRs, they are fed into axial transformers to capture the global dependencies of the features. Our experimental results demonstrate that the proposed PVT-DMHFR outperforms 15 SOTA methods across five public polyp datasets, highlighting its superior generalization and learning capabilities. Specifically, when trained and tested on visible datasets (CVC-ClinicDB and Kvasir-SEG) to assess learning ability, PVT-DMHFR achieves mDic scores of approximately 0.92 and 0.94, respectively. On unseen datasets (CVC-T, ColonDB, and ETIS), used to evaluate generalization capabilities, the PVT-DMHFR achieves mDic scores of 0.897, 0.811, and 0.791, respectively. Furthermore, our MHFRs are versatile and can be easily adapted to process pyramid features in other models by adjusting the channel setting of MHFRs, offering significant potential to enhance deep learning performance in various medical image segmentation tasks. Beyond medical imaging, the DMHFR decoder can also be applied to enhance transformer features in broader medical applications and general computer vision.
Acknowledgement: The authors are thankful to Xiamen Medical and Health Guidance Project and Grant from Guangxi Key Laboratory of Machine Vision and Intelligent Control.
Funding Statement: This work was supported by Xiamen Medical and Health Guidance Project in 2021 (No. 3502Z20214ZD1070). The research was financially supported by a grant from Guangxi Key Laboratory of Machine Vision and Intelligent Control, China (No. 2023B02).
Author Contributions: Study conception and design: Jianuo Huang, data collection: Jianuo Huang, Bohan Lai, Weiye Qiu, Caixu Xu; analysis and interpretation of results: Jianuo Huang, Bohan Lai, Weiye Qiu, Caixu Xu, Jie He; draft manuscript preparation: Jianuo Huang, Jie He. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All publicly available datasets are used in the study.
Ethics Approval: This study utilizes publicly available datasets, all of which have received prior ethical approval. No additional ethical approval was required for this work.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference; 2015 Oct 5–9; Munich, Germany: Springer International Publishing. p. 234–41. [Google Scholar]
2. Fan DP, Ji GP, Zhou T, Chen G, Fu H, Shen J, et al. PraNet: parallel reverse attention network for polyp segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; 2020; Cham: Springer International Publishing. p. 263–73. [Google Scholar]
3. Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J. UNet++: a nested U-Net architecture for medical image segmentation. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018; 2018; Granada, Spain. p. 3–11. [Google Scholar]
4. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016; Las Vegas, NV, USA; p. 770–8. [Google Scholar]
5. Huang H, Lin L, Tong R, Hu H, Zhang Q, Iwamoto Y, et al. UNet 3+: a full-scale connected unet for medical image segmentation. In: ICASSP 2020–2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); 2020; Barcelona, Spain: IEEE. p. 1055–9. [Google Scholar]
6. Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, et al. Attention U-Net: learning where to look for the pancreas. arXiv preprint arXiv:1804.03999. 2018. [Google Scholar]
7. Chen S, Tan X, Wang B, Hu X. Reverse attention for salient object detection. In: Proceedings of the European Conference on Computer Vision (ECCV); 2018; Munich, Germany. p. 234–50. [Google Scholar]
8. Dosovitskiy A. An image is worth 16x16 words: transformers for image recognition at scale. arXiv preprint arXiv:2010.11929. 2020. [Google Scholar]
9. Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, et al. Swin-Unet: Unet-like pure transformer for medical image segmentation. In: European Conference on Computer Vision; 2022; Cham: Springer Nature Switzerland. p. 205–18. [Google Scholar]
10. Zhang Y, Liu H, Hu Q. Transfuse: fusing transformers and CNNs for medical image segmentation. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France; 2021 Sep 27–Oct 1; Strasbourg, France: Springer International Publishing. p. 14–24. [Google Scholar]
11. Wang W, Xie E, Li X, Fan DP, Song K, Liang D, et al. PVT v2: improved baselines with pyramid vision transformer. Comput Vis Media. 2022;8(3):415–24. doi:10.1007/s41095-022-0274-8. [Google Scholar] [CrossRef]
12. Wang Z, Wu Z, Agarwal D, Sun J. MedCLIP: contrastive learning from unpaired medical images and text. arXiv preprint arXiv:2210.10163. 2022. [Google Scholar]
13. Wang W, Xie E, Li X, Fan DP, Song K, Liang D, et al. Pyramid vision transformer: a versatile backbone for dense prediction without convolutions. In: Proceedings of the IEEE/CVF International Conference on Computer Vision; 2021; Montreal, QC, Canada; p. 568–78. [Google Scholar]
14. Xie E, Wang W, Yu Z, Anandkumar A, Alvarez JM, Luo P. SegFormer: simple and efficient design for semantic segmentation with transformers. Adv Neural Inf Process Syst. 2021;34:12077–90. [Google Scholar]
15. Qin Z, Zhang P, Wu F, Li X. FcaNet: frequency channel attention networks. In: Proceedings of the IEEE/CVF International Conference on Computer Vision; 2021; Montreal, QC, Canada; p. 783–92. [Google Scholar]
16. Sasmal P, Iwahori Y, Bhuyan MK, Kasugai K. Active contour segmentation of polyps in capsule endoscopic images. In: 2018 International Conference on Signals and Systems (ICSigSys); 2018; Bali, Indonesia: IEEE. p. 201–4. [Google Scholar]
17. Xia S, Krishnan SM, Tjoa MP, Goh PM. A novel methodology for extracting colon’s lumen from colonoscopic images. J Syst Cybern Inform. 2003;1(2):7–12. [Google Scholar]
18. Gross S, Kennel M, Stehle T, Wulff J, Tischendorf J, Trautwein C, et al. Polyp segmentation in NBI colonoscopy. In: Bildverarbeitung für die Medizin 2009: Algorithmen—Systeme—Anwendungen Proceedings des Workshops vom 22. bis 25. März 2009 in Heidelberg; Heidelberg, Germany: Springer Berlin Heidelberg; 2009. p. 252–6. [Google Scholar]
19. Shi JH, Zhang Q, Tang YH, Zhang ZQ. Polyp-mixer: an efficient context-aware MLP-based paradigm for polyp segmentation. IEEE Trans Circuits Syst Video Technol. 2022;33(1):30–42. doi:10.1109/TCSVT.2022.3197643. [Google Scholar] [CrossRef]
20. Cai L, Wu M, Chen L, Bai W, Yang M, Lyu S, et al. Using guided self-attention with local information for polyp segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; 2022; Cham: Springer Nature Switzerland. p. 629–38. [Google Scholar]
21. Zhang R, Lai P, Wan X, Fan DJ, Gao F, Wu XJ, et al. Lesion-aware dynamic kernel for polyp segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; 2022; Cham: Springer Nature Switzerland. p. 99–109. [Google Scholar]
22. Tomar NK, Jha D, Bagci U, Ali S. TGANet: text-guided attention for improved polyp segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; 2022; Cham: Springer Nature Switzerland. p. 151–60. [Google Scholar]
23. Bui NT, Hoang DH, Nguyen QT, Tran MT, Le N. MEGANet: multi-scale edge-guided attention network for weak boundary polyp segmentation. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision; 2024; Waikoloa, HI, USA; p. 7985–94. [Google Scholar]
24. Akbari M, Mohrekesh M, Nasr-Esfahani E, Soroushmehr SR, Karimi N, Samavi S, et al. Polyp segmentation in colonoscopy images using fully convolutional network. In: 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); 2018; Honolulu, HI, USA: IEEE. p. 69–72. [Google Scholar]
25. Brandao P, Zisimopoulos O, Mazomenos E, Ciuti G, Bernal J, Visentini-Scarzanella M, et al. Towards a computed-aided diagnosis system in colonoscopy: automatic polyp segmentation using convolution neural networks. J Med Robot Res. 2018;3(2):1840002. doi:10.1142/S2424905X18400020. [Google Scholar] [CrossRef]
26. He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. In: Proceedings of the IEEE International Conference on Computer Vision; 2017; Venice, Italy; p. 2961–9. [Google Scholar]
27. Qadir HA, Shin Y, Solhusvik J, Bergsland J, Aabakken L, Balasingham I. Polyp detection and segmentation using mask R-CNN: does a deeper feature extractor CNN always perform better? In: 2019 13th International Symposium on Medical Information and Communication Technology (ISMICT); 2019; Oslo, Norway: IEEE. p. 1–6. [Google Scholar]
28. Ji GP, Chou YC, Fan DP, Chen G, Fu H, Jha D, et al. Progressively normalized self-attention network for video polyp segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; 2021; Cham: Springer International Publishing. p. 142–52. [Google Scholar]
29. Wu L, Hu Z, Ji Y, Luo P, Zhang S. Multi-frame collaboration for effective endoscopic video polyp detection via spatial-temporal feature transformation. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference; 2021 Sep 27–Oct 1; Strasbourg, France: Springer International Publishing. p. 302–12. [Google Scholar]
30. Li C, Liu X, Li W, Wang C, Liu H, Yuan Y. U-KAN makes strong backbone for medical image segmentation and generation. arXiv preprint arXiv:2406.02918. 2024. [Google Scholar]
31. Sun L, Li C, Ding X, Huang Y, Chen Z, Wang G, et al. Few-shot medical image segmentation using a global correlation network with discriminative embedding. Comput Biol Med. 2022;140:105067. doi:10.1016/j.compbiomed.2021.105067. [Google Scholar] [PubMed] [CrossRef]
32. Banik D, Roy K, Bhattacharjee D, Nasipuri M, Krejcar O. Polyp-Net: a multimodel fusion network for polyp segmentation. IEEE Trans Instrum Meas. 2020;70:1–12. doi:10.1109/TIM.2020.3015607. [Google Scholar] [CrossRef]
33. Kim T, Lee H, Kim D. UACANet: uncertainty augmented context attention for polyp segmentation. In: Proceedings of the 29th ACM International Conference on Multimedia; 2021; New York, NY, USA. p. 2167–75. [Google Scholar]
34. Wei J, Hu Y, Zhang R, Li Z, Zhou SK, Cui S. Shallow attention network for polyp segmentation. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference; 2021 Sep 27–Oct 1; Strasbourg, France: Springer International Publishing. p. 699–708. [Google Scholar]
35. Zhao X, Zhang L, Lu H. Automatic polyp segmentation via multi-scale subtraction network. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference; 2021 Sep 27–Oct 1; Strasbourg, France: Springer International Publishing. p. 120–30. [Google Scholar]
36. Xu R, Wang C, Xu S, Meng W, Zhang X. DC-Net: dual context network for 2D medical image segmentation. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference; 2021 Sep 27–Oct 1; Strasbourg, France: Springer International Publishing. p. 503–13. [Google Scholar]
37. Wang J, Huang Q, Tang F, Meng J, Su J, Song S. Stepwise feature fusion: local guides global. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; 2022; Cham: Springer Nature Switzerland. p. 110–20. [Google Scholar]
38. Zhang W, Fu C, Zheng Y, Zhang F, Zhao Y, Sham CW. HSNet: a hybrid semantic network for polyp segmentation. Comput Biol Med. 2022;150:106173. doi:10.1016/j.compbiomed.2022.106173. [Google Scholar] [PubMed] [CrossRef]
39. Dong B, Wang W, Fan DP, Li J, Fu H, Shao L. Polyp-PVT: polyp segmentation with pyramid vision transformers. arXiv preprint arXiv:2108.06932. 2021. [Google Scholar]
40. Rahman MM, Marculescu R. Medical image segmentation via cascaded attention decoding. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision; 2023; Waikoloa, HI, USA; p. 6222–31. [Google Scholar]
41. Ho J, Kalchbrenner N, Weissenborn D, Salimans T. Axial attention in multidimensional transformers. arXiv preprint arXiv:1912.12180. 2019. [Google Scholar]
42. Bhojanapalli S, Chakrabarti A, Glasner D, Li D, Unterthiner T, Veit A. Understanding robustness of transformers for image classification. In: Proceedings of the IEEE/CVF International Conference on Computer Vision; 2021; Montreal, QC, Canada; p. 10231–41. [Google Scholar]
43. He K, Zhang X, Ren S, Sun J. Identity mappings in deep residual networks. In: Computer Vision-ECCV 2016: 14th European Conference; 2016 Oct 11–14; Amsterdam, The Netherlands: Springer International Publishing. p. 630–45. [Google Scholar]
44. Bernal J, Sánchez FJ, Fernández-Esparrach G, Gil D, Rodríguez C, Vilariño F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: validation vs. saliency maps from physicians. Comput Med Imaging Graph. 2015;43:99–111. doi:10.1016/j.compmedimag.2015.02.007. [Google Scholar] [PubMed] [CrossRef]
45. Jha D, Smedsrud PH, Riegler MA, Halvorsen P, De Lange T, Johansen D, et al. Kvasir-seg: a segmented polyp dataset. In: MultiMedia Modeling: 26th International Conference, MMM 2020; 2020 Jan 5–8; Daejeon, Republic of Korea: Springer International Publishing. p. 451–62. [Google Scholar]
46. Vázquez D, Bernal J, Sánchez FJ, Fernández-Esparrach G, López AM, Romero A, et al. A benchmark for endoluminal scene segmentation of colonoscopy images. J Healthc Eng. 2017;2017(1):4037190. doi:10.1155/2017/4037190. [Google Scholar] [PubMed] [CrossRef]
47. Tajbakhsh N, Gurudu SR, Liang J. Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans Med Imaging. 2015;35(2):630–44. doi:10.1109/TMI.2015.2487997. [Google Scholar] [PubMed] [CrossRef]
48. Silva J, Histace A, Romain O, Dray X, Granado B. Toward embedded detection of polyps in wce images for early diagnosis of colorectal cancer. Int J Comput Assist Radiol Surg. 2014;9:283–93. doi:10.1007/s11548-013-0926-3. [Google Scholar] [PubMed] [CrossRef]
49. Milletari F, Navab N, Ahmadi SA. V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV); 2016; Stanford, CA, USA: IEEE. p. 565–71. [Google Scholar]
50. Margolin R, Zelnik-Manor L, Tal A. How to evaluate foreground maps? In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2014; Columbus, OH, USA; p. 248–55. [Google Scholar]
51. Fan DP, Cheng MM, Liu Y, Li T, Borji A. Structure-measure: a new way to evaluate foreground maps. In: Proceedings of the IEEE International Conference on Computer Vision; 2017; Venice, Italy; p. 4548–57. [Google Scholar]
52. Fan DP, Ji GP, Qin X, Cheng MM. Cognitive vision inspired object segmentation metric and loss function. Sci Sin Informationis. 2021;6(6):5. doi:10.1360/SSI-2020-0370. [Google Scholar] [CrossRef]
53. Fan DP, Gong C, Cao Y, Ren B, Cheng MM, Borji A. Enhanced-alignment measure for binary foreground map evaluation. arXiv preprint arXiv:1805.10421. 2018. [Google Scholar]
54. Loshchilov I. Decoupled weight decay regularization. arXiv preprint arXiv:1711.05101. 2017. [Google Scholar]
55. Yang L, Gu Y, Bian G, Liu Y. TMF-Net: a transformer-based multiscale fusion network for surgical instrument segmentation from endoscopic images. IEEE Trans Instrum Meas. 2022;72:1–15. doi:10.1109/TIM.2022.3225922. [Google Scholar] [CrossRef]
56. Duc NT, Oanh NT, Thuy NT, Triet TM, Dinh VS. ColonFormer: an efficient transformer based method for colon polyp segmentation. IEEE Access. 2022;10:80575–86. doi:10.1109/ACCESS.2022.3195241. [Google Scholar] [CrossRef]
57. Chang Q, Ahmad D, Toth J, Bascom R, Higgins WE. ESFPNet: efficient deep learning architecture for real-time lesion segmentation in autofluorescence bronchoscopic video. In: Medical Imaging 2023: Biomedical Applications in Molecular, Structural, and Functional Imaging; 2023; San Diego, CA, USA: SPIE. Vol. 12468. [Google Scholar]
58. Sanderson E, Matuszewski BJ. FCN-transformer feature fusion for polyp segmentation. In: Annual Conference on Medical Image Understanding and Analysis; 2022; Cham: Springer International Publishing. p. 892–907. [Google Scholar]
59. Li Y, Yao T, Pan Y, Mei T. Contextual transformer networks for visual recognition. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2022;45(2):1489–500. doi:10.48550/arXiv.2107.12292. [Google Scholar] [CrossRef]
60. Ouyang D, He S, Zhang G, Luo M, Guo H, Zhan J, et al. Efficient multi-scale attention module with cross-spatial learning. In: ICASSP 2023–2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); 2023; Rhodes Island, Greece: IEEE. p. 1–5. [Google Scholar]
61. Zhang H, Zu K, Lu J, Zou Y, Meng D. EPSANet: an efficient pyramid squeeze attention block on convolutional neural network. In: Proceedings of the Asian Conference on Computer Vision; 2022; Macau SAR, China. p. 1161–77. [Google Scholar]
62. Hu J, Shen L, Sun G. Squeeze-and-excitation networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2018; Salt Lake City, UT, USA. p. 7132–41. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools