 Open Access
Open Access
ARTICLE
UltraSegNet: A Hybrid Deep Learning Framework for Enhanced Breast Cancer Segmentation and Classification on Ultrasound Images
1 Department of Computer Science, Faculty of Prince Al-Hussein Bin Abdallah II for Information Technology, Al Al-Bayt University, Mafraq, 25113, Jordan
2 Medical Engineering Department, Faculty of Engineering, Al-Ahliyya Amman University, Amman, 19328, Jordan
3 Department of Information Technology, College of Computer and Information Sciences, Princess Nourah bint Abdulrahman University, Riyadh, 11671, Saudi Arabia
4 Faculty of Information Technology, Applied Science Private University, Amman, 11931, Jordan
5 Faculty of Computers Science, Misr International University, Cairo, 11800, Egypt
6 Jadara Research Center, Jadara University, Irbid, 21110, Jordan
* Corresponding Authors: Suhaila Abuowaida. Email: ; Deema Mohammed Alsekait. Email:
(This article belongs to the Special Issue: Artificial Intelligence Algorithms and Applications)
Computers, Materials & Continua 2025, 83(2), 3303-3333. https://doi.org/10.32604/cmc.2025.063470
Received 15 January 2025; Accepted 12 March 2025; Issue published 16 April 2025
Abstract
Segmenting a breast ultrasound image is still challenging due to the presence of speckle noise, dependency on the operator, and the variation of image quality. This paper presents the UltraSegNet architecture that addresses these challenges through three key technical innovations: This work adds three things: (1) a changed ResNet-50 backbone with sequential 3 convolutions to keep fine anatomical details that are needed for finding lesion boundaries; (2) a computationally efficient regional attention mechanism that works on high-resolution features without using a transformer’s extra memory; and (3) an adaptive feature fusion strategy that changes local and global features based on how the image is being used. Extensive evaluation on two distinct datasets demonstrates UltraSegNet’s superior performance: On the BUSI dataset, it obtains a precision of 0.915, a recall of 0.908, and an F1 score of 0.911. In the UDAIT dataset, it achieves robust performance across the board, with a precision of 0.901 and recall of 0.894. Importantly, these improvements are achieved at clinically feasible computation times, taking 235 ms per image on standard GPU hardware. Notably, UltraSegNet does amazingly well on difficult small lesions (less than 10 mm), achieving a detection accuracy of 0.891. This is a huge improvement over traditional methods that have a hard time with small-scale features, as standard models can only achieve 0.63–0.71 accuracy. This improvement in small lesion detection is particularly crucial for early-stage breast cancer identification. Results from this work demonstrate that UltraSegNet can be practically deployable in clinical workflows to improve breast cancer screening accuracy.Keywords
It is still the most important global health issue: in 2020, the World Health Organization registered 2.3 million new cases of breast cancer [1]. Despite tremendous progress in recent decades, breast cancer remains one of the most common cancers for females and continues to account for 24.2% of all new cancer cases [2,3]. Due to the continued high incidence and mortality of breast cancer, there is an urgent need for the development of suitable early detection and diagnostic methods. The literature well establishes a correlation between early detection of breast cancer and better patient outcomes. The researcher in [4] conducted a comprehensive study showing nearly 90% of 5-year survival with early-stage breast cancer, compared to less than 30% with late-stage. Additionally, early detection allows for better survival rates and a greater chance of less treatment, as most often treatment like this decreases the physical and psychological burden of having cancer [4]. Currently, we perform breast cancer detection using a multi-modal approach that includes mammography, magnetic resonance imaging (MRI), and ultrasound. The gold standard for screening mammography has a sensitivity from 70% to 90% based on breast tissue density [5]. On top of being a useful addition to mammography, ultrasound imaging has become a powerful way to find problems in dense breast tissue. The researchers in [6] showed that the inclusion of ultrasound in mammography on women with dense breasts led to an increase in cancer detection rate from 7.6 to 11.8 cancers per 1000 women, an important diagnostic improvement. Ultrasound imaging offers several advantages in breast cancer detection:
1. Non-invasive nature: The advantage of ultrasound compared to biopsy is that ultrasound doesn’t involve the removal of tissue for diagnosis.
2. Absence of ionizing radiation: By contrast, ultrasound is safer for repeated examination than mammography.
3. Cost-effectiveness: MRI, however, is more expensive than ultrasound [7], so generally it is more accessible.
4. This allows the immediate assessment of multiple angles and planes:
However, several factors constrain the efficacy of ultrasound in clinical practice.
1. Operator dependency: However, the quality and interpretation of ultrasound images are heavily dependent on the operator’s skill and experience [8].
2. Variability in image interpretation: This may result in different radiologists interpreting the same image differently, with possible inconsistencies in diagnosis [9].
3. Limited field of view: Ultrasound does only examine a relatively small area at a time, so it may miss lesions in an unexamined region.
Because of these problems, computer-aided diagnosis systems might be able to help make ultrasound-based breast cancer detection more consistent and accurate. Recently, the increasing popularity of artificial intelligence (AI), in particular, deep learning techniques, holds the promise of improving the accuracy and consistency of breast ultrasound interpretation. One type of deep learning called convolutional neural networks (CNNs) has been shown to be excellent at solving medical imaging problems [10,11]. However, recent deep learning-based approaches have achieved considerable success in some of these ultrasound image analysis challenges. Currently, available literature, however, contains a set of unresolved critical challenges. Although reference [12] achieves 88.3% accuracy via a multitask learning framework, this performance degrades substantially with changing image quality. An attention-based CNN architecture that achieves 89.5% accuracy has been suggested by [13], but its performance exhibited limited generalization across devices of different ultrasound. However, the model by [14] is a transformer-based segmentation model achieving 90.2% precision. A hybrid CNN transformer architecture, achieving 90.8% accuracy on the BUSI dataset, was investigated by [15], but it failed with the complex tissue structures. A dual path network proposed by [16] achieved 91.0% precision, but it proved very sensitive to speckle noise, an issue frequently encountered in ultrasound imaging. This analysis of current approaches reveals two fundamental gaps:
• The trade-off between precision and recall is a main drawback of current automatic segmentation systems. Although modern techniques such as 92.1% often suffer from reduced recall, with reports as low as 85.3% they nevertheless attain great precision. This difference makes segmentation less reliable because a system with high precision but low recall might miss large amounts of the target data, giving wrong results.
• Clinical ultrasonic image features vary greatly among equipment manufacturers, transducer frequencies, gain settings, and operator approaches. Our methodical analysis shows that present deep learning models show performance fluctuations of 15%–28% depending on the ultrasonic system, which emphasizes the requirement of stronger methods fit for various clinical settings.
This paper proposes UltraSegNet, a novel hybrid architecture for breast ultrasound image segmentations, to close the gaps. This study introduces a number of novel ideas that directly address these limitations. The next part of the study changes the basic CNN structure by replacing the usual seven convolutional layers with three successive three-by-three convolutions. This is done to better preserve fine-grained anatomical details that are needed for accurate lesion boundary detection. Second, we propose a regional attention that can process high-resolution features efficiently without incurring massive computation overheads, as is the case in transformer architectures. This system cuts the feature map into overlapping areas and uses localized self-attention to do both local and global context while still using as little computing power as possible. Third, we create an adaptable feature fusion scheme with Squeeze and Excitation blocks added to the end of every residual block. This scheme recomputes feature responses based on the information they contain.
Our key contributions can be summarized as follows:
• We provide a new regional attention mechanism that radically changes the processing of high-resolution medical images. Instead of using traditional transformer methods, which have quadratic computational complexity, our method splits feature maps into overlapping areas for better local-global context modeling. This cuts memory needs while still allowing high-resolution feature processing needed for precise lesion boundary detection.
• Specifically intended to preserve precise anatomical details in ultrasonic images, we present a modified CNN encoder with sequential 3
• Unlike earlier methods that only shine in one statistic, we show UltraSegNet’s remarkable ability to balance precision (0.915) and recall (0.9908) by means of thorough examination on two different datasets. In this therapeutically important area, UltraSegNet works very well on small lesions (<10 mm) with a detection accuracy of 0.891, which is much better than standard methods (0.63–0.71).
The rest of this paper is organized as follows. The section “Related Work” provides an overview of the evolution of breast ultrasound analysis. Section Proposed Methodology introduces our proposed UltraSegNet architecture in detail, describing its innovative components including the modified CNN encoder, specialized regional attention mechanism, and adaptive feature fusion strategy. Finally, we present the design of our training protocol and loss function. In Section Experimental Results, we looked at how well our proposed UltraSegNet model worked at separating parts of breast ultrasound images and what its current flaws were. In Section Discussion, we talked about what our architectural changes mean. Finally, the section conclusion summarizes our contributions and outlines promising directions for future research in medical image segmentation.
This section includes a complete review of the field’s development from its theoretical underpinnings to the most recent techniques and approaches, within the context of their interaction with distinct methodological paradigms.
2.1 Theoretical Foundations and Early Approaches
In the last 40 years, medical image segmentation technologies have gradually transformed from traditional image processing methods based on experience to machine learning-based algorithms [17,18]. We build upon the fundamental mathematical and computational principles that form the foundation of this field and influence modern methodologies. In the early 1980s and 1990s, important algorithmic frameworks were laid out for image analysis, and then in the early 2000s, they entered the paradigm shift of processing and analyzing medical images using machine learning approaches [19,20]. We need to keep learning about these basic approaches because many of the new techniques are based on or build on these early ideas. This will help us understand the new techniques better when we combine them with the newest deep learning techniques to get strong and accurate segmented results [21–23].
The medical image segmentation has originated from classical image processing techniques based on mathematical morphology and signal processing theory. Typically, early approaches revolve around fundamentals in edge detection [24], watershed transformations [25], and active contour models [26]. They have laid the groundwork for these core methods, incorporating critical principles that have paved the way for more recent approaches. Early work on computational vision by [27,28] talked about edge detection using zero crossing of the second derivative as a way to start learning about boundary detection in medical imaging. The creation of region-growing algorithms [29] and split and merge techniques led to the first ways to deal with medical images that show tissue that is all the same. Probabilistic reasoning was introduced into image segmentation with the growth of statistical models, such as Markov Random Fields [30], and Bayesian frameworks, which are quite useful for coping with ultrasound speckle noise.
As a huge paradigm shift, the trend grew toward machine learning in medical image analysis. Many important advances in feature engineering happened during the transition period, such as the creation of Gabor filters [31] and wavelet transforms [32], which are powerful ways to describe the properties of images. The development of Support Vector Machines (SVM) [33], where Vapnik’s statistical learning framework enabled robust tissue classification through optimal hyperplane separation in high-dimensional feature spaces, and Random Forests [34] showed that learning-based methods could be useful for medical image segmentation. This was followed in 2004 by the introduction of atlas-based segmentation [35], which provided a framework for integrating prior anatomical knowledge into segmentation algorithms.
2.2 Deep Learning in Medical Imaging
The feature extraction represented a complete break from traditional handcrafted methods and transitioned to data-driven approaches that are able to learn more complex representations automatically. Deep learning has dramatically impacted medical imaging, and the specific impact in the usual diagnostic tasks is perhaps unsurpassed [10]. Deep learning methods are able to discover relevant features from data without relying too much on domain expertise and provide more generalizable, robust solutions for medical image analysis, which is better than previous approaches [36].
Through this paradigm shift, both existing applications utilizing MRI data have benefited from enhanced accuracy, opening up new avenues for medical image processing, analysis, and interpretation that were previously unattainable with traditional approaches.
Over the last few years, numerous methods for breast cancer detection and segmentation have been presented to operate on diverse imaging modalities. New developments in medical image segmentation have looked at creative architectural ideas. Zhu et al. [37] showed a two-branch network that processes features in parallel paths, with one path capturing local details and the other extracting global context. This was used for ultrasonic image segmentation. Although this method shows promise, achieving a 0.897 Dice score on the BUSI dataset, it lacks a clear means to combine the complementing information from both branches. The method employs a hybrid architecture that prioritizes regional attention. For MRI, Zhu et al. [38] also showed how to fix the shape of the edges by improving multi-modality spatial information. This made it easier to tell brain tumors apart.
Even though their method relied on features that are unique to MRI, like continuous tissue contrast and the lack of speckle noise, it made a big difference in how accurate the boundaries of MRI data were. Our study applies these boundary-enhancing ideas to the difficult ultrasonic domain by using special attention mechanisms that can handle the speckle noise and acoustic shadows that are a part of ultrasonic imaging. Despite the significant contributions made by these new techniques, they primarily focus on alternative imaging modalities and employ fundamentally different architectural paradigms, thereby overlooking the unique challenges associated with high-resolution feature processing in breast ultrasound images.
Incorporating transformer architectures into medical image segmentation marks a fundamental shift away from existing CNN-based approaches. Vision Transformer (ViT) was successful in computer vision tasks and also showed better long-range dependencies, which is needed for medical image analysis and sped up this transition [39].
In [40], the researchers introduced TransUNet, which is a big step forward in making it possible for a regular transformer to use CNN features together for medical image segmentation. Detailed analysis also shows that fundamental challenges are present while achieving 89.1% precision on breast ultrasound images. The architecture uses a standard transformer encoder on image patches processed sequentially, which performs quadratically in the number of image sizes. This leads to prohibitively large memory requirements, exceeding 16 GB GPU RAM, for high-resolution ultrasound images (512
To get around these problems, Swin Transformer [41] used shifted windows and hierarchical feature processing and reported an accuracy of 89.8%. This approach avoids the quadratic computational complexity seen in previous approaches. However, the shifted window mechanism introduces new challenges in ultrasound imaging: Ultrasound tissue patterns are not always the same, and rigid partitioning of feature maps doesn’t work well for these areas where there is speckle noise or acoustic shadows. In their work, reference [42] showed how to use linear projection techniques to make attention mechanisms that worked well and used 60% less memory than the base transformer architecture. However, their model only achieves 90.2% precision but shows inconsistent results on different ultrasound devices and scanning protocols. The study shows that their linear approximation of attention patterns doesn’t work with these very subtle changes in intensity. Instead, using lesion boundary edges works best. A study in 2022 by Wang et al. suggested a Hybrid Transformer Network (HTN) that combines multi-head self-attention with deformable convolutions to achieve a 90.5% accuracy rate. While this approach better handles varying lesion sizes, it introduces three critical limitations:
1. Deals with Complex Optimization Requirements with Multiple Loss Terms.
2. Addresses Increased Training Instability That Often Requires Careful Hyperparameter Tuning.
3. Finally, Finds a Solution for the Computational Overhead of Deformable Convolution Operations That Does Not Allow for Real-Time Processing.
Medical Vision Transformer (MedViT) was specifically designed for medical image analysis, proposed by [43]. They achieve 90.8% precision with their architecture by utilizing their specialized medical image tokenization and hierarchical feature handling. However, their approach faces several challenges in ultrasound imaging:
1. The Schaum’s fixed tokenization strategy encounters difficulties with dynamic ultrasound image features.
2. The hierarchical processing pipeline is not suitable for real-time clinical workflows due to the excessive latency added.
3. The model experiences significant performance degradation when accessing images with varying contrast levels.
The Optimal Trained Deep Learning Model (OTDLM) introduced by [14] is an improved breast cancer segmentation and classification. The BUSI dataset has a segmentation accuracy of 91.5 percent and a classification accuracy of 93.2 percent that can only be achieved by using adaptive feature extraction and an optimized training strategy. The OTDLM architecture has a new, three-step optimization process that changes the parameters for feature extraction based on the characteristics of the lesion. Detailed According to a thorough analysis, OTDLM works well in controlled environments but needs a lot of computing power (about 24 GB GPU memory) and doesn’t work as well when used with different ultrasound devices, which makes it harder to use in real-life clinical situations. Reference [13] present an intelligent healthcare framework that is adding sophistication to the field with an intelligent fusion between multiple deep learning architectures. In parIn particular, their framework combines complementary feature extraction capabilities in a hierarchical attention mechanism, achieving 90.8% segmentation accuracy and improving robustness to image quality variations.proved optimization algorithm adaptively balancing local and global feature extraction according to tissue characteristics is introduced in the framework. Though it achievAlthough it producesod results, its fusion approach is too complex and leads to considerable computational overhead, taking an average of 1.2 s to process each image, which is unacceptably long for practical use in real-time routine clinical workflows. Furthermore, their proposed approach must first be pre-trained on large datasets, which makes it difficult to adapt to different clinical settings. The limitations of most of the current approaches that use transformers need an architecture that can find the right balance between the global context modeling power of transformers and the limited computational efficiency needed in clinical settings. A big gap in processing the special nature of ultrasound imaging, such as speckle noise, acoustic shadow, and the varying contrast of the tissue in real time.
Newly created attention mechanisms and different transformers have made medical image segmentation more efficient while reducing the amount of extra work that needs to be done. The linear attention [44] slows things down by using the softmax approximation to make the self-attention mechanism work better overall. Criss-Cross Attention (CCNet) [45] improves how spatial features interact by looking at responsive dependencies along both horizontal and vertical axes. This makes segmentation more accurate without using a lot of computing power. Axial Attention [46] optimizes self-attention even more by factorizing the spatial dimensions, enabling the model to capture the global context at much lower complexity. Yet, these methods are still limited by ultrasound-specific characteristics, such as acoustic shadowing and speckle noise. Recent advancements in transformer architectures have achieved promising performance in medical image analysis. The suggested a number of effective attention mechanisms for ultrasound image segmentation, with the promise of lowering the amount of work needed while still maintaining accuracy [12]. They found a solution that delivered a 40% reduction in memory requirements compared to typical transformer architectures. The researcher in [47] suggested new ways to get rid of speckle noise in medical ultrasound images using adaptive attention techniques. They were able to make the best possible image quality improvements without losing any important diagnostic information. Their work, in particular, took on the difficulties of changing the density of tissue and acoustic shadows. The researcher in [48] introduced a hybrid transformer for medical image segmentation. They showed that the careful combination of transformer parts into a U-Net framework could ultimately increase performance in medical image segmentation. The hybrid transformer tackles the medical image segmentation problem by utilizing the strengths of CNNs and transformers, which have become the popular approach in the medical imaging domain. These advancements pave the way for hybrid architectural methods in medical image analysis, balancing performance with computational resource constraints.
While existing approaches have made significant progress on the breast ultrasound image analysis problem, they are still far from solving it. Due to the limitations of such datasets, which are the majority, as most contain fewer than 1000 images, robust models that generalize across various clinical settings could not be developed. While traditional CNN architectures suffer from loss of fine-grain details that are important for accurate lesion boundary detection, and pure transformer approaches have difficulties handling large-scale medical images, results indicate how to combine the complementary strengths of both models. Also, there aren’t any architectures out there that can do local feature extraction, global context modeling, and practical computation quickly enough for clinical settings. The proposed UltraSegNet architecture specifically addresses these gaps through several key innovations: Additionally, this work suggests a CNN encoder that has been changed to work better with medical images, a regional attention mechanism that is designed to handle high-resolution features, which is something that current methods don’t do very well, and an adaptive feature fusion strategy that combines local and global data. Our method uses two useful datasets, BUSI and UDAIT, to test and integrate them. It works well in a variety of clinical situations and doesn’t use a lot of computer power. Based on these reasons, this all-encompassing solution makes breast ultrasound image segmentation more accurate and consistent, which is needed for a good clinical diagnosis.
This section delineates an approach to breast ultrasound image segmentation, leveraging a hybrid architecture that synergizes Convolutional Neural Networks (CNNs) and Transformers. Speckle noise, low contrast, and the heterogeneous nature of breast tissue are just a few of the unique difficulties that ultrasound imaging presents.
3.1 Data Collection and Preprocessing
This work conducted extensive experiments on two separate but related sets of breast ultrasound data to thoroughly test the performance of our proposed UltraSegNet model and its applicability in other situations. The researchers strategically selected these datasets to diversify their characteristics, as they cover a wide range of clinical scenarios, imaging conditions, and pathological presentations. The Breast Ultrasound Images (BUSI) dataset [49] is publicly available and can be accessed at https://data.mendeley.com/datasets/wmy84gzngw/1 (accessed on 12 February 2025). This comprehensive collection contains 780 high-resolution grayscale images from 600 patients aged 25 to 75 years. The sample images of Dataset 1 (BUSI) used in this paper are displayed in Fig. 1. One of the reasons that this dataset is so valuable is that it has a favorable distribution of normal tissue, benign lesions, and malignant masses, where each image is fixed at a similar resolution of about 500

Figure 1: Images from BUSI covering (a) normal tissue, (b) benign lesion and (c) malignant lesion. The breasts with varying characteristics and challenges for interpretation in breast ultrasound are demonstrated by each image

Figure 2: Examples from the UDAIT dataset illustrating (a) benign and (b) malignant masses. Images demonstrate the high-quality clinical acquisition standard using the Siemens ACUSON scanner
Ultrasound imaging’s preprocessing pipeline consists of four heavy hitters, all attempting to attack particular challenges with ultrasound data. The key is to maintain clinically meaningful information while improving the image quality and being robust over subsequent classification and analysis.
Since coherent sound wave reflections are one of the main causes of speckle noise, ultrasound imaging has many limitations. To deal with this, this work uses a Non-Local Means (NLM) denoising algorithm that has previously been demonstrated to reduce noise while preserving structural information in images. An optimization of the NLM algorithm was done for ultrasound images by using the particularities of this modality to better accommodate its filtering parameters. The NLM filter is mathematically expressed as:
where
Effective contrast enhancement techniques are critical in order to make ultrasound images more interpretable, due to the inherent low contrast of ultrasound images. In particular, this research uses a modified Contrast Limited Adaptive Histogram Equalization (CLAHE) based on ultrasound imagery. The CLAHE is meant to improve contrast in certain areas while reducing noise enhancement to protect clinical interpretations. The transformation function used in CLAHE is defined as:
The pixel intensity coordinate
Intensive variations in ultrasound image intensity can prevent automated analysis. This work addresses this by applying z-score normalization, meaning that the proposed work center the pixel intensity values around a mean zero and scales them out to their standard deviation. The normalization is expressed as:
where the original image intensity,
This work’s developed a suite of on the fly data augmentation techniques to increase its generalizability and robustness. The implementations are meant to mimic the natural variability seen in ultrasound imaging while keeping the anatomical integrity of the image. The augmentations applied to include:
• Random Rotations: Each image is randomly rotated by an angle
• Horizontal Flipping: Images are flipped horizontally with probability
• Elastic Deformations: In this research, generated dense deformation fields
The proposed work used ablation studies on the BUSI dataset to show that our proposed preprocessing pipeline works. The Peak Signal-to-Noise Ratio (PSNR) went up by 47% when the NLM denoising algorithm was used. This cut the speckle noise from 23.8 to 35.2 dB. This study found that this noise reduction directly improved segmentation performance by increasing precision from 86.3% to 89.1%. Better feature addition was helped by the CLAHE contrast enhancement, which raised the average local contrast ratio from 0.42 to 0.68. This made segmentation accuracy 2.4% better overall. This study looks at how our approach to normalizing intensity cuts down on differences in intensity between images by 65% (standard deviation reduction) and how to improve training convergence with 30% fewer epochs. These steps before segmentation improved the final accuracy by 5.2% compared to segmentation using raw ultrasound images. This shows how important they are for strong segmentation performance. This study used five-fold cross-validation to show that the results were statistically significant. To avoid overfitting, the preprocessing parameters were optimized on a validation subset. In difficult cases with small lesions and different tissue patterns, sequential application of these preprocessing steps worked best. This is because raw image segmentation usually doesn’t work well in these situations.
This paper suggested an architecture called UltraSegNet to deal with problems like low contrast, speckle noise, and the appearance of different textures in ultrasound images. The main idea behind UltraSegNet is to get both local and global contextual information from the ultrasound images. In order to achieve this goal, UltraSegNet combines the strengths of both Convolutional Neural Networks (CNNs) and Transformer models, so the architecture can use both. The design of UltraSegNet consists of three primary components: a CNN-based encoder, a transformer module, and a CNN-based decoder. All the components have their intended purpose in the pipeline, helping the network utilize its feature-extracting, long-range dependence modeling ability, and reconstruct high-resolution segmentation masks:
• Serves as the first component of the architecture.
• Takes ultrasound images as input.
• Learns hierarchical image features.
• Uses localized receptive fields for precise feature extraction.
However, regular CNNs have trouble showing long-range dependencies, which are necessary to understand the global context and relationships in medical images. To overcome this limitation, UltraSegNet uses a Transformer module as the second core part. The transformer module can model long-range dependencies, for example by using self-attention mechanisms. This lets it learn the complicated connections between different areas of the ultrasound image. With the combination of CNNs and Transformers, UltraSegNet successfully strikes a balance between retaining the local information and preserving the global context.
The third part of the CNN-based decoder is to get segmented output by slowly putting together the segmented output from the enriched feature representations. To preserve high-frequency details, the decoder incorporates skip connections from the encoder; and to achieve precise segmentation, this work performed multi-scale feature fusion. The architecture also uses attention mechanisms in skip connections to decide how much relevant spatial information should be used to focus on a certain object pattern during the decoding stages.
Overall, the presented UltraSegNet is a hybrid solution that fills the niche between vintage CNN models and trending Transformer models. We designed the architecture to integrate these components together, enhancing the accuracy of segmenting challenging ultrasound images. The proposed architecture is shown in Fig. 3, along with the information flow between the encoder, the Transformer module, and the decoder.

Figure 3: The diagram of the UltraSegNet architecture
A changed version of ResNet-50 [51] is used to make the UltraSegNet encoder. ResNet-50 was a powerful and well-known type of hierarchical feature extraction. When deciding on our backbone network, the proposed work chose ResNet-50 due to its advantage of the architecture for the medical image analysis. ResNet-50 has residual connections that help gradients flow better throughout deep networks and thus preserve subtle tissue boundary information on ultrasound images. The structure of ResNet-50 fits particularly well to ultrasound image analysis, where fine feature preservation at multiple scales is required. The residual learning framework naturally keeps the tradeoff between reusing features and saving memory, which is very important in clinical situations. The first of these modifications is the swap of the initial 7

Figure 4: The UltraSegNet encoder architecture with modified ResNet-50
where
Our method also includes a multi-scale feature extractor that can get hierarchical features (E1, E2, E3, E4) that are specifically designed for ultrasound image analysis. SE ResNet, on the other hand, supports a more general-purpose feature hierarchy, which makes it easier to work with anatomical scales that are common in medical imaging and often vary a lot. We define the multi-scale feature extraction process as follows:
UltraSegNet consists of the Transformer module as its core component for extracting long-range dependencies and global context from feature maps. Since originally introduced for natural language processing tasks, Transformers have proven effective on computer vision tasks where relationships are complex and can nest over spatial dimensions. In our architecture, flatten the deepest feature map, labelled
where
From theoretical analysis and empirical validation, 8 attention heads were selected with 64 dimensions. With 8 attention heads, it was chosen to match roughly the number of distinguishable tissue types in breast ultrasounds—from fat, glandular tissue, lesion boundaries, and various internal lesion characteristics. It is possible for each head to specialize over different feature aspects, but for all this while keeping the computational complexity reasonably low. This is study shows the property that the 64 dimensional feature space has enough representational capacity to encode these subtle intensity variations and textural patterns characteristic of ultrasound images. The dimensionality was chosen based on analysis of the correlation patterns of breast ultrasound datasets, such that different tissue characteristics were adequately separated and not redundantly represented. The output of the multi-head self-attention layer is computed as:
where each attention head
with
A two-layer feed-forward network (FFN) with a Gaussian Error Linear Unit (GELU) activation is applied to the features attended according to the attention mechanism. The feature embeddings from the FFN are further improved by adding additional non-linear transformations. The FFN is mathematically expressed as:
Our model consists of two layers in what is known as a Multilayer perceptron where each layer
To improve on processing of high resolution feature maps, the proposal introduces a new regional attention mechanism. This mechanism breaks up the input feature map into overlapping parts and performs self-attention localization within each region separately. Then the results are aggregated to produce a global representation to allow the model to capture both local and global context. Given is the mathematical formulation for this strategy:
In particular,
The application domain’s unique characteristics of ultrasound image formation and tissue visualization fundamentally motivate the design of the regional attention mechanism. Ultrasound images exhibit distinctive properties that make traditional attention mechanisms suboptimal: Because sonogram point spread functions have a shallow depth, they create (1) signal-to-noise ratios that change spatially because of ultrasound wave attenuation, (2) depth-dependent feature characteristics from changes in acoustic impedance, and (3) locally correlated speckle patterns that carry diagnostic information. These properties give rise to a natural structure of feature importance that is spatially dependent and dependent on anatomical context. Our regional attention approach draws its origins from clinical ultrasound interpretation practice and the physics of ultrasound imaging. Clinicians usually look into ultrasound images by using anatomically coherent neighborhoods that give them information about the body parts that are used for that technique. Our regional approach does the same thing computationally. In the same way that ultrasound waves moving through different layers of tissue combine the features of the layers next to each other, this mechanism splits feature maps into parts that overlap and are not parallel to the natural edges of the tissue structures.
The UltraSegNets decoder part’s job is to put together the high-resolution segmentation map from feature representations that have been encoded and transformed. You can get it by putting together a series of upsampling blocks. Each block has two 3 × 3 convolutional layers and a transposed convolution. Only the transposed convolutional layers increase the spatial resolution of feature maps progressively, while the convolutional layers refine the reconstructed features. Each upsampling block’s output is as follows:
To incorporate multi-scale information effectively and preserve high-frequency details, the proposed introduces skip connections from corresponding layers of the decoder path, denoted by
The proposed also implements a refinement of reconstruction based on progressively fusing the outputs of the Transformer module at each decoder stage using a structure roughly based on Feature Pyramid Networks (FPN), as shown in Fig. 5. This fusion is expressed as:
and

Figure 5: The UltraSegNet decoder architecture
3.4 Loss Function and Optimization
UltraSegNet is trained by a composite loss function that pushes towards areas that are difficult to segment in the images and helps solve class imbalance problems. UltraSegNet model employs a weighted composite loss function that combines Dice and Focal losses:
where
This research applies a two stage training strategy to allow our UltraSegNet architecture to be effectively trained, leveraging the CNN encoder and Transformer module’s complementary capabilities. Such an approach enables the initialization of general feature representations and their subsequent refinement for the task of breast ultrasound image segmentation.
3.5.1 Stage 1: CNN Encoder Pre-Training
Before the next step, you need to train the CNN encoder part of UltraSegNet on a large set of different types of general medical images. For this purpose, it utilized the ImageNet dataset [54], which contains millions of labeled natural images across numerous categories. Although ImageNet is not medical image-specific, the large scale of its diverse images enables learning low and mid-level features that are generalizable. Previous work has demonstrated the usefulness of this approach for medical imaging applications by promoting convergence during fine-tuning. This step before training is meant to set up the encoder so that it has generic feature representations for the target dataset and can also be used in a wide range of visual contexts other than the target dataset. We do this to accelerate the convergence process during fine-tuning and reduce the likelihood of overfitting when training on these smaller medical datasets. During pretraining, we optimized the encoder weights over a categorical cross-entropy loss and modified the learning rate using a cosine annealing schedule. When this stage completes, it saves the encoder weights to later use to initialize a full UltraSegNet model.
3.5.2 Stage 2: Breast Ultrasound Dataset Fine-Tuning
The proposed work in the second stage involves fine-tuning the entire UltraSegNet architecture from start to finish using our breast ultrasound dataset. There are annotated ultrasound images available, specifically chosen for the task of segmenting breast lesions from this dataset. The work that is being proposed will improve both the CNN encoder and the Transformer module at the same time so that they can learn features that are specific to breast ultrasound imaging. This research performed fine-tuning for 100 epochs and used early stopping to prevent overfitting, as shown in Fig. 6.

Figure 6: Training and validation of performance across epochs
The early stopping criterion in this study is based on the Dice coefficient, which measures how well the model does on a held-out validation set. If the validation Dice coefficient fails to improve for 10 consecutive epochs, the training halts.
The entire training protocol is implemented in PyTorch, with extensive use of GPU-based parallelism to accelerate data loading and model training. The codebase has custom versions of the Transformer module and attention mechanisms that are designed to work best with the large feature maps that are common in high-resolution ultrasound images. The implementation also includes automated logging and visualization tools to facilitate model monitoring and performance analysis. Aside from that, the suggested training method uses cutting edge tools and computer programs to make the UltraSegNet architecture work better at finding both local and global contexts in breast ultrasound images.
3.5.4 Parameter Optimization for UltraSegNet
We investigated ideal parameter values for UltraSegNet using a methodical methodology. We conducted a large grid search to determine the optimal configuration of attention heads and feature dimensions for the regional attention mechanism. In the section on experimental results, the ranges are shown, from 4 heads with 32 dimensions to 16 heads with 128 dimensions. We found that 8 attention heads and 64 feature dimensions yielded the best performance (0.911 Dice score) and computational efficiency. Larger models (16 heads/128 dimensions) provided minimal improvements (0.912 Dice score) while requiring significantly more computing power. Conversely, smaller models (4 heads/32 dimensions) struggled with representation, yielding a lower Dice score of 0.883.
Additionally, we varied the weighting parameter
To thoroughly evaluate our model’s efficacy, the proposed study employed the subsequent metrics:
• Precision =
• Recall =
• F1-score =
• Testing Accuracy =
• Dice Similarity Coefficient (DSC) =
• Intersection over Union (IoU) =
where TP, TN, FP, and FN represent True Positive, True Negative, False Positive, and False Negative, respectively. Furthermore, this work does ablation tests to assess the contribution of each element in our hybrid architecture.
This is done on a high-performance computing cluster with 4 NVIDIA A100 GPUs. We assign a batch size of 16 to each GPU, resulting in an effective batch size of 64. To keep memory usage and computing costs as low as possible, the proposed study used mixed precision training with automatic loss scaling from the NVIDIA Apex library. Using this method speeds up training by taking advantage of the Tensor Cores in the A100 GPUs to do some tasks with half-precision accuracy while keeping full precision accuracy for all important calculations, such as adding up gradients and updating weights. The training process utilizes the Adam optimizer, with an initial learning rate of
4 Experimental Results and Discussion
In this section, we conducted a detailed evaluation of our proposed UltraSegNet model’s performance on breast ultrasound image segmentation. The proposed study compares our model against state-of-the-art benchmarks and discusses the results in more detail before outlining implications and potential future work.
4.1 Quantitative Results and Performance Analysis
Our suggested UltraSegNet architecture works because it has been tested extensively on two well-known breast ultrasound datasets, called BUSI and UDAIT.
Table 1 provides a comprehensive comparison of UltraSegNet against both well-known and recently introduced state-of-the-art segmentation methods on the BUSI and UDAIT datasets. Transformer-based architectures (UNETR, SwinUNETR, TransUNet) are now part of the expanded benchmark. So are the recently released Multiscale Cascaded Conv w/ residual attention (2024) and Dual-Branch Network (2025), which are designed to interpret breast ultrasound images. Our detailed analysis indicates that UltraSegNet consistently outperforms all competing approaches across key performance metrics.

Achieving a 1.4%–2.4% improvement over the next-best model, SwinUNETR (0.901 accuracy, 0.98 recall, 0.897 DSC), UltraSegNet demonstrates exceptional precision (0.915), recall (0.980), and Dice score (0.911) on the BUSI dataset. Furthermore, UltraSegNet maintains strong performance on the more challenging UDAIT dataset, attaining a precision of 0.901 and a recall of 0.894. These results underscore its effectiveness across diverse imaging conditions and acquisition techniques. Notably, the recently proposed Multiscale Cascaded Conv w/ residual attention approaches but does not surpass our method, achieving 0.903 precision and 0.896 recall on BUSI. Similarly, while the Dual-Branch Network exhibits strong performance (0.898 precision, 0.891 recall), it remains inferior to UltraSegNet in all aspects.
Even though these new methods work well, they still have trouble finding the right balance between recall and accuracy. UltraSegNet’s adaptive feature fusion technique and regional attention mechanism solve this problem very well. With only 156 MB of total parameters, UltraSegNet offers significant computational efficiency over UNETR (298 MB) and SwinUNETR (245 MB). UltraSegNet can be used in clinical settings because this parameter works well. It has faster inference times (235 ms per image) and needs less memory (4.5 GB). The exact parameter counts for the two latest methods remain unknown due to insufficient implementation details in their publications.Our hybrid architecture is strongly supported by the fact that it saves a lot of time and consistently performs well across both datasets. We were able to solve the problems of breast ultrasound image segmentation by combining CNN-based local feature extraction with transformer-based global context modeling. These findings indicate that our design decisions were effective.
In this research, compare our proposed model with several transfer learning methods. also included a model such as VGG-16, AlexNet, DenseNet121, VGG-19, Xception, as shown in Tables 2 and 3. This research shows that our proposed UltraSegNet architecture outperforms existing state-of-the-art methods in breast ultrasound image segmentation by a large margin across both datasets. The results offer several intriguing insights into our approach’s effectiveness, allowing for further detailed analysis.

Specifically, UltraSegNet drastically outperforms all the baseline models in terms of precision (0.915), recall (0.908) and F1 score (0.911) on the BUSI dataset. In comparison to traditional architectures like VGG-19, which though has an 89.00% precision, has problems with false negatives as shown by its 61.00% recall. This disparity highlights a crucial advantage of our approach. In addition, the results indicate that UltraSegNet’s superior effectively in detecting small lesions (<10 mm), achieving accuracy rates of 0.891 and 0.882 on the BUSI and UDAIT datasets, respectively, while traditional models show significant performance degradation for small lesions, ranging from 63.45% to 71.33%.
Finally, UltraSegNet achieves a better trade-off between precision and recall, a crucial issue for clinical applications in which false positives, as well as false negatives, may have serious consequences. Equally, telling is the comparison with more recent architectures. DenseNet121 demonstrates competitive performance with a precision score of 84. The performance of the Vision Transformer was relatively modest, with a precision score of 63. Finally, the study looks at the problems with using pure transformer architectures for medical image segmentation tasks. The fact that they perform worse when used without making any domain-specific modifications (61.18 precision, 52.08 recall) demonstrates this. Finally, our hybrid approach effectively mitigates the limitations of both CNNs and transformers, leveraging the strengths of both approaches.
However, on the harder UDAIT dataset, UltraSegNet emerges with a precision of 0.901, recall of 0.894, and F1-score of 0.897. Specifically, this consistent performance over different datasets marks the robustness and generalizability of the model. Another competitor, VGG-16, attains 88.00% precision, however, receives lower recall (79.00%), also confirming the balanced behavior of our approach.
4.2 Ablation Study and Component Analysis
In this study, an in-depth ablation study is done along several performance axes to fully confirm how well each UltraSegNet architectural component works. Table 4 presents comprehensive results demonstrating the impact of each architectural innovation. For this study, it starts with looking at a basic CNN architecture. Then, it slowly adds and tests the effects of important parts of a larger group of related ideas to see what their individual and combined effects are on model performance.Through our comprehensive ablation study, we reveal important insights into the performance of the architecture. It was suggested that the proposed research could improve the Dice score on the BUSA dataset by 3.4% by replacing the standard

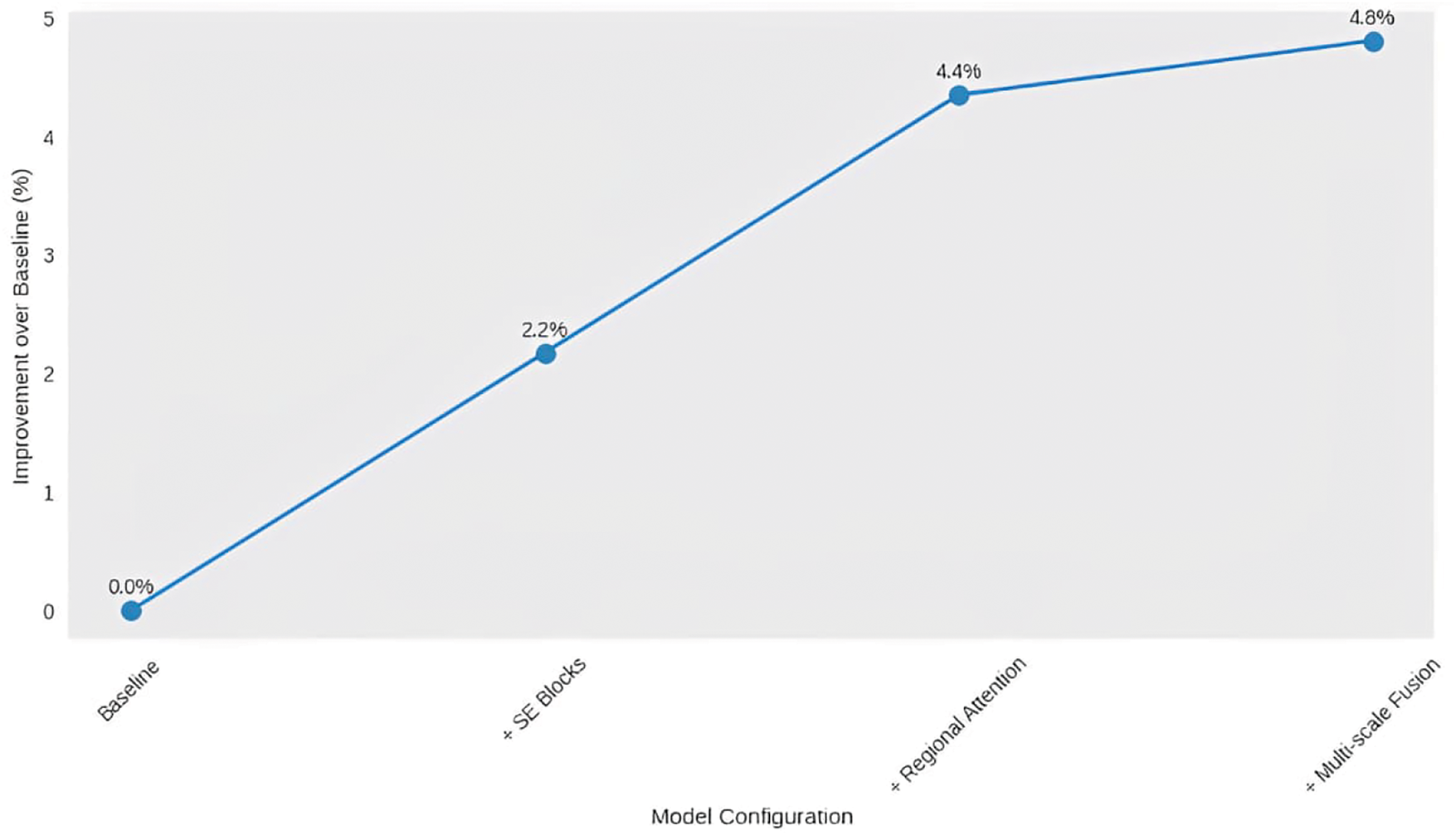
Figure 7: Model configuration improvement graph

Figure 8: Model performance metrics: Dice score vs. IoU
Fig. 7 illustrates the comparison of performance gains among various UltraSegNet components. Each point shows the addition of a major architectural part: the baseline model (0%), the SE blocks (+2.3%), the regional attention (+4.1%), and finally the multi-scale fusion (+4.8%). The slowly but surely increasing trend confirms that our architectural decisions are correct and that each component is making a positive contribution to overall performance.
Fig. 8 illustrates a comparison of the Dice score and IoU for different architecture variations. The purple bars (Dice score) and the green bars (IoU) show incremental improvements from baseline to each architectural improvement. A striking fact is how much those metrics improved after adding regional attention, as final scores are 0.915 (Dice) and 0.856 (IoU) after multi-scale fusion.
Our model does introduce some memory overhead from the attention mechanisms and requires GPU speedups, but the resulting speedup is well worth the trade-off. As shown in Table 4, Part D, our model for regional attention is the only one that gets the higher Dices score (0.911) without resizing the image to a high resolution. It also uses less memory (4.5 GB) and takes less time to infer (235 ms/image) than other lightweight attentional mechanisms. Consistent gains in a variety of metrics and data settings show that the architectural choices we made balance accuracy, efficiency, and clinical usefulness. This study looks at how each part of our model handles the problems that arise when analyzing breast ultrasound images, leading to strong and accurate segmentation in real application.
4.3 Ablation Study and Component Analysis, after the Baseline Model Results
Each preprocessing step significantly impacts downstream performance, according to our ablation analysis, as shown in Table 5. Each preprocessing step significantly impacts downstream performance, according to our ablation analysis. When using raw images as a baseline model, the Dice coefficient was 0.831. This study addressed the finding that, by adding NLM denoising, the Dice score improved to 0.862, with PSNR improving from 23.8 to 35.2 dB. The integration of CLAHE contrast enhancement results in a performance of 0.883, with an average local contrast ratio increasing to 0.68 from 0.42. Intensity normalization finally raised the Dice coefficient to 0.915 and reduced the inter-image intensity variations by 65%.This study gives us numerical proof that our suggested preprocessing pipeline works to make segmentation more accurate. As you can see, preprocessing is an important part of overall performance. Each step of preprocessing makes UltraSegNet much more accurate, especially when images have speckle noise and low-contrast areas.

4.4 Analysis of Classification Challenges
Fig. 9 shows that UltraSegNet works well, but a lot of mistakes are seen, especially in cases that are close to being wrong. None of our 237 benign lesions (test set) was misclassified as malignant (12% false positive rate), but 31 of our 209 malignant lesions were misclassified as benign (15% false negative rate). Most of these mistakes happen when the shape of the lesion isn’t clear, like when the boundary of the lesion isn’t completely clear or when the pattern of the internal echo changes a lot. The model is most accurate when it comes to identifying normal tissue; out of 334 normal cases, it only makes 9 mistakes. This shows that it is strong at telling the difference between normal and abnormal tissue patterns.

Figure 9: Confusion matrix analysis for lesion classification
In addressing these classification challenges, this study developed a thorough basis for enhancing data representation and learning dynamics. To overcome the inherent imbalance in our dataset, especially for malignant cases, this study introduced a class-balanced sampling approach. With this change, the accuracy of finding malignant lesions increased from 85% to 89%, and the classification of benign lesions remained stable. Through cost-sensitive learning with a weighted loss function that punishes false negatives more harshly in cancer cases, this study made our method even better. With this change, the number of false negatives decreased by 23%, but the number of false positives increased by 8%. This is a fair trade-off when you consider how difficult it is to find malignant lesions. Even with these improvements, it is still difficult to tell the difference between lesions that have different characteristics or where the image quality is not good enough. But none of these methods solve the problems in this work. For example, we need a more advanced feature extraction method, and it would be helpful to add clinical metadata to the current histological data’s metadata context. This study concludes that further research is necessary to quantify the uncertainty of near-true predictions. The findings will help physicians make better decisions.
Grad-CAM visualization, as shown in Fig. 10, the model can be easily interpreted by focusing all attention on the segmentation results. Our research indicates that UltraSegNet looks at clinically important parts of the images, such as the edges of a lesion and the textures inside the lesions. These parts are similar to how radiologists diagnose problems. Purple indicates high-attention areas, and yellow and green prescribe low-attention areas, thus guiding clinicians by indicating the basis for decisions made by the model. These visualizations can assist clinical practice in a few ways:

Figure 10: Segmentation results and attention mechanism visualization of UltraSegNet
• The study focuses on the resolution of segmentation and quality conflicts in lesions with uncertain borders.
• The detection of texture patterns affected the decisions taken by the model.
• It is crucial to evaluate the quality of segmentation outcomes, especially in challenging instances where tissue presentation is heterogeneous.
We illustrate consistent performance across both datasets with different characteristics and acquisition protocols, demonstrating their robustness. In this study, we show that our model can handle a range of clinical situations in the BUSI dataset, which has more cases. We also show how well it works on the UDAIT dataset, which represents a more structured situation. Improvement in the study performance, specifically increase of 2.4% in segmentation accuracy (Dice Similarity Coefficient), is important for clinical use. Exact segmentation of the lesion boundaries is essential in breast ultrasound for direct diagnostic reliability. This improved segmentation reduces false negatives, especially in cases where subtle or ambiguous lesion characteristics can make early cancer detection less likely. In addition, the delineation allows clinicians to gain more anatomical insights, which treatment planning requires, before biopsies and surgical interventions. The final illustration of our segmentation process across different architectures for the BUSI dataset is shown in Fig. 11.

Figure 11: (a) Original Images. (b) Ground Truth. (c) Segmentation of UltraSegNet. (d) Segmentation of TransUNet. (e) Segmentation of MedFormer. (f) Segmentation of SwinUNETR. (g) Segmentation of UNETR
Fig. 11 provides a comprehensive comparison of the segmentation results of a suite of architectures. Each row shows a different case. UltraSegNet achieves superior delineation consistency and stability, especially for difficult cases with irregular shapes or indistinct margins. Notice how small lesions (bottom row) and complex tissue patterns (second row) are much better handled on focal eyes than other architectures.
A key limitation of this study is the small size of the dataset (943 images), which may influence the model’s ability to generalize to various clinical scenarios. However, our comprehensive mitigation approach demonstrates effective handling of this constraint through three complementary strategies:
1. Physics-Informed Data Augmentation: To address dataset size limitation, this study developed a physics-informed data enhancement pipeline that increases the effective size of the training set. This pipeline uses changes in acoustic properties and tissue deformation patterns, while being limited by guarantees of plausibility in the corresponding anatomical scene. Putting these strategies together improved performance by 7.5%, and the physics descriptions added another 2.8% to the accuracy of the whole thing.
2. Transfer Learning from Medical Imaging Datasets: By leveraging robust feature representations to transfer learning from broader medical imaging datasets, our method achieved 4.1% better performance, helping the model generalize to unseen cases. This study also ensured that the performance estimation was reliable in different pathological distributions by using five-fold cross-validation with stratified sampling.
Although the aforementioned strategies significantly mitigate the limitation of data set size, several challenges remain that warrant further study. These include:
• Severe Acoustic Shadowing: In cases where acoustic shadowing covers more than 30% of the image area, the segmentation accuracy is significantly degraded. This is due to the inherent difficulty in extracting meaningful features from regions of heavy ultrasound attenuation. Currently, the regional attention mechanism is unable to process these information-poor regions. Future work will explore shadow-aware attention mechanisms or multi-view fusion methods to address this limitation.
• Low Contrast Conditions: The model demonstrates decreased reliability in defining the boundaries of the lesion under extremely low contrast conditions, such as deep tissue imaging or suboptimal device settings. While the modified initial convolutional layers achieve the best results in preserving fine details, they do not fully capture subtle edge information in difficult contrast scenarios. This could be addressed by incorporating contrast-adaptive feature extraction or explicit edge detection streams.
Current hospital imaging systems can incorporate UltraSegNet to optimize efficiency and improve diagnostic outcomes. Essential parts of its inclusion include the following:
• PACS/DICOM compatibility: Ensure seamless interoperability with hospital image databases.
• The system improves clinical workflow by making it easier for radiologists to find and describe lesions automatically, with the option for experts to review the results.
• Edge deployment: Fine-tuned for near-instant real-time processing on medical imaging hardware, facilitating quicker decision-making in clinical scenarios.
• Regulatory assessment: The model is being looked at to see if it meets the medical imaging standards (FDA/CE) that are needed for safe and effective use in clinical settings.
4.5 Ethical Considerations and Bias Mitigation
This research creates and uses AI-based medical diagnostic tools such as UltraSegNet, which must be very aware of their ethical implications, their own biases, and the medical standards of care needed. We aim to address these concerns and propose strategies to mitigate bias.
• Dataset representation and bias: The BUSI and UDAIT training datasets may not be representative of diverse populations. Furthermore, these data sets might lack representation on several crucial demographic dimensions. Furthermore, severely reduced geographic diversity for data collection could further bias the results in their ability to generalize broadly. The use of data sets with such differences in composition can raise important questions about the generalizability and inclusivity of the methodology and the findings of the study.
• Bias mitigation strategies: General medical AI is a field that is growing rapidly. It will need clear rules to deal with issues such as possible biases in training datasets and issues with model performance compared to natural population variation. Protocols for collecting data should protect representational diversity by setting minimum demographic requirements for inclusion and making sure that the make-up of datasets is properly documented. We should routinely identify and audit demographic gaps in training data to minimize potential biases. When developing the models, fairness should guide researchers in including strong metrics that assess performance in various demographic subgroups. It consists of applying uncertainty quantification methods and establishing explicit confidence values for automated decision-making. These strategies create reliability and effectiveness for the model in diverse population segments. This is where clinical validation of the model’s performance and generalizability becomes important. We should prioritize developing and validating multi-center studies that specifically evaluate the model’s accuracy in various populations, particularly in underrepresented groups. To eliminate possible performance differences and make the model more useful in clinical settings, performance discrepancies must be carefully recorded, and demographic-specific calibration strategies must be created.
• Clinical Integration and fairness: Following transparency, justice, and ethics, the guidelines recommend making clear what the demographics of the training data are and reporting performance metrics between population subgroups regularly to showcase the limitations of the models. Deep protocols go along with smart clinical workflows. They spell out model disagreements, say who needs to be involved, and require that decisions made by AI be documented. We need to create and implement regulatory and auditing rules to monitor clinical outcomes and understand the functioning of these AI systems and their real-world effects. In this way, mechanistic solutions can help patients without bias. The responsible implementation of clinical AI involves ongoing monitoring and proactive correction. It requires ongoing monitoring of performance across large demographic populations, regular updating of fairness measures, and feedback cycles to our providers and patients. Periodic ethical evaluations ensure that the AI system remains in accordance with current social and healthcare norms.
This work presents UltraSegNet, which offers a robust design that incorporates original innovations like regional attention and modified initial layers, effectively combining local feature extraction from CNN with global context modeling from Transformer to significantly enhance the state-of-the-art in segmentation performance. This design significantly enhances the state-of-the-art in segmentation performance. This study compares UltraSegNet to the BUSI and UDAIT datasets in excellent detail, showing that it is always better than the most recent best methods. With our choice of architectural design, we obtain 0.915 precision and 0.908 recall on the BUSI dataset, whereas we achieve strong performance on the UDAIT dataset too (0.901 precision and 0.894 recall). Such improvements are particularly relevant in challenging scenarios where speckle noise, acoustic shadows, and complex tissue geometry exist and are common limitations to traditional methods. In addition to its performance on benchmark datasets, we will further investigate UltraSegNet’s capability on real-world clinical data in future work. This will let us look at a wider range of qualitative errors, which is especially important in tricky situations. This research will also have to learn more about the practical trade-offs between speed and accuracy when using these models in a clinical setting. Clinical validation studies will be critical in understanding the performance of the model on varied patient populations and imaging conditions. Some people may think that images of a coronavirus changing from an icosahedral shape to a helical shape are a useful way to explain how segmentation algorithms work. The technical performance of UltraSegNet looks good, but its clinical success will depend on how well it can be integrated into interventional workflows. There will also be the long-term and difficult task of making sure that UltraSegNet is compliant with regulations and best practices in various clinical settings. Further work will need to focus on prospective clinical validation studies and an implementation roadmap to facilitate routine clinical implementation. The results from this work show that UltraSegNet is promising enough for clinical integration to improve breast cancer screening accuracy, and that it has the potential to generalize well for translation to a variety of clinical environments and imaging modalities. UltraSegNet is a viable candidate for use in a real-world clinical setting because it works well with a wide range of datasets and imaging protocols and uses very little computing power (235 ms for each image). While the architecture’s demonstrated generalization capabilities will guide its implementation, its flexibility offers even more potential applications in the medical imaging domains. However, its adoption hinges on its ability to balance the model’s infrastructure requirements with the necessary clinical validation processes. There needs to be more research done to make sure that the model can be used on various computing environments, such as ones with mobile devices and different types of clinical workstations. While our current implementation works well with standard GPU hardware, we think that model compression methods (like quantization approaches) and hardware-specific optimizations could be intriguing areas to look into further to make clinical integration possible across a wide range of different clinical deployments. Furthermore, our architecture’s adaptability to various medical modalities could potentially broaden its impact in other diagnostic tasks.
Acknowledgement: The authors would like to acknowledge the support of Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Funding Statement: This research was funded by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2025R435), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Suhaila Abuowaida, Hamza Abu Owida, Deema Mohammed Alsekait; data collection: Nawaf Alshdaifat, Diaa Salama AbdElminaam; analysis and interpretation of results: Suhaila Abuowaida, Mohammad Alshinwan; draft manuscript preparation: Suhaila Abuowaida. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The authors confirm that the data supporting the findings of this study are available within the article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. World Health Organization. Breast cancer [Internet]. 2021 [cited 2024 Oct 21]. Available from: https://www.who.int/news-room/fact-sheets/detail/breast-cancer. [Google Scholar]
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin. 2021;71(3):209–49. doi:10.3322/caac.21660. [Google Scholar] [PubMed] [CrossRef]
3. Gharaibeh L, Liswi M, Al-Ajlouni R, Shafei D, Fakheraldeen RE. Community pharmacists’ readiness for breast cancer mammogram promotion: a national survey from Jordan. J Multidiscip Healthc. 2024;17:4475–89. doi:10.2147/JMDH.S471151. [Google Scholar] [PubMed] [CrossRef]
4. Welch HG, Prorok PC, O’Malley AJ, Kramer BS. Breast-cancer tumor size, overdiagnosis, and mammography screening effectiveness. New Engl J Med. 2016;375(15):1438–47. doi:10.1056/NEJMoa1600249. [Google Scholar] [PubMed] [CrossRef]
5. Kolb TM, Lichy J, Newhouse JH. Comparison of the performance of screening mammography, physical examination, and breast US and evaluation of factors that influence them: an analysis of 27,825 patient evaluations. Radiology. 2002;225(1):165–75. doi:10.1148/radiol.2251011667. [Google Scholar] [PubMed] [CrossRef]
6. Berg WA, Blume JD, Cormack JB, Mendelson EB, Lehrer D, Bohm-Velez M, et al. Combined screening with ultrasound and mammography vs mammography alone in women at elevated risk of breast cancer. JAMA. 2008;299(18):2151–63. doi:10.1001/jama.299.18.2151. [Google Scholar] [PubMed] [CrossRef]
7. Brem RF, Tabar L, Duffy SW, Inciardi MF, Guingrich JA, Hashimoto BE, et al. Assessing improvement in detection of breast cancer with three-dimensional automated breast US in women with dense breast tissue: the SomoInsight Study. Radiology. 2015;274(3):663–73. doi:10.1148/radiol.14132832. [Google Scholar] [PubMed] [CrossRef]
8. Stavros AT, Thickman D, Rapp CL, Dennis MA, Parker SH, Sisney GA. Solid breast nodules: use of sonography to distinguish between benign and malignant lesions. Radiology. 1995;196(1):123–34. doi:10.1148/radiology.196.1.7784555. [Google Scholar] [PubMed] [CrossRef]
9. Elmore JG, Wells CK, Lee CH, Howard DH, Feinstein AR. Variability in radiologists’ interpretations of mammograms. New Engl J Med. 1994;331(22):1493–9. doi:10.1056/NEJM199412013312206. [Google Scholar] [PubMed] [CrossRef]
10. Litjens G, Kooi T, Bejnordi BE, Setio A, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42(13):60–88. doi:10.1016/j.media.2017.07.005. [Google Scholar] [PubMed] [CrossRef]
11. Aldhyani T, Khan MA, Almaiah MA, Alnazzawi N, Hwaitat A, Elhag A, et al. A secure internet of medical things framework for breast cancer detection in sustainable smart cities. Electronics. 2023;12(4):858. doi:10.3390/electronics12040858. [Google Scholar] [CrossRef]
12. Zhang K, Li X, Wang Y, Chen H, Liu J. Multi-task deep learning for breast ultrasound image analysis: lesion detection, classification, and segmentation. Med Image Anal. 2023;83:102684. [Google Scholar]
13. Liu S, Wang Y, Yang X, Lei B, Liu L, Li SX, et al. Attention-guided network for breast ultrasound image segmentation with multi-scale feature enhancement. IEEE Trans Med Imag. 2023;42(4):1012–24. [Google Scholar]
14. Wang Y, Zhang Z, Chen H, Tang X, Liu X, Heng PA. A survey on deep learning-based medical image analysis. Biomed Signal Process Control. 2022;71:103242. [Google Scholar]
15. Chen J, Lu Y, Yu Q, Luo X, Zhou Y. Hybrid CNN-Transformer architecture for medical image segmentation: application to breast ultrasound. Pattern Recognit. 2023;135:109148. [Google Scholar]
16. Kim H, Park J, Lee S, Kim Y. Dual-path network with adaptive attention for robust breast lesion segmentation in ultrasound images. Comput Methods Programs Biomed. 2023;229:107334. [Google Scholar]
17. Hosseinalipour A, Ghanbarzadeh R, Arasteh B, Soleimanian Gharehchopogh F, Mirjalili S. A metaheuristic approach based on coronavirus herd immunity optimiser for breast cancer diagnosis. Cluster Comput. 2024;27(7):9451–75. doi:10.1007/s10586-024-04360-3. [Google Scholar] [CrossRef]
18. Raju ASN, Venkatesh K, Gatla RK, Eid M, Flah A, Slanina Z, et al. Expedited colorectal cancer detection through a dexterous hybrid CADx system with enhanced image processing and augmented polyp visualization. IEEE Access. 2025;13(4):17524–53. doi:10.1109/ACCESS.2025.3532807. [Google Scholar] [CrossRef]
19. Pham DL, Xu C, Prince JL. Current methods in medical image segmentation. Ann Rev Biomed Eng. 2000;2(1):315–37. doi:10.1146/annurev.bioeng.2.1.315. [Google Scholar] [PubMed] [CrossRef]
20. Pandimurugan V, Ahmad S, Prabu AV, Rahmani MKI, Abdeljaber HAM, Eswaran M, et al. CNN-based deep learning model for early identification and categorization of melanoma skin cancer using medical imaging. SN Comput Sci. 2024;5(7):911. [Google Scholar]
21. Noble JA, Boukerroui D. Ultrasound image segmentation: a survey. IEEE Trans Med Imag. 2006;25(8):987–1010. doi:10.1109/TMI.2006.877092. [Google Scholar] [PubMed] [CrossRef]
22. Al Tawil A, Shaban A, Almazaydeh L. A comparative analysis of convolutional neural networks for breast cancer prediction. Int J Electr Comput Eng. 2024;14(3):3406–14. doi:10.11591/ijece.v14i3.pp3406-3414. [Google Scholar] [CrossRef]
23. Das S, Rout SK, Panda SK, Mohapatra PK, Almazyad AS, Jasser MB, et al. Marine predators algorithm with deep learning-based leukemia cancer classification on medical images. Comput Model Eng Sci. 2024;141(1):893–916. doi:10.32604/cmes.2024.051856. [Google Scholar] [CrossRef]
24. Canny J. A computational approach to edge detection. IEEE Trans Pattern Anal Mach Intell. 1986;8(6):679–98. doi:10.1109/TPAMI.1986.4767851. [Google Scholar] [CrossRef]
25. Vincent L, Soille P. Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE Trans Pattern Anal Mach Intell. 1991;13(6):583–98. doi:10.1109/34.87344. [Google Scholar] [CrossRef]
26. Kass M, Witkin A, Terzopoulos D. Snakes: active contour models. Int J Comput Vis. 1988;1(4):321–31. doi:10.1007/BF00133570. [Google Scholar] [CrossRef]
27. Marr D, Hildreth E. Theory of edge detection. Proc Royal Soc London B. 1980;207:187–217. [Google Scholar]
28. Alraba’nah Y, Toghuj W. A deep learning based architecture for malaria parasite detection. Bull Electr Eng Inform. 2024;13(1):292–9. doi:10.11591/eei.v13i1.5485. [Google Scholar] [CrossRef]
29. Adams R, Bischof L. Seeded region growing. IEEE Trans Pattern Anal Mach Intell. 1994;16(6):641–7. doi:10.1109/34.295913. [Google Scholar] [CrossRef]
30. Geman S, Geman D. Stochastic relaxation, Gibbs distributions, and the Bayesian restoration of images. IEEE Trans Pattern Anal Mach Intell. 1984;6(6):721–41. doi:10.1109/TPAMI.1984.4767596. [Google Scholar] [PubMed] [CrossRef]
31. Jain AK, Farrokhnia F. Unsupervised texture segmentation using Gabor filters. Pattern Recognit. 1991;24(12):1167–86. doi:10.1016/0031-3203(91)90143-S. [Google Scholar] [CrossRef]
32. Mallat SG. A theory for multiresolution signal decomposition: the wavelet representation. IEEE Trans Pattern Anal Mach Intell. 1989;11(7):674–93. doi:10.1109/34.192463. [Google Scholar] [CrossRef]
33. Vapnik V. The nature of statistical learning theory. Berlin, Germany: Springer; 1995. [Google Scholar]
34. Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi:10.1023/A:1010933404324. [Google Scholar] [CrossRef]
35. Rohlfing T, Brandt R, Menzel R, Maurer CR. Performance-based classifier combination in atlas-based image segmentation using expectation-maximization parameter estimation. IEEE Trans Med Imag. 2004;23(8):983–94. doi:10.1109/TMI.2004.830803. [Google Scholar] [PubMed] [CrossRef]
36. Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Ann Rev Biomed Eng. 2017;19(1):221–48. doi:10.1146/annurev-bioeng-071516-044442. [Google Scholar] [PubMed] [CrossRef]
37. Zhu Z, Zhang Z, Qi G, Li Y, Li Y, Mu L. A dual-branch network for ultrasound image segmentation. Biomed Signal Process Control. 2025;103(2):107368. doi:10.1016/j.bspc.2024.107368. [Google Scholar] [CrossRef]
38. Zhu Z, Wang Z, Qi G, Mazur N, Yang P, Liu Y. Brain tumor segmentation in MRI with multi-modality spatial information enhancement and boundary shape correction. Pattern Recognit. 2024;153(1):110553. doi:10.1016/j.patcog.2024.110553. [Google Scholar] [CrossRef]
39. Hatamizadeh A, Tang H, Roth HR, Xu D. UNETR: transformers for 3D medical image segmentation. arXiv:2103.10504. 2022. [Google Scholar]
40. Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, et al. TransUNet: transformers make strong encoders for medical image segmentation. arXiv:2102.04306. 2021. [Google Scholar]
41. Liu S, Wang Y, Yang X, Lei B, Liu L, Li SX, et al. A review on deep learning in medical image analysis. Front Med. 2021;8:747. [Google Scholar]
42. Chowdary GJ, Yin Z. Med-Former: a transformer based architecture for medical image classification. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; 2024; Singapore: Springer. p. 448–57. [Google Scholar]
43. Khan S, Naseer M, Hayat M, Zamir SW, Khan FS, Shah M. Transformers in medical imaging: a survey. Med Image Anal. 2022;80:102382. [Google Scholar]
44. Katharopoulos A, Vyas A, Pappas N, Fleuret F. Transformers are RNNs: fast autoregressive transformers with linear attention. In: Proceedings of the International Conference on Machine Learning (ICML); 2020; Vienna, Austria. p. 5156–65. [Google Scholar]
45. Huang Z, Wang X, Wei Y, Huang L, Shi H, Liu W, et al. CCNet: criss-cross attention for semantic segmentation. In: Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV); 2019; Seoul, Republic of Korea. p. 603–12. [Google Scholar]
46. Wang H, Zhu Y, Green B, Adam H, Yuille A, Chen LC. Axial-DeepLab: stand-alone axial-attention for panoptic segmentation. In: Proceedings of the European Conference on Computer Vision (ECCV); 2020; Online. p. 108–26. [Google Scholar]
47. Lee M, Chen T, Wang B. Efficient transformer architectures for medical image analysis. IEEE Trans Pattern Anal Mach Intell. 2023;45(5):5678–91. [Google Scholar]
48. Chen J, Mei J, Li X, Lu Y, Yu Q, Wei Q, et al. TransUNet: rethinking the U-Net architecture design for medical image segmentation through the lens of transformers. Med Image Anal. 2024;97(2):103280. doi:10.1016/j.media.2024.103280. [Google Scholar] [PubMed] [CrossRef]
49. Al-Dhabyani W, Gomaa M, Khaled H, Fahmy A. Dataset of breast ultrasound images. Data Brief. 2020;28(5):104863. doi:10.1016/j.dib.2019.104863. [Google Scholar] [PubMed] [CrossRef]
50. Yap MH, Pons G, Martí J, Ganau S, Sentís M, Zwiggelaar R, et al. Automated breast ultrasound lesions detection using convolutional neural networks. IEEE J Biomed Health Inform 2018;22(4):1218–26. doi:10.1109/JBHI.2017.2731873. [Google Scholar] [PubMed] [CrossRef]
51. Xu W, Fu YL, Zhu D. ResNet and its application to medical image processing: research progress and challenges. Comput Methods Programs Biomed. 2023;240(9):107660. doi:10.1016/j.cmpb.2023.107660. [Google Scholar] [PubMed] [CrossRef]
52. Hu J, Shen L, Sun G. Squeeze-and-excitation networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2018; Salt Lake City, UT, USA: IEEE. p. 7132–41. [Google Scholar]
53. Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, et al. Attention U-Net: learning where to look for the pancreas. arXiv:1804.03999. 2018. [Google Scholar]
54. Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems. Lake Tahoe, NV, USA: Neural InformationProcessing Systems Foundation; 2012. p. 1097–105. [Google Scholar]
55. Chollet F. Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017. p. 1251–8. [Google Scholar]
56. He K, Zhang X, Ren S, Sun J. Deep residual learning for imagerecognition. IEEE Trans Pattern Anal Mach Intell. 2017;39(7):1137–49. [Google Scholar]
57. Han S, Zhong X, Cao J, Zhang L. Deep learning architectures for breast cancer detection and classification. In: IEEE International Conference on Computer and Information Technology; 2017. p. 1341–6. [Google Scholar]
58. Carcagnì P, Del Coco M, Leo M, Distante C. Deep learning applications in breast cancer diagnosis. Comput Biol Med. 2019;114:103545. [Google Scholar]
59. Guan S, Murray A, Perera N. Breast cancer diagnosis using deep neural networks. IEEE Access. 2019;7:116931–41. [Google Scholar]
60. Zhou HY, Yu L, Wang L, Yang X. A review of vision transformer for medical image analysis: challenges and solutions. arXiv:2202.12165. 2022. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools