 Open Access
Open Access
ARTICLE
BLFM-Net: An Efficient Regional Feature Matching Method for Bronchoscopic Surgery Based on Deep Learning Object Detection
School of Mechanical Engineering, Tianjin University, 135 Yaguan Road, Jinnan District, Tianjin, 300350, China
* Corresponding Author: Kang Kong. Email:
Computers, Materials & Continua 2025, 83(3), 4193-4213. https://doi.org/10.32604/cmc.2025.063355
Received 12 January 2025; Accepted 17 March 2025; Issue published 19 May 2025
Abstract
Accurate and robust navigation in complex surgical environments is crucial for bronchoscopic surgeries. This study purposes a bronchoscopic lumen feature matching network (BLFM-Net) based on deep learning to address the challenges of image noise, anatomical complexity, and the stringent real-time requirements. The BLFM-Net enhances bronchoscopic image processing by integrating several functional modules. The FFA-Net preprocessing module mitigates image fogging and improves visual clarity for subsequent processing. The feature extraction module derives multi-dimensional features, such as centroids, area, and shape descriptors, from dehazed images. The Faster R-CNN Object detection module detects bronchial regions of interest and generates bounding boxes to localize key areas. The feature matching module accelerates the process by combining detection boxes, extracted features, and a KD-Tree (K-Dimensional Tree)-based algorithm, ensuring efficient and accurate regional feature associations. The BLFM-Net was evaluated on 5212 bronchoscopic images, demonstrating superior performance compared to traditional and other deep learning-based image matching methods. It achieved real-time matching with an average frame time of 6 ms, with a matching accuracy of over 96%. The method remained robust under challenging conditions including frame dropping (0, 5, 10, 20), shadowed regions, and variable lighting, maintaining accuracy of above 94% even with the frame dropping of 20. This study presents BLFM-Net, a deep learning-based matching network designed to enhance and match bronchial features in bronchoscopic images. The BLFM-Net shows improved accuracy, real-time performance, and reliability, making a valuable tool for bronchoscopic surgeries.Keywords
The bronchoscopic surgery is crucial for the diagnosis and treatment of pulmonary diseases. Surgeons use bronchoscopes to navigate the complex bronchial tree structure to locate lesions accurately and perform therapeutic interventions. However, the complex anatomy of the bronchial tree presents significant challenges for real-time tracking and precise localization during the surgery. To improve surgical precision and patient safety, image matching techniques are developed for bronchoscopic navigation systems to aid pathway tracking and navigation [1].
Some existing image matching algorithms have been widely used in bronchoscopic navigation for their robustness and efficiency. SIFT (Scale-Invariant Feature Transform) excels in feature extraction under varying conditions, whose application is limited due to its high computational complexity [2]. SURF (Speeded-Up Robust Features) improves processing speed but struggles in complex regions [3], and ORB enhances real-time performance at the expense of higher computational demands [4]. Methods like MSER and KAZE achieve better precision in specific scenarios but are less effective at capturing the complexity of bronchial branching and edge features [5–7]. Optical flow performs motion estimation between frames but is prone to errors caused by occlusion and large displacements, especially in intricate structures like bronchial bifurcations [8]. As analyzed above, the traditional image matching methods face ongoing challenges in handling complex bronchial environments, highlighting the need for more robust and adaptable solutions.
Recently, deep learning-based matching techniques have achieved significant breakthroughs and found pioneering applications in medical image matching and navigation. DeTone et al. [9] proposed a SuperPoint model for keypoint detection and description, enabling efficient and robust matching. Building on this foundation, Sarlin et al. [10] introduced SuperGlue, which optimizes local feature descriptors and geometric relationships to improve inter-frame matching success rates and estimation accuracy. Liu et al. [11] developed a spatially geometric-aware depth estimation network that integrates depth map features with normal vectors and gradient losses, enhancing both matching efficiency and global consistency. Acar et al. [12] applied SuperPoint and SuperGlue to address sparse feature matching challenges in complex endoscopic scenes. Wang et al. [13] presented a 2.5D bronchoscopic tracking method using geometric depth map features to reduce intraoperative noise, but its real-time performance remains limited. Shen et al. [14] proposed a consecutive frame registration loss for bronchoscopic navigation, which incorporates pose, depth map, and temporal consistency constraints, further improving inter-frame matching. Wang et al. [15] employed visual SLAM with restricted 3D point search ranges to improve inter-frame matching accuracy in bronchoscopic scenarios. Liu et al. [16] introduced a joint depth and motion estimation approach for monocular endoscopic image sequences, using a multi-loss framework to enhance feature matching by integrating epipolar constraints and feature descriptor learning, improving robustness in surgical tissue regions with the improved SIFT. However, it relies on high-quality image preprocessing, which can be limiting in poor lighting conditions. Liu et al. [17] also proposed a feature matching method for texture-less endoscopy images, using adaptive gradient-preserving techniques and super pixel-based motion consistency to improve matching accuracy and 3D reconstruction. Yet, the added computational complexity may impact real-time performance in clinical environments. Chu et al. [18] introduced a motion consensus-based feature matching method, combining motion smoothing with affine bilateral regression to refine feature matches and improve 3D reconstruction quality. This method is particularly effective in endoscopic images with specular reflections and deformations, although its computational complexity may limit real-time applicability. Farhat et al. [19] proposed a self-supervised feature matching method for endoscopic images using CNNs and GNNs to model spatial relationships between keypoints, improving matching precision but with potential limitations in real-time applicability due to computational complexity. Lu et al. [20] proposed S2P-Matching, a self-supervised patch-based matching method using Transformers to handle weak textures and large rotations in capsule endoscopic images, improving matching accuracy but with increased computational complexity that may limit real-time performance. Nanehkaran et al. [21] presented a density-based unsupervised approach to the diagnosis of abnormalities in heart patients. In this method, the basic features in the dataset are first selected based on the filter-based feature selection approach. the accuracy of the proposed method for predicting heart patients is approximately 95%, which has improved in comparison with previous methods.
Despite these advancements, the existing methods still face challenges for image matching in bronchoscopic surgery, including insufficient real-time performance, limited robustness in complex bronchial and airway scenarios, and sensitivity to lighting variations. To address these issues, this study introduces BLFM-Net for bronchoscopic surgery, a novel bronchoscopic image matching network that integrates regional feature matching with the FASTER-RCNN object detection framework. Our BLFM-Net introduces key advancements in the following areas.
1. Accurate regional localization: The object detection module is integrated to isolate bronchial tubes and exclude irrelevant areas, significantly improving matching reliability.
2. Optimized regional feature matching: Key regions are emphasized to enhance robustness in complex branching structures, particularly under varying lighting and noise conditions.
3. Improved real-time performance: The regional matching strategy reduces computational cost, making the system suitable for surgical real-time requirements.
In brief, this approach offers a new solution to bronchoscopic navigation challenges, and balances matching accuracy, robustness, and real-time applicability.
2.1 Overall Structure of BLFM-Net
Fig. 1 shows the overall architecture of the BLFM-Net, which is composed of four key modules: the FFA-Net preprocessing module, the Faster R-CNN object detection module, the feature extraction module, and the feature matching module. The FFA-Net preprocessing module (A) is responsible for improving the quality of bronchoscopic video frames by removing haze and segmenting regions of interest. This ensures that the subsequent steps work with clearer and more distinct features. The Faster R-CNN object detection module (B) detects and localizes the bronchial lumen regions. It uses convolutional layers to extract feature maps, which are then processed by the Region Proposal Network (RPN) to identify candidate regions of interest (ROIs) for further analysis. After haze removal and region localization, the feature extraction module (C) extracts important regional features such as center points (fc), area (fa), and Hu moments (fh) from the haze-free frames. These features serve as the foundation for matching across consecutive frames. Finally, the feature matching module (D) computes matching scores by comparing extracted features from different frames. This module first performs detection box matching by using the detection boxes obtained from the Faster R-CNN module and matching them between adjacent frames based on the Intersection over Union (IoU) metric. Once the detection boxes are matched, the module proceeds with feature matching by comparing the region-specific features (such as centroids, areas, and Hu moments) from the matched boxes. The final matching score is computed by evaluating the similarity of these features. This process ensures accurate and efficient feature matching, achieving precise inter-frame alignment.

Figure 1: BLFM-Net architecture: (A) FFA-Net preprocessing module, (B) Faster R-CNN object detection module, (C) feature extraction module, and (D) feature matching module. The gray dashed box illustrates the matching process of the detection boxes between adjacent inter-frames
The preprocessing module in our method employs a simplified version of the Feature Fusion Attention Network (FFA-Net) [22], a deep learning-based, end-to-end image dehazing network designed specifically for single-image dehazing tasks. This network is chosen for its ability to address the prevalent haze effect in bronchoscopic images, which can obscure critical details and pose significant challenges in feature extraction and subsequent analysis. Bronchoscopic images often suffer from haze caused by scattering light, especially in poorly lit or cloudy environments, which reduces the clarity of key anatomical features [23].
To improve the quality of these images, we use a simplified version of FFA-Net, integrating a Channel Attention (CA) module. This integration helps the network focus more effectively on critical channel information—specifically, the features most relevant to enhancing image clarity. The CA module works by applying attention to different feature channels within the network, selectively amplifying those that contain useful information for dehazing and reducing noise in other areas. This adaptation of the original FFA-Net improves its dehazing performance in bronchoscopic images, which often contain subtle and important anatomical details that need to be preserved. The network is trained by minimizing the loss between the dehazed output and the ground truth. The training process involves optimizing the network to remove haze while retaining the fine details of the bronchial structures, ensuring that critical features are enhanced rather than smoothed out. The output from the network is a dehazed image with improved clarity, which is then passed to subsequent modules for feature extraction and matching.
The preprocessing workflow, as shown in Fig. 2, operates as follows: The original bronchoscopic images are input into the network, which processes them through the dehazing pipeline. The final result is a set of dehazed images that exhibit improved contrast and clarity, crucial for the accurate extraction of key features needed for the downstream analysis and tracking tasks in bronchoscopic navigation.

Figure 2: The structure of the preprocessing module in BLFM-Net, integrating FFA-Net with a channel attention (CA) mechanism to enhance haze removal by refining critical channel information
Accurate detection of bronchial lumens is essential for bronchoscopic tracking and localization. Here, the Faster R-CNN model is employed for the object detection module due to its precision and robustness in lumen detection tasks [24]. Fig. 3 presents structure of the object detection module. For the detection of lumens, convolutional layers extract feature maps from the input haze-free images, and the Region Proposal Network (RPN) identifies candidate regions, which are then resized using ROI Pooling to output refined images. These proposals are classified, and their boundaries are refined. Custom anchor box scales and aspect ratios are calibrated to typical bronchial opening sizes, improving detection accuracy and ensuring reliable inputs for feature extraction and matching.

Figure 3: The structure of the object detection module in BLFM-Net, with haze-free images as input and refined detection of bronchial lumens as output
To ensure stable and continuous tracking during bronchoscopic procedures, the proposed BLFM-Net employs a region-based feature extraction method to establish correspondences between consecutive frames for the same bronchial lumen region, thereby maintaining temporal consistency in tracking. Let
Following the preprocessing for image dehazing, bronchial lumen characteristics are extracted using threshold-based segmentation. Each lumen region
2.4.1 Center Point Feature (
The center point feature
where
Then the centroid (cx, cy) of the region is computed as:
where
The area feature
The shape descriptor
where
The normalized central moment
where
The first two Hu moments,
where
These extracted features serve as the foundation for robust and efficient feature matching across consecutive bronchoscopic frames, ensuring the continuity and stability of tracking.
A hierarchical region matching strategy is proposed to achieve efficient and robust feature matching, mainly comprising three stages: box matching, KD-tree-based matching acceleration, and fine-grained candidate region matching.
In the box matching stage, a region-of-interest (ROI)-based strategy is employed to enhance matching efficiency and reduce computational cost. For two consecutive frames t and t + 1, the area of detected bounding boxes are denoted as
where
For each
2.5.2 KD-Tree Matching Acceleration
A KD-tree is utilized to speed up feature point matching within matching boxes, which is a data structure for efficient multi-dimensional searches. Feature points from frames t and t + 1 are organized into array points 1 and points 2 based on their centroid coordinates (cx, cy). The KD-tree is constructed for points 2 to enable fast approximate nearest-neighbor searches, matching each point in points 1 with its closest neighbor in points 2. This approach reduces computational complexity and improves matching efficiency.
Fig. 4 illustrates the impact of box matching strategy and KD-tree-based matching acceleration on the real-time performance. The figure compares the processing time across consecutive frames for the two methods: the original region-based matching (purple curve) and the optimized BLFM-Net with KD-tree (red curve).

Figure 4: Processing time before and after region matching acceleration
Compared to the original Region-based Matching, the optimized BLFM-Net with KD-tree shows a faster processing time, with a markedly smoother and more stable curve. This improvement highlights the effectiveness of the acceleration strategy in reducing computational cost, promoting both speed and stability for real-time feature matching in complex bronchoscopic environments.
2.5.3 Fine-Grained Candidate Region Matching
For each of the matched feature point pairs, a region similarity score
where,
where
where
where
In addition, the weights

In this study, two groups of experiments were designed to verify the stability of the proposed BLFM-Net. The first experiment evaluated feature matching accuracy and real-time performance of BLFM-Net through the comparison with the traditional methods of SURF, ORB, and optical flow, as well as some deep learning-based methods. The second experiment assessed the performance in challenging scenarios of frame dropping.
The dataset for the experiments consists of 5212 bronchoscopic video frames with a resolution of 1920 × 1080, captured by an Olympus endoscope (model: Olympus ESG-400) from the bronchi of a human phantom lung. The preprocessing workflow involves dehazing with the FFA-Net to enhance the features of bronchial lumen regions. The dehazing results are illustrated in Fig. 5.

Figure 5: The dehazing results after applying the FFA-Net on bronchoscopic video frames. The top row shows the original frames, while the bottom row displays the dehazed frames. For each frame, the right-side image represents the extracted feature mask
Altogether 521 frames (about 10%) were randomly extracted from all dehazed video frames and then labeled with Labelme, which were taken as the training dataset to evaluate the performance of the object detection module in BLFM-Net. This dataset was used to train the Faster R-CNN network for lumen detection, in an environment with Windows 10, Python 3.11.8, PyTorch 2.2.1, OpenCV 4.9.0, and CUDA 11.8. The network parameters were set as follows: batch size = 4, and epochs = 100. The loss curve in Fig. 6 shows a reduction in both training and validation losses, with no overfitting observed, indicating effective network training.

Figure 6: Loss curve for lumen region detection
3.2 Matching Results and Hypersensitive Analysis
After the dehazing of the video frames and lumen detection, regional feature extraction and matching shall be performed to complete the feature matching. To validate the effectiveness of BLFM-Net, two consecutive frames were randomly selected from the whole dataset (5212 bronchoscopic video frames) for feature matching. The matching results obtained by BLFM-Net are presented in Fig. 7.

Figure 7: (a) BLFM-Net inter-frame matching results. (b) Feature mask from the matching results
The lighting condition can vary significantly in bronchoscopic examinations, especially when the camera enters darker regions or when the camera angle changes, leading to insufficient or uneven lighting in some areas. These changes can degrade the accuracy of feature matching and hinder tracking, as weak or inconsistent lighting makes it difficult to maintain reliable target detection. Despite these challenges, the proposed method effectively maintains accurate feature matching under variable lighting conditions. As shown in Fig. 8, the proposed method effectively maintains accurate feature matching even in dimly lit regions, as demonstrated by the successful detection and correspondence of key points between Frame1850 and Frame1854. The highlighted region (enclosed in the rectangular box) shows the preserved continuity of the shadowed bronchial passage, ensuring precise and robust matching results. This validates the capability of the algorithm to overcome the limitations posed by uneven lighting conditions.

Figure 8: Feature matching results the under uneven lighting condition in bronchial passage
BLFM-Net is highly sensitive to variations in image quality and lighting conditions, which are common challenges in bronchoscopic procedures due to factors like haze, blur, low resolution, and dynamic lighting changes. Image clarity, which fluctuates due to environmental factors or equipment limitations, directly impacts the method’s accuracy. However, BLFM-Net addresses this by incorporating a preprocessing module (FFA-Net) that removes haze and enhances bronchial features, maintaining accurate feature matching even in degraded images. Additionally, the method adapts to lighting variations, ensuring reliable performance in poorly lit areas or when the camera moves into darker regions, making it highly effective for real-time bronchoscopic navigation under unpredictable conditions, as shown in Fig. 8.
Similarly, BLFM-Net is sensitive to frame skipping and camera movements, which are common in real-time procedures. Frame loss, caused by device limitations or sudden camera shifts, can disrupt feature matching. Nonetheless, BLFM-Net remains resilient, maintaining feature matching accuracy even with dropped frames or motion blur. This capability ensures stable tracking in dynamic environments, which is crucial for successful outcomes in bronchoscopic procedures.
Lastly, BLFM-Net is sensitive to complex anatomical structures, such as intricate bronchial branches and texture-less regions, which make feature extraction challenging for traditional methods. However, its design, incorporating multi-dimensional features like centroids, areas, and Hu moments, allows it to accurately match features even in these challenging regions, ensuring continuous and reliable tracking in complex anatomical environments.
In conclusion, while BLFM-Net demonstrates sensitivity to several factors encountered in bronchoscopic navigation—such as image quality, lighting, frame skipping, camera movement, and complex anatomical structures—its advanced preprocessing and feature matching techniques enable it to overcome these challenges, making it a robust solution for real-time bronchoscopic navigation.
3.3 Comparison of Matching Methods
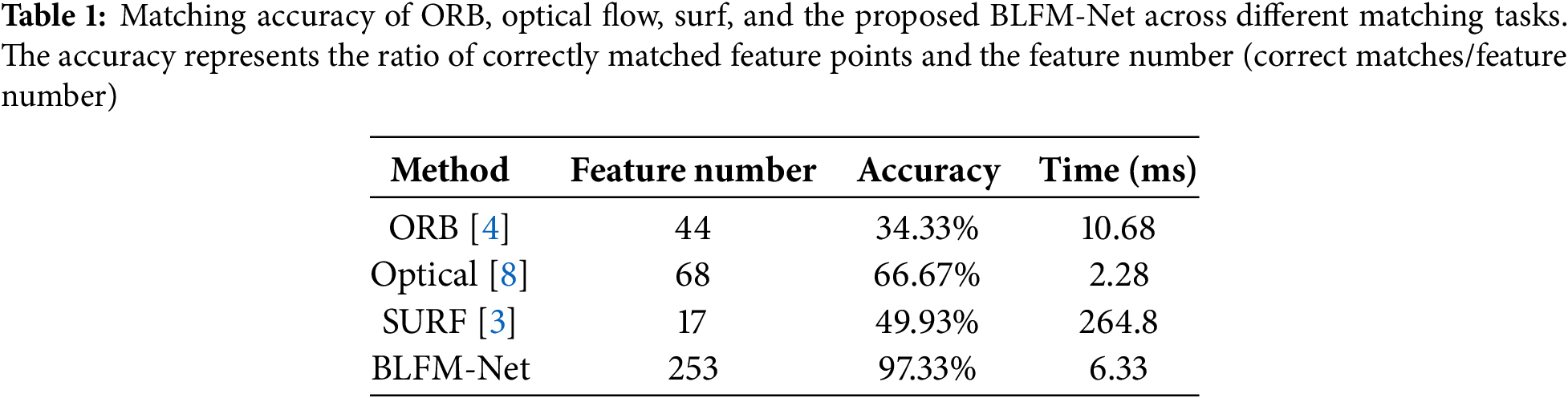
The proposed BLFM-Net was further compared with ORB [4], Optical Flow [8], and SURF [3] to evaluate its matching accuracy and real-time performance. Seven consecutive image frames were selected for feature point matching, and the results of different methods are illustrated in Fig. 9. In the figure, the red points denote feature points in the first frame, the green points indicate feature points in the second frame, and the blue lines represent the matches identified by each method. The matching accuracy of the methods for each frame is summarized in Table 1. The accuracy is presented by the ratio of correctly matched feature points to the feature number, and the ground truth values were obtained through manual calibration. A match is considered correct if the angle deviation between the matched line and the manually calibrated ground truth line is within ±5 degrees. Notably, the proposed method consistently achieves an accuracy above 97%, and the number of extracted feature points significantly surpasses those of other methods.

Figure 9: Comparison of feature point matching results among ORB, optical flow, surf, and the BLFM-Net
To validate the real-time performance and robustness of the proposed algorithm, two frames, Frame2301 and Frame2305, were selected randomly for feature extraction and matching. The results are compared with those of several state-of-the-art deep learning-based image matching methods, including LoFTR (Local Feature Transformer) [26], XoFTR (Cross-Modal Feature Transformer) [27], LightGlue (Lightweight SuperGlue) [28], SuperGlue (SuperPoint + Graph Neural Network) [10], and D2-Net (Dual Disentanglement Network for Brain Tumor Segmentation with Missing Modalities) [29]. Fig. 10 illustrates the matching effect of these methods, highlighting the differences in the number of matches and the visual alignment of feature points. The comparative data are summarized in Table 2.

Figure 10: Feature matching results for the proposed BLFM-Net and several other deep learning-based methods. (a) LoFTR [26]; (b) XoFTR [27]; (c) LightrGlue [28]; (d) SuperGlue [10]; (e) D2-Net [29]; (f) BLFM-Net
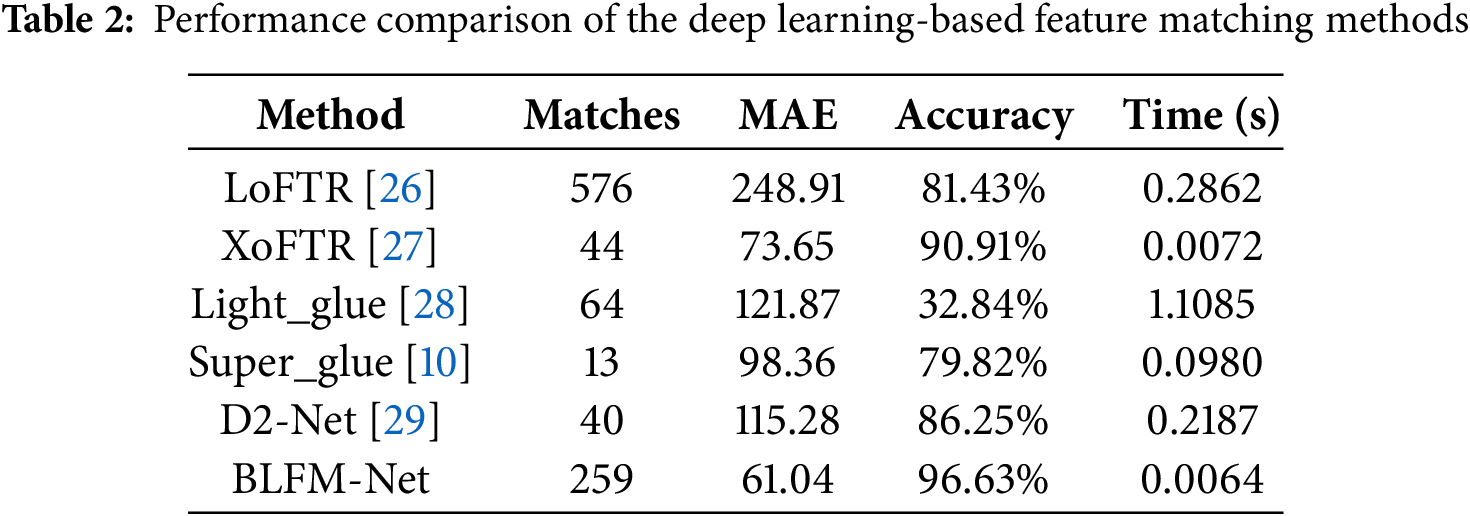
The results in Table 2 demonstrate the superior performance of BLFM-Net compared to other state-of-the-art deep learning-based image matching methods. BLFM-Net achieved the highest accuracy of 96.63%, outperforming all other methods in this comparison, including XoFTR (90.91%) and others, highlighting its precision in feature matching.
Additionally, BLFM-Net demonstrated excellent real-time performance, processing each frame in just 0.0064 s. In comparison, while XoFTR achieved a similar processing time (0.0072 s), its lower accuracy underscores BLFM-Net’s ability to maintain real-time processing without sacrificing precision. In terms of robustness, BLFM-Net showed a significantly low Mean Absolute Error (MAE) of 61.04, indicating reliable and consistent feature matching. Other methods, such as LoFTR (MAE = 248.91) and LightGlue (MAE = 121.87), had notably higher MAE values, suggesting lower robustness, especially in challenging conditions. XoFTR, while competitive with a MAE of 73.65, matched far fewer features (44) compared to BLFM-Net, which successfully matched 259 features. This demonstrates that BLFM-Net not only provides more accurate matches but also captures more relevant features, which is crucial for improving navigation quality in real-time applications.
These results emphasize the optimal balance BLFM-Net achieves between accuracy, computational efficiency, and robustness. While methods like LoFTR, XoFTR, and LightGlue may excel in one aspect, BLFM-Net combines all three, making it particularly well-suited for the dynamic and demanding environments of real-time bronchoscopic navigation. Its ability to consistently achieve high accuracy, handle large numbers of features, and operate in real-time under varying conditions positions BLFM-Net as a leading solution for deep learning-based image matching in medical applications.
3.4 Robustness Evaluation under Frame Dropping Conditions
A frame-dropping experiment was conducted to evaluate the robustness of the proposed method under frame dropping conditions. Frame dropping is genrally caused by interruptions, device vibrations, or lens movement, which often leads to data loss and discontinuities in image sequences, posing significant challenges for feature point matching and tracking. In the experiment, 100 consecutive frames (Frame1000 to Frame1100) were selected, and frame-dropping scenarios were simulated by removing 5, 10, and 20 frames, respectively. The original continuous frames (with 0 dropping frame) served as a control group. Fig. 11 shows the matching results at various dropping intervals, while Fig. 11 presents the number of matched feature points. Even with increased dropping intervals, the proposed method maintains effective feature point extraction and matching, as highlighted by a magnified view for frames 36–44 in Fig. 12.

Figure 11: Matching results under different frame-dropping scenarios (0, 5, 10, and 20 dropping frames)

Figure 12: The number of matched feature points under different dropping frames (0, 5, 10, and 20)
Building on the frame-dropping experiment, tracking accuracy was further evaluated to assess the robustness of the proposed method in maintaining consistent feature matching. Specifically, tracking accuracy is defined as the Euclidean distance between the tracked trajectory and the ground truth, and the ground truth was obtained through manual calibration. Fig. 13 shows the tracking accuracy of BLFM-Net at various dropping frames, where the red dashed lines present values of the ground truth.

Figure 13: Box plot comparison of matching line lengths at different dropping frames (a): 5 dropping frames, (b): 10 dropping frames, (c): 20 dropping frames. The red dashed lines represent the ground truth values
With 5 dropping frames, the method achieved a median error of approximately 962.76, with a 95% confidence interval of [929.64, 998.32], and a matching accuracy of 99.84%. All ground truth values fell within the confidence interval, indicating exceptional performance. With 10 dropping frames, the median error slightly increased to approximately 955.58, with a confidence interval of [920.47, 1012.44], and matching accuracy reduced to 98.19%. With 20 dropping frames, the median error rose to approximately 970, the confidence interval expanded to [917.34, 1036.58], and the matching accuracy decreased to 94.71%, with only 25% of ground truth values falling within the interval.
According to the above results, the tracking performance of the proposed method diminishes as the dropping frame increases, but it still exhibits significant robustness even with 20 dropping frames. Among all scenarios, the case with 5 dropping frames witnessed the best performance of the method, showing both high accuracy and stable tracking. The videos used in this study were recorded at a frame rate of 50 frames per second, and the findings suggest that our method is also well-suited for lower frame rates. Notably, the proposed approach can reliably handle videos with frame rates as low as 5 frames per second, maintaining robust and accurate tracking despite the reduced temporal resolution.
3.5 Evaluation on Simulation of a Fine Transformation
In bronchoscopic scenes, visual feature points are often sparse and unevenly distributed, particularly in texture-less regions of the bronchial tree. In clinical surgery, camera movement and tissue deformation will reduce the number of matches and affect the accuracy of matches. Therefore, we simulated the image scene transformation that might be encountered in clinical surgery to verify the robustness of the proposed method. As shown in Fig. 14, we use the TPS (Thin Plate Spline) image warping method to simulate endoscopic images with general affine transformations. Specifically, three kinds of transformation are included: scale changing (Fig. 14a2), where the scaling transform coefficient is 1.5; contrarotating for 45° (Fig. 14b2); and a random affine transformation (Fig. 14c2). Fig. 14 is an example of the simulations on bronchoscopic image dataset, and the original image is shown in (Fig. 14a1).

Figure 14: Evaluation results of the simulated datasets. On the top left (a1) is the original image. Each row shows an example of the deformation
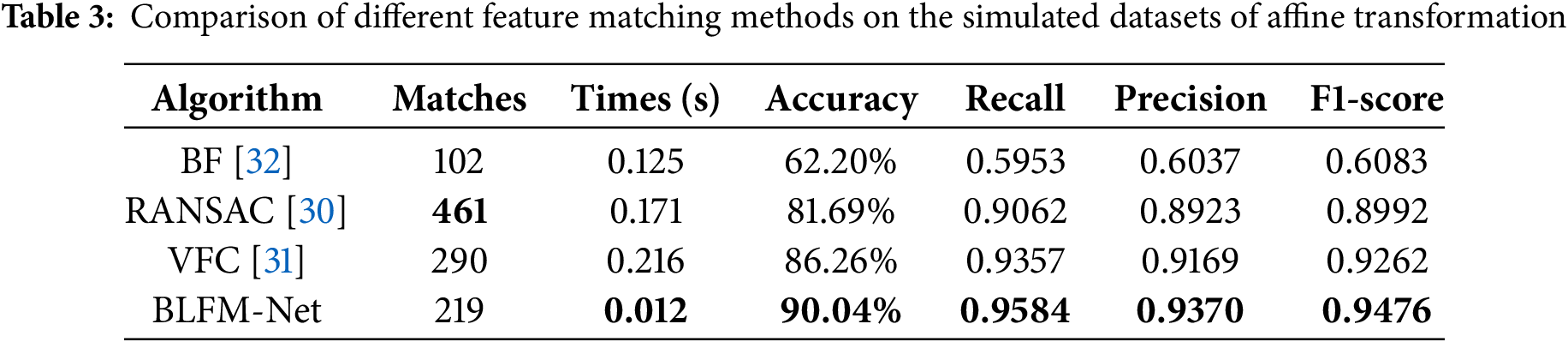
In addition we evaluated the matching ability of the proposed method for the simulated dataset using different matching methods. The ground truth for the exact pixel corresponding to each matching pair is obtained from a known transformation of the simulated dataset. Fig. 14(3) demonstrates the matching results on the paired images. We set the threshold of pixel distance as 10 to distinguish true and false matches. As shown in Table 3, in addition to the number of matches and times obtained from algorithms, we computed the recall, accuracy, precision and F1-score for a general evaluation of their performance. The random sample consensus (RANSAC) [30] and the vector field consensus (VFC) [31] has absolute advantages in the number of matches; however, it exhibits a long matching time. The bilateral functions for global motion modeling (BF) [32] has a short time but exhibits low precision. Our method achieves the highest recall, accuracy, precision and F1-score, which are 90.04%, 0.9584, 0.9370 and 0.9476, respectively. The comprehensive analysis showed that our method has good matching ability in simulated datasets.
The comparison of each matching methods with respect to different indexes is in Fig. 15. There are four groups of evaluation indexes: accuracy, recall, precision and F1-Score. The boxplots indicate the data distribution of each index. Noticeable, although the performance of our method is close to VFC in Table 3, it is more concentrative and reliable. The results also imply that our method is adaptable to different scenes and robust to outliers.

Figure 15: Comparison of different matching methods respect to different index. Each group shows the boxplot of four different methods. The asterisks were the maximums and minimums of each method. In the middle of the boxes, yellow lines marked the median
3.6 Experiments on Clinical Endoscopic Images
We also evaluate the matching ability of the proposed method on clinical endoscopic datasets. In this experiment, we used real-time clinical endoscopic videos of pig bronchi. The clinical data was recorded by the Tianjin University team on 14 April 2024, during a clinical procedure. A full ethical statement is included at the end of the paper. The matching results of the clinical data are shown in Fig. 16. The ground truth was obtained through manual calibration. The average processing time for matching was 0.041 s, and compared to the ground truth, the accuracy for pixel distance errors within 10 pixels reached 97.62%. The number of correctly matched points accounted for 94.44% of the total extracted feature points. These results validate the reliability of the proposed method in clinical applications.

Figure 16: Matches of BLFM-Net on clinical image. (a) Matching result of frame 2486. (b) Matching result of frame 3129
This paper introduces an innovative method named BLFM-Net, which combines the FASTER-RCNN object detection network with regional feature matching, offering significant improvements in accuracy, robustness, and real-time performance for bronchoscopic navigation. By leveraging precise bounding boxes, Hu moments, and KD-Tree-based matching, BLFM-Net efficiently captures both local and global region features while maintaining stable performance under challenging conditions such as complex movements and lighting variations.
For further validation of the effectiveness, BLFM-Net is compared with some traditional image processing methods, such as SURF, ORB, and Optical Flow, and the state-of-the-art deep learning-based approaches, including LoFTR, XoFTR, LightGlue, and SuperGlue. The results demonstrate that BLFM-Net achieves superior accuracy, extracts 2–3 times more feature points than traditional methods, and maintains robustness under complex conditions. Compared to other deep learning-based methods, BLFM-Net achieves a matching accuracy of 96.63% and a processing speed of 0.006 s per frame, showcasing its outstanding balance of accuracy, robustness, and real-time performance. Furthermore, the method remained robust under challenging conditions including frame skipping (0, 5, 10, 20), maintaining accuracy of above 94% even with the frame skipping of 20. Additionally, BLFM-Net was tested in two critical experiments. First, the model was evaluated under simulated affine transformations, including scale changes, counter-rotation, and random affine transformations. The results demonstrated high accuracy and robustness, with a recall rate of 90.04%, accuracy of 0.9584, and F1-score of 0.9476, surpassing methods like RANSAC and VFC in both accuracy and processing time. Second, real-time clinical endoscopic videos of pig bronchi, recorded by the Tianjin University team on 14 April 2024, were used for evaluation. The clinical data showed that BLFM-Net achieved an average processing time of 0.041 s, with 97.62% accuracy for pixel distances within 10 pixels, and 94.44% of the extracted feature points were correctly matched. In conclusion, BLFM-Net offers a robust and efficient solution for real-time bronchoscopic navigation. It excels in navigation and tracking, especially in challenging scenarios involving smooth, textureless regions of the bronchial tree, demonstrating outstanding robustness even in these complex conditions.
The main contributions of this work can be summarized as follows. First, BLFM-Net introduces a novel approach that integrates Faster R-CNN for precise object detection with advanced regional feature matching techniques, enabling accurate and efficient feature extraction. Second, our method demonstrates exceptional real-time performance, processing each frame in just 0.006 s while maintaining 96.63% matching accuracy, making it highly suitable for dynamic and time-sensitive surgical environments. Third, BLFM-Net is robust under challenging conditions such as frame skipping, lighting variations, and complex bronchial structures, maintaining accuracy above 94% even in extreme scenarios. Finally, this method contributes to the advancement of deep learning-based medical image processing, offering a practical and reliable solution for real-time bronchoscopic navigation in clinical applications, thereby improving the precision and safety of bronchoscopic surgeries.
However, BLFM-Net also faces certain limitations. One key limitation is its sensitivity to rapid endoscopic movements, which can lead to a loss of visual information and hinder the accuracy of feature matching. Another challenge is maintaining robustness in environments where there are significant variations in lighting or when the bronchial tree has smooth, textureless regions, which can make it difficult to extract reliable features. Future research can improve the accuracy and robustness of BLFM-Net by integrating endoscopic motion estimation and compensation. Endoscopic movements, such as rapid shifts in camera orientation, often lead to loss of visual information. By incorporating motion estimation, the system can better account for these movements and maintain accurate tracking. This can be achieved by integrating kinematic modeling, which describes the endoscope’s physical motion, with visual features extracted from bronchoscopic images. This fusion of kinematic data and visual information will help mitigate motion blur and frame disruptions, ultimately enhancing the reliability of the navigation system.
In the future, BLFM-Net can be further refined and expanded to enhance its performance and adaptability in clinical applications. The method’s robustness could be improved to address lighting variations, which are common in clinical environments. Enhancing the preprocessing step with techniques such as illumination normalization, light source compensation, or adaptive histogram equalization would increase the system’s resilience under varying lighting conditions. To improve spatial awareness, BLFM-Net could be integrated with 3D imaging technologies or combined with other modalities, such as CT or MRI scans, enabling more accurate navigation through complex 3D bronchial structures. Additionally, integrating BLFM-Net with robotic surgery systems could enhance its capabilities in guiding semi-autonomous or fully autonomous bronchoscopic surgeries, providing more precise real-time interventions. Lastly, incorporating autonomous learning techniques, such as active learning or reinforcement learning, would allow BLFM-Net to continuously improve its performance in real time, reducing the need for manual intervention and making the system more adaptable to various clinical environments. These advancements could enable BLFM-Net to become an even more powerful and versatile tool for real-time bronchoscopic navigation, ultimately improving the precision, safety, and efficiency of bronchoscopic surgeries.
Acknowledgement: The authors would like to thank those who contributed to the article and who support them from Tianjin University and Key Lab for Mechanism Theory and Equipment Design of Ministry of Education for the experience and technical support.
Funding Statement: This study was funded by the National Natural Science Foundation of China (Grant No. 52175028).
Author Contributions: Study conception and design: He Su, Jianwei Gao; data collection: Kang Kong, Jianwei Gao; analysis and interpretation of results: He Su, Jianwei Gao; draft manuscript preparation: He Su, Jianwei Gao. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: The clinical endoscopic images used in this study were obtained from real-time video footage of pig bronchoscopies, recorded on 14 April 2024. All animal experiments were performed in compliance with ethical guidelines and were approved by the Animal Ethics Committee. The procedures followed the principles of the 3Rs (Replacement, Reduction, and Refinement) to ensure humane treatment of the animals involved. The study was conducted under strict supervision to minimize any potential harm or discomfort to the animals. No human data was used in this experiment, and all clinical images were anonymized to protect privacy.
Availability of Data and Materials: Some data are available on request from the authors, which support the findings of this study are available from the corresponding author, Kang Kong, upon reasonable request.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Asano F, Matsuno Y, Shinagawa N, Yamazaki K, Suzuki T, Ishida T, et al. A virtual bronchoscopy navigation system for pulmonary peripheral lesions. Chest. 2006;130(2):559–66. doi:10.1378/chest.130.2.559. [Google Scholar] [PubMed] [CrossRef]
2. Lowe DG. Distinctive image features from scale-invariant keypoints. Int J Comput Vis. 2004;60(2):91–110. doi:10.1023/B:VISI.0000029664.99615.94. [Google Scholar] [CrossRef]
3. Bay H, Ess A, Tuytelaars T, Van Gool L. Speeded-up robust features (SURF). Comput Vis Image Understand. 2008;110(3):346–59. doi:10.1016/j.cviu.2007.09.014. [Google Scholar] [CrossRef]
4. Rublee E, Rabaud V, Konolige K, Bradski G. ORB: an efficient alternative to SIFT or SURF. In: 2011 International Conference on Computer Vision; 2011 Nov 6–13; Barcelona, Spain. p. 2564–71. doi:10.1109/ICCV.2011.6126544. [Google Scholar] [CrossRef]
5. Matas J, Chum O, Urban M, Pajdla T. Robust wide-baseline stereo from maximally stable extremal regions. Image Vis Comput. 2004;22(10):761–7. doi:10.1016/j.imavis.2004.02.006. [Google Scholar] [CrossRef]
6. Alcantarilla PF, Bartoli A, Davison AJ. KAZE features. In: Fitzgibbon A, Lazebnik S, Perona P, Sato Y,Schmid C, editors. Computer vision—ECCV 2012. Berlin/Heidelberg: Springer; 2012. doi:10.1007/978-3-642-33783-3_16. [Google Scholar] [CrossRef]
7. Chien JC, Lee JD, Su E, Li SH. A bronchoscope localization method using an augmented reality co-display of real bronchoscopy images with a virtual 3D bronchial tree model. Sensors. 2020;20(23):6997. doi:10.3390/s20236997. [Google Scholar] [PubMed] [CrossRef]
8. Lucas BD, Kanade T. An iterative image registration technique with an application to stereo vision. In: IJCAI’81: 7th International Joint Conference on Artificial Intelligence; 1981 Aug 24–28; Vancouver, BC, Canada. p. 674–9. [Google Scholar]
9. DeTone D, Malisiewicz T, Rabinovich A. SuperPoint: self-supervised interest point detection and description. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops; 2018 Jun 18–23; Salt Lake City, UT, USA. doi:10.1109/cvprw.2018.00060. [Google Scholar] [CrossRef]
10. Sarlin PE, DeTone D, Malisiewicz T, Rabinovich A. SuperGlue: learning feature matching with graph neural networks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2020 Jun 13–19; Seattle, WA, USA. p. 4937–46. doi:10.1109/cvpr42600.2020.00499. [Google Scholar] [CrossRef]
11. Liu H, Zhang S, Yang Y, Sun L, Song Z, Xu S. A robust pose optimization scheme with spatial geometry awareness for hybrid bronchoscopic navigation. IEEE Trans Instrum Meas. 2024;73:1–10. doi:10.1109/TIM.2024.3488158. [Google Scholar] [CrossRef]
12. Acar A, Lu D, Wu Y, Oguz I, Kavoussi N, Wu JY. Towards navigation in endoscopic kidney surgery based on preoperative imaging. Healthc Technol Lett. 2024;11(2–3):67–75. doi:10.1049/htl2.12059. [Google Scholar] [PubMed] [CrossRef]
13. Wang C, Oda M, Hayashi Y, Kitasaka T, Itoh H, Honma H, et al. Anatomy aware-based 2.5D bronchoscope tracking for image-guided bronchoscopic navigation. Comput Methods Biomech Biomed Eng Imaging Vis. 2022;11(4):1122–9. doi:10.1080/21681163.2022.2152728. [Google Scholar] [CrossRef]
14. Shen M, Gu Y, Liu N, Yang GZ. Context-aware depth and pose estimation for bronchoscopic navigation. IEEE Robot Autom Lett. 2019;4(2):732–9. doi:10.1109/LRA.2019.2893419. [Google Scholar] [CrossRef]
15. Wang C, Oda M, Hayashi Y, Villard B, Kitasaka T, Takabatake H, et al. A visual SLAM-based bronchoscope tracking scheme for bronchoscopic navigation. Int J Comput Assist Radiol Surg. 2020;15(10):1619–30. doi:10.1007/s11548-020-02241-9. [Google Scholar] [PubMed] [CrossRef]
16. Liu S, Fan J, Song D, Fu T, Lin Y, Xiao D, et al. Joint estimation of depth and motion from a monocular endoscopy image sequence using a multi-loss rebalancing network. Biomed Opt Express. 2022;13(7):2707–27. doi:10.1364/BOE.457475. [Google Scholar] [PubMed] [CrossRef]
17. Liu S, Fan J, Ai D, Song H, Fu T, Wang Y, et al. Feature matching for texture-less endoscopy images via superpixel vector field consistency. Biomed Opt Express. 2022;13(7):2247–65. doi:10.1364/BOE.450259. [Google Scholar] [PubMed] [CrossRef]
18. Chu Y, Li H, Li X, Ding Y, Yang X, Ai D, et al. Endoscopic image feature matching via motion consensus and global bilateral regression. Comput Methods Programs Biomed. 2020;190:105370. doi:10.1016/j.cmpb.2020.105370. [Google Scholar] [PubMed] [CrossRef]
19. Farhat M, Chaabouni-Chouayakh H, Ben-Hamadou A. Self-supervised endoscopic image key-points matching. Expert Syst Appl. 2023;213(13):118696. doi:10.1016/j.eswa.2022.118696. [Google Scholar] [CrossRef]
20. Lu F, Zhou D, Chen H, Liu S, Ling X, Zhu L, et al. S2P-matching: self-supervised patch-based matching using transformer for capsule endoscopic images stitching. IEEE Trans Biomed Eng. 2025;72(2):540–51. doi:10.1109/TBME.2024.3462502. [Google Scholar] [PubMed] [CrossRef]
21. Nanehkaran YA, Zhu L, Chen J, Ahmed AM, Zhao S, Dorostkar YN, et al. Anomaly detection in heart disease using a density-based unsupervised approach. Wirel Commun Mob Comput. 2022;2022:6913043. doi:10.1155/2022/6913043. [Google Scholar] [CrossRef]
22. Qin X, Wang Z, Bai Y, Xie X, Jia H. FFA-Net: feature fusion attention network for single image dehazing. In: Proceedings of the AAAI Conference on Artificial Intelligence; 2020 Feb 7–12; New York, NY, USA. doi:10.1609/aaai.v34i07.6865. [Google Scholar] [CrossRef]
23. Luo X, McLeod AJ, Pautler SE, Schlachta CM, Peters TM. Vision-based surgical field defogging. IEEE Trans Med Imaging. 2017;36(10):2021–30. doi:10.1109/TMI.2017.2701861. [Google Scholar] [PubMed] [CrossRef]
24. Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell. 2017;39(6):1137–49. doi:10.1109/TPAMI.2016.2577031. [Google Scholar] [PubMed] [CrossRef]
25. Hu MK. Visual pattern recognition by moment invariants. IRE Trans Inf Theory. 1962;8(2):179–87. doi:10.1109/TIT.1962.1057692. [Google Scholar] [CrossRef]
26. Sun J, Shen Z, Wang Y, Bao H, Zhou X. LoFTR: detector-free local feature matching with transformers. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2021 Jun 22–25; Nashville, TN, USA. p. 8918–27. doi:10.1109/CVPR46437.2021.00881. [Google Scholar] [CrossRef]
27. Tuzcuoğlu Ö., Köksal A, Sofu B, Kalkan S, Alatan AA. XoFTR: cross-modal feature matching transformer. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2024 Jun 17–18; Seattle, WA, USA. p. 4275–86. doi:10.1109/cvprw63382.2024.00431. [Google Scholar] [CrossRef]
28. Lindenberger P, Sarlin PE, Pollefeys M. LightGlue: local feature matching at light speed. In: Proceedings of the IEEE/CVF International Conference on Computer Vision; 2023 Oct 1–6; Paris, France. p. 17581–92. doi:10.1109/ICCV51070.2023.01616. [Google Scholar] [CrossRef]
29. Dusmanu M, Rocco I, Pajdla T, Pollefeys M, Sivic J, Torii A, et al. D2-Net: a trainable CNN for joint detection and description of local features. In: CVPR 2019-IEEE Conference on Computer Vision and Pattern Recognition; 2019 Jun 15–20; Long Beach, CA, USA. p. 8092–101. [Google Scholar]
30. Fischler MA, Bolles RC. Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography. Commun ACM. 1981;24(6):381–95. doi:10.1145/358669.358692. [Google Scholar] [CrossRef]
31. Ma J, Zhao J, Tian J, Yuille AL, Tu Z. Robust point matching via vector field consensus. IEEE Trans Image Process. 2014;23(4):1706–21. doi:10.1109/TIP.2014.2307478. [Google Scholar] [PubMed] [CrossRef]
32. Lin WY, Wang F, Cheng MM, Yeung SK, Torr PHS, Do MN, et al. CODE: coherence based decision boundaries for feature correspondence. IEEE Trans Pattern Anal Mach Intell. 2018;40(1):34–47. doi:10.1109/TPAMI.2017.2652468. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools