 Open Access
Open Access
ARTICLE
An Advanced Medical Diagnosis of Breast Cancer Histopathology Using Convolutional Neural Networks
1 Department of Electrical Engineering, College of Engineering, Jouf University, Sakaka, 72388, Saudi Arabia
2 Laboratory of Biochemistry and Enzymatic Engineering of Lipases, National Engineering School of Sfax, University of Sfax, Sfax, 3038, Tunisia
3 LIPONOV, Biological Engineering Department, HealthTech Industry, Sfax, 3038, Tunisia
4 Department of Electrical Engineering, College of Engineering, University of Hafr Al Batin, Hafr Al Batin, 39524, Saudi Arabia
* Corresponding Author: Ahmed Ben Atitallah. Email:
(This article belongs to the Special Issue: Advanced Medical Imaging Techniques Using Generative Artificial Intelligence)
Computers, Materials & Continua 2025, 83(3), 5761-5779. https://doi.org/10.32604/cmc.2025.063634
Received 20 January 2025; Accepted 10 April 2025; Issue published 19 May 2025
Abstract
Breast Cancer (BC) remains a leading malignancy among women, resulting in high mortality rates. Early and accurate detection is crucial for improving patient outcomes. Traditional diagnostic tools, while effective, have limitations that reduce their accessibility and accuracy. This study investigates the use of Convolutional Neural Networks (CNNs) to enhance the diagnostic process of BC histopathology. Utilizing the BreakHis dataset, which contains thousands of histopathological images, we developed a CNN model designed to improve the speed and accuracy of image analysis. Our CNN architecture was designed with multiple convolutional layers, max-pooling layers, and a fully connected network optimized for feature extraction and classification. Hyperparameter tuning was conducted to identify the optimal learning rate, batch size, and number of epochs, ensuring robust model performance. The dataset was divided into training (80%), validation (10%), and testing (10%) subsets, with performance evaluated using accuracy, precision, recall, and F1-score metrics. Our CNN model achieved a magnification-independent accuracy of 97.72%, with specific accuracies of 97.50% at 40×, 97.61% at 100×, 99.06% at 200×, and 97.25% at 400× magnification levels. These results demonstrate the model’s superior performance relative to existing methods. The integration of CNNs in diagnostic workflows can potentially reduce pathologist workload, minimize interpretation errors, and increase the availability of diagnostic testing, thereby improving BC management and patient survival rates. This study highlights the effectiveness of deep learning in automating BC histopathological classification and underscores the potential for AI-driven diagnostic solutions to improve patient care.Keywords
According to the most recent global cancer statistics, lung cancer was the most commonly diagnosed cancer worldwide in 2022, accounting for 12.4% of all new cases, followed closely by female breast cancer at 11.6%. In women, breast cancer was the most frequently diagnosed cancer and also the leading cause of cancer-related deaths [1]. The most recent WHO data also indicate that BC remains a major global health condition, with the infection being diagnosed in nearly 2.3 million women annually and causing almost 670,000 deaths throughout the world. This situation highlights the role played by the disease in determining the global health burden [2]. BC detection generally combines imaging techniques with histopathological assessments. Predominantly, mammography, ultrasound, and Magnetic Resonance Imaging (MRI) are employed [3–5]. Mammography serves as a primary screening tool but often has high false-positive rates and reduced sensitivity in dense breast tissues [6,7]. Ultrasound acts as an adjunctive tool, particularly beneficial in dense breasts, though its reliability is significantly dependent on the operator’s skill [8]. MRI is recognized for its high sensitivity and is predominantly used to screen individuals at high risk. However, it lacks specificity and incurs higher costs compared to other imaging methods [9]. Despite these modalities, histopathological examination continues to be the definitive method for cancer confirmation following the detection of anomalies via imaging. This method entails the microscopic analysis of biopsied tissue samples [10]. The precision of cancer classification from these samples largely relies on the pathologist’s expertise. Challenges such as pathologist fatigue and a high caseload can lead to diagnostic delays and potential misdiagnoses [11].
Deep learning, and specifically Convolutional Neurol Networks (CNNs), has emerged as a transformative force in medical imaging, offering the potential to automate and enhance the accuracy of diagnostic processes [12,13]. CNNs excel in feature recognition tasks by learning optimal features for classification through training with large labeled datasets. In the context of BC, CNNs can be trained to detect subtle patterns in histopathological images that may be indicative of malignancy. In fact, the integration of the CNNs into the diagnostic workflow for BC helps minimize subjective interpretation errors, thereby improving the accuracy of diagnostic procedures and, consequently, enhancing the care of BC patients.
The BreakHis dataset [14,15], which contains thousands of histopathological images of benign and malignant breast tumors, provides a comprehensive resource for training and testing deep learning models. However, existing approaches often suffer from a dependency on specific magnifications, limiting their generalizability. Our study aims to bridge this gap by developing a magnification-independent CNN model capable of achieving high accuracy across different histopathological image magnifications. The novelty of our work lies in optimizing the CNN architecture to ensure robust feature extraction and classification, outperforming state-of-the-art models in BC classification. This advancement contributes to the early and accurate detection of BC, potentially reducing the mortality rate associated with the disease.
The remainder of the paper is structured as follows: Section 2 covers the literature review. Section 3 details the materials and methods employed in the study, including an in-depth overview of the BreakHis dataset and the CNN model architecture. Section 4 presents the experimental results, addressing both magnification-independent and magnification-dependent binary classification outcomes. Section 5 offers a comparative analysis of different techniques used for BC histopathology classification. Finally, the paper concludes with a summary of findings and recommendations for future research.
Recently, deep learning for BC case classification and detection from histopathological images has gained attention in several studies. The authors of [16] proposed a Deep Manifold Preserving Autoencoder (DMAE) for the classification of BC histopathological images. The DMAE is an autoencoder with the objective of reducing the distance between input and output by maintaining a geometrical data structure. The classifier is implemented using a softmax and fine-tuned in the cascade model with labeled training data. On this point, DMAE outperforms its predecessors, the traditional deep learning and machine learning methods, on the BreaKHis data set, as it can achieve high accuracy in image classification. However, this way seems to be less scalable due to the relative complexity of the architecture and training. The authors in [17] proposed a novel approach to enhance the accuracy of BC histopathology image classification. This approach employs a combination of DenseNet121 and Anomaly Generative Adversarial Networks (AnoGAN) to address the challenge of mislabeled patches in the dataset. DenseNet121 is utilized to extract features from discriminative patches of images, and AnoGAN is used for unsupervised anomaly detection to screen out mislabeled patches effectively. The methodology was tested using the BreaKHis dataset and demonstrated high accuracy, with the best performance showing a 99.13% accuracy rate at 40× magnification. This model performs optimally at 40× and 100× magnifications. However, its effectiveness decreases at 200× and 400× magnifications, requiring modifications for improved performance. In [18], the authors introduced an automatic classification method for BC histopathological images, utilizing deep feature fusion and an enhanced routing mechanism (FE-BiCapsNet). This approach combines the strengths of CNNs and Capsule Networks (CapsNets) through a dual-channel framework to enhance classification accuracy. But the dual-channel feature extraction and the complex feature fusion process result in significant computational demands, leading to increased training and prediction times for FE-BiCapsNet. In [19], the authors discussed the design and evaluation of hybrid architectures for binary classification of BC images using both deep learning techniques for feature extraction and machine learning classifiers. The most effective architecture included the MLP classifier and DenseNet 201 for extracting features. The obtained accuracy was acceptable for the BreakHis datasets with magnification factors. In [20], the authors investigated the effectiveness of handcrafted features and representation learning techniques in BC detection from histopathological images. It contrasts these with pre-trained CNNs and explores feature fusion strategies to enhance detection accuracy. The research involves applying these techniques to two significant datasets, KIMIA Path960 and BreakHis, and demonstrates that feature fusion can significantly improve classification performance compared to individual techniques. But the complexity of combining multiple feature extraction techniques could lead to increased computational demand, which might limit the practical deployment of such models in resource-constrained settings. In [21], the authors introduced the dense residual Dual-Shuffle Attention Network (DRDA-Net) model to advance the classification of BC through histopathological images. Incorporating a channel attention mechanism within its architecture, the model enhances its pattern recognition capabilities. Tested on the BreaKHis dataset across several magnifications, DRDA-Net demonstrates the improvements in the accuracy for cancer detection. But this model is deep due to its densely connected nature, which increases training time and computational cost. In [22], the authors investigated the use of CNNs for classifying histopathological breast images. They presented three types of CNN models: a single CNN model, a fusion of two CNN models, and a fusion of three CNN models. The three CNN models attained an accuracy of 90.10% on the Kaggle dataset. However, the combination of multiple CNN models increases the complexity of model interpretation, which could affect its usability in clinical settings where understanding the model’s decision-making process is crucial. In [23], the authors employed CNNs and transferred learning techniques to classify BC histopathology images. The study aimed to automate the detection process to facilitate faster and more accurate medical diagnoses. The models used include CNNs, VGG16, and ResNet50, which were evaluated on the BreakHis dataset containing images of benign and malignant breast tumors. Data augmentation was applied to enhance model training. The study found that CNNs were the most effective method, achieving a 94% accuracy rate, compared to the lower accuracies of VGG16 and ResNet50. In [24], a deep learning model named BCDecNet was developed. This model features a complex architecture with multiple layers, including convolution layers, activation functions, pooling layers, and fully connected layers, utilizing data from the Kaggle online repository. The BCDecNet demonstrated a performance accuracy rate of 97.33%. However, its complexity and high computational demands may limit its practical application in settings with limited resources. In [25], the authors proposed a breast tumor classification approach using histopathology images from the BreaKHis dataset. In fact, their approach incorporated a custom CNN alongside four pretrained models (MobileNetV3, EfficientNetB0, VGG16, and ResNet50V2) for feature extraction and classification. To enhance performance, they applied Grey Wolf Optimization (GWO) and Modified Gorilla Troops Optimization (MGTO) for hyperparameter tuning. The study highlights the effectiveness of combining deep learning models with metaheuristic optimization for improving diagnostic accuracy in BC classification. In [26], the authors introduced a two-stage hybrid network for BC classification using the BreaKHis dataset. Their method integrates ResNet34 as the initial convolutional network for feature extraction, followed by a Long Short-Term Memory (LSTM) network to capture contextual dependencies. The findings indicate that combining CNNs with LSTM-based contextual learning enhances classification performance in BC histopathology. In [27], the authors presented the CNN approach using the Inception-v3 architecture for the classification of BC images into benign and malignant categories. Experimental results demonstrated over 92% accuracy across all four magnification levels on the BreaKHis dataset, highlighting the effectiveness of CNNs in automating BC histopathological classification.
The BreakHis dataset [14,15] is a collection of histopathological images designed for research in BC analysis. Compiled between January 2014 and December 2014, it consists of 7909 high-resolution microscopic images, including 2480 images of benign tumors and 5429 images of malignant tumors, obtained from 82 patients. This dataset is prepared from tissue biopsies stained with hematoxylin and eosin (H&E) following the standard paraffin process to maintain histological integrity. Digitization of these images ensures high-quality representations for detailed analysis, as illustrated in Fig. 1.
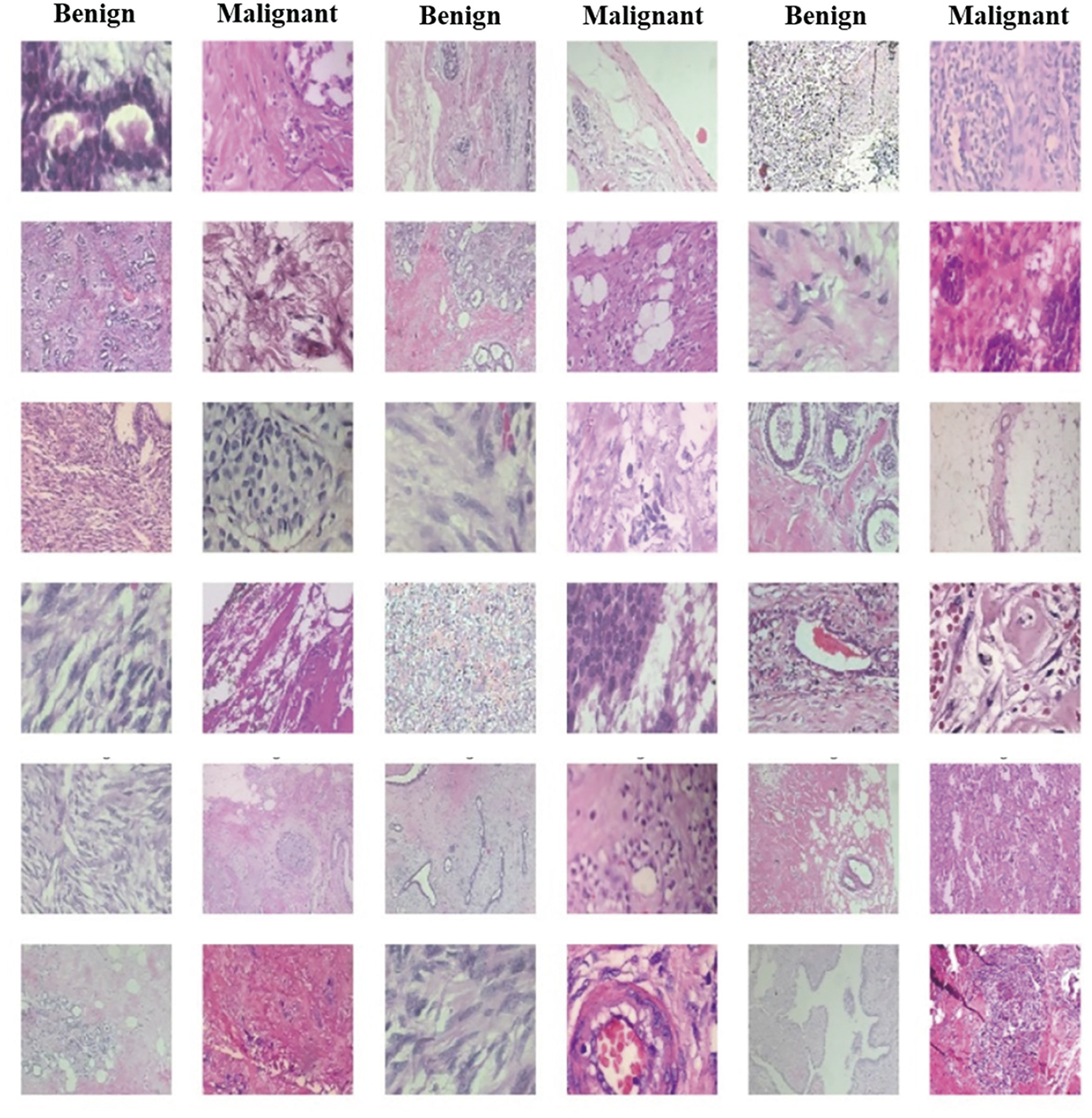
Figure 1: Samples of benign and malignant images from the BreakHis dataset
In this dataset, these images were captured using four different magnification levels (40×, 100×, 200×, and 400×), providing a diverse range of details for examinations. By providing such detailed and diversified data, the BreakHis dataset plays a pivotal role in the ongoing efforts to integrate more sophisticated analytical technologies, like deep learning, into everyday clinical practices, enhancing both the speed and accuracy of cancer diagnostics. In fact, it enables researchers to develop, test, and compare innovative computational methods for BC classification from histopathological images, potentially reducing pathologists’ workload by automatically identifying cancerous regions. This resource is invaluable for enhancing computer-aided diagnosis systems and improving patient outcomes through earlier and more precise detection.
The proposed CNN architecture is designed to improve the diagnostic accuracy for BC by effectively combining feature extraction, abstraction, and classification. By using multiple convolutional and pooling layers, as illustrated in Fig. 2, the network can learn intricate patterns within the data, which is crucial for distinguishing between benign and malignant tumors.
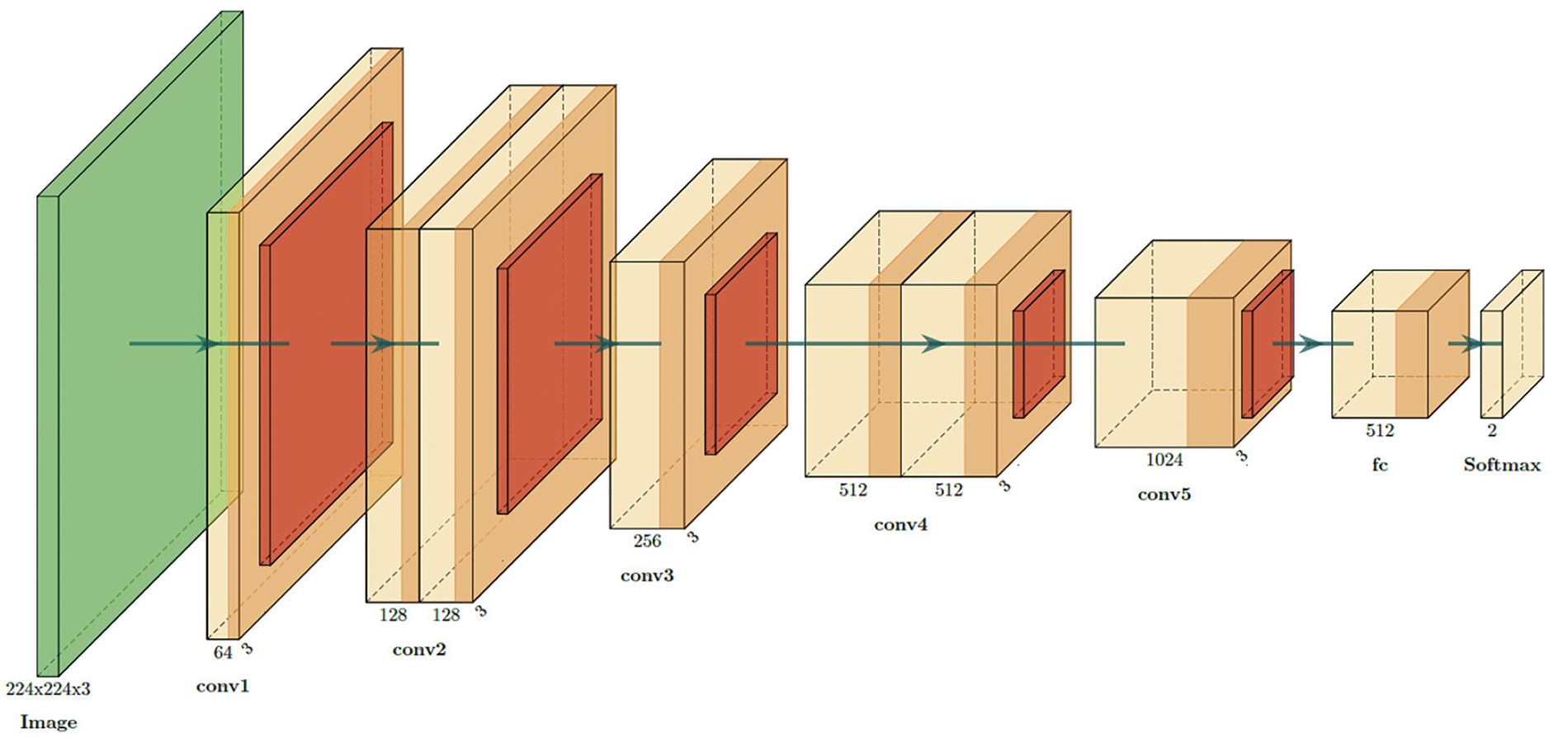
Figure 2: Detailed CNNs architecture diagram for BC classification
In the proposed CNN architecture, the network initiates the process by handling input images sized at 224 × 224 pixels with three color channels (RGB). Initially, the input layer receives images with these dimensions (224 × 224 × 3). The first convolutional layer (conv1) uses 3 × 3 filters to extract 64 feature maps from the input image, reducing the spatial dimensions slightly to 222 × 222. Following this, subsequent convolutional layers (conv2 to conv5) progressively increase the depth of the feature maps from 128 to 1024, allowing the network to capture more complex patterns and features. Each convolutional layer is typically followed by an activation function, which introduces non-linearity. To down-sample the feature maps, MaxPooling layers (MaxPooling1 to MaxPooling5) are employed for each convolution layer. These layers reduce the dimensions while retaining the most significant features, helping to decrease computational complexity and prevent overfitting. Following the convolutional layers, the feature maps are flattened into one vector of 16,384 elements using the flatten layer. Such a vector is then passed through a fully connected (fc) layer of 512 units. It is a dense layer in which the high-level features that were extracted by the convolutional layers are combined into a unified representation. Finally, the output layer contains the dense layer, with 2 units for the two classes: benign and malignant. SoftMax activation was used to generate class probabilities. Table 1 provides a summary of the parameters for the proposed model.
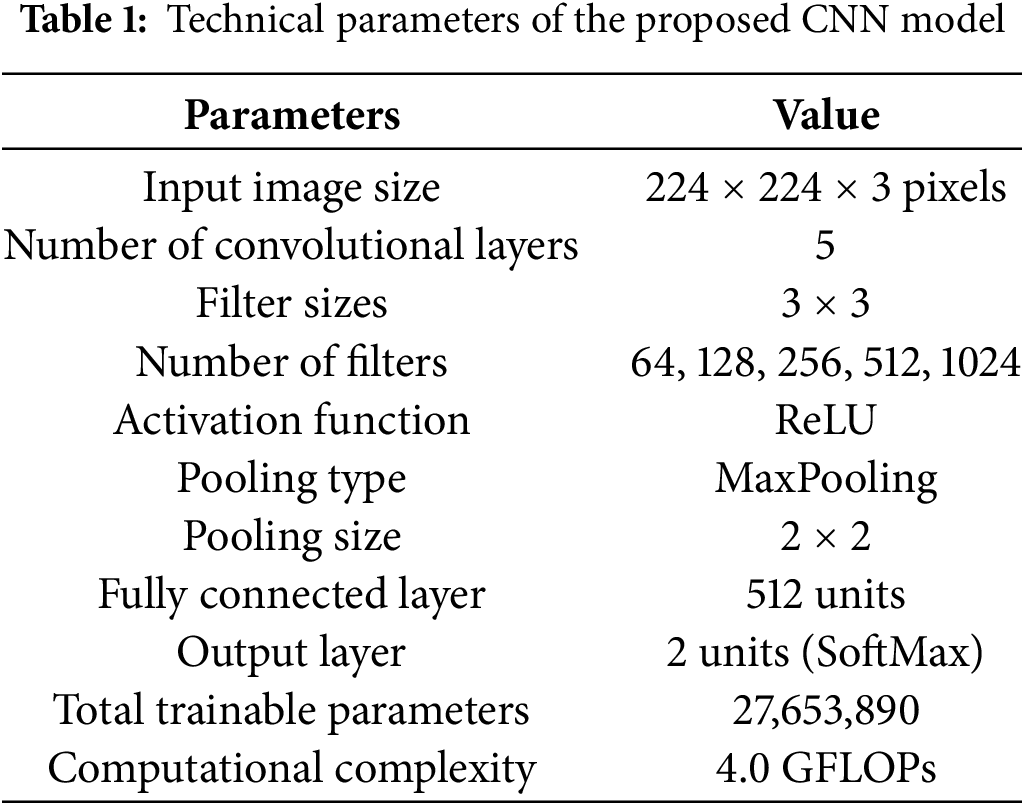
Fig. 3 illustrates the methodology employed in this study, which comprises several key steps: data acquisition, data preprocessing, hyperparameter optimization, model training, evaluation metrics, and data classification. Each step is essential for enhancing the robustness and effectiveness of the CNN model in classifying BC histopathological images from the BreakHis dataset.
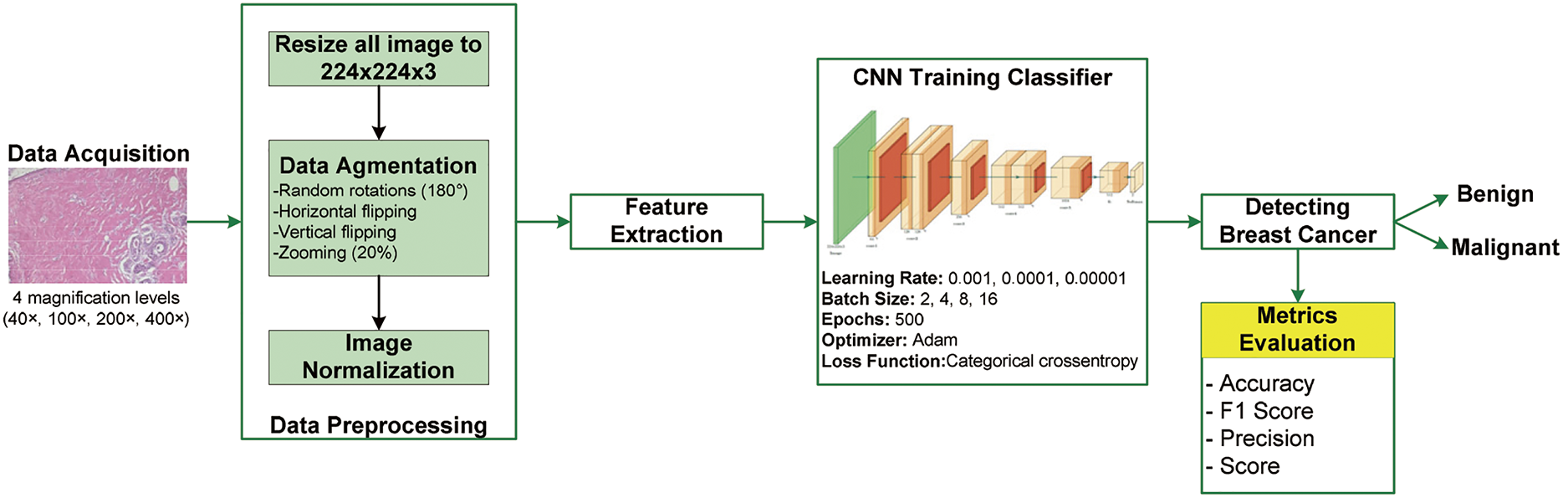
Figure 3: Proposed methodology
We resized input images using a dataset from BreakHis to 224 × 224 pixels in this study. Resizing makes computations efficient and saves training time. Since the dataset contains few images for training the model, the chances of overfitting were high. We applied augmentation techniques using Keras ImageDataGenerator. For that reason, we made random rotations up to 180 degrees, horizontal and vertical flipping, and zooming in up to 20%. Such augmentations guaranteed that our model would be exposed to training on a wide variety of images, thus making it more robust and better at generalizing new, unseen data. Following image augmentation, we converted the color map from RGB to BGR and normalized the pixel values. Normalization is vital as it ensures that the input data values are within a consistent range, enhancing the model’s ability to learn effectively. This task was accomplished by subtracting the mean of the training dataset from each image and then dividing it by the standard deviation, both calculated exclusively from the training dataset. This approach ensures consistent scaling of the input images, which facilitates learning and better performance by the model.
3.3.2 Hyperparameter Optimization and Model Training
In our research, we meticulously adjusted the hyperparameters to improve the performance of our CNN model. We set up the fully connected layer with 512 neurons that were activated by the ReLU function. The output layer used a SoftMax activation function to decide whether an image was benign or malignant. We only used the categorical cross-entropy loss function because it works well for handling outputs from more than one class. We took 80% of the data for training and 20% for validation and testing, making equal parts of it. We have used an Adam optimizer here, which automatically modifies learning rates during adaptive optimization. This technique facilitates fast model convergence. For fine-tuning, we have done experiments using three values of learning rates, such as 0.001, 0.0001, and 0.00001. This experiment was of high importance for grasping how the learning rate influences model performance and is used to determine the best setting for the highest classification accuracy. We monitored the validation accuracy at the end of each epoch, training the model for a total of 500 epochs. We experimented with diverse batch sizes (2, 4, 8, and 16) based on the learning rate to find the best classification performance. By varying the batch sizes, we aimed to balance computational efficiency with the stability of gradient updates. By meticulously configuring these hyperparameters and implementing robust training protocols, we ensured the development of a high-performing model capable of accurately classifying BC from histopathological images.
We utilized the following four widely referred metrics in the literature to measure model performance: accuracy, precision, recall, and F1-score. The latter metrics are widely applicable in any study of whether the model can correctly assign the category of benign or malignant tumors, with the model’s sensitivity towards really positive cases and specificity making the right predictions. Accuracy on (1) says how frequently the model gave a correct prediction in general, regarding both the positive and negative cases. Precision under (2), however, shows how many instances were true when the model predicted them as positive, in relation to the true positives. In other words, recall under (3) shows how effective a model is at capturing all actual positives, representing its suitability in grabbing relevant cases. Conversely, the F1-score in (4) offers a singular metric that harmonizes the measures of precision and recall.
Using these metrics, we can thoroughly evaluate the reliability and effectiveness of our CNN model for BC classification. Moreover, we employed a confusion matrix to analyze the model’s predictions in detail. This matrix distinguishes between True Negatives (TN) and True Positives (TP), which correspond to correctly identified benign and malignant cases, respectively. It also highlights the False Negatives (FN), which indicate malignant cases erroneously classified as benign, and the False Positives (FP), which present benign cases mistakenly identified as malignant. This evaluation was performed on histopathological images from the BreaKHis dataset across various magnifications (40×, 100×, 200×, and 400×).
We implemented our model using TensorFlow and Keras, which are powerful libraries for developing and training deep learning models. The major reasons for selection include their flexibility, ease of use, and comprehensive support of most neural network architectures. In fact, we used the programming language Python, notably known for its simplicity and rich support of machine learning and data science. We developed our project using PyCharm as an integrated development environment (IDE), which played a great role in its effective management, debugging, and organization. Our training was conducted on a computer with an Intel® Core™ i9-13900H processor and NVIDIA GeForce RTX 3050 (4 GB) graphics card. This high-performance processor quickly processed data and performed quick calculations. The dedicated GPU sped up training by a large amount by handling the intensive matrix operations needed for deep learning. To ensure our results were robust, we repeated the experiments multiple times. This procedure helped verify that the model’s performance was consistent and not influenced by anomalies from a single training run.
In this section, we present the results of the proposed CNN model for BC classification using histopathological images from the BreaKHis dataset. The results are categorized into magnification-independent and magnification-dependent (40×, 100×, 200×, and 400×) binary classifications. We divided the dataset into training, validation, and test sets using an 80%, 10%, and 10% split. To find the best setting, we looked at how well the model worked with different learning rates (0.001, 0.0001, and 0.00001) and batch sizes (2, 4, 8, and 16).
4.1 Magnification-Independent Binary Classification
The dataset for the magnification-independent binary classification has 5429 malignant histopathological images and 2480 benign ones. Table 2 presents the classification results, highlighting the impact of varying learning rates (0.001, 0.0001, and 0.00001) and batch sizes (2, 4, 8, and 16) on the model’s diagnostic metrics, such as accuracy, precision, recall, and F1-score. Based on this table, the proposed CNN model works best when the learning rate is 0.0001 and the batch size is 8. It gets an impressive accuracy of 97.72%, a precision of 97.64%, a recall of 99.08%, and an F1-score of 98.35%.
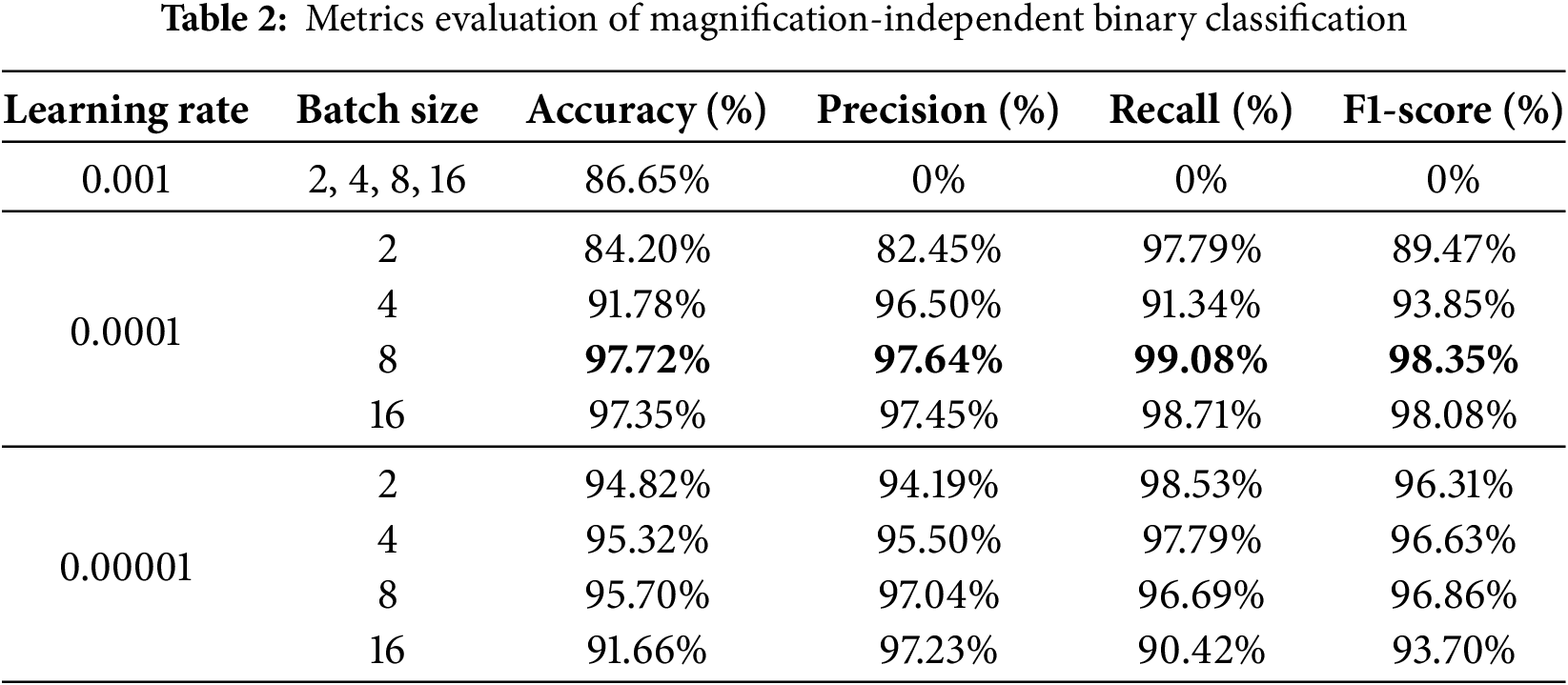
Based on the accuracy and loss curves and the confusion matrix for a learning rate of 0.0001 and a batch size of 8, Fig. 4 lets us see how well our model worked. Fig. 4a displays the training and validation accuracy and loss curves over 500 epochs. These curves show that the model is steadily getting better. The small gap between the training and validation curves indicates that the model is not prone to overfitting and performs well on new data. The confusion matrix in Fig. 4b shows that the model has high rates of TN and TP and low rates of FN and FP. This indicates that the model is accurate and reliable at telling the difference between benign and malignant samples.
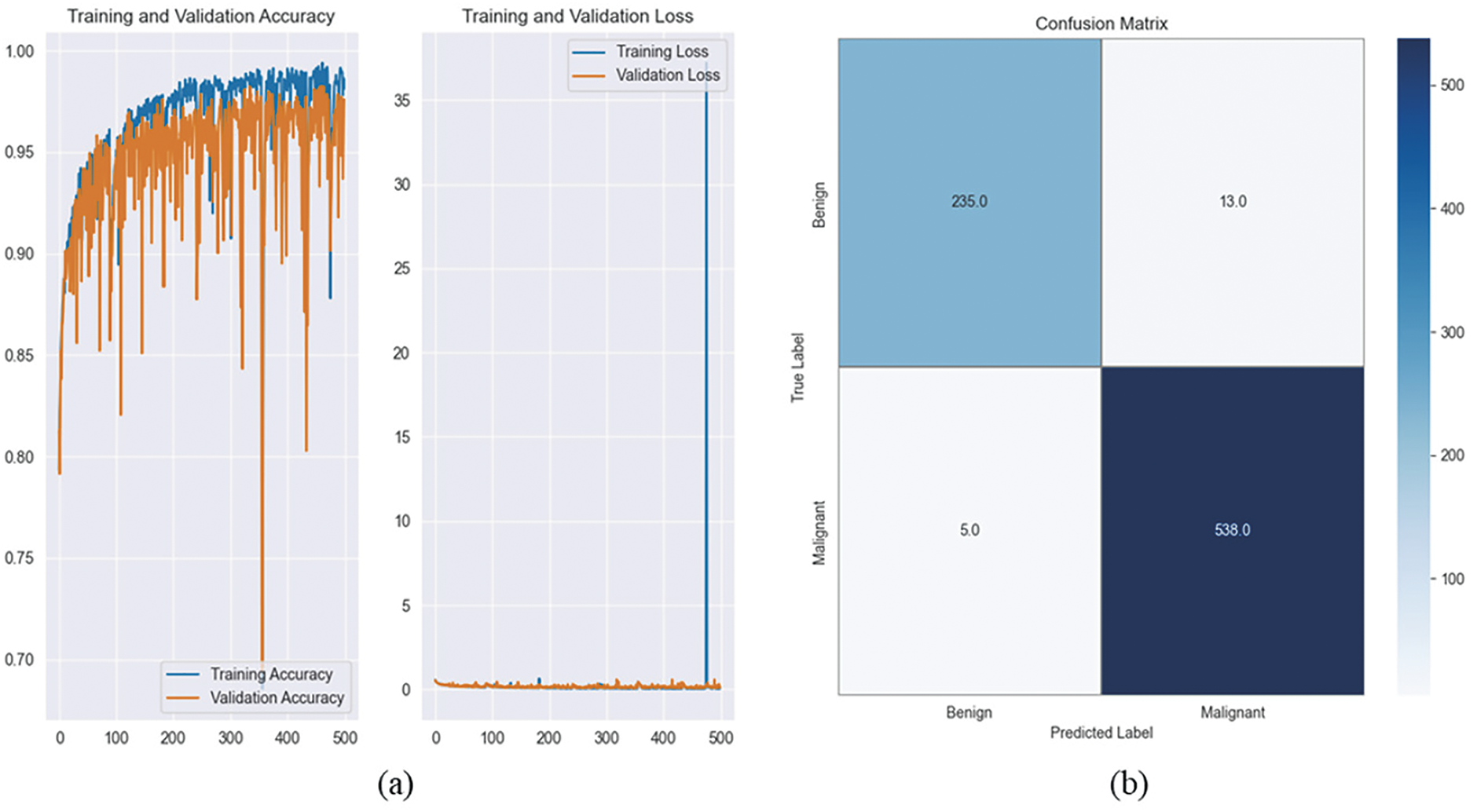
Figure 4: It illustrates the training and validation accuracy, loss curves, and confusion matrix. (a) Training and validation accuracy and loss curves over 500 epochs, illustrating the model’s convergence and stability. The minimal gap between training and validation curves indicates low overfitting and strong generalization performance. (b) Confusion matrix for magnification-independent binary classification, showing high TP and TN rates, with minimal false classifications, demonstrating the model’s effectiveness in distinguishing between benign and malignant breast cancer histopathological images
Fig. 5 presents a comparison of the predicted and actual classifications for 10 images from the BreaKHis dataset. Each subplot displays the true label (0 for benign, 1 for malignant) alongside the model’s prediction, highlighting its effectiveness in distinguishing different breast tissue types. The close match between predicted and actual labels shows how accurate and reliable the CNN model is, which increases its usefulness for diagnostic purposes in the real world.
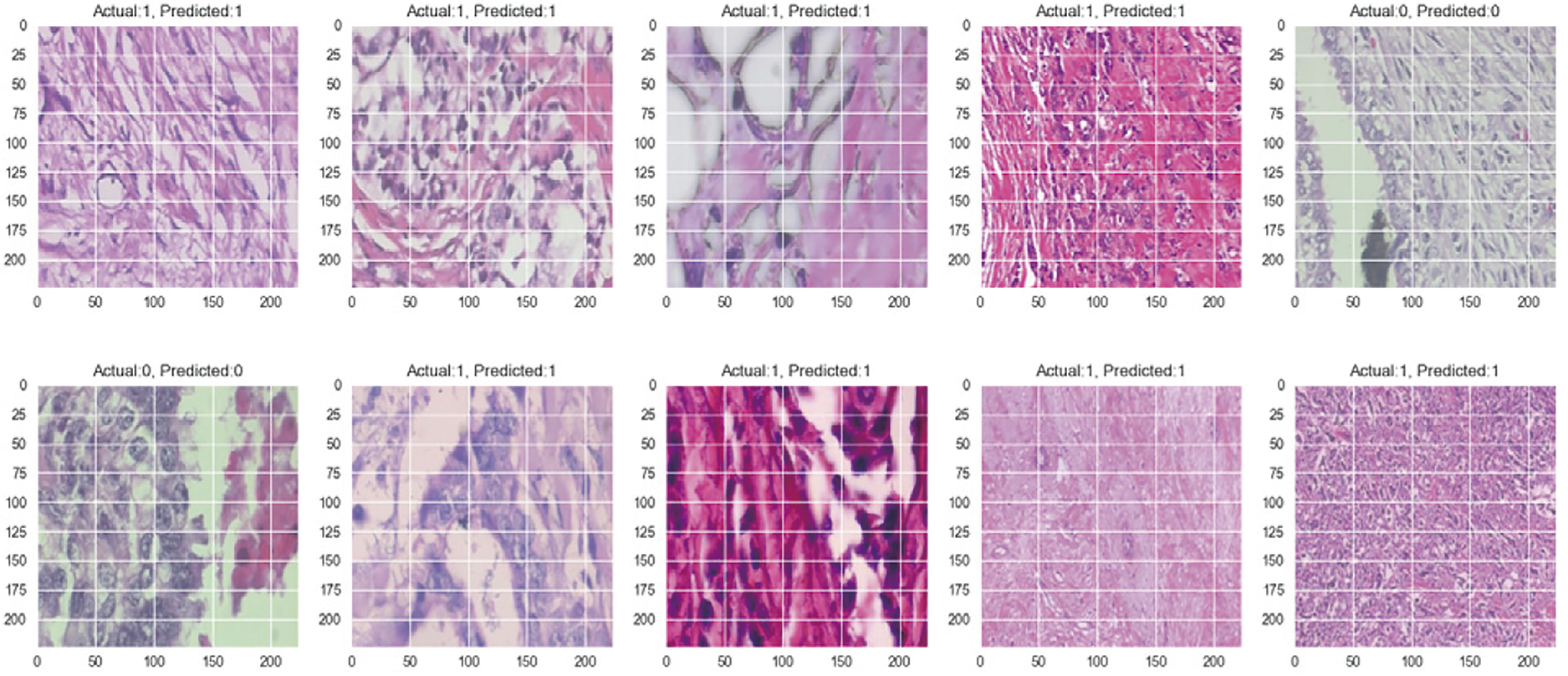
Figure 5: Prediction vs. actual results of 10 images for magnification-independent binary classification
4.2 Magnification-Dependent Binary Classification
The model was further evaluated at specific magnification levels: 40×, 100×, 200×, and 400×. For each magnification, the dataset was split into 80% for training, 10% for validation, and 10% for testing to ensure a balanced evaluation. Table 3 summarizes the performance metrics at each magnification level, demonstrating the impact of different learning rates (0.001, 0.0001, and 0.00001) and batch sizes (2, 4, 8, and 16) on the model’s accuracy, precision, recall, and F1-score. Fig. 6 illustrates the training and validation accuracy, loss curves, and confusion matrix. Fig. 7 provides a comparative visualization of predicted vs. actual classifications for each magnification level.
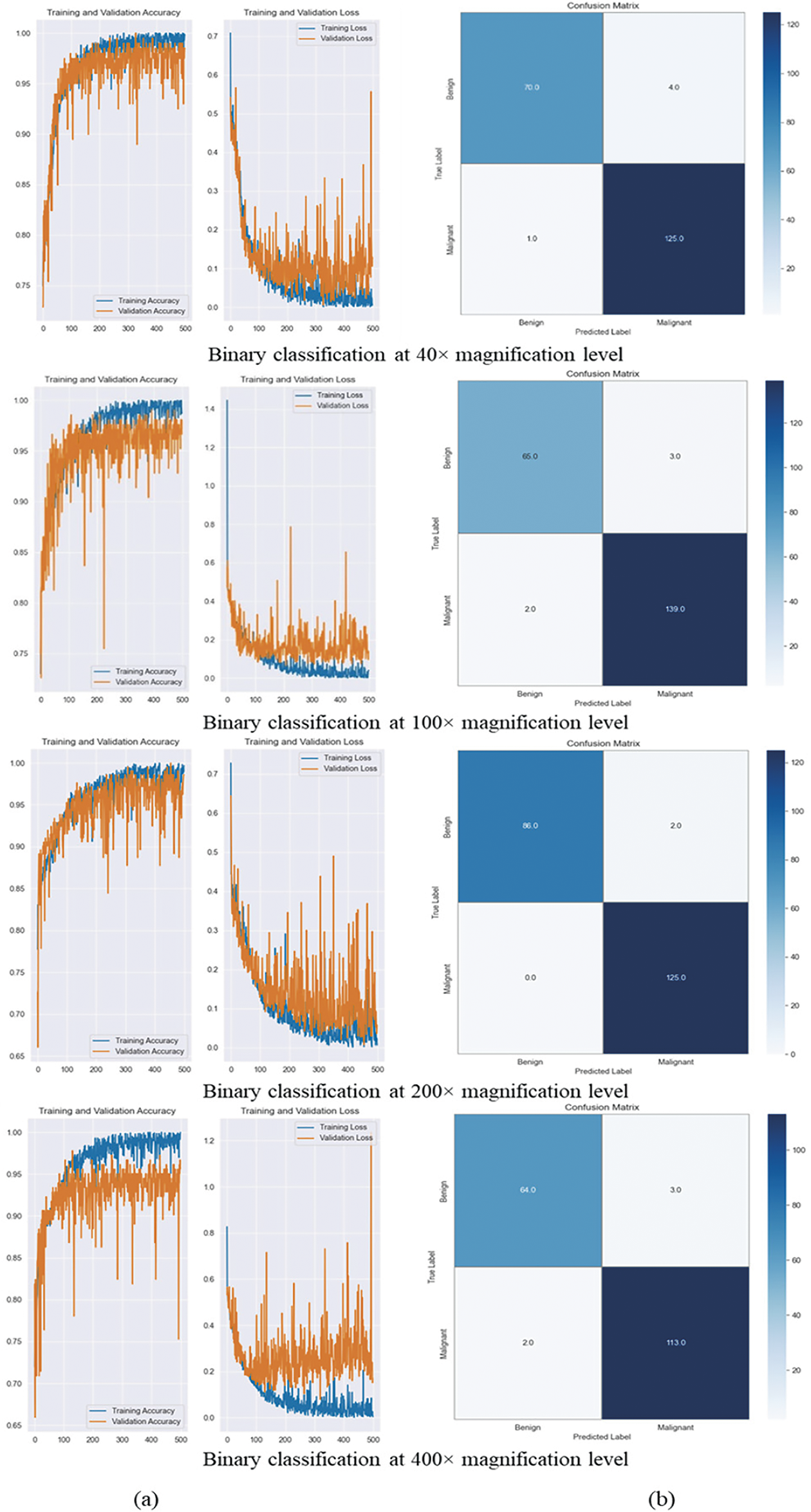
Figure 6: It illustrates the training and validation accuracy, loss curves, and confusion matrix. (a) Training and validation accuracy and loss curves for magnification-dependent binary classification at 40×, 100×, 200×, and 400× magnification levels. The curves indicate stable model convergence across all magnifications, with minimal overfitting. (b) Confusion matrices for each magnification level, illustrating the model’s classification performance. High TP and TN rates confirm the model’s effectiveness in distinguishing benign and malignant histopathological images across different magnifications
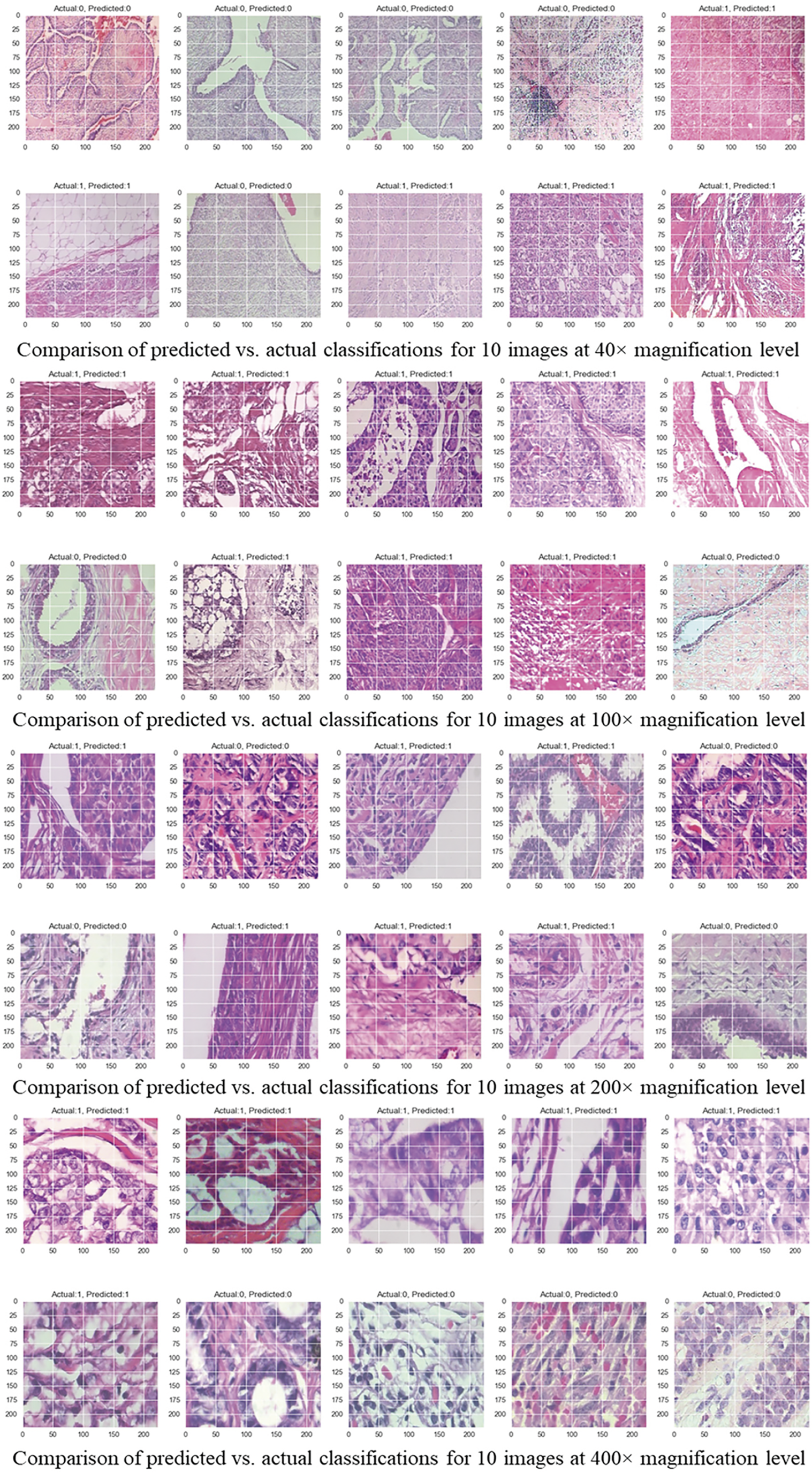
Figure 7: Comparison of predicted vs. actual classifications for 10 images across different magnification levels
As shown in Table 3, we got the best results for classification at 40× magnification with a learning rate of 0.0001 and a batch size of 16. We got a precision of 96.90%, a recall of 99.21%, and an F1-score of 98.04%. There were a total of 1995 images used, with 625 being benign and 1370 being malignant. Further, at 100× magnification, using 2081 images (644 benign, 1437 malignant), the best accuracy of 97.61% was achieved under the same hyperparameter configuration, yielding a precision of 97.89%, a recall of 98.58%, and an F1-score of 98.23%. Moreover, the model did best at classifying images at 200× magnification, with an accuracy of 99.06%, a precision of 98.43%, a recall of 100%, and an F1-score of 99.21%. This score was achieved with 2013 images (623 benign and 1390 malignant) and the best settings of a learning rate of 0.0001 and a batch size of 16. In the end, with a learning rate of 0.0001 and a batch size of 8, the model was able to achieve an accuracy of 97.25% at 400× magnification, with 588 benign images and 1232 malignant images used. It also had a precision of 97.41%, a recall of 98.26%, and an F1-score of 97.84%. These results confirm that the model performs consistently well across different magnifications, with 200× magnification providing the highest accuracy.
Fig. 6a displays the training and validation accuracy and loss curves over 500 epochs. The curves show stable convergence and little overfitting at different levels of magnification. The confusion matrix in Fig. 6b shows that the model has high rates of TP and TN and low rates of FP and FN. This indicates that it is excellent at telling the difference between benign and malignant cases. This finding is supported by Fig. 7, which shows that the predicted and actual labels for 10 images from the BreaKHis dataset are very close at every magnification level.
5.1 Performance Trends across Magnifications
The experimental results confirm the high classification performance of the proposed CNN model across all magnification levels, achieving an overall magnification-independent accuracy of 97.72%, with a peak accuracy of 99.06% at 200× magnification. While the accuracies at 40× (97.50%), 100× (97.61%), and 400× (97.25%) were slightly lower, likely due to the increased complexity of tissue structures, the model still exhibited high classification reliability. We determined these results through systematic experimentation, using a learning rate of 0.0001 and a batch size of 8 or 16. Table 4 provides a summary of the classification performance metrics for each magnification level and magnification-independent classification under the optimal hyperparameter settings for high-accuracy binary classification of histopathology images of BC.
This study underscores the critical role of hyperparameter optimization in enhancing classification accuracy. A learning rate of 0.0001 consistently did better than 0.001 and 0.00001, showing that moderate learning rates help keep models from becoming too perfect while still ensuring stable convergence. Conversely, when the learning rate was set to 0.001, accuracy dropped significantly across all magnifications, confirming that an excessively high learning rate results in unstable training and poor generalization. Furthermore, a batch size of 8 or 16 was found to be the most effective, balancing computational efficiency with stable gradient updates. These results highlight the importance of fine-tuning hyperparameters to enhance the CNN’s ability to extract meaningful features from histopathological images, further demonstrating the effectiveness of deep learning in BC classification. Moreover, the findings emphasize the necessity of dataset diversity and proper hyperparameter selection in maximizing the generalizability and robustness of CNN-based models for histopathological image analysis.
5.2 Comparative Analysis with Existing Methods
Table 5 provides a comprehensive comparison of various techniques used in BC histopathology classification, highlighting their effectiveness and accuracy at different magnification levels. The table includes several references, with each reference corresponding to a specific technique or a combination of techniques employed for classifying BC histopathological images. These techniques range from traditional machine learning methods to advanced deep learning models, such as the DMAE, DenseNet121-AnoGAN, and the FE-BkCapsNet, among others. The primary datasets used in these studies are BreakHis and Kaggle, both providing high-resolution histopathological images essential for training and testing the models. The BreakHis dataset includes images at four magnification levels: 40×, 100×, 200×, and 400×, allowing for a detailed examination of model performance across varying levels of image detail. The table reveals that the accuracy of each technique varies with magnification, indicating the strengths and weaknesses of each model at different levels of detail. For example, DMAE [16] achieves an accuracy of 91.34% at 200× magnification but performs less effectively at 100×, with an accuracy of 87.75%. Similarly, DenseNet121-AnoGAN [17] excels at 40× magnification with 99.13% accuracy but experiences a significant drop to 85.20% at 400×. The models proposed in [18,19,21,26] consistently maintain high accuracy across all magnifications, yet their substantial computational requirements and complex feature fusion processes pose practical challenges. In contrast, the feature fusion approach in [20], which combines CNNs with handcrafted features (CH + ULBP), struggles with lower accuracy across all magnifications. Other deep learning models designed for binary BC classification, such as those in [22–25,27], achieved magnification-independent accuracies of 90.10%, 94%, 97.33%, 93.13%, and 92%, respectively.
The proposed CNN model stands out with its high classification accuracy across all magnifications, achieving an impressive 99.06% accuracy at 200× magnification, suggesting a robust ability to generalize and accurately classify images irrespective of the magnification level. Our model’s magnification-independent accuracy of 97.72% further emphasizes its effectiveness in handling diverse image details, making it a promising tool for practical diagnostic applications. Compared to other models, our CNN model demonstrates superior performance not only in terms of accuracy but also in its ability to maintain high performance consistently across all magnification levels without increasing the model complexity. So, this comparative study highlights the novel aspects of our model:
• Magnification independence: Unlike previous methods that experience accuracy drops at different magnifications, our model maintains high performance across all resolutions, making it more generalizable for clinical applications.
• Optimized feature extraction: Many prior studies use hybrid models (e.g., DenseNet121-AnoGAN, CNNs with LSTM), increasing computational complexity. The proposed CNN model achieves comparable or better accuracy without requiring additional architecture, making it more efficient and reducing both training time and computational costs.
• Balanced performance across metrics: our CNN model achieves high accuracy, precision, recall, and F1-score simultaneously, ensuring reliable and well-rounded classification performance.
While the proposed CNN model demonstrates high classification accuracy across magnification levels, further advancements can be achieved by integrating multimodal data sources, such as histopathological images, genetic data, and clinical information. The combination of these diverse data types has the potential to enhance diagnostic accuracy, improve personalized treatment planning, and provide a more comprehensive understanding of BC.
Our study demonstrates the significant potential of CNNs in improving the accuracy and efficiency of BC histopathology diagnostics. Utilizing the BreaKHis dataset, the proposed CNN model demonstrated high performance across different magnification levels, achieving 97.50% at 40×, 97.61% at 100×, 99.06% at 200×, and 97.25% at 400×, with an overall magnification-independent accuracy of 97.72%. This robust performance underscores the model’s capability to handle diverse image details, making it a valuable tool for practical diagnostic applications. Compared to other models, our CNN model not only excels in accuracy but also ensures reliable performance across all magnifications, addressing a critical need in clinical settings. The adoption of such advanced deep learning techniques in medical diagnostics promises to enhance early detection, reduce diagnostic errors, and ultimately improve patient outcomes. As the field of artificial intelligence in medicine continues to evolve, the integration of CNNs into routine diagnostic workflows could play a transformative role in advancing BC care.
Acknowledgement: Not applicable.
Funding Statement: This work was funded by the Deanship of Graduate Studies and Scientific Research at Jouf University under grant No. (DGSSR-2024-02-01096).
Author Contributions: The authors confirm contribution to the paper as follows: conceptualization: Meshari D. Alanazi and Khaled Kaaniche; methodology: Jannet Kamoun; software: Mohammed Albekairi; validation: Khaled Kaaniche; formal analysis: Ahmed Ben Atitallah and Jannet Kamoun; investigation: Meshari D. Alanazi; resources, Turki M. Alanazi; data curation: Turki M. Alanazi; writing—original draft preparation: Meshari D. Alanazi and Turki M. Alanazi; writing—review and editing: Ahmed Ben Atitallah and Jannet Kamoun; visualization: Mohammed Albekairi; supervision: Ahmed Ben Atitallah; project administration: Khaled Kaaniche; funding acquisition: Ahmed Ben Atitallah. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2024;74(3):229–63. doi:10.3322/caac.21834. [Google Scholar] [PubMed] [CrossRef]
2. Breast cancer [Internet]. Geneva, Switzerland: World Health Organization; 2024 [cited 2024 Mar 13]. Available from: https://www.who.int/news-room/fact-sheets/detail/breast-cancer. [Google Scholar]
3. Sardanelli F, Boetes C, Borisch B, Decker T, Federico M, Gibert FJ, et al. Magnetic resonance imaging of the breast: recommendations from the EUSOMA working group. Eur J Cancer. 2010;46(8):1296–316. doi:10.1016/j.ejca.2010.02.015. [Google Scholar] [PubMed] [CrossRef]
4. Berg WA, Blume JD, Cormack JB, Mendelson EB. Operator dependence of physician-performed whole-breast US: lesion detection and characterization. Radiology. 2006;241(2):355–65. doi:10.1148/radiol.2412051710. [Google Scholar] [PubMed] [CrossRef]
5. Aristokli N, Polycarpou I, Themistocleous S, Sophocleous D, Mamais I. Comparison of the diagnostic performance of magnetic resonance imaging (MRIultrasound and mammography for detection of breast cancer based on tumor type, breast density and patient’s history: a review. Radiography. 2022;28(3):848–56. doi:10.1016/j.radi.2022.01.006. [Google Scholar] [PubMed] [CrossRef]
6. Wang Y, Li Y, Song Y, Chen C, Wang Z, Li L, et al. Comparison of ultrasound and mammography for early diagnosis of breast cancer among Chinese women with suspected breast lesions: a prospective trial. Thorac Cancer. 2022;13(22):3145–151. doi:10.1111/1759-7714.14666. [Google Scholar] [PubMed] [CrossRef]
7. Hussein H, Abbas E, Keshavarzi S, Fazelzad R, Bukhanov K, Kulkarni S, et al. Supplemental breast cancer screening in women with dense breasts and negative mammography: a systematic review and meta-analysis. Radiology. 2023;306(3):e221785. doi:10.1148/radiol.221785. [Google Scholar] [PubMed] [CrossRef]
8. Li X, Zhang L, Ding M. Ultrasound-based radiomics for the differential diagnosis of breast masses: a systematic review and meta-analysis. J Clin Ultrasound. 2024;52(6):778–88. doi:10.1002/jcu.23690. [Google Scholar] [PubMed] [CrossRef]
9. LoDuca TP, Strigel RM, Bozzuto LM. Utilization of screening breast MRI in women with extremely dense breasts. Curr Breast Cancer Rep. 2024;16(1):53–60. doi:10.1007/s12609-024-00525-6. [Google Scholar] [CrossRef]
10. Rana MK, Mahajan MKM. Comprehensive histopathological examination and breast cancer. Ann Pathol Lab Med. 2020;7(1):A28–33. doi:10.21276/apalm.2481. [Google Scholar] [CrossRef]
11. Wang Y, Acs B, Robertson S, Liu B, Solorzano L, Wählby C, et al. Improved breast cancer histological grading using deep learning. Ann Oncol. 2022;33(1):89–98. doi:10.1016/j.annonc.2021.09.007. [Google Scholar] [PubMed] [CrossRef]
12. Arshaghi A, Ashourian M, Ghabeli L. Denoising medical images using machine learning, deep learning approaches: a survey. Curr Med Imagin. 2021;17(5):578–94. doi:10.2174/1573405616666201118122908. [Google Scholar] [PubMed] [CrossRef]
13. Sood T, Bhatia R, Khandnor P. Cancer detection based on medical image analysis with the help of machine learning and deep learning techniques: a systematic literature review. Curr Med Imaging. 2023;19(13):e170223213746. doi:10.2174/1573405619666230217100130. [Google Scholar] [PubMed] [CrossRef]
14. Spanhol FA, Oliveira LS, Petitjean C, Heutte L. A dataset for breast cancer histopathological image classification. IEEE Trans Biomed Eng. 2016;63(7):1455–62. doi:10.1109/TBME.2015.2496264. [Google Scholar] [PubMed] [CrossRef]
15. Benhammou Y, Achchab B, Herrera F, Tabik S. BreakHis based breast cancer automatic diagnosis using deep learning: taxonomy, survey and insights. Neurocomputing. 2020;375(5):9–24. doi:10.1016/j.neucom.2019.09.044. [Google Scholar] [CrossRef]
16. Feng Y, Zhang L, Mo J. Deep manifold preserving autoencoder for classifying breast cancer histopathological images. IEEE/ACM Trans Comput Biol Bioinform. 2020;17(1):91–101. doi:10.1109/TCBB.2018.2858763. [Google Scholar] [PubMed] [CrossRef]
17. Man R, Yang P, Xu B. Classification of breast cancer histopathological images using discriminative patches screened by generative adversarial networks. IEEE Access. 2020;8:155362–77. doi:10.1109/ACCESS.2020.3019327. [Google Scholar] [CrossRef]
18. Wang P, Wang J, Li Y, Li P, Li L, Jiang M. Automatic classification of breast cancer histopathological images based on deep feature fusion and enhanced routing. Biomed Signal Process Control. 2021;65(6):102341. doi:10.1016/j.bspc.2020.102341. [Google Scholar] [CrossRef]
19. Zerouaoui H, Idri A. Deep hybrid architectures for binary classification of medical breast cancer images. Biomed Signal Process Control. 2022;71(1):103226. doi:10.1016/j.bspc.2021.103226. [Google Scholar] [CrossRef]
20. Das R, Kaur K, Walia E. Feature generalization for breast cancer detection in histopathological images. Interdiscip Sci Comput Life Sci. 2022;14(2):566–81. doi:10.1007/s12539-022-00515-1. [Google Scholar] [PubMed] [CrossRef]
21. Chattopadhyay S, Dey A, Singh PK, Sarkar R. DRDA-Net: dense residual dual-shuffle attention network for breast cancer classification using histopathological images. Comput Biol Med. 2022;145(10):105437. doi:10.1016/j.compbiomed.2022.105437. [Google Scholar] [PubMed] [CrossRef]
22. Rafiq A, Chursin A, Awad AW, Rashed AT, Aldehim G, Abdel SN, et al. Detection and classification of histopathological breast images using a fusion of CNN frameworks. Diagnostics. 2023;13(10):1700. doi:10.3390/diagnostics13101700. [Google Scholar] [PubMed] [CrossRef]
23. Chandranegara DR, Pratama FH, Fajrianur S, Putra MRE, Sari Z. Automated detection of breast cancer histopathology image using convolutional neural network and transfer learning. MATRIK J Manaj Tek Inform Dan Rekayasa Komput. 2023;22(3):455–68. doi:10.30812/matrik.v22i3.2803. [Google Scholar] [CrossRef]
24. Zaman R, Shah IA, Ullah N, Khan GZ. A robust deep learning-based approach for detection of breast cancer from histopathological images. In: Cennamo N, editor. Proceedings of the 4th International Electronic Conference on Applied Sciences; 2023 Oct 27–Nov 10; Online. Basel, Switzerland: MDPI. [Google Scholar]
25. Heikal A, El-Ghamry A, Elmougy S, Rashad MZ. Fine tuning deep learning models for breast tumor classification. Sci Rep. 2024;14(1):10753. doi:10.1038/s41598-024-60245-w. [Google Scholar] [PubMed] [CrossRef]
26. Wang G, Jia M, Zhou Q, Xu S, Zhao Y, Wang Q, et al. Multi-classification of breast cancer pathology images based on a two-stage hybrid network. J Cancer Res Clin Oncol. 2024;150(12):505. doi:10.1007/s00432-024-06002-y. [Google Scholar] [PubMed] [CrossRef]
27. Xiao M, Li Y, Yan X, Gao M, Wang W. Convolutional neural network classification of cancer cytopathology images: taking breast cancer as an example. In: Proceedings of the 2024 7th International Conference on Machine Vision and Applications; 2024 Mar 12–14; Singapore. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools