 Open Access
Open Access
REVIEW
A Review of Deep Learning for Biomedical Signals: Current Applications, Advancements, Future Prospects, Interpretation, and Challenges
1 Biomedical Engineering Program, University of Manitoba, Winnipeg, MB R3T 2N2, Canada
2 Department of Electrical and Computer Engineering, University of Manitoba, Winnipeg, MB R3T 2N2, Canada
* Corresponding Author: Zahra Moussavi. Email:
Computers, Materials & Continua 2025, 83(3), 3753-3841. https://doi.org/10.32604/cmc.2025.063643
Received 20 January 2025; Accepted 17 April 2025; Issue published 19 May 2025
Abstract
This review presents a comprehensive technical analysis of deep learning (DL) methodologies in biomedical signal processing, focusing on architectural innovations, experimental validation, and evaluation frameworks. We systematically evaluate key deep learning architectures including convolutional neural networks (CNNs), recurrent neural networks (RNNs), transformer-based models, and hybrid systems across critical tasks such as arrhythmia classification, seizure detection, and anomaly segmentation. The study dissects preprocessing techniques (e.g., wavelet denoising, spectral normalization) and feature extraction strategies (time-frequency analysis, attention mechanisms), demonstrating their impact on model accuracy, noise robustness, and computational efficiency. Experimental results underscore the superiority of deep learning over traditional methods, particularly in automated feature extraction, real-time processing, cross-modal generalization, and achieving up to a 15% increase in classification accuracy and enhanced noise resilience across electrocardiogram (ECG), electroencephalogram (EEG), and electromyogram (EMG) signals. Performance is rigorously benchmarked using precision, recall, F1-scores, area under the receiver operating characteristic curve (AUC-ROC), and computational complexity metrics, providing a unified framework for comparing model efficacy. The survey addresses persistent challenges: synthetic data generation mitigates limited training samples, interpretability tools (e.g., Gradient-weighted Class Activation Mapping (Grad-CAM), Shapley values) resolve model opacity, and federated learning ensures privacy-compliant deployments. Distinguished from prior reviews, this work offers a structured taxonomy of deep learning architectures, integrates emerging paradigms like transformers and domain-specific attention mechanisms, and evaluates preprocessing pipelines for spectral-temporal trade-offs. It advances the field by bridging technical advancements with clinical needs, such as scalability in real-world settings (e.g., wearable devices) and regulatory alignment with the Health Insurance Portability and Accountability Act (HIPAA) and General Data Protection Regulation (GDPR). By synthesizing technical rigor, ethical considerations, and actionable guidelines for model selection, this survey establishes a holistic reference for developing robust, interpretable biomedical artificial intelligence (AI) systems, accelerating their translation into personalized and equitable healthcare solutions.Keywords
Abbreviations
| AI | Artificial Intelligence |
| AHAA | American Heart Association |
| BCI | Brain-Computer Interface |
| BNN | Bayesian Neural Networks |
| CBAM | Convolutional Block Attention Module |
| CNN | Convolutional Neural Network |
| CO | Cardiac Output |
| CP | Conformal Prediction |
| DBN | Deep Belief Networks |
| DL | Deep Learning |
| DNN | Deep Neural Networks |
| DWT | Discrete Wavelet Transform |
| ECG | Electrocardiography |
| EEG | Electroencephalography |
| EIG | Enhanced Integrated Gradients |
| EMG | Electromyography |
| EOG | Electrooculography |
| FDA | Food and Drug Administration |
| FSL | Few-Shot Learning |
| FPR | False Positive Rate |
| GAN | Generative Adversarial Network |
| GDPR | General Data Protection Regulation |
| GPU | Graphics Processing Unit |
| Grad-CAM | Gradient-Weighted Class Activation Mapping |
| GRU | Gated Recurrent Unit |
| IG | Integrated Gradients |
| HA | Hierarchical Attention |
| HAN | Hierarchical Attention Networks |
| HFO | High-Frequency Oscillation |
| HIPAA | Health Insurance Portability and Accountability Act |
| HRV | Heart Rate Variability |
| LELE | Locally Explainable Linear Explanations |
| LIME | Local Interpretable Model-Agnostic Explanations |
| LSTM | Long Short-Term Memory |
| MCBAM-GRU | Multistream Convolutional Block Attention Module-Gate Recurrent Unit |
| MEG | Magnetoencephalography |
| ML | Machine Learning |
| MRI | Magnetic Resonance Imaging |
| NIST | National Institute of Standards and Technology |
| NLP | Natural Language Processing |
| PPG | Photoplethysmography |
| PPV | Positive Predictive Value |
| RNN | Recurrent Neural Network |
| ROC | Receiver Operating Characteristic Curve |
| SHAP | Shapley Additive Explanations |
| ShapAAL | Shapley Attributed Ablation with Augmented Learning |
| SR2CNN | Signal Recognition and Reconstruction Convolutional Neural Network |
| STFT | Short-Time Fourier Transform |
| sEMG | Surface Electromyography |
| SVM | Support Vector Machine |
| ZSL | Zero-Shot Learning |
Deep learning (DL) has become a dominant paradigm for processing biomedical signals because it can extract ostensibly unattainable features through conventional methods [1]. In recent years, there have been an increasing number of publications on the application of DL methods to biomedical signals, which has allegedly led to milestone progress in diagnosis, treatment, and personalized healthcare [2–4]. Different DL methods and techniques, such as convolutional neural networks (CNNs) and recurrent neural networks (RNNs), have been applied to analyze numerous types of biomedical signals, such as electroencephalograms (EEGs), electrocardiograms (ECGs), photoplethysmography (PPGs), electrooculography (EOGs), and electromyography (EMG) signals [2,5,6].
The outcomes of these models are promising, but it is crucial to remember that further validation of their ability to detect all patterns and diseases is needed. This leads to concerns about their reliability in accurate clinical decision-making, and additional evaluations are necessary before they can be widely adopted. However, it has been purportedly used to detect and diagnose several medical conditions, including neuromuscular disorders [7,8], sleep disorders [9,10], epilepsy [11–13], and arrhythmias [2,14–16]. One of the apparent benefits of DL applications in the biomedical signals field is their ability to handle big data. These data became more accessible with the escalation of digital healthcare, and DL models can learn these data to enhance the performance of their outcomes. Moreover, unlabeled data can be used to train DL models, which can be advantageous in scenarios where labeled data are unavailable or costly [17].
The application of DL models for analyzing and interpreting biomedical signals has shown great potential in the medical field. Moreover, these models can provide real-time monitoring of patients’ vital signs and alert clinicians to potential issues. Additionally, these models can analyze large biomedical datasets in depth, identify biomarkers, and predict disease progression, leading to earlier diagnosis and improved treatment outcomes [18]. Thus, these models have the potential to revolutionize healthcare by enabling more personalized and accurate treatment plans. However, further investigations are needed to validate their effectiveness and safety in clinical practice.
Despite the high-performance results achieved by DL models, several challenges are associated with their deployment in biomedical signal processing. The most challenging part is acquiring and preprocessing data and maintaining their quality. Biomedical signals are often corrupted by noise, artifacts, and interference, making it difficult to extract meaningful features [19,20]. Interpreting DL models can be challenging, as they are frequently analyzed as black boxes, making it difficult to comprehend how a particular verdict was reached [18].
Another formidable challenge in deploying DL to biomedical signals is the necessity for specialized hardware and software. However, DL models require copious computational resources and training them can be computationally exorbitant and protracted [21]. Therefore, specialized hardware is needed to perform the training process. One of the well-known hardware components is the graphics processing unit (GPU) [22]. However, their high power consumption can contribute to climate change if not balanced with renewable energy sources or efficiency measures.
Furthermore, specialized DL software tools are required for the design, evaluation, and execution of models. In addition, these models depend strongly on the quality and quantity of the data used to train them. Therefore, data augmentation techniques, such as noise addition and data balancing, enhance the model’s robustness and diminish the possibility of overfitting. Additionally, developing precise and efficient algorithms for data preprocessing, feature extraction, and normalization is crucial for improving the performance of DL models [19,23].
Furthermore, integrating DL models into clinical systems and practice raises ethical and legal concerns about patient data privacy and ownership. In such integration, ensuring that implementation in healthcare is performed ethically and that the benefits outweigh the risks is essential [21]. However, integrating DL models into clinical systems can change healthcare delivery by connecting clinicians with the most recent insights into patient data. These models can analyze large amounts of biomedical data, learn to find patterns and make previously impossible predictions via conventional machine learning (ML) techniques, enabling more accurate diagnoses and personalized treatment plans [18,20]. In addition, the complexity of these models can make it difficult for healthcare professionals to interpret the results generated. Therefore, providing proper data visualization of the outputs and training of DL models and supporting clinicians in verifying that they can effectively use these models in practice are essential [24]. By fully or partially solving these challenges, DL models can be integrated into clinical workflows to enhance patient outcomes and improve the overall quality of healthcare delivery [17,25].
DL models, especially CNNs, RNNs, and GANs, have recently made substantial advancements in analyzing biomedical signals such as ECGs, EEGs, and PPGs. For example, CNNs have achieved high diagnostic accuracy for ECG-based arrhythmia detection, with studies such as those reporting accuracy rates over 98% on datasets such as the MIT-BIH arrhythmia database [26–28]. Similarly, RNNs have shown effectiveness in capturing sequential dependencies in EEG signals for seizure detection, with sensitivity exceeding 90% in specific applications [13,29]. However, the strengths of these models are often balanced by significant challenges, such as dependency on high-quality data and interpretability limitations. For example, [15] noted that CNNs, despite their high accuracy, struggle with noisy data, whereas [30] reported that the “black-box” nature of RNNs poses a barrier to clinical adoption. These studies illustrate that while DL has transformative potential in biomedical signal analysis, these models must overcome critical limitations to achieve real-world clinical viability.
Objectives
This paper covers various DL models and their diverse applications in biomedical signal processing, thoroughly exploring their current successes and the multifaceted challenges they face. Emphasis is placed on addressing the data used, choosing a suitable model for the application, interpreting the model results, and overcoming challenges, as these are central to effective deployment in clinical settings. The paper also examines opportunities to refine these models for more accurate and efficient predictions, explicitly focusing on facilitating early diagnoses and enabling personalized treatments. Ultimately, this review aims to bridge knowledge gaps by highlighting both the potential and limitations of DL in this rapidly evolving domain.
2 Methodology and Bibliometric Analysis
This paper included mainly journal papers covering general DL applications for biomedical signals. We also included journal and conference papers on DL applications in biomedical signals, except that the conference had no extended documents in a journal.
The search for relevant papers in this review encompassed the period up to the mid of 2024. Relevant manuscripts were identified by searching the following databases and search engines:
• Scopus
• Google Scholar
• PubMed
• IEEE
• ScienceDirect
• Taylor & Francis
The selection of these databases is essential because they cover a wide range of scholarly articles to ensure comprehensive coverage of this review. Using keywords as a primary search method helps to locate articles that focus on and are relevant to the topic of this review. Keywords are a vital way to navigate between articles and enhance the visibility of published articles. Fig. 1 shows a graphical representation of the keywords used in this review.
Figure 1: Graphical representation of the keywords covered in this review and their relevance to the scope of this paper
The inclusion criteria for this review were as follows:
• This article proposes the application of DL or the analysis of DL in biomedical signals.
• This article addresses one of the predefined research contributions to DL in biomedical signals.
• This article contributes to the theoretical or practical advancements in DL in biomedical signals.
2.3 Search Keywords and Queries
The selected articles were searched via the following keywords: deep learning, biomedical signals, physiological signals, DL, artificial intelligence (AI), diagnosis, ECG, EEG, EMG, CNN, LSTM, and explainable AI. The following is an example of a search query using the keywords DL AND biomedical signals OR physiological signals AND diagnosis.
To maintain a focus on the most relevant research, articles that did not directly apply DL techniques to one of the biomedical signals were excluded. Our screening methodology was structured in two stages to ensure thorough screening of articles on DL applications in biomedical signals. In the first stage, we assessed the articles based on their titles and abstracts. The selected articles were searched via the keywords mentioned in Section 2.3. In the second stage, we analyzed the full texts of the selected articles from the first stage. In this stage, the full texts of the articles were used to ensure that they met the inclusion criteria. Fig. 2 shows the flowchart of our screening strategy process.
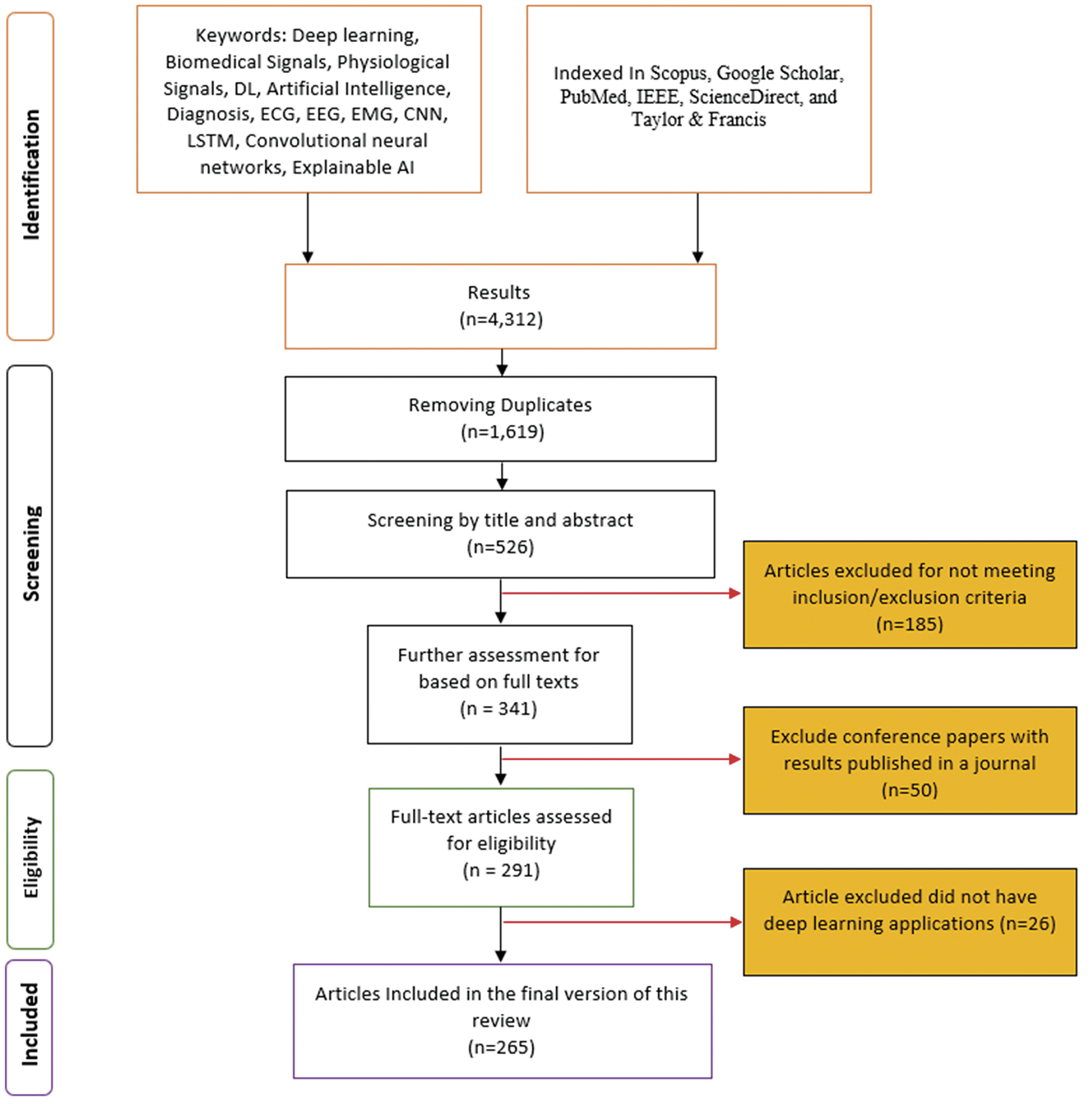
Figure 2: Flowchart showing the multistage screening process for selecting relevant studies
This section provides an in-depth analysis of the research landscape based on bibliometric data. The analysis is organized into several subsections, each focusing on a different aspect of the analyzed data. The study was performed on all included datasets after duplications were removed.
2.5.1 Scientific Production Trends
The analysis of annual publication output reveals a robust increase in scientific production over the past decades. This trend indicates that research interest in this field is growing rapidly, with recent years showing a significant surge in publications. The main findings in this trend analysis are that the dataset shows an increase from approximately 50 publications in the early 2000s to over 400 publications in 2020. A steady growth rate underscores the expanding scope and interest in the field. Fig. 3 shows the number of publications over the years.
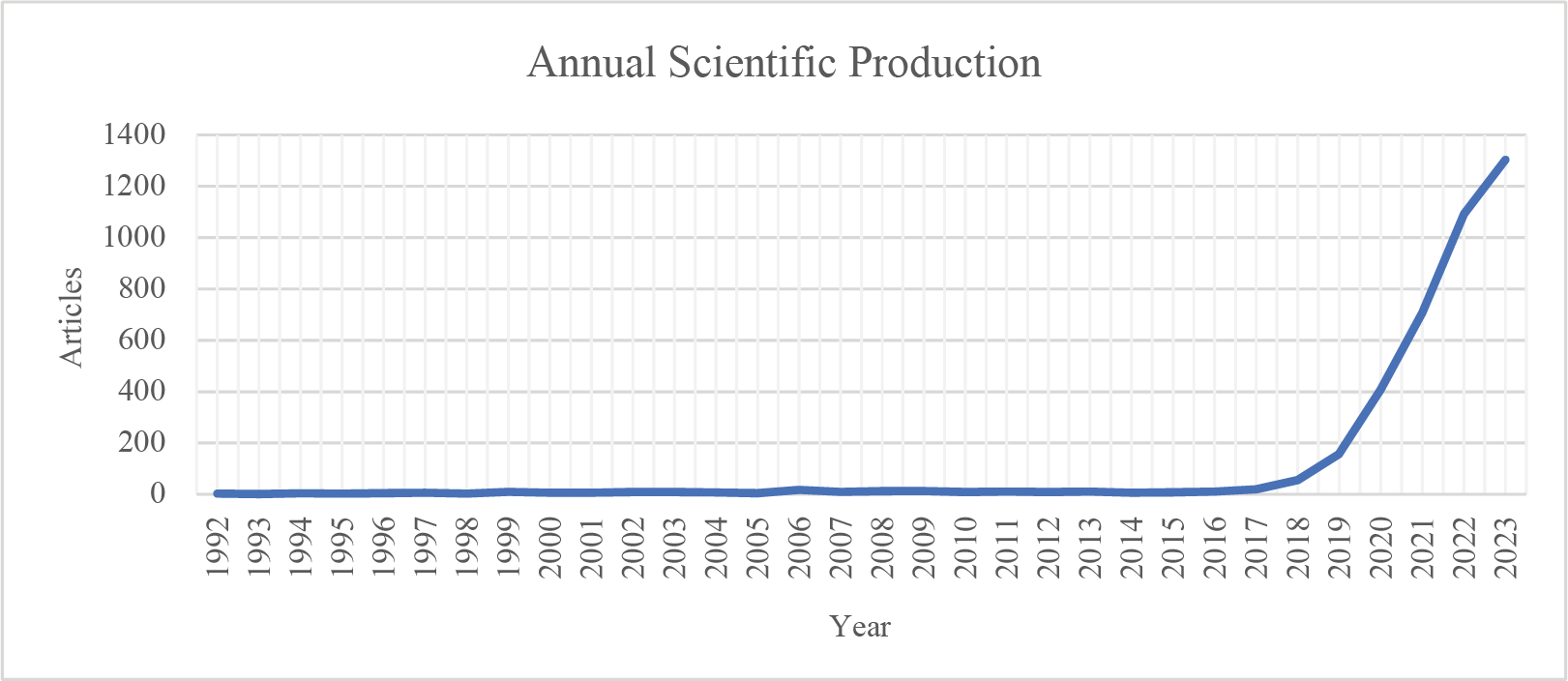
Figure 3: Graph depicting the annual publication counts over time
2.5.2 Analysis of Authors’ Research Output over Time
The analysis identifies the most productive authors and evaluates their scholarly impact. Metrics such as publication counts and citations per publication provide insight into the contributions of key researchers. The key findings are the productivity of various authors over time, spanning from 2018 to mid-2024. Fig. 4 shows the authors’ productivity and impact.
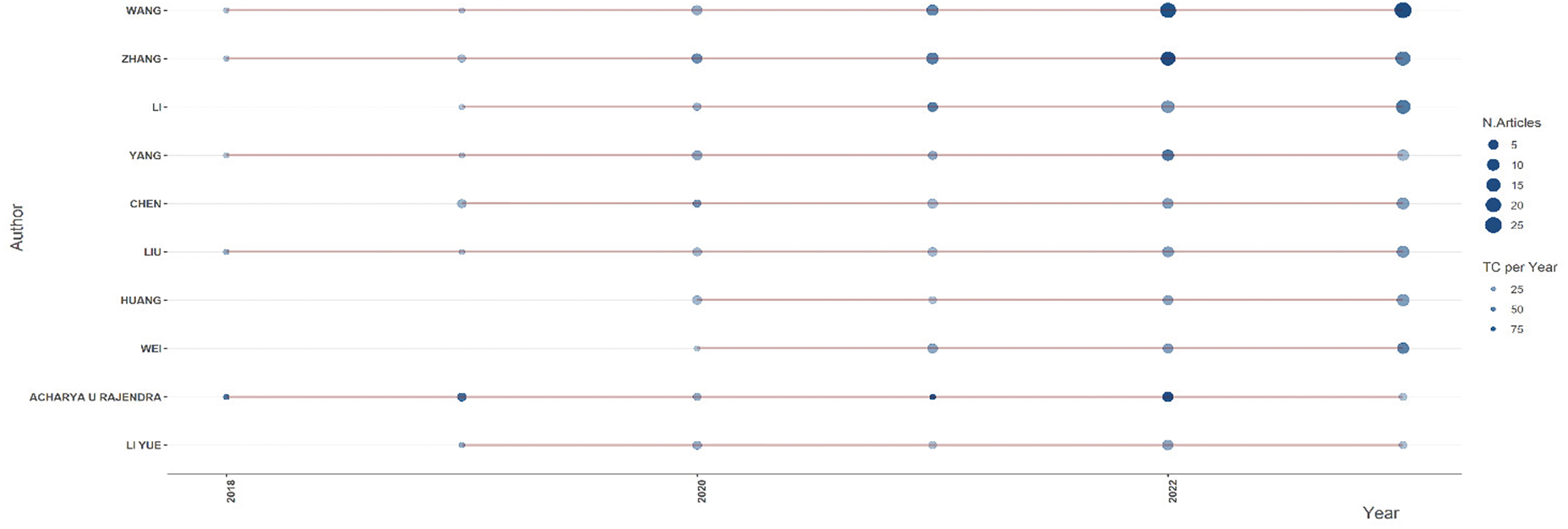
Figure 4: Research productivity and citation impact of authors
The analysis reveals several key observations regarding research productivity and scholarly impact. Authors such as Wang, Zhang, Liu, and Li have demonstrated consistent research output over multiple years, with a noticeable peak in 2022. This indicates their sustained contribution to the field and the increasing volume of their work. The most productive years in terms of the number of published articles appear to be 2022 and 2023, as reflected by the more prominent and darker circles in the visualization. This suggests a surge in research activity during these years, possibly driven by advancements in the field or increased research funding and collaboration. Notably, specific authors, such as Acharya U Rajendra, have comparatively fewer publications but have received significant citations in particular years. This highlights the impact of selected high-quality publications garnering considerable scholarly attention. Overall, the general trend suggests increased research productivity and impact over time, with many authors publishing more frequently in recent years. The findings underscore the growing interest in and expansion of research in this domain, as evidenced by the upward trajectory in both publication count and citation impact.
2.5.3 Journal and Source Analysis
Analyzing publication sources provides valuable insight into the dissemination of research within the field. Identifying the most relevant journals and conference proceedings helps us understand the primary venues for publishing influential studies. Fig. 5 shows the most appropriate sources of publications. The dataset highlights the key sources contributing to the body of knowledge in this domain. The most productive journal is IEEE Access, with 301 publications, demonstrating its role as a leading outlet for research dissemination. Sensors and remote sensing have published 122 and 115 articles, respectively, indicating the strong presence of sensor-related applications in the field. Other significant sources include Biomedical Signal Processing and Control (106 articles), Applied Sciences-Basel (91 articles), and Multimedia Tools and Applications (80 articles), emphasizing the interdisciplinary nature of the research. Additionally, journals such as IEEE Transactions on Geoscience and Remote Sensing (79 articles), Electronics (70 articles), Neurocomputing (60 articles), and IEEE Journal of Biomedical and Health Informatics (48 articles) also contributed significantly to the field. The presence of IEEE journals and high-impact computing journals suggest that the research spans multiple domains, integrating biomedical engineering, signal processing, artificial intelligence, and remote sensing. This distribution of publications across various sources reflects the interdisciplinary growth of the field and underscores the importance of computational and sensor-based methodologies in advancing research. The increasing presence of articles in high-impact journals indicates the field’s maturation and expanding recognition in the broader scientific community.
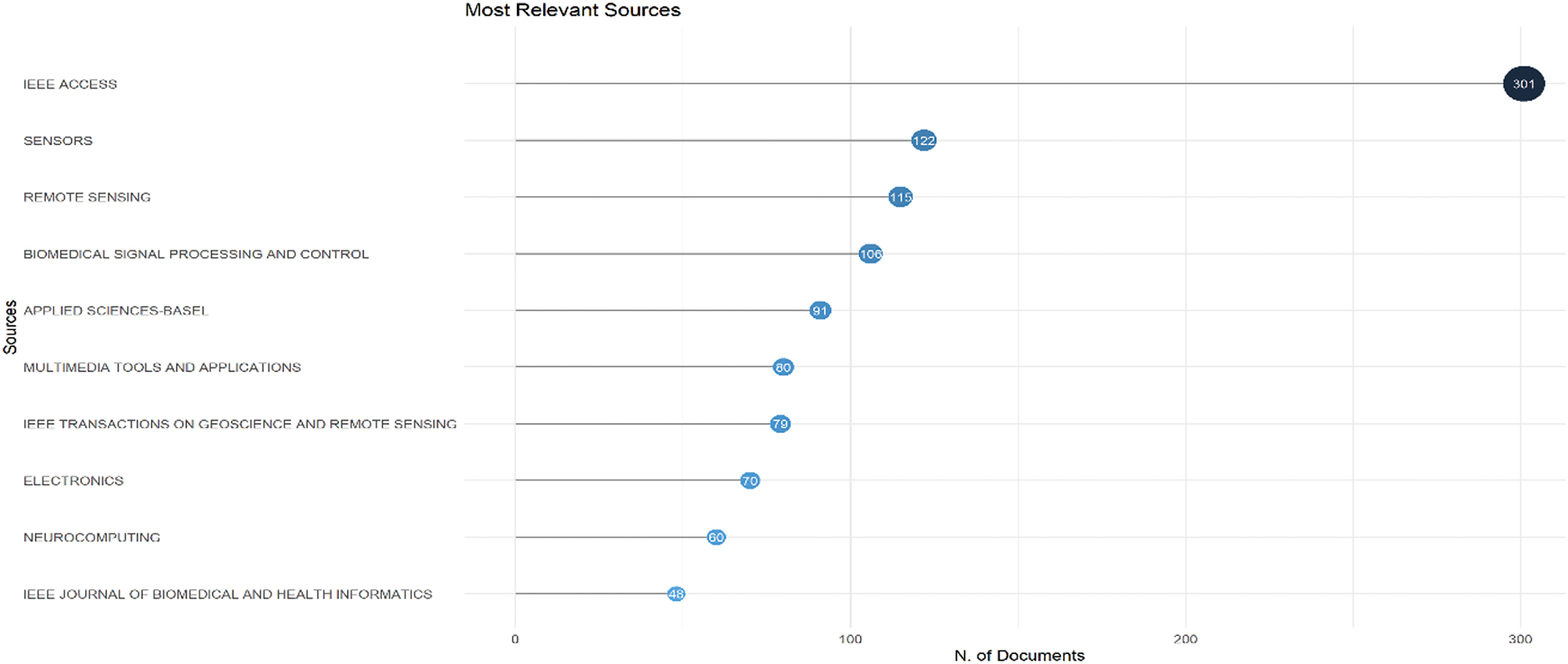
Figure 5: Most relevant sources based on the number of documents published
2.5.4 Country Production Analysis
This section examines countries’ contributions in terms of research citations and publication output, derived from analyzing two datasets: Most Cited Countries and Corresponding Author’s Countries.
• Citation Analysis
The citation dominance of countries is illustrated in the Most Cited Countries chart (scale: 0–40,000 citations). China leads with the highest citation count, occupying the full scale of the chart (≈40,000 citations). The United States (USA), India, South Korea, and Australia follow in descending order, forming the top five most cited countries. Turkey, the United Kingdom, Pakistan, Germany, and Singapore are also prominent, although their citation volumes are lower than those of China. This trend underscores China’s significant influence on global research. Fig. 6 shows the citation analysis per country for the most cited countries.
• Publication Output Analysis
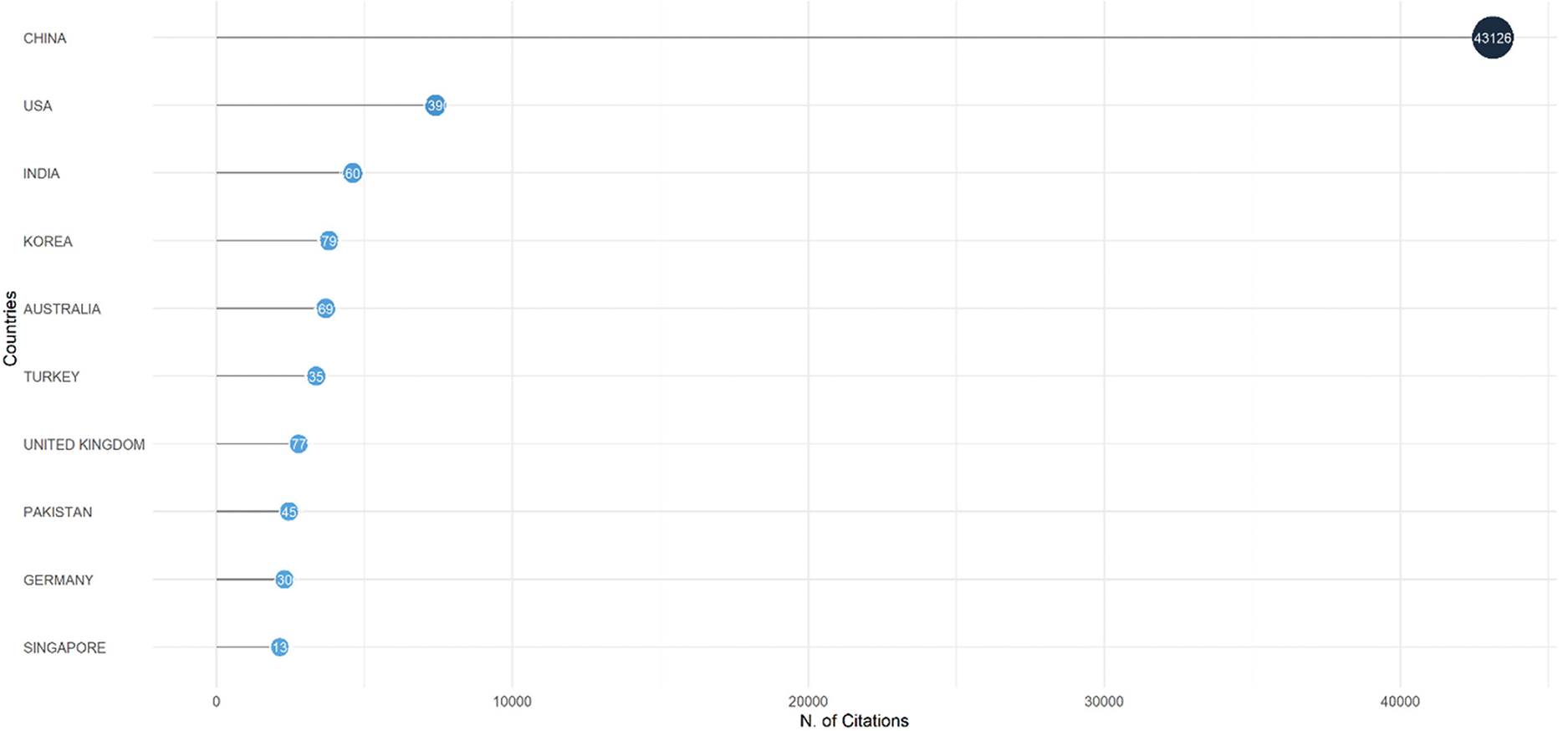
Figure 6: Citation analysis per country for the most cited countries
The Corresponding Author’s Countries dataset highlights the number of documents (scale: 0–2000) contributed by countries, categorized into single-country publications (SCPs) and multiple-country publications (MCPs). China again dominates, producing approximately 2000 documents, followed by India, the USA, South Korea, and Turkey. Countries like Iran, Italy, Saudi Arabia, and Japan appear in the publication rankings. Nevertheless, they are absent from the top-cited list, suggesting a disparity between publication volume and citation impact. European nations such as Germany, Spain, France, Canada, and Brazil contribute moderately. Fig. 7 shows the publication analysis per country.
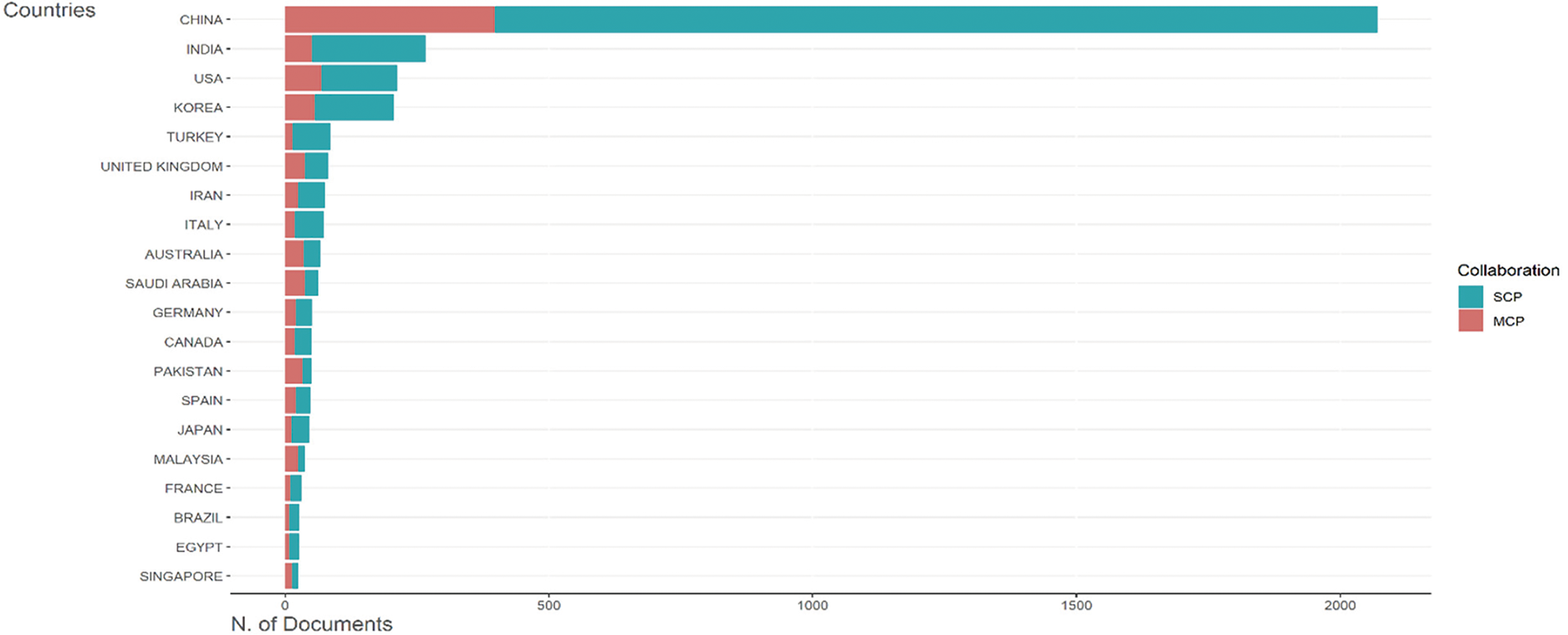
Figure 7: Publication analysis per country
The analysis reveals a stark geographic concentration of research influence. China has emerged as the undisputed leader in citations and publications, reflecting its strategic investments in research and development (R&D), vast academic workforce, and integration into global networks. By contrast, established research hubs such as the USA and Germany maintain strong citation impact despite lower publication volumes relative to China, underscoring their ability to produce high-quality, influential work. Emerging economies, including India, Turkey, and Pakistan, demonstrate growing contributions to global scholarship but face challenges in translating output into citations. This disparity may stem from limited international collaboration, underrepresentation in high-impact journals, or focusing on regionally relevant rather than globally competitive research. Moreover, although their publications are prolific, mid-tier contributors such as Saudi Arabia and Iran lack commensurate citation traction, signaling potential gaps in research visibility or alignment with global priorities. Finally, the dominance of single-country publications (SCPs) across most nations suggests a persistent reliance on domestic expertise and funding. While SCPs strengthen local research ecosystems, the limited prevalence of multiple country publications (MCPs), particularly among emerging economies, highlights untapped opportunities for cross-border partnerships. Such collaboration could enhance the impact of citations, diversify research perspectives, and address global challenges more effectively. These trends underscore the need for policies incentivizing international cooperation while addressing systemic barriers to equitable scholarly recognition.
3 From Traditional Machine Learning to Deep Learning
Machine learning (ML) has become popular in various fields, including biomedical signal processing. Traditional ML methods, such as K-nearest neighbors [31], random forests [32], and gradient boosting [33], have been used for classification [34], regression [35], and clustering tasks [36]. These algorithms require a set of features to be extracted from the input data and then used to train a model. However, the feature engineering process can be time-consuming and requires domain expertise [37,38].
On the other hand, significant attention has been given to DL and its ability to learn features from raw data automatically [39]. DL models, such as CNNs [40], RNNs [41], and deep belief networks (DBNs) [42], have achieved state-of-the-art performance on various tasks, including image and speech recognition, natural language processing (NLP), and biomedical signal processing [39,43].
However, even with the successes achieved by traditional ML methods, the dependency on manual feature engineering has several limitations, particularly in the context of scalability and adaptability [43], and it is also prone to human bias and error [39,43]. This slows the development cycle and limits the model’s generalization ability to new or unforeseen data. In rapidly evolving fields such as biomedical signal processing, where new data types and patterns frequently emerge, the rigidity of traditional feature engineering can hinder the continuous improvement of ML models [3,44,45].
DL addresses these challenges by utilizing neural networks capable of learning directly from raw data, thus automating the feature extraction process. For example, they excel in capturing spatial hierarchies in image data and handling temporal dependencies in sequential data. By removing the need for manual feature engineering, DL models can leverage large datasets to learn more abstract and complex features, leading to improved performance and robustness [46,47].
This shift accelerates the model development process and enhances the potential for discovering novel patterns and insights that human engineers might overlook. Consequently, the transition to DL represents a significant advancement in the field, facilitating more efficient and accurate analysis across diverse applications [43].
Overall, DL has revolutionized the field of AI and has shown significant promise in biomedical applications. With ongoing advancements in this technology and methods, DL is expected to continue to play an essential role in biomedical research and clinical practice. Fig. 8 shows the difference in data flow between ML and deep learning.
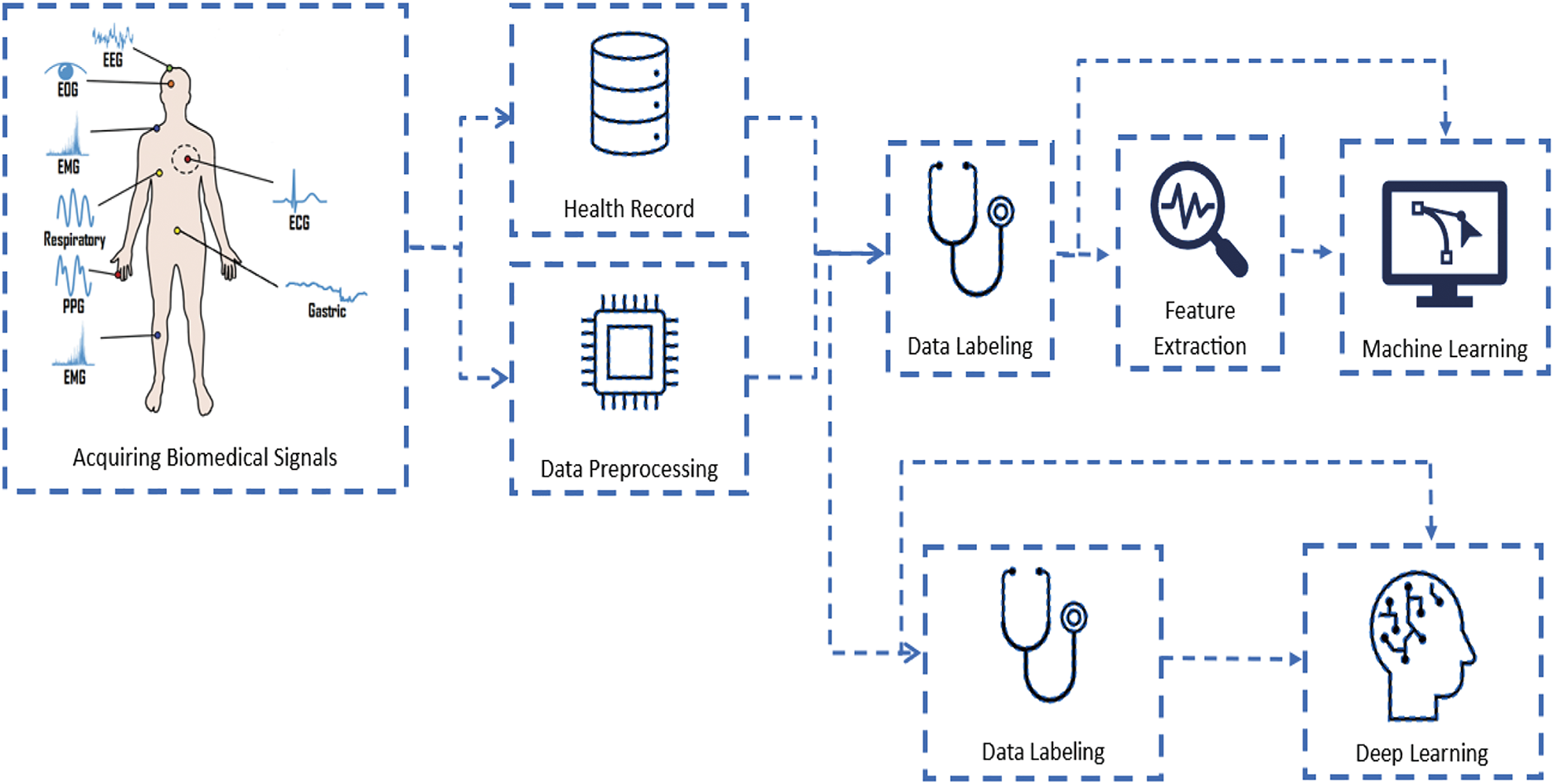
Figure 8: Flow diagram of data from patients to the ML and DL models
ML is a rapidly evolving field that has gained significant attention in recent years because of its ability to learn patterns and make predictions automatically from data. It involves the development of algorithms that can learn from data and improve their performance over time. ML algorithms have been applied in various applications, including image and speech recognition, NLP, and biomedical signal processing [48].
ML has been widely used in biomedical signal processing for classification, regression, and clustering tasks. For example, ML algorithms have been used to classify electroencephalography (EEG) signals into different stages of sleep, predict the development of Alzheimer’s disease via magnetic resonance imaging (MRI) data, and detect abnormal heartbeats in electrocardiography (ECG) signals [49].
Several types of ML algorithms exist, including supervised learning [50], unsupervised learning [51], and reinforcement learning [52]. Supervised learning involves training a model with labeled data, whereas unsupervised learning consists in discovering patterns in unlabeled data. Reinforcement learning involves training a model to make decisions based on a reward system [48,49].
ML algorithms have also been used in biomedical applications, including drug discovery, personalized medicine, and clinical decision support systems. For example, ML algorithms have been used to predict drug interactions and side effects, personalize drug dosages based on patient characteristics, and predict treatment outcomes for various diseases [49]. Fig. 9 shows the block diagram for traditional online and offline detection via ML for biomedical signals.
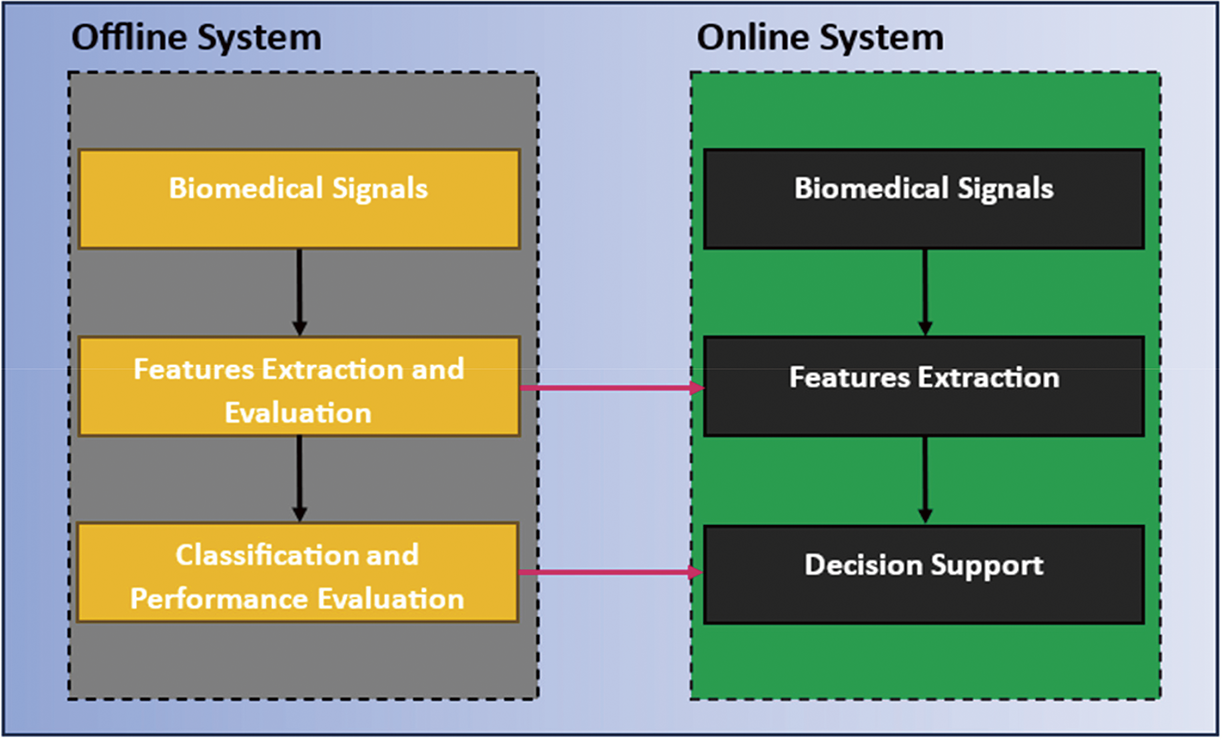
Figure 9: Block diagram of the traditional prediction system based on biomedical signals
Despite the success of ML, several challenges remain. ML algorithms can suffer from overfitting when the utilized model is too complicated [50]. This leads to the learned parameters of the training data not being generalized to the blind testing data. Moreover, the interpretability of complex models can be challenging, as they usually involve too many computations and layers of abstraction [40,53].
ML has shown significant promise in biomedical signal processing and other biomedical applications. With ongoing advancements in technology and methods, ML is expected to continue to play an essential role in biomedical research and clinical practice [54]. Fig. 10 shows the most popular ML algorithms used for biomedical signals.
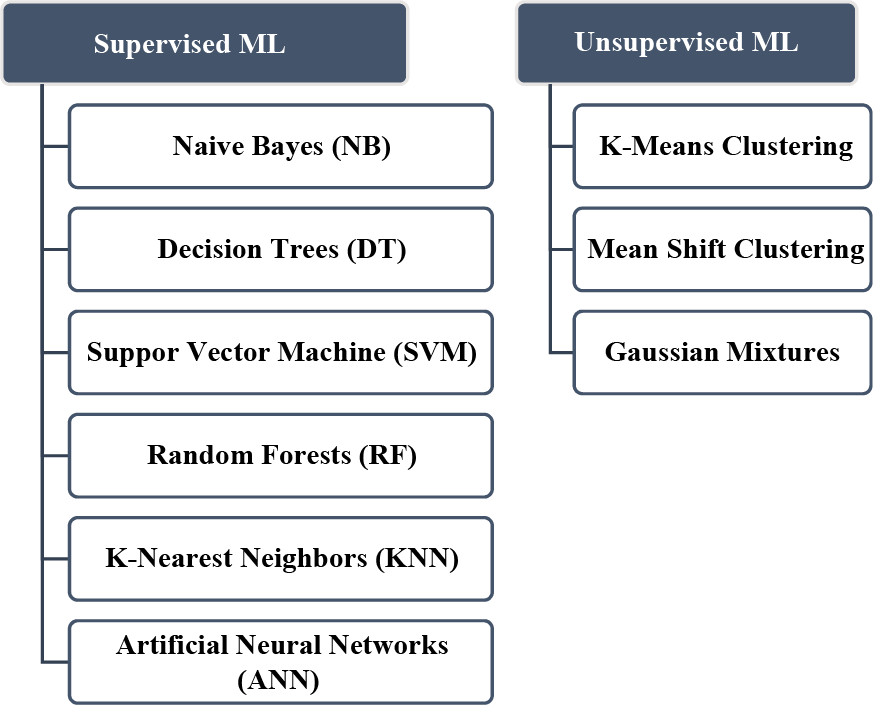
Figure 10: Most popular ML algorithms used in biomedical signals
DL has demonstrated superior capabilities in handling complex and unstructured data. The following subsections discuss the most common DL algorithms.
3.2.1 Convolution Neural Networks (CNNs)
CNNs are a type of DL algorithm that has become increasingly popular for image and signal processing applications. CNNs are inspired by the structure and function of the animal visual cortex and are designed to learn hierarchical representations of pictorial and signal data. Unlike traditional ML algorithms, CNNs automatically know features from raw data rather than requiring manual feature extraction [14,55]. CNNs have been applied to various signal-processing tasks, including image classification, object detection, speech recognition, and biomedical signal processing. In the context of biomedical signal processing, CNNs have been used for EEG analysis, ECG analysis, and MRI analysis [44].
One critical advantage of CNNs is their ability to learn how to extract deep features from data automatically. This can be useful mainly in applications where the signals or features are not well defined or are difficult to detect manually. CNNs are also highly scalable and can be trained on large datasets via parallel processing techniques [1]. Several challenges are associated with training CNNs, including the risk of overfitting and the need for large datasets. Various methods have been developed to address these challenges, such as dropout regularization, data augmentation, and transfer learning [1,40]. In addition to their use in signal processing tasks, CNNs have been applied to various other applications, such as NLP and recommendation systems. DL has achieved state-of-the-art performance in many tasks [40]. DL has emerged as a powerful tool for signal processing tasks, including biomedical signal processing. With ongoing advancements in technology and methods, DL is expected to play an essential role in signal processing and other applications. Fig. 11 shows the general block diagram of CNN.
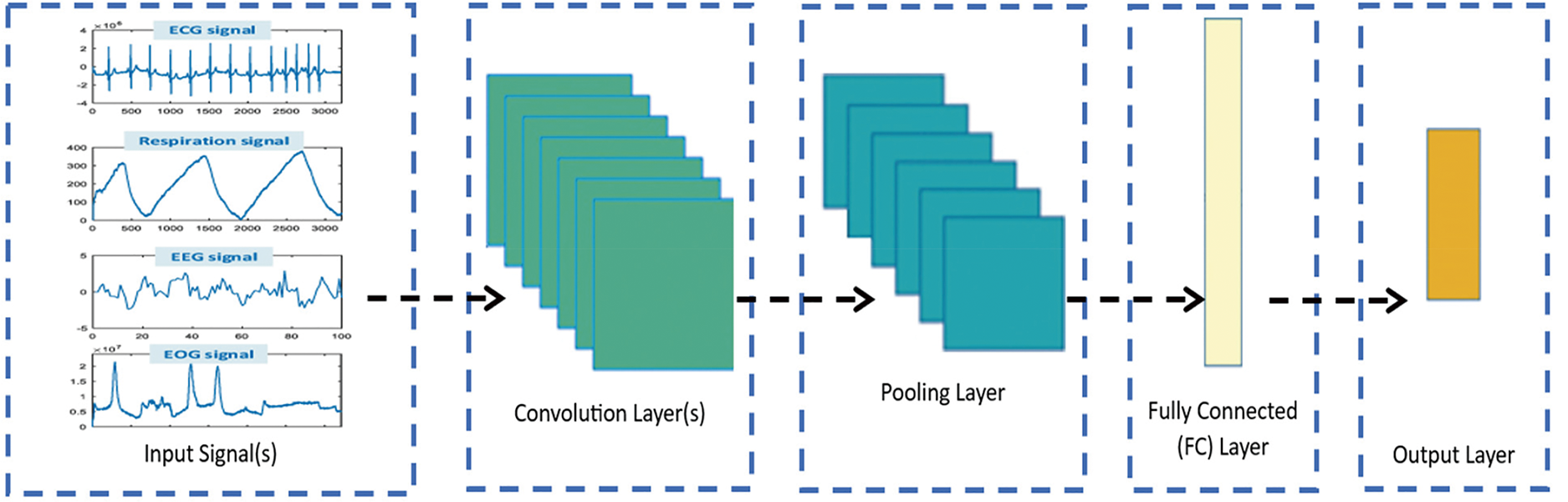
Figure 11: General block diagram of CNN
3.2.2 Autoencoder and Stacked Autoencoder
Autoencoders and stacked autoencoders are powerful DL techniques for biomedical signal-processing tasks. An autoencoder is an unsupervised learning algorithm that can learn how to efficiently represent data via the compressed latent space to reconstruct the data [40,56]. It consists of an encoder network that maps the input data to a lower-dimensional latent representation and a decoder network that reconstructs the original input from the latent representation. In biomedical signals, autoencoders have been used for denoising, dimensionality reduction, and feature extraction [57]. By training an autoencoder on a large set of labeled or unlabeled biomedical signals, it can learn to extract meaningful features that capture essential data characteristics. These learned features can subsequently be used for various downstream tasks, such as classification, anomaly detection, and signal synthesis [39].
Stacked autoencoders, or deep autoencoders or DBNs, are extensions of traditional autoencoders with multiple layers of encoding and decoding units. Stacked autoencoders have shown significant promise in biomedical signal analysis because of their ability to learn complex hierarchical representations. Stacked autoencoders can capture higher-level abstractions and intricate patterns in biomedical signals by adding more layers to the autoencoder architecture. This hierarchical representation learning enables more effective feature extraction, improving performance in various biomedical signal processing applications, such as ECG analysis, EEG decoding, and biomedical image analysis [56,58].
In biomedical signal processing, autoencoders and stacked autoencoders have been widely used. For example, autoencoders have been applied in ECG analysis to denoise signals by learning the underlying noise patterns and reconstructing clean ECG signals. Additionally, stacked autoencoders have been employed for feature extraction in EEG decoding tasks, where hierarchical representations of brain signals that capture discriminative information for classifying different cognitive states or detecting anomalies are known [54]. Moreover, in biomedical image analysis, autoencoders and stacked autoencoders have been utilized for tasks such as feature extraction for disease diagnosis from medical images, image denoising, and image quality through superresolution analysis [58]. Fig. 12 shows the general block diagram of the encoders and decoders.
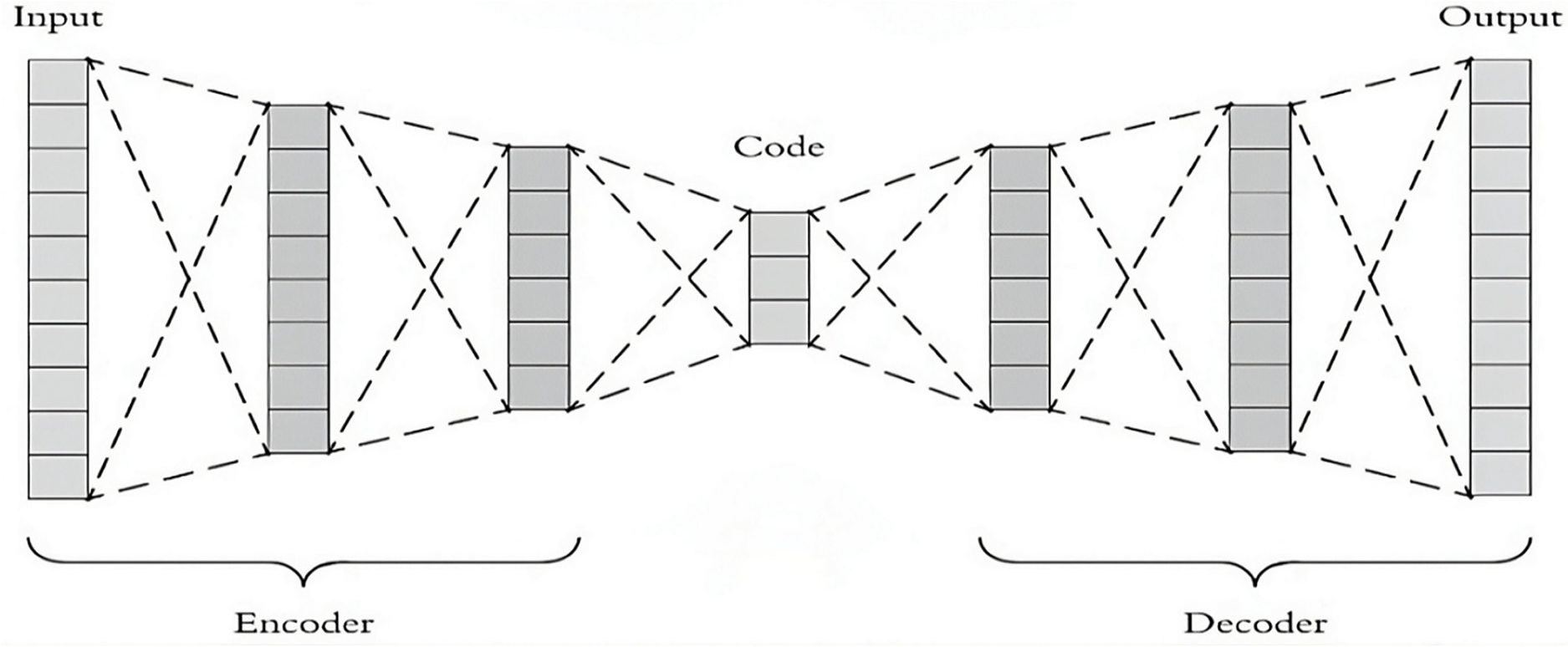
Figure 12: General block diagram of the encoders and decoders
3.2.3 Recurrent Neural Network (RNN)
RNNs have gained significant attention in biomedical signal processing because of their ability to model temporal dependencies and handle sequential data effectively. RNNs are designed to process data sequentially, making them well suited for analyzing biomedical signals, which often exhibit temporal dynamics. RNNs have been successfully applied to various biomedical signal-processing tasks, including ECG, EEG, and speech signal analysis [59,60].
In ECG analysis, RNNs have been used to detect arrhythmias, predict heart rates, and identify abnormalities. By capturing the temporal dependencies in ECG waveforms, RNNs can learn patterns and features crucial for accurately diagnosing and monitoring cardiac conditions. They have shown promising results in detecting various arrhythmias, such as atrial fibrillation, ventricular tachycardia, and heart blocks, contributing to improved clinical decision-making [9,13]. In EEG analysis, RNNs have been applied for brain signal decoding, seizure detection, sleep stage classification, and brain-computer interface (BCI) systems. By modeling the temporal dynamics of EEG signals, RNNs can effectively extract features and patterns that reflect different cognitive states or neurological disorders. They have demonstrated the ability to detect and predict epileptic seizures, classify sleep stages accurately, and enable real-time control in BCI applications [13].
Additionally, RNNs, particularly those with LSTM and GRU components, have been employed for Type-2 diabetes prediction via genomic and tabular data, achieving high performance and showing their effectiveness in predicting chronic diseases from complex datasets [61].
RNNs have also been employed in speech signal analysis for voice recognition, speaker identification, and emotion recognition applications. By considering the temporal context of speech signals, RNNs can capture essential features related to speech dynamics and phonetic patterns [9,62]. They have significantly improved speech recognition accuracy, enabling better speech-based applications in biomedical contexts. Fig. 13 illustrates the distinct architectures of three critical RNN types: the standard RNN, the long short-term memory (LSTM), and the gated recurrent unit (GRU). All three sequential data processes differ in their internal structures and how they handle information flow within the network.
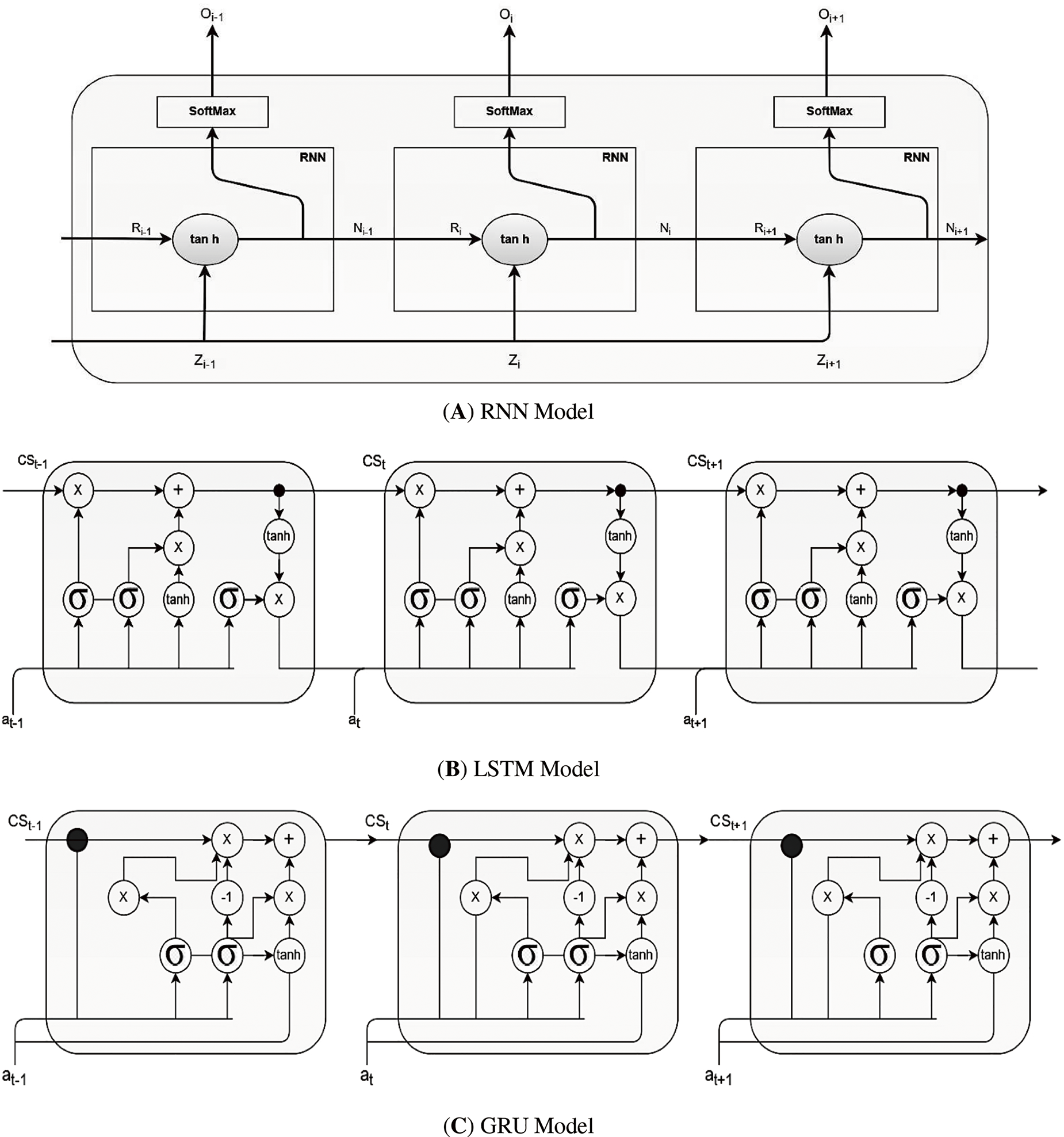
Figure 13: Different types of RNNs
Moreover, when processing sequential biomedical signals such as ECG or EEG data, LSTM networks maintain a cell state that runs through the entire chain of the network, allowing information to persist across time steps. The LSTM cell contains three gates: the forget gate, which determines what data to discard from the cell state; the input gate, which determines what new information to store; and the output gate, which determines what information will be passed to the next step. Fig. 13B illustrates the internal architecture of an LSTM cell, showing how these gates interact to maintain and update the cell state over time. This architecture enables LSTMs to capture long-term dependencies in sequential data, making them particularly effective for arrhythmia detection in ECG signals where temporal patterns are crucial for accurate classification.
The basic RNN unit consists of a single layer, usually an
• Standard RNN
A basic RNN unit consists of a single layer, usually an
where
Despite their simplicity, standard RNNs suffer from the vanishing gradient problem when modeling long-term dependencies. This makes it challenging to retain information over longer sequences.
• Long short-term memory (LSTM)
To mitigate the vanishing gradient problem, LSTMs introduce a cell state and gating mechanisms that regulate the flow of information. An LSTM cell typically has three gates, forget, input, and output gates, which allow the model to retain and utilize information from past inputs effectively [59,60]. The key equations for an LSTM cell are as follows:
where
LSTMs are particularly effective at capturing long-term dependencies in biomedical signals, such as extended ECG waveforms or EEG recordings, by controlling how much past information to keep or forget.
• Gated Recurrent Unit (GRU)
GRUs offer a simplified alternative to LSTMs, using only two gates: update gates and reset gates. These gates regulate the information flow between the previous hidden state and the current candidate state, allowing GRUs to learn long-term dependencies with fewer parameters than LSTMs do [59,60]. The GRU equations are as follows:
where
GRUs often achieve comparable performance to LSTMs on many tasks while being computationally more efficient, making them suitable for real-time biomedical signal analysis and mobile health applications.
3.2.4 Generative Adversarial Network (GAN)
Generative adversarial networks (GANs) have emerged as robust frameworks for generating synthetic data that resemble accurate biomedical signals. GANs consist of two neural networks, a generator and a discriminator, which are trained adversarially. The generator learns to generate synthetic biomedical signals, whereas the discriminator learns to distinguish between accurate and generated signals. GANs have shown promising applications in various biomedical signal-processing tasks, including data augmentation, anomaly detection, and signal synthesis [63]. A GAN consists of three main components or processes:
• Generator
The generator in a GAN architecture serves as the synthetic data producer. It typically takes random noise as input, often samples from a uniform or normal distribution, and transforms this noise into structured data resembling real-world examples [63]. The architecture generally consists of layers designed to process and refine this noise, which may include recurrent layers for capturing temporal patterns, followed by fully connected layers that apply nonlinear activations to shape the output gradually [63]. Regularization techniques such as dropout are commonly employed to prevent overfitting and ensure diversity in the generated outputs. The final layers produce synthetic data, which aims to mimic the statistical properties and characteristics of accurate biomedical signals such as ECG or EEG waveforms [28,64].
• Discriminator
The discriminator functions as the quality assessor in the GAN framework. Its architecture is designed to analyze input data and determine their authenticity. This typically involves layers that extract relevant features from the input, which may include recurrent or convolutional layers depending on the data type, followed by fully connected layers with appropriate activation functions [28,64]. The final output layer usually produces a probability score indicating whether the input is real (from the actual dataset) or synthetic (generated by the generator). The discriminator’s role is crucial, as it provides the feedback mechanism that guides the generator’s learning process [64,65].
• Adversarial training
The core innovation of GANs lies in their adversarial training process. During training, the generator and discriminator engage in a continuous competition where the generator attempts to produce increasingly realistic synthetic data. In contrast, the discriminator strives to better distinguish real from fake data. This dynamic is formalized as a minimax game where the generator minimizes the probability of detection while the discriminator maximizes its classification accuracy [64,65]. Through multiple iterations of this competition, both networks improve iteratively, ultimately reaching an equilibrium where the generator can produce data that are virtually indistinguishable from real examples, and the discriminator cannot reliably differentiate between them [64,65]. The objective function for GANs is as follows:
where:
•
• where
• where
•
A critical application of GANs in biomedical signals is data augmentation. GANs can generate synthetic signals that augment the limited available data, thereby improving the performance and robustness of ML models. By training the generator to mimic the statistical characteristics of natural signals, GANs can generate realistic synthetic data that expand the training dataset, leading to enhanced model generalizability and performance [28,64].
Another application of GANs in biomedical signals is anomaly detection. GANs can be trained on standard, healthy signals to learn their underlying distribution. The discriminator in the GAN is then used as an anomaly detector to identify deviations from the learned normal distribution. This approach has been successfully applied to detect anomalies in various biomedical signals, such as ECGs, electromyograms (EMGs), and EEGs, enabling early detection of abnormalities and improving patient monitoring [64,65].
Furthermore, GANs have been used for signal synthesis in biomedical applications. For example, GANs have been employed to generate synthetic ECG signals that mimic different cardiac conditions, allowing researchers to study and analyze the effects of specific abnormalities without the need for large, diverse clinical datasets [11,65,66]. Synthetic signals generated by GANs can also be used for training and testing DL models, overcoming the challenges of limited or imbalanced datasets. Fig. 14 shows a general block diagram of the GAN.
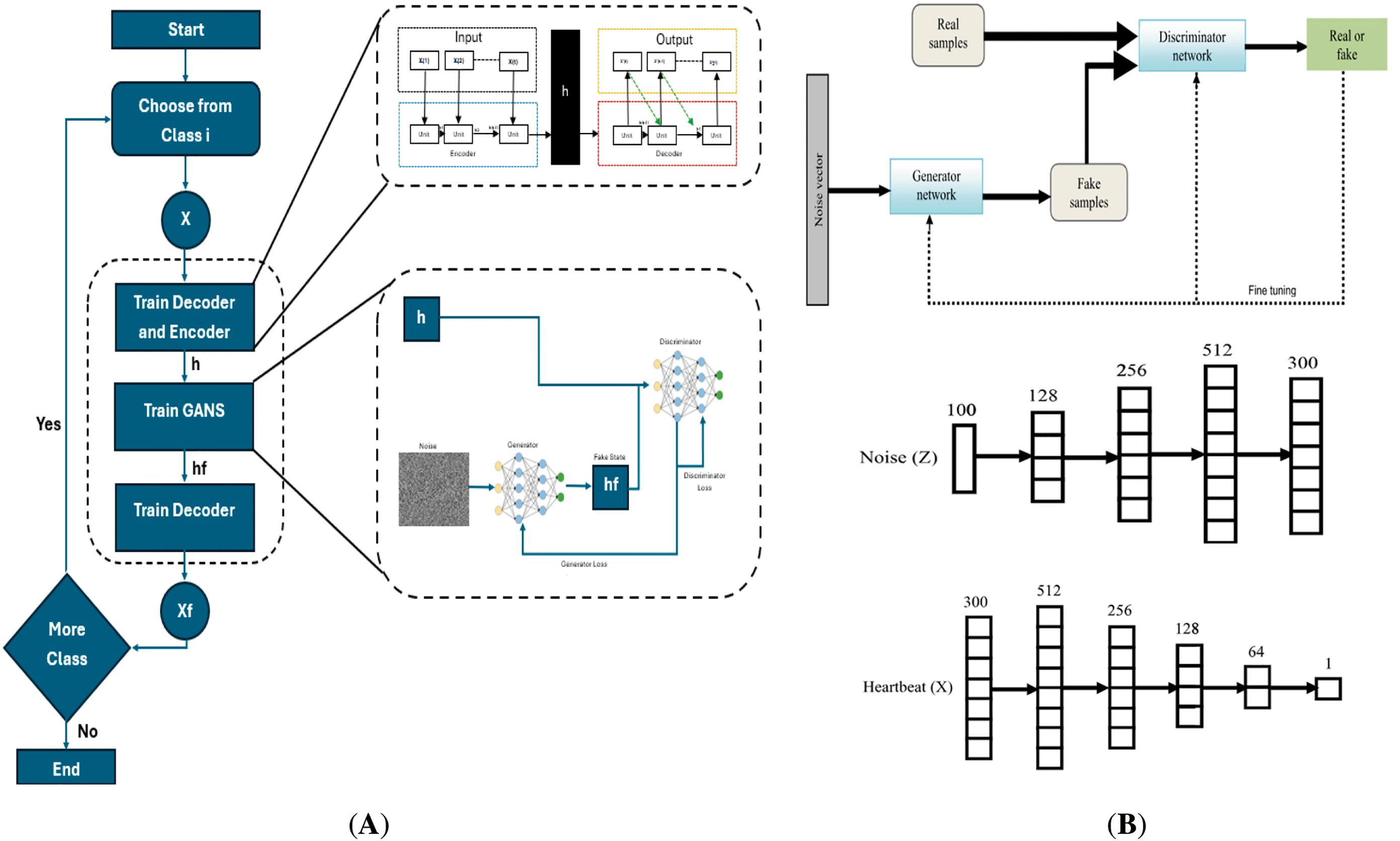
Figure 14: (A) Block diagram of a GAN in real-example, (B) Sample architecture for generator and discriminator networks [67] (CC BY 4.0)
4 DL Applications in Biomedical Signals
DL has revolutionized the field of biomedical signals, offering powerful tools for analyzing and interpreting complex biomedical data. DL models, such as CNNs, RNNs, and GANs, have been extensively applied in various biomedical signal processing and interpretation tasks, including disease diagnosis, anomaly detection, signal classification, and signal synthesis [9,68]. Owing to its ability to efficiently learn representations from complex and unstructured data, DL has remarkably succeeded in extracting meaningful patterns and features from diverse biomedical signals. This section explores different and recent applications of DL in biomedical signals. The following section discusses these topics in detail. Fig. 15 shows the applications of DL in biomedical signals.
Figure 15: DL applications for biomedical signals
4.1 Diagnostic and Disease Detection
This category uses DL algorithms to analyze biomedical signals and diagnose various medical conditions. It includes applications such as ECG arrhythmia classification [2,69], EEG-based seizure detection [10], and speech and voice analysis for diagnosing disorders.
CNNs have been used to analyze ECGs and accurately identify cardiac arrhythmias. RNNs effectively decoded EEG signals to detect epileptic seizures. These models learn to capture intricate patterns and features from signals, enabling accurate disease identification and aiding in early diagnosis and treatment [2,5,25,28,70].
Anomaly detection is another critical area in which DL has made significant contributions. Training DL models on standard, healthy signals, they can learn and identify deviations from the underlying distribution [10,13,71–74]. This approach has been applied to detect anomalies in various biomedical signals, such as detecting abnormal heart rhythms from ECGs or identifying anomalies in brain signals from EEGs. Deep learning-based anomaly detection can enhance patient monitoring systems and enable early detection of critical events [2,5,12,29,74–76].
Signal classification is a fundamental task in biomedical signal processing, and DL models have achieved state-of-the-art performance in this area [40]. CNNs, such as short-time Fourier transform (STFT) and discrete wavelet transform (DWT), have been employed to classify images from biomedical signals [77,78]. To detect specific activities or states, RNNs, such as EEGs or EMGs, have been applied to classify time series signals. DL models can automatically learn discriminative features from signals, enabling accurate and efficient classification [56]. Fig. 16 shows how DL uses inputs from biomedical signals as images.
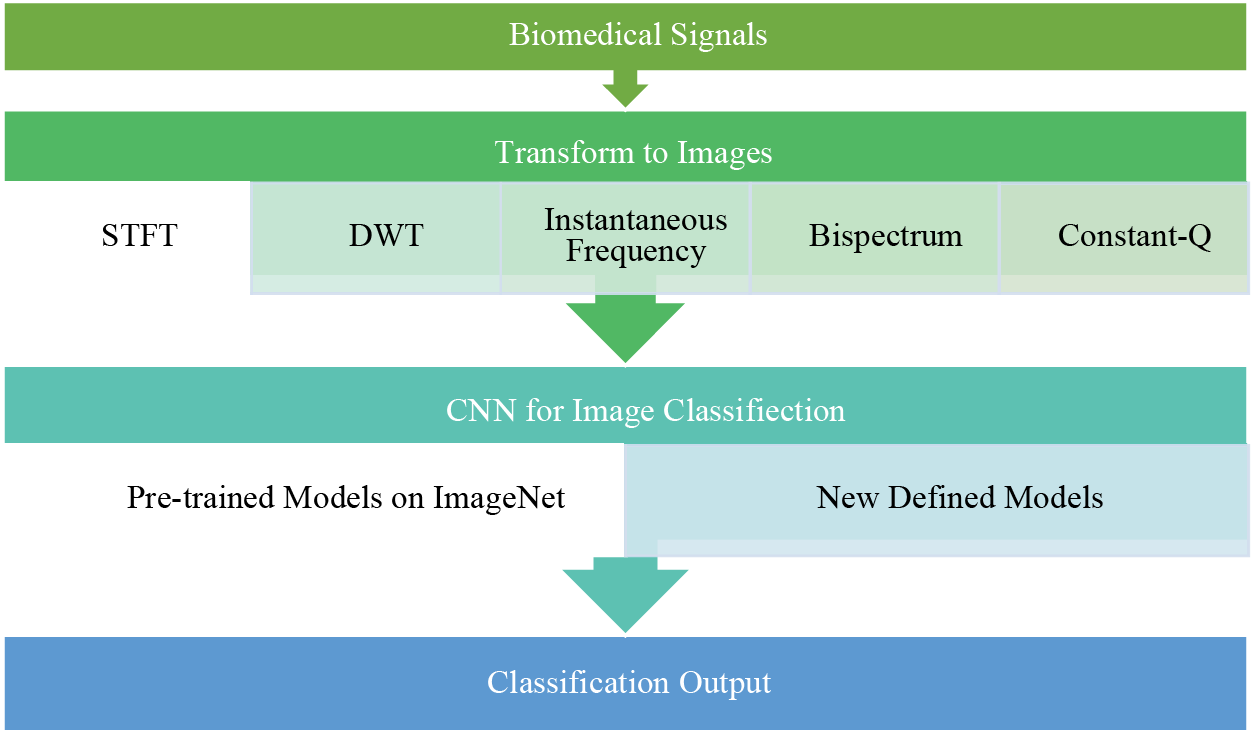
Figure 16: Block diagram using a CNN with image inputs from biomedical signals
4.2 Health Monitoring and Predictive Analytics
DL has shown great potential in using biomedical signals to monitor individuals’ health and predict potential health risks continuously. These signals, including HRV, blood pressure, and glucose levels, offer valuable insights into a person’s physiological state. DL models, such as RNNs and LSTM networks, have been used to analyze temporal patterns in these signals and predict future health events [41,79]. For example, an LSTM-based model effectively predicts hypoglycemic events in patients with type 1 diabetes, providing an early warning system to prevent life-threatening situations [80].
In addition to health monitoring, DL has also been applied to predict disease outcomes and identify potential risk factors based on analyses of longitudinal biomedical data. DL models can identify hidden patterns and complex relationships between signals and patient outcomes by training on large-scale patient data. For example, reference [41] proposed a deep learning-based framework for the early prediction of heart failure-related hospitalization, utilizing electronic health records and physiological signals to achieve high accuracy in forecasting adverse events.
This is a transformative application area for DL in biomedical signals. By harnessing the power of deep neural networks, healthcare professionals can gain valuable insights from continuous health monitoring and make informed decisions for personalized patient care, early disease detection, and risk prediction.
4.3 Biomedical Signal Processing and Enhancement
DL has shown significant potential in enhancing the quality and utility of biomedical signals, which are often corrupted by noise and artifacts that can hinder accurate diagnosis and analysis [81,82]. Deep neural networks have been used to denoise and enhance these signals, improving their fidelity and reliability. For example, a deep learning-based approach using a stacked autoencoder was proposed to effectively denoise electroencephalogram (EEG) signals, reduce artifacts, and improve the accuracy of neurological disorder diagnosis [81]. This represents a critical application area for DL in biomedical signals, where researchers and medical professionals can leverage the capabilities of deep neural networks to effectively denoise, enhance, and augment these signals for improved diagnosis and analysis.
4.4 Data Integration and Multimodal Analysis
To gain comprehensive insights into biomedical signals, DL techniques have shown remarkable potential for combining and analyzing data from multiple sources and modalities. Biomedical research often involves data from various sensors and imaging modalities, and integrating these heterogeneous data can provide a more holistic understanding of complex physiological processes [83,84]. DL models, such as multimodal neural networks and attention mechanisms, have been developed to effectively fuse data from different sources. For example, a multimodal DL framework was proposed that integrated data from ECGs and PPGs to improve the accuracy of heart rate estimation and cardiovascular disease diagnosis [85,86].
DL has also been instrumental in multimodal analysis, combining information from various sources to extract complementary features and facilitate better disease detection and diagnosis. A deep learning-based multimodal analysis approach was developed to diagnose Alzheimer’s disease via structural and functional MRI data, achieving superior performance compared with single-modal analysis [83]. This is an essential application area for DL in biomedical signals, where researchers and clinicians can uncover valuable insights and improve disease detection and diagnosis accuracy and efficiency by integrating data from diverse sources and applying advanced DL techniques.
4.5 Drug Discovery and Therapeutics
This is a rapidly growing field, and DL has been used to accelerate the identification of potential drug candidates and optimize therapeutic strategies. DL techniques have shown great promise in predicting the binding affinity between small molecules and target proteins, which is a crucial step in the design of new drugs [87–89]. CNNs and graph-based DL models have been applied to predict highly accurate protein–ligand interactions. For example, DeepChem, a DL library, was developed to predict the binding affinities of small molecules to protein targets successfully, aiding in virtual screening for potential drug candidates [89].
DL has also significantly optimized drug therapies by analyzing patient data and predicting treatment responses. RNNs and transformer-based models have been applied to electronic health records and genomic data to personalize drug treatment plans and anticipate adverse drug reactions. A DL model was developed that utilized electronic health records to predict patient-specific adverse events, enabling physicians to make informed decisions and reduce the likelihood of harmful drug reactions [90].
This area represents an application for DL in biomedical signals. By using deep neural networks to predict protein–ligand interactions and personalize drug treatments, researchers and clinicians can significantly expedite the drug discovery process and improve patient outcomes in therapeutics.
4.6 Real-Time Monitoring and Decision Support
DL has shown immense potential in providing continuous, real-time analysis of biomedical signals to support clinical decision-making. DL models have been employed to process and analyze streaming data from various sensors and devices, enabling real-time monitoring of patients’ health status and facilitating timely interventions [39,91,92]. For example, deep CNNs have been applied to analyze data from wearable devices, such as smartwatches and fitness trackers, to detect and predict abnormal physiological patterns [92,93].
DL has also provided real-time decision support for critical care settings. By analyzing streaming data from patient monitors and electronic health records, DL models can detect early signs of deterioration and predict adverse events, allowing medical teams to take immediate action. An attention-based DL model was developed for real-time sepsis prediction, achieving high accuracy and sensitivity in identifying patients at risk of septic shock [94].
This topic represents a revolutionary application area for DL in biomedical signals. By leveraging the capabilities of deep neural networks for real-time analysis and prediction, healthcare professionals can receive timely and accurate decision support, leading to improved patient outcomes and more effective healthcare interventions.
4.7 Explainable AI and Interpretability
This is one of the most crucial aspects of applying DL to biomedical signals, where understanding and interpreting model predictions is essential for gaining trust and acceptance from medical professionals. Owing to their complex architecture, DL models are usually considered black boxes and do not understand how decision-making is performed. However, interpretability is vital in biomedical applications to comprehend the rationale behind a model’s predictions and ensure that its recommendations align with medical expertise [95,96].
Recent research has focused on developing explainable DL models that provide insights into the decision-making process. For example, local interpretable model-agnostic explanations (LIMEs) were introduced to explain individual predictions of black-box models such as deep neural networks. Locally explainable linear explanations (LELEs) generate locally interpretable explanations by approximating complex model behavior around a specific instance, allowing medical practitioners to understand why a particular decision was made [97].
Interpretability is especially crucial when deploying DL models for real-world clinical applications. Medical professionals must trust and validate predictions to ensure patient safety and appropriate treatment plans. Researchers have explored methods to make DL models more interpretable by incorporating attention mechanisms and highlighting specific regions in the input data that influence the model’s output. An attention-based DL model was proposed for arrhythmia detection in ECGs, allowing physicians to understand which parts of the ECG signal were most significant in making the diagnosis [96].
Explainable AI is a crucial area of research when applying DL to biomedical signals. Developing methods and techniques that provide transparent and interpretable insights into DL models’ decision-making processes is essential for fostering trust, facilitating validation, and enabling the safe and effective use of DL in real-world clinical settings.
5 Performance Metrics for DL in Biomedical Signals
Assessing the performance of DL models applied to biomedical signals is crucial for evaluating their effectiveness and ensuring their successful application in clinical and research settings [40]. Various performance metrics are commonly used to measure these models’ accuracy, robustness, and generalizability. In this section, we discuss some of the most critical performance metrics and their significance in evaluating the performance of DL models for biomedical signals [38]. These metrics are accuracy, sensitivity (recall), specificity, the F1 score, and the area under the receiver operating characteristic curve (AUC-ROC). The following subsection discusses these metric equations:
Accuracy is one of the most fundamental performance metrics and represents the proportion of correctly classified samples over the total number of samples in the dataset [98]. It is calculated as follows:
where TP (true positives) is the number of correctly classified positive samples, TN (true negatives) is the number of correctly classified negative samples, FP (false positives) is the number of negative samples misclassified as positive, and FN (false negatives) is the number of positive samples misclassified as unfavorable [98].
5.2 Sensitivity (Recall) and Specificity
Sensitivity, also known as recall, measures the ability of a model to identify positive samples correctly. On the other hand, specificity measures the ability to identify negative samples correctly [98]. They are calculated as follows:
High sensitivity is crucial in applications where the cost of false negatives is high, such as in disease diagnosis, whereas high specificity is essential when false positives have severe consequences [98].
The F1 score is the harmonic meaning of precision and recall (sensitivity). It provides a balanced assessment of the model’s performance by considering false positives and negatives [98]. It is calculated as follows:
Precision is the proportion of actual positive samples out of all the predicted positive samples.
5.4 Area Under the Receiver Operating Characteristic Curve (AUC-ROC)
The ROC curve plots the actual positive rate (sensitivity) against the false positive rate (1-specificity) at various classification thresholds. The AUC–ROC metric represents the area under the ROC curve and quantifies the model’s ability to distinguish between positive and negative samples. AUC-ROC values closer to 1 indicate better discriminatory power [98].
5.5 Matthews Correlation Coefficient
The Matthews correlation coefficient (MCC) is a more informative metric for evaluating classification performance, especially in imbalanced datasets. The MCC considers all four confusion matrix components (TP, TN, FP, FN) and produces a value between −1 and 1, where 1 indicates perfect classification, 0 represents random predictions, and −1 signifies complete misclassification. It is calculated as follows:
Unlike accuracy, the MCC remains reliable even when class distributions are skewed, making it a preferred metric for biomedical classification problems.
Balanced accuracy is helpful in imbalanced datasets where standard accuracy may be misleading. It calculates the average sensitivity and specificity, ensuring that both classes contribute equally to the performance measurement. It is defined as:
Balanced accuracy provides a fair assessment when the dataset has a significant class imbalance, making it particularly valuable for biomedical applications such as disease detection.
5.7 Precision (Positive Predictive Value, PPV)
Precision, also known as positive predictive value (PPV), measures the proportion of correctly predicted positive cases among all predicted positive cases. It is defined as:
Positive results are crucial when false positives are costly, such as in cancer screening, where an incorrect positive result can lead to unnecessary biopsies or treatments.
The false positive rate (FPR) measures the proportion of negative samples that were incorrectly classified as positive:
A lower FPR is essential in high-risk applications, such as medical diagnostics, where incorrectly classifying a healthy patient as diseased can lead to unnecessary interventions.
Cohen’s kappa evaluates classification performance by considering agreement beyond what is expected by chance. It is beneficial when working with imbalanced datasets and multiple raters. It is calculated as:
where
A kappa value 1 indicates perfect agreement, whereas 0 indicates random agreement. Cohen’s kappa is especially useful in multiclass classification problems and when comparing multiple models.
5.10 Youden’s Index (J statistic)
Youden’s index provides a single-value measure of a diagnostic test’s performance by considering both sensitivity and specificity:
It ranges from −1 to 1, where 1 indicates perfect classification and 0 suggests no diagnostic ability. Youden’s index is widely used in medical diagnostics to determine the optimal threshold for classification.
The F2 score is a variation of the F1 score that emphasizes recall (sensitivity) more. It is particularly useful in scenarios where false negatives are more costly than false positives, such as critical disease detection.
A higher F2 score is desirable in healthcare applications, where detecting all potential positive cases is more important than minimizing false positives.
5.12 Why Are These Metrics Important?
The choice of performance metrics depends on the specific task and application of deep learning models in biomedical signals. Accuracy is a widely used metric, but more accuracy is needed in imbalanced datasets where the number of positive and negative samples differs significantly. Sensitivity and specificity are essential in scenarios where the cost of false positives or false negatives is asymmetric [40,98]. The F1 score provides a balanced view of the model’s performance and is useful when there is an uneven class distribution. It is essential in applications such as disease diagnosis, where false positives and negatives can have serious consequences, while the F2 score prioritizes recall, which is helpful for disease detection [36,40]. AUC-ROC is particularly valuable in binary classification tasks, as it remains unaffected by the choice of classification thresholds, making it more robust when dealing with imbalanced datasets [40]. The MCC and Cohen’s kappa also provide robust classification performance measures, especially in imbalanced datasets [36,40]. Balanced accuracy ensures that both classes contribute equally, making it more effective than regular accuracy. Youden’s index is particularly valuable in medical diagnostics, as it helps determine the optimal decision threshold [36,40].
5.13 Selecting the Best Metric for Your Problem
The selection of performance metrics depends heavily on the type of task and the application of DL models to biomedical signals. Accuracy is a widely used metric, but greater accuracy is needed for imbalanced datasets where the number of positive and negative samples differs significantly. Sensitivity and specificity are essential in scenarios where the cost of false positives or negatives is asymmetric [40,98].
The F1 score provides a balanced view of the model’s performance and is useful when there is an uneven class distribution. It is essential in applications such as disease diagnosis, where false positives and negatives can have serious consequences [36,40]. AUC-ROC is particularly valuable in binary classification tasks, as it remains unaffected by the choice of classification thresholds, making it more robust when dealing with imbalanced datasets [40].
For imbalanced datasets, metrics such as the MCC, balanced accuracy, Cohen’s kappa, and Youden’s index offer more reliable evaluations than regular accuracy. In medical applications where missing positive cases are costly, the sensitivity, F2 score, and AUC-ROC are more critical than the overall accuracy [36,40]. Conversely, precision and the false positive rate (FPR) should be minimized in applications where false positives carry high consequences. For general classification problems, the F1 score and MCC provide a balanced assessment of model performance [40].
Selecting appropriate performance metrics for evaluating DL models in biomedical signals is crucial for obtaining meaningful insights into their real-world applicability and performance. Researchers and practitioners should carefully consider the specific requirements of their applications and use a combination of metrics to comprehensively assess the model’s capabilities. By doing so, we can advance the field of DL for biomedical signals and promote the adoption of accurate and reliable models in healthcare and related domains [36,99]. Fig. 17 shows the importance of the performance metrics for binary and multiclass scenarios.
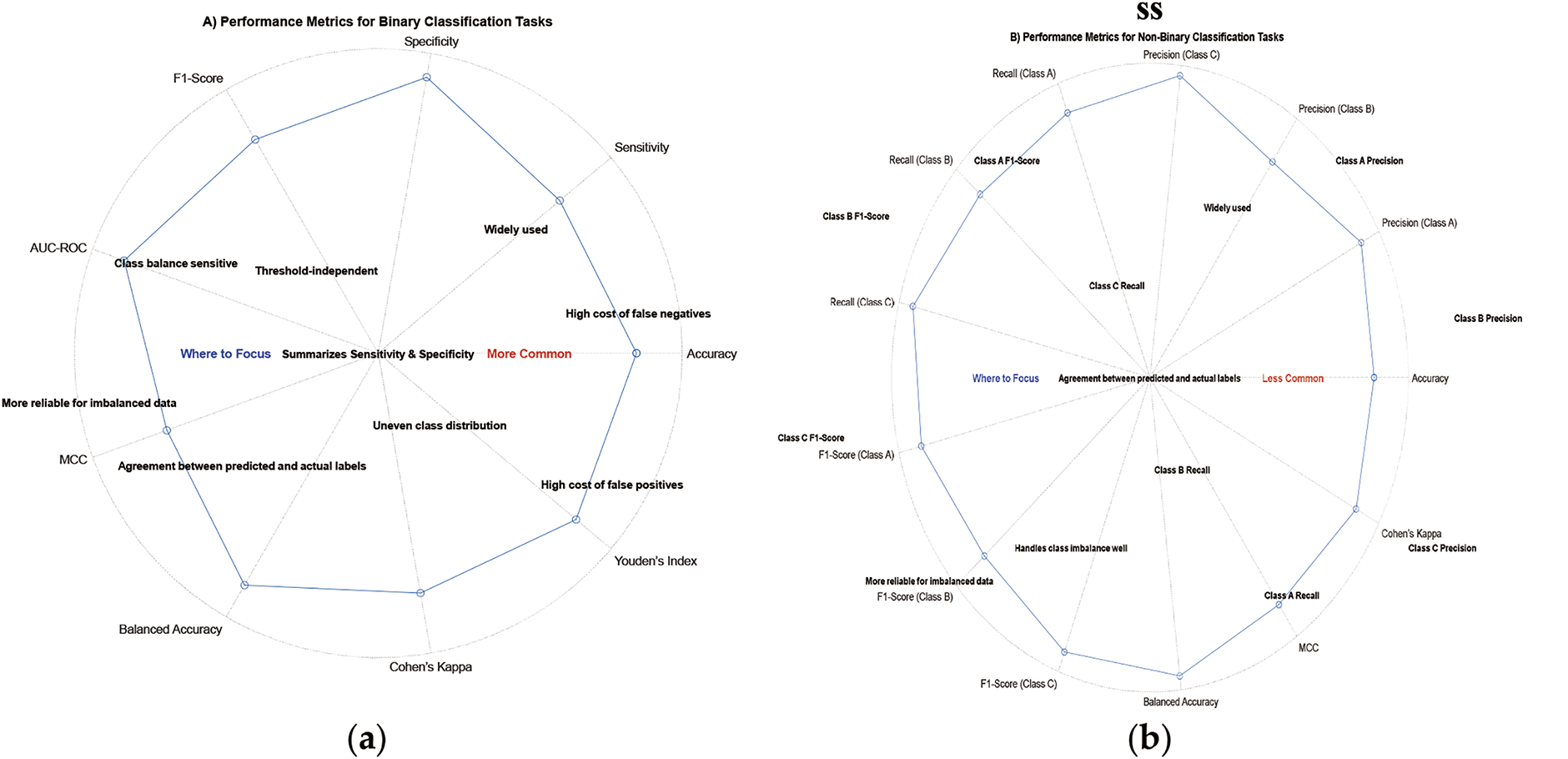
Figure 17: Main metrics for two scenarios: (a) Binary; (b) Multiclass
6 Choosing the Perfect Type of DL for Your Data
DL has emerged as a powerful and versatile approach for analyzing biomedical signals because it can automatically learn complex patterns and representations from raw data. However, with the proliferation of different DL architectures and methodologies, selecting the most appropriate type of DL model for a specific biomedical signal dataset has become crucial. This section discusses various considerations to help researchers and practitioners make informed decisions when choosing the perfect type of DL model for their data.
6.1 Data Characteristics and Scale
The first step in choosing a suitable DL model is thoroughly understanding the data characteristics and scaling. Biomedical signals vary widely, including EEGs for brain activity and ECGs for heart activity. Each type of biomedical signal has a unique data format, temporal or spatial resolution, and noise level [100].
For example, CNNs are well suited for image-like data, such as spectrograms and time-frequency representations of signals, as they effectively capture local patterns and spatial dependencies. On the other hand, RNNs or their variants, such as LSTM networks, are adequate for sequential data such as time series or EEG signals, where temporal dependencies and patterns play crucial roles in understanding the underlying physiology [101].
Additionally, the number of classes in classification tasks is another essential consideration. For multiclass or multilabel classification, models such as CNNs with global pooling layers or transformer-based architectures can be adapted to handle multiple classes effectively.
6.2 Available Data and Annotation
The availability of annotated data plays a crucial role in choosing DL models. For supervised tasks, having a substantial amount of labeled data is necessary for training complex models such as deep neural networks. However, obtaining annotated biomedical signal data can be challenging and time-consuming, especially in medical domains with limited expert annotations [102].
Transfer learning can be a practical solution if labeled data are scarce, where pretrained models are fine-tuned on smaller datasets. Pretrained models trained on large-scale datasets such as ImageNet can capture general patterns often useful for related tasks. Fine-tuning these models on specific biomedical signal data can lead to faster convergence and improved performance [103].
6.3 Model Complexity and Interpretability
The complexity of the DL model should align with the available computational resources and interpretability requirements. While deep models such as transformers achieve state-of-the-art performance in various domains, they are computationally demanding. They may require high-end GPUs or specialized hardware for training and inference [104]. On the other hand, simpler models such as logistic regression or decision trees may offer better interpretability but may sacrifice some predictive performance compared with deep neural networks. For applications where interpretability is critical, researchers may opt for models that allow easier visualization and understanding of the learned features [105].
6.4 Class Imbalance and Performance Metrics
In biomedical signal analysis, class imbalance is a common challenge, where one class may dominate the data distribution while others have fewer samples. Accuracy alone may not be a reliable performance metric for imbalanced biomedical signal datasets. Metrics such as sensitivity (recall), specificity, and the F1 score provide a more comprehensive evaluation of the model’s performance [106].
Specialized loss functions such as focal loss or class-weighted approaches can address class imbalance issues and improve the model’s performance on minority classes. Moreover, data augmentation techniques can help balance the class distribution and enhance the model’s generalizability [107].
6.5 Domain Expertise and Transfer Learning
In medical fields, researchers often have domain-specific knowledge that can guide model design, feature selection, and data preprocessing steps. Incorporating domain expertise can lead to better-informed choices in designing the neural network architecture or selecting relevant features for a specific medical task [108]. Transfer learning from models pretrained on similar tasks or domains can also increase performance, especially when labeled data are limited. Fine-tuning pretrained models on target biomedical signal datasets can help the model leverage knowledge from large-scale datasets and improve generalizability [109].
The choice of DL model also depends on the available computational resources, including GPU capabilities, memory, and processing power. While state-of-the-art models may deliver the best performance, they can be computationally expensive and impractical for resource-constrained environments [110]. Researchers can explore model compression techniques, such as model pruning, quantization, or knowledge distillation, to reduce the model’s size and computational requirements without significantly compromising performance [111].
Overall, selecting the perfect type of DL model for biomedical signal analysis requires careful consideration of the data characteristics, available resources, and performance requirements. By understanding the strengths and limitations of different models, researchers can make informed decisions to achieve optimal results in their specific applications. Fig. 18 shows a tradeoff between the parameters of all the abovementioned considerations.
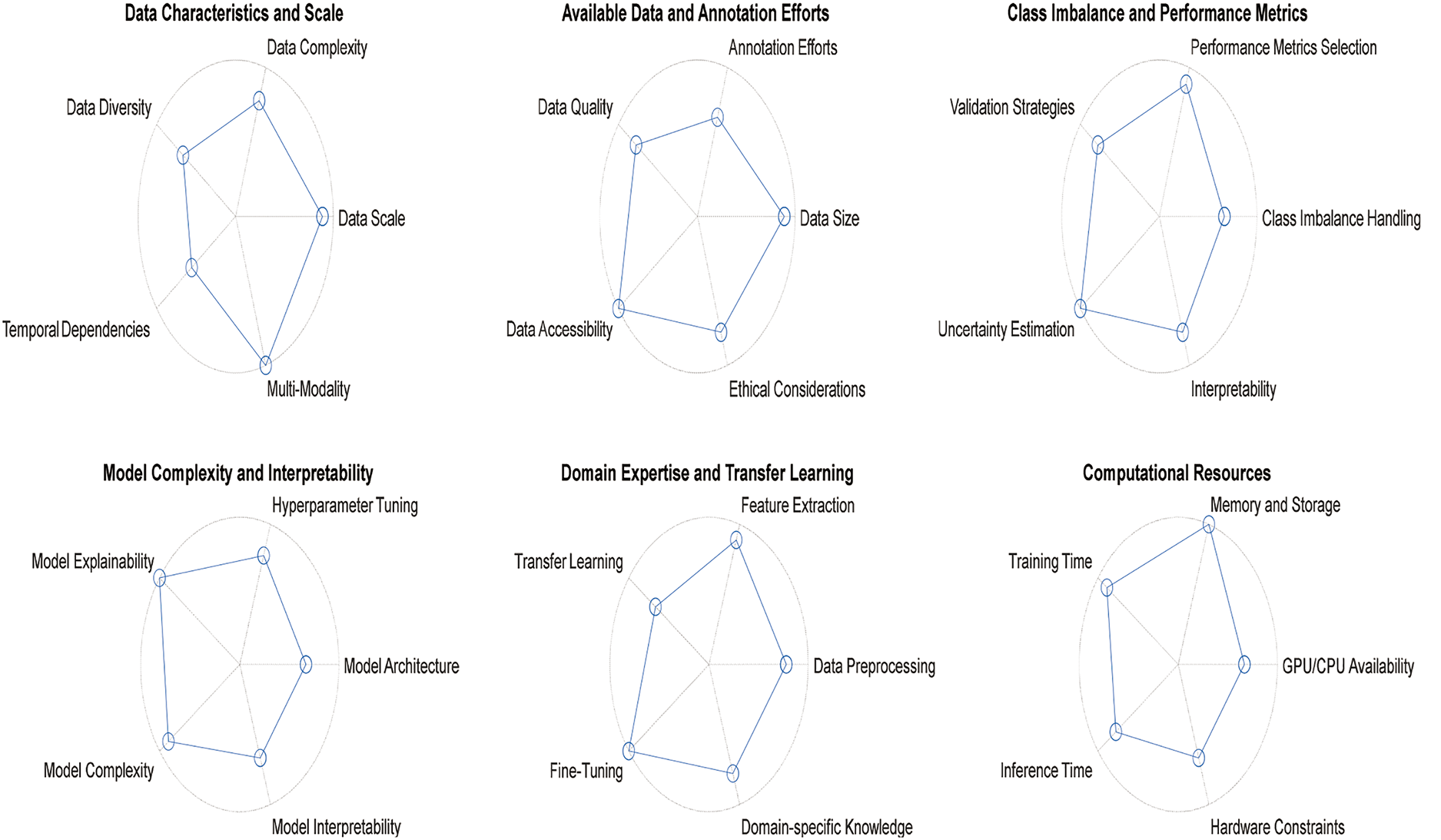
Figure 18: Tradeoff between the parameters of all considerations
While powerful, deep learning models often demand significant computational resources, which can severely restrict their deployment in environments with limited hardware capabilities. In biomedical signal analysis, such constraints are particularly relevant in settings such as mobile health devices, rural clinics with outdated infrastructure, or wearable sensors operating with battery power [110]. These resource-constrained environments face challenges such as insufficient GPU memory, slow processing speeds, and restricted energy budgets, which hinder the use of large-scale models such as transformers or densely connected neural networks. For example, real-time applications such as ECG monitoring on edge devices require models that balance accuracy with low latency. Yet, high-parameter models may fail to meet these requirements even after optimization. This gap between model complexity and practical feasibility underscores the need for lightweight architectures or adaptive frameworks that prioritize efficiency without sacrificing diagnostic reliability [111].
Moreover, the computational burden extends beyond inference, including training and data preprocessing stages. Training state-of-the-art models often requires high-performance computing clusters, which are inaccessible in many research or clinical settings [111]. Even with techniques such as transfer learning or federated learning to mitigate data and resource limitations, the energy consumption and time costs remain prohibitive for continuous operation in low-resource contexts. Consequently, researchers must critically evaluate whether the performance gains of advanced models justify their resource demands or, if more straightforward, specialized architectures such as shallow CNNs or hybrid models offer a more pragmatic solution [110]. Addressing these challenges requires interdisciplinary collaboration to develop hardware-aware algorithms, optimize existing frameworks for deployment, and explore emerging technologies such as neuromorphic computing to bridge the divide between model capability and operational practicality.
7 Methods for Developing and Interpreting DL Models for Biomedical Signals
DL models can be developed via different methodologies, ranging from training models from scratch to leveraging pretrained models through transfer learning. This section discusses several methods for developing DL models, focusing on transfer learning as a practical approach for biomedical signal analysis.
One approach to developing DL models is to train them from scratch. In this method, neural network architecture is designed, initialized with random weights, and trained on the target biomedical signal dataset. While this approach offers complete control over the model architecture and allows for specific customization, it may require a large amount of annotated data and extensive computational resources to achieve competitive performance [112].
Despite their potential challenges, training models from scratch can be suitable when domain-specific expertise suggests that existing pretrained models may not directly apply to the target biomedical signal analysis task [113].
Transfer learning is a powerful technique that leverages knowledge from pretrained models on large-scale datasets and adapts it to new tasks with smaller target datasets. The underlying idea is that lower-level features learned from diverse datasets (e.g., ImageNet) generally apply to various visual recognition tasks [112,114]. By fine-tuning the pretrained model on biomedical signal data, the model can effectively capture relevant patterns and improve performance, even with limited labeled data [115,116].
In transfer learning, there are two main strategies:
Feature Extraction: In this approach, the pretrained model’s convolutional layers are frozen, acting as a feature extractor. The extracted features are fed into a separate classifier for the specific biomedical signal task. This method is effective when the lower-level features learned in the pretrained model are relevant to the target task [40,114–116].
Fine-tuning: Fine-tuning involves using the pretrained model’s lower-level features and adapting the higher-level layers to the target task. During fine-tuning, some or all of the layers in the pretrained model are trainable, allowing the model to adjust its parameters based on the target biomedical signal data [115].
Transfer learning significantly reduces the need for large, annotated datasets, speeds up training, and preserves the knowledge learned from the pretrained model.
7.3 Domain-Specific Model Pretraining
Another method for developing DL models for biomedical signals is domain-specific pretraining. Unlike general-purpose pretraining on large-scale datasets such as ImageNet, domain-specific pretraining focuses on training models on relevant biomedical signal data or related medical datasets [117,118].
Domain-specific pretraining can be helpful when the target biomedical signal dataset differs significantly from generic image datasets. Training the model on more domain-relevant data can be initialized with more task-specific information and may require less fine-tuning on the target task [118].
Transformer networks, initially conceived for NLP, have become a cornerstone of modern DL architecture [119]. Their breakthrough lies in the attention mechanism, which enables the model to focus on crucial elements within a sequence, thereby effectively capturing long-range dependencies. This feature has propelled transformers to the forefront of NLP tasks, including machine translation, sentiment analysis, and question answering, outperforming their predecessors and setting new benchmarks in language understanding and generation [120]. Fig. 19 shows the architecture of the transformer.
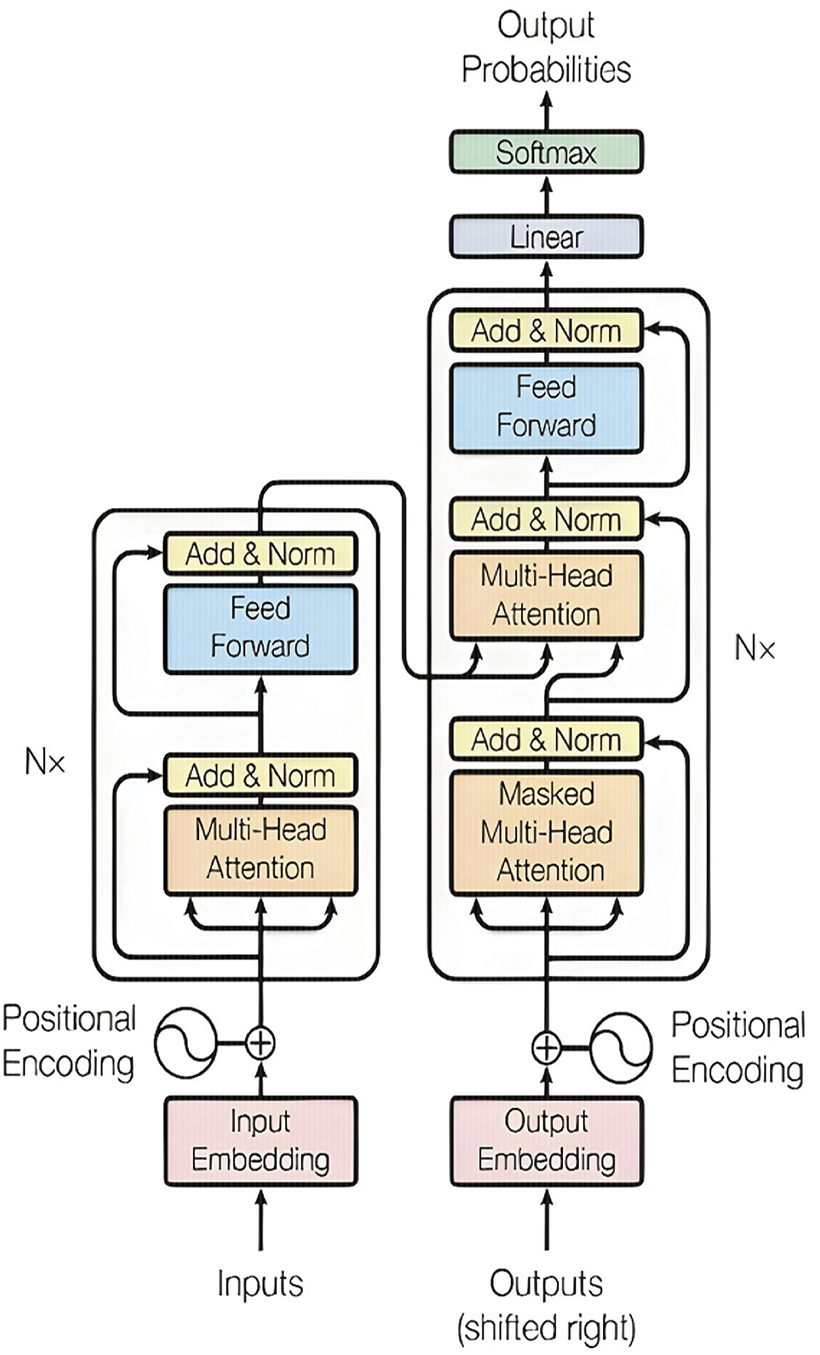
Figure 19: Transformer network architecture [104] (CC BY 4.0)
However, their impact has not been limited to the realm of NLP. Transformer networks have been successfully repurposed for various other domains, including the analysis of biomedical signals. By leveraging their ability to handle sequential data and discern intricate patterns, transformer-based models have shown potential in tasks such as disease diagnosis, anomaly detection, and medical image analysis [119–121]. This adaptability has paved the way for significant advancements in personalized medicine, offering promising avenues for developing innovative diagnostic tools and more effective healthcare solutions. As researchers continue to explore their potential in diverse fields, the integration of transformer networks is expected to foster breakthroughs in biomedical research, ultimately leading to enhanced healthcare practices and a deeper understanding of complex biological systems [121,122].
In the field of biomedical signals, transformer networks have shown potential. For example, a study proposed a constrained transformer network for ECG signal processing and arrhythmia classification [123]. The model combines a CNN and a transformer network to extract temporal information from ECG signals and can perform arrhythmia classification with acceptable accuracy [123]. The transformer network pays more attention to the data’s temporal continuity and captures the data’s hidden deep features well1. Another study proposed a transformer-based high-frequency oscillation (HFO) detection framework for biomedical magnetoencephalography (MEG) one-dimensional signal data [122]. The framework included signal segmentation, virtual sample generation, classification, and labeling. The proposed framework outperformed the state-of-the-art HFO classifiers, increasing the classification accuracy by 7% [122]. Recently a transformer mixture model was used for classification of ECG signals [124]. Fig. 20 shows an example of using transformers for biomedical signals classifications.
Figure 20: Schematic representation of the transformer model for biomedical signals classification [124] (CC BY 4.0)
Hybrid DL models combine the power of two types of DL models. These models have shown great promise in analyzing biomedical signals because of the ability of CNNs to perform spatial feature extraction and capture the temporal dependencies of RNNs, such as LSTM networks [125]. By integrating these models, hybrid models can be used to process complex biomedical signals effectively [126]. One of the most popular hybrid models is the CNN-LSTM hybrid model, which first uses CNN layers to extract features from signals and then employs LSTM layers to analyze the sequential nature of these features, enhancing the model’s ability to detect patterns [127]. Such combinations improve overall performance and enable more robust model interpretation, making hybrid models valuable tools in medical diagnostics and personalized healthcare [125–128]. Fig. 21 shows the general block diagram of hybrid DL models using CNN-LSTM.

Figure 21: General block diagram of the hybrid DL model using CNN-LSTM
7.6 Convolutional Block Attention Module (CBAM)
The convolutional block attention module (CBAM) is a simple yet effective attention module for feedforward CNNs. Given an intermediate feature map, CBAM sequentially infers attention maps along two separate dimensions, channel and spatial; then, the attention maps are multiplied by the input feature map for adaptive feature refinement [129]. Fig. 22 shows a sample block diagram of the CBAM. CBAM has shown potential in the field of biomedical signals. For example, a study experimentally analyzed four attention mechanisms, including CBAM and three CNN architectures, for two representative physiological signal prediction tasks: classification for predicting hypotension and regression for predicting cardiac output (CO) [130]. The CNN models with the spatial attention mechanism performed best in the classification problem, whereas the channel attention mechanism achieved the lowest error in the regression problem [130]. Another study proposed a new sEMG gesture recognition network called the multistream convolutional block attention module-gate recurrent unit (MCBAM-GRU), which is based on sEMG signals [131]. The network is a multistream attention network formed by embedding a GRU module based on CBAM. The experimental results showed that the proposed method obtained excellent performance on the dataset collected in this paper, with a recognition accuracy of 94.1%, achieving advanced performance, with an accuracy of 89.7% on the Ninapro DB1 dataset [131].

Figure 22: Block diagram of CBAM [129] (CC BY 4.0)
7.7 Hierarchical Attention Networks (HANs)
Hierarchical attention networks (HANs) are a type of neural network that applies attention mechanisms at multiple levels of the network hierarchy [132]. The attention mechanism allows the model to focus on specific parts of the input when making predictions, which can be particularly useful when dealing with complex data such as biomedical signals [133].
In biomedical signals, a hierarchical attention (HA) module embedded in HANet captures context information from neighbors of multiple levels, where these neighbors are extracted from a high-order graph [133]. The proposed HA module is robust to the input variance and can be flexibly inserted into existing convolution neural networks [133].
Another study proposed an HA-based capsule model for biomedical document triage [134]. The proposed model employs a hierarchical attention mechanism and capsule networks to capture valuable features across sentences and construct a final latent feature representation for a document. Experimental results have shown that HA mechanisms and capsule networks are helpful in biomedical document triage tasks [134].
Developing DL models for biomedical signal analysis involves carefully selecting methods and considering data availability, computational resources, and performance requirements. Transfer learning and data augmentation are potent techniques for effectively leveraging pretrained models and improving their generalization capabilities even with limited labeled data. Researchers can develop robust and reliable DL models by following a systematic approach and employing suitable methodologies. Fig. 23 shows a quick comparison between the implementation methods mentioned.
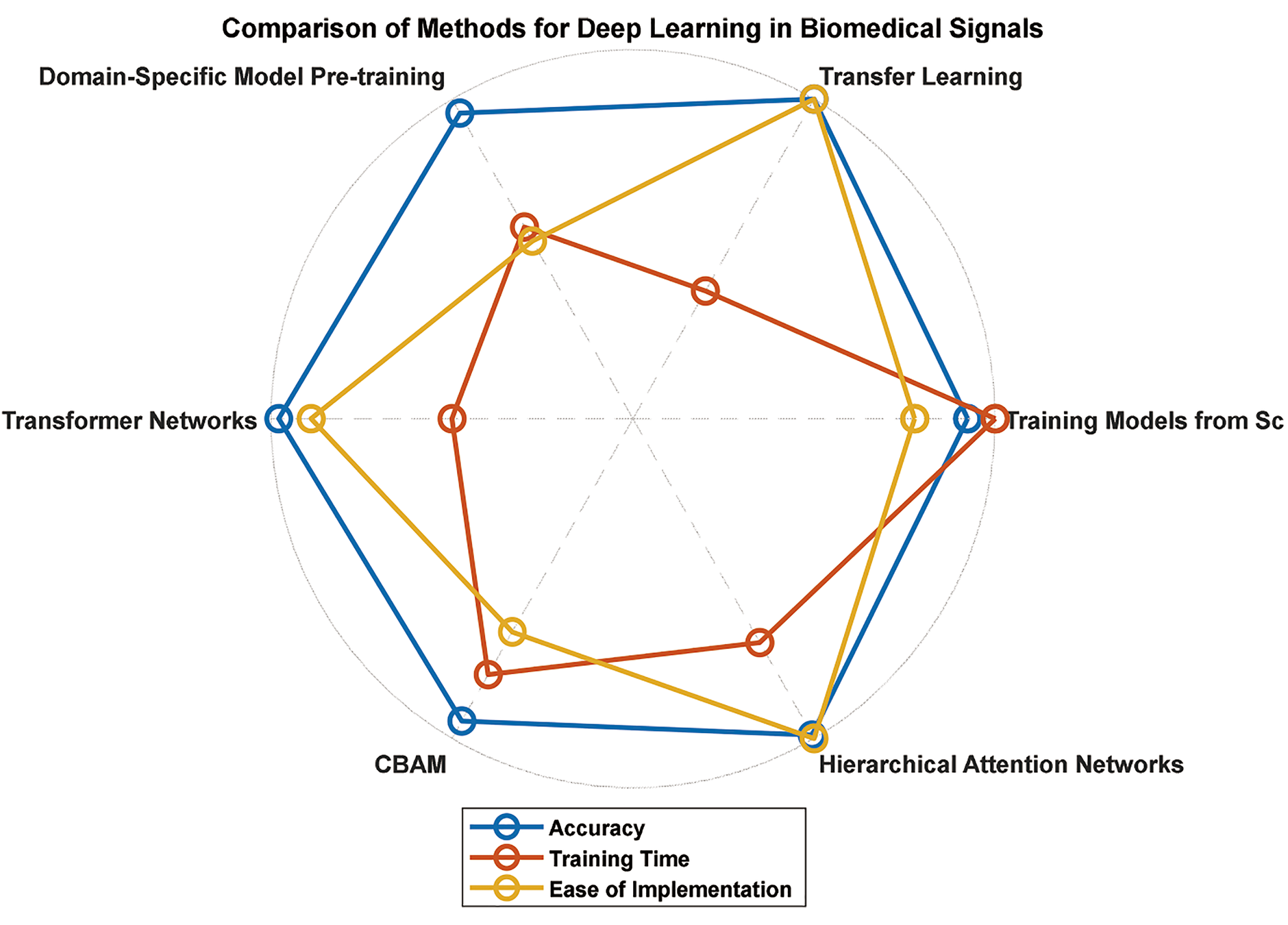
Figure 23: Comparison of DL implementation methods for biomedical signals
7.8 Gradient-Weighted Class Activation Mapping (GradCAM)
Grad-CAM, short for gradient-weighted class activation mapping, has become an indispensable tool in interpretability for CNNs. In essence, it is a powerful means to demystify the decision-making processes of complex models, enhancing their transparency and interpretability [135]. By leveraging the gradients associated with a specific target concept and channelling them back into the last convolutional layer, Grad-CAM excels at generating high-resolution localization maps.
These maps effectively illuminate crucial regions within an input image that contribute significantly to the model’s classification decision, providing a clear and intuitive visualization of the model’s focus [136]. This not only aids in understanding the model’s behavior but also fosters trust and confidence in its predictions, which are essential for applications in fields such as medical imaging, autonomous systems, and beyond. Grad-CAM is a pivotal tool for bridging the gap between the formidable capabilities of CNNs and the need for transparent and interpretable AI systems [135].
In the context of biomedical signals, Grad-CAM can be particularly useful. For example, in a study that aimed to distinguish migraines from EEG signals, researchers converted EEG signals into scalogram images via the continuous wavelet transform method [136]. These scalogram images were then used as inputs to a CNN DL network. Interpretable Grad-CAM images were obtained to support specialists in the diagnosis of migraine. The study revealed a relationship between EEG recordings and migraine disease in terms of frequency components [136].
In addition, there are also advanced AI explainability tools for computer vision, including Grad-CAM, such as PyTorch Grad-CAM. These tools can diagnose model predictions in production or while developing models [135]. They offer a comprehensive collection of pixel attribution methods for computer vision and work with many standard CNNs and vision transformers. The Grad-CAM provides a valuable tool for visualizing and understanding the decision-making processes of DL models, especially in biomedical signal analysis [135]. Fig. 24 shows a sample Grad-CAM output image.
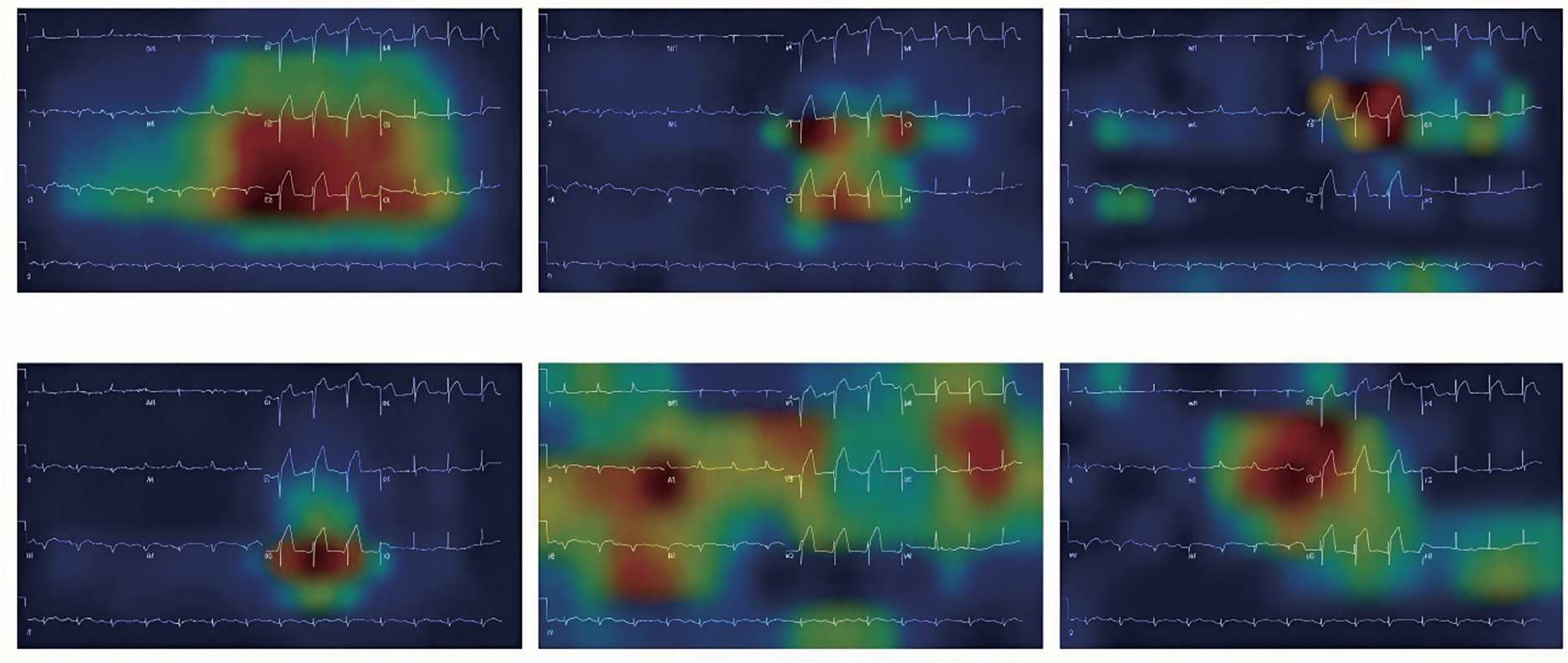
Figure 24: Sample Grad-CAM images for ECG signal classification [137] (CC BY 4.0)
Integrated gradients (IGs) have emerged as critical players in model interpretability, providing a nuanced understanding of how a model arrives at its predictions by attributing them to specific input features. This technique, outlined in the literature [138], offers a valuable means of visualizing the intricate relationship between the input features and the model’s output predictions. In essence, IG is a sophisticated variant of computing gradients, explicitly focusing on the prediction output concerning the input features. By integrating the gradients along a straight path from a baseline to the input, IG assigns importance scores to each feature, elucidating their contributions to the final prediction [138]. This not only aids in comprehending the model’s decision-making process but also facilitates the identification of influential features and their impact on the overall prediction outcome. In the landscape of model interpretation, IG stands as a powerful tool, shedding light on the intricate interplay between input features and model predictions [139].
In the field of biomedical signals, IG has shown potential. For example, in a study that aimed to improve the interpretability of DL models by splicing codes, enhanced integrated gradients (EIG) were introduced [138]. EIG is a method for identifying significant features associated with a specific prediction task. Using RNA splicing prediction as a case study, it was demonstrated that the EIG improves upon the original integrated gradient method and produces informative features [138]. Another study proposed a compensated IG method for a reliable explanation of electroencephalogram (EEG) signal classification [139]. This method does not require a baseline, compensating for the contributions calculated via the IG method at an arbitrary baseline via an example of the Shapley sampling value. The study demonstrated that the contributions obtained via the proposed compensated IG method are more reliable than those obtained via the original IG method [139].
Advanced AI explainability tools for computer vision, including IG, such as TensorFlow IG, are also available. These tools can make model predictions during production or when models are being developed. They offer a comprehensive collection of pixel attribution methods for computer vision and work with many standard CNNs and vision transformers [138,139]. The IG provides a valuable tool for visualizing and understanding the decision-making processes of DL models, especially in biomedical signal analysis.
Shapley values, which originate from cooperative game theory, have proven to be versatile and impactful concepts within the realm of ML [140–142]. Their application extends across various domains, offering a method to equitably distribute the “contribution” of each feature in a model’s prediction. By leveraging the principles of fairness and cooperation, Shapley’s values provide a nuanced understanding of the individual impact of features on a model’s output. This fair attribution of contributions is particularly valuable in scenarios where interpretability and transparency are paramount, allowing stakeholders to grasp the nuanced roles played by each feature in shaping the final prediction [141]. Integrating Shapley values enhances the interpretability of ML models and contributes to building trust and understanding in the decision-making processes of these sophisticated systems. In essence, Shapley’s values bridge cooperative game theory and ML, providing a principled approach to feature attribution and contribution analysis [142].
In the field of biomedical signals, Shapley values have shown potential. For example, Shapley attributed ablation with augmented learning (ShapAAL) was proposed for practical time series sensor data classification [141]. This method demonstrates that a DL algorithm with a suitably selected subset of the seen examples or ablation of the unimportant examples from the given limited training dataset can consistently improve classification performance under augmented training. In ShapAAL, the subset of training examples that contribute positively to a supervised learning setup is derived from the notion of coalition games via Shapley values associated with each of the given inputs’ contributions to the model prediction [141]. Another study used Shapley values to determine the relative importance of input attributes to the result generated by a multivariate molecular diagnostic test for an individual sample or patient [142]. Patient subgroups defined by Shapley value profiles may motivate translational research [142]. Fig. 25 shows an example of the Shapley values used.
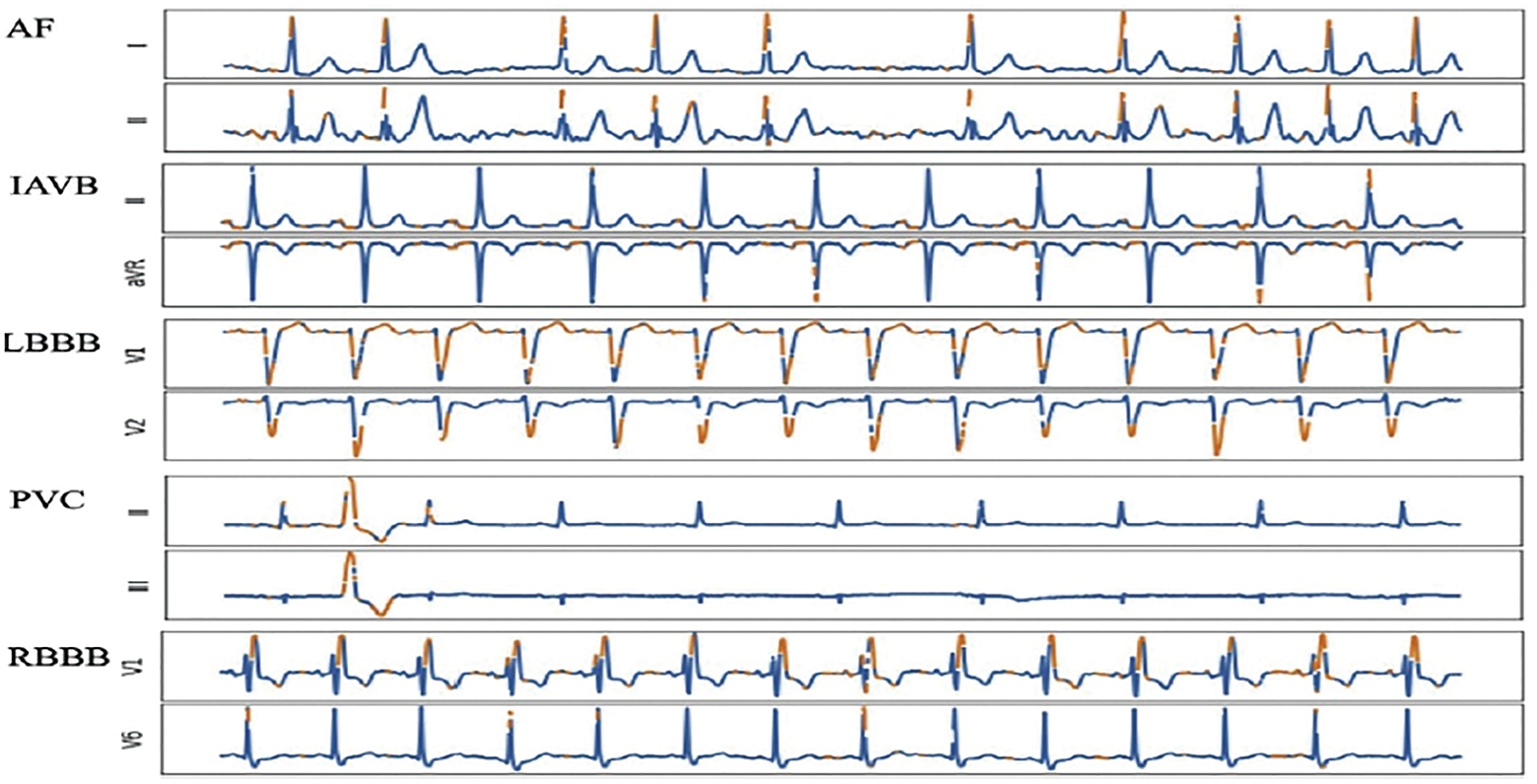
Figure 25: Sample Shapley values for ECG classification for two different rhythms [143] (CC BY 4.0)
In summary, leveraging techniques such as GradCAM, Shapley values, and IG adds interpretability to model outcomes. Researchers aiming to cultivate robust and dependable DL models are encouraged to adopt a systematic approach, integrating appropriate methodologies tailored to the intricacies of biomedical signal data. Fig. 26 illustrates a visual representation comparing these methodologies.
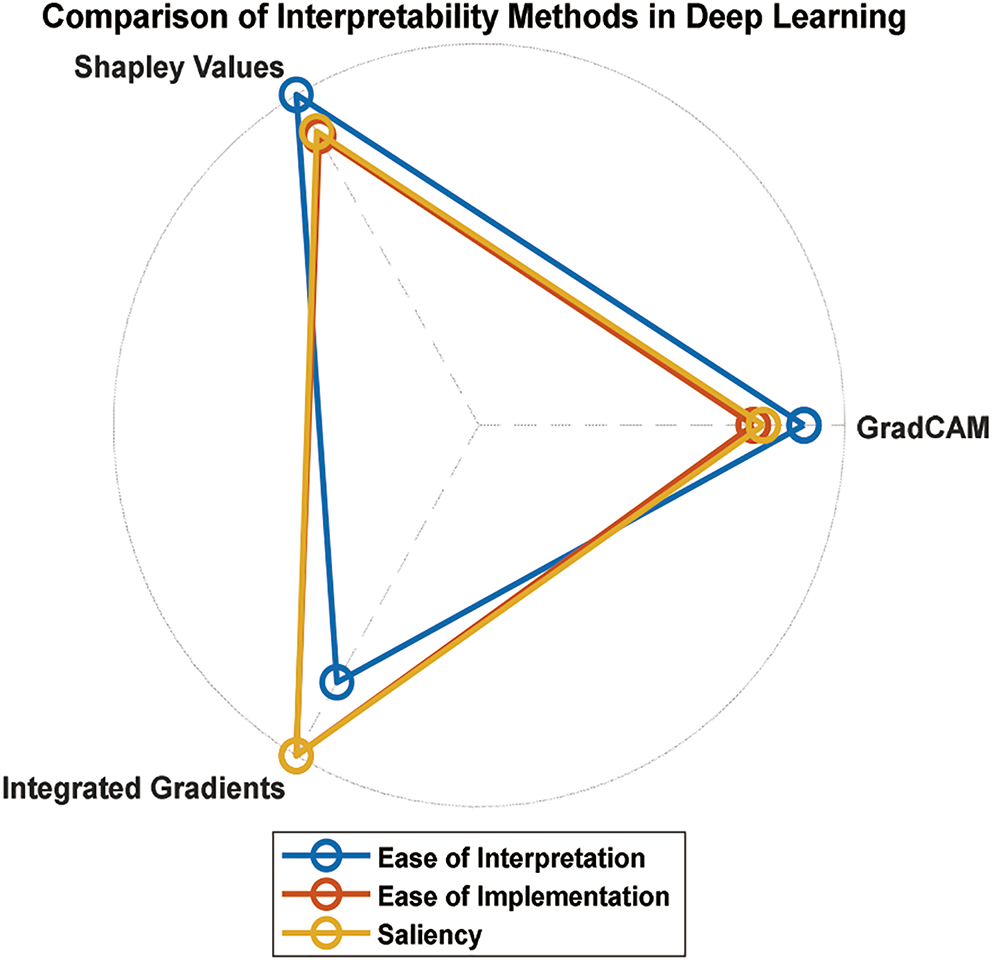
Figure 26: Comparison of methods for interpreting DL in biomedical signals
8 Computational Analysis of Different DL Models
When developing DL models for biomedical signals, computational efficiency is crucial because of the often-limited resources available in medical research and clinical environments [144]. This section evaluates the computational demands of various model architectures discussed in the previous sections, focusing on training time, memory requirements, and performance trade-offs associated with each method [145,146]. Understanding these aspects is essential for selecting models that meet performance criteria while remaining feasible for deployment in real-world healthcare settings.
The computational feasibility of each model depends on the available hardware and performance needs of the application. Transfer learning and CBAM offer efficient solutions for scenarios with limited resources, whereas transformers and hybrid models may be preferable when computational power is available to achieve more nuanced performance [144,147]. By assessing computational requirements alongside model performance, researchers can optimize DL workflows to fit within available resources, promoting the effective deployment of biomedical signal processing applications in clinical and research environments [146,147]. Additionally, as the field progresses, emerging techniques such as model pruning, quantization, and knowledge distillation can further help reduce the computational overhead of DL models.
These techniques aim to simplify complex models without sacrificing performance, making them more accessible for real-time, resource-constrained applications [145]. As such, combining these optimization strategies with the appropriate model architecture could provide a pathway to more scalable and deployable biomedical signal processing solutions. Table 1 compares the computational analysis results of the different methods. By understanding these computational demands, researchers can select model architectures that align with their operational constraints, ensuring an optimal balance between accuracy and resource efficiency.

9 Comparison of Existing Datasets of Biomedical Signals
The availability of numerous biomedical signal datasets has supported the recent rapid growth of DL applications in healthcare. These datasets encompass various signals, such as ECG, EEG, EMG, PPG, and respiratory sounds. These datasets provide a foundation for developing robust models for classifying, predicting, and detecting various health conditions. Table 2 summarizes the most widely used and publicly accessible datasets. Moreover, these datasets provide essential resources for developing and validating DL models in biomedical signal processing, enabling significant advances in various health and diagnostic applications. Each dataset has unique properties, sampling rates, and sample sizes, which cater to specific biomedical signal types and tasks.
Despite the availability of numerous biomedical signal datasets, several limitations hinder their effectiveness in training robust and generalizable DL models. One major limitation is the lack of diversity in many datasets. Most datasets are collected from specific patient populations, often in controlled environments, which limits their applicability to broader or more diverse populations. For example, datasets such as the MIT-BIH Arrhythmia Database primarily include data from adult patients, making it difficult to generalize findings to pediatric or geriatric populations. Additionally, many datasets are imbalanced, with certain classes (e.g., rare diseases or specific arrhythmias) being underrepresented. This imbalance can lead to biased models that perform well on majority classes but poorly in minority classes, which are often clinically significant. Another critical issue is the quality of annotations. While experts meticulously annotate some datasets, others rely on automated or semiautomated labeling, which may introduce errors. Noise and artifacts in the signals, especially in datasets collected in real-world settings (e.g., ICUs or wearable devices), further complicate the training of reliable models. Finally, the size of datasets is often insufficient for training DL models, which typically require large amounts of data to achieve optimal performance. Small datasets increase the risk of overfitting and limit the model’s ability to generalize to new data.
To address these limitations, several strategies can be employed. First, data augmentation techniques, such as synthetic data generation via GANs or time series transformations (e.g., scaling, shifting, or adding noise), can help increase the size and diversity of datasets. Second, collaborative efforts among institutions to create larger, more diverse datasets are essential. Initiatives such as federated learning can enable data sharing while preserving patient privacy, allowing models to be trained on data from multiple sources without transferring sensitive information. Third, improved annotation protocols, including multiple expert annotators, should be adopted to reduce labeling errors and ensure consistency. Additionally, active learning techniques can be used to prioritize the annotation of the most informative data points, reducing the cost and effort of manual labeling. Finally, standardizing data collection protocols across institutions can improve the quality and consistency of datasets, making them more suitable for training generalized models. By addressing these limitations, the biomedical research community can develop more robust datasets that enable the creation of accurate, reliable, and clinically useful DL models.
To further enhance the utility of biomedical signal datasets, providing detailed comparisons of the methodologies employed for their analysis can significantly assist practitioners in selecting the most suitable approaches for their specific tasks. For example, comparing different preprocessing techniques, such as filtering and denoising methods tailored for ECG or EEG signals, can help identify optimal strategies for reducing noise and improving signal quality. Similarly, highlighting the relative performance of feature extraction methods, such as time-domain, frequency-domain, and time-frequency analyses, can provide insights into their effectiveness across various signal types and applications. Additionally, summarizing the strengths and limitations of widely used ML and DL models such as CNNs for spatial signal patterns, RNNs for temporal sequences, or hybrid architectures combining multiple modalities can guide researchers in selecting methods best suited for their datasets. Including benchmarks on publicly available datasets via consistent metrics, such as accuracy, the F1 score, or the area under the curve (AUC), offers a practical reference for evaluating and comparing approaches, fostering better decision-making and promoting innovation in biomedical signal processing.
The issue of overfitting complex models trained on small biomedical datasets represents a significant challenge that requires careful consideration. Complex deep learning architectures, while powerful in their ability to model intricate patterns, are particularly susceptible to overfitting when trained on limited data [173]. This occurs because these models have a high capacity to memorize training examples rather than learn generalizable features, leading to excellent performance on training data but poor generalizability to unseen cases [173]. For example, a deep neural network trained on the relatively small MIT-BIH Arrhythmia Database might achieve near-perfect accuracy on the training set but fail to perform adequately when applied to data from different populations or institutions [174,175]. Researchers should incorporate multiple datasets in their experiments to demonstrate and address this issue, especially when evaluating complex models. The model’s generalizability can be more rigorously assessed by training on one dataset and validating it on another with different characteristics. Additionally, experiments comparing model performance across varying dataset sizes can help identify the minimum data requirements for reliable performance and highlight how overfitting becomes problematic [149]. Techniques such as regularization, dropout, and early stopping should be systematically evaluated in small biomedical datasets to determine their effectiveness in mitigating overfitting while preserving model performance. Through these approaches, the research community can better understand the limitations of complex models when trained on small datasets and develop strategies to enhance their generalization capabilities in real-world clinical settings [176,177].
Several strategies can mitigate the overfitting problem in complex models trained on small biomedical datasets. Regularization techniques such as dropout, L2 regularization, and early stopping have improved generalization by preventing the model from becoming too specialized in the training data. For example, studies on the Sleep-EDF expanded dataset have demonstrated that applying dropout regularization can significantly reduce overfitting in sleep stage classification models [176]. Data augmentation methods, including synthetic data generation via GANs, have also proven effective in expanding dataset size and diversity. Research using PhysioNet Challenge datasets has shown that GAN-augmented data can improve model performance on underrepresented classes [176,177]. Transfer learning represents another promising approach, where models pretrained on larger datasets from related tasks are fine-tuned on smaller target datasets. This approach has been successfully applied in ECG analysis, where models pretrained on the PTB-XL dataset have improved performance when fine-tuned on smaller institutional datasets. By systematically evaluating these techniques across multiple biomedical signal datasets, researchers can develop more robust models that are more generalized to diverse clinical settings [176,177].
10 Handling Small and Unbalanced Data from Biomedical Signals for Deep Learning
Biomedical signal datasets often present challenges related to data size and class imbalance. In this section, we address the issues of small and unbalanced data and discuss strategies to mitigate their impact on DL model performance. Addressing such data constraints is crucial for achieving accurate and robust results in biomedical signal analysis.
10.1 Data Augmentation for Small Datasets
Small biomedical signal datasets can limit the capacity of DL models to learn intricate patterns and may lead to overfitting. Data augmentation is a crucial technique for artificially increasing the dataset size by applying various transformations to the available data. Random rotations, translations, scaling, and flipping can create additional samples, enhancing the generalizability of the model [178].
Because of the unique characteristics of biomedical signals, domain-specific augmentation methods can be applied in addition to conventional data augmentation. For example, in electrocardiogram (ECG) data, using heart rate and morphology variations can augment the dataset and simulate diverse patient conditions [14]. Fig. 27 shows the data flow when the augmentation and nonaugmentation methods are used.
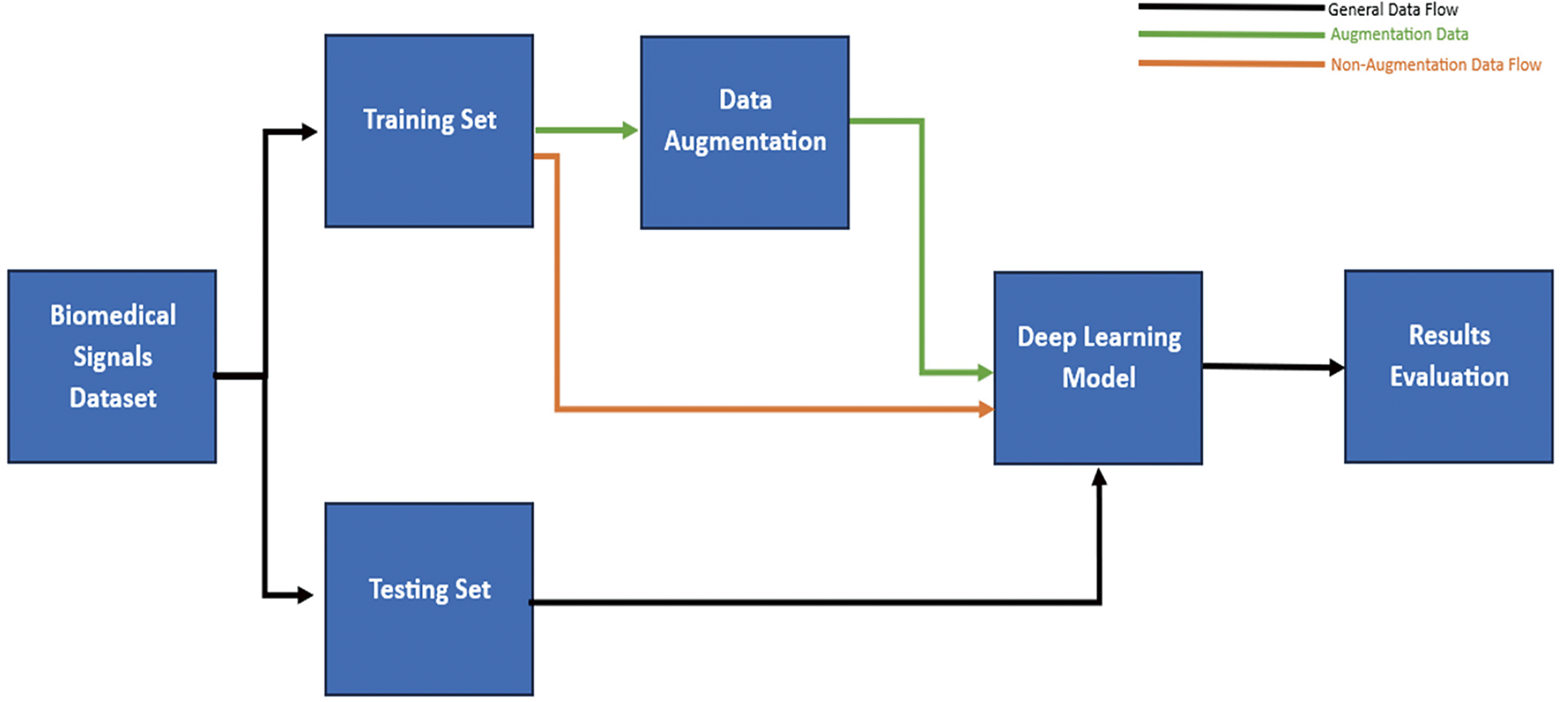
Figure 27: Comparison of methods for interpreting DL in biomedical signals. Data Flow for DL between Augmentation and Nono-augmentation
10.2 Class-Weighted Loss Functions
Class imbalance is common in biomedical signal datasets, where certain classes may have significantly fewer samples than others. In such cases, models trained with conventional loss functions may be biased toward the majority class, leading to poor performance in minority classes [179].
The class-weighted loss functions assign higher weights to the minority classes during training, making the model more focused on these classes. This approach helps balance the impact of imbalanced data on the training process, enabling the model to learn from all types effectively [107].
Ensemble learning is a powerful technique that combines predictions from multiple models to enhance overall performance. In small and imbalanced biomedical signal datasets, ensemble methods can help improve classification accuracy and reduce the risk of overfitting [180].
Ensemble learning increases the model’s robustness and generalization capabilities by training multiple models with different initializations or architectures and combining their outputs through majority voting or weighted averaging [181]. Fig. 28 shows the block diagram of the ensemble model.
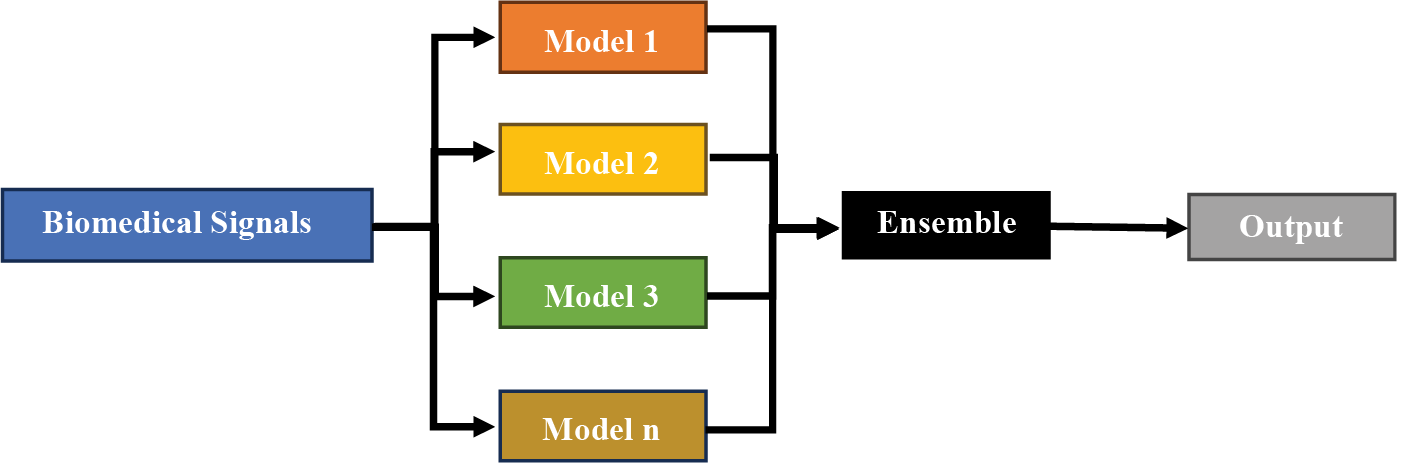
Figure 28: Block diagram of the ensemble model
10.4 Transfer Learning with Pretrained Models
As discussed in the previous section, transfer learning is a practical approach when dealing with small datasets. By leveraging knowledge from pretrained models on large-scale datasets, transfer learning allows the model to benefit from the representations learned from diverse data [102,115]. Compared with training models from scratch, fine-tuning pretrained models on the target biomedical signal data can yield superior performance, especially when the dataset is small or imbalanced [102].
One-class learning can be employed in scenarios where a specific class is rare or only positive samples are available. This approach treats the problem as a binary classification task, where the objective is to distinguish the target class from all other courses combined [182]. One-class learning techniques, such as one-class SVM or isolation forest, are instrumental when dealing with anomalies or rare events in biomedical signal data, where obtaining sufficient samples of the rare class may be challenging [183]. Fig. 29 shows a general block diagram of one-class learning.
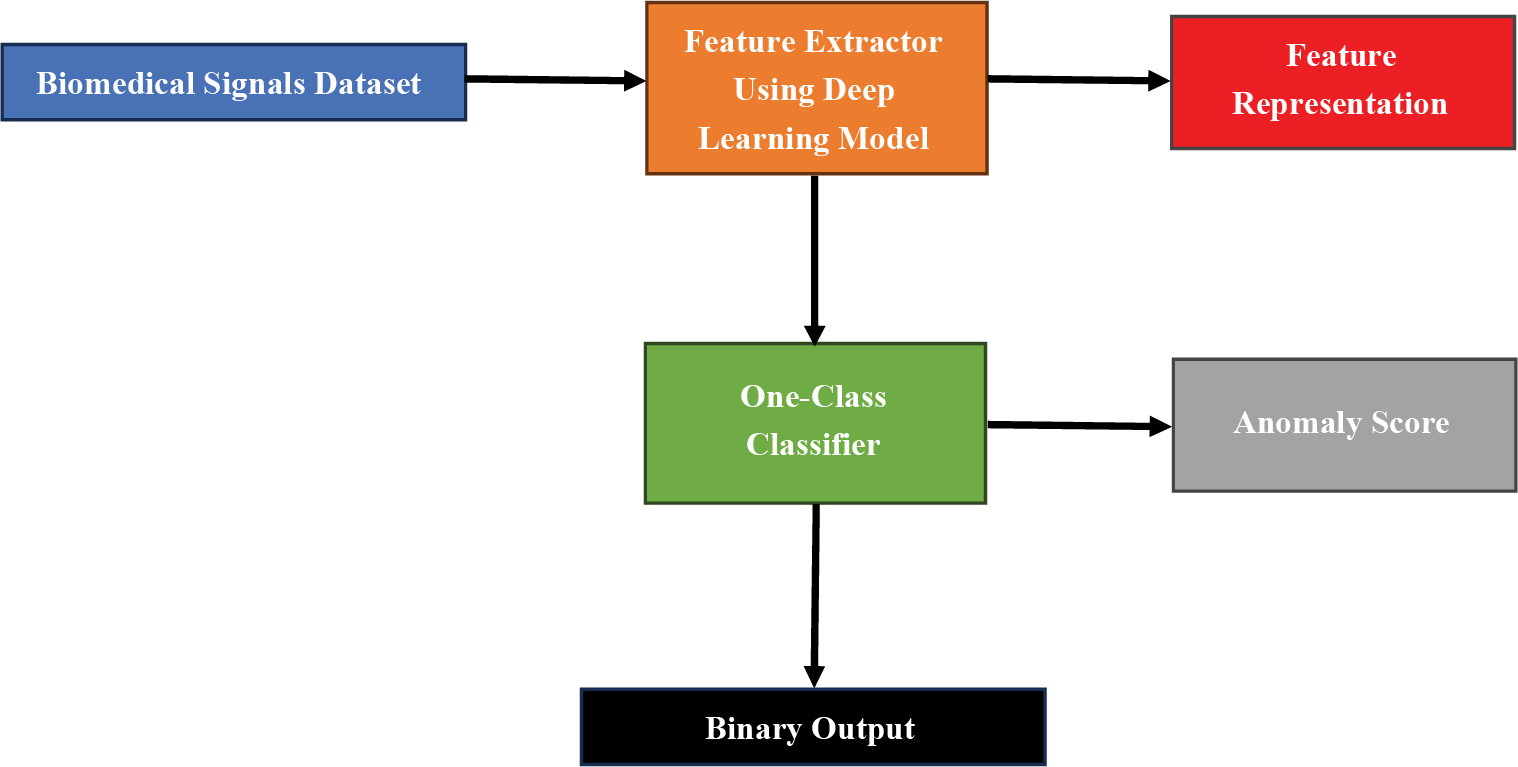
Figure 29: Block diagram of one-class learning
Incorporating these strategies into the DL pipeline for biomedical signal analysis can lead to more accurate, robust, and interpretable models, even with small and unbalanced datasets. By addressing data limitations and biases, researchers can unlock deep learning’s full potential in uncovering valuable insights from biomedical signals.
Few-shot learning (FSL) is an ML technique that aims to design ML models that can adapt to new tasks, given only a few training examples [184,185]. This is particularly useful in biomedical signals, where the data available for each class can often be relatively limited [184]. In biomedical applications, obtaining large and labeled datasets can be challenging because of data privacy concerns, ethical considerations, and the cost and effort involved in data annotation [185]. Therefore, FSL becomes crucial when the model must generalize and quickly make accurate predictions with minimal labeled examples. The ability of FSL to leverage prior knowledge and adapt to new tasks efficiently holds great promise for advancing the capabilities of ML models in the complex and data-limited domain of biomedical signal processing [186].
FSL can effectively handle small and unbalanced biomedical signal data for deep learning. For example, a previous study demonstrated the applicability of FSL for electrocardiogram (ECG) signal classification [185]. The study trained deep CNNs to recognize different heart disease classes with limited examples. Compared with the traditional SoftMax-based classification network, the FSL network has been shown to have greater accuracy in classifying healthy/sick patients [185].
One-shot learning is a concept in ML that aims to make accurate predictions given only a single example of each new class [186,187]. This is especially relevant in biomedical signal analysis, where only one or a few examples of a particular class may be available. Acquiring labeled data for rare medical conditions or specific disease states can be an intricate task in biomedical research. One-shot learning addresses this challenge by emphasizing the importance of leveraging a minimal number of examples to enable the model to discern patterns and characteristics unique to each class [186]. The ability of one-shot learning to draw meaningful insights from limited data is crucial in medical diagnostics, where novel and rare diseases necessitate adaptive and efficient machine-learning approaches for accurate and timely detection. This approach holds significant promise for enhancing the diagnostic capabilities of ML models in biomedical signal analysis [187].
For example, one study utilized one-shot learning to model complex biological systems from biomedical data streams [177]. Despite uncertainty about the data and model, scientists and clinicians can often learn the spatiotemporal dynamics of a complex biological system from just one or a few examples. Given limited exposure to a concept, category, or situation, this innate human ability to make accurate inferences is commonly called one-shot learning [187].
Zero-shot learning (ZSL) is a learning paradigm that recognizes unseen classes during test time; that is, it classifies objects of classes that have not been observed during training [188–190]. This approach is beneficial in biomedical signal analysis, where there are often many potential classes (e.g., types of diseases), but only a few may have sufficient training examples. New diseases and medical conditions continually emerge in the expansive biomedical research landscape, making it challenging to anticipate and gather adequate labeled data for each potential class [189,190]. ZSL addresses this challenge by enabling ML models to generalize their knowledge to classes not part of the training dataset. This capability is crucial for the timely identification and classification of novel diseases or conditions, allowing the model to be extrapolated from its existing knowledge to make informed predictions in a real-world biomedical context where the spectrum of diseases varies and evolves [188].
For example, a study proposed a ZSL framework, signal recognition and reconstruction convolutional neural network (SR2CNN), for signal recognition [190]. The key idea behind the SR2CNN is to learn the representation of the semantic feature space of the signal such that semantic features have more tremendous minimal interclass distances than maximal intraclass distances [190]. The proposed SR2CNN can discriminate signals even if no training data are available for some signal classes. Fig. 30 shows a performance comparison between the few-shot, one-shot, and zero-shot methods.
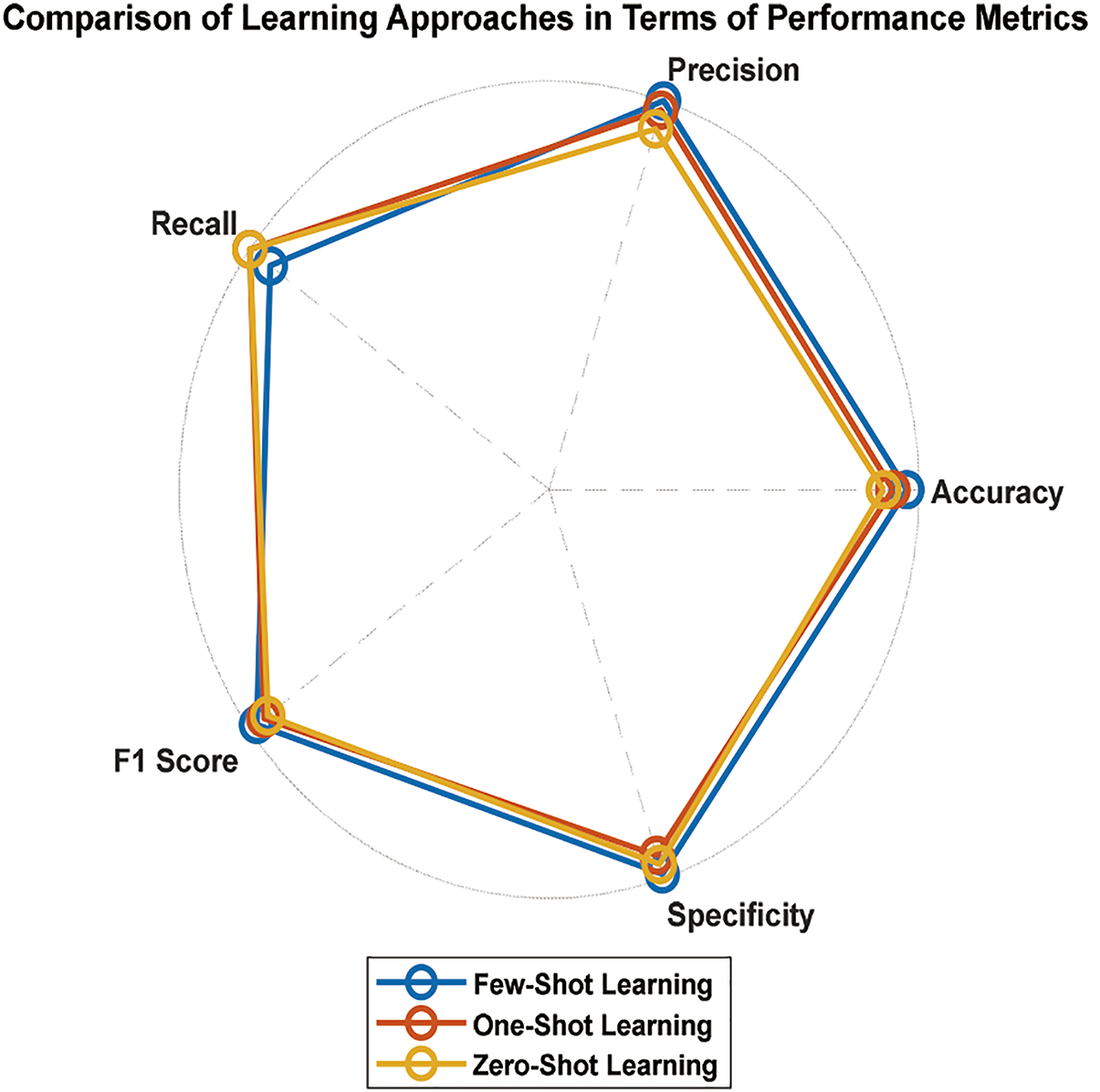
Figure 30: Performance comparison between the few, one, and zero-shot methods
11 Toward Big Data Solutions in Biomedical Signal Analysis
The performance of DL algorithms is enhanced when large amounts of data are used, and DL can learn efficiently from more extensive data [44]. Like other fields, biomedical signal big data present a unique opportunity to unlock the full potential of these powerful algorithms. Big data generally refers to massive, complex, growing datasets that include biomedical signals such as EEGs, ECGs, and EMGs [191]. Fig. 31 shows how the size of biomedical signal data increases over time and the primary application of big data in DL applications in biomedical signals.
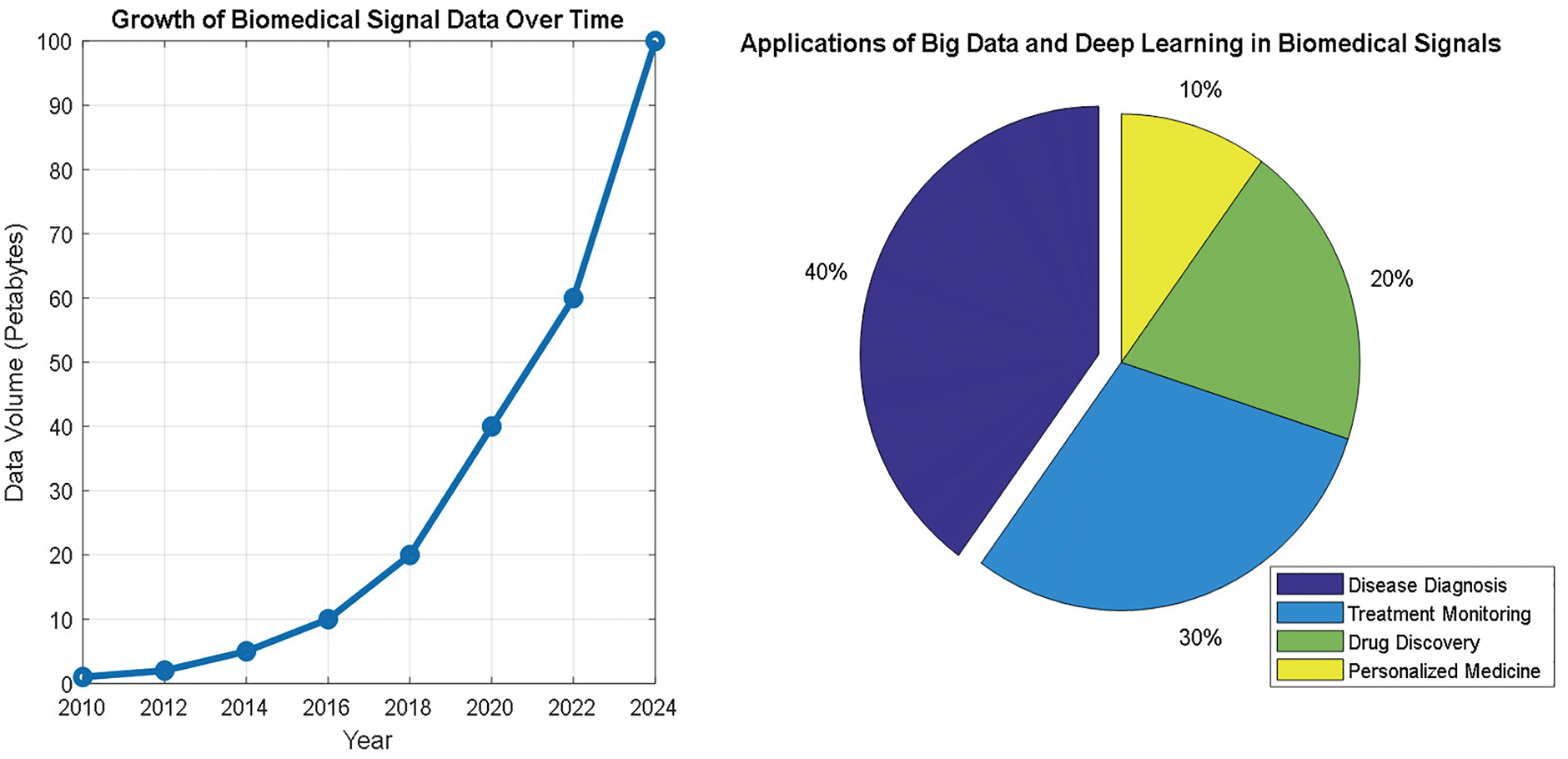
Figure 31: Changes in the size of biomedical signal big data over time and the major applications of big data in DL for biomedical signals
Using big data to train DL models for biomedical signals has different advantages, such as the following:
1. Generalizability: Large biomedical signal datasets enhance the performance of DL models by training them on different patterns and variations in the signal. This leads to more robust predictions of the models by allowing the DL models to generalize the weights to unseen data in real-world scenarios [45]
2. Discovering new and profound features: DL finds and extracts deep patterns within data. With big data, each pattern has enough samples, allowing DL to find distinct features to identify these patterns. This reduces the dependence on manual feature extraction and the time for feature engineering [192].
3. Detecting Rare Events: Biomedical signals can have rare or abrupt anomalies similar to other signals. Using big data increases the opportunity to detect and learn these infrequent events. Therefore, DL models trained on big data have become helpful in identifying these rare events, leading to enhanced diagnoses [191,192].
However, although there are many benefits to using big data in DL for biomedical signals, different challenges still exist. Fig. 32 summarizes these challenges.
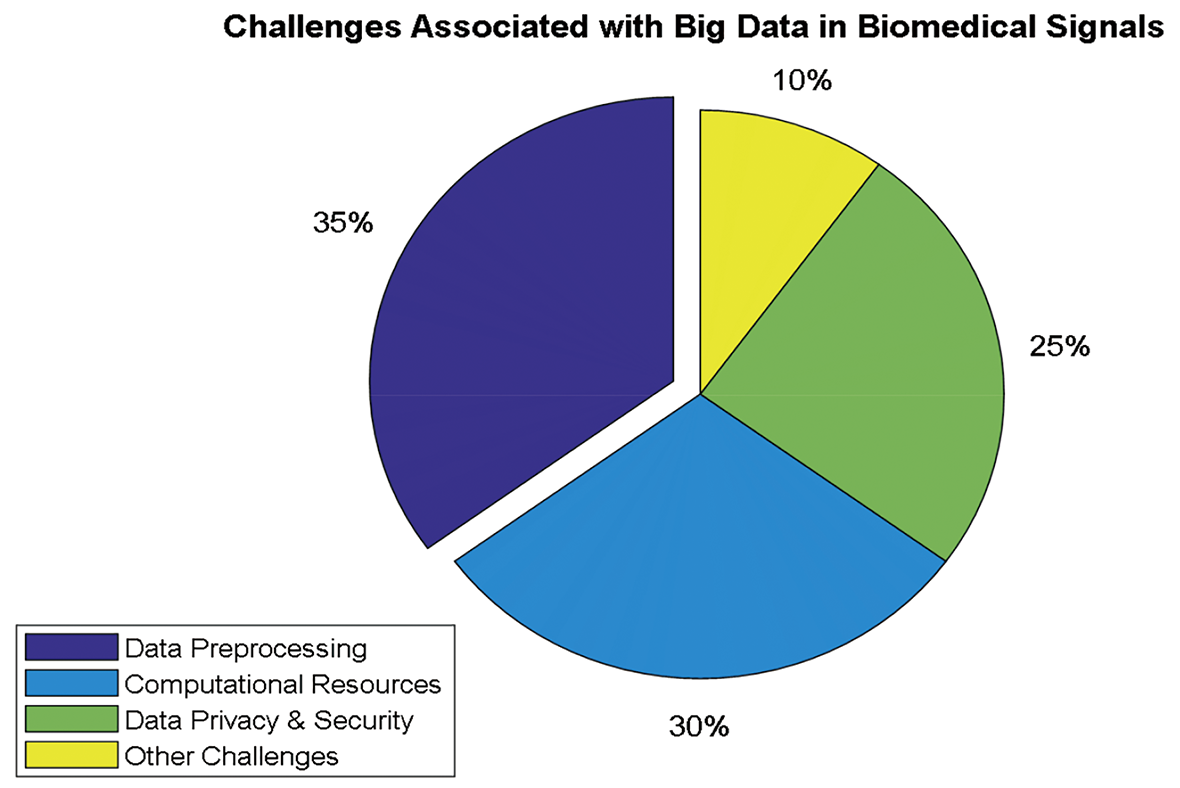
Figure 32: Significant big data challenges in DL for biomedical signals
The significant challenges associated with the use of big data for DL in biomedical signals can be summarized as follows:
1. Data Preprocessing and Management: The massive volume of big data requires the development of robust preprocessing algorithms. These algorithms should be able to deal with noise and reduce noise. Additionally, these algorithms should include data normalization and segmentation techniques to reduce the amount of data. Finally, integrating these algorithms with efficient data storage and retrieval systems is essential for effectively managing the data [191,193].
2. Computational Resources: Using big data to train DL models requires significant computational resources. Since this training will utilize high-performance computing clusters or cloud-based platforms, these platforms have become essential for handling processing requirements [191].
3. Data Privacy and Security: Sharing and distributing medical data, including signals, is very sensitive. Therefore, developing and implementing robust anonymization techniques and secure storage solutions is paramount [193].
Using big data to train DL models offers powerful tools for accessing biomedical signals. Big data can detect different patterns, anomalies, and information. Therefore, researchers can develop more robust models for disease diagnosis, monitoring, and personalized healthcare solutions. Relying on deep learning models on large-scale annotated datasets presents significant challenges in the biomedical domain [191,193]. Acquiring high-quality biomedical signal data requires specialized equipment, controlled environments, and often longitudinal studies that span months or years. This data collection process is financially expensive and resource intensive, requiring coordination among healthcare providers, research institutions, and patients. Furthermore, the ethical considerations surrounding patient privacy and informed consent add complexity that can slow data acquisition efforts [191].
Annotation of biomedical signals represents another substantial bottleneck. Accurate labeling of ECG, EEG, or EMG signals typically requires expertise from medical professionals, who can identify subtle patterns indicative of pathological conditions [191,192]. This manual annotation process is time-consuming and limited by the availability of qualified specialists. For example, annotating rare cardiac arrhythmias in ECG data may require cardiologists to review hours of recordings to identify fleeting anomalies. The cost of such expert annotation can become prohibitive when scaling to big data dimensions, creating a tension between the need for comprehensive training data and practical limitations in dataset creation [191,192].
To mitigate these annotation challenges, researchers are exploring several innovative approaches. Semi-supervised learning techniques leverage labeled and unlabeled data, allowing models to learn from the underlying data distribution while requiring fewer annotated examples. Synthetic data generation via GANs offers another promising avenue, creating realistic biomedical signals that can supplement real datasets while avoiding privacy concerns [192]. Collaborative annotation frameworks that distribute labeling tasks across multiple experts or institutions can help accelerate annotation while maintaining quality standards. While not eliminating the need for annotated data, these approaches can significantly reduce the annotation burden and make large-scale biomedical signal analysis more feasible in resource-constrained research environments [192].
12 Conformal Predictions for DL Models
Conformal prediction (CP) provides a rigorous statistical framework to quantify uncertainty in deep learning (DL) models by producing calibrated prediction sets with guaranteed coverage probabilities [194]. Unlike traditional methods focusing on point estimates, CP ensures reliability under minimal assumptions, making it critical for high-stakes domains such as healthcare, autonomous systems, and financial forecasting [194,195]. By constructing prediction sets rather than single-point estimates, CP guarantees a user-defined coverage probability (e.g., 95%) under the assumption of exchangeability (i.e., the calibration and test data are drawn from the same distribution). This approach is particularly valuable in biomedical applications such as arrhythmia classification, seizure detection, and anomaly segmentation, where model reliability is paramount for clinical decision making [196,197]. Consider a training dataset
and a deep learning model
The nonconformity score is the core component of CP, measuring how unusual a data point is relative to the calibration set. For classification tasks, a common choice for the nonconformity score is based on the predictive probability [197]:
12.1.1 Probability-Based Score
which accounts for the difference between the actual class probability and the next highest class probability.
12.1.3 Negative Log-Likelihood
which emphasizes the calibration of low-probability predictions.
For regression tasks, the typical choices are as follows [198]:
which measures the absolute difference between the actual value and the model’s prediction [185].
12.1.5 Absolute Quantile-Based Scores
where
12.2 Calibration and Quantile Thresholding
After computing the nonconformity scores
This threshold ensures that, under the assumption of exchangeability, the new observation falls within the prediction set with a probability of at least
12.3 Constructing Prediction Sets
For a new test
While CP guarantees coverage, prediction sets may include many classes. Techniques such as top-k conformal prediction or label-conditional thresholds can reduce ambiguity. For example, a prediction set containing multiple possible diagnoses can be presented to clinicians with associated probabilities in medical imaging classification.
The prediction interval is given by:
One-sided intervals or prediction bands can be constructed for time series forecasting. In healthcare applications such as glucose level prediction, these intervals help patients understand possible future values and make informed decisions.
12.4 Integration with DL Architectures
CP can be integrated with deep learning architectures, including CNNs, RNNs, transformer models, and hybrid systems. Notable points include:
• Efficient Computation
Deep models inherently output probabilities or continuous predictions, facilitating direct nonconformity score computation. However, mechanisms such as dropout and batch normalization may impact these scores and the resulting prediction sets [196–198].
• Mondrian Conformal Prediction
For multiclass problems, partitioning the feature space into subgroups (a Mondrian approach) can allow for class-conditional conformal prediction, which addresses heterogeneous uncertainty across classes [197].
• Adaptive Conformal Prediction
In dynamic applications such as real-time monitoring with wearable biosignal devices, adaptive CP methods adjust prediction sets based on incoming data to address nonstationary distributions [198].
12.5 Challenges and Limitations
While CP provides strong theoretical guarantees, several challenges remain:
1. Exchangeability assumption: CP relies on data exchangeability. Recalibration is needed in nonstationary settings (e.g., concept drift in ECG monitoring during different patient activities).
2. Prediction set size: Large prediction sets may hinder clinical usability. Research into compact yet reliable sets remain an open problem.
3. Computational overhead: While minimal during inference, calibration requires a held-out dataset, which may limit its use in low-data regimes. Techniques such as split conformal prediction or jackknife+ can mitigate this.
4. Multitask settings: Extending CP to multitask or multilabel scenarios requires careful design of nonconformity scores that account for label correlations.
Recent research has extended CP to new domains:
• Generative models: Conformalized GANs for reliable sample generation
• Reinforcement learning: CP for uncertainty-aware decision-making in autonomous systems
• Federated learning: CP frameworks that maintain privacy while providing coverage guarantees across distributed datasets
These advances demonstrate the growing relevance of CP in ensuring reliable and trustworthy AI systems across diverse applications.
DL techniques have significantly advanced the processing of biomedical signals; these techniques have shown great potential for extracting meaningful insights from large-scale datasets and improving the accuracy of diagnostic and prognostic models. However, several challenges are associated with applying DL to biomedical signals, as discussed below [106,199].
13.1 Data Acquisition and Quality
One of the primary challenges of DL for biomedical signals is acquiring high-quality data. A large amount of labeled data is crucial for training DL models. However, in many cases, obtaining such data is difficult because of the invasive nature of some medical procedures and the high cost of acquiring high-quality signals. Additionally, biomedical signals are susceptible to various artifacts, including noise, baseline drift, and motion artifacts, which affect their quality [35,106].
Preprocessing is a crucial step in preparing biomedical signals for deep learning. Preprocessing techniques such as filtering, normalization, and artifact removal are necessary to remove unwanted noise and artifacts that affect model performance. However, determining the most appropriate method for preprocessing data can be challenging, and selecting inappropriate methods can lead to poor model performance [200].
13.3 Model Interpretability and Explainability
Interpretability and interpretability are essential in medical applications, as clinicians must understand and trust the models’ decisions. However, DL models are often considered “black boxes,” and understanding how they make decisions can be challenging. Therefore, developing methods to interpret and explain model findings is crucial for adopting DL in clinical practice [201].
13.4 Integration with the Clinical Workflow
Integrating DL models into the clinical workflow can be challenging owing to several factors, including regulatory compliance, data privacy concerns, and the need for models to be user friendly and accessible to clinicians. Additionally, models must be developed and validated in diverse populations to ensure generalizability [202].
13.5 Limited Sample Size and Generalizability
However, DL models require large amounts of data. However, obtaining large datasets may be challenging in some medical applications, such as rare diseases. Additionally, DL models may need to be more balanced with training data, leading to poor generalizability to new datasets. Therefore, developing methods to train DL models effectively on small datasets and improve their generalizability is essential [35,40].
13.6 Hardware and Computational Requirements
However, DL models require significant computational resources for practical training, and deploying them on standard hardware is challenging. Specialized hardware, such as graphical processing units (GPUs), may be necessary for preparing DL models. However, not all institutions have access to such resources, making it challenging to develop and deploy DL models in clinical practice [35,203].
DL techniques require specialized data science, computer science, and mathematics expertise. However, not all medical institutions may have access to such expertise, making it challenging to develop and deploy DL models effectively [106,202].
13.8 Computational Time and Cost
However, DL models can be computationally expensive and require considerable time and resources for training and deployment. Additionally, developing and maintaining DL models can be costly, making it challenging for small medical institutions to adopt these techniques [35,36,106,199].
Regulatory compliance is a significant challenge in DL applications for biomedical signals. The biomedical research and healthcare field is subject to stringent regulatory requirements to ensure patient safety, data privacy, and ethical standards. DL models applied to biomedical signals, such as electrocardiograms, electroencephalograms, or medical imaging data, must adhere to these regulations. Compliance entails navigating complex frameworks such as the Health Insurance Portability and Accountability Act (HIPAA), the General Data Protection Regulation (GDPR), and the Food and Drug Administration (FDA) guidelines. Meeting these standards necessitates meticulous attention to data handling, consent management, secure storage, and rigorous validation of DL models. Striking a balance between innovation and regulatory compliance is crucial to fostering the adoption of DL techniques and maximizing their potential in improving healthcare outcomes while upholding the necessary safeguards [35,39,40,56].
One of the primary challenges in multimodal data fusion for biomedical signal analysis is the complexity of data integration. Biomedical signals often come from various sources, such as ECGs, EEGs, and medical imaging, each with different data formats, scales, and structures. Integrating this diverse information to provide comprehensive insights requires careful data preprocessing and alignment.
Another critical challenge is selecting the appropriate fusion strategy, whether late or early. Late fusion involves training separate DL models for each modality and combining their outputs later through techniques such as averaging or voting. However, early fusion combines modalities at the input level, allowing the model to learn joint representations. The choice of fusion strategy greatly influences the model’s ability to capture and leverage the relationships between different modalities, making it crucial to select the most suitable approach for a specific biomedical signal analysis task.
13.11 Model Uncertainty and Confidence Estimation
Estimating model uncertainty and confidence in DL models for biomedical signal analysis presents several challenges. Bayesian neural networks (BNNs) quantify uncertainty in predictions by incorporating probability distributions over the model’s weights. However, implementing BNNs introduces computational overhead because of the need for multiple samples during inference, which can significantly impact training time and resource requirements.
Moreover, interpreting uncertainty estimates can be challenging in medical decision-making scenarios. Understanding the model’s confidence level in its predictions is crucial for ensuring patient safety and building trust in AI systems. Proper communication and visualization of uncertainty measures are essential to convey the model’s reliability effectively.
13.12 Adversarial Attacks and Defenses
The DL models used in biomedical signal analysis are susceptible to adversarial attacks, where imperceptible perturbations to the input can lead to incorrect predictions. However, generating sufficient adversarial examples for biomedical signal data poses a significant challenge, especially when dealing with limited patient-specific data due to privacy concerns and ethical considerations.
Additionally, adversarial attacks designed for one model might transfer to others, highlighting the importance of building robust defenses that can withstand attacks across various architectures. While effective in enhancing model robustness, adversarial training requires careful tuning and might only partially eliminate the adversarial vulnerability.
13.13 Handling Long Sequences and Temporal Dependencies
Biomedical signals, such as time series data and EEG signals, often exhibit long sequences and temporal dependencies. Incorporating attention mechanisms, memory networks, or transformers to handle these long-term dependencies introduces increased model complexity. Fine-tuning and hyperparameter optimization are crucial for achieving optimal performance while minimizing computational costs.
Data preprocessing is another challenge when dealing with long sequences. Segmentation and padding are commonly used but may introduce artifacts or affect model performance. Maintaining the right balance between sequence handling and data preprocessing is essential to ensure accurate and efficient analysis of extended biomedical signal data.
However, DL models may raise ethical concerns about data privacy, bias, and fairness. Developing methods to address these concerns is crucial to ensure that DL is used responsibly in clinical practice [35,40]. Many examples have appeared because ethics are not considered when applying DL models in the medical field. Moreover, DL models designed to identify high-risk patients for specific diseases can be biased if trained on skewed data. For example 2019, researchers analyzed a healthcare system algorithm to identify patients at high risk for costly hospital readmissions [204]. The model was found to be biased against black patients, potentially leading to unequal allocation of resources and potentially delaying necessary care. This demonstrates how societal biases in healthcare data can be amplified by AI models, leading to discriminatory practices in patient care.
Moreover, DL models often rely on big data, such as sensitive patient data, medical records, and genetic information. If no proper safeguards are implemented, data breaches can occur, compromising patient privacy and potentially causing significant harm. For example, in 2017, the NHS in the UK faced criticism about sharing patient data with DeepMind (a Google subsidiary) without adequate patient consent [205]. This incident raises concerns about the potential for data breaches and unauthorized access to sensitive medical information when DL models are used in clinical settings.
The increasing awareness of ethical concerns surrounding DL in clinical settings has led to the development of AI regulations and acts in various regions to establish ethical frameworks for responsible development and use. Many countries have started establishing rules to guide the development and deployment of AI in medical sectors. The USA and the European Union are the two most important AI regulations. In the USA, to recognize the potential risks associated with AI, the National Institute of Standards and Technology (NIST) is actively developing guidelines for trustworthy AI. These guidelines focus on ensuring fairness, accountability, and transparency throughout the AI development lifecycle [206]. The European Union has taken a more comprehensive approach with the GDPR, which sets strict data privacy regulations for AI development and deployment [207]. This regulation aims to protect individuals’ data rights and ensure responsible data handling practices within the AI field.
Overall, these regulations highlight a growing commitment to harnessing the potential of DL in healthcare while mitigating the associated ethical risks. By prioritizing patient well-being, fairness, and data security, these frameworks pave the way for the responsible and ethical application of DL models in clinical settings.
13.16 Model Deployment and Ethical Considerations
Model deployment in the medical domain requires careful consideration of ethical aspects and patient safety. It must also adhere to regulatory standards and undergo rigorous quality assurance procedures. Every time an AI system is deployed, steps and measures are necessary to ensure the safety and efficacy of DL models in clinical settings [35,40].
13.17 Critical Analysis of Challenges
Data dependency and generalizability remain core challenges across DL applications in biomedical signals. Many studies highlight that DL models, particularly CNNs and GANs, depend heavily on large, clean datasets for high performance. In practice, obtaining such data is difficult because of privacy laws, data variability, and resource constraints. For example, a study by [208] reported that GANs trained on clean, standardized ECG datasets struggled with generalizability when tested on noisy, real-world data, demonstrating an accuracy drop of over 15% [208]. Additionally, models trained on homogeneous demographic datasets can exhibit bias, limiting their applicability across diverse patient populations [13,64] reported that an EEG seizure detection model trained on adult data performed significantly worse when applied to pediatric EEG data, highlighting the urgent need for more representative datasets. These limitations indicate that without addressing data dependency and diversity, the potential of DL in biomedical signals may be unrealized in practical clinical applications.
13.18 Overfitting in Small Datasets
When trained on small biomedical datasets, complex deep learning models face significant overfitting challenges. These models, particularly deep CNNs and RNNs, require substantial data to be generalized effectively. When trained on limited samples, they may memorize noise or idiosyncrasies in the training data rather than learning robust features. For example, studies using the MIT-BIH Arrhythmia Database have shown that complex neural networks can achieve near-perfect training accuracy but perform poorly on external validation sets from different institutions or populations [174]. Similarly, research on sleep stage classification via the Sleep-EDF expanded dataset demonstrated that models trained on small subsets of data exhibited more than 20% accuracy decreases when tested on independent cohorts [174]. Several strategies have been employed to address overfitting, including data augmentation with GANs [175], regularization techniques [149], and transfer learning [177]. Despite these approaches, overfitting remains a critical concern that limits the translation of DL models from research settings to clinical practice.
13.19 Impact of Data Privacy, Security, and Ethical Concerns on Clinical Workflow Integration
Integrating DL into clinical workflows offers significant potential to improve healthcare outcomes, but data privacy, security, and ethical concerns pose substantial challenges. These issues can delay adoption, increase costs, and create resistance among clinicians, ultimately hindering the effective deployment of AI technologies in healthcare settings. Data privacy is a significant barrier, as patients may hesitate to share sensitive health data due to privacy risks, limiting the availability of high-quality datasets for training AI models [35,106]. Strict regulations such as the HIPAA and GDPR further complicate integration by requiring robust data protection measures, which can be resource intensive and time consuming [35,40]. Security risks, such as cyberattacks and data breaches, also threaten the adoption of AI systems, particularly for smaller institutions lacking advanced cybersecurity resources [35,203].
Ethical concerns, including bias and lack of transparency, further impede integration. Models trained on biased datasets can produce discriminatory outcomes, exacerbating healthcare disparities [204]. Many DL models’ “black-box” nature makes it difficult for clinicians to trust or understand their decisions, leading to resistance in adopting these technologies [209]. Additionally, fragmented implementation due to a lack of standardized protocols can limit the scalability and generalizability of AI systems [202]. A combination of technological, policy, and educational strategies is essential to address these challenges. Federated learning and differential privacy can protect data while enabling collaborative model training [210,211]. Explainable AI (XAI) techniques can improve transparency and build trust among clinicians [209]. Policy measures, such as ethical AI guidelines and regulatory sandboxes, can ensure compliance and foster innovation [207]. Training programs for healthcare professionals and patient education can also promote responsible AI adoption [108,202].
In conclusion, addressing data privacy, security, and ethical concerns is critical for integrating DL into clinical workflows. By leveraging technological advancements, implementing robust policies, and fostering collaboration, healthcare institutions can overcome these barriers and harness AI’s full potential to improve patient care.
AI has been instrumental in proposing data-driven solutions to address data privacy concerns. Significant development in this regard has been the field of fair representation learning evolution over the last decade [34,212,213]. This field focuses on three main areas: 1) mitigating bias in AI models to ensure fair treatment across different demographic groups; 2) enhancing interpretability to make AI decision-making processes more transparent and understandable; and 3) developing techniques that allow learning representations from data while preserving privacy, particularly in sensitive domains such as healthcare or finance [214,215]. By advancing fair representation learning, AI researchers and practitioners aim to foster ethical and equitable AI systems that respect individual privacy, minimize biases, and empower users to trust and comprehend the decisions made by intelligent algorithms [34,213]. This multidimensional approach contributes to the responsible deployment of AI technologies in various sectors, promoting inclusivity and ethical considerations in developing and applying intelligent systems [212]. Below are different solutions discussed. These solutions pave the way for more ethical and fair AI systems, ensuring that all can enjoy the benefits of AI without compromising individual privacy and fairness.
Fair machine learning eliminates algorithmic bias in automated decision-making based on ML models [34,212]. The goal is to ensure that decisions made by these models are not unfairly biased toward or against protected subgroups in the population. To achieve fairness, researchers often employ techniques such as adversarial training, reweighting of samples, or adjusting decision boundaries to account for demographic imbalances [216–218]. Moreover, interpretable models contribute to the fairness agenda by allowing stakeholders to understand the factors influencing predictions, enabling the identification and rectification of biased patterns. This approach aligns with the broader movement toward responsible and ethical AI development, emphasizing the importance of addressing biases that may perpetuate societal inequalities [217,218]. In addition to addressing bias, fair representation learning plays a pivotal role in constructing AI models sensitive to privacy concerns. As AI systems increasingly leverage massive amounts of data, particularly in sensitive domains such as healthcare, preserving the privacy of individuals becomes paramount [216,218]. Techniques such as federated learning, differential privacy, and secure multiparty computation enable the extraction of meaningful insights from distributed data sources without compromising the confidentiality of individual records. By incorporating fair representation learning into the development pipeline, AI practitioners aim to balance the need for accurate and practical models and the imperative to safeguard individual privacy, fostering a more responsible and trustworthy landscape for deploying AI technologies [216].
14.2 Fair Representation Learning
In fair representation learning, the aim is to learn latent representations that are informative for a particular task while removing all sensitive factors (e.g., gender or race) contained in the input data [214]. This approach helps in producing fair classifiers via data preprocessing [214]. By disentangling task-relevant information from sensitive attributes, fair representation learning contributes to developing models that can make accurate predictions without reinforcing or perpetuating biases associated with these sensitive characteristics [215,219]. This separation allows for creating models prioritizing fairness, ensuring that decisions are based on factors directly relevant to the task rather than inadvertently incorporating and perpetuating societal prejudices [220,221]. As such, fair representation learning has become a cornerstone in the quest for more equitable and unbiased AI systems.
Dataset bias unlearning, or machine unlearning, is an emerging field that selectively erases learned data from ML models [213]. This process is crucial in cases where learning models are associated with bias, which could result from biased datasets or human mistakes1. Unlearning approaches are used to reduce and eliminate these biases [213]. The significance of dataset bias unlearning lies in its capacity to rectify and adapt models when confronted with evolving and dynamic datasets [222]. As models are exposed to new information, selectively unlearning biased patterns become instrumental in maintaining model fairness and accuracy. This adaptability is especially vital in real-world applications where data distributions may change over time, ensuring that ML systems remain robust, unbiased, and aligned with ethical considerations throughout their operational life cycles [213,222].
Federated learning provides a new and promising technique for developing and training DL models for biomedical signals while preserving medical data privacy [210]. Unlike traditional DL model training, federated learning decentralizes the training process; the trained model’s coefficients and parameters are transferred to the central server, keeping medical data on institution servers or individual devices available [211]. Fig. 33 shows the flowchart of federated learning in a hospital environment.
Figure 33: Federated learning for the hospital environment
In general, federated learning consists of 5 main steps [210,211,223]:
• Model Distribution: The central server receives the model’s parameters from each local server located at each hospital and then distributes the global DL model parameters backward to local servers at the hospitals.
• Local Training: This step includes training a local DL model at each hospital server based on the hospital’s local biomedical signal data (e.g., EEG, ECG). This ensures that the model parameters are only uploaded to the central server, not the patient data and biomedical signals.
• Model Update: The updated local DL model parameters are uploaded to the central server.
• Global Model Aggregation: The central server aggregates the parameters based on the uploaded parameters from all hospital servers. This effectively combines the knowledge gained by each local DL method into one super DL model.
• Improved Global Model: The aggregated model parameters are used to update the global model and are distributed for another round of local training.
Overall, these steps are executed iteratively, allowing the global model to be generalized to all hospital data without compromising patient privacy and by only sharing the knowledge gained by each local model [223]. Additionally, this technique allows the training of robust DL models and enables institutions to gain combined knowledge from diverse datasets. However, federated learning has several issues, such as frequent communication between servers to upload and broadcast data, which can be resource intensive; additionally, the data distributions may vary significantly across institutions, affecting model performance [223].
15 Software Toolkit for Deep Learning
DL has become a key technology in AI and ML, in which sophisticated models are created to process large amounts of data. To facilitate the development and implementation of these models, various software tools have been developed to make DL accessible to researchers, developers, and data scientists [99,224]. Among the most popular algorithms are MATLAB [36], TensorFlow [224], PyTorch [225], Keras [99], Caffe [226], and Apache MXNet [227]. Each of these frameworks offers a range of features and functionalities that make it easier to develop and implement sophisticated DL models.
In addition to these frameworks, other software tools have gained prominence in the DL community. One such tool is MATLAB, which provides various tools and functions for developing and implementing DL models. MATLAB offers a range of DL functions, including neural networks, CNNs, and RNNs, as well as a range of visualization tools for exploring and analyzing DL models [36].
Another critical player in the DL community is Google Brain. Google Brain is an AI research project at Google that aims to develop advanced ML algorithms and technologies. Various tools and frameworks for deep learning, including TensorFlow, one of the world’s most popular DL frameworks, have been developed. Google Brain has also been involved in developing other software tools and technologies for deep learning, such as Keras and TensorFlow.js, which enable DL models to run in the browser [99,224].
Overall, the availability of software tools has played a crucial role in advancing deep learning. TensorFlow, PyTorch, Keras, Caffe, and Apache MXNet are among today’s most popular DL frameworks. Additionally, MATLAB and Google Brain are essential players in the DL community, offering a range of tools and frameworks for developing and implementing DL models. As DL continues to evolve, we expect more advanced software tools to be developed to meet the growing demand for DL models. Fig. 34 shows the main tools for DL model development and design for biomedical signals.
Figure 34: Main tools for developing DL models for biomedical signals
16 Real-World Case Studies: Applications of DL in Biomedical Signal Analysis
In this section, we present real-world case studies that demonstrate the successful application of DL in biomedical signal analysis. These case studies highlight the significant contributions of DL techniques in different medical domains and their potential for improving healthcare outcomes. The studies are categorized based on the algorithm used in the paper.
CNNs are popular in biomedical signal analysis, mainly when signals are represented as images (e.g., spectrograms). CNNs detect spatial features and patterns, making them effective in arrhythmia detection and respiratory sound classification. The robustness of CNNs to noise allows them to focus on relevant patterns, increasing model accuracy even in real-time scenarios. However, CNNs struggle with capturing temporal dependencies, a limitation for sequential data such as ECG and EEG signals. The applications of CNNs in biomedical signals range from arrhythmia classification to EEG artifact removal. Table 3 shows the list of applications.
RNNs and their variant LSTM networks are well suited for analyzing sequential biomedical data such as ECG and EEG, where time dependence is crucial. RNNs capture temporal patterns, effectively detecting cardiac arrhythmia and muscle fatigue. While LSTMs overcome some of the limitations of traditional RNNs by preserving long-term dependencies, they are computationally intensive and may be overfitting with limited data. These models have demonstrated high accuracy across applications, especially in arrhythmia detection. Table 4 shows the list of applications.
16.3 Hybrid Models (CNN-RNN, CNN-LSTM, CNN-SVM)
Hybrid models that combine CNNs with RNNs or LSTMs capture spatial and temporal features in biomedical signals, making them suitable for complex signals requiring multidimensional analysis. The CNN layers extract spatial patterns, whereas the RNN/LSTM layers handle temporal dependencies, enabling robust classification in respiratory sound and EMG analysis tasks. Although computationally intensive and more prone to overfitting, these models are decisive for applications that need pattern recognition and temporal understanding. Combining a CNN with a support vector machine (SVM) allows for effective feature extraction with a CNN followed by robust classification with an SVM, which is beneficial for high-dimensional data. The CNN-SVM approach is helpful in applications with limited labeled data, as SVM helps maximize the decision boundary. However, this combination is computationally intensive, particularly for large datasets. The CNN-SVM is helpful in applications such as muscle fat detection, providing strong classification performance even with small datasets. Table 5 shows the list of applications.
16.4 Empirical Analysis of the Methodologies
CNNs are essential architectures for processing biomedical signals. This analysis evaluates their performance across various applications via research papers applying CNN models to domains. Table 6 Empirical analysis of the CNN models.
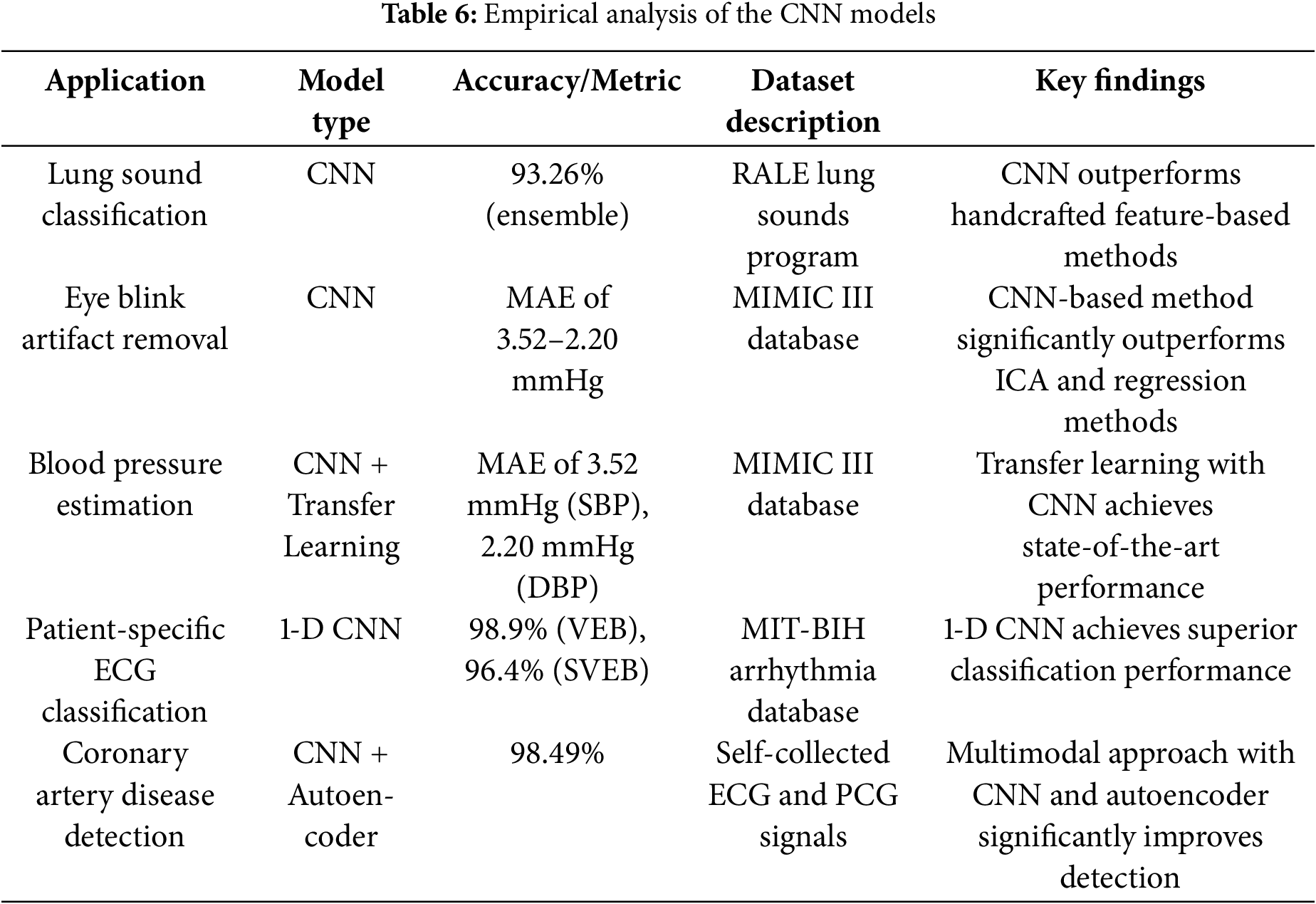
CNNs have emerged as powerful tools in biomedical signal analysis, demonstrating exceptional performance across various healthcare applications. This analysis evaluates the effectiveness of CNNs in different biomedical signal processing tasks based on recent research findings. In lung sound classification, CNNs outperform traditional feature-based methods, achieving an accuracy of 93.26% through automatic feature extraction from raw audio signals. This superior performance suggests that CNNs are particularly well suited for analyzing the complex patterns in lung sound recordings, where traditional handcrafted features may fail to capture the full range of diagnostic information.
For eye blink artifact removal from EEG signals, CNN-based methods show substantial improvements over conventional techniques such as independent component analysis (ICA) and regression. The CNN approach achieves a mean absolute error (MAE) of 3.52 mmHg for systolic blood pressure (SBP) and 2.20 mmHg for diastolic blood pressure (DBP), demonstrating its effectiveness in learning the complex relationships between artifacts and clean signals. A CNN-based transfer learning approach has achieved state-of-the-art performance in blood pressure estimation from photoplethysmography (PPG) signals. This method requires only 50 data samples per person to train accurate personalized models, addressing the challenge of limited patient-specific data while maintaining high accuracy.
For patient-specific ECG classification, one-dimensional CNN architectures have demonstrated exceptional performance. Studies have shown that a simple 1-D CNN can achieve 98.9% accuracy for ventricular ectopic beats and 96.4% for supraventricular ectopic beats on the MIT-BIH arrhythmia database. This performance highlights the effectiveness of CNNs in capturing the temporal patterns and morphological features present in ECG signals. A multimodal CNN approach that combines ECGs, PCGs, and their coupling signals achieves 98.49% coronary artery disease detection accuracy. This architecture demonstrates how combining different data modalities through advanced neural network architectures can significantly enhance diagnostic performance.
These findings demonstrate that CNNs consistently outperform other architectures in biomedical signal analysis because they automatically extract meaningful features from raw data. The effectiveness of these methods in capturing spatial and temporal patterns makes them particularly suitable for complex biomedical signals. While other architectures have strengths, CNNs are the most versatile and practical choice for many biomedical signal processing tasks across diverse healthcare applications.
RNNs, LSTM networks, and GRUs are essential architectures for processing sequential data. This analysis evaluates their performance across various applications via research papers that apply RNN, LSTM, and GRU models to different domains. Table 7 Empirical analysis of the RNN, LSTM, and GRU models.

In biomedical signal analysis tasks, LSTM consistently outperforms traditional RNNs and GRUs in performance. For ECG signal classification via the MIT-BIH arrhythmia database, the LSTM model achieves an accuracy of 99.39%, significantly outperforming standard RNN models. This suggests that the ability of LSTM to capture long-term dependencies in sequential data makes it particularly suitable for ECG analysis. In blood pressure estimation from photoplethysmography signals via the MIMIC II database, the LSTM-based multistage model attains an accuracy of 90.1%, outperforming traditional RNN approaches. For muscle activity detection from surface electromyography (sEMG) signals, LSTM achieves 97% accuracy on simulated data and 90% accuracy on accurate data, clearly outperforming threshold-based methods. In muscle fatigue classification using sEMG signals from 20 participants, the LSTM model reached 95.18% accuracy, surpassing the CNN and SVM models. While the GRU performs well in these medical applications, it generally falls slightly short of the accuracy of the LSTM.
Ensemble models that combine LSTM and the GRU demonstrate the best performance in wearable sensor data for human activity recognition tasks. The LSTM-GRU ensemble model achieves 99.06% accuracy on the UCI-HAR and WISDM datasets, suggesting that combining different RNN architectures can leverage their strengths. The LSTM model alone performs well at 96.61% accuracy, whereas the GRU model achieves 94.06% accuracy. Traditional RNN models perform worse than these specialized RNN variants, indicating that the LSTM and GRU architectures are better suited for capturing the complex patterns in human activity data.
In genomic and tabular data analysis for type 2 diabetes prediction via the PIMA Indian dataset, the LSTM model performs strongly, with an accuracy of 81.8%, outperforming traditional machine learning approaches. This finding suggests that LSTM’s ability to handle long-term dependencies and complex patterns makes it suitable for genomic sequence analysis. Traditional RNN models perform worse than LSTM in this context, further highlighting the advantages of LSTM for such data types.
Based on empirical evidence, LSTM generally outperforms traditional RNNs and GRUs in most applications, particularly in healthcare signal analysis and human activity recognition. While the GRU offers computational efficiency advantages, it typically achieves slightly lower accuracy than LSTM in these tasks. Ensemble approaches combining different RNN architectures can improve performance, but LSTM remains the most consistent and accurate choice for sequential data analysis across diverse domains.
16.4.3 Hybrid Models (CNN-RNN, CNN-LSTM, CNN-SVM)
CNNs have emerged as powerful tools in biomedical signal analysis, with exceptional performance across different applications. Based on recent research findings, this analysis evaluates the effectiveness of hybrid models using CNNs and RNN, LSTM, or SVM in different biomedical signal processing tasks. Table 8 Empirical analysis of the hybrid models.
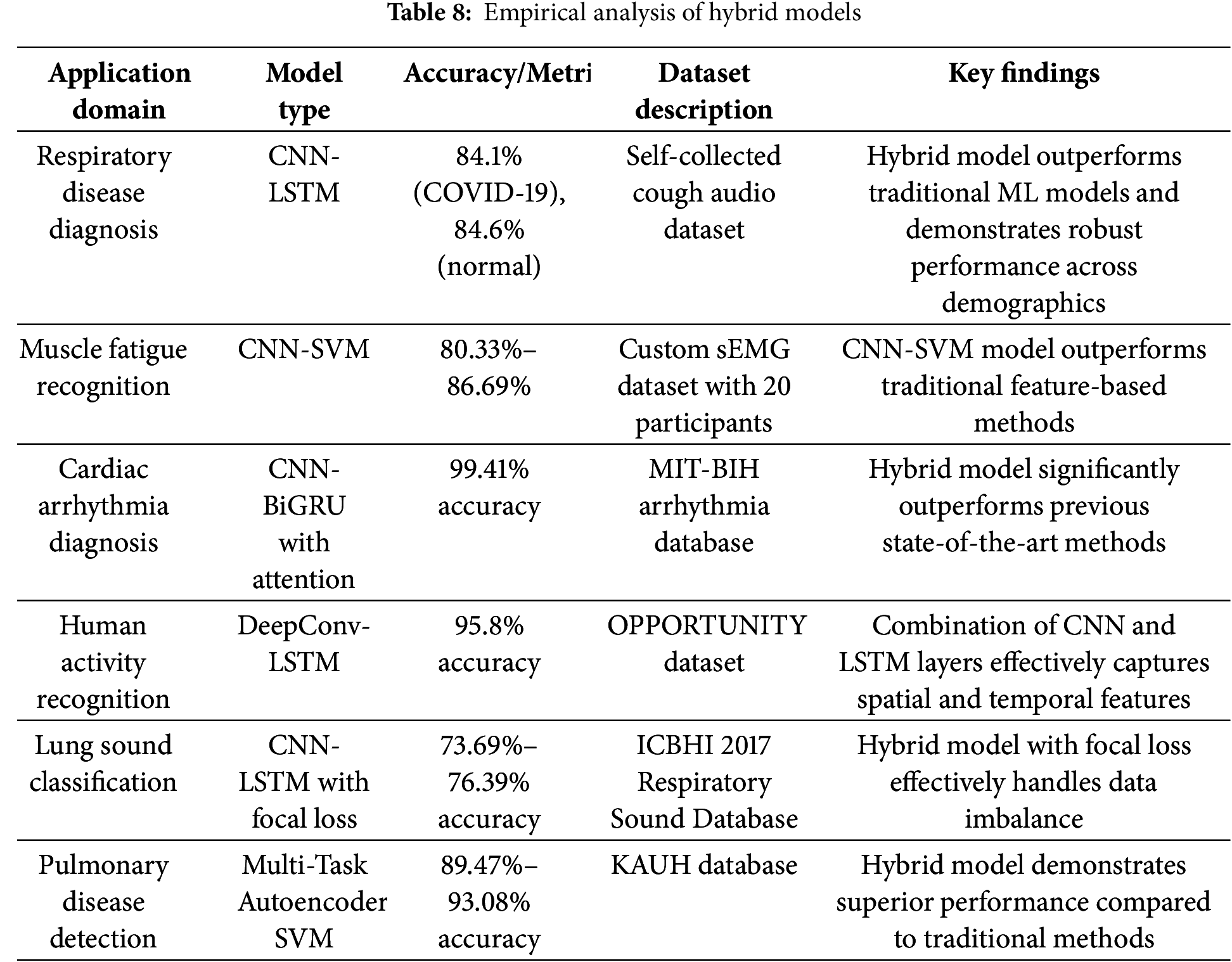
In respiratory disease diagnosis using cough audio signals, hybrid CNN-LSTM models have shown marked effectiveness. The RBF-Net model achieved 84.1% accuracy for COVID-19 detection and 84.6% for standard samples. This hybrid architecture effectively combines CNNs for feature extraction with LSTM layers to capture temporal dependencies in cough audio signals. The model’s robust performance across diverse demographics highlights its potential for real-world applications in respiratory disease screening. A CNN-SVM framework that significantly outperforms traditional feature-based methods was developed for muscle fatigue recognition via surface electromyography (sEMG) signals. Their model achieved classification accuracies ranging from 80.33% to 86.69% on a custom dataset with 20 participants. The CNN component automatically extracted spatial and temporal features from raw sEMG signals, whereas the SVM classifier provided robust classification performance. This approach eliminates the need for manual feature engineering, which is typically time-consuming and requires domain expertise.
A hybrid CNN-BiGRU model with multihead attention achieved 99.41% accuracy on the MIT-BIH arrhythmia database for cardiac arrhythmia diagnosis. This architecture effectively combines CNNs for feature extraction with bidirectional gated recurrent units (BiGRUs) to model temporal dynamics in ECG signals. The multihead attention mechanism further enhances the performance by focusing on the most relevant features for classification. The model significantly outperforms previous state-of-the-art methods, demonstrating the power of combining CNNs with recurrent architectures and attention mechanisms for ECG analysis. For human activity recognition via wearable sensors, a DeepConvLSTM framework outperforms previous approaches by 4% on average on the OPPORTUNITY dataset and 9% on an 18-class gesture recognition task. Their model combined CNN layers for feature extraction with LSTM layers to capture temporal dependencies in sensor data. The architecture demonstrated the effectiveness of deep learning in automatically extracting features from raw sensor data without requiring extensive preprocessing.
A hybrid CNN-LSTM network with focal loss achieved state-of-the-art ICBHI 2017 Respiratory Sound Database results for lung sound classification. Their model effectively handled data imbalance while achieving accuracies ranging from 73.69% to 76.39% across different evaluation strategies. The combination of CNNs for feature extraction and LSTM for temporal modeling, along with the focal loss function, demonstrated superior performance compared with traditional methods. A hybrid multitask autoencoder-SVM model that achieved 89.47% to 93.08% accuracy on the KAUH database was used for pulmonary disease detection from lung sound signals. Their framework combined an autoencoder for unsupervised feature learning with a supervised classifier, optimizing classification accuracy and signal reconstruction and integrating SVM classification with CNN-extracted features enhanced performance, particularly in scenarios with limited training data.
These findings demonstrate that CNNs consistently outperform other architectures in biomedical signal analysis because they automatically extract meaningful features from raw data. The effectiveness of these methods in capturing spatial and temporal patterns makes them particularly suitable for complex biomedical signals. While other architectures have strengths, CNNs are the most versatile and practical choice for many biomedical signal processing tasks across diverse healthcare applications. The hybrid approaches that combine CNNs with other architectures (such as LSTM, attention mechanisms, or SVM) often achieve the highest performance, suggesting that future research should continue to explore these combinations for specific biomedical applications.
16.5 Comparisons of Methodologies
In biomedical signal processing, the choice of DL methodology depends on the nature of the data, the specific task, and the available computational resources. Below, we provide a detailed comparison of the most commonly used methodologies, including CNNs, RNNs, and hybrid models (e.g., CNN-LSTM).
• Strengths
Spatial Feature Extraction: CNNs excel at capturing spatial patterns in data, making them ideal for tasks where the input can be represented as an image (e.g., spectrograms of ECG or EEG signals) [3,36].
• Hierarchical Learning: CNNs automatically learn hierarchical features, starting from low-level patterns (e.g., edges) to high-level abstractions (e.g., arrhythmia patterns in ECG signals) [1,243].
• Robustness to Noise: CNNs are relatively robust to noise, especially when combined with data augmentation techniques such as random cropping or rotation [36,243].
Weaknesses
• Limited Temporal Modeling: CNNs struggle to capture temporal dependencies in sequential data, such as EEG or PPG signals, where the order of data points is crucial [3,36,44].
• High Computational Cost: Training CNNs on large datasets can be computationally expensive, mainly when deep architectures with many layers are used [44].
Best use cases
• ECG Classification: CNNs have been widely used for arrhythmia detection and achieved high accuracy on datasets such as the MIT-BIH Arrhythmia Database [27].
• Image-Based Signal Analysis: CNNs are adequate for tasks where signals are transformed into images, such as time-frequency representations (e.g., spectrograms or scalograms) [2,25].
• Strengths
Temporal Dependency Modeling: RNNs, particularly LSTM and GRU variants, are designed to handle sequential data, making them ideal for tasks such as EEG seizure detection or PPG-based blood pressure estimation [244].
• Memory of Past Inputs: RNNs can remember information from previous time steps, which is crucial for tasks where the context of past data points influences the current prediction [244].
Weaknesses
• Vanishing Gradient Problem: Traditional RNNs suffer from the vanishing gradient problem, which limits their ability to learn long-term dependencies. LSTMs and GRUs mitigate this issue but are still computationally expensive [244].
• Sensitivity to Noise: RNNs can be sensitive to noise in the input data, requiring careful preprocessing and regularization [244].
Best use cases
• EEG Signal Analysis: RNNs are widely used for seizure detection, sleep stage classification, and BCI applications [244].
• Time Series Prediction: RNNs are adequate for tasks such as predicting blood pressure or glucose levels from continuous monitoring data [244].
16.5.3 Hybrid Models (CNN-RNN, CNN-LSTM, CNN-SVM)
• Strengths
Combining Spatial and Temporal Features: Hybrid models, such as CNN-LSTM, leverage the strengths of both CNNs (spatial feature extraction) and RNNs (temporal dependency modeling). This makes them highly effective for tasks that require both spatial and temporal analysis, such as PPG-based blood pressure estimation or respiratory sound classification [26,125].
• Improved Performance: Hybrid models often achieve state-of-the-art performance on complex tasks by capturing both local patterns and long-term dependencies [125,240].
Weaknesses
• Computational Complexity: Hybrid models are computationally intensive and require significant resources for training and inference [26,125].
• Risk of Overfitting: The increased complexity of hybrid models makes them more prone to overfitting, especially when trained on small datasets [26,125].
Best use cases
• Multimodal Signal Analysis: Hybrid models are ideal for tasks that involve multiple types of signals, such as combining ECG and PPG data for cardiovascular disease diagnosis [26,125].
• Complex Temporal Tasks: Tasks such as predicting disease progression or detecting anomalies in long-term monitoring data benefit from the combined strengths of CNNs and RNNs [125].
16.6 Critical Analysis of Current Applications
The studies reviewed here highlight significant advancements in the use of DL for biomedical signal processing, particularly in enhancing diagnostic accuracy and automating feature extraction. Models such as CNNs and RNNs have demonstrated high performance in tasks such as lung sounds classification and wheeze segmentation [229,245], often achieving accuracy levels above 90% on controlled datasets, including PhysioNet’s MIT-BIH Arrhythmia Database and the TUH EEG Corpus [233]. These successes underscore deep learning’s ability to capture complex, nonlinear patterns in biomedical data that are challenging for traditional methods. Despite these achievements, however, the reliability of these models often depends on the availability of high-quality, large-scale datasets, a condition rarely met in clinical practice. Studies have shown that models trained on clean, standardized data can underperform on noisy or inconsistent real-world data, indicating a critical gap in data robustness.
Another common issue is the limited generalizability of these models across diverse patient populations and varying clinical conditions. Many studies rely on homogeneous datasets that lack representation from different demographic or health backgrounds, resulting in models that may perform poorly in broader clinical settings [241]. For example, an ECG model trained on adult data may struggle when applied to pediatric cases because of physiological differences. In contrast, studies have shown that performance declines when models are tested on demographically diverse datasets [30]. DL models, particularly GANs and hybrid models, often demand substantial computational resources, which are typically unavailable in many healthcare settings. This computational complexity restricts real-world applicability, particularly in underresourced medical facilities that lack high-performance computing hardware [30].
Finally, interpretability and ethical concerns further limit the clinical adoption of these DL models. Although some studies have employed tools such as Grad-CAM and SHAP for interpretability [95,105], the practical value of these techniques in clinical contexts remains unclear. DL models are frequently criticized as black-box algorithms, hindering trust among clinicians, who require transparent insights to make informed decisions. Ethical and regulatory considerations also pose challenges, especially when patient privacy and data protection laws such as the GDPR and HIPAA are in play [21,246]. Many studies do not address how to handle these legal requirements, data anonymization, or secure data management, which are essential for real-world applications. As a result, while these models demonstrate promising capabilities, addressing their limitations in data dependency, generalizability, computational demands, interpretability, and ethical compliance will be essential for successful clinical integration.
17 From Lab to Real-Life: Realizing DL in Biomedical Signal Processing
Integrating DL into biomedical signal processing has transformed the field, enabling the transition from theoretical research to practical, life-changing applications. DL models have bridged the gap between laboratory innovation and real-world implementation because of their remarkable ability to analyze complex and noisy biomedical signals. This section explores how these technologies, initially developed in research settings, now underpin tools and systems widely used in the clinical, consumer, and research domains. From wearable devices that monitor heart rhythms to AI-powered diagnostic tools for disease detection, these applications demonstrate the profound impact of DL on patient care, diagnostics, and treatment planning. By examining real-world examples, we highlight the challenges overcome, the factors driving this evolution, and the transformative role of DL in advancing healthcare outcomes.
17.1 Apple Watch–Atrial Fibrillation Detection
The Apple Watch incorporates DL algorithms for atrial fibrillation (AFib) detection via PPG signals. The device processes PPG signals in real time by employing RNNs, identifying irregular heart rhythms indicative of AFib. This feature, validated through large-scale clinical studies such as the Apple Heart Study [247], has received FDA clearance and is now widely used by millions of users for early detection and monitoring of cardiac conditions.
17.2 AliveCor—AI-Powered ECG Interpretation
AliveCor’s KardiaMobile is an FDA-approved personal ECG device in which CNNs detect atrial fibrillation and other arrhythmias. The device pairs with a smartphone application, enabling users to record and analyze real-time ECG data. The CNN model processes signal features and provides instant feedback, empowering users with actionable health insights. This innovation has seen widespread cardiovascular monitoring adoption among clinicians and patients [248].
17.3 Tempus–AI in Precision Oncology
Tempus, a leading health technology company, utilizes DL to analyze genomic and transcriptomic signals alongside clinical data for precision oncology. Tempus AI models predict treatment outcomes and optimize therapy selection for cancer patients. The system integrates DL pipelines into hospitals and research institutions, directly impacting clinical decision-making [249].
17.4 SleepScore Max-Sleep Analysis and Tracking
The SleepScore Max device uses DL algorithms to analyze breathing and movement signals for sleep stage classification. Its noncontact sensors collect respiratory and motion data, which are then processed via hybrid DL models to provide insights into sleep quality and patterns. The product is widely used in consumer markets and research studies for personalized sleep improvement recommendations [250].
17.5 iRhythm-Zio Patch for Cardiac Monitoring
iRhythm Technologies’ Zio Patch is a wearable device for long-term cardiac rhythm monitoring. Powered by DL algorithms, it analyzes ECG data over extended periods, detecting arrhythmias and other abnormalities. The Zio Patch has received FDA approval and is used in clinical practice to diagnose conditions that require continuous monitoring, such as atrial fibrillation and bradycardia [251].
17.6 Bridging the Gap: Advancing Practical Deployment
These real-world applications demonstrate how DL has evolved from research to market-ready products, delivering significant benefits in biomedical signal processing. The transition from academic research to commercial success involves overcoming numerous challenges, including scalability, interoperability, and user acceptance [250]. However, the successful integration of DL technologies into healthcare highlights their potential to revolutionize diagnostics, monitoring, and treatment [248]. The success of these applications in the stock market underscores several critical factors:
1. Regulatory Approvals: Achieving FDA or equivalent clearances validates their reliability and safety. Regulatory approval is a rigorous process that ensures that technology meets stringent standards for efficacy and safety, which is crucial for gaining trust among healthcare providers, investors, and end-users. For example, FDA clearance opens doors to the U.S. market and serves as a benchmark for global markets, enhancing the product’s credibility and market potential [202,207].
2. User-Centric Design: Seamless integration into consumer devices or clinical workflows increased adoption rates. A key aspect of this is the intuitive design that caters to healthcare professionals and patients. For example, wearable devices monitoring vital signs must be easy to use, comfortable, and provide actionable insights without extensive training. Similarly, clinical tools must integrate smoothly with existing electronic health records (EHR) systems to avoid disrupting workflows, increasing their acceptance and utility in real-world settings.
3. Real-World Validation: Extensive testing ensured these tools met real-world demands in diverse environments. Beyond controlled laboratory settings, real-world validation involves testing the technology in various clinical and nonclinical scenarios to ensure robustness and reliability. This includes evaluating performance across patient populations, environmental conditions, and usage patterns. Real-world validation not only demonstrates the technology’s practical applicability but also helps identify and address potential issues before its widespread deployment [248–250].
Moreover, the financial performance of companies leveraging these technologies in the stock market reflects investor confidence in their long-term viability and impact. As these technologies continue to mature, they are likely to drive further innovation, creating new opportunities for growth and improving patient outcomes globally. The convergence of advanced algorithms, regulatory support, and user-friendly design is paving the way for a new era in biomedical signal processing. DL plays a central role in transforming healthcare delivery.
18 Black-Box Challenge: DL Interpretability in Clinical Practice
The “black-box” nature of DL models presents significant challenges for their trust and acceptance in clinical applications. Unlike traditional rule-based systems or simpler machine learning models, DL models often operate as opaque systems where human experts do not easily understand the relationship between input data and output predictions. This lack of transparency raises concerns about reliability, accountability, and safety in healthcare settings where decisions can have life-or-death consequences.
18.1 Critical Need for Interpretability in Healthcare
In clinical practice, interpretability is not merely a desirable feature but also a fundamental requirement. Healthcare providers must understand why a model makes a particular diagnosis or recommendation as follows:
• Validation of the clinical relevance of the predictions
• Identifying potential errors or biases in the model
• Integrate model insights with their professional judgment
• Explain decisions to patients and justify treatment plans
The opacity of DL models creates barriers to these essential clinical processes. A study in 2019 highlighted that while DL models can achieve remarkable accuracy in medical imaging tasks, their inability to provide transparent explanations for predictions remains a significant obstacle to widespread clinical adoption [252].
18.2 Real-World Application Examples Illustrating the Black-Box Challenge
Cardiac monitoring devices that use DL to detect arrhythmias must provide interpretable results for clinicians to trust. A DL model developed for arrhythmia detection from ECG signals has achieved high accuracy but faces resistance in clinical settings because of its lack of transparency regarding which ECG segments or features contribute to its classification [14].
Deep learning models have demonstrated high accuracy in detecting epileptic seizures from EEG signals, with sensitivity exceeding 90% in specific applications [13]. These models analyze complex temporal patterns in EEG data to identify seizure activity. However, they typically output a binary classification (seizure/no seizure) without indicating which specific temporal or spectral features contributed to the prediction. For example, a DL model might flag a seizure event in an EEG recording without specifying whether the detection was based on increased theta activity, decreased alpha waves, or specific spike-and-wave patterns. This lack of transparency makes it difficult for clinicians to validate the model’s decision or to understand potential false positives/negatives.
18.3 Approaches to Enhance Interpretability
To address these challenges, researchers have developed various interpretability techniques:
• Feature importance visualization: Methods such as local interpretability model-agnostic explanations (LIME) and Shapley additive explanations (SHAP) values help identify which input features most influence model predictions [105,253,254]
• Attention mechanisms: These allow models to highlight specific regions of input data that contribute to decisions, which is particularly useful in image and signal analysis [105]
• Prototypal networks: These networks identify representative examples from the training data that are similar to the input being analyzed, providing clinicians with concrete examples to understand model reasoning [253]
DL has revolutionized many fields, and its prospects in biomedical signals hold immense potential for advancing healthcare and improving patient outcomes. By leveraging the power of deep neural networks, researchers and clinicians can extract valuable insights from complex biomedical data, paving the way for more accurate diagnostics, personalized treatments, and innovative healthcare solutions. First, DL algorithms can enhance the analysis and interpretation of biomedical signals, such as ECGs, EEGs, and EMGs. These algorithms can automatically detect patterns, anomalies, and abnormalities in signals, enabling earlier and more accurate detection of diseases. For example, DL models have shown promising results in detecting cardiac arrhythmias and epileptic seizures, allowing timely interventions and reducing the risk of complications.
Second, DL can facilitate the development of intelligent medical devices and wearable technologies. By integrating DL algorithms into these devices, real-time monitoring and analysis of biomedical signals can be performed, providing continuous health monitoring and early warning systems for patients. This can be particularly useful in managing chronic diseases, where regular monitoring is essential. DL can also enable the extraction of valuable biomarkers from signals, aiding in predicting and preventing diseases.
Third, DL can contribute to precision medicine by facilitating the development of personalized treatment strategies. DL algorithms can identify patient-specific patterns and predict treatment outcomes by leveraging large amounts of patient data, including genetic information, medical records, and biomedical signals. This can help clinicians select the most effective therapies, reduce adverse effects, and optimize treatment plans tailored to individual patients.
Furthermore, DL can assist in drug discovery and development processes. DL models can identify potential drug targets, predict drug efficacy, and optimize drug design by analyzing large datasets, including molecular structure, genomics, and clinical trial data. This can significantly accelerate the drug discovery pipeline and lead to the development of novel therapies for various diseases.
Finally, DL can support medical research by enabling the integration and analysis of heterogeneous data sources. Biomedical signals can be combined with other data types, such as imaging data, electronic health records, and omics data, to understand diseases and their underlying mechanisms comprehensively. DL algorithms can integrate and learn from these diverse datasets, leading to discoveries and insights that can advance our understanding of complex diseases and inform the development of innovative treatments.
Despite these promising prospects, challenges remain, including model interpretability and data privacy. The black-box nature of DL models hinders their adoption in critical medical applications where model interpretability is essential. Researchers are actively exploring methods for making DL models more interpretable, fostering trust among healthcare professionals and leading to broader acceptance in clinical settings. Moreover, the sensitive nature of biomedical data demands stringent privacy measures. Federated learning offers a solution by allowing models to be trained across multiple institutions without sharing raw data, preserving patient privacy while benefiting from collective knowledge. Implementing privacy-preserving techniques will be crucial in encouraging healthcare institutions to collaborate and leverage the full potential of DL in biomedical signals.
20 Transforming Healthcare with Deep Learning: Unlocking the Power of Biomedical Signals
DL has emerged as a powerful tool in biomedical signal analysis, exhibiting remarkable success in various applications, such as disease diagnosis, image classification, and predictive modeling. In the future, the prospects of DL in biomedical signals present many exciting opportunities to transform healthcare and medical research [43]. One of the most promising prospects is the advancement of personalized medicine. DL algorithms can learn intricate patterns from diverse biomedical data, enabling the development of tailored treatment strategies for individual patients. This approach can optimize therapeutic outcomes, minimize adverse effects, and improve patient satisfaction. DL has demonstrated the potential to predict drug responses based on genetic information, paving the way for more comprehensive personalized treatment plans [43].
Real-time monitoring and decision support systems are another area where DL holds immense potential. With DL algorithms, wearable devices and implantable sensors can continuously analyze biomedical signals and provide instantaneous feedback to healthcare providers and patients. This real-time monitoring can aid in the early detection of anomalies and prompt intervention, enhancing patient outcomes [255]. Furthermore, integrating DL with medical imaging data and biomedical signals could revolutionize disease diagnosis and prognosis. Multimodal learning techniques can combine information from various sources, providing a more comprehensive view of a patient’s health status. For example, integrating electrocardiogram (ECG) data with medical images improved the accuracy of heart disease diagnosis. This approach can lead to earlier and more accurate diagnoses, allowing timely interventions and improved patient management [256].
Despite these promising prospects, challenges remain, including model interpretability and data privacy. The black-box nature of DL models hinders their adoption in critical medical applications where model interpretability is essential. Researchers are actively exploring methods for making DL models more interpretable, fostering trust among healthcare professionals and leading to broader acceptance in clinical settings [43]. Moreover, the sensitive nature of biomedical data demands stringent privacy measures. Federated learning offers a solution by allowing models to be trained across multiple institutions without sharing raw data, preserving patient privacy while benefiting from collective knowledge. Implementing privacy-preserving techniques will be crucial in encouraging healthcare institutions to collaborate and leverage the full potential of DL in biomedical signals [256].
Overall, the prospects of DL in biomedical signals hold enormous potential to revolutionize healthcare delivery and medical research. The development of personalized medicine, real-time monitoring, multimodal learning, model interpretability, and privacy-preserving approaches is expected to drive the adoption of deep understanding in various clinical applications, ultimately improving patient care and advancing medical knowledge. Fig. 35 illustrates the potential impact of DL in revolutionizing healthcare through biomedical signal analysis. It depicts a futuristic healthcare scenario where deep learning-enabled technologies are pivotal in diagnosis, treatment, and personalized patient care.
Figure 35: Potential impact of DL in revolutionizing healthcare through biomedical signals
21 Advantages and Disadvantages of DL in Biomedical Signals
DL appears to be an emerging tool for practical healthcare applications. It offers numerous benefits but faces significant challenges. Understanding DL strengths and weaknesses is crucial for practical applications in biomedical signals. Below, we outline the key advantages and disadvantages of using DL for biomedical signals.
• High accuracy: DL outperforms ML in processing and analyzing complex biomedical signals when a suitable model is designed and applied.
• Automatic feature extraction: Automatic feature extraction can learn directly from raw data in the time or frequency domain, eliminating manual feature engineering.
• Scalability: The ability to handle large and high-dimensional datasets is essential in fields such as genomics and neuroimaging.
• Versatility: This method can be applied to various tasks, including image-based diagnostics, signal classification, and personalized treatment. Additionally, it can handle multiple inputs simultaneously from different data types.
• Data requirements: Large, labeled datasets are needed, which are often difficult to obtain because of privacy and cost issues.
• Computational demands: Significant computational power and specialized hardware are needed, which can be a barrier for some researchers and institutions.
• Hard interpretability: The model predictions’ “black box” nature makes understanding them in clinical settings challenging. More training is required to understand the model predictions fully.
• Risk of overfitting: Overfitting, especially with small datasets, leads to poor generalization of new data.
• Ethical and privacy concerns: Raise issues related to patient data confidentiality and the ethical use of AI in healthcare.
By examining these advantages and disadvantages, we provide a balanced perspective on using DL in biomedical signals, highlighting the factors that must be addressed for successful integration into healthcare applications.
22 Advancing AI in Healthcare: Strategic Research Directions and Technical Pathways for Enhanced Patient Care
Integrating AI into healthcare systems represents one of our most transformative technological advancements. Since its conceptualization in the 1950s, AI has evolved from simple pattern recognition algorithms to sophisticated machine learning models capable of processing complex medical data. Advances in computing power, the availability of large-scale health datasets, and innovations in deep learning architectures have accelerated this evolution [257,258].
AI applications in healthcare now span a wide range of medical disciplines, including diagnostic imaging, personalized treatment planning, drug discovery, and patient monitoring [259,260]. These technologies have demonstrated remarkable potential for improving diagnostic accuracy, optimizing treatment protocols, and enhancing operational efficiency within healthcare systems. For example, AI-powered imaging analysis has achieved diagnostic performance comparable to that of human experts in some instances, whereas predictive models have enabled early intervention for high-risk patients [261]. Despite these advances, significant challenges remain in translating AI capabilities into widespread clinical practice. Data quality, algorithmic bias, interpretability, and patient privacy hinder adoption [262]. Additionally, there is a critical need for research that addresses the human factors in AI implementation, including clinician trust, workflow integration, and the preservation of the essential human element in patient care [263,264].
This paper identifies specific research directions and technical pathways that can overcome these barriers and accelerate the development of AI systems that truly enhance patient outcomes while supporting healthcare professionals. By focusing on these strategic areas, researchers can contribute to creating AI tools that are technically advanced, clinically practical, and ethically sound. The following subsection discusses and proposes research directions and technical pathways for AI in healthcare.
22.1 Explainable AI (XAI) Development
• More transparent AI models that can justify their diagnostic or treatment recommendations should be created [264]
• Implementing techniques such as layer-wise relevance propagation or attention mechanisms to highlight which patient features influence AI decisions
• Develop standardized evaluation metrics for model interpretability in medical contexts
22.2 Federated Learning Frameworks
• Designing decentralized learning systems that allow AI models to be trained across multiple healthcare institutions without sharing raw patient data
• Implement secure aggregation protocols to protect patient privacy while enabling knowledge sharing
• Creating benchmarking standards to evaluate the performance of federated learning models against centralized approaches
22.3 AI Robustness and Validation
• Establishing rigorous testing protocols for AI models under various clinical scenarios and patient populations
• Development of adversarial training methods to improve model resilience against noisy or manipulated medical data
• Create open-source validation toolkits for healthcare AI that include diverse datasets and performance metrics
22.4 Personalized Treatment Optimization
• Investigate AI-driven adaptive treatment strategies that evolve with patient response
• Explore the integration of multi-omics data (genomics, proteomics, and metabolomics) with clinical data for precision medicine [265]
• Development of AI models that can predict optimal drug combinations for individual patients
22.5 Clinical Workflow Integration
• Study human–AI interaction design principles specifically for healthcare settings
• Creating AI systems that can seamlessly integrate with EHRs and clinical decision support systems
• Investigate the impact of AI-assisted documentation on clinician workload and patient interaction quality [265]
22.6 Health Equity and Bias Mitigation
• Development of AI fairness algorithms that can identify and correct for biases in medical data
• Create benchmark datasets that represent diverse populations to test the generalizability of the AI model
• Establishing guidelines for inclusive AI development that involves multidisciplinary teams, including ethicists and patient representatives
• Research predictive models for identifying patients at risk of developing chronic conditions
• Development of AI-powered interventions for personalized prevention strategies
• Investigate the cost-effectiveness of AI-driven preventive care programs
This paper comprehensively reviews DL in biomedical signal processing, highlighting its current impact and future potential. DL has significantly increased the accuracy of models for analyzing various biomedical signals, enabling more effective extraction of valuable insights. This review demonstrates the versatile applications of DL architectures such as CNNs, RNNs, and GANs across diverse medical domains, from personalized treatment planning to early disease detection.
The study’s findings reveal several essential insights. DL models have consistently outperformed traditional methods in tasks such as arrhythmia detection, seizure identification, and signal classification, with some models achieving accuracy rates exceeding 98% on standard datasets. The superiority of DL approaches is particularly notable in handling complex, high-dimensional data where traditional feature engineering methods fall short. For example, CNNs have proven exceptionally effective in capturing spatial patterns in ECG signals for arrhythmia detection. Moreover, RNNs and their variants have shown remarkable ability to model temporal dependencies in EEG signals for seizure prediction. However, the review also acknowledges significant challenges that must be addressed for effective clinical deployment. These include:
• Data limitations: Most DL models require large, high-quality datasets, often unavailable in clinical settings.
• Computational demands: Specialized hardware remains a barrier for many institutions
• Model interpretability: The “black-box” nature of DL models hinders their clinical acceptance
• Generalizability: Models often perform poorly when applied to diverse patient populations
Future research directions should focus on the following:
1. Development of more efficient DL architectures that require less computational resources while maintaining performance.
2. Robust data augmentation techniques for biomedical signals have been created to address data scarcity.
3. Advancing explainability methods to improve model interpretability and clinical trust.
4. Implementing privacy-preserving techniques such as federated learning to enable multi-institutional collaboration without compromising patient data.
5. Standardizing evaluation frameworks to facilitate meaningful comparisons of different DL approaches.
6. Exploring the integration of multimodal data sources for comprehensive patient assessment.
7. Establishing benchmarks for real-world clinical performance rather than just technical metrics.
The future of this field lies in developing more effective models that can improve patient outcomes through personalized treatment. This paper aims to inspire further research in this field and lead to innovative solutions that can enhance healthcare. Healthcare systems and the well-being of patients and health workers can benefit greatly from AI. However, there are still barriers to applying AI in clinical settings, such as trust, coordination, data and privacy challenges, and patient hesitance. We should design AI systems that put patients first and earn the confidence of health workers in this fantastic technology. AI is not meant to replace health workers but to support them and enhance their lives. After all, people do not want a robot to treat them; they need a human touch, which is vital in every field.
AI and clinicians should collaborate to optimize patient outcomes. This article provides directions for future research and development in AI for healthcare. With the massive amount of data and computing power we currently have, we foresee a growing role for AI and biosensors in clinics that will complement or assist healthcare professionals and ease their burden. Moreover, this review highlights the transformative potential of DL in biomedical signal processing, particularly in improving diagnostic accuracy for conditions detectable through ECG and EEG signals. However, challenges in interpretability, data scarcity, and computational demands persist. For example, while CNNs show high accuracy in arrhythmia detection, they remain constrained by data availability and generalizability across patient populations. Addressing these issues through enhanced data sharing, interpretability frameworks, and efficient model architectures will be pivotal to future advancements in this field.
This review aims to provide researchers and healthcare professionals with a comprehensive view of the recent technologies in this field from both technical and clinical points of view. We explored and discussed ways AI can be applied to biomedical signals to provide diagnoses, predict outcomes and treatments, and monitor patients. Additionally, we focused on the significance of explainable methods in this context. The advantages of using AI in the biomedical signals field include enhanced performance and efficiency in processing data and the ability to develop personalized medical care. However, the main obstacles still need to be overcome, such as providing explainable AI to ensure that the method is trusted by healthcare providers, ensuring that algorithms have no biases, and gathering extensive and high-quality data. Finally, we explored some research inquiries about biomedical signal-based AI systems, including the necessity for improved adaptability across diverse patient groups and the establishment of criteria for assessing AI effectiveness in clinical environments.
Acknowledgement: We acknowledge the support of the NSERC (Natural Sciences and Engineering Research Council of Canada).
Funding Statement: The Natural Sciences and Engineering Research Council of Canada (NSERC) funded this review study.
Author Contributions: The authors confirm their contributions to the paper: Ali Mohammad Alqudah conducted the literature review and wrote the manuscript. Zahra Moussavi contributed to the manuscript by providing editing services and discussing the materials presented. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015;61(3):85–117. doi:10.1016/j.neunet.2014.09.003. [Google Scholar] [PubMed] [CrossRef]
2. Acharya UR, Fujita H, Lih OS, Hagiwara Y, Tan JH, Adam M. Automated detection of arrhythmias using different intervals of tachycardia ECG segments with convolutional neural network. Inf Sci. 2017;405(1):81–90. doi:10.1016/j.ins.2017.04.012. [Google Scholar] [CrossRef]
3. Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19(1):221–48. doi:10.1146/annurev-bioeng-071516-044442. [Google Scholar] [PubMed] [CrossRef]
4. Tobore I, Li J, Liu Y, Al-Handarish Y, Kandwal A, Nie Z, et al. Deep learning intervention for health care challenges: some biomedical domain considerations. JMIR Mhealth Uhealth. 2019;7(8):e11966. doi:10.2196/11966. [Google Scholar] [PubMed] [CrossRef]
5. Attia ZI, Noseworthy PA, Lopez-Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet. 2019;394(10201):861–7. doi:10.1016/S0140-6736(19)31721-0. [Google Scholar] [PubMed] [CrossRef]
6. Xiao L, Luo K, Liu J, Foroughi A. A hybrid deep approach to recognizing student activity and monitoring health physique based on accelerometer data from smartphones. Sci Rep. 2024;14(1):14006. doi:10.1038/s41598-024-63934-8. [Google Scholar] [PubMed] [CrossRef]
7. Zhai X, Jelfs B, Chan RHM, Tin C. Self-recalibrating surface EMG pattern recognition for neuroprosthesis control based on convolutional neural network. Front Neurosci. 2017;11:379. doi:10.3389/fnins.2017.00379. [Google Scholar] [PubMed] [CrossRef]
8. Ghislieri M, Cerone GL, Knaflitz M, Agostini V. Long short-term memory (LSTM) recurrent neural network for muscle activity detection. J Neuroeng Rehabil. 2021;18(1):153. doi:10.1186/s12984-021-00945-w. [Google Scholar] [PubMed] [CrossRef]
9. Goshtasbi N, Boostani R, Sanei S. SleepFCN: a fully convolutional deep learning framework for sleep stage classification using single-channel electroencephalograms. IEEE Trans Neural Syst Rehabil Eng. 2022;30(4):2088–96. doi:10.1109/TNSRE.2022.3192988. [Google Scholar] [PubMed] [CrossRef]
10. Aboalayon K, Faezipour M, Almuhammadi W, Moslehpour S. Sleep stage classification using EEG signal analysis: a comprehensive survey and new investigation. Entropy. 2016;18(9):272. doi:10.3390/e18090272. [Google Scholar] [CrossRef]
11. Xu M, Jie J, Zhou W, Zhou H, Jin S. Synthetic epileptic brain activities with TripleGAN. Comput Math Methods Med. 2022;2022:2841228. doi:10.1155/2022/2841228. [Google Scholar] [PubMed] [CrossRef]
12. Gajic D, Djurovic Z, Gligorijevic J, Di Gennaro S, Savic-Gajic I. Detection of epileptiform activity in EEG signals based on time-frequency and non-linear analysis. Front Comput Neurosci. 2015;9:38. doi:10.3389/fncom.2015.00038. [Google Scholar] [PubMed] [CrossRef]
13. Samee NA, Mahmoud NF, Aldhahri EA, Rafiq A, Muthanna MSA, Ahmad I. RNN and BiLSTM fusion for accurate automatic epileptic seizure diagnosis using EEG signals. Life. 2022;12(12):1946. doi:10.3390/life12121946. [Google Scholar] [PubMed] [CrossRef]
14. Rajpurkar P, Hannun AY, Haghpanahi M, Bourn C, Ng AY. Cardiologist-level arrhythmia detection with convolutional neural networks. arXiv:1707.01836. 2017. [Google Scholar]
15. Salem M, Taheri S, Yuan J. ECG arrhythmia classification using transfer learning from 2-dimensional deep CNN features. In: 2018 IEEE Biomedical Circuits and Systems Conference (BioCAS); 2018 Oct 17–19; Cleveland, OH, USA. p. 1–4. doi:10.1109/BIOCAS.2018.8584808. [Google Scholar] [CrossRef]
16. Lee SH. Natural language generation for electronic health records. npj Digit Med. 2018;1(1):63. doi:10.1038/s41746-018-0070-0. [Google Scholar] [PubMed] [CrossRef]
17. Wang C, Elazab A, Jia F, Wu J, Hu Q. Automated chest screening based on a hybrid model of transfer learning and convolutional sparse denoising autoencoder. Biomed Eng Online. 2018;17(1):63. doi:10.1186/s12938-018-0496-2. [Google Scholar] [PubMed] [CrossRef]
18. Dillen A, Steckelmacher D, Efthymiadis K, Langlois K, De Beir A, Marusic U, et al. Deep learning for biosignal control: insights from basic to real-time methods with recommendations. J Neural Eng. 2022;19(1):011003. doi:10.1088/1741-2552/ac4f9a. [Google Scholar] [PubMed] [CrossRef]
19. Sajno E, Bartolotta S, Tuena C, Cipresso P, Pedroli E, Riva G. Machine learning in biosignals processing for mental health: a narrative review. Front Psychol. 2023;13:1066317. doi:10.3389/fpsyg.2022.1066317. [Google Scholar] [PubMed] [CrossRef]
20. Belo D, Rodrigues J, Vaz JR, Pezarat-Correia P, Gamboa H. Biosignals learning and synthesis using deep neural networks. Biomed Eng Online. 2017;16(1):115. doi:10.1186/s12938-017-0405-0. [Google Scholar] [PubMed] [CrossRef]
21. Murdoch B. Privacy and artificial intelligence: challenges for protecting health information in a new era. BMC Med Ethics. 2021;22(1):122. doi:10.1186/s12910-021-00687-3. [Google Scholar] [PubMed] [CrossRef]
22. Buber E, Diri B. Performance analysis and CPU vs GPU comparison for deep learning. In: 2018 6th International Conference on Control Engineering & Information Technology (CEIT); 2018 Oct 25–27; Istanbul, Turkey. p. 1–6. doi:10.1109/CEIT.2018.8751930. [Google Scholar] [CrossRef]
23. Weissler EH, Naumann T, Andersson T, Ranganath R, Elemento O, Luo Y, et al. The role of machine learning in clinical research: transforming the future of evidence generation. Trials. 2021;22(1):537. doi:10.1186/s13063-021-05489-x. [Google Scholar] [PubMed] [CrossRef]
24. Alzubaidi L, Zhang J, Humaidi AJ, Al-Dujaili A, Duan Y, Al-Shamma O, et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data. 2021;8(1):53. doi:10.1186/s40537-021-00444-8. [Google Scholar] [PubMed] [CrossRef]
25. Alquran H, Alqudah AM, Abu-Qasmieh I, Al-Badarneh A, Almashaqbeh S. ECG classification using higher order spectral estimation and deep learning techniques. Neural Netw World. 2019;29(4):207–19. doi:10.14311/NNW.2019.29.014. [Google Scholar] [CrossRef]
26. Alamatsaz N, Tabatabaei L, Yazdchi M, Payan H, Alamatsaz N, Nasimi F. A lightweight hybrid CNN-LSTM explainable model for ECG-based arrhythmia detection. Biomed Signal Process Control. 2024;90(1):105884. doi:10.1016/j.bspc.2023.105884. [Google Scholar] [CrossRef]
27. Perez Alday EA, Gu A, Shah AJ, Robichaux C, Ian Wong AK, Liu C, et al. Classification of 12-lead ECGs: the PhysioNet/computing in cardiology challenge 2020. Physiol Meas. 2021;41(12):124003. doi:10.1088/1361-6579/abc960. [Google Scholar] [PubMed] [CrossRef]
28. Shan L, Li Y, Jiang H, Zhou P, Niu J, Liu R, et al. Abnormal ECG detection based on an adversarial autoencoder. Front Physiol. 2022;13:961724. doi:10.3389/fphys.2022.961724. [Google Scholar] [PubMed] [CrossRef]
29. Tzallas AT, Tsipouras MG, Fotiadis DI. Automatic seizure detection based on time-frequency analysis and artificial neural networks. Comput Intell Neurosci. 2007;2007(4):80510. doi:10.1155/2007/80510. [Google Scholar] [PubMed] [CrossRef]
30. Yildirim Ö. A novel wavelet sequence based on deep bidirectional LSTM network model for ECG signal classification. Comput Biol Med. 2018;96:189–202. doi:10.1016/j.compbiomed.2018.03.016. [Google Scholar] [PubMed] [CrossRef]
31. Cover T, Hart P. Nearest neighbor pattern classification. IEEE Trans Inf Theory. 1967;13(1):21–7. doi:10.1109/TIT.1967.1053964. [Google Scholar] [CrossRef]
32. Ho TK. Random decision forests. In: Proceedings of 3rd International Conference on Document Analysis and Recognition; 1995 Aug 14–16; Montreal, QC, Canada. p. 278–82. doi:10.1109/ICDAR.1995.598994. [Google Scholar] [CrossRef]
33. Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Statist. 2001;29(5):1189–232. doi:10.1214/aos/1013203451. [Google Scholar] [PubMed] [CrossRef]
34. Sai S, Mittal U, Chamola V, Huang K, Spinelli I, Scardapane S, et al. Machine un-learning: an overview of techniques, applications, and future directions. Cogn Comput. 2024;16(2):482–506. doi:10.1007/s12559-023-10219-3. [Google Scholar] [CrossRef]
35. Rajkomar A, Dean J, Kohane I. Machine learning in medicine. New Engl J Med. 2019;380(14):1347–58. doi:10.1056/NEJMra1814259. [Google Scholar] [PubMed] [CrossRef]
36. Kim P. Matlab deep learning: with machine learning, neural networks and artificial intelligence. 1st ed. New York, NY, USA: Apress Media, LLC; 2017. [Google Scholar]
37. Suganyadevi S, Seethalakshmi V, Balasamy K. A review on deep learning in medical image analysis. Int J Multimed Inf Retr. 2022;11(1):19–38. doi:10.1007/s13735-021-00218-1. [Google Scholar] [PubMed] [CrossRef]
38. Lee JG, Jun S, Cho YW, Lee H, Kim GB, Seo JB, et al. Deep learning in medical imaging: general overview. Korean J Radiol. 2017;18(4):570–84. doi:10.3348/kjr.2017.18.4.570. [Google Scholar] [PubMed] [CrossRef]
39. Ravi D, Wong C, Deligianni F, Berthelot M, Andreu-Perez J, Lo B, et al. Deep learning for health informatics. IEEE J Biomed Health Inform. 2017;21(1):4–21. doi:10.1109/JBHI.2016.2636665. [Google Scholar] [PubMed] [CrossRef]
40. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. doi:10.1038/nature14539. [Google Scholar] [PubMed] [CrossRef]
41. Che Z, Purushotham S, Cho K, Sontag D, Liu Y. Recurrent neural networks for multivariate time series with missing values. Sci Rep. 2018;8(1):6085. doi:10.1038/s41598-018-24271-9. [Google Scholar] [PubMed] [CrossRef]
42. Hinton GE, Osindero S, Teh YW. A fast learning algorithm for deep belief nets. Neural Comput. 2006;18(7):1527–54. doi:10.1162/neco.2006.18.7.1527. [Google Scholar] [PubMed] [CrossRef]
43. Miotto R, Wang F, Wang S, Jiang X, Dudley JT. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform. 2018;19(6):1236–46. doi:10.1093/bib/bbx044. [Google Scholar] [PubMed] [CrossRef]
44. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42(13):60–88. doi:10.1016/j.media.2017.07.005. [Google Scholar] [PubMed] [CrossRef]
45. Li M, Jiang Y, Zhang Y, Zhu H. Medical image analysis using deep learning algorithms. Front Public Health. 2023;11:1273253. doi:10.3389/fpubh.2023.1273253. [Google Scholar] [PubMed] [CrossRef]
46. Somani S, Russak AJ, Richter F, Zhao S, Vaid A, Chaudhry F, et al. Deep learning and the electrocardiogram: review of the current state-of-the-art. Europace. 2021;23(8):1179–91. doi:10.1093/europace/euaa377. [Google Scholar] [PubMed] [CrossRef]
47. Altaheri H, Muhammad G, Alsulaiman M, Amin SU, Ali Altuwaijri G, Abdul W, et al. Deep learning techniques for classification of electroencephalogram (EEG) motor imagery (MI) signals: a review. Neural Comput Appl. 2023;35(20):14681–722. doi:10.1007/s00521-021-06352-5. [Google Scholar] [CrossRef]
48. Kavakiotis I, Tsave O, Salifoglou A, Maglaveras N, Vlahavas I, Chouvarda I. Machine learning and data mining methods in diabetes research. Comput Struct Biotechnol J. 2017;15(7453):104–16. doi:10.1016/j.csbj.2016.12.005. [Google Scholar] [PubMed] [CrossRef]
49. Choubey A, Bhargava Choubey S, Subba Rao SPV. A machine learning algorithm for biomedical signal processing application. In: Machine learning methods for signal, image and speech processing. Aalborg, Denmark: River Publishers; 2021. p. 149–68. [Google Scholar]
50. James G, Witten D, Hastie T, Tibshirani R. An introduction to statistical learning. New York, NY, USA: Springer; 2021. doi:10.1007/978-1-0716-1418-1 [Google Scholar] [CrossRef]
51. Watson DS. On the philosophy of unsupervised learning. Philos Technol. 2023;36(2):28. doi:10.1007/s13347-023-00635-6. [Google Scholar] [CrossRef]
52. Weber T, Racanière S, Reichert DP, Buesing L, Guez A, Rezende DJ, et al. Imagination-augmented agents for deep reinforcement learning. arXiv:1707.06203. 2017. [Google Scholar]
53. Rim B, Sung NJ, Min S, Hong M. Deep learning in physiological signal data: a survey. Sensors. 2020;20(4):E969. doi:10.3390/s20040969. [Google Scholar] [PubMed] [CrossRef]
54. Ganapathy N, Swaminathan R, Deserno TM. Deep learning on 1-D biosignals: a taxonomy-based survey. Yearb Med Inform. 2018;27(1):98–109. doi:10.1055/s-0038-1667083. [Google Scholar] [PubMed] [CrossRef]
55. Craik A, He Y, Contreras-Vidal JL. Deep learning for electroencephalogram (EEG) classification tasks: a review. J Neural Eng. 2019;16(3):031001. doi:10.1088/1741-2552/ab0ab5. [Google Scholar] [PubMed] [CrossRef]
56. Mamoshina P, Vieira A, Putin E, Zhavoronkov A. Applications of deep learning in biomedicine. Mol Pharm. 2016;13(5):1445–54. doi:10.1021/acs.molpharmaceut.5b00982. [Google Scholar] [PubMed] [CrossRef]
57. Min S, Lee B, Yoon S. Deep learning in bioinformatics. Brief Bioinform. 2017;18(5):851–69. doi:10.1093/bib/bbw068. [Google Scholar] [PubMed] [CrossRef]
58. Mahmud M, Kaiser MS, Hussain A, Vassanelli S. Applications of deep learning and reinforcement learning to biological data. IEEE Trans Neural Netw Learn Syst. 2018;29(6):2063–79. doi:10.1109/TNNLS.2018.2790388. [Google Scholar] [PubMed] [CrossRef]
59. Zhang Y, Dong Z, Li S, Cattani C. Deep learning methods for biomedical information analysis. J Ambient Intell Humaniz Comput. 2023;14(5):5293–6. doi:10.1007/s12652-023-04617-6. [Google Scholar] [CrossRef]
60. Petmezas G, Stefanopoulos L, Kilintzis V, Tzavelis A, Rogers JA, Katsaggelos AK, et al. State-of-the-art deep learning methods on electrocardiogram data: systematic review. JMIR Med Inform. 2022;10(8):e38454. doi:10.2196/38454. [Google Scholar] [PubMed] [CrossRef]
61. Srinivasu PN, Shafi J, Krishna TB, Sujatha CN, Praveen SP, Ijaz MF. Using recurrent neural networks for predicting type-2 diabetes from genomic and tabular data. Diagnostics. 2022;12(12):3067. doi:10.3390/diagnostics12123067. [Google Scholar] [PubMed] [CrossRef]
62. Ning Y, He S, Wu Z, Xing C, Zhang LJ. A review of deep learning based speech synthesis. Appl Sci. 2019;9(19):4050. doi:10.3390/app9194050. [Google Scholar] [CrossRef]
63. Xiao C, Choi E, Sun J. Opportunities and challenges in developing deep learning models using electronic health records data: a systematic review. J Am Med Inform Assoc. 2018;25(10):1419–28. doi:10.1093/jamia/ocy068. [Google Scholar] [PubMed] [CrossRef]
64. Habashi AG, Azab AM, Eldawlatly S, Aly GM. Generative adversarial networks in EEG analysis: an overview. J Neuroeng Rehabil. 2023;20(1):40. doi:10.1186/s12984-023-01169-w. [Google Scholar] [PubMed] [CrossRef]
65. Mendez V, Lhoste C, Micera S. EMG data augmentation for grasp classification using generative adversarial networks. In: 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC); 2022 Jul 11–15; Glasgow, Scotland, UK. p. 3619–22. doi:10.1109/EMBC48229.2022.9871625. [Google Scholar] [PubMed] [CrossRef]
66. Sun H, Zhang F, Zhang Y. An LSTM and GAN based ECG abnormal signal generator. In: Advances in artificial intelligence and applied cognitive computing. Cham, Switzerland: Springer; 2021. p. 743–55. doi:10.1007/978-3-030-70296-0_54. [Google Scholar] [CrossRef]
67. Shaker AM, Tantawi M, Shedeed HA, Tolba MF. Generalization of convolutional neural networks for ECG classification using generative adversarial networks. IEEE Access. 2020;8:35592–605. doi:10.1109/ACCESS.2020.2974712. [Google Scholar] [CrossRef]
68. Hershey S, Chaudhuri S, Ellis DPW, Gemmeke JF, Jansen A, Moore RC, et al. CNN architectures for large-scale audio classification. In: 2017 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); 2017 Mar 5–9; New Orleans, LA, USA. p. 131–5. doi:10.1109/ICASSP.2017.7952132. [Google Scholar] [CrossRef]
69. Liang Y, Yin S, Tang Q, Zheng Z, Elgendi M, Chen Z. Deep learning algorithm classifies heartbeat events based on electrocardiogram signals. Front Physiol. 2020;11:569050. doi:10.3389/fphys.2020.569050. [Google Scholar] [PubMed] [CrossRef]
70. Shabaan M, Arshid K, Yaqub M, Jinchao F, Zia MS, Bojja GR, et al. Survey: smartphone-based assessment of cardiovascular diseases using ECG and PPG analysis. BMC Med Inform Decis Mak. 2020;20(1):177. doi:10.1186/s12911-020-01199-7. [Google Scholar] [PubMed] [CrossRef]
71. Ukil A, Bandyoapdhyay S, Puri C, Pal A. IoT healthcare analytics: the importance of anomaly detection. In: 2016 IEEE 30th International Conference on Advanced Information Networking and Applications (AINA); 2016 Mar 23–25; Crans-Montana, Switzerland. p. 994–7. doi:10.1109/AINA.2016.158. [Google Scholar] [CrossRef]
72. Gao Z, Lu G, Yan P, Lyu C, Li X, Shang W, et al. Automatic change detection for real-time monitoring of EEG signals. Front Physiol. 2018;9:325. doi:10.3389/fphys.2018.00325. [Google Scholar] [PubMed] [CrossRef]
73. Bachmann M, Lass J, Hinrikus H. Single channel EEG analysis for detection of depression. Biomed Signal Process Control. 2017;31(1):391–7. doi:10.1016/j.bspc.2016.09.010. [Google Scholar] [CrossRef]
74. Devuyst S, Dutoit T, Stenuit P, Kerkhofs M. Automatic K-complexes detection in sleep EEG recordings using likelihood thresholds. In: 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology; 2010 Aug 31–Sep 4; Buenos Aires, Argentina. p. 4658–61. doi:10.1109/IEMBS.2010.5626447. [Google Scholar] [PubMed] [CrossRef]
75. Chen G, Lu G, Xie Z, Shang W. Anomaly detection in EEG signals: a case study on similarity measure. Comput Intell Neurosci. 2020;2020:6925107. doi:10.1155/2020/6925107. [Google Scholar] [PubMed] [CrossRef]
76. Hassan AR, Subasi A. A decision support system for automated identification of sleep stages from single-channel EEG signals. Knowl Based Syst. 2017;128(7063):115–24. doi:10.1016/j.knosys.2017.05.005. [Google Scholar] [CrossRef]
77. Alqudah AM, Alquran H, Abu Qasmieh I. Classification of heart sound short records using bispectrum analysis approach images and deep learning. Netw Model Anal Health Inform Bioinform. 2020;9(1):66. doi:10.1007/s13721-020-00272-5. [Google Scholar] [CrossRef]
78. Alqudah AM, Qazan S, Al-Ebbini L, Alquran H, Abu Qasmieh I. ECG heartbeatarrhythmias classification: a comparison study between different types of spectrum representation and convolutional neural networks architectures. J Ambient Intell Humaniz Comput. 2022;13(10):4877–907. doi:10.1007/s12652-021-03247-0. [Google Scholar] [CrossRef]
79. Horton WB, Barros AJ, Andris RT, Clark MT, Moorman JR. Pathophysiologic signature of impending ICU hypoglycemia in bedside monitoring and electronic health record data: model development and external validation. Crit Care Med. 2022;50(3):e221–30. doi:10.1097/CCM.0000000000005171. [Google Scholar] [PubMed] [CrossRef]
80. Kim DY, Choi DS, Kim J, Chun SW, Gil HW, Cho NJ, et al. Developing an individual glucose prediction model using recurrent neural network. Sensors. 2020;20(22):E6460. doi:10.3390/s20226460. [Google Scholar] [PubMed] [CrossRef]
81. Leite NMN, Pereira ET, Gurjão EC, Veloso LR. Deep convolutional autoencoder for EEG noise filtering. In: 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2018 Dec 3–6; Madrid, Spain. p. 2605–12. doi:10.1109/BIBM.2018.8621080. [Google Scholar] [CrossRef]
82. Xiong P, Wang H, Liu M, Lin F, Hou Z, Liu X. A stacked contractive denoising auto-encoder for ECG signal denoising. Physiol Meas. 2016;37(12):2214–30. doi:10.1088/0967-3334/37/12/2214. [Google Scholar] [PubMed] [CrossRef]
83. Xu L, Wu X, Chen K, Yao L. Multi-modality sparse representation-based classification for Alzheimer’s disease and mild cognitive impairment. Comput Methods Programs Biomed. 2015;122(2):182–90. doi:10.1016/j.cmpb.2015.08.004. [Google Scholar] [PubMed] [CrossRef]
84. Bhattacharya A, Sadasivuni S, Chao CJ, Agasthi P, Ayoub C, Holmes DR, et al. Multi-modal fusion model for predicting adverse cardiovascular outcome post percutaneous coronary intervention. Physiol Meas. 2022;43(12):124004. doi:10.1088/1361-6579/ac9e8a. [Google Scholar] [PubMed] [CrossRef]
85. Yaacoubi C, Besrour R, Lachiri Z. A multimodal biometric identification system based on ECG and PPG signals. In: Proceedings of the 2nd International Conference on Digital Tools & Uses Congress. Virtual Event, Tunisia; 2020. p. 1–6. doi:10.1145/3423603.3424053. [Google Scholar] [CrossRef]
86. Torres-Soto J, Ashley EA. Multi-task deep learning for cardiac rhythm detection in wearable devices. npj Digit Med. 2020;3(1):116. doi:10.1038/s41746-020-00320-4. [Google Scholar] [PubMed] [CrossRef]
87. Wallach I, Dzamba M, Heifets A. AtomNet: a deep convolutional neural network for bioactivity prediction in structure-based drug discovery. arXiv:1510.02855. 2015. [Google Scholar]
88. Korshunova M, Ginsburg B, Tropsha A, Isayev O. OpenChem: a deep learning toolkit for computational chemistry and drug design. J Chem Inf Model. 2021;61(1):7–13. doi:10.1021/acs.jcim.0c00971. [Google Scholar] [PubMed] [CrossRef]
89. Zhan K, Lu ZH, Zhang YQ. Performance optimization for feature extraction section of DeepChem. In: Algorithms and architectures for parallel processing. Vol. 12452. Cham, Switzerland: Springer; 2020. p. 290–304. doi:10.1007/978-3-030-60245-1_20/COVER. [Google Scholar] [CrossRef]
90. Wilson PM, Philpot LM, Ramar P, Storlie CB, Strand J, Morgan AA, et al. Improving time to palliative care review with predictive modeling in an inpatient adult population: study protocol for a stepped-wedge, pragmatic randomized controlled trial. Trials. 2021;22(1):635. doi:10.1186/s13063-021-05546-5. [Google Scholar] [PubMed] [CrossRef]
91. Nguyen P, Tran T, Wickramasinghe N, Venkatesh S. Deepr: a convolutional net for medical records. IEEE J Biomed Health Inform. 2017;21(1):22–30. doi:10.1109/JBHI.2016.2633963. [Google Scholar] [PubMed] [CrossRef]
92. Harutyunyan H, Khachatrian H, Kale DC, Ver Steeg G, Galstyan A. Multitask learning and benchmarking with clinical time series data. Sci Data. 2019;6(1):96. doi:10.1038/s41597-019-0103-9. [Google Scholar] [PubMed] [CrossRef]
93. Gupta P, Malhotra P, Narwariya J, Vig L, Shroff G. Transfer learning for clinical time series analysis using deep neural networks. J Healthc Inform Res. 2020;4(2):112–37. doi:10.1007/s41666-019-00062-3. [Google Scholar] [PubMed] [CrossRef]
94. Shashikumar SP, Josef CS, Sharma A, Nemati S. DeepAISE—an interpretable and recurrent neural survival model for early prediction of sepsis. Artif Intell Med. 2021;113:102036. doi:10.1016/j.artmed.2021.102036. [Google Scholar] [PubMed] [CrossRef]
95. Zheng L, Lin Y. A multiorder feature tracking and explanation strategy for explainable deep learning. J Intell Syst. 2023;32(1):20220212. doi:10.1515/jisys-2022-0212. [Google Scholar] [CrossRef]
96. Tomita N, Abdollahi B, Wei J, Ren B, Suriawinata A, Hassanpour S. Attention-based deep neural networks for detection of cancerous and precancerous esophagus tissue on histopathological slides. JAMA Netw Open. 2019;2(11):e1914645. doi:10.1001/jamanetworkopen.2019.14645. [Google Scholar] [PubMed] [CrossRef]
97. Lundberg SM, Erion G, Chen H, DeGrave A, Prutkin JM, Nair B, et al. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell. 2020;2(1):56–67. doi:10.1038/s42256-019-0138-9. [Google Scholar] [PubMed] [CrossRef]
98. Al-Issa Y, Alqudah AM. A lightweight hybrid deep learning system for cardiac valvular disease classification. Sci Rep. 2022;12(1):14297. doi:10.1038/s41598-022-18293-7. [Google Scholar] [PubMed] [CrossRef]
99. Gulli A, Pal S. Deep learning with Keras. Birmingham, UK: Packt Publishing Ltd; 2017. [Google Scholar]
100. Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M, et al. Scalable and accurate deep learning with electronic health records. npj Digit Med. 2018;1(1):18. doi:10.1038/s41746-018-0029-1. [Google Scholar] [PubMed] [CrossRef]
101. Ting A, Law J, Lele A, Fang Y, Raychowdhury A. A comparison of CNNs and LSTMs for EEG signal classification. In: 2022 Opportunity Research Scholars Symposium (ORSS); 2022 Apr 27–27; Atlanta, GA, USA. p. 23–6. doi:10.1109/ORSS55359.2022.9806037. [Google Scholar] [CrossRef]
102. Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, et al. CheXNet: radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv:1711.05225. 2017. [Google Scholar]
103. Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, et al. CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison. In: 33rd AAAI Conference on Artificial Intelligence, AAAI 2019, 31st Innovative Applications of Artificial Intelligence Conference, IAAI, 2019 and the 9th AAAI Symposium on Educational Advances in Artificial Intelligence, EAAI 2019; 2019; Honolulu, HI, USA. p. 590–7. doi:10.1609/aaai.v33i01.3301590. [Google Scholar] [CrossRef]
104. Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. In: Advances in neural information processing systems. Cambridge, MA, USA: MIT Press; 2017. p. 5999–6009. [Google Scholar]
105. Lundberg SM, Lee SI. A unified approach to interpreting model predictions. In: Advances in neural information processing systems. Cambridge, MA, USA: MIT Press; 2017. [Google Scholar]
106. Aggarwal R, Sounderajah V, Martin G, Ting DSW, Karthikesalingam A, King D, et al. Diagnostic accuracy of deep learning in medical imaging: a systematic review and meta-analysis. npj Digit Med. 2021;4(1):65. doi:10.1038/s41746-021-00438-z. [Google Scholar] [PubMed] [CrossRef]
107. Lin TY, Goyal P, Girshick R, He K, Dollár P. Focal loss for dense object detection. In: 2017 IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22–29; Venice, Italy. p. 2999–3007. doi:10.1109/ICCV.2017.324. [Google Scholar] [CrossRef]
108. Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25(1):65–9. doi:10.1038/s41591-018-0268-3. [Google Scholar] [PubMed] [CrossRef]
109. Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22(10):1345–59. doi:10.1109/TKDE.2009.191. [Google Scholar] [CrossRef]
110. Cheng Y, Wang D, Zhou P, Zhang T. Model compression and acceleration for deep neural networks: the principles, progress, and challenges. IEEE Signal Process Mag. 2018;35(1):126–36. doi:10.1109/MSP.2017.2765695. [Google Scholar] [CrossRef]
111. Molchanov P, Tyree S, Karras T, Aila T, Kautz J. Pruning convolutional neural networks for resource efficient inference. In: 5th International Conference on Learning Representations, ICLR 2017-Conference Track Proceedings; 2016; Toulon, France. p. 1–17. [Google Scholar]
112. Sabha SU, Assad A, Din NMU, Bhat MR. From scratch or pretrained? An in-depth analysis of deep learning approaches with limited data. Int J Syst Assur Eng Manag. 2024;150(1):106123. doi:10.1007/s13198-024-02345-4. [Google Scholar] [CrossRef]
113. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. In: 3rd International Conference on Learning Representations, ICLR 2015-Conference Track Proceedings; 2015; San Diego, CA, USA. p. 1–14. [Google Scholar]
114. Weimann K, Conrad TOF. Transfer learning for ECG classification. Sci Rep. 2021;11(1):5251. doi:10.1038/s41598-021-84374-8. [Google Scholar] [PubMed] [CrossRef]
115. Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks? Adv Neural Inf Process Syst. 2014;4:3320–8. [Google Scholar]
116. Wu D, Yang J, Sawan M. Transfer learning on electromyography (EMG) tasks: approaches and beyond. IEEE Trans Neural Syst Rehabil Eng. 2023;31:3015–34. doi:10.1109/TNSRE.2023.3295453. [Google Scholar] [PubMed] [CrossRef]
117. Tang Y, Wang X, Harrison AP, Lu L, Xiao J, Summers RM. Attention-guided curriculum learning for weakly supervised classification and localization of thoracic diseases on chest radiographs. In: Machine learning in medical imaging. Cham, Switzerland: Springer International Publishing; 2018. p. 249–58. doi:10.1007/978-3-030-00919-9_29. [Google Scholar] [CrossRef]
118. Shin HC, Roberts K, Lu L, Demner-Fushman D, Yao J, Summers RM. Learning to read chest X-rays: recurrent neural cascade model for automated image annotation. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27–30; Las Vegas, NV, USA. p. 2497–506. doi:10.1109/CVPR.2016.274. [Google Scholar] [CrossRef]
119. Li Z, Li D, Xu C, Wang W, Hong Q, Li Q, et al. TFCNs: a CNN-transformer hybrid network for medical image segmentation. Vol. 13532. In: Artificial neural networks and machine learning–ICANN 2022. Cham, Switzerland: Springer; 2022 Jul. p. 781–92. doi:10.1007/978-3-031-15937-4_65. [Google Scholar] [CrossRef]
120. Yan T, Wan Z, Zhang P. Fully transformer network for change detection of remote sensing images. Vol. 13842. In: Computer vision–ACCV 2022. Cham, Switzerland: Springer; 2023. p. 75–92. doi:10.1007/978-3-031-26284-5_5/COVER. [Google Scholar] [CrossRef]
121. Ma M, Xia H, Tan Y, Li H, Song S. HT-Net: hierarchical context-attention transformer network for medical ct image segmentation. Appl Intell. 2022;52(9):10692–705. doi:10.1007/s10489-021-03010-0. [Google Scholar] [CrossRef]
122. Guo J, Xiao N, Li H, He L, Li Q, Wu T, et al. Transformer-based high-frequency oscillation signal detection on magnetoencephalography from epileptic patients. Front Mol Biosci. 2022;9:822810. doi:10.3389/fmolb.2022.822810. [Google Scholar] [PubMed] [CrossRef]
123. Che C, Zhang P, Zhu M, Qu Y, Jin B. Constrained transformer network for ECG signal processing and arrhythmia classification. BMC Med Inform Decis Mak. 2021;21(1):184. doi:10.1186/s12911-021-01546-2. [Google Scholar] [PubMed] [CrossRef]
124. Mahim SM, Emamul Hossen M, Al Hasan S, Islam MK, Iqbal Z, Alibakhshikenari M, et al. TransMixer-AF: advanced real-time detection of atrial fibrillation utilizing single-lead electrocardiogram signals. IEEE Access. 2024;12(3):143149–62. doi:10.1109/ACCESS.2024.3467181. [Google Scholar] [CrossRef]
125. Lilhore UK, Dalal S, Faujdar N, Margala M, Chakrabarti P, Chakrabarti T, et al. Hybrid CNN-LSTM model with efficient hyperparameter tuning for prediction of Parkinson’s disease. Sci Rep. 2023;13(1):14605. doi:10.1038/s41598-023-41314-y. [Google Scholar] [PubMed] [CrossRef]
126. Wu Y, Tang Q, Zhan W, Li S, Chen Z. Res-BiANet: a hybrid deep learning model for arrhythmia detection based on PPG signal. Electronics. 2024;13(3):665. doi:10.3390/electronics13030665. [Google Scholar] [CrossRef]
127. Kilicarslan S, Celik M, Sahin Ş. Hybrid models based on genetic algorithm and deep learning algorithms for nutritional Anemia disease classification. Biomed Signal Process Control. 2021;63(1):102231. doi:10.1016/j.bspc.2020.102231. [Google Scholar] [CrossRef]
128. Mavaie P, Holder L, Skinner MK. Hybrid deep learning approach to improve classification of low-volume high-dimensional data. BMC Bioinformatics. 2023;24(1):419. doi:10.1186/s12859-023-05557-w. [Google Scholar] [PubMed] [CrossRef]
129. Woo S, Park J, Lee JY, Kweon IS. CBAM: convolutional block attention module. In: Computer vision–ECCV 2018. Cham, Switzerland: Springer International Publishing; 2018. p. 3–19. doi:10.1007/978-3-030-01234-2_1. [Google Scholar] [CrossRef]
130. Park SA, Lee HC, Jung CW, Yang HL. Attention mechanisms for physiological signal deep learning: which attention should we take? In: Medical image computing and computer assisted intervention-MICCAI 2022. Cham, Switzerland: Springer Nature Switzerland; 2022. p. 613–22. doi:10.1007/978-3-031-16431-6_58. [Google Scholar] [CrossRef]
131. Wang S, Huang L, Jiang D, Sun Y, Jiang G, Li J, et al. Improved multi-stream convolutional block attention module for sEMG-based gesture recognition. Front Bioeng Biotechnol. 2022;10:909023. doi:10.3389/fbioe.2022.909023. [Google Scholar] [PubMed] [CrossRef]
132. Li H, Wang T, Zhang M, Zhu A, Shan G, Snoussi H. Hierarchical attention networks for image classification of remote sensing images based on visual Q&A methods. In: 2019 Chinese Automation Congress (CAC); 2019 Nov 22–24; Hangzhou, China. p. 4712–7. doi:10.1109/cac48633.2019.8997347. [Google Scholar] [CrossRef]
133. Huang KH, Yang M, Peng N. Biomedical event extraction with hierarchical knowledge graphs. In: Findings of the Association for Computational Linguistics: EMNLP 2020; Online. Stroudsburg, PA, USA: ACL; 2020. p. 1277–85. doi:10.18653/v1/2020.findings-emnlp.114. [Google Scholar] [PubMed] [CrossRef]
134. Wang J, Li M, Diao Q, Lin H, Yang Z, Zhang Y. Biomedical document triage using a hierarchical attention-based capsule network. BMC Bioinformatics. 2020;21(suppl 13):380. doi:10.1186/s12859-020-03673-5. [Google Scholar] [PubMed] [CrossRef]
135. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. Int J Comput Vis. 2020;128(2):336–59. doi:10.1007/s11263-019-01228-7. [Google Scholar] [CrossRef]
136. Aslan Z. Deep convolutional neural network-based framework in the automatic diagnosis of migraine. Circuits Syst Signal Process. 2023;42(5):3054–71. doi:10.1007/s00034-022-02265-3. [Google Scholar] [CrossRef]
137. Kavak S, Chiu XD, Yen SJ, Chen MY. Application of CNN for detection and localization of STEMI using 12-lead ECG images. IEEE Access. 2022;10:38923–30. doi:10.1109/ACCESS.2022.3165966. [Google Scholar] [CrossRef]
138. Jha A, Aicher JK, Gazzara MR, Singh D, Barash Y. Enhanced integrated gradients: improving interpretability of deep learning models using splicing codes as a case study. Genome Biol. 2020;21(1):149. doi:10.1186/s13059-020-02055-7. [Google Scholar] [PubMed] [CrossRef]
139. Kawai Y, Tachikawa K, Park J, Asada M. Compensated integrated gradients for reliable explanation of electroencephalogram signal classification. Brain Sci. 2022;12(7):849. doi:10.3390/brainsci12070849. [Google Scholar] [PubMed] [CrossRef]
140. Rozemberczki B, Watson L, Bayer P, Yang HT, Kiss O, Nilsson S, et al. The shapley value in machine learning. In: Proceedings of the Thirty-First International Joint Conference on Artificial Intelligence; 2022 Jul 23–29; Vienna, Austria. p. 5572–9. doi:10.24963/ijcai.2022/778. [Google Scholar] [CrossRef]
141. Ukil A, Marin L, Jara AJ. When less is more powerful: shapley value attributed ablation with augmented learning for practical time series sensor data classification. PLoS One. 2022;17(11):e0277975. doi:10.1371/journal.pone.0277975. [Google Scholar] [PubMed] [CrossRef]
142. Roder J, Maguire L, Georgantas R, Roder H. Explaining multivariate molecular diagnostic tests via Shapley values. BMC Med Inform Decis Mak. 2021;21(1):211. doi:10.1186/s12911-021-01569-9. [Google Scholar] [PubMed] [CrossRef]
143. Zhang D, Yang S, Yuan X, Zhang P. Interpretable deep learning for automatic diagnosis of 12-lead electrocardiogram. iScience. 2021;24(4):102373. doi:10.1016/j.isci.2021.102373. [Google Scholar] [PubMed] [CrossRef]
144. Tyagi AK. Computational analysis and deep learning for medical care: principles, methods, and applications. 1st ed. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2021. doi:10.1002/9781119785750. [Google Scholar] [CrossRef]
145. Lac L, Leung CK, Hu P. Computational frameworks integrating deep learning and statistical models in mining multimodal omics data. J Biomed Inform. 2024;152(1):104629. doi:10.1016/j.jbi.2024.104629. [Google Scholar] [PubMed] [CrossRef]
146. Bhandari N, Walambe R, Kotecha K, Khare SP. A comprehensive survey on computational learning methods for analysis of gene expression data. Front Mol Biosci. 2022;9:907150. doi:10.3389/fmolb.2022.907150. [Google Scholar] [PubMed] [CrossRef]
147. Yousef M, Allmer J. Deep learning in bioinformatics. Turk J Biol. 2023;47(6):366–82. doi:10.55730/1300-0152.2671. [Google Scholar] [PubMed] [CrossRef]
148. Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC, Mark RG, et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2000;101(23):E215–20. doi:10.1161/01.cir.101.23.e215. [Google Scholar] [PubMed] [CrossRef]
149. Johnson AEW, Pollard TJ, Shen L, Lehman LH, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3(1):160035. doi:10.1038/sdata.2016.35. [Google Scholar] [PubMed] [CrossRef]
150. Kemp B, Zwinderman AH, Tuk B, Kamphuisen HA, Oberyé JJ. Analysis of a sleep-dependent neuronal feedback loop: the slow-wave microcontinuity of the EEG. IEEE Trans Biomed Eng. 2000;47(9):1185–94. doi:10.1109/10.867928. [Google Scholar] [PubMed] [CrossRef]
151. Sajda P, Gerson A, Müller KR, Blankertz B, Parra L. A data analysis competition to evaluate machine learning algorithms for use in brain-computer interfaces. IEEE Trans Neural Syst Rehabil Eng. 2003;11(2):184–5. doi:10.1109/TNSRE.2003.814453. [Google Scholar] [PubMed] [CrossRef]
152. Blankertz B, Müller KR, Curio G, Vaughan TM, Schalk G, Wolpaw JR, et al. The BCI Competition 2003: progress and perspectives in detection and discrimination of EEG single trials. IEEE Trans Biomed Eng. 2004;51(6):1044–51. doi:10.1109/TBME.2004.826692. [Google Scholar] [PubMed] [CrossRef]
153. Blankertz B, Muller KR, Krusienski DJ, Schalk G, Wolpaw JR, Schlogl A, et al. The BCI competition III: validating alternative approaches to actual BCI problems. IEEE Trans Neural Syst Rehabil Eng. 2006;14(2):153–9. doi:10.1109/TNSRE.2006.875642. [Google Scholar] [PubMed] [CrossRef]
154. Wagner P, Strodthoff N, Bousseljot RD, Kreiseler D, Lunze FI, Samek W, et al. PTB-XL, a large publicly available electrocardiography dataset. Sci Data. 2020;7(1):154. doi:10.1038/s41597-020-0495-6. [Google Scholar] [PubMed] [CrossRef]
155. Shoeb A, Edwards H, Connolly J, Bourgeois B, Treves ST, Guttag J. Patient-specific seizure onset detection. Epilepsy Behav. 2004;5(4):483–98. doi:10.1016/j.yebeh.2004.05.005. [Google Scholar] [PubMed] [CrossRef]
156. Busso C, Bulut M, Lee CC, Kazemzadeh A, Mower E, Kim S, et al. IEMOCAP: interactive emotional dyadic motion capture database. Lang Resour Eval. 2008;42(4):335–59. doi:10.1007/s10579-008-9076-6. [Google Scholar] [CrossRef]
157. Abdelhamid AA, El-Kenawy EM, Alotaibi B, Amer GM, Abdelkader MY, Ibrahim A, et al. Robust speech emotion recognition using CNN+LSTM based on stochastic fractal search optimization algorithm. IEEE Access. 2022;10:49265–84. doi:10.1109/ACCESS.2022.3172954. [Google Scholar] [CrossRef]
158. Shi T, Huang SL. MultiEMO: an attention-based correlation-aware multimodal fusion framework for emotion recognition in conversations. In: Proceedings of the 61st Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). Toronto, ON, Canada. Stroudsburg, PA, USA: ACL; 2023. p. 14752–66. doi:10.18653/v1/2023.acl-long.824. [Google Scholar] [PubMed] [CrossRef]
159. Zhang S, Yang Y, Chen C, Zhang X, Leng Q, Zhao X. Deep learning-based multimodal emotion recognition from audio, visual, and text modalities: a systematic review of recent advancements and future prospects. Expert Syst Appl. 2024;237:121692. doi:10.1016/j.eswa.2023.121692. [Google Scholar] [CrossRef]
160. Alqudah AM, Qazan S, Obeidat YM. Deep learning models for detecting respiratory pathologies from raw lung auscultation sounds. Soft Comput. 2022;26(24):13405–29. doi:10.1007/s00500-022-07499-6. [Google Scholar] [PubMed] [CrossRef]
161. Obeid I, Picone J. The temple university hospital EEG data corpus. Front Neurosci. 2016;10(183):196. doi:10.3389/fnins.2016.00196. [Google Scholar] [PubMed] [CrossRef]
162. Karlen W, Raman S, Ansermino JM, Dumont GA. Multiparameter respiratory rate estimation from the photoplethysmogram. IEEE Trans Biomed Eng. 2013;60(7):1946–53. doi:10.1109/TBME.2013.2246160. [Google Scholar] [PubMed] [CrossRef]
163. Zhang GQ, Cui L, Mueller R, Tao S, Kim M, Rueschman M, et al. The national sleep research resource: towards a sleep data commons. J Am Med Inform Assoc. 2018;25(10):1351–8. doi:10.1093/jamia/ocy064. [Google Scholar] [PubMed] [CrossRef]
164. Chanchaochai N, Cieri C, Debrah J, Ding H, Jiang Y, Liao S, et al. GlobalTIMIT: acoustic-phonetic datasets for the world’s languages. In: Interspeech 2018. Hyderabad, India: ISCA; 2018. p. 192–6. doi:10.21437/Interspeech.2018-1185. [Google Scholar] [CrossRef]
165. Yang J, Soh M, Lieu V, Weber DJ, Erickson Z. EMGBench: benchmarking out-of-distribution generalization and adaptation for electromyography. arXiv:2410.23625. 2024. [Google Scholar]
166. Charlton PH. MESA polysomnography dataset. National Sleep Research Resource; 2024. [cited 2025 Mar 1]. Available from: https://peterhcharlton.github.io/info/datasets/mesa. [Google Scholar]
167. Charlton PH. PPG-DaLiA: a dataset for activity monitoring using photoplethysmography. Physiol Meas. 2018;39(10):105004. [cited 2025 Mar 1]. Available from: https://peterhcharlton.github.io/info/datasets/ppg-dalia. [Google Scholar]
168. Hahn G. PPG Diary: a dataset for continuous PPG monitoring. Physiol Meas. 2019;40(11):115007. [cited 2025 Mar 1]. Available from: https://peterhcharlton.github.io/info/datasets/ppg-diary1. [Google Scholar]
169. Charlton PH, Mariscal Harana J, Vennin S, Li Y, Chowienczyk P, Alastruey J. Modeling arterial pulse waves in healthy aging: a database for in silico evaluation of hemodynamics and pulse wave indexes. Am J Physiol Heart Circ Physiol. 2019 Nov;317(5):H1062–85. doi:10.1152/ajpheart.00218.2019. [Google Scholar] [PubMed] [CrossRef]
170. Charlton PH, Bonnici T, Tarassenko L, Clifton DA, Beale R, Watkinson PJ. An assessment of algorithms to estimate respiratory rate from the electrocardiogram and photoplethysmogram. Physiol Meas. 2016 Apr;37(4):610–26. doi:10.1088/0967-3334/37/4/610. [Google Scholar] [PubMed] [CrossRef]
171. Schmidt P, Reiss A, Duerichen R, Marberger C, Van Laerhoven K. Introducing WESAD, a multimodal dataset for wearable stress and affect detection. In: Proceedings of the 20th ACM International Conference on Multimodal Interaction. Boulder, CO, USA; 2018. p. 400–8. doi:10.1145/3242969.3242985. [Google Scholar] [CrossRef]
172. Clifford G, Liu C, Moody B, Lehman LW, Silva I, Li Q, et al. AF classification from a short single lead ECG recording: the physionet computing in cardiology challenge 2017. In: Computing in Cardiology Conference (CinC); 2017; Rennes, France. p. 1–4. doi:10.22489/cinc.2017.065-469. [Google Scholar] [PubMed] [CrossRef]
173. Subasi A. Practical guide for biomedical signals analysis using machine learning techniques: a MATLAB based approach.Cambridge. MA, USA: Academic Press; 2019. [Google Scholar]
174. Petmezas G, Haris K, Stefanopoulos L, Kilintzis V, Tzavelis A, Rogers JA, et al. Automated atrial fibrillation detection using a hybrid CNN-LSTM network on imbalanced ECG datasets. Biomed Signal Process Control. 2021;63(3):102194. doi:10.1016/j.bspc.2020.102194. [Google Scholar] [CrossRef]
175. Forkan ARM, Khalil I, Atiquzzaman M. ViSiBiD: a learning model for early discovery and real-time prediction of severe clinical events using vital signs as big data. Comput Netw. 2017;113(4):244–57. doi:10.1016/j.comnet.2016.12.019. [Google Scholar] [CrossRef]
176. Del Pup F, Atzori M. Applications of self-supervised learning to biomedical signals: a survey. IEEE Access. 2023;11:144180–203. doi:10.1109/ACCESS.2023.3344531. [Google Scholar] [CrossRef]
177. Kuzmanov I, Ackovska N, Bogadova AM. Transformer models for processing biological signal. In: The 20th International Conference on Informatics and Information Technologies’CIIT 2023. Republic of North Macedonia: Ss Cyril and Methodius University in Skopje, Faculty of Computer Science and Engineering; 2023 Jun. [Google Scholar]
178. LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86(11):2278–324. doi:10.1109/5.726791. [Google Scholar] [CrossRef]
179. Saito K, Ushiku Y, Harada T. Asymmetric tri-training for unsupervised domain adaptation. In: 34th International Conference on Machine Learning, ICML 2017; 2017; Sydney, NSW, Australia. p. 4573–85. [Google Scholar]
180. Dietterich TG. Ensemble methods in machine learning. Vol. 1875. In: Lecture notes in computer science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Berlin/Heidelberg, Germany: Springer; 2000. p. 1–15. doi:10.1007/3-540-45014-9_1/COVER. [Google Scholar] [CrossRef]
181. Längkvist M, Karlsson L, Loutfi A. A review of unsupervised feature learning and deep learning for time-series modeling. Pattern Recognit Lett. 2014;42(5):11–24. doi:10.1016/j.patrec.2014.01.008. [Google Scholar] [CrossRef]
182. Schölkopf B, Platt JC, Shawe-Taylor J, Smola AJ, Williamson RC. Estimating the support of a high-dimensional distribution. Neural Comput. 2001;13(7):1443–71. doi:10.1162/089976601750264965. [Google Scholar] [PubMed] [CrossRef]
183. Liu FT, Ting KM, Zhou ZH. Isolation forest. In: 2008 Eighth IEEE International Conference on Data Mining; 2008 Dec 15–19; Pisa, Italy. p. 413–22. doi:10.1109/ICDM.2008.17. [Google Scholar] [CrossRef]
184. Jiang H, Gao M, Li H, Jin R, Miao H, Liu J. Multi-learner based deep meta-learning for few-shot medical image classification. IEEE J Biomed Health Inform. 2023;27(1):17–28. doi:10.1109/JBHI.2022.3215147. [Google Scholar] [PubMed] [CrossRef]
185. Pałczyński K, Śmigiel S, Ledziński D, Bujnowski S. Study of the few-shot learning for ECG classification based on the PTB-XL dataset. Sensors. 2022;22(3):904. doi:10.3390/s22030904. [Google Scholar] [PubMed] [CrossRef]
186. Tyukin IY, Gorban AN, Alkhudaydi MH, Zhou Q. Demystification of few-shot and one-shot learning. In: 2021 International Joint Conference on Neural Networks (IJCNN); 2021 Jul 18–22; Shenzhen, China. p. 1–7. doi:10.1109/ijcnn52387.2021.9534395. [Google Scholar] [CrossRef]
187. Kalantari J, Mackey MA. One-shot ontogenetic learning in biomedical datastreams. In: Artificial General Intelligence: 10th International Conference, AGI 2017; 2017 Aug 15–18; Melbourne, VIC, Australia. p. 143–53. doi:10.1007/978-3-319-63703-7_14/COVER. [Google Scholar] [CrossRef]
188. Caceres CA, Roos MJ, Rupp KM, Milsap G, Crone NE, Wolmetz ME, et al. Feature selection methods for zero-shot learning of neural activity. Front Neuroinform. 2017;11:41. doi:10.3389/fninf.2017.00041. [Google Scholar] [PubMed] [CrossRef]
189. Mahapatra D, Bozorgtabar B, Ge Z. Medical image classification using generalized zero shot learning. In: 2021 IEEE/CVF International Conference on Computer Vision Workshops (ICCVW); 2021 Oct 11–17; Montreal, BC, Canada. p. 3337–46. doi:10.1109/ICCVW54120.2021.00373. [Google Scholar] [CrossRef]
190. Dong Y, Jiang X, Zhou H, Lin Y, Shi Q. SR2CNN: zero-shot learning for signal recognition. IEEE Trans Signal Process. 2021;69:2316–29. doi:10.1109/TSP.2021.3070186. [Google Scholar] [CrossRef]
191. Karatas M, Eriskin L, Deveci M, Pamucar D, Garg H. Big data for healthcare industry 4.0: applications, challenges and future perspectives. Expert Syst Appl. 2022;200(1):116912. doi:10.1016/j.eswa.2022.116912. [Google Scholar] [CrossRef]
192. Lee J, Liu C, Kim J, Chen Z, Sun Y, Rogers JR, et al. Deep learning for rare disease: a scoping review. J Biomed Inform. 2022;135(2):104227. doi:10.1016/j.jbi.2022.104227. [Google Scholar] [PubMed] [CrossRef]
193. Malin B, Goodman K. Between access and privacy: challenges in sharing health data. Yearb Med Inform. 2018;27(1):55–9. doi:10.1055/s-0038-1641216. [Google Scholar] [PubMed] [CrossRef]
194. Shafer G, Vovk V. A tutorial on conformal prediction. J Mach Learn Res. 2008;9(12):371–421. [Google Scholar]
195. Romano Y, Sesia M, Candès EJ. Classification with valid and adaptive coverage. In: NIPS’20: Proceedings of the 34th International Conference on Neural Information Processing Systems; 2020; Vancouver, BC, Canada. p. 3581–91. [Google Scholar]
196. Gammerman A, Shafer G, Vovk V. Algorithmic learning in a random world. New York, NY, USA: Springer-Verlag; 2005. doi:10.1007/b106715. [Google Scholar] [CrossRef]
197. Lei J, G’Sell M, Rinaldo A, Tibshirani RJ, Wasserman L. Distribution-free predictive inference for regression. J Am Stat Assoc. 2018;113(523):1094–111. doi:10.1080/01621459.2017.1307116. [Google Scholar] [CrossRef]
198. Papadopoulos H, Proedrou K, Vovk V, Gammerman A. Inductive confidence machines for regression. In: Machine learning: ECML 2002. Berlin/Heidelberg: Springer Berlin Heidelberg; 2002. p. 345–56. doi:10.1007/3-540-36755-1_29. [Google Scholar] [CrossRef]
199. Shah P, Kendall F, Khozin S, Goosen R, Hu J, Laramie J, et al. Artificial intelligence and machine learning in clinical development: a translational perspective. npj Digit Med. 2019;2(1):69. doi:10.1038/s41746-019-0148-3. [Google Scholar] [PubMed] [CrossRef]
200. Orphanidou C, Bonnici T, Charlton P, Clifton D, Vallance D, Tarassenko L. Signal-quality indices for the electrocardiogram and photoplethysmogram: derivation and applications to wireless monitoring. IEEE J Biomed Health Inform. 2015;19(3):832–8. doi:10.1109/JBHI.2014.2338351. [Google Scholar] [PubMed] [CrossRef]
201. Anand A, Kadian T, Shetty MK, Gupta A. Explainable AI decision model for ECG data of cardiac disorders. Biomed Signal Process Control. 2022;75:103584. doi:10.1016/j.bspc.2022.103584. [Google Scholar] [CrossRef]
202. Shickel B, Tighe PJ, Bihorac A, Rashidi P. Deep EHR: a survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE J Biomed Health Inform. 2018;22(5):1589–604. doi:10.1109/JBHI.2017.2767063. [Google Scholar] [PubMed] [CrossRef]
203. Chen X, Wang X, Zhang K, Fung KM, Thai TC, Moore K, et al. Recent advances and clinical applications of deep learning in medical image analysis. Med Image Anal. 2022;79:102444. doi:10.1016/j.media.2022.102444. [Google Scholar] [PubMed] [CrossRef]
204. Obermeyer Z, Powers B, Vogeli C, Mullainathan S. Dissecting racial bias in an algorithm used to manage the health of populations. Science. 2019;366(6464):447–53. doi:10.1126/science.aax2342. [Google Scholar] [PubMed] [CrossRef]
205. Powles J, Hodson H. Google DeepMind and healthcare in an age of algorithms. Health Technol. 2017;7(4):351–67. doi:10.1007/s12553-017-0179-1. [Google Scholar] [PubMed] [CrossRef]
206. Gursoy F, Kakadiaris IA. Artificial intelligence research strategy of the United States: critical assessment and policy recommendations. Front Big Data. 2023;6:1206139. doi:10.3389/fdata.2023.1206139. [Google Scholar] [PubMed] [CrossRef]
207. Feldstein S. Evaluating Europe’s push to enact AI regulations: how will this influence global norms? Democratization. 2024;31(5):1049–66. doi:10.1080/13510347.2023.2196068. [Google Scholar] [CrossRef]
208. Xia Y, Wang W, Wang K. ECG signal generation based on conditional generative models. Biomed Signal Process Control. 2023;82(8):104587. doi:10.1016/j.bspc.2023.104587. [Google Scholar] [CrossRef]
209. Majhi B, Kashyap A. Explainable AI-driven machine learning for heart disease detection using ECG signal. Appl Soft Comput. 2024;167(4):112225. doi:10.1016/j.asoc.2024.112225. [Google Scholar] [CrossRef]
210. Guan H, Yap PT, Bozoki A, Liu M. Federated learning for medical image analysis: a survey. Pattern Recognit. 2024;151(3):110424. doi:10.1016/j.patcog.2024.110424. [Google Scholar] [PubMed] [CrossRef]
211. McMahan HB, Moore E, Ramage D, Hampson S, Arcas BAY. Communication-efficient learning of deep networks from decentralized data. arXiv:1602.05629. 2016. [Google Scholar]
212. Wang X, Zhang Y, Zhu R. A brief review on algorithmic fairness. Manag Syst Eng. 2022;1(1):7. doi:10.1007/s44176-022-00006-z. [Google Scholar] [CrossRef]
213. Chen R, Yang J, Xiong H, Bai J, Hu T, Hao J, et al. Fast model debias with machine unlearning. arXiv:2310.12560. 2023. [Google Scholar]
214. Zeng Z, Islam R, Keya KN, Foulds J, Song Y, Pan S. Fair representation learning for heterogeneous information networks. Proc Int AAAI Conf Web Soc Medium. 2021;15:877–87. doi:10.1609/icwsm.v15i1.18111. [Google Scholar] [CrossRef]
215. Jovanović N, Balunović M, Dimitrov DI, Vechev M. FARE: provably fair representation learning with practical certificates. arXiv:2210.07213. 2022. [Google Scholar]
216. Begley T, Schwedes T, Frye C, Feige I. Explainability for fair machine learning. arXiv:2010.07389. 2020. [Google Scholar]
217. Liu LT, Dean S, Rolf E, Simchowitz M, Hardt M. Delayed impact of fair machine learning. In: Proceedings of the 35th International Conference on Machine Learning; 2018 Jul 10–15; Stockholm, Sweden. p. 3150–8. [Google Scholar]
218. Corbett-Davies S, Gaebler JD, Nilforoshan H, Shroff R, Goel S, Nilforoshan H, et al. The measure and mismeasure of fairness. arXiv:1808.00023. 2018. [Google Scholar]
219. Liu J, Li Z, Yao Y, Xu F, Ma X, Xu M, et al. Fair representation learning: an alternative to mutual information. In: Proceedings of the 28th ACM SIGKDD Conference on Knowledge Discovery and Data Mining; 2022; Washington, DC, USA. p. 1088–97. doi:10.1145/3534678.3539302. [Google Scholar] [CrossRef]
220. Creager E, Madras D, Jacobsen J-H, Weis M, Swersky K, Pitassi T, et al. Flexibly fair representation learning by disentanglement. In: Proceedings of the 36th International Conference on Machine Learning; 2019; Long Beach, CA, USA. p. 1436–45. [Google Scholar]
221. McNamara D, Ong CS, Williamson RC. Costs and benefits of fair representation learning. In: Proceedings of the 2019 AAAI/ACM Conference on AI, Ethics, and Society; 2019; Honolulu, HI, USA; 2019. p. 263–70. doi:10.1145/3306618.3317964. [Google Scholar] [CrossRef]
222. Dinsdale NK, Jenkinson M, Namburete AIL. Deep learning-based unlearning of dataset bias for MRI harmonisation and confound removal. Neuroimage. 2021;228(7):117689. doi:10.1016/j.neuroimage.2020.117689. [Google Scholar] [PubMed] [CrossRef]
223. Ben Mansour A, Carenini G, Duplessis A, Naccache D. Federated learning aggregation: new robust algorithms with guarantees. In: 2022 21st IEEE International Conference on Machine Learning and Applications (ICMLA); 2022 Dec 12–14; Nassau, Bahamas. p. 721–6. doi:10.1109/ICMLA55696.2022.00120. [Google Scholar] [CrossRef]
224. Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. TensorFlow: large-scale machine learning on heterogeneous distributed systems. arXiv:1603.04467. 2016. [Google Scholar]
225. Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, et al. PyTorch: an imperative style, high-performance deep learning library. In: Proceedings of the 33rd International Conference on Neural Information Processing Systems; 2019; Red Hook, NY, USA. p. 8026–37. [Google Scholar]
226. Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, et al. Caffe: convolutional architecture for fast feature embedding. In: MM 2014—Proceedings of the 2014 ACM Conference on Multimedia; 2014 Nov 3–7; Orlando, FL, USA. p. 675–8. doi:10.1145/2647868.2654889. [Google Scholar] [CrossRef]
227. Chen T, Li M, Li Y, Lin M, Wang N, Wang M, et al. MXNet: a flexible and efficient machine learning library for heterogeneous distributed systems. arXiv:1512.01274. 2015. [Google Scholar]
228. Kiranyaz S, Ince T, Gabbouj M. Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Trans Biomed Eng. 2016;63(3):664–75. doi:10.1109/TBME.2015.2468589. [Google Scholar] [PubMed] [CrossRef]
229. Bardou D, Zhang K, Ahmad SM. Lung sounds classification using convolutional neural networks. Artif Intell Med. 2018;88(1):58–69. doi:10.1016/j.artmed.2018.04.008. [Google Scholar] [PubMed] [CrossRef]
230. Leitner J, Chiang PH, Dey S. Personalized blood pressure estimation using photoplethysmography: a transfer learning approach. IEEE J Biomed Health Inform. 2022;26(1):218–28. doi:10.1109/JBHI.2021.3085526. [Google Scholar] [PubMed] [CrossRef]
231. Jurczak M, Kołodziej M, Majkowski A. Implementation of a convolutional neural network for eye blink artifacts removal from the electroencephalography signal. Front Neurosci. 2022;16:782367. doi:10.3389/fnins.2022.782367. [Google Scholar] [PubMed] [CrossRef]
232. Sun C, Liu C, Wang X, Liu Y, Zhao S. Coronary artery disease detection based on a novel multi-modal deep-coding method using ECG and PCG signals. Sensors. 2024;24(21):6939. doi:10.3390/s24216939. [Google Scholar] [PubMed] [CrossRef]
233. Mortazavi E, Tarvirdizadeh B, Alipour K, Ghamari M. Deep learning approaches for assessing pediatric sleep apnea severity through SpO2 signals. Sci Rep. 2024;14(1):22696. doi:10.1038/s41598-024-67729-9. [Google Scholar] [PubMed] [CrossRef]
234. Wang J, Sun S, Sun Y. A muscle fatigue classification model based on LSTM and improved wavelet packet threshold. Sensors. 2021;21(19):6369. doi:10.3390/s21196369. [Google Scholar] [PubMed] [CrossRef]
235. Ali NF, Atef M. LSTM multi-stage transfer learning for blood pressure estimation using photoplethysmography. Electronics. 2022;11(22):3749. doi:10.3390/electronics11223749. [Google Scholar] [CrossRef]
236. Batool S, Khan MH, Farid MS. An ensemble deep learning model for human activity analysis using wearable sensory data. Appl Soft Comput. 2024;159(8):111599. doi:10.1016/j.asoc.2024.111599. [Google Scholar] [CrossRef]
237. Petmezas G, Cheimariotis GA, Stefanopoulos L, Rocha B, Paiva RP, Katsaggelos AK, et al. Automated lung sound classification using a hybrid CNN-LSTM network and focal loss function. Sensors. 2022;22(3):1232. doi:10.3390/s22031232. [Google Scholar] [PubMed] [CrossRef]
238. Ordóñez FJ, Roggen D. Deep convolutional and LSTM recurrent neural networks for multimodal wearable activity recognition. Sensors. 2016;16(1):E115. doi:10.3390/s16010115. [Google Scholar] [PubMed] [CrossRef]
239. Saeed T, Ijaz A, Sadiq I, Qureshi HN, Rizwan A, Imran A. An AI-enabled bias-free respiratory disease diagnosis model using cough audio. Bioengineering. 2024;11(1):55. doi:10.3390/bioengineering11010055. [Google Scholar] [PubMed] [CrossRef]
240. Bai X, Dong X, Li Y, Liu R, Zhang H. A hybrid deep learning network for automatic diagnosis of cardiac arrhythmia based on 12-lead ECG. Sci Rep. 2024;14(1):24441. doi:10.1038/s41598-024-75531-w. [Google Scholar] [PubMed] [CrossRef]
241. Wang J, Sun Y, Sun S. Recognition of muscle fatigue status based on improved wavelet threshold and CNN-SVM. IEEE Access. 2020;8:207914–22. doi:10.1109/ACCESS.2020.3038422. [Google Scholar] [CrossRef]
242. Orkweha K, Phapatanaburi K, Pathonsuwan W, Jumphoo T, Rattanasak A, Anchuen P, et al. A framework for detecting pulmonary diseases from lung sound signals using a hybrid multi-task autoencoder-SVM model. Symmetry. 2024;16(11):1413. doi:10.3390/sym16111413. [Google Scholar] [CrossRef]
243. Ansari Y, Mourad O, Qaraqe K, Serpedin E. Deep learning for ECG arrhythmia detection and classification: an overview of progress for period 2017–2023. Front Physiol. 2023;14:1246746. doi:10.3389/fphys.2023.1246746. [Google Scholar] [PubMed] [CrossRef]
244. Chung J, Gulcehre C, Cho K, Bengio Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv:1412.3555. 2014. [Google Scholar]
245. Rocha BM, Pessoa D, Marques A, de Carvalho P, Paiva RP. Automatic wheeze segmentation using harmonic-percussive source separation and empirical mode decomposition. IEEE J Biomed Health Inform. 2023;27(4):1926–34. doi:10.1109/JBHI.2023.3248265. [Google Scholar] [PubMed] [CrossRef]
246. Forcier MB, Gallois H, Mullan S, Joly Y. Integrating artificial intelligence into health care through data access: can the GDPR act as a beacon for policymakers? J Law Biosci. 2019;6(1):317–35. doi:10.1093/jlb/lsz013. [Google Scholar] [PubMed] [CrossRef]
247. Turakhia MP, Desai M, Hedlin H, Rajmane A, Talati N, Ferris T, et al. Rationale and design of a large-scale, app-based study to identify cardiac arrhythmias using a smartwatch: the Apple Heart Study. Am Heart J. 2019;207(12):66–75. doi:10.1016/j.ahj.2018.09.002. [Google Scholar] [PubMed] [CrossRef]
248. Emmett A, Kent B, James A, March-McDonald J. Evaluating the use of the mobile electrocardiogram technology KardiaMobileTM in community settings: an online survey. Nurs Open. 2024;11(6):e2225. doi:10.1002/nop2.2225. [Google Scholar] [CrossRef]
249. Beaubier N, Tell R, Lau D, Parsons JR, Bush S, Perera J, et al. Clinical validation of the tempus xT next-generation targeted oncology sequencing assay. Oncotarget. 2019;10(24):2384–96. doi:10.18632/oncotarget.26797. [Google Scholar] [PubMed] [CrossRef]
250. Lee T, Cho Y, Cha KS, Jung J, Cho J, Kim H, et al. Accuracy of 11 wearable, nearable, and airable consumer sleep trackers: prospective multicenter validation study. JMIR Mhealth Uhealth. 2023;11(5):e50983. doi:10.2196/50983. [Google Scholar] [PubMed] [CrossRef]
251. Barrett PM, Komatireddy R, Haaser S, Topol S, Sheard J, Encinas J, et al. Comparison of 24-hour Holter monitoring with 14-day novel adhesive patch electrocardiographic monitoring. Am J Med. 2014;127(1):95.e11–7. doi:10.1016/j.amjmed.2013.10.003. [Google Scholar] [PubMed] [CrossRef]
252. Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25(1):44–56. doi:10.1038/s41591-018-0300-7. [Google Scholar] [PubMed] [CrossRef]
253. Hicks SA, Isaksen JL, Thambawita V, Ghouse J, Ahlberg G, Linneberg A, et al. Explaining deep neural networks for knowledge discovery in electrocardiogram analysis. Sci Rep. 2021 May;11(1):10949. doi:10.1038/s41598-021-90285-5. [Google Scholar] [PubMed] [CrossRef]
254. Ribeiro MT, Singh S, Guestrin C. Why should I trust you?: explaining the predictions of any classifier. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2016; San Francisco, CA, USA. p. 1135–44. doi:10.1145/2939672.2939778. [Google Scholar] [CrossRef]
255. Komura D, Ishikawa S. Machine learning methods for histopathological image analysis. Comput Struct Biotechnol J. 2018;16:34–42. doi:10.1016/j.csbj.2018.01.001. [Google Scholar] [PubMed] [CrossRef]
256. Lin YM, Gao Y, Gong MG, Zhang SJ, Zhang YQ, Li ZY. Federated learning on multimodal data: a comprehensive survey. Mach Intell Res. 2023;20(4):539–53. doi:10.1007/s11633-022-1398-0. [Google Scholar] [CrossRef]
257. Gupta NS, Kumar P. Perspective of artificial intelligence in healthcare data management: a journey towards precision medicine. Comput Biol Med. 2023;162(1988):107051. doi:10.1016/j.compbiomed.2023.107051. [Google Scholar] [PubMed] [CrossRef]
258. Van den Eynde J, Lachmann M, Laugwitz K-L, Manlhiot C, Kutty S. Successfully implemented artificial intelligence and machine learning applications in cardiology: state-of-the-art review. Trends Cardiovasc Med. 2023;33(5):265–71. doi:10.1016/j.tcm.2022.01.010. [Google Scholar] [PubMed] [CrossRef]
259. Liu Q, Joshi A, Standing JF, van der Graaf PH. Artificial intelligence/machine learning: the new frontier of clinical pharmacology and precision medicine. Clin Pharmacol Ther. 2024;115(4):637–42. doi:10.1002/cpt.3198. [Google Scholar] [PubMed] [CrossRef]
260. van Leeuwen KG, de Rooij M, Schalekamp S, van Ginneken B, Rutten MJCM. How does artificial intelligence in radiology improve efficiency and health outcomes? Pediatr Radiol. 2022;52(11):2087–93. doi:10.1007/s00247-021-05114-8. [Google Scholar] [PubMed] [CrossRef]
261. Alowais SA, Alghamdi SS, Alsuhebany N, Alqahtani T, Alshaya AI, Almohareb SN, et al. Revolutionizing healthcare: the role of artificial intelligence in clinical practice. BMC Med Educ. 2023;23(1):689. doi:10.1186/s12909-023-04698-z. [Google Scholar] [PubMed] [CrossRef]
262. Yin J, Ngiam KY, Teo HH. Role of artificial intelligence applications in real-life clinical practice: systematic review. J Med Internet Res. 2021;23(4):e25759. doi:10.2196/25759. [Google Scholar] [PubMed] [CrossRef]
263. Jiang Y, Wang C, Zhou S. Artificial intelligence-based risk stratification, accurate diagnosis and treatment prediction in gynecologic oncology. Semin Cancer Biol. 2023;96(1):82–99. doi:10.1016/j.semcancer.2023.09.005. [Google Scholar] [PubMed] [CrossRef]
264. Xie Y, Lu L, Gao F, He SJ, Zhao HJ, Fang Y, et al. Integration of artificial intelligence, blockchain, and wearable technology for chronic disease management: a new paradigm in smart healthcare. Curr Med Sci. 2021;41(6):1123–33. doi:10.1007/s11596-021-2485-0. [Google Scholar] [PubMed] [CrossRef]
265. Dave T, Athaluri SA, Singh S. ChatGPT in medicine: an overview of its applications, advantages, limitations, future prospects, and ethical considerations. Front Artif Intell. 2023;6:1169595. doi:10.3389/frai.2023.1169595. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF



 Downloads
Downloads
 Citation Tools
Citation Tools