 Open Access
Open Access
ARTICLE
Multi-Stage Hierarchical Feature Extraction for Efficient 3D Medical Image Segmentation
Department of Electrical and Computer Engineering, Inha University, 100, Inha-Ro, Michuhol-Gu, Incheon, 22212, Republic of Korea
* Corresponding Author: Byeong-Seok Shin. Email:
Computers, Materials & Continua 2025, 83(3), 5429-5443. https://doi.org/10.32604/cmc.2025.063815
Received 24 January 2025; Accepted 18 April 2025; Issue published 19 May 2025
Abstract
Research has been conducted to reduce resource consumption in 3D medical image segmentation for diverse resource-constrained environments. However, decreasing the number of parameters to enhance computational efficiency can also lead to performance degradation. Moreover, these methods face challenges in balancing global and local features, increasing the risk of errors in multi-scale segmentation. This issue is particularly pronounced when segmenting small and complex structures within the human body. To address this problem, we propose a multi-stage hierarchical architecture composed of a detector and a segmentor. The detector extracts regions of interest (ROIs) in a 3D image, while the segmentor performs segmentation in the extracted ROI. Removing unnecessary areas in the detector allows the segmentation to be performed on a more compact input. The segmentor is designed with multiple stages, where each stage utilizes different input sizes. It implements a stage-skipping mechanism that deactivates certain stages using the initial input size. This approach minimizes unnecessary computations on segmenting the essential regions to reduce computational overhead. The proposed framework preserves segmentation performance while reducing resource consumption, enabling segmentation even in resource-constrained environments.Keywords
Accurately segmenting regions of the human body’s anatomical structures, such as organs and bones, is essential for enhancing the accuracy of clinical diagnoses [1] and examinations [2]. However, it remains challenging because the structures vary significantly in size and shape. Such diversity makes it difficult to distinguish boundaries, increasing the likelihood of segmentation errors.
Deep neural networks have brought advancements to medical image segmentation, significantly improving accuracy. U-Net [3], which integrates a symmetrical U-shaped encoder-decoder architecture with skip connections, has become a foundational model for various segmentation models. Numerous studies have continued to develop the U-Net structure [4–6]. Recent studies have combined U-Net with Transformer [7] architectures to enhance global semantic information learning [8–10]. Transformer-based models divide images into sequential patches and leverage self-attention mechanisms to capture global features effectively. However, the model size and computational cost increase rapidly as segmentation accuracy improves. Since 3D-based models experience the exponential increase of resource consumption, it restricts their practical use in real-world applications [11]. These limitations become even more pronounced in resource-constrained environments, restricting the practical applicability of the model [12,13].
Research on lightweight medical image segmentation networks has been proposed, as medical image analysis has extended to resource-constrained environments such as clinical diagnostics [14], telehealth [15], and edge devices [16]. These lightweight networks reduce the cost of medical systems and enable operation in mobile environments. It allows medical professionals to quickly and accurately analyze patients’ image data. Additionally, they facilitate the rapid processing and analysis of medical images in the resource-limited environments encountered in remote areas. It also enhances the efficiency of patient diagnosis and treatment. Moreover, 3D lightweight segmentation models have been proposed to reduce computational resource requirements [17,18]. These methods primarily enhance computational efficiency by reducing the parameters in each network layer. However, since parameters directly influence the network’s capability, decreasing them can lead to performance degradation [19]. Therefore, balancing computational cost and segmentation performance is essential, as excessively simplifying the model may result in a decline in performance. Additionally, these methods struggle to balance global and local features, increasing the risk of errors in multi-scale segmentation [20]. This problem becomes especially prominent when segmenting small and complex structures within the human body due to the regional imbalance between small target objects and large backgrounds. Segmentation models typically allocate a significant portion of their parameters to extract features from the background. Consequently, the influence of the target object on training diminishes, leading to reduced segmentation accuracy. In conclusion, developing a framework that optimally balances computational cost and segmentation performance is essential for ensuring the accurate segmentation of small target objects.
We propose a multi-stage hierarchical architecture that reduces computational resource usage while preserving segmentation accuracy. The proposed architecture comprises a detector and a segmentor. The detector extracts the region of interest (ROI) from medical images and removes background areas. By eliminating unnecessary regions of the input image, the segmentor operates on a more compact input, effectively reducing computational resource consumption. The segmentor is structured as multiple hierarchical stages where each stage handles input of a different size. As the hierarchy of stages goes lower, the depth increases, and more detailed features are extracted from the smaller inputs. If the input size is smaller than the threshold for a specific stage, that stage is deactivated and excluded from training and inference. This stage-skipping approach prevents unnecessary computations on large regions when segmenting small-sized targets. Correspondingly, it effectively reduces computational resource usage while maintaining segmentation performance.
The main contributions of this paper are as follows.
• We introduce a region extraction module within the detector to extract ROIs from input images. By removing unnecessary areas, this module enables the segmentor to achieve segmentation on a compact input.
• We propose a multi-stage hierarchical architecture in the segmentor, where each stage utilizes inputs of different sizes. This architecture optimizes computational efficiency while maintaining segmentation performance by deactivating unnecessary stages using the ROI size.
Section 2 discusses related work on 3D medical image segmentation, including methods to reduce computational resource usage. Section 3 describes the proposed multi-stage hierarchical segmentation model. Section 4 provides comprehensive analyses and interpretations of our experimental results. Finally, Section 5 summarizes our findings.
Various studies have been proposed to enhance the performance of the 3D image segmentation. Çiçek et al. [4] proposed the 3D U-Net architecture to segment 3D medical images. This technique enables high-density 3D image segmentation by leveraging sparsely annotated volumetric data. Milletari et al. [21] proposed a method that captures the relationships between adjacent slices and the overall volumetric context by using a low-resolution 3D image as input. Huang et al. [5] introduced 3D
Several methods have been proposed to reduce computational resources in 3D segmentation. Xie et al. [26] proposed convolutional neural networks with Transformer to mitigate the high computational load and spatial complexity when handling 3D features. It introduces a deformable self-attention mechanism that performs multi-scale and high-resolution feature maps on a limited number of key positions. Perera et al. [27] proposed SegFormer3D, a lightweight 3D medical image segmentation model. It removes complex decoder structures and employs an all-MLP decoder to maintain segmentation performance while reducing resource consumption. Shaker et al. [28] introduced the UNETR++ 3D image segmentation technique, ensuring efficiency by minimizing the parameters and the computational cost. This technique employs dual efficient paired attention blocks to reduce the spatial complexity of attention learning to a linear complexity level. Liao et al. [29] presented LightM-UNet, a lightweight framework integrating Mamba [30] into U-Net. Their technique uses residual vision Mamba layers to extract deep semantic features, decreasing the computational complexity of long-range spatial dependencies as linear. Zhu et al. [20] proposed the lightweight medical image segmentation network to reduce computational resources in semantic segmentation. It integrates a lightweight Transformer into the CNN architecture to optimize multi-scale feature interactions and effectively capture semantic information. Various approaches have been proposed to reduce computational resources during the segmentation of 3D images. However, calculating entire regions while segmenting small and complex structures leads to the excessive and unnecessary consumption of computational resources. Therefore, techniques should be proposed to pre-extract the ROIs and then perform segmentation only within those regions to conserve those resources.
The overall structure of the proposed method shown in Fig. 1a comprises the detector and the segmentor. The detector extracts the ROIs in the 3D image using 2D slices taken from the center of each axis. The segmentor performs segmentation using the extracted ROIs. It is designed as multi-stage hierarchical architecture where each stage handles differently sized input. As the stages move down the hierarchy, their depth increases to extract finer features from smaller input. In addition, the proposed method utilizes a stage-skipping technique that deactivates specific stages if they cannot handle the input size. The training process of the proposed method is as follows. Initially, the detector and segmentor are trained separately, and then the trained modules are combined into a single model. The integrated model is then trained through fine tuning where the detector is frozen and only the segmentor is trained. It enables the segmentor to perform more precisely using the ROI extracted by the detector.
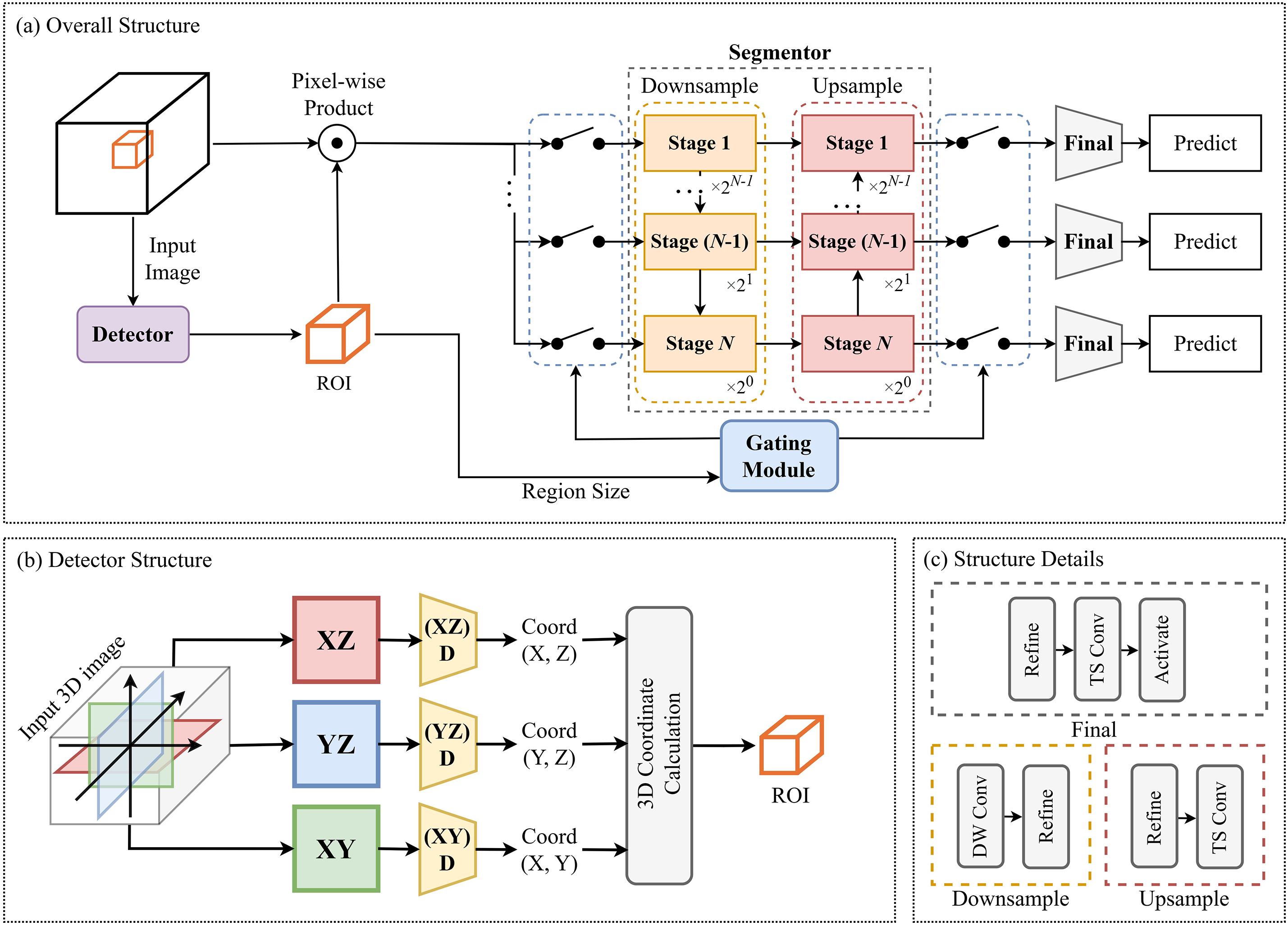
Figure 1: (a) represents an overview of the proposed method. (b) illustrates the structure of the detector that extracts ROIs. (c) shows the detailed structure of the downsample, upsample, and final stages in (a)
The structure of the detector is illustrated in Fig. 1b. The detector calculates the ROI using three 2D slices (
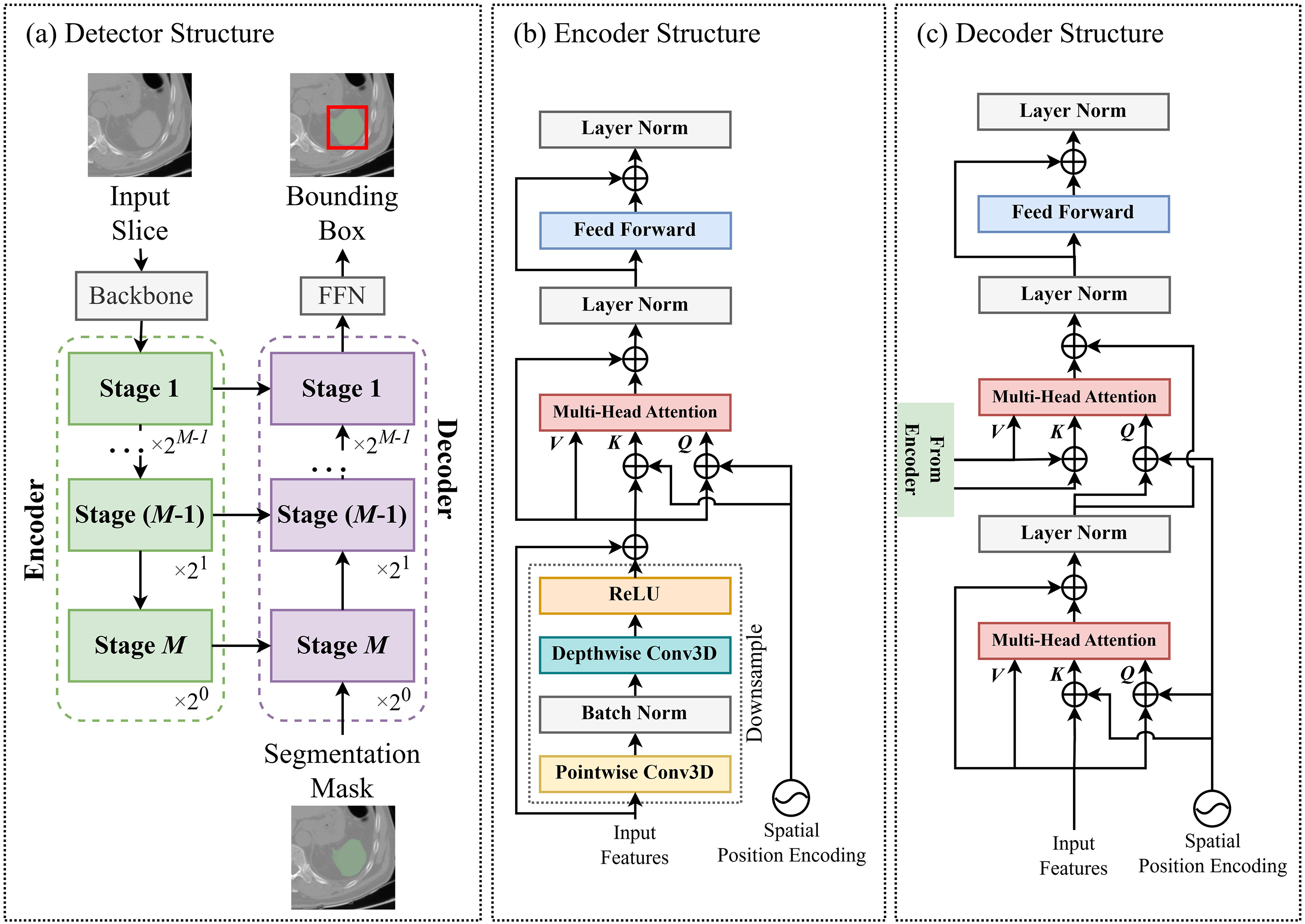
Figure 2: (a) represents the structure of the detector extracting a 2D bounding box from a 2D slice. The detector is divided into the encoder (b) and the decoder (c), each comprising multiple stages (FFN = feed-forward network)
Fig. 2b,c depicts the detailed architectures of the encoder and decoder stages. Initially, a downsampling module is employed to reduce the feature size. It utilizes depth-wise and point-wise 3D convolution layers [33,34], which offer a lower computational load than standard 3D convolution layers. After passing through the downsampling module, multi-scale attention [35] is applied to generate feature maps with the same resolution as the input. Each key and query element consists of pixels from the feature maps, where each query pixel’s reference point is the input feature, itself. We introduce spatial positional embedding to the key and query element, which assigns unique encoding values to different stages to determine each query pixel’s stage location (first to M-th stage). The decoder is composed of two multi-scale attention modules. Similar to the encoder stage, the first multi-scale attention module generates a feature map using the input from the previous decoder stage as each query pixel’s reference point. The second multi-scale attention module utilizes the generated feature map from the previous multi-scale attention module and the feature from the encoder of the same stage index, which is integrated via a skip connection. The encoder’s features serve as the value and key elements, while the feature map produced by the previous multi-scale attention module acts as the query element. Simultaneously, spatial positional embedding, generated in the same manner as in the encoder, is applied.
The segmentor consists of downsampling stages to decrease the feature dimensions and upsampling stages to restore the spatial resolution. N represents the maximum stage index of the segmentor. The segmentor is a multi-stage hierarchical structure, with each stage handling input of different sizes. As the index of each stage increases, the input size computed by each stage is halved. The segmentor employs a stage-skipping technique, deactivating unnecessary stages using the input size to optimize computational efficiency. When the stage index is
The smallest
The input of the segmentor is reshaped into a cube of size
The images used in this experiment were labeled prostate/uterus, spleen, and liver from the abdominal organ segmentation (AMOS) [37] dataset. Additionally, the dataset was categorized according to the proportion of each label present in the image: small for the prostate/uterus (25%), medium for the spleen (50%), and large for the liver (75%). The input images are normalized to [0, 1] and resized to
The outputs from the detector are demonstrated in Fig. 3a. In all cases, the generated ROIs were accurately positioned but were smaller than the ground truth. That leads to errors in certain areas due to the inaccurate ROIs negatively impacting the accuracy of the proposed method combining the detector and segmentor. Further research on loss functions and network architectures should address this issue to minimize errors in the generated ROIs. Fig. 3b displays the outputs from the proposed network, which exhibits reduced accuracy on concave and anisotropic structures in all cases. In an axial slice of the liver, a defect appeared in the shape of the left lobe. The segmentation mask was generated with incorrect locations in the coronal slice of the prostate/uterus. These errors result from misidentifying similar organs when the ROI targets different areas. The segmentor should be refined to solve these problems by preventing the misidentification of similar organs and incorrect segmentation results from the imprecise ROIs. Conversely, in the sagittal slice of the spleen, unnatural shape truncation occurs. This issue is caused by a detector error excluding the correct region occupied by ground truth. That can be resolved by improving the detector’s accuracy. In conclusion, the detector and segmentor in the proposed method are strongly interconnected, and enhancing the network structure of both modules is essential for achieving overall accuracy.
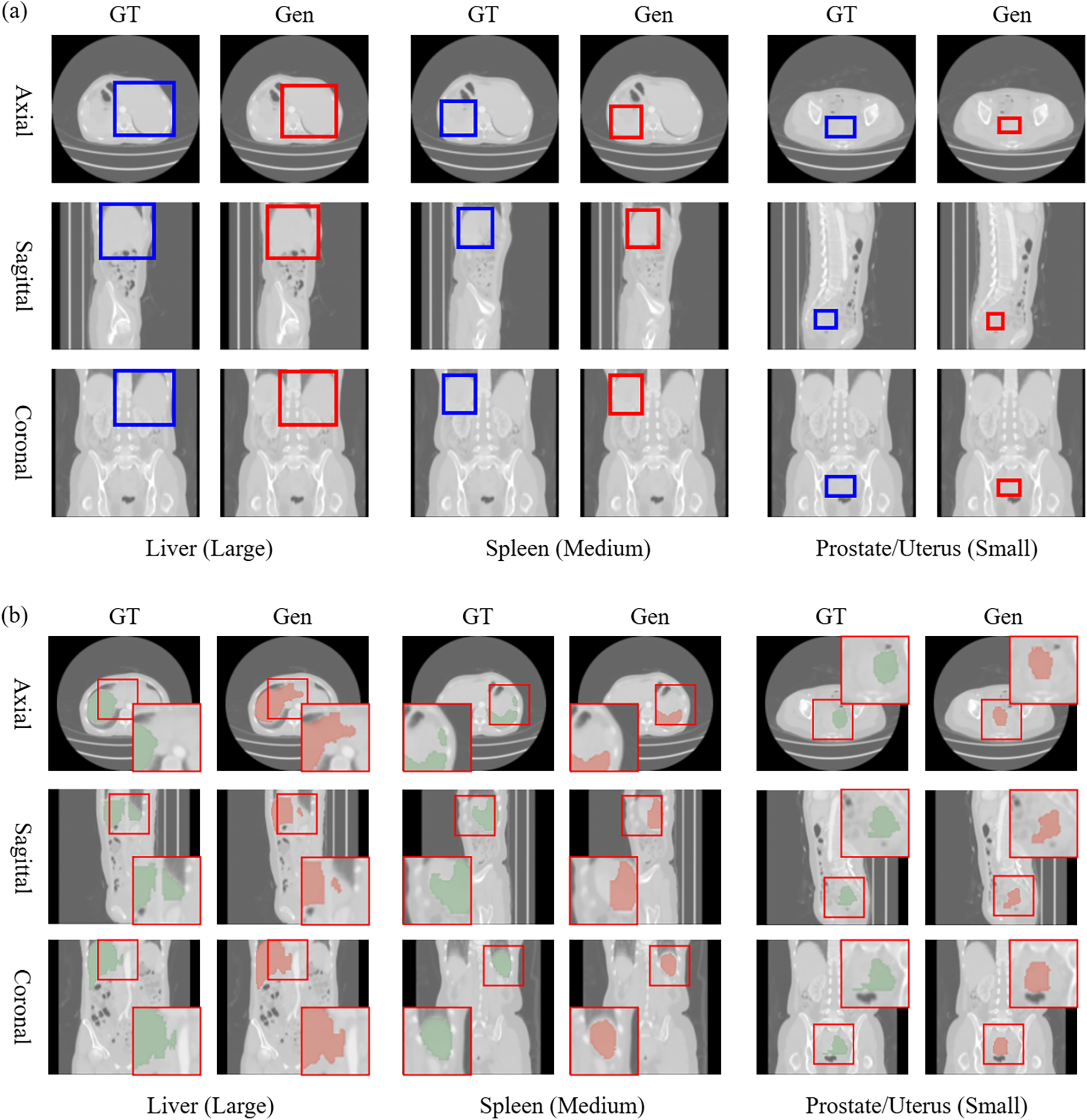
Figure 3: This figure visualizes the results from the detector (a) and the proposed method (b). The blue and red boxes in (a) represent the ground truth (GT) and generated ROI (Gen). The red boxes in (b) reveal magnified areas with significant differences between the generated image and ground truth
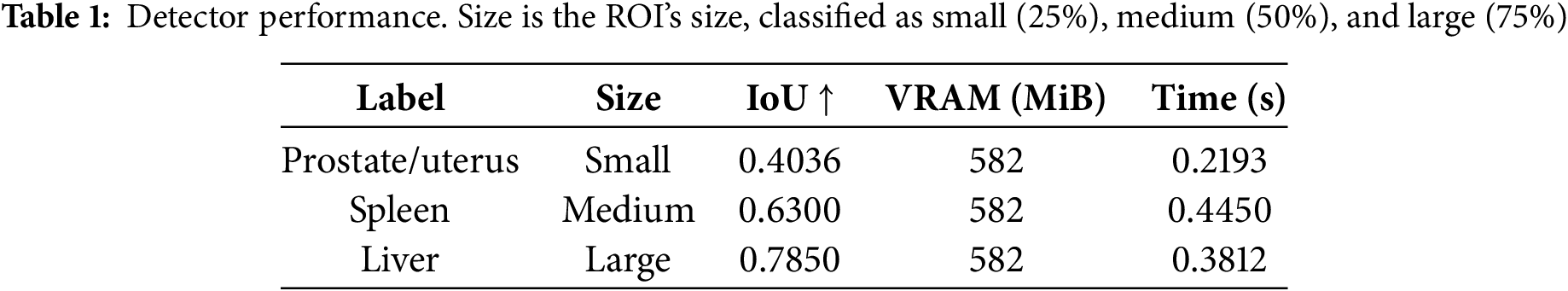
The accuracy was evaluated using the intersection over union (IoU) [38], Dice score [39], and Hausdorff distance (HD95) [40]. GPU memory consumption (video random access memory, VRAM) and iteration time are also measured during inference. The performance of the detector module is revealed in Table 1. The IoU results reveal that the accuracy for the small ROI was 48.59% lower than that for the large ROI. There was no difference in memory consumption because all cases used 2D images at
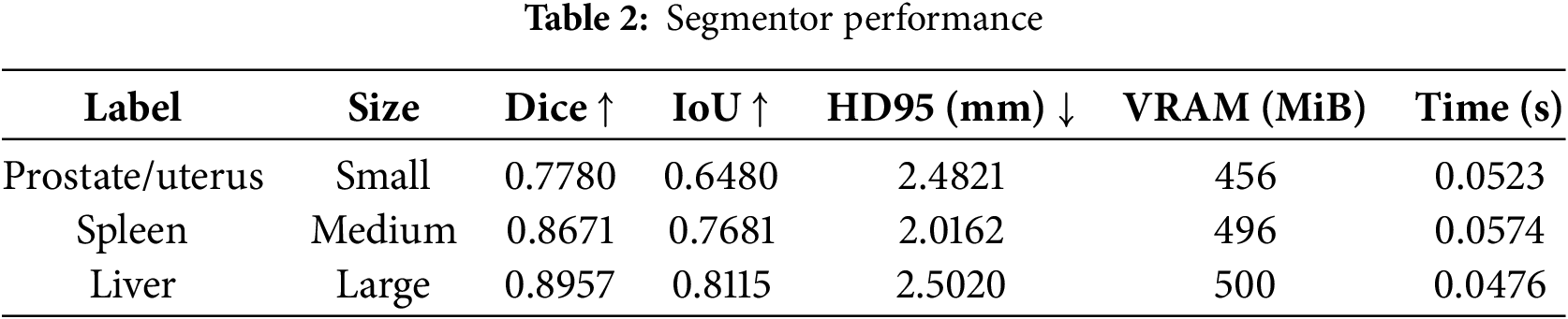
The proposed network’s performance was evaluated using the size of the ROI, as presented in Table 3. Moreover, w/o crop indicates the segmentation of the entire 3D image without a detector, and with crop (our method) applies the detector to generate ROIs for segmentation. Compared to the w/o crop results, the with crop results from the small size and the large size depicted a Dice score of 43.81% lower and 6.45% lower, respectively. Similarly, the IoU results decreased by 51.30% with the small ROI and 11.51% with the large ROI. This result showed that the accuracy of with crop using the detector is lower compared to w/o crop due to the limitations of the 2D-based detector structure. It identifies the ROI in a 3D image using projected 2D slices (axial, sagittal, and coronal) instead of the 3D image. This approach enhances computational efficiency by reducing the input size, which lowers memory consumption and accelerates inference speed. However, it results in accuracy degradation because it fails to consider inter-slice correlations and cannot capture depth across the 3D image. Consequently, regional proposal errors appear in areas with less density variation. This issue becomes more prominent in small regions closely connected with similar complex structures. This accuracy degradation comes from detector inaccuracies, causing the segmentor to receive out-of-distribution (OOD) data. Therefore, accuracy should be improved by fine tuning the newly occurring OOD data. In addition, the proposed method requires at least twice the iteration time and more VRAM than the segmentation of an entire 3D image without a detector. These results stem from the additional detector module.
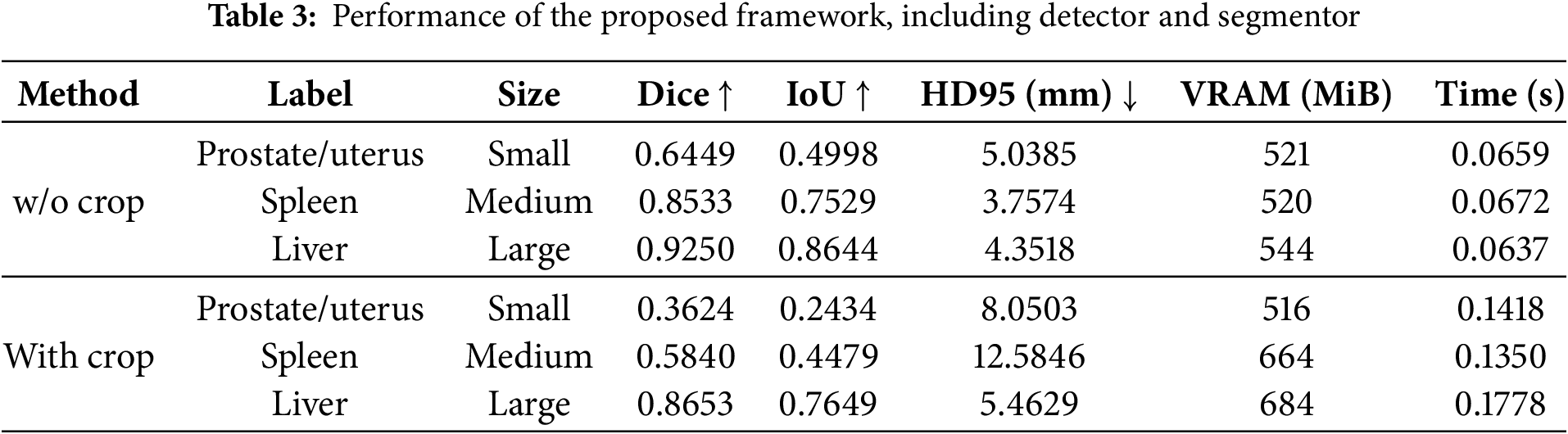
The performance comparison between the proposed framework and state-of-the-art methods is presented in Table 4. We conducted experiments on the proposed method with SwinSMT [23], SegFormer3D [27], and MedNeXt [8]. According to the analysis, MedNeXt achieved the best accuracy across all input sizes regarding the Dice, IoU, and HD95 metrics. However, MedNeXt consumes the highest VRAM compared to other methods. SegFormer3D demonstrated the most efficient VRAM usage across all input sizes. SwinSMT exhibited intermediate results in both accuracy and resource consumption. Although it uses minimal VRAM, the proposed framework showed lower accuracy than other methods at all three input sizes. In particular, the accuracy gap between small and large sizes is noticeable, and further improvements of the small sizes in the detector module are needed to resolve this.
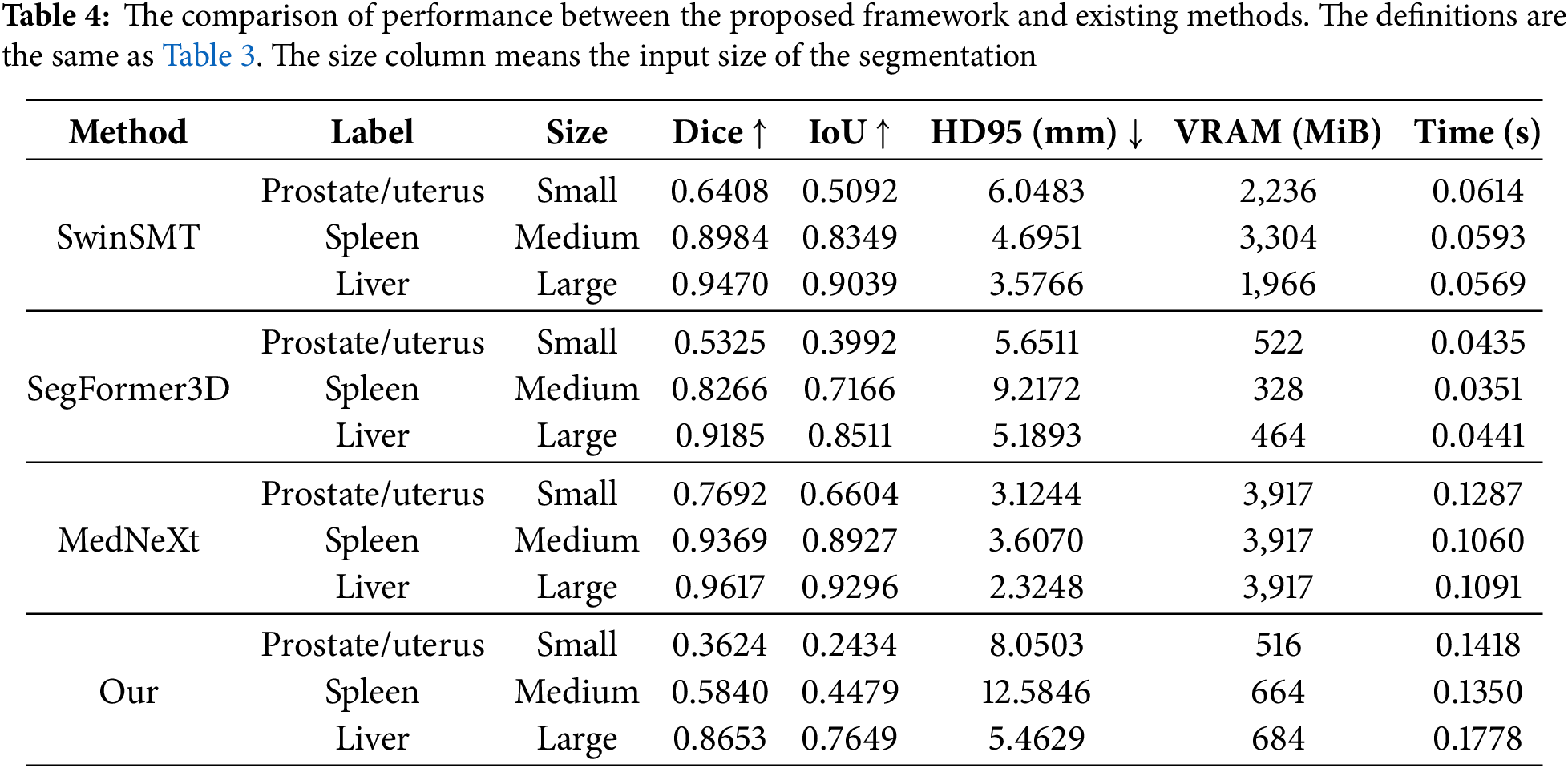
The floating point operations per second (FLOPs) and the number of parameters of the modules in the proposed method are presented in Table 5. The segmentor module with the smaller ROI achieves 85.42% lower FLOPs and 10.32% fewer parameters than the larger ROI. This result demonstrates that dynamically adjusting the network using the size of the proposed regions leads to a substantial improvement in computational efficiency. However, the FLOPs and parameter count of the detector module are 4878.81% and 1841.80% higher than those of the segmentor module. When compared to the existing methods, the segmentor module demonstrated a significant computational cost reduction in FLOPs compared to existing methods, although the number of parameters was higher than those. Meanwhile, the resource consumption of the detector module was similar to or higher than existing methods. The FLOPs of the detector reached 114.49% of SegFormer3D, and the parameters were as high as 3221.57% compared to MedNeXt. These results indicate that the improvements in the segmentor module have a relatively limited impact on the overall efficiency of the entire framework. Additionally, it is observed that the resource consumption in the detector module is significant. Therefore, extending the downsampling approach to reduce the feature size or other optimization techniques are necessary.
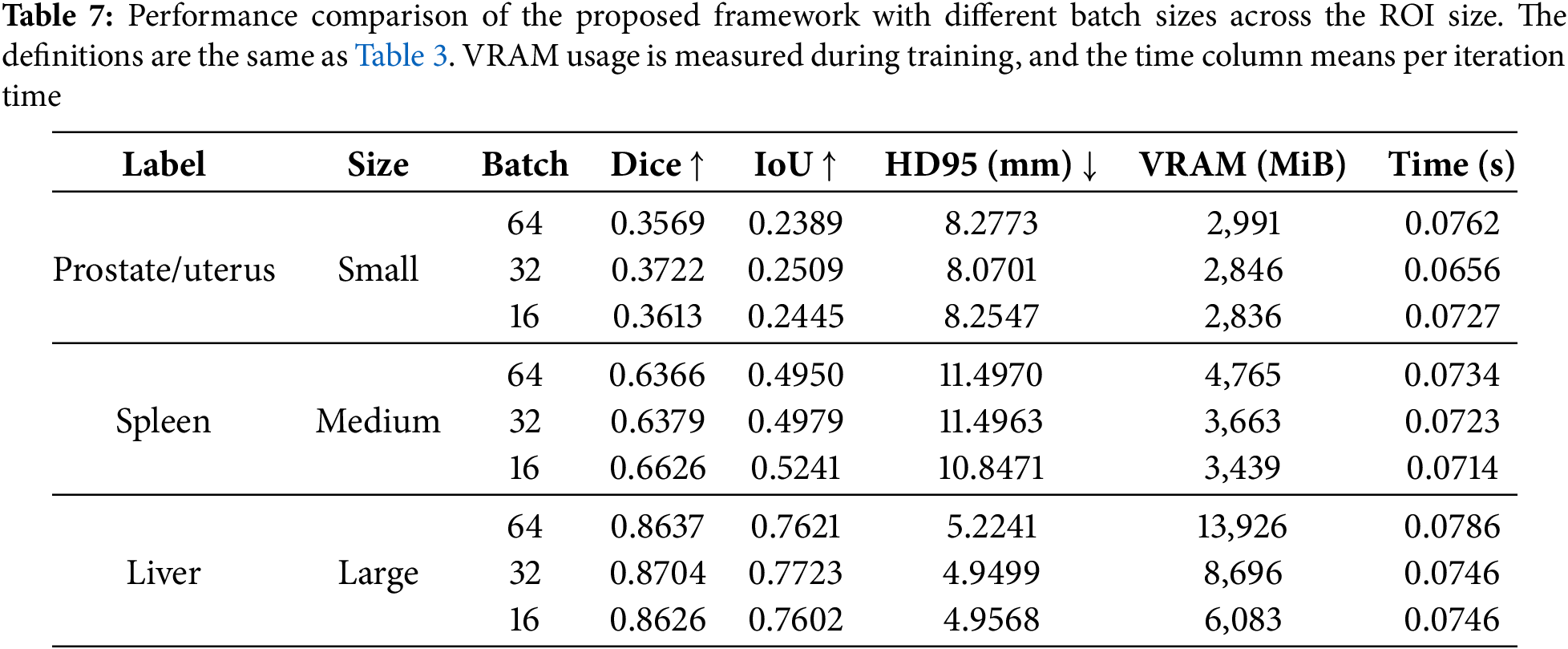
The results of experiments using different learning rates and batch sizes for the proposed method are shown in Tables 6 and 7, respectively. Table 6 presents that the results with small and large ROIs get the highest Dice and IoU scores at a learning rate of
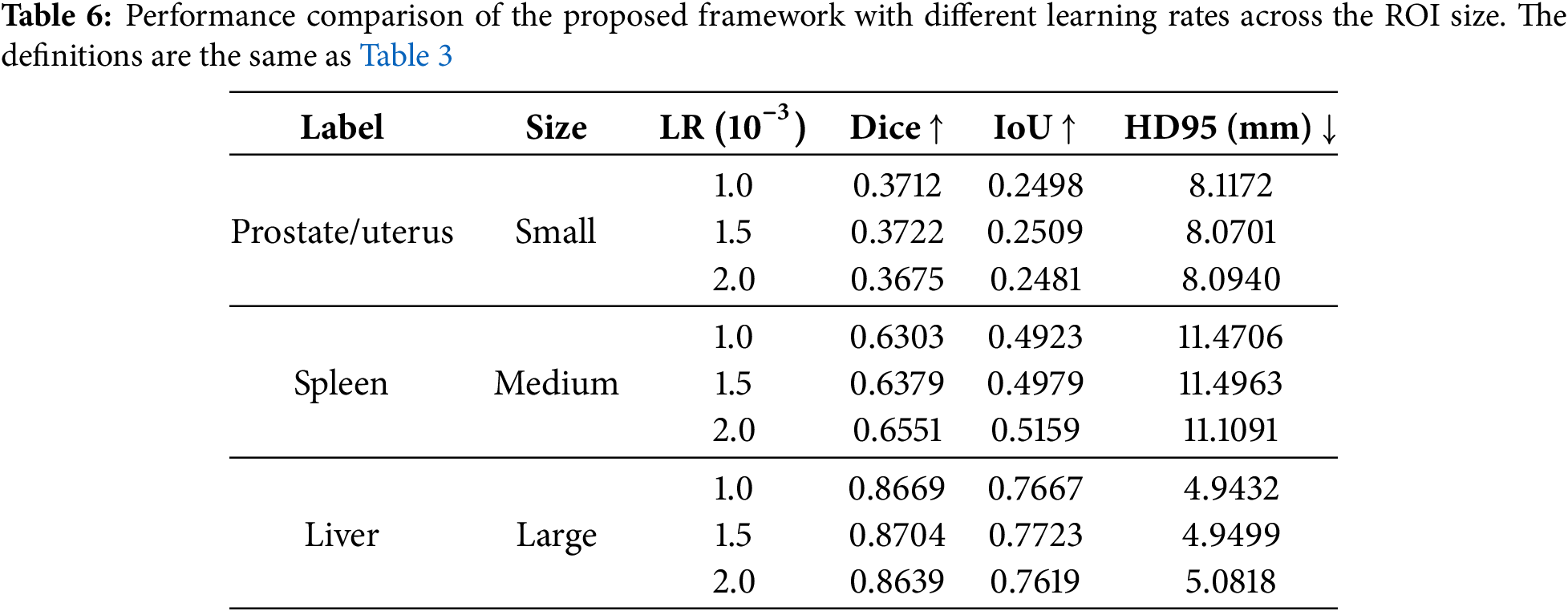
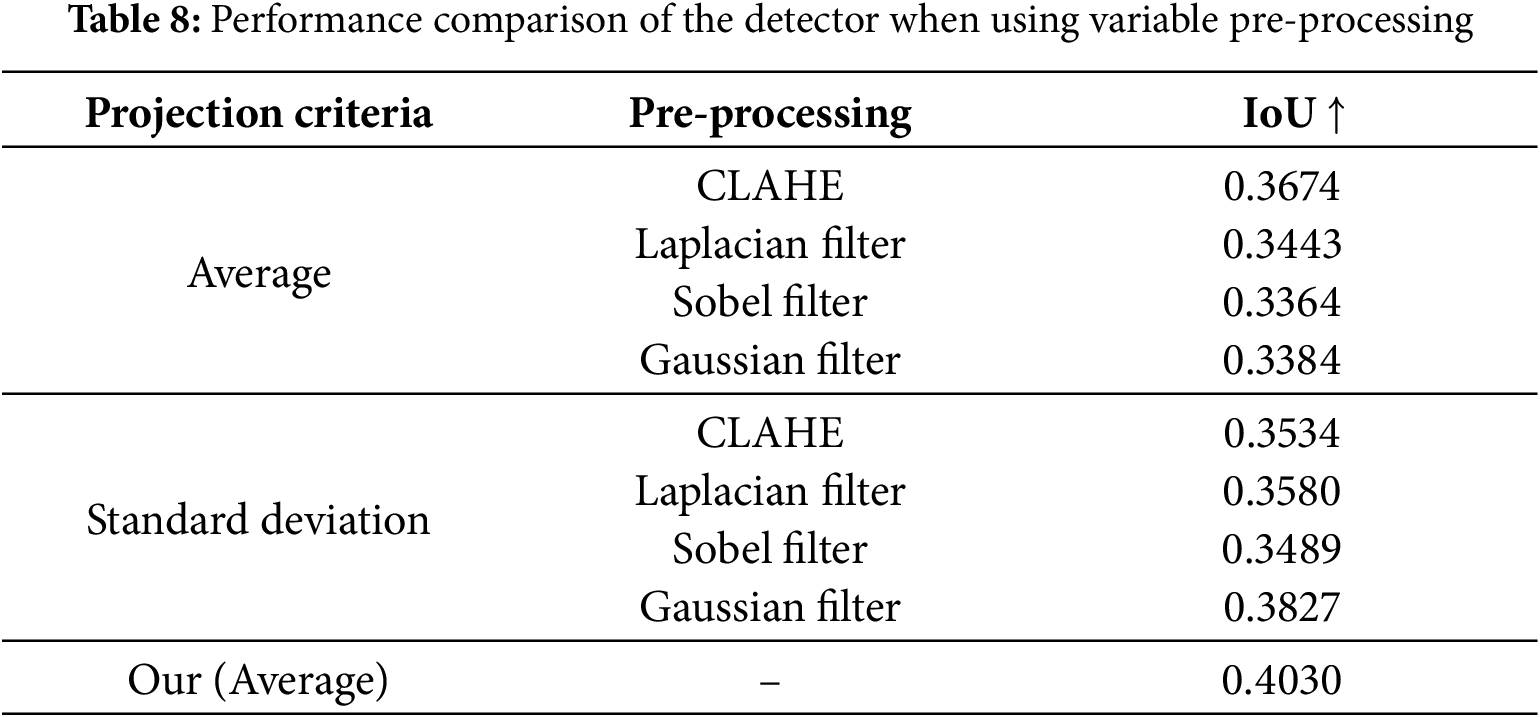
The experimental results for the proposed detector module with different pre-processing methods are presented in Table 8. The experiment was conducted using two types of projections: average and standard deviation projection. CLAHE (Contrast Limited Adaptive Histogram Equalization), Laplacian filter, Sobel filter, and Gaussian filter were applied after projection [41,42]. The projection process converts a 3D image into a 2D image by calculating the average or standard deviation of slices along each axis. Applying CLAHE achieved the highest IoU when using the average as the projection criterion. When the standard deviation was used as the projection criterion, the Gaussian filter yielded the highest IoU. However, in both cases, the accuracy was lower than when no pre-processing was applied. This result shows that applying projection does not directly improve the detector’s accuracy. During average projection, converting a 3D image into a 2D image causes image blurring, making it difficult to distinguish structures with similar intensities as the background, such as muscles and skin. During the standard deviation projection, high-density regions, such as bones, become overly emphasized, hindering the distinction of the outlines of the target object. It is better to use a projection for several sub-regions than a projection for the whole region to solve these problems. Utilizing multiple projected inputs using sub-regions can reduce interference from the background so the detector can recognize the target object more clearly.
This study proposes a multi-stage hierarchical architecture for 3D segmentation that can reduce computational costs while maintaining segmentation accuracy. The proposed structure consists of a detector and a segmentor. The detector extracts the ROIs, and the segmentor processes the input according to the size of the ROI. The stage-skipping mechanism saves computing resources by reducing unnecessary computations while maintaining fine segmentation accuracy. The proposed method can minimize the costs of medical equipment and systems in 3D medical image segmentation while ensuring efficient processing even in resource-constrained environments. It helps minimize environmental constraints in the diagnostic process and facilitates medical services. We plan to enhance network efficiency and accuracy by integrating features extracted from the detector and segmentor. Additionally, we will validate the proposed method on multiple datasets to ensure its robustness. Since the proposed approach is applicable across various imaging modalities (e.g., computed tomography, ultrasound), we also aim to refine the network architecture for modality-specific optimizations, further improving the segmentation performance for each modality.
Acknowledgement: The authors are grateful to all the editors and anonymous reviewers for their comments and suggestions.
Funding Statement: This work was supported by an INHA UNIVERSITY Research Grant.
Author Contributions: The authors’ contributions to this paper are as follows. Study conception and design: Jion Kim, Byeong-Seok Shin; data collection: Jayeon Kim; analysis and interpretation of results: Jion Kim, Jayeon Kim; draft manuscript preparation: Jion Kim, Jayeon Kim; funding acquisition and supervision: Byeong-Seok Shin; manuscript review: Byeong-Seok Shin. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets analyzed during the current study are available in the Zenodo repository, https://doi.org/10.5281/zenodo.7155725.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Addimulam S, Mohammed MA, Karanam RK, Ying D, Pydipalli R, Patel B, et al. Deep learning-enhanced image segmentation for medical diagnostics. Malaysian J Med Biol Res. 2020;7(2):145–52. [Google Scholar]
2. Hossain MS, Rahman MM, Syeed MM, Uddin MF, Hasan M, Hossain MA, et al. DeepPoly: deep learning-based polyps segmentation and classification for autonomous colonoscopy examination. IEEE Access. 2023;11:95889–902. doi:10.1109/ACCESS.2023.3310541. [Google Scholar] [CrossRef]
3. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference; 2015 Oct 5–9; Munich, Germany: Springer. p. 234–41. [Google Scholar]
4. Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Medical Image Computing and Computer-Assisted Intervention-MICCAI 2016: 19th International Conference; 2016 Oct 17–21; Athens, Greece: Springer. p. 424–32. [Google Scholar]
5. Huang C, Han H, Yao Q, Zhu S, Zhou SK. 3D U2-Net: a 3D universal U-Net for multi-domain medical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; 2019; Shenzhen, China: Springer. p. 291–9. [Google Scholar]
6. Ali S, Khurram R, Rehman KU, Yasin A, Shaukat Z, Sakhawat Z, et al. An improved 3D U-Net-based deep learning system for brain tumor segmentation using multi-modal MRI. Multimed Tools Appl. 2024;83:85027–46. doi:10.1007/s11042-024-19406-2. [Google Scholar] [CrossRef]
7. Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Adv Neural Inf Process Syst. 2017;30:1–11. [Google Scholar]
8. Roy S, Koehler G, Ulrich C, Baumgartner M, Petersen J, Isensee F, et al. MedNeXt: transformer-driven scaling of ConvNets for medical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; 2023; Vancouver, BC, Canada: Springer. p. 405–15. doi:10.1007/978-3-031-43901-8_39. [Google Scholar] [CrossRef]
9. Zhao C, Xiang S, Wang Y, Cai Z, Shen J, Zhou S, et al. Context-aware network fusing Transformer and V-Net for semi-supervised segmentation of 3D left atrium. Expert Syst Appl. 2023;214:119105. doi:10.1016/j.eswa.2022.119105. [Google Scholar] [CrossRef]
10. Zhou HY, Guo J, Zhang Y, Han X, Yu L, Wang L, et al. nnFormer: volumetric medical image segmentation via a 3D Transformer. IEEE Trans Image Process. 2023;32:4036–45. doi:10.1109/TIP.2023.3293771. [Google Scholar] [PubMed] [CrossRef]
11. Wu B, Xiao Q, Liu S, Yin L, Pechenizkiy M, Mocanu DC, et al. E2ENet: dynamic sparse feature fusion for accurate and efficient 3D medical image segmentation. Adv Neural Inform Process Syst. 2024;37:118483–512. [Google Scholar]
12. Corral JMR, Civit-Masot J, Luna-Perejón F, Díaz-Cano I, Morgado-Estévez A, Domínguez-Morales M. Energy efficiency in edge TPU vs. embedded GPU for computer-aided medical imaging segmentation and classification. Eng Appl Artif Intell. 2024;127:107298. doi:10.1016/j.engappai.2023.107298. [Google Scholar] [CrossRef]
13. Niepceron B, Nait-Sidi-Moh A, Grassia F. Moving medical image analysis to GPU embedded systems: application to brain tumor segmentation. Appl Artif Intell. 2020;34(12):866–79. doi:10.1080/08839514.2020.1787678. [Google Scholar] [CrossRef]
14. Olatunji AO, Olaboye JA, Maha CC, Kolawole TO, Abdul S. Revolutionizing infectious disease management in low-resource settings: the impact of rapid diagnostic technologies and portable devices. Int J Appl Res Soc Sci. 2024;6(7):1417–32. doi:10.51594/ijarss.v6i7.1332. [Google Scholar] [CrossRef]
15. Ogunsola OO, Olawepo JO, Ajayi O, Osayi E, Akinro YT, Ifechelobi C, et al. AVIVA: a telehealth tool to improve cervical cancer screening in resource-constrained settings. BMJ Glob Health. 2023;8(7):e012311. doi:10.1136/bmjgh-2023-012311. [Google Scholar] [PubMed] [CrossRef]
16. Huang Z, Herbozo Contreras LF, Leung WH, Yu L, Truong ND, Nikpour A, et al. Efficient edge-AI models for robust ECG abnormality detection on resource-constrained hardware. J Cardiovasc Transl Res. 2024;17(4):879–92. doi:10.1007/s12265-024-10504-y. [Google Scholar] [PubMed] [CrossRef]
17. Alwadee EJ, Sun X, Qin Y, Langbein FC. LATUP-Net: a lightweight 3D attention U-Net with parallel convolutions for brain tumor segmentation. Comput Biol Med. 2025;184:109353. doi:10.1016/j.compbiomed.2024.109353. [Google Scholar] [PubMed] [CrossRef]
18. Ruan J, Xiang S, Xie M, Liu T, Fu Y. MALUNet: a multi-attention and light-weight unet for skin lesion segmentation. In: 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2022; Las Vegas, NV, USA: IEEE. p. 1150–6. doi:10.1109/BIBM55620.2022.9995040. [Google Scholar] [CrossRef]
19. Valanarasu JMJ, Patel VM. Unext: mlp-based rapid medical image segmentation network. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; 2022; Singapore: Springer. p. 23–33. doi:10.1007/978-3-031-16443-9_3. [Google Scholar] [CrossRef]
20. Zhu Z, Yu K, Qi G, Cong B, Li Y, Li Z, et al. Lightweight medical image segmentation network with multi-scale feature-guided fusion. Comput Biol Med. 2024;182:109204. doi:10.1016/j.compbiomed.2024.109204. [Google Scholar] [PubMed] [CrossRef]
21. Milletari F, Navab N, Ahmadi SA. V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV); 2016; Stanford, CA, USA: IEEE. p. 565–71. doi:10.1109/3DV.2016.79. [Google Scholar] [CrossRef]
22. Jiang Y, Zhang Y, Lin X, Dong J, Cheng T, Liang J. SwinBTS: a method for 3D multimodal brain tumor segmentation using swin Transformer. Brain Sci. 2022;12(6):797. doi:10.3390/brainsci12060797. [Google Scholar] [PubMed] [CrossRef]
23. Płotka S, Chrabaszcz M, Biecek P. Swin SMT: global sequential modeling in 3D medical image segmentation. arXiv:2407.07514. 2024. doi:10.1007/978-3-031-72111-3. [Google Scholar] [CrossRef]
24. Liu Z, Mao H, Wu CY, Feichtenhofer C, Darrell T, Xie S. A ConvNet for the 2020s. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2022; New Orleans, LA, USA: IEEE. p. 11976–86. [Google Scholar]
25. Zhu Z, Zhang Z, Qi G, Li Y, Li Y, Mu L. A dual-branch network for ultrasound image segmentation. Biomed Signal Process Control. 2025;103:107368. doi:10.1016/j.bspc.2024.107368. [Google Scholar] [CrossRef]
26. Xie Y, Zhang J, Shen C, Xia Y. Cotr: efficiently bridging CNN and transformer for 3D medical image segmentation. In: Medical Image Computing and Computer Assisted Intervention—MICCAI 2021: 24th International Conference; 2021 Sep 27–Oct 1; Strasbourg, France: Springer. p. 171–80. doi:10.1007/978-3-030-87199-4_16. [Google Scholar] [CrossRef]
27. Perera S, Navard P, Yilmaz A. SegFormer3D: an efficient Transformer for 3D medical image segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2024; Seattle, WA, USA: IEEE. p. 4981–8. [Google Scholar]
28. Shaker AM, Maaz M, Rasheed H, Khan S, Yang MH, Khan FS. UNETR++: delving into efficient and accurate 3D medical image segmentation. IEEE Trans Med Imaging. 2024;43(9):3377–90. doi:10.1109/TMI.2024.3398728. [Google Scholar] [PubMed] [CrossRef]
29. Liao W, Zhu Y, Wang X, Pan C, Wang Y, Ma L. LightM-Unet: mamba assists in lightweight UNet for medical image segmentation. arXiv:2403.05246. 2024. doi:10.48550/arXiv.2403.05246. [Google Scholar] [CrossRef]
30. Gu A, Dao T. Mamba: linear-time sequence modeling with selective state spaces. arXiv:2312.00752. 2023. doi:10.48550/arXiv.2312.00752. [Google Scholar] [CrossRef]
31. Zhu X, Su W, Lu L, Li B, Wang X, Dai J. Deformable DETR: deformable transformers for end-to-end object detection. arXiv:2010.04159. 2020. doi:10.48550/arXiv.2010.04159. [Google Scholar] [CrossRef]
32. Lin TY, Dollár P, Girshick R, He K, Hariharan B, Belongie S. Feature pyramid networks for object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017; Honolulu, HI, USA: IEEE. p. 2117–25. doi:10.1109/cvpr.2017.106. [Google Scholar] [CrossRef]
33. Chollet F. Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017; Honolulu, HI, USA: IEEE. p. 1251–8. [Google Scholar]
34. Hua BS, Tran MK, Yeung SK. Pointwise convolutional neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2018; Salt Lake City, UT, USA: IEEE. p. 984–93. [Google Scholar]
35. Tao A, Sapra K, Catanzaro B. Hierarchical multi-scale attention for semantic segmentation. arXiv:2005.10821. 2020. doi:10.48550/arXiv.2005.10821. [Google Scholar] [CrossRef]
36. Pang Y, Liang J, Huang T, Chen H, Li Y, Li D, et al. Slim UNETR: scale hybrid Transformers to efficient 3D medical image segmentation under limited computational resources. IEEE Trans Med Imaging. 2023;43(3):994–1005. doi:10.1109/TMI.2023.3326188. [Google Scholar] [PubMed] [CrossRef]
37. Ji Y, Bai H, Ge C, Yang J, Zhu Y, Zhang R, et al. AMOS: a large-scale abdominal multi-organ benchmark for versatile medical image segmentation. Adv Neural Inform Process Syst. 2022;35:36722–32. [Google Scholar]
38. He K, Gkioxari G, Dollar P, Girshick R. Mask R-CNN. In: Proceedings of the IEEE International Conference on Computer Vision (ICCV); 2017; Venice, Italy: IEEE. [Google Scholar]
39. Eelbode T, Bertels J, Berman M, Vandermeulen D, Maes F, Bisschops R, et al. Optimization for medical image segmentation: theory and practice when evaluating with Dice score or Jaccard index. IEEE Trans Med Imaging. 2020;39(11):3679–90. doi:10.1109/TMI.2020.3002417. [Google Scholar] [PubMed] [CrossRef]
40. Schutze O, Esquivel X, Lara A, Coello CAC. Using the averaged Hausdorff distance as a performance measure in evolutionary multiobjective optimization. IEEE Trans Evol Comput. 2012;16(4):504–22. doi:10.1109/TEVC.2011.2161872. [Google Scholar] [CrossRef]
41. Sharma R, Kamra A. A review on CLAHE based enhancement techniques. In: 2023 6th International Conference on Contemporary Computing and Informatics (IC3I). IEEE; 2023. Vol. 6, p. 321–5. doi:10.1109/IC3I59117.2023.10397722. [Google Scholar] [CrossRef]
42. Soora NR, Vodithala S, Badam JSH. Filtering techniques to remove noises from an image. In: 2022 International Conference on Advances in Computing, Communication and Applied Informatics (ACCAI); 2022; Chennai, India: IEEE. p. 1–9. doi:10.1109/ACCAI53970.2022.9752476. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools