 Open Access
Open Access
ARTICLE
Detection and Classification of Fig Plant Leaf Diseases Using Convolution Neural Network
1 Institute of Computer Science & IT, University of Science & Technology, Bannu, 28100, Pakistan
2 School of Computing, Gachon University, Seongnam, 13120, Republic of Korea
3 Department of Cybersecurity, College of Computer, Qassim University, Buraydah, Saudi Arabia
* Corresponding Authors: Jawad Khan. Email: ; Fahad Alturise. Email:
Computers, Materials & Continua 2025, 84(1), 827-842. https://doi.org/10.32604/cmc.2025.063303
Received 10 January 2025; Accepted 24 March 2025; Issue published 09 June 2025
Abstract
Leaf disease identification is one of the most promising applications of convolutional neural networks (CNNs). This method represents a significant step towards revolutionizing agriculture by enabling the quick and accurate assessment of plant health. In this study, a CNN model was specifically designed and tested to detect and categorize diseases on fig tree leaves. The researchers utilized a dataset of 3422 images, divided into four classes: healthy, fig rust, fig mosaic, and anthracnose. These diseases can significantly reduce the yield and quality of fig tree fruit. The objective of this research is to develop a CNN that can identify and categorize diseases in fig tree leaves. The data for this study was collected from gardens in the Amandi and Mamash Khail Bannu districts of the Khyber Pakhtunkhwa region in Pakistan. To minimize the risk of overfitting and enhance the model’s performance, early stopping techniques and data augmentation were employed. As a result, the model achieved a training accuracy of 91.53% and a validation accuracy of 90.12%, which are considered respectable. This comprehensive model assists farmers in the early identification and categorization of fig tree leaf diseases. Our experts believe that CNNs could serve as valuable tools for accurate disease classification and detection in precision agriculture. We recommend further research to explore additional data sources and more advanced neural networks to improve the model’s accuracy and applicability. Future research will focus on expanding the dataset by including new diseases and testing the model in real-world scenarios to enhance sustainable farming practices.Keywords
The fig tree (Ficus carica), a member of the Moraceae family, is one of the earliest fruit plants cultivated in modern times, originating from the Middle East and Western Asia. Known for its delicious fruit and attractive appearance, fig trees can grow 7–10 m tall and thrive in various soil types and temperatures, provided they have adequate drainage and sunlight. Fig trees are pollinated by specialist wasps to produce the unique syconium fruit, which has upside-down flowers. Figs hold cultural significance, representing fertility and wealth in many civilizations, and are valued for their nutritional benefits and beauty.
Ficus carica tree leaves are affected by many diseases. Farmers must carefully manage this plant because the fruit is nutritious and commercially valuable. Bacteria, viruses, fungi, and environmental factors may cause these diseases. Despite being an important part of agriculture in many countries, fig trees are prone to diseases that reduce their fruit production. Effective fig plant disease therapy requires early leaf disease diagnosis and categorization. This is vital to protecting these trees to quickly identify healthy and bacterially spot-infected leaves. These are possible since fig plants are prone to pathogens. In addition to personal health, they must produce high-quality, commercially and nutritionally valuable figs. Controlling leaf diseases is crucial for agricultural activities in these tree-planted areas [1].
1.1 The Role of Deep Learning in Crop Disease Detection and Classification
Researchers are using deep learning and image processing, particularly convolutional neural networks (CNNs), to identify and categorize crop leaf diseases. This is an advanced technology for agriculture; digitizing leaf images is in the early stages. The images underwent enhancement to collect fundamental data. CNN used the improved image to identify several global endemic agricultural illnesses [2]. Computer vision and machine learning are cutting-edge farming technology to detect plant leaf diseases. This method shows how modern farming technology can boost food security. Computer vision can analyze plant leaf visual data in order to find that the leaf is healthy or affected by the disease. This technology exposes plant diseases faster and more accurately, enabling farmers to detect and treat them earlier, resulting in safer food for end consumers. This technology promotes better crop health and yields [3]. There have been major improvements in computer vision with the rise of novel deep-learning algorithms that can distinguish images. A distinction algorithm that has comprehensively altered image categorization is known as CNN. Unlike previous computer vision algorithms, deep learning systems can categorize images without the help or guidance of a human. Identifying plant diseases is one of the techniques these deep-learning systems may be used. Previously, experts had to detect plant leaf features manually to diagnose diseases. This is a tough skill to get down precisely and easy to screw. These deep-learning systems have the capacity to quickly process a lot of visual data and may recognize patterns in the data that they may use to categorize plant disease. The high-tech, automated images classification method makes disease detection easier and more accurate, making it vital to agricultural and food security [4].
Machine learning (ML) and deep learning (DL) have improved plant disease diagnosis and classifying, identifying plant diseases is faster and easier with leaf images. Recurrent neural networks (RNNs) and convolutional neural networks (CNNs) have long been considered cutting-edge deep learning models [5,6]. Deep learning models can automate plant disease identification. These models quickly and precisely examine hundreds and thousands of plant leaf images. Deep learning technology can accurately recognize visual signals and pieces in images, enabling early sickness diagnosis and classification. Changes that allow people to move swiftly to reduce the effects of disease are crucial for controlling agricultural diseases. They also demonstrate how deep learning and machine learning can detect plant diseases. Most of the Extensive studies have proven their effectiveness [7].
1.2 Diverse Plant Species and Diseases Using Deep Learning
In the past decade, plant diseases have gained attention. Machine learning (ML) and deep learning (DL) have made models effective for more plant varieties than simply basic crops. Typical foods, trees, and ornamentals can now utilize these models. These models may have helped it succeed. Researchers are using ML and DL to tackle plant leaf diseases that affect several species. Powdery mildew, rust, blight, and anthracnose are easy to find using modern technologies. This has changed the focus from traditional farming challenges to comprehensive plant health management. These models correctly identify and classify plant diseases, according to several sources [8,9]. ML and DL algorithms have helped to develop reliable disease identification systems. The most critical challenge in the fig tree preservation system is the early and reliable diagnosis of fig plant diseases such as fig rust, mosaic, and anthracnose, as well as the classification of healthy or sick trees. In addition to being laborious and prone to errors, manual disease classification is also time-consuming. As a result, it is necessary to have an automated system to detect and categorize these disorders promptly while maintaining accuracy. For this purpose, a convolutional neural network (CNN) is used to automatically identify and classify the diseases that affect fig trees through leaves.
This research study contributes the following:
1. To collect a diverse and balanced dataset of healthy and diseased fig tree leaves.
2. To catalog and identify various diseases affecting fig plants through their leaves, including fig rust, fig mosaic, and anthracnose.
3. To apply preprocessing techniques, such as edge detection and segmentation, to enhance the quality of the images in the dataset.
4. To develop a convolutional neural network (CNN) model capable of using the refined dataset to accurately identify and categorize fig leaves as healthy or affected by fig rust, fig mosaic, or anthracnose.
Recent technological advancements in agriculture have enhanced production, efficiency, and environmental sustainability. However, quickly identifying and treating plant diseases remains a challenge, impacting productivity and quality. Fig trees, valued for their fruit and beauty, are susceptible to diseases like fig rust, leaf blight, and anthracnose. Traditional diagnosis methods are laborious, error-prone, and often require specialized knowledge and visual assessment. Consequently, there is a growing need for automated, precise, and personalized solutions.
Ji et al. [10] classified tomato leaf diseases into eleven classes, one of which being the healthy leaf category. First, they employed assessment metrics to compare their model, which was developed via Transfer Learning (TL), to another model that had undergone ablation research to improve its parameters. According to their findings, their model achieved the highest possible accuracy and recall of 95.00% after adding data. After that, the model was made Android and web-based to help tomato growers identify leaf diseases.
Using Deep Convolutional Neural Networks (DCNN), Ngugi et al. [11] evaluated numerous image preprocessing techniques. The researchers tested four preprocessing scenarios utilizing HSI, CMYK, Gaussian, and Median filters and colour models. They sought the best colour models and filters to improve accuracy. They achieved the highest accuracy on Vgg-19 (98.27%), MobileNet-V2 (94.9%), and ResNet-50 (99.5%) by converting RGB to CMYK and utilizing Gaussian Noise and Blur filters. Umamageswari et al. [12] investigated automating mango plant leaf disease detection and classification. They used a wavelet transform to segment photos and a WNN to classify diseases. The 1130 images Plant Village collection provided the high-resolution mango leaf shots. The study team’s WNN model detected mango tree leaf diseases with 98.93% accuracy. Paul et al. [13] used multi-class and multilabel classification to mimic disease recognition in many plant species. This model was created specifically to identify diseases. Images of the leaves of six different plants were collected from various web sources and evaluated. These plants contained tomatoes and apples trees. After analyzing a variety of CNN architectures, they discovered that Xception and DenseNet performed best in multi-label detection of plant diseases. Skip connections, spatial convolutions, and efficient hidden layer connectivity were shown to increase disease classification accuracy in these systems.
Hossain et al. [14] conducted substantial research on deep learning models for plant leaf disease diagnosis, employing a range of models such as Vision Transformer (ViT), DCNN, CNN, RSNSR-LDD, DDN, and YOLO. Along with demonstrating the application of numerous models on various public datasets, they examined the advantages and disadvantages of each model. Mishra et al. [15] tested a CNN to diagnose diseases in tea leaves. Concatenated CNN models Xception, GoogleNet, and Inception-ResNet versions 2 were used. Three of their 4727 images were of Indonesian tea plant diseases and one was of healthy plants. The results found that concatenated CNNs can identify tea leaf diseases with 89.64% accuracy. Kabir et al. [16] constructed a small CNN for plant disease detection. The CNN was built to be lightweight and easy to implement into applications. Eight of the model’s CNN layers are dedicated to feature extraction, while the other four are used as classifiers. The model consists of twelve layers. The algorithm was evaluated on a dataset including 87,867 images and 38 plant disease categories. It worked wonderfully without overfitting. The model achieved an F1-score of 97%, recall of 98%, and accuracy of 97%. It was successful in delivering mobile apps, outperforming other well-known pre-trained models such as InceptionV3 and MobileNetV2. Mustofa et al. [17] created a successful computer vision system for diagnosing diseases in plant leaves. This technique included processes such as denoising, feature extraction with Hu moments, color histograms, and Haralick textures. K-fold validation was used to train many machine learning algorithms on segmented and non-segmented images. These algorithms included Random Forest and Support Vector Machine. The study studied two approaches: using default hyperparameters and optimizing hyperparameters for segmented datasets. As a result, utilizing segmentation increased classification accuracy by 2.19%, while hyperparameter tweaking increased accuracy by another 0.48%. Random Forest’s effectiveness in identifying plant ailments, the highest result was 97.92% accuracy for 40 classes across 10 plant species.
Krisnandi et al. [18] used a hybrid deep learning model to identify and categorize diseases that damage sunflower leaves. Some of these diseases are Verticillium wilt, Downy mildew, Alternaria leaf blight, and Phoma blight. This model uses a stacking ensemble learning technique to integrate VGG-16 with MobileNet to enhance disease diagnosis. To conduct their investigation, scientists gathered 329 images of sunflowers from Google image and divided them into five unique groupings. This dataset was utilized in this work to assess the performance of multiple deep learning models, including the researchers’ hybrid model, to demonstrate the efficacy of combining models for disease categorization. Imanulloh et al. [19] utilized EfficientNet to classify tomato diseases from 18,161 images. They compared U-net to Modified U-net in order to achieve segmentation. Their research included binary, six-class, and ten-class disease classifications. EfficientNet-B7 and B4 performed well in classification tests, with accuracy levels as high as 99.95% in binary classification and 99.89% in ten-class classification. The Modified U-Net obtained an excellent segmentation accuracy of 98.66%. The findings revealed that, when compared to existing approaches, deeper networks and segmented images significantly enhance disease categorization. Kiran et al. [20] used image processing to evaluate the feasibility of identifying two diseases in different wheat cultivars. Images from the field and the lab were both processed. The study included image enhancement, lesion segmentation, and the extraction of 140 features. Support vector machines, BPNN, and RF were then utilized to pick and classify features. Single-variety models performed well (87.18%–100%) when applied to the same variety, but failed badly when applied to various kinds. In contrast, multi-variety models performed brilliantly across a wide range of conditions and kinds, with accuracy levels ranging from 82.05% to 100%. This shows that models trained on a variety of data sets may successfully detect stripe rust and leaf rust in wheat. By utilizing an image-to-image translation model, Malik et al. [21] created a data augmentation technique to improve the collection of sick leaf images. Using attention techniques, they improved the realism of illness textures in the produced images. These synthetic images were contributed to the PlantVillage dataset to evaluate the method. This method improved classification model performance for early plant disease detection. The results suggested that this technique may increase diagnostic accuracy by providing a more balanced and diverse samples.
The literature review reveals a lack of research on detecting and classifying fig tree leaf diseases using machine and deep learning. Most researchers favor deep learning over traditional methods for leaf disease detection due to its superior outcomes. A critical gap is the absence of a dedicated dataset for fig tree leaf diseases like fig rust, fig mosaic, and anthracnose. This research aims to compile a dataset of fig tree leaf images and use Convolutional Neural Network (CNN) models to detect and classify healthy and diseased leaves. By leveraging deep learning techniques, this study seeks to address this gap and develop a specialized tool for fig tree leaf disease identification and classification.
The proposed methodology involves using Convolutional Neural Networks (CNNs) to identify and classify diseases in fig tree leaves. The first step is to collect diverse images of fig tree leaves, ensuring a balanced dataset with healthy and diseased leaves of varying ages, light conditions, and disease intensities. The dataset undergoes extensive pre-processing, including outlier detection, standardization, normalization, and data augmentation, to enhance data quality and model robustness. The processed dataset is then split into training and testing sets. A CNN architecture is designed with convolutional layers for feature extraction, pooling layers for feature refinement, and fully connected layers for accurate classification. After training, the model’s performance is rigorously assessed using metrics such as precision, recall, F1 score, and accuracy. Fig. 1 presents the proposed architecture of our study.
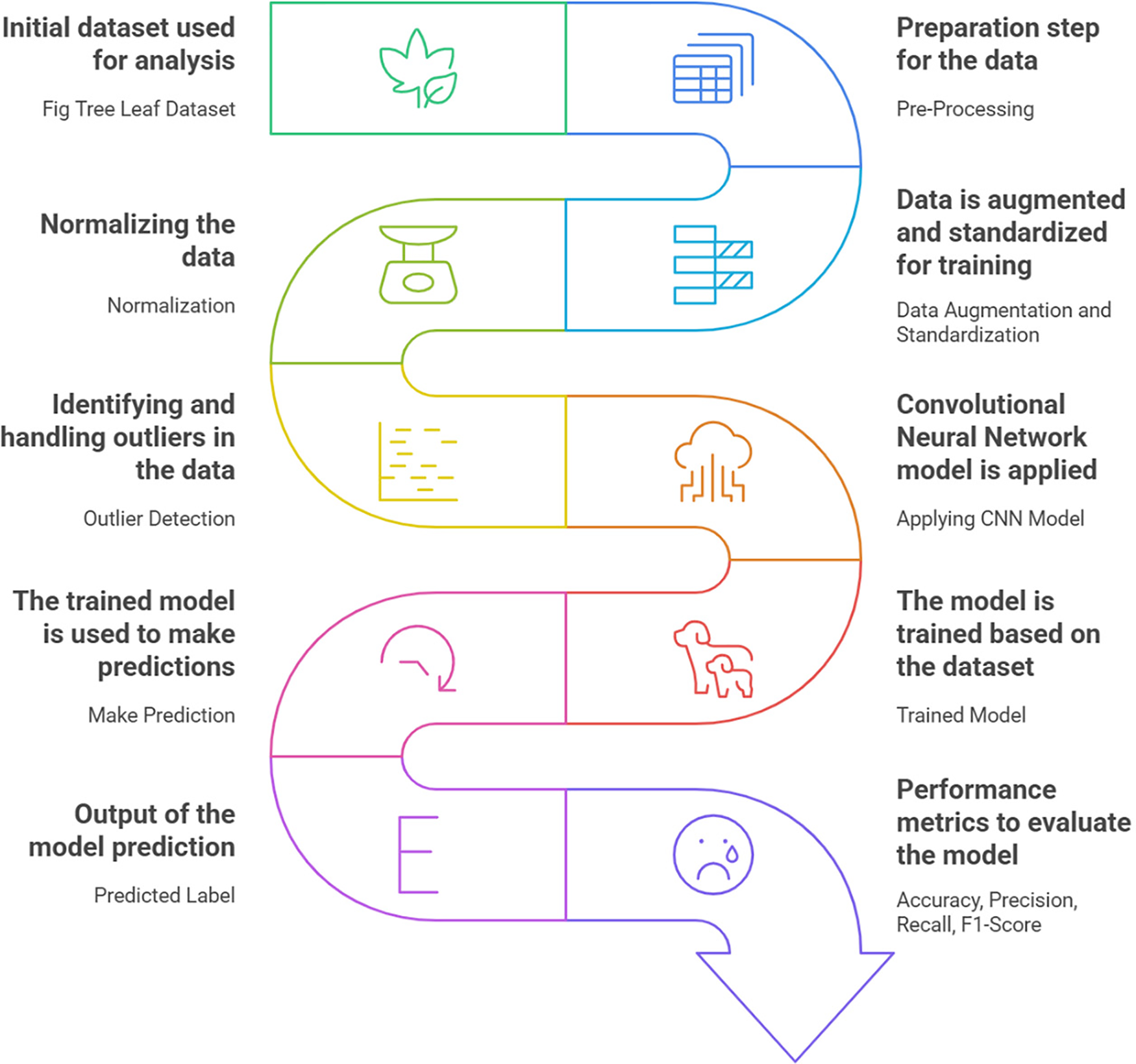
Figure 1: Overall proposed solution
This research study compiles a diverse dataset of Fig Tree leaves, including leaves of various ages, under different light conditions, and with varying disease severity. The dataset includes both healthy leaves and those infected with bacteria. It was collected through fieldwork and photography, adhering to medical and national regulations. Each image was meticulously annotated to ensure high quality. This structured approach aims to create a dataset for detecting and classifying Fig Tree leaf diseases using Convolutional Neural Networks. Fig. 2 shows samples from the dataset.

Figure 2: Fig tree leaf dataset
Image preprocessing optimizations are essential for preparing the Fig tree leaf dataset, improving its quality, and enhancing the model’s performance. Fig. 3 shows the preprocessed dataset images. The following key techniques were applied.

Figure 3: Preprocessed images
To artificially extend the dataset and improve the model’s ability to generalize, we applied several image variations, including rotation, flipping, cropping, and zooming. This step increased the diversity of training samples, preventing the model from overfitting and enabling it to better handle variations in leaf appearance and disease presentation.
Standardization ensures that the images have consistent dimensions, color channels, and pixel value ranges. This uniformity is crucial for speeding up the learning process and helping the model focus on distinguishing healthy leaves from diseased ones. Standardization also allows the model to better interpret variations across different images by eliminating inconsistencies.
Normalizing the pixel values to a specific range (either [0, 1] or [−1, 1]) ensures that the input data remains within a reasonable range, helping to avoid issues such as exploding or vanishing gradients. This normalization step facilitates faster convergence during model training.
Identifying and handling outliers such as data entry mistakes or abnormal disease expressions was critical in preventing the model from learning irrelevant or erroneous information. By cleaning up the data, we ensured that the model could focus solely on relevant and accurate features, improving its performance.
To enhance image quality, edge detection was applied to highlight the boundaries of the fig leaves and any disease spots. Using the Canny edge detector, this technique helped the model focus on important structural features of the leaves, especially around diseased areas, improving the accuracy of disease classification. This step is particularly important when distinguishing between diseases with similar visual features.
Segmentation was used to isolate the leaves from the background and to segment diseased areas from healthy regions. Using techniques such as thresholding and Watershed segmentation, we enhanced the model’s ability to focus specifically on diseased regions, reducing noise from irrelevant parts of the image. This step improved the model’s ability to detect and classify different types of diseases accurately.
These preprocessing techniques together helped improve the dataset’s quality by focusing on the most relevant features, ensuring that the model was trained on well-processed, high-quality images. These optimizations supported the CNN’s ability to accurately detect and diagnose diseases in fig tree leaves, which is essential for effective disease control in fig cultivation.
We scale down the training data to retain a significant amount of Fig Tree diseases for model training and testing. The pre-processed dataset is partitioned into:
Training Data: 80% of the images are used to train the model, helping it learn complex patterns, features, and variants in Fig Tree leaves, thereby establishing the foundational data and reducing learning errors.
Testing Data: The remaining 20% is reserved for testing, allowing the model to be evaluated on unseen data and assessing its ability to generalize learned patterns to new data.
Data visualization interprets and understands data through graphical representations, helping to identify patterns, trends, and outliers. Using tools like matplotlib, seaborn, and plotly, numerical and categorical data are transformed into table, charts, graphs, and maps. Effective visualization supports decision-making and information exchange across various fields.
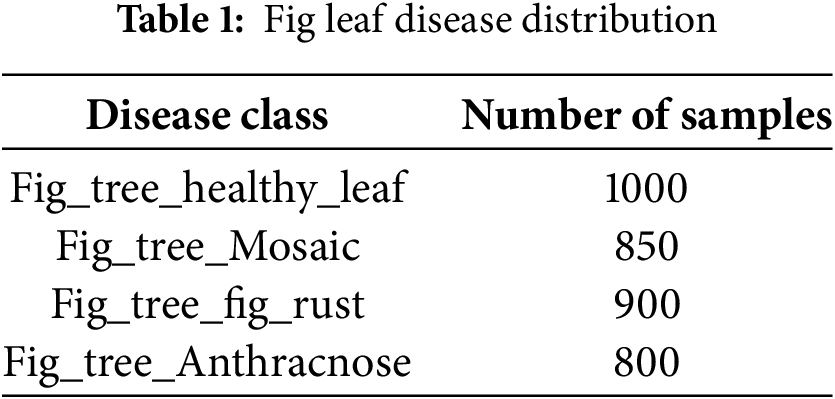
Table 1 presents the number of samples for each disease class in the dataset.
3.5 Applying Convolutional Neural Network (CNN) for Training
To design and train a Convolutional Neural Network (CNN) for identifying and classifying diseases in Fig Tree leaves, we leverage the CNN’s ability to perform image classification tasks. The CNN model includes several layers, each with specific functions:
Convolutional Layers: These are the core of the CNN, extracting micro-features from Fig Tree leaf images using various filters to recognize lines, textures, and patterns. As images pass through multiple convolution layers, the network identifies increasingly complex features to discern the characteristics of Fig Tree diseases. Filters (kernels) are applied to the input image to extract features, using the convolution operation:
Pooling Layers: These layers downsample the image data by reducing its spatial dimensions and summarizing feature presence using methods like max pooling, which takes the maximum value from each patch of pixels. Max pooling reduces the spatial dimensions by selecting the maximum value from each region:
Fully Connected Layers: These layers make the final classification decisions by connecting every neuron to all activations from previous layers, integrating the learned features to determine the disease condition. Features from the previous layers are integrated and classified through a weight matrix:
By intensively designing and modifying the CNN architecture, we created a model tailored for Fig Tree leaf disease detection as shown in Fig. 4. This customized network is fine-tuned to efficiently and accurately learn the unique visual characteristics of diseased leaves. Consequently, our CNN model is both theoretically accurate and practically reliable, effectively differentiating between healthy and diseased leaves, providing valuable information for managing Fig Tree leaf diseases.
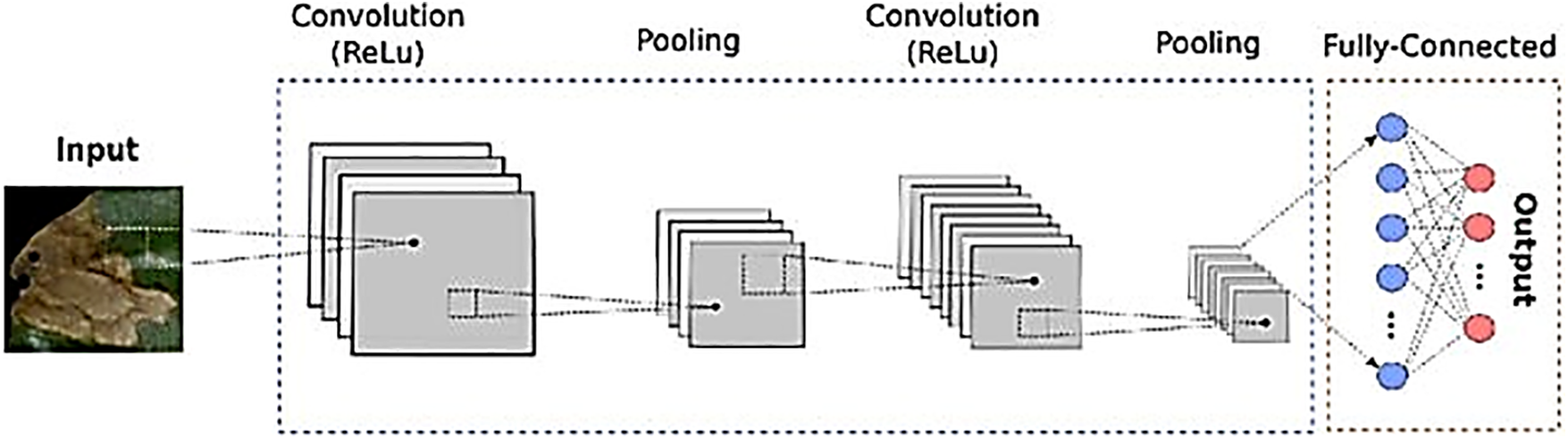
Figure 4: CNN architecture
3.6 Model Performance Evaluation
We are now conducting a robust performance evaluation of our Convolutional Neural Network (CNN) model after completing the training phase. The evaluation uses key metrics to assess the model’s effectiveness in detecting and classifying Fig Tree leaf diseases:
Accuracy: Measures how often the CNN correctly classifies leaves as diseased or healthy.
Precision: Evaluates the model’s accuracy in identifying diseased leaves, particularly minimizing false positives.
Recall: Also known as sensitivity, measures how accurately the model identifies all diseased cases, minimizing false negatives.
F1 Score: Combines precision and recall into a single metric, providing a comprehensive view of the model’s accuracy and balance.
These metrics provide a thorough evaluation of our CNN’s ability to detect and distinguish Fig Tree leaf diseases, validating the model’s accuracy and identifying areas for improvement.
The trained Convolutional Neural Network (CNN) model is now in its final deployment phase, where it predicts whether new Fig Tree leaf images are infected with bacterial spots or healthy. This real-time application will show the model’s effectiveness with new data beyond the training set. Our investigation into detecting and classifying Fig Tree leaf diseases using CNN involved detailed processing stages, from data input to deployment and performance checking. This structured approach ensures accuracy and reliability. Testing the model on new leaf images is the final step, demonstrating its practical applicability for agricultural practitioners and researchers managing plant diseases.
In this section, we discuss our study on detecting and classifying Fig Tree Leaf Disease using a Convolutional Neural Network (CNN). We present the evaluation metrics, training parameters, data augmentation, CNN architecture, and the outcomes of our research. Our model’s performance in detecting plant diseases highlights its significant contribution to agricultural AI. We used Google Colab, built on Jupyter Notebook, for our simulations.
In this research study, we make use of a compilation of libraries with the following objectives:
NumPy: As For efficient numerical operations, arrays, and matrices.
Pandas: For data manipulation and analysis.
Matplotlib.pyplot: For creating visualizations to analyze data and present findings.
TensorFlow: For developing and training our CNN models.
Tensorflow.keras: For simplifying neural network model building.
pathlib and os: For file system path handling and OS-dependent operations.
sklearn.metrics: For evaluating model performance (accuracy, precision, recall).
seaborn: For creating advanced statistical graphics.
These libraries were essential for data processing, model creation, performance testing, and results display.
4.2 Load the Fig Tree Leaf Dataset
In this step, we use the os module to work with the file system and load the Fig Tree leaf dataset. We start by importing os and setting the data path to “/content/Fig Tree Dataset.” Using os.listdir, we list all items in this directory, capturing files and subdirectories. Finally, we iterate over the items and print their names to briefly explore the dataset, ensuring it is well-structured and ready for analysis.
4.3 Data Augmentation Pipeline for Image Processing
We use TensorFlow for our data augmentation pipeline, leveraging the tensorflow.keras module for layers and sequential model building. This pipeline applies several preprocessing steps in order:
Rescaling: Transforms pixel values to a range between 0 and 1.
Random Flip: Randomly flips images horizontally and vertically to increase dataset diversity.
Random Rotation: Randomly rotates images within a range of 0.2 radians (11.5 degrees) to help the model become orientation-invariant.
Random Zoom: Randomly zooms images by 20%, aiding the model in focusing on features at different scales.
These augmentation techniques enhance the model’s reliability and generalizability, improving performance across various image conditions.
4.4 Efficient Image Classification with Tensor Flow Sequential API
We use Tensor Flow, a comprehensive deep learning library, to construct the data augmentation pipeline for improving our image dataset. With Tensor Flow as well as importation of sub modules from tensorflow.keras, we can lay with layers and build models in a sequential manner. This pipeline, which was created using Tensor Flow’s Sequential model, orders a series of preprocessing methods that were created to increase variety of our training dataset and enhance input for the models. Here are the main parts of this pipeline:
Rescaling: Normalizing images pixel values into the [0, 1] range eliminates inconsistencies among images.
Random Flip: Horizontally and vertically flipping a certain percentage of the dataset allows the model to be pseudo-aggregated from different angles.
Random Rotation: Randomly rotation pictures by up to 0.2 radians. The model can gain the ability to achieve perspective invariant data learning object orientation.
Random Zoom: Image data zoomed in or out by a certain percentage. This can improve the model’s ability to understand features of different scales.
Collectively, these augmentation techniques further strengthen the model’s resilience and generalizability to training data, making him more adept at handling scenarios in which the images presented differ greatly from the training images, therefore, providing high prediction power for new data.
4.5 Trained Model Using Convolutional Neural Network (CNN)
We enhance our model training with TensorFlow by incorporating an EarlyStopping callback, which halts training if validation loss doesn’t improve within 5 epochs, with a min_delta of 0.02. The model then restores the best weights based on validation loss. Before training, we optimize the dataset handling:
Training Set: Cached, shuffled, and prefetched using TensorFlow’s AUTOTUNE.
Validation Set: Prefetched, cached, and shuffled.
Training is set for 20 epochs, adjusted based on progress and early stopping. Performance metrics (loss and accuracy) are logged after each epoch. This iterative process, combining early stopping and dataset optimization, ensures a balance between learning efficiency and model performance. To improve testing accuracy and reduce overfitting, we applied data augmentation, cross-validation, and early stopping. Data augmentation techniques such as rotation, flipping, and contrast adjustments helped diversify the training dataset. Cross-validation provided a more reliable evaluation of the model’s performance by assessing it across multiple subsets of the data. Early stopping was implemented to halt training when the validation loss stopped improving, preventing overfitting. These strategies resulted in a significant improvement in testing accuracy, reaching 90.12%. The model achieved a training accuracy of 91.53% and a validation accuracy of 90.12%, demonstrating its ability to accurately classify training images and generalize well to new data, effectively identifying and classifying fig tree leaf diseases. Fig. 5 shows the training and validation accuracy.
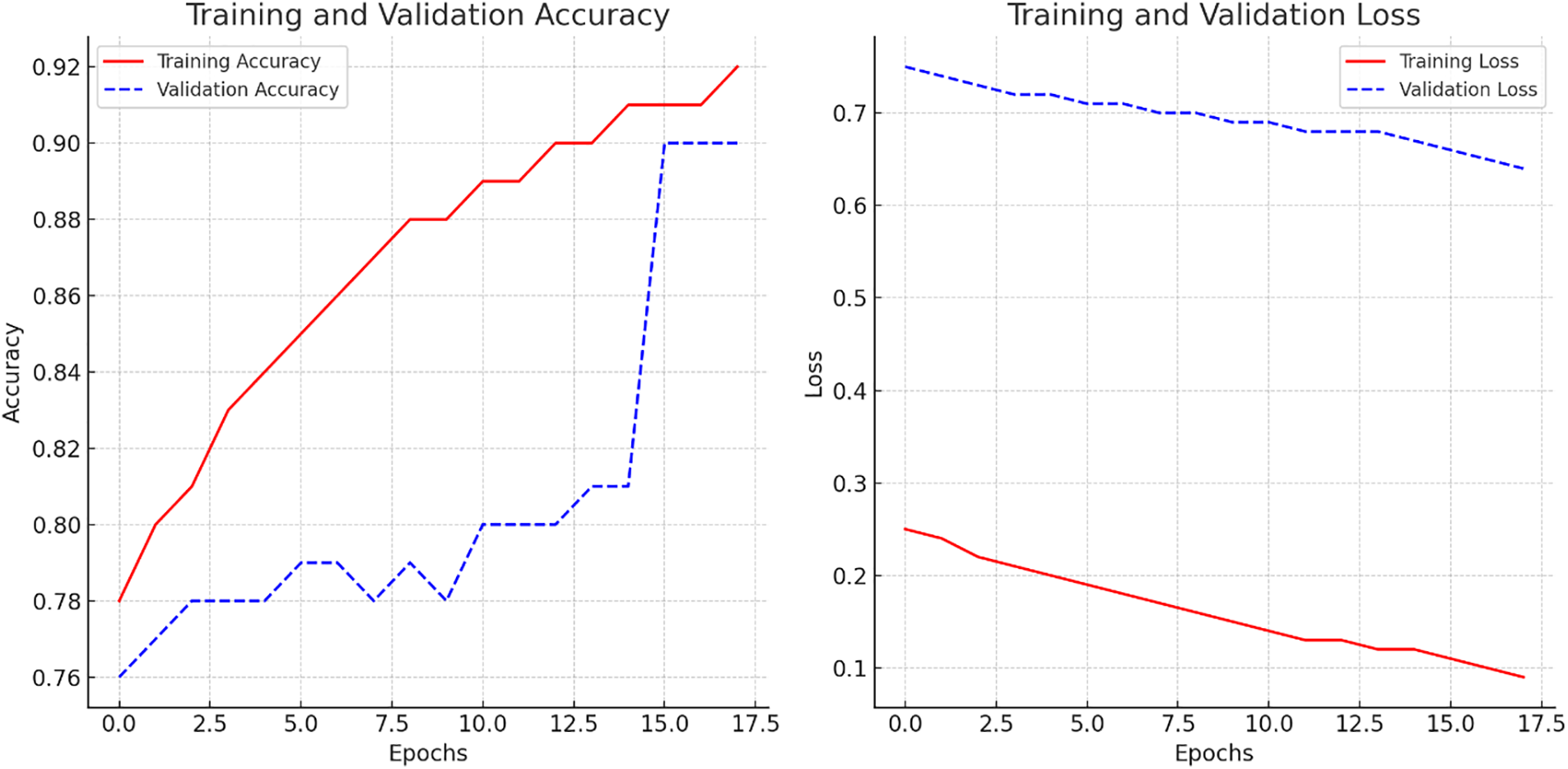
Figure 5: Learning curve of CNN model
4.6 Confusion Matrix of CNN Model
We created a function, plot_advanced_confusion_matrix, to generate a detailed confusion matrix from the model’s predictions. The function predicts labels, extracts true labels, and plots the percent-normalized confusion matrix. Using Seaborn, we plotted and annotated the matrix as a color-coded heatmap, making it easier to interpret. The matrix as given in Fig. 6 displays prediction accuracy for different classes, comparing predicted labels against true labels, and shows how well the CNN model classifies various Fig Tree leaf diseases.
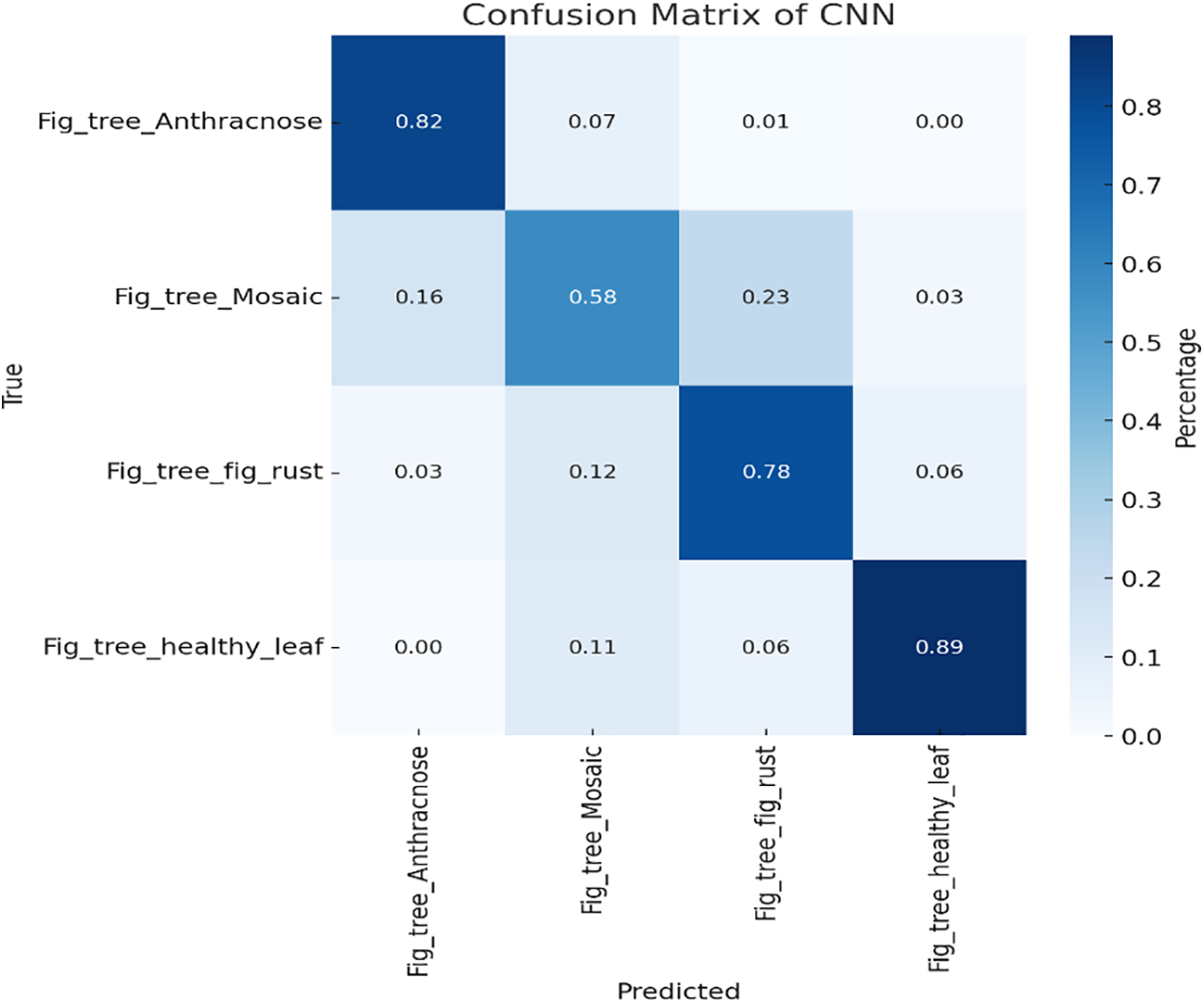
Figure 6: Confusion matrix of CNN model
4.7 Classification Report of CNN Model
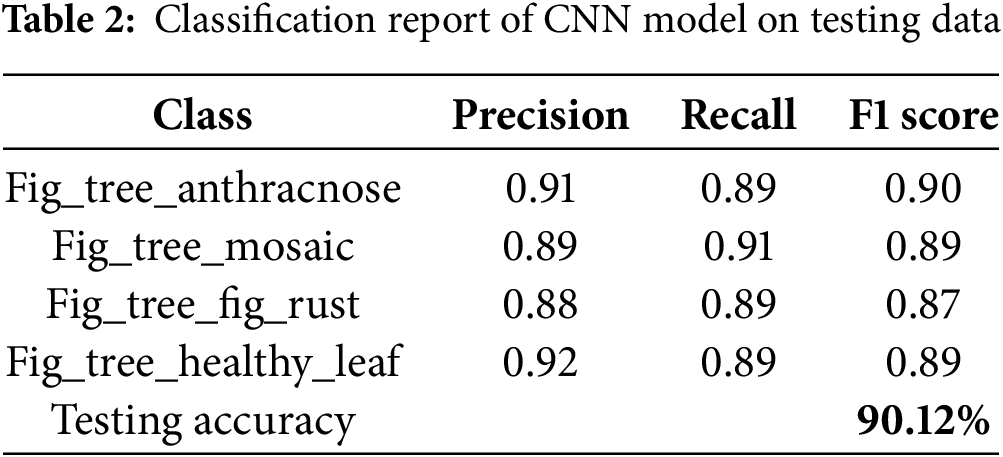
The print classification report function evaluates a CNN model by generating a detailed classification report. It predicts labels, extracts true labels, and uses classification _report from sklearn metrics to calculate precision, recall, F1-score, overall accuracy, and averages. The report, printed for the validation dataset, includes performance metrics for classes: “Fig _tree _Anthracnose”, “Fig _tree _Mosaic”, “Fig _tree _fig_ rust”, and “Fig _tree _healthy _leaf”. With 90.12% accuracy, the model classifies these classes with varying success, indicating its ability to distinguish between different Fig Tree leaf diseases as given in Table 2. This analysis helps guide improvements in model accuracy and reliability.
4.8 Model Prediction Using Grad-Cam Visualization Technique
We use Grad-CAM heatmaps to visualize which parts of validation images the CNN model focuses on when predicting labels. This involves two steps:
1. Generate Heatmaps: Using the make _gradcam _heatmap function, we identify important image areas based on the last convolutional layer’s activations and gradients.
2. Superimpose Heatmaps: The display _grad cam function overlays these heatmaps onto the original images, highlighting critical regions for class predictions.
By shuffling and visualizing the validation set images with heatmaps, we gain insights into model accuracy and areas needing improvement. The predicted output images are shown in Fig. 7.
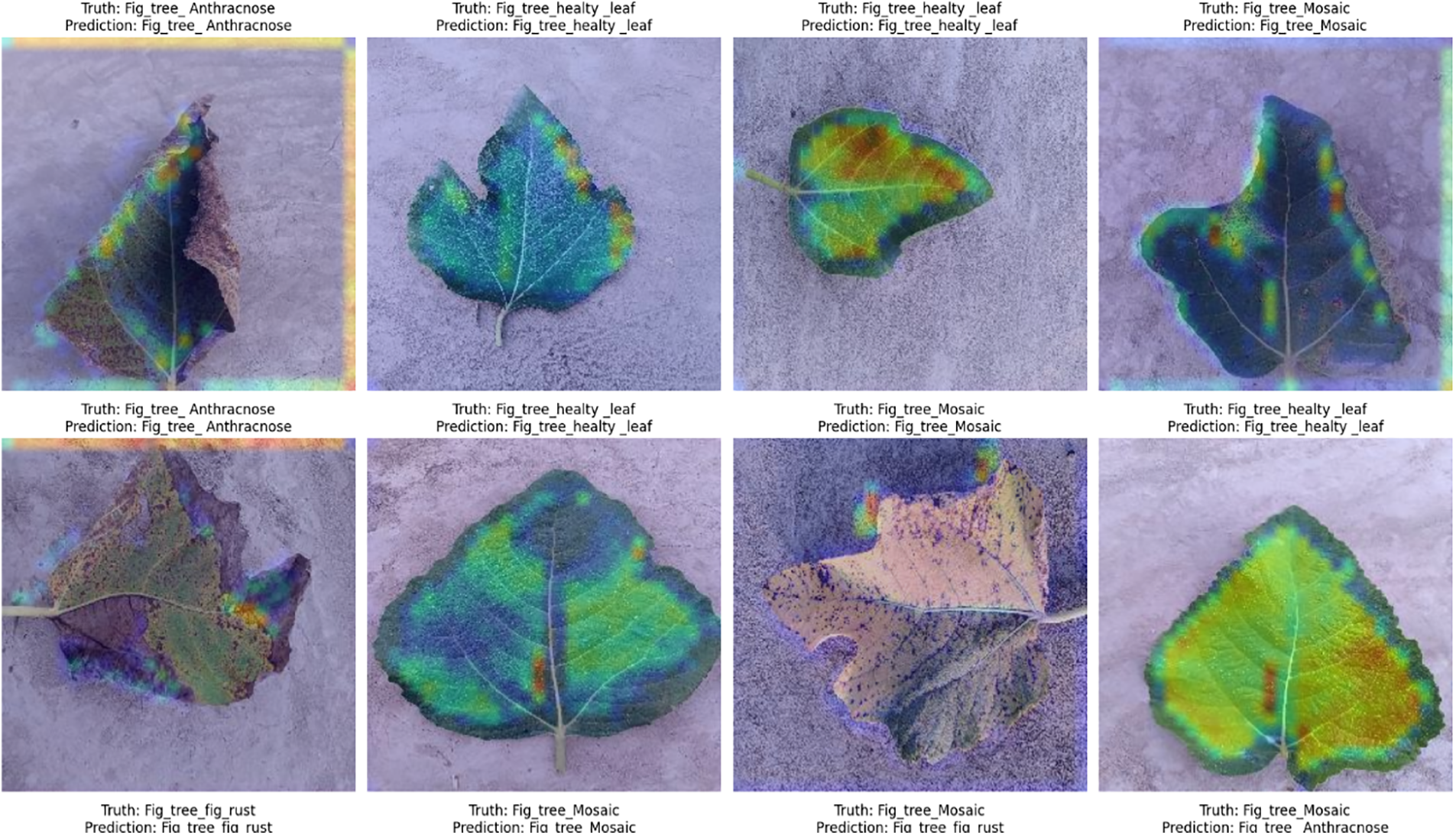
Figure 7: Predicted output
Our research study has contributed to the advancement of the agricultural AI field. We have developed a Convolutional Neural Network model based on TensorFlow to classify the Fig Tree leaf disease into different classes. Data augmentation and early stopping have helped increase the model accuracy and reduce overfitting. The training and validation accuracy were 91.53% and 90.12%, respectively. The Grad-CAM visualizations have shown what the model focuses on in an image and helps in the decision-making process. According to the classification report and confusion matrix, the model performs well in terms of classifying diseases. This method has shown that the generated model could accurately identify leaf diseases and classify them. It also has clear hints to refine the model further and employ other applications in the field of precision agriculture, proving the high potential of CNNs in detecting plant diseases. We suggest expanding the scope of data sources that the model integrates to develop multispectral or hyperspectral imaging, which would substantially enrich the training dataset and potentially increase the accuracy of the disease classification. Additionally, the implementation of more complex neural network architectures, including attention mechanisms or deeper convolutional networks, would extend the model’s learning capabilities. Further steps include an increase in the variety of Fig Tree leaf diseases and environmental conditions, ensuring that the achieved results could be applied on a global scale. Moreover, testing the model in the real-life challenges of agricultural practice would provide essential feedback for further improvement. Finally, advancing in this area of research could result in the creation of an AI-boosted tool for automated disease identification, which would ensure the achievement of precision agriculture and sustainable farming practices.
Acknowledgement: Not applicable.
Funding Statement: The researchers would like to thank the Deanship of Graduate Studies and Scientific Research at Qassim University for financial support (QU-APC-2025).
Author Contributions: Conceptualization, Rahim Khan, Ihsan Rabbi, Umar Farooq; formal analysis, Rahim Khan, Ihsan Rabbi; funding acquisition, Fahad Alturise; investigation, Ihsan Rabbi; methodology, Rahim Khan, Ihsan Rabbi; project administration, Jawad Khan; resources, Ihsan Rabbi; software, Umar Farooq; validation, Rahim Khan, Ihsan Rabbi, Umar Farooq; visualization, Rahim Khan, Umar Farooq; writing—original draft, Rahim Khan, Ihsan Rabbi; writing—reviewing and editing, Jawad Khan, Fahad Alturise, Ihsan Rabbi. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated and analysed during this study are included in this article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
Abbreviations
| I | Input image |
| f | Filter (kernel) |
| O (I, j) | Output feature map |
| P (i, j) | Pooled feature map |
| x | Input vector |
| W | Weight matrix |
| b | Bias term |
| y | Output vector (predicted classes) |
| ti | True label for class iii |
| pi | Predicted probability for class iii |
| η | Learning rate |
| L | Loss function (cross-entropy) |
| RNN | Recurrent Neural Network |
| CNN | Convolutional Neural Network |
| ML | Machine Learning |
| DL | Deep Learning |
| Precision | True Positives/(True Positives + False Positives) |
| Recall | True Positives/(True Positives + False Negatives) |
| F1 Score | Harmonic mean of Precision and Recall |
| Softmax | Activation function |
| Max Pooling | Pooling operation (maximum value) |
References
1. Tsopelas P, Soulioti N, Wingfiield MJ, Barnes I, Marincowitz S, Tjamos EC, et al. Ceratocystis ficicola causing a serious disease of Ficus carica in Greece. Phytopathol Mediterr. 2021;60(2):337–49. doi:10.36253/phyto-12794. [Google Scholar] [CrossRef]
2. Batool A, Hyder SB, Rahim A, Waheed N, Asghar MA, Khan F. Classification and identification of tomato leaf disease using deep neural network. In: 2020 International Conference on Engineering and Emerging Technologies (ICEET); 2020 Feb 22–23; Lahore, Pakistan. p. 1–6. doi:10.1109/iceet48479.2020.9048207. [Google Scholar] [CrossRef]
3. Harakannanavar SS, Rudagi JM, Puranikmath VI, Siddiqua A, Pramodhini R. Plant leaf disease detection using computer vision and machine learning algorithms. Glob Transitions Proc. 2022;3(1):305–10. doi:10.1016/j.gltp.2022.03.016. [Google Scholar] [CrossRef]
4. Mishra S, Sachan R, Rajpal D. Deep convolutional neural network based detection system for real-time corn plant disease recognition. Procedia Comput Sci. 2020;167(11):2003–10. doi:10.1016/j.procs.2020.03.236. [Google Scholar] [CrossRef]
5. Raouhi EM, Lachgar M, Hrimech H, Kartit A. Optimization techniques in deep convolutional neuronal networks applied to olive diseases classification. Artif Intell Agric. 2022;6:77–89. doi:10.1016/j.aiia.2022.06.001. [Google Scholar] [CrossRef]
6. Abd Algani YM, Marquez Caro OJ, Robladillo Bravo LM, Kaur C, Al Ansari MS, Kiran Bala B. Leaf disease identification and classification using optimized deep learning. Meas Sens. 2023;25(1):100643. doi:10.1016/j.measen.2022.100643. [Google Scholar] [CrossRef]
7. Sambasivam G, Opiyo GD. A predictive machine learning application in agriculture: cassava disease detection and classification with imbalanced dataset using convolutional neural networks. Egypt Inform J. 2021;22(1):27–34. doi:10.1016/j.eij.2020.02.007. [Google Scholar] [CrossRef]
8. Agarwal M, Singh A, Arjaria S, Sinha A, Gupta S. ToleD: Tomato leaf disease detection using convolution neural network. Procedia Comput Sci. 2020;167:293–301. doi:10.1016/j.procs.2020.03.225. [Google Scholar] [CrossRef]
9. Ramesh S, Vydeki D. Recognition and classification of paddy leaf diseases using optimized deep neural network with Jaya algorithm. Inf Process Agric. 2020;7(2):249–60. doi:10.1016/j.inpa.2019.09.002. [Google Scholar] [CrossRef]
10. Ji M, Zhang L, Wu Q. Automatic grape leaf diseases identification via UnitedModel based on multiple convolutional neural networks. Inf Process Agric. 2020;7(3):418–26. doi:10.1016/j.inpa.2019.10.003. [Google Scholar] [CrossRef]
11. Ngugi LC, Abelwahab M, Abo-Zahhad M. Recent advances in image processing techniques for automated leaf pest and disease recognition—a review. Inf Process Agric. 2021;8(1):27–51. doi:10.1016/j.inpa.2020.04.004. [Google Scholar] [CrossRef]
12. Umamageswari A, Bharathiraja N, Irene DS. A novel fuzzy C-means based chameleon swarm algorithm for segmentation and progressive neural architecture search for plant disease classification. ICT Express. 2023;9(2):160–7. doi:10.1016/j.icte.2021.08.019. [Google Scholar] [CrossRef]
13. Paul SG, Biswas AA, Saha A, Zulfiker MS, Ritu NA, Zahan I, et al. A real-time application-based convolutional neural network approach for tomato leaf disease classification. Array. 2023;19(3–4):100313. doi:10.1016/j.array.2023.100313. [Google Scholar] [CrossRef]
14. Hossain MI, Jahan S, Al Asif MR, Samsuddoha M, Ahmed K. Detecting tomato leaf diseases by image processing through deep convolutional neural networks. Smart Agric Technol. 2023;5(4):100301. doi:10.1016/j.atech.2023.100301. [Google Scholar] [CrossRef]
15. Mishra S, Ellappan V, Satapathy S, Dengia G, Mulatu BT, Tadele F. Identification and classification of mango leaf disease using wavelet transform based segmentation and wavelet neural network model. Ann Rom Soc Cell Biol. 2021;25(2):1982–9. [Google Scholar]
16. Kabir MM, Ohi AQ, Mridha MF. A multi-plant disease diagnosis method using convolutional neural network. In: Computer vision and machine learning in agriculture. Berlin/Heidelberg, Germany: Springer; 2021. p. 99–111. [Google Scholar]
17. Mustofa S, Munna MMH, Emon YR, Rabbany G, Ahad MT. A comprehensive review on plant leaf disease detection using deep learning. arXiv:2308.14087. 2023. [Google Scholar]
18. Krisnandi D, Pardede HF, Yuwana RS, Zilvan V, Heryana A, Fauziah F, et al. Diseases classification for tea plant using concatenated convolution neural network. Commit Commun Inf Technol J. 2019;13(2):67–77. doi:10.21512/commit.v13i2.5886. [Google Scholar] [CrossRef]
19. Imanulloh SB, Muslikh AR, Setiadi DRIM. Plant diseases classification based leaves image using convolutional neural network. J Comput Theor Appl. 2023;1(1):1–10. doi:10.33633/jcta.v1i1.8877. [Google Scholar] [CrossRef]
20. Kiran SM, Chandrappa DN. Plant leaf disease detection using efficient image processing and machine learning algorithms. J Robot Control (JRC). 2023;4(6):840–8. doi:10.18196/jrc.v4i6.20342. [Google Scholar] [CrossRef]
21. Malik A, Vaidya G, Jagota V, Eswaran S, Sirohi A, Batra I, et al. Design and evaluation of a hybrid technique for detecting sunflower leaf disease using deep learning approach. J Food Qual. 2022;2022:9211700–12. doi:10.1155/2022/9211700. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools