 Open Access
Open Access
ARTICLE
Large Language Model in Healthcare for the Prediction of Genetic Variants from Unstructured Text Medicine Data Using Natural Language Processing
1 Center of Excellence in CyberSecurity (CYBEX), Prince Sultan University, Riyadh, 11586, Saudi Arabia
2 Artificial Intelligence & Data Analytics Lab, CCIS, Prince Sultan University, Riyadh, 11586, Saudi Arabia
3 Department of Mathematical Sciences, College of Science, Princess Nourah Bint Abdulrahman University, P.O. Box 84428, Riyadh, 11671, Saudi Arabia
* Corresponding Author: Faten S. Alamri. Email:
(This article belongs to the Special Issue: Enhancing AI Applications through NLP and LLM Integration)
Computers, Materials & Continua 2025, 84(1), 1883-1899. https://doi.org/10.32604/cmc.2025.063560
Received 17 January 2025; Accepted 30 April 2025; Issue published 09 June 2025
Abstract
Large language models (LLMs) and natural language processing (NLP) have significant promise to improve efficiency and refine healthcare decision-making and clinical results. Numerous domains, including healthcare, are rapidly adopting LLMs for the classification of biomedical textual data in medical research. The LLM can derive insights from intricate, extensive, unstructured training data. Variants need to be accurately identified and classified to advance genetic research, provide individualized treatment, and assist physicians in making better choices. However, the sophisticated and perplexing language of medical reports is often beyond the capabilities of the devices we now utilize. Such an approach may result in incorrect diagnoses, which could affect a patient’s prognosis and course of therapy. This study evaluated the efficacy of the proposed model by looking at publicly accessible textual clinical data. We have cleaned the clinical textual data using various text preprocessing methods, including stemming, tokenization, and stop word removal. The important features are extracted using Bag of Words (BoW) and Term Frequency-Inverse Document Frequency (TFIDF) feature engineering methods. The important motive of this study is to predict the genetic variants based on the clinical evidence using a novel method with minimal error. According to the experimental results, the random forest model achieved 61% accuracy with 67% precision for class 9 using TFIDF features and 63% accuracy and a 73% F1 score for class 9 using Bag of Words features. The accuracy of the proposed BERT (Bidirectional Encoder Representations from Transformers) model was 70% with 5-fold cross-validation and 71% with 10-fold cross-validation. The research results provide a comprehensive overview of current LLM methods in healthcare, benefiting academics as well as professionals in the discipline.Keywords
The Large Language Model self-trains through self-supervised learning and accomplishes this by employing neural networks with billions of parameters and vast amounts of unlabeled textual data. These models are capable of identifying intricate patterns, subtle language differences, and clear logical connections with ease, as evidenced by their extensive training on large internet datasets [1]. LLMs have still performed satisfactorily on other language tasks that necessitate deep learning and extremely large datasets, including translating, summarizing, and analyzing an individual’s emotions [2]. Additionally, the development of these models for future responsibilities has shown significant potential and has yielded novel outcomes across various standards. LLMs have completely revolutionized the use of natural language by enabling the acquisition, analysis, and retrieval of textual material with previously unseen capabilities. This research relies heavily on the Transformer architecture, which Google first used in 2017 for machine translation tasks [3]. To better understand patient’s conditions, make more informed decisions, and streamline administrative processes like scheduling and accounting, models trained on medical literature using NLP might be useful [4,5]. The completely functional artificial language modules in large GPT-3 [6] and GPT-4 [7] models allow them to produce cohesive, human-like material. These are able to comprehend and predict data from various domains, including medical [8]. In addition to their acoustic validation, these models offer novel healthcare uses. Researchers and medical professionals may learn more by applying LLMs to various data sources, including imaging studies, laboratory results, and raw clinical samples [9]. Model validation employing verified datasets, quality control, and descriptive metrics is vital for addressing these challenges and providing dependable and effective application in clinical situations [10]. The LLM supports documented efforts to enhance clinical procedures and treatment alternatives. By evaluating datasets that include genetic and phenotypic variables, the LLM may help in anticipating the severity of diseases [11].
The advancement of LLM presents promising employment prospects in the healthcare sector. This methodology is particularly significant in medicine, as physicians, nurses, and healthcare practitioners encounter substantial amounts of unstructured data daily, including prescriptions, hospital discharge summaries, patient histories, and questionnaires. Historically, comprehending this data has been laborious and susceptible to inaccuracies. LLM facilitates the documentation of these activities, addresses intricate medical inquiries, and enables communication with patients. Also, NLP has enormous potential to transform healthcare because of its capacity to evaluate and arrange the vast amounts of unstructured data that exist in electronic health records, medical records, and other health documents. By extracting vital information, NLP technologies may help physicians make accurate diagnoses and shed light on critical topics, including the course of a disease, the effectiveness of therapy, and risk factors.
However, researchers are adopting novel approaches, including data augmentation and artificial intelligence via the integration of algorithms with clinical data or training through supervised learning. Even though these approaches have potential, they are not yet fully reliable, and further research is necessary to ensure they can meet therapeutic needs globally. Models such as GPT (Generative Pre-Trained Transformer), BERT, or BioBERT extract complex features from medical literature to enhance their ability to provide precise and practical clinical advice. Preserving patient confidentiality, ensuring safety, and avoiding invasiveness are crucial ethical considerations for the widespread use of these tools. It streamlines workflow, improves decision-making, and enables personalized therapy. NLP systems briefly summarize patient histories, treatment protocols, and test results to reduce doctor’s workloads and protect against missing information.
The study makes the following contributions:
• The study presents a novel large language BERT-optimized model for the prediction of Genetic variants using clinical textual data. It also proposed enhanced and adaptive solutions for healthcare settings by fine-tuning the model on medical datasets, resulting in more dependable patient outcomes.
• Natural language processing feature engineering techniques (NLPFET) are used to extract relevant numeric features from the textual data. It will mitigate the influence of less meaningful data and enhance performance across many tasks for the foundation of the RF model.
• To convert unstructured textual data into a structured and suitable form for the large language model, this study used several preprocessing techniques and NLP for the automated extraction and classification of personalized medicine data based on clinical evidence.
• The effect of each preprocessing step is determined and validated by K-fold cross-validation tests utilizing important metrics.
The healthcare sector has gained greatly from LLMs, which enhance the analysis of patient records, medical notes, and medical records. For activities including clinical decision assistance, patient interaction, and health outcome prediction, these models are commonly used. The LLM is utilized in various applications, including predictions, medical education, information extraction, medical documentation, and medical chatbots. The study [12] presented an innovative modular LLM methodology for extracting conceptually relevant characteristics from textual patient intake information. The pipeline excels in feature extraction by meeting the standards for precision and accuracy. The approach reliably identified clinical characteristics from textual data, proving its use across several LLMs. The use of LLM in healthcare has elicited both enthusiasm and concern among people. The use of advanced language models in healthcare might greatly enhance the understanding of clinical terminology and its use in medicine [13]. The advent of transformer architecture and the use of neural networks has radically transformed their understanding and generation of machine language. This evaluation of research offers a thorough examination of LLMs, including their background, development, training methodologies, and various enhancement initiatives. This research illustrates that LLMs are shaping the future of AI and may possess the ability to address intricate problems [14].
Latif and Kim [15] in his work employed the CHARDAT dataset to examine the effectiveness of two extensive language models in producing innovative terminology. The two models that are used are the Bidirectional and Auto-Regressive Transformers (BART) and the Text-To-Text Transfer Transformer. They utilized the ChatGPT technique to get further information from the dataset. ChatGPT could modify English words, but it could not change medical terminology, since it discerned the meanings of words in the dataset according to their context. While medical data may be presented as images, the authors [16] focus only on text-based information. The authors [17] provided a methodology that employed LLM with human expertise to swiftly produce ground truth labels for medical text annotation. Another work indicates that LLMs may accelerate the implementation of tailored NLP solutions by aiding healthcare businesses in optimising the use of unstructured clinical data. Large Language Models may surpass human performance by using several input modalities, including multidimensional visual and numerical data [18].
Peng et al. [19] utilized GatorTronGPT, a generative clinical LLM, using approximately 2 million cases and 277 billion words of clinical literature from 126 departments. The study’s findings about the merits and drawbacks of LLMs may be beneficial for medical research and healthcare. The study [20] produced an annotated dataset for German medical literature with little human intervention. The authors employed GatorTron to conduct a systematic evaluation of five NLP tasks. Clinical idea extraction, medical relation extraction, semantic textual similarity, and medical query answering comprise these tasks [21]. The authors provided an explanation of the construction and deployment of LLM applications in the healthcare industry by utilising examples such as ChatGPT. The objective of this article was to examine the potential advantages and disadvantages of LLMs and their potential to enhance the efficacy and effectiveness of medical research, clinical practice, and education. Although they were not consistently reliable, the results of the use of LLM chatbots in the biological sciences were thrilling in the past [22].
The purpose of research [23] was to evaluate the performance of LLM in comparison to human clinical experts in classifying mental health emergency department patients using terms extracted from a large electronic health record dataset. The primary focus of LLM for postoperative risk prediction using clinical records [24]. Another work proposed by the authors was prediction models for prescription drugs using the MIMIC-IV dataset to improve the analysis of electronic health records [25]. Without further fine-tuning, open weight LLMs may successfully capture patient’s socioeconomic determinants of health, as discussed by the authors of [26].
In the research [27], the authors employed BioBERT for electronic health records, utilized a limited sample size for the tests, and integrated a BERT+BiLSTM+CRF model, which escalates the computational expenses with a reduced number of samples, while also amalgamating several model variations. Additional study [28] employed the pretrained Med-BERT model for the identification of medical records. They utilized a named entity recognition dataset for the studies and did not investigate the preparation processes to enhance performance. Another study [29] only utilized ChatGPT 4 prompts for inquiries regarding medical information and the limits of the chatbot. They examined the advantages and potential impact of ChatGPT in the study; however, no model was proposed. Problems with data consistency and quality could emerge when automatically classifying medical records using LMM models that were built by experts. The practical utility of these models depends on their ability to properly extract relevant clinical data while also being easy to understand. Our model is better at continuous training, textual and variant data, and biomedical touch, which improves the quality of biomedical data, even though other NLP preprocessing pipelines have made important contributions to NLP in the biomedical field. The proposed model links genetic alterations to clinical evidence, explicitly combining classification goals with accuracy.
The authors developed [30] a graph-attentive feature interaction model (CVDLLM) to improve the accuracy of cardiovascular disease diagnoses, which was subsequently refined using the LLM. A novel approach that comprehensively extracts characteristics from ECG(Electrocardiogram) data within its framework was proposed. During the model’s learning phase, a GAT(Graph Attention Networks) subnet was implemented to conduct a systematic analysis of the relationships between inter-lead features. A summary of previous works, highlighting their limitations and research gaps, is shown in Table 1. To address the issues, this study utilized comprehensive preprocessing steps, a natural language processing pipeline, and feature extraction through bag of words and term frequency-inverse document frequency for the RF(Random Forest) model and the proposed LLM model to predict genetic variants from unstructured text medical data, employing various performance metrics.

This section provides the details about the dataset, its preprocessing, and the proposed methodology, as well as the natural language processing techniques. The entire workflow of the proposed study is shown in Fig. 1.
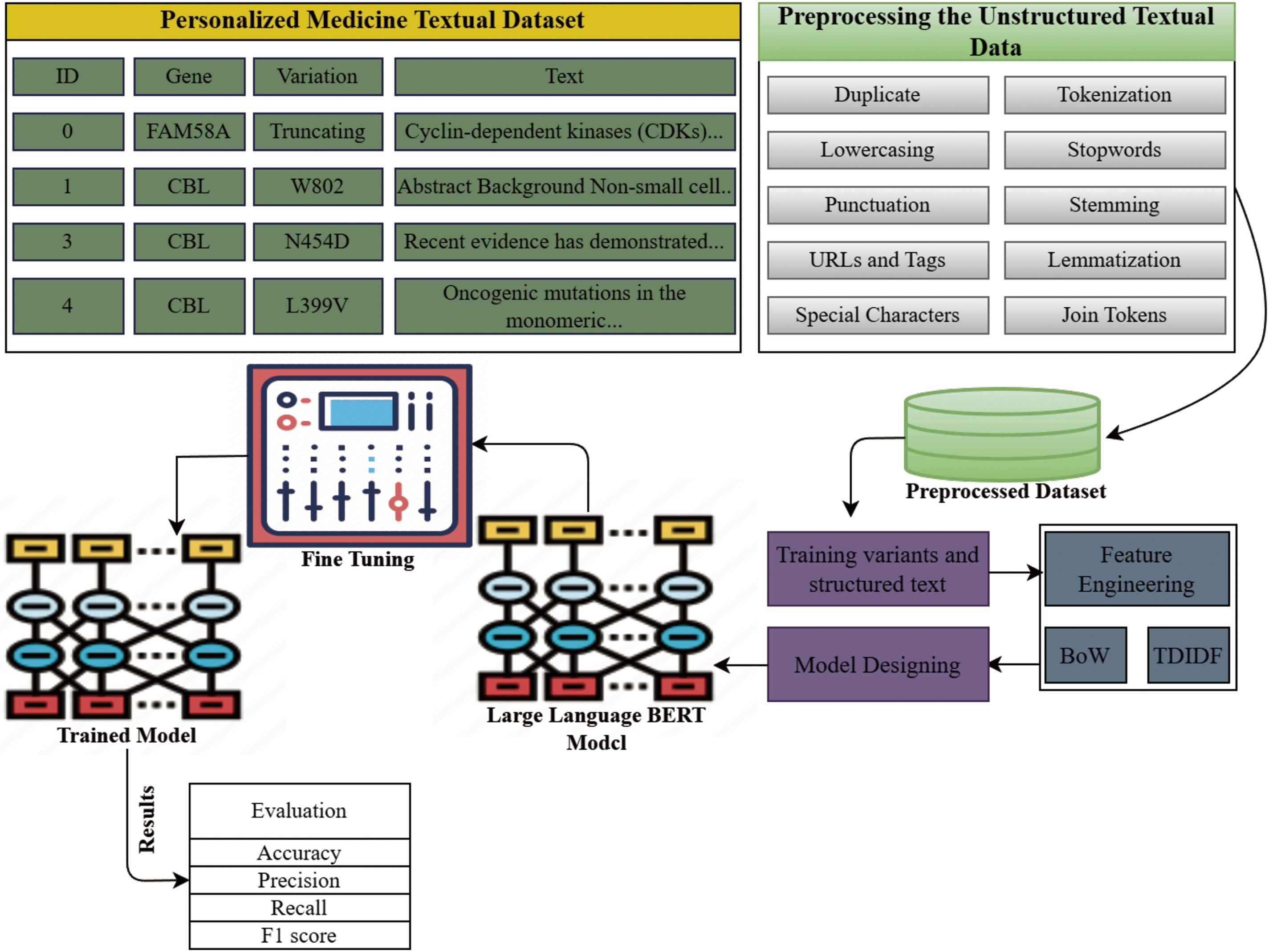
Figure 1: The proposed study workflow for the prediction of genetic variants using Large language BERT model and NLP techniques
3.1 Dataset and Preprocessing Steps
The experiment’s dataset was obtained from the open Kaggle repository. The collection includes nine classifications related to personalised medication for genetic mutations. Text presents the clinical proof, whereas variants in the dataset provide information regarding genetic mutations. The collected data includes unstructured and some useless information, which we need to remove from the textual data for improved model prediction. In the context of data analysis applications, data preparation is of utmost importance since it allows for the removal of unnecessary data, which in turn enhances the classification model learning process and results in increased accuracy. Any data that does not considerably increase the accuracy of the target class prediction is considered data that is considered to be worthless. However, the feature vector decreases, which increases the amount of processing burden. In preparation for encoding, data is cleaned up [31].
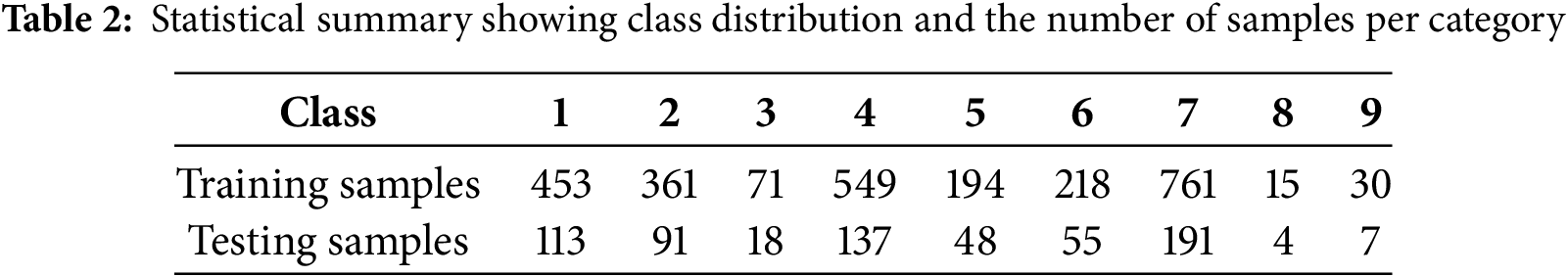
Statistical summary showing class distribution and the number of samples per category is shown in Table 2. We randomly select only 3316 samples for the experiments and split them in an 80:20 ratio, with 80% allocated for training and the remaining 20% for testing the model’s performance. The data encompasses descriptions of genetic mutations, shows ID rows, the genes harboring the mutations, variations, and the associated gene classes. Textual data also encompasses ID and clinical evidence.
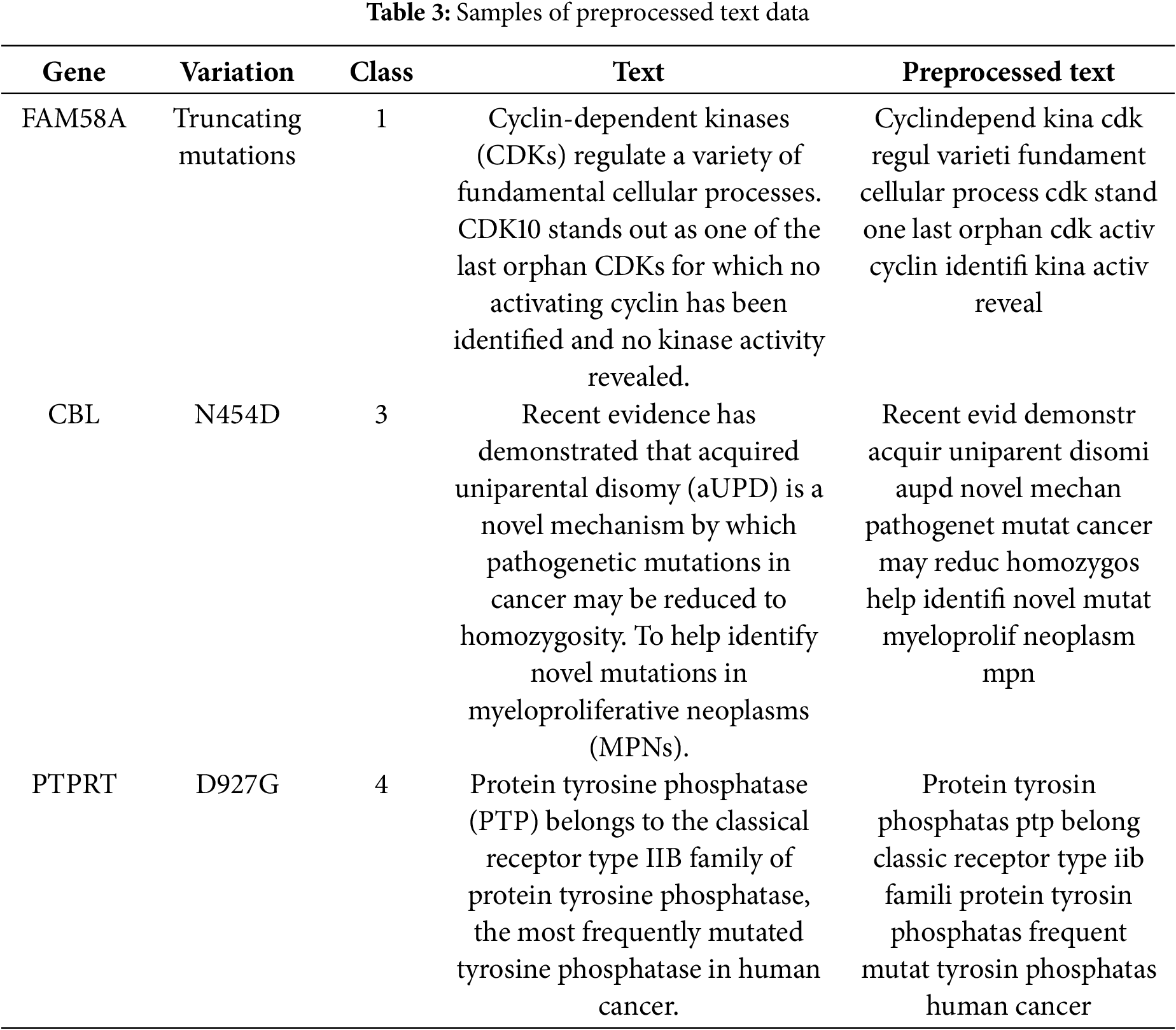
Several steps are taken for preprocessing the unstructured text.
• Lowercase conversion:Lowercasing means changing all the letters in a text to lowercase. In this case, we don’t want the computer to handle the same words in different situations in different ways.
• Punctuation removal:Remove all marks, like periods, commas, exclamation points, emojis, and more, from the text so that it is easier to read and you can focus on the words.
• Stopwords removal:A stopword is a word that doesn’t belong in a phrase and leaving it out doesn’t change what the phrase means. We can use the NLTK library’s stopwords to get rid of stopwords from the text and get a list of word tokens.
• Tokenization:Word tokenisation breaks down a text into its individual words by using spaces, punctuation, or other clues. To do most NLP work, one needs to use word-level tokenisation to process and understand text [32].
• Stemming and lemmatization: Since this is a natural process, the stemmed words that come up may not always be correct language. Lemmatisation is a more advanced method that breaks down a word into its basic form (lemma) by looking at its part of speech and its context. It works better than stems most of the time because it looks at both word meaning and grammar. Table 3 illustrates the preprocessed text data using several techniques.
Feature extraction is a crucial component of NLP. Machine learning systems need this procedure to convert textual input into numerical vectors for utilisation. Term frequency-inverse document frequency (TF-IDF) and bag-of-words (BoW) are the two most recognised methods for vectorising textual data.
BoW technique is a well recognised and straightforward approach to vectorising text data. This entails compiling a list of all distinct terms within a set and then determining the frequency of each word’s occurrence in a text. The result is a vector representation of the text, with each unit denoting the word count inside the text. BoW is a prevalent method for aggregating texts and an effective approach for initiating the vectorisation of text data [33].
TF-IDF is a sophisticated method for vectorising text data since it assesses the significance of words across documents and whole collections. TF-IDF assigns a weight to each word in a text according to its frequency in the text and its rarity throughout the whole corpus. This might enhance the precision of subsequent processes such as categorisation or retrieval by attributing more significance to terms that are crucial for distinguishing texts [34].
Natural language processing derives advantages from the use of large language models. LLM is a broad designation for various types. The BERT deep learning language model aims to improve the effectiveness of NLP tasks. One of its many remarkable features is its ability to examine word connections bidirectionally inside a phrase to account for context. The abbreviation for this method is “Bidirectional Encoder Representations from Transformers.” Encoders, integral components of neural networks, facilitate the simplification of incoming data for machine learning systems. Encoders provide a confidential state vector after processing the provided text. Concealed state vectors are comparable to internal aggregates of values and parameters that provide additional information. Thereafter, the transformer acquires this data payload. By using the aforementioned data, the transformer may provide predictions and conclusions. The proposed large language BERT optimized model is shown in Fig. 2.
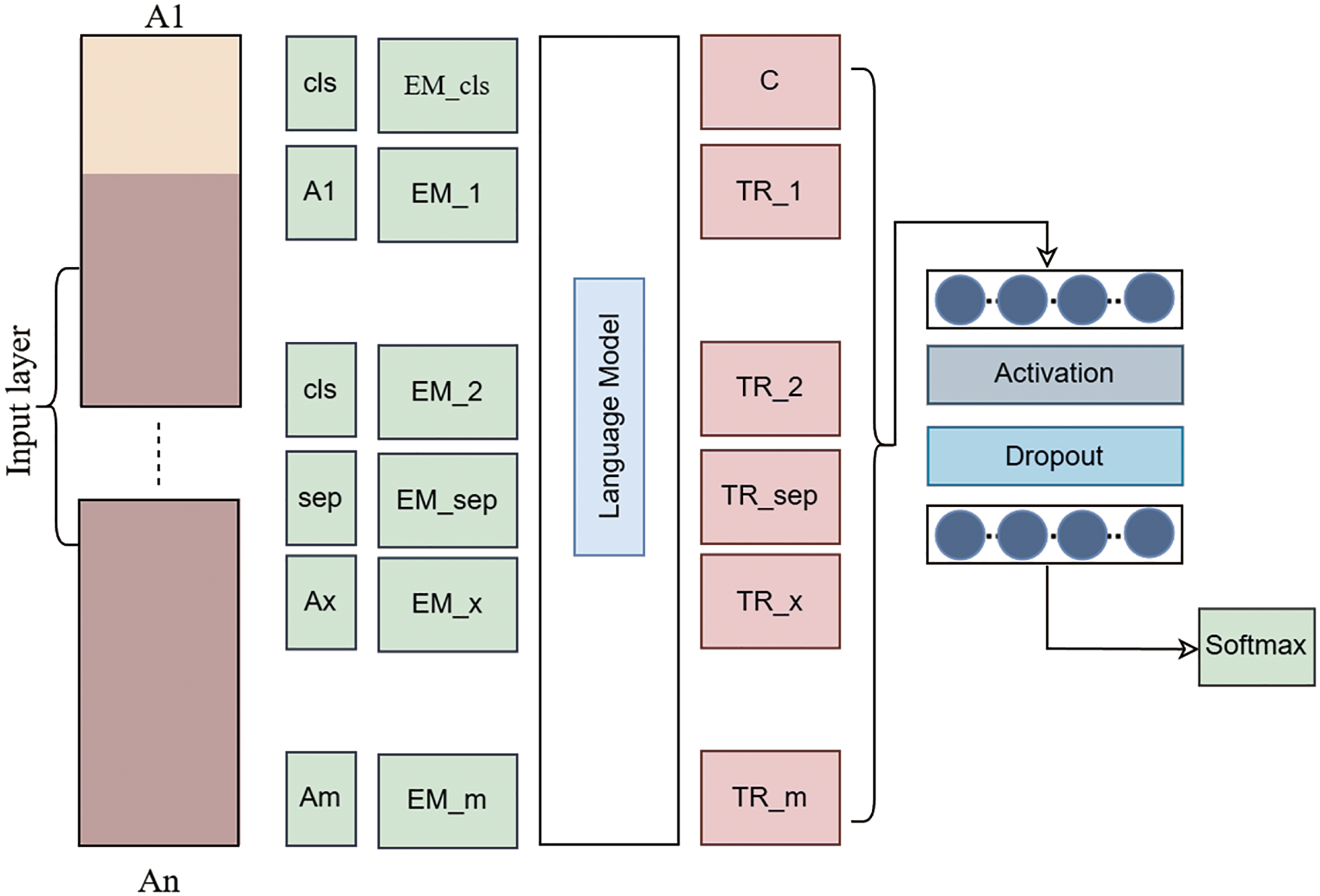
Figure 2: Proposed large language BERT optimized model
A transformer is a deep learning architecture capable of transforming one input into another. Nearly all apps use transformers to process real words. The encoder and decoder constitute the two essential components of a transformer. However, BERT only utilizes components of the generator. The use of BERT for pre-training in language processing is a common approach in the realm of artificial intelligence. A possible use is to improve search engine results by ascertaining context. BERT is the optimal architecture for a wide range of NLP tasks involving words and tokens. It is standard practice to include a [MASK] token that substitutes the words in each word sequence before inputting it into BERT. Subsequently, the model attempts to ascertain the original value of the hidden words by examining the context of the secret terms. The BERT method just assesses concealed value predictions during loss computation. Predicting non-masked words is inconsequential. Thus, the model’s enhanced contextual awareness significantly compensates for its prolonged settling time relative to directed models. BERT is undeniably transformative in the application of machine learning to NLP.
Numerous models, such as the BERT model, which was developed to predict medical text and genetics and served as a foundation for effective classification, were used to supplement this study. BERT is especially helpful for tasks involving technical terms or unstructured data because of its capacity to interpret abstract meanings and complex biomedical language contexts. Even with extremely complicated datasets, it can achieve excellent accuracy because it has been pre-trained on large amounts of data, frequently surpassing more conventional models like pattern recognition and classification. The BERT model does, however, have weaknesses, including a high resource requirement and difficulty in interpretation, which may be detrimental in therapeutic settings where comprehension of the model’s results is crucial. However, conventional machine learning models like random forests, logistic regression, and decision tree models are simple and easy to predict.
This section presents the large language BERT-optimised, fine-tuned model experiments using two important NLP feature engineering techniques, such as bag of words and term frequency and inverse document frequency, for the extraction of numeric features from the medical textual data. We conduct the experiments using various performance metrics.
Predicting genetic polymorphisms and providing specific cancer treatments is personalized medicine. A comma-separated text file (training_variants) describes the training genetic variations. The genetic mutation consists of Identification (ID), Gene (the gene associated with the mutation), Variation (the mutation-induced amino acid modification), and Class (1 to 9). Two methods segment the training_text file, which contains genetic mutation classification text. The authors classify genetic mutations based on ID (clinical evidence) and text. The file test_text contains clinical evidence for genetic mutation classification, separated by double pipes. The genetic mutation database uses ID and text fields to provide clinical information for mutation categorization. Fig. 3 represents the different visualizations of the medical data.
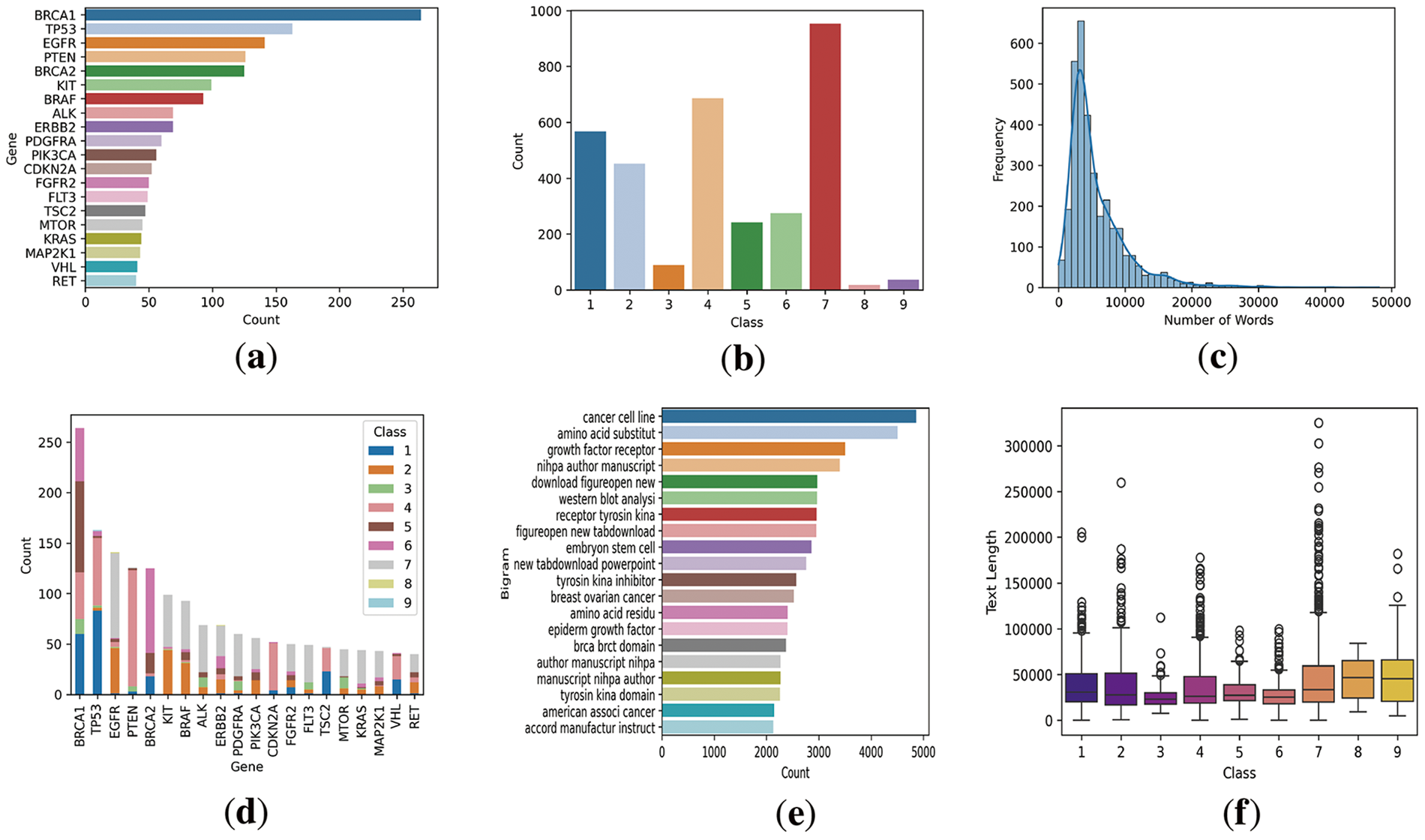
Figure 3: Dataset visualization where (a) presents the top gene counts appeared in the data, (b) presents the number of classes in the data distribution, (c) presents the text length by number of words in the data and its frequency, (d) presents the gene distribution for each class, (e) presents the bi-gram analysis of textual data, (f) presents the textual data length for each class
NLP is a prominent application of WordCloud in the domain of artificial intelligence. We anticipate that by emphasising the most often-used terms in the paragraph, website, social media platform, or discourse, the principal subject of the content will be illuminated. Word clouds serve as a type of data visualisation that illustrates textual information as shown in Fig. 4. The frequency or significance of each word within the sentence determines its prominence in the word cloud. A word cloud allows the visualization of several textual data points. A prevalent use of word clouds is in the analysis of data gathered from social media sites.

Figure 4: Best visualization of the wordclouds extracted from the data
4.2 Performance of the Proposed RF Model Using BoW and TFIDF Features
The selected five features are shown in Fig. 5a, which demonstrates the mean of BoW features along with the classes. Different classes have various means, but Class 9 has the highest BoW mean for cell features. Fig. 5b, which demonstrates the features along with the BoW score. The mutation feature has the highest BoW score in the data. Fig. 5c, which demonstrates the class-wise features and average BoW counts.
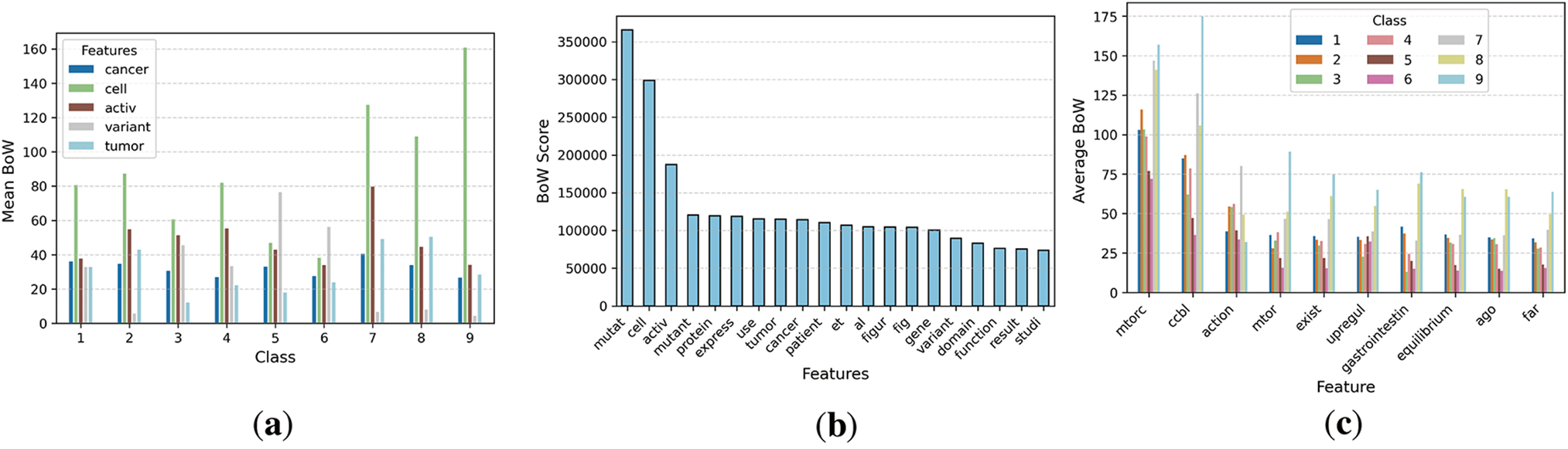
Figure 5: Visualization of BoW features extracted from the dataset
The experimental findings of the proposed model using BoW features are shown in Table 4. Class 9 achieved the most precise findings compared to other classes. Class 2 attained 70% precision and 42% recall; Class 4 earned 65% precision and a 67% F1 score; Class 3 recorded 43% precision and a 38% F1 score; Class 5 reached 40% recall; Class 6 obtained a 68% F1 score; Class 7 realised 64% precision; Class 8 had notably poor performance. The features collected from the Bag of Words approach yielded superior outcomes.
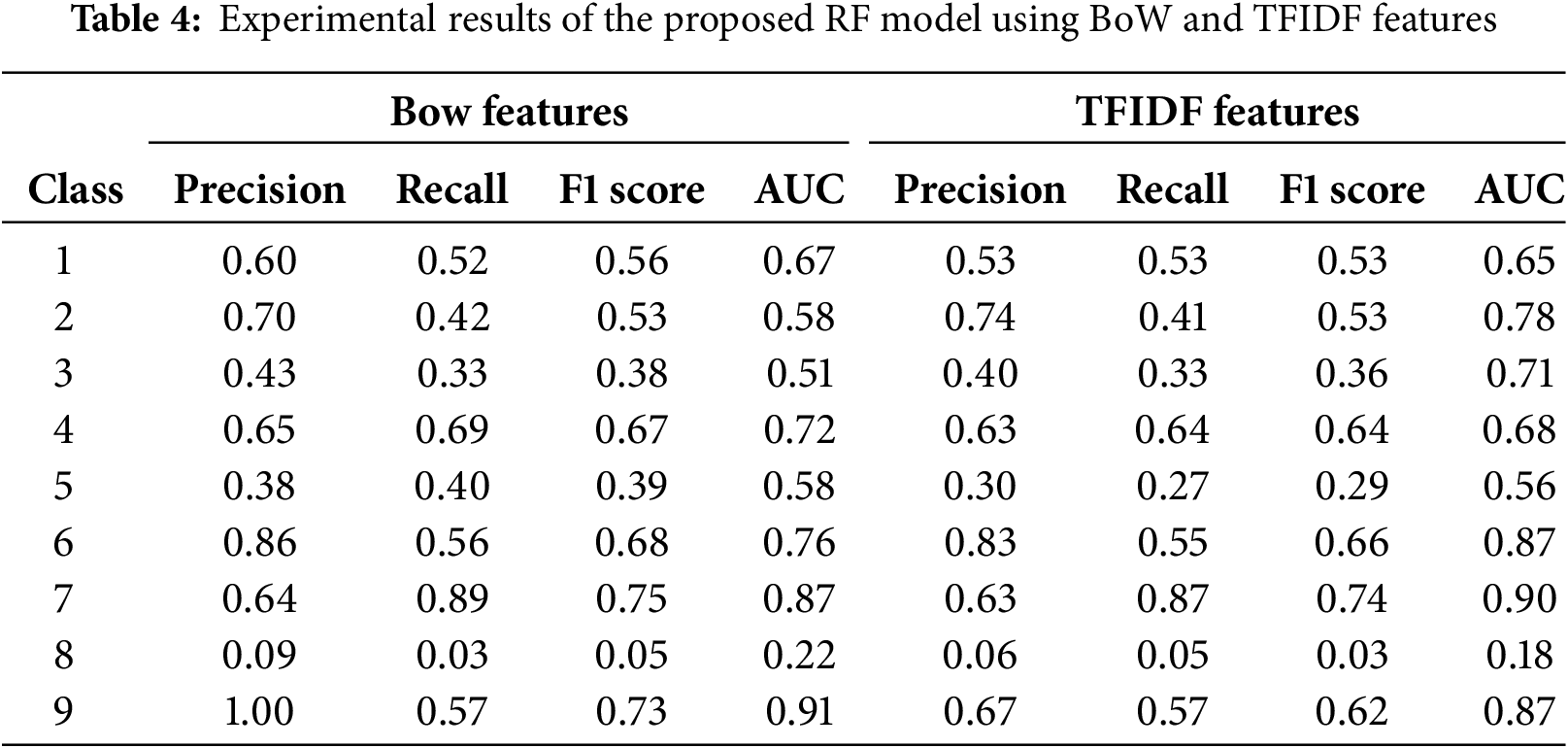
The experimental findings of the proposed model using TFIDF features are also shown in Table 4. Class 9 achieved the most precise findings compared to other classes. Class 2 attained 74% precision and 41% recall; class 3 earned 40% precision and 36% F1 score; class 5 recorded 27% recall; class 6 reached an 66% F1 score; class 7 obtained 63% precision; class 8 had very poor performance. The features retrieved using the TDIDF approach yielded superior results.
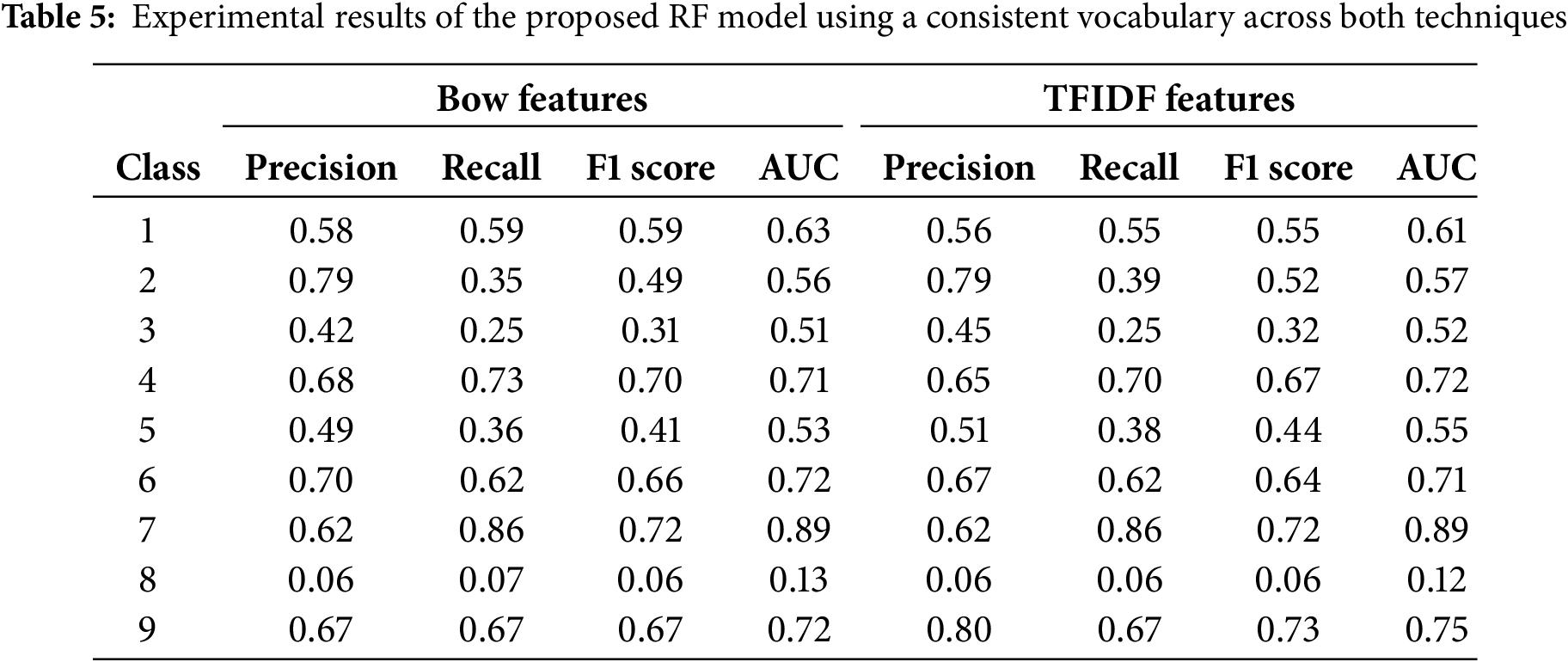
Table 5 presents the experimental results of the proposed RF model using BoW and TFIDF vectorizer with unified vocabulary. We set same frequency counts for both feature extraction techniques.
The five selected features are shown in Fig. 6a, displaying the mean of TFIDF features in relation to the classes. Various classes have distinct means; however, class 5 has the greatest TFIDF mean for variant traits. Fig. 6b illustrates the features besides the TFIDF score. The mutation feature has the greatest TFIDF score in the data, related to BoW. Fig. 6c illustrates the class-specific traits and the average TFIDF numbers.
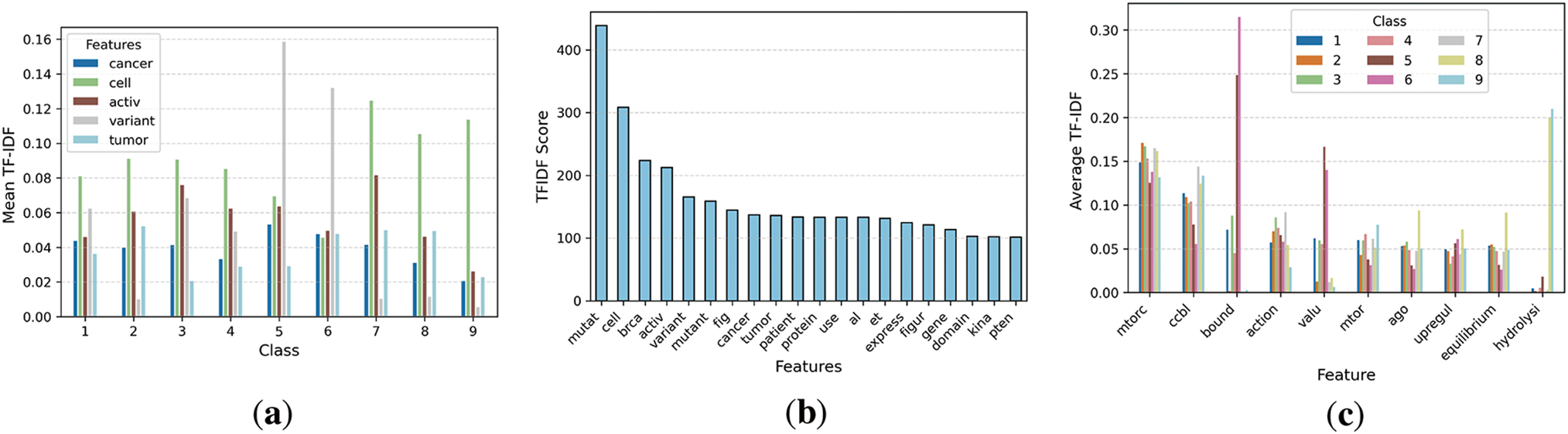
Figure 6: Visualization of TFIDF features extracted from the dataset
4.3 Proposed Large Language BERT Model
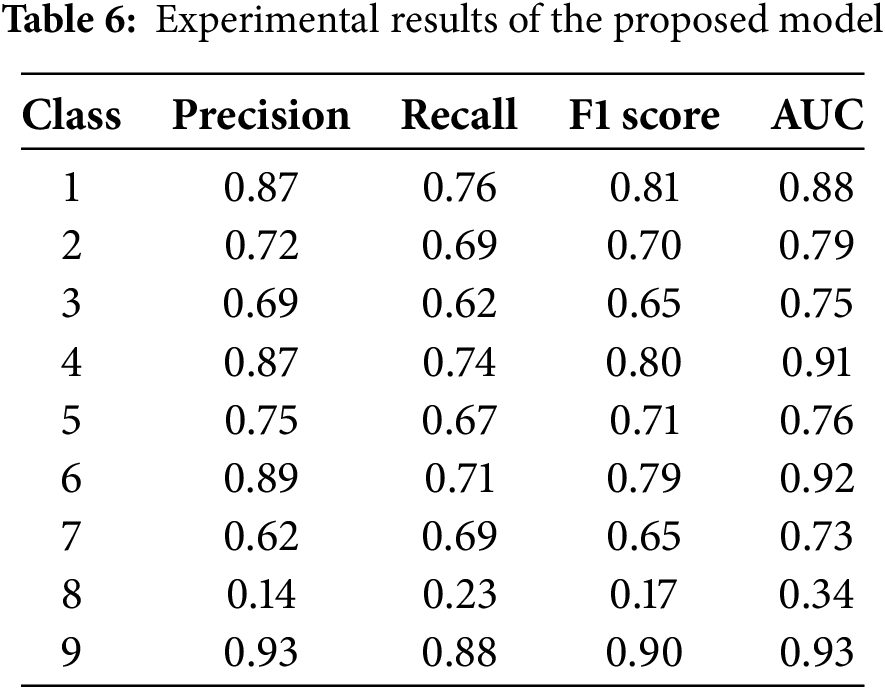
The experimental findings of the proposed model are shown in Table 6. Class 9 achieved the most precise findings compared to other classes. Class 2 attained 87% precision and 76% recall; Class 4 earned 72% precision and a 70% F1 score; Class 3 recorded 69% precision and a 65% F1 score; Class 5 reached 67% recall; Class 6 obtained a 79% F1 score; Class 7 realised 62% precision; Class 8 had notably poor performance. The features collected from the Bag of Words approach yielded superior outcomes.
4.4 Cross Validation Performance
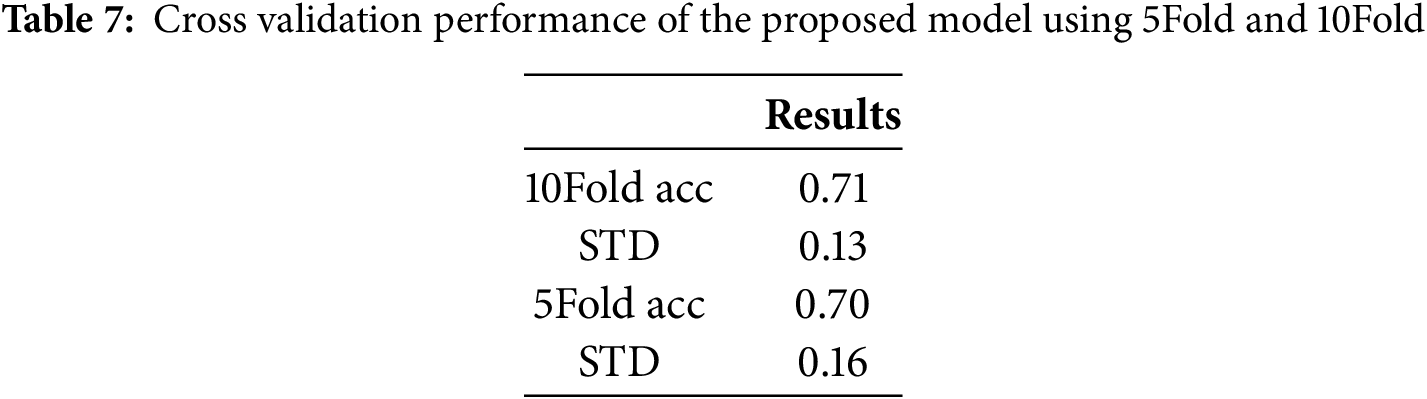
Cross validation performance of the proposed model using 5Fold and 10Fold is presented in Table 7. With cross-validation, the proposed model achieved 71% mean accuracy with 10-fold and 70% accuracy with 5-fold. Also, it achieved a 0.13 standard deviation using 10 folds and 0.16 with 5 folds.
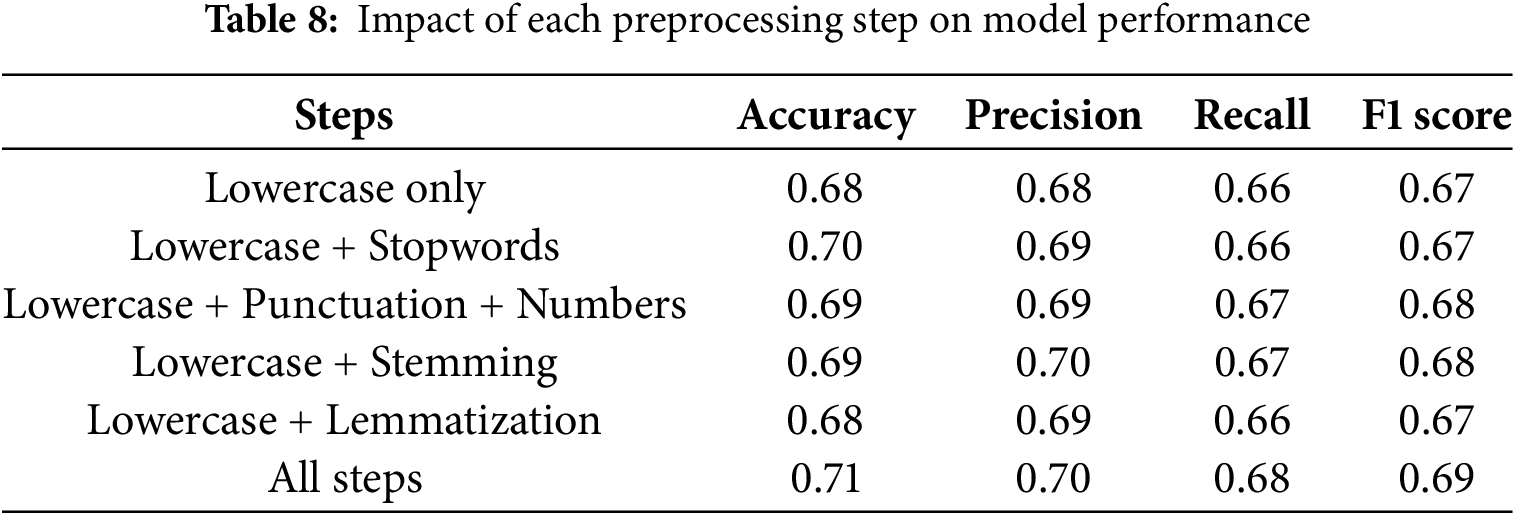
Preprocessing steps affect model performance differently depending on their objective. Lower text reduces contradictions and clarifies vocabulary. Eliminating stopwords keeps text clean, but predictive analytics may lose important data. Table 8 presents the impact of each preprocessing step on model performance. The combination of lowercase and stopwords exerts a greater influence than mere lowercase conversion and lemmatization. Lemmatization, stopwords, and numbers have achieved superior performance compared to lowercase and lemmatized text.
Even though the proposed method performs well technically, a thorough assessment of its therapeutic value and real-world implementation in clinical settings would still be beneficial for this study. This technology reduces manual labor and enhances decision-making by assisting physicians in automatically extracting valuable information from unstructured data, including genetic information, medical presentations, and research publications. They assist in identifying high-risk individuals, developing individualized treatment plans, and matching the right drugs to genetic diseases when integrated with systems like electronic health records. Validating model interpretation, which is crucial in clinical contexts, and managing noisy or missing data are two difficulties that arise during real-world deployment. By concentrating on these areas, the research’s scientific impact will be increased and the gap between AI (artificial intelligence) development and clinical application will be closed. The error analysis per class is shown in Fig. 7. Class 9 has fewer errors, and class 8 has more false positives.
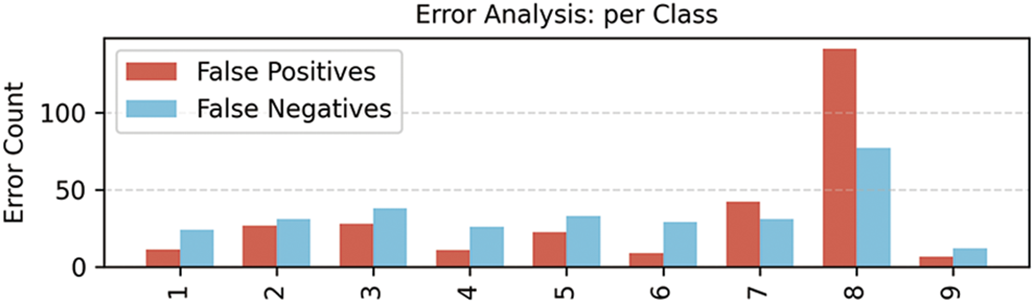
Figure 7: Error analysis made by the proposed model
LLMs attract the interest of many fields as they might revolutionize artificial intelligence capabilities. Training LLMs from genesis using medical datasets or fine-tuning them with generic LLMs could improve their usage in healthcare. The effectiveness of LLM was assessed in this research by examining textual clinical data that was sourced openly. We have used a range of text preparation techniques to clean the clinical textual data. Two feature engineering techniques are BoW and TFIDF, which are used to extract significant features or convert features into numerical formats. The main results show that the RF model reached 63% accuracy and a 73% F1 score for class 9 using Bag of Words features, and 61% accuracy with 67% precision for class 9 using TFIDF features.
The proposed BERT model got 71% accuracy with 10-fold cross-validation and 70% with 5-fold cross-validation. The suggested BERT model achieved 71% accuracy with 10-fold cross-validation and 70% with 5-fold cross-validation. The proposed approach enhances the two-dimensional comprehension of variant sequences, reduces the error rate in predicting genomic variations, and captures intricate impacts that conventional models may ignore. This trend aligns with advancements in NLP, wherein LLMs have surpassed state-of-the-art models in tasks like gene localization and variant prediction. Through extensive pre-training on substantial genomic datasets, our model rapidly acquires the capability to delineate feature sets, hence enhancing its predictive accuracy of behavioral traits relative to current methodologies.
The present research has limitations, since the findings of the proposed model are inadequate, demonstrating poor performance and subpar feature extraction efficacy. Furthermore, this study has limited data that results in less accurate predictions, and there were imbalance issues in the obtained data that caused overfitting. In the future, we will implement efficient feature extraction and selection algorithms to improve accuracy. To address the imbalance issues, we will employ sampling and advanced generative adversarial networks to balance the data and enhance the results. Additionally, we may include more advanced LLM models like ChatGPT-3 and Transformers.
Acknowledgement: This research was supported by Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. The authors would also like to acknowledge the APC support of Prince Sultan University, Riyadh, Saudi Arabia.
Funding Statement: This research was funded by Princess Nourah bint Abdulrahman University and Researchers Supporting Project number (PNURSP2025R346), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Noor Ayesha, Abeer Rashad Mirdad, Amjad R. Khan; data collection: Amjad R. Khan, Faten S. Alamri, Abeer Rashad Mirdad; analysis and interpretation of results: Amjad R. Khan, Muhammad Mujahid, Noor Ayesha; draft manuscript preparation: Amjad R. Khan, Muhammad Mujahid; Supervision: Faten S. Alamri, Amjad R. Khan; project administration: Faten S. Alamri. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Wan M, Safavi T, Jauhar SK, Kim Y, Counts S, Neville J, et al. Tnt-llm: text mining at scale with large language models. In: Proceedings of the 30th ACM SIGKDD Conference on Knowledge Discovery and Data Mining; 2024 Aug 25–29; Barcelona, Spain. p. 5836–47. doi:10.1145/3637528.3671647. [Google Scholar] [CrossRef]
2. Wang Y, Zhang J, Shi T, Deng D, Tian Y, Matsumoto T. Recent advances in interactive machine translation with large language models. IEEE Access. 2024;12:179353–82. doi:10.1109/ACCESS.2024.3487352. [Google Scholar] [CrossRef]
3. Rothman D. Transformers for natural language processing: build innovative deep neural network architectures for NLP with Python, PyTorch, TensorFlow, BERT, RoBERTa, and more. Birmingham, UK: Packt Publishing Ltd.; 2021. [Google Scholar]
4. Radwan A, Amarneh M, Alawneh H, Ashqar HI, AlSobeh A, Magableh AA. Predictive analytics in mental health leveraging LLM embeddings and machine learning models for social media analysis. Int J Web Serv Res (IJWSR). 2024;21(1):1–22. doi:10.4018/IJWSR.338222. [Google Scholar] [CrossRef]
5. Hassan AQ, Al-onazi BB, Maashi M, Darem AA, Abunadi I, Mahmud A. Enhancing extractive text summarization using natural language processing with an optimal deep learning model. AIMS Math. 2024;9(5):12588–609. doi:10.3934/math.2024616. [Google Scholar] [CrossRef]
6. Qazi S, Kadri MB, Naveed M, Khawaja BA, Khan SZ, Alam MM, et al. AI-Driven learning management systems: modern developments, challenges and future trends during the age of ChatGPT. Comput Mater Contin. 2024;80(2):3289–314. doi:10.32604/cmc.2024.048893. [Google Scholar] [CrossRef]
7. Javidan AP, Feridooni T, Gordon L, Crawford SA. Evaluating the progression of artificial intelligence and large language models in medicine through comparative analysis of ChatGPT-3.5 and ChatGPT-4 in generating vascular surgery recommendations. JVS-Vasc Insights. 2024;2:100049. doi:10.1016/j.jvsvi.2023.100049. [Google Scholar] [CrossRef]
8. Rony MA, Islam MS, Sultan T, Alshathri S, El-Shafai W. Medigpt: exploring potentials of conventional and large language models on medical data. IEEE Access. 2024;12:103473–87. doi:10.1109/access.2024.3428918. [Google Scholar] [CrossRef]
9. Sun D, Hadjiiski L, Gormley J, Chan HP, Caoili E, Cohan R, et al. Outcome prediction using multi-modal information: integrating large language model-extracted clinical information and image analysis. Cancers. 2024;16(13):2402. doi:10.3390/cancers16132402. [Google Scholar] [PubMed] [CrossRef]
10. Freyer O, Wiest IC, Kather JN, Gilbert S. A future role for health applications of large language models depends on regulators enforcing safety standards. Lancet Digit Health. 2024;6(9):e662–72. doi:10.1016/S2589-7500(24)00124-9. [Google Scholar] [PubMed] [CrossRef]
11. Shah K, Xu AY, Sharma Y, Daher M, McDonald C, Diebo BG, et al. Large language model prompting techniques for advancement in clinical medicine. J Clin Med. 2024;13(17):5101. doi:10.3390/jcm13175101. [Google Scholar] [PubMed] [CrossRef]
12. Wang L, Ma Y, Bi W, Lv H, Li Y. An entity extraction pipeline for medical text records using large language models: analytical study. J Med Internet Res. 2024;26:e54580. doi:10.2196/54580. [Google Scholar] [PubMed] [CrossRef]
13. Nazi ZA, Peng W. Large language models in healthcare and medical domain: a review. Informatics. 2024;11(3):57. doi:10.3390/informatics11030057. [Google Scholar] [CrossRef]
14. Raiaan MA, Mukta MS, Fatema K, Fahad NM, Sakib S, Mim MM et al. A review on large language models: architectures, applications, taxonomies, open issues and challenges. IEEE Access. 2024;12:26839–74. doi:10.1109/access.2024.3365742. [Google Scholar] [CrossRef]
15. Latif A, Kim J. Evaluation and analysis of large language models for clinical text augmentation and generation. IEEE Access. 2024;12:48987–96. doi:10.1109/access.2024.3384496. [Google Scholar] [CrossRef]
16. Yashwanth YS, Shettar R. Zero and few short learning using large language models for de-identification of medical records. IEEE Access. 2024;12:110385–93. doi:10.1109/ACCESS.2024.3439680. [Google Scholar] [CrossRef]
17. Goel A, Gueta A, Gilon O, Liu C, Erell S, Nguyen LH, et al. Llms accelerate annotation for medical information extraction. Mach Learn Health (ML4H). 2023;225:82–100. [Google Scholar]
18. Yang R, Tan TF, Lu W, Thirunavukarasu AJ, Ting DS, Liu N. Large language models in health care: development, applications, and challenges. Health Care Sci. 2023;2(4):255–63. doi:10.1002/hcs2.61. [Google Scholar] [PubMed] [CrossRef]
19. Peng C, Yang X, Chen A, Smith KE, PourNejatian N, Costa AB, et al. A study of generative large language model for medical research and healthcare. npj Digit Med. 2023;6(1):210. doi:10.1038/s41746-023-00958-w. [Google Scholar] [PubMed] [CrossRef]
20. Frei J, Kramer F. Annotated dataset creation through large language models for non-english medical NLP. J Biomed Inform. 2023;145:104478. doi:10.1016/j.jbi.2023.104478. [Google Scholar] [PubMed] [CrossRef]
21. Yang X, Chen A, PourNejatian N, Shin HC, Smith KE, Parisien C, et al. A large language model for electronic health records. npj Digit Med. 2022;5(1):194. doi:10.1038/s41746-022-00742-2. [Google Scholar] [PubMed] [CrossRef]
22. Thirunavukarasu AJ, Ting DS, Elangovan K, Gutierrez L, Tan TF, Ting DS. Large language models in medicine. Nature Med. 2023;29(8):1930–40. doi:10.1038/s41591-023-02448-8. [Google Scholar] [PubMed] [CrossRef]
23. Cardamone NC, Olfson M, Schmutte T, Ungar L, Liu T, Cullen SW, et al. Classifying unstructured text in electronic health records for mental health prediction models: large language model evaluation study. JMIR Med Inform. 2025;13(1):e65454. doi:10.2196/65454. [Google Scholar] [PubMed] [CrossRef]
24. Alba C, Xue B, Abraham J, Kannampallil T, Lu C. The foundational capabilities of large language models in predicting postoperative risks using clinical notes. npj Digit Med. 2025;8(1):95. doi:10.1038/s41746-025-01489-2. [Google Scholar] [PubMed] [CrossRef]
25. Alghamdi H, Mostafa A. Advancing EHR analysis: predictive medication modeling using LLMs. Inf Syst. 2025;131:102528. doi:10.1016/j.is.2025.102528. [Google Scholar] [CrossRef]
26. Gu B, Shao V, Liao Z, Carducci V, Brufau SR, Yang J, et al. Scalable information extraction from free text electronic health records using large language models. BMC Med Res Methodol. 2025;25(1):23. doi:10.1186/s12874-025-02470-z. [Google Scholar] [PubMed] [CrossRef]
27. Yu X, Hu W, Lu S, Sun X, Yuan Z. BioBERT based named entity recognition in electronic medical record. In: 2019 10th International Conference on Information Technology in Medicine and Education (ITME); 2019 Aug 23–25; Qingdao, China. p. 49–52. doi:10.1109/ITME.2019.00022. [Google Scholar] [CrossRef]
28. Liu N, Hu Q, Xu H, Xu X, Chen M. Med-BERT: a pretraining framework for medical records named entity recognition. IEEE Trans Ind Inform. 2021;18(8):5600–8. doi:10.1109/tii.2021.3131180. [Google Scholar] [CrossRef]
29. Cox A, Seth I, Xie Y, Hunter-Smith DJ, Rozen WM. Utilizing ChatGPT-4 for providing medical information on blepharoplasties to patients. Aesthet Surg J. 2023;43(8):NP658–62. doi:10.1093/asj/sjad096. [Google Scholar] [PubMed] [CrossRef]
30. Qiu X, Wang H, Tan X, Jin Y. CVDLLM: automated cardiovascular disease diagnosis with large-language-model-assisted graph attentive feature interaction. IEEE Trans Artif Intell. 2025. doi:10.1109/tai.2025.3527401. [Google Scholar] [CrossRef]
31. Vijayarani S, Ilamathi MJ, Nithya M. Preprocessing techniques for text mining-an overview. Int J Comput Sci Commun Netw. 2015;5(1):7–16. doi:10.5121/ijcga.2015.5105. [Google Scholar] [CrossRef]
32. Ovalle A, Mehrabi N, Goyal P, Dhamala J, Chang KW, Zemel R, et al. Tokenization mWatters: navigating data-scarce tokenization for gender inclusive language technologies. arXiv:2312.11779. 2023. doi:10.48550/arXiv.2312.11779. [Google Scholar] [CrossRef]
33. Chen S, Li Y, Lu S, Van H, Aerts HJ, Savova GK, et al. Evaluating the ChatGPT family of models for biomedical reasoning and classification. J Am Med Inform Assoc. 2024;31(4):940–8. doi:10.1093/jamia/ocad256. [Google Scholar] [PubMed] [CrossRef]
34. El-Gayar O, Al-Ramahi M, Wahbeh A, Nasralah T, Elnoshokaty A. A comparative analysis of the interpretability of LDA and LLM for topic modeling: the case of healthcare apps [Internet]. 2024 [cited 2025 Apr 29]. Available from: https://scholar.dsu.edu/bispapers/423. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools