 Open Access
Open Access
ARTICLE
A Deep Learning Approach to Classification of Diseases in Date Palm Leaves
1 Department of Biomedical Engineering, KMCT College of Engineering for Women, Kozhikode, 673601, India
2 MCA Department, Federal Institute of Science and Technology, Angamaly, 683577, India
3 ECE Department, National Institute of Technology Calicut, Kattangal, 673601, India
4 Department of Chemistry, College of Science, Princess Nourah bint Abdulrahman University, P.O. Box 84428, Riyadh, 11671, Saudi Arabia
5 Department of Electrical Engineering, College of Engineering, Princess Nourah bint Abdulrahman University, P.O. Box 84428, Riyadh, 11671, Saudi Arabia
* Corresponding Author: Shaeen Kalathil. Email:
Computers, Materials & Continua 2025, 84(1), 1329-1349. https://doi.org/10.32604/cmc.2025.063961
Received 30 January 2025; Accepted 24 April 2025; Issue published 09 June 2025
Abstract
The precise identification of date palm tree diseases is essential for maintaining agricultural productivity and promoting sustainable farming methods. Conventional approaches rely on visual examination by experts to detect infected palm leaves, which is time intensive and susceptible to mistakes. This study proposes an automated leaf classification system that uses deep learning algorithms to identify and categorize diseases in date palm tree leaves with high precision and dependability. The system leverages pretrained convolutional neural network architectures (InceptionV3, DenseNet, and MobileNet) to extract and examine leaf characteristics for classification purposes. A publicly accessible dataset comprising multiple classes of diseased and healthy date palm leaf samples was used for the training and assessment. Data augmentation techniques were implemented to enhance the dataset and improve model resilience. In addition, Synthetic Minority Oversampling Technique (SMOTE) was applied to address class imbalance and further improve the classification performance. The system was trained and evaluated using this dataset, and two of the models, DenseNet and MobileNet, achieved classification accuracies greater than 95%. MobileNetV2 emerged as the top-performing model among those assessed, achieving an overall accuracy of 96.99% and macro-average F1-score of 0.97. All nine categories of date palm leaf conditions were consistently and accurately identified, showing exceptional precision and dependability. Comparative experiments were conducted to assess the performance of the Convolutional Neural Network (CNN) architectures and demonstrate their potential for scalable and automated disease detection. This system has the potential to serve as a valuable agricultural tool for assisting in disease management and monitoring date palm cultivation.Keywords
Date palm trees are of immense cultural and economic importance in many regions of the world, particularly in dry and semidry climates. These trees serve as crucial crops, enhance food security, provide income for farmers, and support various industries including agriculture and food processing. However, leaf diseases in date palms can significantly reduce both the yield and quality [1]. The timely and precise identification of these diseases is vital for maintaining sustainable agricultural practices and implementing effective disease control measures.
This research utilized the “Dataset of Infected Date Palm Leaves for Palm Tree Disease Detection and Classification” by Namoun et al. [1] to tackle this issue. This comprehensive dataset comprises a wide array of leaf images, encompassing multiple disease categories and healthy specimens, captured in diverse environmental settings. This serves as a crucial resource for developing machine learning models capable of classifying infected leaves with high precision and consistency.
Compared with earlier studies on date palm disease detection, such as the work by Al-Shalout and Mansour [2], who applied a deep CNN architecture to a small, manually collected date palm dataset, our methodology diverges from that of Al-Shalout and Mansour [2] in several significant ways. Their research concentrated on a manually assembled, relatively small dataset of date palms, using a single deep CNN model. In contrast, we employed a more varied and extensive dataset that reflected real-world differences, including lighting and background noise. Furthermore, our study investigates and contrasts multiple pre-trained CNN architectures–MobileNet, DenseNet121, and InceptionV3 through transfer learning, allowing for a more thorough assessment of model performance. This expanded scope enhances generalizability and robustness, particularly in practical applications. Furthermore, unlike the DPXception model proposed by Safran et al. [3], which was designed specifically for species classification, our work focuses on disease classification and explores multiple transfer learning architectures (MobileNet, DenseNet121, and InceptionV3), allowing for a richer evaluation of model efficiency and performance trade-offs.
In the realm of general plant disease classification, several reviews demonstrated the success of CNNs on datasets such as PlantVillage [4]. However, these datasets are often collected in controlled settings and do not translate well into real-world environments. By integrating efficient models such as MobileNetV2 with this realistic dataset, our research pushes the boundaries for developing practical plant disease-detection systems for use in the field.
Deep learning models, known as Convolutional Neural Networks (CNNs), have revolutionized image classification by automatically extracting hierarchical features from the input data [5]. Unlike traditional image processing techniques that rely on manually engineered features, CNNs learn directly from raw images, making them highly effective for complex tasks, such as plant leaf classification. Transfer learning with pretrained CNN architectures, such as InceptionV3, DenseNet, and MobileNet, has demonstrated exceptional performance across various domains.
Several review studies, such as those by Kamilaris and Prenafeta-Boldú [6] and Hasan et al. [7] have highlighted the growing role of deep learning, particularly CNNs, in plant disease classification using leaf images.
CNNs have been extensively applied to the detection of diseases in crops. The reader can refer to a review paper for an extensive review of this area [8–12]. As evidenced in studies by Agarwal et al. [13] for tomatoes and Asif et al. [14] for potatoes. These studies have primarily focused on classifying leaf diseases in various plant species. However, the application of deep-learning techniques to date palm leaf disease detection remains relatively unexplored, with a notable scarcity of comprehensive datasets. Prior research has often utilized limited datasets comprising only three or four classes, which fails to capture the full range of conditions in which date palms grow, as noted in studies by Abu-zanona et al. [15].
The proposed method for automated phenotyping of Cymbidium seedlings using 3D point cloud data successfully addressed the issues of tiller segmentation and trait measurement. This approach employs a two-phase segmentation process in line with the natural growth orientation of the plant to accurately extract essential morphological characteristics. This resulted in high precision across various traits including plant height, leaf count, and leaf measurements. Consequently, the need for manual measurements is greatly reduced, while maintaining reliable phenotypic data. Although it is effective in controlled environments, additional testing in diverse real-world settings is required. In summary, this technique offers a scalable and accurate method for automated phenotyping of plants.
Recent developments, such as the Dynamic Feature Network with Point-wise Spatial Attention Network (DFN-PSAN) network, have shown remarkable precision in identifying plant diseases by utilizing deep feature fusion and attention mechanisms, which enhance both interpretability and performance. Similarly, the ITF-WPI model successfully integrates image and text features for pest detection, effectively addressing the challenges of complex agricultural settings [16]. These methods collectively highlight the increasing shift towards intelligent multimodal systems for automated plant analysis and precision farming.
This study marks the first comprehensive investigation of date palm leaf disease detection using an extensive dataset and multiple transfer learning architectures. It not only examines the efficacy of these architectures in classifying date palm leaf diseases but also compares their performance to determine the most suitable architecture for this specific application. By employing a large and diverse dataset, this study addresses the limitations of previous studies and contributes to the field by offering a scalable solution for disease detection in date palms, thereby supporting sustainable agricultural practices.
As illustrated in Fig. 1, the dataset consists of 3089 processed images (totaling 1.6 GB) of infected date palm leaves that have undergone filtering, cropping, augmentation, and classification into specific disease categories [1]. This collection can be employed to develop deep learning models for classifying infected date palm leaves, thereby facilitating early disease prevention in palm trees. The dataset was organized into nine distinct classes:
• class 1 Potassium Deficiency: Encompasses 831 augmented leaf images.
• class 2 Manganese Deficiency: Includes 415 augmented leaf images.
• class 3 Magnesium Deficiency: Contains 566 augmented leaf images.
• class 4 Black Scorch: Comprises 106 augmented leaf images.
• class 5 Leaf Spots: Features 69 augmented leaf images.
• class 6 Fusarium Wilt: Consists of 210 augmented leaf images.
• class 7 Rachis Blight: Incorporates 402 augmented leaf images.
• class 8 Parlatoria Blanchardi: Holds 101 augmented leaf images.
• class 9 Healthy Sample: Encompasses 389 augmented images of healthy date palm leaves.
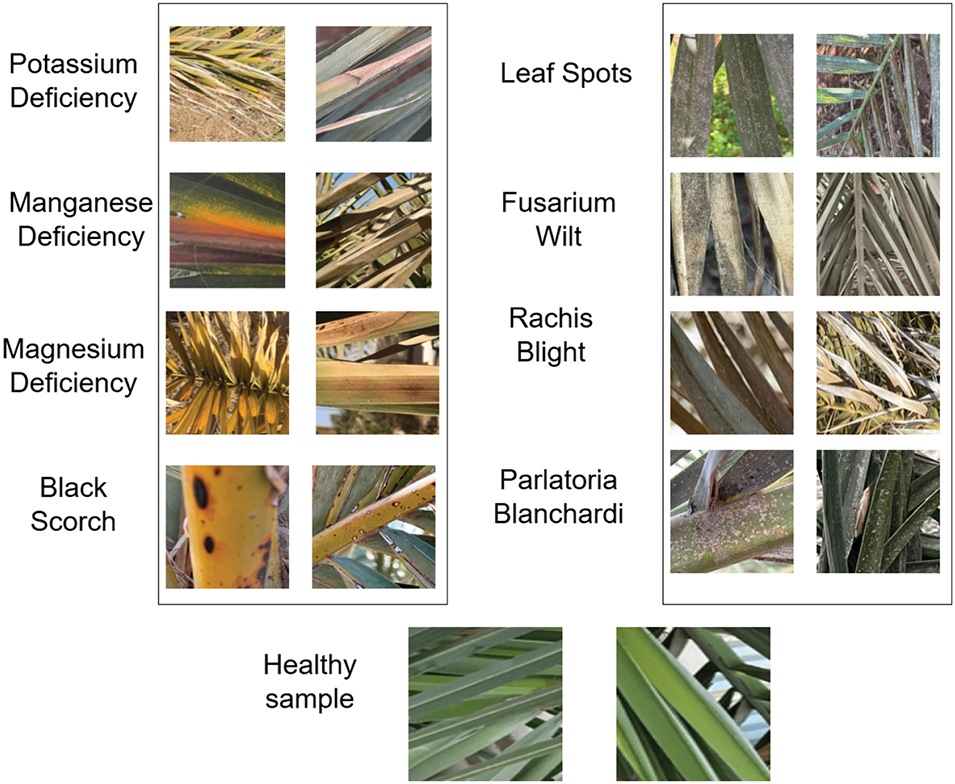
Figure 1: A sample of date palm leaf diseases/disorders and a healthy set
Notably, the dataset exhibited an imbalance with significant variations in sample sizes across different classes. To mitigate this issue, additional data augmentation techniques are implemented during the training process. We employ oversampling using the Synthetic Minority Oversampling Technique (SMOTE) [17].
The dataset employed in this study was previously processed and categorized into images of the diseased and unaffected date palm leaves. All the images were consistently resized to
To prepare the dataset for training, the pixel values were normalized by scaling to the
Finally, the dataset is randomized to prevent the model from detecting unintended patterns based on the inherent data sequence. These minimal modifications to the preprocessed dataset ensured their suitability for the augmentation and subsequent training procedures.
We employed image augmentation techniques using TensorFlow to address the issue of dataset imbalance and enhance the variety of the training data. To achieve a balanced training set and better represent the less frequent categories, approximately 30% of the images underwent augmentation, specifically targeting the minority classes. The augmentation process includes the following modifications [18]:
• Rescaling: Pixel values were standardized to the [0, 1] range by dividing them by 255. This normalization enhances the model training stability and facilitates faster convergence.
• Shear Transformation: A 0.2 shear range was implemented to geometrically alter the images, introducing perspective variations.
• Zooming: Random zoom factors up to 0.2 were applied to images, simulating differences in size and focus.
• Horizontal Flipping: Images were randomly flipped horizontally to create mirrored versions, adding orientation variations.
Dynamic augmentation was employed throughout the training process to create diverse input samples in each batch to prevent overfitting and to enhance the generalizability of the model. These augmentations were implemented with a 20% validation split, to maintain the integrity of the validation set. To address class imbalance, we employed the Synthetic Minority Oversampling Technique (SMOTE) along with image augmentation. Following the extraction of deep features from the images using a feature extractor, SMOTE was applied to the resulting feature vectors. This technique synthetically generates new samples for minority classes by interpolating existing instances, thereby balancing the class distribution without duplicating data. The combination of data augmentation and SMOTE improved the robustness and fairness of the classifier across all the disease categories in the date palm leaf dataset.
SMOTE was utilized at the feature level following the feature extraction. While this helped to balance the class distribution, future research could investigate other methods, such as class-weighted loss functions or generating synthetic images with GANs, to enhance sample diversity.
2.4 Convolutional Neural Networks and Selected Models
Convolutional Neural Networks (CNNs) represent a category of deep learning algorithms specifically engineered for image-related applications, including classification, object identification, and segmentation. These networks utilize convolutional layers to capture spatial characteristics from images, rendering them highly efficient for tasks that require pattern recognition and visual-feature extraction. The layered structure of CNNs enables them to acquire basic features, such as edges and textures, in the initial stages, progressing to more intricate features in the deeper layers.
In this study, we assessed the efficacy of three cutting-edge CNN architectures for categorizing date palm leaf images: InceptionV3, DenseNet121, and MobileNetV2. Each model has distinct advantages in terms of its effectiveness, computational efficiency, and capacity to adapt to various datasets.
To fine-tune the hyperparameters, a grid search was conducted by examining the following parameter ranges for the three models: learning rates of 0.001, 0.0005, and 0.0001, along with batch sizes of 16, 32, and 64.
Fig. 2 illustrates the InceptionV3 model, an advanced deep convolutional neural network known for its distinctive structure incorporating inception blocks. These blocks facilitate multiscale feature extraction within individual layers, thereby achieving an optimal balance between high precision and computational efficiency. In this study, we utilized a pretrained InceptionV3 model initialized with ImageNet weights. To tailor the model to the specific characteristics of the palm-leaf dataset, we unfrozen and fine-tuned the upper layers, thereby enabling the model to acquire domain-specific features effectively.
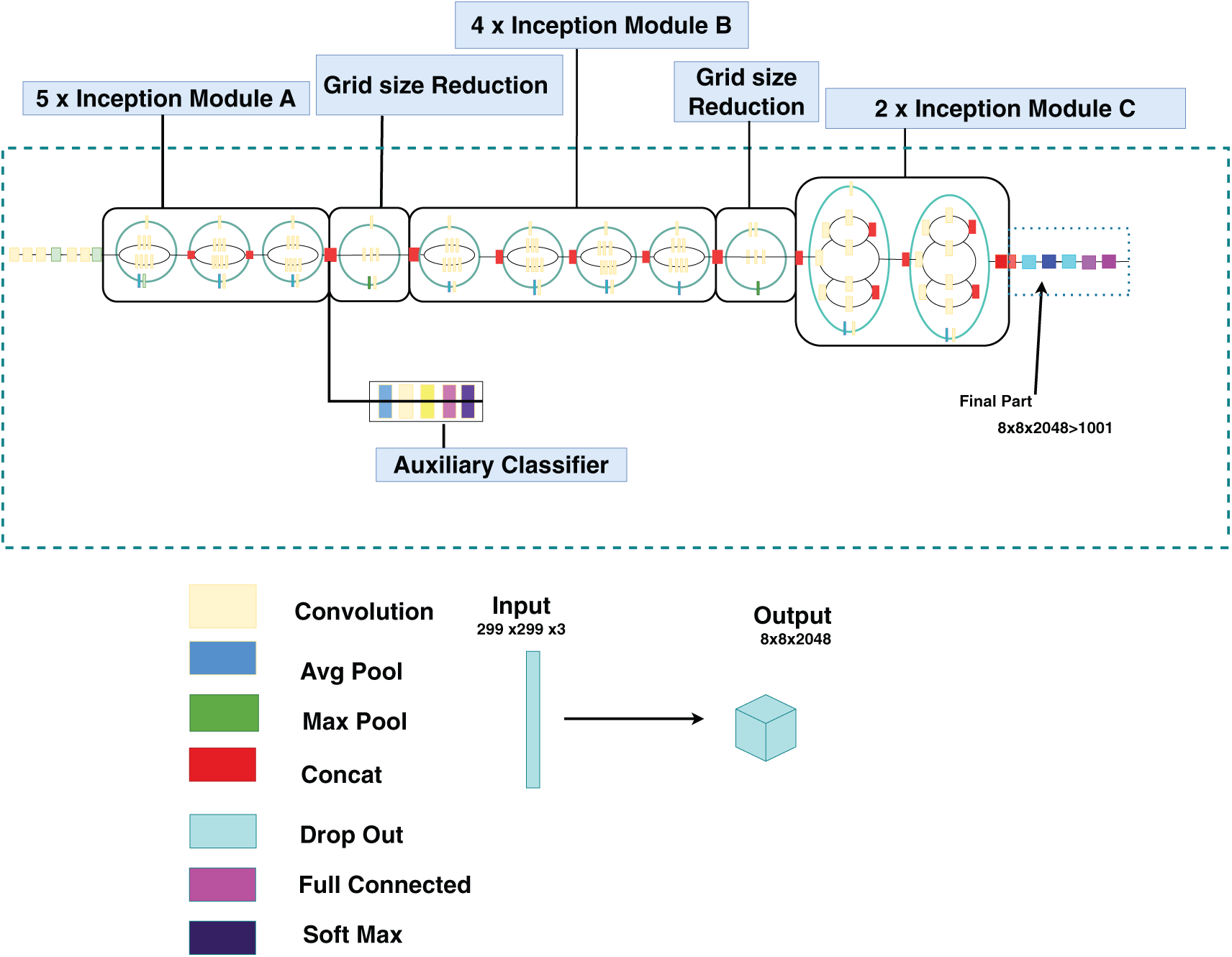
Figure 2: InceptionV3 architecture (adapted from Zorgui et al. [19])
The architecture of the model was modified by removing the last 15 layers of the InceptionV3 network, while maintaining the remaining layers intact. This approach preserves the pre-trained features from the earlier layers while allowing fine-tuning of the dataset using a modified architecture. The following components were integrated into the base model:
• GlobalAveragePooling2D: This layer diminished spatial dimensions while maintaining key features extracted by convolutional layers.
• Dense Layer: A fully connected layer comprising 512 neurons with ReLU activation was introduced to process complex input data representations.
• Dropout Regularization: To mitigate overfitting, a dropout layer with a 0.3 rate was implemented, randomly deactivating neurons during the training phase.
• Output Layer: The final dense layer, consisting of 9 neurons and employing a softmax activation function, was designed for multiclass classification corresponding to the nine disease categories.
For InceptionV3, we removed the following layers:
• Fully Connected Layers: These layers were specific to the original classification task that the model was trained on (e.g., ImageNet). The last fully connected layer, often with softmax activation for classification, is removed.
• Global Average Pooling (GAP) Layer: This layer was removed or modified depending on the task.
After removing these layers, a new fully connected layer is added to match the number of classes for the new task.
To address class imbalance, SMOTE was employed following the feature extraction from the training dataset. This method generates synthetic samples for underrepresented classes and improves the ability of the model to generalize across all categories. The optimal training parameters were determined by hyperparameter tuning. A grid search exploring various learning rates and batch sizes revealed that peak performance was achieved with a learning rate of 0.0005 and a batch size of 16. The performance of the model was assessed using 5-fold Stratified Cross-Validation to ensure a balanced class representation across all folds. The Adam optimizer and categorical cross-entropy loss functions were used for model compilation and training. The model that exhibited the highest validation accuracy during the cross-validation was retained. This comprehensive approach facilitated the effective adaptation of a robust pretrained model to the task of leaf disease classification while minimizing overfitting and addressing class imbalance issues.
DenseNet121 is a sophisticated convolutional neural network architecture that enhances feature utilization through direct connections between each layer and all subsequent layers in a feed-forward manner. This interconnected design maximizes information flow across the layers, reduces the parameter count, and addresses the vanishing gradient problem. In this study, a pretrained DenseNet121 model with ImageNet weights was adapted for the palm leaf dataset. The pre-existing layers were kept frozen to maintain their learned representations, whereas new layers were introduced to tailor the model for a specific task. Fig. 3 illustrates this modified architecture.
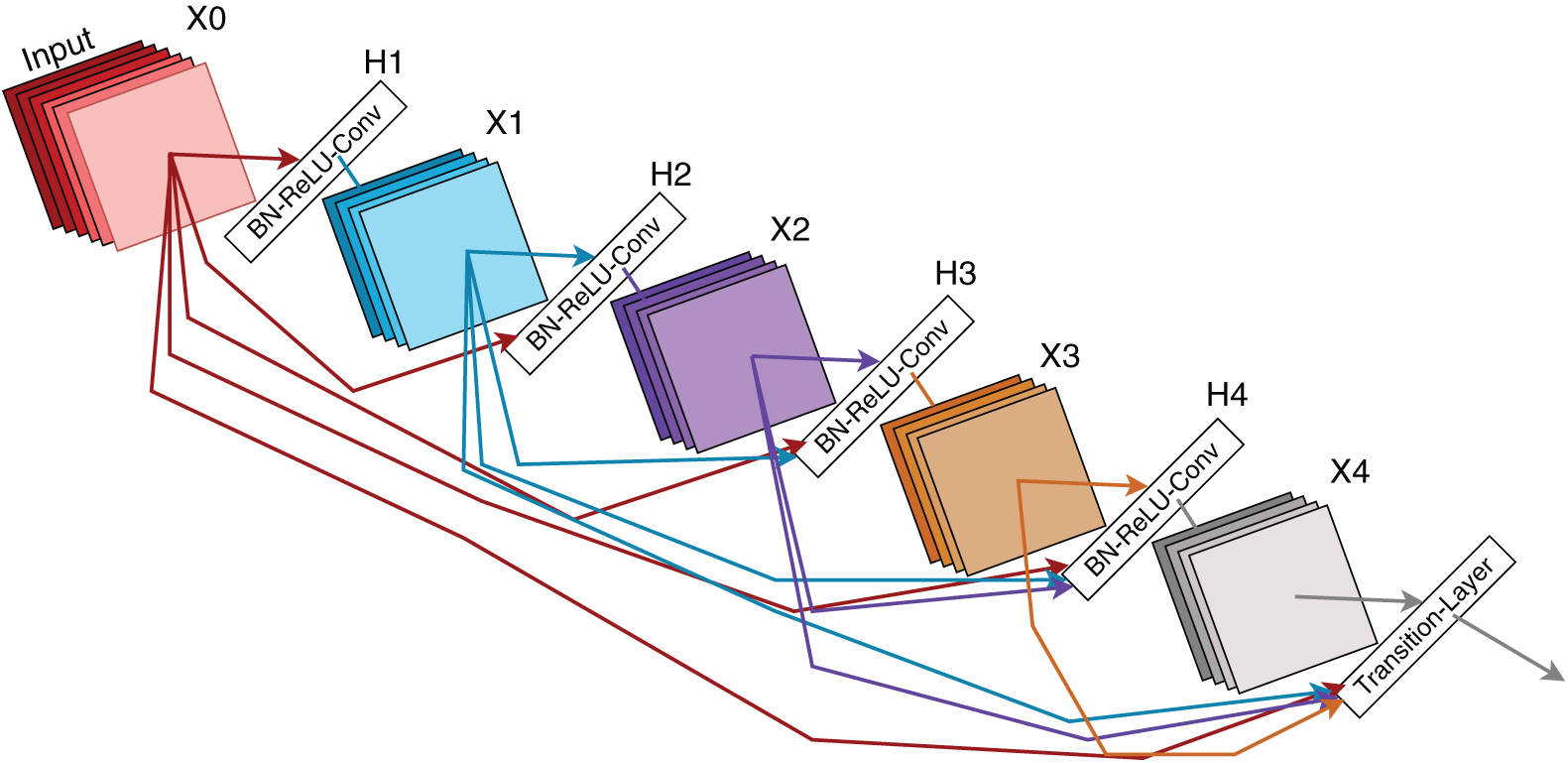
Figure 3: DenseNet121 architecture (adapted from Huang et al. [20])
Architectural Modifications: The following changes were made to the DenseNet121 model:
• A GlobalAveragePooling2D layer was applied to the pre-trained DenseNet121 output, reducing spatial dimensions while preserving key features.
• A Flatten layer transformed the pooled feature maps into a one-dimensional vector.
• A fully connected Dense layer with 512 units and ReLU activation was incorporated to facilitate learning of complex dataset patterns.
• Dropout regularization with a 0.3 rate was implemented after the dense layer to combat overfitting.
• A final Dense layer with 9 units (matching the class count) and a softmax activation function was used for multiclass classification.
For DenseNet121, we removed the following layers:
• Fully Connected Layers: DenseNet has a dense block structure followed by a classifier layer (fully connected layer). The last fully connected layer was removed to suit the new task.
• Classification Layer: The original output layer used for classification was removed and replaced with a new layer that matches the target class count for fine-tuning.
• In some cases, the Global Average Pooling (GAP) Layer at the end was removed depending on the task.
DenseNet uses the growth rate and dense blocks to retain feature reuse; therefore, the focus during fine-tuning is primarily on modifying the final classifier layers to adapt to the new task.
Training Approach:
The DenseNet121-based model was trained using a stratified 5-fold cross-validation approach. Before classification, feature maps from DenseNet121 were extracted and rebalanced using SMOTE to address the class imbalance. Optimal hyperparameters were identified through hyperparameter tuning.
• Learning Rate: 0.0005
• Batch Size: 16
Each fold utilizes an identical architecture compiled using the Adam optimizer and categorical cross-entropy loss. To prevent overfitting, dropout and L2 regularization were employed during training. The class weights were calculated dynamically based on label frequencies to further combat class imbalances.
The model with the best performance across folds was retained for subsequent evaluation. The robustness of the model and its generalization abilities for disease classification were confirmed through various evaluation metrics including accuracy, confusion matrix, and classification reports. The modified DenseNet121 architecture exhibited notable performance enhancements by utilizing its dense connectivity and efficient gradient flow to effectively classify complex visual patterns.
MobileNetV2 is a streamlined CNN architecture designed for deployment in mobile and edge devices. It utilizes depth-wise separable convolutions and linear bottleneck layers to reduce the computational requirements while maintaining high accuracy. MobileNetV2 provides a balanced performance in terms of speed and accuracy, which makes it suitable for swift inference on resource-constrained hardware [21], as illustrated in Fig. 4.
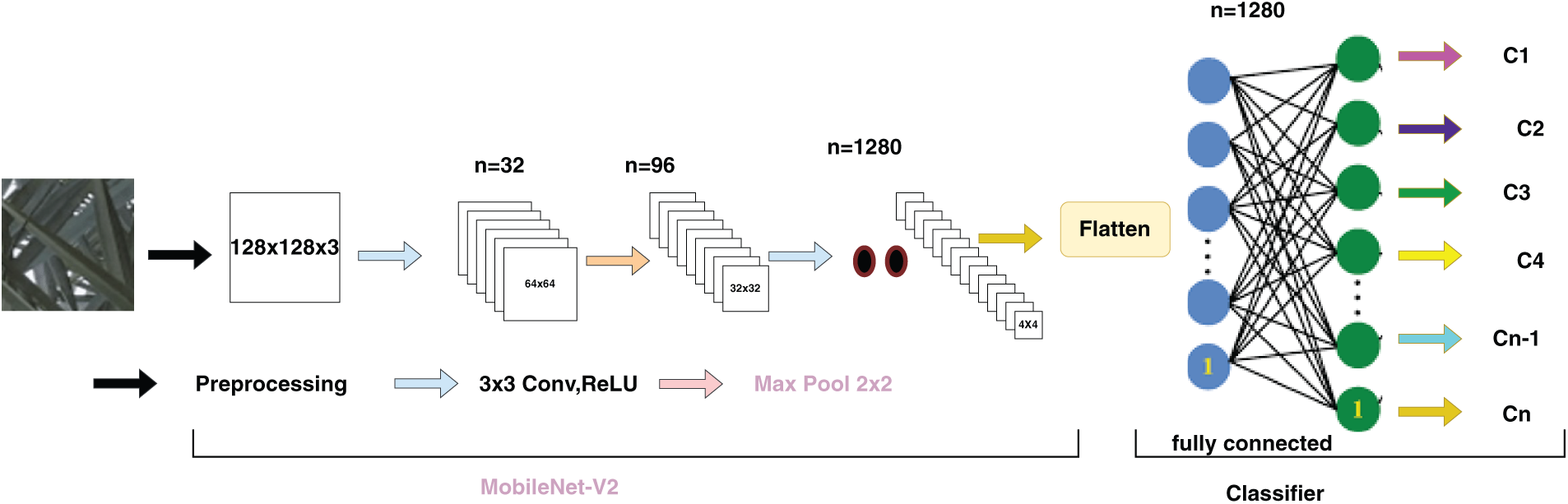
Figure 4: MobileNetV2 architecture (adapted from Golcuk et al. [22])
In our study, we utilized MobileNetV2 as a feature extraction tool, leveraging its pretrained weights from ImageNet. We removed the final layers responsible for classification, retaining only the convolutional foundation to facilitate the transfer learning.
Following feature extraction, a Global Average Pooling (GAP) layer reduces the spatial dimensions of the feature maps, followed by a flattened layer that transforms the pooled features into a one-dimensional array. A fully connected (dense) layer with 512 units and ReLU activation were then applied to learn more intricate patterns from the extracted features.
The SMOTE technique is employed to address the issue of class imbalance. This method generates artificial samples for underrepresented classes using extracted feature vectors, thereby ensuring a balanced distribution across all classes.
For MobileNet, we removed the following layers:
• Fully Connected Layers: Similar to other CNNs, we removed the last fully connected layer(s) and replaced them with a new one designed for the target task.
• Global Average Pooling (GAP) Layer: Although the GAP layer is generally kept for fine-tuning, we adjusted it in some cases to better suit the specific task.
We adopted a 5-fold Stratified Cross-Validation approach to ensure robust performance evaluation across imbalanced classes. The best hyperparameters were obtained through tuning:
• Learning Rate: 0.0005
• Batch Size: 16
The model was trained using the Adam optimizer and categorical cross-entropy loss functions, which are ideal for multiclass classification. The performance was evaluated for each fold using the accuracy, classification reports, and confusion matrices.
The best-performing model across the five folds was saved for final evaluation and deployment. This approach ensures a balanced and generalizable classifier that is capable of robust disease classification across all classes.
MobileNetV2 is designed to be lightweight; therefore, fewer modifications are required for its structure compared with larger models, resulting in fewer layers being removed or adjusted during fine-tuning.
Assessing these models offers valuable insights into their effectiveness in categorizing date palm leaf diseases. Each model underwent fine-tuning and transfer learning processes to adjust its pre-existing weights to a particular dataset, capitalizing on its inherent capabilities for this specific application.
2.5 Model Training and Evaluation
This section describes the setup and methods used for training and assessing the convolutional neural network (CNN) models. This includes model preparation, training protocols, assessment criteria, and technical specifications.
The Adam optimizer was used to compile the models, employing a learning rate of 0.0005 to ensure an effective gradient-based optimization. For effective multiclass classification, categorical cross-entropy is selected as the loss function [23]. The primary metric for evaluating the model performance was accuracy.
The models were trained using batches of 16 samples over 20 epochs, striking a balance between sufficient learning and overfitting. Class imbalance was addressed by applying SMOTE. The model that exhibited the best performance across all folds was retained for subsequent analysis.
The effectiveness of the model was assessed using stratified 5-fold cross-validation, which preserved the class distribution across all the folds. In each iteration, four folds were used for training and the remaining fold served as the validation set. Accuracy served as the primary metric for evaluating performance. Upon the completion of all folds, the average and standard deviation of the validation accuracies were computed to gauge the generalizability of the model. Furthermore, the top-performing model was retained, and its predictions were used to create a classification report and a confusion matrix.
The training framework implemented a specialized technique for preserving the most effective model, despite not explicitly using conventional callback functions such as early stopping and model checkpointing [24]. Throughout the cross-validation process, the system tracked the validation accuracy for each fold and saved the model that exhibited the highest accuracy across all folds for final evaluation. This method ensures that only the most successful model is maintained for eventual deployment, effectively capturing the best performance without relying on the traditional callback mechanisms.
This study was performed using Google Colab’s cloud-based platform, which features an NVIDIA T4 GPU. This powerful graphics processing unit allowed for efficient management of the extensive dataset and enhanced the speed of training the convolutional neural network (CNN) models. By leveraging Colab, researchers were able to conduct experiments smoothly without the need for significant local computing resources, making it an optimal choice for deep-learning investigations and advancements.
These methodologies ensure systematic training and evaluation of the models, resulting in dependable and replicable outcomes.
An 80–20 ratio was employed to separate the dataset into the training and validation portions. The training segment consisted of 2474 images spread across nine classes, while the validation segment included 615 images. This distribution ensured that all the classes were adequately represented in the assessment.
3.2 Model Training Performance Using InceptionV3
The InceptionV3 architecture was trained using 5-fold cross-validation with a learning rate of 0.0005. The training process was aimed at ensuring robust generalization across the different subsets of the dataset. The performance of the model across all folds can be summarized as follows:
• Cross-Validation Accuracies: [91.97%, 92.80%, 95.56%, 93.89%, 96.15%]
• Mean Accuracy: 94.08%
• Standard Deviation of Accuracy: 1.27%
The robustness of the learning ability of the model and generalization is evidenced by the consistent outcomes observed across various folds, suggesting reliable performance.
3.2.1 Validation Classification Report
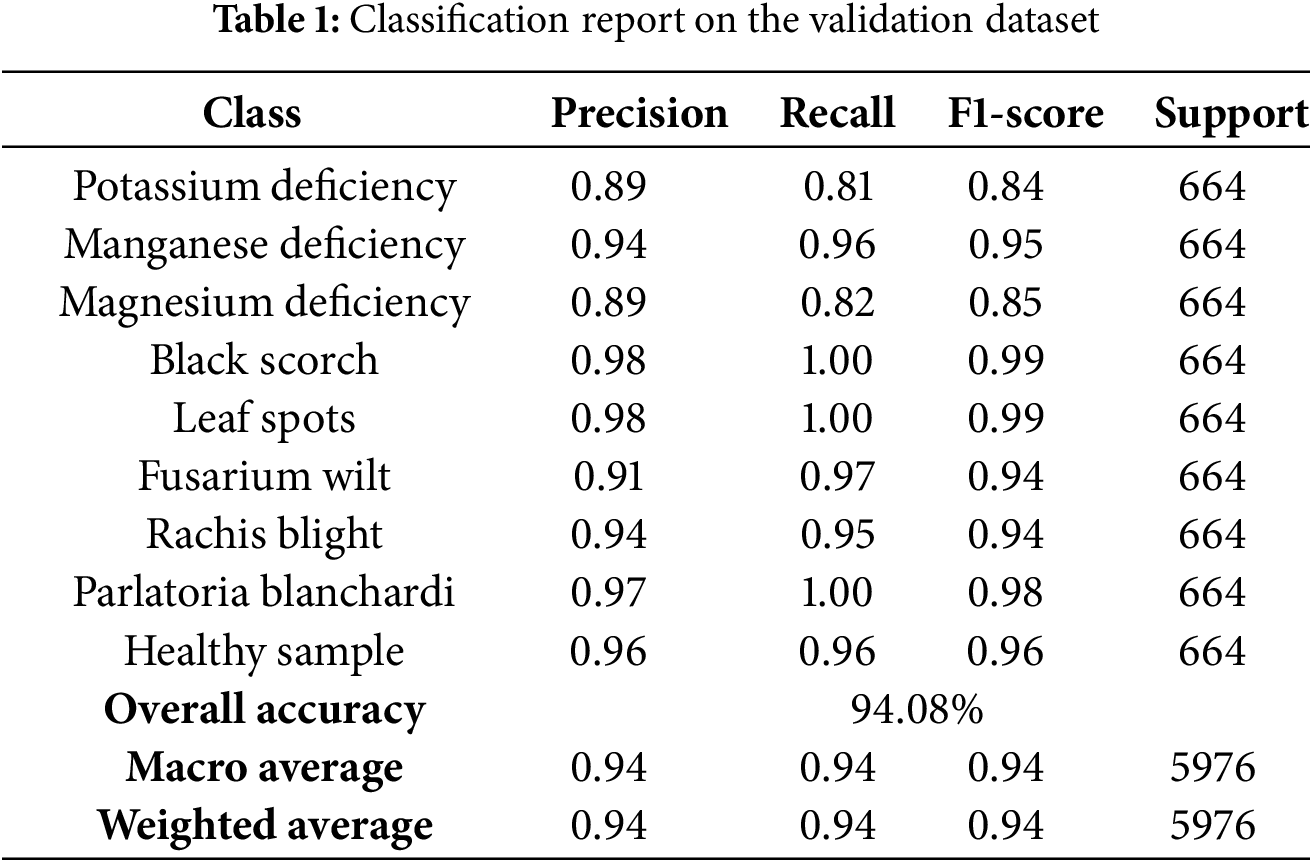
To thoroughly assess the ability of the model to classify, various performance metrics were calculated for each category, including precision, recall, F1-score, and support. A comprehensive classification report derived from the validation data across all folds is presented in Table 1.
3.2.2 Confusion Matrix and ROC curves
Fig. 5a shows a confusion matrix that visually depicts the classification accuracy of the model by comparing the actual and predicted labels across all the nine categories. Furthermore, Fig. 5b shows the ROC curves that offer an in-depth perspective of the capability of the model to differentiate between classes, emphasizing its sensitivity and specificity at different threshold levels.
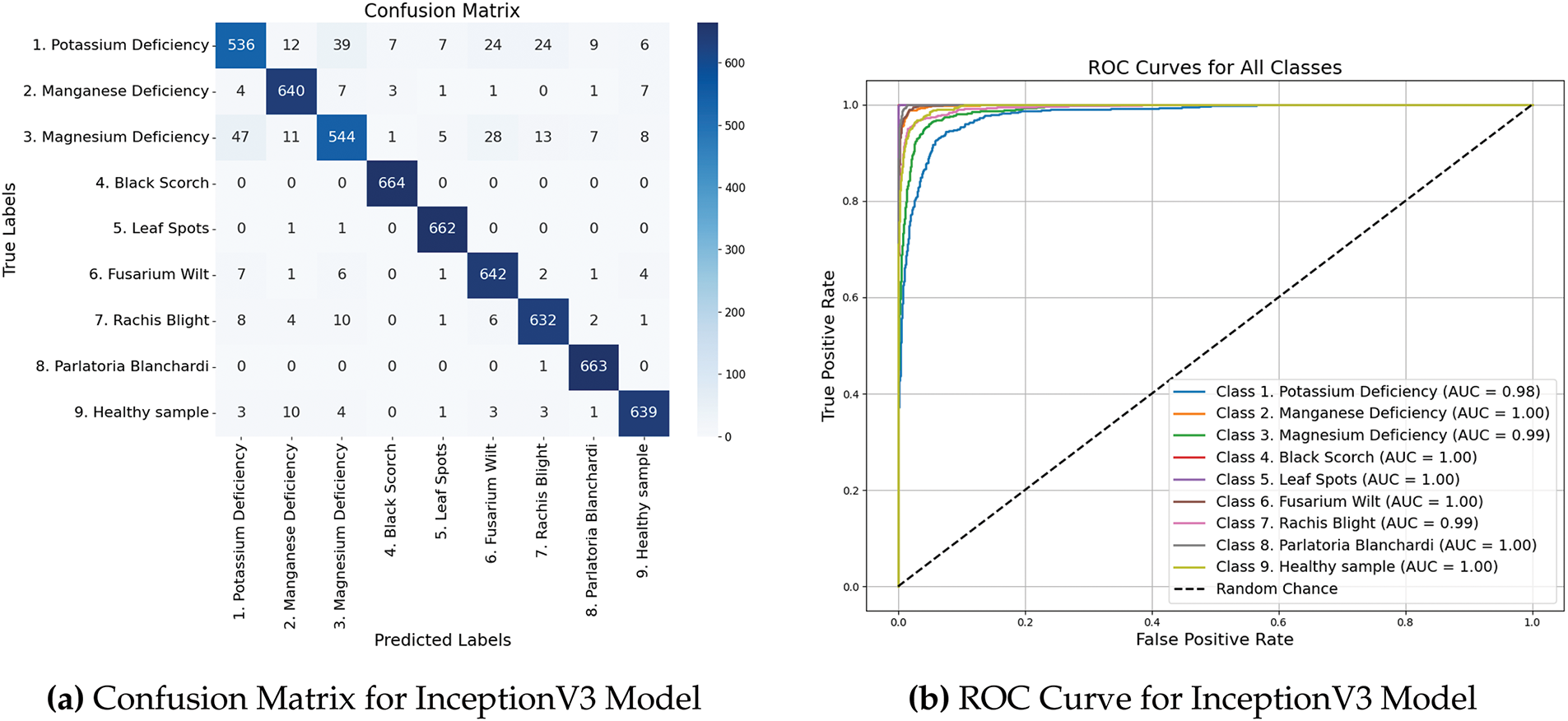
Figure 5: Performance metrics and ROC curves of InceptionV3 model
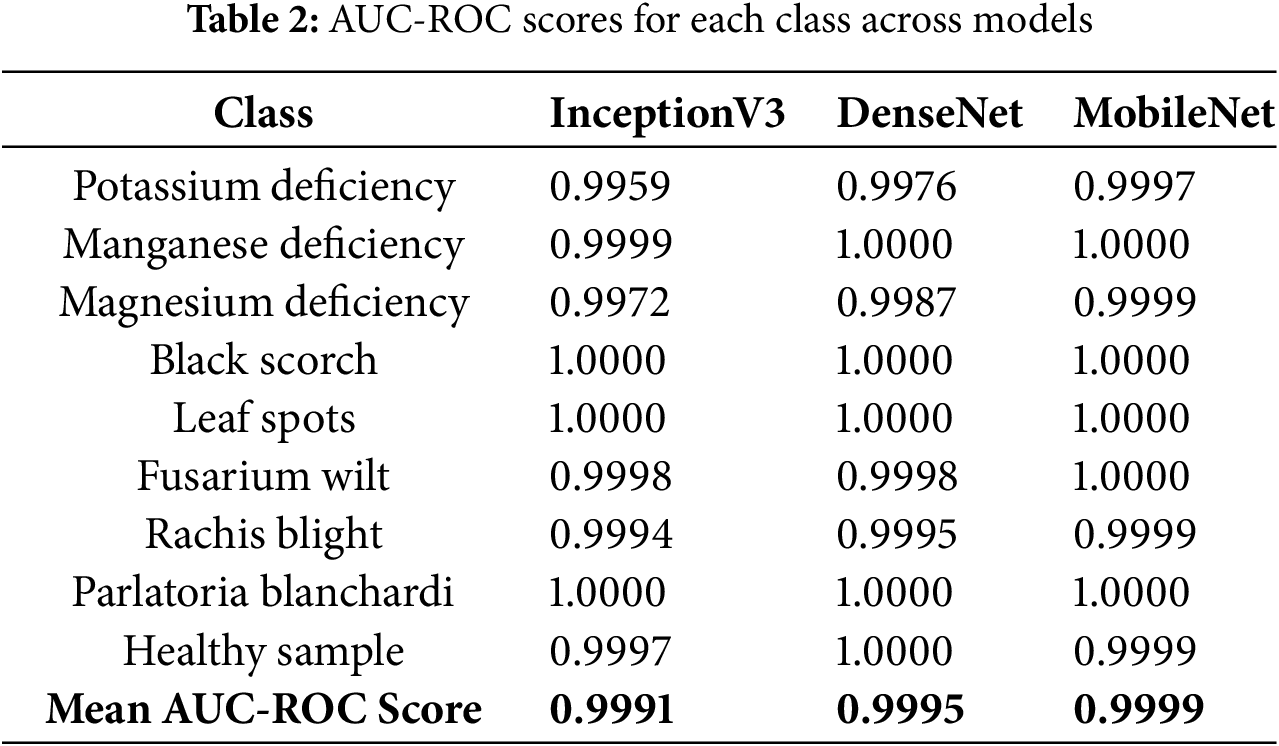
The AUC-ROC scores across all categories remained consistently high, with an average AUC-ROC score of 0.9991, demonstrating excellent category separability. Furthermore, the model attained a Cohen’s Kappa score of 0.9625, indicating a strong correlation between the predicted and actual classifications, which further underscores its dependability. Table 2 presents a comprehensive breakdown of the AUC-ROC scores for each category.
• The InceptionV3 model demonstrated exceptional performance across all categories, with notably high F1-scores exceeding 0.98 in classes such as Black Scorch, Leaf Spots, and Parlatoria Blanchardi.
• Some misclassification was evident in categories like Potassium Deficiency (0.81 recall) and Magnesium Deficiency (0.82 recall), potentially due to visual similarities with other deficiencies or intra-class variations.
• The stability of the model and consistent generalization ability are indicated by the minimal standard deviation observed across the fold accuracies.
• The effectiveness of the model in handling multiclass classification with inter-class variability was confirmed by its balanced performance across the precision, recall, and F1-score metrics.
3.3 Model Training Performance Using DenseNet121
The DenseNet121 model was trained utilizing a 5-fold cross-validation approach. The following accuracies were recorded across various folds during the cross-validation process:
• Fold 1 Accuracy: 95.90%
• Fold 2 Accuracy: 96.40%
• Fold 3 Accuracy: 97.41%
• Fold 4 Accuracy: 97.32%
• Fold 5 Accuracy: 95.06%
Mean Cross-Validation Accuracy: 96.42%
Standard Deviation of Accuracy: 1.25%
These consistently high values reflect the robust learning and generalization capabilities of the model across different data splits.
3.3.1 Validation Classification Report
To assess the effectiveness of the model on the validation set, performance metrics such as precision, recall, F1-score, and support were calculated per class. Table 3 summarizes the results.
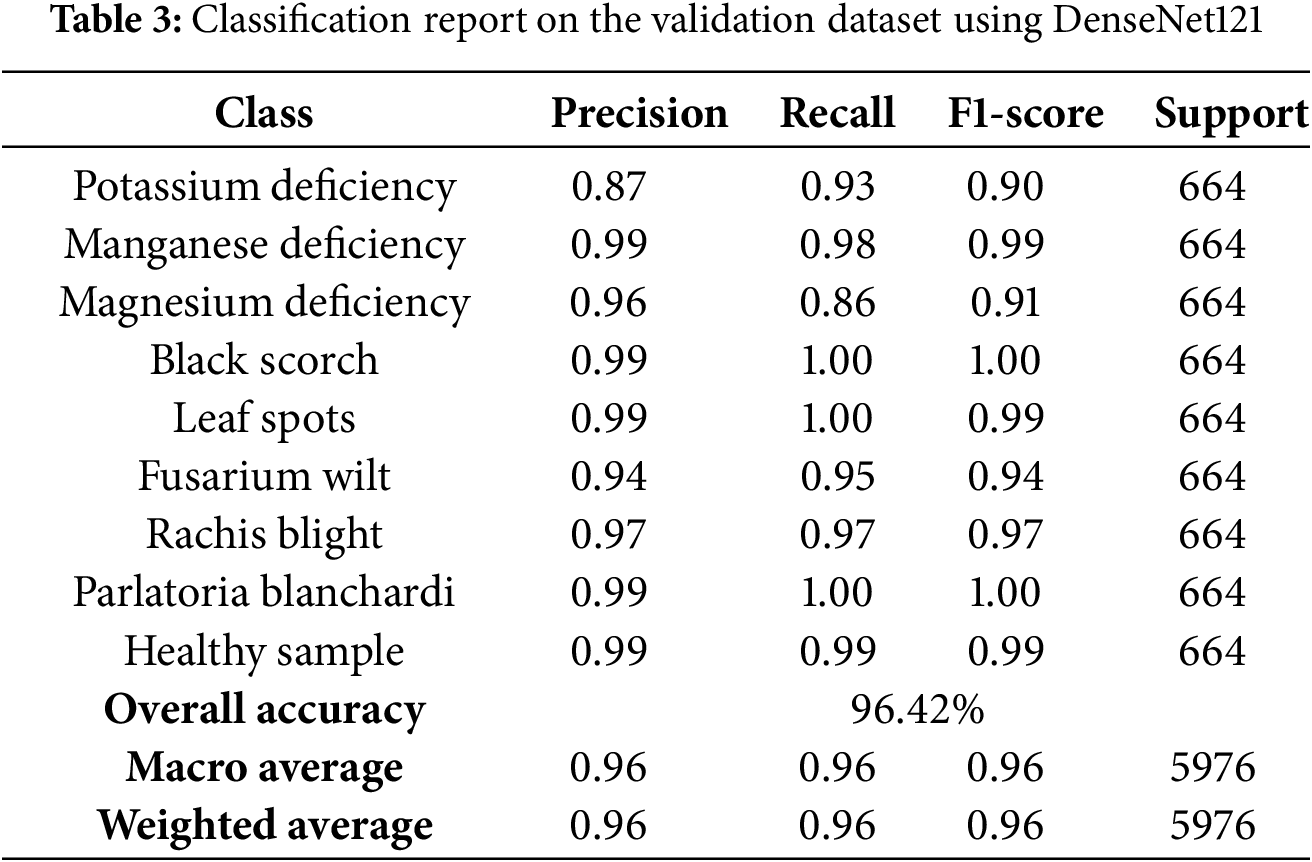
Fig. 6a presents the confusion matrix, which provides a visual representation of the classification behavior of the model across different classes. Fig. 6b shows the ROC curves that offer an in-depth perspective on the capability of the model to differentiate between classes, emphasizing its sensitivity and specificity at different threshold levels.
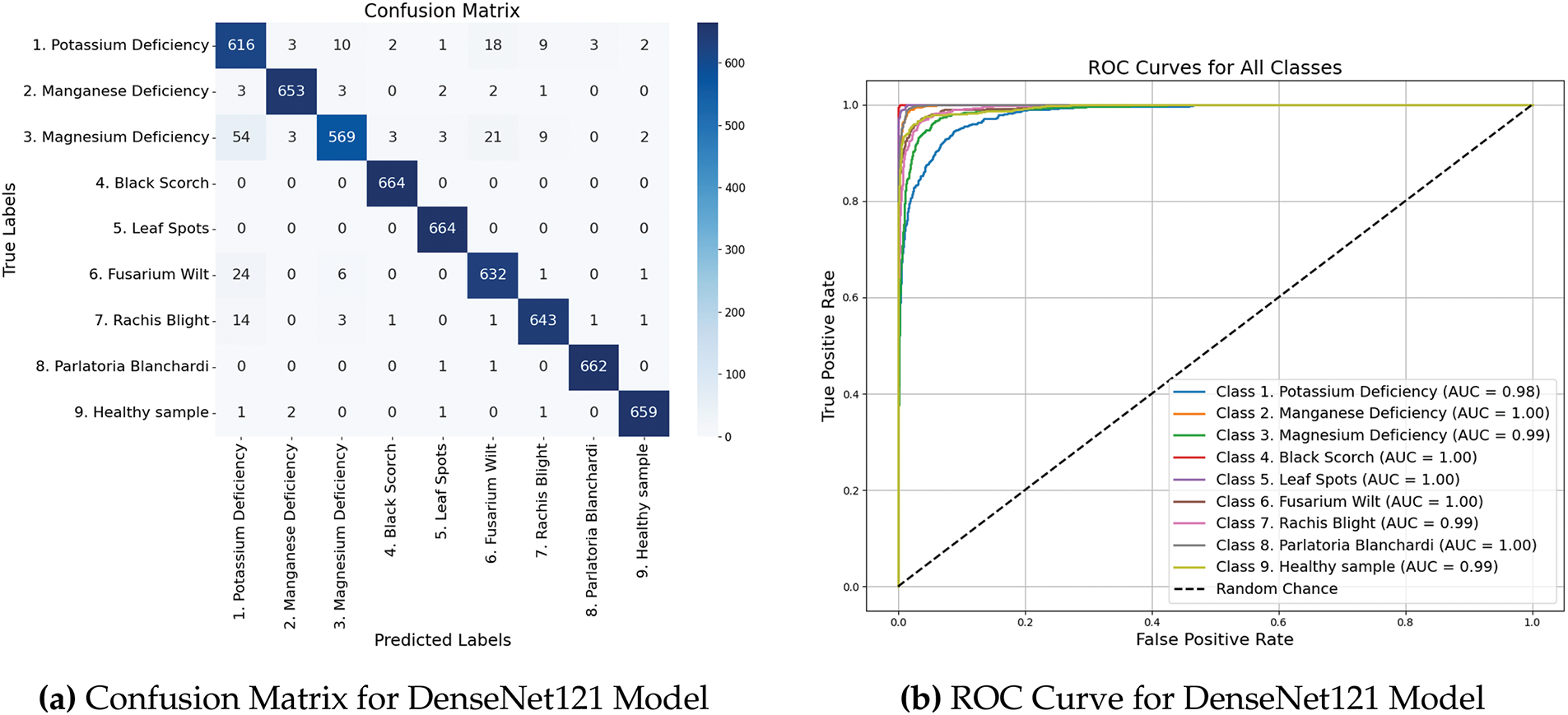
Figure 6: Performance metrics and ROC curves of DenseNet121 model
The AUC-ROC scores across all categories remained consistently high, with an average score of 0.9995, indicating the model’s excellent capability to differentiate between various classes. Moreover, Cohen’s Kappa score of 0.9746 further validated the reliability of the model by showing a strong agreement between the actual and predicted labels. Table 2 summarizes the AUC-ROC scores for each category.
• The DenseNet121 architecture exhibited exceptional results, with a mean cross-validation accuracy of 96.42%. This uniformity across different folds underscores the robustness of the model and its capacity to extract the pertinent features.
• Precision, recall, and F1-scores surpassed 0.90 for all categories, with certain classes such as Black Scorch, Leaf Spots, and Parlatoria Blanchardi attaining near-perfect scores. This highlights the robustness of the model’s ability to differentiate between classes even when faced with class diversity.
• DenseNet121 displayed enhanced generalization capabilities compared to InceptionV3, potentially due to its densely connected structure. This architecture promotes feature reuse and addresses the issue of vanishing gradient.
3.4 Model Training Performance Using MobileNet
A 5-fold cross-validation approach was employed to train the MobileNet. The accuracy results achieved for each fold are as follows:
• Fold 1 Accuracy: 97.32%
• Fold 2 Accuracy: 97.41%
• Fold 3 Accuracy: 97.41%
• Fold 4 Accuracy: 96.07%
• Fold 5 Accuracy: 96.74%
Mean Cross-Validation Accuracy: 96.99%
Standard Deviation of Accuracy: 1.30%
The consistently high values observed across all folds demonstrate the ability of MobileNetV2 to effectively generalize and maintain stability throughout the training process.
3.4.1 Validation Classification Report
Table 4 outlines the performance of the MobileNetV2 model on the validation set, including the class-wise precision, recall, and F1-scores.
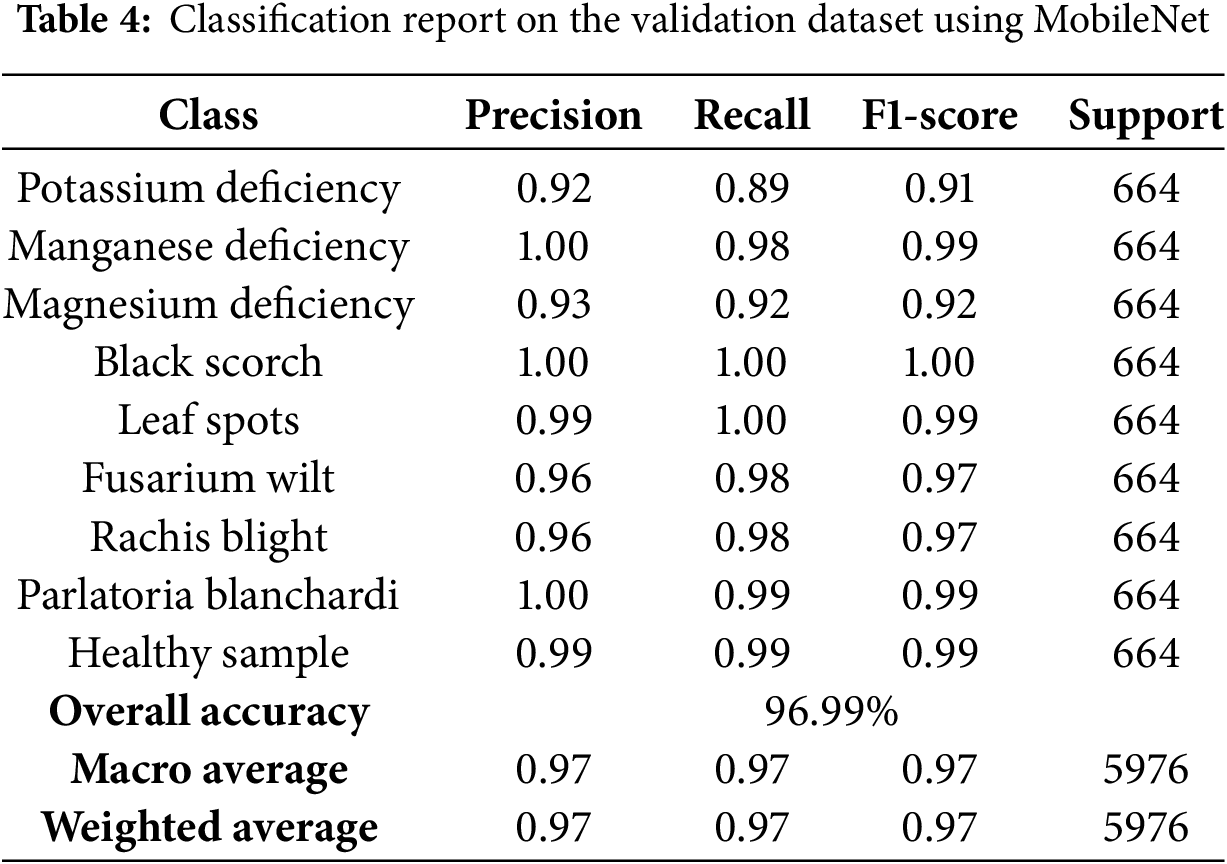
Fig. 7a shows the confusion matrix generated from the validation results, visually representing classification performance of MobileNet. Fig. 7b shows the ROC curves that offer an in-depth perspective on the capability of the model to differentiate between classes, emphasizing its sensitivity and specificity at different threshold levels.
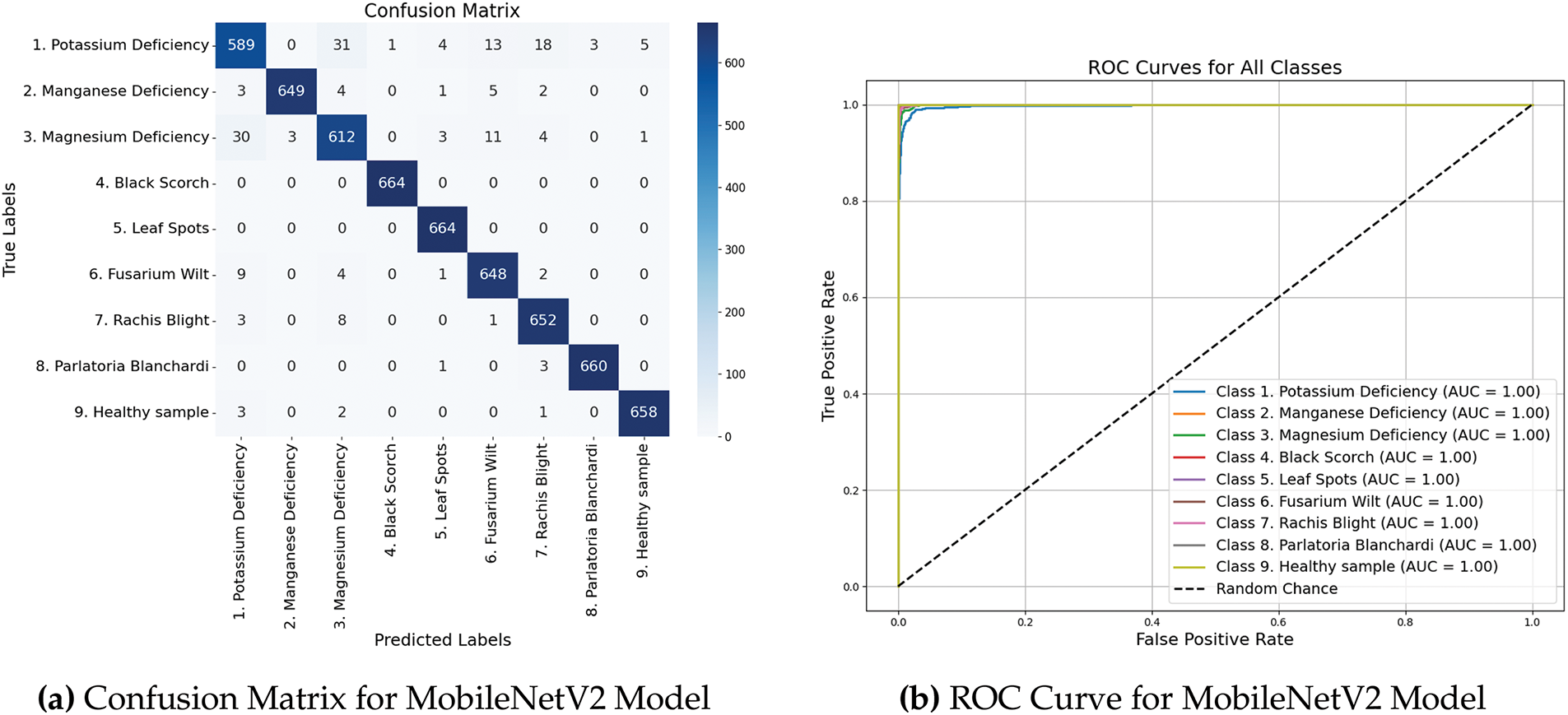
Figure 7: Performance metrics and ROC curves of MobileNetV2 model
The model demonstrated a robust capability to accurately classify various categories, as evidenced by the high AUC-ROC scores across all classes, with an average AUC-ROC score of 0.9999. Additionally, Cohen’s Kappa score of 0.9932 highlights a strong agreement between the predicted and actual labels, underscoring the dependability of the model. Table 2 summarizes the AUC-ROC scores for each class.
• The MobileNetV2 model achieved a strong mean cross-validation accuracy of 96.99%, demonstrating its efficiency and suitability for lightweight, high-accuracy tasks.
• The model maintained consistently high scores across all classes, with particularly impressive results for Black Scorch, Leaf Spots, and Parlatoria Blanchardi classes, achieving nearly perfect metrics.
• MobileNeV2t is well-suited for resource-constrained environments due to its compact architecture while still delivering competitive performance compared to heavier models such as DenseNet121 and InceptionV3.
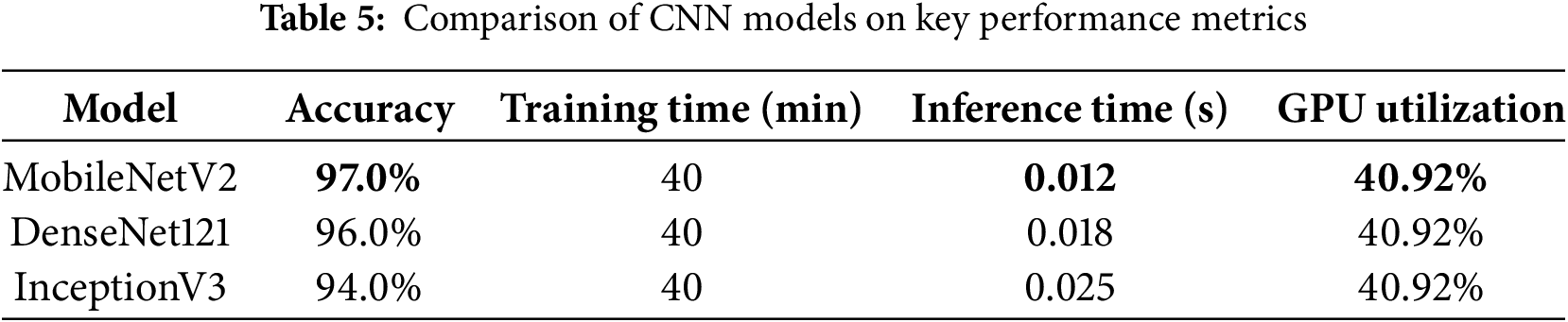
The experimental setup for training MobileNet, DenseNet121, and InceptionV3 was identical, utilizing SMOTE augmentation, 5-fold cross-validation, and consistent hyperparameters. All models underwent training on the same dataset division, utilizing identical hyperparameters, which included batch size of 16 and 20 training epochs. The training process was performed on Google Colab with an NVIDIA T4 GPU to ensure uniform runtime conditions for all models. On average, each model took approximately 40 min to complete training under these standardized conditions. The compact design of MobileNetV2 excelled in terms of speed and memory efficiency, making it ideal for deployment in environments with limited resources. Nevertheless, this streamlined architecture occasionally resulted in a slightly lower accuracy compared with the more intricate DenseNet121 and InceptionV3 models. Table 5 presents a comparison of CNN. Although these latter models demand more computational power, they deliver enhanced classification accuracy. When selecting a model for practical agricultural applications, it is essential to consider the trade-off between the efficiency and precision.
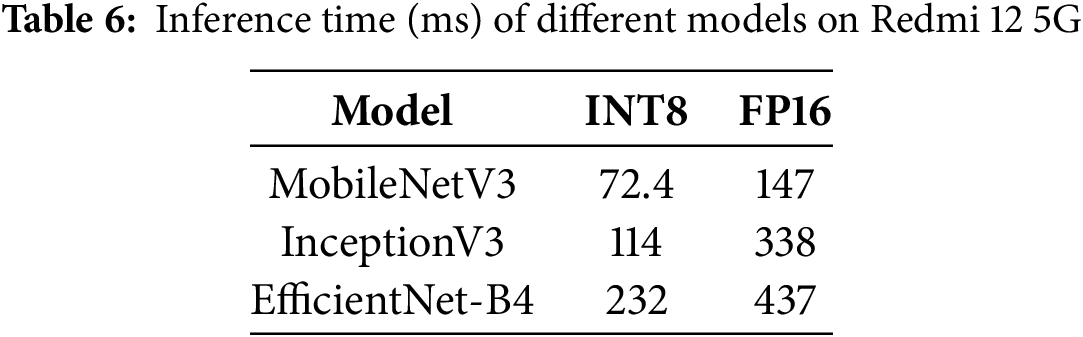
To assess the real-time practicality of various deep learning models, we tested their inference speeds on a Redmi 12 5G smartphone (Qualcomm Snapdragon 4 Gen 2, 4 GB RAM) using AI Benchmark. Since MobileNetV2 was not available in the benchmarking tool, we conducted evaluations using MobileNetV3, which recorded the shortest inference times across different precision formats (INT8, FP16) compared to other architectures, as shown in Table 6. During training, we monitored GPU utilization using built-in TensorFlow/Keras functions in Google Colab. The final GPU utilization check shows 40.92% usage, which indicates that the computational resources are not fully utilized during training. While MobileNetV3 introduces architectural enhancements for improved efficiency, our training experiments with MobileNetV2 indicate that it is also a suitable choice for mobile-based deep learning applications, balancing speed and accuracy effectively.
Heatmap Visualization
To gain a clearer insight into which parts of the input leaf images the model emphasized during classification, we utilized a heatmap visualization method using the MobileNetV2 model, as shown in Fig. 8a. The highlighted regions indicate the focus areas of the model for an accurate classification. This approach helps to pinpoint crucial areas that influence the predictions of the model, thereby improving the interpretability and reliability of the system.
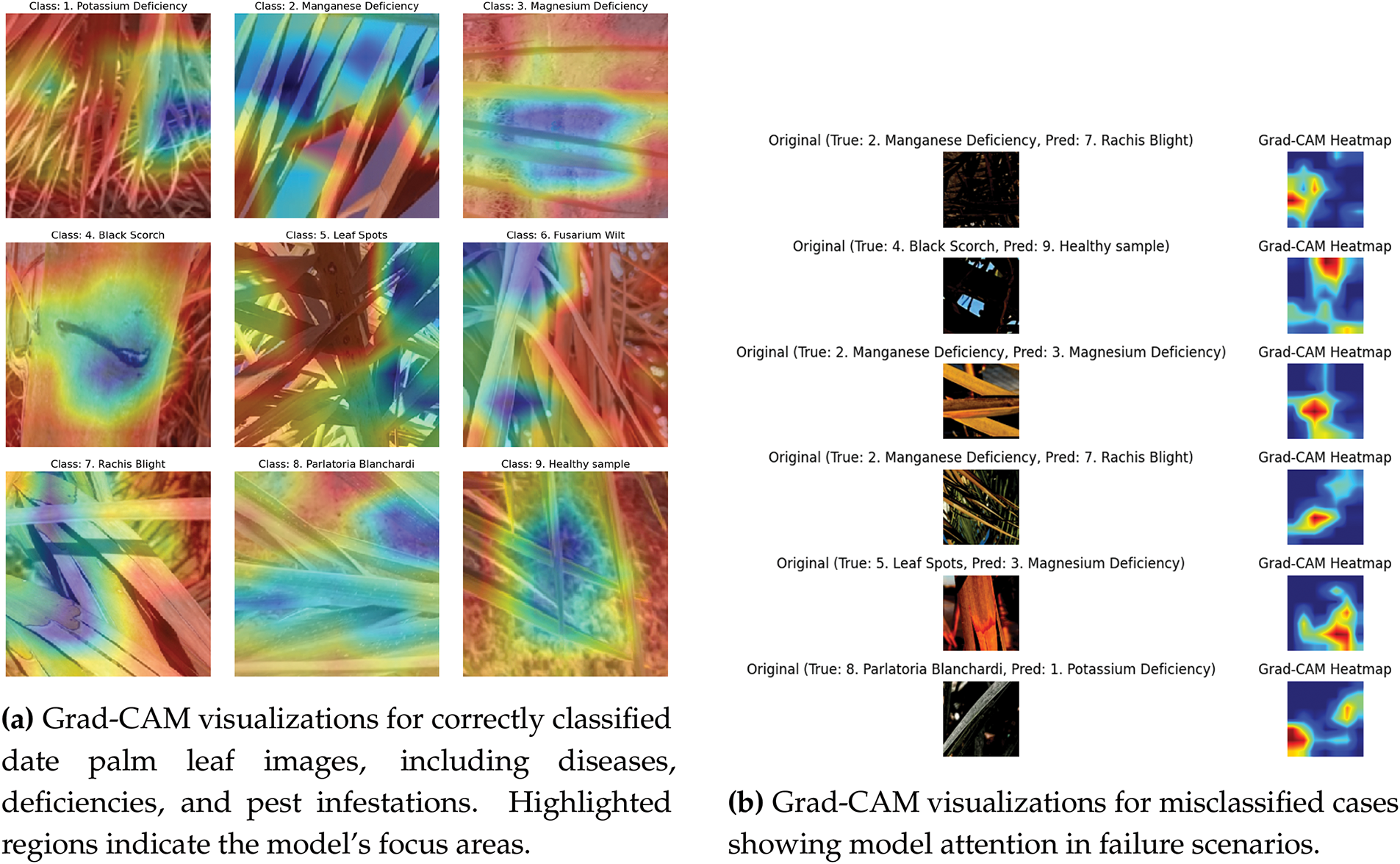
Figure 8: Grad-CAM based attention visualization of the model’s prediction behavior on date palm leaf images
To enhance model interpretability, we analyzed the failure cases using Grad-CAM visualizations, as shown in Fig. 8b. These visualizations revealed that the model occasionally misfocused on non-discriminative regions, leading to misclassification. This analysis provides insights into potential areas for improvement, such as refining feature extraction or incorporating attention mechanisms for better localization of disease-specific patterns.
This research explored the effectiveness of three CNN models, MobileNet, DenseNet121, and InceptionV3, in detecting and categorizing diseases in date palm leaves. Each performance of the model was assessed based on accuracy, precision, recall, computational efficiency, and their capability to manage class imbalance to tackle the dataset’s inherent imbalance, the Synthetic Minority Over-sampling Technique (SMOTE) was utilized to create synthetic samples for underrepresented classes, which significantly enhanced the classification of minority classes and reduced bias. In addition, hyperparameter tuning was conducted for all three models using a grid search, and the optimal batch size and learning rate were identified as 16 and 0.0005, respectively. Cross-validation with 5-fold stratified splitting was performed to assess the generalizability and robustness of the model.
To thoroughly assess model stability, we presented the Mean and Standard Deviation of accuracy across all five folds. This evaluation sheds light on the variability in model performance, highlighting its consistency across various data partitions. By incorporating standard deviation, we provide a more dependable evaluation of the robustness and generalizability of the model.
Although confusion matrices were employed to illustrate classification performance, a more detailed examination of errors showed that Potassium Deficiency and Magnesium Deficiency were sometimes mistaken for one another. This confusion might stem from the visual resemblance of symptoms, such as leaf discoloration, particularly when the lighting conditions vary. Future research should explore the integration of domain-specific visual indicators or expert annotations to enhance the distinction between classes.
Although this study incorporates heatmaps for the MobileNetV2 model to highlight class-specific regions, a more in-depth examination of the focus of the model on disease-related areas could improve its interpretability. The heatmaps produced for the MobileNetV2 model were reviewed by a plant pathologist for expert validation. The specialist confirmed that the highlighted sections in six of the nine images accurately matched the diseased area, and the other three (Manganese Deficiency, Magnesium Deficiency, and Black Scorch) were slightly inaccurate, demonstrating that the model effectively learned pertinent visual features. This validation enhanced the interpretability of the model and its potential application in real-world plant disease diagnostics.
We explored the effects of various fine-tuning methods by evaluating both full fine-tuning and feature-extraction-only techniques. In the feature extraction scenario, the pretrained weights remained unchanged, with only the classifier layers being trained, whereas in full fine-tuning, the entire network underwent updates. Our findings revealed that full fine-tuning achieved slightly better accuracy, but demanded considerably more training time and was more susceptible to overfitting, particularly with smaller datasets. On the other hand, feature extraction strikes a balance between performance and computational efficiency, making it a more suitable option for lightweight deployment.
In our study, MobileNetV2 exhibited a notably lower number of parameters (2.26M) when compared to InceptionV3 (21.80M) and DenseNet121 (7.04M). This makes it a more efficient option for real-time applications, especially for use on mobile devices where computational resources are constrained.
• InceptionV3: Showed strong performance by capturing multi-scale features through inception modules. It effectively balances the accuracy and resource efficiency, making it suitable for mid-range hardware environments.
• DenseNet121: Achieved high precision and recall scores, leveraging its dense connectivity to promote feature reuse and efficient gradient flow. It performed particularly well at detecting subtle disease patterns.
• MobileNet: Outperformed others in computational efficiency while maintaining high accuracy, especially after SMOTE and hyperparameter tuning. The depth-wise separable convolutions enabled real-time performance on resource-constrained devices.
• Computational Demands: While effective, both DenseNet121 and InceptionV3 required considerable training time and memory, making them less ideal for deployment in low-resource settings.
• Minor Class Sensitivity: Although SMOTE reduced class imbalance effects, certain minority classes still exhibited slightly lower performance metrics, indicating room for improvement through advanced data augmentation or ensemble methods.
• Training Time: The use of hyperparameter tuning and cross-validation extended the training duration considerably, especially for complex architectures like InceptionV3.
4.1.3 Practical Applicability Insights
• InceptionV3: Suitable for use cases requiring high accuracy and moderate computational resources, such as centralized agricultural analysis systems.
• DenseNet121: Best suited for scenarios requiring detailed pattern recognition, such as expert-level diagnostics in controlled environments with sufficient GPU support.
• MobileNet: Ideal for real-time disease detection on smartphones or embedded systems, especially in remote agricultural areas where computational resources are limited.
4.2 Training Methodology and Performance Evaluation
The dataset was divided into 80% training set and 20% validation set. SMOTE was applied to balance the training data. All three models underwent hyperparameter tuning, where a batch size of 16 and learning rate of 0.0005 yielded optimal performance. Subsequently, 5-fold cross-validation was used to validate model consistency and generalization. The results confirmed reliable convergence, with MobileNetV2 achieving the highest validation accuracy and lowest inference time, DenseNet121 achieving strong precision and recall, and InceptionV3 maintaining a balance between performance and efficiency.
Although the dataset currently comprises 3089 images, its relatively small size presents challenges for generalization. Future efforts will aim to expand the dataset by gathering real-world data from a range of environments, thereby enhancing the robustness and generalization capabilities of the model.
In this study, a range of deep learning models such as MobileNet and DenseNet121 were assessed for their effectiveness in classifying palm leaf diseases. MobileNetV2 stood out as the best-performing model, achieving a mean cross-validation accuracy of 96.99% and an overall classification accuracy of 97%. Its impressive precision, recall, and F1-scores across all nine disease categories underscore its robustness and efficiency. These outcomes further validate MobileNet’s strong feature extraction capabilities and suitability for real-time, lightweight applications, particularly for mobile and edge devices with limited computational power. Although DenseNet121 also demonstrated strong results, MobileNetV2 offered a superior balance between accuracy and computational efficiency. These findings emphasize the importance of selecting architectures that align with specific deployment needs. To further improve the accuracy and reliability of palm leaf disease detection systems, future research could explore more advanced and cutting-edge architectures, such as
• Vision Transformers (ViT)—for capturing long-range dependencies and global context in images.
• Swin Transformers—providing hierarchical representations and enhanced scalability.
Our model, especially MobileNetV2, is well-suited for real-time deployment in the field because of its lightweight design. Although it is currently utilized for research purposes, future plans will involve creating mobile applications for disease detection. While ViT and Swin Transformers have not yet been evaluated, they hold promise for identifying complex patterns and will be investigated in upcoming projects.
Additionally, incorporating cross-attention mechanisms, self-supervised learning, and hyperparameter tuning, along with the use of larger and more diverse datasets, can significantly enhance the generalizability and effectiveness of future palm leaf disease classification models.
In future research, we intend to test this system in a real-world setting to evaluate its robustness. Moreover, the use of synthetic image generation methods can enhance the generalization. Although MobileNetV2 appears promising for use in devices with limited resources, further investigation is necessary to address real-world deployment issues, such as performance in diverse environments and adaptation to mobile or embedded platforms. As our study used an existing dataset, further investigation may be required on environmental factors such as lighting conditions and background complexity, which may affect real-world deployment.
Acknowledgement: Princess Nourah bint Abdulrahman University Researchers Supporting Project, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. A discussion with Dr. R Praveena, a plant pathologist from the Indian Institute of Spices Research, Kozhikode, was very helpful in resolving some problems with the research.
Funding Statement: This work was funded by the Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2025R821), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Author Contributions: Sameera V Mohd Sagheer and Orwel P V were primarily responsible for the development and implementation of the image classification models and coding aspects of the research. P M Ameer contributed to data analysis and validation. Amal BaQais provided valuable interdisciplinary insights into the biological and chemical characteristics of the plant diseases, ensuring accurate annotation and scientific interpretation. Shaeen Kalathil supervised the overall research direction and manuscript preparation. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The dataset used in this study is publicly available and can be accessed from open data sources.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Namoun A, Alkhodre AB, Abi Sen AA, Alsaawy Y, Almoamari H. Dataset of infected date palm leaves for palm tree disease detection and classification. Data Brief. 2024;57:110933. doi:10.1016/j.dib.2024.110933. [Google Scholar] [PubMed] [CrossRef]
2. Al-Shalout M, Mansour K. Detecting date palm diseases using convolutional neural networks. In: 2021 22nd International Arab Conference on Information Technology (ACIT); 2021 Dec 21–23; Muscat, Oman. p. 1–5. [Google Scholar]
3. Safran M, Alrajhi W, Alfarhood S. DPXception: a lightweight CNN for image-based date palm species classification. Front Plant Sci. 2024;14:1281724. doi:10.3389/fpls.2023.1281724. [Google Scholar] [PubMed] [CrossRef]
4. Abade A, Ferreira PA, de Barros Vidal F. Plant diseases recognition on images using convolutional neural networks: a systematic review. Comput Electron Agric. 2021;185(7):106125. doi:10.1016/j.compag.2021.106125. [Google Scholar] [CrossRef]
5. Jogin M, Mohana, Madhulika MS, Divya G, Meghana G, Apoorva R. Feature extraction using convolution neural networks (CNN) and deep learning. In: 2018 3rd IEEE International Conference on Recent Trends in Electronics, Information & Communication Technology (RTEICT); 2018 May 18–19; Bangalore, India. 2018. p. 2319–23. [Google Scholar]
6. Kamilaris A, Prenafeta-Boldú FX. Deep learning in agriculture: a survey. Comput Electr Agric. 2018;147(2):70–90. doi:10.1016/j.compag.2018.02.016. [Google Scholar] [CrossRef]
7. Hasan RI, Yusuf SM, Alzubaidi L. Review of the state of the art of deep learning for plant diseases: a broad analysis and discussion. Plants. 2020;9(10):1302. doi:10.3390/plants9101302. [Google Scholar] [PubMed] [CrossRef]
8. Bhargava A, Shukla A, Goswami OP, Alsharif MH, Uthansakul P, Uthansakul M. Plant leaf disease detection, classification, and diagnosis using computer vision and artificial intelligence: a review. IEEE Access. 2024;12(4):37443–69. doi:10.1109/ACCESS.2024.3373001. [Google Scholar] [CrossRef]
9. Demilie WB. Plant disease detection and classification techniques: a comparative study of the performances. J Big Data. 2024;11(1):5. doi:10.1186/s40537-023-00863-9. [Google Scholar] [CrossRef]
10. Tandekar D, Dongre S. A review on various plant disease detection using image processing. In: 2023 3rd International Conference on Pervasive Computing and Social Networking (ICPCSN); 2023 Jun 19–20; Salem, India. 2023. p. 552–8. [Google Scholar]
11. Bagga M, Goyal S. Image-based detection and classification of plant diseases using deep learning: state-of-the-art review. Urban Agriculture & Regional Food Systems. 2024;9(1):e20053. doi:10.1002/uar2.20053. [Google Scholar] [CrossRef]
12. Mustofa S, Munna MMH, Emon YR, Rabbany G, Ahad MT. A comprehensive review on plant leaf disease detection using deep learning. arXiv:2308.14087. 2023. [Google Scholar]
13. Agarwal M, Singh A, Arjaria S, Sinha A, Gupta S. ToLeD: tomato leaf disease detection using convolution neural network. Procedia Comput Sci. 2020;167:293–301. doi:10.1016/j.procs.2020.03.225. [Google Scholar] [CrossRef]
14. Asif MKR, Rahman MA, Hena MH. CNN based disease detection approach on potato leaves. In: 2020 3rd International Conference on Intelligent Sustainable Systems (ICISS); 2020; Thoothukudi, India. p. 428–32. [Google Scholar]
15. Abu-zanona M, Elaiwat S, Younis S, Innab N, Kamruzzaman M. Classification of palm trees diseases using convolution neural network. Int J Adv Comput Sci Appl. 2022;13(6):10–14569. doi:10.14569/issn.2156-5570. [Google Scholar] [CrossRef]
16. Dai G, Fan J, Dewi C. ITF-WPI: image and text based cross-modal feature fusion model for wolfberry pest recognition. Comput Electron Agric. 2023;212(2):108129. doi:10.1016/j.compag.2023.108129. [Google Scholar] [CrossRef]
17. Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res. 2002;16:321–57. doi:10.1613/jair.953. [Google Scholar] [CrossRef]
18. Alsaidi M, Jan MT, Altaher A, Zhuang H, Zhu X. Tackling the class imbalanced dermoscopic image classification using data augmentation and GAN. Multimedia Tools Appl. 2024;83(16):49121–47. doi:10.1007/s11042-023-17067-1. [Google Scholar] [CrossRef]
19. Zorgui S, Chaabene S, Bouaziz B, Batatia H, Chaari L. A convolutional neural network for lentigo diagnosis. In: The Impact of Digital Technologies on Public Health in Developed and Developing Countries: 18th International Conference, ICOST 2020; 2020 Jun 24–26; Hammamet, Tunisia. p. 89–99. [Google Scholar]
20. Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition; 2017 Jul 21–26; Honolulu, HI, USA. p. 4700–8. [Google Scholar]
21. Wu D, Zhang Y, Jia X, Tian L, Li T, Sui L, et al. A high-performance CNN processor based on FPGA for MobileNets. In: 2019 29th International Conference on Field Programmable Logic and Applications (FPL); 2019 Sep 8–12; Barcelona, Spain. p. 136–43. [Google Scholar]
22. Golcuk A, Yasar A, Saritas MM, Erharman A. Classification of Cicer arietinum varieties using MobileNetV2 and LSTM. Eur Food Res Technol. 2023;249(5):1343–50. doi:10.1007/s00217-023-04217-w. [Google Scholar] [CrossRef]
23. Gordon-Rodriguez E, Loaiza-Ganem G, Pleiss G, Cunningham JP. Uses and abuses of the cross-entropy loss: case studies in modern deep learning. In: 1st I Can’t Believe It’s Not Better Workshop (ICBINB@NeurIPS 2020); 2020; Vancouver, BC, Canada. [Google Scholar]
24. Bai Y, Yang E, Han B, Yang Y, Li J, Mao Y, et al. Understanding and improving early stopping for learning with noisy labels. Adv Neural Inf Process Syst. 2021;34:24392–403. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools