 Open Access
Open Access
ARTICLE
Enhancing Respiratory Sound Classification Based on Open-Set Semi-Supervised Learning
Department of AI/SW Convergence, Soongsil University, Seoul, 06978, Republic of Korea
* Corresponding Author: Sangjun Lee. Email:
Computers, Materials & Continua 2025, 84(2), 2847-2863. https://doi.org/10.32604/cmc.2025.066373
Received 07 April 2025; Accepted 13 May 2025; Issue published 03 July 2025
Abstract
The classification of respiratory sounds is crucial in diagnosing and monitoring respiratory diseases. However, auscultation is highly subjective, making it challenging to analyze respiratory sounds accurately. Although deep learning has been increasingly applied to this task, most existing approaches have primarily relied on supervised learning. Since supervised learning requires large amounts of labeled data, recent studies have explored self-supervised and semi-supervised methods to overcome this limitation. However, these approaches have largely assumed a closed-set setting, where the classes present in the unlabeled data are considered identical to those in the labeled data. In contrast, this study explores an open-set semi-supervised learning setting, where the unlabeled data may contain additional, unknown classes. To address this challenge, a distance-based prototype network is employed to classify respiratory sounds in an open-set setting. In the first stage, the prototype network is trained using labeled and unlabeled data to derive prototype representations of known classes. In the second stage, distances between unlabeled data and known class prototypes are computed, and samples exceeding an adaptive threshold are identified as unknown. A new prototype is then calculated for this unknown class. In the final stage, semi-supervised learning is employed to classify labeled and unlabeled data into known and unknown classes. Compared to conventional closed-set semi-supervised learning approaches, the proposed method achieved an average classification accuracy improvement of 2%–5%. Additionally, in cases of data scarcity, utilizing unlabeled data further improved classification performance by 6%–8%. The findings of this study are expected to significantly enhance respiratory sound classification performance in practical clinical settings.Keywords
The classification of respiratory sounds is crucial in diagnosing and monitoring respiratory diseases. Abnormal respiratory sounds include crackles and wheezes, such as stridor and rhonchi, which are also considered adventitious lung sounds [1,2]. In this study, we primarily focus on classifying crackles and wheezes.
Diseases associated with crackles include interstitial lung disease, pneumonia, and cognitive heart failure, while wheezes are commonly associated with asthma, chronic obstructive pulmonary disease, and airway obstruction [3]. For diagnosing major respiratory diseases, including asthma, chronic obstructive pulmonary disease (COPD), and pneumonia, auscultation remains a primary and essential diagnostic tool [4–6]. Auscultation is widely used in primary healthcare settings due to its non-invasive nature, cost-effectiveness, and ability to provide immediate results [7,8].
However, classifying respiratory sounds accurately through auscultation is challenging due to its subjective nature and the influence of various factors. The interpretation of auscultation results relies significantly on the experience and expertise of medical professionals. The accuracy of medical students is reported at 60.3%, interns at 53.4%, residents at 68.8%, and fellows at 80.1%, indicating that less-experienced clinicians demonstrate lower auscultation accuracy [3]. There are also variations in accuracy depending on the type of respiratory sound. Compared to normal breath sounds (73.5%) and crackles (72.2%), wheezes (56.3%) and rhonchi (41.7%) pose greater diagnostic difficulties [3]. These difficulties arise from the overlapping frequency spectra and subtle differences between specific respiratory sound patterns. Discerning these minute variations requires substantial clinical experience [9].
Even among experts, diagnostic concordance is not perfect. The agreement rates for crackle and wheeze diagnoses are 82% and 80%, respectively, underscoring the inherent subjectivity and complexity of auscultatory interpretation [3]. The analysis of diagnostic accuracy shows an overall sensitivity of 37% (95% CI: 30–47%) and a specificity of 89% (95% CI: 85–92%), suggesting a high rate of missed diagnoses (false negatives) [3]. This low sensitivity could delay early diagnosis, thereby increasing disease progression and the risk of complications [10]. Several factors influence the accuracy of auscultation, including the patient’s breathing pattern, body position, stethoscope quality, ambient noise, and auscultation site [11]. In noisy environments, such as hospitals, detecting subtle changes in respiratory sounds can be challenging, potentially reducing diagnostic accuracy. Additionally, auscultation can be challenging when patients, such as children or older adults, cannot cooperate effectively [9].
To overcome these limitations, numerous studies have explored the development of respiratory sound classification systems using deep learning techniques. Recent studies have applied various feature extraction methods, such as spectrogram transformation of respiratory sound signals, mel-frequency cepstral coefficients (MFCC) extraction, wavelet transformation, and deep learning-based approaches, to enhance classification performance [4,11]. These feature extraction techniques can utilize information from a wider frequency range than human auditory perception, potentially resulting in improved diagnostic accuracy [4].
Obtaining large-scale labeled datasets remains one of the key challenges in developing deep learning models [12,13]. Experiments assessing respiratory sound classification accuracy and consistency among medical professionals from different specialties showed that pulmonologists achieved higher accuracy than medical professionals from other specialties and medical students. This result underscores the necessity of medical professionals’ involvement in respiratory sound labeling, which is both time-consuming and costly [3,10].
To address the limitations caused by insufficient labeling, semi-supervised learning approaches have recently gained attention [7,14]. Semi-supervised learning is a method that enhances model performance by utilizing a small set of labeled data along with a large volume of unlabeled data, making it highly effective in medical data analysis. Recent studies have applied techniques, such as teacher-student models, consistency regularization, and virtual adversarial training to respiratory sound classification, achieving favorable results [15,16]. Furthermore, research has investigated the use of data augmentation techniques to artificially increase the diversity of constrained labeled datasets [5]. Data augmentation techniques such as time stretching, pitch shifting, and noise injection enhance data variability and improve the model’s generalization ability. Notably, these data augmentation methods are highly effective in enhancing the classification performance of rare respiratory sound patterns and minority classes [4,5,11].
Most conventional semi-supervised learning techniques have been designed based on the closed-set assumption, restricting their capacity to appropriately process new types of respiratory sounds or noise that may arise in practical clinical settings [17,18]. A closed-set setting assumes that during testing, the model will only encounter classes it was exposed to during training. However, previously unseen respiratory sound patterns frequently emerge in practical clinical settings. These new patterns may arise due to unknown environmental noise, diseases, rare conditions, or patient-specific factors. If not adequately addressed, such cases could result in misdiagnosis. In other fields, such as computer vision, open-set semi-supervised learning approaches have been proposed [19–22]; however, research in respiratory sound classification remains in its early stages. Open-set learning enables a model to recognize and appropriately process unknown classes, significantly improving its applicability in practical clinical settings. Distance-based prototype networks can classify samples as known and unknown classes if they exhibit a significant distance in a feature space, making them highly applicable in open-set settings.
This study adopts a distance-based prototype network in the first stage to classify respiratory sounds in an open-set setting. The prototype network is trained using labeled and unlabeled data, after which the prototype representations of the known classes are derived. In the second stage, the distances between unlabeled data points and the known class prototypes are computed. Data points exceeding an adaptive threshold are filtered to form the unknown class, and their prototype is then computed. In the final stage, semi-supervised learning is employed to classify labeled and unlabeled data into known and unknown classes.
Studies on respiratory sound classification in open-set settings are still in their early stages. This section examines prior studies on general open-set audio recognition and classification, as well as image recognition and classification techniques that could be adapted for audio classification. Finally, studies on respiratory sound classification using unsupervised learning are explored.
2.1 Research on Audio Recognition and Classification in Open-Set Settings
An autoencoder-based approach has been proposed to address the problems of few-shot learning and open-set recognition (OSR) in the audio field [23]. The OSR problem involves accurately differentiating unknown classes from known classes when they appear as input. In this study, the autoencoder is trained to learn the internal characteristics of each class, and unknown classes are detected based on the reconstruction error. Recently, deep learning-based approaches have been proposed to address the open-set sound event classification problem [24]. Conventional closed-set classification models cannot detect unknown sounds; however, this study is designed to detect unknown sound events. The approach constructs feature space clusters and utilizes center loss and supervised contrastive loss to enhance the distinction between classes. In the first stage, self-supervised learning is employed to learn features from a large volume of unlabeled data. In the second stage, the model is refined through supervised learning, applying fine-tuning tailored to a specific domain to optimize performance. During this process, clustering techniques encourage known class features to form denser clusters, while ensuring that unknown sounds deviate from these clusters. Finally, the model’s decision head calculates probability distributions, classifying known sounds while detecting unknown sounds as new classes.
2.2 Research on Image Recognition and Classification in Open-Set Settings
OpenMatch [19] is a framework designed to address the open-set semi-supervised learning (OSSL) problem. Conventional FixMatch-based semi-supervised learning [25] approaches exhibit limitations in outlier detection. OpenMatch [19] integrates outlier detection with semi-supervised learning. The model comprises a feature extractor, K-independent class classifiers, and a closed-set classifier. The independent class classifiers detect outliers from each class, while FixMatch [25] generates highly reliable labels from normal data. Soft open-set consistency regularization is introduced to smoothly adjust the decision boundary of the outlier detector, preventing overfitting.
IOMatch [20] proposes a method for addressing the OSSL problem by utilizing outliers in the learning process instead of eliminating them, assuming that the class space of labeled data is a subset of that of unlabeled data. Conventional OOD detection primarily focuses on isolating outliers. In contrast, IOMatch [20] proposes an approach incorporating outliers and normal data. The model integrates distribution alignment (DA), outlier utilization, and self-supervised learning.
A new framework has been proposed to transform the OSSL problem into a closed-set semi-supervised learning (CSSL) problem [21]. Conventional semi-supervised learning methods rely on the closed-set assumption, resulting in performance degradation when OOD data are included in the unlabeled dataset. To address this problem, conventional safe semi-supervised learning (Safe SSL) follows a two-stage process involving OOD detection and data processing; however, this incurs additional data processing costs for OOD data and may conflict with multi-class classification tasks. This study employs a prototype network to model OOD data as a new
SeFOSS [22] is a framework that leverages self-supervision to enhance OSSL. Conventional OSSL approaches assume that OOD data degrades learning performance and thus excludes it, whereas SeFOSS [22] actively utilizes OOD data to enhance learning efficiency. SeFOSS [22] applies self-supervised feature consistency to effectively learn features from the entire unlabeled data set. It utilizes energy-based scoring to effectively distinguish between known and unknown classes. The model enhances the performance of OSR by integrating self-supervised learning, pseudo-labeling, and energy regularization. During training, a trainable linear projection layer reflects feature differences between weak and strong augmentations. This study also utilizes a trainable linear projection layer.
2.3 Research on Respiratory Sound Classification via Unsupervised Learning
In addressing the respiratory sound classification problem, where labeled data is scarce, a semi-supervised learning approach has been introduced to improve model performance [16]. This study employs the Passt [26] model, which has been pre-trained on AudioSet [27], along with the symmetric cross entropy [28] loss function, which is robust to noise, utilizing the PatchMix [29] learning approach. Unlabeled data is processed using a meta pseudo label [30] framework adapted for respiratory sound classification and trained alongside labeled data.
A learning method for evaluating similarity within the same dataset has also been proposed. This approach segments respiratory sounds into multiple sections, identifies recurring patterns, and analyzes similarities between those patterns using a sparse self-relation matrix [13]. This study utilizes HF Lung V1 [31] and COVID-19 [32] datasets as pretraining datasets.
Another approach, graph-based semi-supervised convolutional neural networks (GS-CNNs), has been proposed. It classifies respiratory sounds into normal, crackles, and wheezes by utilizing a limited number of labeled samples and a large number of unlabeled samples [14]. This study constructs a respiratory sound graph (Graph-RS), where labeled and unlabeled samples serve as nodes, capturing relationships among all samples. The information extracted from the Graph-RS is then incorporated into the loss function of a four-layer CNN, constituting the GS-CNNs model.
Although semi-supervised learning has been extensively studied in various domains with abundant unlabeled data but scarce high-quality labeled data, its application to respiratory sound classification remains relatively nascent.
This study proposes a semi-supervised learning method to improve the performance of respiratory sound classification in an environment where a labeled dataset
The proposed method effectively processes OOD data in three stages. In the first stage, a prototype network is trained to obtain prototypes for known classes using labeled and unlabeled data. In the second stage, the trained prototype network calculates the distance between unlabeled data and known class prototypes. Data exceeding the adaptive threshold is filtered out, and the prototype for unknown classes is subsequently calculated. Finally, a trainable projector layer is employed to ensure that the encoder effectively learns even the features of unlabeled respiratory sound data.
3.1 Stage 1: Prototype Network Training
The overall training architecture of the prototype network is shown in Fig. 1. This study utilizes an audio spectrogram transformer (AST) [33], pretrained on AudioSet [27], for the feature extraction of respiratory sounds. AudioSet [27] is a large-scale dataset encompassing thousands of hours of diverse audio events. Pretraining on this dataset enables the model to comprehensively learn audio patterns and features. The AST [33] encoder, utilizing a transformer-based self-attention mechanism, concurrently considers local and global features, thereby efficiently identifying the intricate structure of audio signals. The AST [33] model, pretrained on AudioSet [27], demonstrates excellent generalization performance in transfer learning scenarios and has shown high recognition accuracy, even on small-scale datasets [33].

Figure 1: Stage1: train prototype encoder
The loss function for labeled data is computed based on the distance from the centroid of batch-wise prototypes. In contrast, unlabeled data is processed using a trainable projector layer to apply a consistency loss function. This ensures that the encoder learns the features of labeled and unlabeled data. By training the model to maintain feature consistency across different augmentation methods for the same respiratory sound, meaningful information can be extracted even from unlabeled data [22].
The training process of the proposed method is as follows. As shown in Algorithm 1, labeled and unlabeled data are first transformed from the time domain to the frequency domain using a Mel spectrogram (Line 1). For unlabeled data, both weak and strong augmentations are performed (Line 2), and the AST [33] encoder is utilized to generate embedding vectors (Line 3). The prototype and centroid of each class are computed using the embedding vectors of the labeled data and the labels of the known classes (Line 4).

The loss function is calculated following Algorithm 2, referring to prior research. Considering labeled data

3.2 Stage 2: Generation of OOD Prototypes
The computation of OOD prototypes follows the procedure outlined in Algorithm 3. The prototype network, trained in the previous stage, extracts embedding values from labeled data (Line 1). Based on these, the prototype for each class is computed (Line 2), after which the embedding values of unlabeled data are generated to compute the OOD prototype (Line 3). Next, the distances between the embedding values of unlabeled data and each class prototype are measured (Line 4). Data points exceeding a certain adaptive threshold are identified and used to compute the OOD prototype (Line 5). In the third stage of semi-supervised learning, classification is performed using the class prototypes generated in the second stage alongside the OOD prototypes. The adaptive threshold is computed through the following process: First, for the unlabeled data Xu, the distance from each class prototype is calculated following Eq. (4).
The minimum distance for each sample is then selected. See Eq. (5).
The adaptive threshold for identifying OOD samples is determined according to Eq. (6).

A simple yet effective thresholding strategy is adopted to identify OOD samples, based on the distribution of the minimum distances from each sample to the class prototypes. Specifically, we compute the adaptive threshold as the sum of the mean and standard deviation of the minimum distances. This statistical formulation is commonly used in outlier detection and has been shown effective in prior open-set semi-supervised learning work, such as SeFOSS [22], which similarly employs distribution-based adaptive thresholds to separate pseudo-outliers. The adaptive threshold serves as a criterion for distinguishing OOD data. The mean embedding of
To intuitively understand how the prototype network separates known and unknown samples before semi-supervised learning, we visualized the feature embeddings produced by the student encoder using t-SNE, based on the prototypes trained in Stage 1. As shown in Fig. 2, samples from the known classes form compact clusters around their respective class prototypes. Among the unlabeled data, some samples are located near the known class centroids, indicating similarity to known classes. In contrast, others are positioned far from any prototype, indicating that they may belong to unknown classes.

Figure 2: t-SNE visualization of feature embeddings based on prototypes trained in Stage 1
3.3 Stage 3: Semi-Supervised Learning
The overall structure of semi-supervised learning using the teacher-student model is depicted in Fig. 3, with the detailed training procedure outlined in Algorithm 4. In the proposed learning method, weak and strong augmentations are first applied to unlabeled data. The teacher model predicts labels for data with weak augmentation applied. Cases where the predicted values exceed a predefined threshold are masked and used for UDA [34] computation. The loss function of the teacher model consists of cross-entropy loss, derived from the distance between prototypes and labeled data embedding vectors, along with UDA [34] (Line 2). The student model’s loss function incorporates cross-entropy loss, which utilizes the distance between prototypes and labeled data embedding vectors, and a pseudo-loss function that considers the distance between strongly augmented data embedding vectors and prototypes and the corresponding label information (Line 3). As training progresses, the teacher model generates increasingly accurate pseudo-labels for unlabeled data, which the student model utilizes for training.

Figure 3: Stage3: train semi-supervised learning

The experiments in this study were conducted under the following settings. For respiratory sound classification, the model employed a distance-based prototype network built upon an AST encoder, pre-trained on AudioSet. To ensure efficient learning, the zero redundancy optimizer stage 3 (ZeRO-3) optimization technique, provided by Microsoft’s DeepSpeed library, was employed [35]. ZeRO-3 significantly reduces memory consumption by segmenting optimizer states, gradients, and model parameters across multiple GPUs, thereby enhancing training speed [35]. Regarding batch structure, a larger proportion of unlabeled data was utilized compared to labeled data. This approach reflects practical clinical settings, where labeled data is often scarce, while incorporating various respiratory sound patterns that may arise in open-set settings into learning [36]. The improved memory efficiency of ZeRO-3 enabled training with a larger batch size, allowing for the inclusion of a greater volume of unlabeled data [35].
The datasets used in this study are from ICBHI [37], Fraiwan et al. [38], and HF Lung V2 [39]. The ICBHI [37] dataset comprises 5.5 h of recorded data collected from 126 subjects, encompassing 6898 respiratory cycles. It contains 1864 crackles and 886 wheezes, with 506 cases simultaneously exhibiting both crackles and wheezes. The recordings vary in length from 10 s to 90 s and incorporate background noise at varying levels to simulate practical settings. The labeling files record four key data fields in separate columns: the start and end times of each respiratory cycle and binary indicators (0 or 1) for the presence of crackles and wheezes.
The Fraiwan et al. [38] dataset consists of 112 subjects, including 35 healthy individuals and 77 patients diagnosed with various respiratory diseases. The age range spans 21 to 90 years, with an average age of 50.5 years and a standard deviation of 19.4 years. It comprises 43 females and 69 males, reflecting a broad spectrum of demographic variability. A notable characteristic of this dataset is that respiratory sounds were collected using three different filtering methods: Bell, Diaphragm, and Extended mode. Each filtering method is designed to emphasize different frequency bands, enabling the effective capture of various types of respiratory sounds. The respiratory sounds included in the dataset comprise normal respiratory sounds, crepitations (crackles), wheezes, a combination of crackles and wheezes, and bronchial breath sounds, which are absent from ICBHI [37].
The HF Lung V2 [39] dataset collected data from two different sources. The first subset consists of data from 261 patients recorded in the Taiwan severe emergency and critical care (TSECC) database in 2020. The second subset consists of data collected from 18 patients admitted to the respiratory care ward (RCW) or the respiratory care center (RCC) in northern Taiwan between August 2018 and October 2019. This dataset contains 9765 audio recordings, each 15 s long, including 8457 wheezes, 686 stridor occurrences, 4740 rhonchi, and 15,606 crackles. This dataset also includes stridor and rhonchi, which are not present in ICBHI [37].
The typical frequency range of respiratory sounds is 50–2500 Hz [40]. Crackles are categorized into fine crackles and coarse crackles. Fine crackles last under 5 ms and occur within a frequency range of less than 650 Hz, whereas coarse crackles have a duration of under 30 ms and a frequency range of below 350 Hz [41]. Wheezes have a duration of less than 80 ms and a frequency range of 100–1000 Hz [41]. In this study, respiratory sound augmentation was performed using SpecAugment [42]. After converting into mel spectrograms, augmentation was applied while considering the characteristics of crackles and wheezes. For weak augmentation, the frequency mask was applied once within the range of 125–500 Hz, while the time mask was applied once within 0.8–2.4 s. For strong augmentation, the frequency mask was applied up to twice within the range of 500–1250 Hz, while the time mask was applied up to twice within 2.4–12.8 s. The mel spectrogram was configured with the following parameters: frame length of 400, hop length of 160, FFT length of 512, and 128 mel bins.
This study utilizes the ICBHI Score [37]. This metric was proposed in the ICBHI Challenge and evaluates the classification performance of a model by considering both sensitivity and specificity. This metric specifically employs the mean of the two metrics to mitigate the issue of class imbalance.
Sensitivity represents the proportion of actual positive cases that the model correctly predicts.
Sensitivity represents the proportion of actual positive cases that the model correctly predicts. See Eq. (7).
where
•
•
Specificity represents the proportion of actual negative cases that the model correctly predicts. See Eq. (8).
where
•
•
Finally, the ICBHI Score, denoted as SC in the result tables, is calculated as shown in Eq. (9).
This study used the test data from ICBHI [37] as validation data. The experimental results demonstrate variations in model performance based on varying proportions of training data (100%, 70%, 30%). The results obtained using 100% of the ICBHI [37] training data are presented in Table 1.
The study analyzes how reducing the amount of training data affects the performance compared to this baseline model. When 70% of the ICBHI [37] training data was randomly utilized, the baseline model exhibited an overall performance degradation, as shown in Table 2. However, our proposed method demonstrated a 5%–8% improvement over the PatchMix [29] model in both closed-set and open-set settings.
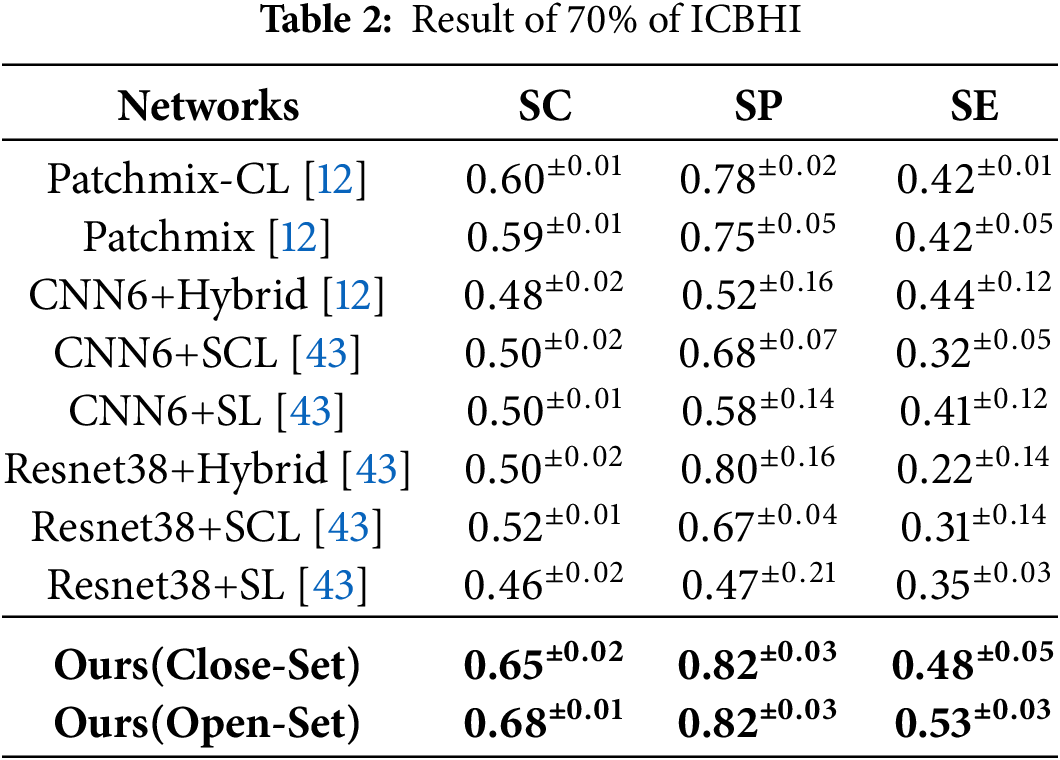
In the closed-set settings, the SC, SP, and SE metrics achieved performance scores of 0.65
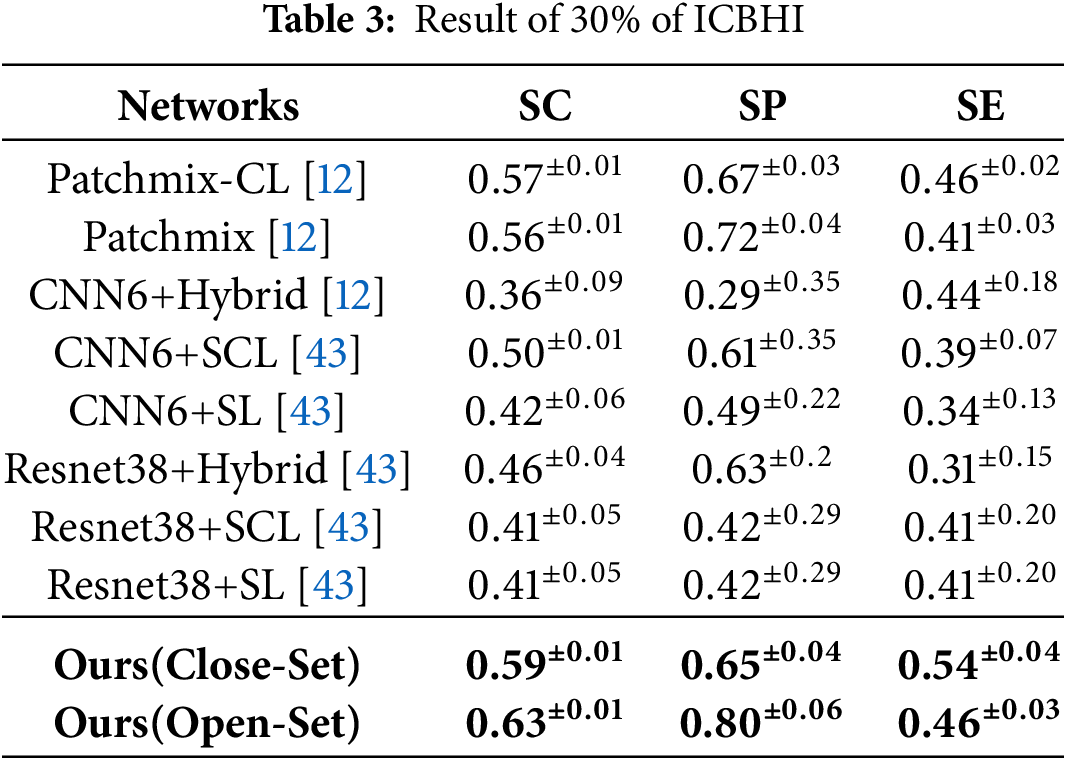
Under the more restricted training conditions of ICBHI [37], the proposed method maintained superior performance compared to existing models, as observed in Table 3, where only 30% of the data was randomly selected for training. Based on the PatchMix-CL [12] model, the proposed method demonstrated a 2% improvement in the closed-set settings and a 6% improvement in the open-set settings. Compared to the PatchMix-CL [12] model trained on 100% of the dataset as shown in Table 1, the proposed method exhibited a 3% performance degradation in the closed-set settings, but a 1% performance improvement in the open-set settings. This indicates that the proposed method can effectively learn with a limited dataset.
An additional analysis of computational resources was conducted to evaluate the model’s efficiency. All metrics were measured using an NVIDIA GeForce RTX 4090. A detailed comparison of model size, FLOPs, and per-sample training and inference time is provided in Table 4. The models referenced in [12] and [43], which follow supervised learning approaches, incur a computational cost of approximately 161.89 GFLOPs and 7 ms–8 ms inference time per sample, based on architectures with around 87 M parameters. A direct comparison may not be entirely appropriate due to differing learning methods. These baselines are trained using supervised learning, whereas the proposed method adopts a three-stage semi-supervised process. The proposed method is trained in three stages: Stage 1—Prototype Network Training, Stage 2—Generation of OOD Prototypes, and Stage 3—Semi-Supervised Learning. This multi-stage process results in a longer training time compared to the baselines. At inference time, the method requires 206.88 GFLOPs and 10.62 ms per sample, with 86.2 M parameters.
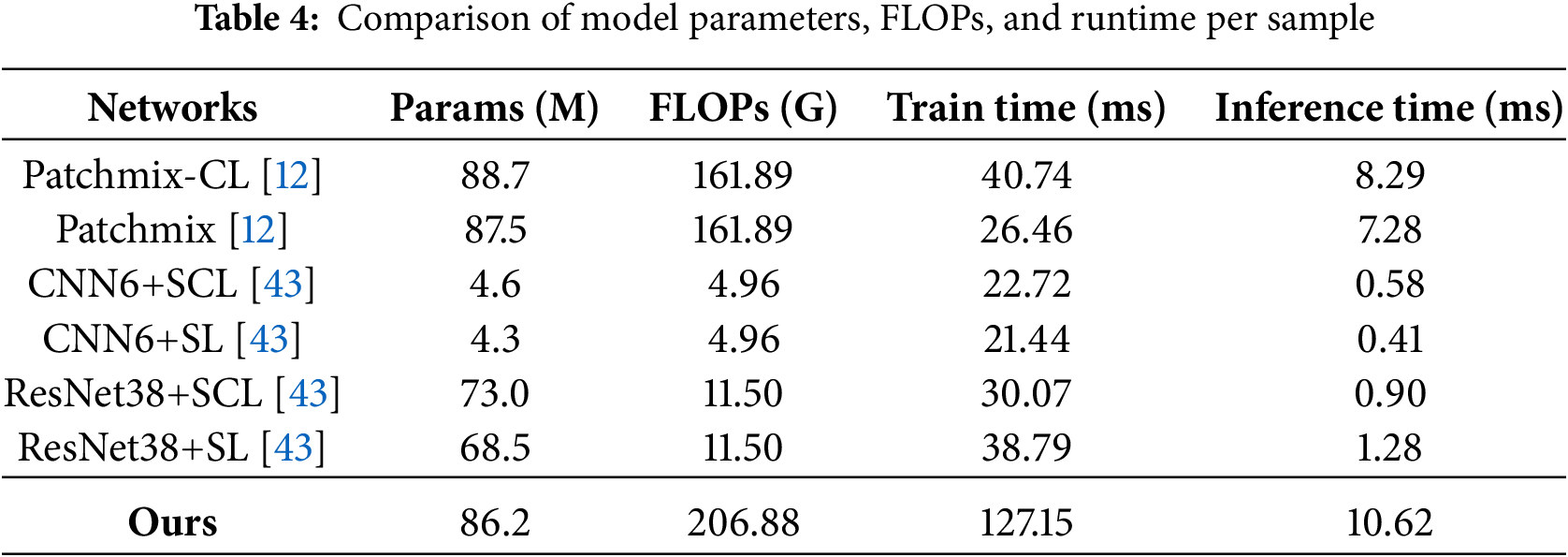
While this represents an approximately 28% increase in inference cost compared to the baselines, the proposed approach demonstrates consistent advantages, with a 2%–5% accuracy improvement over conventional closed-set semi-supervised methods and an additional 6%–8% gain when leveraging unlabeled data in open-set scenarios.
The experimental results demonstrate that the proposed method remains effective when the amount of labeled data is limited. Furthermore, it validates the model’s ability to enhance generalization performance by leveraging unlabeled data from diverse sources. The performance improvements in the open-set settings highlight the model’s robustness in handling the challenges encountered in practical clinical settings.
This study improved the performance of respiratory sound classification by leveraging open-set unsupervised learning based on a distance-based prototype network. In practical clinical settings, new respiratory sound types not included in labeled datasets may emerge. These OOD data significantly degrade the performance of semi-supervised learning models trained under a conventional closed-set setting. This poses a significant challenge in practical clinical settings, where it is essential to account for an open-set setting, as various sounds outside known classes may be introduced. Experimental findings suggest that the proposed method achieved an average classification accuracy improvement of 2%–5% compared to conventional closed-set semi-supervised learning approaches. By classifying OOD data as an unknown class, the model effectively leveraged unlabeled data under conditions with limited labeled data, resulting in a 6%–8% improvement in classification performance. This significantly reduced misclassification errors, which commonly occur in existing models due to their tendency to force all input data into one of the known classes. In the experiment, a part of the unlabeled data was intentionally designated as OOD samples for evaluation. The results demonstrated that the proposed method accurately identifies these OOD samples with high precision. These results indicate that the proposed method can significantly enhance classification accuracy in practical clinical settings, particularly when analyzing respiratory sounds from patients with diverse respiratory diseases. Furthermore, the approach presented in this study is not limited to respiratory sound classification but is also applicable to other fields of medical data analysis. In particular, it plays a crucial role in overcoming the challenges posed by insufficient labeled data and the presence of unknown patterns, which are prevalent in diverse medical diagnostic contexts and practical clinical settings.
Acknowledgement: The authors would like to thank the highly respected editor and reviewers for their valuable suggestions for improving the article.
Funding Statement: This work was supported by Innovative Human Resource Development for Local Intellectualization Program through the Institute of Information & Communications Technology Planning & Evaluation (IITP) grant funded by the Korea government (MSIT) (IITP-2025-RS-2022-00156360).
Author Contributions: Won-Yang Cho: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Writing—original draft preparation, Visualization. Sangjun Lee: Supervision, Writing—review and editing, Project administration, Funding acquisition. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are openly available in the following repositories: ICBHI 2017 Challenge dataset [37] available at https://bhichallenge.med.auth.gr/ICBHI_2017_Challenge (accessed on 12 May 2025), Fraiwan et al. dataset [38] available at https://data.mendeley.com/datasets/jwyy9np4gv/3 (accessed on 12 May 2025), HF Lung V2 dataset [39] available at https://gitlab.com/techsupportHF/HF_Lung_V1_IP (accessed on 12 May 2025).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Sarkar M, Madabhavi I, Niranjan N, Dogra M. Auscultation of the respiratory system. Anna Thorac Med. 2015;10(3):158–68. [Google Scholar] [PubMed]
2. Pramono R, Bowyer S, Rodriguez-Villegas E. Automatic adventitious respiratory sound analysis: a systematic review. PLoS One. 2017;12(5):e0177926. doi:10.1371/journal.pone.0177926. [Google Scholar] [PubMed] [CrossRef]
3. Kim Y, Hyon Y, Jung SS, Lee S, Yoo G, Chung C, et al. Respiratory sound classification for crackles, wheezes, and rhonchi in the clinical field using deep learning. Sci Rep. 2021;11(1):1–11. doi:10.1038/s41598-021-96724-7. [Google Scholar] [PubMed] [CrossRef]
4. Zulfiqar R, Majeed F, Irfan R, Rauf HT, Benkhelifa E, Belkacem AN. Abnormal respiratory sounds classification using deep CNN through artificial noise addition. Front Med. 2021;8:714811. doi:10.3389/fmed.2021.714811. [Google Scholar] [PubMed] [CrossRef]
5. Gairola S, Tom F, Kwatra N, Jain M. RespireNet: a deep neural network for accurately detecting abnormal lung sounds in limited data setting. In: 2021 43rd Annual International Conference of the IEEE Engineering in Medicine&Biology Society (EMBC); 2021 Nov 1–5; Mexico. p. 527–30. [Google Scholar]
6. Celli BR, Fabbri LM, Aaron SD, Agusti A, Brook RD, Criner GJ, et al. Differential diagnosis of suspected chronic obstructive pulmonary disease exacerbations in the acute care setting: best practice. Am J Res Crit Care Med. 2023;207(9):1134–44. doi:10.1164/rccm.202209-1795ci. [Google Scholar] [PubMed] [CrossRef]
7. Chamberlain D, Kodgule R, Ganelin D, Miglani V, Fletcher RR. Application of semi-supervised deep learning to lung sound analysis. In: 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); 2016 Aug 16–20; Orlando, FL, USA. p. 804–7. [Google Scholar]
8. Arts L, Lim EHT, van de Ven PM, Heunks L, Tuinman PR. The diagnostic accuracy of lung auscultation in adult patients with acute pulmonary pathologies: a meta-analysis. Sci Rep. 2020;10(1):7347. doi:10.1038/s41598-020-64405-6. [Google Scholar] [PubMed] [CrossRef]
9. Sfayyih AH, Sabry AH, Jameel SM, Sulaiman N, Raafat SM, Humaidi AJ, et al. Acoustic-based deep learning architectures for lung disease diagnosis: a comprehensive overview. Diagnostics. 2023;13(10):1748. doi:10.3390/diagnostics13101748. [Google Scholar] [CrossRef]
10. Hafke-Dys H, Breborowicz A, Kleka P, Kociński J, Biniakowski A. The accuracy of lung auscultation in the practice of physicians and medical students. PLoS One. 2019;14(8):e0220606. doi:10.1371/journal.pone.0220606. [Google Scholar] [CrossRef]
11. Srivastava A, Jain S, Miranda R, Patil S, Pandya S, Kotecha K. Deep learning based respiratory sound analysis for detection of chronic obstructive pulmonary disease. PeerJ Comput Sci. 2021;7(5):e369. doi:10.7717/peerj-cs.369. [Google Scholar] [PubMed] [CrossRef]
12. Bae S, Kim J, Cho W, Baek H, Son S, Lee B, et al. Patch-mix contrastive learning with audio spectrogram transformer on respiratory sound classification. In: 21st Annual Conference of the International Speech Communication Association (Interspeech 2023); 2023 Aug 20–24; Dublin, Ireland. p. 13459–68. [Google Scholar]
13. Song W, Han J, Deng S, Zheng T, Zheng G, He Y. Joint energy-based model for semi-supervised respiratory sound classification: a method insensitive to distribution mismatch. IEEE J Biomed Health Inform. 2025;29(2):1433–43. doi:10.1109/jbhi.2024.3480999. [Google Scholar] [PubMed] [CrossRef]
14. Lang R, Fan Y, Liu G, Liu G. Analysis of unlabeled lung sound samples using semi-supervised convolutional neural networks. Appl Math Comput. 2021;402(2):126150. doi:10.1016/j.amc.2021.126511. [Google Scholar] [CrossRef]
15. Li Y, Wang X, Liu H, Tao R, Yan L, Ouchi K. Semi-supervised sound event detection with local and global consistency regularization. In: ICASSP 2024—2024 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); 2024 April 14–19; Seoul, Republic of Korea. p. 271–5. [Google Scholar]
16. Cho WY, Lee S. Effective respiratory sound classification based on semi-supervised learning. KIISE Transa Comput Pract. 2024;30(12):669–74. [Google Scholar]
17. Oliver A, Odena A, Raffel CA, Cubuk ED, Goodfellow I. Realistic evaluation of deep semi-supervised learning algorithms. In: Bengio S, Wallach H, Larochelle H, Grauman K, Cesa-Bianchi N, Garnett R, editors. Advances in Neural Information Processing Systems. Vol. 31. Red Hook, NY, USA: Curran Associates, Inc.; 2018. [Google Scholar]
18. Van Engelen JE, Hoos HH. A survey on semi-supervised learning. Mach Learn. 2020;109(2):373–440. doi:10.1007/s10994-019-05855-6. [Google Scholar] [CrossRef]
19. Saito K, Kim D, Saenko K. OpenMatch: open-set semi-supervised learning with open-set consistency regularization. Adv Neural Inform Process Syst. 2021;34:25956–67. [Google Scholar]
20. Li Z, Qi L, Shi Y, Gao Y. IOMatch: simplifying open-set semi-supervised learning with joint inliers and outliers utilization. In: Proceedings of the 2023 IEEE/CVF International Conference on Computer Vision (ICCV); 2023 Oct 1–6; Paris, France. p. 15870–9. [Google Scholar]
21. Ma Q, Gao J, Zhan B, Guo Y, Zhou J, Wang Y. Rethinking safe semi-supervised learning: transferring the open-set problem to a close-set one. In: Proceedings of the 2023 IEEE/CVF International Conference on Computer Vision (ICCV); 2023 Oct 1–6; Paris, France. p. 16370–9. [Google Scholar]
22. Wallin E, Svensson L, Kahl F, Hammarstrand L. Improving open-set semi-supervised learning with self-supervision. In: Proceedings of the 2024 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV); 2024 Jan 3–8; Waikoloa, HI, USA. p. 2356–65. [Google Scholar]
23. Naranjo-Alcazar J, Perez-Castanos S, Zuccarello P, Antonacci F, Cobos M. Open set audio classification using autoencoders trained on few data. Sensors. 2020;20(13):3741. doi:10.3390/s20133741. [Google Scholar] [PubMed] [CrossRef]
24. You J, Wu W, Lee J. Open set classification of sound event. Sci Rep. 2024;14(1):1282. [Google Scholar] [PubMed]
25. Sohn K, Berthelot D, Carlini N, Zhang Z, Zhang H, Raffel CA, et al. FixMatch: simplifying semi-supervised learning with consistency and confidence. In: Advances in Neural Information Processing Systems. Vol. 33; 2020. p. 596–608. [Google Scholar]
26. Koutini K, Schlüter J, Eghbal-zadeh H, Widmer G. Efficient training of audio transformers with patchout. In: Interspeech 2022, 23rd Annual Conference of the International Speech Communication Association; 2022 Sep 18–22; Incheon, Republic of Korea: ISCA. p. 2753–7. doi:10.21437/Interspeech.2022-227. [Google Scholar] [CrossRef]
27. Gemmeke JF, Ellis DP, Freedman D, Jansen A, Lawrence W, Moore RC, et al. Audio set: an ontology and human-labeled dataset for audio events. In: 2017 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); 2017 Mar 5–9; New Orleans, LA, USA. p. 776–80. [Google Scholar]
28. Wang Y, Ma X, Chen Z, Luo Y, Yi J, Bailey J. Symmetric cross entropy for robust learning with noisy labels. In: Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV); 2019 Oct 27–Nov 2; Seoul, Republic of Korea. p. 322–30. [Google Scholar]
29. Hong Y, Chen Y. PatchMix: patch-level mixup for data augmentation in convolutional neural networks. Knowl Inform Syst. 2024;66(7):3855–81. doi:10.1007/s10115-024-02141-3. [Google Scholar] [CrossRef]
30. Pham H, Dai Z, Xie Q, Le QV. Meta pseudo labels. In: Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2021 Jun 20–25; Nashville, TN, USA. p. 11557–68. [Google Scholar]
31. Hsu FS, Huang SR, Huang CW, Huang CJ, Cheng YR, Chen CC, et al. Benchmarking of eight recurrent neural network variants for breath phase and adventitious sound detection on a self-developed open-access lung sound database—HF_Lung_V1. PLoS One. 2021;16(7):e0254134. doi:10.1371/journal.pone.0254134. [Google Scholar] [PubMed] [CrossRef]
32. Xia T, Spathis D, Brown C, Ch J, Grammenos A, Han J, et al. COVID-19 sounds: a large-scale audio dataset for digital respiratory screening. In: Thirty-Fifth Conference on Neural Information Processing Systems Datasets and Benchmarks Track (Round 2); 2021 Dec 6–14; Online. [Google Scholar]
33. Gong Y, Chung YA, Glass J. AST: audio spectrogram transformer. In: Proceedings of Interspeech 2021; 2021 Aug 30–Sep 3; Brno, Czechia. p. 571–5. [Google Scholar]
34. Xie Q, Dai Z, Hovy E, Luong T, Le QV. Unsupervised data augmentation for consistency training. In: Advances in Neural Information Processing Systems (NeurIPS). Vol. 33; 2020. p. 6256–68. [Google Scholar]
35. Rajbhandari S, Ruwase O, Rasley J, Smith S, He Y. Zero-infinity: breaking the GPU memory wall for extreme scale deep learning. In: Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis; 2021 Nov 14–19; Louis, MO, USA. p. 1–14. [Google Scholar]
36. Yu T, Kumar A, Chebotar Y, Hausman K, Finn C, Levine S. How to Leverage Unlabeled Data in Offline Reinforcement Learning. In: International Conference on Machine Learning (ICML); 2022 Jul 17–23; Baltimore, MD, USA. p. 25611–35. [Google Scholar]
37. Rocha BM, Filos D, Mendes L, Vogiatzis I, Perantoni E, Kaimakamis E, et al. A respiratory sound database for the development of automated classification. In: Precision medicine powered by pHealth and connected health. Singapore: Springer; 2018. p. 33–7. [Google Scholar]
38. Fraiwan M, Fraiwan L, Khassawneh B, Ibnian A. A dataset of lung sounds recorded from the chest wall using an electronic stethoscope. Data Brief. 2021;35(1):106913. doi:10.1016/j.dib.2021.106913. [Google Scholar] [PubMed] [CrossRef]
39. Hsu FS, Huang SR, Huang CW, Cheng YR, Chen CC, Hsiao J, et al. A progressively expanded database for automated lung sound analysis: an update. Appl Sci. 2022;12(15):7623. doi:10.3390/app12157623. [Google Scholar] [CrossRef]
40. Reichert S, Gass R, Brandt C, Andrès E. Analysis of respiratory sounds: state of the art. Clin Med Circul Resp Pulmon Med. 2008;2(1):CCRPM.S530. doi:10.4137/ccrpm.s530. [Google Scholar] [PubMed] [CrossRef]
41. Park JS, Kim K, Kim JH, Choi YJ, Kim K, Suh DI. A machine learning approach to the development and prospective evaluation of a pediatric lung sound classification model. Sci Rep. 2023;13(1):1289. doi:10.1038/s41598-023-27399-5. [Google Scholar] [PubMed] [CrossRef]
42. Park DS, Chan W, Zhang Y, Chiu CC, Zoph B, Cubuk ED, et al. SpecAugment: a simple data augmentation method for automatic speech recognition. In: Interspeech 2019; 2019 Sep 15–19; Graz, Austria. p. 2613–7. [Google Scholar]
43. Moummad I, Farrugia N. Pretraining respiratory sound representations using metadata and contrastive learning. In: 2023 IEEE Workshop on Applications of Signal Processing to Audio and Acoustics (WASPAA); 2023; New Paltz, NY, USA. p. 1–5. doi:10.1109/waspaa58266.2023.10248130. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF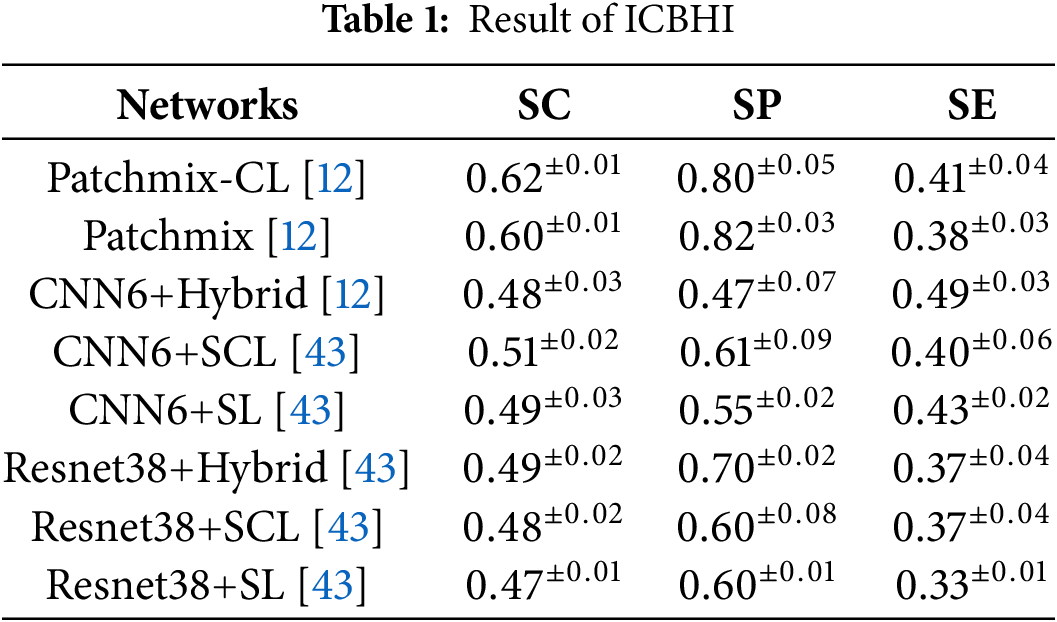
 Downloads
Downloads
 Citation Tools
Citation Tools